Abstract
Telomerase extends chromosome ends by the synthesis of tandem simple-sequence repeats. Studies of minimal recombinant telomerase ribonucleoprotein (RNP) reconstituted in vitro have revealed sequences within the telomerase RNA subunit (TER) that are required to establish its internal template and other unique features of enzyme activity. Here we test the significance of these motifs following TER assembly into telomerase holoenzyme in vivo. We established a method for stable expression of epitope-tagged TER and TER variants in place of wild-type Tetrahymena TER. We found that sequence substitutions in nontemplate regions of TER altered telomere length maintenance in vivo, with an increase or decrease in the set point for telomere length homeostasis. We also characterized the in vitro activity of the telomerase holoenzymes reconstituted with TER variants, following RNA-based RNP affinity purification from cell extracts. We found that nontemplate sequence substitutions imposed specific defects in the fidelity and processivity of template use. These findings demonstrate nontemplate functions of TER that are critical for the telomerase holoenzyme catalytic cycle and for proper telomere length maintenance in vivo.
Telomeres protect authentic chromosome termini from aberrant resection or rearrangement. In most eukaryotes, a stable telomere is composed of a tandem array of simple-sequence repeats assembled into specialized chromatin (37). The overall length of the telomeric simple-sequence repeat tract is maintained in a dynamic balance of sequence loss and addition. To compensate for the inevitable loss from incomplete replication by DNA-dependent DNA polymerases, telomeres recruit the telomerase ribonucleoprotein (RNP) reverse transcriptase in coordination with other DNA replication factors (18). Telomerase copies a defined sequence within its integral RNA subunit (TER) to extend chromosome 3′ ends. Sequence substitutions within the TER template can rapidly impose defects in cell division and viability due to the synthesis of altered repeats (6).
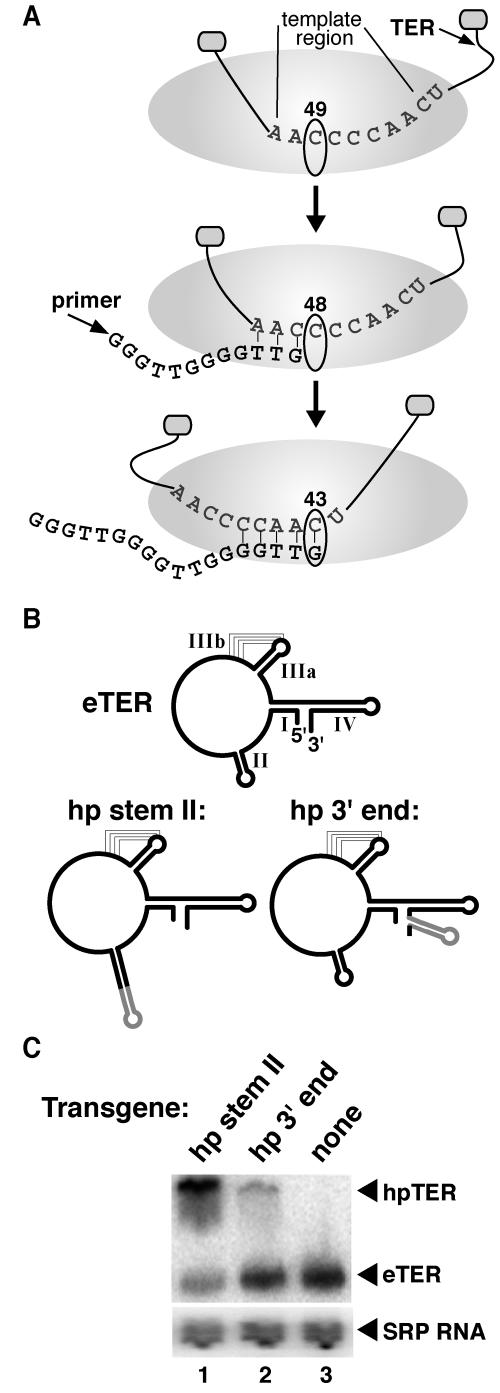
During a catalytic cycle of repeat synthesis, the telomerase active site must proceed through a remarkable number of distinct states of template positioning and pairing (Fig. 1A). Substrates unable to base-pair with the template are extended in alignment with the template 3′ end at its “default” position, defined as the position of template in the absence of a template-primer hybrid (41). For Tetrahymena thermophila TER, default template positioning places C49 in the active site (Fig. 1A, top). As deoxynucleoside triphosphates (dNTPs) are added, a template-product hybrid forms, and successively more 5′ positions of unpaired template must thread into the active site (Fig. 1A, middle). Product synthesis halts at the template 5′ boundary, corresponding to position C43 of T. thermophila TER (Fig. 1A, bottom). Upon eventual dissociation of the template-product hybrid, the liberated template repositions to recover its default placement. Product either dissociates from the enzyme or repositions at the template start for processive repeat addition.
FIG. 1.
Structure and expression of TER. (A) Selected stages of the telomerase catalytic cycle. Internal template boundaries are influenced by TERT-TER interactions that restrict template motion, illustrated here as small ovals. The default positioning of template may favor placement of TER position C49 in the active site (top). With hybrid formation (middle), the template can be pulled through the active site and used to direct dNTP addition until the 5′ boundary is reached at position C43 (bottom). (B) Schematic of eTER and hpTERs. Predicted eTER secondary structure elements are indicated by roman numerals. (C) Total cellular RNA was analyzed by Northern blot hybridization using oligonucleotides complementary to TER and SRP RNA.
Two telomerase holoenzyme subunits have direct roles in catalysis: TER and the telomerase reverse transcriptase protein (TERT). TER domain structure has diverged between ciliates, vertebrates, and yeasts, with the possible exception of a template-adjacent pseudoknot (3, 8). TERT harbors a set of active site motifs shared by all reverse transcriptases and an N-terminal extension conserved only among TERTs (21). The TERT N-terminal extension contains motifs necessary for high-affinity interaction with TER and for recruitment to telomere substrates. Recent studies have exploited rabbit reticulocyte lysate (RRL) for the reconstitution of a minimal Tetrahymena or human telomerase RNP from TERT and TER alone. Using this in vitro system, motifs that influence specific features of catalytic activity have been defined for Tetrahymena TER (23-25, 28, 30, 32, 38) and for human TER as well (7, 9).
Telomerase holoenzyme assembly in vivo could influence TER motif functions previously defined using the minimal RNP assembled in vitro. The holoenzyme contains many subunits in addition to TERT and TER, including an RNA binding protein(s) that alters TER folding (42, 35). Nontemplate substitutions within yeast TERs reconstituted in vivo can alter telomerase catalytic properties, with associated defects in telomere length maintenance (reference 40 and references therein). Here, we investigate nontemplate TER motif functions in the Tetrahymena telomerase holoenzyme reconstituted in vivo. We examined TER motif functions in the holoenzyme catalytic cycle using direct primer elongation activity assays, combined with an RNA-based RNP affinity purification strategy to enrich telomerase holoenzyme from cell extracts. We also determined nontemplate TER motif requirements for telomerase function on its telomere substrates. We find that TER sequence substitutions affect the equilibrium set point for telomere length maintenance in vivo and induce specific changes in the catalytic activity of telomerase holoenzyme in vitro. Our findings demonstrate that nontemplate motifs of Tetrahymena TER have specific roles in telomerase holoenzyme function.
MATERIALS AND METHODS
Cell growth and transformation.
