Abstract
The roles of actin-binding proteins in development and morphogenesis are not well understood. The actin-binding protein UNC-115 has been implicated in cytoskeletal signaling downstream of Rac in Caenorhabditis elegans axon pathfinding, but the cellular role of UNC-115 in this process remains undefined. Here we report that UNC-115 overactivity in C. elegans neurons promotes the formation of neurites and lamellipodial and filopodial extensions similar to those induced by activated Rac and normally found in C. elegans growth cones. We show that UNC-115 activity in neuronal morphogenesis is enhanced by two molecular mechanisms: when ectopically driven to the plasma membrane by the myristoylation sequence of c-Src, and by mutation of a putative serine phosphorylation site in the actin-binding domain of UNC-115. In support of the hypothesis that UNC-115 modulates actin cytoskeletal organization, we show that UNC-115 activity in serum-starved NIH 3T3 fibroblasts results in the formation of lamellipodia and filopodia. We conclude that UNC-115 is a novel regulator of the formation of lamellipodia and filopodia in neurons, possibly in the growth cone during axon pathfinding.
During axon extension in the developing nervous system, the growth cone at the distal tip of a growing axon detects extracellular cues that provide guidance information. These guidance signals result in modulation of the structure and dynamics of the growth cone actin cytoskeleton, which mediates outgrowth and guidance. Dynamic, actin-based plasma membrane protrusions that control growth cone pathfinding include lamellipodia, in which the actin cytoskeleton assumes a cross-linked and branched meshwork, and filopodia, which consist of parallel bundles of actin filaments protruding from the growth cone or lamellipodial margin (15). To understand growth cone pathfinding, it will be necessary to understand the molecules and mechanisms that control the formation of lamellipodia and filopodia in the growth cone. Rac GTPases regulate the actin cytoskeleton (17) and induce the formation of lamellipodia (38), and Rac loss of function or constitutive activation in Drosophila melanogaster and Caenorhabditis elegans leads to axon pathfinding defects (11, 29, 31). In C. elegans, Rac constitutive activation in neurons induces the formation of ectopic neurites and dynamic lamellipodial and filopodial structures, suggesting that Racs may normally mediate the formation of these structures in the growth cone during axon extension (42).
Rac GTPases control cellular morphogenesis in part by interacting with downstream effectors that control the structure and dynamics of the actin cytoskeleton. One cytoskeletal signaling mechanism downstream of Rac involves Pak, LIM kinase, and the actin-severing protein cofilin, which might be involved in dynamic turnover of actin structures necessary for lamellipodial and filopodial extension and retraction (2, 9, 19, 32, 45). Another involves the Arp2/3 complex (34, 46). The Arp2/3 activator WAVE/Scar is present in a complex with Nck (28), Abi1 (22), Sra-1/PIR121 (26, 41), and Kette/Hem2/NAP1 (5, 20, 24). Rac activity stimulates this complex, causing WAVE/Scar to activate Arp2/3 actin nucleation (12, 22, 27). The WAVE/Scar-Arp2/3 complex pathway might be involved in the formation of the actin meshwork of lamellipodia (35, 41, 43).
The UNC-115 family of actin-binding proteins represents a distinct cytoskeletal signaling mechanism downstream of Rac. C. elegans UNC-115 (30), human abLIM (39), and Drosophila UNC-115 (1) each consist of N-terminal LIM domains and a C-terminal villin headpiece domain (VHD) (Fig. 1A). The VHDs from UNC-115 (42), abLIM (39), and the related human protein dematin (4, 36) all bind to actin filaments. In C. elegans, unc-115 mutants display axon pathfinding defects (30), and UNC-115 acts with RAC-2 Rac in axon pathfinding (42). Furthermore, UNC-115 activity is necessary for the formation of ectopic neurites, lamellipodia, and filopodia induced by activated RAC-2, indicating that UNC-115 acts downstream of RAC-2 in neuronal morphogenesis (42). In another study with C. elegans, UNC-115 was shown to act with CED-10 Rac downstream of the UNC-40 guidance receptor in axon pathfinding and neuronal morphogenesis (16).
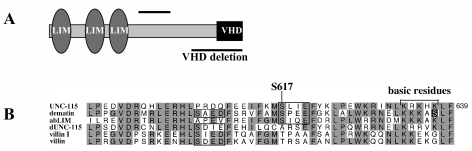
FIG. 1.
The UNC-115 polypeptide and villin headpiece domain. (A) UNC-115 is comprised of three N-terminal LIM domains and a C-terminal VHD. The extent of the VHD deletion is indicated. Bar represents 100 amino acid residues. (B) Alignment of the villin headpiece domains from UNC-115 (GenBank accession number NP509702), human dematin (Q08495), human abLIM (NP006711), Drosophila UNC-115 (dUNC-115) (AAL30429), villin I (NP009058), and villin (A31642) generated by CLUSTALW (3). Residues conserved in at least three out of six molecules are highlighted. Numbering of the UNC-115 residues is based on the long form of UNC-115 encoded by the cDNA with GenBank accession number NP509702. Serine 617 of UNC-115 is indicated, and the conserved putative casein kinase II serine phosphorylation sites are boxed. The serine residue phosphorylated by PKC in the dematin VHD is boxed in black. Also indicated are the conserved basic arginine (R), lysine (K), and histidine (H) residues that, when altered to glutamic acid residues, abolish the actin-binding activity of the UNC-115 VHD.
UNC-115 might directly influence actin cytoskeleton organization and might be involved in the formation of lamellipodial and filopodial structures in the growth cone that mediate growth cone pathfinding. To address this hypothesis, we investigated the role of UNC-115 in neuronal morphogenesis in C. elegans and in actin cytoskeleton organization in cultured fibroblasts. We have found that UNC-115 overactivity in C. elegans neurons and in serum-starved NIH 3T3 fibroblasts induces the formation of lamellipodia and filopodia. Together with previous phenotypic analyses, these data suggest that UNC-115 is a key regulator of lamellipodia and filopodia in the growth cone during axon pathfinding.
MATERIALS AND METHODS
C. elegans genetics and transgenics.
C. elegans was cultured using standard techniques (6, 44). All experiments were performed at 20°C. Wild-type (N2) and unc-115(ky275) mutants were used. Germ line transformation of C. elegans was performed by standard techniques involving injection of a DNA mixture into the syncytial germ line of C. elegans hermaphrodites (33). The DNA mixtures used for injection contained the unc-115 minigenes at 5 ng/μl and the osm-6::gfp transgene (8, 42), used as a cotransformation marker and to score postdeirid neuron (PDE) morphology, at 30 ng/μl.
Construction of the unc-115 minigene and mutants.
Molecular biological and recombinant DNA experiments were performed using standard techniques (40). PCR was used to generate the unc-115 promoter region as well as the unc-115 cDNA region, and primers were used that introduced convenient restriction sites at the ends of the fragments to facilitate cloning. All fragments containing coding regions generated by PCR were sequenced to ensure that no mutations were introduced during the procedure. The unc-115 cDNA::gfp minigene was constructed by cloning the 1.5-kb neuron-specific unc-115 promoter (30) upstream of the unc-115 cDNA fused in frame to gfp at the 3′ end in the pPD95.77 vector (kindly provided by A. Fire) (Fig. 2). The unc-115 cDNA minigene was generated by removing gfp sequences from the unc-115 cDNA::gfp transgene and replacing them with the natural 3′ end of the unc-115 cDNA, including the natural stop codon (Fig. 2). The unc-115 minigenes (gfp tagged and natural 3′ end) were constructed by replacing an internal BglII fragment of the unc-115 cDNA with the same fragment from unc-115 genomic DNA, which included introns 4 to 10 (Fig. 2).
FIG. 2.
Construction of the unc-115 minigene. Boxes represent exons, and lines represent introns or the 5′ promoter region and 3′ downstream region. Open reading frames are represented by black boxes, and 5′ and 3′ untranslated regions are represented by white boxes. The unc-115 cDNA has an SL1 trans-spliced leader at the 5′ end and a poly(A) tail at the 3′ end. The unc-115 cDNA minigene was constructed by placing the 1.5-kb unc-115 promoter upstream of the unc-115 cDNA without SL1 and fusing gfp in frame at the 3′ end of the unc-115 open reading frame. A BglII fragment from the unc-115 genomic region containing introns 4 to 10 was used to replace the equivalent BglII fragment in the unc-115 cDNA minigene to create the unc-115::gfp minigene. Finally, the natural unc-115 stop codon and 3′ untranslated region were used to replace gfp to create the unc-115 minigene. Manipulations described in this work utilized the unc-115::gfp and unc-115 minigenes.
