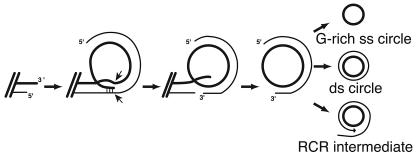Abstract
Recombinational telomere elongation (RTE) known as alternate lengthening of telomeres is the mechanism of telomere maintenance in up to 5 to 10% of human cancers. The telomeres of yeast mutants lacking telomerase can also be maintained by recombination. Previously, we proposed the roll-and-spread model to explain this elongation in the yeast Kluveromyces lactis. This model suggests that a very small (∼100-bp) circular molecule of telomeric DNA is copied by a rolling circle event to generate a single long telomere. The sequence of this primary elongated telomere is then spread by recombination to all remaining telomeres. Here we show by two-dimensional gel analysis and electron microscopy that small circles of single- and double-stranded telomeric DNA are commonly made by recombination in a K. lactis mutant with long telomeres. These circles were found to be especially abundant between 100 and 400 bp (or nucleotides). Interestingly, the single-stranded circles consist of only the G-rich telomeric strand sequence. To our knowledge this is the first report of single-stranded telomeric circles as a product of telomere dysfunction. We propose that the small telomeric circles form through the resolution of an intratelomeric strand invasion which resembles a t-loop. Our data reported here demonstrate that K. lactis can, in at least some circumstances, make telomeric circles of the very small sizes predicted by the roll-and-spread model. The very small circles seen here are both predicted products of telomere rapid deletion, a process observed in both human and yeast cells, and predicted templates for roll-and-spread RTE.
Telomeres are the nucleoprotein structures found at the ends of linear chromosomes (2, 6, 9). Telomeric DNA consists of tandem arrays of short repeats that have G-rich and C-rich strands. The telomeric DNA is divided into a proximal double-stranded region and a more distal single-stranded 3′ overhang (36). This overhang is comprised solely of the G-rich strand and is elongated through the activity of the reverse transcriptase telomerase (53). Telomerase minimally consists of a template RNA that is reverse transcribed onto telomeric ends by the catalytic protein subunit.
Telomeric DNA also associates with a large assemblage of proteins (43, 51). The telomere protein-DNA complex acts as a cap that is dynamic in composition and arrangement. It is the dynamic protein-DNA telomere complex that both controls telomere metabolism and protects telomeres from being recognized as double-stranded breaks. One proposed mechanism for telomere protection is the creation of telomeric secondary structures, such as t-loops, which are stabilized through the action of telomeric DNA binding proteins (19, 49). The maintenance of telomere length is important to the normal functioning of cells.
In most human somatic cells, telomerase activity is very low or absent, so that telomeres gradually shorten, eventually reaching a state which triggers a growth arrest called replicative senescence (15, 40). Mutations or other conditions that bypass senescence produce further telomere shortening and take cells into crisis, a state of high genetic instability and cell death caused by widespread telomere dysfunction (41). In order to survive crisis and become immortal, a means of telomere elongation must arise (14, 39). Most cancer cells are immortal due to the presence of telomerase (40). A subset of cells that emerge from crisis maintain their telomeres through alternate lengthening of telomeres (ALT), a process that involves intertelomeric recombination (3, 16, 38). Studies have shown that telomeres of ALT cells are both highly heterogeneous in size and frequently much longer than telomeres in normal human cells and telomerase-positive cancer cells (3, 4). ALT cells also commonly contain intranuclear structures known as ALT-associated PML bodies, or APBs. APBs contain recombination proteins, telomeric DNA, and replication factor A and have therefore been suggested to play a role in the mechanism of ALT telomere elongation (33, 55). While the structure of telomeric DNA associated with APBs is unknown, extrachromosomal telomeric repeats (ECTR) have recently been extracted from ALT cell lines and have been shown to at least partially exist in circular conformations (7, 52).
