Abstract
The stem cell leukemia (SCL) gene, also known as TAL-1, encodes a basic helix-loop-helix protein that is essential for the formation of all hematopoietic lineages, including primitive erythropoiesis. Appropriate transcriptional regulation is essential for the biological functions of SCL, and we have previously identified five distinct enhancers which target different subdomains of the normal SCL expression pattern. However, it is not known whether these SCL enhancers also regulate neighboring genes within the SCL locus, and the erythroid expression of SCL remains unexplained. Here, we have quantitated transcripts from SCL and neighboring genes in multiple hematopoietic cell types. Our results show striking coexpression of SCL and its immediate downstream neighbor, MAP17, suggesting that they share regulatory elements. A systematic survey of histone H3 and H4 acetylation throughout the SCL locus in different hematopoietic cell types identified several peaks of histone acetylation between SIL and MAP17, all of which corresponded to previously characterized SCL enhancers or to the MAP17 promoter. Downstream of MAP17 (and 40 kb downstream of SCL exon 1a), an additional peak of acetylation was identified in hematopoietic cells and was found to correlate with expression of SCL but not other neighboring genes. This +40 region is conserved in human-dog-mouse-rat sequence comparisons, functions as an erythroid cell-restricted enhancer in vitro, and directs β-galactosidase expression to primitive, but not definitive, erythroblasts in transgenic mice. The SCL +40 enhancer provides a powerful tool for studying the molecular and cellular biology of the primitive erythroid lineage.
One of the central issues facing current biology concerns the molecular mechanisms whereby stem cells give rise to differentiated progeny. Hematopoiesis is the best-characterized stem cell system, and experiments with mice and lower vertebrates have identified a small number of transcription factors, including SCL, LMO-2, and RUNX-1, which are central to the formation and/or behavior of hematopoietic stem cells (HSCs). Tight transcriptional control of these key regulatory genes is frequently critical for their biological functions, but the mechanisms responsible are poorly understood.
The SCL gene encodes a basic helix-loop-helix protein and is expressed in blood, in endothelium, and within specific regions of the central nervous system, a pattern of expression that is highly conserved across vertebrate species from mammals to teleost fish (reviewed in reference 4). Within the hematopoietic system, SCL is expressed in hemangioblasts, HSCs, and a subset of hematopoietic lineages, including both primitive and definitive erythroblasts. Although not required for self-renewal of adult HSCs (9, 46), targeted mutation of the SCL gene has shown that it is essential for the development of all hematopoietic lineages in mice (57, 61) and during murine embryonic stem (ES) cell differentiation (13, 14, 62). However, SCL−/− mouse embryos and ES cells both generate endothelial cells (62, 73), suggesting that SCL is required for lineage commitment to blood cell formation. Consistent with this concept, ectopic expression of SCL during zebra fish development results in excessive formation of hemangioblasts (19). Maintenance of SCL expression is required for normal differentiation along erythroid and megakaryocytic lineages (32, 46), whereas failure to downregulate SCL transcription during T-cell differentiation is associated with T-cell acute lymphoblastic leukemia (T-ALL) (reviewed in reference 4). Current evidence therefore demonstrates that appropriate transcriptional regulation is essential for the biological functions of SCL, and this focuses attention on the mechanisms whereby transcription of SCL itself is initiated and maintained.
Several lines of work have helped define the size of the transcriptional domain necessary for the normal pattern of SCL transcription. A 130-kb human yeast artificial chromosome containing both flanking genes was able to completely rescue the lethal SCL-/- phenotype in mice (66). Consistent with this result, the pufferfish SCL genomic locus gave rise to appropriate expression in transgenic zebra fish (3), and comparisons of SCL flanking genes during vertebrate evolution revealed a limited region of conserved synteny likely to contain regulatory elements responsible for the conserved pattern of SCL expression (21, 24). Analysis of chromatin structure, together with large-scale comparative genomic sequence analysis, has led to identification of SCL enhancers with activity in transfection assays (16, 25) or in transgenic mice (6, 24, 26, 64, 67). These approaches have so far revealed a panel of five enhancers, each of which targets expression to a specific subdomain of the normal pattern of SCL expression and two of which are active in blood and/or endothelial cells.
The +18/19 enhancer is sufficient to direct reporter gene expression to hematopoietic progenitors and endothelial cells during development (64), to the vast majority of long-term-repopulating HSCs from adult bone marrow and fetal liver (65), and to putative hemangioblasts within frog dorsolateral plate mesoderm (26). Expression of an SCL cDNA under the control of this enhancer in transgenic mice selectively rescued the formation of early hematopoietic progenitors in SCL−/− embryos (65), and transgenic mice in which β-geo is driven by this enhancer have been used to identify endoglin as a novel HSC marker (7). Fine mapping demonstrated that the +19 component was sufficient for enhancer activity in transgenic mouse embryos, and biochemical characterization has shown that the +19 element is activated by a novel multiprotein complex containing Fli-1, Elf-1, and GATA-2 (26). These data suggest that this enhancer functions as a nodal point for the integration of signals responsible for establishing the transcriptional program for blood cell development. Interestingly, ES cells in which the +18/19 element was deleted from the endogenous SCL locus were still capable of forming blood cells in vitro and in vivo (23). This observation led to the characterization of a second enhancer (−4 element), which targets expression to endothelial cells and hematopoietic progenitors and which is also bound by Fli-1 and Elf-1 (23).
However, several questions remain. In particular, little is known about the hematopoietic expression of other genes within the SCL locus, and it is unclear whether individual SCL enhancers also regulate the transcription of neighboring genes. Furthermore, the +18/19 and −4 elements target hematopoietic progenitors but not erythroid cells, and yet SCL itself is normally expressed in the primitive and definitive erythroid lineages. As a consequence, we have previously postulated the existence of a separate erythroid enhancer necessary for maintaining SCL expression following erythroid commitment (65). In this paper, we describe the pattern of transcription of SCL and neighboring genes in a panel of hematopoietic cell lines representing multiple lineages. We show that SCL exhibits unexpected coexpression with its downstream neighbor MAP17, suggesting that they share regulatory elements. A systematic survey of histone acetylation throughout the SCL locus resulted in identification of a novel erythroid enhancer 40 kb downstream of SCL exon 1a. This element functions as an erythroid-restricted enhancer in vitro; directs expression to primitive, but not definitive, erythroid cells in vivo; and provides a powerful tool for studying the poorly understood primitive erythroid lineage.
MATERIALS AND METHODS
RT-PCR in human and mouse cells.
Quantitative real-time reverse transcription (RT)-PCR was performed using a 7700 sequence detection system (Applied Biosystems, Foster City, CA) as described by Huntly et al. (34). Primers were located as follows: SIL, from exon 15 to exon 16 in both human and mouse; SCL, from exon 4 to exon 6 for human and from exon 5 to exon 6 for mouse; MAP17, from exon 2 to exon 3 for both human and mouse; SMDR, from exon 1 to exon 2; CYP4A11, from exon 9 to exon 11; Cyp4x1, from exon 8 to exon 9. The sequences of the primers are available from the authors.
The cDNA quality was checked using two control genes: β2-microglobulin (β2 M) and porphobilinogen deaminase (PBGD), as described by Huntly et al. (34). Threshold cycle (CT) values were extracted using the same ΔRn fluorescent threshold of 0.2. In human and mouse cell types, the highest transcript level was obtained for β2M in the EM2 (CT = 14.61) and F4N (CT = 17.53) cell lines. These values were defined as 100% for the human and mouse series of cell lines (Fig. 1B and D), and a CT value of 34 was defined as 0%, that is, no expression detectable (Fig. 1B and D). Transcript levels for other genes and/or cell lines were normalized to these values. The increase was calculated using ΔCT, the difference in CT values between the two genes, and the yield of each PCR primer set was calculated using the following formula: increase (n-fold) = (1 + PCR yield)ΔCT.
