Abstract
Genome stability requires a set of RecQ-Top3 DNA helicase-topoisomerase complexes whose sole budding yeast homolog is encoded by SGS1-TOP3. RMI1/NCE4 was identified as a potential intermediate in the SGS1-TOP3 pathway, based on the observation that strains lacking any one of these genes require MUS81 and MMS4 for viability. This idea was tested by confirming that sgs1 and rmi1 mutants display the same spectrum of synthetic lethal interactions, including the requirements for SLX1, SLX4, SLX5, and SLX8, and by demonstrating that rmi1 mus81 synthetic lethality is dependent on homologous recombination. On their own, mutations in RMI1 result in phenotypes that mimic those of sgs1 or top3 strains including slow growth, hyperrecombination, DNA damage sensitivity, and reduced sporulation. And like top3 strains, most rmi1 phenotypes are suppressed by mutations in SGS1. We show that Rmi1 forms a heteromeric complex with Sgs1-Top3 in yeast and that these proteins interact directly in a recombinant system. The Rmi1-Top3 complex is stable in the absence of the Sgs1 helicase, but the loss of either Rmi1 or Top3 in yeast compromises its partner's interaction with Sgs1. Biochemical studies demonstrate that recombinant Rmi1 is a structure-specific DNA binding protein with a preference for cruciform structures. We propose that the DNA binding specificity of Rmi1 plays a role in targeting Sgs1-Top3 to appropriate substrates.
The RecQ family of DNA helicases is important for maintaining genome integrity. The eukaryotic branch of this family is significant because it includes three human disease proteins (Werner's syndrome protein [WRN], Bloom syndrome protein [BLM], and Rothmund-Thomson syndrome protein [RTS]), in addition to the yeast homologs Sgs1 and Rqh1 from Saccharomyces cerevisiae and Schizosaccharomyces pombe, respectively (10, 16, 22, 56). These proteins are characterized by a 700-amino-acid (700-aa) DNA helicase domain homologous to Escherichia coli RecQ and an equally large N-terminal domain. The N-terminal domain lacks enzyme activity in most of these proteins, but it is known to be functionally important in budding yeast (12, 33, 35). Although SGS1 is not essential for viability, loss of SGS1 results in increased rates of mitotic and meiotic recombination, gross chromosomal rearrangements, and chromosome loss (6, 22, 38, 41, 56, 57, 60). sgs1 mutants also show hypersensitivity to high levels of DNA-damaging agents such as UV light, methylmethane sulfonate (MMS), and hydroxyurea (35, 60). These sensitivities have contributed to the idea that Sgs1 is required to stabilize stalled or collapsed replication forks.
Sgs1, like BLM in humans and Rqh1 in S. pombe, interacts genetically and physically with its cognate DNA topoisomerase III (Top3) (16, 58). Eukaryotic Top3 is a type I enzyme whose activity resembles that of E. coli DNA topisomerase III in that it is most active in unlinking single-strand catenanes (28). Loss of TOP3 results in a profound slow-growth phenotype as well as high levels of recombination and chromosome loss (38, 55). Spontaneously occurring slow-growth suppressors of top3 were originally shown to map to SGS1, and yeast two-hybrid data indicated that the Sgs1 N terminus and Top3 interacted in vivo (16). Physical mapping studies confirmed that Top3 interacts with the N-terminal 100 aa of Sgs1 (4, 14). This interaction, which is conserved in the BLM-TOP3α and Rqh1-Top3 complexes (30, 58), is essential for complementation of sgs1 mutant phenotypes (12, 35, 53, 54). Taken together, these results indicate that Sgs1-Top3 functions as a complex and confirm the idea that the top3 slow-growth phenotype is primarily due to unrestrained Sgs1 DNA helicase activity in the absence of Top3 activity (16).
The RecQ-Top3 complex is needed to complete a late step in homologous recombination (HR), and it may play a specific role in HR events that occur in response to DNA replication damage. Consistent with the DNA damage sensitivity of sgs1 and top3 mutants, yeast strains lacking TOP3 either arrest or delay in G2, suggesting a role in repairing spontaneous S-phase damage (15, 18, 31). SGS1 is required for UV- and MMS-induced heteroalleleic recombination and, like rqh1+, SGS1 has been shown to act in an RAD52-dependent pathway (17, 37, 54). Additional support for a role of Sgs1-Top3 in recombination is provided by genetic suppression studies. Several sgs1-top3 mutant phenotypes appear to result from toxic recombination intermediates, since they are suppressed in strains that are unable to initiate meitotic or mitotic recombination. Of particular relevance is the finding that top3 homozygous diploids are capable of undergoing meiosis as long as recombination is not initiated (15). Similarly, the slow-growth of top3 strains or the synthetic sickness of sgs1 srs2 cells is relieved in cells lacking any of the RAD52 epistasis genes that are required for HR (17, 32, 40, 47).
Recent progress in explaining the molecular mechanism of RecQ-Top3 complexes has come from both genetic and biochemical studies. In yeast, Sgs1 and a second DNA helicase, Srs2, have been shown to control distinct pathways of HR-dependent double-strand break repair. Similar to the increase in sister chromatid exchanges seen in Bloom syndrome cells, sgs1 mutants display an increase in crossover frequency compared to wild-type (wt) cells (23). This result suggests that the normal function of Sgs1-Top3 is to resolve recombination intermediates in a pathway leading to noncrossover products. Complementary in vitro studies with BLM-TOP3α provide a mechanistic explanation for such a pathway. Consistent with the ability of Sgs1 and BLM helicases to branch migrate Holliday junctions (HJs) (3, 25), BLM-TOP3α has been shown to be active in decatenating double HJ-containing substrates. In this reaction, BLM appears to branch migrate double HJs until they collapse into a hemicatenane, which is then a substrate for strand passage by TOP3α (59).
Genes that are redundant with SGS1-TOP3 have been identified by synthetic lethal screens. Newer methodologies such as synthetic genetic arrays (SGA) (51) and synthetic lethal analyis by microarray (43) have been combined with a standard genetic screen (36) to identify over 30 mutations that result in a slow-growth or lethal phenotype in the absence of SGS1. This large number of interactors suggests that SGS1 is a “hub” gene that overlaps multiple pathways (52). Two of these interactors, MUS81 and MMS4, are of particular interest as their loss results in a clear lethal phenotype when combined with sgs1Δ (36). MUS81 and MMS4 encode a heterodimeric structure-specific endonuclease that has been implicated in a late stage of HR (24). The sporulation defect of mus81 diploids is suppressed by eliminating meiotic recombination, like top3 mutants (5, 24), and the lethality of mus81 sgs1 cells is suppressed by eliminating homologous recombination (1, 13). Thus, Sgs1-Top3 and Mus81-Mms4 appear to act in parallel downstream of the initiation of HR.
As an approach to identify genes in the Sgs1-Top3 pathway, we employed a synthetic lethal screen with the synthetic interactor MUS81. While our screen was in progress, the results of SGA screens for both MUS81 and MMS4 were reported (2). Analysis of these candidate genes revealed that one of them, NCE4 (YPL024W), encoded a component of the Sgs1-Top3 complex. Because of its role in controlling genome stability, we hereafter refer to this gene as RMI1 (for RecQ-mediated genome instability).
MATERIALS AND METHODS
Strains and plasmids.
