Abstract
The Cdc14 dual-specificity phosphatases regulate key events in the eukaryotic cell cycle. However, little is known about the function of mammalian CDC14B family members. Here, we demonstrate that subcellular localization of CDC14B protein is cell cycle regulated. CDC14B can bind, bundle, and stabilize microtubules in vitro independently of its catalytic activity. Basic amino acid residues within the nucleolar targeting domain are important for both retaining CDC14B in the nucleolus and preventing microtubule bundling. Overexpression of CDC14B resulted in the formation of cytoplasmic CDC14B and microtubule bundles in interphase cells. These microtubule bundles were resistant to microtubule depolymerization reagents and enriched in acetylated α-tubulin. Expression of cytoplasmic forms of CDC14B impaired microtubule nucleation from the microtubule organization center. CDC14B is thus a novel microtubule-bundling and -stabilizing protein, whose regulated subcellular localization may help modulate spindle and microtubule dynamics in mitosis.
The mammalian spindle midzone or central mitotic spindle formed between the two separating sets of chromatids consists of bundled interdigitating microtubules (MTs) (18). Only at this time of the cell cycle do mammalian cells generate stable and bundled MTs. Numerous proteins required for cell cleavage accumulate at the midzone of mitotic spindles and play important roles for the completion of cytokinesis in several experimental systems (18). Proteins belonging to this category include chromosomal passenger proteins, such as the inner centromere protein (INCENP), Aurora, and survivin; MT motor proteins, such as CHO1/MKLP1 and KLP3A; and the polo family kinases (75). Among these proteins, some play essential roles in regulating density and bundling of the central mitotic spindles and are required for proper formation of the spindle midzone (1, 37, 48, 51, 61, 74). As the cleavage furrow closes, the midbody is formed at the center of intracellular bridging MTs (75). Although the function of the midbody is still not clear, studies from several experimental systems have suggested that the midbody helps coordinate cytokinesis and G1 phase transition (46, 75).
The dual-specificity phosphatase Cdc14 is conserved from yeast to mammals and plays a variety of roles during cell division (64). In budding yeast Saccharomyces cerevisiae, Cdc14 functions primarily to inactivate cyclin-dependent kinase (CDK) activity at the end of mitosis, thereby allowing reentry into G1 phase. Cells that lack Cdc14 arrest with elongated mitotic spindles, separated chromosomes (49), and high levels of Clb-Cdc28 kinase activity (67). Cdc14 inactivates CDK through dephosphorylation of the cyclin-dependent kinase inhibitor Sic1, which is thereby stabilized, and dephosphorylation of Cdh1, which is then able to activate the anaphase-promoting complex, the E3 ubiquitin ligase that degrades the Clb B-type cyclins (24). The low CDK state in G1 phase is essential for the establishment of origins of DNA replication for the ensuing S phase (8). Incomplete inactivation of CDK activity (for example, in cells that lack the CDK inhibitor Sic1) causes massive replication defects and genome instability (30, 43). From G1 until early anaphase, Cdc14 is sequestered in an inactive state in the nucleolus (55, 68). Release of Cdc14 from the nucleolus and exit from mitosis is dictated by the Cdc14 early anaphase release network and mitotic exit network (58, 59). In the fission yeast Schizosaccharomyces pombe, the Cdc14 homolog Clp1/Flp1 appears to function primarily in determining the onset of mitosis (7, 65). In addition to its role in mitotic exit in budding yeast, activation of Cdc14 in early anaphase leads to dephosphorylation of Sli15, which in turn directs the Sli15-Ipl1 (INCENP-Aurora) kinase complex to the anaphase spindle and promotes midzone spindle assembly (45). There is also evidence that Cdc14 helps mediate spindle assembly in Caenorhabditis elegans, where CeCDC-14 is localized to the central spindle and midbody (16). Suppression of CeCDC-14, using RNA-mediated interference, leads to embryonic lethality, primarily due to failure in establishing central mitotic spindles and cytokinesis (16).
In humans, two CDC14 paralogs, CDC14A and CDC14B, have been identified (32). CDC14A dynamically localizes to the interphase centrosome and plays a role in centrosome separation (27, 33). Similar to the budding yeast Cdc14, CDC14A can dephosphorylate Hct1/Cdh1 (3) and INCENP (17). The Xenopus othologs of human CDC14A, called XCdc14α and XCdc14β, are essential for embryonic division and localize to the nucleolus and centrosome, respectively (26). In contrast to CDC14A, CDC14B has not been studied in detail, except that CDC14B associates with nucleoli (27), centrosomes, and intranuclear filaments, and it may be involved in nuclear structure maintenance (39).
Here, we show that the CDC14B protein can bind, bundle, and stabilize MTs in vitro independently of its catalytic activity. We demonstrate that CDC14B localizes to the equator of central spindles in anaphase and to the center of the midbody during telophase and cytokinesis. Overexpression of cytoplasmic CDC14B causes MT bundling and stabilization in interphase cells. Moreover, MT nucleation from the microtubule organization center (MTOC) was delayed in cells with bundled MTs. The regulated localization of CDC14B may therefore directly control spindle assembly and disassembly during mitosis.
MATERIALS AND METHODS
Cell culture and transfection.
U-2OS (ATCC), Saos-2 (ATCC), and 293T (ATCC) cells were cultivated in high-glucose Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin in a 5% CO2 atmosphere at 37°C. Cells were transfected by FuGene 6 (Roche) or Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. After 21-h transfection of Cdc14B plasmids, cells were treated with MT-depolymerizing drugs: nocodazole (10 μg/ml, 2 h; Sigma), vinblastine (1 μM, 4 h; Sigma), or Colcemid (500 ng/ml, 4 h; Sigma).
For the nocodazole washout experiment, U-2OS cells transfected with different Cdc14B-FLAG plasmids were exposed to nocodazole at a final concentration of 10 μg/ml for 2 h. The drug was removed by being rapidly washed three times in warm phosphate-buffered saline (PBS). Cells were fixed at 2, 4, 6, 8, 10, and 12 min after washout in methanol at −20°C and then processed at room temperature for immunofluorescence staining of CDC14B and α-tubulin.
Cloning and site-directed mutagenesis.
Cdc14B open reading frame (ORF) cDNA was amplified by PCR from the Marathon human heart cDNA library (Clontech) using the following primers: forward, 5′-ACTCCCGGGTCCATGAAGCGGAAAAGCGAGC-3′; and reverse, 5′-AGTCCCGGGTTAACGCAAGACTGTTTTAGTCC-3′. The PCR product was digested with SmaI and cloned into the SnaBI site of pMX-pie vector (a kind gift from Gerry Nolan) carrying an N-terminal 6-myc epitope tag. Sequencing analysis of the resulting plasmid confirmed that the Cdc14B cDNA was identical to hCdc14B in GenBank (AF023158.1), except for a T-to-C transition at position 52 from the first ATG. The BamHI/BglII fragment containing the myc-tagged Cdc14B was subcloned into pRSHisGal (MT852) yeast expression vector. The last 68 bp of Cdc14B were deleted from this construct. All of the Cdc14B mutants (C314S, D287A, KKIR29-32AAIA, and KKIR29-32AAIA plus C314S) were generated by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and verified by sequencing analysis. Annealing primers used for site-directed mutagenesis will be provided upon request. Cdc14B expression plasmids were generated by subcloning the entire Cdc14B ORF into pET16b (Novagen), pGEX-3X (Amersham Biosciences), p3xFlag-CMV-14 (Sigma), and pEGFP-N3 (Clontech) vectors. The inducible expression system of Cdc14B was generated by subcloning the C-terminal enhanced green fluorescent protein (EGFP)-tagged Cdc14B into pBI-tet vector (Clontech). Cdc14A ORF cDNA was amplified by PCR from the Marathon human testis cDNA library (Clontech) using the following primers: forward, 5′-CATAGAATTCCATGGCAGCGGAGTCAGGGGAAC-3′; and reverse, 5′-CGCTGCGGCCGCTTAGTAATGAACATATTCAGACTG-3′. The PCR product was cloned into pMXpie. Sequencing analysis of the resulting plasmid confirmed that the Cdc14A cDNA insert was identical to hCdc14A in GenBank (AF122013). The BamHI/NotI fragment containing myc-tagged Cdc14A was subsequently subcloned into pRSHisGal and the entire ORF was subcloned into pET16b. MAP4MTB (MT binding domain, 692 to 1,151 amino acids of MT-associated protein 4 [MAP4]) (22) was amplified from a Marathon human brain cDNA library (Clontech) using the following primers: forward, 5′-GGAGCTCCCACCAAGCCCA-3′; and reverse, 5′-GATGCTTGTCTCCTGGATCTG-3′. It was then cloned into pET16b.