T. thermophila was grown in 2% Proteose Peptone, 0.2% yeast extract, and 10 μM FeCl3 with 250 μg/ml ampicillin and streptomycin and 1.25 μg/ml Fungizone (amphotericin B). All transformations were performed on starved cells by particle bombardment. Transgene integration was selected by disruption of the BTU1 locus. In strain CU522, the β-tubulin encoded at BTU1 confers growth sensitivity to taxol. Because β-tubulin is also encoded by a second gene, disruption of BTU1 is not lethal and can be used as a positive selection for transgene integration (14). Cells were transformed, allowed to recover for 2 to 5 h at 30°C, and then selected in 20 to 30 μM paclitaxel (Sigma). Cells were passaged to progressively higher paclitaxel concentrations for 2 to 3 days until they were growing in 50 μM paclitaxel, after which they were passaged daily in 50 μM paclitaxel for 5 days. Following complete replacement of wild-type chromosomes with the transgene chromosome, transgene expression strains were maintained without selection (42). For disruption of TER, cells were transformed with the same gene disruption vector conferring resistance to paromomycin used in a previous study (33). Cells were brought to maximal selective pressure by culturing in increasing concentrations of paromomycin, maintained at maximal selective pressure for 16 days (∼100 doublings), and then cultured for 8 days (∼50 doublings) in the absence of selection. Growth in the absence of selection provides a test for whether all of the wild-type chromosomes are lost from the macronucleus.
Transformation constructs.
We tagged TER by introduction of a binding site for bacteriophage MS2 coat protein. An optimized version (13) of the hairpin tag (CGUACACCAUCAGGGUACG) was inserted using PCR. The stem II tag location replaced position 28; the 3′ tag location was after position 155. Recombinant hairpin-tagged TER (hpTER) was expressed from a transgene containing TER sequence and 150 bp upstream, including the TER promoter (17). Transcription termination was enforced with a T7 tract in the transcript strand. Transgene expression cassettes were subcloned between the NcoI and BamHI sites of pBICH3 (a gift from Jacek Gaertig, University of Georgia, Athens) to place them between BTU1 5′ and 3′ flanking genomic sequences. TER variants were produced by site-specific mutagenesis. The TER disruption construct has been described elsewhere (33).
Recombinant coat protein.
For affinity purification of telomerase RNPs with tagged TER, we supplemented Tetrahymena extracts with purified, bacterially expressed MS2 coat protein. We used the variant V75E/A81G to reduce the aggregation of coat protein dimers into capsids (27). A construct encoding the MS2 coat protein with V75E and A81G substitutions (a gift from Jamie Williamson, The Scripps Research Institute) was amplified by PCR to append an NdeI site at the 5′ end and a BamHI site at the 3′ end. The resulting fragment was subcloned into pET28 (Novagen) digested with NdeI and BamHI to yield p28MS2. The N-terminal tandem affinity purification (TAP) tag was amplified by PCR to append an NdeI site at the 5′ and 3′ ends. The restriction-digested PCR fragment was cloned into p28MS2 digested with NdeI to yield pET28TAP-MS2. TAP-MS2 coat protein was expressed in BL21(DE3) cells and purified by chromatography on SP-Sepharose (Amersham Pharmacia). Cells were sonicated in buffer containing 20 mM Tris-HCl (pH 8.0), 1 mM MgCl2, and 10% glycerol. Before centrifugation to clear the extract, buffer was adjusted to 60 mM Tris-HCl (pH 7.0), 50 mM NaCl, 1 mM MgCl2, 10% glycerol, and 0.1% NP-40. Chromatography was performed in the same buffer with elution using 0.5 M NaCl.
RNA-based RNP affinity purification.
Extracts were prepared from cells grown at 30°C to mid-log phase (2 × 105 to 4 × 105 cells/ml), harvested, and then resuspended shaking at 30°C in 10 mM Tris-HCl (pH 7.5) for 12 to 17 h. Cells were lysed in T2MG pH 8.5 (20 mM Tris-HCl [pH 8.5], 1 mM MgCl2, 10% glycerol) with protease inhibitors and 15 mM β-mercaptoethanol by addition of 0.2% NP-40. Extract was cleared by centrifugation at 130,000 × g for 1 h. For affinity purification, extracts were adjusted to 50 mM NaCl and incubated with TAP-MS2 coat protein for 1 h at 4°C. Immunoglobulin G (IgG)-agarose (Sigma) was added followed by another 1-h incubation at 4°C. Beads were washed three times in WB (20 mM Tris-HCl [pH 8.0], 2 mM MgCl2, 0.1 M NaCl, 0.1% NP-40, 10% glycerol, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride) and twice in TEV protease elution buffer (10 mM Tris-HCl [pH 8.0], 0.1 M NaCl, 0.1% NP-40, 1 mM dithiothreitol). Elution was accomplished by addition of recombinant TEV protease at room temperature.
RNA, DNA, and activity assays.
RNA was prepared by guanidine thiocyanate-phenol-chloroform extraction. Northern blots were probed with 5′ end-labeled DNA oligonucleotides complementary to the intended RNA target. Genomic DNA isolation was performed as previously described (1). Southern blot assays to assess integration events were performed with random hexamer-primed probes from the genomic region immediately 5′ of the integrated transgene. Southern blots for telomere length analysis were probed with 5′-end-labeled DNA oligonucleotide as previously described (33), using genomic DNA prepared from strains maintained at room temperature and then grown for the same number of doublings at 30°C. Telomerase activity assays were performed in reaction buffer with 50 mM Tris-acetate (pH 8.0), 10 mM spermidine, 5 mM β-mercaptoethanol, 2 mM MgCl2, a 400 μM concentration of each unlabeled nucleotide (dTTP, dATP, and/or ddTTP), 0.3 μM [32P]dGTP (800 Ci/mmol), and 50 nM of primer unless indicated otherwise. Radiolabeled dATP was used at the same specific activity as radiolabeled dGTP (800 Ci/mmol). Purified RNPs that were not eluted from IgG-agarose were assayed after resuspension in T2MG pH 8.5 to make up half of the reaction volume. Reactions were incubated for approximately 45 min at 30°C unless indicated otherwise. Radiolabeled product DNA was analyzed by denaturing gel electrophoresis.
RESULTS
Construction of strains expressing hairpin-tagged TER.
We first developed a method to reconstitute sequence variants of Tetrahymena TER into endogenously assembled telomerase holoenzyme. To facilitate detection and purification of recombinant telomerase RNPs, we tagged TER by introduction of a 19-nucleotide hairpin that serves as a binding site for bacteriophage MS2 coat protein. Affinity chromatography with recombinant coat protein has been used to recover similarly tagged RNAs assembled into RNP in cell extract (13, 20) or Escherichia coli cells (43). The hairpin was inserted in TER at the distal end of stem II or near the 3′ end (Fig. 1B). Stem II was chosen as the internal tag placement site because deletion or interruption of the distal end of the stem has no impact on activity in vitro (28, 32). The 3′ tag was placed immediately preceding the poly-uridine tract added in transcription termination by RNA polymerase III. Transgenes encoding a hpTER were stably integrated into the macronuclear genome by disruption of the nonessential BTU1 locus. In Tetrahymena sp. strain CU522, BTU1 encodes a β-tubulin that slows cell growth in medium supplemented with taxol, and so disruption of BTU1 can be used as a positive selection for transgene integration (14). After complete replacement of BTU1 chromosomes with transgene chromosomes, strains were subsequently grown in medium lacking taxol. Southern blotting confirmed the stable maintenance of the transgene chromosome (data not shown). No difference in growth rate comparing parental and transgene strains was observed.
