Abstract
Efficient assembly of RAG1/2-recombination signal sequence (RSS) DNA complexes that are competent for V(D)J cleavage requires the presence of the nonspecific DNA binding and bending protein HMGB1 or HMGB2. We find that either of the two minimal DNA binding domains of HMGB1 is effective in assembling RAG1/2-RSS complexes on naked DNA and stimulating V(D)J cleavage but that both domains are required for efficient activity when the RSS is incorporated into a nucleosome. The single-domain HMGB protein from Saccharomyces cerevisiae, Nhp6A, efficiently assembles RAG1/2 complexes on naked DNA; however, these complexes are minimally competent for V(D)J cleavage. Nhp6A forms much more stable DNA complexes than HMGB1, and a variety of mutations that destabilize Nhp6A binding to bent microcircular DNA promote increased V(D)J cleavage. One of the two DNA bending wedges on Nhp6A and the analogous phenylalanine wedge at the DNA exit site of HMGB1 domain A were found to be essential for promoting RAG1/2-RSS complex formation. Because the phenylalanine wedge is required for specific recognition of DNA kinks, we propose that HMGB proteins facilitate RAG1/2-RSS interactions by recognizing a distorted DNA structure induced by RAG1/2 binding. The resulting complex must be sufficiently dynamic to enable the series of RAG1/2-mediated chemical reactions on the DNA.
The site-specific V(D)J recombination reaction assembles immunoglobulin and T-cell receptor genes from their separate component gene segments to generate the diverse repertoire of antigen-receptor specificities required by the vertebrate immune system (reviewed in references 7, 13, and 15). Each coding segment within the chromosome is flanked by a recombination signal sequence (RSS) that is recognized by the RAG1/2 V(D)J recombinase. The RSS is composed of conserved heptamer and nonamer elements separated by a spacer region of conserved length (12 or 23 bp) but relatively nonconserved sequence (12 RSS or 23 RSS, respectively). Efficient V(D)J recombination requires that a pair of signals (one 12 and one 23 RSS) be brought together into a synaptic complex with RAG1 and RAG2. In vitro work indicates that the synaptic complex is formed by the binding of the RAG1/2 proteins first to a single RSS followed by the capture of the second RSS by the initial single-site complex (21, 34).
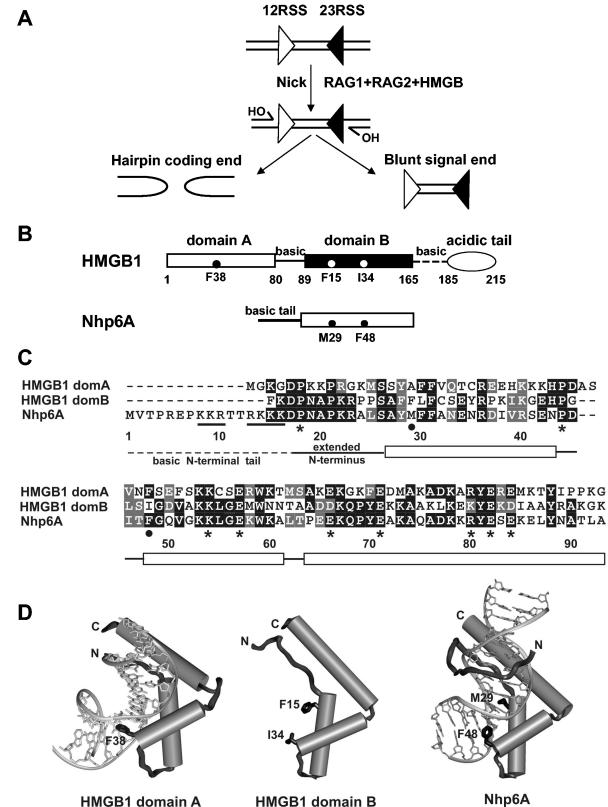
Both the binding of RAG1/2 to a single RSS and formation of the 12/23 RSS synaptic complex have been shown to be strongly enhanced by HMGB1/2 proteins (see references 13 and 15 and references therein). Formation of the RAG1/2-HMGB-RSS complex is followed by the hydrolysis of one strand of DNA, leaving a 3′ hydroxyl group at the border of the coding sequence (Fig. 1A). The free hydroxyl of this nicked DNA then attacks the 5′ phosphate of its base-paired partner on the antiparallel strand, creating a hairpin coding end and a blunt, 5′-phosphorylated signal end (30). These chemical steps are believed to require changes in the structure of DNA within the RSS, particularly over the heptamer region and flanking coding DNA (14). HMGB proteins are ubiquitously present in all eukaryotic organisms and possess one or more structurally conserved HMGB DNA binding domains (for reviews see references 3, 9, 16, 53, and 54). Each domain has 75 to 80 amino acids and consists of an extended strand plus three α-helices that fold into an L-shaped structure (Fig. 1B to D). Members of the sequence-specific class of HMGB proteins, such as LEF1 and SRY, are usually transcription factors whose expression is cell type specific. The nonsequence-specific HMGB proteins, such as HMGB1/2 in mammals and Nhp6A in Saccharomyces cerevisiae, are among the most abundant nonhistone chromatin-associated proteins. Mammalian HMGB1 is found throughout most adult mouse tissues except for regions within the brain (9, 18), whereas HMGB2 is restricted to lymphoid tissues and the testis (44).
FIG. 1.
(A) Outline of the V(D)J recombination reaction. The 12 and 23 RSSs are depicted as triangles with the heptamer at the vertical side of the triangle abutting the coding end. V(D)J recombination is initiated by the combined action of two lymphoid-specific proteins, RAG1 and RAG2. The RAG1-RAG2 complex binds to the RSSs and introduces a double strand break (DSB) at the border of the RSS and the coding DNA. This DSB is generated in two steps. First, a nick is introduced at the 5′ end of the heptamer at the coding segment border, leaving a free 3′ hydroxyl on the coding DNA. In a second step, this 3′ hydroxyl attacks the phosphodiester bond of the opposing strand, leaving a hairpin coding end anda blunt signal end. In vivo (and in vitro with Mg2+ as the divalent metal ion) a 12/23 signal pair is required for hairpin formation. By contrast, in the presence of Mn2+ in vitro, both nicking and hairpinning can proceed on a single RSS (single-site cleavage). Both the binding of RAG1/2 to a single RSS and formation of the 12/23 RSS synaptic complex are greatly enhanced by HMGB1/2 proteins. (B) Schematic representations of full-length rat HMGB1 and yeast Nhp6A. HMGB1 contains domain A (white rectangle), domain B (black rectangle), and an acidic C-terminal domain (oval). There are short basic regions that link domain A and domain B (solid line) and domain B with the C terminus (dashed line). Nhp6A contains a single HMGB domain (white rectangle) and a highly basic amino acid region (solid line) at its N terminus. The amino acids associated with the DNA exit wedge (Phe 38 in HMGB1 domain A, Ile 34 in HMGB1 domain B, and Phe 48 in Nhp6A) and central DNA binding wedge (Phe 15 in HMGB1 domain B and Met 29 in Nhp6A) are marked by black or white (domain B) circles. (C) HMGB protein sequence alignment. Amino acid sequences of the HMGB domains of rat HMGB1 and S. cerevisiae Nhp6A were aligned using Clustal W (UCSD Supercomputer Biology Workbench), and identical and similar residues are shown in black and gray, respectively. The residue numbering and locations of the N-terminal basic tail (dashed line), extended peptide strand at the N-terminal end of the core domain, and the α-helices (rectangles) are with respect to the Nhp6A structure. The locations of the residues comprising the central and exit DNA binding wedges are marked with a solid circle below the sequences. The nine solvent-exposed residues whose chemical character is common between the three proteins, but are not involved in DNA interactions as determined from the DNA complex structures of Nhp6A, HMGB1 domain A, and HMGD, are indicated by asterisks (see the text). (D) The structures of HMGB1 domain A-cisplatin-modified DNA (38), free domain B (43, 60), and Nhp6A-DNA (29). Residue side chains at the central DNA binding wedge (absent in domain A) near the beginning of helix 1 and the DNA exit wedge at the N terminus of helix 2 are denoted in each structure.
Upon binding, HMGB proteins bend the DNA and most preferentially bind to prebent DNA structures, such as cisplatin-cross-linked DNA, bulged DNA, and Holliday junctions (reviewed in references 54, 55, and 67). These proteins often contain highly charged regions outside the conserved HMGB domain, which may vary in their location with respect to the core domain. Mammalian HMGB1 possesses two tandem HMGB domains, referred to as domains A and B, linked by a short basic region (Fig. 1B and C). HMGB1 domain B is followed by a relatively basic region and then 30 contiguous aspartate and glutamate residues. Yeast Nhp6A has a highly basic region at its N terminus but no acidic region. The flanking basic or acidic regions can profoundly influence the DNA binding affinities and functional properties of the HMGB proteins (8, 27, 48, 56, 63). The bent DNA within solved HMGB complexes conforms to the concave surface of the L-shaped fold, creating a wide and shallow minor groove and a highly compressed major groove (Fig. 1D) (28, 29, 35, 37, 38). Localized DNA distortions are also introduced by one or two hydrophobic wedges present on the binding surface of the HMGB protein. These wedges also play important but variable roles for the structure-specific binding and the functional activities of different HMGB proteins (36, 55).
