Abstract
Axin is a central component of the canonical Wnt signal transduction machinery, serving as a scaffold for the β-catenin destruction complex. The related protein Axin2/Conductin, although less extensively studied, is thought to perform similar functions. Loss of Axin causes early embryonic lethality, while Axin2-null mice are viable but have craniofacial defects. Mutations in either gene contribute to cancer in humans. The lack of redundancy between Axin and Axin2 could be due to their different modes of expression: while Axin is expressed ubiquitously, Axin2 is expressed in tissue- and developmental-stage-specific patterns, and its transcription is induced by canonical Wnt signaling. Alternatively, the two proteins might have partially different functions, a hypothesis supported by the observation that they differ in their subcellular localizations in colon epithelial cells. To test the functional equivalence of Axin and Axin2 in vivo, we generated knockin mice in which the Axin gene was replaced with Myc-tagged Axin or Axin2 cDNA. Mice homozygous for the resulting alleles, AxinAx or AxinAx2, express no endogenous Axin but express either Myc-Axin or Myc-Axin2 under the control of the Axin locus. Both AxinAx/Ax and AxinAx2/Ax2 homozygotes are apparently normal and fertile, demonstrating that the Axin and Axin2 proteins are functionally equivalent.
Axin is a central component of the canonical Wnt signal transduction machinery, serving as a scaffold for the β-catenin destruction complex (23, 31, 36, 45, 52). Axin has specific binding sites for many proteins involved in Wnt signal transduction, including β-catenin, glycogen synthase kinase 3 (GSK3), CKI, adenomatous polyposis coli (APC), Dvl, LRP, and protein phosphatase 2A (Fig. 1A) (23, 31). Its key function in this pathway is to bring together β-catenin and the protein kinases CKI and GSK3, thus promoting the phosphorylation and consequent destruction of β-catenin. In the presence of a Wnt signal, this function is overcome, allowing β-catenin to accumulate and enter the nucleus (9, 13). The mechanism by which a Wnt signal leads to the inactivation of the Axin complex is not entirely clear, but it is thought to involve the binding of Axin to the Wnt coreceptor LRP as well as to Dvl (7, 34, 42, 44, 45). This results in the dephosphorylation of Axin, leading to a decrease in its affinity for β-catenin, and in a decrease in the level of Axin (20, 47, 48). Axin also enters the nucleus and appears to play a role in the nuclear-cytoplasmic shuttling of β-catenin (8, 46).
FIG.1.
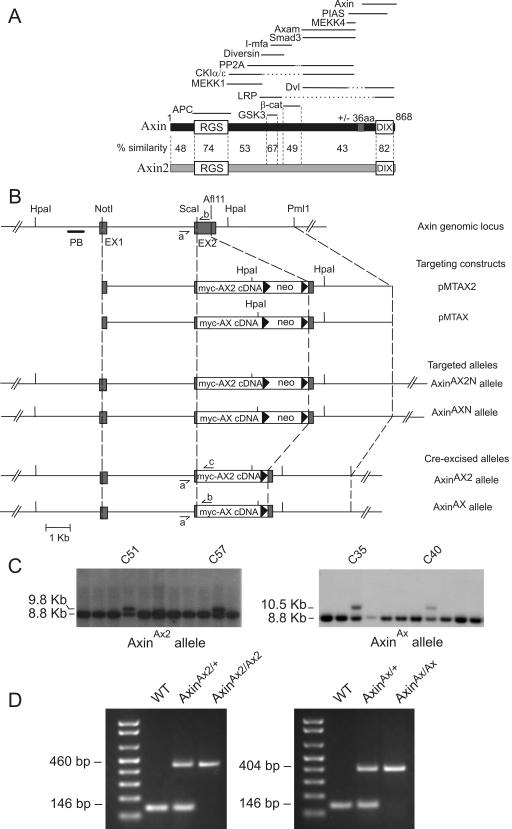
Targeted replacement of the Axin gene with myc-tagged Axin2 cDNA or myc-tagged Axin cDNA. (A) Schematic diagram of Axin and Axin2 proteins and binding partners. Percent similarities between the conserved RGS and DIX domains and the GSK3 and β-catenin (β-cat) binding regions of Axin and Axin2 are indicated, as are those of other less-conserved regions. The solid lines at the top indicate the regions of Axin involved in binding to the indicated proteins (31). Of these, only APC, GSK3, β-catenin, Diversin, and Smad3 are known to bind to Axin2. aa, amino acids; PP2A, protein phosphatase 2A. (B) Schematic diagram of the Axin genomic locus, targeting constructs, and targeted alleles. Exons 1 and 2 (EX1 and EX2) are depicted as grey boxes, and intron sequences are depicted as solid lines. The positions of the restriction enzyme sites and the probe PB are indicated. Small harpoons (⇁ and ↼) show the PCR primers AXL1 (a), MTAXR1 (b), and MTCONR1 (c). (C) Southern blot analysis of G418-resistant colonies after electroporation of ES cells with targeting constructs. Probing with probe PB following digestion of DNA with Hpa1 detected a band of 9.8 kb for the AxinAX2 allele, a band of 10.5 kb for the AxinAX allele, and a band of 8.8 kb for the wild-type allele. (D) Identification of homozygous, heterozygous, and wild-type (WT) AxinAX2 and AxinAX mice by PCR.