The myr::unc-115 minigene constructs were generated by including the coding sequence for the myristoylation site from human c-Src (MGSSKSK) (23) in the 5′ PCR primer used to generate the unc-115 cDNA fragment. Point mutations and deletions were introduced into the unc-115 minigene constructs by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene, La Jolla, CA) and inverse PCR, respectively. For inverse PCR, primers were designed in a tail-to-tail configuration, with sequences to be deleted between the tails. PCR was performed on a circular construct such that a linear fragment was generated lacking the region between the primer tails. Ligation of restriction sites on the tails of the primers was used to recircularize the constructs, and the primers were designed such that the reading frame was maintained upon recircularization. All coding sequences subjected to inverse PCR as well as the junction sites were sequenced to ensure that no PCR-induced mutations were introduced. Nucleotide sequences of all primers and transgenes generated are available upon request.
Residue numbering of UNC-115 is relative to the long form of UNC-115 encoded by the cDNA with GenBank accession number NP509702. The VHD deletion constructs are missing the C-terminal 188 codons (residues 450 to 639) including the villin headpiece domain. The LIM domain deletion constructs lack codons for residues 20 to 243, including all three LIM domains.
Analysis of PDE neuronal morphogenesis defects.
The PDE neuron was visualized using an osm-6::gfp transgene (8, 42), which was included in the construction of extrachromosomal arrays with the unc-115 minigene constructs. osm-6::gfp on its own causes few defects in PDE morphogenesis or development (see Fig. 4) (42). PDE neurons in adult animals of each strain were assayed for the presence of ectopic neurites emanating from the cell body or axon, for veil-like membrane extensions that resemble lamellipodia, and for thin, finger-like membrane extensions that resemble filopodia. The proportions of PDE neurons in animals harboring the different transgenes that displayed ectopic neurites and ectopic lamellipodial and filopodial structures were determined, and the standard error of the proportion for each was calculated. Data from three or more independent lines were pooled and reported in Fig. 4 and Table 1.
FIG. 4.
Neuronal morphogenesis defects induced by unc-115 transgenic expression. The x axis consists of the unc-115 transgenes studied, and the y axis plots the percentages of PDE neurons in transgenic animals displaying morphogenesis defects (white bars, ectopic neurites; black bars, ectopic lamellipodia and filopodia) (see Fig. 3; also see Materials and Methods for scoring). DVHD, C-terminal deletion including the VHD;−VHD, the transgene harbors the VHD point mutations that block F-actin-binding activity; S617D and S617A, transgenes harbor the mutation of serine 617 to aspartic acid or alanine, respectively; DLIM, the transgene harbors the deletion that removes the LIM domains. The asterisk indicates that the percentage of defects in myr::unc-115(S617A) transgenic animals is probably an underrepresentation due to the high proportion of lethality caused by this transgene. Data presented are averages for pooled data generated by scoring three or more independent lines. Error bars, standard errors of the proportions.
TABLE 1.
Rescue of the uncoordinated phenotype of unc-115(ky275) by unc-115 transgenes
| Transgenea | unc-115(ky275) rescueb |
|---|---|
| None | NA |
| unc-115(+) | ++ |
| myr::unc-115 | ++ |
| myr::unc-115(ΔVHD) | − |
| myr::unc-115(−VHD) | − |
| unc-115(S617A) | ++ |
| myr::unc-115(S617A) | ++ |
| unc-115(S617D) | + |
| myr::unc-115(S617D) | + |
| unc-115(ΔLIM) | + |
| myr::unc-115(ΔLIM) | ++ |
All transgenes were scored on a wild-type background. The transgenes were crossed into an unc-115(ky275) background to score the rescue of the Unc phenotype. All transgenes were derivatives of the unc-115 minigene (see Materials and Methods).
Symbols indicate whether the transgene rescued the uncoordinated locomotion phenotype of unc-115(ky275) null mutants (see Materials and Methods). ++, rescue comparable to that of the wild-type unc-115 transgene; +, discernible rescue but not to the extent of the wild-type unc-115 transgene; −, no discernible rescue; NA, not applicable.
For each transgene tested, at least three independent transgenic lines were generated and found to have similar effects. gfp-tagged versions of each transgene were analyzed to ensure that the UNC-115 molecules were expressed at roughly equal levels as determined by green fluorescent protein (GFP) fluorescence and, in the case of the MYR-tagged molecules, that they were localized to the cell margins. Finally, we ensured that each transgene tested was expressed in the PDE neuron as judged by GFP fluorescence.
To determine if the transgenes rescued the Unc phenotype of unc-115(ky275), the arrays were crossed into the unc-115(ky275) background. The uncoordinated locomotion of unc-115(ky275) animals is very strong and 100% penetrant. Furthermore, unc-115(ky275) uncoordinated locomotion is very distinctive: the animals display strong kinking and coiling and are unable to move backward more than 1/2 to 1 body length. A majority (∼90%) of unc-115(ky275) animals harboring the wild-type unc-115 minigene were able to move backward as well as wild-type animals could (∼2 body lengths without kinking and coiling). We used this as a baseline for scoring the abilities of mutant unc-115 transgenes to rescue the Unc of unc-115(ky275) animals. If a majority of transgenic animals displayed good backward movement (∼2 body lengths without kinking and coiling), the transgene was scored as being able to rescue the Unc phenotype as well as the wild-type unc-115 minigene could. If some transgenic animals (<50%) displayed good backward movement, the transgene was scored as providing some rescue but not not as much as the wild-type transgene. If few or no transgenic animals displayed good backward movement, the transgene was scored as providing no rescuing unc-115 activity.
NIH 3T3 fibroblast cell culture and analysis.
An unc-115::egfp construct was generated by placing the unc-115 cDNA open reading frame in the pEGFP-N1 vector (Clontech) fused in frame to egfp. The transfection and serum deprivation protocol used was based on a previously described protocol (18). Briefly, NIH 3T3 cells grown to approximately 50% confluency on polylysine-coated coverslips were transfected by addition of a mixture of the transgene DNA and the Fugene reagent (Roche, Indianapolis, IN) (1 μg of DNA in 3 μl of solution). After transfection, cells were allowed to grow for 6 h and then deprived of serum (serum-containing medium was replaced by Dulbecco's modified Eagle medium). Cells were allowed to grow for another 12 h and then fixed in 3.7% paraformaldehyde. The fixed cells were incubated with 1 μg/ml of rhodamine-labeled phalloidin, washed three times in phosphate-buffered saline, and mounted in 50% glycerin in phosphate-buffered saline for microscopy to visualize GFP and rhodamine fluorescence. For each transgene, at least three transfections were performed and >100 cells were scored to ensure consistency of results. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays for the detection of apoptosis were performed using the In Situ Cell Death Detection kit with tetramethyl rhodamine red (Roche, Indianapolis, IN).
RESULTS
The unc-115 minigene.
unc-115 minigene constructs were generated as a tool to dissect the molecular mechanisms of UNC-115 activity. The 1.5-kb unc-115 upstream promoter region, which is active in all neurons and embryonic neuroblasts (30), was placed upstream of the full-length unc-115 mingene coding region containing unc-115 introns 4 to 10 (Fig. 2). Two types of minigenes were generated: a version with a gfp coding region (7) fused in frame at the 3′ end (the unc-115::gfp minigene) and a version with the natural unc-115 stop codon and 3′ end (the unc-115 minigene) (see Fig. 2 and Materials and Methods). The unc-115::gfp minigene was expressed robustly and consistently in neurons (data not shown). Both the wild-type and the gfp-tagged minigene (Table 1) rescued the uncoordinated locomotion (Unc phenotype) of the unc-115 null mutation unc-115(ky275), indicating that both constructs produced functional UNC-115 molecules.
UNC-115 transgenic expression dominantly disrupts neuronal morphogenesis.