Mammalian HMGB1 efficiently stimulates cleavage by the RAG1/2 proteins, while the yeast HMGB protein Nhp6A exhibits poor but detectable activity, and an unrelated prokaryotic DNA bending protein HU is inactive (57). The poor activity of Nhp6A could be due to (i) the presence of only a single HMGB domain, (ii) an intrinsic difference in the DNA binding properties of Nhp6A and HMGB1, or (iii) the absence of a direct protein-protein interaction between the yeast protein and RAG1/2 that is important for HMGB1 activity in the V(D)J reaction. In support of the last possibility, Aidinis et al. presented evidence that HMGB1 and RAG1 interact in solution and that this interaction requires both HMGB domains (4).
Here we show that individual domains of HMGB1 efficiently assemble cleavage-competent RAG1/2-RSS complexes on naked DNA but that both domains are required for maximal activity when the RSS is associated with a nucleosome. The surface-exposed phenylalanine at the DNA exit wedge of HMGB proteins, a residue critical for recognition of a preexisting DNA kink (19, 61), is essential for stimulating DNA binding and cleavage by RAG1/2. Surprisingly, we find that although Nhp6A efficiently stimulates the binding of RAG1/2 to the RSS, these RAG1/2-RSS complexes do not support DNA cleavage. Thus, the requirements for stimulating binding can be separated from those needed for promoting catalysis. Additional analysis of Nhp6A mutants revealed that those derivatives which bind prebent DNA less stably than the wild type are able to stimulate both RAG1/2-RSS binding and V(D)J cleavage. Taken together, these results identify the functionally important determinants of HMGB proteins for the assembly of catalytically active RAG1/2-RSS complexes and indicate that a dynamic protein-DNA complex is required for RAG-mediated DNA cleavage.
MATERIALS AND METHODS
Construction and purification of HMGB proteins.
The construction of mutant and truncated (Fig. 2A) rat HMGB1 (12, 32) and Nhp6A (5, 29, 62, 63) genes have been described previously. Note that the recombinant deletion forms of HMGB1 used in this study are different from the derivatives obtained by V8 protease digestion employed in an earlier study (39). Recombinant HMGB proteins were typically purified from 2 liters of LB culture after isopropyl-β-d-thiogalactopyranoside induction, using methods similar to those outlined previously (63). Briefly, cells were broken using a French press, and DNA was removed from cleared extracts by precipitation with 0.35% polyethyleneimine (Sigma) in the presence of 0.5 M NaCl. Protein was precipitated with ammonium sulfate (70% saturation) and applied to a column of SP-Sepharose (Pharmacia-Amersham) for Nhp6Ap or phosphocellulose (Whatman P11) for HMGB1 proteins, and the HMGB proteins were eluted by a salt gradient. Fractions containing HMGB proteins were pooled, and contaminants were first removed by precipitation with 2% trichloroacetic acid (TCA), followed by 10% TCA to collect the HMGB proteins. The TCA pellet was rinsed with cold acetone, resuspended in 50 mM HEPES (pH 7.5), 0.1 M NaCl, 1 mM dithiothreitol (DTT), 0.1 M EDTA, 10% glycerol, and subjected to chromatography on an FPLC Mono S and/or a Superdex 75 column. Ion-exchange chromatography on PBE94 (Amersham-Pharmacia) was used to obtain full-length recombinant HMGB1 (1). Recombinant HMGB1 and native HMGB1 purified from calf thymus (39) were indistinguishable with respect to microcircle formation and V(D)J cleavage. In addition, our HMGB1 and Nhp6Ap preparations purified with or without a TCA precipitation step had indistinguishable activities with respect to microcircle formation.
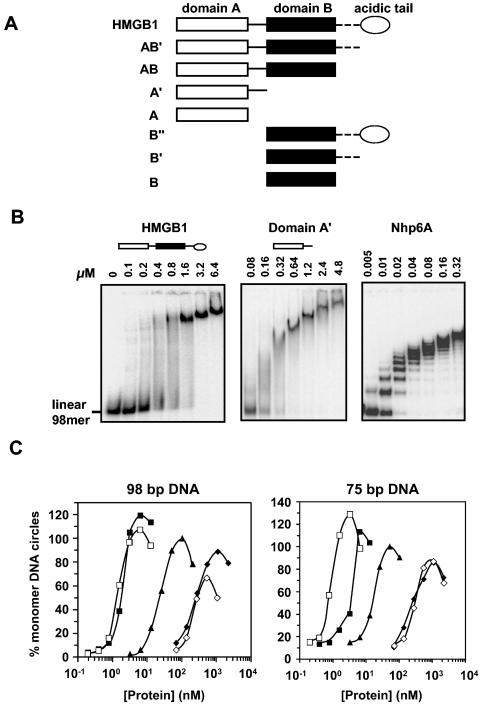
FIG. 2.
DNA binding and bending properties of HMGB1 derivatives. (A) Schematic representation of full-length and truncated derivatives of HMGB1 used in this work. HMGB1 domains are depicted as in Fig. 1B. The amino acid residues present in the derivatives used here are full-length HMGB1 (1 to 215) and domains AB′ (1 to 185), AB (1 to 165), A′ (1 to 88), A (1 to 81), B" (89 to 215), B′ (89 to 185), and B (89 to 165). (B) Gel mobility shift assay for the binding of HMGB1, domain A′, and Nhp6A to a 98-bp linear DNA fragment. The “smeary” patterns of the DNA complexes formed with HMGB1 and domain A′ are suggestive of unstable interactions with DNA. (C) DNA microcircle formation by HMGB1 and its truncated derivatives. Ligation assays on 32P-labeled 98-bp and 75-bp DNA fragments in the presence of different concentrations of HMGB proteins were performed: HMGB1 (filled triangles) and domains AB′ (filled squares), AB (open squares), A′ (filled diamonds), and B′ (open diamonds). The percentage of monomer circles relative to the input DNA was quantitated after exonuclease III digestion and polyacrylamide gel electrophoresis. The percentage of monomer circles formed for truncated HMGB1 proteins was normalized with respect to the full-length HMGB1, whose maximum value was set to 100% and typically represented 30 to 40% of the input DNA. Averaged values from two or three experiments are shown.
HMGB-DNA binding assays.
Gel mobility shift assays on linear substrates were performed essentially as described previously (40, 63). 32P-labeled DNA substrates were purified on polyacrylamide gels after PCRs employing pRJ551-76 (for 98-bp fragments) and pRJ551-53 (for 75-bp fragments), using either end-labeled primers or by internal labeling with [α-32P]dATP as described previously (40, 63). Substrates for microcircles were internally labeled PCR products that were purified using QIAGEN PCR purification columns, digested with 100 units of EcoRI (New England Biolabs) for 3 h at 37°C, extracted with phenol-chloroform, and ethanol precipitated. DNA microcircle ligation assays were performed as described previously (40, 63). Remaining linear DNA substrates were removed by digestion with exonuclease III (New England Biolabs), and the samples were then incubated with proteinase K in the presence of 0.5% sodium dodecyl sulfate followed by extraction with phenol-chloroform. The amount of monomeric microcircles formed was quantitated by phosphorimaging (ImageQuant; Amersham-Pharmacia) after electrophoresis in 7% acrylamide:bisacrylamide (59:1) gels containing 10% glycerol and Tris-borate-EDTA buffer.
For microcircle binding assays, preformed 98-bp 32P-labeled microcircles were prepared using HMGB1 and T4 DNA ligase. Binding reactions were performed under the same conditions as for the linear substrates. For competition assays, sufficient HMGB proteins were incubated with 98-bp microcircles in a 20-μl reaction volume containing 20 mM HEPES (pH 7.5), 40 mM NaCl, 1 mM EDTA, 0.1 mg/ml acetylated bovine serum albumin (BSA) (Roche), and 5% glycerol for 30 min at 30°C to generate at least 50% complexes. From 0 to 3.3 μg sonicated salmon sperm DNA (Sigma) was then added, and the reaction mixtures were incubated an additional 30 min at 30°C and applied to a polyacrylamide gel. For dissociation rate experiments, the complexes were challenged with 55 μg/ml sonicated salmon sperm DNA (Pharmacia), which corresponds to approximately a 1,500-fold mass excess of competitor DNA, and aliquots were loaded onto electrophoresing gels at increasing times. Gels were dried and subjected to phosphorimaging. Dissociation rates reflect the times required for decay of 50% of the starting complex, as measured from the times at which the samples were applied to the gel, and were extrapolated from graphs like those in Fig. 6B.
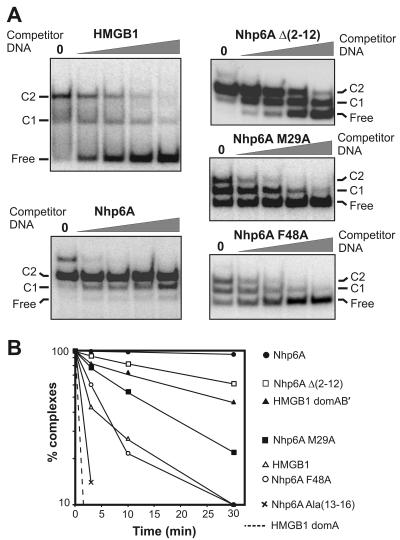
FIG. 6.
