Abstract
Ras GTPases are on/off switches regulating numerous cellular responses by signaling to various effector molecules. In T lymphocytes, Ras can be activated by two Ras exchange factors, SOS and RasGRP1, which are recruited through the adapters Grb2 and LAT and via the second-messenger diacylglycerol (DAG), respectively. Mitogen-activated protein (MAP) kinase phosphorylation patterns induced by active Ras can vary and contribute to distinct cellular responses. The different consequences of Ras activation by either guanine exchange factor are unknown. DAG also recruits and activates the kinase protein kinase Cθ (PKCθ) turning on the Erk MAP kinase pathway, but the biochemical mechanism responsible is unclear. We generated T-cell clones deficient in phorbol myristate acetate (a surrogate for DAG)-induced Ras activation. Analysis of a RasGRP1-deficient Jurkat T-cell clone and RasGRP1 RNA interference in wild-type cells revealed that RasGRP1 is required for optimal, antigen receptor-triggered Ras-Erk activation. RasGRP1 relies on its DAG-binding domain to selectively activate Erk kinases. Activation of Erk correlates with the phosphorylation of threonine residue 184 in RasGRP1. This phosphorylation event requires the activities of novel PKC kinases. Conversely, active PKCθ depends on RasGRP1 sufficiency to effectively trigger downstream events. Last, DAG-PKC-RasGRP1-driven Ras-Erk activation in T cells is a unique signaling event, not simply compensated for by SOS activity.
Proteins of the Ras family are signaling switches that regulate a multitude of cellular processes in various organisms, a noteworthy example being their role in cell division. Ras proteins cycle between inactive GDP-bound and active GTP-bound states (5). Ras is lipid modified and anchored to the membrane. It has intrinsic GTPase activity that is accelerated by GTPase-activating proteins (RasGAPs), which results in the hydrolysis of its GTP into GDP. Strictly regulated Ras activity is fundamental to normal biology. This fact is illustrated by oncogenic mutations resulting in a permanently active Ras in 30% of all metastatic cancers (6).
Active Ras mediates its diverse actions by binding to different effector molecules, such as phosphoinositide 3′ kinase (PI3 kinase), Ral guanine nucleotide dissociation stimulator, Ras interference gene 1, mitogen-activated protein (MAP) kinase kinase kinase 1, and Raf, leading to the activation of divergent signaling pathways (10, 45). In lymphocytes, antigen receptor or phorbol ester stimulation leads to the accumulation of RasGTP (16). It has long been established that introduction of constitutively active Ras in T cells results in induced expression of the early-activation antigen CD69 on the cell surface as well as activation of the transcription factor AP-1 in the nucleus (14). Likewise, engagement of the T-cell receptor (TCR) induces CD69 upregulation, which can be inhibited by dominant negative Ras, emphasizing a crucial role for Ras in TCR-mediated CD69 induction (14).
Illustrative for active Ras acting as a signal transduction branch point is the notion that antigen receptor-triggered signals of different strengths lead to different consequences in developing thymocytes (50). An appropriate signal leads to Ras induction of pathways activating the MAP kinases Erk-1 and -2 (phosphorylated via Raf and MEK-1 and -2), which can result in positive selection of thymocytes, activation of the transcription factor AP-1, and upregulation of the activation marker CD69 (38, 45). Signals that are too strong induce activation of the MAP kinases P38 and JNK-1 and -2 (via MAP kinase kinase kinase 1 and MKK4/7 or MKK3/6) and negative selection.
Originally, Ras activation in lymphocytes was thought to arise from inactivation of RasGAP function, but the biochemical link between antigen receptor ligation and inactivation of RasGAP is still missing (16, 23). The other side of the Ras GTP-GDP cycle is controlled by guanine exchange factors (GEFs), which promote the exchange of GDP for GTP, rendering Ras active. One such GEF expressed in lymphocytes is SOS, named after its homologous Drosophila gene Son of Sevenless. SOS is recruited to the plasma membrane (and brought in close vicinity with Ras) through its polyproline-rich regions that bind to the two SH3 domains in the adapter molecule Grb2 (growth factor receptor-bound protein 2) (43). A crucial signaling event for SOS recruitment and Ras activation is the induced phosphorylation of the adapter LAT (linker for activation in T cells) following TCR stimulation. Tyrosine-phosphorylated LAT functions as a scaffold to allow for recruitment of various SH2 domain-containing signaling molecules, like Grb2, PLCγ1 (phospholipase Cγ1), and Gads-SLP-76 (Grb2-like adapter downstream of Shc-SH2 domain-containing leukocyte protein [76 kDa]) (43). Recruitment and activation of PLCγ leads to the generation of the second messengers diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3) (see Fig. 10).
FIG. 10.
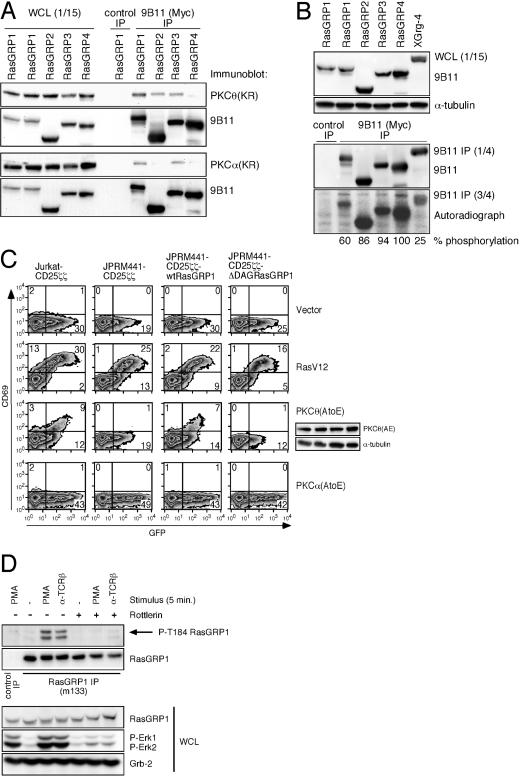
Novel PKC kinases phosphorylate and signal through RasGRP1. (A) Coimmunoprecipitations of Myc-tagged RasGRP-1, -2, -3, and -4 with Express-tagged, kinase-dead PKCθ or PKCα. The indicated constructs were cotransfected into 293T cells. One-fifteenth of the NP-40 lysates was used to verify expression in whole-cell lysates (WCL). The remainder was used for Myc immunoprecipitation (IP). Samples were subjected to Western blotting for Express-tag (PKCθ or PKCα). The same blot was stripped and reprobed for Myc expression (RasGRP's). (B) PKCθ in vitro kinase assay. The indicated constructs were expressed in 293T cells. Following Myc immunoprecipitations, three-fourths of the material was offered as a substrate in an in vitro kinase reaction using purified, active PKCθ. One-fifteenth (whole-cell lysate) and one-fourth (Myc IP) of the material were subjected to Western blotting for Myc expression (RasGRP's). The in vitro kinase assay was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred, and exposed to film. Relative phosphorylation was determined by determining the autoradiograph/Myc expression ratio from the Myc immunoprecipitations. (C) FACS analysis of GFP and induced CD69 expression. The active alleles of RasV12 (10 μg), PKCθ(AE) (20 μg), and PKCα(AE) (20 μg) or the control vector (10 μg) were cotransfected with GFP (10 μg) in the indicated cell lines, and cells were analyzed 40 h later for the amount of CD69 expression. (D) Western blot analysis for phosphorylated RasGRP1. Jurkat T cells were pretreated with DMSO control or Rottlerin and stimulated as indicated. The immunoprecipitated RasGRP1 was analyzed for phosphorylation on threonine 184 using a phospho-specific antibody (51). Backblotting for RasGRP1 identified phospho-RasGRP1 as the top band on the P-T184 blot. The lower band most likely reflects phosphorylated RasGRP3 that can also be immunoprecipitated with the M133 antibody. Analysis of whole-cell lysates demonstrates the proper inducible phosphorylation of Erk kinases, and expression of RasGRP1 and Grb2 indicates equal protein loadings in all samples. Panel D is representative of two independent experiments.
Recruitment of Grb2-SOS to membrane-anchored LAT leads to Ras activation. Also, expression of a membrane-targeted catalytic subunit of SOS is sufficient to activate Ras in lymphocytes (21). However, several lines of research indicate a crucial role for DAG in regulating Ras activation in lymphocytes: (i) TCR-stimulated, SLP-76 deficient, Jurkat T cells reveal impaired PLCγ1 and Ras activation, despite functional membrane recruitment of PLCγ1 and Grb2-SOS (47); (ii) in chicken B cells, B-cell receptor (BCR) stimulation of PLCγ2- and Btk (Burton tyrosine kinase)-deficient, but not of Grb2- and SOS2-deficient, DT40 cells leads to impaired Ras activation (25, 32); and (iii) synthetic DAG analogues, like phorbol myristate acetate (PMA), are efficient activators of Ras in lymphocytes (16, 23). Several labs joined the quest to find a molecule that explained these observations, since Grb2-SOS did not seem involved.
The cloning of Ras guanyl nucleotide-releasing protein 1 (RasGRP1) filled this long-standing void (17). RasGRP1 is the prototype of the RasGRP family and is a Ras exchange factor selectively expressed in T cells and a few other specialized cell types (18). In contrast, SOS proteins are ubiquitously expressed. RasGRP1 contains a DAG-binding domain, a structure resembling a pair of calcium-binding elongation factor (EF) hands, and a catalytic domain consisting of a Ras exchange motif and the CDC25 box. RasGRP1-deficient mice exhibit a severe block in thymocyte development (15). CalDAG-GEF I and RasGRP2 originate from the same RasGRP2 gene through alternative splicing (13, 24). CalDAG-GEF I favors Rap, whereas the longer RasGRP2 can also activate some Ras variants. The dual-specificity (Ras and Rap) GEF RasGRP3 is selectively expressed in B cells (41, 48). RasGRP4 was isolated from mast cells and from acute myeloid leukemia cells (35, 49).