Antibodies and immunofluorescence.
Rabbit anti-CDC14A and anti-CDC14B antibodies (affinity purified with CDC14A and CDC14B peptides, respectively) were purchased from Zymed Laboratories, Inc. To increase the antibody specificity for CDC14B, the antibody was engaged for an additional round of affinity purification using glutathione-S-transferase (GST)-CDC14B fusion protein or CDC14B separated by factor Xa from a GST-CDC14B fusion on a nitrocellulose membrane. The antibody was eluted with a buffer containing 0.75 ml of 4 M MgCl2 and 50 μg/ml bovine serum albumin (BSA). The buffer was then exchanged over an NAP-10 column (Sephadex G25 medium, DNA grade; Pharmacia Biotech) equilibrated with 50 μg/ml BSA in PBS. Each fraction was then tested by immunocytochemical and Western blot analyses. To verify the specificity of the antibody, the CDC14B antibody was either preabsorbed to a GST-CDC14B fusion protein on beads (depleted) or preincubated with glutathione-eluted GST-CDC14B protein in solution (competed). Both the depleted and the competed antibody fractions were used for Western blotting and immunostaining as controls of antibody specificity.
Mouse anti-myc (9E10) and anti β-actin monoclonal antibodies were obtained from Santa Cruz and Cytoskeleton, Inc., respectively. For double immunostaining, rabbit anti-CDC14B and mouse anti α-tubulin (B-5-1-2; Sigma), or mouse anti-acetylated α-tubulin (6-11B-1; Sigma) antibodies were used. For double staining of CDC14B-FLAG transfected cells, mouse anti-FLAG M2 (Sigma) and rabbit anti-α-tubulin (Neo Markers) antibodies were used. Alexa Fluor 568 goat anti-rabbit immunoglobulin G and Alexa Fluor 488 goat anti-mouse immunoglobulin G were purchased from Molecular Probes, Inc.
For immunostaining, U-2OS cells were either grown on coverslips or in slide chambers. We fixed cells in 100% methanol at −20°C, followed by brief extraction with 0.5% NP-40. In some experiments, cells were also fixed with 2.5% paraformaldehyde in PBS for 10 min, followed by 0.1% Triton X-100 extraction at room temperature. Both fixation protocols gave rise to the same staining patterns in this study. Immunofluorescence images were visualized with a Zeiss immunofluorescence microscope and acquired with a charge-coupled device camera using the Photoshop software.
Immunoblotting, recombinant protein production, and phosphatase assay.
U-2OS cell pellets were lysed in buffer 3 (1% NP-40, 0.1 M Tris, pH 8.0, 0.15 M NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). To prepare yeast cell lysates, cells propagating in log phase were collected, and proteins were released by being vortexed in acid-washed glass beads containing 10% trichloroacetic acid. The protein pellets were resuspended in buffer 3, followed by pH adjustment. Forty micrograms of total lysates was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to nitrocellulose membrane with a semidry electrophoresis transfer apparatus (Bio-Rad) and probed with antibodies according to the manufacturer's instructions. Blots were developed using the ECL system (Pharmacia Amersham Biotech).
GST-CDC14B, CDC14B-His, CDC14A-His, and MAP4MTB-His fusion proteins were prepared as described according to the manufacturer's instructions. Briefly, Escherichia coli cells carrying GST- or His-tagged plasmids were induced by 1 mM (final concentration) IPTG (isopropyl-ā-d-thiogalactopyranoside) for 4 h at 30°C, and soluble fractions were isolated with glutathione-Sepharose beads (Amersham Pharmacia) or nitrilotriacetic acid-Ni beads (QIAGEN), respectively. A phosphatase assay was performed at 30°C in a volume of 100 μl containing 15 mM ρ-nitrophenyl phosphate, 50 mM imidazole, pH 6.9, 1 mM dithiothreitol, 1 mM EDTA, and equal amounts of each recombinant protein (∼2 μg). The reaction was terminated by the addition of 400 μl of 0.25 N NaOH, and absorbance was read at an optical density of 405 nm on a spectrophotometer.
In vitro microtubule-binding and -bundling assay.
MT-binding assays were performed with the Microtubule Associated Protein Spin-Down Biochem Assay kit (Cytoskeleton, Inc.). Purified His-tagged CDC14A, CDC14B, and MAP4MTB proteins were exchanged into PEM buffer {80 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8, 1 mM EGTA, and 1 mM MgCl2} using a desalting spin column (Pierce) immediately prior to the assay. In addition, CDC14B preparations were prespun through 50% glycerol cushion-PEM at 100,000 × g for 15 min and the supernatants were used in the binding assays. For each assay, MTs were assembled from 100 μg of pure tubulins (isolated from bovine brain; Cytoskeleton, Inc.) in 20 μl of PEM containing 1 mM GTP and 5% glycerol at 35°C for 20 min and immediately stabilized in 200 μl of warm PEM-20 μM Taxol (paclitaxel). The MTs (4 μM tubulin) were incubated with CDC14Bwt (0.2 μM), CDC14BC314S (0.2 μM), CDC14A (0.5 μM), MAP4MTB (0.5 μM), or BSA (2.3 μM) in a total volume of 50 μl at room temperature for 30 min. The reaction mixtures were then centrifuged through a 50% glycerol cushion-PEM-Taxol at 100,000 × g at 25°C for 15 min. The supernatant and pellet were resolved on 10% SDS-PAGE and the presence of CDC14B, CDC14A, and tubulin was detected by immunoblot analysis. Protein abundance in the control reactions was determined by Coomassie blue staining and densitometry.
For MT-bundling assays, rhodamine-MTs were prepared using a mixture of 20 μg of rhodamine-labeled tubulins (≥99% pure, from bovine brain; Cytoskeleton, Inc.) and 100 μg of unlabeled tubulins as described above. After incubating MTs (0.4 μM tubulin) with CDC14B (0.04 μM), CDC14A (0.03 μM), MAP4MTB (0.1 μM), or MAP2 (0.3 μM, from bovine brain; Cytoskeleton, Inc.) at room temperature for 30 min, the mixtures were immediately dropped onto slides and examined by fluorescence microscopy. Each protein of the same concentration for the MT-bundling assay was incubated with the MTs (0.2 μM tubulin) for electron microscopic analysis. For each reaction mixture, 2 μl was fixed in 60 μl of warm PEM fixative containing 1% glutaraldehyde, 1 mM GTP, and 30% glycerol. Five microliters of the fixed samples was applied to grids with carbon and negatively stained with 2% uranyl formate. All images were recorded at magnifications of ×50,000 or ×70,000 under low-dose conditions with a JEOL 1200 EX II microscope at 100 kV.
Yeast growth conditions.
Conditions for growth of the temperature sensitive cdc14-3 mutant strain were previously described (67). Transformation of yeast cells was performed by the lithium acetate-polyethylene glycol method (13). Transformed cells were selected on His dropoff plates with and without 2% galactose at permissive (25°C) and nonpermissive (37°C) temperatures.
RESULTS
Conservation of CDC14B function.
