Corresponding Author

Key Words: clonal hematopoiesis, dilated cardiomyopathy, genetics, heart failure
During each hematopoietic stem cell (HSC) division, somatic mutations may stochastically arise. Most of the time, these mutations do not lead to a competitive advantage for HSCs. However, a somatic mutation might provide a benefit to the cell, via a loss of balance between self-renewal and differentiation, enhanced resistance against extrinsic insults such as chemotherapy, or protection against inflammation inflicted by mutated progenitor cells thus providing a “competitive” advantage between the mutated clone and non-mutated cells.1 This cell clone can further expand, a phenomenon referred to as clonal hematopoiesis (CH). After further cell differentiation, myeloid and lymphoid cells carrying these clonal hematopoietic driver mutations (CHDMs) enter the circulation and migrate to tissues. When CH occurs in the absence of hematological malignancy, it is referred to as clonal hematopoiesis with indeterminate potential (CHIP).
Previous groundbreaking work established CHIP as a cause of atherosclerotic cardiovascular disease and non-hematological death.2 Myeloid cells (eg, monocytes and macrophages) with CHDMs continuously stimulate an inflammatory response via the inflammasome,1 via histone deacetylase 1 and 2,3 or other mechanisms yet to be elucidated. Since its discovery, the negative impact of CHIP on the prognosis of patients with heart diseases was extensively described: atherosclerotic cardiovascular disease,2 ischemic heart failure,4 and nonischemic heart failure.5 CHIP associates with a 2-fold increase in the risk of cardiac death in patients with nonischemic dilated cardiomyopathy (DCM) over 10-year follow-up.5 The hypothesis is that monocytes with CHDM (eg, DMT3A or TET2) migrate into the myocardium where they stimulate a pro-inflammatory response leading to fibrosis and a reduced systolic function driving an adverse prognosis.6 However, direct evidence of an effect of CHIP on cardiac remodeling in patients with DCM is lacking.
In this issue of JACC: Basic to Translational Science, Inoue et al7 report for the first time on 2 aspects: 1) the effect of CHIP on adverse cardiac remodeling in patients with DCM; and 2) the interaction between CHIP and pathogenic germline variants in DCM-associated genes. DCM was defined as a reduced ejection fraction ≤40% without an apparent cause such as coronary artery disease, uncontrollable hypertension, primary valvular disease, and congenital heart disease. Thus, nondilated left ventricular cardiomyopathy was included in the used definition of DCM. CHIP was detected by whole exome sequencing with an additional panel for deep sequencing of the CHIP driver genes. Only previously published mutations with high coverage were considered as possible CHDMs, if the variant allele frequency was above 2% (corresponding to approximately 4 out of every 100 nucleated blood cells carrying a CHDM). Therefore, relevant mutations with a variant allele frequency ≤2% and previously unpublished mutations may be missed, potentially resulting in an underestimation of the percentage of patients with CHIP.
CHIP was detected in 22 (11%) of the 198 included patients with nonischemic DCM, with most mutations detected in DNMT3A in line with the literature.5 Left ventricular reverse remodeling (LVRR), defined as an absolute increase of ≥10% within 1 year of follow-up, was less prevalent in patients with CHIP compared with patients without CHIP. More specifically, CHIP predicted 73% lower odds of LVRR, even after adjusting for age, sex, heart and kidney function, previous event of heart failure, and germline mutations in DCM-associated genes. To investigate the mechanism of how CHIP might reduce the likelihood of LVRR in DCM, the authors developed an animal model. Mice with a pathogenic germline mutation in Titin (Ttn) exhibited reduced systolic function, increased myocardial inflammation, and greater myocardial fibrosis when they received a bone marrow transplant with cells carrying a pathogenic mutation in additional sex combs-like 1 (ASXL1), which induces myeloproliferation in mice.8 The myocardial inflammation was characterized by increased macrophage infiltration and higher expression of interleukin-1 and interleukin-6, concluding that CHIP may prevent LVRR in patients with DCM by promoting macrophage infiltration in the myocardium, where they stimulate inflammation through interleukin-1 and interleukin-6, eventually leading to fibrosis.
The findings of Inoue et al7 provide novel data on the effect of CHIP on the heart of patients with DCM and might provide an explanation for the observed adverse prognosis. Understanding of the patient population is important for the interpretation and translatability of the results. At baseline, the relatively small cohort of 198 patients already had a median heart failure duration of 18 months. Patients with a left ventricular ejection fraction below 40% after 18 months of treatment are usually less responsive to treatment, partially explaining the low rate of LVRR and high usage of diuretics (62%). The prevalence of pathogenic germline variants was very high (46%), which might be explained by the fact that patients with a genetic variant usually respond less well to standard heart failure medication. However, it has been extensively described that patients with a truncating TTN variant (TTNtv), the most prevalent genetic mutation, respond very well to medication and have a high LVRR. This raises the question whether CHIP might be an important second hit in the pathogenesis of TTNtv-associated DCM, preventing LVRR due to myocardial inflammation. It raises the question whether CHIP might be an important piece of the puzzle to identify those patients who will not respond to standard heart failure therapy. Even further: maybe this is the subgroup of patients with DCM who might benefit from additional anti-inflammatory therapies. These are important research questions that should be addressed in future studies.
