Abstract
Human milk oligosaccharides (HMOs) are a class of structurally diverse and complex unconjugated glycans present in breast milk, which act as selective substrates for several genera of select microbes and inhibit the colonisation of pathogenic bacteria. Yet, not all infants are breastfed, instead being fed with formula milks which may or may not contain HMOs. Currently, formula milks only possess two HMOs: 2′-fucosyllactose (2’FL) and lacto-N-neotetraose (LNnT), which have been suggested to be similarly effective as human breast milk in supporting age-related growth. However, the in vivo evidence regarding their ability to beneficially reduce respiratory infections along with altering the composition of an infant’s microbiota is limited at best. Thus, this review will explore the concept of HMOs and their metabolic fate, and summarise previous in vitro and in vivo clinical data regarding HMOs, with specific regard to 2’FL and LNnT.
Keywords: Human milk oligosaccharides (HMOs), infants, Bifidobacterium, gut microbiota, infant formula, cross-feeding
Introduction: the history of HMOs
Human breast milk is considered the gold standard nutrient source for infants in the early stages of life due to the presence of several remarkable functional ingredients (Van den Abbeele et al., 2019). One group of such ingredients is the human milk oligosaccharides (HMOs). A classification which is given to a group of structurally diverse and complex unconjugated glycans present in human breast milk (Ninonuevo et al., 2006). HMOs were first “discovered” towards the end of the nineteenth century after French biochemist Georges Denigés noted that in addition to lactose, human and bovine milk possessed several other carbohydrate structures with Polonowski and Lespagnol originally terming these unknown fractions as gynolactoses (Polonowski and Lespagnol, 1929, 1931). The levels of HMOs present in breast milk range from 5 to 25 g/L throughout the course of lactation, making the levels of oligosaccharides present in human milk the highest amongst mammalian species. This is over 100 times greater than the oligosaccharide content found in bovine milk, which has been estimated at around 100 mg/L (Robinson, 2019; Zivkovic et al., 2011).
Structural complexity of HMOs
Virtually, all HMOs possess a lactose (Lac) core to which a multitude of different monosaccharide “building blocks,” including galactose (Gal), glucose (Glc), fucose (Fuc), sialic acid (Neu5Ac) and N-acetylglucosamine (GlcNAc), can be attached via the action of specific glycosyltransferases in the presence of α-lactalbumin (Smilowitz et al., 2014). The synthesis of HMOs begins with the enzymatic elongation of lactose (Lac) by either β1-3 or β1-6 linkages of Gal to lacto-N-biose (LNB) or N-acetyllactosamine (LacNAc), respectively. Based on this, HMOs can be classified as either Type-I or Type-II chains. Type-I chain HMOs possess lacto-N-tetraose (LNT), which is lactose coupled to LNB. While Type-II chains, are composed LNT isomer lacto-N-neotetraose (LNnT), and is Lac linked to LacNAc (James et al., 2016). These core HMO structures can then be further elongated and additionally categorised as neutral, fucosylated or sialylated (Plaza-Diaz et al., 2018; Zivkovic et al., 2011). Neutral HMOs possess structures similar to galacto-oligosaccharides (GOSs) containing both Glc and Gal (Barile and Rastall, 2013), and may also accommodate several GlcNAc or LNB units attached via β1-3 and β1-6 linkages (Ayechu-Muruzabal et al., 2018). At this point, Fuc units can be enzymatically attached via α1-2, α1-3 or α1-4 linkages generating fucosylated HMOs. Hereafter, one or more molecules of Neu5Ac may be attached via α2-3 or α2-6 linkages in the presence of sialyl-transferases generating sialylated HMOs (Smilowitz et al., 2014). Figure 1 gives a generalised overview of the complexity and structural diversity of HMOs present in breast milk.
Figure 1.
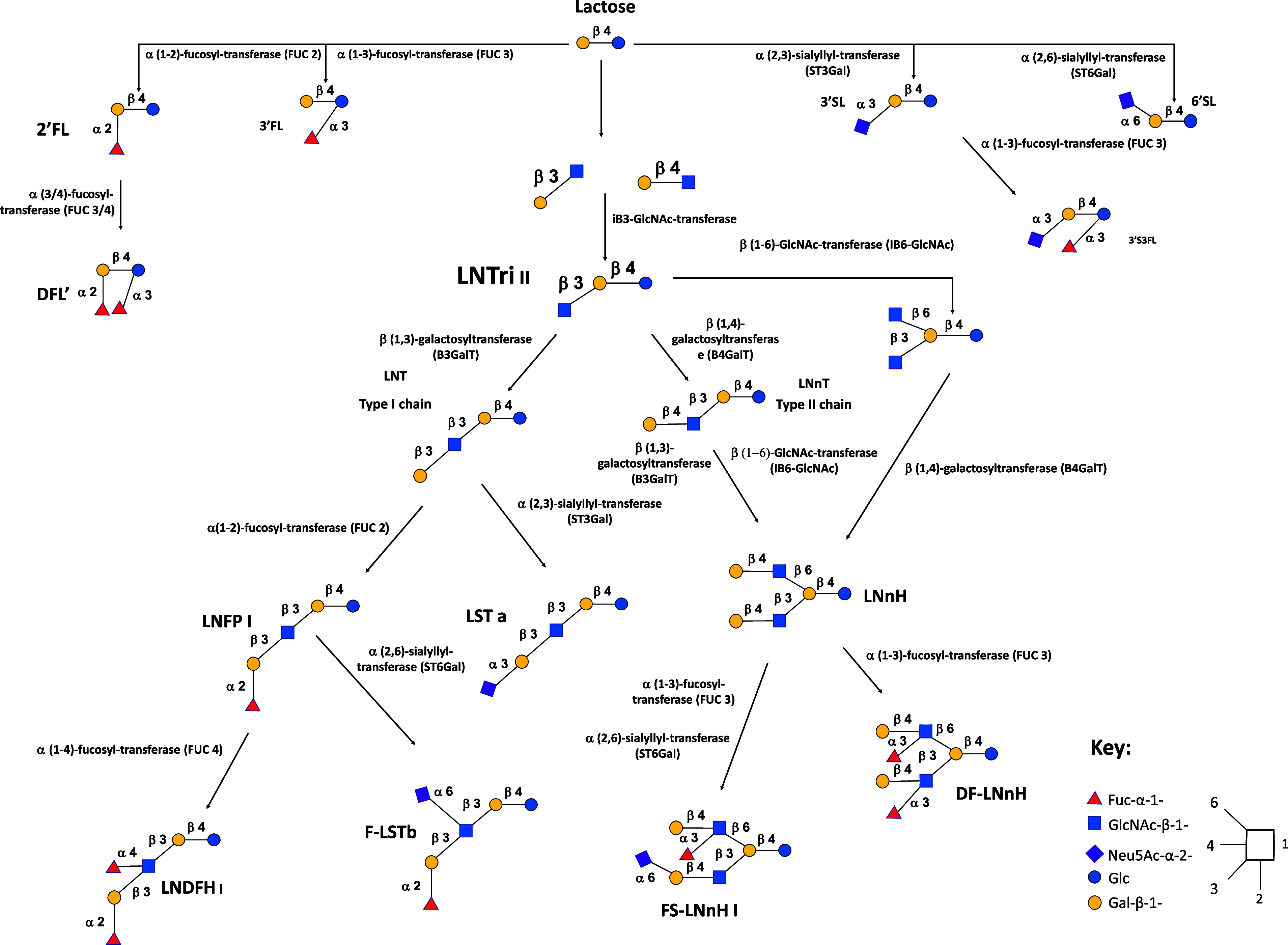
Generalised overview of the complexity and structural diversity of human milk oligosaccharides (HMOs) present in breast milk.
Abbreviations: Neutral HMOs: 2’FL, 2′-fucosyllactose; 3’FL, 3′-fucosyllactose; DFL, difucosyllactose; DF-LNnH, difucosylated lacto-N-neohexaose; LNDFH I, lacto-N-difucohexaitol I; LNFP I, lacto-N-fucopentaose I; LNnH, lacto-N-neohexaose; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetraose. Acidic non-fucosylated HMOs: 3’SL, 3′-sialyllactose; 6’SL, 6′-sialyllactose; LST a, sialyllacto-N-tetraose a. Acidic fucosylated HMOs: 3’S3FL, 3’-Sialyl-3-fucosyllactose; F-LST b, sialylfucosyllacto-N-tetarose b; FS-LNH I, fucosylsiallacto-N-hexose I.
Composition of human milk oligosaccharides in breast milk and factors affecting composition
To date, somewhere in the region of 200 HMOs have been identified in the breast milk of mothers with the most widely recognised HMOs being 2′-fucosyllactose (2’FL), LNT and LNnT (Barile and Rastall, 2013; Egge et al., 1983; Urashima et al., 2018). The composition and concentration of HMOs present in breast milk are highly dependent on several critical factors, including geographical location, ethnicity, length of gestation and secretor status. In general, the levels of HMOs present in breast milk are highest immediately following birth and decrease throughout lactation with the concentration of 2′FL appearing to be highest during the first month postpartum (Thurl et al., 2010, 2017; Xu et al., 2017). Yet, the rates at which HMOs decline are not constant across all HMOs. For example, while 2’FL, difucosyllactose (DFL), lacto-N-fucopentaose-2 (LNFP II), 3’-Sialyllactose (3’SL) and 6’-Sialyllactose (6′SL) all decline in concentration throughout lactation, the rate of decline does not appear to significantly alter during days 30–120 (Gabrielli et al., 2011; Spevacek et al., 2015; Thurl et al., 2010), whereas lacto-N-fucopentaose-1 (LNFP I) appears to decline just 3 days after birth recording a twofold decline by the end of lactation, respectively (Bao et al., 2013; Smilowitz et al., 2013). Other HMOs, including 3′-fucosyllactose (3’FL), appear to increase in concentration throughout lactation by 1.67–1.8-fold (Austin et al., 2019; Gabrielli et al., 2011; Samuel et al., 2019; Smilowitz et al., 2013).
However, a significant percentage of mothers can only synthesise certain HMOs, including 2’FL, dependant on their secretor status. Secretor status is based on the Lewis blood group and depends on the expression of the specific glycosyltransferases, α1-2-fucosyltransferase (FUT2 encoded by the Se gene) and α1-3/4-fucosyltransferase (FUT3 encoded by the Le gene), and can result in marked differences in which HMOs are synthesised (Blank et al., 2012; Hegar et al., 2019). Table 1 summarises these findings.
Table 1.
Human milk oligosaccharide composition of breast milk based on the genetic background of the mother. Source: Vandenplas et al. (2018).
| Gene | Lewis gene + | Lewis gene − |
|---|---|---|
| Secretor gene + | Se+ Le+ | Se+ Le− |
| Secretes all HMOs | Able to secrete 2’FL, 3’FL, LNFP I and LNFP III | |
| Secretor gene − | Se− Le+ | Se− Le− |
| Able to secrete 3’FL, LNFP-II and LNFP III | Able to secrete 3’FL, LNFP III and LNFP V |
However, the Lewis antigen system and secretor status can be described as an over generalisation of an extremely complex situation (Plaza-Diaz et al., 2018) as even when these factors are accounted for, substantial differences in HMO profile can still occur. For example, FUT2 and FUT3 have been shown to compete for several of the same substrates. As a result, differences in levels of expression can alter the synthesis of 2’FL by a “secretor” mother (McGuire et al., 2017; Plaza-Diaz et al., 2018).
Additionally, levels and composition of HMOs present in breast milk may also vary between the mothers of preterm and full-term infants regardless of secretor status. However, reports on this are contradictory with several papers seemingly reporting that HMOs may or may not vary between preterm and term infant mothers (Austin et al., 2019; De Leoz et al., 2012; Kunz and Rudloff, 2017; Thurl et al., 2017).