To study the function of human CDC14B, we set out to determine whether human CDC14B is functionally equivalent to its yeast ortholog, Cdc14, by testing the ability of the human gene to complement a temperature sensitive (TS) cdc14-3 mutant strain of S. cerevisiae (49). Expression of human CDC14B from the inducible GAL1 promoter efficiently restored growth of the cdc14-3 mutant strain at 37°C (Fig. 1A). In contrast, when expressed at a level equivalent to CDC14B, human CDC14A failed to rescue the cdc14-3 strain (Fig. 1A and B). As a control, Clp1p, a Schizosaccharomyces pombe ortholog of Cdc14 also rescued the cdc14-3 defect (65). To determine whether catalytic activity of CDC14B was required for complementation, the conserved cysteine (C314) and aspartate (D287) residues in the active site of CDC14B (15) were mutated to serine and alanine, respectively. Phosphatase assays confirmed that the CDC14BC314S and CDC14BD287A mutant proteins were catalytically inert (Table 1). Neither CDC14BC314S nor CDC14BD287A rescued cdc14-3 lethality at 37°C (Fig. 1A). Human CDC14B is therefore a functional counterpart of the S. cerevisiae Cdc14 phosphatase.
FIG. 1.
(A) Expression of human CDC14B complements cdc14-3 defects at nonpermissive temperatures. The temperature-sensitive mutant cdc14-3 cells containing the indicated plasmids were tested for colony formations on yeast-peptone-dextrose plates with or without (w/o) galactose (Gal) at nonpermissive (37°C) and permissive (25°C) temperatures. Human CDC14BC314S and CDC14BD287A are catalytic “dead” mutants (Table 1) (15). Clp1, a Schizosaccharomyces pombe ortholog of S. cerevisiae Cdc14, was used as a positive control (65). (B) Western blot analysis of Myc-tagged human CDC14A and CDC14B expression using anti-myc antibody (9E10) in cdc14-3 cells transformed with pGAL-Cdc14A (lane 1), pGAL-Cdc14B (lane 2), and vector alone (lane 3). All plasmids were constructed in pRSHisGal (MT852) yeast expression vector.
TABLE 1.
Summary of in vivo and in vitro studies of CDC14B
| Construct | No. (%) of cells, cytoplasmic bundlea | No. (%) of cells, no bundleb | Total no. of cells counted | In vitro MT bindingc | In vitro MT bundlingc | Phosphatase activityd |
|---|---|---|---|---|---|---|
| Cdc14BKKIR | 1,471 (59) | 1,029 (41) | 2,500 | + | + | 1.058 ± 0.012 |
| Cdc14BK&C | 408 (20) | 1,690 (80) | 2,098 | + | + | 0.0003 ± 0.0002 |
| Cdc14Bwt | 202 (14) | 1,298 (86) | 1,500 | + | + | 0.784 ± 0.032 |
| Cdc14BC314S | 63 (4) | 1,436 (96) | 1,499 | + | + | 0.0003 ± 0.0002 |
Twenty-one hours after transfection with the FLAG-tagged constructs indicated above, cells were coimmunostained with anti-FLAG and α-tubulin antibodies. The number of cells containing both cytoplasmic-bundled CDC14B and MT bundles were counted with a fluorescence microscope.
FLAG-tagged CDC14B proteins were found in nucleoli and nuclei.
In vitro MT binding and bundling assays were performed using purified CDC14B His-tagged proteins (see Materials and Methods for details).
The phosphatase activities of purified CDC14B His-tagged proteins were measured using ρ-nitrophenyl phosphate as a substrate. All results represent at least triplicate experiments.
Subcellular localization of CDC14B protein is cell cycle regulated.
In S. cerevisiae, Cdc14 is regulated through sequestration in the nucleolus (55, 68). To investigate whether a similar mechanism operates for human CDC14B, we examined the subcellular localization of CDC14B in U-2OS cells at different stages of the cell cycle. To do so, we developed a highly purified anti-CDC14B antibody by subjecting a commercial affinity-purified anti-CDC14B antibody (Zymed Laboratory, Inc.) to an additional round of purification against a GST-CDC14B fusion protein. Immunoblot analysis demonstrated that the purified antibody recognized a 62-kDa protein in U-2OS cells and that this species was not detected after predepletion or competition of the antibody with GST-CDC14B protein (Fig. 2A, left). This antibody preparation specifically recognized FLAG-tagged CDC14B expressed in transfected U-2OS cells but not FLAG-tagged CDC14A (Fig. 2A, right). Longer exposure of this blot revealed a species at a molecular weight consistent with endogenous CDC14B but not endogenous CDC14A (data not shown). In agreement with a previous report (27), immunostaining of asynchronous U-2OS cells using the affinity-purified antibody revealed that CDC14B localized in nucleoli during interphase and dispersed throughout the cell during prophase and metaphase (Fig. 2B, top). Significantly, we also found that CDC14B accumulated at the spindle midzone in anaphase, at the dense center of the midbody in telophase, and along the intracellular bridge at the termination of cytokinesis (Fig. 2B, top). To affirm the specificity of the CDC14B staining, we showed that the anti-CDC14B antibody preincubated with GST-CDC14B did not reveal any CDC14B signal, regardless of the cell cycle stage (Fig. 2B, panel “compete”). We verified the specificity of midbody staining by colocalization of a midbody protein, AIM-1 (data not shown) (62). Identical subcellular localization of CDC14B was observed with 293T and Saos-2 cells (data not shown).
FIG. 2.
Subcellular localization of human CDC14B protein in U-2OS cells at different stages of the cell cycle. (A) Characterization of anti-CDC14B antibody. (Left) Western blot analysis of endogenous CDC14B protein with anti-CDC14B antibody (original, Zymed Lab, Inc.), affinity-purified anti-CDC14B antibody with GST-CDC14B fusion protein (purified), anti-CDC14B antibody that has been preblocked with GST-CDC14B (compete) and anti-CDC14B antibody that has been predepleted with GST-CDC14B (deplete). (Right) U-2OS cells transiently transfected by FLAG-tagged-Cdc14A (lane 1) and -Cdc14B (lane 2) were analyzed by Western blotting using affinity-purified anti-CDC14B, anti-FLAG antibodies, or anti-CDC14B antibody that had been predepleted with purified GST-CDC14B protein (deplete). (B) U-2OS cells were labeled with affinity-purified antibody against CDC14B (red), monoclonal antibody against α-tubulin (green), and DAPI for DNA (blue). Representative CDC14B localizations in nucleoli, spindle midzone, midbody, and intracellular bridge at different stages of a cell cycle are shown. Note that CDC14B localizes to the equator of central spindle in anaphase, is confined to the dense center of midbody flanked by bundled MTs in telophase, and stretches out along intracellular bridging MTs at the end of cytokinesis. No nucleolar, midzone, or midbody staining was evident when the same anti-CDC14B antibody was preblocked with GST-CDC14B (compete).
The spindle midzone in anaphase contains bundled central MTs and many chromosomal passenger proteins (75). To determine the relationship between CDC14B and bundled central MTs, we performed double immunofluorescence staining using anti-CDC14B and anti-α-tubulin antibodies in U-2OS cells. In anaphase, CDC14B accumulated at the midzone in a region with overlapping MTs, while in telophase, CDC14B localized to the center of the midbody, which was flanked by the bundled MTs (Fig. 2B, bottom). At the end of cytokinesis, when cells were approaching the final stage of separation, CDC14B became dispersed and localized along the intracellular bridge as the bundled MTs became thin and confined within the intracellular bridge (Fig. 2B, bottom).
CDC14B directly binds microtubules in vitro.
The localization of CDC14B in central spindles suggests that CDC14B may interact with MTs. To test this hypothesis, we performed an MT cosedimentation assay using His-tagged CDC14B (CDC14B-His) and Taxol-stabilized MTs assembled from purified tubulins. MTs were incubated with CDC14B-His in solution and then sedimented through a glycerol cushion by centrifugation. For these assays, it was essential that the CDC14B protein be freshly prepared (data not shown). At a tubulin:CDC14B molar ratio of 20:1 (based on the densitometry of the input proteins in Coomassie-stained gels), approximately 60% of the input wild-type CDC14B-His cosedimented with MTs, whereas in the absence of MTs CDC14B-His remained in the supernatant (Fig. 3A). Because the sedimentation efficiency of MTs under these conditions was also approximately 60%, CDC14B was quantitatively bound to the purified MTs in this assay. As controls, after incubation with MTs, His-tagged MAP4MTB (22) was recovered with similar efficiency, while BSA remained exclusively in the supernatant (Fig. 3B, bottom). We then determined whether the interaction with MTs required CDC14B catalytic activity. Similar to wild-type CDC14B, catalytically inactive CDC14BC314S proteins associated efficiently with MTs (Fig. 3A). Taken together, these results demonstrate that CDC14B can directly and specifically bind to MTs in vitro in a manner that appears to be independent of its catalytic activity. As a fraction of CDC14A was recently shown to associate with the spindle poles and the central spindle (17), we tested the possibility that CDC14A might also bind to MTs. Indeed, purified CDC14A-His was able to bind to MTs with a binding efficiency 11-fold higher than that of the background level, although capture was somewhat less efficient than for CDC14B (Fig. 3B). The ability to interact directly with MTs may thus be a general property of the Cdc14 phosphatase family.
