Abstract
Smads 1, 5, and 8 are the intracellular mediators for the bone morphogenetic proteins (BMPs), which play crucial roles during mammalian development. Previous research has shown that Smad1 is important in the formation of the allantois, while Smad5 has been shown to be critical in the process of angiogenesis. To further analyze the BMP-responsive Smads, we disrupted the murine Smad8 gene utilizing the Cre/loxP system. A Smad8 hypomorphic allele (Smad8Δexon3) was constructed that contains an in-frame deletion of exon 3, removing one-third of the MH2 domain and a small portion of the linker region. Xenopus injection assays indicated that this Smad8 deletion allele is still functional but has reduced ventralizing capability compared to the wild type. Although Smad8Δexon3/Δexon3 embryos are phenotypically normal, homozygotes of another hypomorphic allele of Smad8 (Smad83loxP) containing a neomycin cassette within intron 3, phenocopy an embryonic brain defect observed in roughly 22% of Smad1+/− embryos analyzed at embryonic day 11.5. These observations suggest that BMP-responsive Smads have critical functions in the development of the mammalian central nervous system.
Bone morphogenetic proteins (BMPs) are signaling molecules that belong to the transforming growth factor β superfamily, a set of ligands that exert a wide array of biological effects. BMPs were first discovered as the constituent of demineralized bone extracts that could induce the formation of bone and cartilage in rodents (60). Later, BMPs were shown to play important roles in many diverse developmental processes including limb development, axis specification, generation of primordial germ cells, and patterning of the neural tube (37).
To date, there have been eight murine Smad genes identified, with Smads 1, 5, and 8 having been shown to be responsive to BMP signals. Once BMPs are secreted, they bind to type II BMP receptors. These then recruit and phosphorylate type I receptors within intracellular GS (glycine-serine-rich) domains. Type I receptors in turn phosphorylate Smad1, 5, or 8 at the C terminus at a conserved SSXS motif. Smad4, the co-Smad, can then multimerize with the BMP-regulated Smads to facilitate their translocation into the nucleus to either transcriptionally activate or repress target genes (3, 4, 13, 61, 62).
The downstream effects of BMP signaling have been partly elucidated by numerous means, including gene knockout approaches with mice. Smad1 and Smad5 have been previously disrupted in other laboratories. Smad1 null homozygotes die at embryonic day (E10.5) due to defects in the formation of the allantois and placenta (29, 55). Smad5 null homozygotes also die at E10.5 due to angiogenic failure, mesenchymal apoptosis, and other defects (6, 7, 64).
Although the preceding data have been uncovered through gene knockout approaches with mice, additional BMP functions have been revealed through experimental approaches. For example, a number of laboratories have shown that the establishment of dorsal cell fates within the spinal cord is dependent upon the action of BMP signaling (31). For instance, BMP4 and BMP7 proteins can induce the expression of dorsal cell markers when placed in contact with nonneural ectoderm from chicken explants (11). Conversely, specific ablation of the roof plate in mouse embryos using the conditional expression of diphtheria toxin causes a reduction in dorsal cell markers such as BMP7, Msx1, and Zic2 (30). It has been difficult to assess similar roles for specific BMPs in the mouse, probably due to their many redundant functions. However, one example has shown that the deletion of Gdf7, a BMP-related molecule expressed in the dorsal neural tube, results in the loss of the D1A interneuron (32).
Though functional analyses of BMP actions in the murine spinal cord have yielded some insights, there is little known about BMP functions in brain development. Interestingly, BMP2, -4, -5, -6, and -7 are coexpressed within the dorsal medial telencephalon, although single knockouts show either an early embryonic lethality, in the case of BMP2 and -4, or show no neural phenotype, as for BMP5, -6, or -7 (65). However, double knockouts of BMP5 and -7 show defects in neural tube closure, confirming functional redundancies of BMPs within the nervous system (49). Chrd and Nog, antagonists of BMP signaling (46), have also been shown to be important for proper embryonic brain development in the mouse. Chrd−/−; Nog+/− mice display a number of anterior neuroectodermal abnormalities such as cyclopia, holoprosencephaly, and rostral brain reductions. Embryos doubly homozygous for Chrd and Nog display forebrain reductions in addition to dorsoventral and left/right patterning defects (1, 5).
To gain further understanding of BMP-responsive Smads in relation to mammalian development, we disrupted the murine Smad8 gene. Here we report the creation of two hypomorphic alleles for Smad8: an in-frame deletion of exon 3 that causes no observable defects and another that contains a neomycin resistance cassette within intron 3. This latter allele causes a partially penetrant reduction in the embryonic midbrain and hindbrain, which is also exhibited by some Smad1+/− embryos. Our results show that this phenotype is associated with hypercellularity within the dorsal neuroectoderm, although changes in dorsoventral patterning are not seen.
MATERIALS AND METHODS
Construction of the Smad83loxP targeting vector and generation of Smad8Δexon3/Δexon3 mice.
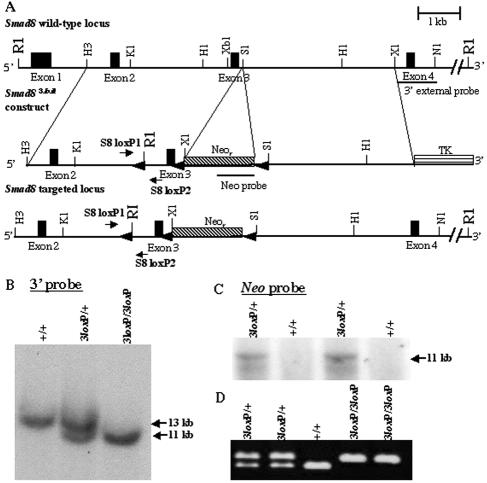
A Smad8 probe containing exons 4 and 5 was used to screen a 129Sv/Ev bacterial artificial chromosome library (43). A 12-kb HindIII fragment of Smad8 spanning exons 2 through 5 was isolated and subcloned into pBluescript (Stratagene, La Jolla, CA). To generate the Smad83loxP construct (Fig. 1A), a 4.2-kb HindIII/SalI subfragment was isolated, filled in with Klenow, and subcloned into the HpaI site of pLoxPII (63). Another 4.3-kb SalI/XhoI arm of homology was subcloned into the SalI site of pLoxPII, which effectively placed a floxed PGK-neo cassette (48) within intron 3. Neomycin was inserted in the forward orientation relative to Smad8 (Fig. 1A). Lastly, a 43-bp loxP site with an engineered flanking EcoRI site was subcloned into an HpaI site within intron 2 of Smad8. TC1 129Sv/Ev embryonic stem (ES) cells (8) were electroporated with 50 micrograms of NotI-linearized DNA and selected with ganciclovir and G418 according to the method of Deng et al. (9). DNA from 160 drug-resistant clones was digested with EcoRI and analyzed by Southern blotting using two probes: a 1-kb 3′ external probe and a neomycin probe (Fig. 1B and C, respectively). Of these ES cell clones, two had been successfully targeted to the Smad8 locus. Correctly targeted clones were injected into C57BL/6 blastocysts, and chimeric mice were generated. Chimeric males were mated with National Institutes of Health (NIH) Black Swiss females, and the offspring were subsequently bred inter se. Once Smad83loxP/3loxP mice were generated, they were bred to EIIA-Cre mice (28) to excise the neomycin cassette for the production of Smad8 floxed and Smad8Δexon3 mice.
FIG. 1.
Targeting the murine Smad8 gene. (A) Partial map of the Smad8 genomic locus containing four of the five exons (top), the Smad83loxP targeting vector (middle), and the targeted Smad8 locus (bottom). (B) Southern blot analysis of tail biopsy DNA confirming correct gene targeting of Smad8. Targeting of the Smad8 locus yields a wild-type 13-kb band and a targeted 11-kb band upon digestion of genomic DNA with EcoRI and hybridization with the 3′ external probe indicated in panel A. (C) Further Southern blot analysis with the same digestion scheme using a neomycin probe confirmed correct targeting. An 11-kb band is detected with a neomycin-specific probe. (D) PCR analysis of tail biopsy DNA extracted from Smad83loxP/3loxP mice. Primers were designed within intron 2 flanking the inserted loxP site (A). The lower and upper bands represent the targeted and wild-type alleles, respectively. R1, EcoRI; H3, HindIII; K1, KpnI; H1, HpaI; Xb1, XbaI; S1, SalI; X1, XhoI; N1, NdeI.