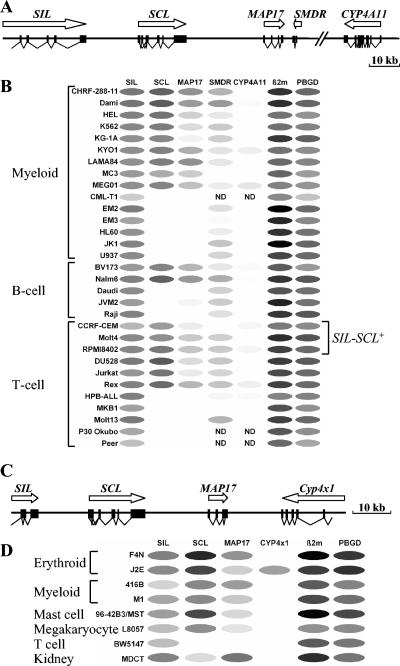
FIG. 1.
Coexpression of SCL and MAP17 in hematopoietic cell lines. (A) Diagram of the human SCL locus. (B) Relative levels of expression of the human SCL and neighboring genes as assessed by quantitative RT-PCR. Expression of β2M in the EM2 cell line is defined as 100% (black oval). Other transcript levels were normalized and are shown as various shades of grey, with white ovals representing 0%. Note the striking correlation between expression of SCL and MAP17. ND, not done. (C) Diagram of the murine SCL locus. (D) Relative levels of expression of murine SCL and neighboring genes. Expression of β2M in F4N is defined as 100% (black oval). Other transcript levels were normalized and are shown as various shades of grey, with white ovals representing 0%.
RT-PCR was performed using an annealing temperature of 56°C for 35 cycles in a 25-μl volume, which contained 0.2 mM deoxynucleoside triphosphate, 1 μM forward primer, 1 μM reverse primer, 10% dimethyl sulfoxide, 1.5 mM MgCl2, and 1 U of AmpliTaq Gold (Applied Biosystems). Details of primers are available on request.
Cell lines and primary tissues.
Cell lines were as described in the DSMZ German cell line bank (http://www.dsmz.de and reference 10), by Green (27), or as follows: CHRF-288-11 (18), Dami (29), KYO-1 (50), MC3 (52), JVM-2 (45), DU.528 (41), Rex (1), MKB-1 (43), P30 Ohkubo (33), J2E (59), L8057 (35), BW5147 (58), RM26 (42), and MDCT (20). All mouse and human cell lines were grown in RPMI 1640 with l-glutamine (Invitrogen, Carlsbad, CA), 10% calf serum (Sigma, St. Louis, MO), and 50 U/ml penicillin-50 μg/ml streptomycin (Invitrogen). Mast cells were cultured from the spleens of (C57BL/6J × CBA/CaJ)F1 adult mice as previously described (28). Cell suspensions were prepared from the thymuses of 6-week-old (C57BL/6J × CBA/CaJ)F1 mice and embryonic day 14.5 (E14.5) livers from (C57BL/6J × CBA/CaJ)F1 embryos by drawing them through an 18-gauge needle.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (17) using anti-diacetylated histone H3, anti-tetra-acetylated histone H4, or nonspecific rabbit immunoglobulin G (IgG) (catalogue numbers 06-599, 06-866, and 12-370, respectively; Upstate, Lake Placid, NY). Detailed protocols are available on request.
Quantitative SyBR green PCR.
Immunoprecipitates were quantified for the genomic content of the SCL locus using SyBR green quantitative PCRs. Primers covered for the mouse a region encompassing the 3′ part of the SIL locus, the full SCL locus, the full MAP17 locus, and a 5′ part of the Cyp4x1 locus. Similarly, a human region encompassing the 3′ part of the SIL locus, the full SCL locus, the full MAP17 locus, and a 5′ part of the CYP4A11 locus was investigated. Primers were tested to reach a sensitivity of 1 pg. The final primer sets are available from the authors. For each final primer set, the yield was calculated. The specificity of the PCR amplification was confirmed by agarose gel electrophoresis. Each mouse primer set was separated on average by 1,012 bp. The minimum and maximum distances between two murine sets were 263 bp and 3,349 bp, respectively. Human primer sets were separated on average by 1,440 bp. The minimum and maximum distances between two human sets were 370 bp and 6,682 bp, respectively. SyBR green PCR was performed on a 7700 sequence detection system (Applied Biosystems), and the CT values were extracted. A ΔCT value was calculated as the difference between the CT value of the rabbit IgG immunoprecipitate and that of the acetylated histone H3 or H4 immunoprecipitate. The enrichment of the immunoprecipitated DNA fragment was calculated using the following formula: increase (n-fold) = (1 + PCR yield)ΔCT.
Stable transfection assays.
The simian virus 40 (SV40) “promoter alone” luciferase reporter construct (SV40/luc) was the pGL2-Promoter plasmid (Promega, Southampton, United Kingdom). The thymidine kinase (TK) “promoter alone” luciferase construct was made by inserting the human herpesvirus TK minimal promoter (equivalent to bases 121 to 252 in GenBank sequence V00470) as a BglII-to-HindIII fragment into the pGL2-Basic plasmid (Promega). The SCL +40 region was amplified by PCR from a genomic DNA template extracted from mouse strain 129-derived ES cells using a proofreading polymerase (Extensor Hi-Fidelity Mix; ABgene, Epsom, United Kingdom). Restriction sites were engineered on either end (BglII and SalI for the SV40 and TK promoter plasmids; SalI and KpnI for the −0.9E3 luciferase plasmid). In each case, the +40 region (equivalent to bases 98649 to 102310 of AJ297131) was inserted into the BamHI/SalI sites situated 3′ of the luciferase gene.
The −0.9E3 “promoter alone” construct (−0.9E3/luc) contained a 3.8-kb mouse genomic DNA fragment extending from 0.9 kb upstream of the SCL transcription start site at exon 1a to exon 3 (corresponding to bases 59449 to 63258 of AJ297131) (64). This fragment was inserted into the BamHI/NheI sites of the pBluescriptII plasmid (Stratagene, La Jolla, CA), with the NheI-to-SalI fragment of pGL2-Basic containing the luciferase gene coding region and SV40 poly(A) (Promega) fused downstream of exon 3 (64). In this case, the +40 region was inserted into the SalI/KpnI sites situated 3′ of the SV40 poly(A) sequence to create the −0.9E3/luc/+40 plasmid.
For in vitro transfection assays of enhancer activity, 107 log-phase growing cells were electroporated (220 V; 900 μF) with 10 μg of linearized luciferase test plasmid and 2 μg of linearized pPGK-neo pA selection plasmid (kindly provided by Lorraine Robb, The Walter and Eliza Hall Institute for Medical Research, Melbourne, Australia). Immediately after electroporation, the cells were divided evenly into four independent pools. After 24 h, the neomycin analogue G418 (Invitrogen, Paisley, United Kingdom) was added to a final concentration of 0.3 mg/ml (F4N and J2E) or 0.8 mg/ml (416B and BW5147). Neomycin-resistant pools were derived 1 to 2 weeks following electroporation, and luciferase assays were performed on extracts from 106 cells per pool essentially as described previously (5). Each experiment was repeated at least three times per cell line. The results are expressed as luciferase activity (relative light units), where the mean of the four pools of the relevant “promoter alone” transfected cells was normalized to 1 and the activities in each of the four pools transfected with the +40-containing plasmid were compared.
Generation and analysis of transgenic mice.
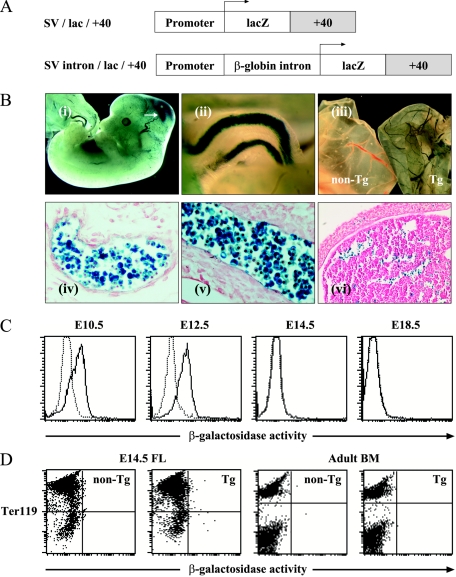
The SV/lac/+40 transgene was constructed from the −0.9E3/lacZ plasmid (64) in the pBluescriptII backbone. It contains the SV40 minimal promoter from pGL2-Promoter (inserted as a BglII/HindIII fragment, after removal of the −0.9E3 promoter as a SpeI/NheI fragment and religation of the remaining vector) and the lacZ coding region and SV40 poly(A), both derived from the −0.9E3/lacZ plasmid. As with the −0.9E3/luc/+40 plasmid, the 3.7-kb +40 region was inserted into the SalI/KpnI sites 3′ of the SV40 poly(A) to create the SV/lac/+40 transgene vector. The SV intron/lac/+40 transgene is equivalent to this, with the exception that it also contained the rabbit β-globin intron from the pSG5 vector (Stratagene; equivalent to bases 395 to 967 of GenBank sequence AF013258) inserted between the SV40 minimal promoter and the lacZ coding region.