FIG. 3.
CDC14B and CDC14A are MT-binding proteins. CDC14Bwt-His, CDC14BC314S-His, or CDC14A-His proteins were incubated in the presence (+) or absence (−) of Taxol-stabilized MTs and then subjected to a glycerol cushion centrifugation. Pellets (P) and supernatants (S) were collected and separated by SDS-PAGE. The presence of CDC14B, CDC14A, and MTs were detected by Western blot analysis (A and B), and the input proteins were quantified on a Coomassie-stained gel (A and B). The input molar ratio of CDC14Bwt-His or CDC14BC314S-His to MT (tubulin) was 1:20 the input molar ratio of CDC14A-His to tubulin was 1:8. Note that there are two bands in the CDC14A-His input (B) but only the lower one is CDC14A determined by Western blot analysis. MAP4MTB-His and BSA were used as positive and negative controls, respectively (Coomassie-stained gel) (B, bottom).
Cytoplasmic CDC14B has microtubule-bundling activity in vivo.
The direct association of CDC14B with MTs in vitro prompted us to investigate a possible role of CDC14B in MT network regulation in vivo. For this purpose, we transfected FLAG-tagged wild-type CDC14B (CDC14Bwt-FLAG) into U-2OS cells and then immunostained cells with anti-FLAG antibody. As reported previously (27), the majority of the transfected cells exhibited colocalization of CDC14B with interphase nucleoli and nuclei (Table 1). However, in 14% of the cells, CDC14Bwt-FLAG also appeared to be bundled as perinuclear rings or MT-like bundled structures. Double immunostaining with anti-FLAG and anti-α-tubulin antibodies revealed that the CDC14Bwt-FLAG bundles partially colocalized with MT networks (Fig. 4A, panels a to f). To further investigate possible physical interactions between CDC14B and MT networks, we generated a CDC14B mutant that predominantly localizes in the cytoplasm, where MT networks are normally assembled. The first 54 amino acids of CDC14B contain a potential nucleolar targeting domain, the removal of which results in relocalization of the majority of CDC14B protein into the cytoplasm (27). Within this domain, amino acid residues 7 to 32 (7RRSSWAAAPPCSRRCSSTSPGVKKIR32) contain a potential bipartite nuclear localization sequence (K/R2-X10-20-K/R3; in boldface type in the sequence above) (9). To abrogate nuclear localization, we thus mutated the three basic residues at positions 29, 30, and 32 to alanines (CDC14BKKIR29-32AAIA, abbreviated as CDC14BKKIR). In comparison with CDC14Bwt-FLAG transfected cells, the number of cytoplasmic CDC14B bundles in CDC14BKKIR-FLAG-transfected cells was significantly increased (t test; P < 0.01) (Table 1). All of the cytoplasmic CDC14BKKIR-FLAG protein appeared in rings or MT-like bundles and partially colocalized with MTs (Fig. 4A, panels a to f). This pattern of CDC14B bundles resembled that of other MT-associated proteins, which often form bundled rings and irregular filaments when overexpressed in cells (2, 29, 34, 57, 63, 70, 72). CDC14BKKIR-FLAG did not appear to colocalize with cytoskeletal intermediate filaments or actins by double immunostaining with anti-FLAG and anti-vimentin (VIM-3B4; Chemicon) antibodies, or with anti-FLAG antibody and fluorescein-conjugated phalloidin (Molecular Probes, Inc.) (data not shown). These results suggest that the 29KKIR32 motif restrains the ability of CDC14B to form cytoplasmic bundles, presumably through nucleolar and/or nuclear sequestration of CDC14B.
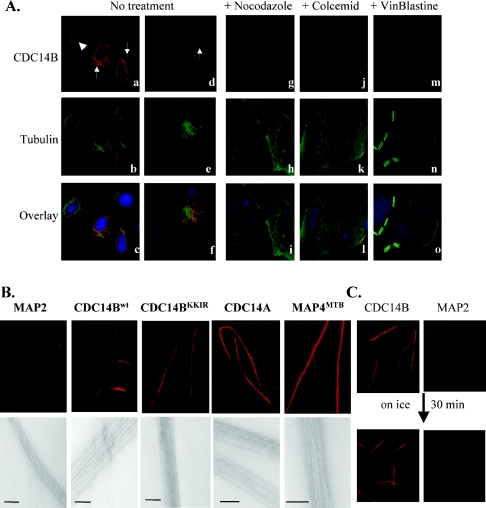
FIG. 4.
Overexpressed CDC14B proteins form cytoplasmic bundles and stabilize MT network. (A) Immunofluorescence staining of U-2OS cells transfected with CDC14Bwt- and CDC14BKKIR-FLAG (see also Table 1). Twenty-one hours after transfection, cells were fixed and immunostained with anti-FLAG and α-tubulin antibodies (No treatment). Representative images of CDC14B cytoplasmic bundles or perinuclear rings (arrows) and nucleolar or nuclear expression (arrowhead) are shown (a and d). CDC14BK&C- and CDC14BC314S-FLAG proteins showed similar bundling structures. However, the ratio of bundles versus nucleolar expression differed (see Table 1 for details). All cytoplasmic CDC14B bundles partially colocalized with the bundled MTs (c and f). After nocodazole (10 μg/ml; 2 h) and Colcemid (500 ng/ml; 4 h) treatment, MT structures were completely disrupted in the nontransfected cells (h and, k). High dose of vinblastine (1 μM; 4 h) resulted in the formation of tubulin-vinblastine paracrystals (n). In contrast, in cells expressing cytoplasmic CDC14B bundles, MTs remained bundled (h, k, and n) and colocalized with bundled CDC14Bs (i, l, and o). Red, CDC14B; green, tubulin; and blue, DNA. (B) In vitro MT-bundling assay. The MT-bundling activities of CDC14B and CDC14A were examined by incubating purified CDC14Bwt-His, CDC14BKKIR-His, or CDC14A-His proteins with rhodamine-labeled MTs. Each reaction mixture was then applied to slides for examination under fluorescence (top) and electron microscopes (bottom). MAP4MTB-His (22) and MAP2 proteins (23) were used as positive and negative controls, respectively. Note that MTs in the presence of CDC14Bwt-His, CDC14BKKIR-His, and CDC14A-His are long and thick in comparison with MTs in MAP2 reaction (top). High magnification (×50,000) of MTs showed that CDC14Bwt-His, CDC14BKKIR, CDC14A-His, and MAP4MTB-His caused extensive MT bundles (bottom). Scale bar, 200 nm. (C) CDC14B stabilizes MTs in vitro. Rhodamine-labeled MTs were incubated with purified CDC14Bwt-His or MAP2 for 30 min at room temperature. Subsequently, the reaction mixtures were placed on ice for 30 min, followed by fluorescence microscopic examination. Cold-resistant MT bundles were also observed when CDC14BKKIR-His, CDC14BC314S-His, and CDC14BK&C-His proteins were used.