The hpTER transgene strains should transcribe TER from the transgene and from the endogenous gene locus. Previous studies using plasmid vectors to overexpress template variants of Tetrahymena TER demonstrated that recombinant TER must compete with endogenous TER (eTER) for accumulation (44). This competition is likely to arise from the limiting expression of Tetrahymena p65, a telomerase holoenzyme protein required for TER stability (42). To investigate the relative accumulation of hpTER and eTER in the coexpression strains, we performed Northern blot hybridization. Probing with an oligonucleotide fully complementary to eTER and both hpTERs revealed that either hpTER could accumulate (Fig. 1C, top). The stem II-tagged hpTER was more abundant than eTER in total RNA samples, potentially due to a slightly elevated level of TER transcription from the transgene context. The 3′-tagged hpTER was less abundant than eTER, revealing a disadvantage in RNP assembly or stability. As expected, total TER accumulation remained constant relative to the signal recognition particle (SRP) RNA loading control (Fig. 1C, bottom). Based on these results, subsequent studies utilized the stem II hpTER backbone.
Functional complementation of eTER by the hpTER transgene.
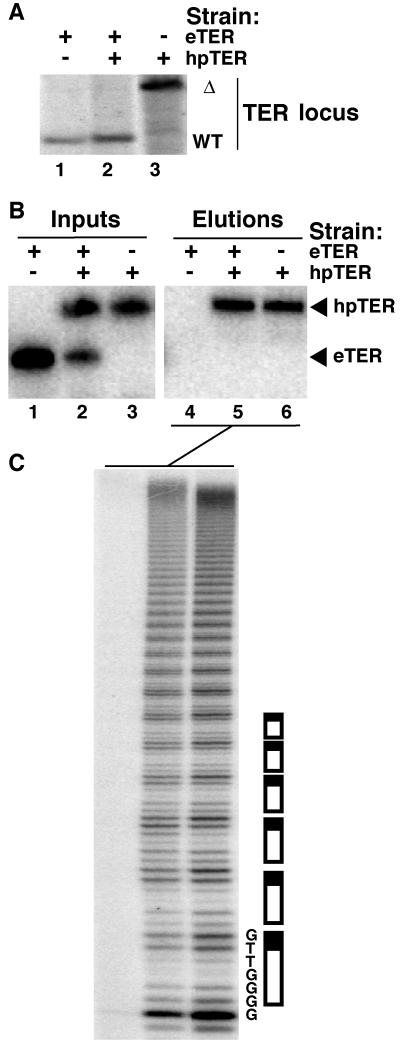
The TER locus is essential in Tetrahymena (33). We performed TER gene disruption in the hpTER transgene strain to determine whether hpTER could fulfill the essential TER function(s). An expression cassette conferring paromomycin resistance was integrated into the TER locus to perform gene disruption, as described previously (33). Cells were grown in medium supplemented with paromomycin to maximize the replacement of TER chromosomes by gene disruption chromosomes bearing the selectable marker. In cells expressing the hpTER transgene, the TER gene disruption cassette could substitute the macronuclear copies of the wild-type TER locus to completion (Fig. 2A and additional data not shown). This outcome demonstrates that hpTER fulfills the essential functions of eTER.
FIG. 2.
Function of hpTER. (A) Genomic DNA was digested with HindIII and analyzed by Southern blotting from the parental strain (lane 1), the hpTER strain coexpressing eTER (lane 2), or the hpTER strain with TER gene disruption (lane 3). The hybridization probe had identical homology with endogenous and disrupted TER loci, which were distinguishable by restriction fragment length as indicated. (B and C) Cell extracts normalized for total protein concentration (inputs) were used to purify hpTER-tagged telomerase RNP (elutions). For TER analysis by Northern blot hybridization in panel B, inputs represent ∼5% of the total amount of cell extract used for binding. Aliquots of the elution mixtures were assayed for activity in panel C with the primer (G4T2)3 in reaction mixtures with 0.6 μM [32P]dGTP. The first seven dNTP additions are indicated along with a schematic of sequential repeat additions.
RNA-based telomerase RNP affinity purification.
To accurately study the in vitro properties of endogenously assembled telomerase, partial purification is needed to remove nonspecific inhibitors present in crude cell extract. We supplemented cell extract with purified, bacterially expressed MS2 coat protein fused to an N-terminal TAP tag (36). Coat protein was allowed to bind hpTER in extract followed by recovery on IgG-agarose using the protein A domains of the TAP tag. Bound RNP was eluted by protease cleavage at a site positioned between the coat protein and protein A domains. We examined the specificity of affinity purification using extracts from strains expressing eTER, eTER plus hpTER, or hpTER alone (Fig. 2B). Both forms of TER could be detected in the input extracts (lanes 1 to 3), but only hpTER was recovered by affinity purification (lanes 4 to 6). We assayed the purified samples for direct primer extension activity using radiolabeled dGTP and dTTP (Fig. 2C). The enrichment of telomerase catalytic activity paralleled the recovery of hpTER. No background binding of hpTER or telomerase activity to IgG-agarose was detected in mock purifications from extracts without added coat protein (data not shown). Notably, even when eTER and hpTER were coexpressed, only hpTER was recovered in the affinity-purified material (Fig. 2B, compare lanes 2 and 5). Some eTER should have copurified with hpTER if more than one TER were incorporated per RNP particle. Instead, the affinity purification results suggest that TER is monomeric in an endogenously assembled telomerase holoenzyme RNP.
Importantly, the purified hpTER-tagged telomerase RNP possessed catalytic activity features diagnostic of the Tetrahymena telomerase holoenzyme. The minimal Tetrahymena telomerase RNP assembled in vitro from TERT and untagged TER has a low repeat addition processivity that is also dependent on allosteric activation by dGTP (16). With minimal recombinant enzyme, activity assays with <1 μM dGTP support predominantly single-repeat synthesis. In contrast, telomerase holoenzyme RNP assembled in vivo with eTER (12) or hpTER (Fig. 2C) displayed a much higher repeat addition processivity in reactions with <1 μM dGTP. This high repeat addition processivity results from protein-DNA “anchor site” interactions that are not reconstituted in the minimal RNP (10).
Reconstitution of hpTER variants.
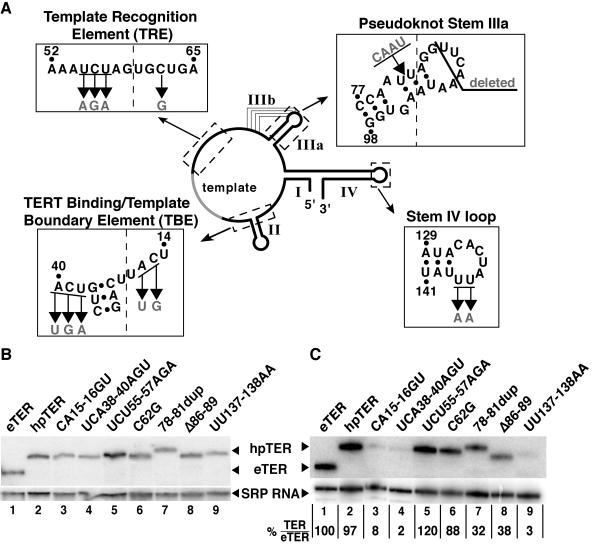
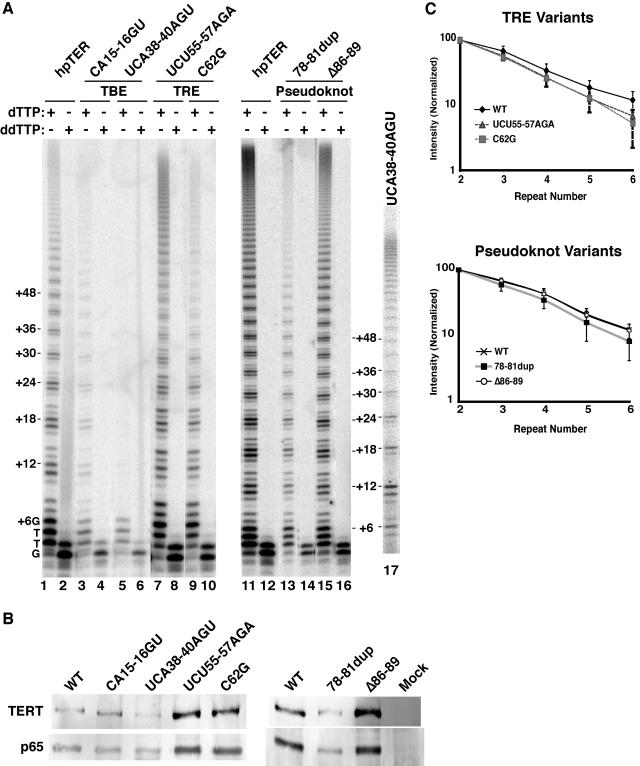
The hpTER transgene expression strategy described above was used to reconstitute TER sequence variants into the endogenous telomerase holoenzyme context in vivo. Seven TER variants (Fig. 3A) were selected based on the studies described below. We verified that in vitro reconstitution of hpTER variants recapitulated the expected catalytic activity defects (described below; see Fig. 4).
