Abstract
The mitogen-activated protein kinase (MAPK) phosphatase 1 (MKP-1) is an immediate-early gene comprised of a dual-specificity phosphatase domain and a noncatalytic NH2 terminus. Here, we show that the NH2 terminus of MKP-1, containing the cdc25 homology domains A (CH2A) and B (CH2B), mediates MKP-1 nuclear targeting and modulates MAPK-mediated gene expression. An LXXLL motif which is known to mediate protein-protein interactions with nuclear-targeted hormone receptors was identified proximal to the CH2A domain of MKP-1. The NH2 terminus alone of MKP-1 containing this LXXLL motif was sufficient to direct nuclear targeting, and mutating this motif to LXXAA resulted in the exclusion of MKP-1 from the nucleus. We found that the LXXLL motif proximal to the CH2A domain was present in other nuclear-localized MKPs but was absent in MKPs that localized to the cytoplasm. These data suggest that this LXXLL motif confers nuclear targeting properties to the MKPs. The NH2 terminus of MKP-1 was also found to inhibit the activation of the serum response element (SRE) by preventing MAPK-mediated phosphorylation of the regulatory serine 383 residue on Elk-1. Moreover, we show that MKP-1 plays a major role in the attenuation of serum-induced SRE activity, since MKP-1 null fibroblasts exhibited enhanced SRE activity in response to serum compared with wild-type fibroblasts. The NH2 terminus of MKP-1, when reconstituted into MKP-1 null fibroblasts to levels similar to endogenous MKP-1 following serum stimulation, reduced serum-mediated SRE activity. Collectively, these data reveal novel roles for the NH2 terminus of MKP-1 in nuclear targeting and transcriptional regulation.
The mitogen-activated protein kinase (MAPK) phosphatases (MKPs) are members of a family of dual-specificity phosphatases that inactivate the MAPKs in order to regulate cell proliferation, development, apoptosis, differentiation, and inflammation (3, 11, 14, 21, 22, 42). To date, 11 members of the classical MKP family have been identified, all of which inactivate the MAPKs by dephosphorylation of the regulatory threonine and tyrosine residues on MAPK. The MKPs share some basic structural similarities. The NH2 terminus of the MKPs is the most variable among this family (3, 21, 22), but they typically contain two conserved regions that exhibit similarities to the cdc25 phosphatase called the CH2 domain. The function of the CH2 domain, however, remains to be defined. All MKPs share strong amino acid sequence identity within their protein tyrosine phosphatase (PTP) domain that is defined by the signature catalytic motif [C(X)5R] at the COOH terminus (3, 42). Resolution of the crystal structure of the MKPs reveals that their PTP domain shares features similar to that of the VH1-related phosphatase by adopting a shallow catalytic cleft (14). Of the 11 MKP family members, MKP-1 (CL100/Erp/3CH134) represents the archetypal MKP, containing two CH2 domains designated CH2A and CH2B and a conserved PTP domain (5, 6, 23, 24, 31).
MKP-1 exhibits substrate specificity towards the inactivation of the stress-activated MAPKs, p38 MAPK and JNK, followed by the growth factor-activated extracellular-regulated kinase (Erk) (3, 15, 16, 21, 22, 34). The substrate specificity for the MKPs towards the MAPKs has been attributed to the ability of MAPK to interact with the modular docking surface that is located between the CH2A and CH2B domains of the MKPs (4). The MAPK-docking domain of the MKPs is composed of clusters of both basic and hydrophobic residues. A combination of biochemical and structural evidence strongly suggests that this docking surface defines the molecular determinants required for the interaction between the MKPs and MAPKs (12, 37, 41). With regards to MKP-1, the first cluster of basic residues are critical for Erk and, partially, p38 MAPK binding (34, 37, 41). In addition, the hydrophobic cluster and the second basic cluster of MKP-1 appear critical for the association with JNK and p38 MAPK (37, 41). Binding of MKP-1 to p38 MAPK, JNK, and Erk enhances its catalytic activity (1, 18, 34). Similarly, MAPK binding to the common MAPK interaction motif on MKP-3 induces a conformational change in the COOH terminus of the phosphatase that also enhances its catalytic activity (4, 13, 35). However, not all interactions between the MKPs and the MAPKs enhance MKP catalysis (7, 38, 40), suggesting that additional regions within the MKPs contribute to the regulation of MAPK dephosphorylation.
Subcellular localization is an important regulatory feature of the MKPs. Some MKPs are localized to the nucleus, such as MKP-1, MKP-2 (hVH-2 and TYP-1), PAC-1, and B23 (hVH-3) (3, 14, 42), whereas other MKPs localize to the cytoplasm, such as MKP-3 (Pyst1), Pyst2, MKP-4 (Pyst-3), MKP-5, M3/6 (hVH-5), and MKP-7 (3, 14, 42). Despite the fact that the subcellular distribution of many of the MKPs is known, the mechanisms regulating their localization remain poorly defined. MKP-1 is localized to the nucleus, yet it contains no classical nuclear localization sequence (NLS). It has been suggested that the two basic clusters of amino acids found between the CH2A and CH2B domains on the MKP [RRRAK-(X14)-RXR] serve to target it to the nucleus (7). How MKP-1, as well as other nuclearly localized MKPs, localizes to the nucleus remains to be identified.
Following activation, a fraction of the MAPKs translocate to the nucleus to regulate gene expression by phosphorylating transcription factors. For example, the serum response element (SRE) is regulated by MAPK-dependent pathways through the combined actions of the serum response factor (SRF) and the ETS domain transcription factor, Elk-1. It is well established that activation of Elk-1 is dependent upon the integrity of MAPK to phosphorylate Elk-1 in the nucleus (47). Thus, the localization of MKP-1 to the nucleus serves to inactivate this subcellular pool of MAPKs in order to provide discrete control of the temporal kinetics of Elk-1-mediated activity and ultimately gene expression. Because of the existence of other MKPs which also localize to the nucleus, it is not known whether MKP-1 plays a critical role in negatively regulating MAPK-mediated gene expression. Earlier work using MKP-1-deficient fibroblasts demonstrated that the effects of MKP-1 can be subserved by other MKPs (9). Therefore, the role of MKP-1 in the regulation of transcriptional activation remains to be fully defined.
Here we show that the NH2 terminus of MKP-1 is necessary and sufficient for directing MKP-1 nuclear targeting. Specifically, an LXXLL motif proximal to the CH2A domain was identified to be required for the localization of MKP-1 to the nucleus. We also identified that the NH2 terminus of MKP-1 was capable of attenuating growth factor-mediated activation of the serum response element by preventing phosphorylation and activation of the transcription factor Elk-1. We propose that these observations may represent a general mechanism for the nuclear targeting and the control of gene expression in other immediate-early responsive MKP family members.
MATERIALS AND METHODS
Cells and cell culture.
293 and COS-7 cell lines were cultured at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS; Sigma, St. Louis, MO), 1 mM sodium pyruvate (Invitrogen), 50 U/ml penicillin (Sigma), and 50 μg/ml streptomycin (Sigma). Serum deprivation medium contained DMEM, 0.1% FBS, 1 mM sodium pyruvate, 50 U/ml penicillin, and 50 μg/ml streptomycin.
Generation of GFP-MKP-1 fusion proteins.
To generate full-length green fluorescent protein (GFP)-MKP-1, MKP-1 cDNA was PCR amplified with oligonucleotides to introduce 5′ SmaI and 3′ XhoI restriction sites. The sequences of all the oligonucleotide primers can be found in the supplemental material. The purified PCR products were subcloned to the carboxyl terminus of enhanced GFP (EGFP) in pWay21 (a generous gift from Thomas Hughes, Montana State University) using SmaI and XhoI restriction sites, resulting in an in-frame GFP-MKP-1 fusion designated pWay-EGFP-MKP-1 (GFP-MKP-1). Nucleotide sequences corresponding to amino acids 1 to 46, 47 to 136, and 1 to 136 were PCR amplified and subcloned into pWay21-EGFP using SmaI and XhoI, except for the PCR product representing amino acids 1 to 46, which was subcloned using SmaI and NotI to generate pWay21-EGFP-MKP-11-46 (GFP-MKP-11-46), pWay21-EGFP-MKP-147-136 (GFP-MKP-147-136), and pWay21-EGFP-MKP-11-136 (GFP-MKP-11-136), respectively. PCR products for pWay21-GFP- MKP-147-367 (GFP-MKP-147-367) and pWay21-GFP-MKP-1137-367 (GFP-MKP-1137-367) were generated by amplifying with nucleotides corresponding to amino acids 47 to 367 and 137 to 367 and subcloning into pWay21-EGFP using SmaI and XhoI. The GFP-MKP-1Δ47-136 fusion protein was generated by using pWay21-EGFP-MKP-1 as a template to delete amino acids 47 to 136 of MKP-1. Oligonucleotides designed towards amino acids 46 and 137 were used to generate a GFP-MKP-1Δ47-136 construct in which residues 47 to 136 of MKP-1 were deleted. Site-directed mutagenesis (Stratagene, La Jolla, CA) was employed to introduce a triple point mutation in the pWay21-EGFP-MKP-1 vector to generate pWay-EGFP-MKP-1(ASA) [GFP-MKP-1 (ASA)]. First, arginine 53 was mutated to alanine to generate pWay-EGFP-MKP-1(R53A). This mutant was used as a template to mutate arginine 54 to serine. Finally, in the context of an alanine 53, serine 54 double mutant, arginine 55 was mutated to alanine, resulting in a triple mutant. Site-directed mutagenesis was also used to generate GFP-MKP-1 containing a leucine 16-to-alanine 16 (L16A), GFP-MKP-1 leucine 17-to-alanine 17 (L17A), and GFP-MKP-1 leucine 16 and 17-to-alanine 16 and 17 (L16A/L17A) mutant. GFP-MKP-1 was used as a template to generate pWay-EGFP-MKP-1(L16A) [GFP-MKP-1(L16A)] and pWay-EGFP-MKP-1(L17A) [GFP-MKP-1(L17A)]. GFP-MKP-1(L16A) was used as a template to generate the pWay-EGFP-MKP-1(L16A/17A) [GFP-MKP-1(L16A/17A)] double mutant. All expression constructs were confirmed using automated DNA sequencing.