Stability of HMGB complexes formed on 98-bp microcircles. (A) Nhp6A, HMGB1, and Nhp6A mutants were bound to 32P-labeled 98-bp microcircles and then challenged with increasing amounts (0, 0.12, 0.37, 1.1, and 3.3 μg) of sonicated salmon sperm DNA in 20-μl reaction mixtures. After an additional 30-min incubation, the samples were subjected to native polyacrylamide gel electrophoresis. The positions of the unbound (Free) 98-bp microcircles and complexes (C) containing one and two molecules of the HMGB protein are indicated on the left. A small amount of an unstable third complex is present in the Nhp6A sample without competitor DNA. (B) Dissociation kinetics of HMGB complexes formed on 98-bp microcircles after addition of 55 μg/ml sonicated salmon sperm DNA. The relative number of complexes (percentage of the total DNA) formed by each HMGB protein prior to the addition of competitor DNA was set to 100, and the relative numbers of complexes remaining at each time point were scaled accordingly. The symbols for the HMGB1 and Nhp6A derivatives plotted are given to the right of the graph.
RAG-RSS complex assembly and DNA cleavage assays. (i) Proteins.
The core RAG2 protein was produced from vaccinia virus infection of HeLa cells as described previously (30). Core RAG1 proteins (either with a histidine tag or fused to the maltose binding protein) were produced as described previously (23, 30).
(ii) Single-site cleavage.
Plasmids12RSS/TP3 and 23RSS/TP2 (25) served as the sources for the DNA substrates for single-site cleavage. 32P-labeled substrate DNA was prepared as described previously (25). The single-site cleavage reactions were performed in a 20-μl final volume containing 1.5 ng labeled substrate DNA, 20 ng His-tagged core RAG1, 20 ng core RAG2, 1% BSA, 21 mM HEPES (pH 7.5), 1.7 mM DTT, 62.5 mM K-glutamate, 2 mM MnCl2, and 20 ng fragmented and boiled herring sperm DNA. The use of Mn2+ as the divalent metal ion permits cleavage in the presence of only a single RSS. Reaction mixtures were incubated for 2 h at 30°C. Reaction products were separated on a 6% denaturing acrylamide:bisacrylamide (19:1) gel, visualized by autoradiography, and quantified with a phosphorimager using ImageQuant software.
(iii) Nucleosome cleavage assays.
Nucleosome cleavage assays were carried out essentially as described previously (25, 26). Nucleosomes were assembled by the salt dilution method using bulk acetylated histones and a 152-bp DNA template (12/TP3 or 23/TP2) and then purified through a 5 to 30% glycerol gradient. Equal amounts of free DNA and nucleosomes (using equal 32P activities equivalent to 1 ng) were used in 20-μl cleavage reaction mixtures as described previously (25, 26).
(iv) Gel shift assay.
Electrophoretic mobility shift assay reactions were carried out as previously described (34) using Ca2+ as the divalent metal ion to allow the RAG proteins to bind to but not cleave their substrate DNA. In brief, binding reaction mixtures in a final volume of 10 μl contained 0.75 ng labeled substrate DNA, 10 ng maltose-binding protein-core RAG1, 10 ng core RAG2, 20 mM HEPES (pH 7.5), 55 mM K-glutamate, 2 mM CaCl2, 1.6 mM DTT, 1% glycerol, 1% BSA, and 0.3 ng herring sperm DNA. The amount of HMGB protein present in each reaction mixture is indicated in the figure legends. Binding was allowed to proceed at 30°C for 30 min, and the products separated on a 5% polyacrylamide gel (29:1) (0.5× Tris-borate-EDTA and 5% glycerol) at 250 V for 3 h.
RESULTS
DNA binding activities of truncated HMGB1 proteins.
To address the functional importance of different segments of HMGB1 in V(D)J recombination, a set of truncated derivatives (Fig. 2A) was purified. Initially, the ability of these proteins to bind and bend DNA independently of the RAG proteins was evaluated. Although, HMGB1 does not form sufficiently stable complexes with linear DNA to generate discrete bands in native polyacrylamide gels (Fig. 2B), it does form discrete complexes with DNA microcircles (41, 58). On 98-bp DNA circles, two complexes are formed with an apparent Kd of 4.5 nM (e.g., see Fig. 6A). Each of the truncated versions of HMGB1 also formed two complexes on the prebent DNA, with the exception of domains B and B", which exhibit only very weak DNA interactions at high protein concentrations (Table 1). The individual HMGB domains containing their C-terminal basic regions (A′ and B′) displayed only a modest decrease in equilibrium binding affinity to 98-bp microcircles when compared to the didomain HMGB1 proteins (Table 1). The stimulatory effect of the basic residues present on the C-terminal end of each domain on DNA binding was much greater for domain B than domain A, as the affinity of the domain B′ peptide for DNA microcircles is over 50 times greater than the domain B peptide.
TABLE 1.
DNA binding properties of HMGB1 and Nhp6A mutants
| HMGB protein | Kd lineara (nM) | Kd circlea (nM) | Kcircleb (nM) | Half-lifec (min) |
|---|---|---|---|---|
| HMGB1 | ND | 4.5 | 24 | 3 |
| Domains AB′ | ND | 7.5 | 2.0 | 26 |
| Domains AB | ND | 4.5 | 1.5 | 26 |
| Domain A′ | ND | 15 | 230 | <2 |
| Domain A | ND | 65 | 668 | <2 |
| Domain A F38A | ND | 120u | NF | ND |
| Domain B′ | ND | 15 | 210 | <2 |
| Domain B | ND | 1,020u | 2414 | ND |
| Domain B" | ND | ND | 3482 | ND |
| Nhp6A | 10 | 1.5 | 35 | 300 |
| Δ(2-12) | 20u | 2.0 | 55 | 41 |
| Δ(2-16) | 4,100u | 530u | 2400 | ND |
| Ala(13-16) | 108u | 7.5 | 600 | <2 |
| R23A R36A | 105u | 9.0 | 146 | <2 |
| M29A | 21 | 2.5 | 223 | 12 |
| F48A | 19 | 14 | NF | 4 |
| M29A F48A | 15 | 13 | NF | 4 |
Equilibrium dissociation constants for binding to linear or circular 98-bp DNA as measured by gel mobility shift assays. Values for binding of HMGB1 or its derivatives to linear DNA or domain B" binding to microcircular DNA were not determined (ND) since they do not form discrete complexes (e.g., see Fig. 2B). Nhp6A data were obtained as part of this work or are from references 5, 29, and 63. u, complexes appeared unstable as reflected by smeary bands after electrophoresis.
The concentration of protein needed to generate 50% of the maximal levels of 98-bp monomer circles in a 30-min ligation reaction. The maximal levels of microcircles formed were similar among all of the active HMGB proteins, with 30 to 40% of input linear DNA converted into microcircles. NF, 98-bp microcircles are not formed. Average values from two or more experiments were determined as part of this work, or values are from references 5, 29, and 63.
Rate of dissociation of complexes preassembled on 98-bp microcircles after addition of 55 μg/ml competitor DNA. Averaged values were extrapolated from two or more experiments and represent the time (min) required for decay of 50% of the starting number of complexes. ND, not determined due to very poor binding.
DNA bending activities of the HMGB1 derivatives were evaluated using microcircle formation assays (39, 41). Ninety-eight- and 75-bp DNA fragments containing EcoRI cohesive ends were incubated with different amounts of HMGB proteins in the presence of DNA ligase, and the percentage of monomeric DNA circles was determined (Fig. 2C). Although each of the derivatives was active in forming microcircles, the protein concentrations needed for reaching half-maximal levels of circle formation (Kcircle) differed by up to three orders of magnitude (Fig. 2C and Table 1). Our preparations of HMGB1 derivatives containing both domains were more active than the single-domain derivatives, as has been noted earlier (17). Microcircle formation by domains A′ and B′ was very similar, but domain B without the C-terminal basic region (derivative B) and especially domain B" containing both the basic and acidic C-terminal extensions exhibited low activity. The 10-fold-higher Kcircle measured for full-length HMGB1 relative to HMGB1 domain AB′ presumably reflects inhibition by the acidic region at the C terminus (22, 27, 48).
Stimulation of RAG1/2-RSS binding and DNA cleavage by truncated HMGB1 derivatives.
While RAG1/2 alone can bind to an individual RSS, HMGB1 enhances this binding, promoting efficient formation of a complex that migrates more slowly than the RAG1/2-RSS complex formed in the absence of HMGB1 (for examples, see references 45 and 57 and Fig. 3A). We found that the individual HMGB domains are sufficient to stimulate formation of high levels of RAG1/2 complexes on either the 12 or 23 RSS (Fig. 3A), although more domain B′ than domain A′ protein is required to generate an equivalent amount of the slower-migrating RAG1/2-HMGB-RSS complex (Fig. 3A). HMGB1 does not require the presence of the C-terminal charged extensions to form the RAG1/2-HMGB-RSS complex. Derivatives AB′, AB, A, and B all are capable of forming high levels of slower-migrating complexes (Fig. 3A and data not shown). The domain B" derivative was ineffective, consistent with its very poor DNA binding and microcircle formation activities (not shown).
