Abstract
Posttranslational modification by SUMO elicits a repressive effect on many transcription factors. In principle, sumoylation may either influence transcription factor activity on promoters, or it may act indirectly by targeting the modified factors to specific cellular compartments. To provide direct experimental evidence for the above, not necessarily mutually exclusive models, we analyzed the role of SUMO modification on the localization and the activity of the orphan nuclear receptor LRH-1. We demonstrate, by using fluorescence resonance energy transfer (FRET) and fluorescence recovery after photobleaching (FRAP) assays, that sumoylated LRH-1 is exclusively localized in promyelocytic leukemia protein (PML) nuclear bodies and that this association is a dynamic process. Release of LRH-1 from nuclear bodies correlated with its desumoylation, pointing to the pivotal role of SUMO conjugation in keeping LRH-1 in these locations. SUMO-dependent shuttling of LRH-1 into PML bodies defines two spatially separated pools of the protein, of which only the soluble, unmodified one is associated with actively transcribed target genes. The results suggest that SUMO-PML nuclear bodies may primarily function as dynamic molecular reservoirs, controlling the availability of certain transcription factors to active chromatin domains.
The selectivity of gene expression is mainly controlled by the limited ability of transcription factors to access the genome. Besides alterations of chromatin structure, other processes, such as the positioning of genes within the nucleus and compartmentalization of the proteins that regulate their expression, have been implicated in the pathways determining the specificity of gene activation (9, 15, 30). Covalent attachment of the small-ubiquitin-related modifier (SUMO) to proteins has been characterized as a modification with diverse effects, including targeting of the substrates to specific nuclear territories (17, 42, 46, 52). For example, sumoylation is required for the targeting of the cytoplasmic nuclear import factor RanGAP1 to the nuclear pore complex or to mitotic spindles and kinetochores in dividing cells (24, 32, 33). Furthermore, under conditions that allow SUMO conjugation, several proteins, including transcription factors, have been found to localize in nuclear speckles (38, 40, 41), although direct evidence for the identity of the proteins at the nuclear speckles as SUMO-modified ones is still missing.
These sites of accumulations often coincide with the nuclear bodies associated with promyelocytic leukemia protein nuclear bodies (PML-NBs). PML-NBs are nuclear matrix-associated structures, ranging in size from 0.2 to 1 μm (7, 52). Besides PML, several other proteins accumulating in these structures have been identified. These include SUMO-1, Sp100, Daxx, Rb, BLM, CBP, and p53, suggesting roles in DNA replication and repair, cell cycle control, apoptosis, and transcription (1, 6, 23, 29, 50, 51). SUMO modification of several of the above components, including the PML protein itself, is important for their recruitment to the PML-NBs and consequently for the proper formation of the nuclear domain (52). Although several lines of evidence suggest that PML-NBs may play a role in the regulation of gene expression (3, 5, 47, 52), the relationship between SUMO modification-dependent compartmentalization of transcription factors into these structures and their function on target genes is less well understood.
To address this question, we studied the role of SUMO modification on the localization and the activity of the orphan nuclear receptor LRH-1 (for liver receptor homologue 1; also known as FTF and CPF). LRH-1 plays a crucial role in liver development during early embryogenesis in controlling cell proliferation and renewal of intestinal crypt cells and in the regulation of cholesterol-bile acid homeostasis in adult hepatocytes (8, 12, 20, 31, 36). Here we show that LRH-1 is reversibly modified by SUMO-1 in vitro and in vivo. We present evidence for a SUMO-dependent sequestration of LRH-1 into PML-NBs, which precludes its access to active chromatin domains.
MATERIALS AND METHODS
Plasmids and antibodies.
The open reading frame of the human LRH-1 cDNA was subcloned into pCDNA-myc (Invitrogen) and pEYFP (Clontech) vectors. Site-directed mutagenesis was performed with the GeneEditor kit (Promega). Glutathione S-transferase (GST)-SUMO-1, GST-Ubc9, GST-PIASxα, and pCMV-Flag-PIASxα plasmids were provided by N. Kotaja and J. Palvimo, GST-SAE1/2 was provided by R. Hay, GST-RanBP2 was provided by F. Melchior, and pSG-PML-III and green fluorescent protein (GFP)-PML-III were provided by H. Will. pMT-HA-SUMO-1, pECFP-SUMO-1, pEYFP-SUMO-1, and pCMV-Flag-SuPr-1 were generated by amplifying the open reading frame of SUMO-1 missing the last four amino acids and the entire open reading frame of SuPr1 by PCR, followed by ligation into pMT-HA, pECFP, pEYFP, and pCMV-Flag vectors, respectively. All constructs were verified by DNA sequencing.
Anti-SUMO-1 (FL-101), anti-myc (9E10 and A-14), antihemagglutinin (anti-HA; Y-11), anti-PML, and anti-Cy3-labeled PML (PG-M3) were from Santa Cruz Biotechnology; anti-Pol-II (8WG16) was from Covance; anti-histone 3 (ab8580) was from Abcam; and anti-Flag (M2) and anti-Lamin B were from Sigma. A polyclonal antibody against LRH-1 was raised by immunization of New Zealand White rabbits with bacterially expressed recombinant human LRH-1 protein. In Western blots and immunofluorescence assays horseradish peroxidase-conjugated anti-rabbit and anti-mouse immunoglobulin G (Jackson Laboratories) and anti-rabbit or anti-mouse AlexaFluor568 or AlexaFluor488 (Molecular Probes) were used as secondary antibodies, respectively.
siRNA-mediated knockdown of SUMO-1.
Double-stranded small interfering RNA (siRNA) for targeting human SUMO-1 and control siRNA containing scrambled sequence were purchased from Santa Cruz Biotechnology (sc-29498 and sc-37007). HepG2 cells were transfected with the siRNAs at 100 nM final concentrations by using the jetSI-ENDO kit (Polyplus). Extract preparations and immunofluorescence analyses were performed 72 h after transfection.
Immunoprecipitations, Western blots and in vitro SUMO conjugation assays.
Cells were lysed by resuspension in lysis buffer containing 1.72% sodium dodecyl sulfate (SDS), 50 mM Tris-HCl (pH 6.7), 10% glycerol, 0.33% NP-40, 0.33% sodium deoxycholate, 10 mM N-ethylmaleimide (NEM), 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin/ml, and 10 μg of E64/ml, followed by mild sonication and centrifugation at 12,000 × g. The extracts were either analyzed directly on Western blots or, after a 17-fold dilution with a buffer containing 50 mM HEPES (pH 7.9), 5% glycerol, 0.1% NP-40, 150 mM NaCl, 0.1 mM EDTA, 10 mM NEM, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin/ml, and 10 μg of E64/ml, subjected to immunoprecipitation as described previously (45). Chromatin immunoprecipitation (ChIP) assays were performed as described previously (21, 44) except for the cross-linking step. For the latter, mouse livers were perfused successively with phosphate-buffered saline (PBS), 1% formaldehyde, and 0.125 M glycine for 10 min. Cross-linked nuclei were purified by centrifugation through a sucrose gradient as described previously (45). In re-ChIP assays, after the protein G-Sepharose beads from the primary immunoprecipitation were washed, bound complexes were eluted from the beads by incubation with 10 mM dithiothreitol at 37°C for 30 min. After a 50-fold dilution with 1× sonication buffer, the eluates were subjected to immunoprecipitation with the second antibody. Reporter assays were performed as described previously (28).