Yeast strains are described in Table 1. Strain construction, growth, and transformation were carried out by standard procedures (45). Unless otherwise indicated, cells were grown at 30°C in 1% yeast extract-2% peptone-2% dextrose (YPD) media. PCR-mediated gene disruptions replaced complete open reading frames (ORFs) with the indicated antibiotic resistance marker as previously described (19). Yeast strain NJY1906 was constructed by integrative transformation of W303-1a with the following plasmids: pNJ1569, pNJ2565, and pNJ7134. This placed different C-terminal epitope tags on the chromosomal loci of Sgs1, Top3, and Rmi1, respectively. To test the functionality of these tagged genes, we crossed NJY1906 to JMY381 (a mus81Δ strain). After sporulation and tetrad dissection, we obtained mus81Δ segregants at the expected frequency (50% of all spores), and among these were all combinations of tagged genes, including those with three tags.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| W303-1a | MATaade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 | 50a |
| W303-1b | MATα ade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 | 50a |
| JMY372 | W303-1a top3-2::HIS3 | This study |
| JMY376 | W303-1b mms4-10::KAN | This study |
| JMY381 | W303-1b mus81-10::KAN | This study |
| JMY361 | W303-1b slx1-10::TRP1 | 36 |
| NJY506 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 sgs1-11::KAN | This study |
| NJY518 | MATα ade2-1 ade3::hisG ura3-1 his3-11,15 lys2 trp1-1 can1-100 slx4-11::KAN | This study |
| NJY540 | K1875 sgs1-11::loxP | 36 |
| SIY777 | W303-1b slx8-10::KAN | This study |
| NJY1236 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 lys2 trp1-1 can1-100 top3-2::HIS3 sgs1-3::TRP1 | This study |
| JMY1452 | W303-1a slx1-11::HIS3 rad55::LEU2 | This study |
| JMY1455 | MATα ade2-1 ade3::hisG ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-10 slx5-10::TRP1 rad51::HIS3 | This study |
| JMY1457 | MATα ade2-1 ade3::hisG ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 mms4-10::KAN rad55::LEU2 | This study |
| JMY1459 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 mus81-10::KAN rad51::HIS3 | This study |
| VCY1515 | W303-1b slx4-11::KAN rad54::HIS3 | This study |
| JMY1699 | W303-1b slx5-11::HGR | This study |
| K1875 | MATaade2-1 ura3-52 his4-260 leu2-3,112 trp1-H3 lys2-ΔBX-CAN1-LYS2 can1 rRNA gene::ADE2 rRNA gene::URA3 | 27 |
| NJY1906 | W303-1a SGS1-FLAG::LEU2 TOP3-V5::TRP1 RMII-HA::HIS3 | This study |
| JMY1918 | W303-1a rmi1-10::KAN | This study |
| NJY1959 | W303-1a SGS1-FLAG::LEU2 TOP3-V5::TRP1 rmi1-10::KAN | This study |
| JMY1960 | MATaade2-1 ura3-1 his3-11,15 lys2 trp1-1 can1-100 top3-2::HIS3 rmi1-10::KAN | This study |
| JMY1961 | W303-1b top3-2::HIS3 rmi1-10::KAN sgs1-3::TRP1 | This study |
| JMY1962 | W303-1b top3-2::HIS3 rmi1-10::KAN::loxP sgs1-3::TRP1 | This study |
| JMY1963 | MATaade2-1 ura3-1 his3-11,15 leu2-3,112 lys2 trp1-1 can1-100 rmi1-10::KAN sgs1-3::TRP1 | This study |
| JMY1964 | W303-1a rmi1-11::HGR | This study |
| NJY1975 | W303-1b sgs1-11::KAN TOP3-V5::TRP1 RMI1-HA::HIS3 | This study |
| NJY1977 | W303-1b SGS1-FLAG::LEU2 top3::loxP RMI1-HA::HIS3 | This study |
| JMY1996 | K1875 rmi1-11::HGR | This study |
| JMY1997 | K1875 rmi1-11::HGR sgs1-20::HGR | This study |
| JMY2012 | W303-1b esc2-10::KAN | This study |
The epitope-tagging vectors pNJ1569, pNJ2565, and pNJ7134 were constructed by fusing the Sgs1, Top3, and Rmi1 ORFs to 3× FLAG, V5, or 3× hemagglutinin (HA) coding regions, respectively. Fusions replaced the natural stop codons with a NotI linker encoding the three amino acids (GGR) between the ORF and the epitope tag. The plasmids pNJ1569, pNJ2565, and pNJ7134 are based on the integrating plasmids pRS405, pRS404, and pRS403 (48), respectively, and were targeted for site-specific integration by restriction enzyme digestion within the respective ORF prior to transformation. The T7 expression plasmids pKR1566, pSAS402, and pNJ7135 are based on pET11a (50) and express the C-terminal epitope-tagged Sgs1-V5, Top3-V5, and Rmi1-3xHA, respectively. The double expression plasmid pAM1567 expresses Sgs1-V5 and untagged Top3. To create double expression plasmids, the T7 promoter and ORF of pSAS402 were subcloned on a BstEII/BamHI fragment into the BstEII/BglII sites of pNJ7135 to create pCS7141. Similarly, the BstEII/BamHI fragment of pNJ1566 was inserted into the BstEII/BglII site of pNJ7135 to create pNJ1572. A triple expression plasmid (Sgs1-V5, Top3, and Rmi1-3xHA) was constructed by moving the BstEII/BamHI fragment of pAM1567 into the BstEII/BglII sites of pNJ7135 to create pNJ1571. To express His6-Rmi1-3xHA, the His6 encoding region of pET28a was moved on an NcoI-NdeI fragment and inserted upstream of the Rmi1 ORF of pNJ7135 to create pNJ7138.
Yeast extracts, immunoprecipitations (IPs), and gel filtration chromatography.
Total yeast cell extracts were prepared essentially as described previously (46). Cells from 2 liters of culture were washed in water followed by a wash in 2× extraction buffer (100 mM HEPES [pH 7.4], 400 mM sodium acetate, 100 mM magnesium acetate, 1 mM EDTA, 20% glycerol, 1 mM sodium orthovanadate, 0.1% 2-mercaptoethanol) plus the following protease inhibitors: pepstatin, 10 μg/ml; leupeptin, 5 μg/ml; benzamidine, 10 mM; bacitracin, 100 μg/ml; aprotinin, 20 μg/ml; and sodium metabisulfite, 10 mM. The cells were packed into a syringe and extruded through an 18-gauge needle into liquid nitrogen. The frozen material was then placed in ground dry ice and ground in a coffee grinder for 3 min. This mixture was placed on ice and when the remaining dry ice had sublimed, the cells were resuspended in 1 volume of cold 2× extraction buffer plus protease inhibitor. The insoluble material was pelleted by centrifugation at high speed in a microcentrifuge for 10 min at 4°C, and the soluble portion was taken as extract. Typical extracts contained 10-mg/ml protein, which were aliquoted and stored at −80°C.
IPs were performed at 4°C essentially as previously described (35). Two milligrams of total protein was incubated for 1 h with 40 μl protein G-Sepharose beads conjugated to anti-FLAG mouse monoclonal antibodies (Sigma) or with 1 μl of anti-HA (5 μg/μl; Roche) or anti-V5 (1 μg/μl; Invitrogen) monoclonal antibodies. Thirty microliters of protein A-Sepharose beads (Amersham-Pharmacia) was added to each sample, followed by rocking for 1 h. The immune complexes were then washed three times with 1 ml of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 1% [vol/vol] NP-40, 0.5% [wt/vol] deoxycholate, 0.1% [wt/vol] sodium dodecyl sulfate [SDS]. Bound proteins were resuspended in Laemmli buffer and resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Following SDS-PAGE, the gels were transferred to nitrocellulose membranes and treated with either anti-FLAG (Sigma), anti-V5, or anti-HA as the primary antibody at a 1:10,000 dilution. Blots were then treated with anti-mouse horseradish peroxidase-conjugated secondary antibody (1:10,000; Gibco-BRL) and developed with chemiluminescence reagents (Pierce), prior to capturing the image on a chemiluminescence camera (Fujifilm LAS3000).
Superose 6 HR10/30 chromatography was carried out with an AKTA fast-performance liquid chromatography system (GE Healthcare). The running buffer was buffer A (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.01% [vol/vol] NP-40, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol [DTT]) containing 150 mM NaCl but lacking glycerol. Approximately 400 μg protein was fractionated at a flow rate of 0.4 ml/min. Fractions were collected, precipitated with trichloroacetic acid, and analyzed by immunoblotting as described above.
Expression and purification of recombinant Rmi1.
