Abstract
Changes in chromatin structure are a key aspect in the epigenetic regulation of gene expression. We have used a lac operator array system to visualize by light microscopy the effect of heterochromatin protein 1 (HP1) α (HP1α) and HP1β on large-scale chromatin structure in living mammalian cells. The structure of HP1, containing a chromodomain, a chromoshadow domain, and a hinge domain, allows it to bind to a variety of proteins. In vivo targeting of an enhanced green fluorescent protein-tagged HP1-lac repressor fusion to a lac operator-containing, gene-amplified chromosome region causes local condensation of the higher-order chromatin structure, recruitment of the histone methyltransferase SETDB1, and enhanced trimethylation of histone H3 lysine 9. Polycomb group proteins of both the HPC/HPH and the EED/EZH2 complexes, which are involved in the heritable repression of gene activity, are not recruited to the amplified chromosome region by HP1α and HP1β in vivo targeting. HP1α targeting causes the recruitment of endogenous HP1β to the chromatin region and vice versa, indicating a direct interaction between the two HP1 homologous proteins. Our findings indicate that HP1α and HP1β targeting is sufficient to induce heterochromatin formation.
Packing of the eukaryotic genome into higher-order chromatin structures is tightly related to gene expression (reviewed in references 17 and 37). Changes in chromatin structure are a key element in epigenetic gene control. Transcriptional activation is associated with large-scale chromatin decondensation, whereas silencing is related to condensation (2, 31, 36, 51, 52). The molecular mechanisms that establish and maintain functionally distinct higher-order chromatin states in the interphase nucleus are poorly understood (8, 42). Posttranslational modifications of histones have been directly linked with transcriptional regulation and with changes in chromatin structure (reviewed in references 22 and 24). Such modifications alter the interactions between histones and DNA and between chromatin-associated proteins. The “histone code” apparently defines functionally distinct chromatin domains of different transcriptional states (15, 21, 35, 49). Many key questions concerning the role of histone modifications regulating chromatin structure and gene expression are unsolved.
A number of heterochromatin-associated proteins, including heterochromatin protein 1 (HP1) and polycomb group (PcG) proteins, which all are involved in epigenetic silencing, share a common evolutionarily conserved domain, the chromodomain (CD), which is responsible for the association with chromatin (4, 9, 19, 24, 48). HP1 also contains a chromoshadow domain (CSD), which mediates protein-protein interactions, including homodimerization (1). Moreover, HP1 also binds to DNA and linker histones through its hinge domain (HD), positioned between the CD and the CSD. This HD may also be involved in targeting HP1 to heterochromatin through an RNA binding activity (30). A link between histone modifications and the formation of a repressive chromatin state is suggested by the finding that the methylation of histone H3 at lysine 9 (H3K9) by histone methyltransferase (HMT) SU(VAR)3-9 establishes a binding site for HP1.
In mammals, three isoforms of HP1 that show different subnuclear distributions, i.e., HP1α, HP1β, and HP1γ, have been identified. HP1α is found mainly in pericentromeric heterochromatin. HP1β and, to a lesser extent, HP1γ are found in pericentromeric domains but are also present at dispersed euchromatic sites throughout the nucleoplasm (9, 18, 24, 29, 56). PcG-mediated gene silencing has often been compared with HP1-induced heterochromatin gene silencing. However, PcG-mediated gene silencing and HP1-mediated gene silencing require distinct sets of proteins, and it is still unclear whether they use similar mechanisms (13). At least two distinct PcG complexes, the HPC/HPH and EED/EZH2 complexes, exist (reviewed in reference 25). The EED/EZH2 complex contains HMT activity capable of methylating H3K27 and, to a much lesser extent, H3K9; little evidence exists suggesting that enzymatic activities are associated with the HPC/HPH complex, although interactions have been demonstrated between SU(VAR)3-9 and the mammalian homologue HPC2 in overexpression studies (38, 46). The physiological consequences of HP1-associated proteins are still uncertain (34, 47).
In the present study, we used a lac operator array-based system (36) in mammalian cells to target HP1α and HP1β to chromatin in living cells. To investigate the effect of HP1 targeting on large-scale chromatin structure, we used the CHO-derived cell line RRE_B1, containing large amplified genomic domains in an extended, often fibrillar conformation, suggesting a euchromatin-like structure. We confirmed that proteins involved in gene silencing and known to associate with condensed chromatin (such as HP1α and HP1β, trimethylated H3K9 [tri-MeH3K9], and the HMT SETDB1) are not present at the amplified chromosome region. We analyzed the resulting changes in large-scale chromatin structure, site-specific histone modifications, and recruitment of other chromatin proteins. In vivo HP1 targeting to the lac operator-containing amplified chromosome region, as an enhanced green fluorescent protein (EGFP)-tagged lac repressor (EGFP-lacR) fusion, caused local condensation of large-scale chromatin structure, recruitment of HMT SETDB1, and enhancement of tri-MeH3K9. We examined whether PcG proteins are involved in the HP1-induced processes of gene silencing and chromatin condensation by analyzing the distributions of various proteins of the HPC/HPH and EED/EZH2 complexes after HP1 targeting. We showed that PcG proteins from neither complex are recruited to the amplified chromosome region by HP1 targeting to that region. Furthermore, targeting of HP1α and HP1β to the chromosome array causes the recruitment of endogenous HP1β and HP1α, respectively, indicating a direct interaction between the two HP1 forms. Our results show that HP1α and HP1β targeting to a defined chromatin region is sufficient to induce heterochromatin formation in that region.
MATERIALS AND METHODS
Construction of plasmids.
A plasmid expressing an EGFP-lacR-simian virus 40 nuclear localization signal (NLS) fusion under the control of the F9-1 promoter, called p3′SS-EGFP-dimer lac repressor (52), was used as the basis for these studies. This plasmid contains a mutant form of the lac repressor which forms dimers rather than tetramers due to a five-amino-acid deletion in the C terminus (36). The N terminus of the lac repressor was replaced with a proline-3→ tyrosine mutant from pAFS135 to produce a tightly binding lac repressor. The resulting plasmid, p3′SS EGFP-dimer tb lac repressor, then was modified as described previously to generate EGFP-lacR-AscI-NLS (NYE4) (36). HP1α and HP1β were amplified by PCR with primers which contain AscI sites in frame. The PCR products were digested with AscI and ligated into the AscI-digested EGFP-lacR-AscI-NLS (NYE4) vector to create HP1α or HP1β C-terminal in-frame fusions with the EGFP-lacR-AscI-NLS (NYE4) vector. All regions of constructs that had undergone PCR were sequenced to ensure fidelity. The eight lac operator repeat-containing pGL3 luciferase reporter plasmid was generated by removing an XbaI-XhoI fragment containing eight lac operator repeats from pPS-8.1 (43) and ligating this fragment into the pGL3-Control luciferase vector (Promega, Madison, WI).
Cell culture, transfection, and luciferase reporter assay.