In vitro sumoylation reactions were performed by using 1 μl of in vitro-translated, 35S-labeled LRH-1 in a 15-μl reaction mixture containing 50 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 1 mM dithiothreitol, and 2 mM ATP, 50 ng of GST-SUMO-1, 25 ng of GST-SAE1/2, 25 ng of GST-UBC9, and 1 μg of GST-RanBP2, or GST-PIAS-1, or GST-PIASxα. After incubation at 30°C for 1 h, the reaction products were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Nuclear matrix isolation was carried out essentially as described in references 22 and 37, except that all buffers were supplemented with protease inhibitor cocktail (Roche), 10 mM NEM, and 10 μg of E64/ml. Briefly, after being washed with PBS, the cells were resuspended in CSK buffer (see below) and incubated for 3 min on ice. After centrifugation for 3 min at 5,000 × g, the supernatant (Nucleocytoplasm) was separated, and the pellet was resuspended in CSK buffer containing 1 mg of RNase-free DNase/ml. After incubation at 37°C for 15 min, the reaction mix was adjusted with ammonium sulfate to 0.25 M final concentration and centrifuged again. The supernatant (chromatin fraction) was saved, and the pellet was extracted with 2 M NaCl. Complete removal of histones and DNA was verified by SDS-PAGE and agarose gel electrophoresis. The final insoluble pellet (nuclear matrix) was resuspended in SDS sample buffer. Equal proportions of each fraction were separated by SDS-PAGE and analyzed in Western blots.
Quantitative assessment of Western blot signals was performed with a Fujifilm LAS-1000 luminescent image analyzer.
Immunofluorescence and confocal microscopy.
Cells were plated on poly-d-lysine-coated glass coverslips and transfected with various combinations of expression vectors by using the PolyPlus transfection reagent. At 24 to 36 h after transfection the cells were either used for live cell imaging or fixed with 2% paraformaldehyde. Immunostainings were performed as described previously (28). Fluorescence images were obtained on Zeiss Axioscope 2 Plus microscope outfitted with a Bio-Rad Radiance 2100 laser scanning system and Lasersharp-2000 imaging software.
For fluorescence resonance energy transfer (FRET) analysis the following filter settings were used. Cyanfluorescent protein (CFP) fluorescence was observed by using argon laser 457-nm excitation and 488/10 band-pass emission filters, whereas yellow fluorescent protein (YFP) fluorescence was observed by using argon laser 514-nm excitation and 570LP band-pass emission filters. Photobleaching of YFP fluorescence was achieved by irradiation of the selected cells for 1 min with the 514-nm excitation filter at maximum intensity. Sequential images before or after photobleaching were collected, and pixel intensities of the different regions were analyzed by using NIH Image-J software. FRET efficiency was expressed as the percent increase of prebleach CFP fluorescence in the individual areas compared to that observed after YFP photobleaching.
For fluorescence recovery after photobleaching (FRAP) analysis, the coverslips were placed in PBS and observed by using argon laser 514-nm excitation and 545/40 band-pass emission filters for YFP and argon laser 488-nm excitation and 515/30 band-pass emission filters for GFP. An approximately 1-μm2 spot was bleached for 2 s at 50 to 60% intensity setting of the appropriate excitation laser, and serial images were collected over a 60-s period. Fluorescence intensities of the bleached areas from five independent experiments were normalized to out-of-focus contribution and are expressed as a percentage of the measured prebleach intensities.
In situ permeabilization experiments were performed by treating the cells with 0.5% Triton X-100 containing CSK buffer (10 mM PIPES [pH 6.8], 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA) for 1 min at room temperature. Permeabilization was stopped by fixing the cells with 2% paraformaldehyde or by extraction with lysis buffer.
RESULTS
LRH-1 is modified by SUMO-1 in vitro and in vivo.
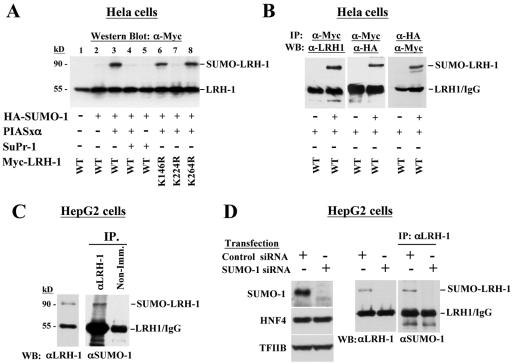
The LRH-1 protein has three conserved consensus sumoylation sites (ΨKXE) at amino acid positions 146, 224, and 264. In order to demonstrate that LRH-1 can be modified by SUMO, we performed Western blot analysis with lysates from HEK293 cells, which were transfected with HA-tagged SUMO-1 and myc-tagged wild type, or SUMO consensus-site mutant forms of LRH-1. A slower-migrating LRH-1 species (∼90 kDa) was observed only when LRH-1 was coexpressed with SUMO-1 and the known SUMO E3 ligase PIASxα (25) (Fig. 1A, lane 3). The fact that this band corresponds to SUMO-modified LRH-1 is demonstrated by its dependence on the coexpression of SUMO-1 and PIASxα (Fig. 1A, lanes 1 to 3) and by its disappearance upon coexpression of SuPr-1 (Fig. 1A, lane 4), which is a known SUMO protease (3). Furthermore, we could detect a band with the same mobility in myc-LRH-1 immunoprecipitates with an HA antibody recognizing HA-tagged SUMO-1 and in HA-SUMO-1 immunoprecipitates by using a myc antibody recognizing myc-tagged LRH-1 (Fig. 1B). When the same analyses were performed with the different sumoylation consensus site LRH-1 mutants, the slower-migrating band disappeared only in the case of K224R, suggesting that lysine 224 of LRH-1 is the major SUMO conjugation site (Fig. 1A, lanes 6 to 8).
FIG. 1.