FIG. 3.
(A) Stimulation of RAG1/2-HMGB-RSS complex assembly by mammalian HMGB1 and its truncated derivatives. Complex assembly with the 12 RSS substrate was analyzed using fixed amounts of RAG1/2 protein and increasing amounts of the indicated HMGB derivative. Reactions were carried out in the presence of Ca2+ to permit binding, but not cleavage, to occur, and mixtures were electrophoresed on native polyacrylamide gels. The amounts of HMGB protein per lane were as follows: HMGB1, 0, 0.05, 0.1, 0.2, 0.4, 0.8, 1.5, 3.1, 6.2, and 12.5 ng; domains AB′, AB, A′, A, and B′, 0.05 to 12.5 ng; domain B, 0, 1.5, 3.2, 6.2, 12.5, 25, 50, and 100 ng. (B) Stimulation of RAG1/2-mediated cleavage and hairpin formation by HMGB1 and its truncated derivatives. Cleavage of a 32P-labeled 150-bp DNA fragment containing a 12 RSS was analyzed in the presence of a fixed concentration of RAG1/2 and increasing amounts of the HMGB protein. The amounts of HMGB protein per lane were as follows: HMGB1, 0, 0.012, 0.05, 0.2, 0.8, 3.2, 12.5 ng; domains AB′ and AB, 0.05, 0.2, 0.8, 3.2, 12.5, and 50 ng; domains A′, A, B′, and B, 0.2, 0.8, 3.2, 12.5, 50, and 100 ng. Reaction products were separated on 6% denaturing polyacrylamide gels. HP, hairpin; S, 12 RSS DNA substrate; N, nicked DNA.
The ability of the truncated HMGB1 proteins to stimulate RAG1/2-mediated cleavage of substrates containing a single RSS was also assessed. HMGB1 and all the truncated derivatives (with the exception of domain B") strongly stimulated RSS cleavage, as measured by the accumulation of nicked and hairpinned products of both the 12 RSS (Fig. 3B) and 23 RSS (not shown) over the very low basal activity seen in the absence of HMGB1. Domains A and A′ stimulated cleavage at concentrations similar to those of full-length HMGB1 or domain AB′ or AB, while the domain B derivatives required more protein (see legend of Fig. 3). We conclude that both isolated domains are effective in stimulating assembly of RAG1/2-HMGB-RSS complexes that mediate V(D)J cleavage but that the derivatives of domain B have a lower specific activity than those of domain A.
Stimulation of RAG1/2 activity on nucleosomal substrates by HMGB1 derivatives.
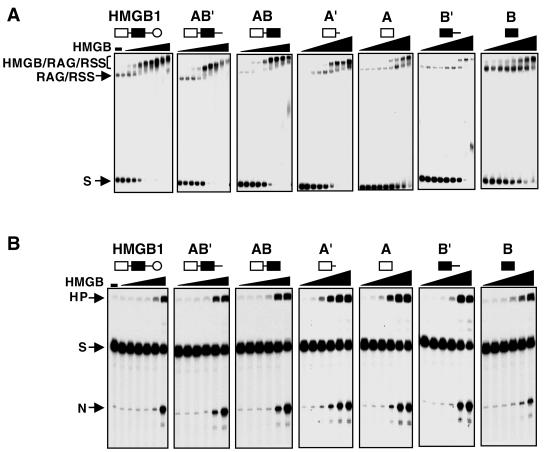
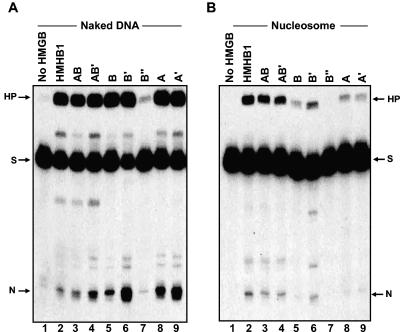
Previous studies have shown that nucleosomal substrates are refractory to V(D)J cleavage. This inhibition can be relieved in part through acetylation of the nucleosomes and the addition of HMGB1, particularly if the RSS is positioned near the entry/exit points of the nucleosome (25, 26). The ability of different HMGB1 derivatives to support cleavage of nucleosomal DNA was tested using two different substrates: 12TP3 and 23TP2 (see Materials and Methods and references 25 and 26). In these constructs, the 12 RSS and 23 RSS are positioned approximately 30 bp and 20 bp away from the dyad, respectively. Acetylated histones were assembled on 12TP3 and 23TP2, and cleavage in the presence of different HMGB1 derivatives was assessed in comparison to cleavage of naked 12TP3 and 23TP2 DNA. In the context of a nucleosome, the single HMGB domains were less efficient at stimulating RAG1/2 cleavage than full-length HMGB1 or the derivatives containing both domains (Fig. 4). Thus, unlike cleavage of naked DNA, both HMGB domains appear to be required for efficient RAG1/2 activity in the context of chromatin.
FIG. 4.
Stimulation of V(D)J cleavage by HMGB1 derivatives on nucleosomal versus naked 23 RSS substrates. (A) Naked DNA substrate. A 32P-labeled 150-bp DNA fragment (23TP2) (25) containing a 23 RSS was incubated with RAG1 and RAG2 either without HMGB protein (lane 1) or with 100 ng of the indicated HMGB1 derivatives (lanes 2 to 9). (B) Nucleosomal substrate. Nucleosomes assembled from bulk-acetylated histones as described previously (26) were challenged for cleavage in the presence of RAG1 and RAG2 either without HMGB (lane 1) or with 100 ng of each of the indicated HMGB1 derivatives (lanes 2 to 9).
Yeast Nhp6A supports RAG1/2-HMGB-RSS complex assembly but not RAG1/2-mediated cleavage.
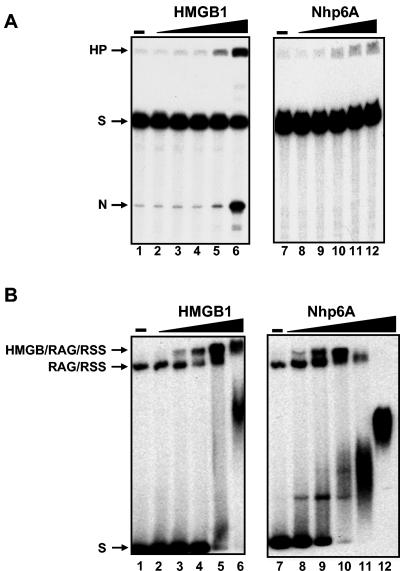
Yeast Nhp6A contains a single HMGB domain whose amino acid residues are 26% and 48% identical to the mammalian HMGB1 domains A and B, respectively (Fig. 1C). Addition of Nhp6A did little to stimulate RAG1/2-mediated hairpin formation at a 12 (not shown) or 23 (Fig. 5A) RSS, consistent with earlier results of van Gent et al. (57). Surprisingly, even though cleavage was poorly stimulated by Nhp6A, the yeast protein was efficient at assembling RAG1/2-HMGB-RSS complexes (Fig. 5B). Thus, Nhp6A cooperates with RAG1/2 to assemble RAG1/2-Nhp6A-RSS complexes, but these complexes are only minimally catalytically competent.
FIG. 5.
Nhp6A promotes efficient assembly of RAG1/2-RSS complexes but supports minimal V(D)J cleavage. (A) V(D)J cleavage reactions performed in the presence of increasing amounts of Nhp6A and HMGB1 on a 23 RSS substrate. The 23 RSS substrate was incubated with RAG1 and RAG2 either without HMGB protein (lanes 1 and 7) or with HMGB1 (0.05, 0.2, 0.8, 3.2, and 12.5 ng in lanes 2 to 6, respectively) or Nhp6A (0.2, 0.8, 3.2, 12.5, and 50 ng in lanes 8 to 12, respectively). HP, hairpin; S, substrate; N, nicked DNA. (B) RAG1/2-HMGB-23 RSS complex formation in the presence of increasing amounts of Nhp6A or HMGB1. The 23 RSS substrate was incubated with RAG1 and RAG2 either without HMGB protein (lanes 1 and 7) or with increasing amounts of HMGB1 (0.04, 0.2, 1, 5, and 25 ng in lanes 2 to 6, respectively) or Nhp6A (0.04, 0.2, 1, 5, and 25 ng in lanes 8 to 12, respectively).
Surprisingly, the yeast protein efficiently assembled RAG1/2-HMGB-RSS complexes at low concentrations; at higher concentrations, complex formation was inhibited, presumably due to the strong binding and coating of the probe by Nhp6A (Fig. 5B). We conclude that Nhp6A cooperates with RAG1/2 to assemble RAG1/2-Nhp6A-RSS complexes but these complexes exhibit poor DNA cleavage and hairpin formation.