The RRE_B1 cell line was derived in three steps (43). First, pSV2-DHFR-8.32, containing the dihydrofolate reductase (DHFR) cDNA transgene and 256 direct repeats of the lac operator, was modified by adding a 225-bp Rev response element (RRE) to the 3′ untranslated region by blunt-end ligation with the HpaI restriction site, generating the pSV2-DHFR-8.32-RRE plasmid. pSV2-DHFR-8.32-RRE was transfected by using Lipofectamine (GIBCO, Breda, The Netherlands) into CHO DG44 cells, containing a double knockout of DHFR. At 3 days after transfection, selection for stable transformants was done by growth in Ham's F-12 medium without hypoxanthine or thymidine but supplemented with 10% dialyzed fetal bovine serum (HyClone Labs, Logan, Utah). Second, a mixed population of stable transformants was subjected to gene amplification with stepwise increases of methotrexate up to a final concentration of 10 μM. Third, cells were subcloned from this gene-amplified population. Many of these subclones, including the RRE_B1 clone, showed unusually extended, often fibrillar conformations of the amplified chromosome region, suggesting a euchromatin-like structure. Osteosarcoma cells (U2OS) and NIH/3T3 mouse fibroblasts were cultured in Dulbecco minimal essential medium containing 10% fetal bovine serum. All cells were cultured at 37°C in a 5% CO2 atmosphere.
Transfection on coverslips was performed with FuGENE6 reagent (Roche, Indianapolis, Ind.) according to the manufacturer's instructions with 500 ng of DNA and 6 μl of FuGENE6 reagent per ml of culture medium. Fresh medium was added at 24 h after transfection. At 48 h after transfection, cells were rinsed with phosphate-buffered saline (PBS) and used for immunofluorescence labeling or for transcription assay measurements. Transfections for luciferase reporter assays were done with 500 ng of eight lac operator repeat-containing pGL3 luciferase vector, 500 ng of pSV/β-Gal construct (Promega) as an internal reference reporter, and 500 ng of effector plasmid (i.e., EGFP-lacR, EGFP-lacR-HP1α, or EGFP-lacR-HP1β) combined with 48 μl of FuGENE6 reagent per 25-cm2 culture flask. Cells were harvested and lysed at 48 h posttransfection. A luciferase reporter gene-targeted repression assay was performed as described previously (6, 44, 53).
Immunofluorescence labeling.
Immunofluorescence labeling was performed as described previously (54). Briefly, cells were fixed for 10 min at 4°C with 2% (wt/vol) formaldehyde in PBS. After fixation, cells were permeabilized with 0.5% (wt/vol) Triton X-100 in PBS for 5 min and incubated in PBS containing 100 mM glycine for 10 min. Subsequently, cells were incubated for 1 h at 37°C with primary antibodies diluted in PBS containing 0.5% (wt/vol) bovine serum albumin and 0.1% (wt/vol) gelatin (PBG) (Sigma, St. Louis, MO). The following primary antibodies were used: rabbit anti-dimethylated H3K9 (di-MeH3K9) (Upstate, Milton Keynes, United Kingdom) (33), rabbit anti-branched(4×)-MeH3K9 peptide (23), rabbit anti-tri-MeH3K9 (9), rabbit anti-SETDB1 (45), mouse anti-HP1α and anti-HP1β (Euromedex; Mundolsheim, France) (34), and rabbit anti-BMI and mouse anti-RING, anti-HPC2, anti-EED, and anti-EZH2 (16). After several washes with PBS, cells were incubated with the appropriate secondary antibodies: Cy3-conjugated donkey anti-rabbit antibody, donkey anti-mouse antibody, or donkey anti-rat antibody (Jackson, West Grove, PA). Secondary antibodies were diluted in PBG. Incubations were performed for 1 h at room temperature. Cells were rinsed with PBS at room temperature, and DNA staining was performed with 0.4 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) (33258; Sigma) in PBS. Slides were mounted in Vectashield (Brunschwig, Burlingame, CA). Slides were stored at 4°C and analyzed within 24 h. As a control, the primary antibody was omitted.
Confocal scanning laser microscopy.
All experiments were performed at least two times in duplicate. For each experiment, 10 nuclei were visualized and at least 5 nuclei were imaged. Images were recorded by using a Zeiss LSM 510 (Zeiss, Jena, Germany) confocal laser scanning microscope equipped with a ×100/1.23 NA oil immersion objective. We used an argon laser at 488 nm in combination with a helium-neon laser at 543 nm to excite green and red fluorochromes simultaneously. Emitted fluorescence was detected with a 505- to 530-nm band-pass filter for the green signal and a 560-nm long-pass filter for the red signal. Pairs of images were collected simultaneously in the green and red channels. Three-dimensional (3D) images were scanned at 512 by 512 by 32 voxels (sampling rates, 49 nm lateral and 208 nm axial).
Image analysis.
Image analysis was performed with a Huygens system 2 software package (Scientific Volume Imaging, Hilversum, The Netherlands). For a semiquantitative analysis of the spatial relationship between the relative spatial distributions of components in dually labeled cells, we made line scans. The signal intensities of the two labels are plotted along a line through the nucleus. The line scan analysis method provides a rapid and straightforward way to determine to what extent two components colocalize.
To analyze quantitatively changes in large-scale chromatin structure induced by HP1 targeting, we applied novel 3D image analysis tools. 3D images of the amplified region in control cells and in HP1-targeted cells were acquired (30 images of each type). A number of attributes of the 3D structure were determined by using two independent methods. In the first method, a 3D region of interest, i.e., the EGFP-labeled chromosome region, was defined. The images were Gaussian filtered to reduce noise with a size of 20 pixels (sigma value is 250 nm). The area of interest was the set of voxels that had an intensity higher than 25% of the maximum found in the filtered image. For selection of the region of interest of a control sample and of an HP1-targeted sample, see Fig. 3A and C and Fig. 3B and D, respectively. For each set, we recorded the size of the selected region (in μm3) and the average magnitude of the intensity gradient of the region. To determine the latter, the original data in the selected region were filtered by using a Gaussian filter of size 8 (sigma value is 100 nm) to smooth out high frequencies, such as noise. The intensity gradient was calculated in the filtered data for the voxels in the region of interest by using a gradient magnitude filter (41). The displacements used were 8 in the x and y directions and 2 in the z direction. For filtering used to obtain the average magnitude of the intensity gradient of the selected region of a control sample and of an HP1-targeted sample, see Fig. 3A and E and Fig. 3B and F, respectively. In the second 3D image analysis method, a difference-of-Gaussian (DoG) approximation of the Laplacian method (28) was used to determine distinct spots within the EGFP-labeled chromosomal domain (distinct spots were determined by grouping labeled voxels which shared a face with another labeled voxel). The data were filtered by using a Gaussian filter with sigma values of 75 nm and 250 nm. A threshold value of 15% of the possible maximum value was used. For the selection of distinct spots within the EGFP-labeled chromosome region of a control sample and of an HP1-targeted sample, see Fig. 3A and G and Fig. 3B and H, respectively. Using the second method, we assessed the number of distinct spots within the chromosomal domain and the average size of these spots (in μm3). For the occurrence of the measured values in histograms, see Fig. 4; in addition, a Gaussian approximation of the distributions is shown by the curve.
