Abstract
BID, a proapoptotic BCL-2 family member, plays an essential role in the tumor necrosis factor alpha (TNF-α)/Fas death receptor pathway in vivo. Activation of the TNF-R1 receptor results in the cleavage of BID into truncated BID (tBID), which translocates to the mitochondria and induces the activation of BAX or BAK. In TNF-α-activated FL5.12 cells, tBID becomes part of a 45-kDa cross-linkable mitochondrial complex. Here we describe the biochemical purification of this complex and the identification of mitochondrial carrier homolog 2 (Mtch2) as part of this complex. Mtch2 is a conserved protein that is similar to members of the mitochondrial carrier protein family. Our studies with mouse liver mitochondria indicate that Mtch2 is an integral membrane protein exposed on the surface of mitochondria. Using blue-native gel electrophoresis we revealed that in viable FL5.12 cells Mtch2 resides in a protein complex of ca. 185 kDa and that the addition of TNF-α to these cells leads to the recruitment of tBID and BAX to this complex. Importantly, this recruitment was partially inhibited in FL5.12 cells stably expressing BCL-XL. These results implicate Mtch2 as a mitochondrial target of tBID and raise the possibility that the Mtch2-resident complex participates in the mitochondrial apoptotic program.
Programmed cell death, or apoptosis, is critical for both the development and maintenance of tissues. Caspases, a family of cysteine proteases, are the major executioners of the apoptotic process (24), whereas the BCL-2 protein family members are the major regulators of this process (5). The mechanisms by which the BCL-2 proteins regulate cell death are unknown, although it is believed that their function depends mostly on their ability to modulate the release of proteins from the intermembrane space (IMS) of the mitochondria. The BCL-2 family includes both proapoptotic (e.g., BAX) and antiapoptotic (e.g., BCL-2) proteins, which possess up to four conserved BCL-2 homology (BH) domains (BH1 to -4). A subset of the proapoptotic proteins is the BH3-only group of proteins (e.g., BID).
Two major apoptotic pathways, intrinsic and extrinsic, have been identified. The cell-intrinsic apoptotic pathway involves activation of proapoptotic BCL-2 family members, which induce the permeabilization of the outer mitochondrial membrane (OMM), resulting in the release of cytochrome c (Cyt c) and other IMS proteins (29).
In the extrinsic pathway, apoptosis is initiated through activation of members of the tumor necrosis factor (TNF)/Fas receptor family (26). Once engaged by ligand, these receptors initiate the formation of the death-inducing signaling complex (DISC), which leads to activation of caspase-8. Activated caspase-8 can initiate both a cascade of caspases and the cleavage of the BID protein. Cleavage of cytosolic BID at Asp59 yields a p15 C-terminal truncated fragment (tBID) that translocates to the mitochondria (13, 15, 16). Targeting of tBID to mitochondria induces the activation of BAX and BAK in a BH3-dependent manner, resulting in the release of Cyt c (7, 30). That BID is an essential component of the extrinsic death pathway was demonstrated in Bid-deficient mice, which were resistant to Fas and TNF-α-induced hepatocellular apoptosis (33, 35).
BAX and BAK are the keys to the “apoptotic lock” of mitochondria, since Bax, Bak double-knockout mouse embryonic fibroblasts are resistant to multiple apoptotic stimuli, as well as to tBID and several other BH3-only molecules (31, 36). Activation of BAX and BAK involves their oligomerization and integration into the cell membrane (6, 8, 10, 12). In vitro, recombinant BAX forms channels in artificial membranes, enabling the passage of large macromolecules through the membrane (14, 20, 23). In addition, it has been hypothesized that BAX forms lipidic pores, based on its ability to produce a “growing” pore in pure lipid bilayers, which results in membrane breakage (2). Thus, the tBID-induced formation of BAX/BAK oligomers, which form nonselective channels/lipidic pores serves as an attractive model for the release of IMS proteins such as Cyt c. Alternatively, BAX and BAK could regulate the activity of preexisting channels rather than form channels themselves. In this regard, it has been proposed that altered conductance of existing channels would eventually lead to mitochondrial swelling and the nonspecific rupture of the OMM (3, 25).
The mitochondrial carrier protein (MCP) family comprises a variety of proteins that catalyze the exchange of substrates across the IMM (27). All MCP family members are relatively small proteins, with a molecular mass ranging from 28 to 34 kDa. Comparison of the amino acid sequences of the different carriers has shown that they are made up of three tandem repeats, each about 100 amino acids in length and known as the mitochondrial carrier domain (MCD). Each repetitive element contains two hydrophobic stretches that are of sufficient length to span the membrane as α-helices, separated by an extensive hydrophilic region. Based on this information, it was proposed that the overall structure of this protein family consists of six transmembrane α-helices, in which both the N and the C termini of the proteins are facing the IMS and the three long hydrophilic segments (connecting the two transmembrane regions of each domain) are facing the matrix (17). From the functional characterization of several different carriers that were reconstituted in liposomes, it would appear that the mitochondrial carriers are not only similar in structure but also in function and are characterized by a common kinetic mechanism (17).
We previously demonstrated that the addition of TNF-α to FL5.12 cells results in the formation of a 45-kDa tBID cross-linkable mitochondrial complex (11). In the present study, we describe the purification of the 45-kDa complex and demonstrate that it represents an association between tBID and mitochondrial carrier homolog 2 (Mtch2). Mtch2 is a conserved protein that has one MCD. Using blue-native gel electrophoresis, we demonstrate that in viable FL5.12 cells, Mtch2 resides in a large protein complex; moreover, TNF-α leads to the recruitment of both tBID and BAX to this complex. It is important to note that this recruitment was partially inhibited in FL5.12 cells stably expressing BCL-XL. These results implicate Mtch2 as a mitochondrial target of tBID and raise the possibility that the Mtch2-resident complex participates in tBID- and BAX-induced apoptosis.
MATERIALS AND METHODS
Cell lines.
FL5.12, an interleukin-3 (IL-3)-dependent murine early hematopoietic cell line, was maintained in Iscove medium containing 10% fetal bovine serum supplemented with 10% WEHI-3B conditioned medium as a source of IL-3. FL5.12-BCL-XL cells are FL5.12 cells that stably express mouse BCL-XL (13). To induce apoptosis by means of TNF-α, FL5.12 and FL5.12-BCL-XL cells were treated with recombinant mouse TNF-α (40 ng/ml; Sigma) and cycloheximide (CHX; 1 μg/ml; Sigma) for 5 h. 293T, an embryonic kidney cell line; 293T-T-Rex, a 293T stable clone expressing the tetracycline repressor (Invitrogen); and HeLa, a human cervical adenocarcinoma cell line were maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum. Mouse embryonic fibroblasts (MEFs) were prepared from 11- to 13-day-old embryos and maintained in Iscove medium containing 10% fetal bovine serum. Bax, Bak double-knockout MEFs were a generous gift from Stanley J. Korsmeyer (Dana-Farber Cancer Institute).
tBID expression plasmids.
Wild-type p15 tBID was amplified by PCR from wild-type p22 BID (28). The pcDNA3 plasmid containing a hemagglutinin (HA) epitope tag was ligated in frame to the tBID PCR product under the cytomegalovirus promoter to create the HA-tBID plasmid. To create the pT-Rex-HA-tBID-inducible expression vector, HA-tBID was amplified by using PCR from pcDNA3-HA-tBID. The PCR product was then ligated to the pCA14T-Rex vector to create pT-Rex-HA-tBID.
Cloning of the human Mtch2 gene and construction of Mtch2 expression plasmids.
Single-stranded oligo(dT)-primed cDNA prepared from total RNA of HeLa cells was used as a template for PCR amplification of the human Mtch2 cDNA. The following primers were used for the PCR: 5′-GGGAATTCATGGCGGACGCGGCCAG-3′ (sense primer) and 5′-CCGGCTCCAATTAACATTTTCAGGTCAC-3′ (antisense primer). The PCR product was digested with EcoRI/BamHI and subcloned into the EcoRI/BamHI sites of pcDNA3.1/myc/His (Invitrogen). The sequence of human Mtch2 was confirmed by DNA sequencing. To create the Mtch2-green fluorescent protein (GFP) chimera, the human Mtch2 PCR product was ligated in-frame to the N terminus of EGFP in the pEGFP plasmid (Clontech).
Transient-transfection system and adenovirus infections.
Transient transfections were performed either by means of the calcium phosphate method (9) or with Lipofectamine 2000 (Gibco-BRL). For the experiments involving tBID adenovirus infection, we produced an adenovirus vector for the constitutive expression of tBID, as previously described (21). Virus preparations were made from freeze-thaw lysis of the cells, and virus titers were performed on 293T cells. For infection, cells were generally seeded at 70 to 80% confluence. Cells were infected with a multiplicity of infection of 50 in a minimal culture medium volume for 1 h; the medium volume was then increased until the cells were harvested.
Viewing Mtch2-GFP, tBID, and mitochondria by means of confocal microscopy.
