Abstract
During the mitotic cell cycle, microtubule depolymerization leads to a cell cycle arrest in metaphase, due to activation of the spindle checkpoint. Here, we show that under microtubule-destabilizing conditions, such as low temperature or the presence of the spindle-depolymerizing drug benomyl, meiotic budding yeast cells arrest in G1 or G2, instead of metaphase. Cells arrest in G1 if microtubule perturbation occurs as they enter the meiotic cell cycle and in G2 if cells are already undergoing premeiotic S phase. Concomitantly, cells down-regulate genes required for cell cycle progression, meiotic differentiation, and spore formation in a highly coordinated manner. Decreased expression of these genes is likely to be responsible for halting both cell cycle progression and meiotic development. Our results point towards the existence of a novel surveillance mechanism of microtubule integrity that may be particularly important during specialized cell cycles when coordination of cell cycle progression with a developmental program is necessary.
In the course of gamete production, a specialized cell division called meiosis creates four haploid cells from one diploid progenitor. Many aspects of cell cycle regulation are similar during proliferative mitotic growth and meiosis, but the different division pattern of meiosis requires modification of the mitotic cell cycle machinery to fit the needs of the meiotic differentiation program. During the meiotic cell cycle, DNA is replicated once and then separated twice during meiosis I and meiosis II without an intervening S phase. In addition, a prolonged G2 phase (meiotic prophase) separates premeiotic DNA replication from the first meiotic division. During meiotic prophase, homologous chromosomes align, and meiotic recombination creates the links between homologs that are necessary for proper meiosis I chromosome segregation. After completion of prophase, homologous chromosomes are segregated first during meiosis I, followed by the segregation of sister chromatids during meiosis II.
Concurrent with the chromosomal events, cells progress through an intricate developmental program that culminates in the production of highly specialized cell types, such as sperm and egg, or spores in budding yeast. For gamete formation to occur successfully, it is essential that the meiotic cell cycle and the developmental program are tightly coupled via molecular interactions that we are only beginning to understand. Mutations that uncouple meiotic cell cycle progression from spore formation emphasize how important these interactions are. For example, cells that fail to decrease cyclin-dependent kinase (CDK) activity cannot disassemble the meiosis I spindle, but other aspects of the meiotic cell cycle and the developmental program (spore production) continue, leading to the formation of defective gametes (5, 36).
The complex transcriptional program that underlies gametogenesis appears to be one key level of control that couples the meiotic cell cycle to gamete development. Stage-specific expression of crucial meiotic regulators controls most meiotic processes, including meiotic recombination, formation of the synaptonemal complex (SC), meiosis I chromosome segregation, and spore wall formation. Factors required for the respective cell cycle stage and the corresponding developmental genes are coordinately up-regulated within characteristic transcriptional waves, creating a link between cell cycle and development (7, 8, 44). For example, nutrient limitation that provides the signal for entry into the meiotic cell cycle is relayed through the transcription factor Ime1 (17), which is not only essential for initiating cell cycle entry but also responsible for inducing the transcription of genes required for the early metabolic program of gametogenesis. It does so by interacting with the transcriptional modulator Ume6 that coordinates expression of at least 80 loci involved in metabolic, as well as early and middle meiotic, gene functions (68). Similarly, at the end of meiotic prophase, the transcription factor Ndt80 induces not only expression of genes essential for entry into meiosis I but also that of genes important for prospore wall assembly (7, 8, 44).
Several meiotic events are monitored by surveillance mechanisms known as checkpoints. Checkpoints halt cell cycle progression until the event that is surveyed has been completed, thereby ensuring that the cell cycle, as well as developmental events, occur in an ordered manner. Typically, the transcription factors required to initiate the next phase of the meiotic cell cycle are under direct checkpoint control. For example, the transcription factor NDT80 is a direct target of the recombination (or pachytene) checkpoint (49, 63), which prevents entry into meiosis I and prospore wall assembly until the DNA damage created during meiotic recombination is repaired. Thus, both the initiation of prospore wall assembly and entry into meiosis I are coupled to the completion of meiotic prophase.
The integrity of the microtubule cytoskeleton is also monitored by surveillance mechanisms. In mitotic cells, perturbations of the microtubule cytoskeleton lead to unattached kinetochores, which cause activation of the spindle assembly checkpoint (32, 56, 57). The spindle checkpoint component Mad2 binds to and inhibits the anaphase-promoting complex or cyclosome (APC/C), a crucial activator of chromosome segregation (20). This inhibition causes a cell cycle arrest in metaphase until the spindle defects have been repaired. Disruption of the microtubule cytoskeleton during the meiotic cell cycle by exposure to the microtubule-depolymerizing drug benomyl (methyl 1-[butylcarbamoyl]-2-benzimidazolecarbamate) has also been reported to cause a transient delay in metaphase I that, much like in the mitotic cell cycle, is dependent upon the checkpoint component Mad2 (56).
Here, we report that severe disruption of the yeast microtubule cytoskeleton after premeiotic DNA replication leads to a G2 rather than a metaphase I arrest, with low protein levels of cyclin Clb3 and incompletely synapsed chromosomes. Whole-genome expression profiling revealed that this arrest is accompanied by a substantial change in the meiotic gene expression program. In particular, genes essential for meiotic recombination, cell cycle progression, or spore formation and/or maturation are not expressed at wild-type levels. Down-regulation of meiotic transcripts also occurs when microtubules are destabilized by culturing cells at 10°C. Finally, we demonstrate that the G2 arrest caused by microtubule depolymerization is independent of the checkpoints controlling spindle assembly and meiotic recombination. Our results indicate that the transcriptional response to microtubule perturbations serves to bring both the meiotic developmental program and the cell cycle to a halt. Our data also point towards the existence of a novel mechanism of microtubule integrity surveillance that coordinates the meiotic cell cycle with spore development.