Ras activation has also been reported to occur at the endoplasmic reticulum and Golgi membranes, in addition to the plasma membrane (12). Overexpression experiments have demonstrated that among all the Ras activators, RasGRP1 and -3 are unique in their ability to localize to the Golgi apparatus (4, 9). Calcium signals were reported to be essential for the translocation of RasGRP1 to the Golgi apparatus (4, 9). Interestingly, the same calcium signals induce localization of CAPRI (Ca2+-promoted Ras inactivator) to the plasma membrane, where it inactivates Ras (4, 27). Thus, spatial localization of Ras activation may very well be an important aspect and explain the need for different Ras-activating proteins in the same cell.
Protein kinase Cδ (PKCδ), -ɛ, -η, and -θ are members of the novel PKC subfamily that are Ca2+ independent and contain a DAG-binding C1 domain (2). PKC molecules were long thought to be the link between DAG and Ras activation. However, the exact PKCθ-Ras signaling mechanism is unclear and seems to be cell type specific, i.e., dominant negative Ras (N17) blocks PKC-induced Erk activation in some but not all cell systems (28). PKCθ is highly expressed in T cells and is the only member that translocates to the immunological synapse, like RasGRP1 (3, 11). Direct activity of PKCθ towards Ras activation has not been established, but TCR cross-linking does result in the phosphorylation and activation of Raf (and Erk) in a PKC-dependent manner (39). Like active Ras, active PKCθ leads to CD69 upregulation, and conversely, CD69 upregulation is impaired in PKCθ-deficient T cells or when T cells express dominant negative PKCθ (2, 40). Thymocyte development in PKCθ knockout mice is, however, grossly normal (40).
Here we use a RasGRP1-deficient T-cell line and RasGRP1 RNA interference (RNAi) to study the role of RasGRP1 activation and show that novel PKC kinases phosphorylate RasGRP1 at threonine residue 184 in TCR-triggered T cells, resulting in activation of the Ras-Erk-CD69 pathway. PKCθ-RasGRP1 signals are functionally unique since an inactive PKCθ-RasGRP1 pathway is not compensated for by LAT-Grb2-SOS signaling.
MATERIALS AND METHODS
Cell lines, stimulations, and inhibitors.
Clone numbers of cell lines are as follows: Jurkat E6-1 clone 1 (37), Jurkat-CD25ζζ#35, and JPRM441-CD25ζζ#1. Jurkat T cells, derived cell lines, and 293T cells were grown as described before (37, 42). Stable cell lines were selected with Zeocin (200 μg/ml; Invitrogen) or G418 (Genetecin, 2 mg/ml; Gibco BRL). Human CD4 T cells were purified according to the manufacturer's instructions (RosetteSep; StemCell Technologies). Stimulations were carried out at 37°C with PMA at 25 ng/ml unless mentioned otherwise, ionomycin (Calbiochem) at 1 μM, a 1:1,000 dilution of the C305 supernatant recognizing TCRβ, and mouse anti-human CD25 antibody (ICN Biochemicals) at 1 μg/ml, with goat anti-mouse antibody (Jackson Laboratories) at 10 μg/ml. Cells were preloaded for 1 h with PKC inhibitor Rottlerin (20 μM; Calbiochem) or Ro 31-8220 (1 μM; Alexis Biochemicals) before stimulations.
Generation of EMS mutants.
Jurkat E6-1 clone 1 cells (100 × 106) were treated with 200 μg/ml methanesulfonic acid ethyl ester (EMS; Sigma) for 24 h, washed, and cultured for 5 days prior to being screened. Jurkat PMA response mutant (JPRM) clones were generated as described in Results though magnetic cell sorting (MACS) depletion using anti-CD69-fluorescein isothiocyanate (FITC; BD Biosciences) and anti-FITC beads (Miltenyi Biotec) according to Miltenyi's instructions. In short, 100 × 106 cells were (i) washed in fluorescence-activated cell sorter (FACS) buffer, (ii) resuspended in 1 ml FACS buffer with 40 μl CD69-FITC (1 h at 4°C), (iii) washed twice, (iv) resuspended in 1 ml FACS buffer with 50 μl anti-FITC beads (1 h at 4°C), (v) washed twice, and (vi) loaded on a MACS depletion column; (vii) then the column was washed four times. Steps iv to vii were repeated twice, and the CD69-negative cells were eluted from the column and expanded in culture until the next round of selection.
Plasmids.
EcoRI/EcoRV-digested SRα-Tac/ζ/ζ (Weiss lab inventory) was cloned into EcoRI/Pme-digested pEF4MycHisA to create pEF4-CD25ζζ. pAWneo3/Myc-HisA, -B, and -C were created by ligating a SpeI/PmeI fragment from pEF6Myc-HisA-C (Invitrogen) into XbaI-blunted, BamHI-digested pAWneo3. RasGRP1 was amplified by PCR using the primers 5′-CGACGCGTCCGCGGCCATGGGAACCCTG-3′ and 5′-TCTGTCGACAGAACAGTCACCCTGCTC-3′ and cloned into MluI/SalI-digested pAWneo3/Myc-HisB. ΔDAGRasGRP1-pAWneo3/Myc-HisB (MluI/SalI) was created as a three-point ligation using the additional primers 5′-GAAGGATCCAGGAAAGCCCAGGCCCAGCTT-3′ and 5′-TTTGGATCCAAGAAGCGAGCCAAGAACCCA-3′. Additional Myc-tagged versions of full-length RasGRP-1, -2, -3, and -4 were generated through PCR and cloned into pEF6MycHisA (Invitrogen) using NotI/XbaI and the respective primer combinations: 5′-GGCGGCCGCGGATCCGCGGCCATGGGAACCCTG-3′ and 5′-CCTCTAGAAGAACAGTCACCCTGCTCCATTTG-3′, 5′-CGCGGCCGCAGCCATGGCAGGCACCCTGGAC-3′ and 5′-CCTCTAGACAAGTGGATGTCAAACACCCCATC-3′, 5′-GGCGGCCGCATAACCATGGGATCAAGTGGCCTTG-3′ and 5′-CCTCTAGAGCCATCCTCACCATCCTGTCTGGTC-3′, and 5′-GGCGGCCGCGCGGGAAGCATGAACAGAAAAGAC-3′ and 5′-CCTCTAGAGGAATCCGGCTTGGAGGATGCAGTTG-3′. XGrg-4 encodes Myc-tagged Xenopus Grg-4 (36).
FACS analysis.
FACS assays were carried out as described before (37) using phycoerythrin (PE)- and allophycocyanin-conjugated CD69, PE-CD25, and PE-CD3 (all BD Biosciences).
Transfections.
Jurkat and derived cell lines were transfected in 0.3 ml of RPMI 1640, 10% fetal calf serum, and glutamine, without penicillin-streptomycin, with a Gene Pulser electroporator (Bio-Rad) set at 250 V and 960 μF and 293T cells as described before (42).
Western blot analysis and immunoprecipitations.
Western blot analysis of 1% NP-40 lysates were performed as described before (37). A total of 1 × 106 cell equivalents were analyzed per sample with antibodies (Abs) for the following proteins: A176 Ab for RasGRP1 (18), Thr204/Tyr204 for phospho-p44/42 MAP kinase, P38 MAP kinase Ab, 9B11 Ab for Myc (Cell Signalling), Ab, active JNK, active p38 Ab (Promega), C-16 and C-14 for ERK-1 and -2, F3 and D2 Ab for JNK-1 and -2 (Santa Cruz Biotechnology); α-tubulin Ab (Sigma), PKCθ Ab, (BD Transduction Laboratories), Express Ab (Invitrogen), SOS1 Ab, 4G10 Ab for phospho-Y (Upstate Biotechnologies). M133 antibody was developed by injection of recombinant full-length rat RasGRP1 into BALB/c mice. The M133 antibody recognizes human, rat, and mouse RasGRP1. The P-T184 antibody was developed as described by Zheng et al. (51). Immunoprecipitations were performed as described before (37) for RasGRP1 (30 μg of M133) (51), Grb2 (with 10 μg of Grb2 C23 [Santa Cruz] cross-linked to protein A Sepharose with dimethyl pimelimidate [Pierce]), or the control (using purified rabbit immunoglobulin G; Pierce). Coimmunoprecipitations of 20 μg transfected Myc-tagged RasGRP and 20 μg transfected kinase dead PKCθ or -α were performed on 500 μl NP-40 lysates of the transfected 10-cm plate with 293T cells using 5 μg of 9B11 or control antibody (purified mouse immunoglobulin G2b; ICN) bound to protein G-Sepharose (Pharmacia). Proteins were visualized using Western Lightning chemiluminescence reagent plus (Perkin Elmer) and a Kodak image station 440CF, and Kodak ID Image Analysis software 3.5 was used to quantify expression levels.
RNA isolation, Northern blot analysis, and RT-PCR.