UNC-115 is a key regulator of axon pathfinding and might act by controlling morphogenetic changes in the growth cone that mediate growth cone outgrowth and steering. To test if UNC-115 regulates neuronal morphogenesis, we assayed the effects of UNC-115 transgenic expression on neuronal development. Transgenic expression can often lead to overexpression and overactivity. We assayed PDE neuron morphology in wild-type animals harboring the unc-115 minigene. The PDEs are a bilaterally symmetric pair of neurons located laterally in the posterior of the animal in the postdeirid ganglion (47) (Fig. 3A). The PDEs extend a single dendrite dorsally, and a single unbranched axon ventrally to the ventral nerve cord (VNC), where the axon bifurcates and extends anteriorly and posteriorly in the VNC (Fig. 3A). The PDE is a useful model for neuronal morphogenesis studies because it has a relatively simple morphology and because the PDE cell body and axon can be unambiguously distinguished from other neurons via osm-6::gfp expression (8, 42). The 1.5-kb unc-115 promoter used to construct the unc-115 minigene is expressed in the PDEs, as is the osm-6::gfp transgene, which was used to visualize PDE morphology in all of these experiments (see Materials and Methods).
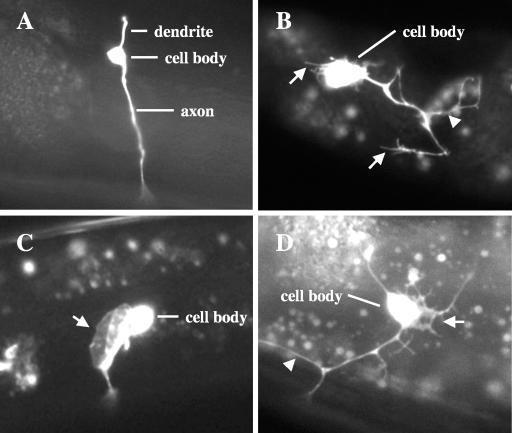
FIG. 3.
PDE neuronal morphogenesis defects induced by MYR::UNC-115. All micrographs are of PDE neurons from young adult animals with osm-6::gfp expression. (A) A wild-type PDE neuron. The dendrite, the cell body, and a single unbranched axon that extends ventrally to the ventral nerve cord are visible. (B and C) PDE neurons from animals expressing MYR::UNC-115. (B) Multiple filopodial structures (arrows) extend from the cell body and neurites, and ectopic neurites are visible (arrowhead). The PDE axon fails to extend to the ventral nerve cord. (C) A large lamellipodial extension from the cell body is indicated by an arrow. The PDE axon extends normally to the ventral nerve cord (out of focus). (D) A PDE neuron from an animal expressing constitutively activated RAC-2(G12V). An ectopic neurite is indicated by an arrowhead, and the arrow indicates a lamellipodial extension with multiple filopodial extensions.
Animals harboring the unc-115::gfp transgene displayed defects in PDE neuron morphogenesis, including formation of ectopic neurites and ectopic plasma membrane extensions from the cell bodies and axons that resembled lamellipodia and filopodia found on C. elegans growth cones (Fig. 3B and C) (25). The ectopic structures induced by UNC-115 were observed in adult animals past the time of normal extension of the PDE growth cone. Even though UNC-115 induced ectopic neuronal structures, in most cases the PDE axon extended to the ventral cord, indicating that UNC-115 did not completely perturb axon outgrowth and guidance. However, PDE axons were sometimes misguided and wandered laterally before reaching the VNC (data not shown). Of PDE neurons with transgenic UNC-115 expression, 9% displayed ectopic neurites and 6% displayed ectopic lamellipodia and filopodia (Fig. 4). Expression of the 1.5-kb promoter alone fused to gfp did not cause these defects (data not shown). While PDE morphology was scored in adult animals, defects in PDE morphogenesis caused by unc-115 expression were observed earlier in larval development, when the PDEs first became visible by osm-6::gfp expression (mid- to late second larval stage).
Membrane-targeted UNC-115 is an activated molecule.
We have previously shown that constitutively active Rac molecules, including RAC-2, cause dominant effects in the PDE neurons (42), including induction of ectopic neurites and lamellipodial and filopodial structures that are dynamic over time (Fig. 3D). The UNC-115-induced structures resembled those induced by activated RAC-2 (compare Fig. 3B and C with Fig. 3D). Previous experiments also indicated that UNC-115 is an actin-binding protein that mediates signaling downstream of the RAC-2 GTPase during neuronal morphogenesis (42). Possibly, UNC-115 is activated in response to Rac signaling during axon pathfinding and neuronal morphogenesis. Like all Rac GTPases, RAC-2 accumulates at the plasma membrane by a prenylation event mediated by a C-terminal CAAX box (42). UNC-115 contains no obvious membrane-targeting sequences, and UNC-115::GFP from the unc-115::gfp minigene showed no discernible plasma membrane localization (Fig. 5A), consistent with previous observations of UNC-115::GFP from the full-length genomic fusion to gfp (30).
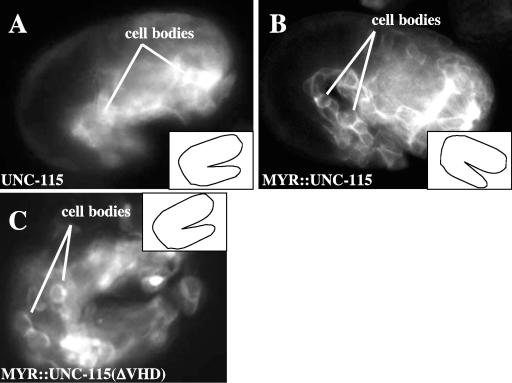
FIG. 5.
MYR::UNC-115 accumulates at cell margins and causes lethality with neuronal morphogenesis defects. Inset line drawings represent the orientation of the 1.5-fold embryo in each micrograph. (A) A 1.5-fold embryo with UNC-115::GFP expression in embryonic blastomeres. UNC-115::GFP was distributed uniformly in the cytoplasm of the cells, and the cell margins are not visible. (B) MYR::UNC-115::GFP accumulated at the plasma membranes of blastomeres of a 1.5-fold embryo, highlighting the margins between the cells. (C) MYR::UNC-115(ΔVHD) expression in a 1.5-fold embryo. Despite lacking the VHD, MYR::UNC-115(ΔVHD) localized to the plasma membrane, highlighting the cell margins in a manner similar to full-length MYR::UNC-115::GFP.
To test if membrane localization is important for UNC-115 function, we generated membrane-targeted unc-115 and unc-115::gfp minigenes containing the myristoylation sequence (Myr) from c-Src fused in frame at the 5′ end of the unc-115 open reading frame (23) (see Materials and Methods). The c-Src Myr sequence directs covalent attachment of a myristoyl moiety which inserts into cellular membranes including the plasma membrane (23) and has been used to analyze membrane localization of heterologous signaling molecules (16, 37). The myr::unc-115::gfp transgene is predicted to produce a full-length UNC-115 molecule with an N-terminal Myr sequence and C-terminal GFP. Indeed, MYR::UNC-115::GFP accumulated at the margins of embryonic blastomeres in transgenic animals (Fig. 5B), demonstrating that the Myr sequence drives MYR::UNC-115::GFP to the plasma membrane. MYR::UNC-115::GFP also accumulated at the plasma membranes of neurons in larval and adult animals (data not shown).
To determine the effects of MYR::UNC-115 on neuronal morphogenesis, we analyzed PDE neurons of animals harboring the myr::unc-115 transgene. We found that MYR::UNC-115 expression caused dominant defects in PDE morphogenesis similar to those caused by wild-type UNC-115 expression, including ectopic neurite formation and induction of lamellipodia and filopodia. However, the dominant effect of MYR::UNC-115 was much stronger than that of wild-type UNC-115 (Fig. 4). Of PDEs from animals harboring the myr::unc-115 transgene, 48% displayed ectopic neurites and 29% displayed ectopic lamellipodial and filopodial structures compared to 9% and 6%, respectively, for wild-type UNC-115. In most cases MYR::UNC-115 did not perturb normal PDE axon outgrowth (Fig. 3C) or cause uncoordinated locomotion. However, the outgrowth of some PDE axons (10%) was affected (Fig. 3B). MYR::UNC-115 caused similar defects in other neurons, including the VD/DD motor neurons and the amphid and phasmid sensory neurons (data not shown), indicating that the effects of MYR::UNC-115 are not specific to the PDE neuron.