Yeast cells have constitutively active telomerase and normally maintain their telomeres within a fixed size range (17). However, deletion of telomerase in yeast cells leads to telomere shortening and coincident growth senescence (27, 29, 42). Although most cells eventually die from this senescence, some postsenescence survivors emerge. These survivors, which are dependent upon the recombination gene RAD52 (26, 28), elongate their telomeres through recombinational telomere elongation (RTE), a process that appears to be analogous to human ALT. In Saccharomyces cerevisiae, there are two pathways of RTE, each of which produces a distinctly different elongation pattern. Type I survivors have amplification of the long Y′ subtelomeric elements and short extensions of the telomeric TG(1-3) repeats (10, 26, 46). Type II survivors lack subtelomeric amplification and are characterized by long telomeric TG(1-3) extensions which are heterogeneous in length (10, 46). Both type I and type II survivors of Saccharomyces require RAD52 as well as type-specific sets of recombination genes. Type I survivors require RAD51, RAD54, and RAD57, while type II survivors require components of the MRX complex, RAD59, and the helicase SGS1 (10, 12, 21, 24, 45).
Only type II survivors arise when the telomerase RNA gene (TER1) is deleted from Kluveromyces lactis cells (28). Inspection of telomeric sequence from K. lactis ter1-Δ survivors derived from cells with telomeres composed of basal wild-type and terminal phenotypically silent mutant repeats revealed repeating patterns containing both types of repeats (35). Most or all telomeres within a clone shared a single pattern, but patterns varied between survivor clones. These data led us to propose the roll-and-spread model, which proposes that the first elongated telomere is made within a cell by rolling circle replication around a circle as small as 100 bp and that the sequence of this long telomere is then spread by gene conversion to most or all other telomeres (35). Consistent with this model, K. lactis cells can utilize transformed telomeric circles of 1.6 kb and 100 nucleotides (nt) to elongate their telomeres (34, 35). Other recent work has confirmed that the sequence of one elongated telomere spreads at very high efficiency to all other telomeric ends during survivor formation (49a). In this work we show that the unusually small telomeric circles proposed by the roll-and-spread model can be made by recombination in a long telomere mutant of K. lactis.
MATERIALS AND METHODS
Yeast strains.
The strain 7B520 (ura3-1 his2-2 trp1) originally described by Wray et al. (54) was used as wild type in this study. The telomerase RNA gene mutant ter1-16T is a mutant of 7B520 originally described by Underwood et al. (50). The double mutant ter1-16T rad52Δ was created by mating of the single mutant ter1-16T with TAQ-STU-19 (ura3 his2-2 rad52Δ) also containing one chromosome with a subtelomeric insert of the S. cerevisiae URA3 gene (30; M. J. McEachern and S. Iyer, unpublished data). Diploids were selected for on plates lacking both histidine and uracil and sporulated. Tetrad dissection was performed, and each spore was screened for RAD52 and telomere length phenotype by Southern blot analysis (50). The TAQ-STU-19 parental strain is not fully isogenic with 7B520.
Southern blotting.
Yeast genomic DNA (cut or uncut) was run on 1% Sea Kem LE agarose gel (Cambrex Bio Science Rockland Inc., Rockland, ME) or 4% Gene Pure 3:1 agarose gel (ISC Bioexpress, Kaysville, UT) and then transferred onto Hybond N+ membrane (Amersham Biosciences, Piscataway, NJ). Southern blots were hybridized and washed at 47°C or 50°C with γ-32P-labeled probes. Probes were either Klac1-25 G-strand telomeric probe (5′-ACGGATTTGATTAGGTATGTGGTGT-3′) or the KC25-1 C-stranded telomeric probe (3′-ACACCACATACCTAATCAAATCCGT-5′). All hybridizations were carried out in the presence of 500 mM Na2HPO4 and 7% sodium dodecyl sulfate (SDS) (11). All washes were done in 100 mM Na2HPO4 and 2% SDS.
Two-dimensional (2D) gel analysis.
Genomic DNA from ter1-16T (uncut or exonuclease I treated) was run at 75 V for 6 h on a 4% nondenaturing Gene Pure 3:1 agarose gel containing 0.6 μg/ml chloroquine. These gels were then soaked in 0.5× Tris-borate-EDTA (TBE) containing 3 μg/ml chloroquine. The gels were then rotated 90° and run in the second dimension for 6 h at 75 V. Both dimensions were run in 0.5× TBE running buffer with chloroquine concentrations equal to that of the gel.
Isolation of extrachromosomal telomeric DNA.
Uncut genomic DNA from ter1-16T was run on 0.8% agarose gels at 30 V for 90 min. DNA migrating below a 500-bp linear marker was cut from the gel. This DNA was electro-eluted from the gel fragments at 90 V for 1 h while enclosed by 12,000 to 14,000 MWCO Spectra/Por dialysis tubing (Spectrum Laboratories Incorporated, Rancho Dominguez, CA). Solutions containing eluted DNA were concentrated by using the Microcon model YM-10 as directed by the manufacturer (Amicon Bioseparations, Raleigh, NC).
