Abstract
In 1997, an epizootic in Taiwan, Province of China, was caused by a type O foot-and-mouth disease virus which infected pigs but not cattle. The virus had an altered 3A protein, which harbored a 10-amino-acid deletion and a series of substitutions. Here we show that this deletion is present in the earliest type O virus examined from the region (from 1970), whereas substitutions surrounding the deletion accumulated over the last 29 years. Analyses of the growth of these viruses in bovine cells suggest that changes in the genome in addition to the deletion, per se, are responsible for the porcinophilic properties of current Asian viruses in this lineage.
Foot-and-mouth disease (FMD) is a highly contagious disease of cloven-hoofed animals, notably cattle, pigs, and sheep. Presence of FMD in a country severely limits its trade in animals and animal products due to international regulations designed to limit spread of the disease. FMD virus (FMDV) is a positive-stranded RNA virus belonging to the Aphthovirus genus in the family Picornaviridae. The FMDV RNA genome contains a single long open reading frame encoding a polypeptide that is cleaved to mature polypeptide products by virally encoded proteinases. In addition to four capsid proteins (VP1 to -4), the RNA-dependent RNA polymerase (3D), and three proteinases (L, 2A, and 3C), the FMDV genome encodes multiple mature polypeptides and intermediate cleavage products (20). The mature polypeptides include 3A, a highly conserved protein of 153 amino acids (aa) in most viruses examined to date. Changes in 3A have been associated with altered host range in the hepatoviruses (10, 11, 14, 15), rhinoviruses (12), and enteroviruses (13). Among the aphthoviruses, a deletion in 3A has been associated with bovine attenuation of egg-adapted FMDVs, which were developed and used as FMD vaccines in South America (9). In addition, we recently identified a similar deletion in the 3A coding region (3) which is associated with the porcinophilic properties of the virus that caused an outbreak of FMD in Taiwan in 1997 (3, 5). Interestingly, these deletions are all found in a region of the FMDV 3A protein that does not have a homologue in any of the 3A proteins of the well-characterized picornaviruses. In fact, all picornaviruses examined to date have 3A proteins less than 100 aa in length, except for equine rhinitis B virus, which has an extended 3A (110 aa) (25). Additionally, FMDV is unique among the picornaviruses in that it encodes three copies of 3B (VPg) (20), the viral genome-bound protein, which serves as a primer for picornavirus genome replication (17).
The source of the 3A deletions in egg-adapted FMDVs is unclear, but the fact that a deletion arose independently during serial passage of two different serotypes (9) suggests that mutations in 3A may reflect a common method of altering the host range of FMDV. The 3A deletion that we detected in the Taiwan virus (3) could have arisen as a result of viral adaptation to new environmental conditions and/or hosts. In particular, a virus with this deletion could have been selected from a wild-type virus by an enhanced ability to spread among the porcine populations in Asia. Alternatively, a virus with this deletion could have been derived from a live-attenuated vaccine strain of FMDV, which later became established in the field.