FIG. 3.
Expression of hpTER variants. (A) Sequence substitutions are depicted within boxes expanded from the schematic. Nucleotide positions are numbered in the eTER context. (B and C) RNA was prepared from equal numbers of cells (B) or from cell extracts (C) and analyzed by Northern blot hybridization. In panel C, levels of hpTER relative to eTER are indicated after normalization to SRP RNA as a loading control.
FIG. 4.
In vitro reconstitution of TERT and hpTER in RRL. Two microliters of TERT protein expression reaction mixture was combined with 1 pmol of TER prior to RNP assembly for 15 min at 30°C, as done previously (25, 28, 35). Samples were then diluted 10-fold into T2MG buffer to provide half of the final activity assay reaction volume. Assay mixtures contained 5.6 μM dGTP and 400 μM dTTP for optimal activity of the minimal recombinant RNP (16). (A and B) Reconstituted wild-type TER (lanes 1), wild-type hpTER (lanes 2), and hpTER variants were assayed using either 250 nM of the telomeric repeat primer (GT2G3)3 (A) or 250 nM of (TG)8T2G3 (B), a primer optimal for the minimal recombinant RNP. (C) Reconstituted wild-type TER (lanes 1 and 2), wild-type hpTER (lanes 3 and 4), and the hpTER variant UCA38-40AGU (lanes 5 and 6) were assayed using 250 nM (TG)8T2G3 in reaction mixtures without (-) or with (+) 400 μM dATP. The dATP-dependent change in product profile for the TBE variant enzyme occurs due to synthesis past the normal template 5′ boundary to copy TER position U42 (aberrant template position +7).
The only nontemplate sequence conserved among all ciliate TERs is the GUCA motif 5′ of the template (29, 31). For T. thermophila TER, this GUCA sequence (positions 37 to 40) and a nearby CAUU sequence (positions 15 to 18) comprise the core of the TERT binding/template 5′ boundary element (TBE) (Fig. 3A). This motif interacts with TERT with high affinity in vitro (25) and blocks the active site from copying past the intended template 5′ end (2, 24). Consistent with a physiological role for the TBE in TERT interaction, CA15-16 and other local residues become protected from chemical modification by assembly as RNP in vivo (45). We constructed hpTER TBE variants testing the substitutions CA15-16GU and UCA38-40AGU. Both substitutions affect both TBE functions in vitro, but CA15-16GU more severely reduces TERT binding while UCA38-40AGU more strongly compromises fidelity at the template 5′ boundary (24).
TER sequence immediately 3′ of the template is conserved among Tetrahymena species RNAs (31). Results from reconstitution assays in vitro suggest that this region, termed the template recognition element (TRE) (Fig. 3A), functions to position adjacent 5′ sequence in the active site (32). The conserved, template-adjacent residues UCU55-57 can be cross-linked directly at short range to TERT translated in RRL (23), and nearby C62 becomes protected from modification when assembled into RNP in vivo (45). We constructed both TRE substitutions UCU55-57AGA and C62G as hpTER variants. The substitution UCU55-57AGA reduces the repeat addition processivity of minimal recombinant Tetrahymena telomerase RNP, while the substitution C62G strongly inhibits its activity overall (25, 28) (Fig. 4A), both consistent with defects in positioning the template.
Pseudoknot folding has been proposed in all TERs (8). Tetrahymena TER variants with an unpaired pseudoknot stem IIIb did not efficiently reconstitute telomerase holoenzyme when assembled in vivo in competition with eTER (15). In assays of the minimal recombinant RNP, unpairing or removal of either pseudoknot stem reduced repeat addition processivity and activity overall (23, 28, 38). We constructed two hpTER variants in the pseudoknot region (Fig. 3A): a duplication of positions 78 to 81 (78-81dup) and a deletion of nucleotides 86 to 89 (Δ86-89). The 78-81dup variant mimics a TER allele that arose spontaneously in continuous log-phase growth, coexpressed with wild-type TER (1). Continuous growth at 30°C promotes progressive telomere lengthening, which in turn appears to select for cells that acquire stable, short telomeres (26, 1). The 4-nucleotide, position 78 to 81 duplication was predicted to destabilize the pseudoknot structure. The Δ86-89 variant removes residues in the stem IIIa loop that accomplish stem IIIb pairing, which should prevent formation of the predicted pseudoknot entirely. Both pseudoknot sequence substitutions reconstituted as hpTER variants in vitro reduced recombinant Tetrahymena telomerase RNP repeat addition processivity and activity overall (Fig. 4A).
We also constructed an hpTER variant in the stem IV loop. The loop nucleotides UU137-138 are absolutely conserved in TERs of all Tetrahymena species examined (29, 31). RRL reconstitution studies revealed that Tetrahymena TER residues UU137-138 can be cross-linked to TERT directly at short range (23). Substitutions of these and other stem IV loop residues reduced catalytic activity and the processivity of nucleotide addition within a single round of repeat synthesis (23, 38). We constructed the hpTER stem IV loop variant UU137-138AA (Fig. 3A). In the minimal recombinant RNP, this variant reconstituted a telomerase enzyme with very low overall activity and a defect in nucleotide addition processivity (Fig. 4B).
Endogenous accumulation and extract stability of the hpTER variants.
Transgenes encoding the hpTER variants were used to disrupt and completely replace BTU1, followed by disruption and complete replacement of TER using the methods described above. All strains expressing hpTER variants with or without coexpression of eTER doubled at the same rate as the parental strain at 30°C (data not shown). We quantified hpTER accumulation by Northern blot hybridization using total RNA isolated from equal numbers of cells expressing only eTER (Fig. 4B, lane 1), hpTER (lane 2), or an hpTER variant (lanes 3 to 9). When normalized to SRP RNA as a loading control, each hpTER variant accumulated to the same level as eTER accumulation in wild-type cells. Cell lysis and extract production allowed some differences in the levels of hpTER variants to develop (Fig. 4C). In particular, hpTER variants substituted in the TBE (lanes 3 to 4) or the stem IV loop (lane 9) were lost over time in extract relative to the level of SRP RNA. These sequence substitutions affect some sites of TERT contact defined for the minimal RNP reconstituted in RRL (23, 25).