Generation of MKP-1-deficient MEFs.
Mice containing a disruption within exon 2 of MKP-1 (MKP-1−/−) were derived by implantation of heterozygotic (MKP+/−) 129Sv/C57BL6 embryos (9) obtained from Bristol-Myers Squibb (Princeton, NJ) into surrogate mothers. Mouse embryonic fibroblasts (MEFs) from the wild type (MKP-1+/+) and MEFs containing a homozygotic deletion for MKP-1 (MKP-1−/−) were derived from embryonic day 12 to embryonic day 14 embryos. Isolated fibroblasts were resuspended and cultured at 37°C, 5% CO2 in MEF growth medium containing DMEM supplemented with 10% FBS, 50 U/ml penicillin, and 50 μg/ml streptomycin. Either primary or spontaneously immortalized MEFs were used for further experiments.
Confocal imaging of GFP-MKP-1 fusion proteins.
COS-7 cells were cultured on coverslips and transiently transfected with the indicated GFP fusion constructs using either calcium phosphate DNA precipitation or FuGENE-6 (Roche Molecular Biochemicals, Indianapolis, IN) according to the manufacturer's instructions. The cells were fixed with 4% paraformaldehyde (Sigma) and mounted on microscope slides. GFP was visualized on a Zeiss LSM 510 META confocal microscope, and images were analyzed using LSM510 version 3.2 software (Zeiss, Germany).
Measurement of Elk-1 activation and SRE-mediated gene expression.
Elk-1 luciferase activity was measured using the luciferase assay system kit from Promega (Madison, WI) according to the manufacturer's instructions. 293 cells were cotransfected with 0.5 μg Elkc-Gal4, 0.5 μg 5X-Gal4-luciferase, 50 ng pRL-Renilla, and 3 μg of the various GFP-MKP-1 expression plasmids using calcium phosphate DNA precipitation. The cells were serum deprived for 18 to 24 h prior to stimulation with 10% FBS for 4 h. Luciferase and Renilla activities were determined, and luciferase values were normalized to Renilla activities. Data are expressed as the fold change relative to unstimulated vector control transfectants. SRE-mediated activity was performed by cotransfecting 5XSRE-luciferase (0.25 μg) and pRL-Renilla (50 ng) along with 3 μg GFP, GFP-MKP-1, or GFP-MKP-11-136. Immortalized MKP-1 MEFs were cotransfected with 5XSRE-luciferase (0.25 to 0.5 μg) and pRL-Renilla (25 ng) along with 4 μg GFP, 1 μg GFP-MKP-1, and 3 μg GFP or 1 μg GFP-MKP-11-136, and 3 μg GFP with FuGENE-6 (Roche) according to the manufacturer's instructions. The cells were serum deprived for 18 to 24 h prior to stimulation with 10% FBS for 4 h. Primary MEFs were transfected with 1 μg 5XSRE-luciferase and 50 ng pRL-Renilla using FuGENE-6.
Analysis of Erk, p38 MAPK, JNK, and Elk-1 phosphorylation.
To determine the phosphorylation status of Erk, 293 cells were transiently transfected with 1 μg pCG-HA-Erk2 and 6 μg of the indicated GFP-MKP-1 expression plasmids using Lipofectamine 2000 (Invitrogen). Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed in 1 ml of NP-40 lysis buffer (1.0% NP-40, 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 1 μg/ml pepstatin A, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 2 mM sodium orthovanadate [Na3VO3], and 50 mM sodium fluoride [NaF]) for 30 min at 4°C. Cell lysates were clarified by centrifugation at 4°C at 20,800 × g for 15 min. Protein concentration was determined using Coomassie protein reagent (Pierce, Rockford, IL). Approximately 1 to 2 mg of cell lysates was precleared with Pansorbin (Calbiochem) for 10 min. The precleared lysates were incubated with 5 μg of antihemagglutinin (anti-HA) monoclonal antibody (Roche Molecular Biochemicals, Indianapolis, IN) overnight at 4°C. The immune complexes were collected on protein A-Sepharose and washed three times with 1 ml of ice-cold NP-40 lysis buffer (containing 2 mM Na3VO3 and 50 mM NaF) followed by washing with 1 ml of ice-cold STE buffer (150 mM NaCl, 25 mM Tris-HCl [pH 8], and 1 mM EDTA). The immune complex was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were transferred to Immobilon-P membranes (Millipore, Bedford, MA) and immunoblotted with anti-phospho-Erk1/2 antibodies (New England BioLabs). The immunoblot was reprobed with anti-Erk2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA). p38 MAPK phosphorylation was determined by transiently transfecting 293 cells with 7 μg of the indicated plasmids using Lipofectamine 2000. 293 cells were transfected with 0.5 μg Elk-1-Flag (provided by Andrew Sharrocks, University of Manchester, Manchester, England) and 3 μg of the indicated GFP-MKP-1 expression plasmids in order to assess Elk-1 phosphorylation. Serum-deprived cells were either left untreated or treated with 10% FBS for 30 min. MEFs were stimulated for 1 h with 10% FBS. MKP-1−/− MEFs were transiently transfected with either pWay21 (4 μg), GFP-MKP-1 (0.5 to 4 μg), or GFP-MKP-11-136 (0.5 to 4 μg) plus an equivalent amount of pWay21 to achieve a total of 4 μg cDNA. Cells were washed twice with ice-cold PBS and lysed in sample buffer. Cell lysates were resolved by SDS-PAGE, transferred to Immobilon-P membranes, and subjected to immunoblotting with anti-phospho p38 MAPK (New England BioLabs), anti-p38 MAPK (Santa Cruz), phospho-Elk-1 (New England BioLabs), anti-Flag (Sigma), anti-MKP-1 (SC-1102; Santa Cruz), and anti-GFP antibodies (Molecular Probes, Eugene, OR). Primary antibodies were detected using peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (Amersham Pharmacia Biotechnology).
Assessment of MKP-1-MAPK interactions.
293 cells were transiently transfected with either 6 μg pCMV-Flag-p38 MAPK or 6 μg pCG-HA-Erk2 and with 6 μg of the indicated GFP-MKP-1 plasmids. Cells were lysed as described, and 1 mg of protein lysate was precleared with Pansorbin and incubated with 5 μg of anti-HA or anti-Flag antibodies overnight at 4°C. The immunocomplexes were resolved by SDS-PAGE, and immunoblot analyses with anti-GFP, anti-p38 MAPK, and anti-Erk antibodies were performed.
RESULTS
The NH2 terminus of MKP-1 is necessary and sufficient for nuclear targeting.