LRH-1 is modified by SUMO-1 in vivo. (A) Extracts of HEK-293 cells transfected with the indicated expression vectors were analyzed in Western blots with a myc tag-specific antibody to detect modified and unmodified LRH-1. (B) myc-LRH-1 and HA-SUMO-1 proteins were immunoprecipitated with αmyc or αHA antibody, and the presence of LRH-1 and SUMO-1 in the immunoprecipitates was detected in Western blots by using αLRH-1, αmyc and αHA antibodies as indicated. (C) Endogenous LRH-1 from C3A-HepG2 cells was detected in Western blot by an αLRH-1 antibody (left panel) or immunoprecipitated with αLRH-1 antibody and analyzed in Western blots with αSUMO-1 antibody. As a control, nonimmune (Non-Imm.) antiserum was used for immunoprecipitation. (D) C3A-HepG2 cells were transfected with SUMO siRNA and control siRNA as indicated. Cell lysates prepared 72 h after transfection were analyzed in Western blots with antibodies recognizing SUMO-1, HNF4, TFIIB, and LRH-1 (left and middle panels). Part of the extracts was immunoprecipitated with αLRH-1 antibody and analyzed in Western blots with αSUMO-1 antibody (panel at right).
In order to examine the modification of endogenous LRH-1 proteins, we performed Western blot analysis with nuclear extracts from C3A-HepG2 cells. A slower-migrating αLRH-1-reactive band (∼90 kDa) was observed in addition to the major 55-kDa species, which corresponds to unmodified LRH-1 protein. The identity of the 90-kDa band as SUMO-conjugated LRH-1 was confirmed by Western blot analysis of anti-LRH-1 immunoprecipitates with an αSUMO-1 antibody (Fig. 1C). Additional evidence for the sumoylation of endogenous LRH-1 was provided by the analysis of cells transfected with SUMO-1 siRNA. In SUMO-1-specific siRNA-transfected cells endogenous SUMO-1 was efficiently knocked down and the slower-migrating band in simple Western blots stained with anti-LRH-1, or in LRH-1 immunoprecipitates probed with anti-SUMO-1, disappeared (Fig. 1D). The specificity of the assay was confirmed by the unaltered expression of LRH-1, or HNF-4 and TFIIB, which were used as additional controls (Fig. 1D).
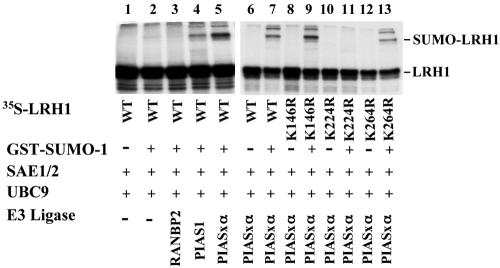
Next, we tested whether LRH-1 could also be a substrate for SUMO modification in vitro. We used an in vitro sumoylation assay using purified recombinant GST-SUMO-1 protein, E1 (SAE1/2), E2 (Ubc9) enzymes, and various E3 ligases (RANBP2, PIAS1, and PIASxα.). As substrates, we used in vitro-translated 35S-labeled wild-type LRH-1 or its different SUMO consensus site mutant derivatives. We could detect SUMO attachment to radiolabeled LRH-1 only when SUMO-1, SAE1/2, Ubc9 and PIAS1, or PIASxα ligases were present in the reaction (Fig. 2, lanes 4 and 5). As expected, unlike the K146R or the K264R mutants, the K224R mutant form of LRH-1 was not modified in vitro (Fig. 2, lanes 8 to 13). Collectively, these results suggest that LRH-1 can be modified by SUMO-1 on lysine 224 by the PIAS family of E3 ligases and that sumoylated LRH-1 is a substrate for SuPr-1, which can hydrolyze the SUMO moiety from the protein.
FIG. 2.
Modification of LRH-1 by SUMO-1 in vitro. 35S-labeled wild-type LRH-1 and its mutant forms were synthesized in vitro and incubated in a reaction reconstituted with the indicated recombinant proteins. The reaction products were separated by electrophoresis in SDS-10% polyacrylamide gels and visualized by autoradiography.
Sumoylated LRH-1 is localized in discrete nuclear dots.
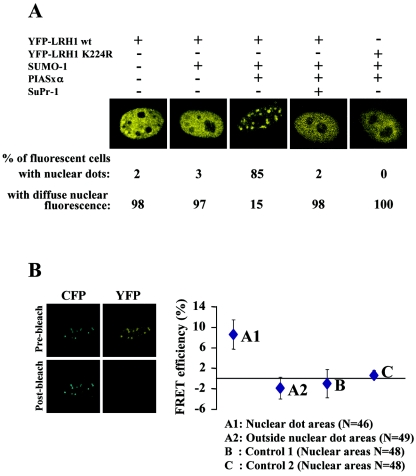
To study the subnuclear localization of LRH-1, we expressed a YFP-LRH-1 fusion protein in HeLa cells. A diffuse nuclear staining was observed (Fig. 3A). However, when the wild-type YFP-LRH-1 was coexpressed with SUMO-1 and PIASxα, which, as judged by Western blot analysis leads to the sumoylation of ca. 50% of the protein (data not shown), a substantial part of the fluorescence signal was accumulated in dot-like structures in the majority of the cells. No such accumulation was observed with the K224R mutant protein or in SuPr-1-expressing cells, suggesting that the observed relocalization of LRH-1 was dependent on sumoylation (Fig. 3A). Although such SUMO-dependent subnuclear redistribution has been observed with other transcription factors (9, 38, 40), the question of whether the proteins accumulating at the nuclear dots correspond to SUMO-modified ones has not been directly addressed before. To this end, we performed FRET after photobleaching assays (25), with CFP-tagged SUMO-1 and YFP-tagged LRH-1 proteins. FRET is based on the ability of a higher-energy donor fluorophore (CFP) to transfer energy to a lower-energy acceptor molecule (YFP), causing sensitized fluorescence of the acceptor and simultaneous quenching of the donor fluorescence, when the two molecules are in close proximity. If FRET occurs, photobleaching of YFP should lead to an increase in the emission of the CFP protein. An average of 8% increase in CFP emission was observed only at the nuclear dots but not in the areas outside them (Fig. 3B, A1 and A2 datum set). The significance of this increase is substantiated by the absence of FRET signal in control experiments where either YFP-LRH-1 K224R was coexpressed with CFP-SUMO-1 and PIASxα, or YFP-LRH-1wt was coexpressed with CFP-SUMO-1 without PIASxα (Fig. 3B, B and C datum set). Because FRET occurs only when the donor and acceptor molecules are within 50 to 100 Å of each other (25), these results can be interpreted either as a true molecular interaction between wtLRH-1 and SUMO-1 or as an interaction of LRH-1 with other sumoylated proteins. The latter scenario can be excluded by the absence of FRET signal in any of the randomly selected nuclear locations under conditions where LRH-1 sumoylation does not occur (e.g., in YFP-LRH-1 K224R-transfected cells or in cells not transfected with PIASxα). Therefore, we conclude that SUMO-modified LRH-1 is localized solely at the nuclear dot areas, pointing to the existence of two populations of LRH-1 molecules (sumoylated and nonsumoylated), which are distributed into distinct nuclear territories.
FIG. 3.