Expression of the unc-115 promoter alone or of MYR::GFP, which accumulated at the cell margins as did MYR::UNC-115::GFP, did not cause morphological defects in neurons, indicating that the defects were not caused by the unc-115 promoter sequence or by the myr sequence (data not shown). The myr::unc-115 transgene rescued the uncoordinated locomotion of unc-115 null mutants (Table 1), suggesting that the MYR::UNC-115 molecule is an activated molecule rather than a dominant-negative molecule. Indeed, MYR::UNC-115 caused the formation of neurites, lamellipodia, and filopodia in an unc-115(ky275) mutant background (data not shown). We have previously shown that unc-115 is required in motor neurons for coordinated locomotion and that unc-115 mutants display motor neuron axon pathfinding defects (30). Together, these data indicate that the neurites, lamellipodia, and filopodia induced by MYR::UNC-115 are possibly the results of unregulated UNC-115 activity and that UNC-115 plasma membrane translocation might activate the molecule.
The effects of MYR::UNC-115 require the UNC-115 actin-binding domain.
The neurites, lamellipodia, and filopodia induced by MYR::UNC-115 might be the results of the effects of UNC-115 on the actin cytoskeleton. The VHD of UNC-115 binds to actin filaments. We next tested if the UNC-115 VHD was required for the dominant effects of MYR::UNC-115. The coding region for the C-terminal 188 residues including the VHD was deleted from the myr::unc-115 transgene (Fig. 1A). We found that the VHD-deleted construct caused far fewer defects in PDE morphogenesis than full-length MYR::UNC-115 (Fig. 4). Furthermore, the VHD-deleted construct failed to rescue the uncoordinated locomotion of unc-115(ky275) mutants (Table 1). While the VHD deletion reduced the effect of MYR::UNC-115, the GFP-tagged VHD deletion molecule still accumulated at cell margins (Fig. 5C), indicating that the VHD-mutant molecules were expressed and localized to the plasma membrane.
To examine the role of the VHD in greater detail, we tested the role of conserved basic residues in the C terminus of the VHD required for actin binding (Fig. 1B) (14, 42). Point mutations that changed the conserved basic VHD residues to glutamic acid residues, which abolishes actin filament binding of the UNC-115 VHD (42), were introduced into the myr::unc-115 transgenes. We found that these mutations significantly reduced the ectopic neurites induced by MYR::UNC-115 but did not reduce formation of lamellipodia and filopodia (Fig. 4). Further, the mutant molecule accumulated at the plasma membrane (data not shown). These results indicate that the actin-binding site consisting of the conserved basic residues in the VHD is required for the full effect of MYR::UNC-115 and might contribute to neurite formation. However, a region encompassed by the VHD deletion but left unaltered by the point mutations might mediate formation of lamellipodia and filopodia. In fact, the VHD-containing protein dematin has two F-actin-binding sites, one in the VHD and another in the N-terminal “undefined” region (4, 30). Possibly, UNC-115 also contains a second actin-binding site which is unaltered by the VHD point mutations but is removed by the VHD deletion. While the VHD point mutations did not completely eliminate the dominant effects of MYR::UNC-115, they did abolish the rescue of unc-115(ky275) uncoordination (Table 1).
Mutations in the putative serine phosphorylation site in the UNC-115 villin headpiece domain affect UNC-115 function.
Phosphorylation regulates the activity of the VHD-containing protein dematin. Phosphorylation of the dematin VHD by protein kinase C (PKC) blocks the actin-bundling activity but not the F-actin-binding activity of the molecule (4, 21). While this PKC site is not conserved in the abLIM and UNC-115 VHDs (Fig. 1B), abLIM is phosphorylated in vivo (39). Thus, phosphorylation might also control the activities of abLIM and UNC-115. A comparison of the UNC-115 polypeptide sequence with abLIM and dematin revealed a conserved casein kinase II serine phosphorylation site (serine 617 [S617] in UNC-115) in the VHD regions of these molecules (Fig. 1B). The position of this site was not conserved in Drosophila UNC-115 (dUNC-115) or in the other VHD-containing proteins villin and villin I. However, a similar putative serine phosphorylation site was found in dUNC-115, villin, and villin I, as well as in dematin, 11 residues N-terminal to the S617 site in UNC-115 (Fig. 1B). To test if the putative S617 phosphorylation site is important for UNC-115 function, we altered the S617 residue in both the wild-type unc-115 minigene and the myr::unc-115 minigene and assayed the effects of these transgenes on PDE morphogenesis and unc-115(ky275) rescue.
We first changed S617 to an aspartic acid residue (S617D) to mimic the negative charge introduced by phosphorylation at the site. UNC-115(S617D) showed levels of PDE neuronal defects similar to those with wild-type UNC-115 (Fig. 4). However, the S617D mutation greatly reduced the effects of membrane-targeted MYR::UNC-115, including ectopic neurites, lamellipodia, and filopodia (Fig. 4). Both S617D mutant molecules were expressed, as determined by the GFP tag, and the Myr-tagged molecules accumulated at the plasma membrane (data not shown). While the S617D mutation reduced the dominant effects of MYR::UNC-115, it did not completely abolish unc-115 activity; both unc-115(S617D) and myr::unc-115(S617D) partially rescued the Unc phenotype of unc-115(ky275) animals (Table 1).
We next determined the effect of altering S617 to alanine, which might block the ability of this site to be phosphorylated. In transgenic animals, UNC-115(S617A)::GFP was expressed and was located throughout the cytoplasm and processes of neurons and embryonic blastomeres (data not shown). UNC-115(S617A) had little effect on PDE morphogenesis compared to wild-type UNC-115 (Fig. 4). However, unc-115(S617A) efficiently rescued the uncoordinated locomotion of unc-115(ky275) animals (Table 1), indicating that a functional molecule was produced.
The S617A mutation had a strong effect when included in the myr-tagged transgene. Animals harboring the myr::unc-115(S617A) transgene gave rise to an average of 35% dead embryos and larvae with severe morphological defects. The dead larvae displayed incomplete elongation and a lumpy body shape (Fig. 6A and B). Furthermore, some dead embryos displayed defects in tissue organization, possibly in gastrulation and epiboly (data not shown). In arrested larvae expressing GFP-tagged MYR::UNC-115(S617A), we observed neurons with severely defective morphology. Figure 6C shows VD and DD motor neurons (47) from an animal with myr::unc-115(S617A)::gfp expression. The cell bodies failed to migrate to their final positions at the ventral nerve cord and stayed in a lateral position; moreover, the cell bodies were greatly enlarged, with extensive lamellipodial and filopodial protrusions, so that they were morphologically unrecognizable as neurons. Membrane accumulation of myr::unc-115(S617A)::gfp was visible in these cells (Fig. 6C). We speculate that MYR::UNC-115(S617A)-induced lethality is caused by expression in cells other than neurons, possibly in the hypodermal cells required for embryonic and larval morphogenesis or the pharyngeal cells necessary for feeding. While the 1.5-kb unc-115 promoter is active primarily in neurons and we cannot detect gfp expression in nonneuronal tissues, low levels of expression of the transgenes in other cells remains a possibility.
FIG. 6.
Lethality and neuronal morphogenesis defects induced by MYR::UNC-115(S617A). (A) Differential interference contrast image of a wild-type L1 larva. Note the smooth body shape. (B) Differential interference contrast image of an L1 larva with myr::unc-115(S617A)::gfp expression. The animal displayed irregular and lumpy body morphology, incomplete elongation, and uncoordinated locomotion. Arrows point to body morphology abnormalities (a lump near the head and a knobbed tail). (C) An L2 larval animal with full-length MYR::UNC-115(S617A)::GFP expression in motor neurons. These neurons, of the VD/DD class, had severely defective morphology. Instead of migrating ventrally after birth and extending neurites, the neurons displayed a spread morphology accompanied by large membrane extensions that resembled lamellipodial and filopodial extensions (arrows). MYR::UNC-115(S617A)::GFP accumulated at the margins of these cells (indicated by arrowheads).
We scored PDE morphogenesis defects in animals that survived to adulthood (Fig. 4). Seventeen percent of PDE neurons displayed ectopic neurites, and 12% displayed lamellipodia and filopodia. This is a lower precentage of defects than that for myr::unc-115 without the S617A mutation. Possibly, the high proportion of lethality is masking the true extent of defects in the PDE neuron (i.e., we were unable to score PDEs for the most severely affected animals, which died).
myr::unc-115(S617A) rescued the uncoordinated phenotype of unc-115(ky275) in animals that survived to adulthood (Table 1). While animals harboring the myr::unc-115(S617A) transgene were sometimes uncoordinated, unc-115(ky275) animals displayed a very strong and characteristic kinking and coiling uncoordination which was qualitatively distinct from the milder myr::unc-115(S617A)-induced uncoordination (see Materials and Methods). These data suggest that MYR::UNC-115(S617A) can compensate for a loss of endogenous UNC-115 activity and that the molecule does not have a dominant-negative effect. MYR::UNC-115(S617A)::GFP showed expression and accumulation at the cell margins (Fig. 6C and data not shown), indicating that the molecule was expressed and properly localized.