Furthermore, the sizeable heterogeneity in the levels and composition of HMOs detected in breast milk may be in part due to differences in analytical techniques used in studies, including high-performance liquid chromatography mass spectrometry (HPLCMS) and HPLC time-of-flight mass spectrometry, high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD), as well as differences in the number of preparation steps taken (van Leeuwen, 2019; Zivkovic et al., 2011). A further complication is the lack of analytical standards needed in order to quantitatively assess the concentration of HMOs present (Wicinski et al., 2020). As a result, it could be argued that determining the average composition and quantity of HMOs in breast milk, based on current published information, is not achievable due to too many confounding variables. Furthermore, to reiterate (Thurl et al., 2017), to be able to determine the average composition of HMOs in breast milk, a worldwide multicentre study following the same protocols would be required. However, even if the average HMO content of breast milk was determined, what relevance this would have regarding clinical significance remains uncertain.
Human milk oligosaccharides, health benefits and clinical data
It is well documented that breastfeeding is highly associated with several health benefits, including improved growth rate, lower prevalence of respiratory, intestinal and urinary infections (Li et al., 2014) and lower incidence of allergies and autoimmune conditions, with some of this being put down to the presence of HMOs in breast milk (Doherty et al., 2018; Triantis et al., 2018).
The structural nature of HMOs renders them resistant to digestive enzymes found within the GI tract (Garrido et al., 2012a) with >90 per cent of HMOs reaching the colon intact. HMOs likely function as prebiotics, stimulating the growth of beneficial bacteria, including bifidobacteria (Barile and Rastall, 2013; Bode, 2009). HMOs can also act as soluble decoys, preventing the adhesion of pathogenic bacteria to cell surface receptors due to their resemblance to the glycans found on the surface of epithelial cells in the intestinal tract (Wicinski et al., 2020). In addition, the formation of the short-chain fatty acids (SCFAs) butyrate, acetate and propionate from saccharolytic fermentation plays vital roles in the activation and differentiation of immune cells and may also reduce the risk of infections and allergies (Ayechu-Muruzabal et al., 2018; Kumari and Kozyrskyj, 2017). Additionally, it has been shown in infants that a lack of bifidobacteria, particularly those associated with HMO degradation and utilisation, is associated with increases in systemic inflammation and immune dysfunction (Henrick et al., 2021). Furthermore, supplementation of 2’FL in combination with B. pseudocatenulatum MP80 was associated with changes in gene expression of both anti-inflammatory and pro-inflammatory markers in the cecum of healthy mice, whereas in mice with dextran sulphate sodium (DSS)-induced colitis, supplementation of 2’FL in combination with B. pseudocatenulatum MP80 resulted in attenuations of bodyweight loss, along with a reduction in DSS-induced immune cell infiltration, increase in colon length and disrupted mucosal architecture along with preventing a reduction in occludin expression (Heiss et al., 2021).
However, not all infants are breastfed, with many being fed formula milk which may or may not be supplemented with HMOs. The first two HMOs to be commercially produced were 2’FL and LNnT, and while these compounds are referred to as HMOs, they are not sourced from human milk, but are produced by microbial fermentation using strains of E. coli and yeasts which have been genetically modified (Sprenger et al., 2017).
Several studies have suggested that infants fed HMO-supplemented formula milk present lower risks of parent-reported bronchitis and respiratory infections, along with reduced use of antibiotics, less waking at nights and improved age-appropriate growth (Marriage et al., 2015; Puccio et al., 2017). In a study on the 2’FL, solely or in combination with Bifidobacterium longum subsp. infantis (Bi-26), on cognitive and structural development in young pigs (Sutkus et al., 2022), the authors noted that synbiotic administration of 2’FL and Bi-26 had several interactive effects on microstructural brain components; however, it appeared to have no effect on memory. Preclinical in vitro and in vivo studies propose that both 2′FL and LNnT promote the growth of several Bifidobacterium and Bacteroides species, strains, and subspecies including B. longum subsp. infantis, Bacteroides fragilis, and Bacteroides vulgatus, amongst others (Marcobal et al., 2010; Yu et al., 2013).
However, while the importance of breast milk on infant health outcomes is well supported, there remains a great deal unknown regarding the efficacy of HMOs added to infant formulas on infant health outcomes. Furthermore, it has been demonstrated that several species, strains and subspecies of microorganisms found within the gut microbiota, including Bifidobacterium adolescentis and Bifidobacterium animalis, do not grow well on HMOs, including 2′FL and LNnT on their own (Lawson et al., 2020; LoCascio et al., 2010; Marcobal et al., 2010; Sela et al., 2012; Xiao et al., 2010). This suggests that the composition of gut microbiota may be key if formula milks containing 2’FL and LNnT are to be effectively utilised. Therefore, the rest of this review will focus on the effects that HMOs have on the composition of the infant gut microbiota both in vitro and in vivo and will attempt to determine the metabolic fate of HMOs.
The metabolic fate of human milk oligosaccharides: infant and rodent studies
It was established as early as the 1970s that HMOs are present in the urine of expecting mothers as soon as 8 weeks (Hallgren et al., 1977) and can be detected in the mammary glands using 13C isotopes (Dotz et al., 2014), suggesting that HMOs circulate throughout maternal serum. Hirschmugl et al. (2019) investigated individual and temporal variations in the composition and levels of HMOs in maternal serum throughout the course of pregnancy. In this study, serum samples were collected from healthy pregnant woman throughout the course of gestation at weeks 10–14 (Visit 1), 20–24 (Visit 2) and 30–35 (Visit 3) and at the time of admission to delivery at the Department of Obstetrics, Medical University of Graz, Graz, Austria. In total, 16 HMOs [2′FL, 3’FL, DF’, 3′SL, 6′SL, LNT, LNnT, LNFP I, II, and III, LST a, b, and c, LNDFH, LNH and disialyllacto-N-tetraose (DSLNT)] were detected in serum with the authors reporting a steady increase in the presence of HMOs, in particular, fucosylated HMOs in circulation throughout the course of pregnancy.
Based on this, one could speculate that HMOs may undergo maternal-to-foetal transport. Indeed, there are data suggesting that several HMOs, including 2′FL, 3’FL, DFL and 6′SL, were present in amniotic fluid (Wise et al., 2018). Likewise, it has also been demonstrated, using an ex vivo experimental model involving isolated placental cotyledons perfused with 2’FL, that 2’FL was able to cross the placenta (Hirschmugl et al., 2019). Furthermore, given that HMOs function as signalling molecules and that similar glycan structures can act as receptors in cells and tissues (Bhargava et al., 2012), it is likely that HMOs may also contribute towards the development and functioning of immune (Plaza-Diaz et al., 2018) and endothelial cells (Donovan and Comstock, 2016) of infants in utero. However, since foetal circulation and cord blood are not accessible during pregnancy, these findings should be interpreted with a great deal of care (Hornef and Penders, 2017; Walker et al., 2017).
To date, the ability of HMOs to end up in the urine of infants has been extensively studied using 13C-labelled glycans along with several analytical techniques, including MALDI-TOF-MS, HPAEC-PAD, nano-LC–MS and MALDI FT-ICR MS. The results of these studies suggest that fully and partially intact HMOs, including 2’FL, 3’SL, 6’SL, LNT and LNnT, can be present in the urine of infants, albeit in far lower concentrations than that found in milk at just 4 per cent, with secretor/non-secretor status massively impacting on which HMOs are detected (Borewicz et al., 2020; De Leoz et al., 2013; Dotz et al., 2014, 2015; Goehring et al., 2014).
Given that HMOs can end up in the urine of infants, HMOs can clearly be absorbed through the epithelial cells of the gastrointestinal tract and enter the bloodstream (Dotz et al., 2014; Rudloff et al., 2012). To date, the potential for HMOs to enter circulation has been extensively studied in animal models. In one study, conducted in rats fed both mixed and isolated HMOs, including 2’FL, it was noted that when mixed HMOs were ingested, 2’FL was detected in circulation 30 min later in a dose-dependent manner, reaching a maximum at 60 min (Vazquez et al., 2017), whereas in rats fed isolated HMOs, levels of 2’FL detected in circulation similarly increased in concentration over time, again in a dose-dependent manner. However, they did not reach a maximum peak within the 4-h sampling period. In contrast, Jantscher-Krenn et al. (2013) reported that only 3’SL, along with very few other HMOs, was detected in the serum of rats. The discrepancies in findings between these studies probably result from biological differences in the metabolism of HMOs due to the ages of rats used in each respective study.
The potential for HMOs to enter into the circulation and plasma of infants was investigated by Ruhaak et al. (2014) in 13 full-term infants using solid-phase extraction followed by an analysis by nHPLC-PGC-chip-TOF-MS. In total, 15 oligosaccharides, including LNT, LDFP, LNFT, 3’SL, 6’SL, 3′sialyllactosamine (3’SLN), 6′sialyllactosamine (6’SLN), LNFP III and 2’FL, were detected in the plasma of infants, albeit in lower concentrations than levels found in breast and formula milk, with over a 10-fold variation in LNT being recorded between partially breastfed and formula-fed infants. Interestingly, an unknown isomer of SLN was found. Given SLN is usually derived from bovine milk (McGrath et al., 2016; Ruhaak et al., 2014) and the relative abundances detected in the infants of this study far exceed those found in bovine milk (Fong et al., 2011). From this, one could theorise that new SLNs detected in the circulation of infants may have been produced via interactions between milk, milk by-products and bacterial glycosidases (Lis-Kuberka and Orczyk-Pawilowicz, 2019).
Moreover, using isotopically labelled standards, coupled with ultra-performance liquid chromatography and HPLC, Goehring et al. (2014) investigated the presence of HMOs in both breastfed and formula-fed infants. The authors reported the detection of several HMOs, including 2′FL, 3’FL and LNnT, in the plasma of breastfed but not formula-fed infants, again albeit in far lower levels. A similar finding was reported by Radzanowski et al. (2013), who documented that the concentration of HMOs present in infant plasma was substantially less (3′SL: 0.10–0.78 mg/L; 6′SL: 0.05–0.68 mg/L; 2′FL: 0–2.25 g/L) compared with breast milk (3′SL: 54.3–225 mg/L; 6′SL: 29.3–726 mg/L; 2′FL: 0–3.8 g/L) at just 0.1 per cent, respectively.
The degradation and transportation of HMOs
As previously discussed, virtually, all HMOs possess a lactose core to which a multitude of different monosaccharide “building blocks,” including Gal, Glc, Fuc, sialic acid (Neu5Ac) and N-acetylglucosamine (GlcNAc), can be attached via the action of specific glycosyltransferases in the presence of α-lactalbumin (Smilowitz et al., 2014). In order to stimulate the fructose 6-phosphate phosphoketolase-dependent glycolytic pathway more commonly termed the bifid shunt active in bifidobacteria, these complex milk glycans must be degraded (Pokusaeva et al., 2011). The mechanisms by which HMOs undergo degradation can be characterised as intracellular (transport-dependant) and extracellular (glycosidase-dependent). Both mechanisms require the use of specific ATP (adenosine triphosphate)-binding cassette (ABC) transporters to either import intact or processed glycans inside the bacterial cell (Garrido et al., 2011) with the most common strains of bifidobacteria (B. breve, B. longum and Bifidobacterium bifidum) preferring to utilise specific mechanisms of action (Sakanaka et al., 2020). See Figure 2.
Figure 2.