To view Mtch2-GFP and mitochondria, HeLa cells were transfected with Mtch2-GFP and were incubated with 100 nM MitoTracker Red (MTR; Molecular Probes) at 37°C for 30 min prior to fixation. To view tBID and mitochondria, HeLa cells were transfected with tBID in the presence of the broad caspase inhibitor zVAD-fmk (50 μM; Biomol Research Laboratories), prestained with MTR, fixed, and immunostained with anti-BID Abs. To view Mtch2-GFP and tBID, HeLa cells were transfected with both Mtch2-GFP and tBID in the presence of zVAD-fmk (50 μM), fixed, and immunostained with anti-BID Abs. The coverslips were mounted with elvanol, and the cells were viewed under a Nikon fluorescence microscope at a magnification of ×400. Pictures were obtained with a 1310 digital camera (DVC). Confocal microscopy was performed by using an Axiovert 100 TV microscope (Zeiss, Oberkochen, Germany) attached to the Bio-Rad Radiance 2000 laser scanning system (Bio-Rad) and operated by LaserSharp software.
Preparation of mitochondria from cultured cells.
Cells were suspended in isotonic HIM buffer (200 mM mannitol, 70 mM sucrose, 1 mM EGTA, 10 mM HEPES [pH 7.5]) and homogenized by using either a polytron homogenizer (Brinkmann Instruments) at a setting 6.5 for 10 s or by passing them 20 times through a 25-gauge (0.5- by 16-mm) needle. Nuclei and unbroken cells were removed by centrifugation at 120 × g for 5 min. The supernatant was centrifuged at 10,000 × g for 10 min to collect the mitochondrion-enriched fraction and the supernatant (cytosol).
Crude mitochondria were further purified according to the method of Da Cruz et al. (4). Briefly, 5 to 10 mg of crude mitochondria was suspended in 2.5 ml of HIM buffer and loaded on a discontinuous nycodenz gradient. The gradient was prepared by stepwise layering of 3.5 ml of a 22.5% Nycodenz gradient (Sigma) and 5.6 ml of a 9.5% nycodenz solution in a 12-ml SW45 Ti centrifuge tube. The nycodenz solutions themselves were prepared by diluting a 36% nycodenz solution with HIM buffer. The gradient was centrifuged (141,000 × g at 4°C for 1 h), and the pellet was obtained was resuspended in 10 ml of HIM and centrifuged twice more (100,000 × g at 4°C for 10 min each) to obtain the final pellet.
BN-PAGE.
Blue-native polyacrylamide gel electrophoresis (BN-PAGE) was performed according to the method of Schagger (22). Briefly, purified mitochondria from FL5.12 cells either not treated or treated with TNF-α-CHX for 5 h were solubilized with 0.3% n-dodecyl β-d-maltoside (DM). After high-speed centrifugation (217,000 × g at 4°C for 20 min), solubilized complexes were mixed with Coomassie blue G-250 prepared in 750 mM γ-caproic acid so that the concentration of the dye was 1/4 of the DM. The complexes were resolved on a 5 to 13% gradient native gel. In order to separate the complexes into their individual components, lanes from the native gels were incubated while shaking with buffer containing 50 mM Tris (pH 8.8), 6 M urea, 30% glycerol, 2% sodium dodecyl sulfate (SDS), 1% dithiothreitol, and a trace of bromophenol blue at room temperature for 2 h. The lanes were horizontally laid on a 8 to 20% gradient SDS gel to separate the native complexes into their individual protein constitutes.
Mitochondrial import, alkali/urea extraction, and proteinase K treatment.
In vitro mitochondrial import assays, urea extraction and proteinase K treatment were performed as previously described (8). Briefly, mitochondria were isolated from mouse liver that was homogenized in HIM buffer, with two up-and-down strokes in a glass homogenizer. The homogenate was centrifuged at 600 × g for 10 min. The supernatant was centrifuged at 7,000 × g for 15 min. The pellet containing mitochondria was then resuspended in HIM and recentrifuged at 600 × g for 10 min. The supernatant was centrifuged at 7,000 × g for 15 min, and the mitochondrial pellet was resuspended in MRM-S (250 mM sucrose, 10 mM HEPES, 1 mM ATP, 5 mM succinate, 0.08 mM ADP, 2 mM K2HPO4 [pH 7.4]).
For mitochondrial import experiments, mitochondria were incubated with the cytosolic fraction of 293T cells (containing HA-tBID) at 30°C for 30 min. At the end of the reaction, mitochondria were centrifuged, and the pellet and supernatant fractions were solubilized in Laemmli sample buffer and then analyzed by Western blotting. For alkali extraction, the mitochondrial pellet was resuspended in 0.1 M Na2CO3 (pH 11.5). For urea extraction, the mitochondrial pellet was resuspended in 8 M urea. In both instances, mitochondria were incubated at 4°C for 30 min, followed by high-speed centrifugation (75,000 × g) for 10 min. The resulting pellet and supernatant were solubilized in Laemmli sample buffer and analyzed by Western blotting. For proteinase K treatment, the mitochondrial pellet was resuspended in SEM (250 mM sucrose, 10 mM 3-(N-morpholino)propanesulfonic acid-KOH, 2.5 mM EDTA), together with 0.1 or 1 μg of proteinase K/ml, and then incubated at 4°C for 20 min. The reaction was stopped with 1 mM phenylmethylsulfonyl fluoride, and the mitochondria were centrifuged at 10,000 × g for 10 min, resuspended in HIM containing 1 mM phenylmethylsulfonyl fluoride, and recentrifuged at 10,000 × g for 10 min. To perform import experiments with mitochondria pretreated with proteinase K, the mitochondrial pellet was resuspended in MRM-S buffer prior to the experiment.
Cross-linking and dissociation of the cross-link bond.
Sulfo-BSOCOES, i.e., sulfo-bis[2-(sulfosuccinimidooxy-carbonyloxy)ethyl]sulfone (Pierce), from a 10-fold stock solution was added to obtain a final concentration of 10 mM. The cross-linker was added either to the mitochondrion-enriched fraction suspended in isotonic HIM buffer or to purified mouse liver mitochondria suspended in MRM-S. After incubation at room temperature for 30 min, the cross-linker was quenched by the addition of 1 M Tris-HCl (pH 7.5) to a final concentration of 20 mM. After quenching, samples were lysed and analyzed by Western blotting with the indicated Ab. Dissociation of the cross-link bond was performed according to the manufacturer's instructions. Briefly, the cross-linked product was incubated in alkaline conditions (0.1 M Na2CO3; pH 11) at 37°C for 2 h.
Purification of the HA-tBID cross-linked complex.
One thousand plates (10 cm) of 293T cells were transiently transfected with pcDNA3-HA-tBID. At 18 h posttransfection, cells were harvested and subcellularly fractionated by differential centrifugation, as described above. The mitochondrion-enriched heavy membrane fractions were treated with sulfo-BSOCOES at a final concentration of 10 mM. After incubation at room temperature for 30 min, the cross-linker was quenched by the addition of 1 M Tris-HCl (pH 7.5) to a final concentration of 20 mM.
After quenching, the membrane fraction was separated from the soluble fraction by centrifugation and lysed in Laemmli sample buffer without reducing agents. The resulting lysate was diluted in binding buffer (20 mM Tris [pH 7.5], 0.1 M NaCl, 0.1 mM EDTA) to reach a final concentration of 0.2% SDS. The diluted lysate was incubated for 16 h with 5 mg of anti-HA monoclonal Abs coupled to agarose beads (Roche), followed by extensive washing of the beads with binding buffer containing 0.05% Tween 20. The material that remained bound to the beads was eluted by incubation with 1 ml (1 mg/ml) of HA peptide at 37°C for 15 min. Elution was repeated twice more, and the three eluents were pooled and concentrated by using a Centricon tube with a 3K cutoff (Amicon). The concentrated material was loaded onto a single lane, separated by SDS-PAGE, and then stained with Coomassie blue.
In gel proteolysis and mass spectrometry analysis.
The stained protein bands or spots in the gel were cut with a clean razor blade. The proteins in the gel were then reduced with 10 mM dithiothreitol and modified with 100 mM iodoacetamide in 10 mM ammonium bicarbonate. The gel pieces were treated with 50% acetonitrile in 10 mM ammonium bicarbonate to remove the stain from the proteins; the gel pieces were then dried. The dried gel pieces were rehydrated with 10% acetonitrile in 10 mM ammonium bicarbonate containing ca. 0.1 μg of trypsin per sample. The gel pieces were then incubated overnight at 37°C, and the resulting peptides were recovered with 60% acetonitrile with 0.1% trifluoroacetate. The tryptic peptides were resolved by reversed-phase chromatography on 0.1-by-300-mm fused silica capillaries (100-μm inner diameter; J&W) filled with porous R2 (Perspective). The peptides were then eluted by using a 80-min linear gradient of 5 to 95% acetonitrile with 0.1% acetic acid in water at a flow rate of 1 μl/min. The liquid from the column was electrosprayed into an ion-trap mass spectrometer (LCQ, Finnegan, San Jose, CA).