SUMO-modified LRH-1 is localized in discrete nuclear dots. (A) HeLa cells were transfected with the indicated expression vectors and observed by confocal microscopy. Representative images of cells from several experiments are shown. (B) HeLa cells were transfected with pECFP-SUMO-1, pEYFP-LRH-1, and pCMV-Flag-PIASxα. Sequential CFP and YFP fluorescence images were recorded before and after 1 min of photobleaching of YFP fluorescence by 514-nm laser line. FRET efficiency was expressed as the percent increase of prebleach CFP fluorescence after YFP photobleaching in the nuclear dot areas (A1) and similar-sized areas outside the nuclear dots (A2). In the control experiment B, the cells were cotransfected with pECFP-SUMO-1, pEYFP-LRH-1 K224R, and pCMV-Flag-PIASxα, whereas in the control experiment C, the cells were cotransfected with pECFP-SUMO-1 and pEYFP-LRH-1 without the PIASxα expression vector. The differences in fluorescence intensity in similar-sized nuclear areas were calculated as described above. The graph shows mean values and standard errors from the indicated number (N) of measurements, which were obtained from six to eight different cells.
The sites where sumoylated LRH-1 accumulates correspond to PML-containing nuclear bodies.
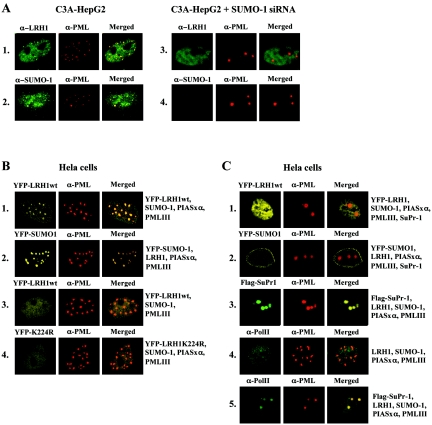
In order to further characterize the identity of the nuclear domains where sumoylated LRH-1 accumulates, we performed colocalization assays with PML, which is known to be concentrated in similar speckles in a sumoylation-dependent manner (3, 13). In C3A-HepG2 cells endogenous LRH-1 and SUMO-1 were detected throughout the nucleoplasm and at small speckles, which largely overlapped with endogenous PML-containing nuclear dots (Fig. 4A1 and 2). When endogenous SUMO-1 expression was knocked down, LRH-1 disappeared from the nuclear speckles, whereas PML was concentrated in two to four large nuclear aggregates (Fig. 4A3 and 4).
FIG. 4.
SUMO-modified LRH-1 is localized in PML-containing nuclear bodies. (A) Untransfected and SUMO-siRNA-transfected C3A-HepG2 cells were stained with αLRH-1 (rows 1 and 3) and αSUMO-1 (rows 2 and 4). (B and C) HeLa cells, which were transfected with the expression vectors indicated at the right, were observed by direct fluorescence of YFP or immunofluorescence staining with anti-Flag or anti-RNA Pol II antibodies (left panels). In all cases PML-NBs were detected in the same field by immunofluorescence staining with a TRITC (tetramethyl rhodamine isothiocyanate)-conjugated PML antibody (middle panels). Merged images are depicted in the right panels.
The correlation between sumoylation of LRH-1 and its colocalization with PML was further corroborated by the analysis of HeLa cells transfected with various combinations of expression vectors that mimic the conditions under which SUMO modification of the ectopically expressed YFP-LRH-1 can occur (transfection with wtLRH-1, PIASxα, and SUMO-1 [Fig. 4B1]) or cannot occur (transfection with wtLRH-1 and SUMO-1 without PIASxα [Fig. 4B3] or transfection with LRH-1 K224R with PIASxα and SUMO-1 [Fig. 4B4]). As expected, colocalization of YFP-LRH-1 with PML in the NBs could be detected in a SUMO-1- and PIASxα-dependent manner (Fig. 4B1). Under the same conditions YFP-SUMO-1 also colocalized with PML (Fig. 4B2). Overexpression of SuPr-1 resulted in a diffuse nuclear distribution of YFP-LRH-1 and YFP-SUMO-1, with some accumulation of the latter at the nuclear periphery (Fig. 4C1 and 2). Although this could be consistent with the idea that desumoylation of LRH-1 leads to its release from PML-NBs, we note that SuPr-1 action might not be selective because sumoylation of the PML protein itself is important for the integrity of nuclear bodies (3, 13, 34). In agreement with a previous study (3), we observed that SuPr-1 overexpression resulted in the disruption of PML-NBs and that PML was relocated in two to four large, abnormally shaped aggregates (Fig. 4C1 to C3). Interestingly, in addition to the diffuse nuclear fraction, a clear concentration of endogenous RNA polymerase II was detected in these SuPr-1-containing and SUMO-deficient structures, suggesting that they may represent novel PML deposit sites functionally different from those of PML-NBs, which contain sumoylated proteins but do not accumulate RNA polymerase-II (Fig. 4C4 and 5). Because of the high density of RNA polymerase II in these SuPr-1-generated territories, it is tempting to speculate that they may represent hot spots of active transcription for several genes. However, the diffuse nucleoplasmic distribution of LRH-1 in SuPr-1-overexpressing cells suggests that the SuPr-1-mediated induction of LRH-1 activity (see below) is more likely a consequence of the release of LRH-1 from PML-NBs and the concomitant increase of the free LRH-1 pool than of its relocation to the above RNA polymerase II-dense nuclear regions.
FIG. 5.
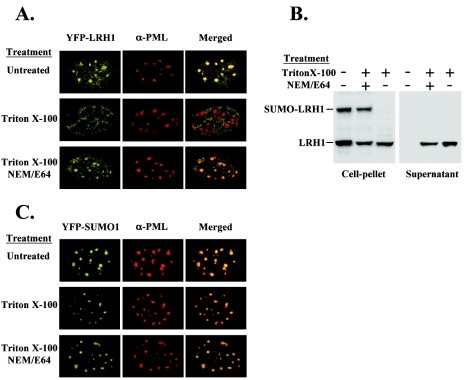
Release of LRH-1 from PML-NBs correlates with the loss of SUMO moiety. (A) HeLa cells were transfected with YFP-LRH-1, HA-SUMO, PIASxα, and PML-III expression vectors. (C) HeLa cells were transfected with YFP-SUMO-1, myc-LRH-1, PIASxα, and PML-III expression vectors. The cells were incubated in a 0.5% Triton X-100-containing buffer for 1 min before fixation or extraction. Triton X-100 treatments were performed in the presence or absence of 10 mM NEM and 10 μg of E64 isopeptidase inhibitors/ml. Confocal microscopic images (A and C) and Western blot analysis of extracts with αLRH-1 antibody (B) are shown.
Dynamics of SUMO-dependent association of LRH-1 with PML-NBs.