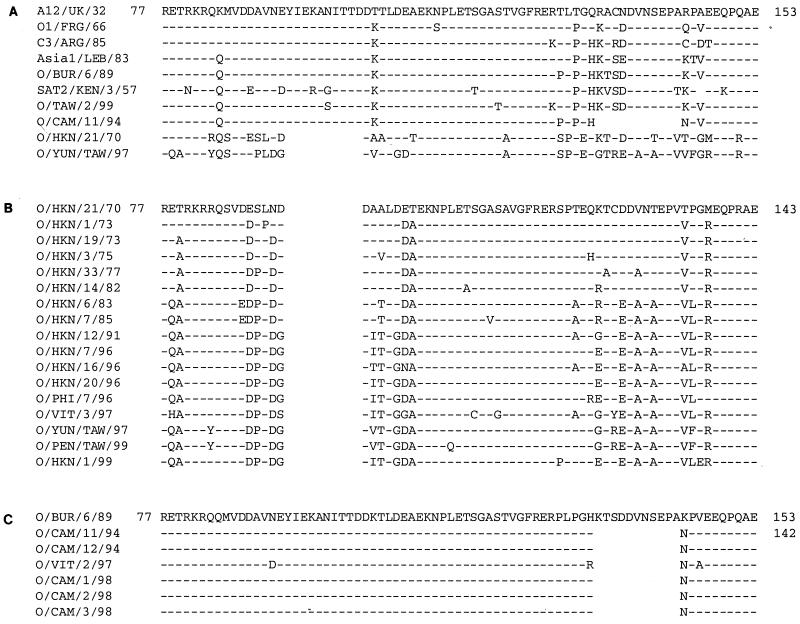
To investigate the source of the deleted and mutated segments of the 3A coding region of the Taiwan virus (referred to here as O/YUN/TAW/97), we undertook analyses of the 3A coding regions of 28 type O isolates from Southeast Asia (Table 1). For these studies, we selected viruses from several Asian countries based on analyses of VP1 nucleic acid sequences (N. J. Knowles and A. R. Samuel, unpublished data) which demonstrated that the virus causing the 1997 outbreak in Taiwan was related to a number of viruses obtained from Hong Kong and the Philippines. Interestingly, these viruses were also related to an FMDV strain that caused an outbreak in Moscow in 1995, which had been traced to a Chinese pork shipment (V. V. Drygin, personal communication, 1995). In addition, our analyses included isolates which had been obtained from pigs and cattle and deposited with the OIE/FAO World Reference Laboratory for FMD (WRLFMD) at the same time. These analyses revealed that the first half of the 3A coding region, which encodes an N-terminal hydrophilic domain and a hydrophobic domain capable of binding to membranes, was highly conserved among all viruses examined (results not shown). However, there were substantial alterations identified in the latter half of the 153-codon 3A coding region (Table 1; Fig. 1). These data show that the deletion previously found in the 3A coding region of O/YUN/TAW/97 (codons 93 to 102; hereafter referred to as the 93–102 deletion) is also present in O/HKN/21/70, the oldest virus in the WRLFMD collection from this region of the world. Other viruses examined from this region had full-length 3A coding regions, and a third group contained a different deletion, spanning codons 133 through 143 of 3A (133–143 deletion) (Table 1; Fig. 1).
TABLE 1.
Designation, origin, and 3A phenotype of FMD viruses examined in this study
| Virus designation | Geographic origina | Date of collection (day/mo/yr) | Host species | Deletionb | Sequence accession no.c | Reference or source |
|---|---|---|---|---|---|---|
| O1/FRG/66 | Kaufbeuren, Germany | 1966 | Bovine | None | X00871 | 8 |
| A10/ARG/61 | Argentina | 1961 | Bovine | None | X00429 | 4 |
| A12/UK/119/32 | Brasted, Kent, United Kingdom | 29/12/1932 | Bovine | None | M10975 | 19 |
| A22/USSR/65 | Azerbaijan | 1965 | Bovine | None | X74812 | —d |
| C1/SPA/70 | Santa Pau, Spain | 02/1970 | Porcine | None | AJ133357 | 24 |
| C3/ARG/85 | Marcos Juárez, Córdoba, | 12/1984 | Bovine | None | AJ007347 | 6 |
| Argentina | ||||||
| SAT2/KEN/3/57 | Kenya | 1957 | Bovine | None | AJ251473 | — |
| Asia1/LEB/83 | Kfar Kela, Lebanon | 1983 | Bovine | None | AJ294931, AJ295004 | This work |
| O/HKN/21/70 | Hang Tau, Sheung Shiu, N. T., | 13/03/1970 | Porcine | 93–102 | AJ294911, AJ294984 | This work |
| Kowloon, Hongkong | ||||||
| O/HKN/1/73 | Lantau Island, N. T., Kowloon, | 01/1973 | Porcine | 93–102 | AJ294912, AJ294985 | This work |
| Hong Kong | ||||||
| O/HKN/19/73 | Ha Cheung Sha, Lantau Island, | 19/06/1973 | Bovine | 93–102 | AJ294913, AJ294986 | This work |
| N.T., Kowloon, Hong Kong | ||||||
| O/HKN/1/75 | Shiu Hum Tsuen, Sun Tin, Yuen | 9/12/1974 | Bovine | None | AJ294914, AJ294987 | This work |
| Long, N.T., Kowloon, Hong | ||||||