While we have no direct evidence of phosphorylation at S617, these phosphomimetic mutations suggest that phosphorylation of S617 might repress UNC-115 activity. Interestingly, the S617A mutation was effective only in the membrane-targeted MYR::UNC-115 molecule, suggesting that UNC-115 must be at the plasma membrane to be activated by the S617A mutation.
The UNC-115 LIM domains are required for UNC-115 function.
Previous studies indicate that the UNC-115 LIM domains mediate localization to cell margins in nonneuronal cells (30). Furthermore, expression of the mouse abLIM LIM domains alone leads to dominant defects in neuronal development (13). To determine the role of the LIM domains in UNC-115 activity, we constructed unc-115 transgenes in which coding regions for the LIM domains were deleted and assayed the effects of these transgenes on PDE development. Deletion of the LIM domains from the wild-type unc-115 minigene and the myr::unc-115 transgene reduced their effects on neuronal morphogenesis (Fig. 4). GFP-tagged versions of both molecules were expressed, and MYR::UNC-115(ΔLIM)::GFP accumulated at cell margins (data not shown). While the LIM domains were required for the full activity of activated UNC-115 molecules, the LIM domain deletion did not completely abrogate UNC-115 activity: both UNC-115(ΔLIM) and MYR::UNC-115(ΔLIM) rescued the uncoordinated phenotype of unc-115(ky275) mutants (Table 1).
If the role of the LIM domains were simply to mediate UNC-115 plasma membrane translocation, we would expect that membrane targeting by the Myr sequence could fully restore the ectopic neurites, lamellipodia, and filopodia induced by the activated molecule. The fact that the LIM domain-deleted version of the Myr-tagged molecule caused less-severe defects than the intact version indicates that this is not the case. The LIM domains are likely to play a role in UNC-115 regulation outside of plasma membrane localization.
UNC-115 alters cell morphology and actin cytoskeleton structure in fibroblasts.
The above data suggest that activated UNC-115 can induce ectopic lamellipodial and filopodial structures in neurons. Possibly, UNC-115 is a key regulator of the actin cytoskeleton involved in formation of lamellipodia and filopodia. To test if UNC-115 can induce bona fide lamellipodia and filopodia, we expressed C. elegans UNC-115 fused to enhanced GFP (EGFP) in serum-starved NIH 3T3 fibroblasts and assayed actin cytoskeleton structure and cell morphology. As expected, untransfected or egfp-transfected serum-starved cells displayed a spread morphology with obvious actin stress fibers, antiparallel bundles of actin filaments that traverse the cell and provide stability and adhesion (Fig. 7A, B, and C). These cells also displayed a thin layer of actin around the cortex of the cell that defines the cell margin. Cells transfected with unc-115::egfp frequently (68%) displayed a rounded morphology compared to untransfected cells or egfp-transfected cells (Fig. 8D, E, and F). Furthermore, nearly all of the unc-115::egfp-transfected cells with a rounded morphology exhibited large conglomerates of F-actin at the cell periphery (Fig. 7E) not seen in untransfected or egfp-transfected cells. Accompanying peripheral actin accumulation in unc-115::egfp-transfected cells was a loss of stress fibers (Fig. 7D). At a lower frequency (2%), cells expressing UNC-115::EGFP displayed plasma membrane extensions that resembled lamellipodia and filopodia (Fig. 7G, H, and I). These data suggest that UNC-115::GFP caused loss of stress fibers and accumulation of F-actin in bundles at the cell periphery and, at a lower frequency, induced the formation of lamellipodia and filopodia.
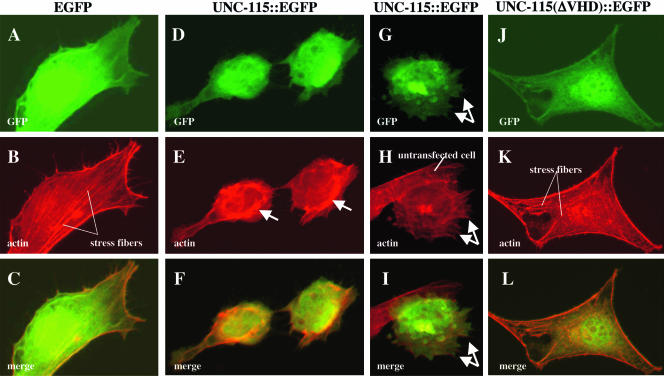
FIG. 7.
UNC-115 causes actin rearrangement in cultured fibroblasts. All panels are micrographs of serum-starved NIH 3T3 fibroblasts taken at the same plane of focus. (Top panels) GFP fluorescence; (center panels) the actin cytoskeleton visualized by rhodamine-phalloidin fluorescence; (bottom panels) merged images. (A through C) Cell transfected with egfp; (D through I) cells transfected with unc-115::egfp; (J through L) cell transfected with unc-115(ΔVHD)::egfp lacking the actin-binding villin headpiece domain. Arrows in panel E indicate abnormal peripheral actin bundles. Arrows in panels G, H, and I indicate lamellipodial and filopodial structures. An untransfected cell indicated in panel H shows normal fibroblast morphology (spread appearance with stress fibers and no lamellipodia or filopodia).
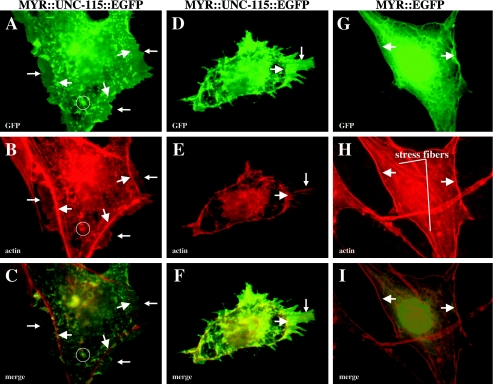
FIG. 8.
MYR::UNC-115 induces lamellipodia and filopodia in cultured fibroblasts. (A, D, and G) GFP fluorescence; (B, E, and H) rhodamine-phalloidin staining to visualize the actin cytoskeleton; (C, F, and I) merged images. Large arrows point to F-actin at the cell cortex that defines the cell margin. Small arrows in panels A, B, and C point to lamellipodia that extend past the cell margin. Small arrows in panels D, E, and F point to a lamellipodial protrusion with bundled actin filaments that extend into filopodia. A cytoplasmic punctum of MYR::UNC-115::EGFP is circled in panels A, B, and C.
UNC-115::EGFP was distributed throughout the cytoplasm and was not strictly colocalized with F-actin (Fig. 7B). Similarly, in transgenic C. elegans, UNC-115::GFP was not localized to any particular cellular structure but was found throughout the cytoplasm and processes of neurons (Fig. 5A). The fact that UNC-115::EGFP did not strictly colocalize with F-actin suggests that the association of UNC-115 with actin is not constitutive and might be under cellular control.
In C. elegans neurons, MYR::UNC-115 strongly induced lamellipodial and filopodial structures. To determine if MYR::UNC-115 had a similar effect in starved fibroblasts, we transfected cells with MYR::UNC-115::EGFP, which accumulated in part on the plasma membrane. MYR::UNC-115::EGFP also accumulated in cytoplasmic conglomerates not observed with wild-type UNC-115 (Fig. 8B). Actin was not strictly colocalized with these cytoplasmic structures (Fig. 8A, B, and C). These structures are likely to be membranous organelles, such as endosomal vesicles or the endoplasmic reticulum, at which the Myr site of Src can also cause accumulation (37). Cells transfected with MYR::UNC-115::EGFP often (24%) displayed striking morphological changes, including formation of lamellipodia and filopodia (Fig. 8), similar to the rare events noted with UNC-115::EGFP. In many cases, cells retained a spread morphology with some stress fibers but also displayed lamellipodia, veil-like protrusions that extended past the marginal actin border, not seen in untransfected cells (Fig. 8A, B, and C). Some MYR::UNC-115::EGFP-transfected cells displayed a rounded morphology with few or no stress fibers (4%). These cells often had multiple lamellipodial protrusions containing bundles of actin filaments that extended into filopodial protrusions (Fig. 8D, E and F).