Electron microscopy.
Gel-isolated low-molecular-weight K. lactis DNA was incubated with 20 μg/ml T4 gene product 32 (gift of Nancy Nossal, National Institutes of Health, Bethesda, MD) for 5 min in a buffer containing 10 mM HEPES (pH 7.5) and 1 mM EDTA. The samples were treated with 0.6% glutaraldehyde on ice for 10 min and chromatographed over a 2.5-ml BioGel A-1.5 M column (Bio-Rad, Hercules, CA). Fractions containing DNA and DNA-protein complexes were prepared for electron microscopy (18) and examined on an FEI Tecnai 12 instrument (Eindhoven, The Netherlands). Images were captured using a Gatan Ultrascan US4000SP digital camera (Gatan, Pleasanton, CA), and molecule dimensions were determined using Gatan Digital Micrograph 3.0 software. Images for publication were captured on sheet film and digitized using ACT-1 software (Nikon, Tokyo, Japan) and a Nikon SMZ1000 stereoscope. Brightness and contrast were adjusted using Adobe Photoshop (Adobe Systems, San Jose, CA).
RESULTS
2D gel analysis of extrachromosomal telomeric DNA from ter-16T cells.
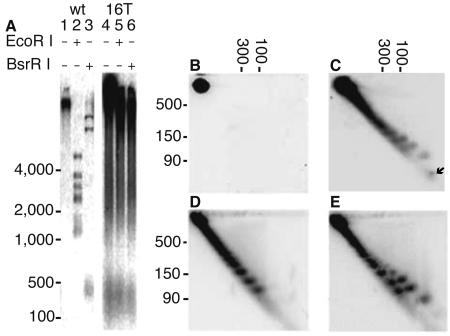
The long telomere mutant ter1-16T has extremely elongated telomeres relative to the wild type (Fig. 1A, compare lanes 2 and 3 with 5 and 6) (50). Also apparent in the ter1-16T sample is the ECTR, which can be seen running under 1 kb in lane 1 of Fig. 1A. This ECTR is especially abundant between the 100- and 500-bp linear markers (Fig. 1A, lane 4). Based on the unit size of repeating patterns in the telomeres of postsenescence survivors (35), we predicted that circles at least as little as 100 bp can sometimes be formed in vivo by a senescent ter1-Δ cell. However, ter1-Δ cells at various stages of senescence do not produce amounts of ECTR that are detectable by Southern blotting (data not shown) (28). This is not surprising, as they are presumably produced only rarely in this mutant.
FIG. 1.
Gel analysis of ECTR from ter1-16T. (A) Southern blot of 1% 1D agarose gels hybridized to C-strand telomere probe. Cut and uncut genomic DNA from TER1 (lanes 1 to 3) and the ter1-16T mutant (lanes 4 to 6) show abundance of small telomeric material running under 500 bp. (B to E) Southern blots of 2D 4% agarose gels hybridized to either the G-strand or C-strand telomeric probes. (B) Uncut DNA from rad52 ter1-16T hybridized to the C-strand probe shows that the double mutant produces no ECTR. The spot seen represents chromosomal telomeric signal. Similar results were obtained with the G-strand probe (data not shown). (C) Uncut ter1-16T DNA run with 66-nt control oligo (arrow), hybridized with the C-strand telomeric probe. Two ladders of partially separated spots are visible. (D) Exo I-treated ter1-16T genomic DNA hybridized to the G-strand telomeric probe. (E) Same membrane shown in panel D hybridized to the C-strand telomeric probe. Molecular masses of double-stranded linear control fragments are shown in base pairs.
Previous work done on telomerase RNA template gene (TER1) mutants has shown that mutants within and near the Rap1p binding domain produce extremely long telomeres and abundant ECTR (22, 23, 29, 50); however, the nature of the ECTR in these mutants was not previously investigated. We therefore selected a representative long telomere mutant that produces abundant ECTR, ter1-16T (Fig. 1A), for detailed analysis. This mutant contains a 1-bp change in the Rap1p binding region of the telomerase RNA template (50). Consistent with the hypothesis that decreased Rap1p binding at ter1-16T telomeres causes its telomeric phenotype, both the long telomere phenotype and presence of abundant ECTR are partially suppressed when Rap1p is overexpressed in ter1-16T (50).