Genotype analysis.
Smad83loxP mice were genotyped for the presence of the loxP site within intron 2 of Smad8 using PCR (Fig. 1D). Forward and reverse primers were designed flanking the loxP site, S8 loxP1 (5′-CACAGAAGGAGAGAGGGACGG-3′) and S8 loxP2 (5′-TGGATACAGGTTGCCTCACAG-3′). Mice bearing Smad8Δexon3 alleles were genotyped using the Smad8 loxP1 primer and S8R (5′-CTATCCTGTCTCACAGCCCTC-3′) located 3′ to the neomycin cassette. Smad1 genotyping was performed as previously described (29).
Histological analysis and in situ hybridization.
E11.5 embryos were dissected free from the uterus and extraembryonic membranes and fixed in 4% paraformaldehyde with rocking at 4°C overnight. The following day, the embryos were dehydrated through an ethanol series, embedded in paraffin, sectioned at 7 μm, and stained with hematoxylin and eosin. Radioactive in situ hybridization was performed using standard procedures. 35S-labeled probes were prepared for Pax3 (22), Pax6 (56), Foxa2 (27), and Shh (17).
Neurofilament antibody staining.
E11.5 embryos were stained with the 2H3 neurofilament antibody developed by T. M. Jessell and T. Dodd and obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa, Iowa City, IA.
BrdU cellular proliferation assay.
Pregnant mice were subcutaneously injected with 5-bromo-2′-deoxyuridine (BrdU) at 10 μg per gram body weight. After 3 hours, the mice were sacrificed and embryos were prepared as described above. A BrdU in situ detection kit (BD Pharmingen, San Diego, CA) was used to perform immunohistochemical staining against BrdU according to the manufacturer's instructions. Two abnormal Smad1+/− embryos and wild-type siblings were evaluated by analysis under high magnification of the number of stained cells within the dorsal neural tube. Three random areas within the dorsal neural tube were analyzed, and mitotic indexes were calculated. Results were shown to be statistically significant with a P value of <1.0 × 10−4.
Xenopus injection assay.
The Smad8 wild-type (WT) or Smad8 truncated (TD) cDNAs were cloned into the EcoRI site of the pCS2+ vector from a Smad8 expression vector (42). A Smad8Δexon3 cDNA was generated from an inverse PCR using the Smad8 expression vector as a template and polyacrylamide gel electrophoreis-purified primers 5′-CAG ACA GTC CCT ATC AAC ACT CAG GTG TG CAT TTG TAC TAC GTT GGG G-3′ and 5′-CCC CAA CGT AGT ACA AAT GCA CAC CTG AGT GTT GAT AGG GAC TGT CTG-3′. The Smad8Δexon3 cDNA was sequenced to verify the absence of mutations and was then cloned into the EcoRI site of the pCS2+ vector. The Smad8Δexon4,5 cDNA vector was constructed by amplifying the first three exons of Smad8 and cloning this fragment into the EcoRI site of the pCS2+ vector. Each of these constructs was linearized with NotI and used as a template to produce mRNA by in vitro transcription using the mMessage mMachine kit (Ambion, Austin, TX). Xenopus embryos were prepared and cultured as previously described (18). One nanogram of Smad8 and derivative mRNAs was injected into the marginal zone of 4-cell-stage embryos. To control for the injection process, β-galactosidase mRNA was injected into the marginal zone of control embryos. A total of 60 Xenopus blastulae were injected with the above mRNAs and phenotypically scored utilizing the Xenopus dorso-anterior index system (26), and the results were analyzed using an unpaired t test.
Transfections and reporter assays.
All culture media and reagents used in this study were acquired from Invitrogen (CA). Bl6 cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum supplemented with 60 units/ml penicillin and 60 μg/ml streptomycin. All cell lines were grown in 5% CO2, 95% humidity at 37°C. For transfections into various cell lines, SuperFect reagent (QIAGEN, Hilden, Germany) was used according to the manufacturer's instructions. Cells were seeded into 12-well dishes at 70 to 80% confluence 24 h before transfection. The amount of DNA per transfection was kept consistent by the addition of an empty expression vector, pCS2+. Cells were harvested 36 h after transfection, rinsed with 1× phosphate-buffered saline, and lysed in 200 μl of reporter lysis buffer (Promega, Madison, WI). Lysates were frozen on dry ice and thawed in a 37°C water bath. For luciferase and β-galactosidase reporter assays, kits were purchased from Promega and used according to the manufacturer's instructions. Experiments were performed in triplicate, and results were normalized to β-galactosidase values. Error bars represent the deviations from the results of at least three independent experiments.
RESULTS
Targeted disruption of the murine Smad8 gene.
A targeting vector was constructed such that exon 3 of the Smad8 gene was flanked by loxP sites (48), while a neomycin resistance cassette (neo) was introduced into intron 3 of Smad8 (Fig. 1A). The neo cassette within the targeting vector, driven by the PGK promoter, is in the same transcriptional orientation as Smad8 (Fig. 1A). This allele of Smad8 will be subsequently referred to as Smad83loxP.
After electroporation and selection with G418 and ganciclovir, ES cell clones were screened by Southern blot analysis (Fig. 1B and C). Correctly targeted clones were injected into C57BL/6 blastocysts to generate chimeric mice. Smad83loxP/+ mice were genotyped for the presence of the loxP site within intron 2 using PCR (Fig. 1D).
The Smad8Δexon3 allele functions as a hypomorph in Xenopus injection assays.
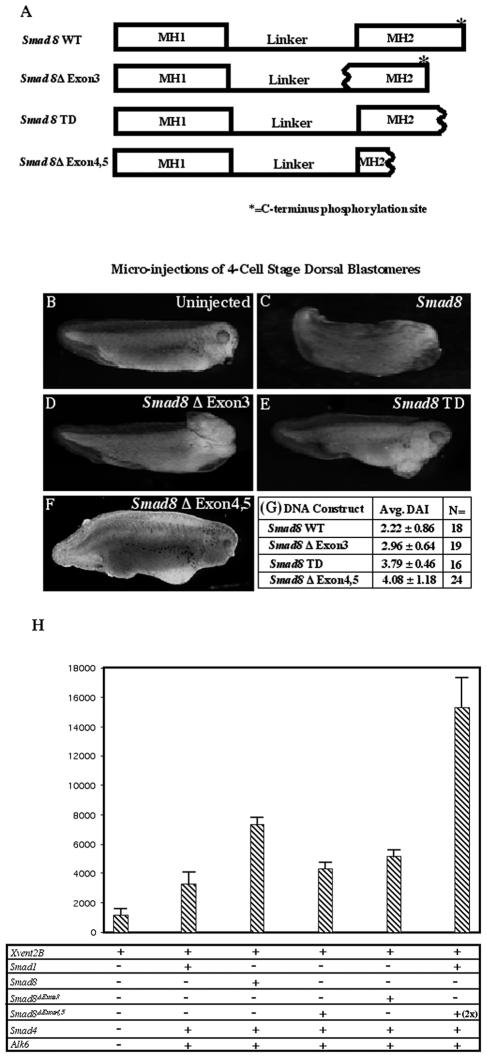
To elucidate the functional consequence(s) of deleting exon 3 from Smad8, we utilized an in vivo assay with Xenopus laevis. Prior research has shown that Xsmad1, Xsmad5, and Xsmad8 mRNAs are all capable of ventralizing Xenopus tadpoles when injected into the dorsal side of 4-cell-stage embryos, although some homologs of the latter fail to do so (39, 41, 47, 50, 52). Here we show that mouse Smad8 is similarly capable of ventralizing Xenopus embryos. Four Smad8 mRNAs were injected into Xenopus embryos: full-length Smad8 (Smad8 WT), Smad8 lacking exon three (Smad8Δexon3), Smad8 lacking exons four and five (Smad8Δexon4,5), and a Smad8 cDNA lacking the C-terminal phosphorylation site (Smad8 TD) (42) (Fig. 2A). A qualitative analysis utilizing the dorso-anterior index (DAI) indicated that Smad8 WT mRNA had a ventralizing effect on Xenopus embryos compared to uninjected controls (Fig. 2B and C), while Smad8 TD had an attenuated effect (Fig. 2E). The latter result was expected given the fact that a similar Smad1 mutant behaves as a dominant negative (45). Although Smad8Δexon3 was able to ventralize Xenopus tadpoles, it had a reduced effect compared to wild-type Smad8 (P = 5.2 × 10−3) (Fig. 2D). This suggests that the Smad8Δexon3 allele could act as a hypomorph rather than as a null mutation of Smad8 in the mouse. The mRNA made by the Smad83loxP allele, Smad8Δexon4,5, was tested similarly. Interestingly, this allele was also able to weakly ventralize Xenopus embryos (P < 1.0 × 10−4) (Fig. 2F). This was quite surprising, as the Smad8Δexon4,5 mutation deletes roughly 75% of the MH2 domain and would be expected to disable the protein. Indeed, a similar allele of Smad2, in which the MH2 domain had been deleted, was nonfunctional in Xenopus assays (34). DAI scores for the various Smad8 alleles are shown in Fig. 2G.