To determine whether CDC14B catalytic activity is required for its cytoplasmic translocation and MT bundling, we generated a double mutant by changing the active cysteine into serine (C314S) in a CDC14BKKIR mutant (CDC14BKKIR29-32AAIA&C314S, abbreviated as CDC14BK&C). Approximately 20% of CDC14BK&C-FLAG-transfected cells exhibited cytoplasmic CDC14B bundles, which partially colocalized with MT bundles (Table 1). The percentage of cells with bundles appeared to vary somewhat among cells transfected with different Cdc14B constructs, ranging from 59% (CDC14BKKIR), 20% (CDC14BK&C), and 14% (CDC14Bwt) to 4% (CDC14BC314S) (Table 1). Since the extent of cytoplasmic CDC14BK&C localization was about twofold less than that of CDC14BKKIR (t test; P < 0.005), it is possible that Cdc14B catalytic activity is required for Cdc14B release from the nucleoli and/or its maintenance in the cytoplasm. In all cases, however, each of the CDC14B variants showed very similar bundled structures and colocalization with MTs, regardless of catalytic activity. These findings suggest that the catalytic activity of CDC14B is not required for induction of MT bundles in the cytoplasm or for localization of CDC14B within these MT bundles. Similar results were also obtained when wild-type or mutant CDC14B was expressed as GFP fusions from a doxycycline-regulatable promoter (see Table S1 in the supplemental material). We noticed that the CDC14Bwt-EGFP cells also formed intranuclear filamentous structures and associated with centrosomes, depending on the level of protein expression (see Fig. S1 in the supplemental material) in agreement with a recent report (39).
MT-bundling activity of CDC14B.
The collective evidence that CDC14B localizes to midzone central spindles (Fig. 2B), directly binds to MTs in vitro (Fig. 3), and causes MT bundling in vivo (Fig. 4A, panels a to f) suggests that CDC14B might be a bona fide MT-bundling protein. We therefore examined the effects of CDC14B in an in vitro MT-bundling assay based on the visualization of MTs from rhodamine-labeled tubulins. Purified CDC14Bwt-His or CDC14BKKIR-His proteins were incubated with rhodamine-MTs, and the reaction mixtures were examined immediately by fluorescence microscopy. CDC14B stimulated the formation of thick and long MTs (Fig. 4B, top), comparable to the MTs bundled by the MAP4MTB-His protein used as a positive control (22). A similar result was obtained when CDC14A-His was used in the MT-bundling assay (Fig. 4B, top). In contrast, MTs were thin and short in the presence of the MAP2 protein, which is unable to support MT bundling in vitro (5, 28). Thin and short MTs were also observed when BSA was used as a negative control (data not shown). To confirm that the thick and long MTs were indeed MT bundles, we examined the reaction mixtures by electron microscopy. Compared with a single MT seen in the presence of MAP2, ordered arrays of multiple MTs were observed in the presence of CDC14Bwt-His, CDC14BKKIR-His, CDC14A-His, and MAP4MTB-His (positive control) proteins (Fig. 4B, bottom). CDC14BKKIR-bundled MTs often formed tighter bundles and contained more MTs than CDC14Bwt-bundled MTs, suggesting that the 29KKIR32 sequence may partially impair MT bundling, in addition to serving as a nuclear localization signal. All of the CDC14B proteins that bound to MTs also appeared to support MT bundling in vitro, even those mutants devoid of phosphatase activity (Table 1). The results obtained in this purified system suggest that both CDC14A and CDC14B have intrinsic MT-bundling activity.
To examine whether the MT-bundling activity of CDC14B plays a role in MT regulation, we first tested MT stability by treating U-2OS cells with MT depolymerization reagents. Twenty-one hours after transfection with various Cdc14B-FLAG constructs (Table 1), cells were treated with nocodazole (10 μg/ml; 2 h), Colcemid (500 ng/ml; 4 h) or vinblastine (1 μM; 4 h) and then double labeled with anti-FLAG and anti-α-tubulin antibodies. In nontransfected cells, nocodazole and Colcemid completely disrupted MT networks (Fig. 4A, panels h and k), while treatment with vinblastine induced multiple vinblastine-tubulin paracrystals (56) (Fig. 4A, panel n). However, in cells bearing cytoplasmic CDC14B bundles, the MT network structures were refractory to disruption by each of the drugs tested (Fig. 4A, panels h, k, and n). Importantly, in the drug-treated cells the stabilized MTs colocalized exclusively with cytoplasmic CDC14B (Fig. 4A, panels i, l, and o), in comparison with the partial colocalization of MTs with CDC14B in untreated cells (Fig. 4A, panel c). To test whether CDC14B could directly stabilize MTs independently of any other MT-associated proteins, we examined the effect of CDC14B on MT cold sensitivity in the in vitro-purified system. Most mammalian MTs are cold sensitive and undergo rapid disassembly at low temperatures (69). When MTs were premixed with CDC14B-His protein and incubated on ice for 30 min, there was little or no depolymerization (Fig. 4C). In contrast, in the presence of MAP2, most MTs disassembled upon incubation at the low temperature (Fig. 4C).
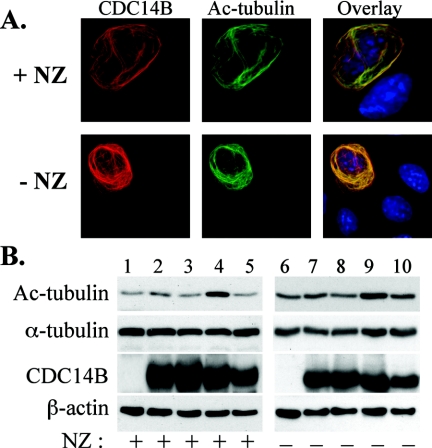
CDC14B stabilizes microtubules that are enriched in acetylated α-tubulins.
Acetylated α-tubulin accumulates in stable MTs (44, 47, 52, 60, 71). To further verify the MT stabilization by CDC14B, we examined the tubulin acetylation status of the drug-resistant MTs. When nocodazole-treated cells expressing CDC14B bundles were examined using an antibody specific to acetylated α-tubulin, we found that the nocodazole-resistant MTs were enriched with acetylated tubulins and completely colocalized with the CDC14B bundles (Fig. 5A, top). Colocalization of acetylated α-tubulin and CDC14B was also detected in the absence of nocodazole treatment (Fig. 5A, bottom). Immunoblot analysis revealed that acetylated tubulins accumulated in Cdc14BKKIR-FLAG-transfected cells (Fig. 5B, lanes 4 and 9), regardless of nocodazole treatment. Because the majority of the ectopically expressed CDC14Bwt, CDC14BK&C, and CDC14BC314S proteins were constrained to the nucleoplasm and nucleoli (Table 1), we were not able to detect accumulation of acetylated tubulin in these cells (Fig. 5B, lanes 2, 3, 5, 7, 8, and 10). These findings strengthen the suggestion that CDC14B is an MT-stabilizing protein.
FIG. 5.
Effect of cytoplasmic CDC14B bundles on α-tubulin acetylation. (A) Colocalization of acetylated tubulins and CDC14B bundles in the presence (+ NZ, 10 μg/ml and 2 h) or absence (− NZ) of nocodazole treatment. (B) Expression of cytoplasmic CDC14B leads to accumulation of acetylated tubulins. After transient transfection, U-2OS cells were incubated with (+) or without (−) nocodazole for 2 h. Cell lysates were resolved on 10% SDS-PAGE and Western blotting was performed by using anti-acetylated tubulin, α-tubulin, FLAG, and β-actin antibodies, respectively. Mock transfection (lanes 1 and 6), transfection with CDC14BK&C-FLAG (lanes 2 and 7), CDC14BC314S-FLAG (lanes 3 and 8), CDC14BKKIR-FLAG (lanes 4 and 9), and CDC14Bwt-FLAG (lanes 5 and 10).
Cytoplasmic CDC14B delays microtubule nucleation from MTOCs.
MTs emanating from MTOCs in interphase cells can rapidly reorganize to form a bipolar mitotic spindle during cell division. The dynamic behavior of these MTs can be analyzed by measuring the kinetics of MT nucleation from MTOCs (12, 47). Analysis of MTs recovered after nocodazole treatment revealed that while MT nucleation from MTOCs still occurred, these events were considerably delayed in cells bearing cytoplasmic CDC14B bundles. Two minutes after nocodazole washout, nontransfected cells exhibited long MTs radiating from MTOCs (Fig. 6, top right panel), whereas no obvious MTOC could be identified in the cytoplasmic CDC14B expressing cells (Fig. 6, top left panel). Similar results were observed at 4, 6, 8, and 10 min after nocodazole washout (data not shown). Twelve minutes after washout, the majority of nontransfected cells developed normal MT arrays (Fig. 6, bottom), as described by others (47, 57). At this time point, however, only a small fraction of cells bearing cytoplasmic CDC14B bundles had initiated MT nucleation from MTOCs (Fig. 6, bottom, right). Despite impairment of MT nucleation by cytoplasmic CDC14B, we did not observe any alteration of centrosome copy numbers by immunostaining for γ-tubulin (data not shown). These results suggest that alteration of CDC14B subcellular localization can alter MT dynamics and that CDC14B-mediated MT stabilization may delay MT nucleation.