2D gel analysis was employed to investigate the structure of ECTR made by ter1-16T. Two ladders of telomeric DNA migrating faster than 500 bp were seen on 2D Southern blots hybridized to a C-strand telomeric probe (Fig. 1C and E). One of these ladders migrated faster than the other in high concentrations of the intercalating agent chloroquine. This faster-moving ladder hybridized only to the C-stranded telomeric probe (Fig. 1C and E). In contrast, the slower-moving ladder hybridized to both telomeric strand probes (compare Fig. 1D and E). We therefore conclude that one ladder is double-stranded ECTR and the other ladder is single-stranded ECTR specifically composed of the G-rich strand of K. lactis telomeric DNA. The single-stranded ladder is resistant to the single-strand-specific exonuclease I under conditions where a single-stranded control oligonucleotide is completely digested away (Fig. 1E and data not shown). This indicated that the molecules in the single-stranded ladder of ECTR do not contain nuclease-accessible ends (Fig. 1E).
Neither ladder of spots was visible in a ter1-16T rad52Δ double mutant (Fig. 1B) in spite of the fact that long telomeres persisted (50). This finding indicates that the creation of both the single-stranded and double-stranded small ECTR species is a recombination-driven process. Because homologous recombination is a precise process, we further conclude that the discreet spots in each ladder represent forms differing by integral numbers of the 25-bp (nt) telomeric repeat.
Electron microscopy of ECTR from ter1-16T.
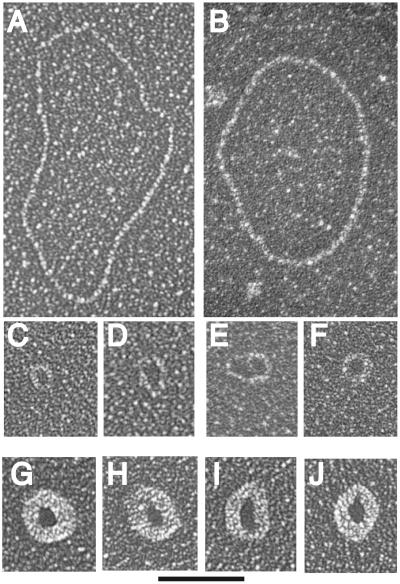
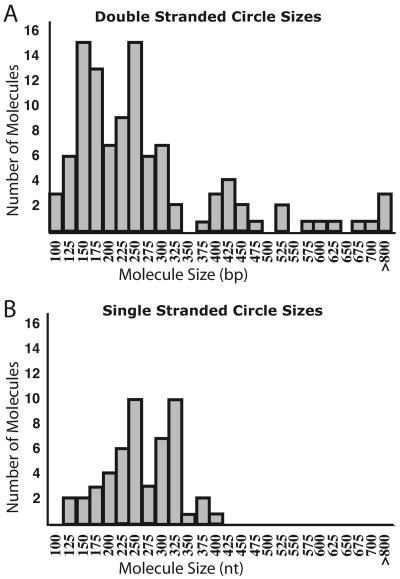
We next examined the small ECTR from ter1-16T by electron microscopy. Uncut genomic DNA from ter1-16T was run a short distance on a 0.8% agarose gel. Material running ahead of a 500-bp linear marker was extracted and purified. The extracted ECTR was treated with the single-strand binding protein T4 gene product 32 and visualized by electron microscopy (Fig. 2). Greater than 75% of the ter1-16T ECTR molecules visualized were circular. Of the circular DNA molecules, approximately 60% were double stranded and 40% were single stranded. Fifty-one single-stranded circles ranging in size from 125 to 400 nt and 106 double-stranded circles ranging in size from 100 to 1,600 bp were measured, and their distribution is shown in Fig. 3. Previously, telomeric circles have been referred to as t-circles (7, 47). The number of small t-circles counted may have been underestimated due to the difficulty involved in visualizing DNA circles near or below 100 bp (nt) in size. Ninety percent of all circles measured were under 400 bp (nt) in size, consistent with the migration patterns of this DNA as seen on both 1D and 2D gels. Among the circles examined, we found a few surprisingly large circles. It is not clear why these larger circles migrated faster than the 500-bp linear marker. Limitations of digital molecular measurement did not allow precise sizing of circles. Both the double- and single-stranded circles are expected to be multiples of the 25-bp increment of the K. lactis telomeric repeat, as indicated by the incremental nature of the telomeric ladders visualized on 2D Southern blots and by their dependence on homologous recombination. Our results demonstrate that tiny telomeric circles can be readily generated by telomeric recombination in at least one class of mutant with long dysfunctional telomeres.