The major product of the Axin gene is a protein of 832 or 868 amino acids (depending on alternative splicing) containing two highly conserved domains (52): the RGS domain, which encompasses the binding site for APC (16, 18), and the DIX domain, a region of homology with Dvl proteins that is implicated in the binding of Axin to Dvl as well as in homodimerization (10, 17, 22, 24, 29, 40). (The open reading frame of Axin cDNA potentially encodes proteins of 956 and 992 amino acids, but proteins of this length have not been detected, and the major protein begins at codon Met-125 of the original sequence.) Mutant embryos lacking Axin die at embryonic day 9.5 (E9.5) with abnormalities including truncation of the forebrain, neural tube defects, and embryonic axis duplications (14, 35). Axin is expressed ubiquitously during embryogenesis, and the presence of axis duplications in its absence is thought to be a consequence of the abnormal accumulation of β-catenin, mimicking a Wnt signal, in the early embryo.
Axin2 (also known as Conductin) is 44% identical to Axin and shares the RGS and DIX domains (Fig. 1A) (5, 39, 49) as well as the binding sites for β-catenin, GSK3, Diversin, and Smad3 (Fig. 1A) (5, 11, 39, 49). While Axin2 has been studied less extensively than Axin, most data suggest that they are similar in function. Thus, when overexpressed in cultured cells, both proteins reduce the levels of β-catenin and the expression of Wnt target genes, and when expressed in frog embryos, both inhibit the development of dorsal structures (5, 19, 37, 49, 52). However, they are not fully redundant in vivo, as Axin2 is clearly unable to compensate for the absence of Axin in Axin-null embryos (52). Furthermore, humans with heterozygous germ line AXIN2 mutations have familial tooth agenesis (25), while mice homozygous for loss of Axin2 have skull abnormalities (51), indicating that Axin does not fully substitute for the lack of Axin2. In addition, deletions or mutations in AXIN or AXIN2 have been observed in a number of types of tumors, including colorectal cancer and hepatocellular carcinoma, indicating that each gene is a tumor suppressor and that neither gene can replace the other for this function (30, 38, 43).
One reason for the lack of full redundancy may be the different expression patterns of Axin and Axin2. While Axin mRNA is ubiquitous (52), Axin2 is expressed in tissue- and developmental-stage-specific patterns, due (at least in part) to its transcriptional upregulation by canonical Wnt signaling (4, 21, 27). Thus, Axin2 potentially constitutes a negative-feedback mechanism regulating the response to Wnt signals (21, 27, 32, 50). In the mouse embryo, the inability of Axin2 to compensate for the lack of Axin might be explained simply by the fact that Axin2 is not expressed in every cell. However, the two proteins might also differ significantly in function, a possibility supported by their different subcellular localizations in at least some cell types (3). If the functional differences were subtle, they might not be detected using assays that involve overexpression; since the normal level of Axin is extremely low (26), the abnormally high levels of expression in these assays could mask a functional difference.
To address the question of functional redundancy in a more physiological way, we generated knockin mice in which the Axin gene was replaced with an Axin2 cDNA sequence. This gene replacement strategy is similar to those used successfully to compare the functions of other paralogous gene pairs (6, 12, 15, 33, 41). The main advantage of this approach over in vitro assays or other in vivo functional assays (e.g., transgenic rescue) is that targeting of a cDNA to the appropriate genetic locus results in a normal pattern and level of expression. In animals homozygous for the resulting allele, AxinAx2, there will be no Axin, but Axin2 will be expressed ubiquitously from the Axin locus at a physiological level, as well as in its normal tissue-specific pattern from the unmodified Axin2 locus. Thus, the total level of Axin plus Axin2 expression in any cell type should be unchanged from that of the wild type. The ability of these mice to develop and survive normally thus tests the capacity of Axin2 to replace Axin in vivo.