| Kong | ||||||
| O/HKN/3/75 | Ping Shan, Yuen Long, N.T., | 23/12/1974 | Porcine | 93–102 | AJ294915, AJ294988 | This work |
| Kowloon, Hong Kong | ||||||
| O/HKN/33/77 | Tin Ping Shan, Sheung Shui, | 1977 | Porcine | 93–102 | AJ294916, AJ294989 | This work |
| N.T., Kowloon, Hong Kong | ||||||
| O/HKN/14/82 | Hei Ling Chau Island, N.T., | 25/02/1982 | Porcine | 93–102 | AJ294917, AJ294990 | This work |
| Kowloon, Hong Kong | ||||||
| O/HKN/17/82 | Pok Wai, San Tin, N.T., Hong | 19/03/1982 | Bovine | None | AJ294918, AJ294991 | This work |
| O/HKN/6/83 | Pokfulam, Hong Kong Island | 18/12/1982 | Bovine | 93–102 | AJ294919, AJ294992 | This work |
| O/HKN/7/85 | Ma On Kong, Pat Heung, N.T., | 25/01/1985 | Porcine | 93–102 | AJ294920, AJ294993 | This work |
| Kowloon, Hong Kong | ||||||
| O/BUR/2/89 | Hlegu, Burma | 10/06/1989 | Bovine | None | AJ294904, AJ29477 | This work |
| O/BUR/6/89 | Tharyawaddy, Burma | 02/07/1989 | Porcine | None | AJ294905, AJ294978 | This work |
| O/HKN/12/91 | Sek Kwu Chau, N.T., Kowloon, | 26/11/1991 | Porcine | 93–102 | AJ294921, AJ294994 | This work |
| Hong Kong | ||||||
| O/CAM/11/94 | Mong Russey District, | 09/1994 | Bovine | 133–143 | AJ294906, AJ294979 | This work |
| Battambang Province, Cambodia | ||||||
| O/CAM/12/94 | Mong Russey District, | 09/1994 | Porcine | 133–143 | AJ294907, AJ294980 | This work |
| Battambang Province, Cambodia | ||||||
| O/HKN/7/96 | Government abattoir, Hong Kong | 06/02/1996 | Bovinee | 93–102 | AJ294922, AJ294995 | This work |
| O/HKN/16/96 | Takwuling, N.T., Kowloon, Hong Kong | 29/03/1996 | Porcine | 93–102 | AJ294923,AJ294996 | This work |
| O/HKN/20/96 | Hong Kong | 17/04/1996 | Bovinee | 93–102 | AJ294924, AJ294997 | This work |
| O/PHI/7/96 | Mahabang Parang, Angono, | 1996 | Porcine | 93–102 | AJ294926, AJ294999 | This work |
| Philippines | ||||||
| O/VIT/2/97 | Vietnam | 1997 | Bovine | 133–143 | AJ294929, AJ295002 | This work |
| O/VIT/3/97 | Vietnam | 1997 | Porcine | 93–102 | AJ294930, AJ295003 | This work |
| O/YUN/TAW/97 | Yunlin Prefecture, Taiwan, POC | 04/1997 | Porcine | 93–102 | AF308157 | 3 |
| O/CAM/1/98 | Borset, Kg. Speu, Cambodia | 05/01/1998 | Porcine | 133–143 | AJ294908, AJ294981 | This work |
| O/CAM/2/98 | Kg. Speu, Cambodia | 05/01/1998 | Bovine | 133–143 | AJ294909, AJ294982 | This work |
| O/CAM/3/98 | Kg. Speu, Cambodia | 05/01/1998 | Bovine | 133–143 | AJ294910, AJ294983 | This work |
| O/HKN/1/99 | Mong Tseng Tsuen, Yuen Long, | 05/01/1999 | Porcine | 93–102 | AJ294925, AJ294998 | This work |
| N.T., Kowloon Hong Kong | ||||||
| O/PEN/TAW/99 | Penghu Island, Taiwan, POC | 02/1999 | Porcine | 93–102 | AJ294928, AJ295001 | This work |
| O/TAW/2/99 | Kinmen Island, Taiwan, POC | 06/1999 | Bovine | None | AJ294927, AJ295000 | This work |
Most were obtained from the WRLFMD as low-passage-number stocks isolated in primary bovine thyroid cell cultures or in cultures of the swine kidney cell line IB-RS-2. Other isolates studied included a virus obtained from Yunlin Prefecture, Taiwan, in April 1997 (O/YUN/TAW/97; referred to as OTai in reference 3) and an isolate obtained from Penghu Island Prefecture, Taiwan, in February 1999 (O/PEN/TAW/99). N.T., New Territories; POC, Province of China; Kg., Kampong.