FIG. 2.
Visualization of DNA circles isolated from K. lactis ter1-16T. (A to F) Electron micrographs of double-strand DNA circles. Circle sizes are 1,580, 1,285, 155, 230, 265, and 190 bp, respectively, for panels A to F. (G to J) Electron micrographs of single-strand DNA circles bound by T4 gene 32 single-strand DNA binding protein. Circle sizes are 360, 365, 400, and 330 nt for panels G to J, respectively. Samples were directly mounted onto thin carbon-coated foils and rotary shadowcast with tungsten (see Materials and Methods). Images are shown in negative contrast. Solid bar is equivalent to 200 bp (A to F) or 235 nt (G to J).
FIG. 3.
Size distribution of t-circles. (A) Double-stranded t-circle sizes. (B) Single-stranded t-circle sizes. For both panels bars represent t-circles ranging from 12.5 bp (nt) below and above the size indicated. Ninety percent of the circles measured fell below 400 bp (nt).
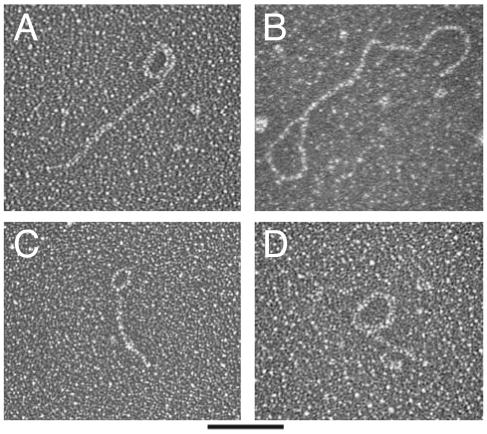
In the course of examining the telomeric DNA sample containing circles, we also visualized several double-stranded circles with double-stranded tails (Fig. 4). These structures were not abundant in the sample, representing less than 1% of the total number of molecules visualized. Because this sample was not subjected to any cross-linking, noncovalent structures were likely not preserved (20). The small ECTR from ter1-16T may therefore have contained many more circle-with-tail type structures that were lost in sample preparation. There are several possible explanations for these structures. They could represent protective t-loops (19). Alternatively, they could be intramolecular strand invasions that occurred as a result of telomere uncapping. If this happened on extrachromosomal linear pieces, either end of the fragment might be involved in the strand invasion. Finally, the structures could be rolling circle replication (RCR) intermediates. Larger structures of this sort have been shown to be intermediates of RCR in a variety of systems (1, 10).
FIG. 4.
Visualization of circle-with-tail structures present in K. lactis ter1-16T ECTR. (A to D) Electron micrographs of molecules resembling rolling circle replication intermediates. Estimated sizes of circular and tail portions of molecules shown in panels A to D were 230 and 570, 430 and 730, 140 and 240, and 300 and 210 bp, respectively. Molecules were prepared as for Fig. 2. Bar, 200 bp.
DISCUSSION
A major prediction of the roll-and-spread model is that senescing K. lactis cells lacking telomerase will occasionally be able to generate small circles of telomeric DNA that can trigger RTE (35). The t-circles at least as small as 100 bp (nt) would need to be produced by such cells to account for the repeating patterns in the elongated telomeres of postsenescence survivors (35). Our results here demonstrate that at least some K. lactis cells with dysfunctional telomeres can readily use homologous recombination to produce remarkably small single- and double-stranded t-circles.
The production of t-circles by recombination can likely be caused by a variety of telomere capping defects. All K. lactis mutants with extremely long telomeres produce abundant ECTR (50). These include mutations, such as ter1-16T, that produce immediate telomere elongation as a result of Rap1p binding defects, other TER1 template mutations that cause delayed elongation and do not disrupt Rap1p binding, and a mutation in the gene encoding the telomeric cap protein Stn1 (22, 50; S. Iyer, A. Chadha, and M. J. McEachern, submitted for publication). Evidence that these different mutants have telomere capping defects includes highly elevated subtelomeric gene conversion rates and the presence of extensive single-stranded DNA specifically of the G-rich strand (50; Iyer et al., submitted). Preliminary evidence from 2D gel analysis shows that ECTR from representatives of each of these classes of mutants contains telomeric ladders similar to those from the ter1-16T mutant (S. Iyer and M. J. McEachern, unpublished data) (data not shown). Taken together, the presence of ECTR ladders in all classes of long telomere mutants suggests that t-circle formation is a general consequence of telomere uncapping.