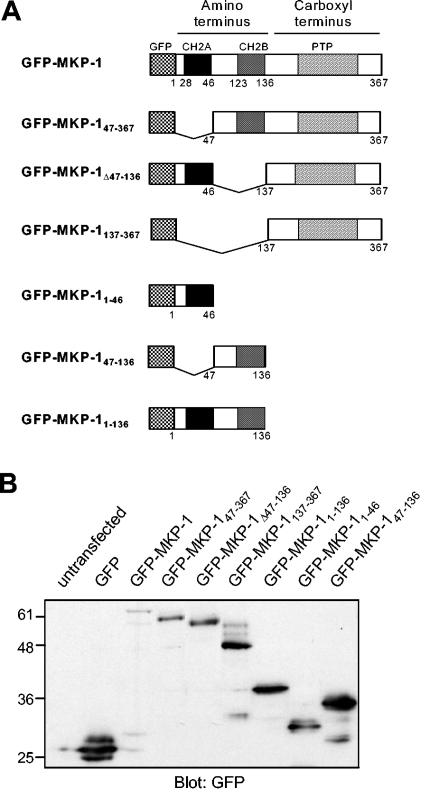
MKP-1 consists of tandem CH2 domains (CH2A and CH2B) at the NH2 terminus and a PTP domain at the COOH terminus. In order to identify the region within MKP-1 that is required for targeting it to the nucleus, we generated NH2-terminal GFP fusions of wild-type MKP-1 (GFP-MKP-1) and MKP-1 containing deletions of amino acids 1 to 46 (GFP-MKP-147-367), 47 to 136 (GFP-MKP-1Δ47-136), and 1 to 136 (GFP-MKP-1137-367). In addition, GFP fusions representing MKP-1 amino acids 1 to 46 (GFP-MKP-11-46), 47 to 136 (GFP-MKP-147-136), and 1 to 136 (GFP-MKP-11-136) were constructed (Fig. 1A). Wild-type and mutant MKP-1-GFP fusion proteins were transiently transfected into COS-7 cells, and immunoblotting of cell lysates prepared from these transfectants with anti-GFP antibodies confirmed that the wild type and the MKP-1 mutants generated proteins of the expected molecular mass (Fig. 1B).
FIG. 1.
Generation of GFP-MKP-1 fusion proteins. (A) Schematic representation of the GFP fusion proteins of MKP-1 used in this study. (B) COS-7 cells were either left untransfected or were transfected with expression vectors encoding the GFP-MKP-1 fusion proteins shown in panel A. Cell lysates were prepared and subjected to SDS-PAGE followed by immunoblotting with anti-GFP antibodies.
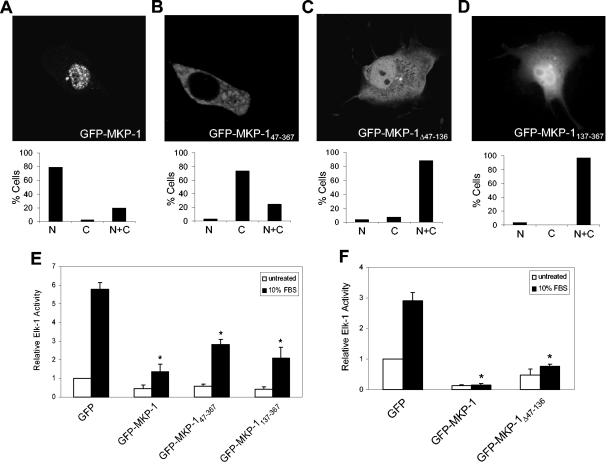
Next, COS-7 cells were transiently transfected with GFP-MKP-1, GFP-MKP-147-367, GFP-MKP-1Δ47-136, or GFP-MKP-1137-367 in order to determine their subcellular localization (Fig. 2). Visualization and expression of GFP was determined by confocal microscopy. As a control, COS-7 cells were transiently transfected with GFP alone, which was equally distributed between the cytoplasm and nucleus (data not shown). As expected, GFP-MKP-1 was localized predominantly within the nucleus (Fig. 2A). In contrast, GFP-MKP-147-367 was almost entirely excluded from the nucleus, and it accumulated in the cytoplasm (Fig. 2B). In contrast, GFP-MKP-1Δ47-136 and GFP-MKP-1137-367 were equally localized in both the nucleus and cytoplasm (Fig. 2C and D). The GFP-MKP-1Δ47-136 fusion protein is ∼54 kDa (Fig. 1A), exceeding the 40- to 50-kDa limit for passive diffusion (19, 32) and suggesting that amino acids 47 to 136 also contribute to the accumulation of MKP-1 in the nucleus. GFP-MKP-1137-367 is ∼49 kDa, and its subcellular localization in both the cytoplasm and nucleus likely reflects its ability to passively diffuse throughout the cell. These results demonstrate that the first 136 amino acids of MKP-1 are required for its nuclear targeting and accumulation.
FIG. 2.
The NH2 terminus of MKP-1 is required for nuclear localization. (A to D) COS-7 cells were transiently transfected with GFP-MKP-1 (A), GFP- MKP-147-367 (B), GFP-MKP-1Δ47-136 (C), or GFP-MKP-1137-367 (D). Confocal imaging was used to visualize GFP expression. The graphs below each photomicrograph in panels A to D represent the quantitation from a representative experiment in which >100 GFP-positive cells were analyzed for each condition. Shown is the percentage of cells in which GFP fluorescence was predominately nuclear (N), cytoplasmic (C), or both (N+C). (E and F) 293 cells were transiently transfected with the indicated GFP-MKP-1 expression plasmids plus Elk-1c-GAL4, 5XGAL4-luciferase, and pRL-Renilla. Cells were serum deprived and restimulated with 10% FBS. Elk-1 activation was measured as described in Materials and Methods. Data are representative of the mean ± standard error of the mean from three to four separate experiments. *, P < 0.05.
In Fig. 2, we show that GFP-MKP-147-367, GFP-MKP-1Δ47-136, and GFP-MKP-1137-367 failed to localize exclusively to the nucleus. It was conceivable that deletion of the NH2 terminus of MKP-1 perturbed protein folding, resulting in nonspecific cytoplasmic accumulation. To rule out this possibility, we asked whether the NH2-terminal truncations of MKP-1 were capable of dephosphorylating MAPK and, thus, inhibiting MAPK-mediated gene expression. We utilized the Elk-1 system to test this, since MAPK phosphorylates Elk-1, leading to its activation (33, 47). If these MKP-1 truncation mutants were appropriately folded and active as PTPs, they should retain their catalytic activity, dephosphorylate MAPK, and inhibit Elk-1. When GFP-MKP-1 was transiently overexpressed in 293 cells, as expected it inhibited the ability of serum to stimulate Elk-1 activation compared to GFP-alone control transfectants (Fig. 2E and F). We also found that serum-induced Elk-1 activation was inhibited when GFP-MKP-147-367, GFP-MKP-1137-367, or GFP-MKP-1Δ47-136 was overexpressed in 293 cells (Fig. 2E and F). These data suggest that failure of the NH2-terminal MKP-1 truncations to correctly localize is unlikely to be due to a global disruption of MKP-1 protein folding but rather to a specific loss of targeting.
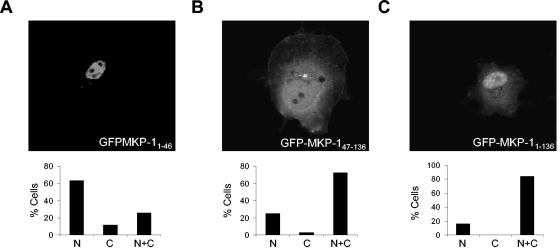
To test the sufficiency of the NH2 terminus of MKP-1 to direct heterologous targeting of GFP to the nucleus, we analyzed the localization of GFP fused either to amino acids 1 to 46 (GFP-MKP-11-46), 47 to 136 (GFP-MKP-147-136), or 1 to 136 (GFP-MKP-11-136) of MKP-1 (Fig. 3). When transiently expressed in COS-7 cells, GFP-MKP-11-46 localized predominantly in the nucleus (Fig. 3A). GFP-MKP-147-136 was distributed between the cytoplasm and nucleus (Fig. 3B), whereas GFP-MKP-11-136 accumulated predominantly in the nucleus but could also be detected, albeit at lower levels, in the cytoplasm (Fig. 3C). It is conceivable that GFP-MKP-11-136 contains sequences which may influence the nuclear targeting properties of amino acids 1 to 46 (see Discussion). Nevertheless, these data demonstrate that amino acids 1 to 46 are necessary and sufficient to direct nuclear targeting (Fig. 2B and A), whereas amino acids 46 to 136 contribute to nuclear accumulation (Fig. 2C).
FIG. 3.
The NH2 terminus of MKP-1 is sufficient for nuclear targeting. GFP-MKP-11-46 (A), GFP-MKP-147-136 (B), and GFP-MKP-11-136 (C) were transiently transfected into COS-7 cells, and confocal imaging was performed to visualize for GFP. The graphs below each photomicrograph in panels A to C represent the quantitation from a representative experiment in which >100 GFP-positive cells were analyzed for each condition. Shown are the percentages of cells in which GFP fluorescence was either predominately nuclear (N), cytoplasmic (C), or both (N+C).
An LXXLL motif proximal to the CH2A domain of MKP-1 directs nuclear targeting.