MATERIALS AND METHODS
Generation of AxinAX2 and AxinAX mice.
Targeting vectors pMTAX2 and pMTAX were constructed for the knockin of myc-Axin2 cDNA and myc-Axin cDNA into the Axin locus. Using pBluescriptIISK (Stratagene) as the backbone, a series of fragments was subcloned into the polylinker. First, a 4.7-kb KpnI-ScaI fragment of the mouse Axin gene (containing intron 1 and parts of exons 1 and 2, including the exon 2 splice acceptor) was inserted into the KpnI site. Next, a 3.2-kb AfIII-PmlI fragment (containing the exon 2 splice donor and part of intron 2) was added to the XbaI site, and a floxed PGK-neo cassette was added to the adjacent SpeI site. Finally, mouse Axin and Axin2 cDNAs with N-terminal Myc tags were excised from the vector pCS2-MT (10) using ClaI and NotI, their ends were blunted, and the blunt-ended cDNAs were cloned into the ClaI site of pBluescriptIISK. Axin cDNA sequences started at codon Met125 of the published sequence (52), and Axin2 sequences started at Met1.
The targeting vectors were linearized with NotI and electroporated into CSL3 embryonic stem (ES) cells, which were selected with 0.35 mg/ml G418. DNA was digested with HpaI and screened by Southern blotting with a 479-bp probe, PB (Fig. 1B), generated by PCR of genomic DNA with primers 5′-CTTCTAATGGTATGAGGCTG-3′ and 5′-GCATCTGCACTTGCCATCTAC-3′. The targeting frequencies were 5/150 for pMTAX2 and 7/180 for pMTAX. ES cell clones were microinjected into C57BL/6J host blastocysts, and the PGK-neo cassette was excised by mating chimeric males to a β-actin-Cre transgenic line (28).
PCR genotyping.
The following primers were used for genotyping. AXL1 (5′-GGACCACCTTTCCTAATCCTTG-3′) and MTAXR1 (5′-AACCCTGCTCCTGGACATTC-3′) amplify the wild-type (146-bp) and the AxinAX (404-bp) alleles, while AXL1 in combination with MTCONR1 (5′-TGGGATCTGAAGGAGAGTCAC-3′) detects the AxinAX2 allele (460 bp) at an annealing temperature of 56.5°C.
Immunoblot analysis of embryonic tissue.
Embryos were dissected at E10.5 and homogenized as described previously (10). The anti-myc antibody (Ab-1) was from Calbiochem, and the two anti-Axin polyclonal antibodies were provided by David Virshup (antiserum DV, raised against full-length mouse Axin) and Francois Fagotto (antiserum FF, raised against amino acids 406 to 685 of mouse Axin).
RESULTS AND DISCUSSION
To generate mice in which Axin is replaced by Axin2, we inserted a mouse Axin2 cDNA in place of exon 2 of the Axin gene (Fig. 1B). The resulting AxinAx2 allele cannot encode Axin but should express Myc-tagged Axin2 from the Axin locus. As a control, we generated a second allele, AxinAx, in which Axin cDNA was inserted into the Axin locus. Although Axin normally encodes two isoforms that differ by the presence or absence of 36 amino acids (52), a cDNA can encode only a single isoform, and we used form 1 Axin, which lacks the 36 amino acids. There is only one known isoform of Axin2.
Correctly targeted ES cell lines were obtained (Fig. 1B and C) and used to generate germ line chimeric mice. Heterozygotes for both alleles appeared normal and were mated to produce homozygotes, which were identified by Southern blotting and PCR (Fig. 1). AxinAx2/Ax2 and AxinAx/Ax homozygotes were found in the expected proportions and also appeared normal. Both sexes were fertile and had an apparently normal life span. No premature deaths, obvious behavioral defects, or overt tumors were observed among approximately 50 AxinAx2 homozygotes over a period of 18 months. Histological analysis on a number of organs (liver, lungs, kidney, small and large intestines, stomach, heart, and spleen), including several where Wnt signaling is known to be important for development, revealed no abnormalities (data not shown). Of course, we cannot rule out the possibility that mice expressing only Axin2 have subtle defects yet to be detected.