The transgene fragment was separated from the plasmid vector by electrophoresis of the NotI-digested plasmid and purified using a QIAquick gel extraction kit (QIAGEN, Crawley, United Kingdom). Transgene DNA was diluted in the supplied EB buffer at 4 μg/ml and spun through an Ultrafree MC sterile centrifuge filter unit (Millipore, Watford, United Kingdom) before injection into the pronuclei of (C57BL/6J × CBA/CaJ)F1 fertilized mouse embryos. The injected embryos were transferred into the oviducts of C57BL/6J pseudopregnant recipient females. The resulting transgenic founders were bred to nontransgenic (C57BL/6J × CBA/CaJ)F1 mice to generate progeny lines. For analysis of embryos or tissues, timed matings between transgenic males and nontransgenic (C57BL/6J × CBA/CaJ)F1 females were performed. The morning on which a vaginal plug was observed was designated E0.5 of gestation. PCR-based genotyping of mice and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining of whole-mount embryos were performed as described previously (47).
Flow cytometric analysis of β-galactosidase activity.
Single-cell suspensions of embryonic peripheral blood and fetal liver were prepared and analyzed for β-galactosidase activity as described previously (49), except that no red cell lysis was done. The data acquisition and analysis were carried out on a FACScalibur using CellQuest software (BD Biosciences Immunocytometry Systems, San Jose, CA). Phycoerythrin-conjugated rat anti-mouse Ter119 and rat IgG2b isotype control antibodies were purchased from BD Biosciences Pharmingen (San Diego, CA). A forward scatter gate excluded debris, whereas dead cells were excluded by propidium iodide (Sigma, St. Louis, MO) uptake. For peripheral blood analyses, between 0.5 × 104 and 2 × 104 viable cells were examined. Similarly, between 2 × 104 and 10 × 104 viable fetal liver cells were analyzed.
RESULTS
Coexpression of SCL and MAP17 in hematopoietic cells.
The genomic organizations of the human and mouse SCL loci are shown in Fig. 1A and C. SCL is flanked by SIL and MAP17 in both species and also in chickens (22, 26). A novel gene called SMDR (SCL-MAP17 downstream RNA) was identified by expressed sequence tag analysis downstream of MAP17 in the human, but not the mouse, sequence. SMDR consists of two exons (225 bp and 472 bp) separated by an intron of 429 bp. No major open reading frame is present, suggesting that SMDR may encode a regulatory or structural RNA. A cytochrome p450 gene cluster lies telomeric to SMDR in humans and to MAP17 in mice. However, the first gene in the human cluster (CYP4A11) is not orthologous to the first gene in the murine cluster (Cyp4x1), an observation that reflects genomic rearrangement within the gene cluster following divergence of humans and mice (24).
In order to clarify the transcriptional activities of genes within the SCL locus in different hematopoietic lineages, quantitative RT-PCR was used to measure transcript levels of SCL and its neighboring genes, together with β2M and PBGD as widely expressed control genes. The results obtained using a large panel of human hematopoietic cell lines are shown in Fig. 1B. SIL was expressed in all cell lines, and there was a significant correlation between the levels of SIL and PBGD (r = 0.77; n = 31). SMDR was also widely expressed, with transcripts detectable in 25 of 28 lines tested. By contrast, CYP4A11 was expressed only at very low levels in 10 out of 29 cell lines. These results are consistent with previous data suggesting that SIL is widely expressed (8, 36) and that CYP4A11 expression is largely restricted to the liver and kidney (2).
MAP17 has been reported to be expressed mainly in renal-proximal tubules (40) and in the skin (37). However, in hematopoietic cells we observed an unexpected and striking concordance between the levels of transcripts derived from SCL and MAP17 (r = 0.80; n = 31). All 17 cell lines expressing SCL also expressed MAP17, and 13 out of the 14 cell lines that were negative for SCL were also negative for MAP17 (Fig. 1B). The only discrepancy observed was in the B-cell line JVM-2, which was negative for SCL but in which very low levels of MAP17 were detected. Levels of SCL transcripts were higher than those derived from MAP17 in all 17 SCL-expressing cell lines (mean difference, 234-fold; median difference, 34-fold; range, 3- to 1,744-fold).
Similar results were obtained using a smaller panel of mouse cell lines (Fig. 1D). SIL was expressed in all eight cell lines, whereas Cyp4x1 was expressed in only a single cell line (J2E). Of seven hematopoietic cell lines, six expressed SCL and all six also expressed MAP17, albeit at a lower level. By contrast, the MDCT line (derived from renal tubule cells) expressed 162-fold more MAP17 transcripts than SCL transcripts. Thus, all 23 human or mouse hematopoietic cell lines that expressed SCL also expressed MAP17, and in each case, SCL transcripts were more abundant than MAP17 transcripts. These results demonstrate that SCL and MAP17 lie within a common transcriptional domain and suggest the existence of regulatory elements that act on both genes in hematopoietic cells.
Identification of a putative SCL enhancer downstream of MAP17.
In order to identify additional enhancers that might contribute to the regulation of SCL or MAP17, a systematic analysis of histone acetylation was performed using several different cell types. PCR primers were designed at approximately 1-kb intervals throughout the SCL locus, and ChIP was then used to assess the acetylation of histones H3 and H4 over a 90-kb region of the murine locus from SIL intron 14 to Cyp4x1 intron 8. Figure 2 summarizes the results obtained with an antibody directed to diacetylated histone H3. A similar pattern of results was obtained with an antibody directed to tetra-acetylated histone H4 (data not shown).
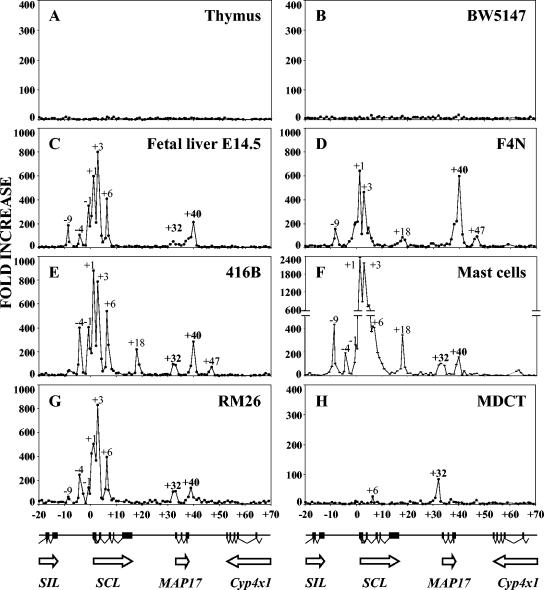
FIG. 2.
Chromatin immunoprecipitation of acetylated histone H3 throughout the murine SCL locus. A diagram of the murine SCL locus is shown beneath each set of four panels. Quantitative PCR was performed using primers located approximately every 1 kb throughout a 90-kb region. The increase over background was calculated by comparison with immunoprecipitates obtained using rabbit IgG. Peaks of H3 acetylation are labeled according to their positions relative to the first nucleotide of SCL exon la. (A) Adult mouse thymus; (B) BW5147 T-cell line; (C) embryonic day 14.5 fetal liver; (D) F4N erythroid cell line; (E) 416B myeloid progenitor cell line; (F) adult spleen mast cells; (G) RM26 myeloid cell line; (H) MDCT kidney cell line.