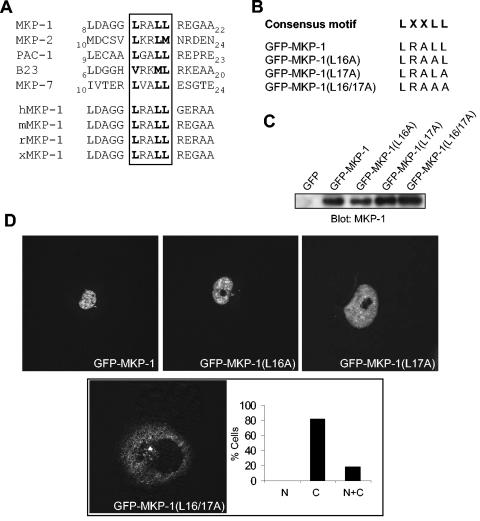
To further define the molecular basis for the ability of the first 46 amino acids of MKP-1 to direct nuclear targeting, we inspected the amino acid sequence of the NH2 terminus of MKP-1. Notably, MKP-1 does not contain a classical NLS within amino acids 1 to 46. We therefore compared the amino acid sequence that corresponds to the NH2 terminus of nuclear-targeted MKPs in order to identify any conserved sequence motifs. A sequence alignment of the NH2 termini of MKP-1, MKP-2, PAC-1, and the nuclear-shuffling MKP-7 revealed, with the exception of B23, the presence of a conserved LXXLL motif within the first 25 amino acids (Fig. 4A). This LXXLL motif in MKP-1 is also found in human, mouse, rat, and Xenopus laevis MKP-1 sequences (Fig. 4A). In contrast, the cytoplasmically localized MKPs, MKP-3, MKP-4, MKP-5, and M3/6, do not contain an LXXLL motif within this region (29, 30, 38, 43). The LXXLL motif describes an α-helix that is responsible for mediating protein-protein interactions between many nuclear receptor coactivators and nuclear hormone receptors (17). These observations raised the possibility that the LXXLL motif may be responsible for targeting MKP-1 to the nucleus.
FIG. 4.
The LXXLL motif proximal to the CH2A domain is required for MKP-1 nuclear targeting. (A) Sequence alignment of the NH2 terminus of MKP-1, MKP-2, PAC-1, B23, and MKP-7 reveals a conserved LXXLL motif. The LXXLL motif is conserved in human, mouse, rat, and Xenopus (xCL100/xMKP-1) MKP-1. (B) Sequences of the LXXLL MKP-1 mutants. (C) The LXXLL motif mutants shown in panel B were expressed transiently in COS-7 cells along with GFP-MKP-1. Lysates were resolved by SDS-PAGE, and GFP expression was detected by immunoblotting with an anti-MKP-1 antibody. (D) GFP-MKP-1 or the MKP-1-LXXLL mutants were transiently expressed in COS-7 cells, and confocal imaging for the expression of GFP was performed. The inset in panel D shows a quantitative analysis of the GFP fluorescence distribution of the GFP-MKP-1(L16/17A) mutant analyzed as described in the legend for Fig. 2.
The LXXLL motif identified in MKP-1 is positioned ∼10 amino acids proximal to the start of the CH2A domain (Fig. 1A and 4A). To test the possibility that this LXXLL sequence confers nuclear targeting to MKP-1, we mutated the last two leucines to alanines (LXXLL→LXXAA) in order to disrupt the functionality of this motif (17, 20, 27). Additionally, single (LRAAL and LRALA) and double leucine-to-alanine mutants (LRAAA) within the LXXLL motif were generated (Fig. 4B). Transient transfection of GFP-MKP-1(L16A), GFP-MKP-1(L17A), and GFP-MKP-1(L16/17A) in COS-7 cells indicated that these mutants were appropriately expressed compared with MKP-1 (Fig. 4C). Visualization for GFP in COS-7 cells transfected with these expression plasmids showed that both GFP-MKP-1(L16A) and MKP-1(L17A) were targeted to the nucleus (Fig. 4D). Remarkably, when expressed in COS-7 cells GFP-MKP-1(L16/17A) was almost completely redistributed from the nucleus to the cytoplasm (Fig. 4D). These results demonstrated that the LXXLL motif is required for the localization of MKP-1 to the nucleus.
MKP-1 nuclear accumulation is mediated by its MAPK-binding domain.
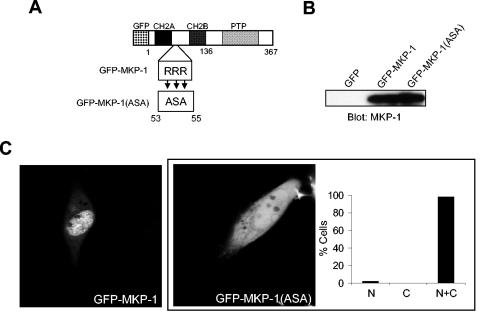
Previously, it was shown that mutating the first basic cluster in MKP-2 abolishes MAPK binding (34) and disrupts MKP-2 nuclear accumulation (7). Moreover, the basic cluster in MKP-1 has been suggested to be involved in targeting MKP-1 to the nucleus (7, 30, 43). However, the basic cluster found within GFP-MKP-147-136 was not sufficient to target MKP-1 to the nucleus (Fig. 3B). Nevertheless, deletion of this domain prevented the accumulation of MKP-1 to the nucleus (Fig. 2C), suggesting that the basic cluster is involved in MKP-1 nuclear accumulation. To specifically investigate the contribution of the MAPK-docking motif for MKP-1 localization, the triple arginine cluster that constitutes this first basic cluster was mutated to alanine, serine, and alanine within the context of GFP-MKP-1 to obtain GFP-MKP-1(ASA) (Fig. 5A and B). Confocal microscopic imaging for the detection of GFP showed that when transfected into COS-7 cells GFP-MKP-1(ASA) was equally distributed between the cytoplasm and nucleus (Fig. 5C). In contrast, GFP-MKP-1 was again entirely localized to the nucleus (Fig. 5C). These data demonstrate that the first basic cluster within MKP-1 contributes to its accumulation within the nucleus.
FIG. 5.
The MAPK-binding site on MKP-1 contributes to nuclear accumulation. (A) Schematic representation of the basic cluster (RRR) on MKP-1 that resides between the CH2A and CH2B domains. This RRR basic cluster was mutated to ASA in the context of GFP-MKP-1 to generate GFP-MKP-1(ASA). (B) Expression of GFP-MKP-1(ASA) was confirmed by transient transfection into COS-7 cells followed by immunoblot analysis using anti-MKP-1 antibodies. (C) Confocal imaging for GFP expression in COS-7 cells transiently expressing GFP, GFP-MKP-1, or GFP-(MKP-1)ASA proteins. The inset in panel C shows a graphical representation of the GFP fluorescence distribution of the GFP-MKP-1(ASA) mutant analyzed as described in the legend for Fig. 2.
The NH2 terminus of MKP-1 regulates Elk-1 independently of MAPK activity.
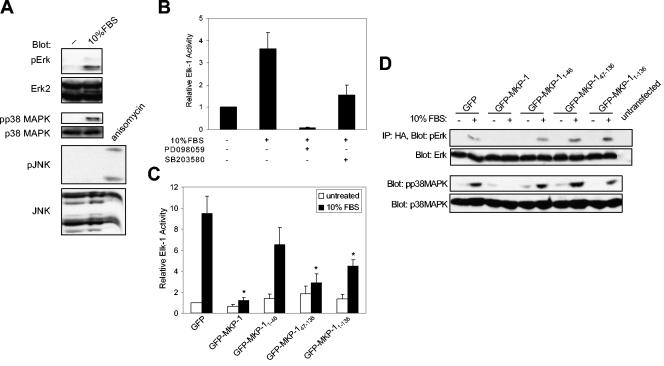
The region between the CH2A and CH2B domains of the NH2 terminus of MKP-1 has been shown to bind directly to the MAPKs through a common docking motif that is also shared by Elk-1 (1, 39). We hypothesized that MAPK-mediated phosphorylation of its substrates in the nucleus could be inhibited in a competitive manner by the NH2 terminus of MKP-1 binding to and preventing MAPK access to its substrates in the nucleus. Such a mechanism for MAPK-mediated gene inactivation would occur independently of the PTP domain. Since one of the major substrates for the MAPKs in the nucleus is the transcription factor Elk-1 and since phosphorylation of Elk-1 by the MAPKs is required for its activation (33, 47), we set out to test whether the NH2 terminus of MKP-1 could modulate MAPK-mediated Elk-1 activity. Because Elk-1 can be activated by Erk, p38 MAPK, and JNK, we first determined which of these MAPK family members participated in Elk-1 activation in response to serum in 293 cells. When 293 cells were stimulated with serum, Erk and p38 MAPK, but not JNK, were activated (Fig. 6A). Next, we tested the contribution of p38 MAPK and Erk to stimulate Elk-1 activation (Fig. 6B). Pretreatment with either the Erk-specific or p38 MAPK-specific inhibitors, PD098059 or SB203580, respectively, inhibited serum-induced Elk-1 activation compared to control (Fig. 6B). Thus, only p38 MAPK and Erk participate in the phosphorylation and activation of Elk-1 in 293 cells in response to serum, indicating that these MAPKs are the primary targets of MKP-1 in this system.