FIG. 2.
The Smad8Δexon3 allele acts as a hypomorph in Xenopus laevis. (A) Schematic representations of Smad8 WT, Smad8Δexon3, Smad8Δexon4,5, and Smad8 TD constructs used for microinjections into four-cell-stage embryos. Wavy lines represent deletion breakpoints. Smad8Δexon3 lacks one-third of the MH2 domain and a small portion of the linker region. Smad8Δexon4,5 lacks the majority of the MH2 domain. Smad8 TD lacks the C-terminal phosphorylation site. (B) Uninjected embryos develop normally. (C) Microinjection of Smad8 WT mRNA into dorsal blastomeres ventralizes Xenopus tadpoles. (D) Microinjection of Smad8Δexon3 into dorsal blastomeres also ventralizes Xenopus tadpoles but to an intermediate extent. (E) Smad8 TD lacks the majority of the Smad8 ventralizing capability. (F) Microinjection of Smad8Δexon4,5 into dorsal blastomeres also ventralizes Xenopus tadpoles but to a lesser extent. Note, β-galactosidase mRNA was coinjected with Smad8Δexon4,5 mRNA in the image shown in panel F. (G) Average (Avg.) DAI scores and numbers of embryos (N) areshown. (H) Results from a luciferase reporter assay utilizing the Xvent2B promoter driving luciferase and cDNAs for the following constructs: Smad1, Smad8, Smad4, Alk-6, Smad8Δexon3, and Smad8Δexon4,5.
Our Xenopus assays demonstrated that we had created two novel hypomorphic alleles of Smad8. Although these alleles were not null, we suspected that they might be invaluable, since null Smad1 and Smad5 mutants die during early embryogenesis (6, 7, 29, 55, 64). We therefore hypothesized that an analysis of hypomorphic Smad8 alleles would allow for further dissection of BMP functions throughout mammalian development.
Smad8Δexon3/Δexon3 mice are viable and fertile.
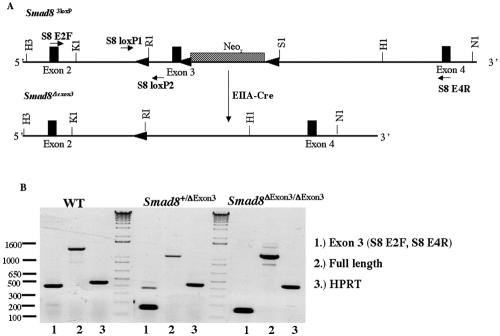
Smad83loxP/3loxP mice were mated with EIIA-Cre mice, which zygotically express Cre in a mosaic fashion (28). This has previously been shown to efficaciously create excision products from alleles with multiple loxP sites (24) and was successful in creating Smad8Δexon3 mice (Fig. 3A). The F1 generation, resulting from the cross between an EIIA-Cre male and a Smad83loxP/3loxP female, produced mosaic offspring, which carried all of the expected excision alleles. F1 progeny carrying the Smad8Δexon3 allele were mated to wild-type females, and F2 mice were genotyped again for the Smad8Δexon3 allele and allowed to interbreed. This intercross resulted in Smad8Δexon3/Δexon3 mice that were healthy, fertile, and represented at normal Mendelian ratios (data not shown) and that have life spans of approximately 2 years, equivalent to their wild-type siblings. To confirm that exon 3 was absent from the Smad8 transcript, reverse transcription (RT)-PCR was performed, demonstrating its complete deletion in Smad8Δexon3/Δexon3 mice (Fig. 3B). Additionally, the junction between exons 2 and 4 was sequenced and shown to be in frame with the rest of the Smad8 transcript, as expected (data not shown). The Smad8Δexon3 mutation should result in deletion of amino acids 224 to 298, deleting part of the linker as well as one-third of the MH2 domain of Smad8. Real-time PCR was used to ascertain the level of Smad8 transcript in Smad8Δexon3/Δexon3 mice showing no difference compared to wild-type siblings (data not shown). Therefore, deletion of exon 3 does not appear to affect the stability or steady-state level of the Smad8 mRNA.
FIG. 3.
Cre mediates the excision of exon 3 and neo in Smad83loxP/3loxP mice. (A) Schematic diagram of the Smad83loxP allele undergoing the Cre-mediated deletion of neo and exon 3 from the Smad8 locus. (B) RT-PCR analysis illustrating that exon 3 has been excised from Smad8 in Smad8Δexon3/Δexon3 mice. Lanes 1 show PCR products amplified by primers (S8 E2F and S8 E4R) that flank exon 3 and give a product of ∼400 bp. The 150-bp band seen in the Smad8Δexon3/+ and Smad8Δexon3/Δexon3 samples represent the regions that flank exon 3. Lanes 2 show PCR products of full-length Smad8. As a positive control, hypoxanthine phosphoribosyltransferase (HPRT) was used and is shown in lanes 3. Restriction enzyme abbreviations are given in the legend to Fig. 1.
Phenotypic Smad83loxP/3loxP and Smad1+/− embryos exhibit partially penetrant midbrain and hindbrain reductions.
The presence of an intronic neomycin resistance cassette within a gene can cause aberrant splicing, resulting in a neo fusion transcript, due to the presence of strong cryptic splice acceptor and donor sites within the neo cassette (38). When neomycin is inserted in the forward orientation relative to gene transcription, the frequency of aberrant splicing is much less than if it is placed in the reverse orientation (40). Indeed, hypomorphic alleles have been created through the intronic placement of neo (38).
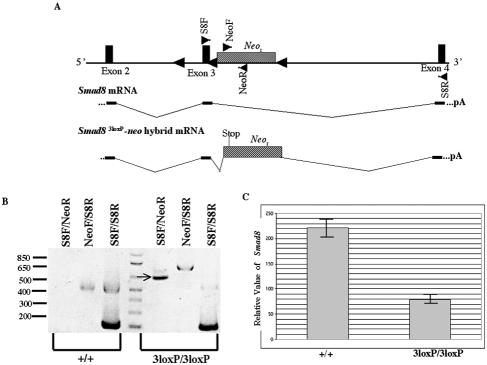
We therefore tested whether the neo cassette could disrupt expression of the Smad8 gene. Our analysis shows that a Smad8-neo fusion mRNA can be detected in the Smad83loxP/3loxP mice (Fig. 4A and B). Sequencing of this Smad8-neo fusion product demonstrates that neo was spliced in between exons 3 and 4, utilizing a previously described splice site 5 base pairs upstream of the initiating ATG codon (38). This aberrant splicing causes neo to be out of frame with the Smad8 transcript, resulting in a premature stop codon, which ultimately allows for the synthesis of a truncated Smad8 protein lacking roughly three-fourths of the MH2 domain. Xenopus assays have suggested that this protein retains partial function (Fig. 2F). To determine the level of wild-type Smad8 transcript in Smad83loxP/3loxP mice, we employed real-time PCR technology. Our analysis has shown that Smad8 is reduced in the adult Smad83loxP/3loxP brain (Fig. 4C), confirming that the Smad83loxP allele is hypomorphic (data not shown).
FIG. 4.