As senescing ter1-Δ mutants also experience very high rates of telomeric and subtelomeric recombination (30; Topcu et al., submitted), they are also likely to form t-circles, at least occasionally. However, several factors may act to drastically limit t-circle formation in senescing ter1-Δ cells relative to the levels seen in ter1-16T. The most obvious is telomere length, which is very short in senescing ter1-Δ cells and very long in ter1-16T and other mutants known to have abundant ECTR. Additionally, the relative amounts of single-stranded 3′ overhangs might also be very different. Another difference is the apparent nature of the respective capping defects. The telomeres of ter1-Δ cells are prone to initiating recombination only when they drop below ∼100 bp in length (49a). It may therefore be the case that most telomeres in senescing ter1-Δ cells become recombination prone at sizes too short to form t-circles via intratelomeric recombination. If ter1-Δ cells only rarely make circles, their formation may be the rate-limiting step of RTE. In marked contrast, the mutants that produce abundant ECTR presumably have perpetual capping defects and are therefore likely to continuously be prone to circle formation and other recombinational processes.
We suggest that all the ECTR structures seen and described above may form from a common recombination product. The model in Fig. 5 proposes that a 3′ telomeric overhang strand invades a more internal region of the telomeric repeat array. Subsequent nucleolytic processing of this structure could give rise to a partially single-stranded t-circle. Degradation of the partial C strand or helicase activity could lead to the creation of a totally single-stranded circle comprised of the G-rich telomeric strand. Extension of the partial duplex strand by a DNA polymerase could lead to a double-stranded circle or, with continued DNA synthesis, to a rolling circle replication intermediate. This model for ECTR production is similar to a model proposed by Lustig and colleagues for telomere rapid deletion (TRD), a process which can lead to the sudden loss of up to thousands of base pairs from a telomere in yeast or human cells (5). TRD is largely RAD52 dependent in S. cerevisiae, where it was suggested to arise from resolution of an equivalent intramolecular strand invasion structure into a shortened telomere and a t-circle (25). TRD and t-circle production may therefore be one and the same.
FIG. 5.
Model of t-circle creation. Intramolecular strand invasion of the G-rich strand 3′ telomeric tip and internal telomeric C-rich strand. Cutting of both telomeric strands and ligation of the G-rich strand creates a partially double-stranded circular intermediate. This initial product could be degraded to leave a single G-rich stranded circle, filled in to create a double-stranded circle, or used as a template for RCR.
Intramolecular strand invasion to form a t-loop structure has been proposed to be a protective mechanism that helps provide the capping function of telomeres (19). t-loops have been observed at telomeres of mammals (19), protozoans (31, 32), plants (8), and chickens (37). While t-loops have not been identified in vivo on yeast chromosomes, they have been visualized on in vitro models of telomeres from the yeast Schizosaccharomyces pombe (48). Their formation appears to be promoted by certain telomere binding proteins (44, 49). There are two general possibilities for the relationship between a protective t-loop that contributes to telomere capping and an intramolecular strand invasion of the telomeric end that results from defective telomere capping and which may be the precursor to t-circles. The first is that the two structures are fundamentally the same at the DNA level. In this scenario, a t-loop is kept from being processed by the nucleases and polymerases that would normally accompany a recombination event by the complex of telomere proteins. Presumably, this would include the double-stranded telomeric binding protein Rap1p in K. lactis. Disruption of this protective complex could therefore directly lead to a t-loop behaving as a recombination intermediate. The alternative possibility is that t-loops are not identical to a recombination intermediate at the DNA level. Conceivably, for example, t-loops could form through non-Watson-Crick base interactions. In this case, the t-loop DNA structure could be intrinsically incompatible with recombination and might need to be disrupted before a telomeric end could initiate a recombination event.