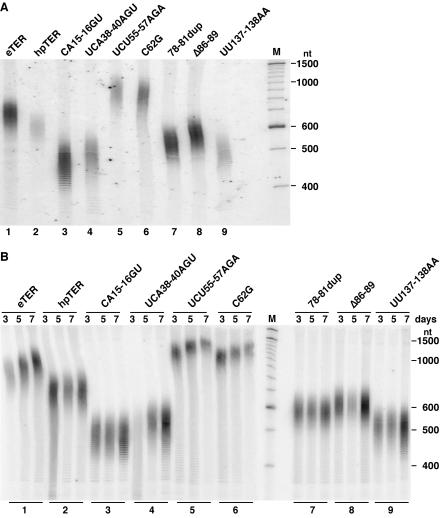
In vivo function of the hpTER variants. Using the hpTER strains lacking eTER expression, we could determine the impact of TER sequence substitutions on telomere length maintenance. Genomic DNA prepared from these strains was digested with HindIII to release an identical ∼360-bp subtelomeric sequence from half of the macronuclear chromosome ends, representing both ends of the amplified, palindromic rRNA chromosome. Digested DNA was resolved by denaturing gel electrophoresis and probed with an oligonucleotide complementary to the subtelomeric region (Fig. 5A). The length of telomeric repeat tracts can be compared by subtraction of the ∼360 bp between the first telomeric repeat and the nearest HindIII site.
FIG. 5.
Telomere length maintenance by telomerase holoenzymes reconstituted with hpTER variants. Genomic DNA was digested with HindIII, resolved by denaturing polyacrylamide gel electrophoresis, and assayed by Southern blotting for telomeric restriction fragments containing the rRNA chromosome subtelomeric region. End-labeled DNA marker (M) lengths are indicated at right. (A) Cells were grown for a fixed number of doublings at 30°C. (B) Cells were grown in continuous log-phase culture at 30°C with aliquots harvested for analysis at days 3, 5, and 7. All strains doubled at similar rates.
Macronuclear telomeric repeat tracts in the parental strain expressing eTER averaged ∼325 bp (Fig. 5A, lane 1). Telomeres in the otherwise-unsubstituted hpTER strain averaged ∼250 bp (lane 2), which serves as the reference length for changes imposed by sequence substitutions in the hpTER variants (lanes 3 to 9). Pseudoknot region substitutions reduced average telomere length to ∼160 to 210 bp (lanes 7 and 8). The TBE and stem IV loop substitutions brought telomere length to under 150 bp (lanes 3, 4, and 9), representing a loss of half of the wild-type telomeric repeat tract. Strikingly, TRE substitutions increased telomere length to twofold above that in wild-type cells (lanes 5 and 6). Hybridization with a probe complementary to the other strand of subtelomeric sequence confirmed that the lengths of both strands were affected in parallel (data not shown). Analysis of bulk telomeres using a telomeric-repeat probe for hybridization also produced similar results (data not shown).
The long-term viability of the hpTER variant strains suggested that their macronuclear telomeres must be stably maintained. To demonstrate this directly, we passaged the parental and hpTER strains as continuous log-phase cultures at 30°C. Under these conditions, macronuclear telomeres in wild-type cells experience a progressive lengthening (26). Telomere length was monitored by hybridization to the rRNA chromosome subtelomeric region as described above, using genomic DNA harvested after 3, 5, and 7 days of continuous culture (Fig. 5B). Cells expressing eTER (lane set 1) or either of the hpTER TRE variants (lane sets 5 and 6) gradually gained telomeric repeats with time in culture, as expected. Cells with initial telomere lengths shorter than the parental strain stably maintained their telomeres, showing less increase in length over time (lane sets 2 to 4 and 7 to 9). We conclude that nontemplate sequence variants of Tetrahymena TER alter the equilibrium set point for telomere length maintenance.
Catalytic activity of telomerase holoenzymes with hpTER variants.
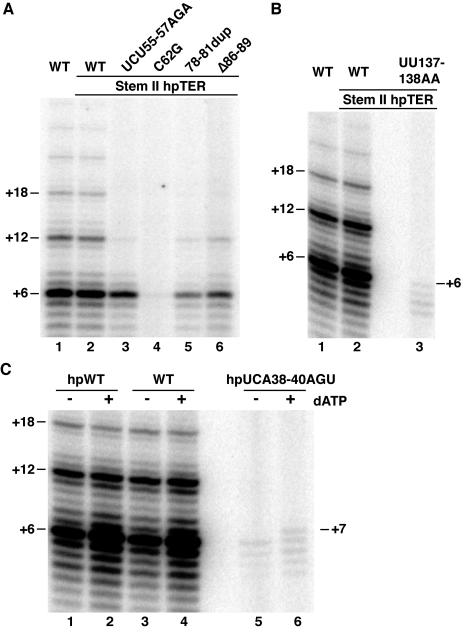
We next examined holoenzyme catalytic activity differences imposed by the hpTER sequence substitutions (Fig. 6A). We performed telomerase RNP purification from cell extracts as described above. Affinity purification of telomerase RNP assembled on each hpTER variant enriched full-length TER (data not shown) and proportionally recovered TERT and other telomerase holoenzyme proteins such as p65 (Fig. 6B). Eluted samples were assayed in a processive repeat synthesis reaction with dGTP and dTTP (Fig. 6A, odd lanes) and in a nonprocessive repeat synthesis reaction with dGTP and ddTTP (even lanes). Activity purified from cells expressing the UU137-138AA variant was difficult to detect, due at least in part to the instability of this hpTER in cell extract. For the other hpTER RNPs, we quantified relative specific activity using the product intensities of the nonprocessive repeat synthesis reaction; these reaction products are concentrated in a smaller region of the gel and thus more accurately corrected for background. We normalized activity assay product intensities by quantifying hpTER recovery using an aliquot of each purified sample for Northern blot hybridization (data not shown). After normalization, the variant hpTER RNPs had activity levels within ∼3-fold of the wild-type hpTER control in vitro.
FIG. 6.
Catalytic activity of telomerase holoenzymes containing hpTER variants. All wild-type (WT) samples refer to hpTER with no additional sequence substitutions. (A) Eluted samples were assayed for activity as indicated with the primer (GT2G3)3, which is elongated by addition of the sequence GTTG to reach the template 5′ end at position +6. The product ladder is annotated to indicate complete repeat additions. Lane 17 is another activity assay using RNP with the TBE variant UCA38-40AGU in dTTP; this relatively long exposure revealed repeat addition processivity. (B) TERT and p65 were detected in purified samples by immunoblotting using rabbit polyclonal antibodies raised and affinity purified against a TERT peptide or full-length p65 (42). Neither protein was recovered by the control affinity purification from a cell extract lacking hpTER (mock). Note that differences in recovery do not reflect relative differences in RNP stability or purification yield because extracts were not normalized for hpTER prior to purification. (C) Successive complete-repeat addition products from parallel assays of wild-type or TRE and pseudoknot substitution hpTER RNPs were quantified for intensity, normalized for specific activity, and plotted to compare repeat addition processivities.
We evaluated holoenzyme repeat addition processivity by quantifying the intensities of successive complete-repeat addition products within a single reaction mixture and then normalizing for their content of radiolabeled dGTP (4). In the Tetrahymena minimal telomerase RNP context, substitutions in the TRE and pseudoknot reduce repeat addition processivity (23, 25, 28). In the holoenzyme context, substitutions in the TRE decreased repeat addition processivity by a small but detectable margin, while substitutions in the pseudoknot had no detectable impact (Fig. 6C). We could not detect a defect for the pseudoknot variants, even when we monitored product synthesis over time using pulse-chase methods (data not shown). High telomerase holoenzyme repeat addition processivity independent of an intact TRE or pseudoknot is consistent with the proposed significance of protein-DNA “anchor site” interactions in governing enzyme-product interaction.