FIG. 6.
The NH2 terminus of MKP-1 inhibits MAPK-mediated Elk-1 activation. (A) Serum-deprived 293 cells were either left untreated or were restimulated with 10% FBS for 30 min. Protein lysates were resolved by SDS-PAGE and immunoblotted with anti-phospho-specific antibodies to Erk1/2 (pErk), p38 MAPK (pp38 MAPK), and JNK (pJNK). Immunoblots were probed with anti-Erk, p38 MAPK, and JNK antibodies. Anisomycin-treated cells (5 μM anisomycin for 1 h) served as a positive control for JNK activation. (B) 293 cells were cotransfected with the Elk-1 reporter genes, and Elk-1 activation was measured as described in the legend for Fig. 2. 293 cells were pretreated with either dimethyl sulfoxide, PD098059 (5 μM), or SB203580 (10 μM) prior to stimulation with 10% FBS for 4 h. Data represent the means ± standard errors of the means (SEM) of three separate experiments. (C) The indicated GFP-MKP-1 fusion proteins (Fig. 1A) were expressed with the Elk-1 reporter genes in 293 cells, and Elk-1 activity was determined either in the absence or presence of 10% FBS stimulation. The data are represented as the mean ± SEM change in Elk-1 activation from four separate experiments performed in triplicate. *, P < 0.05. (D) 293 cells were cotransfected with the indicated GFP-MKP-1 fusion proteins along with HA-Erk2. Transfectants were serum deprived and were then either left untreated or restimulated with 10% FBS for 30 min. Anti-HA-Erk2 immunoprecipitates were immunoblotted with phospho-Erk1/2 antibodies (upper panel). The immunoblot was reprobed with anti-Erk1/2 antibodies (lower panel). Whole-cell lysates were prepared and analyzed for p38 MAPK activation using anti-phospho-p38 MAPK (upper panel) and p38 MAPK (lower panel) antibodies.
To determine whether the NH2 terminus of MKP-1 could prevent Erk/p38 MAPK-mediated activation of Elk-1 in the nucleus, we overexpressed GFP-MKP-11-46, GFP-MKP-147-136,and GFP-MKP-11-136, all of which lack the PTP domain, in 293 cells and measured Elk-1 activation in response to serum (Fig. 6C). We found that serum increased Elk-1 activation by approximately ninefold in GFP control transfectants, and this induction was inhibited upon expression of GFP-MKP-1 (Fig. 6C). Significantly, expression of GFP-MKP-11-136 and GFP-MKP-147-136 inhibited serum-induced Elk-1 activation (Fig. 6C). Expression of the GFP-MKP-11-46 domain did not significantly inhibit Elk-1 activation, although it was consistently reduced (Fig. 6C). These data suggest that the NH2 terminus of MKP-1 is not only important for nuclear targeting (Fig. 2 to 5) but also may contribute to the termination of MAPK-mediated gene expression in the nucleus.
Next, we examined if the upstream activators of Elk-1 in 293 cells, namely Erk and p38 MAPK, were affected when the NH2 terminus of MKP-1 was overexpressed. Serum stimulation of quiescent 293 cells increased both Erk and p38 MAPK activation in GFP controls, and this activity was inhibited upon the expression of GFP-MKP-1 (Fig. 6D). However, expression of the GFP-MKP-11-46, GFP-MKP-147-136, or GFP-MKP-11-136 domain did not suppress either serum-induced Erk or p38 MAPK activation (Fig. 6D). These data show that the NH2 terminus of MKP-1 does not provide any trans-acting catalytic properties to facilitate either Erk or p38 MAPK dephosphorylation, strengthening the argument that the NH2 terminus of MKP-1 can regulate MAPK-mediated transcription independently of MAPK activation.
Attenuation of MAPK-mediated Elk-1 Ser 383 phosphorylation and SRE activity by the MKP-1 NH2 terminus.
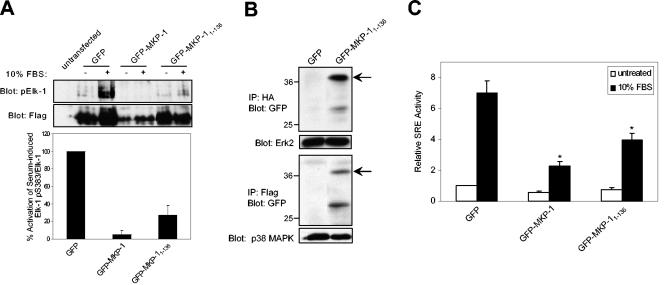
To further define the mechanism by which the NH2 terminus of MKP-1 inhibited serum-induced Elk-1 activation, we assessed the phosphorylation status of Elk-1 serine 383. Phosphorylation of this residue by MAPK is required for Elk-1 activation (33). Therefore, if our hypothesis is correct, the inhibition of Elk-1 activation by the NH2 terminus of MKP-1 should block the ability of either Erk and/or p38 MAPK to phosphorylate Ser 383 on Elk-1. We tested this by performing cotransfection experiments with the NH2 terminus of MKP-1 and an epitope-tagged expression vector for Elk-1 (Flag-Elk-1) followed by immunoblotting using a phospho-specific antibody to Ser 383 of Elk-1 (Fig. 7A). In 293 cells, endogenous phospho-Elk-1 levels were undetectable (Fig. 7A). However, when Elk-1 was overexpressed in 293 cells in response to serum stimulation, it was detectably phosphorylated on Ser 383 (Fig. 7A). Ser 383 phosphorylation, however, was inhibited completely upon expression of GFP-MKP-1 (Fig. 7A), and expression of GFP-MKP-11-136 was also able to inhibit serum-induced Ser 383 phosphorylation of Elk-1 by ∼70% (Fig. 7A). These data demonstrate that the NH2 terminus of MKP-1 can inhibit Elk-1-mediated phosphorylation by either Erk and/or p38 MAPK, thereby suppressing Elk-1 activation in response to serum.
FIG. 7.
The NH2 terminus of MKP-1 inhibits SRE-mediated activation by preventing Elk-1 phosphorylation. (A) Serum-deprived 293 cells expressing the indicated GFP-MKP-1 fusion proteins along with Flag-Elk-1 were stimulated with 10% FBS for 30 min. Whole-cell lysates prepared from these transfectants were resolved by SDS-PAGE and immunoblotted with either anti-phospho-Elk-1 (Ser 383) or anti-Flag antibodies. The graph below represents the means ± standard errors of the means (SEM) of three separate experiments performed as described above, in which densitometric analyses were determined from phospho-Elk-1 and Elk-1 immunoblots. The phospho-Elk-1/Elk-1 ratio as a percentage of serum-induced Elk-1 phosphorylation upon GFP-MKP-1 and GFP-MKP-11-136 expression relative to GFP is shown. (B) 293 cells were transfected with HA-Erk2 (upper panel) or Flag-p38 MAPK (lower panel) with either GFP control or GFP-MKP-11-136. HA-Erk2 and Flag-p38 MAPK immune complexes were resolved and immunoblotted using anti-GFP antibodies. The lower panels represent controls for Erk (upper panel) and p38 MAPK (lower panel) using anti-Erk2 and p38 MAPK, respectively. (C) 293 cells were cotransfected with 5XSRE-luciferase reporter along with pRL-Renilla and GFP, GFP-MKP-1, and GFP-MKP-11-136. Transfectants were serum deprived and were either left untreated or restimulated with 10% FBS for ∼4 h. SRE luciferase activity was normalized to Renilla units and expressed as the relative SRE activity. The data shown are representative of the means ± SEM from four separate experiments. *, P < 0.05.
The MAPK-binding site on MKP-1 is similar to the MAPK-docking motif on Elk-1, raising the possibility that the NH2 terminus of MKP-1 could physically associate with MAPK to prevent Ser 383 phosphorylation on Elk-1. Therefore, we tested whether the NH2 terminus of MKP-1 complexes with either Erk and/or p38 MAPK. When either Erk2 or p38 MAPK was transiently transfected into 293 cells along with GFP-MKP-11-136, we found that both Erk and p38 MAPK complexed with GFP-MKP-11-136 but not with GFP alone (Fig. 7B). Similar results have been reported by others (41). These data support the interpretation that the NH2 terminus of MKP-1 interacts with Erk and p38 MAPK, providing a potential mechanism for how it can prevent MAPK from phosphorylating Ser 383 on Elk-1.