Evidence for the role of desumoylation on the release of LRH-1 from PML-NBs was provided by in situ permeabilization experiments. Our rationale was that if the accumulation of SUMO-LRH-1 at PML-NBs has a “storage” function, then decreasing the intranuclear concentration of LRH-1 might mobilize the molecules located in these compartments. To this end, we treated the cells with 0.5% Triton X-100 for 1 min and, after fixation, examined the distribution of YFP-LRH-1, YFP-SUMO-1, and PML proteins. Under these mild permeabilization conditions, ca. 50% of the LRH-1 protein leaked out of the nuclei, whereas the overall morphology of the cells was preserved. As shown in Fig. 5A, YFP-LRH-1 was cleared from the nuclear dots in Triton X-100-treated cells and was localized mainly at the surrounding areas. On the other hand, under the same conditions, PML and YFP-SUMO-1 remained concentrated in discrete nuclear speckles (Fig. 5A and C). Importantly, when the detergent treatment was performed in the presence of the isopeptidase inhibitors NEM and E64, which prevent the cleavage of SUMO from modified proteins (14), a substantial proportion of YFP-LRH-1 fluorescence remained associated with the PML-containing nuclear dots (Fig. 5A). Parallel Western blot analysis revealed the loss of SUMO moiety from LRH-1 in Triton X-100-permeabilized cells and its retention when the treatment was performed in the presence of isopeptidase inhibitors (Fig. 5B). These results suggest that lowering the nuclear concentration of proteins leads to desumoylation and the release of LRH-1 from the nuclear speckles, while a large portion of SUMO molecules, which may correspond to certain proteins that retained SUMO-1, cleaved SUMO-1, or free SUMO-1, are retained in PML-NBs.
If the observed isopeptidase inhibitor-sensitive desumoylation and release of LRH-1 is mediated by the activation of the SuPr-1 family of enzymes, the results of the above experiment are in an apparent contradiction with those obtained in SuPr-1-overexpressing cells, where PML-NBs were disrupted. We have to assume that the observed desumoylation of LRH-1 is either not selective and is accompanied by at least a partial disruption of the PML-NBs or that it is mediated by other, specific isopeptidases that do not cleave the SUMO moiety from PML. We favor the first scenario, which is also supported by the less-compact appearance of PML dots and the partial loss of SUMO-1 from PML-NBs in Triton X-100-permeabilized cells (Fig. 5A and C). We also note that high levels of SuPr-1 expression over a long period of time may lead to more dramatic effects compared to those exerted by the activation of the endogenous protein. In either case, however, the results clearly demonstrate that desumoylation of LRH-1 leads to its dissociation from the PML-NBs.
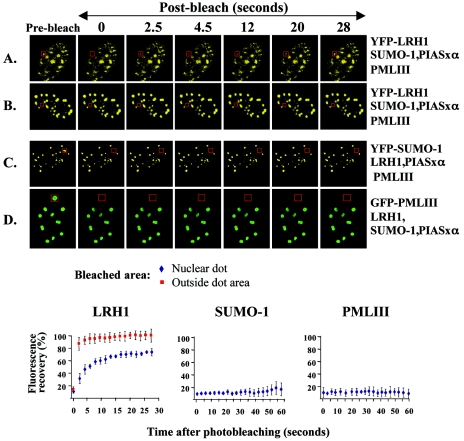
The kinetics of LRH-1 association with PML-NBs was evaluated by FRAP experiments. In cells expressing YFP-LRH-1, YFP-SUMO-1, or GFP-PML-III, individual nuclear dot areas and areas outside of the dots were photobleached by using a 2-s pulse, and the kinetics of the recovery of fluorescence intensity at the bleached spots were measured. Recovery of YFP-LRH-1 fluorescence in nuclear dot areas reached equilibrium at ca. 75% of the prebleach intensity within 15 s, pointing to the existence of a highly mobile and a relatively immobile population of the protein (Fig. 6A). In areas outside the nuclear speckles almost full recovery of YFP-LRH-1 fluorescence was observed within 2.5 s (Fig. 6B). The existence of an immobile fraction in the nuclear speckles and the difference in residence time between the two territories (dots and outside dot areas) provide further support for the idea that PML-NBs may function as molecular reservoirs of nuclear LRH-1. In agreement with a previous report, PML protein residing at NBs was found to be immobile (Fig. 6D), which is consistent with the nature of structural proteins that contribute to the integrity of NBs (6). Interestingly, however, SUMO-1 was also found to be a relatively immobile protein (Fig. 6C). This is most likely due to the sumoylation of immobile PML and other structural components (e.g., SP100) of the NBs, without precluding the possibility that NB-associated SUMO-1 stably resides in these structures as part of the sumoylation machinery in reserve for modification of incoming protein substrates.
FIG. 6.
Dynamic association of LRH-1 with PML-NBs. HeLa cells were transfected with the expression vectors indicated at the right, and expression was detected by direct fluorescence of YFP or GFP in live cells. After targeted photobleaching of the boxed areas corresponding to nuclear dots (A, C, and D) or areas outside the nuclear dots (B), serial images were collected every 2.5 s. Images from one experiment at selected time points are shown. The graphs at the bottom represent fluorescence recovery curves for the bleached areas. The results are presented as mean values and standard errors of the percentages of the postbleach fluorescence intensities at the irradiated areas relative to the prebleach intensity from five independent experiments.
Taken together, these results suggest that association of LRH-1 with PML-NBs is a dynamic process, which involves rapid sumoylation and desumoylation-driven exchange of the majority of LRH-1 protein residing in these structures.
Sumoylation negatively regulates LRH-1 transcriptional activity by excluding it from active chromatin domains.