Whether the telomeres of K. lactis form t-loops is not currently known. The somewhat surprising characteristic of the ter1-16T mutant with its very long telomeres to preferentially accumulate t-circles of very small sizes might indicate that normal K. lactis telomeres preferentially form tightly folded loop-back or t-loop structures within their normal length of 350 to 600 bp. Failure to form a properly folded structure has been suggested to be responsible for the relatively abrupt loss of protection against recombination that occurs when K. lactis telomeres drop to below ∼100 bp in length (49a).
What role, if any, the t-circles play in telomere function in ter1-16T cells is unknown. The presence of long telomeres and the absence of ECTR in rad52 ter1-16T cells clearly indicate that small t-circles are not required for the telomere elongation in this mutant (50) (Fig. 1). However, it remains possible that recombination, possibly including the copying of t-circles, does contribute to the formation of long telomeres in ter1-16T cells. The presence of telomeres composed of another type of mutant telomeric repeat (the delayed elongation “Kpn” repeats [22, 23, 29, 50]) leads to pronounced telomere elongation even in the absence of telomerase (49a).
Recent evidence indicates that t-circles contribute to RTE in other organisms in addition to K. lactis. Telomeric “nanocircles” as small as 36 nt can be utilized in vitro for rolling circle replication by a variety of DNA polymerases (20). The sudden emergence of elongated telomeric repeat tracts in type II survivors of S. cerevisiae has been suggested to be triggered by rolling circle copying of t-circles (45). Double-stranded t-circles are abundantly produced from the mitochondrial telomeres of several yeast species, including Candida parapsilosis, and have been proposed to be required for normal mitochondrial telomere maintenance (48). Recently, t-circles have been isolated from human ALT cells (7, 52). Xenopus laevis has also been found to produce t-circles during part of its normal embryonic development (13). The presence of t-circles in this broad group of organisms opens up the possibility that RTE based on rolling circle replication is a phylogenetically diverse phenomenon.
Acknowledgments
We thank Gary Fielding for his help with figure preparations.
Grants from both the National Institutes of Health (CGM61645 and GM31819) and the Ellison Foundation have supported this work. J.D.G. is an Ellison Senior Scholar. Cindy Groff-Vindman is supported through a training grant from the National Institutes of Health (5T32GMOO&10330).
REFERENCES
- 1.Backert, S. 2002. R-loop-dependent rolling-circle replication and a new model for DNA concatemer resolution by mitochondrial plasmid mp1. EMBO J. 21:3128-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, S. M., and E. H. Goodwin. 2004. DNA and telomeres: beginnings and endings. Cytogenet. Genome Res. 104:109-115. [DOI] [PubMed] [Google Scholar]
- 3.Bryan, T. M., A. Englezou, L. Dalla-Pozza, M. A. Dunham, and R. R. Reddel. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271-1274. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, T. M., A. Englezou, J. Gupta, S. Bacchetti, and R. R. Reddel. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14:4240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bucholc, M., Y. Park, and A. J. Lustig. 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cech, T. R. 2004. Beginning to understand the end of the chromosome. Cell 116:273-279. [DOI] [PubMed] [Google Scholar]
- 7.Cesare, A. J., and J. D. Griffith. 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 24:9948-9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesare, A. J., N. Quinney, S. Willcox, D. Subramanian, and J. D. Griffith. 2003. Telomere looping in P. sativum (common garden pea). Plant J. 36:271-279. [DOI] [PubMed] [Google Scholar]
- 9.Chan, S. R., and E. H. Blackburn. 2004. Telomeres and telomerase. Philos. Trans. R. Soc. London B 359:109-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, H., and D. A. Sinclair. 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. USA 98:3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, S., and M. Mechali. 2002. Formation of extrachromosomal circles from telomeric DNA in Xenopus laevis. EMBO Rep. 3:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong, Y. S., W. E. Wright, and J. W. Shay. 2002. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 66:407-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristofalo, V. J., A. Lorenzini, R. G. Allen, C. Torres, and M. Tresini. 2004. Replicative senescence: a critical review. Mech. Ageing Dev. 125:827-848. [DOI] [PubMed] [Google Scholar]
- 16.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447-450. [DOI] [PubMed] [Google Scholar]
- 17.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 18.Griffith, J. D., and G. Christiansen. 1978. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu. Rev. Biophys. Bioeng. 7:19-35. [DOI] [PubMed] [Google Scholar]