Many immediate-early genes, such as c-fos, contain within their promoter an SRE (44). The SRE mediates a diverse array of growth factor and stress-responsive stimuli by forming a complex with the SRF and Elk-1 to direct transcriptional activation (47). We therefore asked whether the NH2 terminus of MKP-1 could inhibit SRE activity mediated by endogenous SRF/Elk-1 complexes. 293 cells were cotransfected with 5XSRE-luciferase and either vector, GFP-MKP-1, or GFP-MKP-11-136. Upon serum stimulation of 293 cell transfectants, SRE-mediated luciferase activity increased by up to sevenfold, and this was inhibited upon expression of GFP-MKP-1 (Fig. 7C). Expression of GFP-MKP-11-136 was also capable of significantly inhibiting serum-induced SRE-luciferase activity, although to a lesser extent than with GFP-MKP-1 (Fig. 7C). Together, these results imply that the NH2 terminus of MKP-1 can function to inhibit endogenous SRE-mediated gene expression by preventing Erk/p38 MAPK-mediated Ser 383 phosphorylation of Elk-1.
Critical role for MKP-1 in SRE-mediated gene expression.
Our data suggest a mechanism whereby upon activation, the NH2 terminus of MKP-1 functions independently of the PTP domain to attenuate SRE-mediated gene expression by physically disrupting MAPK-mediated Elk-1 Ser 383 phosphorylation. If this model is correct, then the NH2 terminus of MKP-1 alone should be less efficient at inhibiting SRE-mediated gene expression than wild-type MKP-1. Indeed, we observed that this is the case under conditions when GFP-MKP-1 and GFP-MKP-11-136 were overexpressed (Fig. 7A and C). Additionally, we wished to determine whether the NH2 terminus of MKP-1 could inhibit SRE-mediated gene transcription under conditions which represented more closely the endogenous levels of MKP-1 expression.
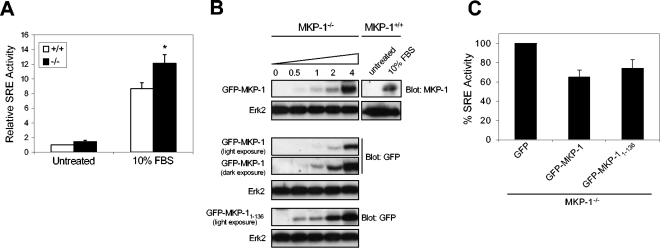
We therefore sought to reexpress GFP-MKP-1 and GFP-MKP-11-136 in MKP-1-deficient MEFs to levels that are similar to endogenous MKP-1 in order to test the ability of the reconstituted MKP-1-deficient cells to attenuate normal SRE-mediated activity. First, we derived MEFs from wild-type (MKP-1+/+) and MKP-1-deficient (MKP-1−/−) mice (9) and then tested whether loss of MKP-1 resulted in enhanced serum-induced SRE activity. We found that MKP-1−/− MEFs exhibited ∼30% higher levels of serum-induced SRE activity in response to serum than MKP-1+/+ MEFs (Fig. 8A). These data indicate that MKP-1 plays an important role in the inactivation of serum-induced SRE-mediated gene activity.
FIG. 8.
MKP-1 and its NH2 terminus are important for the inactivation of SRE-mediated transcriptional activity. (A) Primary wild-type MKP-1 (MKP-1+/+) and MKP-1 null (MKP-1−/−) MEFs were serum deprived for 48 h and restimulated with 10% FBS for ∼4 h. SRE luciferase activity was normalized to Renilla units and expressed as the relative fold SRE activity. The data shown are representative of the means ± standard errors of the means (SEM) from five separate experiments. *, P < 0.05. (B) Immortalized MKP-1−/− MEFs were transiently transfected with the indicated amounts of MKP-1 expression plasmid, or MKP-1+/+ MEFs were left untreated or were stimulated with 10% FBS for 1 h. Cells were lysed, resolved by SDS-PAGE, and immunoblotted with anti-MKP-1 antibodies. Expression of the GFP fusion proteins was determined by immunoblotting cell lysates with anti-GFP antibodies. (C) MKP-1−/− MEFs were transfected with 5XSRE-luciferase reporter along with pRL-Renilla and either GFP, GFP-MKP-1, or GFP-MKP-11-136. Cells were stimulated with 10% FBS for ∼4 h, and SRE luciferase activity was normalized to Renilla units. Results are shown as percent SRE activity relative to the GFP control and represent the means ± SEM from four separate experiments.
Next, in order to show that the ability of the GFP-MKP-11-136 domain to inhibit SRE activity was not simply a consequence of overexpression, we established conditions under which GFP-MKP-11-136, when introduced into MKP-1−/− MEFs, could be expressed to levels equivalent to that of endogenous MKP-1 following serum stimulation. We determined the titers of GFP-MKP-1 and GFP-MKP-11-136 cDNA transfected into MKP-1−/− MEFs that would achieve levels of expression that were similar to that of endogenous MKP-1 following serum stimulation. In Fig. 8B we show that ∼4 μg of GFP-MKP-1 expression plasmid is sufficient to express GFP-MKP-1 to levels similar to that of endogenous MKP-1 following serum stimulation, as determined by densitometric analyses (data not shown). Based upon the transfection efficiency of ∼30 to 40% in these cells, as measured by GFP expression, we estimated that ∼2 μg GFP-MKP-1 cDNA was required to achieve levels of MKP-1 similar to that of the endogenous protein. Unfortunately, these MKP-1 antibodies only recognize the carboxyl terminus of MKP-1 and, thus, we were unable to detect GFP-MKP-11-136 in order to perform this analysis. Therefore, we compared the expression levels of GFP-MKP-1 and GFP-MKP-11-136 using anti-GFP antibodies in these MKP-1−/− MEFs. When GFP-MKP-1 and GFP-MKP-11-136 were expressed in MKP-1−/− MEFs, we found that GFP-MKP-1 was expressed to ∼3-fold-lower levels than with GFP-MKP-11-136 when normalized for transfection efficiency in these MKP-1−/− MEFs (Fig. 8C). Because we estimated that ∼2 μg MKP-1 cDNA was equivalent to that of endogenous MKP-1 and that the GFP-MKP-11-136 was expressed to levels up to ∼3-fold higher, we estimated that when corrected for transfection efficiency ∼1 μg GFP-MKP-11-136 cDNA would be sufficient to represent expression levels similar to that of endogenous MKP-1. Therefore, GFP-MKP-11-136 (1 μg) was transfected into MKP-1−/− MEFs which were subsequently stimulated with serum. We found that under these conditions, GFP-MKP-11-136 was capable of inhibiting serum-induced SRE activity by ∼25% compared with the GFP vector control (Fig. 8C). We also observed that 0.5 μg GFP-MKP-11-136 also inhibited SRE activity when transfected into MKP-1−/− MEFs (data not shown). Importantly, when MKP-1 (1 μg) was transfected into MKP-1−/− MEFs we found that it too was capable of inhibiting serum-induced SRE activity to similar levels as with GFP-MKP-11-136 (Fig. 8C). Taken together, these data support the interpretation that the NH2 terminus of MKP-1 contributes to the suppression of SRE-mediated activity.
DISCUSSION
Although MKP-1 nuclear localization has been well documented (22), the structural determinants that mediate its nuclear targeting have not been identified. The role of the NH2 terminus in MKP nuclear localization has been suggested previously in experiments performed on MKP-2 and MKP-7 (7, 26). In the present study, we found that the LXXLL motif prior to the CH2A domain of MKP-1 is critical for its nuclear targeting. Either deletion of the LXXLL motif (GFP-MKP-147-367) or point mutations of the LXXLL motif within wild-type MKP-1 [GFP-MKP-1(L16A/17A)] resulted in the exclusion of MKP-1 from the nucleus. Although amino acids 1 to 46 (GFP-MKP-11-46) were sufficient to direct heterologous targeting of GFP to the nucleus, we noted that the entire NH2 terminus (GFP-MKP-11-136) was not as effectively targeted to the nucleus. There are several plausible explanations for this result. The first explanation is that in the context of amino acids 1 to 136, the LXXLL motif may not be sufficiently accessible in order for it to exert its nuclear targeting properties. The second explanation is that amino acids 1 to 136 contain a basic cluster which contributes to MAPK binding. It is conceivable that this basic cluster binds MAPKs that are in the cytosol, hindering complete nuclear localization of this fragment.