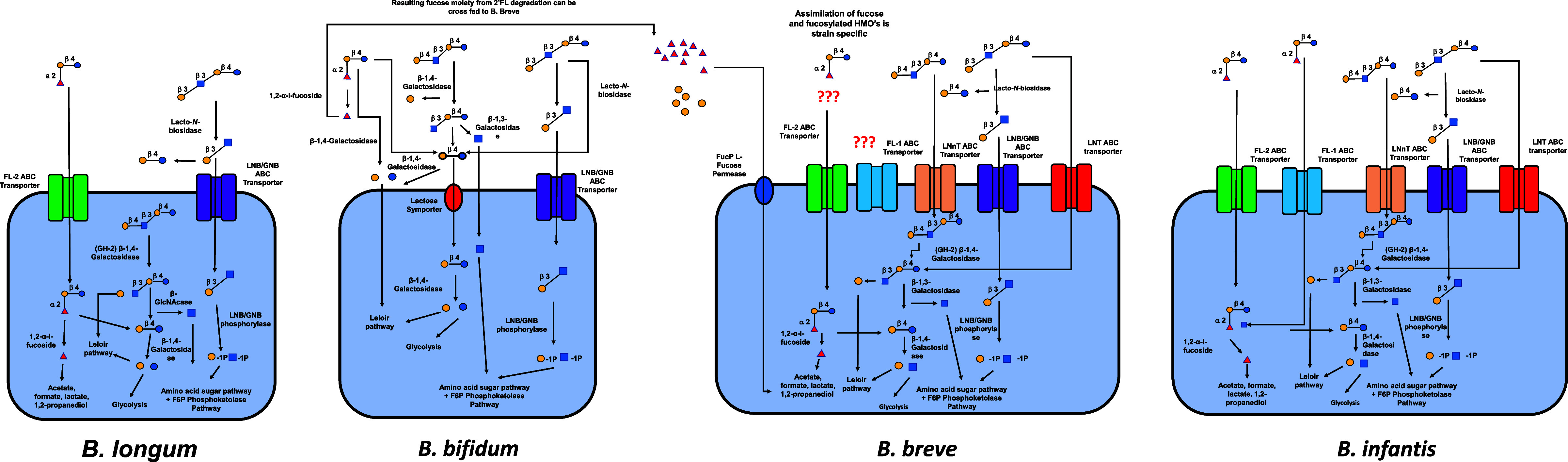
Intracellular and extracellular degradation of the three main human milk oligosaccharides (2’FL, LNT and LNnT) and resulting metabolites by four common species of Bifidobacterium (Bifidobacterium longum, B. bifidium, B. breve and B. longum) and selective fermentation pathways. Abbreviations: ABC, ATP-binding cassette; FL, fucosyllactose; GNB, galacto-N-biose; LNB, lacto-N-biose; LNnT, lacto-N-neotetraos.
Fucosidases and fucosylated human milk oligosaccharide transporters
To hydrolyse fucosidic-linked HMOs, two distinct glycoside hydrolase (GH) families, GH95 and GH29, are required for the degradation of specific fucosidic linkages (Sakanaka et al., 2019). While both GH95 and GH29 can target the 1,2-, 1,3- and 1-4-α-l-fucosides, GH29 displays a higher affinity towards both 1,3- and 1,4-α-l-fucosides, whereas GH95 displays a higher preference towards the hydrolysis of 1,2-α-l-fucosides (Matsuki et al., 2016; Shani et al., 2022; Zeuner et al., 2018). The transporters responsible for the uptake of fucosyllactose (FL’) were first discovered in B. longum subsp. infantis ATCC 15697T (Sela et al., 2008). B. longum subsp. infantis ATCC 15697T possesses two paralogous FL’ transporters which share up to 60 per cent of the same solute-binding proteins (SBPs), suggesting that there is some degree of overlap in their ability to transport various HMOs (Sakanaka et al., 2020). This overlap was demonstrated in a recent study conducted by Sakanaka et al. (2019) with the authors concluding that FL transporter-1 was only able to import low molecular weight HMOs, including 2′FL and 3’FL. In contrast, FL transporter-2 was able to import not only 2′FL and 3’FL, but also LDFT and LNFP I. Yet, the ability of different species and strains of bifidobacteria to express FL’ transporters is not identical. As research by Garrido et al. (2016) and Matsuki et al. (2016) revealed, there was a remarkable difference in the ability of B. longum subsp. infantis and B. bifidum to utilise FL’ due to the presence of different intracellular and extracellular ABC-type transporters (K02025 and K02026).
Outside of Bifidobacterium spp., GH29 and GH95 have also been detected in Roseburia inulinivorans and GH29 detected in Akkermansia muciniphila, enabling it to cleave the α1-2-fucosyl linkage to Gal (Kostopoulos et al., 2020).
Lacto-N-biosidase and LNB transporter
LNT is one of the most abundant neutral HMOs present in breast milk and is hydrolysed by lacto-N-biosidase (lnb), resulting in the formation of LNB and Lac (Yamada et al., 2017). The specificity of lnb present in Bifidobacterium also varies remarkably between species (Sakanaka et al., 2020). On this basis, lnb present in B. bifidum is classified as GH20, whereas in B. longum, lnb is categorised as GH136. The differences being that GH136 requires an additional chaperonin for proper protein folding and is also capable of accepting sialyllacto-N-tetraose a (LST a) in addition to LNT (Sakurama et al., 2013; Yamada et al., 2017).
In B. bifidum, degradation of HMOs to LNB and monosaccharides begins extracellularly with the hydrolysis of LNFP I, II and lacto-N-difucohexaose I, II (LNDFH I, II) to LNT and Fuc with the aid of additional fucosidases (Marcobal and Sonnenburg, 2012). LNB is then hydrolysed from LNT by Lnb and then transported inside of the cell leaving any remaining fucosyl residues behind. Lnb phosphorylase then converts LNB into its respective monosaccharides Gal and GlcNAc, which then undergo further assimilation (Zivkovic et al., 2011). In B. longum, the degradation of LNT seemingly follows similar principles to that of B. bifidum with LNB being extracellularly hydrolysed from LNT. However, unlike B. bifidum, the hydrolysis of LNB from LNFP I by B. longum occurs intracellularly, meaning that no Fuc residues are left behind (Xiao et al., 2010; Yamada et al., 2017). In contrast to both B. bifidum and B. longum, the degradation of LNT and LNnT to LNB by B. longum subsp. infantis occurs entirely intracellularly with LNB constituent monosaccharides Gal and GlcNAc entering selective fermentation pathways (Ozcan and Sela, 2018). More recently, the presence of GH136 has been discovered in Roseburia with the degradation of fuscoylated pentose and hexose HMOs said to occur extracellularly (Pichler et al., 2020).
Sialidase
For microorganisms to be able to utilise sialic acids, they must possess the necessary sialidases required to hydrolyse the α-2,3 and α-2,6 linkages of sialylated HMOs (Kiyohara et al., 2011; Zivkovic et al., 2011). In bifidobacteria, the most common sialidases are classified as GH33 with each member of the GH33 family exhibiting preference for specific sialic acid linkages. For example, in B. bifidum, the sialidase present is categorised as SiaBb2 and exhibits a preference for α-2,3 linkages, whereas the sialidase present in B. longum subsp. infantis is classified as NanH2, but unlike SiaBb2 appears to exhibit an equal preference for both α-2,3 and α-2,6 linkages (Juge et al., 2016).
LNT β-1,3-galactosidase
In bifidobacteria, β-1,3-galactosidases are categorised as GH42 and GH35 and are responsible for cleaving HMOs possessing β-1,3 Gal linkages, displaying hydrolytic ability for both Type-I and Type-II chains with highest activity being displayed on LNT, followed by Lac, LNB and LNnT (James et al., 2016; Yoshida et al., 2012).
β-1,4-galactosidase
Several other glycosidases for the assimilation of HMOs exist, including β-1,4-galactosidases (Zeuner et al., 2019). In B. bifidum and B. breve, the β-1,4-galactosidases belong to the GH2 family and are categorised as BbgIII and LacZ2 and LacZ6, respectively, and are responsible for the hydrolysis of HMOs possessing Lac and Type-II chains (James et al., 2016; Yoshida et al., 2012). This mechanism of HMOs degradation is particularly prominent in B. bifidum and functions extracellularly cleaving LNnT at its Galβ-1,4 residue liberating Gal and LNT. Thereafter, LNT is further hydrolysed producing GlcNAc and lactose with lactose undergoing additional hydrolysis resulting in the formation of Glc and Gal, respectively (James et al., 2016; Miwa et al., 2010).
β-D-hexosaminidases
N-acetyl-β-D-hexosaminidases are another set of hydrolytic enzymes, which belong to the GH20 family with three enzymes seemingly responsible for the hydrolysis of HMOs. N-acetyl-β-D-hexosaminidases have been detected in B. longum subsp. infantis (Garrido et al., 2012b) with these GHs being categorised as Blon_2355 and Blon_0732 and Blon_0459. Blon_2355 displays a preference for GlcNAc β1-3 Gal linkages, whereas Blon_0732 and Blon_0459 can additionally release GlcNAc from branched HMOs characterised by GlcNAc β1-6 Gal linkages (Garrido et al., 2012b). Additionally, N-acetyl-β-D-hexosaminidases have been discovered in B. bifidum JCM 1254, categorised as BbhI and BbhII, with BbhI being shown to hydrolyse lacto-N-triose II into GlcNAc and lactose, respectively (Miwa et al., 2010).
It is clear that different species, strains and subspecies of Bifidobacterium possess several different mechanisms for the assimilation of HMOs and it is likely that substantial differences in rates of consumption and fermentation occur.
The in vitro assimilation and consumption of HMOs
Differences in the consumption behaviour between different microorganisms found within in the gut microbiota have been assessed in vitro using either pooled or singular HMOs as sole carbon sources. To date, bifidobacteria are the most widely studied microorganisms, both singular and in combination, in relation to their ability to utilise HMOs due to their predominance in the breastfed infant gut microbiota. Yet, it is not only bifidobacteria that have the ability to degrade HMOs, several other genera within the gut, including Bacteroides, Roseburia and Akkermansia amongst others, appear to play a role in HMO utilisation.
Single cultures
As mentioned, bifidobacteria are one of the most well-documented microorganisms in the gut regarding HMO consumption. In one study, conducted by Gotoh et al. (2018), the authors aimed to determine the ability of various strains of bifidobacteria to utilise HMOs, including 2’FL. The four strains of B. bifidum tested (JCM1254, JCM7004, TMC3108 and TMC3115) were isolated from either infant faecal samples or obtained from other researchers at the Riken Bioresource centre and subjected to in vitro assays using GAM broth and with sugar (HMOs) analysis and concentration being analysed in the spent media. There was a large amount of Fuc and Lac in the supernatants of all four B. bifidum strains assimilating both HMOs and Lac by 15 h, with the degradation of 2’FL beginning even before cells entered the exponential phase. This suggests that each strain of B. bifidum possessed the necessary 1,2-α-fucosidase and 1-3- and 1-4-α-fucosidases required for the degradation of fucosylated HMOs. Furthermore, the fermentation of 2’FL appeared to occur rapidly and extracellularly which may have important implications regarding the utilisation of HMOs within bifidobacterial communities and subsequent microbial diversity (Sakanaka et al., 2020).
Garrido et al. (2015) also examined the ability of several strains of B. longum subsp. infantis and B. bifidum utilised from breastfed infants to utilise HMOs, including 2’FL, 3’FL, 6’SL and LNT, as sole carbon sources. All B. longum subsp. infantis strains displayed excellent growth on all HMOs, while the ability of various B. bifidum strains was highly variable with several strains showing little growth on pooled HMOs. These findings are similar to those of Gotoh et al. (2018), who recorded that the growth of B. bifidum strains (JCM1254, TMC3108 and TMC3115) resulted in higher cell biomass compared with B. bifidum strain JCM7004. There is clearly a remarkable variability in the ability of Bifidobacterium strains, even of the same species, to utilise HMOs with several species, strains and subspecies appearing to be better adapted than others (Sakanaka et al., 2020).