Template 5′ boundary definition.
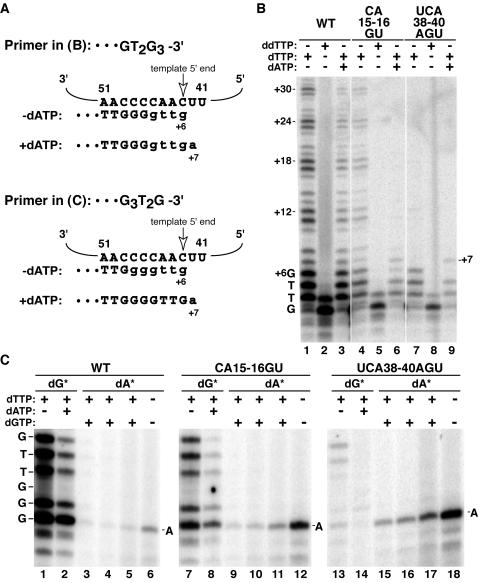
TBE sequence substitutions that reduce high-affinity TERT-TER interactions in RRL allow the minimal RNP to copy past the normal 5′ template boundary (24). We tested the fidelity of the holoenzyme 5′ template boundary using RNPs endogenously assembled on hpTER variants, including the TBE substitutions CA15-16GU and UCA38-40AGU. In assays with dGTP and either dTTP or ddTTP, only the correct template residues could be copied (Fig. 7A and B, lanes 1 and 2, 4 and 5, and 7 and 8). In assays with dGTP and dTTP alone, the TBE variant holoenzymes could generate a product ladder indicative of repeat addition processivity (Fig. 6A, lanes 3 and 17). If assay mixtures contained dGTP, dTTP, and dATP, however, the TBE variant holoenzymes generated a product profile distinct from that of the wild-type enzyme (Fig. 7B, compare lanes 3, 6, and 9). In the presence of dATP, template use by the TBE variant holoenzymes extended beyond the normal 5′ boundary to include U42 as an aberrant +7 position of the template. This dATP-dependent change in product synthesis was observed only for the TBE variants, whether reconstituted in vitro or in vivo (Fig. 4C and additional data not shown).
FIG. 7.
Template boundary definition by the TBE. (A) Schematic of products produced in panels B and C by enzymes with TBE substitutions. TER positions 41 to 51 and primer sequence are shown in uppercase letters; nucleotides added during product synthesis are shown in lowercase letters. (B) Eluted samples were assayed for activity as indicated with the primer (GT2G3)3, which was elongated by addition of GTTG to reach the template 5′ boundary at position +6. The product ladder is annotated to indicate complete repeat additions. Synthesis past the normal template 5′ boundary to copy TER position U42, indicated as +7 product, was detected in reactions with dATP. (C) Eluted samples were assayed for activity as indicated with 1 μM primer (G3T2G)3, which can be elongated by dGTP addition at template position +1 or by dATP addition at aberrant template position +7. Either dGTP (dG*) or dATP (dA*) was radiolabeled. The dG* reaction mixtures contained 0.3 μM radiolabeled dGTP and 1 μM unlabeled dGTP, with 400 μM dTTP and dATP if indicated. Reactions in lanes 3 to 5, 9 to 11, and 15 to 17 contained, respectively, 0.3, 0.6, and 1.3 μM radiolabeled dATP mixed with unlabeled dATP to provide a total of 1.3 μM; these reaction mixtures also contained 0.3 μM dGTP and 400 μM dTTP. Reaction mixtures for lanes 6, 12, and 18 contained 1.3 μM radiolabeled dATP as the only nucleotide substrate.
To confirm incorporation templated by the aberrant +7 position U42, we performed activity assays using dATP as the radiolabeled nucleotide. A primer was employed that could align at either the 3′ or 5′ end of the template (Fig. 7A, bottom). In control reactions that contained only radiolabeled dGTP and dTTP, all enzymes elongated the primer in alignment with the template 3′ end (Fig. 7C, lanes 1, 7, and 13). In reactions that contained radiolabeled dATP (lanes 3 to 6, 9 to 12, and 15 to 18), primer extension could instead occur in alignment with the template 5′ end to copy position U42. Strikingly, the TBE variant holoenzymes relatively efficiently utilized radiolabeled dATP for primer elongation (Fig. 7C, lanes 6, 12, and 18), even in the presence of unlabeled dGTP as competitor (lanes 3 to 5, 9 to 11, and 15 to 17). The extent of difference between wild-type and TBE variant holoenzymes is most evident in the comparison of product intensities from reactions with radiolabeled dGTP versus radiolabeled dATP (Fig. 7C, first and last lanes of each set). We note that the UCA38-40AGU substitution imposed a more severe template boundary defect than the CA15-16GU substitution when compared in the context of holoenzyme RNPs reconstituted in vivo (Fig. 7) or minimal RNPs reconstituted in vitro (24). These results establish TER TBE function in defining the 5′ template boundary in endogenously assembled telomerase holoenzyme.
Efficient use of the template 3′ end.
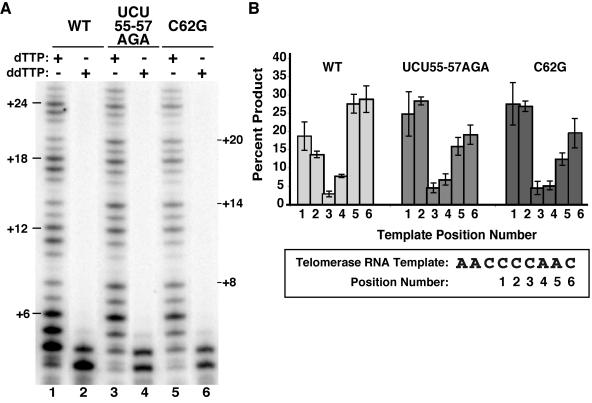
The predominant products of Tetrahymena telomerase assayed in vitro end with the permutation -TTG-3′, reflecting copying of the entire template (Fig. 1A). We noticed that telomerase holoenzymes with TRE substitutions reproducibly generated more product representing only one to two dGTP additions at the template 3′ end (Fig. 6A, lanes 7 and 9, and 8A). This change in product profile was observed only for the TRE variants and was evident in every repeat of the product ladder for every primer tested, indicative of a position-specific defect in template copying. We quantified the amount of product accumulated after synthesis at each position of the template by averaging values from multiple repeats of a product ladder (Fig. 8B [from reactions in A, lanes 1, 3, and 5]) from several independent activity assays (additional data not shown). Each product intensity was normalized for its relative content of radiolabeled dGTP and then expressed as a percentage of the total product within a repeat. In the Fig. 8 reactions, template position +2 products of the wild-type enzyme were 14% of the total, while for RNPs with UCU55-57AGA or C62G substitutions they accumulated to twice this level, at 28% or 27%, respectively.
FIG. 8.
A TRE role in use of the template 3′ end. (A) Eluted samples were assayed for activity as indicated with the primer (GT2G3)3. The product ladder is annotated to indicate sequential repeat additions to template position +6 or products from synthesis to template position +2. (B) The intensity of each product within the first five repeats in odd lanes of panel A was adjusted for specific activity and then normalized as the percentage of total product within the repeat. A schematic of the template position numbers is shown.