LXXLL sequences describe a core protein-protein-interacting motif, and the amino acids surrounding this motif dictate binding specificity (27). Although the LXXLL motif has not previously been ascribed to perform nuclear targeting directly, it is conceivable that this sequence could mediate the interaction with other NLS-containing proteins. It is well established that the LXXLL motif mediates protein-protein interactions with nuclear hormone receptors (17). Thus, MKP-1 may utilize nuclear hormone receptors as well as coactivators and corepressors as chaperones in order to attain its localization to the nucleus. This nuclear receptor chaperone-like mechanism involving the LXXLL motif of the nuclear hormone receptor, Dax-1, has been reported. The interaction between the LXXLL motifs on Dax-1 and the NLS-containing protein Ad4BP (SF-1) has been shown to be critical for targeting Dax-1 to the nucleus (20). Significantly, we identified that the LXXLL motif also is present within sequences of other nuclear-targeted MKPs, such as MKP-2 and PAC-1, as well as the nuclear shuffling protein MKP-7 (26, 28). We suggest, given the strong sequence conservation of the LXXLL motif among the nuclear-localized MKPs, that these proteins may be targeted to the nucleus in a similar manner.
The conserved basic cluster RRRAK-(X14)-RGR, which is located between the CH2A and CH2B domains on MKP-1, has also been suggested to provide nuclear targeting to MKP-1 (7, 30, 43). However, our results indicate that this basic cluster is not sufficient to target MKP-1 to the nucleus. Instead, the basic cluster appears to provide MKP-1 with the property that retains it in the nucleus. Deletion of either the domain that contains the basic clusters (GFP-MKP-1Δ47-136) or point mutations of the basic cluster [GFP-MKP-1(ASA)] results in the distribution of MKP-1 between the nucleus and cytoplasm. This subcellular distribution is similar to the localization found when the basic cluster of MKP-2 is mutated (7). In addition, GFP-MKP-147-136, which contains this basic cluster, fails to target MKP-1 to the nucleus, directly demonstrating that this domain alone does not function as a nuclear targeting sequence. The basic cluster of MKP-1, and possibly other nuclear-targeted MKPs, may function in a cooperative manner with the LXXLL motif to target and subsequently retain these MKPs to the nucleus. Thus, two mechanisms for the nuclear localization of MKP-1 may exist, nuclear targeting followed by nuclear accumulation.
The MAPK-binding site on MKP-1 is similar to the MAPK-docking motif on the MAPK substrate, Elk-1 (1, 12, 37). Therefore, the NH2 terminus of MKP-1 may compete with Elk-1 for MAPK binding, thus physically preventing MAPK from phosphorylating its downstream substrates. We found that the NH2 terminus of MKP-1 when overexpressed failed to affect MAPK activation, but it inhibited the ability of MAPK to phosphorylate Elk-1 on serine 383, the critical site for Elk-1 activation (25). The consequences of this phosphorylation at serine 383 within the C terminus result in the relief of an inhibitory intramolecular interaction within Elk-1 that enhances its DNA binding through conformational modulation of the N terminus (48). Consistent with this, the NH2 terminus of MKP-1 inhibited SRE-mediated gene expression. These data suggest that the NH2 terminus of MKP-1 can inhibit MAPK-mediated phosphorylation of Elk-1 by competing out MAPK for its target substrate, Elk-1. It could be argued that these effects are due solely to the overexpression of the NH2 terminus of MKP-1. To address this issue, we reintroduced either GFP-MKP-1 or GFP-MKP-11-136 into fibroblasts lacking MKP-1 to levels that were similar to endogenous MKP-1 when induced by serum stimulation. These data showed that MKP-1 and GFP-MKP-11-136 were both capable of suppressing serum-induced SRE activity at levels close to endogenous MKP-1 expression following serum stimulation. A significant finding from these studies is that we show that MKP-1 is important for regulating SRE gene activity, because in MKP-1-deficient cells SRE-mediated gene activation in response to serum is enhanced. Thus, MKP-1 plays a major role in the downregulation of SRE-mediated gene regulation. Consistent with this we have shown that MKP-1-deficient fibroblasts show hyperactivation of p38 MAPK and JNK, but not Erk, in response to serum stimulation (J. J. Wu and A. M. Bennett, unpublished observations). These results provide evidence for a novel mechanism of MAPK-mediated gene regulation by MKP-1 that occurs independently of its catalytic activity. MKP-1 is an immediate-early gene whose expression becomes rapidly induced in response to a variety of growth factor and stress stimuli (2, 8, 10, 23, 36). The transcriptional induction of MKP-1 occurs following activation of the MAPKs, and it has been shown that both p38 MAPK and Erk can stimulate MKP-1 gene expression (2, 8, 10, 23, 36). Thus, once expressed in the cytoplasm, MKP-1 is targeted to the nucleus, and as its level of expression increases its NH2 terminus may serve to compete for MAPK binding to Elk-1. This event may serve to attenuate MAPK-mediated gene expression via Elk-1 in two ways. First, by displacing MAPK from Elk-1, MAPK can no longer continue to phosphorylate and thus activate Elk-1. Second, the NH2 terminus of MKP-1 can now direct MAPK dephosphorylation via the PTP domain.
This working model predicts a mechanism of MKP-1-mediated inhibition of gene expression that occurs independently of its PTP domain and may represent a common regulatory feature of the MKPs that exhibit immediate-early gene activation properties. The idea that the NH2 terminus of MKPs contributes to the inhibition of MAPK-mediated gene expression may be an important mechanism for how MK-STYX may regulate MAPK-mediated signaling. The STYX family of PTPs contains a point mutation in the PTP domain rendering them catalytically inactive (45, 46). MK-STYX is a STYX family member that contains an NH2 terminus similar to that of MKP-1 and, thus, although catalytically inactive may function to regulate MAPK-mediated gene expression by competing for MAPK substrates. Taken together, these data reveal new roles for MKP-1 and, more broadly, our data provide novel mechanisms for how the MKP family may regulate subcellular localization as well as MAPK-mediated gene expression.
Supplementary Material
Acknowledgments
We thank members of the Bennett lab for comments and helpful discussions during the course of this study. We also thank Michael Nathanson and Barbara Ehrlich for their helpful discussions and review of this work. We are grateful to Thomas Hughes for providing the pWay21 plasmid and Andrew Sharrocks for the Elk-1 reagents.
This work was supported by a Burroughs Wellcome Foundation New Investigators Award and NIH grant and P01-DK-57751 to A.M.B.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bardwell, A. J., M. Abdollahi, and L. Bardwell. 2003. Docking sites on mitogen-activated protein kinase (MAPK) kinases, MAPK phosphatases and the Elk-1 transcription factor compete for MAPK binding and are crucial for enzymic activity. Biochem. J. 370:1077-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brondello, J.-M., A. Brunet, J. Pouyssegur, and F. R. McKenzie. 1996. The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J. Biol. Chem. 272:1368-1376. [DOI] [PubMed] [Google Scholar]
- 3.Camps, M., A. Nichols, and S. Arkinstall. 2000. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 14:6-16. [PubMed] [Google Scholar]
- 4.Camps, M., A. Nichols, C. Gillieron, B. Antonsson, M. Muda, C. Chabert, U. Boschert, and S. Arkinstall. 1998. Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science 280:1262-1264. [DOI] [PubMed] [Google Scholar]
- 5.Charles, C. H., A. S. Abler, and L. F. Lau. 1992. cDNA sequence of a growth-inducible immediate early gene and characterization of its encoded protein. Oncogene 7:187-190. [PubMed] [Google Scholar]
- 6.Charles, C. H., H. Sun, L. F. Lau, and N. K. Tonks. 1993. The growth factor-inducible immediate-early gene 3CH134 encodes a protein-tyrosine phosphatase. Proc. Natl. Acad. Sci. USA 90:5292-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, P., D. Hutter, X. Yang, M. Gorospe, R. J. Davis, and Y. Liu. 2001. Discordance between the binding affinity of mitogen-activated protein kinase subfamily members for MAP kinase phosphatase-2 and their ability to activate the phosphatase catalytically. J. Biol. Chem. 276:29440-29449. [DOI] [PubMed] [Google Scholar]