Mass spectrometry was performed in the positive ion mode, utilizing a repetitively full mass spectrometry scan, followed by collision-induced dissociation (CID) of the most dominant ion selected from the first mass spectrometry scan. The mass spectrometry data was compared to simulated proteolysis and CID of the proteins in the NR-NCBI database by using Sequest software (J. Eng, University of Washington, Seattle, and J. Yates, Finnegan, San Jose, CA). The amino terminal of the protein was sequenced on Peptide Sequencer 494A (Perkin-Elmer) according to the manufacturer's instructions.
Western blot analysis and Abs.
Proteins were size fractionated by SDS-PAGE and then transferred to polyvinylidene difluoride membranes (Immun-Blot; Bio-Rad). Western blots were developed by use of the enhanced chemiluminescence reagent (Amersham). Purified recombinant histidine-tagged murine BID was used as an immunogen to generate polyclonal anti-mBID Abs. Protein A purified anti-mBID Abs were used for Western blotting. Additional Abs included anti-mBAX Ab (651), anti-BCL-XL Ab (Santa Cruz), anti-myc MAb (9E10; Santa Cruz), and anti-HA MAb (3F10; Roche).
Anti-Mtch2 Abs were prepared by subcutaneously injecting each of the peptides (aa92-105 [Ab1]), aa109-127 [Ab2], and aa273-288 [Ab3]) conjugated to KLH (Imject Maleimide-activated mcKLH from Pierce, according to the manufacturer's protocol) in several places on the backs of three white female rabbits. Two weeks after the injection, 200 μg of booster prepared with incomplete Freund adjuvant (Sigma) was injected. Three weeks after boosting, blood was collected and incubated at room temperature for 2 h before being stored at 4°C overnight to enable clotting of the red blood cells. After the clot was removed, the serum was centrifuged for 30 min at 1,500 × g. The supernatant which resulted was then divided into aliquots and stored at −20°C.
RESULTS
Formation of the 45-kDa tBID complex requires an ∼30-kDa surface-exposed mitochondrial protein, other than BAX or BAK.
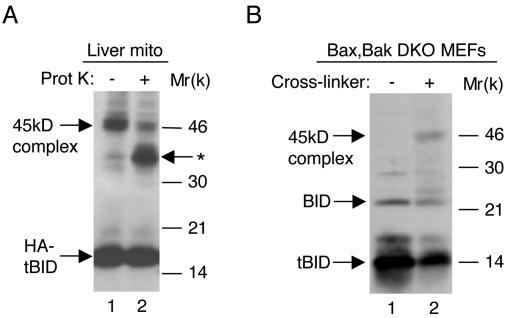
In a previous study, we proposed that the TNF-α-induced 45-kDa complex represents a tBID homotrimer (11). However, further studies in our laboratory involving in vitro mitochondrial import experiments suggested that this might not be the case. In these studies, we initially found that the 45-kDa tBID complex is formed by incubating purified, intact mouse liver mitochondria with a cytosolic extract prepared from 293T cells transfected with hemagglutinin (HA)-tagged tBID (Fig. 1A, lane 1). To determine whether the formation of this complex required the presence of a surface-exposed mitochondrial protein, mitochondria were pretreated with proteinase K. This pretreatment led to a significant decrease in the intensity of the 45-kDa band and the appearance of a new, ∼35-kDa BID-immunoreactive band (Fig. 1A, lane 2). These results suggest that pretreatment of naive mitochondria with proteinase K cuts off an exposed ∼10-kDa region from an ∼30-kDa mitochondrial protein, resulting in an ∼20-kDa fragment that cross-links with p15 tBID to form an ∼35-kDa complex. Thus, the 45-kDa BID-immunoreactive band most likely represents a complex between tBID and an ∼30-kDa surface-exposed mitochondrial protein.
FIG. 1.
Formation of the 45-kDa tBID complex requires an ∼30-kDa surface-exposed mitochondrial protein other than BAX or BAK. (A) Formation of the 45-kDa tBID complex requires an ∼30-kDa surface-exposed mitochondrial protein. Mouse liver mitochondria were pretreated with proteinase K and then incubated with HA-tBID as described in Materials and Methods. At the end of the reaction, mitochondria were treated with the sulfo-BSOCOES cross-linker and lysed, and equal amounts of protein (20 μg per lane) were subjected to SDS-PAGE, followed by Western blot analysis with anti-BID antibodies. ✽, a new ∼35-kDa BID-immunoreactive band that appears as a result of proteinase K pretreatment. (B) BAX and BAK are not part of the 45-kDa BID immunoreactive complex. Bax, Bak DKO MEFs were infected with tBID adenoviruses for 3 h. The mitochondrion-enriched fraction was treated with either sulfo-BSOCOES (+) or with dimethyl sulfoxide (−), lysed, and analyzed as described for panel A.
BAX and BAK are known to be essential downstream effectors of tBID, since Bax, Bak double-knockout (DKO) MEFs are resistant to tBID-induced apoptosis (31, 36). It was previously demonstrated that tBID heterodimerizes with BAX or BAK to induce their oligomerization in the mitochondrial membrane (7, 30). Therefore, in the present study we sought to determine whether BAX and/or BAK were part of the 45-kDa complex or played an essential role in its formation. Accordingly, Bax, Bak DKO MEFs were infected with adenoviruses containing tBID (Ad-tBID). The heavy membrane fraction was then prepared and subsequently either not treated or treated with cross-linker. Western blot analysis indicated that the 45-kDa cross-linked complex is formed in heavy membranes prepared from these cells (Fig. 1B). Thus, these results indicate that BAX and BAK are not part of this complex and are not essential to its formation.
HA-tBID forms a 45-kDa complex with Mtch2, a previously uncharacterized 33-kDa mitochondrial protein.
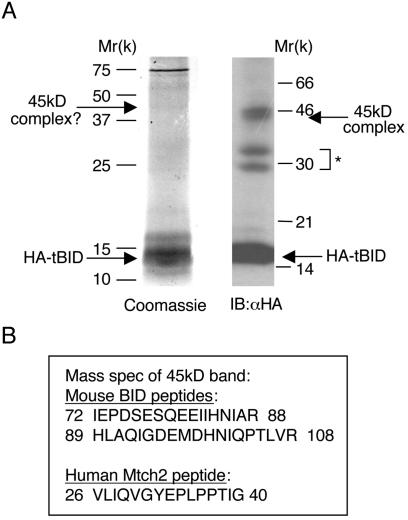
We previously demonstrated that HA-tBID is capable of forming a 45-kDa complex in 293T cells (11). We took advantage of this finding to purify the HA-tBID complex and identify its components. The mitochondrion-enriched heavy membrane fraction prepared from 293T cells transfected with HA-tBID was treated with cross-linker and then lysed. The mitochondrial lysate was then incubated with anti-HA antibodies coupled to agarose beads, and the HA-tBID monomers/complexes were eluted with an HA peptide (see Materials and Methods). Our initial results indicated that the anti-HA antibody column efficiently captured the 45-kDa complex and that the HA peptide was efficient in releasing it from the column (Fig. 2A, right lane).
FIG. 2.
HA-tBID forms an ∼45-kDa complex with Mtch2, a previously uncharacterized 33-kDa mitochondrial protein. (A) Purification of a 45-kDa tBID cross-linked complex from 293T cells transfected with HA-tBID. Purification of this complex is described in Materials and Methods. The eluted material from the anti-HA antibody affinity column was loaded onto a single lane and separated by SDS-PAGE, followed by Coomassie blue staining (left panel). The top arrow marks the band suspected to be the 45-kDa band that was cut out of the gel and analyzed by mass spectrometry. A small portion of the eluted material was also taken for Western blot analysis with anti-HA Abs (right lane). ✽, additional BID immunoreactive bands that may represent additional tBID cross-linked complexes. (B) Mass spectrometry results of the 45-kDa band. Sequences of the two peptides from the BID protein and the single peptide from the Mtch2 protein that resulted from the analysis are shown.
Next, we significantly increased the amount of the starting material for purification of the complex and eventually reached a point at which an ∼45-kDa band, which we suspected to represent the 45-kDa-tBID complex, was visible as a faint band by means of Coomassie blue staining (Fig. 2A, left lane). In this gel, HA-tBID was clearly visible as an ∼15-kDa band (confirmed by mass spectrometry). Mass spectrometry analysis of the ∼45-kDa band revealed that it included two peptides from tBID and one peptide from a 33.4-kDa human protein known as Mtch2 (mitochondrial carrier homolog 2) (Fig. 2B).
Mtch2 is a conserved protein with a MCD.
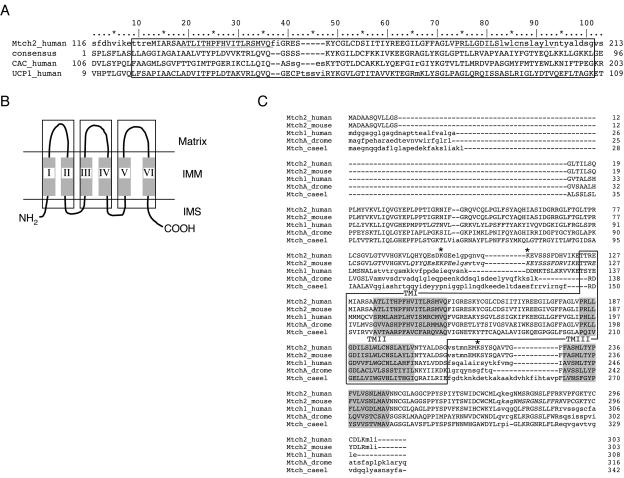
Mitochondrial carrier homolog 2 is an uncharacterized protein first identified in a screen for human cDNAs (34). It was named after the conserved MCD it contains (Fig. 3A and 3B; see also the Introduction). This 100-residue domain is composed of two transmembrane (TM) domains, which are connected by a linker region. Mitochondrial carrier proteins (MCPs) have three mitochondrial carrier domains, whereas Mtch2 appears to have only one (Fig. 3B and C, respectively).