FIG. 6.
Expression of cytoplasmic CDC14B bundle delays MT nucleation from microtubule organization center. Twenty-one hours after transfection with CDC14BKKIR-FLAG, cells were exposed to the MT depolymerization drug nocodazole (10 μg/ml) for 2 h and then washed out of nocodazole, followed by 2-, 4-, 6-, 8-, and 12-min incubation in warm fresh culture medium. Representative results from 2- and 12-min time points are shown. Transfected CDC14B proteins and MTs were visualized by being stained with anti-FLAG (left) and α-tubulin (right) antibodies. Two minutes after nocodazole washout, long MTs were nucleated from MTOCs in nontransfected cells (2 min; arrowhead). In contrast, MTs remained bundled and no obvious MTOC appeared in cells with CDC14B bundles at the 2-min time point. Twelve minutes after nocodazole washout, all of the nontransfected cells appeared to be repopulated by relative normal MT arrays. In contrast, only a small portion of CDC14B-bundled cells just started to nucleate MTs from semiprominent MTOCs (arrowhead).
DISCUSSION
Human CDC14B is the functional ortholog of budding yeast Cdc14, as judged by its ability to complement yeast strains defective in Cdc14 function and the analogous spatial regulation evident in both yeast and human cells. In budding yeast, Cdc14 is sequestered in the nucleolus prior to anaphase through binding to its nucleolar inhibitor Net1/Cfi1 (55, 68). Release of Cdc14 from the nucleolus in the late stages of mitosis allows the dephosphorylation of substrates and subsequent exit from mitosis (67). We suggest that a similar nucleolar sequestration mechanism may in part serve to prevent CDC14B from directly interacting with interphase MTs in human cells. Failure to prevent this interaction will have dire effects on MT networks and dynamics. As there is no obvious metazoan homolog of Cfi1/Net1, it has yet to be determined how sequestration of CDC14B in the nucleolus is controlled. However, the potential bipartite NLS containing the 29KKIR32 sequence within CDC14B could interact with importin α and β in a manner similar to that of other MT-associated proteins, such as NuMA and TPX2 (25). Alternatively, it is possible that this motif may mediate an interaction of CDC14B with an unknown nucleolar anchor protein(s). Regardless of the precise mechanism, reimport of CDC14B into the nucleus and/or nucleolus after mitosis must be necessary to prevent disruption of nascent MT networks in G1 phase.
The cell cycle-dependent subcellular localization of CDC14B to spindle structures resembles that of other midzone-midbody-associated proteins, for example, PRC1, CHO1, and PTP-BL (20, 36, 37). Sequence analysis of CDC14B did not reveal any consensus repetitive motifs commonly observed in the MT binding domains of other MAPs, such as Tau, MAP2, and MAP4 (10, 41), suggesting that CDC14B may contain novel MT binding motifs. The induction of drug- and cold-resistant MT bundles associated with CDC14B overexpression indicates that CDC14B can regulate MT stability. Many other MAPs such as Tau, MAP2, and NuSAP are capable of stabilizing MTs both in vivo and in vitro (31, 50, 60). During anaphase, central overlapping MTs become bundled to form the midzone central spindle (35). Successful completion of cytokinesis depends on the timely and spatially regulated assembly of the midzone (6, 14, 73). Because CDC14B localizes to the midzone and can bundle and/or stabilize MTs, CDC14B may be involved in the regulation of central spindle assembly and mitotic MT dynamics. Indeed, genetic evidence suggests an analogous function for the C. elegans Cdc14 ortholog (16). While the manuscript was under review, Higuchi and Uhlmann demonstrated that yeast Cdc14 indeed functions to stabilize mitotic MT dynamics and suggested that downstream targets of Cdc14 are involved in this process (21). Thus, MT-binding and -stabilizing activity may be a general and conserved feature of all Cdc14 family members. A recent study indicates that CDC14B may have an unexpected role in maintenance of nuclear structure (39). In agreement with this suggestion, we found that a portion of CDC14B was localized to centrosomes when expressed at a low level but to intranuclear filaments and cytoplasmic bundles when expressed at a high level from a doxycycline-regulatable promoter (see Fig. S1 in the supplemental material). CDC14B may thus play a number of roles in mitosis, including spindle and centrosome regulation and control of nuclear architecture. At this point, genetic evidence in support of this notion in mammalian cells is lacking, in part because small interfering RNA and short hairpin RNA approaches fail to completely eliminate CDC14B protein and cause little or no defect in tissue culture cells (reference 39 and data not shown). Genetic analysis of CDC14B function must await generation of CDC14B null cell lines or CDC14B knockout mice. Interestingly, ectopic expression of PLK1, Cdc5-like polo-like kinase 1 (38), in CDC14B knockdown cells led to severe nuclear deformation and cytokinesis defects (39). This synthetic genetic interaction between PLK1 and CDC14B may in part reflect the fact that the two proteins regulate cytokinesis in the same pathway, in agreement with our finding that CDC14B may be involved in the regulation of central spindle formation.
Our observation that acetylated α-tubulin accumulates in MT bundles of CDC14B transfected cells is consistent with the fact that overexpression of other MT-associated proteins, such as MAP1B, MAP2, or TAU, increases MT stability and α-tubulin acetylation (60). In several experimental systems, acetylated α-tubulins are most abundant in stable MTs but absent from more dynamic MT structures (44, 47, 52, 60, 71). Concordantly, we find that CDC14B protein normally localizes in the spindle midzone and the center of the midbody, precisely when acetylated tubulins accumulate in the stable MTs (47). It is possible that under physiological conditions, CDC14B participates in the establishment and/or maintenance of MT stabilization in the spindle midzone either through directly stabilizing and bundling MTs and/or through modulating tubulin acetylation. Recently, it has been suggested that CDC14B dephosphorylates a tubulin deacetylase, SIRT2, which in turn triggers SIRT2 ubiquitination and degradation (11, 42). It is conceivable that CDC14B exerts its effects on MT stabilization in part through SIRT2 degradation and consequent accumulation of acetylated tubulins. However, we cannot rule out the possibility that CDC14B stabilizes MTs and that these stabilized MTs accumulate acetylated tubulins, as suggested by Palazzo et al. in drug-induced tubulin acetylation experiments (44).
Cytoskeletal MTs undergo continuous assembly and disassembly during the cell cycle and development (12, 19, 54). We have found that inappropriate localization of CDC14B to the cytoplasm delays MT nucleation from MTOCs. The timely regulated exit of CDC14B from the nucleolus during the cell cycle may well be critical to avoid disruption of MT dynamics. Recently, Saito et al. demonstrated that CDC14 promotes cell cycle arrest in various tissues of C. elegans, possibly through regulating CKI-1 protein stability (53). It is possible that CDC14 participates in both cell cycle regulation (53) and microtubule organization (16) in C. elegans. Indeed, constitutive expression of cytoplasmic CDC14B causes MT bundling and stabilization and arrests cells in G1 (our unpublished observations). Overexpression of the MT-associated protein MAP4 also stabilizes MTs and significantly delays G1/S transition (40), while the anticancer agent Taxol induces MT hyperpolymerization and blocks untransformed cells in late G1 through an as-yet-uncharacterized MT-dependent G1 checkpoint mechanism (66).