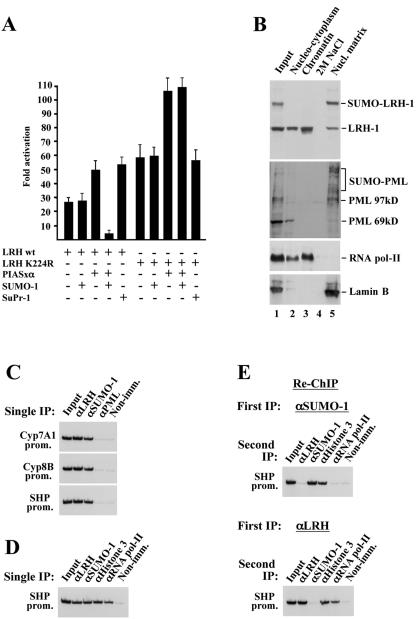
The regulated SUMO-dependent shuttling of LRH-1 into PML-NBs defines two spatially sequestered pools of the protein with apparently different roles in transcription. Therefore, we analyzed the functional consequence of sumoylation on the transcriptional activity of LRH-1 and on its recruitment to target genes in the context of chromatin. In transient-transfection assays the sumoylation-deficient mutant of LRH-1 exhibited an approximately twofold increased transactivation capacity (Fig. 7A). A similar level of increase in wtLRH-1-mediated transcription was observed in SuPr1-transfected cells, suggesting that sumoylation elicits a repressive effect on the transcriptional activity of LRH-1. Although in our Western blot assays (Fig. 1) we could not observe sumoylated LRH-1 in cells transfected with wtLRH-1 alone, the different transactivation potential of wild-type and mutant LRH-1 may be explained by assuming that some of the overexpressed wild-type protein became sumoylated by endogenous SUMO-1 and PIASxα which, due to the high turnover of the modification, limited detection. To clarify this, we performed additional assays by coexpressing LRH-1 with PIASxα and SUMO-1. Interestingly, coexpression of PIASxa alone with either wild-type and the K224R mutant of LRH-1 increased their transactivation potential (Fig. 7A), suggesting that PIASxα can function as a coactivator for LRH-1 independent of sumoylation. However, when SUMO-1 was coexpressed (conditions when sumoylated LRH-1 can be detected by Western blot analysis), an eightfold decrease in wtLRH-1-induced promoter activity was observed. Importantly, no such repression was evident in LRH-K224R-induced transcription (Fig. 7A). In principle, a repressive effect in reporter assays could be the result of a direct action of SUMO-1 on LRH-1 properties or an indirect effect brought about the modification of coactivators (e.g., p300) (19) or corepressors (e.g., HDAC) (11, 26, 49), which may modulate LRH-1 activity. Although the results obtained with the sumoylation-deficient LRH mutant argue against the latter possibility, in either of the above cases SUMO is presumed to act on promoter-bound LRH-1-containing transcription complexes. An alternative possibility is that sumoylated LRH-1 cannot access its target genes because it is spatially sequestered to nuclear locations from which active chromatin domains are excluded. Because, as shown above, sumoylated LRH-1 is localized in PML-NBs, one way to distinguish between these possibilities is to determine whether sumoylated LRH-1 and/or PML can associate with chromatin and LRH-1-regulated target genes.
FIG. 7.
SUMO-modified LRH-1 is not associated with transcriptionally active LRH-1 target genes in vivo. (A) Sumoylation has a repressive effect on the transactivation potential of LRH-1. HeLa cells were transfected with the SHP-luc reporter containing the 500-bp proximal promoter region of the SHP gene in front of firefly luciferase cDNA, together with the indicated expression vectors. The bars represent mean values and standard errors of normalized luciferase activities from three independent experiments and are expressed as the fold increase over the values obtained from cells transfected with the reporter alone. (B) C3A-HepG2 cells were sequentially extracted with Triton X-100 (Nucleocytoplasm), DNase I, and (NH4)2SO4 (Chromatin), and 2 M NaCl. Equal aliquots of each fraction and the solubilized pellet (Nucl. Matrix) were subjected to SDS-PAGE and, after being blotted to nitrocellulose membranes, were stained with the indicated antibodies. (C and D) ChIP assays were performed with the indicated antibodies by using cross-linked, soluble chromatin from mouse livers. LRH-1 target promoters (CYP7A1, CYP8B, and SHP genes) in the immunoprecipitates were detected by radioactive PCR. (E) αSUMO-1 (top panel) or αLRH-1-immunoprecipitated (bottom panel) complexes (1st IP) were eluted from the protein G-Sepharose beads and, after dilution, were subjected to a second immunoprecipitation step with the indicated antibodies (2nd IP) and analyzed by radioactive PCR.
To this end, we first performed a nuclear protein fractionation procedure as described in references 22 and 37. In the first fractionation step, soluble nucleocytoplasmic proteins are removed by extraction with Triton X-100. Chromatin and associated proteins are then released by DNase I digestion and extraction by 0.25 M ammonium sulfate. After being washed with 2 M NaCl, the remaining insoluble material in the pellet is composed of nuclear matrix proteins (22, 37). Equal amounts of the different fractions were analyzed by Western blot analysis, with antibodies to LRH-1, PML, RNA polymerase (Pol) II, and lamin B (Fig. 7B). As expected, RNA Pol II was distributed mainly in the “chromatin” fraction and to a lesser extent in the “nucleocytoplasmic” fraction. Lamin B and sumoylated PML were detected only at the “nuclear matrix” fraction. These results verify that the different fractions are essentially pure or that only minimal cross-contamination occurred during the procedure. The majority of unmodified LRH-1 was detected in the detergent-soluble and chromatin fractions. Importantly, SUMO-modified LRH-1 was solely observed in the final insoluble fraction (Fig. 7B), indicating that sumoylated LRH-1 is associated with the nuclear matrix but not with DNA. Although the above data argue against the possibility that sumoylated LRH-1 is targeted to gene regulatory regions, they do not entirely exclude it. It is still possible that, due to its tight association with components of the nuclear matrix, chromatin-bound SUMO-LRH-1 could not be liberated by DNase treatment or that the DNase-I-released protein was somehow desumoylated.
Therefore, we addressed the issue by using an independent approach. We performed ChIP assays with soluble chromatin prepared from mouse liver cells that were cross-linked in their native tissue environment. We analyzed LRH-1, SUMO-1, and PML occupancy on three well-characterized LRH-1-regulated genes (Cyp7A1, Cyp8B, and SHP). As expected, LRH-1 occupancy was observed at the promoter regions of all three genes (Fig. 7C and D). Interestingly, however, positive ChIP signals could be detected in anti-SUMO-1-immunoprecipitated chromatin, but not in that obtained with anti-PML. To determine whether the observed SUMO-1 signal corresponds to sumoylated LRH-1 and to find a correlation between SUMO-1 occupancy and the activation state of the genes, we performed sequential ChIP assays. In these experiments SUMO-1-containing, or LRH-1-containing chromatin purified by the first immunoprecipitation was eluted from the protein G-Sepharose beads and subjected to a second immunoprecipitation with antibodies recognizing SUMO-1, LRH-1, histone 3, or RNA Pol II. As shown in Fig. 7E, histone H3, but not LRH-1 or RNA Pol II, could be detected on the SUMO-1-associated SHP promoter. Furthermore, SUMO-1 was absent in LRH-1-occupied chromatin, which contained histone H3 and RNA Pol II. This finding clearly demonstrates that sumoylated LRH-1 is not associated with its target genes. Because RNA Pol II recruitment is a molecular indicator of the activation state of genes, the results point to the existence of two populations of hepatocytes or alleles at any given time: one with active promoters, occupied by unmodified LRH-1 and RNA Pol II, and another with inactive promoters, occupied by an unidentified sumoylated protein(s) but lacking LRH-1 and RNA Pol II.