FIG. 3.
Mtch2 is a conserved protein with a mitochondrial carrier domain. (A) Human Mtch2 sequence (NCBI gi 7657347) was searched against the NCBI Conserved Domain Database (CDD; version 1.60) by using the RPS-BLAST program (1). Positions 128 to 192 of human Mtch2 were similar to the mitochondrial carrier protein domain (CDD 9082) with an e-value of 2−8 (53.8 bits). Uppercase letters denote the aligned regions in the query and the domain multiple alignment. The two transmembrane regions predicted by the PHD server (18) are underlined in human Mtch2. Below the human Mtch2 sequence are the consensus and multiple alignment of the MCDs taken from the two proteins with known function that were most similar to human Mtch2. The NCBI gi codes of the sequences are as follows: human carnitine/acylcarnitine translocase (CAC), 3914023s; and human mitochondrial uncoupling protein 1 (UCP1), 1351353. The conserved MCD is marked by a box. (B) Graphic display of the topology of the MCP family members in the inner membrane of mitochondria. All MCP family members have three MCDs (marked by boxes), each composed of two transmembrane regions connected by a linker region. (C) Multiple sequence alignment of Mtch2 and closely related proteins. Only residues in uppercase letters are confidently aligned. The NCBI gi codes of the sequences are as follows: Mtch2_human, 7657347; Mtch2_mouse, 5815347 (93% identity); Mtch1_human, 6995989 (48% identity); MtchA_drome (Drosophila melanogaster mitochondrial carrier homolog A), 5815351 (38% identity); and Mtch_caeel (Caenorhabditis elegans), 17533979 (28% identity). The three transmembrane regions predicted by the PHD server (18) are shaded, and the single MCD is marked by a box. ✽, lysines 100, 111, and 219, which we propose to be the sites that are cleaved by proteinase K. The peptides used to generate the antibodies (aa92-105 [Ab1], aa109-127 [Ab2], and aa273-288 [Ab3]) appear in italics.
Mtch2 has several close relatives, which together form a “sister family” separate from that of the MCPs. In chordates, the Mtch family includes two members (Mtch1 and Mtch2) in mammals and birds, whereas fish and tunicates apparently have only one Mtch2 ortholog. In invertebrates, Caenorhabditis elegans has one Mtch protein, and Drosophila melanogaster has two. It is hypothesized that all Mtch family members have a single MCD and contain three transmembrane domains, in which the first and second are part of the mitochondrial carrier domain (Fig. 3C). Moreover, all members of this family can be aligned across nearly their entire length. Mtch proteins should therefore be considered a distinct subgroup of proteins related to MCPs.
Exogenous Mtch2 and tBID associate in mitochondria.
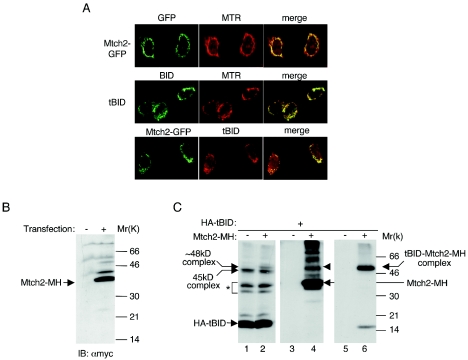
We have cloned the human Mtch2 gene from total RNA prepared from HeLa cells, and initially fused it to green fluorescent protein (GFP) to confirm that it localizes to mitochondria. HeLa cells transfected with the Mtch2-GFP chimeric protein were incubated with MTR (to label mitochondria) and were analyzed by means of confocal microscopy. These studies demonstrated that a major part of the chimeric molecules colocalized with MTR, confirming their mitochondrial localization (Fig. 4A, top panel). We have also transfected HeLa cells with either tBID alone or tBID combined with Mtch2-GFP and performed overlays of tBID/MTR and tBID/Mtch2-GFP (Fig. 4A, middle and bottom panels, respectively). These studies demonstrate that Mtch2 colocalizes with tBID at the mitochondria.
FIG. 4.
Mtch2-myc-His and HA-tBID associate in mitochondria. (A) Mtch2-GFP and tBID colocalize at mitochondria. The top panels show HeLa cells transfected with human Mtch2-GFP were stained with MTR, fixed, and analyzed by using a confocal fluorescence microscope. The green areas (left panel) are Mtch2-GFP, the red areas (middle panel) are mitochondria stained with MTR, and the right panel is a merge image of the two previous panels. The middle panels show HeLa cells transfected with mouse tBID in the presence of the broad caspase inhibitor zVAD-fmk (50 μM) that were prestained with MTR, fixed, immunostained with anti-BID Abs, and analyzed as described above. The green areas (left panel) are tBID, the red areas (middle panel) are mitochondria stained with MTR, and the right panel is a merge image of the two previous panels. The bottom panels show HeLa cells transfected with both human Mtch2-GFP and mouse tBID that were fixed, immunostained with anti-BID Abs, and analyzed as described above. The green areas (left panel) are Mtch2-GFP, the red areas (middle panel) are tBID, and the right panel is a merge image of the two previous panels. (B) Expression of Mtch2-myc-His in 293T cells. The human Mtch2 cDNA product was cloned into pcDNA3.1-Myc-His (Invitrogen) and transiently transfected into 293T cells. At 18 h posttransfection the cells were harvested, lysed, and analyzed by Western blotting with anti-myc Abs. 293T cells transfected (+) or nontransfected (−) are shown. (C) HA-tBID and Mtch2-MH associate in mitochondria. 293T-T-Rex cells (Invitrogen) were transiently transfected with pT-Rex-HA-tBID, with or without the addition of pcDNA3.1-Mtch2-MH. At 21 h posttransfection (doxycycline was added for the last 5 h), mitochondria were prepared from these cells and treated with the sulfo-BSOCOES cross-linker. After treatment, the mitochondria were lysed, and equal amounts of protein (20 μg per lane) were subjected to SDS-PAGE, followed by Western blot analysis with anti-HA Abs (lanes 1 and 2). Alternatively, mitochondria were lysed and loaded onto Ni columns, and the eluted proteins were subjected to SDS-PAGE, followed by Western blot analysis with either anti-myc Abs (lanes 3 and 4) or anti-HA Abs (lanes 5 and 6). ✽, additional BID immunoreactive bands that may represent additional tBID cross-linked complexes. The arrow (lane 4) marks the Mtch2-MH band, and the arrowhead (lane 4) marks the band that represents the ∼48-kDa complex (HA-tBID/Mtch2-MH complex).
Next, we fused Mtch2 to a Myc-His epitope (Mtch2-MH), which we predicted would migrate as an ∼36-kDa protein, and expressed the chimeric protein in 293T cells. Western blot analysis with anti-myc antibodies detected a single major band of the expected size (Fig. 4B).
To assess whether tBID interacts with Mtch2-MH at the mitochondria, we utilized 293T cells in which expression of tBID was controlled by tetracycline. 293T-T-REx cells were transfected with pT-Rex-HA-tBID in the presence or absence of Mtch2-MH and, at 16 h posttransfection, doxycycline was added for 5 h. The mitochondrion-enriched heavy membrane fraction prepared from these cells was treated with cross-linker, lysed, and then analyzed by Western blotting with anti-HA or anti-myc antibodies. Western blot analysis with anti-HA antibodies detected a new ∼48-kDa band in cells expressing both proteins but not in cells expressing only HA-tBID (Fig. 4C, compare lanes 1 and 2).
To determine whether this band indeed represents a complex between HA-tBID and Mtch2-MH, the mitochondrial lysates were loaded onto Ni columns and the eluted proteins were separated by SDS-PAGE, followed by Western blot analysis with either anti-myc (Fig. 4C, lanes 3 and 4) or anti-HA (Fig. 4C, lanes 5 and 6) antibodies. As expected, Mtch2-MH bound to the Ni column and was detected in the eluted material by using anti-myc antibodies (Fig. 4C, lane 4). Western blot analysis with anti-HA antibodies indicated that the ∼48-kDa band that appeared in lanes 2 and 4 represents a complex between HA-tBID and Mtch2-MH (Fig. 4C, lane 6).
Mtch2 is an integral membrane protein exposed on the surface of mitochondria.