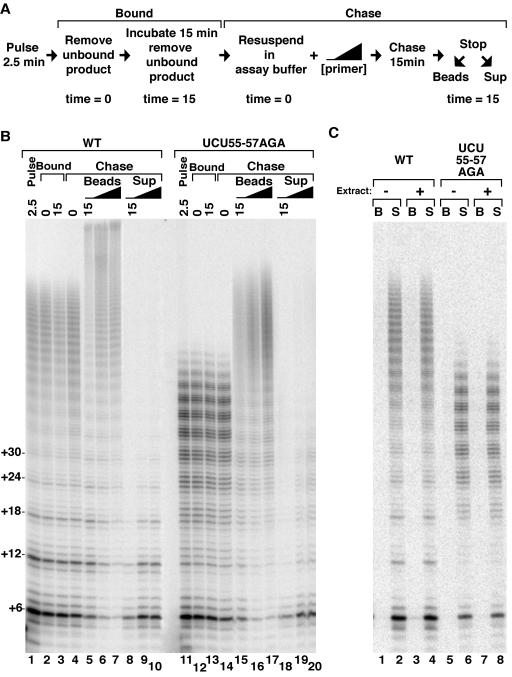
A defect in nucleotide addition processivity specific to the template 3′ end could result from enhanced product dissociation or reduced efficiency of template use. To distinguish between these possibilities, we performed pulse-chase activity assays with wild-type and TRE variant holoenzymes still bound to IgG-agarose (summarized in Fig. 9A). After an initial 2.5-min pulse of product synthesis, the immobilized enzyme and any bound products were recovered from the activity assay and resuspended in activity assay buffer without substrates. Following an additional 15-min incubation to allow product dissociation, immobilized enzyme and any bound products were again recovered. To our surprise, even the aberrantly accumulated, incomplete-repeat products of the TRE variant holoenzyme remained bound to RNP (Fig. 9B, lanes 1 to 3 and 11 to 13). As a control, we verified that purified product DNA did not associate with IgG-agarose under similar incubation conditions (Fig. 9C). These results argue against premature product dissociation as the basis for the nucleotide addition processivity defect.
FIG. 9.
A kinetic defect imposed by substitution of the TRE. (A and B) RNPs were immobilized on IgG-agarose using TAP-tagged coat protein. Bead-bound enzymes were assayed with the primer (GT2G3)3, 400 μM dTTP, and 0.6 μM radiolabeled and 1 μM unlabeled dGTP for 2.5 min at 30°C. Reactions were stopped (lanes 1 and 11), and bound products were retained after washes to remove unbound primer and product (lanes 2 and 12). After an additional 15-min incubation at 30°C, beads were again washed to detect stably bound products (lanes 3 and 13). Extension of bound products was then assayed by addition of unlabeled 1 μM dGTP and 400 μM dTTP (lanes 4 and 14, time zero) in the absence or presence of 1 μM or 5 μM of the primer (G3T2G)3 as depicted by the black triangle. Chase reactions were halted after 15 min at 30°C followed by separation of bead-associated (lanes 5 to 7 and 15 to 17) or released (lanes 8 to 10 and 18 to 20) products. (C) IgG-agarose was preincubated without (-) or with (+) cell extract and washed, followed by addition of telomerase products purified after a 2.5-min activity assay. Samples were incubated for 20 min at 30°C followed by separation of products that became bead associated (B, odd lanes) or remained in solution (S, even lanes).
The aberrant products of the TRE variant holoenzyme could be enzyme bound as arrested, dead-end complexes or could be paused in elongation as the consequence of a new rate-limiting step in the catalytic cycle. To distinguish between these alternatives, washed enzyme-product complexes were returned to a chase reaction with unlabeled dNTPs (summarized in Fig. 9A). All bound products were elongated by a chase in the absence or presence of competitor primer (Fig. 9B, lanes 4 to 10 and 14 to 20). In the chase interval, for a given enzyme, elongated products that remained bound to RNP or that dissociated shared the same profile of product 3′ permutation (compare lanes 5 to 7 with 8 to 10 or lanes 15 to 17 with 18 to 20). This result implies that product dissociation is not biased to occur at a specific step of the catalytic cycle. Taken together, the findings above reveal that TRE substitution imposes a kinetic defect in use of the template 3′ end. The kinetic nature of this defect was also evident in the much more pronounced accumulation of incomplete-repeat products in the resolved region of the gel at short reaction times (Fig. 9, 2.5 min) versus long reaction times (Fig. 6, 45 min, and additional time course data not shown).
DISCUSSION
Telomerase holoenzyme assembly using tagged TER.
We created Tetrahymena strains expressing individual variants of epitope-tagged TER. A hairpin tag inserted at the distal end of stem II allowed the rapid and gentle, RNA-based affinity purification of active telomerase RNP. Affinity purification from cell extracts of strains coexpressing eTER and wild-type hpTER (Fig. 2), or eTER and any hpTER variant (data not shown), failed to reveal any suggestion of endogenous TER dimerization. These studies, along with previous studies of Tetrahymena telomerase RNPs reconstituted in vitro and in vivo (5, 42), demonstrate that telomerase catalytic activity and repeat addition processivity do not require the cooperative effort of TERT or TER multimers.
Fidelity of the template 5′ boundary.
TBE substitutions reduce the fidelity of template 5′ boundary definition in Tetrahymena telomerase RNPs reconstituted in vitro (2, 24) and in vivo (above). The synthesis of mutant telomeric repeats in vivo can impact Tetrahymena telomere length maintenance, chromosome segregation, and cell growth (22, 34, 44). Tetrahymena strains expressing the TBE variant hpTERs grew normally, however, suggesting that altered telomeric repeats were not abundant or were tolerated. We could not detect mutant telomeric repeat synthesis in vivo using Southern blot hybridization with oligonucleotides complementary to putative altered-repeat sequences (data not shown). Strains expressing an hpTER with TBE substitution did harbor significantly shorter telomeres than strains expressing eTER or wild-type hpTER. These atypically short telomeres could arise from the removal of aberrant repeats by the nucleolytic cleavage activity of telomerase or another enzyme (11, 19). Atypically short telomeres could also arise from a reduction in telomerase repeat addition processivity: relaxed template 5′ boundary fidelity may indicate relaxed strain on the template-product hybrid at the template 5′ end, which would be accompanied by reduced product dissociation.
Pseudoknot function.
TERs from all species have been predicted to form a pseudoknot pairing (8). Pseudoknot region mutations of yeast TERs expressed in vivo or mammalian TERs expressed in vivo or in vitro can reduce telomerase catalytic activity and telomere length maintenance (reference 39 and references therein). Findings here indicate that Tetrahymena TER sequence substitutions in the pseudoknot region do not necessarily affect telomerase holoenzyme catalytic activity assayed in vitro. The Tetrahymena telomerase holoenzyme has a mechanism of repeat addition processivity different from that of the minimal RNP, dependent on holoenzyme protein-DNA interactions (10). This difference can account for the reduced impact of pseudoknot region substitutions on the repeat addition processivity of the holoenzyme versus the minimal RNP. Curiously, Tetrahymena TER pseudoknot formation is also not required for stable telomere length maintenance in vivo, albeit at a homeostasis slightly different from wild type. Future studies could test whether impaired pseudoknot formation imposes phenotypes that would not be evident during vegetative growth, for example in the process of telomerase-mediated chromosome healing during new macronuclear development.
Use of the template 3′ end.