The differences in the ability of Bifidobacterium to utilise HMOs were also demonstrated by Bunesova et al. (2016), who tested several strains of Bifidobacterium, including B. longum subsp. infantis, B. longum subsp suis BSM11–5 and B. bifidum and Bifidobacterium kashiwanohense isolated from the stool samples of 6-month infants. There was substantial variability in the ability of bifidobacterial strains and subspecies to utilise HMOs with B. longum subsp. infantis being able to utilise 2′-FL, 3′-FL, 3′-SL and LNnT. B. bifidum BSM28-1 was able to utilise 6’SL in addition to 2′-FL, 3′-FL, 3′-SL and LNnT, while B. longum subsp. suis BSM11-5 and B. kashiwanohense strains all grew in the presence of 2′FL and 3’FL. Several strains and subspecies of bifidobacteria, however, including B. bifidum DSM20215 and B. breve DSM 20213, were unable to utilise HMOs to any real degree, with Bifidobacterium pseudolongum not being able to utilise HMOs at all. Furthermore, all B. longum strains and subspecies tested, including B. longum subsp. suis BSM11-5 and B. longum subsp. infantis DSM 20088, were able to utilise the resulting Fuc moiety, albeit with a varying degree of success with B. kashiwanohense not being able to metabolise Fuc at all. This adds to the evidence that the composition of the gut microbiota appears to be critical if HMOs are to be effectively utilised.
Ward et al. (2007) also investigated the capability of Bifidobacterium to effectively degrade 2’FL and utilise the resulting Fuc and sialic acid moiety with B. longum bv. infantis ATCC 15697 achieving both the highest growth rate and the highest Fuc consumption amongst the majority of species and strains of Bifidobacterium tested, whereas B. breve ATCC 1570 was only able to achieve intermediate levels of growth with only moderate Fuc usage and B. adolescentis and B. bifidum ATCC 15696 exhibiting no growth on either Fuc or sialic acids. These findings are similar to those recorded by Garrido et al. (2015), who noted that while B. longum subsp. infantis ATCC 15697 exhibited excellent growth on both 2’FL and 3’FL, B. bifidum JCM 7004 only exhibited moderate growth on 2’FL, 3’FL and 6’SL with B. animalis JCM 10602 exhibiting no growth, on HMOs at all. Accordingly, Locascio et al. (2007) reported that B. adolescentis ATCC 15703 presented little-to-no sign of growth in the presence of several HMOs, whereas Xiao et al. (2010) found that B. adolescentis ATCC 15704 and 15705 appeared to be unable to utilise HMOs including LNB. This infers that B. adolescentis ATCC 15703, 15704 and 15705 lack the glycosidases and transporters required to assimilate HMOs (LoCascio et al., 2010).
The ability of the B. longum subsp. infantis ATCC 15697 to effectively utilise HMOs can be put down to the presence of five distinct gene clusters (Sela et al., 2008). In the genomic sequencing of B. longum subsp. infantis ATCC 15697 a 43-kb gene cluster dedicated to HMO import encoding 21 genes was identified with one of its four loci encompassing all of the necessary sialidases, fucosidases, galactosidases and hexosaminidases required for transporting and metabolising HMOs (LoCascio et al., 2007, 2009; Sela et al., 2008). Several other strains and subspecies of bifidobacteria, including B. longum subsp. longum DJO10AB and B. adolescentis ATCC 15703, possess fewer than 11 genes where the lack of SBPs results in an inability to effectively utilise HMOs (Lee et al., 2008; LoCascio et al., 2009).
It is not just Bifidobacterium which possesses the ability to utilise and exhibit growth on HMOs. In addition to Bifidobacterium, several strains of Bacteroides, including B. fragilis and B. vulgatus, are also known prominent consumers of HMOs (Marcobal et al., 2010). Of these, two strains of B. fragilis ATCC2585 can effectively utilise a full range of HMOs, albeit displaying a preference for non-fucosylated HMOs, recording an overall consumption between 25 and 90 per cent. These findings are substantially higher than B. vulgatus ATCC8482, which can also utilise a full range of HMOs, again displaying a preference for fucosylated HMOs. This strain displayed much lower consumption rates of total HMOs than B. fragilis ATCC2585 at 16–40 per cent, respectively (Marcobal et al., 2010), the difference being the presence of Fuc-specific GH95 and GH29 glycoside hydrolases.
Furthermore, in contrast to Bifidobacterium and Bacteroides, several strains of Enterobacteriaceae, including EC1000, EC11775, EC29425 and SD13313, appear to be unable to utilise several HMOs, including 2‘FL and 6′SL while displaying limited growth on LNnT. However, these strains could also readily utilise Glc, maltodextrin and GOS as a sole carbon source in pure cultures (Hoeflinger et al., 2015).
Mixed culture/faecal inoculum
The ability of 2’FL to alter the composition of the gut microbiota has been investigated using an in vitro Simulator of Human Intestinal Microbial Ecosystem (SHIME) model using faecal samples from 6-month-old infants had been exclusively formula-fed (Van den Abbeele et al., 2019). The authors noted that 2’FL increased the relative abundance of bifidobacteria and butyrate-producing bacteria, shifting the distribution of Bifidobacterium spp. from B. bifidum towards B. adolescentis: an interesting finding given that, as previously discussed, B. adolescentis appears to be unable to utilise whole HMOs. This likely indicates that B. adolescentis can exploit products of the degradation of HMOs by other microbial community members (Thongaram et al., 2017).
Increases in the concentration of acetate and butyrate were seen in both parts of the distal and proximal colon of the SHIME model, with levels of propionate displaying a more rapid increase in the distal part of the colon upon 2’FL dosing. This is consistent with Vester Boler et al. (2013), who reported that 2’FL was rapidly fermented upon inoculation with mixed faecal cultures. However, the bifidogenic effect of 2’FL appeared to be donor-specific with increases in numbers of bifidobacteria only being observed in the proximal colon of one donor. In another, donor increases in bifidobacteria were observed in both the proximal and distal colon.
Additionally, the authors reported that increases in the concentration of acetate and butyrate detected were seen in both parts of the distal and proximal colon of the SHIME model with levels of propionate recording a more rapid increase in the distal part of the colon upon 2’FL dosing. Thus, from these results, it suggests that the microbiota dependence of individual rates of fermentation of 2’FL is likely to be highly variable between subjects (Marcobal and Sonnenburg, 2012). Yet, the supplementation of 2’FL in this study was undertaken at 2 g/L, approximately twice the concentration of 2‘FL found in formula milks currently for sale on the market (SMA Nutrition, 2020), indicating that the results generated by this study may not give a fair reflection of what might transpire in real life.
In another study conducted by Salli et al. (2019) using faecal samples from healthy infants aged below 1 year, the effects of 2’FL on the composition and metabolites of the infant microbiota were investigated using a semi-continuous colon simulator and was compared against GOSs with Lac as a control. Changes in microbial composition and metabolites were measured via 16S RNA amplicon sequencing and gas chromatography. From the results, it was noted that 2’FL recorded similar increases in numbers of total bacteria, Firmicutes and Actinobacteria (including bifidobacteria) compared to GOS, but 2’FL was unable to match GOS in reductions of numbers of Proteobacteria. Furthermore, levels of SCFAs and lactic acid produced by 2’FL were only half compared with those of GOS, suggesting that at least in this regard GOS results in a greater generation of metabolites associated with beneficial health outcomes than supplementation of 2’FL on its own.
Interestingly, Li et al. (2012) noted that in the in vitro fermentation of piglet faeces, LNnT recorded the largest increases in levels of acetate and butyrate compared with FOS, GOS/polydextrose mixture and pooled HMOs, whereas pooled HMOs and FOS recorded higher levels of propionate and lactate. Furthermore, both pooled HMOs and LNnT were able to stimulate changes in the microbial composition, including increasing numbers of total bacteria, Bifidobacterium, Lactobacillus, B. vulgatus and Clostridium cluster XIVa along with resulting in reductions in Clostridium cluster IV, suggesting that both pooled and single HMOs can drive beneficial changes in microbial composition. However, despite differences being detected in microbial composition, both pooled HMOs and LNnT appeared to be no more effective in stimulating changes in microbial composition compared with both FOS and the GOS/polydextrose mixture, respectively.
The metabolic by-products and fermentation characteristics of prebiotics, including GOS, 2’FL, LNnT, 6’SL, high-performance inulin (HP) and gum arabic, were investigated by Vester Boler et al. (2013) using faecal samples isolated from both breast and formula-fed infants using an in vitro fermentation model. From the results, it was noted that the rates of fermentation of prebiotics differed significantly between breastfed and formula-fed infants inocula. For example, formula-fed inocula generated higher levels of acetate compared with breastfed inocula (P < 0.001) with 6’SL producing the largest concentration of acetate after 12-h fermentation. However, acetate production also varied over time between substrates with GOS generating large quantities of acetate regardless of diet. Butyrate appeared to be less affected by substrate or diet, but was higher in formula-fed inocula compared with breastfed inocula overall (P < 0.01); however, no differences were detected at any individual time point. Conversely, propionate was affected by diet, but more so by substrate and time with 6’SL producing large amounts of propionate after 12 h of fermentation. Moreover, the fermentation of 2’FL seemingly levelled off after 6 h, further indicating that 2’FL likely undergoes rapid fermentation upon inoculation (Salli et al., 2019). Finally, regarding microbial composition, numbers of bifidobacteria increased, whereas numbers of E. coli and Clostridium perfringens decreased regardless of the substrate used.
While using a pH-controlled in vitro fermentation model involving faecal donors from healthy, Irritable bowel syndrome (IBS) and ulcerative colitis patients, the most noticeable changes in gut microbiota composition were in Bifidobacterium (P < 0.01), while supplementation of 2’FL also resulted in increased numbers of Eubacterium rectale and Clostridium coccoides after 8- and 24-h fermentation [P < 0.01 (8 h) and P < 0.05 (24 h)] in healthy and [P < 0.01 (8 and 24 h)] IBD-ulcerative colitis donors. Significant increases in Roseburia at 8 h fermentation were seen in both healthy and IBS but not inflammatory bowel disease (IBD)-ulcerative colitis donors. Interestingly, in both IBD-ulcerative colitis and IBS patients but not healthy donors, there were significant increases in Atopobium cluster at 8 and 24 h (P < 0.01; Ryan et al., 2021).
These results further add to the evidence that the ability of HMOs and 2’FL to stimulate changes in the microbiota and its resulting metabolites appears to be highly specific and restricted to certain species, strains and subspecies of microbes as well as the initial composition of the gut microbiota (Gotoh et al., 2018; Sakanaka et al., 2020; Yu et al., 2013).
Cross-feeding: a strategy to ensure dominance?
As previously discussed, the gut microbiota, in particular bifidobacteria, have developed several strategies to colonise and dominate the microbiota of an infant’s gut with some strains, species and subspecies, displaying better potential than others. Interestingly, to help drive the colonisation of the gut, Bifidobacterium, Bacteroides as well as several other genera, including Akkermansia, Anaerostipes and Roseburia, have developed another strategy based on cross-feeding. Strains and species of Bifidobacterium and Bacteroides that are not able to utilise whole HMOs can feed on metabolites resulting from exploitation of HMOs by other species and strains (White et al., 2014).
In breastfed infants, B. bifidum is said to make up of over 10 per cent of the total number of bifidobacteria present within their gut (Sakanaka et al., 2019). When B. bifidum is in abundance, the corresponding microbiota appears to follow suit with higher numbers of several other bifidobacterial species and strains also being recorded (Tannock et al., 2013). The potential for B. bifidum to act as cross-feeders for other members of the Bifidobacterium genus was noted by Asakuma et al. (2011), who documented that B. bifidum left several HMO components, including Fuc and Gal in spent media, indicating that extracellular degradation had occurred and suggesting that non-HMOs utilising species/subspecies may be able to exploit these monosaccharide moieties (Kitaoka, 2012).
Additionally, using faecal suspensions isolated from infants, children and adults in a mucin-based medium supplemented with HMOs, Egan et al. (2014) and Gotoh et al. (2018) recorded the ability of several species and strains of bifidobacteria to grow in the presence and absence of B. bifidum. The overall findings of these studies suggest that in faecal suspensions possessing B. bifidum, the numbers of several bifidobacteria species and strains, including B. longum 105-A and B. breve UCC2003, increased; strains not known to effectively utilise whole HMOs to any real extent. It seems that B. bifidum is likely to be a prominent player in the establishment of the microbiota in early life (Kitaoka, 2012).