DISCUSSION
LRH-1 is an orphan member of the nuclear receptor superfamily, which plays a pivotal role in early endodermal development, whereas in adults it controls the expression of enzymes involved in lipid and bile acid homeostasis (12, 16, 20, 31, 36, 39). In addition, LRH-1 has also been implicated in cell proliferation via cross talk with the β-catenin signaling pathway (8). The functioning of LRH-1 in such diverse biological processes highlights the importance of identification and exploration of potential signaling pathways that may modulate its activity.
We demonstrate here that LRH-1 is a direct substrate for the SUMO conjugation machinery and that sumoylation can modulate its activity. LRH-1 is reversibly modified by SUMO-1 at a single lysine residue, located at the hinge domain of the protein. Several lines of evidence described here suggest that SUMO modification-mediated inhibition of LRH-1 activity is due to its spatial sequestration from active chromatin domains. First, we demonstrate that sumoylated LRH-1 is localized in PML-containing nuclear bodies but not in nuclear territories outside them. Second, sumoylated LRH-1 is not associated with chromatin and its actively transcribed target genes. Third, desumoylation, which leads to the dissociation of LRH-1 from PML-NBs, increases its transcriptional activity.
Various functions have been attributed to PML-NBs, including a role in transcriptional regulation (52). Although the precise mechanism by which PML-NBs regulate gene expression is unknown, accumulating evidence on various transcription factors has led to the proposal of speculative models, which assume that PML-NBs may function either as “storage sites,” which can titrate the concentration of soluble factors in the nucleus, or as sites of specific posttranslational modifications (e.g., sumoylation), or as scaffolds into which transcription complexes assemble (52). Our results provide experimental evidence for links between the proposed functions by demonstrating a dynamic, sumoylation-dependent association of LRH-1 with the PML-NBs. The sumoylation machinery at these sites may play an operational role in the supply of active LRH-1 for transcription, as demonstrated by the finding that the release of LRH-1 from PML-NBs requires enzymatic removal of the SUMO moiety. Although the mode of activation of SUMO proteases in response to nuclear protein concentration is not known, it is clear that their function is central to the establishment of a dynamic equilibrium between the two pools of the LRH-1 protein: the one localized at PML-NBs and the other an unmodified soluble pool situated outside the PML-NBs. In this way, SUMO-mediated compartmentalization may play a role in regulating the actual nuclear concentration of the protein available for transcription in addition to or more likely in combination with the pathways that control the production and degradation of LRH-1.
The PML protein itself is also subject to SUMO modification, which is required for the proper formation of the nuclear body (3, 13, 35, 53). Interestingly, global desumoylation of proteins brought about by overexpressing a SUMO protease or by knocking down endogenous SUMO-1 expression results in relocalization of PML into a small number of nuclear structures, which unlike the regular PML-NBs accumulate RNA Pol II. These novel PML deposit sites may therefore represent hot spots of active transcription of genes although, because LRH-1 was not concentrated in these territories, it is unlikely that they contribute to the desumoylation-mediated activation of LRH-1 target genes. Nevertheless, the observation illuminates the plasticity of specific territories within the nuclear environment, where proteins can accumulate in response to different stimuli and possibly exercise distinct biological functions. Consistent with this notion is the recent demonstration of association of PML with nucleoli in response to specific stress signals (2).
SUMO modification of transcription factors is becoming increasingly recognized as an important pathway regulating gene expression (10, 27, 34, 38, 40, 41, 48). Several models have been put forward as possible mechanism(s) for SUMO modification-dependent transcriptional effects, including competition with other modifications, modulation of interactions with DNA and other proteins on the promoters, or the above-mentioned “sequestration model,” whereby sumoylation leads to the localization of the transcription factor to subnuclear domains where it cannot access its target genes (4, 17, 18, 42, 46, 52). Although the models described above are not necessarily mutually exclusive, probably no generally applicable mechanism exists and, depending on the modified factor, sumoylation may elicit its transcriptional effects by several means. This scenario is corroborated by our sequential ChIP assays, which detected SUMO-1 or LRH-1 on the promoters but not the two proteins together. The results are consistent with the idea that two populations of hepatocytes or promoter alleles exist at any given time: one with active promoters, occupied by unmodified LRH-1 and RNA Pol II, and another, with inactive promoters occupied by unidentified sumoylated protein(s), which lack LRH-1 and RNA Pol II. This points to the parallel involvement of a direct, LRH-1 modification-independent function of SUMO-1 at the promoters. The recent finding that histone H4 and, to a lesser extent, histones H2A, H2B, and H3 can be modified by SUMO (43), which at least in transfected reporters mediates gene silencing, is in line with our detection of histones but not of an active RNA Pol II-containing preinitiation complex on the SUMO-associated promoter (Fig. 7 and data not shown). Although our results do not provide definite evidence for the presence of sumoylated nucleosomes on the repressed promoters, these results clearly show that sumoylated proteins (histones or others) can be associated with chromatin, which, as indicated by the absence of PML ChIP signals on these regulatory regions, most likely reside outside the PML-NBs. In this respect we note that a significant amount of SUMO-1 fluorescence signal can be detected throughout the nucleoplasm, outside of the PML-NBs (Fig. 3, 5, and 6). SUMO modification of some of these proteins may affect preinitiation complex formation at the promoters by recruiting corepressor activities. Examples for such direct repression have been provided by studies on Elk-1 and histones, whose sumoylation at promoters can facilitate the recruitment of HDAC or HP-1 repressor proteins (43, 49).
Our results on LRH-1 point to a fundamentally different control mechanism for the set of transcription factors that are modified by SUMO at PML-NBs. Sumoylation of these factors leads to their “storage” at PML-NBs, and thus they become spatially sequestered from active chromatin. SUMO-dependent compartmentalization of LRH-1 is a reversible and highly dynamic process, which may play a fine-tuning role in controlling the availability of this factor to chromatin. PML-NBs, where sumoylated LRH-1 accumulates, are not regarded as sites of active transcription since they are devoid of DNA, but nascent RNAs and transcriptionally active genomic loci are concentrated in their immediate periphery (31, 47). In light of this and the evidence provided here, we propose that sumoylation may act as a “modus operandi” for the formation of dynamic reservoirs of transcription factors in the vicinity of active chromatin domains. Shuttling of transcription factors between these nuclear domains may represent an important control mechanism of gene expression.
Acknowledgments
We are indebted to R. Hay, F. Melchior, N. Kotaja, J. Palvimo, and H. Will for providing reagents; N. Katrakili for technical assistance; N. Tavernarakis for help with confocal microscopy; and P. Hatzis for helpful comments on the manuscript.
This study was supported by grants from EU (QLRT-2000-01513 and LSHG-2004-502950) and from GSRT (01ED509).