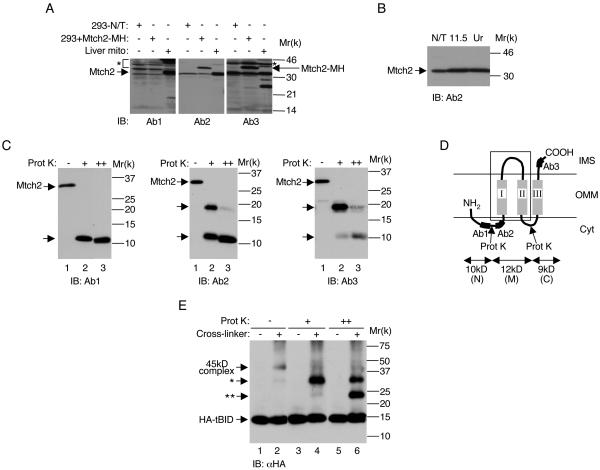
To further characterize the biochemical properties of endogenous human and mouse Mtch2, we raised three polyclonal Abs to three separate peptides in mouse Mtch2 (aa92-105 [Ab1], aa109-127 [Ab2], and aa273-288 [Ab3]; these peptides appear in italics in Fig. 3C). The first two mouse peptides are very similar to their human counterparts, and the third mouse peptide is identical to the human peptide.
We next examined the ability of these antibodies to recognize Mtch2 by using Western blot analysis of mouse and human cells. All three antibodies recognized endogenous mouse Mtch2 from liver mitochondria, whereas only Ab2 and Ab3 recognized human Mtch2, exogenously expressed in 293T cells (Fig. 5A). In addition, Ab3 and, to a lesser extent, Ab2 also recognized endogenous human Mtch2 in 293T cells.
FIG. 5.
Mtch2 is an integral membrane protein exposed on the surface of mitochondria. (A) Characterization of anti-Mtch2 antibodies. 293T cells (293-N/T), 293T cells transfected with Mtch2-MH (293+Mtch2-MH), and mouse liver mitochondria (Liver mito) were lysed and analyzed by Western blotting with the indicated anti-Mtch2 antibodies. ✽, cross-reactive bands. (B) Mtch2 is an integral membrane protein. Mouse liver mitochondria were left untreated (N/T) or treated with 0.1 M Na2CO3 (pH 11.5) or with 8 M urea (Ur). After treatment, the membranes were separated from the soluble fraction by centrifugation, and the membrane fractions were analyzed by Western blotting with anti-Mtch2 antibodies (Ab2). (C) Proteinase K cleaves Mtch2 into three fragments. Mouse liver mitochondria were either left untreated (−) or treated with a low (0.1 μg/ml; +) or a high (1 μg/ml; ++) concentration of proteinase K, lysed, size fractionated by SDS-PAGE, and analyzed by Western blotting with the indicated anti-Mtch2 antibodies. The arrows mark the resulting ∼20- and ∼10-kDa fragments. (D) A graphic display of the predicted topology of Mtch2 in the outer membrane of mitochondria. The MCD is marked by a box, and the “Ab” labelings indicate the location of the peptides in Mtch2 that were used for generating Ab1, Ab2, and Ab3. The predicted proteinase K cleavage sites are marked by arrows, and the resulting three fragments are labeled as follows: N, ∼10-kDa N-terminal fragment; M, ∼12-kDa middle fragment; and C, ∼9-kDa C-terminal fragment. (E) tBID seems to interact with the C fragment of Mtch2. Mouse liver mitochondria were either left untreated (−) or pretreated with a low (+) or a high (++) concentration of proteinase K and incubated with HA-tBID as described in Materials and Methods. At the end of the reaction, mitochondria were treated with the BSOCOES cross-linker and lysed, and equal amounts of protein (20 μg per lane) were subjected to SDS-PAGE, followed by Western blot analysis with anti-HA antibodies. ✽, ∼35-kDa tBID-Mtch2 complex; ✽✽, ∼24-kDa tBID-Mtch2 complex.
To assess whether Mtch2 is an integral membrane protein, intact mouse liver mitochondria were treated with either alkali or urea, two treatments which release all except for the most integral membrane proteins. After treatment, the membranes were separated from the soluble fractions by centrifugation and the membrane fractions were analyzed by Western blotting with Ab2. The results demonstrate that Mtch2 is largely resistant to both alkali and urea extraction (Fig. 5B), indicating that it is an integral membrane protein.
Next, we used mouse liver mitochondria to examine the sensitivity of Mtch2 to proteinase K, since the results presented in Fig. 1 suggested that the 45-kDa complex represents an association between tBID and a mitochondrial protein that is sensitive to proteinase K. Treatment of mitochondria with a low concentration of proteinase K resulted in the cleavage of Mtch2 into several fragments: an ∼10-kDa fragment recognized by Ab1 (Fig. 5C, left panel, lane 2), an ∼20-kDa fragment, and an ∼10-kDa fragment recognized by Ab2 (middle panel, lane 2), and an ∼20-kDa fragment and an ∼10-kDa fragment recognized by Ab3 (right panel, lane 2). Treatment of mitochondria with a 10-fold-higher concentration of proteinase K resulted in a decrease in the intensity of the ∼20-kDa fragment and in an increase in the intensity of the ∼10-kDa fragment recognized by Ab2 and Ab3 (Fig. 5C, middle and right panels, lane 3).
Taken together, these results lead us to propose that proteinase K cleaves Mtch2 at two sites (Fig. 5D): at lysine 100 or 111 (positioned in a hydrophilic loop between the peptides used to generate Ab1 and Ab2; the exact position of these lysines is marked in Fig. 3C) and at lysine 219 (positioned in a hydrophilic loop connecting TMII and TMIII). Cleavage at these two sites results in the generation of three fragments: an ∼10-kDa N-terminal fragment (N), an ∼12-kDa middle fragment (M), and an ∼9-kDa C-terminal fragment (C). Thus, lysines 100/111 and 219 (and the areas surrounding them) are most likely exposed on the surface of mitochondria, and therefore all three TM domains are likely to span the outer mitochondrial membrane (Fig. 5D).
The ∼10-kDa reduction in size of Mtch2 after treatment with the low concentration of proteinase K is similar to the reduction in size (from ∼45- to ∼35-kDa) that we observed with the tBID cross-linked complex (Fig. 1A and 5E, compare lanes 2 and 4). Thus, tBID cross-links to the middle plus the C-terminal fragment and not to the N-terminal fragment. To determine whether tBID cross-links to both the middle and C-terminal fragments or only to one of them, mitochondria were pretreated with a 10-fold-higher concentration of proteinase K (which results in the generation of separate middle and C-terminal fragments; see Fig. 5C) prior to the addition of HA-tBID and cross-linker. Pretreatment with the higher concentration of proteinase K led to a decrease in the intensity of the ∼35-kDa cross-linked band and the corresponding appearance of a new HA-immunoreactive band, which migrated slightly below the 25-kDa marker (Fig. 5E, lane 6). These results suggest that tBID cross-links to the C-terminal fragment (15 + 9 = 24 kDa) rather than the middle fragment (15 + 12 = 27 kDa) of Mtch2.
The 45-kDa complex formed in TNF-α-treated FL5.12 cells is composed of tBID and Mtch2.
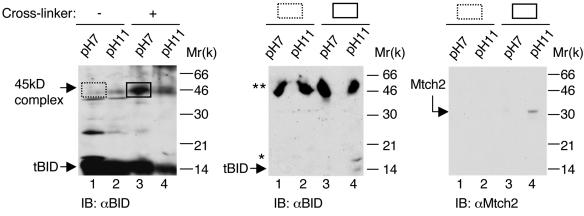
Previously, we have demonstrated that the 45-kDa cross-linked complex appears in TNF-α-treated, but not in untreated (−TNF-α), FL5.12 cells (11). To assess whether the 45-kDa band detected in TNF-α-treated FL5.12 cells indeed represents a complex between endogenous tBID and Mtch2, we took advantage of the fact that the cross-linker used in these experiments, BSOCOES, can be reversed in alkaline conditions. Accordingly, FL5.12 cells were treated with TNF-α-CHX for 5 h, and the heavy membrane fraction was either not treated or treated with cross-linker, followed by incubation in either neutral (pH 7) or alkaline (pH 11) conditions. At the end of the incubation, the membranes were lysed and analyzed by Western blotting with anti-BID antibodies. Figure 6 (left panel) shows that alkaline conditions significantly reduced the intensity of the 45-kDa band.
FIG. 6.
The 45-kDa complex formed in TNF-α-treated FL5.12 cells is composed of tBID and Mtch2. (Left panel) Alkaline conditions significantly reduce the intensity of the 45-kDa band. Mitochondria from FL5.12 cells treated with TNF-α-CHX for 5 h were either not treated (−) or treated (+) with the cross-linker BSOCOES, followed by incubation in either neutral (pH 7) or alkaline (pH 11) conditions (see Materials and Methods). At the end of the incubation, the membranes were lysed and analyzed by Western blot with anti-BID antibodies. (Middle and right panels) The 45-kDa band represents a complex between tBID and Mtch2. The experiment presented in the left panel was repeated, and the gel sections corresponding to the position of the 45-kDa complex in lanes 1 and 3 (left panel; marked by boxes) were excised and incubated in either neutral or alkaline conditions. The sections were then layered onto a 15% denaturing gel, and the proteins were resolved by SDS-PAGE and analyzed by Western blot with either anti-BID Abs (middle panel) or anti-Mtch2/Ab2 Abs (right panel). ✽, an additional BID immunoreactive band that might represent a modified form of tBID; ✽✽, cross-reactive substances that might have been released from the gel sections.