As a more stringent test, we crossed AxinAx2 mice to those carrying the AxinTg1-null allele (52), thus generating AxinAx2/Tg1 compound heterozygotes, in which the level of Axin2 should be only half that in AxinAx2/Ax2 homozygotes. If Axin2 were less active than Axin at performing their shared functions, the AxinAx2/Tg1 compound heterozygotes might reveal a phenotypic defect not seen in AxinAx2/Ax2 homozygotes. However, these mice were indistinguishable from the AxinAx2/Ax2 or AxinAx/Ax homozygotes.
The only minor defect we detected, both in AxinAx2/Ax2 and AxinAx/Ax homozygotes, was a reduction of 11 to 14% in birth weight compared to that of wild-type littermates. AxinAx2/Ax2 mice were 11% smaller than wild-type mice at birth (P = 0.06), 12% smaller at 14 days (P = 0.06), and 12% smaller at 28 days (P = 0.03). AxinAx/Ax mice were 14% smaller than wild-type mice at birth (P = 0.001), 12% smaller at 14 days (P = 0.003), and 11% smaller at 28 days (P = 0.06). However, by 6 weeks of age, the difference was only 5 to 6% and was not statistically significant (P = 0.55 for AxinAx2/Ax2 mice and P = 0.32 for AxinAx/Ax mice). As the AxinAx2 and AxinAx alleles had the same effect, this does not reflect a functional difference between Axin and Axin2 but rather a property of both knockin alleles. One possibility was that these alleles might express too little or too much Axin/Axin2. To compare the amounts of protein encoded by the wild-type and AxinAx alleles, we performed immunoblotting with AxinAx/+ heterozygous embryos and an anti-Axin antiserum that detects both Myc-tagged and endogenous Axin. Myc-tagged Axin is larger than endogenous Axin, and the two bands are easily resolved (Fig. 2A). This analysis showed that the AxinAx allele encodes Myc-Axin at the same level as endogenous Axin, ruling out this explanation for the low birth weight. An alternative possibility is that the Myc tag, present in both AxinAx and AxinAx2 alleles, causes this transient defect.
FIG. 2.
Expression of Axin2 and Axin in AxinAX2 and AxinAX mice. (A) Protein lysates from wild-type (WT) and AxinAx/+ heterozygous embryos were probed with anti-Axin antibodies. The heterozygotes expressed equal amounts of Myc-tagged Axin from the AxinAX allele and wild-type Axin from the endogenous allele, showing that the AxinAX allele is expressed at a normal level. (B, C, and D) Protein lysates from wild-type, AxinAX/AX, and AxinAX2/AX2 homozygous embryos were probed with anti-Myc antibody (B) or with two different anti-Axin antisera (C and D). In wild-type embryos, the anti-Axin antibodies detected endogenous Axin (∼110 kDa), while the anti-Myc antibody detected only background bands. In AxinAX/AX embryos, both the anti-Axin and the anti-Myc antibodies detected Myc-tagged Axin (∼130 kDa), while endogenous Axin was absent. In AxinAX2/AX2 embryos, the anti-Axin antibodies detected only background bands also seen in wild-type embryos, while the anti-Myc antibody detected Myc-tagged Axin2. Anti-Axin antiserum FF, a gift from François Fagotto, was raised against amino acids 406 to 685 of mouse Axin, while anti-Axin antiserum DV, a gift from David Virshup, was raised against full-length mouse Axin. Molecular size markers (in kilodaltons) are noted at the left of blots.
Although our targeting strategy precluded the expression of any functional Axin from the AxinAx2 locus (since the initiation codon and exon 2, which encodes the essential RGS domain, were deleted), we wanted to test this at the protein level. We therefore performed immunoblotting with anti-myc and anti-Axin antisera on extracts of AxinAx2 embryos (Fig. 2B to D). This confirmed that the AxinAx2/Ax2 homozygotes expressed Myc-tagged Axin2 and lacked endogenous Axin.