REFERENCES
- 1.Alcalay, M., L. Tomassoni, E. Colombo, S. Stoldt, F. Grignani, M. Fagioli, L. Szekely, K. Helin, and P. G. Pelicci. 1998. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell. Biol. 18:1084-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernardi, R., P. P. Scaglioni, S. Bergmann, H. F. Horn, K. H. Vousden, and P. P. Pandolfi. 2004. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 6:665-672. [DOI] [PubMed] [Google Scholar]
- 3.Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. [DOI] [PubMed] [Google Scholar]
- 4.Boggio, R., R. Colombo, R. T. Hay, G. F. Draetta, and S. Chiocca. 2004. A mechanism for inhibiting the SUMO pathway. Mol. Cell 16:549-561. [DOI] [PubMed] [Google Scholar]
- 5.Boisvert, F. M., M. J. Hendzel, and D. P. Bazett-Jones. 2000. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 148:283-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boisvert, F. M., M. J. Kruhlak, A. K. Box, M. J. Hendzel, and D. P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botrugno, O. A., E. Fayard, J. S. Annicotte, C. Haby, T. Brennan, O. Wendling, T. Tanaka, T. Kodama, W. Thomas, J. Auwerx, and K. Schoonjans. 2004. Synergy between LRH-1 and β-catenin induces G1 cyclin-mediated cell proliferation. Mol. Cell 15:499-509. [DOI] [PubMed] [Google Scholar]
- 9.Carmo-Fonseca, M. 2002. The contribution of nuclear compartmentalization to gene regulation. Cell 108:513-521. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarti, S. R., R. Sood, S. Nandi, and G. Nucifora. 2000. Posttranslational modification of TEL and TEL/AML1 by SUMO-1 and cell-cycle-dependent assembly into nuclear bodies. Proc. Natl. Acad. Sci. USA 97:13281-13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colombo, R., R. Boggio, C. Seiser, G. F. Draetta, and S. Chiocca. 2002. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 3:1062-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Castillo-Olivares, A., J. A. Campos, W. M. Pandak, and G. Gil. 2004. The role of alpha1-fetoprotein transcription factor/LRH-1 in bile acid biosynthesis: a known nuclear receptor activator that can act as a suppressor of bile acid biosynthesis. J. Biol. Chem. 279:16813-16821. [DOI] [PubMed] [Google Scholar]
- 13.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localization. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 14.Eloranta, J. J., and H. C. Hurst. 2002. Transcription factor AP-2 interacts with the SUMO-conjugating enzyme UBC9 and is sumolated in vivo. J. Biol. Chem. 277:30798-30804. [DOI] [PubMed] [Google Scholar]
- 15.Emerson, B. M. 2002. Specificity of gene regulation. Cell 109:267-270. [DOI] [PubMed] [Google Scholar]
- 16.Fayard, E., J. Auwerx, and K. Schoonjans. 2004. LRH-1: an orphan nuclear receptor involved in development, metabolism, and steroidogenesis. Trends Cell Biol. 14:250-260. [DOI] [PubMed] [Google Scholar]
- 17.Gill, G. 2003. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 13:108-113. [DOI] [PubMed] [Google Scholar]
- 18.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046-2059. [DOI] [PubMed] [Google Scholar]
- 19.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11:1043-1054. [DOI] [PubMed] [Google Scholar]
- 20.Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, P. R. Maloney, T. M. Willson, and S. A. Kliewer. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517-526. [DOI] [PubMed] [Google Scholar]
- 21.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10:1467-1477. [DOI] [PubMed] [Google Scholar]
- 22.He, D. C., J. A. Nickerson, and S. Penman. 1990. Core filaments of the nuclear matrix. J. Cell Biol. 110:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodges, M., C. Tissot, K. Howe, D. Grimwade, and P. S. Freemont. 1998. Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am. J. Hum. Genet. 63:297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joseph, J., S. H. Tan, T. S. Karpova, J. G. McNally, and M. Dasso. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karpova, T. S., C. T. Baumann, L. He, X. Wu, A. Grammer, P. Lipsky, G. L. Hager, and J. G. McNally. 2003. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 209:56-70. [DOI] [PubMed] [Google Scholar]
- 26.Kirsh, O., J. S. Seeler, A. Pichler, A. Gast, S. Muller, E. Miska, M. Mathieu, A. Harel-Bellan, T. Kouzarides, F. Melchior, and A. Dejean. 2002. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ktistaki, E., and I. Talianidis. 1997. Modulation of hepatic gene expression by hepatocyte nuclear factor 1. Science 277:109-112. [DOI] [PubMed] [Google Scholar]
- 29.LaMorte, V. J., J. A. Dyck, R. L. Ochs, and R. M. Evans. 1998. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA 95:4991-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 31.Lu, T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6:507-515. [DOI] [PubMed] [Google Scholar]
- 32.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 33.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller, S., M. Berger, F. Lehembre, J. S. Seeler, Y. Haupt, and A. Dejean. 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 275:13321-13329. [DOI] [PubMed] [Google Scholar]
- 35.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pare, J. F., D. Malenfant, C. Courtemanche, M. Jacob-Wagner, S. Roy, D. Allard, and L. Belanger. 2004. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J. Biol. Chem. 279:21206-21216. [DOI] [PubMed] [Google Scholar]
- 37.Reyes, J. C., C. Muchardt, and M. Yaniv. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross, S., J. L. Best, L. I. Zon, and G. Gill. 2002. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10:831-842. [DOI] [PubMed] [Google Scholar]
- 39.Sablin, E. P., I. N. Krylova, R. J. Fletterick, and H. A. Ingraham. 2003. Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Mol. Cell 11:1575-1585. [DOI] [PubMed] [Google Scholar]
- 40.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sapetschnig, A., G. Rischitor, H. Braun, A. Doll, M. Schergaut, F. Melchior, and G. Suske. 2002. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 21:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell. Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 43.Shiio, Y., and R. N. Eisenman. 2003. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100:13225-13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 45.Soutoglou, E., B. Viollet, M. Vaxillaire, M. Yaniv, M. Pontoglio, and I. Talianidis. 2001. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 20:1984-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verger, A., J. Perdomo, and M. Crossley. 2003. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 4:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, J., C. Shiels, P. Sasieni, P. J. Wu, S. A. Islam, P. S. Freemont, and D. Sheer. 2004. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 164:515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, S. H., E. Jaffray, R. T. Hay, and A. D. Sharrocks. 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 12:63-74. [DOI] [PubMed] [Google Scholar]
- 49.Yang, S. H., and A. D. Sharrocks. 2004. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13:611-617. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Y., and Y. Xiong. 1999. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3:579-591. [DOI] [PubMed] [Google Scholar]
- 51.Zhong, S., P. Hu, T. Z. Ye, R. Stan, N. A. Ellis, and P. P. Pandolfi. 1999. A role for PML and the nuclear body in genomic stability. Oncogene 18:7941-7947. [DOI] [PubMed] [Google Scholar]
- 52.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]
- 53.Zhong, S., P. Salomoni, S. Ronchetti, A. Guo, D. Ruggero, and P. P. Pandolfi. 2000. Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. J. Exp. Med. 191:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]