Rmi1 protein was expressed as a His6-Rmi1-3xHA fusion protein from plasmid pNJ7138, which was transformed into E. coli BL21(DE3)-RIL cells (Stratagene). Freshly transformed colonies were pooled and grown by being shaken in 1 liter of LB media containing 0.1 mg/ml ampicillin at 37°C to an optical density at 600 nm of 0.4. Expression of the recombinant protein was induced by addition of isopropyl-1-thio-d-galactopyranoside to a final concentration of 0.4 mM for 2 h at 37°C. Induced cells were pelleted and resuspended in 100 ml buffer A containing 50 mM NaCl. Cell suspensions were incubated with 1 mg/ml lysozyme for 15 min at 4°C and then sonicated three times for 1 min each with a Branson sonifier 450 microtip at setting 4 and a 60% duty cycle. The lysate was centrifuged at 15,000 rpm in an SS34 rotor at 4°C for 15 min. The insoluble pellet was resuspended in 200 ml TEK buffer (100 mM Tris-HCl [pH 8.7], 200 mM KCl, 1 mM EDTA, 1 mM DTT) using sonication to disrupt aggregates and centrifuged as above. This pellet was washed as above in TEK containing 0.1% NP-40, followed by a wash in TEK, and it was then solubilized in 50 ml TEK containing 10 mM DTT and 8 M urea. After centrifugation, the soluble extract was dialyzed over a 24-h period against a gradient of TEK buffer containing 8 M to 0.25 M urea. The dialyzate was centrifuged, and the supernatant was dialyzed against N buffer (25 mM Tris-HCl [pH 8.0], 500 mM NaCl, 0.01% [vol/vol] NP-40, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM DTT) containing 10 mM imidazole. The proteins were then batch bound to 5 ml Ni Pro-Bond resin (Invitrogen). After 3 h, the resin was poured into a column and washed with 10 column-volumes of N buffer containing 10 mM imidazole. The column was then successively eluted with 6 half-column volumes of N buffer containing 50, 100, 200, and 500 mM imidazole. Peak Rmi1 proteins levels were found in the 200 mM fractions. The Rmi1 eluate was dialyzed against buffer A containing 200 mM NaCl and stored at −80°C. Protein concentrations were determined by Bradford reagent (Bio-Rad) using bovine serum albumin (BSA) as a standard. Rmi1 protein was estimated to be at least 95% pure as judged by Coomassie blue staining.
Electrophoretic mobility shift assay (EMSA) and DNA substrates.
The specified amounts of Rmi1 protein were incubated with 50 fmol 32P-labeled DNA substrate in a final volume of 25 μl containing 25 mM HEPES (pH 7.5), 50 mM NaCl, 0.2 mM dithiothreitol, 0.1% NP-40, 1.0 mg/ml BSA, and 5.8% glycerol at 25°C for 45 min. Loading dye was added to a final concentration of 8% glycerol and 0.25% bromophenol blue. Gel electrophoresis was modified from a previously described method (34). After the gel was loaded, the sample was electrophoresed at 10 V/cm through a 7.5% polyacrylamide gel (29:1 acrylamide:bis) in 12.5 mM Tris-HCl (pH 8.7), 55 mM glycine, and 1.5 mM EDTA at 4°C. The gel was fixed in 50% ethyl alcohol-10% acetic acid for 15 min, dried, and visualized with a Molecular Dynamics PhosphorImager.
DNA substrates were prepared from deoxyoligonucleotides (oligonucleotides) of approximately 50 nucleotides as previously described (24). Specific substrates were assembled as follows: linear double-stranded DNA (dsDNA), oligonucleotides 892 and 897; Y, oligonucleotides 888 and 891; 3′ flap, oligonucleotides 888, 891, and 994; 5′ flap, oligonucleotides 888, 891 and 992; pseudoreplication fork, oligonucleotides 888, 891, 992 and 994; Holliday junction, oligonucleotides 892, 893, 894, and 895.
UV cross-linking assay.
To identify proteins cross-linked to substrate DNA, 30 ng of purified Rmi1 was incubated in a final volume of 25 μl with 20,000 cpm of the indicated 32P-labeled substrate DNA at 25°C for 15 min under the following conditions: 25 mM HEPES (pH 7.5), 50 mM NaCl, 0.2 mM DTT, 0.1% NP-40, 0.1 mg/ml BSA, and 5.8% glycerol. Binding reaction mixtures were cross-linked with UV light at a dose of 1,000 J/m2 in a Stratalinker (Stragene) and incubated at 4°C with either 1 μl anti-HA or anti-V5 antibodies for 1 h in 200 μl RIPA buffer. Protein A-Sepharose resin (Amersham) was added to each reaction mixture, and the mixture was rotated at 4°C for 1 h and washed three times with 1 ml RIPA buffer. The washed beads were resuspended in Laemmli loading buffer, and the bound products were either loaded directly for gel electrophoresis without being heated or boiled 5 min before being loaded. The products were resolved on a 15% SDS-PAGE gel, fixed, dried, and visualized as above.
RESULTS
rmi1 mutants display the same spectrum of synthetic lethality as sgs1 strains.
Using genome-wide SGA screens, Brown and colleagues identified four genes, in addition to SGS1 and TOP3, that are required for viability in strains lacking MUS81 or MMS4 (2). Among these interacting genes, RMI1 and ESC2 were considered candidates for the SGS1-TOP3 pathway because they displayed many of the same interactions that were reported for sgs1 and top3 mutants (52). To test this idea, we reasoned that if these genes were in the SGS1-TOP3 pathway, then esc2 and rmi1 mutations should exhibit synthetic lethality with mutations in SLX1, -4, -5, and -8, which were isolated in the same SGS1 synthetic lethal screen as MUS81-MMS4 (36). After the appropriate diploid strains were sporulated and microdissected, their meiotic products were analyzed. As shown in Fig. 1A, we obtained viable esc2 slx1 double mutants. Because these double mutants were obtained at the expected frequency and lacked any significant growth defect, we eliminated ESC2 from further analysis. In the case of RMI1, however, we were unable to isolate any rmi1 slx1 double-mutant spore clones that were either healthy or slow growing (Fig. 1B and Table 2). This strong synthetic lethal phenotype is typical of the interactions between sgs1 and the slx mutations (36). A more stringent test of whether RMI1 functions in the SGS1-TOP3 pathway relates to the specificity of its synthetic interaction with mus81 and the observation that sgs1 mus81 synthetic lethality is dependent on HR. In agreement with Bellaoui et al. (2), we were unable to isolate any viable rmi1 mus81 or rmi1 mms4 double mutants (Fig. 1C and D); however, we did obtain rmi1 mus81 rad51 triple mutants, as well as rmi1 mms4 rad55 strains at about the expected frequency (Fig. 1C and D and Table 2). As with sgs1 mus81 rad51 strains (13), these rmi1 mus81 rad51 triple mutants displayed a slow-growth phenotype, indicating that RMI1 or MUS81 must have some function independent of RAD51. Lastly, rmi1 mutants displayed synthetic-lethal interactions with SLX4 and SLX5 and, like sgs1 mutants, these synthetic lethal interactions were not suppressed by loss of rad54 or rad51, respectively (Fig. 1E and F and Table 2). A summary of these results (Table 2) reveals a precise overlap between the synthetic lethal interactions of RMI1 and SGS1 with respect to MUS81, MMS4, SLX1, SLX4, SLX5, and SLX8, as well as the role of HR in mediating the lethality with MUS81 and MMS4 (1, 13, 36).
FIG. 1.
Synthetic lethal interactions with rmi1 identify a potential member of SGS1-TOP3 pathway. Diploid strains were isolated from the indicated crosses and sporulated. Following sporulation, the spores were microdissected and allowed to germinate for 3 days at 30°C. A tetrad consists of one vertical column. (A) Strains: JMY2012 × JMY1452. (B) Strains: JMY1918 × JMY361. (C) Strains: JMY1964 × JMY1459. (D) Strains: JMY1964 × JMY1457. (E) Strains: JMY1964 × VCY1515. (F) Strains: JMY1918 × JMY1455. Spore clones marked “r” contain the indicated rad mutation.
TABLE 2.