Next, we repeated this experiment, and the gel sections corresponding to the position of the 45-kDa complex in lanes 1 and 3 (marked by boxes in Fig. 6, left panel) were excised and incubated in either neutral or alkaline conditions. The sections were then layered onto a 15% denaturing gel, and the proteins were resolved by SDS-PAGE. Western blot analysis with either anti-BID antibodies (Fig. 6, middle panel) or anti-Mtch2 antibodies (Ab2; Fig. 6, right panel) indicated that tBID and Mtch2 appear exclusively in the gel section that represents mitochondria treated with cross-linker and incubated in alkaline conditions. These results strongly suggest that the endogenous 45-kDa complex formed in TNF-α-treated FL5.12 cells is composed of tBID and Mtch2.
Mtch2 resides in an ∼185-kDa resident mitochondrial complex, and activation with TNF-α leads to the recruitment of tBID to this complex.
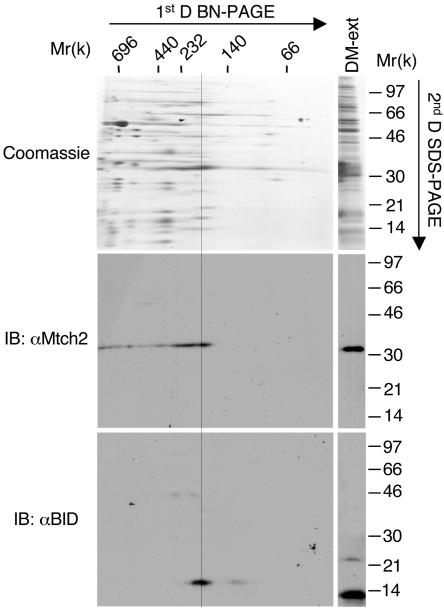
Next, we utilized BN-PAGE (22) as an additional approach to verify the interaction between endogenous tBID and Mtch2 in the mitochondria of FL5.12 cells signaled to die by TNF-α. This method has been shown to be a powerful means for purifying the respiratory chain complexes. For these studies, we utilized mitochondria purified from FL5.12 cells either not treated or treated with TNF-α-CHX for 5 h. Purified mitochondria were solubilized with DM and centrifuged, and the solubilized complexes were mixed with Coomassie blue G-250. The resulting complexes were resolved on a 5 to 13% gradient gel under nondenaturing conditions (Fig. 7, marked by “1st D,” and the horizontal molecular weight markers). These native complexes were separated into their individual components by placing a native gel slice horizontal as a stack above an SDS-denaturing gel (marked by “2nd D”). A Coomassie blue stain of this gel revealed the individual protein constituents of each native complex (Fig. 7, top panel).
FIG. 7.
Mtch2 resides in an ∼185-kDa resident mitochondrial complex, and activation with TNF-α leads to the recruitment of tBID to this complex. FL5.12 cells were grown in the presence of TNF-α-CHX for 5 h. Mitochondria were prepared and solubilized with 0.3% DM. DM-solubilized protein complexes were resolved on a 5 to 13% gradient BN gel (1st D BN-PAGE). Lanes from the BN gel were layered onto an 8 to 20% gradient denaturing gel, and the proteins were resolved by SDS-PAGE and stained with Coomassie blue (top panel; 2nd D SDS-PAGE). The stained gel was then transferred to a PVDF membrane and immunoblotted with either anti-Mtch2/Ab2 Abs (middle panel) or anti-BID Abs (bottom panel). DM-ext (top panel, right lane) marks DM solubilized protein complexes directly loaded onto the SDS-gel. The line marks the comigrating positions of Mtch2 and tBID.
The stained gel was then transferred to a PVDF membrane and immunoblotted with either anti-Mtch2 or anti-BID antibodies. In mitochondria prepared from viable FL5.12 cells, Mtch2 was detected as a smear that ranged from ca. 185 to 230 kDa in the native gel (see below), whereas tBID, as expected, was not detected. Strikingly, in mitochondria prepared from TNF-α-treated FL5.12 cells, tBID appeared in two spots, and one of them comigrated with one of the Mtch2 spots (Fig. 7, middle and bottom panels). Both tBID and Mtch2 were detected on the second-dimension gel at higher molecular masses than expected, since the migration of proteins from the horizontal native gel slice to the SDS-gel takes longer than the migration of soluble proteins that are directly loaded onto the SDS-gel (compare the positions of the tBID and Mtch2 spots in the large blots to the tBID and Mtch2 bands in the “DM-ext” lanes [Fig. 7, middle and bottom panels]). These results strongly suggest that Mtch2 resides in a relatively large mitochondrial complex in viable FL5.12 cells and that activation with TNF-α leads to the recruitment of tBID to this complex.
Activation by TNF-α also recruits BAX to the Mtch2-resident complex.
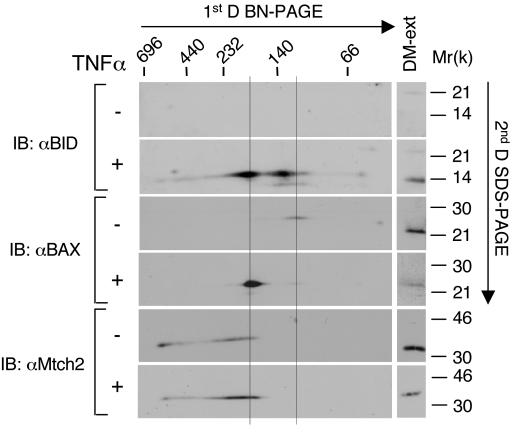
To identify additional components of the ca. 185-kDa complex, individual protein spots that aligned with Mtch2 and tBID were excised from second-dimension gels and sequenced by mass spectrometry. By sequencing two of the spots (one at ∼35 kDa and the other at 25 kDa), we identified peptides that are identical to mouse Mtch2 and mouse BAX, respectively (data not shown).
To confirm the presence of BAX in this complex, we performed the BN experiment again with FL5.12 cells either not treated or treated with TNF-α-CHX and then probed the second-dimension blots with anti-BAX, anti-BID, and anti-Mtch2 antibodies. In mitochondria prepared from viable FL5.12 cells, BAX was detected in a single spot that migrated at ∼115 kDa in the native gel, whereas Mtch2 was detected as a smear that ranged from ∼185 to 230 kDa in the native gel (Fig. 8). Strikingly, in TNF-α-treated FL5.12 cells the position of BAX was seen to shift to a single spot that comigrated with one of the two tBID spots at ca. 185 kDa in the native gel (Fig. 8). On the other hand, TNF-α had no significant effect on the position of Mtch2. Thus, it appears that TNF-α leads to the recruitment of both tBID and BAX to the Mtch2-resident complex.
FIG. 8.
Activation by TNF-α also recruits BAX to the Mtch2-resident complex. FL5.12 cells were either not treated (−) or treated (+) with TNF-α-CHX for 5 h. Mitochondria were prepared from the cells and processed as described in Fig. 7. Western blot analysis was performed with anti-BID (top two panels), anti-BAX (middle two panels), and anti-Mtch2 (bottom two panels) Abs. The left line is placed in the same position in which it is placed in Fig. 7, and it marks the comigrating positions of tBID, BAX, and Mtch2. The right line marks the position of BAX in viable FL5.12 cells.
BCL-XL partially inhibits the recruitment of both BAX and tBID to the Mtch2-resident complex.
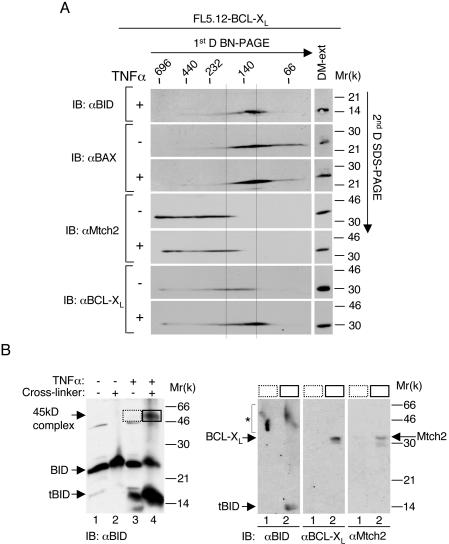
We previously demonstrated that FL5.12 cells that stably express BCL-XL (FL5.12-BCL-XL) do not release Cyt c from the mitochondria in response to TNF-α (13). Since the recruitment of tBID and BAX to the Mtch2 complex might be related to Cyt c release, we utilized these cells to assess whether the presence of BCL-XL affects this recruitment. In viable FL5.12-BCL-XL cells, BAX was detected as a smear that ranged from ca. 66 to 185 kDa in the native gel, whereas BCL-XL was detected as a smear that ranged from ca. 115 to 230 kDa in the native gel and partially comigrated with the Mtch2 smear (Fig. 9). Strikingly, in TNF-α-treated FL5.12-BCL-XL cells, the position of BAX did not shift, and tBID now appeared as a smear that comigrated with BAX (Fig. 9). On the other hand, the addition of TNF-α did induce a shift in the position of BCL-XL, which now comigrated with BAX and tBID. Thus, overexpression of BCL-XL inhibits the recruitment of both tBID and BAX to the Mtch2-resident complex.