It was reported in a single ecosystem that 2′FL derived metabolites from B. pseudocatenulatum strains LH9, LH11, LH13 and LH14 supported the growth of several non-HMOs utilising strains including B. longum LH12. However, B. longum subsp. infantis LH23 2′FL degradation products did not support the growth of B. breve (LH21 and LH24), respectively. Additionally, with B. longum LH206 2′FL conditioned media, increases in numbers of all strains of B. longum subsp. infantis and B. pseudocatenulatum tested were seen. This indicates that the metabolism of 2′FL by B. infantis LH206 may generate a wider variety of growth-promoting compounds (Lawson et al., 2020).
It has been documented in a co-culture experiment that Anaerostipes cacae was able to utilise monosaccharides, as well as lactate and acetate, resulting from HMOs degradation by B. Infantis (Chia et al., 2021). Roseburia spp. were able to grow in the presence of A. muciniphila, whereas in pure culture Roseburia spp. showed little-to-no sign of growth (Pichler et al., 2020). The ability of Bacteroides to act as primary degraders of HMOs was demonstrated in mice-fed sialylated HMOs, including 3’SL and 6’SL, when a marked increase in Enterobacteriaceae was seen. This led to an exacerbation of the pro-inflammatory response (Huang et al., 2015).
Additionally, in antibiotic-treated germ-free mice infected with either Salmonella typhimurium or C. difficile, it has been demonstrated that S. typhimurium was able to access both Fuc and sialic acid and C. difficile was able to readily utilise sialic acid as a result of breakdown of host carbohydrates by Bacteroides thetaiotaomicron (Ng et al., 2013). However, while the expansion of enteric bacterial pathogens via the utilisation of HMOs is sometimes seen in vitro in co-cultures or using in vivo mechanistic disease state rodent models, in the highly complex ecosystem in the human gut, this has never been reported.
Not all microorganisms found within the gut can participate in cross-feeding due to intracellular metabolism of polysaccharides/glycans. The inability of specific bifidobacteria to act as cross-feeders was demonstrated by Garrido et al. (2016). B. longum SC596 exhibited excellent growth on both Type-I and Type-II chain HMOs, albeit displaying a preference for fucosylated HMOs; however, no monosaccharides from HMOs degradation were detected in the medium. As the degradation of HMOs by B. longum SC596 appears to be similar to that of B. longum subsp. infantis ATCC 15697, which uses intracellular metabolism (Garrido et al., 2013), this suggests that B. longum SC596 cannot partake in the cross-feeding of other microorganisms.
These results suggest that the mutualistic behaviour which exists between microorganisms found in the gut likely influences the rates at which metabolites such as SCFAs are generated (Comstock, 2009). From this, one could conclude that the degree to which this mutualistic behaviour exists between microorganisms found in the gut not only increases the diversity of the gut microbiota, but is maybe one of the most critical characteristics in helping to shape a flexible, healthy ecosystem (Gotoh et al., 2018).
The influence of human milk oligosaccharides on infant microbiota composition in vivo
As previously discussed, the ability of HMOs to alter microbial composition in infants has been studied extensively using in vitro test conditions, but less so in vivo with only a limited number of studies being undertaken to date.
In a proof-of-concept study (De Leoz et al., 2015), serial faecal samples were collected from two vaginally born infants. Infant A was breastfed directly from birth, whereas infant B received formula supplementation for 4 days from days 2–6 and then was solely breastfed thereafter. Faecal samples were collected twice per week for the first month, twice per month in the second month and once or twice per month thereafter. Microbial compositions were analysed via 16S rRNA sequencing. The results demonstrated that after an initial rise in non-HMO-consuming bacteria, including Enterobacteriaceae and Staphylococcaeae, large shifts in microbial composition from non-HMO-consuming bacteria to HMO-consuming bacteria Bacteroidaceae and Bifidobacteriaceae were seen. Yet, large differences were seen between both donors whereby week 13 Bifidobacterium spp. dominated in infant A, and levels of most faecal HMOs dropped dramatically, whereas by week 14, Bacteroides spp. were most dominant in infant B.
Borewicz et al. (2019, 2020) aimed to correlate the HMOs in breast milk with changes in faecal microbiota composition, analysed via Illumina HiSeq 16S rRNA gene sequencing, in healthy 2-, 4-, 6- and 12-week-old breastfed infants. Unsurprisingly, the ability of infants to utilise HMOs, including 2’FL, was associated with differences in the faecal microbiota composition, with those infants possessing relatively high abundances of Bifidobacterium 418, 614 and 681 and Lactobacillus 744 (FDR < 0.05) reporting higher rates of 2’FL consumption. Additionally, infants who recorded higher consumptions rates of LNT and LNnT, LNFP III, LNFP II and lacto-N-hexaose (LNH) possessed significantly higher relative abundances of Bifidobacterium OTUs 418, 406, 643, 658, 423, 1335 and 597 and Bacteroides 144 (FDR < 0.05). Moreover, the degradation of sialylated HMOs 3′SL, 6′SL, LST a, LST b and LST c appeared to be more highly associated with Bacteroides; a finding confirming those reported by Yu et al. (2013), who demonstrated using in vitro models that B. fragilis, B. vulgatus and B. thetaiotaomicron could utilise 3’SL and 6’SL as sole carbon sources.
Interestingly, Borewicz et al. (2019, 2020) also recorded that lactobacilli appeared to thrive in the presence of 2′FL, DFL, LNDFH I, LNT, LNnT and LNFP II. This is fascinating given that it has been shown repeatedly in several in vitro studies that lactobacilli appear to be unable to utilise HMOs (Schwab and Ganzle, 2011; Ward et al., 2006). This suggests that lactobacilli might be able to thrive in the infant’s gut via cross-feeding, scavenging any resulting metabolites, including Fuc and lactose from the extracellular degradation of HMOs (Zuniga et al., 2018). This likely infers that the degradation of HMOs strongly correlates with the microbiota and specifically with the relative abundance of the phylotypes Bifidobacterium, Bacteroides and Lactobacillus present within an infant’s gut.
Moreover, in a randomised, double-blind, multicentre clinical trial, healthy full-term infants (aged 0–14 days) were fed infant formula with no added HMOs (control), or the same formula with the addition of 2’FL and LNnT for a timeframe of 6 months. Thereafter, all infants were fed the same non-HMO-containing infant formula (Berger et al., 2020). Results were analysed against a breastfed reference group with changes in microbial community types being analysed at 3 and 12 months via 16S rRNA gene sequencing. The results indicated that, compared with the breastfed reference group, the HMO-containing formula stimulated increases in Bifidobacterium, albeit to a lower degree than the reference breastfed group. Levels of Escherichia were, however, significantly lower in the HMO-containing formula group compared with the control group and were similar to those in the breastfed group. Numbers of Peptostreptococcaceae were far higher in the control and HMO-containing formula group compared with the breastfed group. Yet, at 12 months, no differences were detected between the two formula groups. This suggests that the supplementation of infants with HMO-containing formulae may offset some of the ill effects associated with not breastfeeding from birth. However, this study is not without limitation. First, only two faecal samples were collected: one at 3 months and one at 12 months. Second, no data were collected on day-care attendance and when solid foods were introduced (weaning) which, due to the effects these factors have on the microbial composition (McBurney et al., 2019), may have introduced biases into the results. Consequently, further investigation into this area would be highly beneficial to determine the true effects that both 2’FL and LNnT have on altering the composition of healthy infants in vivo.
In another study, differences in gut microbiota composition between caesarean and vaginally born babies of α1-2 fucosylated secreting mothers were conducted by Tonon et al. (2021). In this study, faecal microbiota composition from caesarean and vaginally born infants was analysed by 16S rRNA gene sequencing and qPCR with results being stratified by secretor status. The authors concluded that levels of Bifidobacterium were similar between caesarean and vaginally born infants of secretor mothers. Yet, there were differences between caesarean and vaginally born infant microbiotas with the caesarean born infants from secretors possessing higher amounts of Kluyvera and Veillonella. Vaginally born infants from secretor mothers possessed higher amounts of Bacteroides. This further adds to the evidence that mode of delivery may likely impact on proliferation of the gut microbiota and HMO utilisation.
In addition to healthy infant’s, the effects of HMOs on the gut microbiota and health outcomes also been studied in preterm infants. In one study, 12 premature infants were randomised into two groups – one group containing formula and increasing doses of short-chain GOSs (degree of polymerisation < 8) and the other group receiving formula + HMOs (Underwood et al., 2014). The authors noted that relative abundances of clostridia increased with increasing doses of HMOs. The authors also noted that there were trends towards increase of γ-proteobacteria over time/dose in preterm infants feed formula + HMOs.
Additionally, in a study involving preterm infants with necrotising enterocolitis (NEC), it was noted that infants with NEC possessed higher levels of Proteobacteria and lower levels of Actinobacteria at phylum levels, along with lower relative abundances of B. longum and higher relative abundances of Enterobacter cloacae. The authors also noted that the composition of breast milk, specifically lower concentration of DSLNT, was associated with the likelihood of developing NEC (Masi et al., 2021). While, similarly, Underwood et al. (2015) concluded that preterm infants of non-secretor mothers possessed higher levels of Proteobacteria and lower levels of Firmicutes, secretor mothers possess specific fucosylated HMOs including LDFT and LNFP V, which may be associated with lower levels of Enterobacteriaceae and potentially protective effect against pathogens associated with NEC. These results potentially infer that the composition of HMOs present in breast milk may be a contributing factor towards the development of NEC in preterm infants.
Moreover, in another study conducted in healthy rats, Chleilat et al. (2020) investigated the effects that the supplementation of 2′FL and 3′SL either together or on their own had on microbial composition compared to a non-HMOs control. In general, all HMO-fortified diets altered gut microbiota composition. However, larger increases in Bifidobacterium spp. were recorded in the 2′FL group compared with the 3′SL-fortified group (P = 0.03). Additionally, A. muciniphila numbers were significantly lower in 3′SL + 2′FL group compared with the control (P < 0.01), respectively.
In a study involving mice supplemented with the 2’FL and 2’FL consuming strain B. pseudocatenulatum MP80, it was noted that 2’FL created an environment that allowed B. pseudocatenulatum MP80 to thrive, along with finding that 2’FL increased the Bifidobacteriaceae relative abundance, as well as resulting in higher log ratios of Bacteroidaceae and Bifidobacteriaceae relative to Lachnospiraceae and Ruminococcaceae amplicon sequencing variant (P = 0.003).
These results help to explain the large variability in the presence and levels of HMOs detected in the faecal samples of infants, even when secretor status is considered, with virtually no HMOs being detected in faecal samples of several infants, whereas, in the faecal samples of other infants, there was a strong presence of non-fucosylated HMOs, suggesting the presence of the fucoside-utilising microorganisms needed to degrade fucosylated HMOs (Asakuma et al., 2011). In faecal samples of several other infants, large quantities of LNnT were detected with LNnT not being detected in others. However, despite these differences, a common characteristic amongst nearly all infants used in these studies was what appeared to be the presence of several new, non or partially intact HMOs and HMOs by-products in faecal samples with the majority of new HMOs detected displaying a high proportion of HexNAc (Davis et al., 2016; De Leoz et al., 2013; Dotz et al., 2015).
Thus, while results seemingly imply that the supplementation of 2’FL and LNnT may contribute towards a positive shift in the composition of the gut microbiota, in reality, the make-up of the infant’s microbiota is shaped through several often complex and interacting factors from birth, including mode of delivery (vaginal vs. c-section), feeding practices (breast vs. bottle feeding) and age at which the introduction of complex dietary substrates occurs (weaning). The use of gut microbiome altering medications and supplements, namely antibiotics and probiotics (Bertelsen et al., 2016; McBurney et al., 2019), will also have an impact. Consequently, the degradation of HMOs will differ greatly depending on the relative abundance of specific species, strains and subspecies of microorganisms present within an individual infant’s gut microbiome. More detailed analysis is needed for infant microbiota prior to supplementation in such studies. The supplementation of 2’FL and LNnT in infant formula milk may be of little-to-no benefit to many infants as several infants especially those who were never breastfed may not possess the necessary microorganisms and thus glycosidases and transporters needed to effectively utilise these specific HMOs.