To ensure that these effects were due to UNC-115 activity and not the myristoylation sequence, we transfected cells with MYR::EGFP. These cells resembled nontransfected cells; they displayed a spread morphology and stress fibers, and they lacked lamellipodial and filopodial protrusions (Fig. 8G, H, and I). MYR::EGFP accumulated at the cell membrane and in cytoplasmic networks distinct in appearance from the cytoplasmic puncta observed in MYR::UNC-115::EGFP-expressing cells (compare Fig. 8B and H). MYR::EGFP also accumulated around the nucleus. The distinct patterns of MYR::EGFP and MYR::UNC-115::EGFP could be due to interactions of UNC-115 with other molecules that restrict its localization to puncta.
To determine if the actin-binding VHD of UNC-115 is required for its morphological effects, we transfected cells with wild-type and Myr-tagged UNC-115(ΔVHD)::EGFP, which lacks the VHD and other C-terminal residues (Fig. 1A). While both VHD deletion transgenes were expressed at levels comparable to those of their intact counterparts, as determined by GFP fluorescence (compare Fig. 7D and J), the morphological change induced by these transgenes was dramatically reduced: UNC-115::EGFP caused 68% (n = 137) rounded morphology compared to 41% (n = 97) for UNC-115(ΔVHD)::EGFP, and MYR::UNC-115::EGFP caused 24% (n = 157) formation of lamellipodia and filopodia compared to 13% (n = 184) for MYR::UNC-115(ΔVHD)::EGFP. Furthermore, the morphological change induced by the VHD-deleted constructs was less severe than that induced by the intact molecules. Figure 7J, K, and L show a cell transfected with UNC-115(ΔVHD)::EGFP that resembled those transfected with EGFP alone. The cell displayed a spread morphology and stress fibers. These data suggest that the effects of UNC-115 in starved fibroblasts are in part mediated by the C-terminal region containing the VHD. The fact that the effects were not completely blocked by the C-terminal deletion suggests that other regions of UNC-115 can participate in actin rearrangement and formation of lamellipodia and filopodia.
It is possible that the observed actin rearrangements and morphological changes were due to UNC-115::EGFP-induced programmed cell death rather than being a direct effect of UNC-115::EGFP on cytoskeletal organization. We assayed programmed cell death in transfected cells using the TUNEL assay (see Materials and Methods) and found no effect (data not shown), indicating that UNC-115::EGFP did not induce programmed cell death. Our results with serum-starved fibroblasts indicate that UNC-115 can induce the formation of lamellipodia and filopodia when targeted to the plasma membrane.
DISCUSSION
The roles of actin-binding proteins, including those of the UNC-115/abLIM family, in development and morphogenesis are not well understood. While UNC-115 has been shown to affect axon pathfinding downstream of Rac signaling (30, 42), very little is known about the cellular and developmental mechanisms of UNC-115 activity in axon pathfinding. Results reported here provide the first evidence of the cellular role of the UNC-115/abLIM family of actin-binding proteins in developmental morphogenesis. Our results indicate that UNC-115 abLIM is a new regulator of formation of lamellipodia and filopodia. We showed that membrane-targeted UNC-115 dominantly induces ectopic neurites and lamellipodial and filopodial structures in neurons that resemble lamellipodia and filopodia in C. elegans growth cones (25). The actin-binding domain and LIM domains of UNC-115 were shown to be necessary for these effects. Furthermore, we showed that C. elegans MYR::UNC-115 induces lamellipodia and filopodia in NIH 3T3 fibroblasts. Together with previous results that show that unc-115 mutants have defects in axon pathfinding and that UNC-115 acts downstream of Rac GTPase signaling, these findings suggest that UNC-115 directs cytoskeletal change in the growth cone during axon pathfinding that lead to the formation of growth cone lamellipodia and filopodia, which underlie axon outgrowth and guidance.
Membrane targeting activates UNC-115.
Our results indicate that plasma membrane targeting of UNC-115 activates the molecule, as MYR::UNC-115 dominantly induces the formation of ectopic neurites and lamellipodial and filopodial structures. Three lines of evidence indicate that MYR::UNC-115 is activated and is not a dominant-negative molecule: (i) the defects caused by MYR::UNC-115 do not resemble unc-115 loss-of-function mutations, which cause some ectopic neurite formation but do not display ectopic lamellipodia and filopodia (30); (ii) MYR::UNC-115 expression rescues the Unc phenotype of the null unc-115(ky275) mutation, which would not be expected if MYR::UNC-115 were a dominant-negative molecule; and (iii) deletion of the actin-binding villin headpiece domain abolishes the dominant effects of the molecule, indicating that actin-binding activity is required for the dominant effects of MYR::UNC-115 in C. elegans. While MYR::UNC-115 caused the formation of ectopic neurites, lamellipodia, and filopodia, it did not strongly affect the outgrowth of the normal PDE axon or cause uncoordinated locomotion. This suggests that MYR::UNC-115 does not interfere with endogenous UNC-115 in PDE axon extension and that overactivity of UNC-115 results in ectopic neuronal structures.
It is possible that the effects of MYR::UNC-115 do not reflect the endogenous role of the molecule in neuronal development. For example, MYR::UNC-115 might ectopically affect a process in which wild-type UNC-115 is not involved. Several lines of evidence suggest that this is not the case and that MYR::UNC-115 reflects endogenous UNC-115 activity. First, overexpression of wild-type UNC-115 caused defects similar to those induced by MYR::UNC-115 but at a lower frequency (Fig. 4). Second, the defects induced by MYR::UNC-115 were similar to those induced by activated Rac in neurons, and UNC-115 was previously shown to mediate some of the effects of activated Rac (42). Third, forms of MYR::UNC-115 missing specific domains (the VHD and the LIM domains) were less active than full-length MYR::UNC-115 (Fig. 4), indicating that MYR::UNC-115 is not nonspecifically perturbing an ectopic process. Finally, MYR::UNC-115 rescued the uncoordinated locomotion of unc-115(ky275) animals (Table 1), indicating that MYR::UNC-115 can compensate for loss of endogenous UNC-115 activity. Together, these results suggest that MYR::UNC-115 can compensate for loss of endogenous UNC-115 activity and drive formation of ectopic neurites, lamellipodia, and filopodia because of overactivity.
Possibly, other molecules present only at the plasma membrane interact with UNC-115 and activate it upon translocation to the membrane. The LIM domains are required for UNC-115 activity even when UNC-115 is driven to the plasma membrane, indicating that the LIM domains might have a role in UNC-115 function besides mediating plasma membrane translocation.
Mutations at serine 617 modulate MYR::UNC-115 activity.
We have found that mutation of S617 alters the activity of UNC-115 in the neuronal morphogenesis and rescue assays. S617 is part of a predicted casein kinase II phosphorylation site in the VHD of UNC-115 that is conserved in the VHDs of human abLIM and dematin. We found that mutation of S617 to aspartic acid (S617D), which can mimic the negative charge of phosphorylation of the site, reduced the ability of MYR::UNC-115 to form neurites, lamellipodia, and filopodia. Furthermore, we found that alteration of S617 to an alanine (S617A) in MYR::UNC-115, which eliminates the potential of the site to be phosphorylated, led to lethality and severe neuronal morphogenesis defects, consistent with the idea that the S617A mutation is an activating mutation. While we have no direct biochemical evidence that S617 is subject to phosphorylation, these data suggest that phosphorylation of S617 represses UNC-115 activity. Furthermore, these studies indicate that MYR::UNC-115 activity can be modulated, suggesting that MYR::UNC-115 is not nonspecifically perturbing an ectopic process and that UNC-115 activity might be under cellular control.