Our results show that Axin2, when expressed under the control of Axin regulatory sequences, can carry out all the essential functions of Axin during development as well as in adult life. If Axin and Axin2 are functionally identical, why have the two genes been conserved during evolution? One explanation is that the total level of Axin-like protein (Axin plus Axin2) needs to be elevated in certain cells; Axin provides a basal level in all cells (52), while Axin2, which is induced by Wnt/β-catenin signaling, is regulated to provide elevated levels where needed (4, 21, 27, 32, 50). Additional insight into this question derives from the analysis of a null-mutant allele (lacZ insertion) of Axin2 (32). Mice lacking Axin2 are viable and fertile but display skull malformations resembling craniosynostosis in humans, due to the premature fusion of cranial sutures (51). Thus, in the presence of a normal complement of Axin alleles, Axin2 is dispensable for most developmental processes, although it is important for postnatal skull development. To further examine the relationship between Axin and Axin2, mice with various combinations of AxinTg1- and Axin2lacZ-null alleles were examined (B. Jerchow and W. Birchmeier, personal communication). Double homozygotes lacking both Axin and Axin2 died much earlier (by E6.5) than those lacking only Axin, indicating that endogenous Axin2 can partially compensate for the absence of Axin in early embryogenesis. Furthermore, while mice with only one wild-type Axin allele (AxinTg1/+) are normal (52), the further removal of Axin2 (AxinTg1/+ and Axin2lacZ/lacZ) resulted in severe brain and craniofacial abnormalities at birth. Thus, one Axin allele is sufficient in the presence of Axin2 but not in its absence. Overall, these findings argue that when Axin is absent or reduced, Axin2 can partially compensate for its developmental functions. Axin2 does not fully compensate because it is not ubiquitously expressed. However, in the AxinAx2 allele, where the pattern of Axin2 expression is altered to resemble that of Axin, it can replace the functions of Axin.
The functional equivalence of the Axin and Axin2 proteins during mouse development implies that any amino acids that are not conserved between the two proteins are not required for their function. Similarly, any interactions with other proteins that are not shared by Axin and Axin2 are unlikely to be important. Axin interacts directly with at least 17 other proteins (Fig. 1A) (31), but only a small subset of these has been tested for interaction with Axin2. We suggest that one way to evaluate the importance of these multiple protein-protein interactions for the functions of Axin would be to test if they interact similarly with Axin2; those that fail are unlikely to be required for the developmental functions of Axin.
Our results also call into question the significance of the differences in subcellular locations observed for Axin and Axin2. Anderson et al. (3) previously reported that in normal colon epithelial cells, Axin was found in several locations—diffusely in the nucleus, along cell membranes, and often in the cytoplasm—while Axin2 was uniformly expressed in the nucleus. In adenomatous polyps, Axin was strongly cytoplasmic while Axin2 remained nuclear. Given our results, it is unlikely that these differences in localization reflect important differences in the functions of the two proteins. While other studies have shown previously that Axin shuttles between the nucleus and the cytoplasm (8, 46), it is very unlikely that all of the functions of Axin and Axin2 can be carried out in the nucleus. Therefore, the apparent nuclear localization of Axin2 probably does not reflect its major site of action. Perhaps a low level of the protein in the cytoplasm or at the cell surface, below the level of detection by immunostaining, is sufficient to carry out its essential cytoplasmic functions. The equivalence of Axin and Axin2 proteins, despite their different subcellular localizations, is reminiscent of the finding that APC1 and APC2 are functionally redundant in the fly, although these proteins also display different intracellular locations (1, 2).
The insertion of form 1 Axin cDNA into the Axin locus (in the AxinAx allele), while primarily intended as a control for the AxinAx2 allele, provided some useful information. First, it showed that the long isoform of Axin, form 2, is not required for normal development. On the other hand, the transient growth defects observed in both AxinAx and AxinAx2 homozygotes might be due to the lack of form 2 Axin or to the absence of the upstream Axin coding sequences. Second, the normal development of AxinAx/Ax homozygotes suggests that this targeting strategy can be used to efficiently generate new mutant alleles of Axin, in which specific domains or amino acids are altered. This approach is currently being used to examine the importance in vivo of several conserved sequences believed to play important roles in the functions of Axin.
Acknowledgments
We thank Zaiqi Wu for excellent technical assistance, Victor Lin for the CSL3 ES cell line, and Francois Fagotto and David Virshup for the anti-Axin antisera.
This work was supported by grant HD-44265 from the NIH to F.C.