RNA was isolated and Northern blots were carried out as described before (37). Hybridizations were performed using a 1,500-bp RasGRP1 3′ XhoI/EcoRI fragment, 350-bp TCRα HindIII/PvuII and 360-bp TCRβ BglII/StuI fragments encompassing the constant region, and T-to-A cloned PCR fragments of TCRζ (959 bp, primers 5′-CCAGGGGATTTCACCACTCAAAGGCC-3′ and 5′-AGCAGAGCAGAGAGCGTTTTCCATCC-3′), of CD3ɛ (629 bp, primers 5′-TATTCTGGCCTGAATCAGAGACGC-3′ and 5′-GCTGAGGAGCCAGAGCACATATACGG-3′), and of beta-actin (540 bp, primers 5′-GTGGGCCGCTCTAGGCACCAA-3′ and 5′-CTCTTTGATGTCACGCACGATTTC-3′). First-strand cDNA was produced using a synthesis kit from Amersham, and RasGRP1-specific (33 cycles, primers 5′-TCCTTCTGTGTGATGGACAAAGAC-3′ and 5′-CTGTGGGAGCTACTGGGTTCTT-3′), RasGRP2-specific (34 cycles, primers 5′-TACCTCAGCGCCTTTGGGGACCTC-3′ and 5′-CAGAGAGAAGCTGAAGGCGCGGTG-3′), RasGRP3-specific (37 cycles, primers 5′-TTGGATTCCTTCTGTGTTCTGGAC-3′ and 5′-GGCTTCCAGGCAGTGACCCATGAC-3′), RasGRP4-specific (40 cycles, primers 5′-CCACGCCAGGGGAGAGGATCCTTC-3′ and 5′-TCCTGCATCGCCCTTGGCCCCTGG), and HPRT-specific (30 cycles, primers 5′-AACGTCTTGCTCGAGATGTG-3′ and 5′-AGCAAGCTTGCGACCTTGAC-3′) reverse transcription (RT)-PCRs were carried out.
Ras activation assays.
Activation of Ras was analyzed by a RasGTP pull-down assay (with 15 μl of Raf-1 Ras binding domain (RBD) agarose and 10 × 106 cells for each time point) according to the manufacturer's instructions (Upstate).
Luciferase assays.
Jurkat and JPRM441 cells were transfected with 20 μg of an AP-1 luciferase reporter construct and 2 μg of pEF6-LacZ as a transfection control, and luciferase assays were performed as described before (42). All luciferase assay data were normalized to β-galactosidase numbers.
Measurement of intracellular Ca2+ levels.
Jurkat, Jurkat-CD25ζζ, and JPRM441-CD25ζζ cells were loaded with the Ca2+ indicator Fluo-3-AM and analyzed as described before (26). Cells were preloaded with anti-CD25 or medium for 30 min on ice and washed. Cells were warmed to 37°C for 5 min prior to stimulation by C305 or goat anti-mouse antibody and analyzed by FACS.
Kinase assays.
For in vitro kinase assays, Myc immunoprecipitations were performed, and protein G-Sepharose beads with the antibody-protein complex were washed once in kinase buffer and resuspended in 37.6 μl kinase buffer (25 mM Tris [pH 7.5], 5 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM MgCl2, and 200 μM ATP as described by Perez et al. [33]). Next, 1.4 μl active PKCθ (Upstate) and 10 μCi γ-ATP (Amersham) were added and the mix was incubated for 30 min at 37°C. Products were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane, and exposed to film.
RNAi.
RASGRP1 RNAi constructs were generated using BglII/HindIII-digested pTER plasmid (44) and the following oligonucleotides: for RasGRP1-RNAi274, 5′-GATCCCGTCATGCTGACCATGCACCTTCAAGAGAGGTGCATGGTCAGCATGACTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGTCATGCTGACCATGCACCTCTCTTGAAGGTGCATGGTCAGCATGACGG-3′; for RasGRP1-RNAi363, 5′-GATCCCTTCACCAGGACTTTGCCTGTTCAAGAGACAGGCAAAGTCCTGGTGAATTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAATTCACCAGGACTTTGCCTGTCTCTTGAACAGGCAAAGTCCTGGTGAAGG-3′; for RasGRP1-RNAi492, 5′-GATCCCGGGTGAGGAGTTACATTGCTTCAAGAGAGCAATGTAACTCCTCACCCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGGGTGAGGAGTTACATTGCTCTCTTGAAGCAATGTAACTCCTCACCCGG-3′; for RasGRP1-RNAi1419, 5′-GATCCCACACGTCCAGAGGATGGTGTTCAAGAGACACCATCCTCTGGACGTGTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAACACGTCCAGAGGATGGTGTCTCTTGAACACCATCCTCTGGACGTGTGG-3′; for RasGRP1-RNAi1503, 5′-GATCCCGATTGCTGCGAGTTTTCCATTCAAGAGATGGAAAACTCGCAGCAATCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGATTGCTGCGAGTTTTCCATCTCTTGAATGGAAAACTCGCAGCAATCGG-3′; and for RasGRP1-RNAi2174, 5′-GATCCCAGCGGGCTTTTGTCAAGTGTTCAAGAGACACTTGACAAAAGCCCGCTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGCGGGCTTTTGTCAAGTGTCTCTTGAACACTTGACAAAAGCCCGCTGG-3′. Jurkat cells were transfected with RasGRP1-RNAi1503 and a human CD16/CD7 expression construct. Four days later, trans-fected cells were MACS purified using anti-CD16 FITC (CalTag) and anti-FITC beads (Miltenyi Biotec) and analyzed as described before. PKCθ small interfering RNA (siRNA) transfection was performed using the same transfection protocol and purification strategy as for RasGRP1 RNAi with PKCθ siRNA 802-822 (Ambion) as previously described by Ishaq et al. (22).
RESULTS
Generation of mutant Jurkat T-cell clones defective in diacylglycerol-mediated Ras activation.
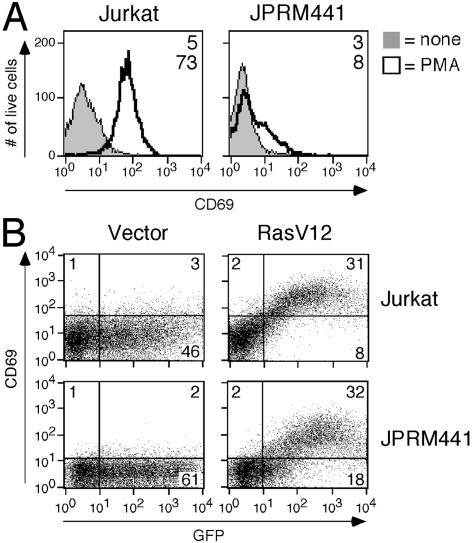
In order to identify signaling molecules that activate Ras in T cells in response to the second-messenger DAG, we designed the following selection strategy for the Jurkat T-cell leukemic line E6-1. First, these cells were randomly mutagenized using EMS. Subsequently, they were selected for their inability to upregulate the activation marker CD69, a Ras-dependent event, in response to a synthetic DAG analogue PMA (50 ng/ml). After three rounds of selection using antibody-coated bead depletion of CD69-positive cells, mutant cells were subcloned by limiting dilution. Seven hundred JPRMs were individually reanalyzed by FACS for their inability to upregulate CD69 expression in response to PMA. One hundred twenty-five clones were identified as true phenotypic mutants (for a typical example, see Fig. 1A).
FIG. 1.
Screening strategy for the isolation of Jurkat T-cell clones with impaired DAG-induced Ras activation. (A) FACS analysis of induced CD69 expression on Jurkat or JPRM441 cells following overnight PMA stimulation. Numbers indicate the mean fluorescent intensity. (B) FACS analysis of GFP and induced CD69 expression on Jurkat and JPRM441 cells, 40 h after transient cotransfection with GFP (10 μg) and vector or RasV12 (10 μg). Numbers indicate the percentage of live cells in each quadrant. All experiments in Fig. 1 and all subsequent figures are representative examples of at least three independent experiments unless indicated otherwise.
We were primarily interested in JPRM lines that contained a defect in the most proximal part of the PMA-Ras-MAP kinase-CD69 pathway, i.e., PMA leading to Ras activation. To examine if the distal part of the pathway, i.e., Ras-MAP kinase-CD69, was still functional in our JPRM clones, we transiently introduced an active allele of Ras, RasV12, which leads to CD69 upregulation in wild-type Jurkat T cells (Fig. 1B). Out of 85 JPRM clones (45 severe- and 40 mild-phenotype mutants), 82 JPRM lines had an intact Ras-MAP kinase-CD69 pathway (for a typical example, see Fig. 1B). We also noticed a decreased basal level of CD69 in unstimulated JPRM441 cells (Fig. 1A and B). Cell fusion assays using polyethylene glycol divided the JPRM clones into five complementation groups (data not shown).
We predicted RasGRP1 and PKCθ to be affected candidate genes in our JPRM clones, and indeed, the largest complementation group expressed RasGRP1 at a severely reduced level. None of the 125 JPRM clones expressed decreased levels of PKCθ protein (data not shown). JPRM441 is a typical clone from the largest complementation group and expresses roughly 1/10 of the RasGRP1 protein produced by wild-type Jurkat T cells but expresses normal levels of PKCθ, Erk-1 and -2, P38, and JNK-1 and -2 (Fig. 2A, 3B, and data not shown). All the clones belonging to this complementation group exhibit the same amount of reduced RasGRP1 expression (data not shown). The RasGRP1 defect is at the transcriptional level, as we were unable to detect RasGRP1 mRNA in JPRM441 by Northern blot analysis (Fig. 2B).
FIG. 2.
Isolation of a RasGRP1-deficient Jurkat T-cell clone. (A) Western blot analysis of RasGRP1 expression in Jurkat and JPRM441 whole-cell lysates (WCL) or RasGRP1 immunoprecipitations (m133 IP). The expression was quantified and set to a relative 100% expression level in wild-type Jurkat cells. (B) Northern blot analysis of RasGRP1, TCRα, TCRβ, TCRζ, and CD3ɛ gene expression in Jurkat (lanes 1) and JPRM441 (lanes 2) cell lines. RNA levels are indicated by β-actin hybridization of the same blots. (C) RT-PCR analysis of RasGRP-1, -2, -3, and -4 gene expression on 1:10 serial dilutions of Jurkat and JPRM441 cDNAs and on cDNA derived from human peripheral blood mononuclear cells, and a panel of lymphoid and nonlymphoid cell lines (cell type): lane 1, peripheral blood mononuclear cells; lane 2, peer (γδ T cell); lane 3, RAJI (B cell); lane 4, K562 (myeloid cell); lane 5, U937 (myeloid cell); lane 6, RPMI8226 (multiple myeloma cell); and lane 7, HELA (cervix carcinoma). RT-PCR for HPRT gene expression demonstrates equal RNA levels.