Aberrant splicing of neo within Smad8 results in a Smad8-neo fusion transcript. (A) Schematic diagram of the Smad83loxP allele (top) with primer pairs represented as triangles within exons (S8F and S8R) or in the neo cassette (NeoF and NeoR). The Smad8 transcript is depicted (middle) showing exons 2, 3, and 4. The Smad8-neo hybrid transcript found in the Smad83loxP/3loxP mice is spliced in between exons 3 and 4, causing neo to be out of frame with the Smad8 transcript, which results in a premature stop codon within neo. (B) RT-PCR analysis of the Smad8-neo hybrid transcript, with primer pairs shown on top and the indicated sample cDNA shown below. The PCR product using primer pair S8F-NeoR in the Smad83loxP/3loxP sample indicated by an arrow was sequenced and shown to be a Smad8-neo hybrid transcript. (C) Relative level of Smad8 expression as determined by real-time PCR.
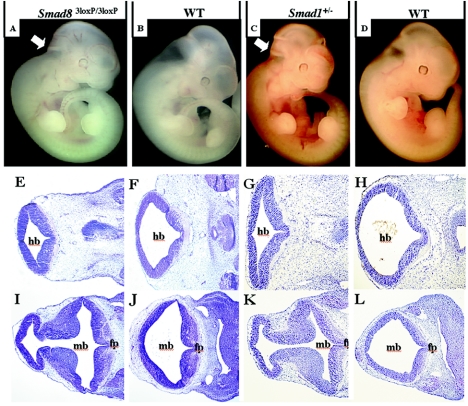
To test whether the reduction in Smad8 levels had an effect on normal development, we observed 45 embryos at E11.5 from Smad83loxP/3loxP intercrosses and found midbrain and hindbrain reductions in 11% of them (Fig. 5A and B). This phenotype was not observed in Smad8Δexon3/Δexon3 embryos, suggesting that it is specifically associated with the Smad83loxP allele. There exists the possibility that neo might be affecting the expression of neighboring genes, causing this phenotype indirectly. An analysis of the annotated mouse genome through the Ensembl Mouse Genome Browser revealed a single gene in close proximity to Smad8, RFXAP, an important regulatory gene associated with the major histocompatibility complex class II molecules (15). To address the possibility that the intronic neo was affecting genes nearby Smad8 and causing the observed phenotype, we performed semiquantitative RT-PCR on wild-type and phenotypic Smad83loxP/3loxP embryos and found expression levels for RFXAP to be consistent between the two (data not shown).
FIG. 5.
Phenotypic Smad83loxP/3loxP and Smad1+/− embryos display reductions in the hindbrain and midbrain. Genotypes of pictured embryos and sections are shown at the top. (A to D) Whole mount pictures of phenotypic Smad83loxP/3loxP(A), littermate control (B), phenotypic Smad1+/− (C), and littermate control (D) embryos at E11.5. Note that the hindbrain and midbrain are significantly reduced both in phenotypic Smad83loxP/3loxP and Smad1+/− embryos as indicated by arrows. (E to L) Frontal histological sections within the neural tube show reduced space and hypercellularity within the midbrain of phenotypic Smad83loxP/3loxP (I) and phenotypic Smad1+/− (K) embryos at E11.5 compared to wild-type siblings (J and L, respectively). More caudal histological sections show the same diminished space within the hindbrain of abnormal Smad83loxP/3loxP (E) and abnormal Smad1+/− (G) embryos compared to wild-type siblings (F and H, respectively). mb, hb, and fp inbdicate midbrain, hindbrain, and floor plate, respectively.
More intriguingly, we have also identified a partially penetrant lethality in Smad1 heterozygotes due to previously uncharacterized midbrain/hindbrain defects (Fig. 5C and D). Analyzing intercrosses between Smad1+/− and wild-type mice, we have observed a statistically significant reduction (P < 0.01) in the number of Smad1+/− mice at weaning (Table 1). Although there were no apparent defects in 40 Smad1+/− embryos at E9.5 (data not shown), roughly 22% of the E11.5 Smad1+/− embryos displayed midbrain and hindbrain reductions (Table 1).
TABLE 1.
Phenotype and genotype analysis of Smad1+/− × WT crosses
| Age | No. with phenotype of Smad1+/−
|
No. observed with genotype (no. expected)
|
Total no. of mice | ||
|---|---|---|---|---|---|
| Normal | Brain reduction | +/+ | +/− | ||
| E11.5 | 32 | 9 | 37 (39) | 41 (39) | 78 |
| Postnatal | NAa | NA | 102 (80) | 58 (80) | 160 |
NA, not applicable.
There remains the possibility that the aberrant Smad8 transcript produced in Smad83loxP/3loxP embryos acts as a dominant inhibitor of Smad1 signaling, thereby recapitulating the nervous system defect observed in Smad1 heterozygotes. However, the cDNA produced by the Smad83loxP mutants (Smad8Δexon4,5) was injected into Xenopus laevis embryos and was shown to possess very weak ventralizing activity (Fig. 2F). This result is indicative of a hypomorphic and not an antimorphic allele. To confirm this, transcriptional reporter assays were carried out utilizing the BMP-responsive Xvent2B promoter (23) driving the luciferase reporter gene. The Xvent2B promoter exhibited some endogenous activity (Fig. 2H, lane 1) which was increased roughly twofold with the addition of Smad1, Smad4, and a constitutively activated receptor, Alk6 (Fig. 2H, lane 2), in agreement with experiments done previously (23). However, we are now able to demonstrate that Smad8 also transactivates the Xvent2B promoter to the same extent as or possibly to a greater extent than Smad1 (Fig. 2H, lane 3.) The Smad8Δexon4,5 cDNA was also capable of transactivating the Xvent2B promoter but to a lesser degree than wild-type Smad8 (Fig. 2H, lane 4), as was the Smad8Δexon3 allele (Fig. 2H, lane 5). To test whether the Smad8Δexon4,5 allele could function in an antimorphic fashion and reduce the signaling of the other BMP-regulated Smads, it was expressed alongside wild-type Smad1, to determine whether it could restrict Smad1-mediated activation. However, the Smad8Δexon4,5 protein did not interfere with Smad1-mediated transcription (Fig. 2H, lane 6).
Abnormal Smad1+/− embryos exhibit increased cellular proliferation in the dorsal region of the neural tube.
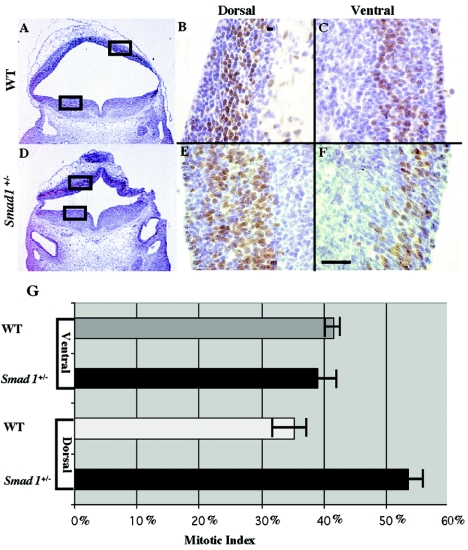
Histological analysis reveals that the observed neuroectodermal phenotype in abnormal Smad1 heterozygous embryos is associated with hypercellularity within the dorsal region of the neural tube (Fig. 5E to L). To address the cause of this hypercellularity, we examined the level of cellular proliferation within the neural tube of abnormal Smad1+/− embryos using BrdU (Fig. 6A and D). Higher numbers of labeled cells were found in the dorsal midbrain of Smad1+/− embryos than in that of their wild-type siblings (Fig. 6B and E). Interestingly, the number of labeled cells within the ventral neural tube was consistent between Smad1+/− and wild-type mice (Fig. 6C and F). Quantitation of the BrdU incorporation showed a 20% increase in the mitotic index on the dorsal side of the neural tube in abnormal Smad1 heterozygotes compared to the wild type (Fig. 6G). The results were similar in the hindbrain, although cellular proliferation was normal in the spinal cord (data not shown). This indicates that increased cellular proliferation contributes to the observed hypercellularity. Interestingly, this increase appears to be specific to the dorsal region of the midbrain and hindbrain, as no increase in the rate of cellular proliferation was seen in the ventral region of the brain. The level of cellular proliferation was also analyzed in Smad83loxP/3loxP embryos utilizing Ki-67 and phospho-H3 as markers, although we were unable to detect any significant changes between mutant and wild-type embryos. Abnormal Smad1+/− and Smad83loxP/3loxP embryos were examined for apoptosis within the neural tube, but no changes were seen (data not shown). We suspect that the Smad83loxP/3loxP embryos do exhibit increased cellular proliferation in the dorsal brain, as evidenced by the observed hypercellularity (Fig. 5E and I). There is more tissue but no increase in cell death. It may be that Ki-67 and phospho-H3 antibodies are insufficient to reveal this increased cellular proliferation, indeed they did not detect proliferative changes in the abnormal Smad1+/− embryos.