Nhp6A forms more stable complexes with DNA than does HMGB1.
What properties of yeast Nhp6A as compared to the mammalian HMGB1 single-domain derivatives might account for the different effects of these HMGB proteins on V(D)J cleavage? Both HMGB1 and Nhp6A promote the ligation of 98-bp microcircles, with Kcircle values of 24 and 35 nM, respectively (Table 1); similar Kcircle values for HMGB1 (17 nM) and Nhp6A (38 nM) were also obtained for 75-bp microcircle formation. Moreover, both Nhp6A and HMGB1 bind to 98-bp DNA microcircles with similar apparent equilibrium dissociation constants (Table 1). However, the different behaviors of Nhp6A and HMGB1 complexes with linear DNA segments upon polyacrylamide gel electrophoresis implies that Nhp6A forms more stable DNA complexes than HMGB1 (Fig. 2B) (40).
To directly evaluate the stabilities of DNA complexes containing Nhp6A and HMGB1, complexes assembled on 98-bp microcircles were challenged with increasing amounts of sonicated salmon sperm DNA. Two Nhp6A molecules associate very tightly with the 98-bp microcircle substrates and are resistant to addition of up to ∼5,000-fold mass excess of competitor DNA (Fig. 6A). By contrast the two full-length HMGB1 molecules readily dissociate from the microcircles upon addition of increasing amounts of competitor DNA.
To further quantify the differences in stability of the HMGB-DNA microcircle complexes, the kinetics of dissociation were measured after addition of 55 μg/ml competitor DNA. Complexes formed with wild-type Nhp6A on 98-bp DNA microcircles decay extremely slowly, with an estimated half-life of about 5 h (Fig. 6B; Table 1). By contrast, 50% of the complexes formed with full-length HMGB1 dissociated within 3 min, and essentially all of the complexes formed with the individual domain A′ and B′ peptides dissociate within 3 min (Fig. 6B; Table 1). Interestingly, HMGB1 derivatives containing both domains but lacking the acidic C-terminal tail (HMGB1 domains AB′ and AB) are markedly more stable than the full-length protein, with half-lives of 26 min (Fig. 6B; Table 1). However, they remain less stable than complexes formed with wild-type Nhp6A.
Mutations in Nhp6A that decrease complex stability on microcircles enhance V(D)J cleavage.
The results described above indicate that a key difference between the properties of DNA complexes formed with Nhp6A and HMGB1 and its derivatives is their stability. This led us to consider whether Nhp6A may be assembling RAG1/2-HMGB-RSS complexes that are insufficiently dynamic to support DNA catalysis. We therefore asked if Nhp6A proteins containing mutations that destabilize their interaction with DNA were more effective at stimulating V(D)J cleavage. The basic N-terminal tail of Nhp6A, which wraps around the major groove on the opposite side of DNA from the HMGB core domain (Fig. 1D), is largely responsible for its high-affinity binding to linear DNA (29, 63). Nhp6A Δ(2-12) is missing the first set of basic residues (Lys 8, Lys 9, and Arg 10; Fig. 1C). This mutant is able to bind to and promote ligation of 98-bp and 75-bp microcircles as efficiently as wild-type Nhp6A (Table 1). Nhp6A Δ(2-12) is much more sensitive to challenge with competitor DNA than wild-type Nhp6A and exhibits a dissociation rate from 98-bp microcircles that is similar to HMGB1 domains AB and AB′ (Fig. 6A and B; Table 1). Nhp6A Δ(2-16) has the entire N-terminal basic arm region deleted. It binds poorly to linear DNA but is able to form unstable complexes on 98-bp microcircles and promote ligation of 98-bp microcircles at high protein concentrations (Table 1) (this work and reference 63). Whereas wild-type Nhp6A stimulates RAG1/2 cleavage of 12 and 23 RSS substrates poorly, reactions performed with either Nhp6A Δ(2-12) or Δ(2-16) generate substantial amounts of hairpins (Fig. 7A and D), while supporting complex formation at levels equivalent to wild-type Nhp6A (Fig. 7B and C). Similar results were obtained in coupled-cleavage assays; whereas Nhp6A stimulated a small amount of coupled cleavage at the 12 and 23 RSS sites, the Δ(2-12) and Δ(2-16) derivatives were much more effective (data not shown).
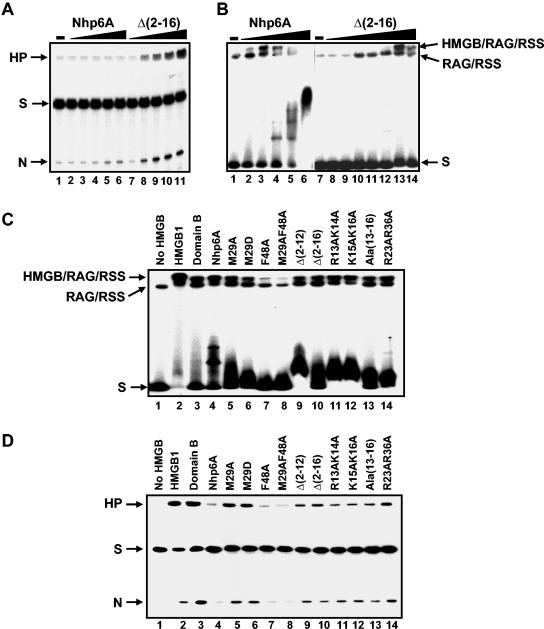
FIG. 7.
Effects of mutations in Nhp6A on RAG1/2-HMGB-RSS complex formation and V(D)J cleavage. (A) Cleavage at a 12 RSS in the presence of increasing amounts of Nhp6A and the Δ(2-16) mutant derivative. The 12 RSS substrate was incubated with RAG1 and RAG2 without HMGB (lane 1), RAG1 and RAG2 with Nhp6A (1, 2.5, 10, 50, and 100 ng in lanes 2 to 6, respectively), and Δ(2-16) (10, 20, 40, 50, and 100 ng in lanes 7 to 11, respectively). HP, hairpin; S, substrate; N, nicked DNA. (B) Both Nhp6A and Nhp6A Δ(2-16) support RAG1/2-RSS complex formation. The 12 RSS substrate was incubated with RAG1 and RAG2 without HMGB (lane 1 and lane 7), RAG1 and RAG2 with Nhp6A (0.04, 0.2, 1, 5, and 25 ng in lanes 2 to 6, respectively), and Nhp6A Δ(2-16) (0.04, 0.2, 1, 5, 25, 50, and 100 ng in lanes 8 to14, respectively). (C) RAG1/2-HMGB-12 RSS complex formation. A 150-bp 12 RSS substrate was incubated with RAG1 and RAG2 without HMGB (lane 1) or with HMGB1 (25 ng), HMGB1 domain B (50 ng), Nhp6A (1 ng), or Nhp6A mutant M29A (5 ng), M29D (5 ng), F48A (1 ng), M29A F48A (1 ng), Δ(2-12) (5 ng), Δ(2-16) (50 ng), K13A K14A (5 ng), K15A K16A (5 ng), Ala(13-16) (5 ng), or R23AR36A (5 ng). The amount of each protein added represents the optimal concentration for maximum RAG1/2-HMGB-RSS complex formation. (D) Single-site V(D)J cleavage. A 150-bp 32P-labeled 12 RSS substrate was incubated with RAG1 and RAG2 without HMGB or with HMGB1 (50 ng), HMGB1 domain B (50 ng), Nhp6A (2.5 ng), or Nhp6A mutant M29A (50 ng), M29D (50 ng), F48A (50 ng), M29A F48A (50 ng), Δ(2-12) (50 ng), Δ(2-16) (50 ng), R13A K14A (50 ng), K15A K16A (50 ng), Ala(13-16) (50 ng), or R23A R36A (50 ng). The amount of each protein added represents the optimal concentration for maximum nick and hairpin formation.
Specific amino acid residue substitutions that destabilize Nhp6A-DNA interactions also result in enhanced RSS cleavage by RAG1/2. Nhp6A Ala(13-16) has the lysines and arginine in the second basic patch of the N-terminal segment (Fig. 1C) replaced with alanines. These substitutions result in a strong decrease in binding affinity, with most complexes formed on 98-bp microcircles dissociating within 3 min of competitor addition (Fig. 6B; Table 1). Like the N-terminal deletions, Nhp6A Ala(13-16) promotes enhanced cleavage and hairpin formation at the 12 and 23 RSS (Fig. 7C and D and not shown). Alanine substitutions at residues Arg 13 plus Lys 14 or Lys 15 plus Lys 16 have an intermediate defect in DNA binding (62) and, likewise, display moderately enhanced cleavage and hairpin formation by RAG1/2 (Fig. 7D). The R23A R36A double mutant contains substitutions of residues within the HMGB core domain which directly interact with DNA from the minor groove side (29). This Nhp6A derivative binds linear DNA with a 10-fold-lower affinity, requires four times the wild-type amount of protein to promote equivalent microcircle formation, and dissociates from 98-bp microcircles with a <3-min half-life (Table 1) (5). Nevertheless, R23A R36A facilitates binding and DNA catalysis by RAG1/2 with the 12 and 23 RSS substrates (Fig. 7C and D and not shown). Thus, a variety of mutations in the HMGB core or arm of Nhp6A that destabilize DNA binding result in enhanced RAG1/2-mediated catalysis of DNA relative to wild-type Nhp6A.