The S617A mutation was effective only in the context of MYR::UNC-115 and had no effect when introduced into wild-type UNC-115. It is possible that UNC-115 must be at the plasma membrane in order for S617 dephosphorylation to activate the molecule. Other molecules that interact with dephosphorylated UNC-115 might be present at the plasma membrane. Furthermore, the S617A mutation did not cause UNC-115::GFP to be localized at the plasma membrane (data not shown), indicating that S617 phosphorylation and membrane translocation are independent events. One potential model is that when UNC-115 is translocated to the plasma membrane, a phosphatase present at the plasma membrane dephosphorylates UNC-115, allowing it to interact with other molecules at the plasma membrane to drive formation of lamellipodia and filopodia. It is also possible that the S617A mutation causes MYR::UNC-115 to interfere with a process in which endogenous UNC-115 is not involved. However, MYR::UNC-115(S617A) and UNC-115(S617A) were both able to rescue the Unc phenotype of unc-115(ky275) animals, indicating that they can compensate for loss of endogenous UNC-115 activity and that their activities might reflect wild-type UNC-115 activity.
MYR::UNC-115 stimulates formation of lamellipodia and filopodia.
Our results show that UNC-115 activity can induce the ectopic formation of neurites and lamellipodial and filopodial structures in neurons. Lamellipodia and filopodia form in part due to rearrangements of the actin cytoskeleton. Lamellipodia consist of a peripheral meshwork of cross-linked and branched actin filaments at the leading edge of a migrating cell or growth cone, whereas filopodia consist of parallel bundles of actin filaments that protrude from the periphery of a migrating cell or growth cone (10, 15). Taking our results together with previous studies showing that unc-115 mutants have defects in axon pathfinding, we suggest that UNC-115 is involved in actin rearrangements underlying the formation of lamellipodia and filopodia in the growth cone during axon pathfinding.
The C terminus of UNC-115 including the actin-binding VHD was required for induction of neurites, lamellipodia, and filopodia. However, we found that specific alteration of the known actin-binding residues in the UNC-115 VHD reduced ectopic neurite formation but had little effect on the formation of lamellipodia and filopodia, suggesting that UNC-115 might contain additional actin-binding sites and/or serve as a scaffold for the recruitment of other actin-binding proteins.
We have found that transgenic UNC-115 expression in serum-starved NIH 3T3 cells caused morphological changes. Wild-type UNC-115 caused formation of peripheral actin conglomerates at the expense of stress fibers and a low frequency of formation of lamellipodia and filopodia, and MYR::UNC-115 strongly induced lamellipodia and filopodia. While formation of lamellipodia and filopodia was more robust with MYR::UNC-115::EGFP, these structures were formed at a lower frequency with wild-type UNC-115::EGFP, indicating that formation of lamellipodia and filopodia is not due to an ectopic activity mediated by the MYR sequence but indeed reflects a role of wild-type UNC-115 activity. The different effects of UNC-115 and MYR::UNC-115 could simply be due to differing levels of activity of the two molecules. Alternatively, they might reveal different activities of the molecules. Possibly, UNC-115 in the cytoplasm inhibits stress fiber formation and UNC-115 at the plasma membrane induces formation of lamellipodia and filopodia. The fact that the C terminus of UNC-115 containing the actin-binding VHD was required for the full effect suggests that UNC-115 might directly interact with actin filaments to control morphogenesis. However, UNC-115 molecules with the C-terminal deletion retained some ability to induce morphological change, suggesting that UNC-115 might contain additional regions that participate in actin rearrangement and lamellipodial and filopodial formation. In any case, the interaction of UNC-115 with actin might be under cellular regulation, as UNC-115::EGFP and MYR::UNC-115::EGFP distribution was not strictly coincident with actin in transfected fibroblasts.
The VHD-containing molecule dematin, which contains two actin-binding sites, causes actin cross-linking and bundling (4), and we show evidence here that UNC-115 might have more than one actin-interacting domain (the VHD point mutations did not completely abolish the morphogenetic ability of activated UNC-115 in C. elegans neurons, whereas deletion of the C-terminal region did). While previous studies show that the point mutations abolish F-actin binding (42), it is possible that the second actin-binding site was not detected in this assay. The second binding site might not be entirely included in the C-terminal deleted region.
UNC-115 and Rac signaling in axon pathfinding.
Previous results show that UNC-115 is required for proper axon pathfinding and that UNC-115 acts with Rac signaling in C. elegans axon pathfinding. Furthermore, UNC-115 activity is necessary for the formation of lamellipodial and filopodial structures induced by activated RAC-2 in C. elegans neurons, indicating that UNC-115 acts downstream of RAC-2 (30, 42). Results reported here show that UNC-115 can induce the formation in C. elegans neurons of lamellipodial and filopodial structures that are similar in appearance to the structures induced by activated Racs (compare Fig. 3B and C with Fig. 3D). In starved fibroblasts, Rac induces the formation of lamellipodia but little or no formation of filopodia. Our results indicate that UNC-115 induces both lamellipodia and filopodia in starved fibroblasts. Possibly, UNC-115 acts downstream of Rac signaling to mediate formation of lamellipodia and downstream of a distinct signaling network to mediate formation of filopodia (e.g., Cdc42). In C. elegans neurons, Rac activity might induce both lamellipodia and filopodia by regulating the activity of UNC-115. Furthermore, UNC-115 might serve as a scaffold to recruit other actin-modulating complexes involved in formation of lamellipodia and filopodia. Considered together, the data presented here suggest a model in which UNC-115 might be locally recruited to the plasma membrane by activated Rac and subsequently dephosphorylated on serine 617. Dephosphorylated UNC-115 then might interact with other molecules at the plasma membrane to induce formation of lamellipodia and filopodia by reorganization of the actin cytoskeleton.
Acknowledgments
We thank M. Buechner, K. Neufeld, R. Ward, and members of the Lundquist lab for critical reading of the manuscript and discussions; K. Neufeld, S. Quackenbush, and J. Rovnak for assistance with cell culture and transfection; E. Struckhoff and S. Mariano for technical assistance; and A. Fire for gfp vectors.
Some C. elegans strains used in this work were provided by the C. elegans Genetics Center, funded by the National Center for Research Resources. This work was supported by NIH grant NS40945 and NSF grant IBN93192 to E.A.L.
REFERENCES
- 1.Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, G. G. Sutton, J. R. Wortman, M. D. Yandell, Q. Zhang, L. X. Chen, R. C. Brandon, Y. H. Rogers, R. G. Blazej, M. Champe, B. D. Pfeiffer, K. H. Wan, C. Doyle, E. G. Baxter, G. Helt, C. R. Nelson, G. L. Gabor, J. F. Abril, A. Agbayani, H. J. An, C. Andrews-Pfannkoch, D. Baldwin, R. M. Ballew, A. Basu, J. Baxendale, L. Bayraktaroglu, E. M. Beasley, K. Y. Beeson, P. V. Benos, B. P. Berman, D. Bhandari, S. Bolshakov, D. Borkova, M. R. Botchan, J. Bouck, P. Brokstein, P. Brottier, K. C. Burtis, D. A. Busam, H. Butler, E. Cadieu, A. Center, I. Chandra, J. M. Cherry, S. Cawley, C. Dahlke, L. B. Davenport, P. Davies, B. de Pablos, A. Delcher, Z. Deng, A. D. Mays, I. Dew, S. M. Dietz, K. Dodson, L. E. Doup, M. Downes, S. Dugan-Rocha, B. C. Dunkov, P. Dunn, K. J. Durbin, C. C. Evangelista, C. Ferraz, S. Ferriera, W. Fleischmann, C. Fosler, A. E. Gabrielian, N. S. Garg, W. M. Gelbart, K. Glasser, A. Glodek, F. Gong, J. H. Gorrell, Z. Gu, P. Guan, M. Harris, N. L. Harris, D. Harvey, T. J. Heiman, J. R. Hernandez, J. Houck, D. Hostin, K. A. Houston, T. J. Howland, M. H. Wei, C. Ibegwam, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa, H., S. Wakatsuki, A. Ishii, K. Moriyama, Y. Sasaki, K. Ohashi, Y. Sekine-Aizawa, A. Sehara-Fujisawa, K. Mizuno, Y. Goshima, and I. Yahara. 2001. Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat. Neurosci. 4:367-373. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., and D. J. Lipman. 1990. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. USA 87:5509-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azim, A. C., J. H. Knoll, A. H. Beggs, and A. H. Chishti. 1995. Isoform cloning, actin binding, and chromosomal localization of human erythroid dematin, a member of the villin superfamily. J. Biol. Chem. 270:17407-17413. [DOI] [PubMed] [Google Scholar]