FIG. 6.
Phenotypic Smad1+/− embryos display increased cellular proliferation in the dorsal region of the neural tube. A histological section stained for BrdU shows a greater number of proliferating cells on the dorsal side of the neural tube in abnormal Smad1 heterozygotes (D and E) than the wild type (A and B). The boxes in panels A and D indicate the areas enlarged in panels B, C, E, and F. On the ventral side of the neural tube, the number of proliferating cells is equal between Smad1+/− (F) and wild-type embryos (C). Quantification of this data shows that there are 20% more proliferating cells on the dorsal side of the neural tube in the Smad1+/− embryo than in the wild-type embryo (G).
In an effort to determine the cause of the increased cellular proliferation in the dorsal region of the midbrain and hindbrain, immunohistochemistry was performed to compare the levels of important cell cycle regulatory proteins between wild-type and Smad1+/− mutant embryos. This analysis revealed no discernible difference between wild-type and mutant embryos in expression of the cyclin-dependent kinase inhibitor proteins p21 (WAF1/CIP1) and p27 (Kip1) or cyclin D1 (data not shown).
Neurofilament staining of abnormal Smad1+/− embryos reveals relatively normal cranial nerve development.
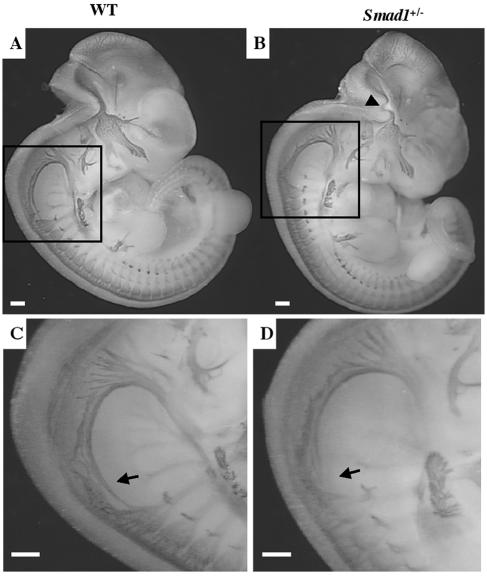
The early hindbrain is segmented into discrete units termed rhombomeres from which signals emanate that will orchestrate the development of the nervous system (58). During hindbrain patterning, positional information along the anterior/posterior axis is laid down that allows for the expansion of the peripheral nervous system (54). We therefore wanted to know whether the hindbrain defect observed in abnormal Smad1 heterozygotes caused a disruption in cranial nerve development. To analyze the anatomy of differentiating neurons in abnormal Smad1 heterozygotes, we visualized the nervous system at E11.5 by neurofilament staining (12). Careful examination of neurofilament-labeled embryos failed to reveal any drastic changes in cranial nerve development (Fig. 7A and B). Examination of the hindbrain in the phenotypic Smad1+/− embryos revealed a curved cranial nerve that is likely to be a secondary effect caused by the abnormal architecture of the hindbrain (Fig. 7B) and not a specific result of Smad1 disruption. However, abnormal Smad1+/− embryos exhibited a clearly truncated spinal accessory nerve (Fig. 7C, D), which is more likely to be a direct result of Smad1 haploinsufficiency.
FIG. 7.
Abnormal Smad1+/− embryos exhibit a truncated spinal accessory nerve. Whole-mount micrograph of a wild-type embryo (A) and a phenotypic Smad1 heterozygous embryo (B) at E11.5 stained with neurofilament antibody. The Smad1+/− hindbrain contains a warped nerve, most likely a secondary effect due to a reduced hindbrain (arrowhead). Higher magnification of the Smad1+/− embryo revealed a shortened spinal accessory neuron (XI) (D) compared to the wild type (C).
Examination of genes involved in dorsoventral patterning of the neural tube in phenotypic Smad1+/− and Smad83loxP/3loxP embryos.
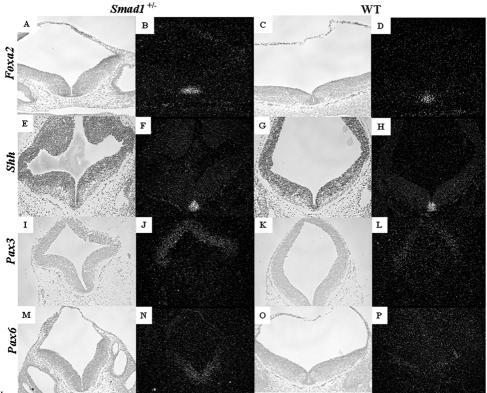
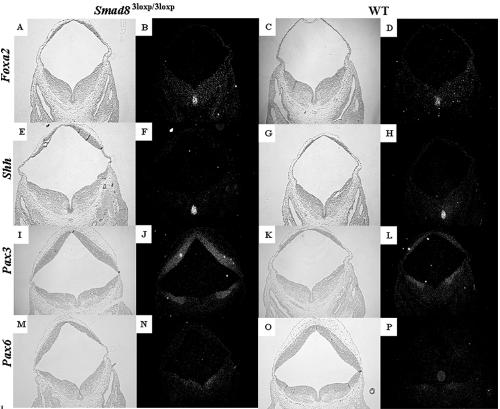
A well-known activity of the BMP family is the patterning of the embryonic neural tube, in which BMPs are expressed dorsally and are thought to promote the formation of dorsal cell fates (16). We therefore undertook an analysis of genes critical in dorsoventral neural tube patterning. Foxa2 (Hnf3β) is expressed in the floor plate and ventral regions of the neural tube in wild-type, Smad83loxP/3loxP, and Smad1+/− embryos (Fig. 8A to D and 9A to D) and is required for the formation of ventral cell types (2). Foxa2 was expressed at a similar level and in a similar domain in the wild-type, Smad1+/−, and Smad83loxP/3loxP phenotypic embryos (Fig. 8A to D and 9A to D), showing that ventral cell fates were not expanded in the Smad1+/− and Smad83loxP/3loxP phenotypic embryos. Similar results were seen for Shh (Fig. 8E to H and 9E to H), another ventrally expressed gene (17). Pax3, a dorsally expressed transcription factor (19, 36), was expressed in its correct domain, although it was upregulated in phenotypic Smad1 heterozygous and Smad83loxP/3loxP embryos compared to wild-type siblings (Fig. 8I to L and 9I to L). This suggests that the defect seen in the midbrain and hindbrain of the Smad1+/− and Smad83loxP/3loxP embryos was not the result of change in the dorsoventral axis within the neural tube. Interestingly, Pax6 was also upregulated in phenotypic Smad1 heterozygous and Smad83loxP/3loxP embryos (Fig. 8M to P and 9M to P); however, its expression domain was not expanded and was confined to the intermediate and ventral regions of the neural tube (57).
FIG. 8.
Examination of genes involved in dorsoventral patterning of the neural tube in phenotypic Smad1+/− embryos. Genes that are responsible for ventralizing the neural tube such as Foxa2 (A to D) and Shh (E to H) are expressed at similar levels and similar domains between a phenotypic Smad1 heterozygous embryo (A and B) and a wild-type embryo (C and D). Bright-field images are shown to the left of the dark-field images. Pax3 (I to L) and Pax6 (M to P), genes expressed in the dorsal and ventrolateral regions of the neural tube, respectively, show a significant increase in the Smad1+/− embryo (I, J, M, and N) compared to the control embryo (K, L, O, and P).
FIG. 9.
Examination of genes involved in dorsoventral patterning of the neural tube in phenotypic Smad83loxP/3loxP embryos. Genes that are responsible for ventralizing the neural tube such as Foxa2 (A to D) and Shh (E to H) are expressed at similar levels and similar domains between a phenotypic Smad8 homozygous embryo (A and B) and a wild-type embryo (C and D). Bright-field images are shown to the left of the dark-field images. Pax3 (I to L) and Pax6 (M to P), genes expressed in the dorsal and ventrolateral regions of the neural tube, respectively, show a significant increase in the Smad83loxP/3loxP embryos (I, J, M, and N) compared to the control embryo (K, L, O, and P).