FIG. 3.

Quantitative image analysis methods. For quantitative analysis of the chromatin structure, 3D images of the chromosomal array of control (A, C, E, and G) or HP1-targeted (B, D, F, and H) cells were analyzed. A number of attributes were tested by two independent methods (see Materials and Methods). (A and B) Illustration of the quantitative selection methods for an optical section of the chromosomal array of a control sample (A) and an HP1β-targeted sample (B). (C and D) Illustration of the selection of a 3D region of interest, i.e., the EGFP-labeled chromosome region of an optical section of a control sample (C) and an HP1β-targeted sample (D). (E and F) Illustration of the filtering used to obtain the intensity gradient of the selected region of an optical section of a control sample (E) and an HP1β-targeted sample (F). (G and H) Illustration of the determined distinct spots within the EGFP-labeled chromosomal domain of an optical section of a control sample (G) and an HP1-targeted sample (H). Nuclei in panels A to H are on the same scale. Bar, 0.9 μm.
FIG. 4.
Image analysis of the chromatin structure of the chromosomal array after HP1β targeting. 3D images of RRE_B1 cells transfected with EGFP-lacR-HP1β and EGFP-lacR (control) fusion constructs were collected 48 h after transfection A number of attributes of the control (A, C, E, and G) and HP1β-targeted (B, D, F, and H) data sets were tested by two independent methods (see Materials and Methods). The first method involved the selection of a 3D region of interest, i.e., the labeled chromosome domain (A to D), to analyze the size of the defined region (in μm3) (A and B) and the average magnitude of the intensity gradient over the region (C and D). The second method involved the determination of distinct spots within the labeled chromosomal domain (E to H) to determine the number of distinct spots (E and F) and the average size of these spots (in μm3) (G and H) within the labeled chromosomal domain. The data are presented as histograms of the occurrence of the measured values and Gaussian approximations of the distributions.
RESULTS
Functionality of HP1 fusion constructs.
We made fusion constructs consisting of EGFP, the lac repressor domain, and HP1α or HP1β. The ability to silence gene expression in a chromatin environment of the constructs was tested with a luciferase reporter assay. To this end, we constructed a pGL3 luciferase vector (Promega) containing eight lac operator binding sites. Transiently transfected U2OS cells with the EGFP-lacR-HP1α, or the EGFP-lacR-HP1β fusion construct and the eight lac operator repeat-containing luciferase vector caused 20-fold repression of luciferase activity compared with activity of the EGFP-lacR control constructs (Fig. 1A), showing that the targeted HP1 fusion efficiently silences gene activity. Normal behavior of the HP1 fusions was further established by showing that in NIH/3T3 mouse fibroblasts they localized, as expected, in pericentromeric heterochromatin domains, identified as DAPI-dense regions (Fig. 1B to E). These results indicate that the HP1 constructs are functional with respect to their ability to repress gene activity and to properly localize expressed HP1.
FIG. 1.
Functionality of the HP1 fusions. (A) U2OS cells were transfected with the pSV/β-Gal construct, the eight lac operator repeat-containing luciferase vector, and the test constructs EGFP-lacR, EGFP-lacR-HP1α, and EGFP-lacR-HP1β. At 48 h after transfection, the cells were harvested and lysed to measure luciferase activity relative to β-galactosidase activity. Bars 1, 2, and 3 represent relative luciferase activities of test constructs EGFP-lacR, EGFP-lacR-HP1α, and EGFP-lacR-HP1β, respectively. Values are the mean ± standard error of six independent experiments. (B to E) NIH/3T3 fibroblasts were transiently transfected with EGFP-lacR-HP1α (B and C) and EGFP-lacR-HP1β (D and E) fusion constructs, fixed, and stained with DAPI 48 h after transfection. In cells transfected with EGFP-lacR-HP1α, the distribution of exogenous HP1α (C) occurred in the domains intensely stained with DAPI (B). Similarly, in cells transfected with EGFP-lacR-HP1β, the distribution of exogenous HP1β (E) occurred in the domains intensely labeled with DAPI (D). Bars, 2.8 μm.
HP1 targeting induces local condensation of chromatin.
To investigate the effect of HP1 targeting on large-scale chromatin structure we used the CHO-derived cell line RRE_B1, containing large amplified genomic domains (several tens of Mb) in an extended, often fibrillar conformation, suggesting a euchromatin-like structure (43). According to unpublished data from the Belmont laboratory (see also reference 20), essentially all multicopy plasmid insertions appear to form condensed structures, a property which is most likely related to the phenomenon of transgene silencing. This is in contrast to the behavior of transgene arrays created in a multistep process by stable transfection followed by gene amplification, which leads to copies of transgene insertions being flanked by hundreds to thousands of kb of coamplified genomic DNA. These gene-amplified transgene arrays show a range of conformations from condensed to unusually extended, which presumably depends on which coamplified genomic regions are present. Based on unpublished light and electron microscopy data, the lac operator array of the RRE_B1 clone appears to be more extended than even bulk chromatin. We performed several tests on this cell line to determine that proteins involved in gene silencing and known to associate with condensed chromatin (such as HP1α and HP1β, tri-MeH3K9, and the HMT SETDB1) are not present at the amplified chromosome region. We clearly do not observe any of these endogenous proteins present at the amplified chromosome region (these results are shown in Fig. S1 in the supplemental material). We selected the RRE_B1 clone for further studies as we believed that any effects of HP1 in terms of causing large-scale chromatin condensation would be particularly obvious given the starting “open” conformation of the RRE_B1 amplified chromosome region. Therefore, these cells constitute a useful system to investigate euchromatin-heterochromatin transitions. EGFP-lacR-HP1 fusion constructs were transiently transfected into RRE_B1 cells, whereas the EGFP-lacR fusion construct is used as a control. 3D images were collected 48 h after transfection. In control cells, transfected with EGFP-lacR, the amplified domain occurs as a fibrous structure that may extend over long distances in the nucleus (Fig. 2A and C). Expression of the EGFP-lacR-HP1α fusion, thereby inducing HP1α targeting to the amplified chromosome region, caused chromatin condensation of that region (Fig. 2B and D). The same results were obtained for the HP1β fusion and after transfection of the HP1α and HP1β constructs together (data not shown). After targeting of the HP1α or HP1β fusion the extended and diffusely labeled chromatin domain condensed to a string of strongly labeled subdomains. The chromatin structure after HP1 targeting appeared as a collection of a small number of distinct intensely labeled round spots, ∼0.4 μm in diameter, well above the 0.2-μm diameter of diffraction limited point sources. These data suggest both a local condensation of large-scale chromatin structure and a coalescence of lac operator-tagged regions into a smaller number of larger condensed regions.
FIG. 2.