Summary of rmi1 genetic interactions
| Strain crossed to rmi1 mutant | No. of viable spores/no. expected | No. of viable double mutants/no. expecteda | No. of viable “rad” triple mutants/no. expecteda |
|---|---|---|---|
| slx1 | 29/36 | 0/7 | NA |
| slx4 | 41/56 | 0/13 | NA |
| slx4 rad54 | 77/96 | 0/7 | 0/10 |
| mms4 | 38/52 | 0/14 | NA |
| mms4 rad55 | 48/56 | 0/6 | 2/2 |
| mus81 | 29/44 | 0/10 | NA |
| mus81 rad51 | 62/76 | 0/11 | 8/9 |
| slx5 | 65/92 | 0/25 | NA |
| slx5 rad51 | 59/80 | 0/14 | 0/7 |
| slx8 | 24/36 | 0/10 | NA |
The number of mutants expected for a given genotype is inferred from the genotype of surviving spores. NA, not applicable.
sgs1 suppresses rmi1 phenotypes.
Epistasis analysis was used to test whether RMI1 functioned in the SGS1-TOP3 pathway. An sgs1 top3 rmi1 heterozygous diploid was sporulated, and tetrads were microdissected and analyzed. As previously observed (16), top3 spore clones formed small colonies compared to sgs1 cells, and sgs1 top3 clones were almost as large as sgs1 clones (Fig. 2A). We observed that rmi1 spore clones were also smaller than sgs1, although in general not as small as top3. Surprisingly, sgs1 rmi1 spore clones were about the same size as sgs1 colonies, indicating that sgs1 is epistatic to rmi1 with respect to its slow-growth phenotype (Fig. 2A). This conclusion is confirmed by streaking for single colonies (Fig. 2B), by spot dilution assays (Fig. 2C), and by growth rate measurements in liquid culture (Table 3). In liquid cultures, the rmi1 strain exhibited a doubling time (DT) of 156 min, while the DT of an sgs1 rmi1 strain approximated that of an sgs1 strain (120 min). In addition, the sgs1 top3 rmi1 triple mutants (DT, 129 min) grow nearly as well as sgs1 strains, confirming that sgs1 is epistatic to both top3 and rmi1 (Fig. 2C; Table 3).
FIG. 2.
Loss of SGS1 suppresses rmi1 growth and DNA damage phenotypes. (A) Spores from the indicated cross (JMY1918 × NJY1236) were microdissected in vertical columns and allowed to germinate for 3 days at 30°C. The sizes of the spore clones reflect the strains' relative growth rates. (B) The indicated haploid strains were streaked on YPD plates to single colonies and allowed to grow for 3 days at 30°C to demonstrate their relative growth rates and the accumulation of suppressor mutations. (C) The indicated haploid strains were replica plated in 1:10 serial dilutions on YPD plates or YPD plates containing 0.012% MMS and incubated for 3 days at the indicated temperatures. Strains shown in panels B and C: W303-1a, JMY1918, JMY1963, NJY506, JMY1961, JMY1962, JMY1960, and NJY372.
TABLE 3.
Doubling times of selected mutantsa
| Genotype | Doubling time (min) | Relative rate |
|---|---|---|
| Wild type | 108 | 1.0 |
| sgs1 | 120 | 1.1 |
| rmi1 | 186 | 1.7 |
| sgs1 rmi1 | 120 | 1.1 |
| top3 | 273 | 2.5 |
| top3 sgs1 | 144 | 1.3 |
| rmi1 top3 | 240 | 2.2 |
| rmi1 top3 sgs1 | 129 | 1.2 |
Strains are W303-1a, NJY506, JMY1918, NJY372, NJY1236, JMY1960, and JMY1962.
As shown in Fig. 2C, epistasis with sgs1 extends to additional phenotypes. The rmi1 and top3 strains show a slight temperature-sensitive growth defect as well as sensitivity to MMS (7). The temperature and MMS sensitivity of rmi1 or rmi1 top3 double mutants is suppressed in sgs1 backgrounds (Fig. 2C), although in both cases growth was not fully restored to sgs1 levels. Thus, RMI1 appears to have roles independent of SGS1. This phenotype is reminiscent of TOP3, which has also been observed to have SGS1-independent functions (42). It is also interesting that rmi1 mutants resemble top3 strains in that sporulation is compromised in each of these backgrounds (55). Homozygous rmi1 diploids display a sporulation frequency of only 1% compared with 35% for wt cells.
In contrast to sgs1 mutations, loss of RMI1 had no significant effect on the temperature or MMS sensitivity of top3 strains (Fig. 2C), nor did it affect the slow growth of top3 spore clones (Fig. 2A). These results indicate that top3 is epistatic to rmi1. Following continued growth, however, the effect of rmi1 on top3 growth was less clear. The rmi1 top3 strains gave rise to fast-growing suppressors at a high frequency (data not shown), and in liquid cultures their growth rate (DT, 240 min) was slightly faster than that of top3 (DT, 273 min) (Table 3). Therefore, it appears that spontaneous suppressors may be responsible for the slightly faster growth rate of rmi1 top3 cells in liquid media compared to their initial growth rate as spore clones.
Because of its presumed role in the SGS1-TOP3 pathway, we tested whether RMI1 was required for genome stability. Genome stability was assayed by measuring recombination rates in a strain carrying two selectable markers (ADE2 and URA3) that were independently integrated at the rRNA genes, as well as a third marker (CAN1) integrated at LYS2 (27). These markers are most often lost by means of excision recombination leading to ade2, ura3, or can1 phenotypes. An increased rate of ADE2 marker loss was immediately apparent from the elevated frequency of red (ade2) colonies and red sectors in rmi1 colonies (Fig. 3, compare WT and rmi1 panels). The frequency of red colonies and sectors was reduced in rmi1 sgs1 strains (Fig. 3), consistent with sgs1 suppression. Table 4 quantifies the URA3 and CAN1 recombination data. Compared to wt cells, the rmi1 mutants displayed a 28-fold (URA3) or 21-fold (CAN1) increase in marker excision rate. In each case, these large increases in recombination are reduced to sgs1 levels in the rmi1 sgs1 background. We conclude that loss of RMI1 results in genome instability that is suppressed by eliminating SGS1.
FIG. 3.
rmi1 mutants display a hyperrecombination phenotype that is suppressible by sgs1. The indicated strains were taken from selective media lacking adenine, grown nonselectively for 50 generations, plated for single colonies, and allowed to grow for 3 days before the colonies were photographed. Each strain contains a single copy of ADE2 integrated in the rRNA genes and cells turn from white (ADE2) to red (ade2) as the marker is lost through recombination. Two panels (low and high magnifications) are shown for each strain. Strains are WT, K1875; rmi1, JM1996; sgs1, NJY540; and rmi1 sgs1, JMY1997.
TABLE 4.
rmi1 hyperrecombination is suppressed by loss of SGS1a
| Genotype |
URA3
|
CAN1
|
||
|---|---|---|---|---|
| Loss rate at RDN1b | Fold increase | Loss rate at LYS2 | Fold increase | |
| wt | 1.3 × 10−7 | 1 | 6.6 × 10−8 | 1 |
| sgs1 | 9.6 × 10−7 | 4.8 | 42 × 10−8 | 6.4 |
| rmi1 | 36 × 10−7 | 28 | 140 × 10−8 | 21 |
| rmi1 sgs1 | 3.2 × 10−7 | 2.5 | 30 × 10−8 | 4.5 |
Strains are K1875, NJY540, JMY1996, and JMY1997.
Marker loss rates (per cell per generation) were calculated by the method of the median as previously described (49).
Rmi1 binds Sgs1 and Top3 in vivo and in vitro.