As a dual-specificity phosphatase, CDC14B executes its functions in part through dephosphorylation of proteins that are phosphorylated by proline-directed kinases (15, 27). The spectrum of Cdc14 substrates encompasses a number of proteins important for CDK regulation. A spindle-associated function has been established for Cdc14 in yeast, as it has been demonstrated that dephosphorylation of Sli15 by Cdc14 directs Sli15-Ipl1, an INCENP/Aurora kinase-like complex, to the spindle midzone in a step important for central spindle assembly (45). Since INCENP is phosphorylated prior to mitosis in higher organisms (4), the intrinsic MT-stabilizing and -bundling properties of CDC14B may serve to direct CDC14B to the central spindle where it coordinates central spindle assembly with INCENP dephosphorylation. Indeed, CDC14A localizes to the central spindle and can dephosphorylate mitotic INCENP (17); furthermore, we demonstrated here that CDC14A can bind and bundle MTs in vitro. Given that both CDC14A and CDC14B are present in the central spindle, it will be important to determine whether the two phosphatases act in synergy to regulate INCENP dephosphorylation and central spindle formation. In addition to the catalytic functions of Cdc14 in spindle dynamics, we have found that CDC14B can unexpectedly bind, stabilize, and bundle MTs independently of its phosphatase activity. Thus, CDC14B may directly stabilize and organize the central spindle independently of any intermediate substrate. To our knowledge, this is also the first instance in which the biological function of a phosphatase has been segregated from its intrinsic phosphatase activity. Elucidation of the mechanisms that regulate CDC14B and the means whereby it alters MT dynamics will undoubtedly be crucial for understanding mitotic transition in the normal and cancer cell cycles.
Supplementary Material
Acknowledgments
We thank Lea Harrington and Angelika Amon for critical reading of the manuscript and Ashley Davis for technical support regarding the in vitro MT-bundling assay. We also thank Angelika Amon for the cdc14-3 strain and Kathy Gould for the pGAL-clp1 plasmid.
Y.W. acknowledges the support of the Laboratory Directed Research and Development Program (LDRD), Oak Ridge National Laboratory, and the Office of Biological and Environmental Research, U.S. Department of Energy, under contract DE-AC05-00OR22725 with UT-Battelle, LLC. H.P.C. is a postdoctoral fellow supported by LDRD. M.T. is supported by grants from the Canadian Institutes of Health Research and holds a Canada Research Chair in Functional Genomics and Bioinformatics.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, R. R., A. A. Tavares, A. Salzberg, H. J. Bellen, and D. M. Glover. 1998. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 12:1483-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlow, S., M. L. Gonzalez-Garay, R. R. West, J. B. Olmsted, and F. Cabral. 1994. Stable expression of heterologous microtubule-associated proteins (MAPs) in Chinese hamster ovary cells: evidence for differing roles of MAPs in microtubule organization. J. Cell Biol. 126:1017-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bembenek, J., and H. Yu. 2001. Regulation of the anaphase-promoting complex by the dual specificity phosphatase human Cdc14a. J. Biol. Chem. 276:48237-48242. [DOI] [PubMed] [Google Scholar]
- 4.Bolton, M. A., W. Lan, S. E. Powers, M. L. McCleland, J. Kuang, and P. T. Stukenberg. 2002. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell 13:3064-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgin, K. E., B. Ludin, J. Ferralli, and A. Matus. 1994. Bundling of microtubules in transfected cells does not involve an autonomous dimerization site on the MAP2 molecule. Mol. Biol. Cell 5:511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, L. G., and Y. L. Wang. 1996. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol. Biol. Cell 7:225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cueille, N., E. Salimova, V. Esteban, M. Blanco, S. Moreno, A. Bueno, and V. Simanis. 2001. Flp1, a fission yeast orthologue of the s. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 114:2649-2664. [DOI] [PubMed] [Google Scholar]
- 8.Diffley, J. F., and K. Labib. 2002. The chromosome replication cycle. J. Cell Sci. 115:869-872. [DOI] [PubMed] [Google Scholar]
- 9.Dingwall, C., and R. A. Laskey. 1998. Nuclear import: a tale of two sites. Curr. Biol. 8:R922-R924. [DOI] [PubMed] [Google Scholar]
- 10.Doll, T., M. Meichsner, B. M. Riederer, P. Honegger, and A. Matus. 1993. An isoform of microtubule-associated protein 2 (MAP2) containing four repeats of the tubulin-binding motif. J. Cell Sci. 106:633-639. [DOI] [PubMed] [Google Scholar]
- 11.Dryden, S. C., F. A. Nahhas, J. E. Nowak, A. S. Goustin, and M. A. Tainsky. 2003. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol. Cell. Biol. 23:3173-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dustin, P. 1984. Microtubules. Springer-Verlag, New York, N.Y.
- 13.Gietz, R. D., and R. A. Wood. 2002. Transformation of yeast by lithium acetate/single stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 14.Glotzer, M. 2003. Cytokinesis: progress on all fronts. Curr. Opin. Cell Biol. 15:684-690. [DOI] [PubMed] [Google Scholar]
- 15.Gray, C. H., V. M. Good, N. K. Tonks, and D. Barford. 2003. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 22:3524-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruneberg, U., M. Glotzer, A. Gartner, and E. A. Nigg. 2002. The CeCDC-14 phosphatase is required for cytokinesis in the Caenorhabditis elegans embryo. J. Cell Biol. 158:901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruneberg, U., R. Neef, R. Honda, E. A. Nigg, and F. A. Barr. 2004. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166:167-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guertin, D. A., S. Trautmann, and D. McCollum. 2002. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66:155-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gundersen, G. G., S. Khawaja, and J. C. Bulinski. 1989. Generation of a stable, posttranslationally modified microtubule array is an early event in myogenic differentiation. J. Cell Biol. 109:2275-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann, L., T. Dittmar, and K. S. Erdmann. 2003. The protein tyrosine phosphatase PTP-BL associates with the midbody and is involved in the regulation of cytokinesis. Mol. Biol. Cell 14:230-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higuchi, T., and F. Uhlmann. 2005. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 433:171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iida, J., T. J. Itoh, H. Hotani, K. Nishiyama, H. Murofushi, J. C. Bulinski, and S. Hisanaga. 2002. The projection domain of MAP4 suppresses the microtubule-bundling activity of the microtubule-binding domain. J. Mol. Biol. 320:97-106. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, T. J., and H. Hotani. 1994. Microtubule-stabilizing activity of microtubule-associated proteins (MAPs) is due to increase in frequency of rescue in dynamic instability: shortening length decreases with binding of MAPs onto microtubules. Cell Struct. Funct. 19:279-290. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen, P., and M. Tyers. 1999. Altered states: programmed proteolysis and the budding yeast cell cycle. Curr. Opin. Microbiol. 2:610-617. [DOI] [PubMed] [Google Scholar]
- 25.Kahana, J. A., and D. W. Cleveland. 2001. Cell cycle. Some importin news about spindle assembly. Science 291:1718-1719. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser, B. K., M. V. Nachury, B. E. Gardner, and P. K. Jackson. 2004. Xenopus Cdc14α/β are localized to the nucleolus and centrosome and are required for embryonic cell division. BMC Cell Biol. 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser, B. K., Z. A. Zimmerman, H. Charbonneau, and P. K. Jackson. 2002. Disruption of centrosome structure, chromosome segregation, and cytokinesis by misexpression of human Cdc14A phosphatase. Mol. Biol. Cell 13:2289-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, H., L. I. Binder, and J. L. Rosenbaum. 1979. The periodic association of MAP2 with brain microtubules in vitro. J. Cell Biol. 80:266-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koonce, M. P., J. Kohler, R. Neujahr, J. M. Schwartz, I. Tikhonenko, and G. Gerisch. 1999. Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J. 18:6786-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lengronne, A., and E. Schwob. 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G1. Mol. Cell 9:1067-1078. [DOI] [PubMed] [Google Scholar]