HP1-induced local compaction of the chromatin structure of the amplified chromosome region. RRE_B1 cells were transfected with EGFP-lacR-HP1α or EGFP-lacR (control) fusion constructs. 3D images were collected 48 h after transfection of large numbers of cells. 3D images were recorded; the images shown represent individual midnuclear optical sections. (A) The chromosomal array in a control nucleus transfected with EGFP-lacR occurs as an extended fibrillar structure. (B) After HP1 targeting to the chromosomal array, chromatin occurred as a row of distinct beads. The circumference of the nucleus in panels A and B is shown by the white line. (C and D) Enlargements of the chromosomal arrays in panels A and B, respectively. Bars, A and B, 0.9 μm; C and D, 0.4 μm.
Quantitative analysis of changes in chromatin structure.
There is significant variability in the appearance of the chromatin domains in a population of identical cells. This is possibly related to cell cycle effects and/or differences in expression levels of the transfected fusion constructs. In addition, the actual size of the amplified chromosome region may vary due to genetic instability of this cell line. We have quantitatively and objectively analyzed the changes in large-scale chromatin structure that were induced by HP1 targeting (Fig. 3 and 4). Similar data were obtained for HP1α or HP1β separately and in combination (the results of HP1β targeting are shown in Fig. 4). 3D images were acquired of the amplified chromosomal region in control cells and of cells after targeting of HP1β (30 sets of each). We used two methods to quantify differences in chromatin structure (Fig. 3A and B in control and HP1β targeted cells, respectively). In the first method a 3D region of interest is defined, i.e., the labeled chromosomal domain, to analyze the size of the defined region and the average magnitude of the gradient of the intensity over the region. Figure 3 illustrates the selection of the region of interest (Fig. 3C and D for a control sample and an HP1β-targeted sample, respectively) and the filtering to obtain the average magnitude of intensity gradient of the selected region (Fig. 3E and F for a control sample and an HP1β-targeted sample, respectively). The second 3D method uses a DoG approximation of the Laplacian method (28) to determine the number of distinct EGFP-labeled spots within the labeled chromosomal domain and the average size of these spots (distinct spots are determined by grouping EGFP-labeled voxels which share a face with another EGFP-labeled voxel). Figure 3 illustrates the selection of distinct spots within the labeled chromatin domain (G and H of a control and HP1β targeted sample, respectively). Results of the quantitative analysis of the changes in chromatin structure upon HP1 targeting are plotted as histograms in Fig. 4.
Using the region-of-interest method, both the distributions of size (in μm3) (Fig. 4A and B) and magnitude of intensity gradient (Fig. 4C and D) of the defined region are significantly different between control cells and cells transfected with the HP1 constructs. The average magnitude of intensity gradient defines the more gradual or steep change of the intensity over the chromatin region. Following HP1 targeting, the size of the array is smaller and the intensity gradient steeper. t tests show that the differences in control versus HP1 targeted cells regarding size and average magnitude of intensity gradient of the chromatin region are highly significant (P = 0.0006 and 0.004, respectively). Similarly, using the second method, the number of distinct spots within the chromosomal domain (Fig. 4E and F) and the size of distinct spots (in μm3) within the chromosome domain (Fig. 4G and H) are significantly different (P = 0.002 and 0.02, respectively) in control cells versus HP1 targeted cells. Upon HP1 targeting more distinct spots are detectable, whereas the average size of the spots is larger. Taken together HP1 recruitment causes a contraction of the amplified chromosome region into a smaller size, consisting of a more spot-like appearance, with more distinct spots of larger size and also a very steep change in intensity indicating a more compact structure. The results of the quantitative analysis support the conclusion that targeting of HP1 to the amplified chromosome region induces condensation of a number of subdomains in the amplified region.
HP1 targeting induces recruitment of SETDB1 and tri-MeH3K9.
Methylation of H3K9 creates a binding site for the HP1 CD resulting in the assembly of a repressive protein complex. Propagation of a repressive chromatin state is postulated to occur by HP1-mediated recruitment of additional HMTs, resulting in histone methylation of neighboring nucleosomes (14). We investigated the distribution of HMT SETDB1 and of di- and tri-MeH3K9 after HP1 targeting. SETDB1 is member of a subclass of SET proteins that methylates H3K9 predominantly in euchromatic regions, thereby stimulating the binding of HP1 (45). We performed immunofluorescent labeling, using antibodies against di-MeH3K9, tri-MeH3K9 (9), or branched(4×)-MeH3K9 peptide (26) and an antibody against SETDB1 (45). The distribution of endogenous di- and tri-MeH3K9 and endogenous SETDB1 was analyzed in control cells that had been transfected with EGFP-lacR compared to cells transfected with EGFP-lacR-HP1.
Labeling with anti-di-MeH3K9 antibody showed a diffuse distribution throughout the nucleoplasm (Fig. 5A). Labeling with the anti-branched(4×)-MeH3K9 antibody and with the anti-tri-MeH3K9 also showed a distribution over many small sites throughout the nucleoplasm, as well as in larger domains (Fig. 6A and C), reproducing previous results with these antibodies (9, 23). The di-MeH3K9 antibody is known to display high affinity for di-MeH3K9, while showing minor cross-reactivity with di-MeH3K4 and tri-MeH3K27 (39). It displays a rather uniform labeling that is underrepresented in pericentromeric heterochromatin domains (39). The antibody against branched(4×)-MeH3K9 peptide is described to have a high affinity for tri-MeH3K9 and to significantly cross-react with di- and trimethylation of several H3 lysine positions, and was therefore renamed as “multi-methyllysine” antiserum (39). This antibody selectively decorates silenced chromatin regions at pericentromeric heterochromatin and the inactive X chromosome (39). The tri-MeH3K9 antibody is known to display a very similar staining pattern as the anti-branched(4×)-MeH3K9 antiserum (9, 39).
FIG. 5.
Spatial relationship between the distribution of di-MeH3K9 and the amplified chromosome region after HP1 targeting. RRE_B1 cells transfected with EGFP-lacR (A to D) or a combination of EGFP-lacR-HP1α and EGFP-lacR-HP1β (E to G) were fixed after 48 h and immunofluorescently labeled with an antibody against di-MeH3K9. Panels B and C and panels F and G are enlargements of the chromosomal arrays shown in panels A and E, respectively. The green signal shows the EGFP-tagged chromosomal array, and the red signal shows the distribution of immunolabeled di-MeH3K9. 3D images were recorded; the images shown represent individual midnuclear optical sections. Panels D and H show line scans through the nuclei shown in panels A and E, respectively; the position of the line scan is shown by the white horizontal line. Nuclei in panels A and E are on the same scale. Bar, 1 μm.
FIG. 6.