Acknowledgement
Peter Jackson would like to thank Prof Bob Rastall and Dr Anisha Wijeyesekera for their support and feedback in writing this manuscript.
Disclosure statement
No potential conflicts of interest were reported by the authors.
Research transparency and reproducibility
The opinions/views contained within this article are those of the authors.
Author contributions
Conceptualisation: P.P.J.J, R.A.R and A.W; Formal analysis: P.P.J.J.; Funding acquisition: R.A.R.; Investigation: P.P.J.J.; Methodology: all authors; Project administration: R.A.R.; Supervision: R.A.R and A.W.; Visualisation: all authors; Writing – original draft: P.P.J.J.; Writing – review and editing: all authors. P.P.J.J. drafted and revised all manuscript drafts following feedback from co-authors R.A.R and A.W.
Funding
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
References
- Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J and Kitaoka M (2011) Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. The Journal of Biological Chemistry 286(40), 34583–34592. 10.1074/jbc.M111.248138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S, De Castro CA, Sprenger N, Binia A, Affolter M, Garcia-Rodenas CL, Beauport L, Tolsa JF and Fumeaux CJF (2019) Human milk oligosaccharides in the milk of mothers delivering term versus preterm infants. Nutrients 11(6), 1282. 10.3390/nu11061282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayechu-Muruzabal V, van Stigt AH, Mank M, Willemsen LEM, Stahl B, Garssen J and van’t Land B (2018) Diversity of human milk oligosaccharides and effects on early life immune development. Frontiers in Pediatrics 6, 239. 10.3389/fped.2018.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao YW, Chen C and Newburg DS (2013) Quantification of neutral human milk oligosaccharides by graphitic carbon high-performance liquid chromatography with tandem mass spectrometry. Analytical Biochemistry 433(1), 28–35. 10.1016/j.ab.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile D and Rastall RA (2013) Human milk and related oligosaccharides as prebiotics. Current Opinion in Biotechnology 24(2), 214–219. 10.1016/j.copbio.2013.01.008 [DOI] [PubMed] [Google Scholar]
- Berger B, Porta N, Foata F, Grathwohl D, Delley M, Moine D, Charpagne A, Siegwald L, Descombes P, Alliet P, Puccio G, Steenhout P, Mercenier A and Sprenger N (2020) Linking human milk oligosaccharides, infant fecal community types, and later risk to require antibiotics. mBio 11(2). 10.1128/mBio.03196-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen RJMP, Jensen ETMPHP and Ringel-Kulka TMDMPH (2016) Use of probiotics and prebiotics in infant feeding. Best Practice & Research: Clinical Gastroenterology 30(1), 39–48. 10.1016/j.bpg.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Bhargava P, Li CL, Stanya KJ, Jacobi D, Dai LL, Liu SH, Gangl MR, Harn DA and Lee CH (2012) Immunomodulatory glycan LNFPIII alleviates hepatosteatosis and insulin resistance through direct and indirect control of metabolic pathways. Nature Medicine 18(11), 1665. 10.1038/nm.2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank D, Dotz V, Geyer R and Kunz C (2012) Human milk oligosaccharides and Lewis blood group: Individual high-throughput sample profiling to enhance conclusions from functional studies. Advances in Nutrition 3(3), 440S–449S. 10.3945/an.111.001446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L (2009) Human milk oligosaccharides: Prebiotics and beyond. Nutrition Reviews 67(11), S183–S191. 10.1111/j.1753-4887.2009.00239.x [DOI] [PubMed] [Google Scholar]
- Borewicz K, Gu F, Saccenti E, Hechler C, Beijers R, de Weerth C, van Leeuwen SS, Schols HA and Smidt H (2020) The association between breastmilk oligosaccharides and faecal microbiota in healthy breastfed infants at two, six, and twelve weeks of age. Scientific Reports, 10(1), 4270. 10.1038/s41598-020-61024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borewicz K, Gu FJ, Saccenti E, Arts ICW, Penders J, Thijs C, van Leeuwen SS, Lindner C, Nauta A, van Leusen E, Schols HA and Smidt H (2019) Correlating infant fecal microbiota composition and human milk oligosaccharide consumption by microbiota of 1-month-old breastfed infants. Molecular Nutrition & Food Research 63(13), 1801214. 10.1002/mnfr.201801214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunesova V, Lacroix C and Schwab C (2016) Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiology 16. 10.1186/s12866-016-0867-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia, L. W., Mank, M., Blijenberg, B., Bongers, R. S., van Limpt, K., Wopereis, H., Tims, S., Stahl, B., Belzer, C., & Knol, J. (2021). Cross-feeding between Bifidobacterium infantis and Anaerostipes caccae on lactose and human milk oligosaccharides. Beneficial Microbes, 12(1), 69‑83. 10.3920/bm2020.0005 [DOI] [PubMed] [Google Scholar]
- Chleilat F, Klancic T, Ma K, Schick A, Nettleton JE and Reimer RA (2020) Human milk oligosaccharide supplementation affects intestinal barrier function and microbial composition in the gastrointestinal tract of young Sprague Dawley rats. Nutrients 12(5), 1532. 10.3390/nu12051532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock LE (2009) Importance of glycans to the host-Bacteroides mutualism in the mammalian intestine. Cell Host & Microbe 5(6), 522–526. 10.1016/j.chom.2009.05.010 [DOI] [PubMed] [Google Scholar]
- Davis JCC, Totten SM, Huang JO, Nagshbandi S, Kirmiz N, Garrido DA, Lewis ZT, Wu LD, Smilowitz JT, German JB, Mills DA and Lebrilla CB (2016) Identification of oligosaccharides in feces of breast-fed infants and their correlation with the gut microbial community. Molecular & Cellular Proteomics 15(9), 2987–3002. 10.1074/mcp.M116.060665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leoz MLA, Gaerlan SC, Strum JS, Dimapasoc LM, Mirmiran M, Tancredi DJ, Smilowitz JT, Kalanetra KM, Mills DA, German JB, Lebrilla CB and Underwood MA (2012) Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. Journal of Proteome Research 11(9), 4662–4672. 10.1021/pr3004979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leoz MLA, Wu S, Strum JS, Ninonuevo MR, Gaerlan SC, Mirmiran M, German JB, Mills DA, Lebrilla CB and Underwood MA (2013) A quantitative and comprehensive method to analyze human milk oligosaccharide structures in the urine and feces of infants. Analytical and Bioanalytical Chemistry 405(12), 4089–4105. 10.1007/s00216-013-6817-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leoz, M. L. A., Kalanetra, K. M., Bokulich, N. A., Strum, J. S., Underwood, M. A., German, J. B., Mills, D. A., & Lebrilla, C. B. (2014). Human Milk Glycomics and Gut Microbial Genomics in Infant Feces Show a Correlation between Human Milk Oligosaccharides and Gut Microbiota: A Proof-of-Concept Study. Journal of Proteome Research, 14(1), 491‑502. 10.1021/pr500759e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty AM, Lodge CJ, Dharmage SC, Dai X, Bode L and Lowe AJ (2018) Human milk oligosaccharides and associations with immune-mediated disease and infection in childhood: A systematic review. Frontiers in Pediatrics 6, 91. 10.3389/fped.2018.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan SM and Comstock SS (2016) Human milk oligosaccharides influence neonatal mucosal and systemic immunity. Annals of Nutrition and Metabolism 69, 42–51. 10.1159/000452818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotz V, Rudloff S, Blank D, Lochnit G, Geyer R and Kunz C (2014) 13C-labeled oligosaccharides in breastfed infants’ urine: Individual-, structure- and time-dependent differences in the excretion. Glycobiology 24(2), 185–194. 10.1093/glycob/cwt099 [DOI] [PubMed] [Google Scholar]
- Dotz V, Rudloff S, Meyer C, Lochnit G and Kunz C (2015) Metabolic fate of neutral human milk oligosaccharides in exclusively breast-fed infants. Molecular Nutrition & Food Research 59(2), 355–364. 10.1002/mnfr.201400160 [DOI] [PubMed] [Google Scholar]
- Egan, M., O’Connell Mtherway, M., Kilcoyne, M., Kane, M., Joshi, L., Ventura, M., & van Sinderen, D. (2014). Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiology, 14(1). 10.1186/s12866-014-0282-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egge H, Dell A and Von Nicolai H (1983) Fucose containing oligosaccharides from human milk. I. Separation and identification of new constituents. Archives of Biochemistry and Biophysics 224(1), 235–253. 10.1016/0003-9861(83)90207-2 [DOI] [PubMed] [Google Scholar]
- Fong B, Ma K and McJarrow P (2011) Quantification of bovine milk oligosaccharides using liquid chromatography-selected reaction monitoring-mass spectrometry. Journal of Agricultural and Food Chemistry 59(18), 9788–9795. 10.1021/jf202035m [DOI] [PubMed] [Google Scholar]
- Gabrielli O, Zampini L, Galeazzi T, Padella L, Santoro L, Peila C, Giuliani F, Bertino E, Fabris C and Coppa GV (2011) Preterm milk oligosaccharides during the first month of lactation. Pediatrics 128(6), E1520–E1531. 10.1542/peds.2011-1206 [DOI] [PubMed] [Google Scholar]
- Garrido D, Barile D and Mills DA (2012a) A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Advances in Nutrition 3(3), 415S–421S. 10.3945/an.111.001586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Dallas DC and Mills DA (2013) Consumption of human milk glycoconjugates by infant-associated bifidobacteria: Mechanisms and implications. Microbiology 159, 649–664. 10.1099/mic.0.064113-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Kim JH, German JB, Raybould HE and Mills DA (2011) Oligosaccharide binding proteins from Bifidobacterium longum subsp infantis reveal a preference for host glycans. PLoS One 6(3). 10.1371/journal.pone.0017315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, Lemay DG, Ugalde JA, German JB, Lebrilla CB and Mills DA (2016) A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp longum SC596. Scientific Reports 6, 35045. 10.1038/srep35045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S, Lemay DG, Sela DA, German JB and Mills DA (2015) Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Scientific Reports 5, 13517. 10.1038/srep13517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S and Mills DA (2012b) Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp infantis. Anaerobe 18(4), 430–435. 10.1016/j.anaerobe.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring KC, Kennedy AD, Prieto PA and Buck RH (2014) Direct evidence for the presence of human milk oligosaccharides in the circulation of breastfed infants. PLoS One 9(7), e101692. 10.1371/journal.pone.0101692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh A, Katoh T, Sakanaka M, Ling YW, Yamada C, Asakuma S, Urashima T, Tomabechi Y, Katayama-Ikegami A, Kurihara S, Yamamoto K, Harata G, He F, Hirose J, Kitaoka M, Okuda S and Katayama T (2018) Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Scientific Reports 8, 13958. 10.1038/s41598-018-32080-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgren P, Lindberg BS and Lundblad A (1977) Quantitation of some urinary oligosaccharides during pregnancy and lactation. Journal of Biological Chemistry 252(3), 1034–1040. 10.1016/S0021-9258(19)75202-8 [DOI] [PubMed] [Google Scholar]
- Heiss, B. E., Ehrlich, A. M., Maldonado-Gomez, M. X., Taft, D. H., Larke, J. A., Goodson, M. L., Slupsky, C. M., Tancredi, D. J., Raybould, H. E., & Mills, D. A. (2021). Bifidobacterium catabolism of human milk oligosaccharides overrides endogenous competitive exclusion driving colonization and protection. Gut Microbes, 13(1). 10.1080/19490976.2021.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegar B, Wibowo Y, Basrowi RW, Ranuh RG, Sudarmo SM, Munasir Z, Atthiyah AF, Widodo AD, Supriatmo, Kadim M, Suryawan A, Diana NR, Manoppo C and Vandenplas Y (2019) The role of two human milk oligosaccharides, 2’-fucosyllactose and lacto-N-neotetraose, in infant nutrition. Pediatric Gastroenterology Hepatology & Nutrition 22(4), 330–340. 10.5223/pghn.2019.22.4.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrick, B. M., Rodriguez, L., Lakshmikanth, T., Pou, C., Henckel, E., Arzoomand, A., Olin, A., Wang, J., Mikes, J., Tan, Z., Chen, Y., Ehrlich, A. M., Bernhardsson, A. K., Mugabo, C. H., Ambrosiani, Y., Gustafsson, A., Chew, S., Brown, H. K., Prambs, J., … Brodin, P. (2021). Bifidobacteria-mediated immune system imprinting early in life. Cell, 184(15), 3884–3898.e11. 10.1016/j.cell.2021.05.030 [DOI] [PubMed] [Google Scholar]
- Hirschmugl B, Brandl W, Csapo B, van Poppel M, Kofeler H, Desoye G, Wadsack C and Jantscher-Krenn E (2019) Evidence of human milk oligosaccharides in cord blood and maternal-to-fetal transport across the placenta. Nutrients 11(11), 2640. 10.3390/nu11112640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflinger JL, Davis SR, Chow J and Miller MJ (2015) In vitro impact of human milk oligosaccharides on Enterobacteriaceae growth. Journal of Agricultural and Food Chemistry 63(12), 3295–3302. 10.1021/jf505721p [DOI] [PubMed] [Google Scholar]
- Hornef M and Penders J (2017) Does a prenatal bacterial microbiota exist? Mucosal Immunology 10(3), 598–601. 10.1038/mi.2016.141 [DOI] [PubMed] [Google Scholar]