REFERENCES
- 1.Ahmed, Y., A. Nouri, and E. Wieschaus. 2002. Drosophila Apc1 and Apc2 regulate Wingless transduction throughout development. Development 129:1751-1762. [DOI] [PubMed] [Google Scholar]
- 2.Akong, K., B. M. McCartney, and M. Peifer. 2002. Drosophila APC2 and APC1 have overlapping roles in the larval brain despite their distinct intracellular localizations. Dev. Biol. 250:71-90. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, C. B., K. L. Neufeld, and R. L. White. 2002. Subcellular distribution of Wnt pathway proteins in normal and neoplastic colon. Proc. Natl. Acad. Sci. USA 99:8683-8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aulehla, A., C. Wehrle, B. Brand-Saberi, R. Kemler, A. Gossler, B. Kanzler, and B. G. Herrmann. 2003. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4:395-406. [DOI] [PubMed] [Google Scholar]
- 5.Behrens, J., B. A. Jerchow, M. Wurtele, J. Grimm, C. Asbrand, R. Wirtz, M. Kuhl, D. Wedlich, and W. Birchmeier. 1998. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science 280:596-599. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard, M., P. Pfeffer, and M. Busslinger. 2000. Functional equivalence of the transcription factors Pax2 and Pax5 in mouse development. Development 127:3703-3713. [DOI] [PubMed] [Google Scholar]
- 7.Cliffe, A., F. Hamada, and M. Bienz. 2003. A role of Dishevelled in relocating Axin to the plasma membrane during wingless signaling. Curr. Biol. 13:960-966. [DOI] [PubMed] [Google Scholar]
- 8.Cong, F., and H. Varmus. 2004. Nuclear-cytoplasmic shuttling of Axin regulates subcellular localization of beta-catenin. Proc. Natl. Acad. Sci. USA 101:2882-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eastman, Q., and R. Grosschedl. 1999. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr. Opin. Cell Biol. 11:233-240. [DOI] [PubMed] [Google Scholar]
- 10.Fagotto, F., E. Jho, L. Zeng, T. Kurth, T. Joos, C. Kaufmann, and F. Costantini. 1999. Domains of Axin involved in protein-protein interactions, Wnt pathway inhibition, and intracellular localization. J. Cell Biol. 145:741-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuhashi, M., K. Yagi, H. Yamamoto, Y. Furukawa, S. Shimada, Y. Nakamura, A. Kikuchi, K. Miyazono, and M. Kato. 2001. Axin facilitates Smad3 activation in the transforming growth factor β signaling pathway. Mol. Cell. Biol. 21:5132-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng, Y., W. Whoriskey, M. Y. Park, R. T. Bronson, R. H. Medema, T. Li, R. A. Weinberg, and P. Sicinski. 1999. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell 97:767-777. [DOI] [PubMed] [Google Scholar]
- 13.Giles, R. H., J. H. van Es, and H. Clevers. 2003. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta 1653:1-24. [DOI] [PubMed] [Google Scholar]
- 14.Gluecksohn-Schoenheimer, S. 1949. The effects of a lethal mutation responsible for duplications and twinning in mouse embryos. J. Exp. Zool. 110:47-76. [DOI] [PubMed] [Google Scholar]
- 15.Hanks, M., W. Wurst, L. Anson-Cartwright, A. B. Auerbach, and A. L. Joyner. 1995. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science 269:679-682. [DOI] [PubMed] [Google Scholar]
- 16.Hart, M. J., R. de los Santos, I. N. Albert, B. Rubinfeld, and P. Polakis. 1998. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr. Biol. 8:573-581. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, W., L. Zeng, and F. Costantini. 1999. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J. Biol. Chem. 274:3439-3445. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda, S., S. Kishida, H. Yamamoto, H. Murai, S. Koyama, and A. Kikuchi. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 17:1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh, K., V. E. Krupnik, and S. Y. Sokol. 1998. Axis determination in Xenopus involves biochemical interactions of axin, glycogen synthase kinase 3 and beta-catenin. Curr. Biol. 8:591-594. [DOI] [PubMed] [Google Scholar]