FIG. 9.
BCL-XL partially inhibits the recruitment of both BAX and tBID to the Mtch2-resident complex. (A) FL5.12-BCL-XL cells were either not treated (−) or treated (+) with TNF-α-CHX for 5 h. Mitochondria were prepared from the cells and processed as described in Fig. 7. Western blot analysis was performed with anti-BID, anti-BAX, anti-Mtch2, and anti-BCL-XL Abs. The two lines are placed in the same position in which they are placed in Fig. 8. (B) tBID forms a complex with either BCL-XL or Mtch2 in TNF-α-treated FL5.12-BCL-XL cells. For the left panel, FL5.12-BCL-XL cells were grown in the presence or absence of TNF-α-CHX for 5 h. Mitochondria were prepared from the cells and either not treated (−) or treated (+) with the BSOCOES cross-linker. At the end of the incubation, the membranes were lysed and analyzed by Western blotting with anti-BID Abs. For the right panels, the experiment presented in the left panel was repeated, and the gel sections corresponding to the position of the 45-kDa band in lanes 3 and 4 (left panel; marked by boxes) were excised, incubated in alkaline conditions, and later resolved as described for Fig. 6. Western blot analysis was performed with anti-BID Abs (left panel), anti-BCL-XL Abs (middle panel), or anti-Mtch2 Abs (right panel). ✽, cross-reactive substances that might have been released from the gel sections.
To confirm these results, we performed cross-linking experiments with mitochondria prepared from FL5.12-BCL-XL cells either not treated or treated with TNF-α-CHX. Surprisingly, a 45-kDa BID-immunoreactive band was detected in TNF-α-treated FL5.12-BCL-XL cells treated with cross-linker (Fig. 9B, left panel). To assess whether this band represented a complex between tBID (15 kDa) and BCL-XL (30 kDa), we repeated this experiment, and the gel sections corresponding to the position of the 45-kDa band in lanes 3 and 4 (marked by boxes in Fig. 9B, left panel) were excised, incubated in alkaline conditions, and later resolved as described in Fig. 6. Western blot analysis with either anti-BID antibodies or anti-BCL-XL antibodies indicated that tBID and BCL-XL appear exclusively in the gel section that represents mitochondria treated with cross-linker and incubated in alkaline conditions (Fig. 9B, right panels). To assess whether this 45-kDa gel section might also contain a complex comprised of tBID and Mtch2, we performed Western blot analysis with anti-Mtch2 antibodies and found that Mtch2 was also released from this gel section (Fig. 9B, right panels). Taken together, these results suggest that the 45-kDa BID-immunoreactive band detected in TNF-α-treated FL5.12-BCL-XL cells represents two complexes: one composed of tBID and BCL-XL and the other composed of tBID and Mtch2. Thus, overexpression of BCL-XL only partially inhibits the recruitment of tBID to the Mtch2-resident complex.
DISCUSSION
BCL-2 family members are major regulators of the apoptotic process. The mechanisms by which these proteins regulate cell death are largely unknown, although it is thought that their function depends mostly on their ability to modulate the release of proteins from the intermembrane space of the mitochondria.
Many approaches have been used in the past to identify proteins that interact with BCL-2 family members. These approaches included coimmunoprecipitation from detergent-lysed cells, yeast two-hybrid screening, expression cloning, purification of protein complexes using native conditions, and the use of chemical cross-linkers. Many interacting proteins, mainly members of the BCL-2 family, have been identified by using these approaches. We believe that the cross-linking and the native purification approaches are preferable to the other approaches, since BCL-2 family members are (or become) associated with membranes. Therefore, their interactions with other membrane proteins are likely to depend upon the presence of an intact membrane.
In a previous study, we used the cross-linking approach to demonstrate that the addition of TNF-α to FL5.12 cells resulted in the formation of a 45-kDa tBID mitochondrial complex (11). Based on a series of experiments that included coimmunoprecipitation of epitope-tagged proteins, cross-linking of endogenous/exogenous proteins, fluorescence resonance energy transfer analysis of chimeric molecules, and enforced dimerization using the FKBP12/FK1012 system, we originally proposed that the 45-kDa complex represents a tBID homotrimer.
In the present study, we demonstrate that in fact the 45-kDa complex represents an association between tBID and Mtch2, a novel and previously uncharacterized protein. Mtch2 might participate in tBID-induced apoptosis, since we could show, using a native purification approach, that in apoptotic cells, tBID is recruited together with BAX to a relatively large resident complex that includes Mtch2. Furthermore, we demonstrate that overexpression of BCL-XL, which inhibits the TNF-α-induced release of Cyt c, partially inhibits the recruitment of both tBID and BAX to this resident complex.
Mitochondrial carrier homolog 2 has several close relatives, which together form a “sister family” separate from the other MCPs (Fig. 3). All Mtch family members contain a single MCD, three transmembrane domains, and can be aligned across almost their entire length (Fig. 3). Mtch proteins are thus a distinct subgroup of proteins related to MCPs.
The function(s) of Mtch2 and its relatives are unknown. Mtch2 does not seem to be a proapoptotic protein, since we found that its overexpression in 293T or in HeLa cells had no effect on cell viability (data not shown). On the other hand, it was previously reported that human presenilin-1-associated protein (PSAP, also known as human Mtch1, a protein with 48% identity to human Mtch2; see Fig. 3) localizes to mitochondria and induces Cyt c release, caspase activation, and apoptosis when overexpressed in 293T cells (32). Thus, Mtch1 is a proapoptotic protein acting at the mitochondria.
What might be the function(s) of Mtch2 and its relatives? As described above, the MCP family comprises proteins that reside in the mitochondria and catalyze the transport of metabolites across the IMM (17). Thus, one hypothesis would be that Mtch proteins are carrier proteins directly involved in the transport of metabolites. On the other hand, since these proteins contain only one and not three MCDs, it is possible that they are involved in transferring signals (via protein-protein interactions) rather than metabolites (via carrier activity). The fact that Mtch2 is an integral membrane protein, which is likely to be exposed on both sides of the OMM (Fig. 5), suggests that it and perhaps other members of the same family play a role in transferring metabolites/signals across the OMM.
In the present study, we used two different approaches to demonstrate that endogenous Mtch2 and tBID associate in TNF-α-treated FL5.12 cells. First, we reversed the cross-linker connection of the 45-kDa complex and found that tBID and Mtch2 were released from the complex (Fig. 6). Second, by using the BN-PAGE technique we found that tBID comigrates with Mtch2 in the native gel (Fig. 7).
Furthermore, we utilized the BN-PAGE technique to demonstrate, for the first time, that tBID and BAX coreside in the mitochondrial membrane of apoptotic cells (Fig. 8). The fact that tBID interacts with Mtch2 and that BAX coresides with both proteins in the native gel suggests that Mtch2 might also be involved in tBID-induced activation of BAX. The fact that we could not capture the interaction between tBID and BAX using cross-linkers (11) suggests that this interaction is a transient “touch-and-go” interaction, as previously proposed for tBID and BAK (30). Since Mtch2 is an integral membrane protein that is exposed on the surface of mitochondria, it is tempting to speculate that it might act to stably anchor and/or correctly position tBID in the membrane to enable the transient activation of BAX. In this respect, it was previously demonstrated that BAX oligomerization requires the presence of both tBID and a mitochondrial protein that is sensitive to proteinase K (19). The fact that we could show that a tBID BH3 mutant (tBID-G94E) is capable of forming the 45-kDa complex (11) but does not interact with BAX (28) suggests that tBID could interact simultaneously with both Mtch2 and BAX.
BAX is a downstream effector of tBID, and its presence is essential for tBID-induced apoptosis (31). It is well established that tBID induces the dimerization/oligomerization of BAX (7); however, it is still not clear whether these oligomers can indeed form channels in the mitochondrial membrane of intact cells. Alternatively, BAX could regulate the activity of preexisting channels and/or carriers rather than form channels itself. Thus, it is possible that the role played by tBID involves the recruitment of BAX to the Mtch2 resident complex, which contains a channel/carrier that is activated by BAX.
Our BN-PAGE studies also revealed that overexpression of BCL-XL inhibits the recruitment of tBID and BAX to the Mtch2 resident complex (Fig. 9). Inhibition of tBID is due to a direct interaction with BCL-XL, since TNF-α induces the formation of a cross-linked complex between tBID and BCL-XL in FL5.12-BCL-XL cells. Interestingly, a similar interaction between tBID and BCL-XL also occurs in TNF-α-treated FL5.12 parent cells (data not shown), but in these cells the recruitment of tBID and BAX to the Mtch2 resident complex is not affected (Fig. 8). Thus, at low levels of BCL-XL the recruitment to the Mtch2 complex is not inhibited. On the other hand, at high levels of BCL-XL, the recruitment to the Mtch2 complex is inhibited, but this inhibition seems to be partial since we could still detect an interaction between tBID and Mtch2 (Fig. 9). Nevertheless, the fact that FL5.12-BCL-XL cells do not release Cyt c in response to TNF-α (13) suggests that this partial inhibition of the recruitment of tBID and BAX to the Mtch2-resident complex is sufficient to inhibit Cyt c release.