- Huang YL, Chassard C, Hausmann M, von Itzstein M and Hennet T (2015) Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nature Communications 6, 8141. 10.1038/ncomms9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K, Motherway MO, Bottacini F and van Sinderen D (2016) Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Scientific Reports 6, 38560. 10.1038/srep38560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantscher-Krenn E, Marx C and Bode L (2013) Human milk oligosaccharides are differentially metabolised in neonatal rats. British Journal of Nutrition 110(4), 640–650. 10.1017/S0007114512005727 [DOI] [PubMed] [Google Scholar]
- Juge N, Tailford L and Owen CD (2016) Sialidases from gut bacteria: A mini-review. Biochemical Society Transactions 44, 166–175. 10.1042/BST20150226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka M (2012) Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Advances in Nutrition 3(3), 422S–429S. 10.3945/an.111.001420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H and Yamamoto K (2011) An exo-alpha-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology 21(4), 437–447. 10.1093/glycob/cwq175 [DOI] [PubMed] [Google Scholar]
- Kostopoulos, I., Elzinga, J., Ottman, N., Klievink, J. T., Blijenberg, B., Aalvink, S., Boeren, S., Mank, M., Knol, J., de Vos, W. M., & Belzer, C. (2020). Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro. Scientific Reports, 10(1). 10.1038/s41598-020-71113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M and Kozyrskyj AL (2017) Gut microbial metabolism defines host metabolism: An emerging perspective in obesity and allergic inflammation. Obesity Reviews 18(1), 18–31. [DOI] [PubMed] [Google Scholar]
- Kunz C and Rudloff S (2017) Compositional analysis and metabolism of human milk oligosaccharides in infants. Intestinal Microbiome: Functional Aspects in Health and Disease 88, 137–147. 10.1159/000455398 [DOI] [PubMed] [Google Scholar]
- Lawson MAE, O’Neill IJ, Kujawska M, Javavdi SG, Wijeyesekera A, Flegg Z, Chalklen L and Hall LJ (2020) Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME Journal 14(2), 635–648. 10.1038/s41396-019-0553-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, Weimer B and O’Sullivan DJ (2008) Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9, 247. 10.1186/1471-2164-9-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Bauer LL, Chen X, Wang M, Kuhlenschmidt TB, Kuhlenschmidt MS, Fahey GC and Donovan SM (2012) Microbial composition and in vitro fermentation patterns of human milk oligosaccharides and prebiotics differ between formula-fed and sow-reared piglets. Journal of Nutrition 142(4), 681–689. 10.3945/jn.111.154427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RW, Dee D, Li CM, Hoffman HJ and Grummer-Strawn LM (2014) Breastfeeding and risk of infections at 6 years. Pediatrics 134, S13–S20. 10.1542/peds.2014-0646D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis-Kuberka J and Orczyk-Pawilowicz M (2019) Sialylated oligosaccharides and glycoconjugates of human milk. The impact on infant and newborn protection, development and well-being. Nutrients 11(2), 306. 10.3390/nu11020306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio RG, Desai P, Sela DA, Weimer B and Mills DA (2010) Broad conservation of milk utilization genes in Bifidobacterium longum subsp infantis as revealed by comparative genomic hybridization. Applied and Environmental Microbiology 76(22), 7373–7381. 10.1128/AEM.00675-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA and German JB (2007) Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. Journal of Agricultural and Food Chemistry 55(22), 8914–8919. 10.1021/jf0710480 [DOI] [PubMed] [Google Scholar]
- LoCascio RG, Ninonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB and Mills DA (2009) A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microbial Biotechnology 2(3), 333–342. 10.1111/j.1751-7915.2008.00072.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi, A. C., Embleton, N. D., Lamb, C. A., Young, G., Granger, C. L., Najera, J., Smith, D. P., Hoffman, K. L., Petrosino, J. F., Bode, L., Berrington, J. E., & Stewart, C. J. (2020). Human milk oligosaccharide DSLNT and gut microbiome in preterm infants predicts necrotising enterocolitis. Gut, 70(12), 2273‑2282. 10.1136/gutjnl-2020-322771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB and Mills DA (2010) Consumption of human milk oligosaccharides by gut-related microbes. Journal of Agricultural and Food Chemistry 58(9), 5334–5340. 10.1021/jf9044205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A and Sonnenburg JL (2012) Human milk oligosaccharide consumption by intestinal microbiota. Clinical Microbiology and Infection 18, 12–15. 10.1111/j.1469-0691.2012.03863.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriage BJ, Buck RH, Goehring KC, Oliver JS and Williams JA (2015) Infants fed a lower calorie formula with 2’ FL show growth and 2’ FL uptake like breast-fed infants. Journal of Pediatric Gastroenterology and Nutrition 61(6), 649–658. 10.1097/MPG.0000000000000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, Ogawa E, Kodama H, Yamamoto K, Yamada T, Matsumoto S and Kurokawa K (2016) A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nature Communications 7, 11939. 10.1038/ncomms11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MI, Davis C, Fraser CM, Schneeman BO, Huttenhower C, Verbeke K, Walter J and Latulippe ME (2019) Establishing what constitutes a healthy human gut microbiome: State of the science, regulatory considerations, and future directions. Journal of Nutrition 149(11), 1880–1893. 10.1093/jn/nxz154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath BA, Fox PF, McSweeney PLH and Kelly AL (2016) Composition and properties of bovine colostrum: A review. Dairy Science & Technology 96(2), 133–158. 10.1007/s13594-015-0258-x [DOI] [Google Scholar]
- McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice AM, Kvist LJ, Otoo GE, Brooker SL, Price WJ, Shafii B, Placek C, Lackey KA, Robertson B, Manzano S, Ruiz L, Rodriguez JM, Pareja RG and Bode L (2017) What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. American Journal of Clinical Nutrition 105(5), 1086–1100. 10.3945/ajcn.116.139980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M, Horimoto T, Kiyohara M, Katayama T, Kitaoka M, Ashida H and Yamamoto K (2010) Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology 20(11), 1402–1409. 10.1093/glycob/cwq101 [DOI] [PubMed] [Google Scholar]
- Ninonuevo MR, Park Y, Yin HF, Zhang JH, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm R and Lebrilla CB (2006) A strategy for annotating the human milk glycome. Journal of Agricultural and Food Chemistry 54(20), 7471–7480. 10.1021/jf0615810 [DOI] [PubMed] [Google Scholar]
- Ng, K. M., Ferreyra, J. A., Higginbottom, S. K., Lynch, J. B., Kashyap, P. C., Gopinath, S., Naidu, N., Choudhury, B., Weimer, B. C., Monack, D. M., & Sonnenburg, J. L. (2013). Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature, 502(7469), 96‑99‑. 10.1038/nature12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan, E., & Sela, D. A. (2018). Inefficient Metabolism of the Human Milk Oligosaccharides Lacto-N-tetraose and Lacto-N-neotetraose Shifts Bifidobacterium longum subsp. infantis Physiology. Frontiers in Nutrition, 5. 10.3389/fnut.2018.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler, M. J., Yamada, C., Shuoker, B., Alvarez-Silva, C., Gotoh, A., Leth, M. L., Schoof, E., Katoh, T., Sakanaka, M., Katayama, T., Jin, C., Karlsson, N. G., Arumugam, M., Fushinobu, S., & Abou Hachem, M. (2020). Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nature Communications, 11(1). 10.1038/s41467-020-17075-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza-Diaz J, Fontana L and Gil A (2018) Human milk oligosaccharides and immune system development. Nutrients 10(8), 1038. 10.3390/nu10081038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K, Fitzgerald GF and van Sinderen D (2011) Carbohydrate metabolism in Bifidobacteria. Genes and Nutrition 6(3), 285–306. 10.1007/s12263-010-0206-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polonowski, M. and Lespagnol A. (1931). “Sur deux nouveaux sucres du lait de femme, le gynolactose etl’allolactose.” Compte Rendu de l’ Academie de Science. 192(1319). [Google Scholar]
- Polonowski, M. and Lespagnol A. (1929). “Sur la nature glucidique de la substance lévogyre du lait defemme.” Bulletin de la Societe de Chimie Biologique. 101, (61–63). [Google Scholar]
- Puccio G, Alliet P, Cajozzo C, Janssens E, Corsello G, Sprenger N, Wernimont S, Egli D, Gosoniu L and Steenhout P (2017) Effects of infant formula with human milk oligosaccharides on growth and morbidity: A randomized multicenter trial. Journal of Pediatric Gastroenterology and Nutrition 64(4), 624–631. 10.1097/MPG.0000000000001520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzanowski GG, Garrett PN, Li X and Anita M (2013) Short-chain milk oligosaccharide levels in human milk and infant plasma. FASEB Journal 27, 629.16. 10.1096/fasebj.27.1_supplement.629.16 [DOI] [Google Scholar]
- Robinson RC (2019) Structures and metabolic properties of bovine milk oligosaccharides and their potential in the development of novel therapeutics. Frontiers in Nutrition 6, 50. 10.3389/fnut.2019.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak LR, Stroble C, Underwood MA and Lebrilla CB (2014) Detection of milk oligosaccharides in plasma of infants. Analytical and Bioanalytical Chemistry 406(24), 5775–5784. 10.1007/s00216-014-8025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudloff, S., & Kunz, C. (2012). Milk Oligosaccharides and Metabolism in Infants. Advances in Nutrition, 3(3), 398S–405S. 10.3945/an.111.001594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, J. J., Monteagudo-Mera, A., Contractor, N., & Gibson, G. R. (2021). Impact of 2′-Fucosyllactose on Gut Microbiota Composition in Adults with Chronic Gastrointestinal Conditions: Batch Culture Fermentation Model and Pilot Clinical Trial Findings. Nutrients, 13(3), 938. 10.3390/nu13030938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao J, Kitaoka M and Katayama T (2020) Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: Prevalence of the gene set and its correlation with Bifidobacteria-rich microbiota formation. Nutrients 12(1), 71. 10.3390/nu12010071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanaka M, Hansen ME, Gotoh A, Katoh T, Yoshida K, Odamaki T, Yachi H, Sugiyama Y, Kurihara S, Hirose J, Urashima T, Xiao JZ, Kitaoka M, Fukiya S, Yokota A, Lo Leggio L, Abou Hachem M and Katayama T (2019) Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Science Advances 5(8). 10.1126/sciadv.aaw7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurama H, Kiyohara M, Wada J, Honda Y, Yamaguchi M, Fukiya S, Yokota A, Ashida H, Kumagai H, Kitaoka M, Yamamoto K and Katayama T (2013) Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. Journal of Biological Chemistry 288(35), 25194–25206. 10.1074/jbc.M113.484733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salli K, Anglenius H, Hiryonen J, Hibberd AA, Ahonen I, Saarinen MT, Tiihonen K, Maukonen J and Ouwehand AC (2019) The effect of 2’-fucosyllactose on simulated infant gut microbiome and metabolites; a pilot study in comparison to GOS and lactose. Scientific Reports 9, 13232. 10.1038/s41598-019-49497-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel TM, Binia A, de Castro CA, Thakkar SK, Billeaud C, Agosti M, Al-Jashi I, Costeire MJ, Marchinr G, Martinez-Costa C, Picaud JC, Stiris T, Stoicescu SM, Vanpee K, Domellof M, Austin S and Sprenger N (2019) Impact of maternal characteristics on human milk oligosaccharide composition over the first 4 months of lactation in a cohort of healthy European mothers. Scientific Reports 9, 11767. 10.1038/s41598-019-48337-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab C and Ganzle M (2011) Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiology Letters 315(2), 141–148. 10.1111/j.1574-6968.2010.02185.x [DOI] [PubMed] [Google Scholar]
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM and Mills DA (2008) The genome sequence of Bifidobacterium longum subsp infantis reveals adaptations for milk utilization within the infant microbiome. Proceedings of the National Academy of Sciences of the United States of America 105(48), 18964–18969. 10.1073/pnas.0809584105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Garrido D, Lerno L, Wu SA, Tan KM, Eom HJ, Joachimiak A, Lebrilla CB and Mills DA (2012) Bifidobacterium longum subsp infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Applied and Environmental Microbiology 78(3), 795–803. 10.1128/AEM.06762-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani, G., Hoeflinger, J. L., Heiss, B. E., Masarweh, C. F., Larke, J. A., Jensen, N. M., Wickramasinghe, S., Davis, J. C., Goonatilleke, E., El-Hawiet, A., Nguyen, L., Klassen, J. S., Slupsky, C. M., Lebrilla, C. B., & Mills, D. A. (2022). Fucosylated Human Milk Oligosaccharide Foraging within the Species Bifidobacterium pseudocatenulatum Is Driven by Glycosyl Hydrolase Content and Specificity. Applied and Environmental Microbiology, 88(2), 1–12. 10.1128/aem.01707-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMA Nutrition. (2020). SMA® ADVANCED FOLLOW-ON MILK from 6 months + data card.