HP1 targeting induces SETDB1 recruitment and enhanced tri-MeH3K9 expression. RRE_B1 cells transfected with EGFP-lacR (A to D and I to L) or EGFP-lacR-HP1β (E to H and M to P) were, after 48 h, immunofluorescently labeled with an antibody against branched(4×)-MeH3K9 (A to D and E to H) or against SETDB1 (I to L and M to P). The green signal shows the EGFP-tagged chromosomal array, and the red signal shows the distribution of immunolabeled tri-MeH3K9 or SETDB1. Panels B and C, panels F and G, panels J and K, and panels N and O are enlargements of the chromosomal array shown in panels A, E, I, and M, respectively. 3D images were recorded; the images shown represent individual midnuclear optical sections. Panels D, H, L, and P show line scans through the nuclei shown in panels A, E, I, and M, respectively; the position of the line scan is shown by the white arrow. The double-headed arrows indicate the sizes of the tri-MeH3K9 or SETDB1 signal (red) and the EGFP-tagged chromosomal array signal (green). Nuclei in panels A, E, I, and M are on the same scale. Bar, 1 μm.
Using any of these three antibodies, a low level of di- and tri-MeH3K9 was observed in the amplified chromosomal array in control cells after EGFP-lacR transfection (Fig. 5A, C, and D and Fig. 6A, C, and D). After HP1β targeting, labeling with the anti-branched(4×)-MeH3K9 (Fig. 6E to H) and anti-tri-MeH3K9 antibody (data not shown) was significantly enhanced in the amplified chromosome region. Domains with enhanced tri-MeH3K9 (red) colocalized with the amplified domains to which the HP1β is targeted (green) (Fig. 6E to G), as demonstrated by line scans (Fig. 6H). Very similar results were obtained after targeting HP1α, followed by labeling with anti-branched(4×)-MeH3K9 or anti-tri-MeH3K9 antibody (data not shown). No increase in labeling with the anti-di-MeH3K9 antibody was observed after HP1α or HP1β targeting, and also not after targeting HP1α and HP1β simultaneously (Fig. 5E, G, and H).
SETDB1 is known to localize predominantly in nuclear regions that show HP1 labeling outside the pericentromeric domains (45). In control RRE_B1 cells, transfected with the EGFP-lacR construct, SETDB1 is present at low levels in the amplified chromosomal region (Fig. 6I to K). After targeting HP1α, HP1β, or HP1α and HP1β simultaneously, SETDB1 specifically accumulates in the amplified domain (Fig. 6M to P). The colocalization of anti-SETDB1 signal and the EGFP-tagged amplified chromosome region (Fig. 6M to P) upon HP1 targeting is similar as the observed colocalization of anti-tri-MeH3K9 and anti-branched(4×)-MeH3K9 signal and the EGFP-tagged chromosomal array after HP1 targeting. To rule out that the observed enrichment upon HP1 targeting is simply giving a more prominent signal as a consequence of a higher DNA concentration of the amplified chromosome region, we carefully analyzed a significant number of cells in 3D, also performing line scan analysis demonstrating that endogenous levels of tri-MeH3K9 and SETDB1 are not correlated with the EGFP signal of the amplified chromosome region in the control cells. Moreover, the enrichment of the signal observed after anti-tri-MeH3K, anti-branched(4×)-MeH3K9 and anti-SETDB1 labeling at the amplified chromosome region upon HP1 targeting compared to the low levels in the rest of the nucleus is not related to the level of DAPI DNA staining at the amplified chromosome region compared to the DAPI signal in the rest of the nucleus (data not shown). Our results convincingly show that targeting of HP1α, HP1β or a combination of HP1α and HP1β, to the amplified region in RRE_B1 cells causes recruitment of SETDB1 and enhanced tri-MeH3K9. Of interest, the anti-tri-MeH3K9 and anti-branched(4×)-MeH3K9 signal (Fig. 6E to H; see also Fig. S1E to H in the supplemental material) that we observe upon HP1 targeting encompasses but appears significantly larger than the EGFP-tagged amplified chromosome region. Crude measurements of this observation revealed that the tri-MeH3K9 and SETDB1 signals extend approximately 300 to 500 nm beyond the signal of the amplified chromosome region (Fig. 6E to H; see also Fig. S1G and H in the supplemental material).
HP1α and HP1β recruit each other but not PcG proteins.
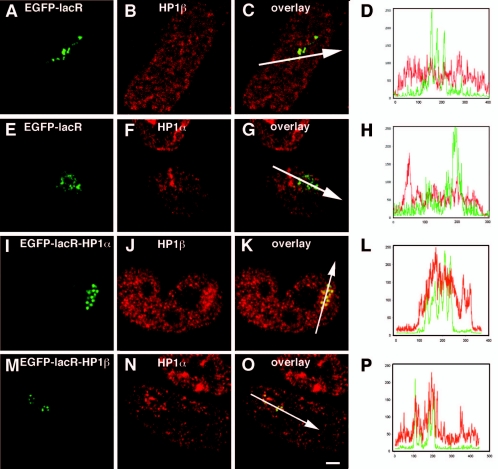
We wondered whether targeting of HP1 to the amplified chromosome region would recruit endogenous HP1. In control cells, transfected with EGFP-lacR, endogenous HP1β and HP1α were present only at low levels in the amplified regions Fig. 7A to D and E to H, respectively. To analyze the recruitment of endogenous HP1 proteins after HP1 targeting, cells were transfected with the EGFP-lacR-HP1α construct and the distribution of endogenous HP1β was determined by immunofluorescent labeling (Fig. 7I to L). Also the reverse experiment was carried out transfecting the cells with the EGFP-lacR-HP1β construct and analyzing the distribution of endogenous HP1α (Fig. 7M to P). Significant recruitment of endogenous HP1α and HP1β was observed after HP1 targeting (Fig. 7J to K and Fig. 7N to O), as was confirmed by line scans through the amplified chromosome region (Fig. 7L and P). The signal of the endogenous recruited HP1α upon HP1β targeting (and vice versa) is less obvious expanding beyond the amplified chromosome region as observed for the tri-MeH3K9 and SETDB1. Crude measurements show that the recruited HP1 signal outside the amplified chromosome region is in the range of 200 nm, which limits the light microscopy resolution (Fig. 7I to P; see also Fig. S1E and F in the supplemental material).
FIG. 7.
HP1 targeting induces the recruitment of endogenous HP1. RRE_B1 cells transfected with EGFP-lacR (A to H), EGFP-lacR-HP1α (I to L), or EGFP-lacR-HP1β (M to P) were, after 48 h, immunofluorescently labeled with an antibody against HP1β (A to D and I to L) or against HP1α (E to H and M to P). 3D images were recorded; the images shown represent individual midnuclear optical sections. The green signal shows the EGFP-lacR-tagged chromosomal array, and the red signal shows the distribution of immunolabeled endogenous HP1. Panels D and H show line scans through the nuclei shown in panels C and G, respectively, and panels L and P show line scans through the nuclei shown in panels K and O, respectively; the position of the line scan is shown by the white arrow. Nuclei in panels A to C, E to G, I to K, and M to O are on the same scale. Bar, 1.4 μm.