In the thymus and the T-cell line BW5147, neither of which expresses SCL, acetylation was at background levels throughout the SCL locus (Fig. 2A and B). By contrast, a broadly similar pattern of increased histone acetylation was observed in the vicinity of the SCL and MAP17 genes in five other hematopoietic cell types (Fig. 2C to G). The SCL promoter region and the body of the SCL gene were associated with several peaks of acetylation that correspond to regions previously shown to have promoter or enhancer activity, including SCL promoter 1a and promoter 1b, together with regions located +3 and +7 kb downstream of the beginning of SCL exon 1a (25). Upstream of SCL, most of the hematopoietic cell lines showed acetylation peaks at −4 and −9 kb. These are likely to correspond, respectively, to a previously described endothelial or hematopoietic enhancer at −4 kb (23, 67) and to a DNase I-hypersensitive site at −9 kb, the function of which remains obscure (21, 25, 67). An acetylation peak located between SCL and MAP17 at approximately +18 kb was observed in F4N cells (erythroid), 416B cells (early myeloid), and primary mast cells (Fig. 2D to F). This peak corresponds to the +18/19 enhancer previously shown to target expression to hemangioblasts and to their hematopoietic and endothelial progeny (26, 64, 65). In addition, a peak at +32 kb colocalized with the putative MAP17 promoter and was the major peak observed in the MDCT renal tubule cell line (Fig. 2H), which expresses high levels of MAP17. Smaller peaks at +32 kb were seen in several hematopoietic cell types (Fig. 2C, E, F, and G), consistent with activity of the MAP17 promoter in some hematopoietic cell types.
These results, therefore, did not identify new candidate enhancers within the SCL gene or between it and its immediate flanking genes. However, downstream of MAP17, two acetylation peaks were of particular interest. A peak at +40 kb was identified in all five SCL-expressing hematopoietic cell types, two of which also exhibited a smaller peak at +47 kb. The presence of the +40 and +47 kb peaks was not associated with Cyp4x11 expression, since F4N, 416B, and RM26 do not express detectable levels of Cyp4x11 transcripts (Fig. 1D and data not shown). Moreover, neither peak was necessary for MAP17 expression, since they were not found in the MDCT cell line. The presence of both peaks, therefore, correlated with SCL expression, suggesting that they might represent previously unrecognized SCL enhancers.
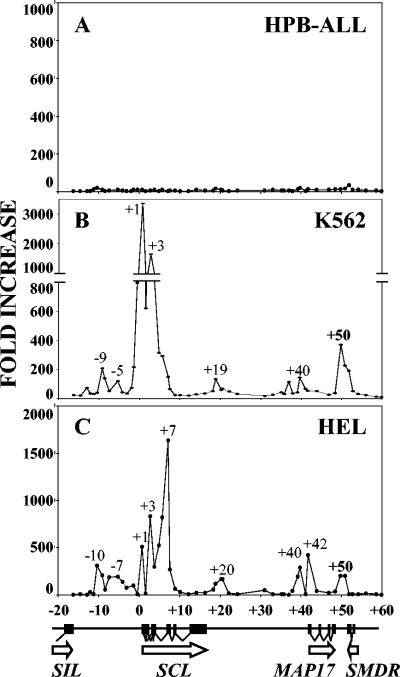
Additional ChIP experiments were performed to investigate whether similar acetylation patterns were observed downstream of MAP17 in human cells (Fig. 3). Experiments were performed using an SCL-negative T-cell line (HPB-ALL) and two primitive myeloid lines (K562 and HEL), both of which express SCL and exhibit erythroid potential. No peaks of acetylation were evident in HPB-ALL, whereas both K562 and HEL exhibited an acetylation peak at +50 kb, a region that corresponds to the mouse +40 region. These results are consistent with the presence of a conserved element that regulates SCL transcription in hematopoietic cells.
FIG. 3.
Chromatin immunoprecipitation of acetylated histone H3 throughout the human SCL locus. A diagram of the human SCL locus is shown at the bottom. Quantitative PCR was performed using primers located approximately every 1 kb throughout an 80-kb region. The increase was calculated by comparison with immunoprecipitates obtained using rabbit IgG. Peaks of H3 acetylation are labeled according to their positions relative to SCL exon 1a. (A) HPB-ALL T-cell line; (B) K562 myeloid-erythroid progenitor cell line; (C) HEL myeloid-erythroid cell line.
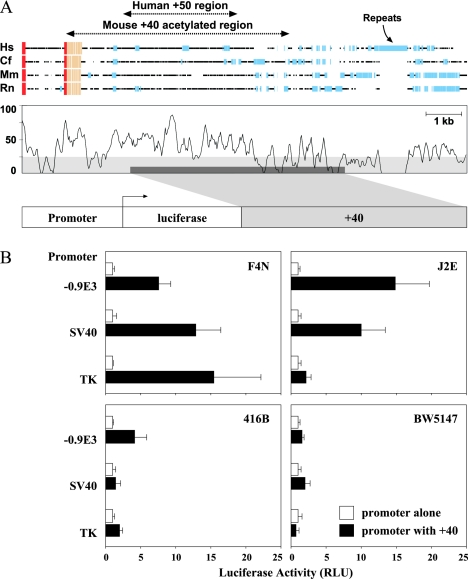
The +40 region functions as an erythroid enhancer in vitro.
A four-way comparison of the human, dog, mouse, and rat sequences downstream of MAP17 is shown in Fig. 4A. The positions of the mouse +40 and human +50 acetylation peaks are indicated and were found to span a region containing several peaks of high sequence homology. A 3.7-kb fragment of mouse DNA spanning the +40 acetylation peak was cloned into a luciferase reporter plasmid containing either the −0.9E3 SCL promoter cassette (64), an SV40 promoter, or a TK promoter. As shown in Fig. 4B, the 3.7-kb fragment increased luciferase transcription from all three promoters in the erythroid cell line F4N and from two of the three promoters in a second erythroid line, J2E. By contrast, little or no activity was observed in primitive myeloid cells (416B) or in a T-cell line (BW5147). These data demonstrate that the +40 region functions as an enhancer in erythroid cells in vitro. Interestingly, 416B cells did contain an acetylation peak at +40 (Fig. 2E), suggesting that the enhancer may be in an open chromatin configuration but that 416B cells lack one or more critical transcription factors necessary for enhancer function.
FIG. 4.
The +40 region is conserved and functions as an erythroid enhancer in vitro. (A) Schematic representation of a four-way sequence alignment of the MAP17 gene downstream region in human (Hs), dog (Cf), mouse (Mm), and rat (Rn) showing peaks of conserved sequence. Red boxes, coding exons; beige boxes, untranslated exons; blue boxes, repeat sequences. The 3.7-kb fragment incorporated into luciferase reporter constructs is shown at the bottom. (B) Stable transfection of hematopoietic cell lines with luciferase reporter constructs. Cell lines representing erythroid (F4N and J2E), myeloid progenitor (416B), or T-lymphoid (BW5147) cell types were transfected with reporter constructs containing various promoters (−0.9E3 SCL promoter, SV40 minimal promoter, or thymidine kinase promoter) with or without the +40 region. The histograms represent the mean (and standard deviation) luciferase activities (in relative light units [RLU]) of four independent pools normalized to the mean result obtained with a construct containing the corresponding promoter alone. The results shown were obtained in a single representative experiment. The same pattern of results was observed in at least three independent experiments for each cell line.
The +40 region directs expression to primitive, but not definitive, erythroid cells in vivo.
In order to assess the activity of the +40 enhancer in vivo, four transgenic mouse lines were generated in which β-galactosidase activity was controlled by the 3.7-kb +40 enhancer. Three lines were generated using a construct containing an intron from the rabbit β-globin gene (SV intron/lac/+40) and one with a construct lacking this intron (SV/lac/+40) (Fig. 5A).
FIG. 5.