FIG. 3.
Restored induction of CD69 in stimulated JPRM441 cells reconstituted with wild-type RasGRP1 cDNA. (A) Strategy to mimic TCR signaling in JPRM441 through cross-linking of the introduced chimeric CD25ζζ protein. A FACS analysis for CD25 demonstrates equal levels of expression of CD25ζζ in the stable clones Jurkat-CD25ζζ and JPRM441-CD25ζζ. (B) Western blot analysis of RasGRP1 expression in stable cell lines expressing wild-type RasGRP1 cDNA or ΔDAGRasGRP1 cDNA in JPRM441-CD25ζζ, yielding the indicated cell lines. Western blot analyses of PKCθ, Erk-1 and -2, and P38 expression demonstrate equal levels of these four proteins in all the lines. WCL, whole-cell lysates. (C) The indicated cell lines were left unstimulated or were stimulated overnight with PMA or soluble anti-CD25 and goat anti-mouse antibodies and analyzed by FACS for induced CD69 expression. Numbers in the histograms indicate mean fluorescent intensities.
RT-PCR analysis for transcripts of RasGRP1, -2, -3, and -4 genes confirmed the RasGRP1 Northern blot result, with JPRM441 expressing approximately 50-fold-reduced RasGRP1 mRNA levels (Fig. 2C). There was no apparent altered expression of the other three RasGRP family members in JPRM441, suggesting that there is no compensatory mechanism at the level of gene expression. The RasGRP2 RT-PCR indicates the expression level of the Rap-specific GEF, since we were able to amplify only that shorter form of RasGRP2 from Jurkat or JPRM441 cDNA (data not shown). Notably, the expression pattern of the various RasGRP genes in Jurkat T cells closely resembled that of human peripheral blood mononuclear cells, typically containing more than 70% T cells (Fig. 2C, lane 1). In contrast, other lymphoid and nonlymphoid cell types displayed different patterns of expressed RasGRP genes (Fig. 2C).
RasGRP1 deficiency causes impaired CD69 upregulation in both PMA- and TCR-stimulated JPRM441.
FACS analysis for cell surface markers revealed that the RasGRP1 mutant clone JPRM441 did not express a TCR/CD3 complex on the surface. Western blot analysis following immunoprecipitations demonstrated that this mutant clone does not express any (intracellular) TCRα or TCRβ protein (data not shown). Last, Northern blot analysis established that JPRM441 cells do not express any TCRα mRNA and low levels of TCRζ mRNA. JPRM441 cells expressed only low levels of the immature 1.0-kb D→J TCRβ transcript and no detectable mature 1.3-kb TCRβ transcripts (Fig. 2B). The reduced transcript levels were evident in all tested clones that belonged to the same complementation group as JPRM441. In addition, the reduced levels of TCRα and -β transcripts were also found in independent JPRM clones from some of the other groups that complemented the JPRM441 line. These results suggest that the downregulation of TCRα and -β transcripts is not due to an additional mutation in JPRM441 but rather a result of impaired DAG-Ras signaling. Reconstitution of JPRM441 with wild-type RasGRP1 did not lead to reexpression of these transcripts in the 208, 240, or 247 clones. However, transient transfection of RasGRP1-RNAi-1503 plasmid in wild-type Jurkat cells led to a 20% reduction in the surface expression of CD3 in the transfected cells 4 days after transfection, whereas control pTER plasmid did not (data not shown). We are currently investigating the underlying mechanism responsible for this regulation of TCRα and -β transcript levels.
In order to trigger JPRM441 cells with a TCR-like stimulus, we next generated stable transfectants expressing a chimeric CD25ζζ molecule, consisting of the extracellular portion of human CD25 and the transmembrane region and intracellular portion of murine TCRζ (Fig. 3A). A second generation of stable lines was obtained by expressing full-length wild-type RasGRP1 or ΔDAGRasGRP1 into JPRM441-CD25ζζ. ΔDAGRasGRP1 results in the deletion of the DAG-binding C1 domain (amino acids 542 to 591). Stable clones expressing levels of RasGRP1 protein similar to those observed in wild-type Jurkat cells were selected (Fig. 3B).
To test whether the deficiency in RasGRP1 was the true cause of decreased PMA-induced CD69 expression in JPRM441, we analyzed the panel of cell lines depicted in Fig. 3C. Indeed, the impaired CD69 induction by PMA was specifically restored in the two wild-type RasGRP1-reconstituted JPRM441-CD25ζζ clones, but not in the two clones expressing ΔDAGRasGRP1. In addition, we observed that cross-linking of CD25ζζ induced high levels of CD69 on the surfaces of Jurkat-CD25ζζ cells as cross-linking of the TCR does, but not on that of JPRM441-CD25ζζ cells. Again, the defect in CD69 upregulation was restored only by reconstitution with wild-type RasGRP1 and not with ΔDAGRasGRP1. RasGRP1 is therefore also a critical component in the TCR-induced Ras-MAP kinase-CD69 pathway.
Impaired activation of Ras and the MAP kinases Erk-1 and -2 upon phorbol ester stimulation of RasGRP1-deficient cells.
We next examined the activation of Ras by the DAG surrogate PMA in JPRM441 cells, the signaling event that prompted us to perform this study. As depicted in Fig. 4A, activation of Ras upon stimulation with PMA was impaired in JPRM441, yet not absent.
FIG. 4.
Defective PMA-Ras-Erk signaling due to RasGRP1 deficiency. (A) Resting and PMA-stimulated Jurkat and JPRM441 cells were analyzed for the extent of Ras activation at the indicated time points by a GST-Raf pull-down assay. Bands were quantified, and the percentage of Ras activation was obtained through normalization to total Ras expression levels. (B) Western blot analysis of PMA-induced activation of Erk-1 and -2 in Jurkat and JPRM441 cells. The mean levels of phosphorylation of Erk-1 (P-Erk1) and -2 and the standard deviations from three independent experiments were calculated using normalization for α-tubulin expression and are indicated in the box. These mean levels of activation were also plotted as relative percentages for the various time points. (C) Western blot analysis of PMA- and ionomycin-induced (ion.) phosphorylation of P38 or PMA-induced phosphorylation of JNK-1 and -2 in Jurkat and JPRM441 cells. The same blot was subjected to α-tubulin Western blot analysis as a loading control. (D) Luciferase assay for the transcriptional activity of AP-1 in resting or PMA-stimulated Jurkat and JPRM441 cells. The bar graph represents the mean luciferase values and standard deviations from three independent assays.
Ras functions as a branch point and can lead to the activation of three classes of MAP kinases: Erk-1 and -2, P38, and JNK-1 and -2 (45). We observed that phosphorylation of Erk-1 and -2 was delayed, reduced, and more transient in PMA-stimulated RasGRP1-deficient JPRM441 cells (Fig. 4B). Importantly, these three defects were restored in wild-type RasGRP1 (wtRasGRP1)- but not in ΔDAGRasGRP1-reconstituted cells (Fig. 5). Expression of ΔDAGRasGRP1 in JPRM441 even further decreased the phosphorylation levels compared to those in nonreconstituted JPRM441, suggesting a dominant negative effect. wtRasGRP1#208 and ΔDAGRasGRP1#11 are representative clones and were selected for all further presented experimentation. The majority of the assays were verified in the other clones and the results were in agreement with those obtained with wtRasGRP1#208 and ΔDAGRasGRP1#11 (data not shown).
FIG. 5.
PMA-MAPK signaling in RasGRP1-reconstituted cells. Results of an analysis of the PMA-induced phosphorylation of Erk-1 and -2 or JNK-1 and -2 and the PMA- and ionomycin-induced (ion.) phosphorylation of P38 in Jurkat-CD25ζζ, JPRM441-CD25ζζ, and wtRasGRP1- or ΔDAGRasGRP1-reconstituted JPRM441-CD25ζζ cells are shown. The mean levels of phosphorylation of Erk-1 and -2 and the standard deviations from three independent experiments were calculated using normalization for α-tubulin expression and are indicated in the box.
We next analyzed how impaired, transient activation of Erk kinases in RasGRP1-deficient cells impacts cumulative signals that are received in the nucleus. Luciferase assays for the activation of the transcription factor AP-1, which, like overnight upregulation of CD69, measures cumulative MAP kinase input following PMA stimulation, demonstrated a 75% reduction of activity in JPRM441 cells (Fig. 4D).
The deficiency in RasGRP1 expression did, however, not affect P38 phosphorylation upon PMA and ionomycin stimulation (Fig. 4C and 5). Phosphorylation of JNK-1 and -2 was reduced in PMA-stimulated JPRM441 cells, but it was not restored in any of the three wtRasGRP1-reconstituted clones (Fig. 4C, 5, and data not shown). The defect in activation of JNK kinases may be the result of an additional mutation generated by the EMS treatment. In support of this explanation, RasGRP1 RNAi introduction in wild-type Jurkat cells did not affect PMA-induced phosphorylation of JNK-1 and -2 (data not shown).
Normal proximal TCR signaling and calcium mobilization in TCR-stimulated JPRM441 cells.