- 20.Jho, E., S. Lomvardas, and F. Costantini. 1999. A GSK3beta phosphorylation site in axin modulates interaction with beta-catenin and Tcf-mediated gene expression. Biochem. Biophys. Res. Commun. 266:28-35. [DOI] [PubMed] [Google Scholar]
- 21.Jho, E., T. Zhang, C. Domon, C.-K. Joo, J.-N. Freund, and F. Costantini. 2002. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22:1172-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julius, M. A., B. Schelbert, W. Hsu, E. Fitzpatrick, E. Jho, F. Fagotto, F. Costantini, and J. Kitajewski. 2000. Domains of Axin and Disheveled required for interaction and function in Wnt signaling. Biochem. Biophys. Res. Commun. 276:1162-1169. [DOI] [PubMed] [Google Scholar]
- 23.Kikuchi, A. 1999. Modulation of Wnt signaling by Axin and Axil. Cytokine Growth Factor Rev. 10:255-265. [DOI] [PubMed] [Google Scholar]
- 24.Kishida, S., H. Yamamoto, S.-I. Hino, S. Ikeda, M. Kishida, and A. Kikuchi. 1999. DIX domains of Dvl and Axin are necessary for protein interactions and their ability to regulate β-catenin stability. Mol. Cell. Biol. 19:4414-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lammi, L., S. Arte, M. Somer, H. Jarvinen, P. Lahermo, I. Thesleff, S. Pirinen, and P. Nieminen. 2004. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am. J. Hum. Genet. 74:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, E., A. Salic, R. Kruger, R. Heinrich, and M. W. Kirschner. 2003. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 1:E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung, J. Y., F. T. Kolligs, R. Wu, Y. Zhai, R. Kuick, S. Hanash, K. R. Cho, and E. R. Fearon. 2002. Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J. Biol. Chem. 277:21657-21665. [DOI] [PubMed] [Google Scholar]
- 28.Lewandoski, M., E. N. Meyers, and G. R. Martin. 1997. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harbor Symp. Quant. Biol. 62:159-168. [PubMed] [Google Scholar]
- 29.Li, L., H. Yuan, C. D. Weaver, J. Mao, G. H. Farr III, D. J. Sussman, J. Jonkers, D. Kimelman, and D. Wu. 1999. Axin and Frat1 interact with Dvl and GSK, bridging Dvl to GSK in Wnt-mediated regulation of LEF-1. EMBO J. 18:4233-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, W., X. Dong, M. Mai, R. S. Seelan, K. Taniguchi, K. K. Krishnadath, K. C. Halling, J. M. Cunningham, C. Qian, E. Christensen, P. C. Roche, D. I. Smith, and S. N. Thibodeau. 2000. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat. Genet. 26:146-147. [DOI] [PubMed] [Google Scholar]
- 31.Luo, W., and S. C. Lin. 2004. Axin: a master scaffold for multiple signaling pathways. Neurosignals 13:99-113. [DOI] [PubMed] [Google Scholar]
- 32.Lustig, B., B. Jerchow, M. Sachs, S. Weiler, T. Pietsch, U. Karsten, M. van de Wetering, H. Clevers, P. M. Schlag, W. Birchmeier, and J. Behrens. 2002. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 22:1184-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malynn, B. A., I. M. de Alboran, R. C. O'Hagan, R. Bronson, L. Davidson, R. A. DePinho, and F. W. Alt. 2000. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 14:1390-1399. [PMC free article] [PubMed] [Google Scholar]
- 34.Mao, J., J. Wang, B. Liu, W. Pan, G. H. Farr III, C. Flynn, H. Yuan, S. Takada, D. Kimelman, L. Li, and D. Wu. 2001. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol. Cell 7:801-809. [DOI] [PubMed] [Google Scholar]
- 35.Perry, W. L. I., T. J. Vasicek, J. J. Lee, J. M. Rossi, L. Zeng, T. Zhang, S. M. Tilghman, and F. Costantini. 1995. Phenotypic and molecular analysis of a transgenic insertional allele of the mouse Fused locus. Genetics 141:321-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polakis, P. 2000. Wnt signaling and cancer. Genes Dev. 14:1837-1851. [PubMed] [Google Scholar]