Changes in spinal cord gene expression in phenotypic Smad1+/− embryos.
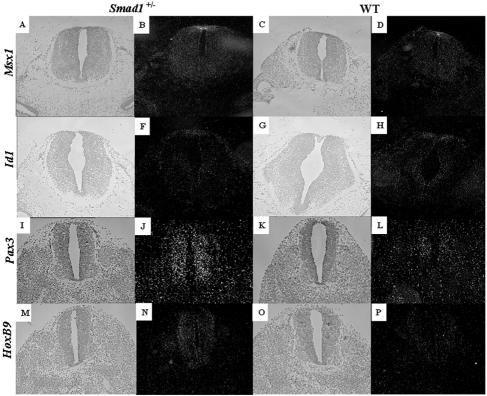
The effects of BMPs in assigning dorsal fate identities within the spinal cord has been studied far more extensively than their dorsalizing capabilities in the brain. One specialized compartment within the dorsal neural tube that requires BMP signals for its propagation and maintenance is the roof plate. This structure is localized at the dorsal midline and formed from glial cells derived from the lateral neural plate (31). Two roof plate markers which have been shown to be responsive to BMP signals, Id1 and Msx1 (20, 35), were analyzed in phenotypic Smad1+/− embryos for the existence of a roof plate. Msx1 and Id1 were both expressed in the roof plate and at similar levels between phenotypic Smad1+/− embryos and their wild-type siblings (Fig. 10A to H). These data confirm that the midbrain/hindbrain defect observed in phenotypic Smad1+/− embryos is not due to the absence of a roof plate.
FIG. 10.
Analysis of spinal cord markers in phenotypic Smad1+/− embryos. BMP-responsive genes that are expressed within the roof plate such as Id1 (A to D) and Msx1 (E to H) are expressed at similar levels and similar domains between a phenotypic Smad1 heterozygous embryo (A, B, E, and F) and a wild-type embryo (C, D, G, and H). Bright-field images are shown to the left of the dark-field images. Pax3 (I to L) and HoxB9 (M to P), genes expressed within the spinal cord, show a significant increase in the Smad1+/− embryo (I, J, M, and N) compared to the control embryos (K, L, O, and P).
Given the increased expression of Pax3 in the midbrain and hindbrain of the Smad1+/− and Smad83loxP/3loxP embryos, we were curious if its expression was normal in the spinal cord. Indeed, Pax3 was upregulated in the dorsal spinal cord of the Smad1+/− (Fig. 9I to L) and Smad83loxP/3loxP mutants (data not shown). Of note, the expression of Pax3 appears to increased ventrally in the spinal cord (Fig. 9I to L, compare panels J and L), suggesting an increase in dorsal cell fates in the spinal cord. However, an examination of other genes and proteins located along the dorsal/ventral axis in the spinal cord, including Shh, Foxa2, Ncam, and Isl-1, revealed no differences, nor were there differences in cellular proliferation (data not shown). While examining the anterior/posterior axis of the neural tube in abnormal Smad1+/− embryos, the expression of HoxB9, a gene expressed in the spinal cord (51), was examined. Surprisingly, its expression was elevated in abnormal Smad1 heterozygous embryos compared to their wild-type littermates (Fig. 9M to P). Therefore, although there was no obvious phenotype in the spinal cords of the affected Smad1+/− embryos, there were clearly changes in gene expression.
DISCUSSION
Here we have targeted the murine Smad8 gene, creating two distinct alleles of Smad8. One allele, in which exon 3 has been deleted, causes no overt phenotype in the mouse, although it displays reduced ventralizing activity in Xenopus. It is probable that some Smad8 functions have become impaired in Smad8Δexon3/Δexon3 mice; however, the consequences of this are undetectable under gross examination. Thus, it can be inferred that exon 3 of Smad8 is dispensable for normal mouse development. The second allele, which contains a neo cassette within intron 3, has a 65% reduction of wild-type RNA due to aberrant splicing of neo within the Smad8 transcript. In addition, it creates a truncated product with diminished activity and therefore represents a hypomorphic allele. Intriguingly, we have observed a partially penetrant neuroectodermal defect in 11% of homozygotes of the Smad83loxP allele as well as in 22% of Smad1 heterozygous embryos.
The Smad1 and Smad8 knockouts display an unusual nervous system phenotype. One aspect observed only in phenotypic Smad1 heterozygous embryos is an increase in cellular proliferation only observed on the dorsal side of the neural tube. This is not due to differences in the expression pattern of Smad1 throughout the neural tube, since it has a ubiquitous expression pattern early in development (10, 55) (M. Hester and M. Weinstein, unpublished data). The increased cellular proliferation is interesting, since application of BMPs to explants of the lateral telencephalon inhibits cell proliferation in vitro (21) and reduction of Smad1 activity would presumably result in decreased BMP signaling.
An increase in Pax3 expression is also observed in the dorsal mid- and hindbrain of the abnormal Smad1 and Smad8 mutants, while Pax6 expression appears to be elevated ventrally. Using chicken neural explants, it has been shown that increased BMP signaling elevates the level of Pax3 and Pax7 as well as the expression of other dorsal neuroectodermal genes (33, 53). From these experiments, it would be expected that phenotypic Smad83loxP/3loxP and Smad1+/− embryos would exhibit a decrease in Pax3 expression. Presumably, the decrease in BMP signaling seen in the affected Smad1+/− or Smad83loxP/3loxP embryos is sufficiently subtle that Pax3 expression might be modulated differently than seen previously. In addition, experiments using chicken neural explants increase the level of BMP signaling to nonphysiological levels.
Another anomaly observed in phenotypic Smad1 heterozygotes was a shortened spinal accessory nerve (X). Interestingly, it has been shown recently that mutations in the transcription factor Nkx2.9 cause a shortened spinal accessory nerve (44); otherwise Nkx2.9−/− mice are viable and fertile and do not display any behavioral phenotype. Semiquantitative PCR analysis using RNA from phenotypic Smad1+/− embryonic brains revealed no significant difference in levels of Nkx2.9 compared to the wild type (M. Hester and M. Weinstein, unpublished data).
Although BMPs have been shown to dorsalize the embryonic neural tube (25, 31), the phenotypic Smad1 and Smad8 embryos did not appear to have abnormal dorsoventral patterning in the brain, in that morphologically distinct floor plates and roof plates were evident in the mutants (Fig. 5E to L). This has been confirmed by normal expression patterns of Shh, Foxa2 (Fig. 8 and 9), Msx1, Id1, NCAM, and Isl-1 (Hester and Weinstein, unpublished), genes involved in brain development and expressed within narrow dorsoventral boundaries. Although there were increases in the level of Pax3 and Pax6 in the abnormal Smad1 and Smad8 embryos, the expression domains of these genes did not appear to be changed in the brain (Fig. 8 and 9), further arguing against expansion or reduction of dorsal cell fates within the brain. Interestingly, there appears to be increased expression of Pax3 ventrally in the spinal cord, suggesting an increase in dorsal cell fates. However, our data suggest that this did not come at the expense of ventral cell fates, nor were any differences seen in the expression of other neuroectodermal genes. This might require further investigation.
Interestingly, another group has constructed a similar Smad1 knockout allele but has not observed any phenotype associated with Smad1 heterozygotes (55). A likely explanation for this discrepancy is the presence of modifiers within the different strain backgrounds utilized in these studies (29, 55). The frequency at which Smad1 heterozygotes (which are on a mixed 129/NIH Black Swiss background) exhibit the observed abnormality can be increased by breeding onto 129Sv/Ev or decreased by breeding onto NIH Black Swiss (Hester and Weinstein, unpublished). Indeed, reductions in the viability of Smad1 heterozygous mice on a mixed 129Sv/Ev and NIH Black Swiss background were seen in another laboratory (29). This suggests that there are strain-specific modifiers that affect Smad1 function.