Even though RasGRP1 activity is reportedly distal to DAG generation, we wanted to examine the proximal signaling events that are triggered by TCR ligation in JPRM441 cells. TCR stimulation of JPRM441-CD25ζζ cells is modeled by cross-linking of CD25ζζ on these cells, as in Fig. 3C. Cross-linking of anti-TCRβ or CD25ζζ on Jurkat-CD25ζζ cells and CD25ζζ on JPRM441-CD25ζζ cells induced very similar levels of tyrosine-phosphorylated proteins, including p36 (LAT), p56 (Lck), p70 (ZAP-70), and p76 (SLP-76) (Fig. 6A). We also tested the ability of JPRM441-CD25ζζ cells to generate a calcium flux upon CD25ζζ cross-linking. As demonstrated in Fig. 6B, RasGRP1-deficient cells were capable of generating a normal calcium flux. The failure to express RasGRP1 or a possible unrelated mutation caused by the EMS treatment therefore does not compromise the ability of JPRM441 cells to phosphorylate many of the proteins in the TCR pathway or to flux calcium following CD25ζζ stimulation.
FIG. 6.
TCR-induced tyrosine-phosphorylated proteins and calcium mobilization. (A) Induced tyrosine phosphorylation patterns upon TCRβ or CD25ζζ cross-linking in the indicated cell lines by 4G10 Western blot analysis are shown. Positions of known tyrosine-phosphorylated proteins are indicated. Equal protein loadings are indicated by α-tubulin expression on the same blot. (B) Measurement of intracellular Ca2+ levels using the indicator Fluo-3-AM in Jurkat, Jurkat-CD25ζζ, and JPRM441-CD25ζζ cells. Arrows indicate the administration of C305 cross-linking TCRβ or anti-CD25 cross-linking CD25ζζ.
RasGRP1 is required for the activation of Erk-1 and -2 following TCR stimulation.
Activation of the Ras-Erk pathway following engagement of the TCR is a crucial signaling event for positive selection of developing thymocytes (38). The current literature suggests that there are at least two distinct pathways leading to TCR-induced activation of Ras: one that involves the formation of a trimolecular phospho-LAT-Grb2-SOS complex resulting in the recruitment of SOS to the membrane, where it can activate Ras. A second pathway requires the generation of the second-messenger DAG, leading to the recruitment of molecules with a DAG-binding C1 domain, like PKC and RasGRP. Indirectly, this pathway also relies on LAT, i.e., through LAT-dependent PLCγ recruitment and activation (43). Based on our CD69 assays, we hypothesized that the RasGRP1 deficiency could lead to some degree of impaired activation of Erk kinases by a TCR-like stimulus. Indeed, activation of Erk-1 and -2 following cross-linking of CD25ζζ on JPRM441-CD25ζζ cells was delayed, reduced, and less sustained, compared to the efficient and sustained phosphorylation in Jurkat-CD25ζζ cells (Fig. 7A). Direct analysis of Ras activation using the Raf-glutathione S-transferase pull-down assay demonstrated impaired CD25ζζ-triggered Ras activation in JPRM441-CD25ζζ cells (data not shown). Again, the defect in phosphorylation of Erk-1 and -2 was specifically restored by the expression of a full-length molecule of RasGRP1, but not by a molecule that lacks the DAG-binding C1 domain (Fig. 7B).
FIG. 7.
Impaired TCR-Erk signal transduction in RasGRP1-deficient cells. (A) Western blot analysis of activation of Erk-1 and -2 by cross-linking CD25ζζ on Jurkat-CD25ζζ and JPRM441-CD25ζζ cells. The mean levels of phosphorylation of Erk-1 (P-Erk1) and -2 and the standard deviations from three independent experiments were calculated and are plotted as in Fig. 4B. (B) Analysis of anti-CD25-induced phosphorylation of Erk-1 and -2 in Jurkat-CD25ζζ, JPRM441-CD25ζζ, and wtRasGRP1- or ΔDAGRasGRP1-reconstituted JPRM441-CD25ζζ cells. The mean levels of phosphorylation of Erk-1 and -2 and the standard deviations from three independent experiments were calculated usingnormalization for α-tubulin expression and are indicated in the box. (C) Western blot analysis of SOS1 and phospho-p36 (LAT) coimmunoprecipitating (IP) with Grb2 in the indicated cell lines following CD25ζζ cross-linking. An analysis of whole-cell lysates (WCL) indicates equal levels of expression or phosphorylation of the three proteins in the two cell lines.
The defect observed after CD25ζζ cross-linking was at least as severe as the impairment seen using PMA as a stimulus (compare Fig. 7A with 4B). This is remarkable given the fact that the RasGRP1 deficiency does not affect Grb2 and SOS1 recruitment to phosphorylated LAT upon CD25ζζ stimulation (Fig. 7C). The Ras-activating pathway involving the formation of a trimolecular phospho-LAT-Grb2-SOS complex is thus intact in RasGRP1-deficient JPRM441 cells but does not compensate for the defect in the second, DAG-dependent, Ras-activating pathway. Similarly, SOS cannot compensate in RasGRP1-deficient thymocytes (15).
The deficiency in RasGRP1 expression did not affect P38 phosphorylation following TCR stimulation (data not shown). Phosphorylation of JNK-1 and -2 was reduced in TCR-stimulated JPRM441 cells, but it was not restored in any of the three wtRasGRP1-reconstituted JPRM441 clones (data not shown).
Optimal, phorbol ester-induced activation of Ras and the kinases Erk-1 and -2 requires both RasGRP1 and the activity of novel PKC family members.
The JPRM441 line is not completely deficient for both Ras and Erk kinase activation upon PMA stimulation. Intriguingly, introduction of a RasGRP1 molecule lacking the DAG-binding C1 domain did not simply fail to restore PMA-induced Erk activation but also decreased this signaling event in JPRM441 cells further. This result suggests that the ΔDAGRasGRP1 molecule acts as a dominant negative allele and implies that the residual phosphorylation of Erk is generated through residual RasGRP1 or through a RasGRP(-like) molecule. We postulated that the ΔDAGRasGRP1 molecule competes with the remaining RasGRP proteins for signals coming from another PMA-responsive element in this pathway and hypothesized that this element was PKC.
To address this question, we performed the following experiments in the absence or presence of Rottlerin, a relative selective inhibitor of novel PKC family members, including PKCθ (20). Administration of this inhibitor completely abolished the residual activation of Ras and of Erk kinases in JPRM441 cells that were stimulated with PMA (Fig. 8A and B). In RasGRP1-sufficient, wild-type Jurkat T cells, the strong activation of Ras and Erk kinases that normally occurs upon PMA treatment was nearly abolished by Rottlerin treatment (Fig. 8A and B). The same results were obtained with Ro 31-8220, an inhibitor of both classical and novel PKC family members (46; data not shown). In contrast, Go 6976, an inhibitor specific to classical PKC family members (29), did not have such dramatic effects (data not shown). The effect of Rottlerin was not limited to the Jurkat T-cell line. Purified human CD4 T cells from peripheral blood (99% CD3+, 89% CD4+, 0% CD19+) that express RasGRP1 protein (data not shown) were equally sensitive to the inhibitor, which blocked all PMA-induced phosphorylation of Erk-1 and -2 (Fig. 8C). Optimal activation of the DAG-Ras-Erk pathway therefore requires not only sufficient levels of RasGRP1 but also the activity of novel PKC kinase molecules, suggesting a connection between these two DAG-responsive signaling proteins.
FIG. 8.
Both RasGRP1 and novel PKC family members are required for PMA-Ras-Erk signaling. (A) Jurkat and JPRM441 cells were treated with DMSO control or with the PKC inhibitor Rottlerin for 1 h. Resting and PMA-stimulated Jurkat and JPRM441 cells were analyzed for the extent of Ras activation. Bands were quantified, and the percentages of Ras activation were obtained through normalization to total Ras expression levels. (B) Following Rottlerin or control treatment, the phosphorylation of Erk-1 (P-Erk1) and -2 was determined by Western blot analysis of PMA-induced or resting Jurkat and JPRM441 cells. Activation of the two kinases was quantified using normalization for α-tubulin expression on the same blot. Numbers are the means and standard deviations from three independent experiments. (C) Purified human CD4 T cells were pretreated with Rottlerin or with DMSO control and analyzed for the amount of PMA-induced phosphorylation of Erk-1 and -2 as described for panel B.
Novel PKC kinase signals are responsible for residual activity in the PMA-Erk pathway in RasGRP1-null cells.
To address the residual PMA-Ras-Erk signal in JPRM441 cells, we designed RasGRP1 RNAi constructs to target RasGRP1 transcripts, avoiding random mutagenesis. We selected six target sequences using Ambion's selection criteria (www.ambion.com/techlib/misc/siRNA_finder.html), cloned the annealed oligonucleotides in pTER, and tested the indicated RNAi constructs by cotransfection of 293T cells with Myc-tagged RasGRP1 (Fig. 9A). RasGRP1-RNAi1503 yielded efficient and reproducible knock-down of RasGRP1 levels and was used for further experimentation. Western blot analysis of the purified, transfected pool of wild-type Jurkat and mutant JPRM441 cells indicated that this RNAi construct resulted in RasGRP1 expression levels of 16% and 2% of normal Jurkat levels in the respective cell lines (Fig. 9C). Analysis of PMA-induced phosphorylation of Erk in the same cells demonstrated the following. (i) Wild-type Jurkat cells transfected with RasGRP1 RNAi respond very similarly to RasGRP1-deficient JPRM441 cells (Fig. 9D, top). These experiments validate the results obtained with our mutant cell line. (ii) As in JPRM441, Rottlerin treatment combined with RasGRP1 RNAi transfection abolished the residual phosphorylation of Erk. In cells expressing only 2% of normal RasGRP1 levels, PMA still induced Erk phosphorylation to a modest extent, but this was blocked by Rottlerin (Fig. 9D, bottom). These results indicate the presence of another Ras activator that is still responsive to DAG and is regulated by novel PKC kinases.
FIG. 9.