The genetic results described above suggest that Rmi1 may physically interact with Sgs1 and/or Top3 in yeast. To test this idea, we placed a different C-terminal epitope tag on each single-copy gene in yeast to create strain NJY1906. The SGS1-FLAG, TOP3-V5, and RMI1-HA alleles present in strain NJY1906 were shown to be functional based on the fact that MUS81 is dispensable in this background (see Materials and Methods). Following preparation of a whole-cell extract, we immunoprecipitated each protein under stringent RIPA washing conditions and detected coprecipitating proteins by immunoblotting. When Sgs1 was immunoprecipitated from extracts of strain NJY1906 with anti-FLAG antibody, both Top3 and Rmi1 coprecipitated, albeit with weak signals (Fig. 4A, lane 1). Consistent with this result, reciprocal IPs of either Top3 or Rmi1 coprecipitated the remaining two proteins but with more intense signals (Fig. 4A, lanes 2 and 3). For example, when Top3 was immunoprecipitated, both Sgs1 and Rmi1 strongly coprecipitated. The more intense Sgs1 signal obtained from Top3 IPs was previously observed (14) and is thought to be due to an excess of Top3 over Sgs1 protein. These results are consistent with two possibilities. Either Sgs1, Top3, and Rmi1 exhibit a set of pairwise interactions or they exist as a heteromeric complex of the three proteins.
FIG. 4.
Rmi1 binds stably to Sgs1-Top3 in yeast. (A) Unless deleted, each strain contains epitope-tagged versions of Sgs1, Top3, and Rmi1 proteins. Cell extracts were prepared from the indicated strains and subjected to immunoprecipitation with the appropriate antibody as follows: S, Sgs1 IP (α-FLAG); T, Top3 IP (α-V5); R, Rmi1 IP (α-HA). IPs were then immunoblotted to detect Sgs1 (top), Top3 (middle), and Rmi1 (bottom). Strains: WT, NJY1906; Δsgs1, NJY1975; Δtop3, NJY1977; and Δrmi1, NJY1959. (B) Extract from NJY1906 was fractionated by Superose 6 gel filtration chromatography. Fractions with the indicated retention volumes (in milliliters) were precipitated and subjected to immunoblotting to detect Sgs1, Top3, or Rmi1, as indicated. Size markers (top of panel) indicate retention volumes for the indicated molecular mass standards in kilodaltons.
To test the genetic requirements for these interactions, we performed co-IPs from strains in which one of the genes had been deleted. Top3 and Rmi1 coprecipitated from extracts lacking Sgs1 (Fig. 4A, lanes 4 to 6), indicating that Top3 and Rmi1 form a stable complex even in the absence of Sgs1. In contrast, Sgs1 IPs coprecipitated little or no Rmi1 in the absence of Top3 (Fig. 4A, lane 7), and more noticeably, Rmi1 IPs failed to coprecipitate Sgs1 in these extracts (Fig. 4A, lane 9). Both Sgs1 and Rmi1 proteins were present in extracts made from top3 cells, although Rmi1 abundance may have been reduced compared to that of the wt (Fig. 4A, lane 9). This result indicates that Top3 is required for an optimal Sgs1-Rmi1 interaction in yeast. Similar to this result, Sgs1 IPs coprecipitated little or no Top3 in the absence of Rmi1 (Fig. 4A, lane 10); in the reciprocal test, Top3 IPs failed to coprecipitate Sgs1 in this background (Fig. 4A, lane 11). Given that Sgs1 and Top3 proteins were stable in the rmi1 mutant, we conclude that Rmi1 is required for optimal Sgs1-Top3 interaction in vivo. The above data are most simply explained by a model in which Sgs1, Top3, and Rmi1 form a heteromeric complex in which the binding of the Rmi1-Top3 subcomplex to Sgs1 is codependent.
The question of whether Sgs1, Top3, and Rmi1 exist in a single heteromeric complex was tested directly by gel filtration chromatography of NJY1906 extracts. Following fractionation on a Superose 6 column, individual fractions were immunoblotted for the presence of the three epitope-tagged proteins. As shown in Fig. 4B, the three proteins coeluted in the high-molecular-weight (high-MW) region of the profile, with the peak of elution judged to be 8.4 ml. This elution volume corresponds to a native molecular mass of approximately 2.5 MDa, which is somewhat larger than the previously estimated values for native Sgs1-Top3, Rqh1-Top3, or recombinant BLM (14, 26, 30). The coelution of these proteins provides the strongest evidence that they form a heteromeric complex in yeast. In addition to this result, we note the presence of much weaker Top3 and Rmi1 signals eluting with retention volumes of about 16.5 ml. These signals suggest the presence of minor amounts of Top3-Rmi1 dimer, separate from Sgs1.
To test whether Rmi1 interacts directly with the other two subunits, we expressed epitope-tagged Sgs1, Top3, and Rmi1 in E. coli and analyzed them by co-IP. Bacterial extracts were prepared from cells expressing Rmi1 and Top3, and these proteins were individually immunoprecipitated. As shown in Fig. 5A and B, a portion of each protein coimmunoprecipitated with the other. This indicates that Rmi1 and Top3 interact stably in the absence of other yeast proteins. Extracts were also prepared from cells expressing Rmi1 and Sgs1, and these proteins were individually immunoprecipitated. Under these conditions, Rmi1 was found to communoprecipitate with Sgs1 (Fig. 5C). Consistent with this result, Sgs1 communoprecipitated with Rmi1 in the reciprocal experiment (Fig. 5D); however, the signal was significantly weaker in this direction. This difference is due at least in part to the degradation of Sgs1 in this system. We consistently observed a low abundance of full-length Sgs1 protein (relative molecular mass, 220 kDa) compared to truncated products (Fig. 5D and data not shown). The interaction between Rmi1 and Sgs1 was also observed when all three proteins were coexpressed (Fig. 5E and F). In this case, full-length Sgs1 was associated with Rmi1 IPs. Taken together, these results indicate that Rmi1 can bind directly to Sgs1 and Top3 when overexpressed in E. coli.
FIG. 5.
Rmi1 binds Top3 and Sgs1 in recombinant form. Cell lysates were prepared from bacterial strains expressing the indicated epitope-tagged proteins in the left column (Extract). Extracts were immunoprecipitated with the indicated antibodies, washed three times in RIPA buffer, and immunoblotted with either anti-HA (left) or anti-V5 (right). (A and B) Rmi1-HA and Top3-V5 were coexpressed from plasmid pCS7141. (C and D) Rmi1-HA and Sgs1-V5 were coexpressed from plasmid pNJ1572. (E and F) Sgs1-V5, untagged Top3, and Rmi1-HA were coexpressed from plasmid pNJ1571. (G) Rmi1-HA was expressed alone from plasmid pNJ7135. EXT is a control representing 1/20 of the amount of extract subjected to IP. ΔSgs1-V5 indicates the position of a 150-kDa breakdown product of full-length Sgs1-V5. Molecular mass markers are shown to the left of each blot in kilodaltons.
Rmi1 is a conserved protein.
The Rmi1 protein has a predicted molecular mass of 26.9 kDa and a pI of 10.3. The charged nature of the protein may explain why it migrates anomalously on SDS-PAGE with a relative molecular mass of 35 kDa. Analysis of its amino acid sequence revealed no obvious protein motifs using the PROSITE, Pfam, or eMOTIF programs. BLAST analysis using Rmi1 as query identified homologous proteins of similar size in at least seven fungal species. BLAST analysis using the more highly conserved C-terminal region of these proteins as a query identified numerous metazoan homologs, including two each in Caenorhabditis elegans and humans. In general, the metazoan homologs were about twice the size of the fungal proteins. Multiple sequence alignment of several homologs revealed a region of limited sequence conservation corresponding to the C terminus of yeast Rmi1 (Fig. 6). Within this domain, there were only isolated invariant residues such as those represented by the yeast L150, G174, and K176 residues. Although amino acid sequence analysis did not provide significant information as to its function, the conservation of Rmi1 homologs in multiple species suggests that it may play an important role in RecQ-Top3 function in higher cells.
FIG. 6.
Amino acid sequence alignment of Rmi1 homologs. Eight homologous sequences, listed by genus and species, were aligned using the Multalign program (9). Identical (reverse type) or conserved residues (shaded) were identified with BoxShade 3.21. The four metazoan proteins have C-terminal extensions of about 300 aa that are not shown. GenBank sequence numbers are as follows: S. cerevisiae, 6325233; Kluyveromyces lactis, 49642738; Neurospora crassa, 28919965; S. pombe, 1177350; Gallus gallus, 53136712; Homo sapiens, 13376427; C. elegans, 25150267; Aradopsis thaliana, 42573433.
Rmi1 is a structure-specific DNA binding protein.