The +40 region targets expression to primitive erythroblasts in vivo. (A) Constructs used to generate transgenic mice. (B) Whole-mount (i to iii) and histological (iv to vi) sections of SV intron/lac/+40 transgenic mice (line 4092). (i) Lateral view of an E12.5 embryo. Note β-galactosidase expression in blood vessels and in the midbrain (arrow). (ii) High-power view of umbilical vessels from an E12.5 embryo. (iii) Yolk sacs from E13.5 nontransgenic (non-Tg) and transgenic (Tg) embryos. (iv) Sagittal section through E12.5 yolk sac blood island. (v) E12.5 umbilical vessel. (vi) E12.5 fetal liver. (C) Flow cytometric analysis (FACS) of β-galactosidase activity in circulating erythroid cells of SV intron/lac/+40 transgenic embryos (line 4092). Data from transgenic animals (solid lines) are shown superimposed on those from nontransgenic littermates (dashed lines). Similar results were obtained in at least three mice of each genotype for each time point. Similar data were also obtained at E12.5 and E14.5 in two additional independent transgenic lines (lines 4102 and 4274). (D) FACS analysis of β-galactosidase activity and Ter119 erythroid lineage marker expression in E14.5 fetal liver and adult bone marrow from SV intron/lac/+40 transgenic (Tg) animals (line 4092) and nontransgenic (non-Tg) littermates. The fetal liver results shown are representative of at least three embryos of each genotype, whereas the bone marrow data represent experiments in which at least four mice of each genotype (lines 4092, 4102, and 4274) were analyzed.
Similar patterns of β-galactosidase staining were observed in three of the four transgenic lines (SV intron/lac/+40 lines 4092 and 4102; SV/lac/+40 line 4274), with only ectopic expression in the remaining line. In E12.5 to E13.5 embryos from these three lines, β-galactosidase activity was observed in the midbrain and in blood vessels of the embryo (Fig. 5B, i to iii). Close inspection of whole-mount embryos suggested that the vascular staining reflected β-galactosidase expression in hematopoietic cells within the lumen of the vessels (Fig. 5B, ii). This was confirmed by analysis of histological sections, which showed that the vast majority of hematopoietic cells in the yolk sac blood islands (Fig. 5B, iv) and within the lumen of blood vessels (Fig. 5B, v) expressed β-galactosidase. By contrast, β-galactosidase activity was detected in only a minority of cells in E12.5 livers (Fig. 5B, vi).
During mouse development, circulating erythroid cells are initially generated by a transient cohort of primitive erythroid progenitors originating in the yolk sac between E7 and E9 (55). The resultant primitive erythroid cells are large and nucleated and undergo semisynchronous differentiation, with eventual enucleation while circulating in the bloodstream (15, 39). Smaller enucleated definitive erythrocytes begin to emerge from the fetal liver at E12.5 and thereafter rapidly become the predominant cell type in the circulation.
To further investigate the ability of the +40 element to target the primitive and definitive erythroid lineages, β-galactosidase expression was assessed by flow cytometric analysis (FACS) of peripheral blood cells obtained from three independent lines of transgenic mice at various time points. At E12.5 (and at earlier time points), over 90% of circulating erythroid cells are nucleated primitive erythroblasts, a figure which drops to approximately 4% by E14.5, mainly as a result of a dramatic influx of enucleated definitive erythrocytes into the circulation (39). As shown for line 4092 in Fig. 5C, at E10.5 and E12.5, most circulating erythroid cells expressed β-galactosidase, whereas by E14.5 (and E18.5), virtually all circulating erythroid cells lacked β-galactosidase expression. A similar loss of β-galactosidase expression between E12.5 and E14.5 was seen in lines 4102 and 4274 (data not shown). These results demonstrate that the +40 enhancer is active in circulating primitive erythroblasts. The lack of β-galactosidase expression in enucleated definitive erythrocytes also suggested that the +40 enhancer was unlikely to be active in definitive erythroblasts immediately prior to their enucleation. In order to investigate whether the +40 enhancer was active in definitive erythroblasts, FACS analysis of E14.5 liver was performed. The vast majority (>90%) of liver cells at this time point are Ter119+ definitive erythroblasts, but virtually none of these expressed detectable β-galactosidase in multiple embryos obtained from two independent transgenic lines (lines 4092 and 4274) (Fig. 5D and data not shown). Similarly, Ter119+ erythroid cells in adult bone marrow also lacked detectable expression of β-galactosidase (Fig. 5D). Taken together, these data demonstrate that the +40 enhancer targets β-galactosidase expression to primitive, but not definitive, erythroblasts.
DISCUSSION
In this paper, we describe two new findings. First, we demonstrate unexpected coexpression of SCL and its downstream neighbor MAP17 in hematopoietic cells and thus define a functional transcriptional domain flanked upstream by SIL and downstream by the cytochrome p450 locus. Second, a survey of histone acetylation across the SCL locus has allowed us to identify a new SCL enhancer that directs expression to primitive, but not definitive, erythroblasts in transgenic mice.
Coexpression of SCL and MAP17.
Coexpression of SCL and MAP17 in hematopoietic cells was unexpected, since MAP17 has been reported to be expressed primarily in renal tubular epithelium and in the skin (37, 40). Coregulation of neighboring genes has been reported recently in Caenorhabditis elegans (63), Drosophila (68), and human (44, 60), but in most cases the mechanisms responsible are obscure. Shared enhancers represent one possibility (69), although in the case of the SCL locus none of the SCL enhancers identified so far are able to direct expression to the renal tubular epithelium or to the skin, tissues known to express MAP17. SCL transcripts were more abundant than MAP17 transcripts in all 23 human or murine hematopoietic cell lines representing multiple lineages, implying that any shared enhancer would be likely to interact preferentially with the SCL promoter.
In addition to their coregulation in hematopoietic cells, SCL is expressed in specific regions of the central nervous system and MAP17 is expressed in the renal tubular epithelium and skin. It therefore seems likely that the two genes are controlled by enhancers specific for each gene, as well as by one or more shared regulatory elements. Potential candidates for a shared hematopoietic regulatory element include the three SCL enhancers known to target hematopoietic cells, the +18/19 enhancer (26, 64), the SCL −4 enhancer (23), and the +40 enhancer described in this paper, together with three peaks of sequence homology upstream of MAP17 (24), which may represent additional enhancers.
The boundaries of the SCL-MAP17 transcriptional domain identified here are consistent with previous comparative analyses of vertebrate SCL loci (21). The pattern of SCL expression is highly conserved between mammals, chickens, and zebra fish (19). Comparative genomic sequence analysis has identified a region of conserved synteny that extends in mammals from the 3′ end of the SIL gene to the beginning of the cytochrome p450 locus and which would be expected to contain all regulatory elements responsible for the conserved pattern of SCL expression (21). This region of conserved synteny corresponds to the SCL-MAP17 transcriptional domain described here. In mammals, the latter is flanked upstream by the SIL gene, which is ubiquitously expressed, and downstream by the cytochrome p450 locus, which is not expressed in hematopoietic cells. It is not yet clear whether the functional boundaries of the SCL-MAP17 domain reflect the presence of insulator elements, the specificity of promoters for particular enhancers, or the operation of other mechanisms.
Identification of an SCL erythroid enhancer.
Previous studies have identified two SCL enhancers active in hematopoietic progenitors and endothelium, but neither enhancer was able to direct expression to primitive or definitive erythroid cells (23, 64, 67). Since SCL is expressed in both primitive and definitive erythroblasts (12, 28), we had postulated the existence of a distinct erythroid enhancer (65). In this paper, we describe a systematic survey of histone acetylation throughout 90 kb of the SCL locus, which led to the identification of a previously unrecognized enhancer with erythroid specificity in vitro and in transgenic mice. This enhancer is situated between MAP17 and the cytochrome p450 gene cluster and, in the mouse sequence, lies 40 kb downstream of SCL exon 1a. In addition to its erythroid activity, several other lines of evidence suggest that the +40 enhancer regulates SCL transcription. The murine +40 acetylation peak (or the equivalent human +50 peak) was present in all seven SCL-expressing cell lines studied. Where tested, cell types containing the +40 (or +50) acetylation peak did not express Cyp4x1 or Cyp4A11, suggesting that the +40 enhancer does not regulate these cytochrome p450 genes. In addition, our data suggest that the +40 enhancer is not necessary for MAP17 transcription, since the +40 enhancer acetylation peak was not found in MAP17-expressing MDCT cells derived from renal tubule epithelium.