Novel PKC kinases signal to a RasGRP1-like molecule in RasGRP1-null cells. (A) Immunoblotting for Myc expression in Myc-tagged RasGRP1-transfected 293T cells in combination with the indicated RNAi constructs. (B) Immunoblotting for Express expression in Express-tagged PKCθ(KR)-transfected 293T cells in combination with the indicated siRNA construct. (C) Western blot analysis for RasGRP1, and Grb2 of MACS-purified Jurkat and JPRM441 cells transfected with the indicated constructs. Numbers indicate the relative percentages of RasGRP1 expression. (D) The indicated purified, transfected pools were preincubated with DMSO control or Rottlerin, stimulated as indicated, and analyzed for Erk phosphorylation by Western blot analysis. Numbers indicate the relative levels of phosphorylated Erk-1 (P-Erk1) and -2, obtained by normalizing for protein loading through Grb2. Panel D is representative of two independent experiments.
Novel PKC kinases phosphorylate and signal to RasGRP1 to activate the Ras-Erk-CD69 pathway.
To address a possible PKCθ-RasGRP1 connection, we first examined whether the two proteins could interact with each other. To this end, 293T cells were transfected with Myc-tagged RasGRP and Express-tagged, kinase-dead PKC constructs. Immunoprecipitations of the Myc-tagged material followed by Western blot analysis of RasGRP (Myc) and PKC (Express) expression demonstrated that PKCθ coimmunoprecipitated with RasGRP-1, -2, and -3 (Fig. 10A). PKCα was able to interact with only RasGRP-1 and -3. Only kinase-dead forms of PKC-θ or -α and not active PKC alleles were consistently coimmunoprecipitated with the RasGRP species (data not shown).
Some kinase-dead enzymes function as traps for their substrates, supporting the possibility that PKC may phosphorylate RasGRP1. To explore this, we performed in vitro kinase assays to establish whether RasGRP proteins are potential PKC substrates. We produced Myc-tagged RasGRP-1, -2, -3, and -4 and an irrelevant control XGrg-4 of similar sizes in 293T cells (Fig. 10B). Following immunoprecipitations of the Myc-tagged material, one-fourth of the Myc-tagged material was subjected to Western blotting to determine the level of production and three-fourths was used as a substrate in a kinase reaction with [γ-32P]ATP and purified, active PKCθ. Active PKCθ was able to in vitro phosphorylate all four RasGRP proteins but had very little effect (25%, 32P-signal/protein expressed) on the XGrg-4 control (Fig. 10B).
The decreased Ras activation in PMA-stimulated RasGRP1-deficient cells, the elimination of any PMA-Ras activation by Rottlerin, and the fact that RasGRP proteins are potential PKC substrates led us to investigate a possible PKCθ-RasGRP1-Erk-CD69 pathway. To this end, we transiently cotransfected our panel of cell lines with green fluorescent protein (GFP) and either empty vector or active RasV12, active PKCθ(AE), or active PKCα(AE) and analyzed the level of induced CD69 expression. As seen before, RasV12 led to strong CD69 expression in all four lines regardless of the RasGRP1 status. Active PKCθ(AE) led to a more moderate CD69 induction (Fig. 10C). The induction, however, was critically dependent on both sufficient levels of RasGRP1 protein and the presence of the DAG-binding domain in RasGRP1 (Fig. 10C). The active allele of PKCθ was expressed at similar levels in the four lines (Fig. 10C). When expressed at higher levels, active PKCθ did lead to some CD69 induction in JPRM441-CD25ζζ and JPRM441-CD25ζζΔDAGRasGRP1 cells, presumably signaling through the residual 10% RasGRP1 or another molecule present in these cells. However, the induced CD69 was never comparable to the level observed in wild-type Jurkat or JPRM441-CD25ζζwtRasGRP1 cells, and these higher PKCθ(AE) levels also caused severe cytotoxicity (data not shown). The RasGRP1-dependent induction of CD69 by active PKCθ was relatively specific since we did not observe any CD69 induction with an active allele of PKCα [expressed at levels higher than those of PKCθ(AE) (data not shown)], even though kinase-dead PKCα could interact with RasGRP1 in 293T cells. To address the specific actions of PKCθ by means other than the Rottlerin inhibitor, we used PKCθ siRNA as described before (22). The siRNA effectively reduced the expression of transfected PKCθ in 293T cells (Fig. 9B). Unfortunately, the same siRNA reduced endogenous PKCθ levels in Jurkat T cells to only 66% of the level in control transfected cells (data not shown). This moderate reduction of PKCθ levels in Jurkat T cells did not affect their ability to activate Erk kinases in response to PMA (data not shown). The same results were obtained when PKCθ was targeted by RNAi-802 transcribed from the pTER plasmid (data not shown).
Last, very recently it was reported that RasGRP1 and RasGRP3 are phosphorylated on a conserved threonine residue in the catalytic region in stimulated T or B cells (1, 51). We studied whether TCR or PMA stimulation of Jurkat cells also resulted in the phosphorylation of RasGRP1. The phosphorylation of threonine 184 in RasGRP1 is induced by both stimuli and is inhibited by Rottlerin treatment (Fig. 10D). Importantly, this phosphorylation event correlates well with the induced phosphorylation of the downstream Erk kinases. These experiments indicate that the phosphorylation of RasGRP1 by novel PKC kinases, including PKCθ, is a crucial signaling event required for optimal Ras-Erk activation.
DISCUSSION
We have demonstrated here that RasGRP1 is critical for the phorbol ester- and TCR-induced activation of Ras and Erk-1 and -2, leading to upregulation of CD69 in Jurkat T cells. However, RasGRP1 was dispensable for the activation of P38 or JNK-1 and -2 in Jurkat T cells. RasGRP1 fulfills a unique function since the impaired TCR-induced activation of this pathway in RasGRP1-deficient cells cannot be compensated for by the RasGEF SOS. Last, we demonstrate a functional PKCθ-RasGRP1 pathway in T cells, providing a mechanism for previous observations that PKC family members are able to activate the Ras-Raf-Erk-AP-1 signaling pathway (Fig. 11).
FIG. 11.
Model of TCR-induced Ras-Raf-Erk activation in T lymphocytes. T lymphocytes express two RasGEF molecules, SOS and RasGRP1, and hypothetically can achieve Ras activation through two different pathways. One pathway, (1), utilizes SOS, involving the formation of a phospho-LAT-Grb2-SOS complex to bring SOS in proximity with Ras-GDP. The second pathway, (2), requires generation of DAG by PLCγ. DAG recruits RasGRP1 and PKCθ to the membrane through their C1 domains. Possibly, DAG can also recruit molecules Y (e.g., PKCη) and X (RasGRP3). Novel PKC kinases phosphorylate RasGRP1 on threonine 184 (or RasGRP3 on threonine 133) and other residues, likely leading to increased GEF activity of RasGRP1. RasGRP1 subsequently converts Ras-GDP to active Ras-GTP. Disruption of the PKCθ-RasGRP1 pathway results in impaired TCR-induced Ras and Erk activation, while formation of the phospho-LAT-Grb2-SOS complex is unaffected. RasGRP1-induced Ras activation is therefore unique and cannot be compensated for by SOS. MHC, major histocompatibility complex.
Our findings with the mutant JPRM441 T-cell line are in agreement with studies of RasGRP1-deficient mice. Thymocytes in these mice are developmentally blocked at the CD4+ CD8+ double-positive stage because of a defect in positive selection that arises from impaired Erk activation (15, 34). As in our mutant cell line, these thymocytes are capable of activating P38 (34). In contrast, Grb2 haploinsufficiency results in normal Erk but attenuated P38 and JNK activation as well as in impaired negative selection (19). So, although both RasGEFs activate Ras following TCR stimulation, RasGRP1 seems to preferentially couple to Erk activation, whereas SOS may play a greater role in P38 and JNK activation. SOS also has a Rac GTPase-binding Dbl homology domain, which is not present in RasGRP1. These differences may offer an explanation for two different Ras-activating pathways in thymocytes (50).
Judging from the high incidence of activating mutations in the Ras gene in various types of cancer, the regulation of Ras activity is of fundamental importance. Two distinct types of RasGEFs (RasGRP1 and SOS) may provide more control over the regulation of Ras activity. Do the two pathways in T lymphocytes each simply contribute 50% of the Ras activation? In RasGRP1-deficient cells, Erk activation was more severely impaired upon TCR stimulation than upon PMA stimulation, but recruitment of Grb2-SOS to LAT following CD25ζζ cross-linking appears to occur normally in JPRM441-CD25ζζ cells. Why does SOS not compensate for the loss of RasGRP1? It is possible that T lymphocytes at different stages of development make different uses of either a DAG-dependent RasGRP1 pathway or a DAG-independent SOS pathway. For example, RasGRP1-deficient thymocytes early in development display no apparent defect in pre-TCR signaling, a process that is known to depend on Ras activation (15). Alternatively, Ras activation at different cellular locations may be achieved by the different GEFs (4, 9, 12). We are currently investigating the hierarchy and cross talk between the two pathways in T lymphocytes.
The PKC inhibitor Rottlerin blocked the activation of both Ras and Erk-1 and -2 in PMA-stimulated mutant JPRM441 T cells, a phenomenon that had been observed in T lymphoblasts as well (23). One could argue that residual Erk activation in RasGRP1-deficient cells stems from novel PKCs acting directly on Raf. However, others have shown that activation of Raf by PKC requires RasGTP to recruit Raf (28). The fact that Rottlerin blocks Ras activation itself strongly suggests that RasGRP1 is the relevant activator but that it requires the activity of PKCθ or another novel PKC kinase to do so.