The distribution of the HPC/HPH PcG proteins BMI, RING, HPC2 and EED/EZH2 PcG proteins EED and EZH2 in RRE_B1 control cells (transfected with EGFP-lacR) occurs diffusely throughout the nucleoplasm (data not shown). After transfection with EGFP-lacR-HP1 (either HP1α, HP1β or HP1α and HP1β simultaneously), no accumulation of any of these PcG proteins was observed at the amplified chromosome regions (Fig. 8). Figure 8A to O show the distribution of BMI, RING, HPC2, EED and EZH2, respectively, upon immunolabeling 48 h after transfection with the EGFP-lacR-HP1β construct. Our results show that targeting of HP1α and HP1β at the amplified chromosomal region does not recruit proteins of the HPC/HPH or EED/EZH2 PcG complex suggesting that these PcG proteins are not involved in the changes in chromatin structure induced by HP1.
FIG. 8.
HP1 targeting does not recruit endogenous PcG proteins, BMI, RING, HPC2, EED, or EZH2 to the amplified chromosome region. RRE_B1 cells transfected with EGFP-lacR-HP1β were, after 48 h, immunofluorescently labeled with antibodies against the HPC/HPH and EED/EZH2 PcG complexes: anti-BMI (A to C), anti-RING (D to F), anti-HPC2 (G to I), anti-EED (J to L), and anti-EZH2 (M to O). 3D images were recorded; the images shown represent individual midnuclear optical sections. The green signal shows the EGFP-tagged chromosomal array (A, D, G, J, and M), and the red signal shows the distribution of the immunolabeled PcG proteins (B, E, H, K, and N). Panels C, F, I, L, and O show overlays of panels A and B, panels D and E, panels G and H, panels J and K, and panels M and N, respectively. Nuclei in panels A to O are on the same scale. Bar, 1.2 μm.
DISCUSSION
HP1α and HP1β are nonhistone chromatin proteins that are involved in heterochromatin formation and gene silencing (11, 12, 27, 47, 53). Still, the actual molecular mechanisms by which these proteins mediate transcriptional repression and whether this repression is linked to changes in chromatin packing remains unclear, despite clear links between HP1 targeting and histone methylation (4, 9, 19, 26). In the present study, we report on the ability of HP1α and HP1β to induce changes in mammalian large-scale chromatin structure in vivo. We have targeted EGFP-tagged HP1 to an amplified chromosome region in CHO cells, using the lac repressor-lac operator system developed by the Belmont group (5, 52). We show that HP1 targeting to a defined chromatin region resulted in local chromatin condensation and enrichment of HMT SETDB1 and tri-MeH3K9 at the amplified chromosome region. Interestingly, targeting of the HP1α fusion construct recruited endogenous HP1β, and vice versa. In contrast, no components of the HPC/HPH and EED/EZH2 PcG group protein complexes, which are involved in epigenetic silencing, were associated with targeted HP1.
The amplified chromosome region of the RRE_B1 cells contains the DFHR gene adjacent to an array of 256 lac operator sites plus an unknown amount of coamplified genomic DNA. The total amplified chromosome region is several tens of Mb in size (36) and it occurs as a fibrous, irregular open structure, as can be visualized after targeting EGFP-lacR (Fig. 2A and C). Targeting of HP1α or HP1β to the amplified chromosome region as an EGFP-lacR-HP1 fusion causes chromatin condensation. HP1 targeting induces large-scale changes in chromatin structure, changing it from the fibrous open state to a more condensed structure, consisting of multiple compact subdomains (Fig. 2B and D). To objectively and quantitatively analyze such structural transitions we have developed novel software that allows one to interrogate the 3D images using a wide variety of structural parameters. Several of such parameters confirmed the large-scale chromatin condensation upon HP1 targeting (Fig. 3 and 4). These results show that targeting of HP1α or of HP1β is sufficient to induce chromatin condensation.
Heterochromatin formation is known to involve MeH3K9 (9). Subsequently, MeH3K9 creates a binding site for HP1, which in turn is able to bind HMT (4, 9, 19, 26). In this way, heterochromatin formation may spread along the chromatin fiber (14). Furthermore, multiple HMTs methylate the same lysine and lysine can be mono-, di-, or trimethylated. This raises the question how the activities of different enzymes are regulated and whether methylation by different enzymes has different consequences. Different enzymes might be targeted to different chromosomal domains and/or genes. Alternatively, different enzymes might have different preferences toward non-, mono-, and dimethylated substrates. SU(VAR)3-9 HMT is specific for tri-MeH3K9 at constitutive heterochromatin, whereas G9a HMT, a dominant di-MeH3K9, and minor monomethylated H3K9 (mono-MeH3K9) most likely functions at facultative heterochromatin. The remaining mono-MeH3K9 may be attributed to euchromatic histone methyltransferase 1, an HMT with significant homology to G9a (7, 40, 50, 55). Concerning ESET/SETDB1 it can be a di- or tri-MeH3K9 in vivo (45). Together these observations suggest that specific histone methyltransferases direct mono- and di-MeH3K9 to silent domains in euchromatin, whereas the bulk of tri-MeH3K9 is directed to pericentric regions typically associated with constitutive heterochromatin. In this context, our in vivo data show that in the absence of HP1, chromatin of the amplified region is hypotrimethylated at H3K9 and contains low levels of endogenous HP1α, HP1β, and SETDB1, whereas targeting of HP1α or HP1β recruits HMT SETDB1 and induces tri-MeH3K9, which is recognized by anti-tri-MeH3K9 and anti-branched(4×)-MeH3K9 antibodies. In contrast, the level of di-meH3K9 did not increase upon HP1α or HP1β targeting to the amplified chromosome region. di-MeH3K9 is observed throughout the nucleoplasm, whereas tri-MeH3K9 is enriched in pericentromeric heterochromatin (39). Our data underline the current view that defined histone methyltransferases are responsible for adding or subtracting histone lysine methylation degrees (mono- versus di- versus tri-), thereby effecting distinct downstream events.
In line with our observations in mammalian cells, Li et al. (24) showed in Drosophila that HP1 targeting as a lacR-HP1 fusion is able to induce gene silencing in a euchromatin locus. They showed that HP1-induced silencing appeared to function downstream of the SU(VAR)3-9-induced di-MeH3K9, as they observed no increased di-MeH3K9 upon HP1 targeting and no suppression of gene silencing or reduction in HP1 levels in a SU(VAR)3-9 mutant background. These results are consistent with our observations, since we also find that HP1 targeting causes no enrichment of di-MeH3K9. In our present study we show that HP1 targeting leads to enrichment of tri-MeH3K9 as well as SETDB1 at the targeted locus. In this mammalian system, HP1 therefore is sufficient to directly or indirectly induce tri-MeH3K9 modifications, presumably by recruitment of SETDB1. In more recent work by the Wallrath group (10) they demonstrated that HP1 induced gene silencing and nucleosomal restriction enzyme accessibility were similar at reporter genes positioned 1.9 or 3.7 kb downstream of lac operator repeats in wild-type flies, indicating that the HP1 effect can spread over these distances. Moreover, they showed that the HP1 effect in the SU(VAR)3-9 mutant background is minimally effected at 1.9 kb but is eliminated at 3.7 kb, suggesting that HP1-mediated silencing can operate both in a SU(VAR)3-9-dependent and -independent way. Apparently, the short-range HP1 effect might function independent of SU(VAR)3-9 induced histone methylation, but the long-range silencing effects of HP1 might in fact depend on tri-MeH3K9. This would argue for a functional role of the SU(VAR)3-9 recruitment and increased histone methylation for long-range silencing. It is intriguing that upon HP1 targeting in our assay we observe a tri-MeH3K9, SETDB1 (and less clear for endogenous HP1) signal which encompasses but appears significantly larger than the EGFP-tagged amplified chromosome region, suggesting a long-range propagation of increased histone methylation. These studies clearly illustrate that the role of HP1 at a given locus depends on the presence of interacting partners at that locus. Ayyanathan et al. (3) demonstrated in mammalian cells using a synthetic hormone regulated KRAB repression domain that HP1, HMT SETDB1 and MeH3K9 are enriched at the expressed transgene. In this system they observe silencing of the transgene and recruitment of the transgene close to HP1 rich regions assuming the induction of a compact chromatin structure. The observations made with this regulated recruitment of HP1 are in agreement with our findings. In this context a very recent study of the McNally group (32) shows similar light microscopical observations of changes in the large-scale chromatin structure of natural chromosome domains consisting of MMTV tandem arrays when they are either transcriptionally active or less active.