- 19.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 20.Hartig, J. S., and E. T. Kool. 2004. Small circular DNAs for synthesis of the human telomere repeat: varied sizes, structures and telomere-encoding activities. Nucleic Acids Res. 32:e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, P., P. F. Lester, D. Maddison, R. L. Borts, R. H. Hickson, I. D. Louis, E. J. 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11:125-129. [DOI] [PubMed] [Google Scholar]
- 22.Krauskopf, A., and E. H. Blackburn. 1996. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383:354-357. [DOI] [PubMed] [Google Scholar]
- 23.Krauskopf, A., and E. H. Blackburn. 1998. Rap1 protein regulates telomere turnover in yeast. Proc. Natl. Acad. Sci. USA 95:12486-12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, B., and A. J. Lustig. 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10:1310-1326. [DOI] [PubMed] [Google Scholar]
- 26.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 27.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 28.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822-1834. [DOI] [PubMed] [Google Scholar]
- 29.McEachern, M. J., and E. H. Blackburn. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376:403-409. [DOI] [PubMed] [Google Scholar]
- 30.McEachern, M. J., and S. Iyer. 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7:695-704. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Jordan, J. L., G. A. Cross, T. de Lange, and J. D. Griffith. 2001. t-loops at trypanosome telomeres. EMBO J. 20:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murti, K. G., and D. M. Prescott. 1999. Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl. Acad. Sci. USA 96:14436-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nabetani, A., O. Yokoyama, and F. Ishikawa. 2004. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J. Biol. Chem. 279:25849-25857. [DOI] [PubMed] [Google Scholar]
- 34.Natarajan, S., C. Groff-Vindman, and M. J. McEachern. 2003. Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot. Cell 2:1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Natarajan, S., and M. J. McEachern. 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22:4512-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neidle, S., and G. N. Parkinson. 2003. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 13:275-283. [DOI] [PubMed] [Google Scholar]
- 37.Nikitina, T., and C. L. Woodcock. 2004. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 166:161-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddel, R. R., and T. M. Bryan. 2003. Alternative lengthening of telomeres: dangerous road less travelled. Lancet 361:1840-1841. [DOI] [PubMed] [Google Scholar]
- 39.Scheel, C., and C. Poremba. 2002. Telomere lengthening in telomerase-negative cells: the ends are coming together. Virchows Arch. 440:573-582. [DOI] [PubMed] [Google Scholar]
- 40.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33:787-791. [DOI] [PubMed] [Google Scholar]
- 41.Shay, J. W., and W. E. Wright. 2004. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis [Online.] doi:10.1093/carcin/bgh296. [DOI] [PubMed]
- 42.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 43.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 44.Stansel, R. M., T. de Lange, and J. D. Griffith. 2001. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20:5532-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 46.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomaska, L., M. J. McEachern, and J. Nosek. 2004. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. 567:142-146. [DOI] [PubMed] [Google Scholar]
- 48.Tomaska, L., J. Nosek, A. M. Makhov, A. Pastorakova, and J. D. Griffith. 2000. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 28:4479-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomaska, L., S. Willcox, J. Slezakova, J. Nosek, and J. D. Griffith. 2004. Taz1 binding to a fission yeast model telomere: formation of telomeric loops and higher order structures. J. Biol. Chem. 279:50764-50772. [DOI] [PubMed] [Google Scholar]
- 49a.Topcu, Z., K. Nickles, C. Davis, and M. J. McEachern. 2005. Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis. Proc. Natl. Acad. Sci. USA 102:3348-3353. [DOI] [PMC free article] [PubMed]
- 50.Underwood, D. H., C. Carroll, and M. J. McEachern. 2004. Genetic dissection of the Kluyveromyces lactis telomere and evidence for telomere capping defects in TER1 mutants with long telomeres. Eukaryot. Cell 3:369-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vega, L. R., M. K. Mateyak, and V. A. Zakian. 2003. Getting to the end: telomerase access in yeast and humans. Nat. Rev. Mol. Cell Biol. 4:948-959. [DOI] [PubMed] [Google Scholar]
- 52.Wang, R. C., A. Smorgorzewska, and T de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119:335-368. [DOI] [PubMed] [Google Scholar]
- 53.Wei, C., and M. Price. 2003. Protecting the terminus: t-loops and telomere end-binding proteins. Cell. Mol. Life Sci. 60:2283-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wray, L. V., Jr., M. M. Witte, R. C. Dickson, and M. I. Riley. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175-4179. [PubMed] [Google Scholar]