- 37.Sakanaka, C., J. B. Weiss, and L. T. Williams. 1998. Bridging of beta-catenin and glycogen synthase kinase-3beta by Axin and inhibition of beta-catenin-mediated transcription. Proc. Natl. Acad. Sci. USA 95:3020-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh, S., Y. Daigo, Y. Furukawa, T. Kato, N. Miwa, T. Nishiwaki, T. Kawasoe, H. Ishiguro, M. Fujita, T. Tokino, Y. Sasaki, S. Imaoka, M. Murata, T. Shimano, Y. Yamaoka, and Y. Nakamura. 2000. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat. Genet. 24:245-250. [DOI] [PubMed] [Google Scholar]
- 39.Schwarz-Romond, T., C. Asbrand, J. Bakkers, M. Kühl, H.-J. Schaeffer, J. Huelsken, J. Behrens, M. Hammerschmidt, and W. Birchmeier. 2002. The ankyrin repeat protein Diversin recruits Casein kinase Iɛ to the β-catenin degradation complex and acts in both canonical Wnt and Wnt/JNK signaling. Genes Dev. 16:2073-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smalley, M. J., E. Sara, H. Paterson, S. Naylor, D. Cook, H. Jayatilake, L. G. Fryer, L. Hutchinson, M. J. Fry, and T. C. Dale. 1999. Interaction of Axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF-dependent transcription. EMBO J. 18:2823-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suda, Y., J. Nakabayashi, I. Matsuo, and S. Aizawa. 1999. Functional equivalency between Otx2 and Otx1 in development of the rostral head. Development 126:743-757. [DOI] [PubMed] [Google Scholar]
- 42.Tamai, K., X. Zeng, C. Liu, X. Zhang, Y. Harada, Z. Chang, and X. He. 2004. A mechanism for Wnt coreceptor activation. Mol. Cell 13:149-156. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi, K., L. R. Roberts, I. N. Aderca, X. Dong, C. Qian, L. M. Murphy, D. M. Nagorney, L. J. Burgart, P. C. Roche, D. I. Smith, J. A. Ross, and W. Liu. 2002. Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene 21:4863-4871. [DOI] [PubMed] [Google Scholar]
- 44.Tolwinski, N. S., M. Wehrli, A. Rives, N. Erdeniz, S. DiNardo, and E. Wieschaus. 2003. Wg/Wnt signal can be transmitted through arrow/LRP5,6 and Axin independently of Zw3/Gsk3beta activity. Dev. Cell 4:407-418. [DOI] [PubMed] [Google Scholar]
- 45.Tolwinski, N. S., and E. Wieschaus. 2004. Rethinking WNT signaling. Trends Genet. 20:177-181. [DOI] [PubMed] [Google Scholar]
- 46.Wiechens, N., K. Heinle, L. Englmeier, A. Schohl, and F. Fagotto. 2004. Nucleo-cytoplasmic shuttling of Axin, a negative regulator of the Wnt-beta-catenin pathway. J. Biol. Chem. 279:5263-5267. [DOI] [PubMed] [Google Scholar]
- 47.Willert, K., S. Shibamoto, and R. Nusse. 1999. Wnt-induced dephosphorylation of Axin releases beta-catenin from the Axin complex. Genes Dev. 13:1768-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto, H., S. Kishida, M. Kishida, S. Ikeda, S. Takada, and A. Kikuchi. 1999. Phosphorylation of Axin, a Wnt signal negative regulator, by glycogen synthase kinase-3beta regulates its stability. J. Biol. Chem. 274:10681-10684. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto, H., S. Kishida, T. Uochi, S. Ikeda, S. Koyama, M. Asashima, and A. Kikuchi. 1998. Axil, a member of the Axin family, interacts with both glycogen synthase kinase 3β and β-catenin and inhibits axis formation of Xenopus embryos. Mol. Cell. Biol. 18:2867-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan, D., M. Wiesmann, M. Rohan, V. Chan, A. B. Jefferson, L. Guo, D. Sakamoto, R. H. Caothien, J. H. Fuller, C. Reinhard, P. D. Garcia, F. M. Randazzo, J. Escobedo, W. J. Fantl, and L. T. Williams. 2001. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/β-catenin signaling is activated in human colon tumors. Proc. Natl. Acad. Sci. USA 98:14973-14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, H.-M. I., B. Jerchow, T.-J. Sheu, B. Liu, F. Costantini, J. E. Puzas, W. Birchmeier, and W. Hsu. 2005. The role of Axin2 in calvarial morphogenesis and craniosynostosis. Development 132:1995-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng, L., F. Fagotto, T. Zhang, W. Hsu, T. J. Vasicek, W. L. I. Perry, J. J. Lee, S. M. Tilghman, B. M. Gumbiner, and F. Costantini. 1997. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 90:181-192. [DOI] [PubMed] [Google Scholar]