The phenotype seen in the Smad1+/− and Smad83loxP/3loxP embryos is not completely penetrant, a phenomenon that has been observed in other knockouts of BMP signaling factors, including Bmp4 (14) and Chrd (1). For example, Bmp4 homozygous and heterozygous embryos display partially penetrant phenotypes, indicating that the level of Bmp4 is critical for proper development. Bmp4−/− embryos die between E6.5 and 9.5, with the majority arresting at the egg cylinder stage due to their inability to gastrulate. The embryos that survive past E6.5 have either unorganized or truncated posterior axes as well as defects in extraembryonic mesoderm formation (59). Interestingly, Bmp4 heterozygotes also display a range of phenotypes that is dependent upon genetic background. On a mixed 129Sv/Ev and C57BL/6 background, there is no reduction in viability or any significant phenotype associated with Bmp4 heterozygosity. However, when the Bmp4 mutation is backcrossed onto the C57BL/6 genetic background, 25% of the heterozygotes die within 3 weeks of birth, exhibiting incompletely penetrant phenotypes such as cystic kidney, polydactyly, microphthalmia, and craniofacial abnormalities (14). Genetic background has also been shown to alter the penetrance of phenotypes observed in Chrd knockout mice (1). It is therefore not surprising that the phenotypes we have observed in Smad1+/− and Smad83loxP/3loxP embryos are not completely penetrant and that the Smad1+/− phenotype is strain dependent.
In summary, we have created hypomorphic alleles for Smad8, one of which can function at 35% of the level of wild-type Smad8. Interestingly, homozygotes for this allele display a partially penetrant phenotype that phenocopies a hindbrain/midbrain reduction we have observed in Smad1+/− embryos at E11.5. Numerous laboratories have shown the importance of BMPs and related family member molecules in nervous system development, but we have for the first time shown the functional importance for their mediators, Smad1 and Smad8, in embryonic brain development. Future studies that delete Smad1 or Smad8 specifically from the embryonic brain utilizing brain-specific Cre strains will be invaluable, allowing the dissection of their functions in brain development.
Acknowledgments
We thank Natarajan Muthusamy for performing the microinjections of blastocysts to make chimeric Smad8 mice and Robert Lechleider for generously providing the Smad1 mutant mice. We also thank Hiroshi Shibuya for kindly providing the Smad8 expression vector and Christoph Plass for obtaining the bacterial artificial chromosome used for generating the Smad8 knockout construct. We are also indebted to Susan Cole and members of the Weinstein laboratory for critical input on this paper.
REFERENCES
- 1.Anderson, R. M., A. R. Lawrence, R. W. Stottmann, D. Bachiller, and J. Klingensmith. 2002. Chordin and noggin promote organizing centers of forebrain development in the mouse. Development 129:4975-4987. [DOI] [PubMed] [Google Scholar]
- 2.Ang, S. L., and J. Rossant. 1994. HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78:561-574. [DOI] [PubMed] [Google Scholar]
- 3.Attisano, L., and S. Tuen Lee-Hoeflich. 2001. The Smads. Genome Biol. 2:REVIEWS3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attisano, L., and J. L. Wrana. 2002. Signal transduction by the TGF-beta superfamily. Science 296:1646-1647. [DOI] [PubMed] [Google Scholar]
- 5.Bachiller, D., J. Klingensmith, C. Kemp, J. A. Belo, R. M. Anderson, S. R. May, J. A. McMahon, A. P. McMahon, R. M. Harland, J. Rossant, and E. M. De Robertis. 2000. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature 403:658-661. [DOI] [PubMed] [Google Scholar]
- 6.Chang, H., D. Huylebroeck, K. Verschueren, Q. Guo, M. M. Matzuk, and A. Zwijsen. 1999. Smad5 knockout mice die at mid-gestation due to multiple embryonic and extraembryonic defects. Development 126:1631-1642. [DOI] [PubMed] [Google Scholar]
- 7.Chang, H., A. Zwijsen, H. Vogel, D. Huylebroeck, and M. M. Matzuk. 2000. Smad5 is essential for left-right asymmetry in mice. Dev. Biol. 219:71-78. [DOI] [PubMed] [Google Scholar]
- 8.Deng, C., A. Wynshaw-Boris, F. Zhou, A. Kuo, and P. Leder. 1996. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84:911-921. [DOI] [PubMed] [Google Scholar]
- 9.Deng, C. X., A. Wynshaw-Boris, M. M. Shen, C. Daugherty, D. M. Ornitz, and P. Leder. 1994. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 8:3045-3057. [DOI] [PubMed] [Google Scholar]
- 10.Dick, A., W. Risau, and H. Drexler. 1998. Expression of Smad1 and Smad2 during embryogenesis suggests a role in organ development. Dev. Dyn. 211:293-305. [DOI] [PubMed] [Google Scholar]
- 11.Dickinson, M. E., M. A. Selleck, A. P. McMahon, and M. Bronner-Fraser. 1995. Dorsalization of the neural tube by the non-neural ectoderm. Development 121:2099-2106. [DOI] [PubMed] [Google Scholar]
- 12.Dodd, J., S. B. Morton, D. Karagogeos, M. Yamamoto, and T. M. Jessell. 1988. Spatial regulation of axonal glycoprotein expression on subsets of embryonic spinal neurons. Neuron 1:105-116. [DOI] [PubMed] [Google Scholar]
- 13.Ducy, P., and G. Karsenty. 2000. The family of bone morphogenetic proteins. Kidney Int. 57:2207-2214. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, N. R., G. E. Winnier, L. K. Hargett, J. J. Schrick, A. B. Fogo, and B. L. Hogan. 1997. Haploinsufficient phenotypes in Bmp4 heterozygous null mice and modification by mutations in Gli3 and Alx4. Dev. Biol. 188:235-247. [DOI] [PubMed] [Google Scholar]
- 15.Durand, B., P. Sperisen, P. Emery, E. Barras, M. Zufferey, B. Mach, and W. Reith. 1997. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 16:1045-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebendal, T., H. Bengtsson, and S. Soderstrom. 1998. Bone morphogenetic proteins and their receptors: potential functions in the brain. J. Neurosci. Res. 51:139-146. [DOI] [PubMed] [Google Scholar]
- 17.Echelard, Y., D. J. Epstein, B. St-Jacques, L. Shen, J. Mohler, J. A. McMahon, and A. P. McMahon. 1993. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75:1417-1430. [DOI] [PubMed] [Google Scholar]
- 18.el-Hodiri, H. M., and M. Perry. 1995. Interaction of the CCAAT displacement protein with shared regulatory elements required for transcription of paired histone genes. Mol. Cell. Biol. 15:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epstein, J. A. 2000. Pax3 and vertebrate development. Methods Mol. Biol. 137:459-470. [DOI] [PubMed] [Google Scholar]
- 20.Evans, S. M., and T. X. O'Brien. 1993. Expression of the helix-loop-helix factor Id during mouse embryonic development. Dev. Biol. 159:485-499. [DOI] [PubMed] [Google Scholar]
- 21.Furuta, Y., D. W. Piston, and B. L. Hogan. 1997. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 124:2203-2212. [DOI] [PubMed] [Google Scholar]
- 22.Goulding, M. D., G. Chalepakis, U. Deutsch, J. R. Erselius, and P. Gruss. 1991. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 10:1135-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henningfeld, K. A., S. Rastegar, G. Adler, and W. Knochel. 2000. Smad1 and Smad4 are components of the bone morphogenetic protein-4 (BMP-4)-induced transcription complex of the Xvent-2B promoter. J. Biol. Chem. 275:21827-21835. [DOI] [PubMed] [Google Scholar]
- 24.Holzenberger, M., C. Lenzner, P. Leneuve, R. Zaoui, G. Hamard, S. Vaulont, and Y. L. Bouc. 2000. Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucleic Acids Res. 28:E92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jessell, T. M. 2000. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1:20-29. [DOI] [PubMed] [Google Scholar]