Codons deleted from the 153-codon 3A coding region.
For sequences determined in this work, RNA extracted from virus stocks (in some cases following amplification in BHK cells) using standard techniques was used to prepare cDNA using mouse mammary tumor virus reverse transcriptase (Promega or Life Technologies) and random hexamers. Virus-specific cDNA fragments were amplified with Taq polymerase (Promega or Sigma) and pairs of oligonucleotide primers using PCR and standard techniques (22). Primer pairs were located in the 2C, 3A, 3B, and 3C regions or in the VP3/2B region of the genome. PCR products were sequenced using asymmetric amplification with Cy5-labeled oligonucleotide primers and resolved on a ALFexpress II system (Amersham Pharmacia Biotech), or by using asymmetric amplification with Big-dye terminators (ABI), and resolved on an ABI 373 sequencer.
—, no literature citation available.
Animal imported from mainland China.
FIG. 1.
Alignment of predicted amino acid sequences corresponding to codons 77 to 153 of the 3A coding region of selected viruses. Sources of the sequence data are listed in Table 1. (A) Comparison of the sequences of selected deleted genomes to those of prototype strains of FMDV. (B) Comparison of sequences of O/HKN/21/70 genotype viruses, which display the deletion of codons 93 to 102. (C) Comparison of sequences of O/CAM/11/94 genotype viruses, which display the deletion of codons 133 to 143. In each panel, viruses are shown in chronological order with the predicted amino acids shown in one-letter code. Dashes indicate identity with oldest virus in the group; spaces represent deleted codons.
Among the viruses in the O/HKN/21/70 lineage, most were isolated from pigs (Table 1), consistent with our previous studies showing that the 3A coding region of O/YUN/TAW/97 could confer bovine growth restriction on genetically engineered viruses (3). In our survey, we studies 6 of 27 type O bovine isolates obtained from bovine samples submitted to the WRLFMD by Hong Kong between 1970 and 1999 (461 type O viruses were isolated from porcine samples submitted from Hong Kong during this period). Of these six isolates, four were of the O/HKN/21/70 genotype, somewhat surprising in light of the above-mentioned studies (3). The bovine isolates from this lineage were obtained only from Hong Kong; none of the viruses obtained from cattle from Cambodia or Vietnam during this time period contained the 93–102 deletion.