Our study focused on the in situ changes in large-scale chromatin structure upon targeting HP1 to a chromosomal array. We show for the first time that direct targeting of HP1α and HP1β causes the formation of a condensed large-scale chromatin structure in combination with enrichment of tri-MeH3K9 (not di-MeH3K9), SETDB1 and the other HP1 homologue as visualized in vivo at the single cell level. Recently, Janicki et al. (20) developed an inducible system, also based on the lac operator/repressor arrays, to correlate changes in chromatin structure with transcriptional activation. They showed that the 200-copy transgene array occurs as a condensed MeH3K9 locus, where HP1 is bound. Upon transcriptional activation and hence decondensation of the chromatin structure of the array, both HP1 and MeH3K9 disappear from the locus. In the present study, we show the reverse process in vivo, i.e., large-scale chromatin condensation, enhanced tri-MeH3K9, and recruitment of histone methyltransferase SETDB1 upon direct HP1α or HP1β targeting.
Together, our observations indicate that HP1 plays a key role in the formation of a condensed chromatin state in mammalian cells. However, recent experiments by us, using a dominant-negative approach for overexpressing truncated HP1 that lacks a functional chromodomain, showed that HP1α or HP1β can be competed out of the mouse chromocenters without changing their compact chromatin structure (G. Mateos-Langerak et al., unpublished). These data indicate that HP1 is not required for maintaining the heterochromatin state. The structure of HP1, containing CD, CSD, and HD, allows it to bind to a variety of proteins (27). Our data indicate that components from the other class of silencing factors, the PcG proteins, are not recruited by HP1α or HP1β. Moreover, HP1 forms are able to interact with each other via their CSDs. Our in vivo data support the notion that this interaction is probably responsible for the recruitment of HP1α by HP1β and vice versa.
In summary, our in vivo results show for the first time that HP1α and HP1β are able to induce heterochromatin formation and concomitant large-scale changes in chromatin structure, involving HMT SETDB1 and tri-MeH3K9 and the recruitment of endogenous HP1. Combining our observations with those of others suggests that the transition to heterochromatin can be triggered by HP1 as well as by tri-MeH3K9. Together, these processes seem to form a self-propagating loop of events, resulting in spreading of the heterochromatin state along the chromatin fiber. To further unravel the mechanism of HP1 in gene regulation, it will be important to characterize the interacting components at a given genomic site.
Supplementary Material
Acknowledgments
We are grateful to P. B. Singh, T. Jenuwein, D. C. Schultz, and A. P. Otte for kindly providing us with the antibodies used in the present study. Confocal microscopy was performed at the Centre for Advanced Microscopy. We gratefully thank E. M. M. Manders for expert assistance.
This work was supported by ALW-NWO PULS and VIDI grants to P. J. V. (project numbers PULS/33-98l/805-48011 and VIDI2003/03921/ALW/016.041.311).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aasland, R., and A. F. Stewart. 1995. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 23:3168-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, K., R. Mahr, B. Bjorkroth, and B. Daneholt. 1982. Rapid reformation of the thick chromosome fiber upon completion of RNA synthesis at the Balbiani ring genes in Chironomus tentans. Chromosoma 87:33-48. [DOI] [PubMed] [Google Scholar]
- 3.Ayyanathan, K., M. S. Lechner, P. Bell, G. G. Maul, D. C. Schultz, Y. Yamada, K. Tanaka, K. Torigoe, and F. J. Rauscher. 2003. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 17:1855-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 5.Belmont, A. S. 2001. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 11:250-257. [DOI] [PubMed] [Google Scholar]
- 6.Bunker, C. A., and R. E. Kingston. 1996. Activation domain-mediated enhancement of activator binding to chromatin in mammalian cells. Proc. Natl. Acad. Sci. USA 93:10820-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, R., Wang, L., Wang, H., Xia, L. Erdjument-Bromage, H., Tempst, P., Jones, R., and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 8.Chambeyron, S., and W. A. Bickmore. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowell, I. G., R. Aucott, S. K. Mahadevaiah, P. S. Burgoyne, N. Huskisson, S. Bongiorni, G. Prantera, L. Fanti, S. Pimpinelli, R. Wu, D. M. Gilbert, W. Shi, R. Fundele, H. Morrison, P. Jeppesen, and P. B. Singh. 2002. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111:22-36. [DOI] [PubMed] [Google Scholar]
- 10.Danzer, J. R., and L. L. Wallrath. 2004. Mechanisms of HP1-mediated gene silencing in Drosophila. Development 131:3571-3580. [DOI] [PubMed] [Google Scholar]
- 11.Eissenberg, J. C., T. C. James, D. M. Foster-Hartnett, T. Hartnett, V. Ngan, and S. C. Elgin. 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 87:9923-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Festenstein, R., S. SharghiNamini, M. Fox, K. Roderick, M. Tolaini, T. Norton, A. Saveliev, D. Kioussis, and P. Singh. 1999. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat. Genet. 23:457-461. [DOI] [PubMed] [Google Scholar]
- 13.Fischle, W., Y. M. Wang, S. A. Jacobs, Y. C. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 bv polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grewal, S. I. S., and D. Moazed. 2003. Heterochromatin and epigenetic control of gene expression. Science 301:798-802. [DOI] [PubMed] [Google Scholar]
- 15.Hall, I. M., G. D. Shankaranarayana, K. I. Noma, N. Ayoub, A. Cohen, and S. I. S. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 16.Hamer, K. M., R. G. Sewalt, J. L. Den Blaauwen, T. Hendrix, D. P. Satijn, and A. P. Otte. 2002. A panel of monoclonal antibodies against human polycomb group proteins. Hybrid Hybridomics 21:245-252. [DOI] [PubMed] [Google Scholar]
- 17.Horn, P. J., and C. L. Peterson. 2002. Chromatin higher order folding—wrapping up transcription. Science 297:1824-1827. [DOI] [PubMed] [Google Scholar]
- 18.Horsley, D., A. Hutchings, G. W. Butcher, and P. B. Singh. 1996. M32, a murine homologue of Drosophila heterochromatin protein 1 (HP1), localises to euchromatin within interphase nuclei and is largely excluded from constitutive heterochromatin. Cytogenet. Cell Genet. 73:308-311. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295:2080-2083. [DOI] [PubMed] [Google Scholar]