The activity of the +40 enhancer in primitive, but not definitive, erythroblasts was particularly striking. We considered the possibility that this pattern of expression might represent an artifact of the β-galactosidase reporter gene, since it has been reported that the β-galactosidase cassette interferes with the ability of the β-globin locus control region to confer position-independent expression (30), and it has been suggested that the β-galactosidase sequence may act as a nucleation point for the formation of heterochromatin in definitive erythroid cells (71). However, several groups have shown that a variety of erythroid enhancers, including the β-globin locus control region (30, 31), the α-globin −40 element (56, 70), and the EKLF −950 promoter (77), can direct expression of β-galactosidase to definitive erythroid cells in transgenic mice. Instead, we favor the alternative explanation that the 3.7-kb fragment containing the +40 region exhibits intrinsic specificity for the primitive erythroid lineage. This is a remarkable attribute, since most of the erythroid enhancers that have been characterized during embryonic development are active in both primitive and definitive erythroblasts (30, 70, 77). However, despite their many similarities, primitive and definitive erythropoieses differ in several characteristics, including cell size, types of hemoglobin accumulated, and location during embryonic development (39, 55). The two lineages also utilize distinct types of progenitors (38) and are differentially affected by targeted mutations of several transcription factor genes (48, 53, 72, 75, 76). These observations suggest that at least some features of the transcriptional environment are different in the two erythroid lineages and are consistent with the existence of regulatory elements displaying selective activity in one lineage but not the other. The 3.7-kb +40 region contains conserved potential binding sites for multiple transcription factors, including members of the GATA, Ets, basic helix-loop-helix, and homeobox families (data not shown). Refinement of a core enhancer region, together with subsequent detailed biochemistry and functional analyses, will begin to unravel the molecular basis for the biological activity of this unusual element.
The results presented here demonstrate that the +40 element is sufficient to direct expression in transgenic mice to primitive, but not definitive, erythroblasts. However, our data do not exclude the possibility that the +40 region may be necessary for expression in definitive erythroblasts, perhaps in collaboration with one or more additional elements. This scenario would be consistent with our observation that the +40 region is acetylated in F4N cells, which were generated by retroviral infection of neonatal mice and are therefore thought to represent definitive erythroid progenitors (11). Alternatively, expression of SCL in the definitive erythroid lineage may reflect the independent activity of a separate SCL enhancer.
It has been reported that an enhancer approximately 3.7 kb upstream of the GATA-1 erythroid promoter also directs β-galactosidase expression to primitive, but not definitive, erythroid cells (54), although a second group has found that the same element can target β-galactosidase to both cell types (74). The explanation for this discrepancy is not clear. Moreover, the reported specificity of the GATA-1 −3.7 element for the primitive erythroid lineage requires promoter-proximal sequences, as well as the −3.7 element itself (51). By contrast, the SCL +40 enhancer can distinguish between primitive and definitive erythroblasts in the context of a heterologous SV40 promoter.
The primitive erythroid lineage has been difficult to study because of its transient nature during embryogenesis and because the growth factor requirements of its progenitors are ill defined. The SCL +40 enhancer provides a powerful tool for dissecting the molecular and cellular biology of this poorly understood lineage.
Acknowledgments
This work was supported by The Wellcome Trust, the Leukemia Research Fund, the Association pour la Recherche sur le Cancer (ARC), and the Fondation de France (Comité Leucémie).
REFERENCES
- 1.Acuto, O., R. E. Hussey, K. A. Fitzgerald, J. P. Protentis, S. C. Meuer, S. F. Schlossman, and E. L. Reinherz. 1983. The human T cell receptor: appearance in ontogeny and biochemical relationship of alpha and beta subunits on IL-2 dependent clones and T cell tumors. Cell 34:717-726. [DOI] [PubMed] [Google Scholar]
- 2.Baker, J. R., S. Satarug, R. J. Edwards, M. R. Moore, D. J. Williams, and P. E. Reilly. 2003. Potential for early involvement of CYP isoforms in aspects of human cadmium toxicity. Toxicol. Lett. 137:85-93. [DOI] [PubMed] [Google Scholar]
- 3.Barton, L. M., B. Gottgens, M. Gering, J. G. Gilbert, D. Grafham, J. Rogers, D. Bentley, R. Patient, and A. R. Green. 2001. Regulation of the stem cell leukemia (SCL) gene: a tale of two fishes. Proc. Natl. Acad. Sci. USA 98:6747-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begley, C. G., and A. R. Green. 1999. The SCL gene: from case report to critical hematopoietic regulator. Blood 93:2760-2770. [PubMed] [Google Scholar]
- 5.Bockamp, E. O., F. McLaughlin, A. M. Murrell, B. Gottgens, L. Robb, C. G. Begley, and A. R. Green. 1995. Lineage-restricted regulation of the murine SCL/TAL-1 promoter. Blood 86:1502-1514. [PubMed] [Google Scholar]
- 6.Chapman, M. A., F. J. Charchar, S. Kinston, C. P. Bird, D. Grafham, J. Rogers, F. Grutzner, J. A. Marshall Graves, A. R. Green, and B. Gottgens. 2003. Comparative and functional analyses of LYL1 loci establish marsupial sequences as a model for phylogenetic footprinting. Genomics 81:249-259. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Z., M. Li, D. de Graaf, S. Monti, B. Gottgens, M. J. Sanchez, E. S. Lander, T. R. Golub, A. R. Green, and H. F. Lodish. 2002. Identification of endoglin as a functional marker that defines long-term repopulating hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 99:15468-15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collazo-Garcia, N., P. Scherer, and P. D. Aplan. 1995. Cloning and characterization of a murine SIL gene. Genomics 30:506-513. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, D. J., M. A. Hall, L. J. Van Stekelenburg, L. Robb, S. M. Jane, and C. G. Begley. 2004. SCL is required for normal function of short-term repopulating hematopoietic stem cells. Blood 103:3342-3348. [DOI] [PubMed] [Google Scholar]
- 10.Drexler, H. G. 2000. The leukemia-lymphoma cell line factsbook. Academic Press, London, United Kingdom.