How does PKC activity regulate RasGRP1-induced Ras activation? We show that kinase-dead PKCθ can bind to RasGRP-1, -2, and -3 when expressed in 293T cells. In addition, active PKCθ can phosphorylate RasGRP family members in vitro, and the introduction of kinase-active PKCθ resulted in efficient CD69 upregulation only in the presence of sufficient wild-type RasGRP1. These data indicate that PKCθ may play a critical role in activating RasGRP1 via phosphorylation. Evidence for a similar pathway exists in B cells (41). In BCR-stimulated DT40 chicken B cells, RasGRP3 is necessary to couple PLCγ2 to Ras activation (32). Metabolically labeled Ramos B cells revealed BCR-triggered phosphorylation of RasGRP3 (41). Very recently it has been demonstrated that RasGRP3 is inducibly phosphorylated on threonine 133 in antigen receptor-stimulated B cells (1, 51). Similarly, RasGRP1 is phosphorylated on threonine 184 in TCR-stimulated T cells (51). In our Jurkat clone, we find that phosphorylation of RasGRP1 on threonine 184 is inhibited by Rottlerin treatment and that this phosphorylation event very strongly correlates with activation of the Erk kinases (Fig. 10D). Together with our other findings, these findings suggest that RasGRP1 and RasGRP3 couple antigen receptor signaling to Ras activation in either T cells or B cells and provide a biochemical link between PKC kinases and Ras activation. Given the location of the conserved threonine residue in the catalytic region, phosphorylation could very well influence GEF activity.
The question of which PKC family member is responsible for the phosphorylation of RasGRP-1 or -3 is still difficult to answer. Originally, it was thought that PKCδ was the enzyme responsible for RasGRP3 phosphorylation. In addition, kinase-dead PKCα, a classical form, or Go 6976, an inhibitor of classical PKC enzymes, did not influence the mobility shift in RasGRP3 in PMA-stimulated CHO or LN229 cells (7). However, in PKCδ-deficient DT40 B cells, Ras activation is unaffected (1). Moreover, treatment of B cells with inhibitors of classical or novel PKC family members suggested that both may play a role in RasGRP3 phosphorylation (51). We find that inhibition of novel PKC kinases by Rottlerin abrogated the phosphorylation of RasGRP1 and the downstream activation of Erk kinases. In our studies, Go 6976 did not substantially affect the PMA-induced phosphorylation of Erk kinases in Jurkat T cells, yet the same inhibitor has been reported to block the phosphorylation of RasGRP1 at threonine 184 (51).
Eleven serine residues and threonine residue 184 are predicted by the NetPhos 2.0 server (http://www.cbs.dtu.dk/services/NetPhos/) to be possible phosphorylation sites in both human and mouse RasGRP1. The functional analysis of mutated serine and threonine residues that match PKC phosphorylation consensus sequences is complicated since the results thus far suggest that more than one residue in RasGRP1 are involved and not all the signaling input is transduced by threonine 184 (Zheng et al. [51] and data not shown).
PKCθ is expressed at high levels in T cells and is the only member reported to date to translocate to the immunological synapse, where it may interact with RasGRP1, which has also been reported to translocate to the synapse (3, 11). Given the cellular location and our results presented here, PKCθ is a very likely candidate to phosphorylate RasGRP1 in T cells. An apparent disconnect in a PKCθ-RasGRP1 pathway is the difference between the phenotypes of mice deficient for either signaling molecule. RasGRP1-deficient thymocytes are blocked in development at the double-positive stage, whereas PKCθ-deficient thymocytes are normal (15, 40). However, it has been noticed that Erk kinases can be activated in PKCθ−/− T cells by plate-bound/immobilized anti-TCR antibodies through the activity of classical PKC family members but that wild-type T cells do not require classical PKC family members for Erk activation (Hanne Ostergaard, personal communications). Very recently it has been reported that PKCη expression is induced upon positive selection of thymocytes (30, 31). Strikingly, in PKCθ-deficient double-positive thymocytes, PKCη is expressed at higher levels than in wild-type cells and it translocates to the immunological synapse as an apparent compensatory mechanism for the loss of PKCθ (Nicholas Gascoigne, personal communication). It is therefore very likely that classical PKC isotypes function to compensate for the loss of PKCθ during positive selection. PKCθ-deficient cells are thus not ideal to study RasGRP phosphorylation. Likewise, chemical inhibitors may have some unwanted cross-reactivity. Additional experiments, possibly through selective RNAi of the various PKC family members, will be required to definitively identify the kinases that phosphorylate the various residues in the four RasGRP molecules.
In summary, the second-messenger DAG plays a unique and pivotal role in T-lymphocyte Ras activation by recruiting both PKCθ and RasGRP1. Our studies demonstrate that these act in concert in regulating Ras activation, yet we cannot rule out possible roles for other molecules like PKCη and RasGRP3 in T lymphocytes. Upon recruitment through C1 domains, PKCθ phosphorylates RasGRP1, resulting in Ras activation. Active Ras can then recruit Raf. PKCθ may subsequently exert a second function by phosphorylating Raf, leading to downstream Erk activity (Fig. 11). We have isolated several JPRM mutants that are able to complement the JPRM441 clone but are nevertheless impaired for PMA-induced Ras activation. These JPRM lines could harbor mutations for additional molecules involved in DAG-mediated Ras activation. The superfamily of DAG-binding proteins also contains PKD, Chimaerin, and Munc-13 signaling proteins that may well contribute to the PKCθ-RasGRP1-Ras pathway described here (8). Further research is required to analyze these possible additional pathways that are triggered by DAG generation.
Acknowledgments
We express our gratitude to Warner Greene for the PKCθA148E and -K409R constructs, Stacey Stang for preparation of anti-RasGRP1 antibodies, and Yong Zheng for preparation of the P-T184 antibody. We are grateful to Hanne Ostergaard and Nicholas Gascoigne for communicating unpublished results. We thank the members of the Weiss lab for their suggestions and comments and Rene Bernards and Hans Bos for continuous support.
J.R. is grateful for grants from The Netherlands Organization for Scientific Research (NWO) and the Dutch Cancer Society (KWF). This work was supported in part by a grant from the NCI (CA 72531).
REFERENCES
- 1.Aiba, Y., M. Oh-hora, S. Kiyonaka, Y. Kimura, A. Hijikata, Y. Mori, and T. Kurosaki. 2004. Activation of RasGRP3 by phosphorylation of Thr-133 is required for B cell receptor-mediated Ras activation. Proc. Natl. Acad. Sci. USA 101:16612-16617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman, A., N. Isakov, and G. Baier. 2000. Protein kinase Ctheta: a new essential superstar on the T-cell stage. Immunol. Today 21:567-573. [DOI] [PubMed] [Google Scholar]
- 3.Altman, A., and M. Villalba. 2003. Protein kinase C-theta (PKCtheta): it's all about location, location, location. Immunol. Rev. 192:53-63. [DOI] [PubMed] [Google Scholar]
- 4.Bivona, T. G., I. Perez De Castro, I. M. Ahearn, T. M. Grana, V. K. Chiu, P. J. Lockyer, P. J. Cullen, A. Pellicer, A. D. Cox, and M. R. Philips. 2003. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature 424:694-698. [DOI] [PubMed] [Google Scholar]
- 5.Bos, J. L. 1998. All in the family? New insights and questions regarding interconnectivity of Ras, Rap1 and Ral. EMBO J. 17:6776-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos, J. L. 1989. ras oncogenes in human cancer: a review. Cancer Res. 49:4682-4689. [PubMed] [Google Scholar]
- 7.Brodie, C., R. Steinhart, G. Kazimirsky, H. Rubinfeld, T. Hyman, J. N. Ayres, G. M. Hur, A. Toth, D. Yang, S. H. Garfield, J. C. Stone, and P. M. Blumberg. 2004. PKCdelta associates with and is involved in the phosphorylation of RasGRP3 in response to phorbol esters. Mol. Pharmacol. 66:76-84. [DOI] [PubMed] [Google Scholar]
- 8.Brose, N., A. Betz, and H. Wegmeyer. 2004. Divergent and convergent signaling by the diacylglycerol second messenger pathway in mammals. Curr. Opin. Neurobiol. 14:328-340. [DOI] [PubMed] [Google Scholar]
- 9.Caloca, M. J., J. L. Zugaza, and X. R. Bustelo. 2003. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J. Biol. Chem. 278:33465-33473. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, S. L., R. Khosravi-Far, K. L. Rossman, G. J. Clark, and C. J. Der. 1998. Increasing complexity of Ras signaling. Oncogene 17:1395-1413. [DOI] [PubMed] [Google Scholar]
- 11.Carrasco, S., and I. Merida. 2004. Diacylglycerol-dependent binding recruits PKCtheta and RasGRP1 C1 domains to specific subcellular localizations in living T lymphocytes. Mol. Biol. Cell 15:2932-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu, V. K., T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Johnson II, A. D. Cox, and M. R. Philips. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343-350. [DOI] [PubMed] [Google Scholar]