- 26.Kao, K. R., and R. P. Elinson. 1988. The entire mesodermal mantle behaves as Spemann's organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev. Biol. 127:64-77. [DOI] [PubMed] [Google Scholar]
- 27.Lai, E., V. R. Prezioso, W. F. Tao, W. S. Chen, and J. E. Darnell, Jr. 1991. Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 5:416-427. [DOI] [PubMed] [Google Scholar]
- 28.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lechleider, R. J., J. L. Ryan, L. Garrett, C. Eng, C. Deng, A. Wynshaw-Boris, and A. B. Roberts. 2001. Targeted mutagenesis of Smad1 reveals an essential role in chorioallantoic fusion. Dev. Biol. 240:157-167. [DOI] [PubMed] [Google Scholar]
- 30.Lee, K. J., P. Dietrich, and T. M. Jessell. 2000. Genetic ablation reveals that the roof plate is essential for dorsal interneuron specification. Nature 403:734-740. [DOI] [PubMed] [Google Scholar]
- 31.Lee, K. J., and T. M. Jessell. 1999. The specification of dorsal cell fates in the vertebrate central nervous system. Annu. Rev. Neurosci. 22:261-294. [DOI] [PubMed] [Google Scholar]
- 32.Lee, K. J., M. Mendelsohn, and T. M. Jessell. 1998. Neuronal patterning by BMPs: a requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev. 12:3394-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liem, K. F., Jr., G. Tremml, and T. M. Jessell. 1997. A role for the roof plate and its resident TGFbeta-related proteins in neuronal patterning in the dorsal spinal cord. Cell 91:127-138. [DOI] [PubMed] [Google Scholar]
- 34.Liu, Y., M. H. Festing, M. Hester, J. C. Thompson, and M. Weinstein. 2004. Generation of novel conditional and hypomorphic alleles of the Smad2 gene. Genesis 40:118. [DOI] [PubMed] [Google Scholar]
- 35.MacKenzie, A., L. Purdie, D. Davidson, M. Collinson, and R. E. Hill. 1997. Two enhancer domains control early aspects of the complex expression pattern of Msx1. Mech. Dev. 62:29-40. [DOI] [PubMed] [Google Scholar]
- 36.Mansouri, A. 1998. The role of Pax3 and Pax7 in development and cancer. Crit. Rev. Oncog. 9:141-149. [DOI] [PubMed] [Google Scholar]
- 37.Mehler, M. F., P. C. Mabie, D. Zhang, and J. A. Kessler. 1997. Bone morphogenetic proteins in the nervous system. Trends Neurosci. 20:309-317. [DOI] [PubMed] [Google Scholar]
- 38.Meyers, E. N., M. Lewandoski, and G. R. Martin. 1998. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 18:136-141. [DOI] [PubMed] [Google Scholar]
- 39.Miyanaga, Y., I. Torregroza, and T. Evans. 2002. A maternal Smad protein regulates early embryonic apoptosis in Xenopus laevis. Mol. Cell. Biol. 22:1317-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagy, A. 2000. Cre recombinase: the universal reagent for genome tailoring. Genesis 26:99-109. [PubMed] [Google Scholar]
- 41.Nakayama, T., M. A. Snyder, S. S. Grewal, K. Tsuneizumi, T. Tabata, and J. L. Christian. 1998. Xenopus Smad8 acts downstream of BMP-4 to modulate its activity during vertebrate embryonic patterning. Development 125:857-867. [DOI] [PubMed] [Google Scholar]
- 42.Nishita, M., N. Ueno, and H. Shibuya. 1999. Smad8B, a Smad8 splice variant lacking the SSXS site that inhibits Smad8-mediated signalling. Genes Cells 4:583-591. [DOI] [PubMed] [Google Scholar]
- 43.Osoegawa, K., M. Tateno, P. Y. Woon, E. Frengen, A. G. Mammoser, J. J. Catanese, Y. Hayashizaki, and P. J. de Jong. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10:116-128. [PMC free article] [PubMed] [Google Scholar]
- 44.Pabst, O., J. Rummelies, B. Winter, and H. H. Arnold. 2003. Targeted disruption of the homeobox gene Nkx2.9 reveals a role in development of the spinal accessory nerve. Development 130:1193-1202. [DOI] [PubMed] [Google Scholar]
- 45.Pera, E. M., A. Ikeda, E. Eivers, and E. M. De Robertis. 2003. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 17:3023-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasai, Y., and E. M. De Robertis. 1997. Ectodermal patterning in vertebrate embryos. Dev. Biol. 182:5-20. [DOI] [PubMed] [Google Scholar]
- 47.Sater, A. K., H. M. El-Hodiri, M. Goswami, T. B. Alexander, O. Al-Sheikh, L. D. Etkin, and J. Akif Uzman. 2003. Evidence for antagonism of BMP-4 signals by MAP kinase during Xenopus axis determination and neural specification. Differentiation 71:434-444. [DOI] [PubMed] [Google Scholar]
- 48.Sauer, B., and N. Henderson. 1988. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 85:5166-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solloway, M. J., and E. J. Robertson. 1999. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development 126:1753-1768. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki, A., C. Chang, J. M. Yingling, X. F. Wang, and A. Hemmati-Brivanlou. 1997. Smad5 induces ventral fates in Xenopus embryo. Dev. Biol. 184:402-405. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki, M., Y. Mizutani-Koseki, Y. Fujimura, H. Miyagishima, T. Kaneko, Y. Takada, T. Akasaka, H. Tanzawa, Y. Takihara, M. Nakano, H. Masumoto, M. Vidal, K. Isono, and H. Koseki. 2002. Involvement of the Polycomb-group gene Ring1B in the specification of the anterior-posterior axis in mice. Development 129:4171-4183. [DOI] [PubMed] [Google Scholar]
- 52.Thomsen, G. H. 1996. Xenopus mothers against decapentaplegic is an embryonic ventralizing agent that acts downstream of the BMP-2/4 receptor. Development 122:2359-2366. [DOI] [PubMed] [Google Scholar]
- 53.Timmer, J. R., C. Wang, and L. Niswander. 2002. BMP signaling patterns the dorsal and intermediate neural tube via regulation of homeobox and helix-loop-helix transcription factors. Development 129:2459-2472. [DOI] [PubMed] [Google Scholar]
- 54.Trainor, P. A., and R. Krumlauf. 2000. Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat. Rev. Neurosci. 1:116-124. [DOI] [PubMed] [Google Scholar]
- 55.Tremblay, K. D., N. R. Dunn, and E. J. Robertson. 2001. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development 128:3609-3621. [DOI] [PubMed] [Google Scholar]
- 56.Walther, C., and P. Gruss. 1991. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 113:1435-1449. [DOI] [PubMed] [Google Scholar]
- 57.Warren, N., and D. J. Price. 1997. Roles of Pax-6 in murine diencephalic development. Development 124:1573-1582. [DOI] [PubMed] [Google Scholar]
- 58.Wilkinson, D. G., and R. Krumlauf. 1990. Molecular approaches to the segmentation of the hindbrain. Trends Neurosci. 13:335-339. [DOI] [PubMed] [Google Scholar]
- 59.Winnier, G., M. Blessing, P. A. Labosky, and B. L. Hogan. 1995. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9:2105-2116. [DOI] [PubMed] [Google Scholar]
- 60.Wozney, J. M., V. Rosen, A. J. Celeste, L. M. Mitsock, M. J. Whitters, R. W. Kriz, R. M. Hewick, and E. A. Wang. 1988. Novel regulators of bone formation: molecular clones and activities. Science 242:1528-1534. [DOI] [PubMed] [Google Scholar]
- 61.Wrana, J. L. 2000. Crossing Smads. Sci. STKE 2000:RE1. [DOI] [PubMed] [Google Scholar]
- 62.Wrana, J. L., and L. Attisano. 2000. The Smad pathway. Cytokine Growth Factor Rev. 11:5-13. [DOI] [PubMed] [Google Scholar]
- 63.Xu, X., C. Li, L. Garrett-Beal, D. Larson, A. Wynshaw-Boris, and C. X. Deng. 2001. Direct removal in the mouse of a floxed neo gene from a three-loxP conditional knockout allele by two novel approaches. Genesis 30:1-6. [DOI] [PubMed] [Google Scholar]
- 64.Yang, X., L. H. Castilla, X. Xu, C. Li, J. Gotay, M. Weinstein, P. P. Liu, and C. X. Deng. 1999. Angiogenesis defects and mesenchymal apoptosis in mice lacking SMAD5. Development 126:1571-1580. [DOI] [PubMed] [Google Scholar]
- 65.Zhao, G. Q. 2003. Consequences of knocking out BMP signaling in the mouse. Genesis 35:43-56. [DOI] [PubMed] [Google Scholar]