- 11.Dube, S. K., I. B. Pragnell, N. Kluge, G. Gaedicke, G. Steinheider, and W. Ostertag. 1975. Induction of endogenous and of spleen focus-forming viruses during dimethylsulfoxide-induced differentiation of mouse erythroleukemia cells transformed by spleen focus-forming virus. Proc. Natl. Acad. Sci. USA 72:1863-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elefanty, A. G., C. G. Begley, L. Hartley, B. Papaevangeliou, and L. Robb. 1999. SCL expression in the mouse embryo detected with a targeted lacZ reporter gene demonstrates its localization to hematopoietic, vascular, and neural tissues. Blood 94:3754-3763. [PubMed] [Google Scholar]
- 13.Endoh, M., M. Ogawa, S. Orkin, and S. Nishikawa. 2002. SCL/tal-1-dependent process determines a competence to select the definitive hematopoietic lineage prior to endothelial differentiation. EMBO J. 21:6700-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faloon, P., E. Arentson, A. Kazarov, C. X. Deng, C. Porcher, S. Orkin, and K. Choi. 2000. Basic fibroblast growth factor positively regulates hematopoietic development. Development 127:1931-1941. [DOI] [PubMed] [Google Scholar]
- 15.Fantoni, A., A. De la Chapelle, and P. A. Marks. 1969. Synthesis of embryonic hemoglobins during erythroid cell development in fetal mice. J. Biol. Chem. 244:675-681. [PubMed] [Google Scholar]
- 16.Fordham, J. L., B. Göttgens, F. McLaughlin, and A. R. Green. 1999. Chromatin structure and transcriptional regulation of the stem cell leukaemia (SCL) gene in mast cells. Leukemia 13:750-759. [DOI] [PubMed] [Google Scholar]
- 17.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fugman, D. A., D. P. Witte, C. L. A. Jones, B. J. Aronow, and M. A. Lieberman. 1990. In vitro establishment and characterization of a human megakaryoblastic cell line. Blood 75:1252-1261. [PubMed] [Google Scholar]
- 19.Gering, M., A. R. Rodaway, B. Gottgens, R. K. Patient, and A. R. Green. 1998. The SCL gene specifies hemangioblast development from early mesoderm. EMBO J. 17:4029-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gesek, F. A., and P. A. Friedman. 1992. Mechanism of calcium transport stimulated by chlorothiazide in mouse distal convoluted tubule cells. J. Clin. Investig. 90:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottgens, B., L. M. Barton, M. A. Chapman, A. M. Sinclair, B. Knudsen, D. Grafham, J. G. Gilbert, J. Rogers, D. R. Bentley, and A. R. Green. 2002. Transcriptional regulation of the stem cell leukemia gene (SCL)—comparative analysis of five vertebrate SCL loci. Genome Res. 12:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottgens, B., L. M. Barton, J. G. Gilbert, A. J. Bench, M. J. Sanchez, S. Bahn, S. Mistry, D. Grafham, A. McMurray, M. Vaudin, E. Amaya, D. R. Bentley, and A. R. Green. 2000. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 18:181-186. [DOI] [PubMed] [Google Scholar]
- 23.Gottgens, B., C. Broccardo, M.-J. Sanchez, S. Deveaux, G. Murphy, J. Gothert, E. Kotsopoulou, S. Kinston, L. Delaney, S. Piltz, L. M. Barton, W. N. Erber, C. G. Begley, K. Frampton, and A. R. Green. 2004. The SCL +18/19 stem cell enhancer is not required for hematopoiesis: identification of a 5′ bifunctional hematopoietic-endothelial enhancer regulated by Fli-1 and Elf-1. Mol. Cell. Biol. 24:1870-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottgens, B., J. G. Gilbert, L. M. Barton, D. Grafham, J. Rogers, D. R. Bentley, and A. R. Green. 2001. Long-range comparison of human and mouse SCL loci: localized regions of sensitivity to restriction endonucleases correspond precisely with peaks of conserved noncoding sequences. Genome Res. 11:87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottgens, B., F. McLaughlin, E. O. Bockamp, J. L. Fordham, C. G. Begley, K. Kosmopoulos, A. G. Elefanty, and A. R. Green. 1997. Transcription of the SCL gene in erythroid and CD34 positive primitive myeloid cells is controlled by a complex network of lineage-restricted chromatin-dependent and chromatin-independent regulatory elements. Oncogene 15:2419-2428. [DOI] [PubMed] [Google Scholar]
- 26.Gottgens, B., A. Nastos, S. Kinston, S. Piltz, E. C. Delabesse, M. Stanley, M. J. Sanchez, A. Ciau-Uitz, R. Patient, and A. R. Green. 2002. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21:3039-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green, A. R. 1992. Transcription factors, translocations and hematological malignancies. Blood Rev. 6:118-124. [DOI] [PubMed] [Google Scholar]
- 28.Green, A. R., J. Visvader, T. Lints, R. Harvey, and C. G. Begley. 1992. SCL is co-expressed with GATA-1 in hemopoietic cells but is also expressed in developing brain. Oncogene 7:653-660. [PubMed] [Google Scholar]
- 29.Greenberg, S. M., D. S. Rosenthal, T. A. Greeley, R. Tantravahi, and R. I. Handen. 1988. Characterization of a new megakaryocytic cell line: the Dami cell. Blood 72:1968-1977. [PubMed] [Google Scholar]
- 30.Guy, L. G., R. Kothary, Y. DeRepentigny, N. Delvoye, J. Ellis, and L. Wall. 1996. The β-globin locus control region enhances transcription of but does not confer position-independent expression onto the lacZ gene in transgenic mice. EMBO J. 15:3713-3721. [PMC free article] [PubMed] [Google Scholar]
- 31.Guy, L. G., R. Kothary, and L. Wall. 1997. Position effects in mice carrying a lacZ transgene in cis with the beta-globin LCR can be explained by a graded model. Nucleic Acids Res. 25:4400-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall, M. A., D. J. Curtis, D. Metcalf, A. G. Elefanty, K. Sourris, L. Robb, J. R. Gothert, S. M. Jane, and C. G. Begley. 2003. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc. Natl. Acad. Sci. USA 100:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirose, M., K. Minato, K. Tobinai, M. Ohira, T. Ise, S. Watanabe, M. Shimoyama, M. Taniwaki, and T. Abe. 1982. A novel pre-T cell line derived from acute lymphoblastic leukemia. Gann 73:600-605. [PubMed] [Google Scholar]
- 34.Huntly, B. J. P., A. J. Bench, E. Delabesse, A. G. Reid, J. Li, M. Scott, L. Campbell, J. Byrne, E. Pinto, A. Brizzard, D. Niedermeiser, E. P. Nacheva, F. Guilhot, M. Deininger, and A. R. Green. 2002. Derivative chromosome 9 deletions in chronic myeloid leukemia: poor prognosis is not associated with loss of ABL-BCR expression, elevated BCR-ABL levels or karyotypic instability. Blood 99:4547-4553. [DOI] [PubMed] [Google Scholar]
- 35.Ishida, Y., J. Levin, G. Baker, P. E. Stenberg, Y. Yamada, H. Sasaki, and T. Inoue. 1993. Biological and biochemical characteristics of murine megakaryoblastic cell line L8057. Exp. Hematol. 21:289-298. [PubMed] [Google Scholar]
- 36.Izraeli, S., T. Colaizzo-Anas, V. L. Bertness, K. Mani, P. D. Aplan, and I. R. Kirsch. 1997. Expression of the SIL gene is correlated with growth induction and cellular proliferation. Cell Growth Differ. 8:1171-1179. [PubMed] [Google Scholar]
- 37.Jaeger, C., B. M. Schaefer, R. Wallich, and M. D. Kramer. 2000. The membrane-associated protein pKe#192/MAP17 in human keratinocytes. J. Investig. Dermatol. 115:375-380. [DOI] [PubMed] [Google Scholar]
- 38.Kennedy, M., M. Firpo, K. Choi, C. Wall, S. Robertson, N. Kabrun, and G. Keller. 1997. Identification of a common precursor for primitive erythropoiesis and definitive hematopoiesis. Nature 386:488-493. [DOI] [PubMed] [Google Scholar]
- 39.Kingsley, P. D., J. Malik, K. A. Fantauzzo, and J. Palis. 2004. Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood 104:19-25. [DOI] [PubMed] [Google Scholar]
- 40.Kocher, O., P. Cheresh, and S. W. Lee. 1996. Identification and partial characterization of a novel membrane-associated protein (MAP17) up-regulated in human carcinomas and modulating cell replication and tumor growth. Am. J. Pathol. 149:493-500. [PMC free article] [PubMed] [Google Scholar]
- 41.Kurtzberg, J., S. H. Bigner, and M. S. Hershfield. 1985. Establishment of the DU.528 human lymphohemopoietic stem cell line. J. Exp. Med. 162:1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange, C., C. Kaltz, K. Thalmeier, H. J. Kolb, and R. Huss. 1999. Hematopoietic reconstitution of syngeneic mice with a peripheral blood-derived, monoclonal CD34−, Sca-1+, Thy-1(low), c-kit+ stem cell line. J. Hematother. Stem Cell Res. 8:335-342. [DOI] [PubMed] [Google Scholar]