In summary, the present study demonstrates that tBID forms a stable interaction with a resident mitochondrial protein in cells signaled to die by TNF-α. The fact that Mtch2 resides in a resident mitochondrial complex and that tBID and BAX are recruited to this complex, whereas BCL-XL inhibits this recruitment, strongly suggests that Mtch2 and possibly other proteins in this complex are participating in the tBID-BAX death pathway.
Acknowledgments
We are grateful to David Wallach and Michael Forte for helpful comments on the manuscript.
This study was supported in part by the Israel Science Foundation, Israel Cancer Research Fund, Minerva Stiftung, the Y. Leon Benoziyo Institute for Molecular Medicine, the Willner Family Center for Vascular Biology, and Stanley Chais. M.S. is a recipient of the David Aftalion fellowship, and H.N. is a recipient of the Sara Lee Schupf fellowship. A.G. is the incumbent of the Armour Family Career Development Chair of Cancer Research.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basanez, G., J. C. Sharpe, J. Galanis, T. B. Brandt, J. M. Hardwick, and J. Zimmerberg. 2002. Bax-type apoptotic proteins porate pure lipid bilayers through a mechanism sensitive to intrinsic monolayer curvature. J. Biol. Chem. 277:49360-49365. [DOI] [PubMed] [Google Scholar]
- 3.Bernardi, P., V. Petronilli, F. Di Lisa, and M. Forte. 2001. A mitochondrial perspective on cell death. Trends Biochem. Sci. 26:112-117. [DOI] [PubMed] [Google Scholar]
- 4.Da Cruz, S., I. Xenarios, J. Langridge, F. Vilbois, P. A. Parone, and J. C. Martinou. 2003. Proteomic analysis of the mouse liver mitochondrial inner membrane. J. Biol. Chem. 278:41566-41571. [DOI] [PubMed] [Google Scholar]
- 5.Danial, N. N., and S. J. Korsmeyer. 2004. Cell death: critical control points. Cell 116:205-219. [DOI] [PubMed] [Google Scholar]
- 6.Desagher, S., A. Osen-Sand, A. Nichols, R. Eskes, S. Montessuit, S. Lauper, K. Maundrell, B. Antonsson, and J. C. Martinou. 1999. Bid-induced conformational change of Bax is responsible for mitochondrial cytochrome c release during apoptosis. J. Cell Biol. 144:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eskes, R., S. Desagher, B. Antonsson, and J. C. Martinou. 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 20:929-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goping, I. S., A. Gross, J. N. Lavoie, M. Nguyen, R. Jemmerson, K. Roth, S. J. Korsmeyer, and G. C. Shore. 1998. Regulated targeting of BAX to mitochondria. J. Cell Biol. 143:207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths, G. J., L. Dubrez, C. P. Morgan, N. A. Jones, J. Whitehouse, B. M. Corfe, C. Dive, and J. A. Hickman. 1999. Cell damage-induced conformational changes of the pro-apoptotic protein Bak in vivo precede the onset of apoptosis. J. Cell Biol. 144:903-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grinberg, M., R. Sarig, Y. Zaltsman, D. Frumkin, N. Grammatikakis, E. Reuveny, and A. Gross. 2002. tBID homooligomerizes in the mitochondrial membrane to induce apoptosis. J. Biol. Chem. 277:12237-12245. [DOI] [PubMed] [Google Scholar]
- 12.Gross, A., J. Jockel, M. C. Wei, and S. J. Korsmeyer. 1998. Enforced dimerization of BAX results in its translocation, mitochondrial dysfunction, and apoptosis. EMBO J. 17:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross, A., X. M. Yin, K. Wang, M. C. Wei, J. Jockel, C. Milliman, H. Erdjument-Bromage, P. Tempst, and S. J. Korsmeyer. 1999. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J. Biol. Chem. 274:1156-1163. [DOI] [PubMed] [Google Scholar]
- 14.Kuwana, T., M. R. Mackey, G. Perkins, M. H. Ellisman, M. Latterich, R. Schneiter, D. R. Green, and D. D. Newmeyer. 2002. Bid, bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111:331-342. [DOI] [PubMed] [Google Scholar]
- 15.Li, H., H. Zhu, C. J. Xu, and J. Yuan. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94:491-501. [DOI] [PubMed] [Google Scholar]
- 16.Luo, X., I. Budihardjo, H. Zou, C. Slaughter, and X. Wang. 1998. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94:481-490. [DOI] [PubMed] [Google Scholar]
- 17.Palmieri, F. 1994. Mitochondrial carrier proteins. FEBS Lett. 346:48-54. [DOI] [PubMed] [Google Scholar]
- 18.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5:1704-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roucou, X., S. Montessuit, B. Antonsson, and J. C. Martinou. 2002. Bax oligomerization in mitochondrial membranes requires tBid (caspase-8-cleaved Bid) and a mitochondrial protein. Biochem. J. 368:915-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito, M., S. J. Korsmeyer, and P. H. Schlesinger. 2000. BAX-dependent transport of cytochrome c reconstituted in pure liposomes. Nat. Cell Biol. 2:553-555. [DOI] [PubMed] [Google Scholar]
- 21.Sarig, R., Y. Zaltsman, R. C. Marcellus, R. Flavell, T. W. Mak, and A. Gross. 2003. BID-D59A is a potent inducer of apoptosis in primary embryonic fibroblasts. J. Biol. Chem. 278:10707-10715. [DOI] [PubMed] [Google Scholar]
- 22.Schagger, H. 2001. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65:231-244. [DOI] [PubMed] [Google Scholar]
- 23.Schendel, S. L., M. Montal, and J. C. Reed. 1998. Bcl-2 family proteins as ion-channels. Cell Death Differ. 5:372-380. [DOI] [PubMed] [Google Scholar]
- 24.Shi, Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9:459-470. [DOI] [PubMed] [Google Scholar]
- 25.Vander Heiden, M. G., and C. B. Thompson. 1999. Bcl-2 proteins: regulators of apoptosis or of mitochondrial homeostasis? Nat. Cell Biol. 1:E209-E216. [DOI] [PubMed] [Google Scholar]
- 26.Varfolomeev, E. E., and A. Ashkenazi. 2004. Tumor necrosis factor: an apoptosis JuNKie? Cell 116:491-497. [DOI] [PubMed] [Google Scholar]
- 27.Walker, J. E., and M. J. Runswick. 1993. The mitochondrial transport protein superfamily. J. Bioenerg. Biomembr. 25:435-446. [DOI] [PubMed] [Google Scholar]
- 28.Wang, K., X. M. Yin, D. T. Chao, C. L. Milliman, and S. J. Korsmeyer. 1996. BID: a novel BH3 domain-only death agonist. Genes Dev. 10:2859-2869. [DOI] [PubMed] [Google Scholar]
- 29.Wang, X. 2001. The expanding role of mitochondria in apoptosis. Genes Dev. 15:2922-2933. [PubMed] [Google Scholar]
- 30.Wei, M. C., T. Lindsten, V. K. Mootha, S. Weiler, A. Gross, M. Ashiya, C. B. Thompson, and S. J. Korsmeyer. 2000. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14:2060-2071. [PMC free article] [PubMed] [Google Scholar]
- 31.Wei, M. C., W. X. Zong, E. H. Cheng, T. Lindsten, V. Panoutsakopoulou, A. J. Ross, K. A. Roth, G. R. MacGregor, C. B. Thompson, and S. J. Korsmeyer. 2001. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292:727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu, X., Y. C. Shi, W. Gao, G. Mao, G. Zhao, S. Agrawal, G. M. Chisolm, D. Sui, and M. Z. Cui. 2002. The novel presenilin-1-associated protein is a proapoptotic mitochondrial protein. J. Biol. Chem. 277:48913-48922. [DOI] [PubMed] [Google Scholar]
- 33.Yin, X. M., K. Wang, A. Gross, Y. Zhao, S. Zinkel, B. Klocke, K. A. Roth, and S. J. Korsmeyer. 1999. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature 400:886-891. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Q. H., M. Ye, X. Y. Wu, S. X. Ren, M. Zhao, C. J. Zhao, G. Fu, Y. Shen, H. Y. Fan, G. Lu, M. Zhong, X. R. Xu, Z. G. Han, J. W. Zhang, J. Tao, Q. H. Huang, J. Zhou, G. X. Hu, J. Gu, S. J. Chen, and Z. Chen. 2000. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 10:1546-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao, Y., S. Li, E. E. Childs, D. K. Kuharsky, and X. M. Yin. 2001. Activation of pro-death Bcl-2 family proteins and mitochondria apoptosis pathway in tumor necrosis factor-alpha-induced liver injury. J. Biol. Chem. 276:27432-27440. [DOI] [PubMed] [Google Scholar]
- 36.Zong, W. X., T. Lindsten, A. J. Ross, G. R. MacGregor, and C. B. Thompson. 2001. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 15:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]