- 20.Janicki, S. M., T. Tsukamoto, S. E. Salghetti, W. P. Tansey, R. Sachidanandam, K. V. Prasanth, T. Ried, Y. Shav-Tal, E. Bertrand, R. H. Singer, and D. L. Spector. 2004. From silencing to gene expression: real-time analysis in single cells. Cell 116:683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 22.Lachner, M., and T. Jenuwein. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286-298. [DOI] [PubMed] [Google Scholar]
- 23.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 24.Li, Y. H., J. R. Danzer, P. Alvarez, A. S. Belmont, and L. L. Wallrath. 2003. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 130:1817-1824. [DOI] [PubMed] [Google Scholar]
- 25.Lund, A. H., and M. Lohuizen. 2004. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 16:239-246. [DOI] [PubMed] [Google Scholar]
- 26.Maison, C., D. Bailly, A. H. F. M. Peters, J. P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329-334. [DOI] [PubMed] [Google Scholar]
- 27.Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell. Biol. 5:296-304. [DOI] [PubMed] [Google Scholar]
- 28.Marr, D., and E. Hildrith. 1980. Theory of edge detection. Proc. R. Soc. London B 207:187-217. [DOI] [PubMed] [Google Scholar]
- 29.Minc, E., V. Allory, H. J. Worman, J. C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108:220-234. [DOI] [PubMed] [Google Scholar]
- 30.Muchardt, C., M. Guilleme, J. S. Seeler, D. Trouche, A. Dejean, and M. Yaniv. 2002. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1 alpha. EMBO Rep. 3:975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller, W. G., D. Walker, G. L. Hager, and J. G. McNally. 2001. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 154:33-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller, W. G., D. Rieder, G. Kreth, C. Cremer, Z. Trajanoski, and J. G. McNally. 2004. Generic features of tertiary chromatin structure as detected in natural chromosomes. Mol. Cell. Biol. 24:9359-9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, A. L., M. OuladAbdelghani, J. A. Ortiz, E. Remboutsika, P. Chambon, and R. Losson. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7:729-739. [DOI] [PubMed] [Google Scholar]
- 35.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 36.Nye, A. C., R. R. Rajendran, D. L. Stenoien, M. A. Mancini, B. S. Katzenellenbogen, and A. S. Belmont. 2002. Alteration of large-scale chromatin structure by estrogen receptor. Mol. Cell. Biol. 22:3437-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Brien, T. P., C. J. Bult, C. Cremer, M. Grunze, B. B. Knowles, J. Langowski, J. McNally, T. Pederson, J. C. Politz, A. Pombo, G. Schmahl, J. P. Spatz, and R. van Driel. 2003. Genome function and nuclear architecture: from gene expression to nanoscience. Genome Res. 13:1029-1041. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Burgos, L., A. H. Peters, S. Opravil, M. Kauer, K. Mechtler, and T. Jenuwein. 2004. Generation and characterization of methyl-lysine histone antibodies. Methods Enzymol. 376:234-254. [DOI] [PubMed] [Google Scholar]
- 40.Peters, A. H. F. M., S. Kubicek, K. Mechtler, R. J. O'Sullivan, A. A. H. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. A. Martens, and T. Jenuwein. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12:1577-1589. [DOI] [PubMed] [Google Scholar]
- 41.Pratt, E. K. 1991. Digital image processing, p. 497-508. John Wiley & Sons, Inc., San Francisco, Calif.
- 42.Ragoczy, T., A. Telling, T. Sawado, M. Groudine, and S. T. Kosak. 2003. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 11:513-525. [DOI] [PubMed] [Google Scholar]
- 43.Robinett, C. C., A. Straight, G. Li, C. Willhelm, G. Sudlow, A. Murray, and A. S. Belmont. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satijn, D. P. E., M. J. Gunster, J. van der Vlag, K. M. Hamer, W. Schul, M. J. Alkema, A. J. Saurin, P. S. Freemont, R. Van Driel, and A. P. Otte. 1997. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol. Cell. Biol. 17:4105-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz, D. C., K. Ayyanathan, D. Negorev, G. G. Maul, and F. J. Rauscher. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16:919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sewalt, R. G. A. B., M. Lachner, M. Vargas, K. M. Hamer, J. L. denBlaauwen, T. Hendrix, M. Melcher, D. Schweizer, T. Jenuwein, and A. P. Otte. 2002. Selective interactions between vertebrate polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of polycomb group proteins. Mol. Cell. Biol. 22:5539-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh, P. B., and S. D. Georgatos. 2002. HP1: facts, open questions, and speculation. J. Struct. Biol. 140:10-16. [DOI] [PubMed] [Google Scholar]
- 48.Singh, P. B., J. R. Miller, J. Pearce, R. Kothary, R. D. Burton, R. Paro, T. C. James, and S. J. Gaunt. 1991. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 19:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 50.Tachibana, M., Sugimoto, K., Nozaki, M., Ueda, J., Ohta, T., Ohki, M., Fukuda, M., Takeda, N., Kato, H., and Y. Shinkai. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 Lysine 9 methylation. Genes Dev. 16:1779-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsukamoto, T., N. Hashiguchi, S. M. Janicki, T. Tumbar, A. S. Belmont, and D. L. Spector. 2000. Visualization of gene activity in living cells. Nat. Cell Biol. 2:871-878. [DOI] [PubMed] [Google Scholar]
- 52.Tumbar, T., G. Sudlow, and A. S. Belmont. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Vlag, J., J. L. den Blaauwen, R. G. A. B. Sewalt, R. van Driel, and A. P. Otte. 2000. Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J. Biol. Chem. 275:697-704. [DOI] [PubMed] [Google Scholar]
- 54.Verschure, P. J., I. Van Der Kraan, J. M. Enserink, M. J. Mone, E. M. Manders, and R. Van Driel. 2002. Large-scale chromatin organization and the localization of proteins involved in gene expression in human cells. J. Histochem. Cytochem. 50:1303-1312. [DOI] [PubMed] [Google Scholar]
- 55.Wang, H., W. An, R. Cao, L. Xia, H. Erdjument-Bromage, B. Chatton, P. Tempst, R. G. Roeder, and Y. Zhang. 2003. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12:475-487. [DOI] [PubMed] [Google Scholar]
- 56.Wreggett, K. A., F. Hill, P. S. James, A. Hutchings, G. W. Butcher, and P. B. Singh. 1994. A mammalian homologue of Drosophila heterochromatin protein 1 (Hp1) is a component of constitutive heterochromatin. Cytogenet. Cell Genet. 66:99-103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.