- 5.Bogdan, S., and C. Klambt. 2003. Kette regulates actin dynamics and genetically interacts with Wave and Wasp. Development 130:4427-4437. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 8.Collet, J., C. A. Spike, E. A. Lundquist, J. E. Shaw, and R. K. Herman. 1998. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics 148:187-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dan, C., A. Kelly, O. Bernard, and A. Minden. 2001. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J. Biol. Chem. 276:32115-32121. [DOI] [PubMed] [Google Scholar]
- 10.Dickson, B. J. 2002. Molecular mechanisms of axon guidance. Science 298:1959-1964. [DOI] [PubMed] [Google Scholar]
- 11.Dickson, B. J. 2001. Rho GTPases in growth cone guidance. Curr. Opin. Neurobiol. 11:103-110. [DOI] [PubMed] [Google Scholar]
- 12.Eden, S., R. Rohatgi, A. V. Podtelejnikov, M. Mann, and M. W. Kirschner. 2002. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418:790-793. [DOI] [PubMed] [Google Scholar]
- 13.Erkman, L., P. A. Yates, T. McLaughlin, R. J. McEvilly, T. Whisenhunt, S. M. O'Connell, A. I. Krones, M. A. Kirby, D. H. Rapaport, J. R. Bermingham, D. D. O'Leary, and M. G. Rosenfeld. 2000. A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron 28:779-792. [DOI] [PubMed] [Google Scholar]
- 14.Friederich, E., K. Vancompernolle, C. Huet, M. Goethals, J. Finidori, J. Vandekerckhove, and D. Louvard. 1992. An actin-binding site containing a conserved motif of charged amino acid residues is essential for the morphogenetic effect of villin. Cell 70:81-92. [DOI] [PubMed] [Google Scholar]
- 15.Gallo, G., and P. C. Letourneau. 2004. Regulation of growth cone actin filaments by guidance cues. J. Neurobiol. 58:92-102. [DOI] [PubMed] [Google Scholar]
- 16.Gitai, Z., T. W. Yu, E. A. Lundquist, M. Tessier-Lavigne, and C. I. Bargmann. 2003. The netrin receptor UNC-40/DCC stimulates axon attraction and outgrowth through Enabled and, in parallel, Rac and UNC-115/AbLIM. Neuron 37:53-65. [DOI] [PubMed] [Google Scholar]
- 17.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 18.Heo, W. D., and T. Meyer. 2003. Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell 113:315-328. [DOI] [PubMed] [Google Scholar]
- 19.Hing, H., J. Xiao, N. Harden, L. Lim, and S. L. Zipursky. 1999. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell 97:853-863. [DOI] [PubMed] [Google Scholar]
- 20.Hummel, T., K. Leifker, and C. Klambt. 2000. The Drosophila HEM-2/NAP1 homolog KETTE controls axonal pathfinding and cytoskeletal organization. Genes Dev. 14:863-873. [PMC free article] [PubMed] [Google Scholar]
- 21.Husain-Chishti, A., W. Faquin, C. C. Wu, and D. Branton. 1989. Purification of erythrocyte dematin (protein 4.9) reveals an endogenous protein kinase that modulates actin-bundling activity. J. Biol. Chem. 264:8985-8991. [PubMed] [Google Scholar]
- 22.Innocenti, M., A. Zucconi, A. Disanza, E. Frittoli, L. B. Areces, A. Steffen, T. E. Stradal, P. P. Di Fiore, M. F. Carlier, and G. Scita. 2004. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 6:319-327. [DOI] [PubMed] [Google Scholar]
- 23.Kamps, M. P., J. E. Buss, and B. M. Sefton. 1985. Mutation of NH2-terminal glycine of p60src prevents both myristoylation and morphological transformation. Proc. Natl. Acad. Sci. USA 82:4625-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura, T., Y. Kitamura, K. Yonezawa, N. F. Totty, I. Gout, K. Hara, M. D. Waterfield, M. Sakaue, W. Ogawa, and M. Kasuga. 1996. Molecular cloning of p125Nap1, a protein that associates with an SH3 domain of Nck. Biochem. Biophys. Res. Commun. 219:509-514. [DOI] [PubMed] [Google Scholar]
- 25.Knobel, K. M., E. M. Jorgensen, and M. J. Bastiani. 1999. Growth cones stall and collapse during axon outgrowth in Caenorhabditis elegans. Development 126:4489-4498. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, K., S. Kuroda, M. Fukata, T. Nakamura, T. Nagase, N. Nomura, Y. Matsuura, N. Yoshida-Kubomura, A. Iwamatsu, and K. Kaibuchi. 1998. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem. 273:291-295. [DOI] [PubMed] [Google Scholar]
- 27.Kunda, P., G. Craig, V. Dominguez, and B. Baum. 2003. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13:1867-1875. [DOI] [PubMed] [Google Scholar]
- 28.Li, W., J. Fan, and D. T. Woodley. 2001. Nck/Dock: an adapter between cell surface receptors and the actin cytoskeleton. Oncogene 20:6403-6417. [DOI] [PubMed] [Google Scholar]
- 29.Lundquist, E. A. 2003. Rac proteins and the control of axon development. Curr. Opin. Neurobiol. 13:384-390. [DOI] [PubMed] [Google Scholar]
- 30.Lundquist, E. A., R. K. Herman, J. E. Shaw, and C. I. Bargmann. 1998. UNC-115, a conserved protein with predicted LIM and actin-binding domains, mediates axon guidance in C. elegans. Neuron 21:385-392. [DOI] [PubMed] [Google Scholar]
- 31.Luo, L. 2000. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1:173-180. [DOI] [PubMed] [Google Scholar]
- 32.Meberg, P. J. 2000. Signal-regulated ADF/cofilin activity and growth cone motility. Mol. Neurobiol. 21:97-107. [DOI] [PubMed] [Google Scholar]
- 33.Mello, C., and A. Fire. 1995. DNA transformation. Methods Cell Biol. 48:451-482. [PubMed] [Google Scholar]
- 34.Miki, H., and T. Takenawa. 2002. WAVE2 serves a functional partner of IRSp53 by regulating its interaction with Rac. Biochem. Biophys. Res. Commun. 293:93-99. [DOI] [PubMed] [Google Scholar]
- 35.Nozumi, M., H. Nakagawa, H. Miki, T. Takenawa, and S. Miyamoto. 2003. Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J. Cell Sci. 116:239-246. [DOI] [PubMed] [Google Scholar]
- 36.Rana, A. P., P. Ruff, G. J. Maalouf, D. W. Speicher, and A. H. Chishti. 1993. Cloning of human erythroid dematin reveals another member of the villin family. Proc. Natl. Acad. Sci. USA 90:6651-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reuther, G. W., J. E. Buss, L. A. Quilliam, G. J. Clark, and C. J. Der. 2000. Analysis of function and regulation of proteins that mediate signal transduction by use of lipid-modified plasma membrane-targeting sequences. Methods Enzymol. 327:331-350. [DOI] [PubMed] [Google Scholar]
- 38.Ridley, A. J., H. F. Paterson, C. L. Johnston, D. Diekmann, and A. Hall. 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70:401-410. [DOI] [PubMed] [Google Scholar]
- 39.Roof, D. J., A. Hayes, M. Adamian, A. H. Chishti, and T. Li. 1997. Molecular characterization of abLIM, a novel actin-binding and double zinc finger protein. J. Cell Biol. 138:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Steffen, A., K. Rottner, J. Ehinger, M. Innocenti, G. Scita, J. Wehland, and T. E. Stradal. 2004. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Struckhoff, E. C., and E. A. Lundquist. 2003. The actin-binding protein UNC-115 is an effector of Rac signaling during axon pathfinding in C. elegans. Development 130:693-704. [DOI] [PubMed] [Google Scholar]
- 43.Suetsugu, S., D. Yamazaki, S. Kurisu, and T. Takenawa. 2003. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell. 5:595-609. [DOI] [PubMed] [Google Scholar]
- 44.Sulston, J., and J. Hodgkin. 1988. Methods, p. 587-606. In W. B. Wood (ed.), The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Symons, M. 2000. Adhesion signaling: PAK meets Rac on solid ground. Curr. Biol. 10:R535-R537. [DOI] [PubMed] [Google Scholar]
- 46.Welch, M. D., and R. D. Mullins. 2002. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18:247-288. [DOI] [PubMed] [Google Scholar]
- 47.White, J. G., E. Southgate, J. N. Thomson, and S. Brenner. 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. 314:1-340. [DOI] [PubMed] [Google Scholar]