The DNA exit wedge generated by Nhp6A Phe 48 is required for assembly of RAG1/2-HMGB-RSS complexes.
Nhp6A possesses two hydrophobic wedges that insert into the base stack via the minor groove and distort the structure of the bound DNA (5, 29). Met 29 is the most important residue of the wedge, located near the center of the binding site, and Phe 48 specifies the wedge at one of the DNA exit sites (Fig. 1D). Mutants with alanine substitutions at either of these residues remain capable of forming complexes on linear DNA, but nuclear magnetic resonance analysis has revealed that both residues appear important in binding site selection (29). Phe 48 is uniquely required for recognition of the kinked DNA structure induced by cisplatin cross-linking (61). Both M29A and F48A are compromised for bending DNA, as revealed by the failure of either mutant to form 75-bp microcircles; however, M29A remains capable of forming 98-bp microcircles. Both mutants form complexes on 98-bp microcircles that are much less stable than that formed by wild-type Nhp6A but more stable than those formed by individual HMGB1 domains in the presence of competitor DNA (Fig. 6; Table 1).
The two Nhp6A wedge mutants have opposite properties with respect to the V(D)J reaction. Nhp6A F48A is unable to assemble RAG1/2-HMGB-RSS complexes (Fig. 7C) and thus also fails to promote V(D)J cleavage (Fig. 7D). By contrast, Nhp6A M29A (or Nhp6A M29D) supports RAG1/2-RSS complex formation and stimulates V(D)J cleavage relative to wild-type Nhp6A (Fig. 7C and D). A mutant containing alanine substitutions at both wedges (Nhp6A M29A F48A) behaves like Nhp6A F48A with respect to RAG1/2-HMGB-RSS complex formation and cleavage. Likewise, F48A and M29A F48A mutants failed to support coupled cleavage, but reactions performed with Nhp6A M29A or M29D exhibited enhanced cleavage at the 12 and 23 RSS sites relative to wild-type Nhp6A (data not shown). These results indicate that the Phe 48 wedge is of critical importance for initial assembly of RAG1/2-HMGB-RSS complexes. The enhanced catalytic activity of RAG1/2-HMGB-RSS complexes formed with Nhp6A M29A (and M29D) relative to wild-type Nhp6A presumably reflects the less stable binding properties exhibited by the mutant (Fig. 6A and B; Table 1).
Requirement for Phe 38 of HMGB1 domain A for RAG1/2-RSS complex formation.
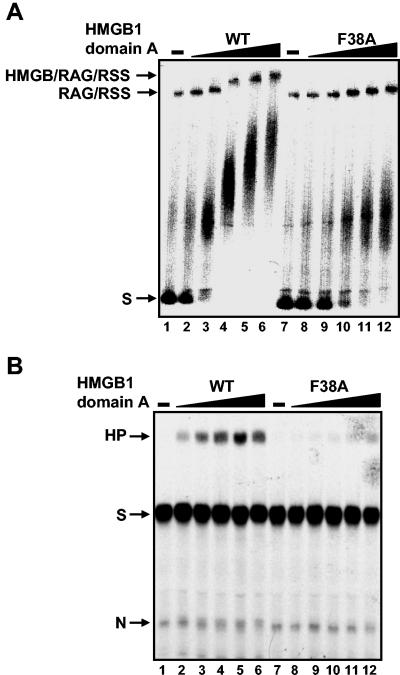
Phe 38 in HMGB1 domain A corresponds to Phe 48 of Nhp6A. We therefore asked whether Phe 38 in the domain A peptide was similarly required for the assembly of RAG1/2-RSS complexes. Domain A peptide containing the F38A mutation failed to promote ligation of 98-bp microcircles, although it still was able to form unstable complexes with preformed 98-bp microcircles (Table 1). Similarly, domain A F38A does not promote assembly of RAG1/2 complexes on the 23 RSS, and therefore, as expected, no stimulation of cleavage by the mutant HMGB1 derivative is detected (Fig. 8).
FIG. 8.
Phe 38 of HMGB1 domain A is required for the stimulation of complex formation and V(D)J cleavage. (A) Complex formation with a 23 RSS in the presence of increasing concentrations of HMGB1 domain A (WT) (0.04, 0.2, 1, 5, and 25 ng in lanes 2 to 6, respectively) and the HMGB1 domain A mutant F38A (0.04, 0.2, 1, 5, and 25 ng in lanes 8 to 12, respectively) is shown. No HMGB protein was included in the reactions in lanes 1 and 7. (B) V(D)J cleavage in the presence of wild-type HMGB1 domain A and HMGB1 domain A mutant F38A. The 23 RSS substrate was incubated with RAG1 and RAG2 either without HMGB (lane 1 and lane 7), with HMGB1 domain A (WT) (25, 50, 100, 200, and 400 ng in lanes 2 to 6, respectively), or with domain A mutant F38A (25, 50, 100, 200, and 400 ng in lanes 8 to 12, respectively). HP, hairpin; S, substrate; N, nicked DNA.
DISCUSSION
Previous in vitro studies on the V(D)J reaction have established that HMGB1 or HMGB2 potentiate RAG1/2 binding to the RSS (4, 10, 45, 57; also see references 13 and 15 and references therein). In this report we show that either of the two HMGB DNA binding domains of HMGB1 can independently promote efficient assembly of catalytically competent complexes on naked DNA in vitro. However, the didomain structure of HMGB1 is required for maximum activity when the RSS substrate is wrapped in a nucleosome. Analysis of mutants of the yeast HMGB protein Nhp6A uncovered two important features of HMGB proteins for stimulation of V(D)J recombination. First, the HMGB protein must interact with bent DNA in a relatively dynamic manner in order for the complexes to support DNA nicking and hairpin formation. This feature may reflect sequential changes in DNA conformations that are likely to be required as the RAG1/2-HMGB-RSS complex proceeds from initial binding through the two chemical steps leading to the formation of the hairpin product. Second, the hydrophobic wedge at the DNA exit site, which is specified in Nhp6A by Phe 48, is a critical feature that is required to promote the initial step of RAG1/2-RSS binding. Phe 48 in Nhp6A, as well as its counterpart in HMGB1 domain A (Phe 38), is known to be a critical determinant in the recognition of kinked DNA (19, 38, 61), and its importance is consistent with HMGB proteins recognizing or stabilizing a distorted DNA structure within the RAG1/2-RSS complex (4). We discuss the biochemical properties of HMGB proteins with respect to their stimulation of the V(D)J recombination reaction below.
Relationship between the DNA binding properties of resected derivatives of mammalian HMGB1 and their support of RAG1/2-RSS complex assembly and cleavage activity. (i) HMGB1 domain A versus domain B.
The rat HMGB1 DNA binding domains (A and B) are 29% identical and 50% similar to each other (Fig. 1C). Our recombinant versions of domains A′ and B′ containing their C-terminal basic ends function very similarly with respect to their abilities to form as well as to selectively bind to DNA microcircles. The similarity in DNA bending efficiencies between domains A′ and B′ observed here contrasts with some other studies where domain B, with its two intercalating wedges, has been found to exhibit greater bending activity (46, 52, 64). However, we note that the recombinant form of domain B used here lacks the lysine-rich linker region (amino acids 85 to 88; TKKK) separating domains A and B, which has been reported to enhance DNA binding (17, 46, 47). Our minimal-domain peptides that lack the basic residues at their C-terminal ends show pronounced differences in bending and binding to microcircular DNA; domain A requires less protein than domain B for equivalent activity. We conclude that both HMGB1 domains are effective at bending DNA but that flanking basic residues enhance their activities, particularly for the B domain.
The DNA binding and bending properties of the isolated domains largely parallel their activities in the V(D)J reaction on naked DNA. Whereas each of the isolated domain constructs are able to efficiently promote RAG1/2-RSS assembly and V(D)J cleavage, the B′- and B-domain peptides require more protein than the respective A′- and A-domain peptides for optimal activity. However, the ability to bend DNA, as evaluated by microcircle formation, is not the sole determinant of HMGB function in the V(D)J reaction, since the domain A′ peptide behaved nearly indistinguishably from the didomain derivatives on naked DNA, even though 10- or 100-fold more domain A′ was required to generate equivalent numbers of microcircles compared to full-length HMGB1 or domains AB/AB′, respectively. As elaborated further below, the differences that we observe with the minimal domains are consistent with the view that domain A of HMGB1 is more active than domain B in interacting with prebent and structured DNA (19, 22, 52, 59) and that a localized DNA disruption exists within the RAG1/2-HMGB-RSS assembly (4, 10, 11, 42).
(ii) The C-terminal acidic tail.