To characterize Rmi1's biochemical function we expressed Rmi1-HA in E. coli as a fusion to an N-terminal six-histidine tag. Following induction, the expressed protein was found almost entirely in the pellet fraction, regardless of induction temperature (data not shown). This material was solubilized in urea, refolded by gradient dialysis, and then purified by Ni-affinity chromatography. As shown in Fig. 7A, the purified protein (calculated molecular mass, 33.1 kDa) migrated at a relative molecular mass of about 40 kDa by SDS-PAGE. Because of its potential role in DNA metabolism, we tested Rmi1 for DNA binding activity. An EMSA was used to detect the binding of Rmi1 to single-stranded DNA (ssDNA) or dsDNA (Fig. 7B). Relatively high concentrations of Rmi1 were capable of shifting a small portion of these probes to a position near the top of the gel. With increasing protein concentrations, these signals migrated progressively more slowly until they were unable to enter the gel. This progressive retardation suggests that Rmi1 either binds to multiple sites on the probe or aggregates at high protein concentrations. It should be noted that even at maximum protein concentrations Rmi1 was unable to bind a significant fraction of either of these substrates. In contrast, Rmi1 bound well to a HJ substrate. Compared to dsDNA, HJ binding could be detected at a 10-fold-lower protein concentration and the substrate was quickly saturated (Fig. 7B, right). To compare the relative affinity of Rmi1 to dsDNA or HJ substrates, we performed a competition experiment in which Rmi1 protein was incubated with radiolabeled HJ probe together with increasing amounts of unlabeled competitor DNA. Whereas 5 pmol of cold HJ was sufficient to compete about 50% binding (Fig. 7C, lane 7), a 30-fold-higher level of cold dsDNA had little or no effect on the binding Rmi1 to a HJ (Fig. 7C, lane 20). The effectiveness of competition by dsDNA was not improved by using a larger competitor of 90 bp (data not shown). This analysis was extended by using a variety of unlabeled branched structures as competitor. The resulting data were quantified and are presented in Fig. 7D. Branched DNA by itself is not sufficient for Rmi1 binding, since a simple branched molecule, a Y structure, was unable to compete significantly with HJ binding. Single-stranded DNA was similarly unable to compete (data not shown). As expected, unlabeled HJ DNA was an effective competitor, although a pseudoreplication fork structure was only threefold-less effective than the HJ. Two related but less-complex structures, the 3′ flap and 5′ flap, were intermediate in their effectiveness. These structures were 10-fold-less effective competitors than HJ DNA but were significantly better than the Y structure. Taken together, these results indicate that Rmi1 is a structure-specific DNA binding protein with highest affinity for HJ DNA.
FIG. 7.
Rmi1 is a structure-specific DNA binding protein. (A) Three micrograms of purified Nce 4 and the indicated MW markers were subjected to 17% SDS-PAGE and Coomassie blue staining. (B) A total of 20,000 cpm (50 fmol) of the indicated radiolabeled DNA probe was incubated with 0, 0.125, 0.25, 0.5, 1.0, 2.0, 4.0, or 8.0 μg of Rmi1 prior to electrophoresis as described in Materials and Methods. (C) Competition experiments were set up as in standard EMSAs except that 0, 0.05, 0.15, 0.5, 1.5, 5, 15, 50, or 150 pmol of the indicated competitor DNAs were premixed with 50 fmol of labeled HJ probe prior to the addition of Rmi1. Neither competitor nor protein was added to mock reaction mixtures (lanes 1 and 11), which were used to identify the free probe. (D) Competition experiments were performed as in the results shown in panel C but included Y (closed triangles), 5′ flap (open squares), 3′ flap (open circles), and pseudoreplication fork (closed circles), in addition to linear dsDNA (closed squares) and HJ (closed diamonds) competitors. The fraction of free and bound probe was determined by quantifying the signal from phosphorimager scans with IP LabGel software. For each competitor, the percentage of initial HJ binding (obtained in the absence of unlabeled DNA) is plotted as a function of competitor DNA input. Substrates are presented graphically, with arrowheads indicating 3′ ends and filled dots as 5′ ends.
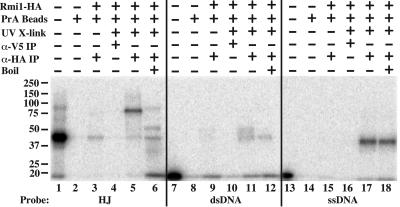
As an alternative method of measuring Rmi1 DNA binding activity, we used a UV cross-linking assay. This assay not only allows the use of lower concentrations of protein and DNA but is capable of detecting protein-DNA interactions that are too weak to be seen by EMSA, which requires stable DNA binding (44). Rmi1 protein was incubated with radiolabeled DNA probes, UV irradiated, and purified away from unbound probe by IP. Protein-DNA complexes were then detected by SDS-PAGE and phosphorimaging. Compared to dsDNA, the HJ probe was efficiently cross-linked to Rmi1 and resulted in a signal that migrated at approximately 80 kDa (Fig. 8, lane 5). This size is expected from the sum of the relative migrations of unbound HJ probe (40 kDa) (Fig. 8, lane 1) and Rmi1-HA (40 kDa). The 40-kDa signal in lane 5 is most likely due to cross-linking to low amounts of contaminating ssDNA (see below). To confirm these assignments, the sample was boiled prior to SDS-PAGE. This resulted in the release of some free ssDNA (relative molecular mass, 20 kDa), a more intense 40-kDa band, and some higher-molecular-mass bands that may represent Rmi1 protein cross-linked to multiple strands of the HJ (Fig. 8, lane 6). Rmi1 bound poorly to dsDNA under these conditions and produced two weak bands at 40 and 50 kDa (Fig. 8, lane 11). The 50-kDa band appears to be due to dsDNA because it collapses upon being boiled to a 40-kDa band, which is consistent with its cross-linking to one strand of ssDNA. Surprisingly, Rmi1 bound most strongly to ssDNA by this assay (Fig. 8, lane 17), and all of the signal resulting from this reaction was resistant to boiling. We conclude that Rmi1 interacts preferrentially with ssDNA in a UV cross-linking assay. This result suggests that Rmi1 may interact weakly or transiently with the nucleotide bases of ssDNA, even though it may not be observed by EMSA. Such an activity is reminiscent of the monomeric ssDNA binding domains of RPA, which cross-link to ssDNA as monomers but must be tandemized to bind ssDNA stably (44). The ability of Rmi1 to interact with ssDNA may play a role in recognizing or creating ssDNA for strand passing by Sgs1-Top3.
FIG. 8.
UV cross-linking detects Rmi1 interactions with HJ and ssDNA substrates. The indicated radiolabeled probes (20,000 cpm) were incubated without (lanes 2, 8, and 14) or with (lanes 3 to 6, 9 to 12, and 15 to 18) 30 ng Rmi1-HA and either treated with UV (lanes 4 to 6, 10 to 12, 16 to 18) or mock treated (lanes 3, 9, and 15). The reactions were then processed for immunoprecipitation with nonspecific (anti-V5; lanes 4, 10, and 16) or specific (anti-HA; lanes 3, 5, 6, 9, 11, 12, 15, 17, and 18) antibodies under RIPA buffer conditions. The immunoprecipitates were resuspended in Laemmli buffer and either boiled (lanes 6, 12, and 18) or loaded directly (lanes 1 to 5, 7 to 11, and 13 to 17) for 15% SDS-PAGE. Approximately 1/20 of the free probe used per binding reaction mixture was loaded in lanes 1, 7, and 13. PrA Beads, protein A beads used for IP.
DISCUSSION
Synthetic lethal screens are powerful tools for identifying overlapping pathways in yeast. Recent global screens using functional genomics approaches have provided a wealth of data from which interaction maps can be assembled (43, 51, 52). Our laboratory previously identified six mutants that gave a strong synthetic lethal phenotype when combined with mutations in SGS1 (36), and it seemed likely that a search in the “reverse” direction could identify genes in the SGS1-TOP3 pathway. In this effort, the global SGA screens against deletions of MUS81 and MMS4 provided an exceptionally powerful resource (2). Not only did these two SGA screens approach saturation, but the interactors were likely to be valid since they were directed against two genes known to encode a single enzyme. Of the two candidates we tested, rmi1 mutants gave the expected result when crossed to the previously untested strong SGS1 synthetic-lethal interactors (SLX1, SLX4, SLX5, and SLX8). The link between RMI1 and SGS1-TOP3 became even more probable given the requirement for HR in rmi1 mus81 synthetic lethality.