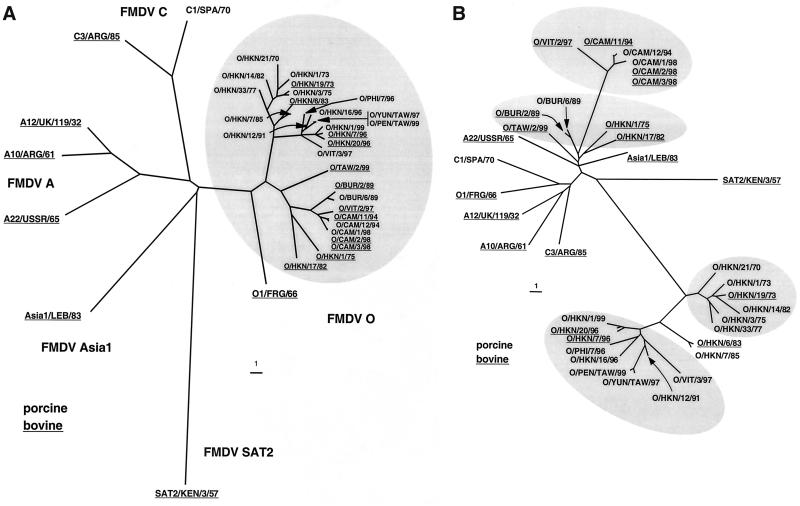
A detailed examination of the alterations in the coding capacity of the 3A coding region of the O/HKN/21/70 lineage between 1970 and 1999 revealed that the region encoding aa 77 to 143 accumulated a large number of substitutions (Fig. 1B). These substitutions fell into two regions, one surrounding the site of the 93–102 deletion and the other in the region of the deletion observed in O/CAM/11/94 and related viruses (Fig. 1). Dendrograms prepared from either the VP1 (Fig. 2A) or 3A (Fig. 2B) coding region showed similar (although not identical [see below]) clusterings of viruses with the 93–102-deleted 3A genotype, suggesting that these two regions coevolved over the time period rather than being exchanged by recombination.
FIG. 2.
Dendrograms showing relationships among Asian serotype O viruses and prototype strains of FMDV. Relationships were determined based on nucleotide differences of the sequences encoding the capsid protein VP1 (A) or nonstructural protein 3A (B). The bars represent the distances as percentage differences (7). For VP1, 633 to 648 nucleotides were used for the analyses; in the case of 3A, 426 to 459 nucleotides were used. Sources of the sequence data are listed in Table 1. Distance matrices were calculated using a program written by N. J. Knowles; all pairwise comparisons were performed giving each base substitution equal statistical weight (ambiguities were ignored). Phylogenetic trees were constructed using the neighbor-joining algorithm of Saitou and Nei (23) as implemented in the program NEIGHBOR, part of the PHYLIP package (7). Trees were drawn using the program TreeView 1.6 (16).
Four of the five Asian viruses that had a full-length 3A coding region were obtained from cattle (Table 1). These included the recently identified bovine-virulent virus isolated from a sub clinically infected animal on the Taiwanese island of Kinmen (O/TAW/2/99). The dendrogram generated from VP1 sequence data (Fig. 2A) shows that these full-length 3A viruses cluster with the viruses containing the 133–143 deletion (from Vietnam and Cambodia). Based on these data, it appears the 133–143 deletion must have arisen more recently than the 93–102 deletion found in the O/HKN/21/70 group of viruses. Specifically, the dendrograms based on VP1 nucleotide sequence data show that the viruses with this deletion, O/CAM/11/94, O/CAM/12/94, O/CAM/1/98, O/CAM/2/98, O/CAM/3/98, and O/VIT/2/97, are closely related to the BUR/89 viruses, which contain full-length 3A coding regions (Fig. 1C and 2A). Unlike the O/HKN/21/70 lineage viruses, the O/CAM/11/94 virus group did not undergo significant mutation in the last half of the 3A coding region between 1994 and 1998 (Fig. 1C). Rather, there were essentially no changes in 3A surrounding the deletion over this time frame. Furthermore, unlike the O/HKN/21/70 viruses, there did not appear to be an association of this deletion with a particular host species, although the number of viruses identified in this group was quite low.
Analysis of the dendrogram generated from the 3A sequence data shows that these viruses can be subdivided into four groups. One group contains viruses with a full-length 3A coding region, another consists of viruses with the 133–143 deletion, and the last two groups contain viruses with the 93–102 deletion, one with the viruses from the 1970 to 1982 and a second including viruses obtained in the 1990s (Fig. 2B).
Determination of serotype O virus host-range in primary lingual keratinocyte cultures.
To obtain a relevant in vitro model to study FMD, we developed a system based on the known predilection of the virus for the epidermis of the tongues of livestock animals. To this end, we adapted a method of producing human dermal keratinocytes to isolate keratinocytes from the tongues of slaughtered cattle and pigs (based on the method of Barlow and Pye [2] and data not shown) and used these cells to evaluate the ability of selected viruses to replicate in swine and bovine cells. Using these cells, we were able to demonstrate that genetically engineered viruses expressing the 3A coding region of O/YUN/TAW/97 replicated 10- to 100-fold more in keratinocytes of swine origin than those of bovine origin (results not shown), consistent with our earlier studies (3).