The C-terminal acidic tail has generally been found to inhibit binding of HMGB1 derivatives to naked DNA, and our results are consistent with this view (22, 27, 48). Nevertheless, full-length HMGB1 appears no less efficient at promoting RAG1/2 interactions on RSS DNA than domain A or didomain derivatives lacking the C-terminal tail. As others also have noted, the single domain B" peptide containing the acidic region exhibits very poor DNA binding and bending activities, and consequently it appears essentially inactive in the V(D)J reaction. Recent nuclear magnetic resonance and cross-linking experiments have provided evidence that the acidic region can specifically interact with residues within both domains and the connecting basic linker segment, which presumably inhibits DNA binding (22, 24).
The acidic region in the context of the didomain structure has been reported to facilitate HMGB1 interactions with nucleosomes, possibly by interacting with the histone H3 N terminus (56). Bonaldi et al. have reported that the presence of the acidic region on HMGB1 enhances nucleosome sliding by the ACF (ISWI) remodeling complex (8). We find that full-length HMGB1 is somewhat more efficient in promoting RSS cleavage by RAG1/2 on nucleosomal substrates than the didomain derivatives without the acidic region, suggesting that the acidic region could have a modest beneficial role in V(D)J recombination in vivo.
Nhp6A mutants reveal parameters important for assembly of catalytically competent RAG1/2-RSS complexes.
The sequence and structure of the HMGB domain of Nhp6A are more closely related to HMGB1 domain B (48% amino acid residue identity, 1.2-Å root mean square deviation between peptide backbones) than domain A (26% amino acid residue identity, 2.2-Å root mean square deviation between peptide backbones) (Fig. 1C) (5, 29). Nhp6A binds DNA much more stably than any of the HMGB1 derivatives, primarily because of its N-terminal basic arm. We find that the greater affinity for DNA does not improve RAG1/2-RSS complex formation. Nhp6A promotes efficient RAG1/2-RSS complex formation at protein levels that are equivalent to HMGB1 domain A′ or the didomain constructs. However, the stable DNA binding by Nhp6A strongly inhibits DNA cleavage by the RAG1/2-Nhp6A-RSS complexes. Mutations within the N-terminal tail or within the HMGB core that destabilize binding and thus lead to Nhp6A proteins whose DNA binding properties more closely mimic those of the HMGB1 derivatives result in corresponding increases in V(D)J cleavage. These findings suggest that a dynamic association of the HMGB protein with the RAG1/2-RSS complex is critical for the DNA cleavage steps. The HMGB protein is not released from the complex, however, since HMGB1 has been shown to directly enhance hairpin formation in experiments utilizing prenicked substrates and to remain with the complex after the hairpin has been formed (20, 33, 34, 49, 50, 57).
Nhp6A contains two hydrophobic wedges at the DNA binding surface, but only one, Phe 48, is critical for stimulating RAG1/2-RSS assembly. This residue corresponds to Phe 38 in domain A, which we also show to be essential for its ability to promote RAG1/2-RSS assembly, and to Ile 34 in domain B (Fig. 1D). The importance of Phe 48 and the dispensability of the Met 29 wedge parallel the roles of these two residues in selective binding by Nhp6A to cisplatin adducts on DNA (61). Consistent with its moderately poorer activity in the V(D)J reaction, HMGB1 domain B has been found to be less selective for binding to cisplatin adducts than domain A, in part because of the isoleucine in place of a phenylalanine at the DNA exit wedge (19, 22). Cisplatin forms an intrastrand cross-link at the N7 position of adjacent guanines, generating a kink in the duplex DNA that compresses the major groove (51). An X-ray structure of mammalian HMGB1 domain A bound to a DNA duplex containing a cisplatin cross-link revealed that the Phe 38 intercalates between the cross-linked guanines on the minor groove side (Fig. 1D) (19, 38).
Aidinis et al. have provided evidence for bending of the RSS sites by RAG1/2 by circular permutation analysis of the electrophoretic migrations of RAG1/2-12 or 23 RSS complexes, even in the absence of HMGB1 (4). This finding, combined with the importance of the phenylalanine wedge, suggests that the HMGB protein may be recognizing a RAG1/2-induced DNA distortion between the nonamer and heptamer regions. Binding of the HMGB proteins may then stabilize the bent structure, thus enhancing complex formation. Consistent with this model, both protein-DNA cross-linking and ethylation interference experiments have suggested that HMGB1 is located in the intervening DNA segment. Ethylation interference experiments point to the HMGB protein contacting DNA near the 5′ end of the nonamer on the opposite side of the duplex from the bound RAG1/2 (33, 49). There are also several lines of evidence suggesting that unpairing of DNA strands, particularly over the heptamer region, may be important for the DNA cleavage/hairpin formation steps (10 and reviewed in reference 14). HMGB proteins induce considerable untwisting of DNA along with bending, which may further contribute to their stimulation of RAG1/2 function.
Comparison of HMGB1 stimulation of RAG1/2 and other DNA binding proteins.
The DNA binding domain of RAG1 has been proposed to contain a helix-turn-helix motif that is similar to the Tc1/mariner family of transposases (6). Interestingly, a member of the Tc1/mariner family of transposons, Sleeping Beauty, has also been shown to require HMGB1 for transposition in human cells (66). HMGB1 also facilitates binding of a number of transcription factors with structurally diverse DNA binding domains (2, 54). As is the case for RAG1/2 binding, a single domain from HMGB1 has been found to be sufficient in stimulating DNA binding of some of these regulatory proteins, including the Oct1/2 and HOXD9 homeodomains and Rta (31, 32, 65, 68). However, in at least one case, the cooperative binding of the basic leucine zipper protein Zebra to adjacent sites within the regulatory region of an Epstein-Barr virus promoter, both linked domains of HMGB1 are required (12). In some situations, there is evidence that HMGB1 interacts in solution with its binding partner, implying that direct protein-protein interactions, in addition to effects on DNA architecture by HMGB1, appear to contribute to binding cooperativity (reviewed in (2, 54). On the other hand, stable ternary complexes containing HMGB1 and DNA have not been observed upon polyacrylamide gel electrophoresis in several systems. Therefore, the HMGB protein appears to function in some contexts only transiently to chaperone the transcription factor to its DNA binding site.
Aidinis et al. reported that RAG1/2 and HMGB1 interact in solution in experiments employing immobilized RAG1/2 or HMGB1 (4). Both domains A and B were required in order to observe this interaction, leading the authors to propose that both domains are required for HMGB1 stimulation of RAG1/2 activity. Our finding that either isolated HMGB1 domain or unstable binding mutants of yeast Nhp6A are effective in promoting the assembly of catalytically competent RAG1/2-HMGB-RSS complexes suggests that a specific RAG1/2-HMGB interaction may be secondary to an effect of HMGB on DNA architecture. Only nine solvent-exposed residues, which have similar chemical characteristics but are not directly involved in DNA binding, are common between the three HMGB domains (Fig. 1C).
Acknowledgments
This work was supported by USPHS grants GM38509 to R.C.J. and GM48026 to M.A.O.