The rmi1 and top3 mutants are similar in that both show slow growth, DNA damage sensitivity, and hyperrecombination phenotypes. This similarity suggested that these genes lie in the same genetic pathway. Analysis of the rmi1 top3 double mutant confirmed this idea that top3 was epistatic to rmi1 with respect to temperature and MMS sensitivity. With respect to the slow-growth phenotype, RMI1 and TOP3 appear to function in the same genetic pathway downstream of SGS1, since the rmi1 top3 growth defect is completely suppressed in the sgs1 background. Although it is possible that these two genes have separate functions downstream of SGS1, the physical interaction between Rmi1 and Top3 suggests that they cooperate to perform a single function that is not entirely eliminated by the loss of one protein alone. In this case, Top3 would appear to have the more important role, since its phenotypes are more severe. An alternative possibility is that Rmi1 is simply a negative regulator of Sgs1 DNA helicase activity. In this case, the rmi1 slow-growth phenotype might due to excess Sgs1 DNA helicase activity, which is known to inhibit growth in otherwise-wt cells (35). This excess Sgs1 helicase activity would exacerbate the top3 phenotype in top3 rmi1 cells and be suppressible by the loss of SGS1. This hypothesis predicts that extra copies of SGS1 should be tolerated in cells overexpressing Rmi1.
Several lines of evidence indicate that Sgs1, Top3, and Rmi1 form a heteromeric complex. Coimmunoprecipitation experiments in yeast revealed that there are at least pairwise interactions between all three proteins, and the resistance to 0.1% SDS in these IPs indicates that the interactions are robust. We observed no requirement for Sgs1 in forming the Rmi1-Top3 complex in either yeast or bacteria. On the other hand, mutations in Rmi1 or Top3 appeared to compromise or eliminate the binding of its partner to Sgs1. Since the Sgs1-Top3-Rmi1 and Top3-Rmi1 complexes are resistant to these stringent conditions, we must conclude that the stability of the Sgs1-Rmi1 and Sgs1-Top3 subcomplexes is diminished compared to that of the wt. Interestingly, a decrease in the stability of the Sgs1-Top3 complex may contribute to the rmi1 phenotype. It has been suggested that Sgs1-Top3 or BLM-TOP3α must form a complex to function (12, 35, 53, 58), so that reducing or altering the binding of Top3 to Sgs1 in rmi1 cells could explain its top3-like phenotype. If this were true in the extreme, however, one would expect rmi1 phenotypes to be identical to top3. Although their phenotypes are very similar, it is clear that rmi1 mutants do not grow as poorly as top3 cells at either 30 or 37°C. Top3 may therefore retain a partial interaction with Sgs1 in the rmi1 mutant. This idea is consistent with the pairwise interactions observed with Sgs1 with recombinant proteins. In this case, the stability of the pairwise interactions between Sgs1-Rmi1 or Sgs1-Top3 appears to result from the high protein concentrations obtained with recombinant systems (4, 14). But in yeast, where these proteins are less abundant, the ability to form these pairwise interactions may be reduced in the absence of the third protein. This suggests that the rmi1 phenotype may be suppressed, at least partially, by overexpressing Top3. As described below, however, we do not think that the sole function of Rmi1 is to serve as a linker between Sgs1 and Top3.
The most compelling evidence for concluding that Sgs1, Top3, and Rmi1 function as a complex comes from gel filtration chromatography. The coelution of these proteins at a high native MW indicates that the complex is more than a simple heterotrimer. Based on the hexameric structure of recombinant BLM (26), a number of models for Sgs1 are possible. These models range from a hexamer of Sgs1 plus one Rmi1-Top3 dimer to a hexamer of Sgs1-Top3-Rmi1 trimers. The challenge still remains to purify the complex to homogeneity to determine its native subunit structure. The identification of the third subunit in the complex may help accomplish this with a recombinant system.
In our search for a biochemical function for Rmi1, we were guided by previous studies. In addition to its 3′-5′ DNA helicase activity, Sgs1, like other RecQ and hexameric DNA helicases, is capable of branch-migrating HJs (3, 8, 25). Genetically, Sgs1-Top3 and BLM-TOP3α function to suppress crossover formation; in vitro, the BLM-TOPα complex has been shown to decatenate double HJ-containing structures (23, 47, 59). These studies implicate Sgs1-Top3 in binding to or interacting with DNA in a structure-specific manner. Using a standard EMSA, we found that Rmi1 bound a number of DNA structures, although it preferred HJs. We were surprised by the requirement for high protein concentrations in this assay, but this probably reflects a low abundance of active protein in our recombinant preparations. It was also unexpected that the protein-DNA complexes migrated near the top of the gel and in the well. This migration may be due to multiple specific binding events or to protein aggregation, although we suspect it is the latter. Purified Rmi1 may aggregate simply because it is separated from its other two subunits. Similar to Rmi1, both Sgs1 and Top3 are largely insoluble when expressed at high levels in E. coli.
Competition experiments confirmed Rmi1's preference for HJs. This binding specificity seems consistent with the fact that pseudoreplication forks competed better than flap structures, which were better than ssDNA, linear dsDNA, or Y structures. DNA binding by Rmi1 requires more than simple branched DNA, as in the Y structure, but also requires that the substrate have multiple duplex arms. The binding of Rmi1 to HJs is consistent with the suspected role of Sgs1 in regulating crossover formation, as well as the known activity of Sgs1 DNA helicase on HJ substrates. We suggest that the structure-specific DNA binding of Rmi1 helps target Sgs1-Top3 to substrates such as single or double HJs. A prediction of this model is that Rmi1 would stimulate the HJ binding or branch-migration activity of Sgs1-Top3 in vitro.
In vitro studies of eukaryotic DNA topoisomerase III indicate that it is most active in strand-passing reactions, such as decatenation of ssDNA catenanes, and relatively poor at relaxing negatively supercoiled DNA. These activities reflect the fact that Top3 binds ssDNA well but has a low affinity for dsDNA substrates. The high temperatures required for superhelical relaxation are presumably needed to melt the substrate DNA to allow the Top3 to gain access to ssDNA (28, 29). Bacterial DNA topoisomerase III exhibits similar activities (11, 21, 39) and, in a catenation assay dependent on bacterial RecQ, it has been shown that eukaryotic Top3 can replace bacterial Top3 (20). This suggests that bacterial RecQ may provide Top3 with access to a substrate that it would be unable to act on by itself. Yeast Sgs1 and/or Rmi1 may perform a similar role.
The cross-linking of Rmi1 to ssDNA suggests that it may have an affinity for ssDNA bases which could be important for Top3 strand-passing activity. If Sgs1-Top3 acts at hemi-catenanes that result from the collapse of two HJs, then Top3 may use Rmi1 to help recognize and pass these ssDNA strands. This model predicts that Rmi1 may distort or melt hemicatenated DNA to reveal ssDNA. In addition to the above approaches that can be taken to determine the biological function of Rmi1, it may be informative to test whether Rmi1 alters these DNA structures and whether it stimulates the decatenation activity of Top3.
Acknowledgments
We gratefully acknowledge Steve Anderson, Marc Gartenberg, Hiroshi Hiasa, and Janet Huang for advice and suggestions. We are also grateful to Grant Brown for communicating results prior to publication.
This work was supported by NIH grant GM071268.