- 43.Matsuo, Y., T. Tomiyasu, F. Shirakawa, U. Yamashita, K. Sagawa, M. M. Yokoyama, and J. Minowada. 1989. Establishment and characterization of a clonal human T-cell line, MKB-1 derived from a patient with acute myeloblastic leukemia. Hum. Cell 2:423-429. [PubMed] [Google Scholar]
- 44.Megy, K., S. Audic, and J. M. Claverie. 2003. Positional clustering of differentially expressed genes on human chromosomes 20, 21 and 22. Genome Biol. 4:P1. [DOI] [PubMed] [Google Scholar]
- 45.Melo, J. V., V. Brito-Babapulle, L. Foroni, D. S. Robinson, L. Luzzatto, and D. Catovsky. 1986. Two new cell lines from B-prolymphocytic leukaemia: characterization by morphology, immunological markers, karyotype and Ig gene rearrangement. Int. J. Cancer 38:531-538. [DOI] [PubMed] [Google Scholar]
- 46.Mikkola, H. K., J. Klintman, H. Yang, H. Hock, T. M. Schlaeger, Y. Fujiwara, and S. H. Orkin. 2003. Hematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421:547-551. [DOI] [PubMed] [Google Scholar]
- 47.Miles, C., M.-J. Sanchez, A. Sinclair, and E. Dzierzak. 1997. Expression of the Ly-6E. 1 (Sca-1) transgene in adult hematopoietic stem cells and the developing mouse embryo. Development 124:537-547. [DOI] [PubMed] [Google Scholar]
- 48.Mucenski, M. L., K. McLain, A. B. Kier, S. H. Swerdlow, C. M. Schreiner, T. A. Miller, D. W. Pietryga, W. J. Scott, Jr., and S. S. Potter. 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65:677-689. [DOI] [PubMed] [Google Scholar]
- 49.Ogilvy, S., A. G. Elefanty, J. Visvader, M. L. Bath, A. W. Harris, and J. M. Adams. 1998. Transcriptional regulation of vav, a gene expressed throughout the hematopoietic compartment. Blood 91:419-430. [PubMed] [Google Scholar]
- 50.Ohkubo, T., T. Kamamoto, K. Kita, A. Hiraoka, Y. Yoshida, and H. Uchino. 1985. A novel Ph1 chromosome positive cell line established from a patient with chronic myelogenous leukemia in blastic crisis. Leuk. Res. 9:921-926. [DOI] [PubMed] [Google Scholar]
- 51.Ohneda, K., R. Shimizu, S. Nishimura, Y. Muraosa, S. Takahashi, J. D. Engel, and M. Yamamoto. 2002. A minigene containing four discrete cis elements recapitulates GATA-1 gene expression in vivo. Genes Cells 7:1243-1254. [DOI] [PubMed] [Google Scholar]
- 52.Okabe, M., Y. Kunieda, S. Nakane, M. Kurosawa, T. Itaya, W. R. Vogler, M. Shoji, and T. Miyazaki. 1995. Establishment and characterization of a new Ph1-positive chronic myeloid leukemia cell line MC3 with trilineage phenotype and an altered p53 gene. Leuk. Lymphoma 16:493-503. [DOI] [PubMed] [Google Scholar]
- 53.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 54.Onodera, K., S. Takahashi, S. Nishimura, J. Ohta, H. Motohashi, K. Yomogida, N. Hayashi, J. D. Engel, and M. Yamamoto. 1997. GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc. Natl. Acad. Sci. USA 94:4487-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palis, J., S. Robertson, M. Kennedy, C. Wall, and G. Keller. 1999. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126:5073-5084. [DOI] [PubMed] [Google Scholar]
- 56.Pondel, M. D., J. A. Sharpe, S. Clark, L. Pearson, W. G. Wood, and N. J. Proudfoot. 1996. Proximal promoter elements of the human zeta-globin gene confer embryonic-specific expression on a linked reporter gene in transgenic mice. Nucleic Acids Res. 24:4158-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Porcher, C., W. Swat, K. Rockwell, Y. Fujiwara, F. W. Alt, and S. H. Orkin. 1996. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86:47-57. [DOI] [PubMed] [Google Scholar]
- 58.Ralph, P. 1973. Retention of lymphocyte characteristics by myelomas and theta +-lymphomas: sensitivity to cortisol and phytohemagglutinin. J. Immunol. 110:1470-1475. [PubMed] [Google Scholar]
- 59.Rapp, U. R., J. L. Cleveland, T. N. Fredrickson, K. L. Holmes, H. C. Morse III, H. W. Jansen, T. Patschinsky, and K. Bister. 1985. Rapid induction of hemopoietic neoplasms in newborn mice by a raf(mil)/myc recombinant murine retrovirus. J. Virol. 55:23-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reymond, A., V. Marigo, M. B. Yaylaoglu, A. Leoni, C. Ucla, N. Scamuffa, C. Caccioppoli, E. T. Dermitzakis, R. Lyle, S. Banfi, G. Eichele, S. E. Antonarakis, and A. Ballabio. 2002. Human chromosome 21 gene expression atlas in the mouse. Nature 420:582-586. [DOI] [PubMed] [Google Scholar]
- 61.Robb, L., N. J. Elwood, A. G. Elefanty, F. Köntgen, R. Li, L. D. Barnett, and C. G. Begley. 1996. The SCL gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 15:4123-4129. [PMC free article] [PubMed] [Google Scholar]
- 62.Robertson, S. M., M. Kennedy, J. M. Shannon, and G. Keller. 2000. A transitional stage in the commitment of mesoderm to hematopoiesis requiring the transcription factor SCL/tal-1. Development 127:2447-2459. [DOI] [PubMed] [Google Scholar]
- 63.Roy, P. J., J. M. Stuart, J. Lund, and S. K. Kim. 2002. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418:975-979. [DOI] [PubMed] [Google Scholar]
- 64.Sanchez, M., B. Gottgens, A. M. Sinclair, M. Stanley, C. G. Begley, S. Hunter, and A. R. Green. 1999. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult hematopoietic progenitors. Development 126:3891-3904. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez, M. J., E. O. Bockamp, J. Miller, L. Gambardella, and A. R. Green. 2001. Selective rescue of early hematopoietic progenitors in Scl−/− mice by expressing Scl under the control of a stem cell enhancer. Development 128:4815-4827. [DOI] [PubMed] [Google Scholar]
- 66.Sinclair, A. M., A. J. Bench, A. J. Bloor, J. Li, B. Gottgens, M. L. Stanley, J. Miller, S. Piltz, S. Hunter, E. P. Nacheva, M. J. Sanchez, and A. R. Green. 2002. Rescue of the lethal scl−/− phenotype by the human SCL locus. Blood 99:3931-3938. [DOI] [PubMed] [Google Scholar]
- 67.Sinclair, A. M., B. Gottgens, L. M. Barton, M. L. Stanley, L. Pardanaud, M. Klaine, M. Gering, S. Bahn, M. Sanchez, A. J. Bench, J. L. Fordham, E. Bockamp, and A. R. Green. 1999. Distinct 5′ SCL enhancers direct transcription to developing brain, spinal cord, and endothelium: neural expression is mediated by GATA factor binding sites. Dev. Biol. 209:128-142. [DOI] [PubMed] [Google Scholar]
- 68.Spellman, P. T., and G. M. Rubin. 2002. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spitz, F., F. Gonzalez, and D. Duboule. 2003. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113:405-417. [DOI] [PubMed] [Google Scholar]
- 70.Sutherland, H. G., D. I. Martin, and E. Whitelaw. 1997. A globin enhancer acts by increasing the proportion of erythrocytes expressing a linked transgene. Mol. Cell. Biol. 17:1607-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tewari, R., N. Gillemans, A. Harper, M. Wijgerde, G. Zafarana, D. Drabek, F. Grosveld, and S. Philipsen. 1996. The human β-globin locus controls region confers an early embryonic erythroid-specific expression pattern to a basic promoter driving the bacteria lacZ gene. Development 122:3991-3999. [DOI] [PubMed] [Google Scholar]
- 72.Tsai, F.-Y., G. Keller, F. C. Kuo, M. Weiss, J. Z. Chen, M. Rosenblatt, F. W. Alt, and S. H. Orkin. 1994. An early hemopoietic defect in mice lacking the transcription factor GATA-2. Nature 371:221-226. [DOI] [PubMed] [Google Scholar]
- 73.Visvader, J. E., Y. Fujiwara, and S. H. Orkin. 1998. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 12:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vyas, P., M. A. McDevitt, A. B. Cantor, S. G. Katz, Y. Fujiwara, and S. H. Orkin. 1999. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development 126:2799-2811. [DOI] [PubMed] [Google Scholar]
- 75.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93:3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weiss, M. J., G. Keller, and S. H. Orkin. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1− embryonic stem cells. Genes Dev. 8:1184-1197. [DOI] [PubMed] [Google Scholar]
- 77.Xue, L., X. Chen, Y. Chang, and J. J. Bieker. 2004. Regulatory elements of the EKLF gene that direct erythroid cell-specific expression during mammalian development. Blood 103:4078-4083. [DOI] [PubMed] [Google Scholar]