REFERENCES
- 1.Adachi, Y., S. Mizuno, and M. Yoshida. 1990. Efficient large-scale purification of non-histone chromosomal proteins HMG1 and HMG2 by using Polybuffer-exchanger PBE94. J. Chromatogr. 530:39-46. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, A., and D. G. Schatz. 1997. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell 89:43-53. [DOI] [PubMed] [Google Scholar]
- 3.Agresti, A., and M. E. Bianchi. 2003. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13:170-178. [DOI] [PubMed] [Google Scholar]
- 4.Aidinis, V., T. Bonaldi, M. Beltrame, S. Santagata, M. E. Bianchi, and E. Spanopoulou. 1999. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol. 19:6532-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allain, F. H., Y. M. Yen, J. E. Masse, P. Schultze, T. Dieckmann, R. C. Johnson, and J. Feigon. 1999. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 18:2563-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee-Basu, S., and A. D. Baxevanis. 2002. The DNA-binding region of RAG 1 is not a homeodomain. Genome Biol. 3:interactions 1004.1-1004.4. [Online.] http://genomebiology.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassing, C. H., W. Swat, and F. W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl.):S45-S55. [DOI] [PubMed] [Google Scholar]
- 8.Bonaldi, T., G. Langst, R. Strohner, P. B. Becker, and M. E. Bianchi. 2002. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 21:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciubotaru, M., and D. G. Schatz. 2004. Synapsis of recombination signal sequences located in cis and DNA underwinding in V(D)J recombination. Mol. Cell. Biol. 24:8727-8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuomo, C. A., C. L. Mundy, and M. A. Oettinger. 1996. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 16:5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellwood, K. B., Y. M. Yen, R. C. Johnson, and M. Carey. 2000. Mechanism for specificity by HMG-1 in enhanceosome assembly. Mol. Cell. Biol. 20:4359-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 14.Gellert, M. 2002. V(D)J recombination, p. 705-729. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 15.Gellert, M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 16.Grasser, K. D. 2003. Chromatin-associated HMGA and HMGB proteins: versatile co-regulators of DNA-dependent processes. Plant Mol. Biol. 53:281-295. [DOI] [PubMed] [Google Scholar]
- 17.Grasser, K. D., S. H. Teo, K. B. Lee, R. W. Broadhurst, C. Rees, C. H. Hardman, and J. O. Thomas. 1998. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1). Eur. J. Biochem. 253:787-795. [DOI] [PubMed] [Google Scholar]
- 18.Guazzi, S., A. Strangio, A. T. Franzi, and M. E. Bianchi. 2003. HMGB1, an architectural chromatin protein and extracellular signalling factor, has a spatially and temporally restricted expression pattern in mouse brain. Gene Expr. Patterns 3:29-33. [DOI] [PubMed] [Google Scholar]
- 19.He, Q., U. M. Ohndorf, and S. J. Lippard. 2000. Intercalating residues determine the mode of HMG1 domains A and B binding to cisplatin-modified DNA. Biochemistry 39:14426-14435. [DOI] [PubMed] [Google Scholar]
- 20.Hiom, K., and M. Gellert. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell 1:1011-1019. [DOI] [PubMed] [Google Scholar]
- 21.Jones, J. M., and M. Gellert. 2002. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 21:4162-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung, Y., and S. J. Lippard. 2003. Nature of full-length HMGB1 binding to cisplatin-modified DNA. Biochemistry 42:2664-2671. [DOI] [PubMed] [Google Scholar]
- 23.Kim, D. R., Y. Dai, C. L. Mundy, W. Yang, and M. A. Oettinger. 1999. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 13:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knapp, S., S. Muller, G. Digilio, T. Bonaldi, M. E. Bianchi, and G. Musco. 2004. The long acidic tail of high mobility group box 1 (HMGB1) protein forms an extended and flexible structure that interacts with specific residues within and between the HMG boxes. Biochemistry 43:11992-11997. [DOI] [PubMed] [Google Scholar]
- 25.Kwon, J., A. N. Imbalzano, A. Matthews, and M. A. Oettinger. 1998. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol. Cell 2:829-839. [DOI] [PubMed] [Google Scholar]
- 26.Kwon, J., K. B. Morshead, J. R. Guyon, R. E. Kingston, and M. A. Oettinger. 2000. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell 6:1037-1048. [DOI] [PubMed] [Google Scholar]
- 27.Lee, K. B., and J. O. Thomas. 2000. The effect of the acidic tail on the DNA-binding properties of the HMG1,2 class of proteins: insights from tail switching and tail removal. J. Mol. Biol. 304:135-149. [DOI] [PubMed] [Google Scholar]
- 28.Love, J. J., X. Li, D. A. Case, K. Giese, R. Grosschedl, and P. E. Wright. 1995. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376:791-795. [DOI] [PubMed] [Google Scholar]
- 29.Masse, J. E., B. Wong, Y. M. Yen, F. H. Allain, R. C. Johnson, and J. Feigon. 2002. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 323:263-284. [DOI] [PubMed] [Google Scholar]
- 30.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 31.McKinney, K., and C. Prives. 2002. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol. Cell. Biol. 22:6797-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsouras, K., B. Wong, C. Arayata, R. C. Johnson, and M. Carey. 2002. The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol. Cell. Biol. 22:4390-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo, X., T. Bailin, S. Noggle, and M. J. Sadofsky. 2000. A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res. 28:1228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mundy, C. L., N. Patenge, A. G. Matthews, and M. A. Oettinger. 2002. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol. 22:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, E. C., V. B. Zhurkin, J. M. Louis, G. Cornilescu, and G. M. Clore. 2001. Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol. 312:481-499. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, F. V. T., and M. E. Churchill. 2000. Nonsequence-specific DNA recognition: a structural perspective. Structure Fold Des. 8:R83-R89. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, F. V. T., R. M. Sweet, and M. E. Churchill. 1999. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 18:6610-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohndorf, U. M., M. A. Rould, Q. He, C. O. Pabo, and S. J. Lippard. 1999. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature 399:708-712. [DOI] [PubMed] [Google Scholar]
- 39.Paull, T. T., M. J. Haykinson, and R. C. Johnson. 1993. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 7:1521-1534. [DOI] [PubMed] [Google Scholar]
- 40.Paull, T. T., and R. C. Johnson. 1995. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem. 270:8744-8754. [DOI] [PubMed] [Google Scholar]
- 41.Pil, P. M., C. S. Chow, and S. J. Lippard. 1993. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc. Natl. Acad. Sci. USA 90:9465-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramsden, D. A., J. F. McBlane, D. C. van Gent, and M. Gellert. 1996. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 15:3197-3206. [PMC free article] [PubMed] [Google Scholar]
- 43.Read, C. M., P. D. Cary, C. Crane-Robinson, P. C. Driscoll, and D. G. Norman. 1993. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 21:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronfani, L., M. Ferraguti, L. Croci, C. E. Ovitt, H. R. Scholer, G. G. Consalez, and M. E. Bianchi. 2001. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein HMGB2. Development 128:1265-1273. [DOI] [PubMed] [Google Scholar]
- 45.Sawchuk, D. J., F. Weis-Garcia, S. Malik, E. Besmer, M. Bustin, M. C. Nussenzweig, and P. Cortes. 1997. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J. Exp. Med. 185:2025-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stros, M. 1998. DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J. Biol. Chem. 273:10355-10361. [PubMed] [Google Scholar]
- 47.Stros, M. 2001. Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry 40:4769-4779. [DOI] [PubMed] [Google Scholar]
- 48.Stros, M., J. Stokrova, and J. O. Thomas. 1994. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 22:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson, P. C. 2002. Fine structure and activity of discrete RAG-HMG complexes on V(D)J recombination signals. Mol. Cell. Biol. 22:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson, P. C. 2002. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 22:7790-7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahara, P. M., A. C. Rosenzweig, C. A. Frederick, and S. J. Lippard. 1995. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature 377:649-652. [DOI] [PubMed] [Google Scholar]
- 52.Teo, S. H., K. D. Grasser, and J. O. Thomas. 1995. Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex-determining factor SRY. Eur. J. Biochem. 230:943-950. [DOI] [PubMed] [Google Scholar]
- 53.Thomas, J. O. 2001. HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans. 29:395-401. [DOI] [PubMed] [Google Scholar]
- 54.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 55.Travers, A. 2000. Recognition of distorted DNA structures by HMG domains. Curr. Opin. Struct. Biol. 10:102-109. [DOI] [PubMed] [Google Scholar]
- 56.Ueda, T., H. Chou, T. Kawase, H. Shirakawa, and M. Yoshida. 2004. Acidic C-tail of HMGB1 is required for its target binding to nucleosome linker DNA and transcription stimulation. Biochemistry 43:9901-9908. [DOI] [PubMed] [Google Scholar]
- 57.van Gent, D. C., K. Hiom, T. T. Paull, and M. Gellert. 1997. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 16:2665-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webb, M., D. Payet, K. B. Lee, A. A. Travers, and J. O. Thomas. 2001. Structural requirements for cooperative binding of HMG1 to DNA minicircles. J. Mol. Biol. 309:79-88. [DOI] [PubMed] [Google Scholar]
- 59.Webb, M., and J. O. Thomas. 1999. Structure-specific binding of the two tandem HMG boxes of HMG1 to four-way junction DNA is mediated by the A domain. J. Mol. Biol. 294:373-387. [DOI] [PubMed] [Google Scholar]
- 60.Weir, H. M., P. J. Kraulis, C. S. Hill, A. R. Raine, E. D. Laue, and J. O. Thomas. 1993. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 12:1311-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong, B., J. E. Masse, Y. M. Yen, P. Giannikoupolos, J. Feigon, and R. C. Johnson. 2002. Binding to cisplatin-modified DNA by the Saccharomyces cerevisiae HMGB protein Nhp6A. Biochemistry 41:5404-5414. [DOI] [PubMed] [Google Scholar]
- 62.Yen, Y. M., P. M. Roberts, and R. C. Johnson. 2001. Nuclear localization of the Saccharomyces cerevisiae HMG protein NHP6A occurs by a RAN-independent nonclassical pathway. Traffic 2:449-464. [DOI] [PubMed] [Google Scholar]
- 63.Yen, Y. M., B. Wong, and R. C. Johnson. 1998. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J. Biol. Chem. 273:4424-4435. [DOI] [PubMed] [Google Scholar]
- 64.Yoshioka, K., K. Saito, T. Tanabe, A. Yamamoto, Y. Ando, Y. Nakamura, H. Shirakawa, and M. Yoshida. 1999. Differences in DNA recognition and conformational change activity between boxes A and B in HMG2 protein. Biochemistry 38:589-595. [DOI] [PubMed] [Google Scholar]
- 65.Zappavigna, V., L. Falciola, M. Helmer-Citterich, F. Mavilio, and M. E. Bianchi. 1996. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 15:4981-4991. [PMC free article] [PubMed] [Google Scholar]
- 66.Zayed, H., Z. Izsvak, D. Khare, U. Heinemann, and Z. Ivics. 2003. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31:2313-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zlatanova, J., and K. van Holde. 1998. Binding to four-way junction DNA: a common property of architectural proteins? FASEB J. 12:421-431. [PubMed] [Google Scholar]
- 68.Zwilling, S., H. Konig, and T. Wirth. 1995. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 14:1198-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]