Viruses containing full-length 3A coding region or the coding region with the 133–143 deletion (Table 1; Fig. 1) replicated well in keratinocytes obtained from both pigs and cattle (Fig. 3). However, the Taiwan virus isolate obtained from pigs in 1997 and the closely related viruses obtained from pigs in Vietnam and Hong Kong (Fig.1 and 2; Table 1) grew approximately 10- to 100-fold better in the swine-derived keratinocyte cultures than in cultures prepared from bovine tissue (Fig. 3). Somewhat surprisingly, the viruses from the 93–102-deleted 3A lineage isolated before O/HKN/6/83 grew very well in the bovine cells, with the first virus in this lineage, O/HKN/21/70, growing equally well in bovine and swine cells (Fig. 3). These results indicate that the 93–102 deletion in this region of 3A cannot, alone, account for the inability of O/YUN/TAW/97 to replicate in bovine cells. In addition, these results suggest that other mutations in the genome of this virus lineage, possibly generated in response to the deletion of aa 93 to 102, may be responsible for the species specificity of O/YUN/TAW/97.
FIG. 3.
Growth of selected viruses in bovine ( ) and swine (■) keratinocytes. Representative data show replication of the indicated viruses in keratinocytes of the species indicated, expressed relative to side-by-side replication of vCRM8 (a genetically engineered virus which is highly virulent in bovine and swine [1, 21]). The bar and extended bar represent the results of two independent values obtained in the same experiment. Nomenclature and host of origin for viruses are given in Table 1; underlined viruses were isolated from bovine samples, and all other isolates were from swine (Table 1). For the replication assays, monolayer cultures of bovine or swine keratinocytes derived from lingual epithelial tissue (prepared using a modification of a method for developed for cultivating keratinocytes from human skin biopsies [2]) were incubated with virus for 1 h at 37°C with a multiplicity of infection of 10 (based on plaque assay in BHK cells). At the end of the adsorption period, the inoculum was removed and the cell layers were rinsed twice with ice-cold 145 mM NaCl, 25 mM morpholineethanesulfonic acid (pH 5.5) to remove residual virus particles; the cells were then rinsed with BME containing 1% fetal calf serum and 25 mM HEPES (pH 7.4) and incubated in 1 ml of this medium for 6 h at 37°C. The cultures were frozen at −70°C, and the titer (PFU per milliliter) was determined on BHK cells (18).
) and swine (■) keratinocytes. Representative data show replication of the indicated viruses in keratinocytes of the species indicated, expressed relative to side-by-side replication of vCRM8 (a genetically engineered virus which is highly virulent in bovine and swine [1, 21]). The bar and extended bar represent the results of two independent values obtained in the same experiment. Nomenclature and host of origin for viruses are given in Table 1; underlined viruses were isolated from bovine samples, and all other isolates were from swine (Table 1). For the replication assays, monolayer cultures of bovine or swine keratinocytes derived from lingual epithelial tissue (prepared using a modification of a method for developed for cultivating keratinocytes from human skin biopsies [2]) were incubated with virus for 1 h at 37°C with a multiplicity of infection of 10 (based on plaque assay in BHK cells). At the end of the adsorption period, the inoculum was removed and the cell layers were rinsed twice with ice-cold 145 mM NaCl, 25 mM morpholineethanesulfonic acid (pH 5.5) to remove residual virus particles; the cells were then rinsed with BME containing 1% fetal calf serum and 25 mM HEPES (pH 7.4) and incubated in 1 ml of this medium for 6 h at 37°C. The cultures were frozen at −70°C, and the titer (PFU per milliliter) was determined on BHK cells (18).