TRE substitutions in TER reconstituted with TERT in vitro impose defects in activity and repeat addition processivity (25, 28). A subset of the same TRE variants reconstituted in vivo yielded enzymes with reduced repeat addition processivity and a kinetic defect in nucleotide addition specific to the template 3′ end. Disruption of the TRE, particularly with the template-adjacent substitution UCU55-57AGA, seems likely to affect the positioning of the 3′ end of the template and/or the hybrid of the template 3′ end with product. The same molecular defect could impact the product profile of the minimal and holoenzyme RNPs differently, because only the holoenzyme can maintain strong, anchor site interactions with product released from the template. Reduced formation or stability of DNA hybrid at the template 3′ end could inhibit the activity and repeat addition processivity of the minimal RNP by enhancing product dissociation; this same defect could inhibit the nucleotide addition processivity of the holoenzyme by delaying elongation of a stably bound product. Regardless of the exact mechanism, it is fascinating to consider why substitution of the TRE leads to telomere elongation rather than telomere shortening. If telomerase dissociation from the telomere is not dependent on synthesis to the template 5′ end, in keeping with our observations for dissociation from product in vitro (Fig. 9), holoenzymes with TRE substitutions would leave an altered permutation of the 3′ overhang. This in turn could influence the balance of telomere length homeostasis.
Acknowledgments
This work was supported by NIH grant GM54198.
We thank Jacek Gaertig for strain CU522 and plasmid pBICH3, Jamie Williamson for MS2 coat protein plasmid, Bobby Hogg for purified TAP-MS2 coat protein, the Lemaux lab for generous use of their biolistic transformation system, and Martin Gorovsky and members of the Collins laboratory, including Keren Witkin and Catherine O'Connor, for advice and discussions on the manuscript.
REFERENCES
- 1.Ahmed, S., H. Sheng, L. Niu, and E. Henderson. 1998. Tetrahymena mutants with short telomeres. Genetics 150:643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Autexier, C., and C. W. Greider. 1995. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 9:2227-2239. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn, E. H. 2000. The end of the (DNA) line. Nat. Struct. Biol. 7:847-850. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, T. M., K. J. Goodrich, and T. R. Cech. 2000. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 275:24199-24207. [DOI] [PubMed] [Google Scholar]
- 5.Bryan, T. M., K. J. Goodrich, and T. R. Cech. 2003. Tetrahymena telomerase is active as a monomer. Mol. Biol. Cell 14:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, S. R., and E. H. Blackburn. 2004. Telomeres and telomerase. Philos. Trans. R. Soc. London B 359:109-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. L., and C. W. Greider. 2003. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 22:304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J. L., and C. W. Greider. 2004. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends Biochem. Sci. 29:183-192. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. L., and C. W. Greider. 2003. Template boundary definition in mammalian telomerase. Genes Dev. 17:2747-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, K. 1999. Ciliate telomerase biochemistry. Annu. Rev. Biochem. 68:187-218. [DOI] [PubMed] [Google Scholar]
- 11.Collins, K., and C. W. Greider. 1993. Nucleolytic cleavage and non-processive elongation catalyzed by Tetrahymena telomerase. Genes Dev. 7:1364-1376. [DOI] [PubMed] [Google Scholar]
- 12.Collins, K., and C. W. Greider. 1995. Utilization of ribonucleotides and RNA primers by Tetrahymena telomerase. EMBO J. 14:5422-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, R., Z. Zhou, and R. Reed. 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell 5:779-787. [DOI] [PubMed] [Google Scholar]
- 14.Gaertig, J., Y. Gao, T. Tishgarten, T. G. Clark, and H. W. Dickerson. 1999. Surface display of a parasite antigen in the ciliate Tetrahymena thermophila. Nat. Biotechnol. 17:462-465. [DOI] [PubMed] [Google Scholar]
- 15.Gilley, D., and E. H. Blackburn. 1999. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc. Natl. Acad. Sci. USA 96:6621-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy, C. D., C. S. Schultz, and K. Collins. 2001. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem. 276:4863-4871. [DOI] [PubMed] [Google Scholar]
- 17.Hargrove, B. W., A. Bhattacharyya, A. M. Domitrovich, G. M. Kapler, K. Kirk, D. E. Shippen, and G. R. Kunkel. 1999. Identification of an essential proximal sequence element in the promoter of the telomerase RNA gene of Tetrahymena thermophila. Nucleic Acids Res. 27:4269-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington, L. 2003. Biochemical aspects of telomerase function. Cancer Lett. 194:139-154. [DOI] [PubMed] [Google Scholar]
- 19.Jacob, N. K., K. E. Kirk, and C. M. Price. 2003. Generation of telomeric G strand overhangs involves both G and C strand cleavage. Mol. Cell 11:1021-1032. [DOI] [PubMed] [Google Scholar]
- 20.Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff, and M. J. Moore. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8:426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelleher, C., M. T. Teixeira, K. Forstemann, and J. Lingner. 2002. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27:572-579. [DOI] [PubMed] [Google Scholar]
- 22.Kirk, K. E., B. P. Harmon, I. K. Reichardt, J. W. Sedat, and E. H. Blackburn. 1997. Block in anaphase chromosome separation caused by a telomerase template mutation. Science 275:1478-1481. [DOI] [PubMed] [Google Scholar]
- 23.Lai, C. K., M. C. Miller, and K. Collins. 2003. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell 11:1673-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, C. K., M. C. Miller, and K. Collins. 2002. Template boundary definition in Tetrahymena telomerase. Genes Dev. 16:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, C. K., J. R. Mitchell, and K. Collins. 2001. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21:990-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larson, D. D., E. A. Spangler, and E. H. Blackburn. 1987. Dynamics of telomere length variation in Tetrahymena thermophila. Cell 50:477-483. [DOI] [PubMed] [Google Scholar]
- 27.LeCuyer, K. A., L. S. Behlen, and O. C. Uhlenbeck. 1995. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry 34:10600-10606. [DOI] [PubMed] [Google Scholar]
- 28.Licht, J. D., and K. Collins. 1999. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev. 13:1116-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingner, J., L. L. Hendrick, and T. R. Cech. 1994. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 8:1984-1998. [DOI] [PubMed] [Google Scholar]
- 30.Mason, D. X., E. Goneska, and C. W. Greider. 2003. Stem-loop IV of Tetrahymena telomerase RNA stimulates processivity in trans. Mol. Cell. Biol. 23:5606-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCormick-Graham, M., and D. P. Romero. 1995. Ciliate telomerase RNA structural features. Nucleic Acids Res. 23:1091-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, M. C., and K. Collins. 2002. Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl. Acad. Sci. USA 99:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, M. C., and K. Collins. 2000. The Tetrahymena p80/p95 complex is required for proper telomere length maintenance and micronuclear genome stability. Mol. Cell 6:827-837. [DOI] [PubMed] [Google Scholar]
- 34.Petcherskaia, M., J. M. McGuire, J. M. Pherson, and K. E. Kirk. 2003. Loss of cap structure causes mitotic defect in Tetrahymena thermophila telomerase mutants. Chromosoma 111:429-437. [DOI] [PubMed] [Google Scholar]
- 35.Prathapam, R., K. L. Witkin, C. M. O'Connor, and K. Collins. 2005. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 12:252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 37.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 38.Sperger, J. M., and T. R. Cech. 2001. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry 40:7005-7016. [DOI] [PubMed] [Google Scholar]
- 39.Theimer, C. A., C. A. Blois, and J. Feigon. 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell 17:671-682. [DOI] [PubMed] [Google Scholar]
- 40.Tzfati, Y., Z. Knight, J. Roy, and E. H. Blackburn. 2003. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev. 17:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, H., and E. H. Blackburn. 1997. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 16:866-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witkin, K. L., and K. Collins. 2004. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 18:1107-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Youngman, E. M., J. L. Brunelle, A. B. Kochaniak, and R. Green. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117:589-599. [DOI] [PubMed] [Google Scholar]
- 44.Yu, G., J. D. Bradley, L. D. Attardi, and E. H. Blackburn. 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344:126-132. [DOI] [PubMed] [Google Scholar]
- 45.Zaug, A. J., and T. R. Cech. 1995. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA 1:363-374. [PMC free article] [PubMed] [Google Scholar]