- 13.Clyde-Smith, J., G. Silins, M. Gartside, S. Grimmond, M. Etheridge, A. Apolloni, N. Hayward, and J. F. Hancock. 2000. Characterization of RasGRP2, a plasma membrane-targeted, dual specificity Ras/Rap exchange factor. J. Biol. Chem. 275:32260-32267. [DOI] [PubMed] [Google Scholar]
- 14.D'Ambrosio, D., D. A. Cantrell, L. Frati, A. Santoni, and R. Testi. 1994. Involvement of p21ras activation in T cell CD69 expression. Eur. J. Immunol. 24:616-620. [DOI] [PubMed] [Google Scholar]
- 15.Dower, N. A., S. L. Stang, D. A. Bottorff, J. O. Ebinu, P. Dickie, H. L. Ostergaard, and J. C. Stone. 2000. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat. Immunol. 1:317-321. [DOI] [PubMed] [Google Scholar]
- 16.Downward, J., J. D. Graves, P. H. Warne, S. Rayter, and D. A. Cantrell. 1990. Stimulation of p21ras upon T-cell activation. Nature 346:719-723. [DOI] [PubMed] [Google Scholar]
- 17.Ebinu, J. O., D. A. Bottorff, E. Y. Chan, S. L. Stang, R. J. Dunn, and J. C. Stone. 1998. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science 280:1082-1086. [DOI] [PubMed] [Google Scholar]
- 18.Ebinu, J. O., S. L. Stang, C. Teixeira, D. A. Bottorff, J. Hooton, P. M. Blumberg, M. Barry, R. C. Bleakley, H. L. Ostergaard, and J. C. Stone. 2000. RasGRP links T-cell receptor signaling to Ras. Blood 95:3199-3203. [PubMed] [Google Scholar]
- 19.Gong, Q., A. M. Cheng, A. M. Akk, J. Alberola-Ila, G. Gong, T. Pawson, and A. C. Chan. 2001. Disruption of T cell signaling networks and development by Grb2 haploid insufficiency. Nat. Immunol. 2:29-36. [DOI] [PubMed] [Google Scholar]
- 20.Gschwendt, M., H. J. Muller, K. Kielbassa, R. Zang, W. Kittstein, G. Rincke, and F. Marks. 1994. Rottlerin, a novel protein kinase inhibitor. Biochem. Biophys. Res. Commun. 199:93-98. [DOI] [PubMed] [Google Scholar]
- 21.Holsinger, L. J., D. M. Spencer, D. J. Austin, S. L. Schreiber, and G. R. Crabtree. 1995. Signal transduction in T lymphocytes using a conditional allele of Sos. Proc. Natl. Acad. Sci. USA 92:9810-9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishaq, M., G. DeGray, and V. Natarajan. 2003. Protein kinase C theta modulates nuclear receptor-corepressor interaction during T cell activation. J. Biol. Chem. 278:39296-39302. [DOI] [PubMed] [Google Scholar]
- 23.Izquierdo, M., J. Downward, J. D. Graves, and D. A. Cantrell. 1992. Role of protein kinase C in T-cell antigen receptor regulation of p21ras: evidence that two p21ras regulatory pathways coexist in T cells. Mol. Cell. Biol. 12:3305-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawasaki, H., G. M. Springett, S. Toki, J. J. Canales, P. Harlan, J. P. Blumenstiel, E. J. Chen, I. A. Bany, N. Mochizuki, A. Ashbacher, M. Matsuda, D. E. Housman, and A. M. Graybiel. 1998. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc. Natl. Acad. Sci. USA 95:13278-13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurosaki, T. 1999. Genetic analysis of B cell antigen receptor signaling. Annu. Rev. Immunol. 17:555-592. [DOI] [PubMed] [Google Scholar]
- 26.Lin, J., A. Weiss, and T. S. Finco. 1999. Localization of LAT in glycolipid-enriched microdomains is required for T cell activation. J. Biol. Chem. 274:28861-28864. [DOI] [PubMed] [Google Scholar]
- 27.Lockyer, P. J., S. Kupzig, and P. J. Cullen. 2001. CAPRI regulates Ca(2+)-dependent inactivation of the Ras-MAPK pathway. Curr. Biol. 11:981-986. [DOI] [PubMed] [Google Scholar]
- 28.Marais, R., Y. Light, C. Mason, H. Paterson, M. F. Olson, and C. J. Marshall. 1998. Requirement of Ras-GTP-Raf complexes for activation of Raf-1 by protein kinase C. Science 280:109-112. [DOI] [PubMed] [Google Scholar]
- 29.Martiny-Baron, G., M. G. Kazanietz, H. Mischak, P. M. Blumberg, G. Kochs, H. Hug, D. Marme, and C. Schachtele. 1993. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J. Biol. Chem. 268:9194-9197. [PubMed] [Google Scholar]
- 30.Mick, V. E., T. K. Starr, T. M. McCaughtry, L. K. McNeil, and K. A. Hogquist. 2004. The regulated expression of a diverse set of genes during thymocyte positive selection in vivo. J. Immunol. 173:5434-5444. [DOI] [PubMed] [Google Scholar]
- 31.Niederberger, N., L. K. Buehler, J. Ampudia, and N. R. Gascoigne. 20. January 2005. Thymocyte stimulation by anti-TCR-β, but not by anti-TCR-α, leads to induction of developmental transcription program. J. Leukoc. Biol. 10.1189/jlb.1004608. [DOI] [PubMed]
- 32.Oh-hora, M., S. Johmura, A. Hashimoto, M. Hikida, and T. Kurosaki. 2003. Requirement for Ras guanine nucleotide releasing protein 3 in coupling phospholipase C-gamma2 to Ras in B cell receptor signaling. J. Exp. Med. 198:1841-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez, O. D., D. Mitchell, G. C. Jager, S. South, C. Murriel, J. McBride, L. A. Herzenberg, S. Kinoshita, and G. P. Nolan. 2003. Leukocyte functional antigen 1 lowers T cell activation thresholds and signaling through cytohesin-1 and Jun-activating binding protein 1. Nat. Immunol. 4:1083-1092. [DOI] [PubMed] [Google Scholar]
- 34.Priatel, J. J., S. J. Teh, N. A. Dower, J. C. Stone, and H. S. Teh. 2002. RasGRP1 transduces low-grade TCR signals which are critical for T cell development, homeostasis, and differentiation. Immunity 17:617-627. [DOI] [PubMed] [Google Scholar]
- 35.Reuther, G. W., Q. T. Lambert, J. F. Rebhun, M. A. Caligiuri, L. A. Quilliam, and C. J. Der. 2002. RasGRP4 is a novel Ras activator isolated from acute myeloid leukemia. J. Biol. Chem. 277:30508-30514. [DOI] [PubMed] [Google Scholar]
- 36.Roose, J., M. Molenaar, J. Peterson, J. Hurenkamp, H. Brantjes, P. Moerer, M. van de Wetering, O. Destree, and H. Clevers. 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395:608-612. [DOI] [PubMed] [Google Scholar]
- 37.Roose, J. P., M. Diehn, M. G. Tomlinson, J. Lin, A. A. Alizadeh, D. Botstein, P. O. Brown, and A. Weiss. 2003. T cell receptor-independent basal signaling via Erk and Abl kinases suppresses RAG gene expression. PLoS Biol. 1:E53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sebzda, E., S. Mariathasan, T. Ohteki, R. Jones, M. F. Bachmann, and P. S. Ohashi. 1999. Selection of the T cell repertoire. Annu. Rev. Immunol. 17:829-874. [DOI] [PubMed] [Google Scholar]
- 39.Siegel, J. N., R. D. Klausner, U. R. Rapp, and L. E. Samelson. 1990. T cell antigen receptor engagement stimulates c-raf phosphorylation and induces c-raf-associated kinase activity via a protein kinase C-dependent pathway. J. Biol. Chem. 265:18472-18480. [PubMed] [Google Scholar]
- 40.Sun, Z., C. W. Arendt, W. Ellmeier, E. M. Schaeffer, M. J. Sunshine, L. Gandhi, J. Annes, D. Petrzilka, A. Kupfer, P. L. Schwartzberg, and D. R. Littman. 2000. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature 404:402-407. [DOI] [PubMed] [Google Scholar]
- 41.Teixeira, C., S. L. Stang, Y. Zheng, N. S. Beswick, and J. C. Stone. 2003. Integration of DAG signaling systems mediated by PKC-dependent phosphorylation of RasGRP3. Blood 102:1414-1420. [DOI] [PubMed] [Google Scholar]
- 42.Tomlinson, M. G., L. P. Kane, J. Su, T. A. Kadlecek, M. N. Mollenauer, and A. Weiss. 2004. Expression and function of Tec, Itk, and Btk in lymphocytes: evidence for a unique role for Tec. Mol. Cell. Biol. 24:2455-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomlinson, M. G., J. Lin, and A. Weiss. 2000. Lymphocytes with a complex: adapter proteins in antigen receptor signaling. Immunol. Today 21:584-591. [DOI] [PubMed] [Google Scholar]
- 44.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitmarsh, A. J., and R. J. Davis. 1996. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74:589-607. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson, S. E., P. J. Parker, and J. S. Nixon. 1993. Isoenzyme specificity of bisindolylmaleimides, selective inhibitors of protein kinase C. Biochem. J. 294:335-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yablonski, D., M. R. Kuhne, T. Kadlecek, and A. Weiss. 1998. Uncoupling of nonreceptor tyrosine kinases from PLC-gamma1 in an SLP-76-deficient T cell. Science 281:413-416. [DOI] [PubMed] [Google Scholar]
- 48.Yamashita, S., N. Mochizuki, Y. Ohba, M. Tobiume, Y. Okada, H. Sawa, K. Nagashima, and M. Matsuda. 2000. CalDAG-GEFIII activation of Ras, R-ras, and Rap1. J. Biol. Chem. 275:25488-25493. [DOI] [PubMed] [Google Scholar]
- 49.Yang, Y., L. Li, G. W. Wong, S. A. Krilis, M. S. Madhusudhan, A. Sali, and R. L. Stevens. 2002. RasGRP4, a new mast cell-restricted Ras guanine nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Identification of defective variants of this signaling protein in asthma, mastocytosis, and mast cell leukemia patients and demonstration of the importance of RasGRP4 in mast cell development and function. J. Biol. Chem. 277:25756-25774. [DOI] [PubMed] [Google Scholar]
- 50.Yun, T. J., and M. J. Bevan. 2001. The Goldilocks conditions applied to T cell development. Nat. Immunol. 2:13-14. [DOI] [PubMed] [Google Scholar]
- 51.Zheng, Y., H. Liu, J. Coughlin, J. Zheng, L. Li, and J. C. Stone. 18. January 2005. Phosphorylation of RasGRP3 on threonine 133 provides a mechanistic link between PKC and RAS signaling systems in B cells. Blood Epub. [DOI] [PubMed]