The two bovine viruses with the 93–102 deletion genotype that were isolated in the late 1990s (O/HKN/7/96 and O/HKN/20/96) displayed an intermediate growth phenotype in bovine keratinocytes, suggesting that a porcine-derived virus may have readapted to a bovine host. Although we have no direct evidence for this phenomenon, it is clear that most of the type O viruses deposited at the WRLFMD from Hong Kong during this time period were from pigs (see above). Thus, if passage through cattle could select altered sequences permitting replication in this host, it is also possible that propagation in bovine cells in vitro could result in acquisition of new sequences not present in the field. From this standpoint, it is worth noting that many of the early samples used in this study were propagated initially in bovine thyroid cell cultures (results not shown). However, our initial attempts to adapt porcinophilic viruses to grow in bovine keratinocytes were not successful (results not shown), indicating that the observed ability of the early O/HKN/21/70 genotype porcine-derived viruses to grow in bovine keratinocytes was not a result of their in vitro manipulations prior to our analyses.
Although we analyzed only a few of the Asian serotype O viruses present in the WRLFMD collection, most type O viruses with full-length 3A coding regions were obtained from cattle (these include the virus causing an outbreak in Taiwan in 1999, which is related to the viruses responsible for the outbreaks in the Republic of Korea and Japan in March of 2000 and in eastern Russia and Mongolia in April of 2000). On the other hand, viruses with the 3A 133–143 deletion, which were all obtained from Vietnam and Cambodia, were isolated from both cattle and pigs.
Current studies are aimed at attempting to utilize reverse genetics to study which regions of the genome interact with 3A and determine the biochemical basis by which these genetic alterations produce the altered host range in vivo and in cell culture. These studies may help to elucidate how new viruses with altered pathogenesis and host range emerge and are likely to yield important information concerning the replicative cycle of FMDV in infected cells and animals.
Acknowledgments
We thank the Organization for Economic Cooperation and Development and the National Research Initiative Competitive Grants program, USDA/CSREES, for financial support. This work was partially supported by the Agricultural Research Service, USDA, and the U.K. Ministry of Agriculture, Fisheries and Food (MAFF), grant 2911.
We thank J. Lubroth, Foreign Animal Disease Diagnostic Laboratory, NVSL, APHIS, USDA, PIADC, for supplying the Penghu Island isolate. We thank V. V. Drygin, All Russian Research Institute for Animal Health, Vladimir, Russia, for supplying information on the FMDV strain which caused the 1995 outbreak in Moscow.
REFERENCES
- 1.Almeida M R, Rieder E, Chinsangaram J, Ward G, Beard C, Grubman M J, Mason P W. Construction and evaluation of an attenuated vaccine for foot-and-mouth disease: difficulty adapting the leader proteinase-deleted strategy to the serotype O1 virus. Virus Res. 1998;55:49–60. doi: 10.1016/s0168-1702(98)00031-8. [DOI] [PubMed] [Google Scholar]
- 2.Barlow Y, Pye R J. Keratinocyte culture. In: Pollard J W, Waljer J M, editors. Methods in molecular in biology. 5. Animal cell culture. Clifton, N.J: Humana Press; 1989. pp. 83–98. [Google Scholar]
- 3.Beard C W, Mason P W. Genetic determinants of altered virulence of Taiwanese foot-and-mouth disease virus. J Virol. 2000;74:987–991. doi: 10.1128/jvi.74.2.987-991.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll A R, Rowlands D J, Clarke B E. The complete nucleotide sequence of the RNA coding for the primary translation product of foot and mouth disease virus. Nucleic Acids Res. 1984;12:2461–2472. doi: 10.1093/nar/12.5.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn C S, Donaldson A I. Natural adaption to pigs of a Taiwanese isolate of foot-and-mouth disease virus. Vet Rec. 1997;141:174–175. doi: 10.1136/vr.141.7.174. [DOI] [PubMed] [Google Scholar]
- 6.Escarmis C, Carrillo E C, Ferrer M, Arriaza J F, Lopez N, Tami C, Verdaguer N, Domingo E, Franze-Fernandez M T. Rapid selection in modified BHK-21 cells of a foot-and-mouth disease virus variant showing alterations in cell tropism. J Virol. 1998;72:10171–10179. doi: 10.1128/jvi.72.12.10171-10179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), 3.5c ed. Department of Genetics, University of Washington, Seattle, Wash. (Distributed by author.)