REFERENCES
- 1.Bastin-Shanower, S. A., W. M. Fricke, J. R. Mullen, and S. J. Brill. 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23:3487-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellaoui, M., M. Chang, J. Ou, H. Xu, C. Boone, and G. W. Brown. 2003. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 22:4304-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, R. J., J. L. Keck, and J. C. Wang. 1999. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol. 289:235-248. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, R. J., M. F. Noirot-Gros, and J. C. Wang. 2000. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 275:26898-26905. [DOI] [PubMed] [Google Scholar]
- 5.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates III, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 6.Chakraverty, R. K., and I. D. Hickson. 1999. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays 21:286-294. [DOI] [PubMed] [Google Scholar]
- 7.Chang, M., M. Bellaoui, C. Boone, and G. W. Brown. 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. USA 99:16934-16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Constantinou, A., M. Tarsounas, J. K. Karow, R. M. Brosh, V. A. Bohr, I. D. Hickson, and S. C. West. 2000. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey, S., C. S. Han, S. A. Ramer, J. C. Klassen, A. Jacobson, A. Eisenberger, K. M. Hopkins, H. B. Lieberman, and G. A. Freyer. 1998. Fission yeast rad12+ regulates cell cycle checkpoint control and is homologous to the Bloom's syndrome disease gene. Mol. Cell Biol. 18:2721-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiGate, R. J., and K. J. Marians. 1988. Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 263:13366-13373. [PubMed] [Google Scholar]
- 12.Duno, M., B. Thomsen, O. Westergaard, L. Krejci, and C. Bendixen. 2000. Genetic analysis of the Saccharomyces cerevisiae Sgs1 helicase defines an essential function for the Sgs1-Top3 complex in the absence of SRS2 or TOP1. Mol. Gen. Genet. 264:89-97. [DOI] [PubMed] [Google Scholar]
- 13.Fabre, F., A. Chan, W. D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99:16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fricke, W. M., V. Kaliraman, and S. J. Brill. 2001. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 276:8848-8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangloff, S., B. de Massy, L. Arthur, R. Rothstein, and F. Fabre. 1999. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 18:1701-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 18.Goodwin, A., S. W. Wang, T. Toda, C. Norbury, and I. D. Hickson. 1999. Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res. 27:4050-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmon, F. G., R. J. DiGate, and S. C. Kowalczykowski. 1999. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell 3:611-620. [DOI] [PubMed] [Google Scholar]
- 21.Hiasa, H., R. J. DiGate, and K. J. Marians. 1994. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 269:2093-2099. [PubMed] [Google Scholar]
- 22.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 23.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaliraman, V., J. R. Mullen, W. M. Fricke, S. A. Bastin-Shanower, and S. J. Brill. 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15:2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karow, J. K., A. Constantinou, J. L. Li, S. C. West, and I. D. Hickson. 2000. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 97:6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karow, J. K., R. H. Newman, P. S. Freemont, and I. D. Hickson. 1999. Oligomeric ring structure of the Bloom's syndrome helicase. Curr. Biol. 9:597-600. [DOI] [PubMed] [Google Scholar]
- 27.Keil, R. L., and A. D. McWilliams. 1993. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics 135:711-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, R. A., and J. C. Wang. 1992. Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J. Biol. Chem. 267:17178-17185. [PubMed] [Google Scholar]
- 29.Kim, Y. C., J. Lee, and H. S. Koo. 2000. Functional characterization of Caenorhabditis elegans DNA topoisomerase III alpha. Nucleic Acids Res. 28:2012-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laursen, L. V., E. Ampatzidou, A. H. Andersen, and J. M. Murray. 2003. Role for the fission yeast RecQ helicase in DNA repair in G2. Mol. Cell. Biol. 23:3692-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maftahi, M., C. S. Han, L. D. Langston, J. C. Hope, N. Zigouras, and G. A. Freyer. 1999. The top3+ gene is essential in Schizosaccharomyces pombe and the lethality associated with its loss is caused by Rad12 helicase activity. Nucleic Acids Res. 27:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maftahi, M., J. C. Hope, L. Delgado-Cruzata, C. S. Han, and G. A. Freyer. 2002. The severe slow growth of Δsrs2 Δrqh1 in Schizosaccharomyces pombe is suppressed by loss of recombination and checkpoint genes. Nucleic Acids Res. 30:4781-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyajima, A., M. Seki, F. Onoda, M. Shiratori, N. Odagiri, K. Ohta, Y. Kikuchi, Y. Ohno, and T. Enomoto. 2000. Sgs1 helicase activity is required for mitotic but apparently not for meiotic functions. Mol. Cell. Biol. 20:6399-6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohammad, M., R. D. York, J. Hommel, and G. M. Kapler. 2003. Characterization of a novel origin recognition complex-like complex: implications for DNA recognition, cell cycle control, and locus-specific gene amplification. Mol. Cell. Biol. 23:5005-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mullen, J. R., V. Kaliraman, and S. J. Brill. 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154:1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullen, J. R., V. Kaliraman, S. S. Ibrahim, and S. J. Brill. 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157:103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray, J. M., H. D. Lindsay, C. A. Munday, and A. M. Carr. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17:6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myung, K., A. Datta, C. Chen, and R. D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 39.Nurse, P., C. Levine, H. Hassing, and K. J. Marians. 2003. Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J. Biol. Chem. 278:8653-8660. [DOI] [PubMed] [Google Scholar]
- 40.Oakley, T. J., A. Goodwin, R. K. Chakraverty, and I. D. Hickson. 2002. Inactivation of homologous recombination suppresses defects in topoisomerase III-deficient mutants. DNA Repair (Amsterdam) 1:463-482. [DOI] [PubMed] [Google Scholar]
- 41.Onoda, F., M. Seki, A. Miyajima, and T. Enomoto. 2001. Involvement of SGS1 in DNA damage-induced heteroallelic recombination that requires RAD52 in Saccharomyces cerevisiae. Mol. Gen. Genet. 264:702-708. [DOI] [PubMed] [Google Scholar]
- 42.Onodera, R., M. Seki, A. Ui, Y. Satoh, A. Miyajima, F. Onoda, and T. Enomoto. 2002. Functional and physical interaction between Sgs1 and Top3 and Sgs1-independent function of Top3 in DNA recombination repair. Genes Genet. Syst. 77:11-21. [DOI] [PubMed] [Google Scholar]
- 43.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35:277-286. [DOI] [PubMed] [Google Scholar]
- 44.Philipova, D., J. R. Mullen, H. S. Maniar, C. Gu, and S. J. Brill. 1996. A hierarchy of SSB protomers in replication protein A. Genes Dev. 10:2222-2233. [DOI] [PubMed] [Google Scholar]
- 45.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Rouse, J., and S. P. Jackson. 2000. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 19:5801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shor, E., S. Gangloff, M. Wagner, J. Weinstein, G. Price, and R. Rothstein. 2002. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162:647-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spell, R. M., and S. Jinks-Robertson. 2004. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 262:3-12. [DOI] [PubMed] [Google Scholar]
- 50.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 50a.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 51.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 52.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 53.Ui, A., Y. Satoh, F. Onoda, A. Miyajima, M. Seki, and T. Enomoto. 2001. The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol. Genet. Genomics 265:837-850. [DOI] [PubMed] [Google Scholar]
- 54.Ui, A., M. Seki, H. Ogiwara, R. Onodera, S. Fukushige, F. Onoda, and T. Enomoto. 2005. The ability of Sgs1 to interact with DNA topoisomerase III is essential for damage-induced recombination. DNA Repair (Amsterdam) 4:191-201. [DOI] [PubMed] [Google Scholar]
- 55.Wallis, J. W., G. Chrebet, G. Brodsky, M. Rolfe, and R. Rothstein. 1989. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58:409-419. [DOI] [PubMed] [Google Scholar]
- 56.Watt, P. M., I. D. Hickson, R. H. Borts, and E. J. Louis. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watt, P. M., E. J. Louis, R. H. Borts, and I. D. Hickson. 1995. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81:253-260. [DOI] [PubMed] [Google Scholar]
- 58.Wu, L., S. L. Davies, P. S. North, H. Goulaouic, J. F. Riou, H. Turley, K. C. Gatter, and I. D. Hickson. 2000. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275:9636-9644. [DOI] [PubMed] [Google Scholar]
- 59.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]
- 60.Yamagata, K., J. Kato, A. Shimamoto, M. Goto, Y. Furuichi, and H. Ikeda. 1998. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA 95:8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]