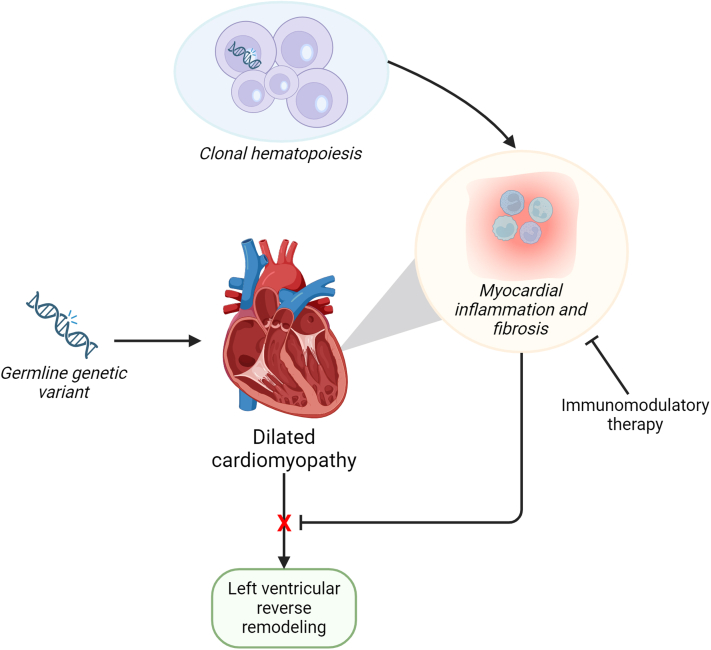
The interaction between pathogenic germline variants and CHIP was focused on LVRR (Figure 1). It remains the question whether CHIP also leads to an earlier disease onset and more severe presentation in those patients with a germline variant. The animal study demonstrates that Ttn-mutated mice with CHIP have a worse systolic function compared with Ttn-mutated mice without CHIP, but it does not compare Ttn-mutated mice with CHIP and wild-type mice with CHIP. Although mutations in DNMT3A are the most prevalent driver mutations in patients, the mouse work focuses on ASXL1 but CHDMs in ASXL1 are rare in patients with DCM. A study focusing on the effect of DNMT3A CHDMs on top of germline mutations in DCM-associated genes would be extremely interesting.
Figure 1.
CH Predicts Adverse Cardiac Remodeling
Clonal hematopoiesis (CH) driver mutations increase myocardial inflammation via macrophage inflammasome, interleukin-1, and interleukin-6 expression. Patients with dilated cardiomyopathy (DCM) and CH have a lower chance of reverse cardiac remodeling, independent of germline mutation in DCM-associated genes. Immunomodulatory therapy may ultimately improve cardiac remodeling in DCM.
In summary, Inoue et al7 are the first to report on the effect of CHIP on LVRR in patients with DCM. Their findings suggest that CHIP can be a biomarker for myocardial inflammation, fibrosis, and poor recovery in patients not completely responding to heart failure therapy. Future trials could use CHIP to select patients who would best benefit from additional anti-inflammatory therapy.
Funding Support and Author Disclosures
The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
References
- 1.Sikking M.A., Stroeks S., Waring O.J., et al. Clonal hematopoiesis of indeterminate potential from a heart failure specialist's point of view. J Am Heart Assoc. 2023;12 doi: 10.1161/JAHA.123.030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaiswal S., Natarajan P., Silver A.J., et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q., Zhao K., Shen Q., et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–393. doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorsheimer L., Assmus B., Rasper T., et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol. 2019;4:25–33. doi: 10.1001/jamacardio.2018.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sikking M.A., Stroeks S., Henkens M., et al. Clonal hematopoiesis has prognostic value in dilated cardiomyopathy independent of age and clone size. JACC Heart Fail. 2024;12:905–914. doi: 10.1016/j.jchf.2023.06.037. [DOI] [PubMed] [Google Scholar]
- 6.Sano S., Oshima K., Wang Y., et al. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1β/NLRP3 inflammasome. J Am Coll Cardiol. 2018;71:875–886. doi: 10.1016/j.jacc.2017.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue S., Ko T., Shindo A., et al. Association between clonal hematopoiesis and left ventricular reverse remodeling in nonischemic dilated cardiomyopathy. JACC Basic Transl Sci. 2024;9(8):956–967. [Google Scholar]
- 8.Uni M., Masamoto Y., Sato T., et al. Modeling ASXL1 mutation revealed impaired hematopoiesis caused by derepression of p16Ink4a through aberrant PRC1-mediated histone modification. Leukemia. 2019;33:191–204. doi: 10.1038/s41375-018-0198-6. [DOI] [PubMed] [Google Scholar]