- 8.Forss S, Strebel K, Beck E, Schaller H. Nucleotide sequence and genome organization of foot-and-mouth disease virus. Nucleic Acids Res. 1984;12:6587–6601. doi: 10.1093/nar/12.16.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giraudo A T, Beck E, Strebel K, de Mello P A, La Torre J L, Scodeller E A, Bergmann I E. Identification of a nucleotide deletion in parts of polypeptide 3A in two independent attenuated aphthovirus strains. Virology. 1990;177:780–783. doi: 10.1016/0042-6822(90)90549-7. [DOI] [PubMed] [Google Scholar]
- 10.Graff J, Kasang C, Normann A, Pfisterer-Hunt M, Feinstone S M, Flehmig B. Mutational events in consecutive passages of hepatitis A virus strain GBM during cell culture adaptation. Virology. 1994;204:60–68. doi: 10.1006/viro.1994.1510. [DOI] [PubMed] [Google Scholar]
- 11.Graff J, Normann A, Feinstone S M, Flehmig B. Nucleotide sequence of wild-type hepatitis A virus GBM in comparison with two cell culture-adapted variants. J Virol. 1994;68:548–554. doi: 10.1128/jvi.68.1.548-554.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinz B A, Vance L M. Sequence determinants of 3A-mediated resistance to enviroxime in rhinoviruses and enteroviruses. J Virol. 1996;70:4854–4857. doi: 10.1128/jvi.70.7.4854-4857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lama J, Sanz M A, Carrasco L. Genetic analysis of poliovirus protein 3A: characterization of a non-cytopathic mutant virus defective in killing Vero cells. J Gen Virol. 1998;79:1911–1921. doi: 10.1099/0022-1317-79-8-1911. [DOI] [PubMed] [Google Scholar]
- 14.Lemon S M, Murphy P C, Shields P A, Ping L H, Feinstone S M, Cromeans T, Jansen R W. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991;65:2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morace G, Pisani G, Beneduce F, Divizia M, Pana A. Mutations in the 3A genomic region of two cytopathic strains of hepatitis A virus isolated in Italy. Virus Res. 1993;28:187–194. doi: 10.1016/0168-1702(93)90135-a. [DOI] [PubMed] [Google Scholar]
- 16.Page R D. Tree View: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 17.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 18.Rieder E, Bunch T, Brown F, Mason P W. Genetically engineered foot-and-mouth disease viruses with poly(C) tracts of two nucleotides are virulent in mice. J Virol. 1993;67:5139–5145. doi: 10.1128/jvi.67.9.5139-5145.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson B H, Grubman M J, Weddell G N, Moore D M, Welsh J D, Fischer T, Dowbenko D J, Yansura D G, Small B, Kleid D G. Nucleotide and amino acid sequence coding for polypeptides of foot-and-mouth disease virus type A12. J Virol. 1985;54:651–660. doi: 10.1128/jvi.54.3.651-660.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P H, editors. Field's virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 609–654. [Google Scholar]
- 21.Sa-Carvalho D, Rieder E, Baxt B, Rodarte R, Tanuri A, Mason P W. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J Virol. 1997;71:5115–5123. doi: 10.1128/jvi.71.7.5115-5123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Toja M, Escarmis C, Domingo E. Genomic nucleotide sequence of a foot-and-mouth disease virus clone and its persistent derivatives. Implications for the evolution of viral quasispecies during a persistent infection. Virus Res. 1999;64:161–171. doi: 10.1016/s0168-1702(99)00089-1. [DOI] [PubMed] [Google Scholar]
- 25.Wutz G, Auer H, Nowotny N, Grosse B, Skern T, Kuechler E. Equine rhinovirus serotypes 1 and 2: relationship to each other and to aphthoviruses and cardioviruses. J Gen Virol. 1996;77:1719–1730. doi: 10.1099/0022-1317-77-8-1719. [DOI] [PubMed] [Google Scholar]