- 8.Cook, S. J., J. Beltman, K. A. Cadwallader, M. McMahon, and F. McCormick. 1997. Regulation of mitogen-activated protein kinase phosphatase-1 expression by extracellular signal-related kinase-dependent and Ca2+-dependent signal pathways in Rat-1 cells. J. Biol. Chem. 272:13309-13319. [DOI] [PubMed] [Google Scholar]
- 9.Dorfman, K., D. Carrasco, M. Gruda, C. Ryan, S. A. Lira, and R. Bravo. 1996. Disruption of the erp/mkp-1 gene does not affect mouse development: normal MAP kinase activity in ERP/MKP-1-deficient fibroblasts. Oncogene 13:925-931. [PubMed] [Google Scholar]
- 10.Duff, J. L., M. B. Marrero, W. G. Paxton, C. H. Charles, L. F. Lau, K. E. Bernstein, and B. C. Berk. 1993. Angiotensin II induces 3CH134, a protein tyrosine phosphatase, in vascular smooth muscle cells. J. Biol. Chem. 268:26037-26040. [PubMed] [Google Scholar]
- 11.English, J., G. Pearson, J. Wilsbacher, J. Swantek, M. Karandikar, S. Xu, and M. H. Cobb. 1999. New insights into the control of MAP kinase pathways. Exp. Cell Res. 253:255-270. [DOI] [PubMed] [Google Scholar]
- 12.Enslen, H., and R. J. Davis. 2001. Regulation of MAP kinases by docking domains. Biol. Cell 93:5-14. [DOI] [PubMed] [Google Scholar]
- 13.Farooq, A., G. Chaturvedi, S. Mujtaba, O. Plotnikova, L. Zeng, C. Dhalluin, R. Ashton, and M. M. Zhou. 2001. Solution structure of ERK2 binding domain of MAPK phosphatase MKP-3: structural insights into MKP-3 activation by ERK2. Mol. Cell 7:387-399. [DOI] [PubMed] [Google Scholar]
- 14.Fauman, E., and M. Saper. 1996. Structure and function of the protein tyrosine phosphatases. Trends Biochem. Sci. 21:413-417. [DOI] [PubMed] [Google Scholar]
- 15.Franklin, C. C., and A. S. Kraft. 1997. Conditional expression of the mitogen-activated protein kinase (MAPK) phosphatase MKP-1 preferentially inhibits p38 MAPK and stress-actviated protein kinase in U397 cells. J. Biol. Chem. 272:16917-16923. [DOI] [PubMed] [Google Scholar]
- 16.Franklin, C. C., S. Srikanth, and A. S. Kraft. 1998. Conditional expression of mitogen-activated protein kinase phosphatase-1, MKP-1, is cytoprotective against UV-induced apoptosis. Proc. Natl. Acad Sci. USA 95:3014-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 18.Hutter, D., P. Chen, J. Barnes, and Y. Liu. 2000. Catalytic activation of mitogen-activated protein (MAP) kinase phosphatase-1 by binding to p38 MAP kinase: critical role of the p38 C-terminal domain in its negative regulation. Biochem. J. 352:155-163. [PMC free article] [PubMed] [Google Scholar]
- 19.Kaffman, A., and E. K. O'Shea. 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15:291-339. [DOI] [PubMed] [Google Scholar]
- 20.Kawajiri, K., T. Ikuta, T. Suzuki, M. Kusaka, M. Muramatsu, K. Fujieda, M. Tachibana, and K. Morohashi. 2003. Role of the LXXLL-motif and activation function 2 domain in subcellular localization of Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1). Mol. Endocrinol. 17:994-1004. [DOI] [PubMed] [Google Scholar]
- 21.Keyse, S. M. 1998. Protein phosphatases and the regulation of MAP kinase activity. Cell. Dev. Biol. 9:143-152. [DOI] [PubMed] [Google Scholar]
- 22.Keyse, S. M. 2000. Protein phosphatases and the regulation of mitogen-activated protein kinase signalling. Curr. Opin. Cell Biol. 12:186-192. [DOI] [PubMed] [Google Scholar]
- 23.Keyse, S. M., and E. A. Emslie. 1992. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature 359:644-647. [DOI] [PubMed] [Google Scholar]
- 24.Kwak, S. P., D. J. Hakes, K. J. Martell, and J. E. Dixon. 1994. Isolation and characterization of a human dual specificity protein-tyrosine phosphatase gene. J. Biol. Chem. 269:3569-3604. [PubMed] [Google Scholar]
- 25.Li, Q. J., S. H. Yang, Y. Maeda, F. M. Sladek, A. D. Sharrocks, and M. Martins-Green. 2003. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 22:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda, K., H. Shima, M. Watanabe, and K. Kikuchi. 2001. MKP-7, a novel mitogen-activated protein kinase phosphatase, functions as a shuttle protein. J. Biol. Chem. 276:39002-39011. [DOI] [PubMed] [Google Scholar]
- 27.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misra-Press, A., C. S. Rim, H. Yao, M. S. Roberson, and P. J. S. Stork. 1995. A novel mitogen-activated protein kinase phosphatase. J. Biol. Chem. 270:14587-14596. [DOI] [PubMed] [Google Scholar]
- 29.Muda, M., U. Boschert, R. Dickinson, J.-C. Martinou, I. Martinou, M. Camps, W. Schlegel, and S. Arkinstall. 1996. MKP-3, a novel cytosolic protein-tyrosine phosphatase that exemplifies a new class of mitogen-activated protein kinase phosphatase. J. Biol. Chem. 271:4319-4326. [DOI] [PubMed] [Google Scholar]
- 30.Muda, M., U. Boschert, A. Smith, B. Antonsson, C. Gillieron, C. Chabert, M. Camps, I. Martinou, A. Ashworth, and S. Arkinstall. 1997. Molecular cloning and functional characterization of a novel mitogen-activated protein kinase phosphatase, MKP-4. J. Biol. Chem. 272:5141-5151. [DOI] [PubMed] [Google Scholar]
- 31.Noguchi, T., R. Metz, L. Chen, M.-G. Mattei, D. Carrsaco, and R. Bravo. 1993. Structure, mapping, and expression of erp, a growth factor-inducible gene encoding a nontransmembrane protein tyrosine phosphatase, and effect of ERP on cell growth. Mol. Cell. Biol. 13:5195-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlenstedt, G. 1996. Protein import into the nucleus. FEBS Lett. 389:75-79. [DOI] [PubMed] [Google Scholar]
- 33.Sharrocks, A. D. 2001. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2:827-837. [DOI] [PubMed] [Google Scholar]
- 34.Slack, D. N., O. M. Seternes, M. Gabrielsen, and S. M. Keyse. 2001. Distinct binding determinants for Erk2/p38α and JNK MAP kinases mediate catalytic activation and substrate selectivity of map kinase phosphatase-1. J. Biol. Chem. 276:16491-16500. [DOI] [PubMed] [Google Scholar]
- 35.Stewart, A. E., S. Dowd, S. M. Keyse, and N. Q. McDonald. 1999. Crystal structure of the MAPK phosphatase Pyst1 catalytic domain and implications for regulated activation. Nat. Struct. Biol. 6:174-181. [DOI] [PubMed] [Google Scholar]
- 36.Sun, H., C. H. Charles, L. F. Lau, and N. K. Tonks. 1993. MKP-1 (3CH134), an intermediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell 75:487-493. [DOI] [PubMed] [Google Scholar]
- 37.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2:110-116. [DOI] [PubMed] [Google Scholar]
- 38.Tanoue, T., T. Moriguchi, and E. Nishida. 1999. Molecular cloning and characterization of a novel dual specificity phosphatase, MKP-5. J. Biol. Chem. 274:19949-19956. [DOI] [PubMed] [Google Scholar]
- 39.Tanoue, T., and E. Nishida. 2003. Molecular recognitions in the MAP kinase cascades. Cell. Signalling 15:455-462. [DOI] [PubMed] [Google Scholar]
- 40.Tanoue, T., T. Yamamoto, R. Maeda, and E. Nishida. 2001. A Novel MAPK phosphatase MKP-7 acts preferentially on JNK/SAPK and p38 alpha and beta MAPKs. J. Biol. Chem. 276:26629-26639. [DOI] [PubMed] [Google Scholar]
- 41.Tanoue, T., T. Yamamoto, and E. Nishida. 2002. Modular structure of a docking surface on MAPK phosphatases. J. Biol. Chem. 277:22942-22949. [DOI] [PubMed] [Google Scholar]
- 42.Theodosiou, A., and A. Ashworth. 2002. MAP kinase phosphatases. Genome Biol. 3:reviews3009.1-reviews3009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theodosiou, A., A. Smith, C. Gillieron, S. Arkinstall, and A. Ashworth. 1999. MKP5, a new member of the MAP kinase phosphatase family, which selectively dephosphorylates stress-activated kinases. Oncogene 18:6981-6988. [DOI] [PubMed] [Google Scholar]
- 44.Treisman, R. 1995. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 14:4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wishart, M. J., J. M. Denu, J. A. Williams, and J. E. Dixon. 1995. A single mutation converts a novel phosphotyrosine binding domain into a dual-specificity phosphatase. J. Biol. Chem. 270:26782-26785. [DOI] [PubMed] [Google Scholar]
- 46.Wishart, M. J., and J. E. Dixon. 1998. Gathering STYX: phosphatase-like form predicts functions for unique protein-interaction domains. Trends Biochem. Sci. 23:301-306. [DOI] [PubMed] [Google Scholar]
- 47.Yang, S.-H., A. D. Sharrocks, and A. J. Whitmarsh. 2003. Transcriptional regulation by the MAP kinase signaling cascades. Gene 320:3-21. [DOI] [PubMed] [Google Scholar]
- 48.Yang, S. H., P. Shore, N. Willingham, J. H. Lakey, and A. D. Sharrocks. 1999. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 18:5666-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.