- Smilowitz JT, Lebrilla CB, Mills DA, German JB and Freeman SL (2014) Breast milk oligosaccharides: Structure-function relationships in the neonate. Annual Review of Nutrition 34, 143–169. 10.1146/annurev-nutr-071813-105721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilowitz JT, O’Sullivan A, Barile D, German JB, Lonnerdal B and Supsky CM (2013) The human milk metabolome reveals diverse oligosaccharide profiles. Journal of Nutrition 143(11), 1709–1718. 10.3945/jn.113.178772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevacek AR, Smilowitz JT, Chin EL, Underwood MA, German JB and Slupsky CM (2015) Infant maturity at birth reveals minor differences in the maternal milk metabolome in the first month of lactation. Journal of Nutrition 145(8), 1698–1708. 10.3945/jn.115.210252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger N, Lee LY, De Castro CA, Steenhout P and Thakkar SK (2017) Longitudinal change of selected human milk oligosaccharides and association to infants’ growth, an observatory, single center, longitudinal cohort study. PLoS One 12(2), e0171814. 10.1371/journal.pone.0171814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutkus LT, Joung S, Hirvonen J, Jensen HM, Ouwehand AC, Mukherjea R, Donovan SM and Dilger RN (2022) Influence of 2′-fucosyllactose and Bifidobacterium longum subspecies infantis supplementation on cognitive and structural brain development in young pigs [original research]. Frontiers in Neuroscience 16, 860368. 10.3389/fnins.2022.860368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW, Lawley B, Munro K, Pathmanathan SG, Zhou SJ, Makrides M, Gibson RA, Sullivan T, Prosser CG, Lowry D and Hodgkinson AJ (2013) Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Applied and Environmental Microbiology 79(9), 3040–3048. 10.1128/aem.03910-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongaram T, Hoeflinger JL, Chow J and Miller MJ (2017) Human milk oligosaccharide consumption by probiotic and human-associated bifidobacteria and lactobacilli. Journal of Dairy Science 100(10), 7825–7833. 10.3168/jds.2017-12753 [DOI] [PubMed] [Google Scholar]
- Thurl S, Munzert M, Boehm G, Matthews C and Stahl B (2017) Systematic review of the concentrations of oligosaccharides in human milk. Nutrition Reviews 75(11), 920–933. 10.1093/nutrit/nux044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurl S, Munzert M, Henker J, Boehm G, Muller-Werner B, Jelinek J and Stahl B (2010) Variation of human milk oligosaccharides in relation to milk groups and lactational periods. British Journal of Nutrition 104(9), 1261–1271. 10.1017/s0007114510002072 [DOI] [PubMed] [Google Scholar]
- Tonon KM, Morais TB, Taddei CR, Araujo HB, Abrao A, Miranda A and de Morais MB (2021) Gut microbiota comparison of vaginally and cesarean born infants exclusively breastfed by mothers secreting alpha 1-2 fucosylated oligosaccharides in breast milk. PLoS One 16(2), e0246839. 10.1371/journal.pone.0246839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantis V, Bode L and van Neerven RJJ (2018) Immunological effects of human milk oligosaccharides. Frontiers in Pediatrics 6, 190. 10.3389/fped.2018.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood MA, Gaerlan S, De Leoz MLA, Dimapasoc L, Kalanetra KM, Lemay DG, German JB, Mills DA and Lebrilla CB (2015) Human milk oligosaccharides in premature infants: Absorption, excretion, and influence on the intestinal microbiota. Pediatric Research 78(6), 670–677. 10.1038/pr.2015.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood MA, Kalanetra KM, Bokulich NA, Mirmiran M, Barile D, Tancredi DJ, German JB, Lebrilla CB and Mills DA (2014) Prebiotic oligosaccharides in premature infants. Journal of Pediatric Gastroenterology and Nutrition 58(3), 352–360. 10.1097/mpg.0000000000000211 [DOI] [PubMed] [Google Scholar]
- Urashima T, Hirabayashi J, Sato S and Kobata A (2018) Human milk oligosaccharides as essential tools for basic and application studies on galectins. Trends in Glycoscience and Glycotechnology 30(172), SE51–SE65. 10.4052/tigg.1734.1SE [DOI] [Google Scholar]
- Van den Abbeele P, Duysburgh C, Vazquez E, Chow J, Buck R and Marzorati M (2019) 2’-fucosyllactose alters the composition and activity of gut microbiota from formula-fed infants receiving complementary feeding in a validated intestinal model. Journal of Functional Foods 61, 103484. 10.1016/j.jff.2019.103484 [DOI] [Google Scholar]
- van Leeuwen SS (2019) Challenges and pitfalls in human milk oligosaccharide analysis. Nutrients 11(11), 2684. 10.3390/nu11112684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenplas Y, Berger B, Carnielli VP, Ksiazyk J, Lagstrom H, Luna MS, Migacheva N, Mosselmans JM, Picaud JC, Possner M, Singhal A and Wabitsch M (2018) Human milk oligosaccharides: 2-fucosyllactose (2-FL) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients 10(9), 1161. 10.3390/nu10091161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester Boler, B. M., Rossoni Serao, M. C., Faber, T. A., Bauer, L. L., Chow, J., Murphy, M. R., & Fahey, G. C. (2013). In Vitro Fermentation Characteristics of Select Nondigestible Oligosaccharides by Infant Fecal Inocula. Journal of Agricultural and Food Chemistry, 61(9), 2109‑2119. 10.1021/jf305056f [DOI] [PubMed] [Google Scholar]
- Vazquez E, Santos-Fandila A, Buck R, Rueda R and Ramirez M (2017) Major human milk oligosaccharides are absorbed into the systemic circulation after oral administration in rats. British Journal of Nutrition 117(2), 237–247. 10.1017/s0007114516004554 [DOI] [PubMed] [Google Scholar]
- Walker RW, Clemente JC, Peter I and Loos RJF (2017) The prenatal gut microbiome: Are we colonized with bacteria in utero? Pediatric Obesity 12, 3–17. 10.1111/ijpo.12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RE, Ninonuevo M, Mills DA, Lebrilla CB and German JB (2006) In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Applied and Environmental Microbiology 72(6), 4497–4499. 10.1128/aem.02515-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RE, Ninonuevo M, Mills DA, Lebrilla CB and German JB (2007) In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Molecular Nutrition & Food Research 51(11), 1398–1405. 10.1002/mnfr.200700150 [DOI] [PubMed] [Google Scholar]
- White BA, Lamed R, Bayer EA and Flint HJ (2014) Biomass utilization by gut microbiomes. Annual Review of Microbiology 68, 279–296. 10.1146/annurev-micro-092412-155618 [DOI] [PubMed] [Google Scholar]
- Wicinski M, Sawicka E, Gebalski J, Kubiak K and Malinowski B (2020) Human milk oligosaccharides: Health benefits, potential applications in infant formulas, and pharmacology. Nutrients 12(1), 266. 10.3390/nu12010266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise A, Robertson B, Choudhury B, Rautava S, Isolauri E, Salminen S and Bode L (2018) Infants are exposed to human milk oligosaccharides already in utero. Frontiers in Pediatrics 6, 270. 10.3389/fped.2018.00270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao JZ, Takahashi S, Nishimoto M, Odamaki T, Yaeshima T, Iwatsuki K and Kitaoka M (2010) Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Applied and Environmental Microbiology 76(1), 54–59. 10.1128/aem.01683-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Davis JCC, Goonatilleke E, Smilowitz JT, German JB and Lebrilla CB (2017) Absolute quantitation of human milk oligosaccharides reveals phenotypic variations during lactation. Journal of Nutrition 147(1), 117–124. 10.3945/jn.116.238279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada C, Gotoh A, Sakanaka M, Hattie M, Stubbs KA, Katayama-Ikegami A, Hirose J, Kurihara S, Arakawa T, Kitaoka M, Okuda S, Katayama T and Fushinobu S (2017) Molecular insight into evolution of symbiosis between breast-fed infants and a member of the human gut microbiome Bifidobacterium longum. Cell Chemical Biology, 24(4), 515–524. 10.1016/j.chembiol.2017.03.012 [DOI] [PubMed] [Google Scholar]
- Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K and Kumagai H (2012) Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22(3), 361–368. 10.1093/glycob/cwr116 [DOI] [PubMed] [Google Scholar]
- Yu ZT, Chen C and Newburg DS (2013) Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 23(11), 1281–1292. 10.1093/glycob/cwt065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuner B, Muschiol J, Holck J, Lezyk M, Gedde MR, Jers C, Mikkelsen JD and Meyer AS (2018) Substrate specificity and transfucosylation activity of GH29 alpha-L-fucosidases for enzymatic production of human milk oligosaccharides. New Biotechnology 41, 34–45. 10.1016/j.nbt.2017.12.002 [DOI] [PubMed] [Google Scholar]
- Zeuner B, Teze D, Muschiol J and Meyer AS (2019) Synthesis of human milk oligosaccharides: Protein engineering strategies for improved enzymatic transglycosylation. Molecules 24(11), 2033. 10.3390/molecules24112033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic AM, German JB, Lebrilla CB and Mills DA (2011) Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America 108, 4653–4658. 10.1073/pnas.1000083107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga M, Monedero V and Yebra MJ (2018) Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium species. Frontiers in Microbiology 9, 1917. 10.3389/fmicb.2018.01917 [DOI] [PMC free article] [PubMed] [Google Scholar]