- 31.Lewis, S. A., I. E. Ivanov, G. H. Lee, and N. J. Cowan. 1989. Organization of microtubules in dendrites and axons is determined by a short hydrophobic zipper in microtubule-associated proteins MAP2 and tau. Nature 342:498-505. [DOI] [PubMed] [Google Scholar]
- 32.Li, L., B. R. Ernsting, M. J. Wishart, D. L. Lohse, and J. E. Dixon. 1997. A family of putative tumor suppressors is structurally and functionally conserved in humans and yeast. J. Biol. Chem. 272:29403-29406. [DOI] [PubMed] [Google Scholar]
- 33.Mailand, N., C. Lukas, B. K. Kaiser, P. K. Jackson, J. Bartek, and J. Lukas. 2002. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell Biol. 4:317-322. [DOI] [PubMed] [Google Scholar]
- 34.Mandelkow, E. M., O. Schweers, G. Drewes, J. Biernat, N. Gustke, B. Trinczek, and E. Mandelkow. 1996. Structure, microtubule interactions, and phosphorylation of tau protein. Ann. N. Y. Acad. Sci. 777:96-106. [DOI] [PubMed] [Google Scholar]
- 35.Mastronarde, D. N., K. L. McDonald, R. Ding, and J. R. McIntosh. 1993. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 123:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matuliene, J., and R. Kuriyama. 2002. Kinesin-like protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol. Biol. Cell 13:1832-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mollinari, C., J. P. Kleman, W. Jiang, G. Schoehn, T. Hunter, and R. L. Margolis. 2002. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157:1175-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mundt, K. E., R. M. Golsteyn, H. A. Lane, and E. A. Nigg. 1997. On the regulation and function of human polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem. Biophys. Res. Commun. 239:377-385. [DOI] [PubMed] [Google Scholar]
- 39.Nalepa, G., and J. W. Harper. 2004. Visualization of a highly organized intranuclear network of filaments in living mammalian cells. Cell Motil. Cytoskeleton 59:94-108. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen, H. L., S. Chari, D. Gruber, C. M. Lue, S. J. Chapin, and J. C. Bulinski. 1997. Overexpression of full- or partial-length MAP4 stabilizes microtubules and alters cell growth. J. Cell Sci. 110:281-294. [DOI] [PubMed] [Google Scholar]
- 41.Noble, M., S. A. Lewis, and N. J. Cowan. 1989. The microtubule binding domain of microtubule-associated protein MAP1B contains a repeated sequence motif unrelated to that of MAP2 and tau. J. Cell Biol. 109:3367-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.North, B. J., B. L. Marshall, M. T. Borra, J. M. Denu, and E. Verdin. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11:437-444. [DOI] [PubMed] [Google Scholar]
- 43.Nugroho, T. T., and M. D. Mendenhall. 1994. An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol. Cell. Biol. 14:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palazzo, A., B. Ackerman, and G. G. Gundersen. 2003. Cell biology: tubulin acetylation and cell motility. Nature 421:230. [DOI] [PubMed] [Google Scholar]
- 45.Pereira, G., and E. Schiebel. 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302:2120. [DOI] [PubMed] [Google Scholar]
- 46.Piel, M., J. Nordberg, U. Euteneuer, and M. Bornens. 2001. Centrosome-dependent exit of cytokinesis in animal cells. Science 291:1550-1553. [DOI] [PubMed] [Google Scholar]
- 47.Piperno, G., M. LeDizet, and X. J. Chang. 1987. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J. Cell Biol. 104:289-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powers, J., O. Bossinger, D. Rose, S. Strome, and W. Saxton. 1998. A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 8:1133-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pringle, J. R., and L. H. Hartwell. 1981. The Saccharomyces cerevisiae cell cycle, p. 97-142. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Raemaekers, T., K. Ribbeck, J. Beaudouin, W. Annaert, M. Van Camp, I. Stockmans, N. Smets, R. Bouillon, J. Ellenberg, and G. Carmeliet. 2003. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. 162:1017-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raich, W. B., A. N. Moran, J. H. Rothman, and J. Hardin. 1998. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell 9:2037-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robson, S. J., and R. D. Burgoyne. 1989. Differential localisation of tyrosinated, detyrosinated, and acetylated alpha-tubulins in neurites and growth cones of dorsal root ganglion neurons. Cell Motil. Cytoskeleton 12:273-282. [DOI] [PubMed] [Google Scholar]
- 53.Saito, R. M., A. Perreault, B. Peach, J. S. Satterlee, and S. van den Heuvel. 2004. The CDC-14 phosphatase controls developmental cell-cycle arrest in C. elegans. Nat. Cell Biol. 6:777-783. [DOI] [PubMed] [Google Scholar]
- 54.Saxton, W. M., D. L. Stemple, R. J. Leslie, E. D. Salmon, M. Zavortink, and J. R. McIntosh. 1984. Tubulin dynamics in cultured mammalian cells. J. Cell Biol. 99:2175-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed, Z. W. Chen, J. Jang, H. Charbonneau, and R. J. Deshaies. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244. [DOI] [PubMed] [Google Scholar]
- 56.Smith, C. D., and X. Zhang. 1996. Mechanism of action cryptophycin. Interaction with the Vinca alkaloid domain of tubulin. J. Biol. Chem. 271:6192-6198. [DOI] [PubMed] [Google Scholar]
- 57.Smith, D. S., M. Niethammer, R. Ayala, Y. Zhou, M. J. Gambello, A. Wynshaw-Boris, and L. H. Tsai. 2000. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2:767-775. [DOI] [PubMed] [Google Scholar]
- 58.Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108:207-220. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan, M., and F. Uhlmann. 2003. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 5:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takemura, R., S. Okabe, T. Umeyama, Y. Kanai, N. J. Cowan, and N. Hirokawa. 1992. Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J. Cell Sci. 103:953-964. [DOI] [PubMed] [Google Scholar]
- 61.Terada, Y. 2001. Role of chromosomal passenger complex in chromosome segregation and cytokinesis. Cell Struct. Funct. 26:653-657. [DOI] [PubMed] [Google Scholar]
- 62.Terada, Y., M. Tatsuka, F. Suzuki, Y. Yasuda, S. Fujita, and M. Otsu. 1998. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 17:667-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Togel, M., G. Wiche, and F. Propst. 1998. Novel features of the light chain of microtubule-associated protein MAP1B: microtubule stabilization, self interaction, actin filament binding, and regulation by the heavy chain. J. Cell Biol. 143:695-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trautmann, S., and D. McCollum. 2002. Cell cycle: new functions for Cdc14 family phosphatases. Curr. Biol. 12:R733-R735. [DOI] [PubMed] [Google Scholar]
- 65.Trautmann, S., B. A. Wolfe, P. Jorgensen, M. Tyers, K. L. Gould, and D. McCollum. 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11:931-940. [DOI] [PubMed] [Google Scholar]
- 66.Trielli, M. O., P. R. Andreassen, F. B. Lacroix, and R. L. Margolis. 1996. Differential Taxol-dependent arrest of transformed and nontransformed cells in the G1 phase of the cell cycle, and specific-related mortality of transformed cells. J. Cell Biol. 135:689-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers, and A. Amon. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2:709-718. [DOI] [PubMed] [Google Scholar]
- 68.Visintin, R., E. S. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818-823. [DOI] [PubMed] [Google Scholar]
- 69.Wallin, M., and E. Stromberg. 1995. Cold-stable and cold-adapted microtubules. Int. Rev. Cytol. 157:1-31. [DOI] [PubMed] [Google Scholar]
- 70.Waterman-Storer, C. M., S. Karki, and E. L. Holzbaur. 1995. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1). Proc. Natl. Acad. Sci. USA 92:1634-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Webster, D. R., and G. G. Borisy. 1989. Microtubules are acetylated in domains that turn over slowly. J. Cell Sci. 92:57-65. [DOI] [PubMed] [Google Scholar]
- 72.Weisshaar, B., T. Doll, and A. Matus. 1992. Reorganisation of the microtubular cytoskeleton by embryonic microtubule-associated protein 2 (MAP2c). Development 116:1151-1161. [DOI] [PubMed] [Google Scholar]
- 73.Wheatley, S. P., and Y. Wang. 1996. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell Biol. 135:981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams, B. C., M. F. Riedy, E. V. Williams, M. Gatti, and M. L. Goldberg. 1995. The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 129:709-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeitlin, S. G., and K. F. Sullivan. 2001. Animal cytokinesis: breaking up is hard to do. Curr. Biol. 11:R514-R516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.