MATERIALS AND METHODS
Strains.
Strains used are described in Table 1 and are derivatives of SK1 (24). CLB3-3HA::kanMX6 and mad2Δ::kanMX6 were constructed by a one-step gene replacement method (34). mek1::kanMX4 and swe1::kanMX4 were PCR amplified from the corresponding knockout strains in the Saccharomyces deletion collection (13) and introduced into SK1. REC8-3HA and URA3::tetO were described in reference 27 and LEU2::pURA3-tetR-GFP::tetO::HIS3 was described in reference 37. rad50::URA3 was described in reference 6.
TABLE 1.
Strains
| Strain | Relevant genotype |
|---|---|
| A727 | MATa/α ho::LYS2/ho::LYS2 lys2/lys2 his4X/his4X leu2::hisG/leu2::hisG ura3/ura3 |
| A1771 | MATa/α rad50::URA3/rad50::URA3 |
| A1972 | MATa/α REC8-3HA::URA3/REC8-3HA::URA3 |
| A4563 | MATa/α CLB3-3HA::kanMX6/+ |
| A4704 | MATa/α swe1Δ::kanMX4/swe1Δ::kanMX4, CLB3-3HA::kanMX6/+ |
| A4838 | MATa/α mek1Δ::kanMX4/mek1Δ::kanMX4 |
| A4843 | MATa/α mad2::kanMX/mad2::kanMX, CLB3-3HA::kanMX6/+ |
| A4967 | MATa/α URA3::tetOx224 (Chr.V)/+ LEU2::pURA3-tetR-GFP/LEU2::pURA3-tetR-GFP::tetO:: HIS3 (Chr.III) |
| A5009 | MATa/α URA3::tetOx224 (Chr.V)/URA3::tetOx224 (Chr.V) LEU2::pURA3::tetR-GFP/LEU2:: pURA3:: tetR-GFP |
| A5779 | MATa/α tub2-150/tub2-150 CLB3-3HA::kanMX6/+ |
| A12927 | MATa/α mad2::kanMX/mad2::kanMX mek1Δ::kanMX4/mek1Δ::kanMX4 CLB3-3HA::kanMX6/+ |
Sporulation conditions.
Cells were grown to saturation in YPD (yeast extract-peptone plus 2% glucose) for 24 h, diluted into YPA (yeast extract-peptone plus 2% potassium acetate) at an optical density at 600 nm of 0.3 and grown overnight (16 h) at 30°C. Cells were washed with sterilized water the next day and resuspended in SPO medium (0.3% potassium acetate; pH = 7.0) at an optical density at 600 nm of 1.9 to induce sporulation. Cells were sporulated at 25°C or 10°C as indicated. Sporulation medium containing benomyl was always prepared freshly on the day of the experiment, following the directions in reference 56. Briefly, dimethyl sulfoxide (DMSO; Sigma-Aldrich) or benomyl (30 mg/ml stock in DMSO; Sigma-Aldrich) was dissolved in near-boiling SPO medium to avoid precipitation of benomyl. The medium was then allowed to slowly cool to room temperature. At the time of drug treatment, cells were filtered and immediately resuspended in the medium containing benomyl or DMSO.
Immunofluorescence and meiotic spreads.
Unless noted otherwise, 200 cells were scored for each time point. Indirect immunofluorescence on whole cells was carried out as described previously (65). Rat α-tubulin antibody (Oxford Biotechnology) and fluorescein isothiocyanate-conjugated α-rat antibody (Jackson ImmunoResearch) were diluted at 1:200 and 1:100, respectively. Meiotic spreads were performed as described previously (42). α-Zip1 antibody (kindly provided by N. Kleckner) and fluorescein isothiocyanate-conjugated α-rabbit antibody (Jackson ImmunoResearch) were diluted at 1:150 and 1:50, respectively. Immunofluorescence samples were analyzed with a Zeiss Axioplan 2 microscope.
Immunoprecipitation and kinase assays.
H1 kinase assays were performed as described previously (2). Briefly, Clb3-3HA protein was immunoprecipitated from 50 μl crude extract (250 μg total protein) using α-hemagglutinin (α-HA) antibody (Babco) and immunoglobulin G (IgG) Sepharose (Amersham Pharmacia). Sepharose beads were washed extensively with NP-40 buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40) and 25 mM MOPS (morpholinepropanesulfonic acid [pH 7.0]). Beads were preincubated with 6 μl buffer HBII (25 mM MOPS, 15 mM MgCl2, 5 mM EGTA, and 1 mM dithiothreitol, supplemented with protease and phosphatase inhibitors) for 15 min, before the addition of 10 μl kinase reaction mixture (25 mM MOPS, 2 mg/ml histone H1, 0.2 mM ATP) containing 50 nCi [γ-32P]ATP. Kinase reactions were allowed to proceed for 15 min at 25°C before they were stopped by the addition of 10 μl 3× sodium dodecyl sulfate loading buffer. Samples were separated on a 15% sodium dodecyl sulfate-acrylamide gel, fixed in 10% methanol-10% acetic acid, dried, and analyzed by autoradiography.
Northern blot analysis.
Northern blot analysis was performed as described previously (2). Total RNA was purified by phenol extraction and ethanol precipitation. A total of 10 μg RNA was loaded per lane and separated on a 1.1% agarose gel containing 6% formaldehyde and 40 mM MOPS (pH 7.0). Gels were blotted in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH = 7.0]) onto Hybond-XL membranes (Amersham Biosciences). Blots were probed overnight with randomly 32P-labeled DNA probes typically spanning ∼1 kb of the respective open reading frame.
cRNA target synthesis and microarray hybridization.
Frozen yeast cell pellets stored at −80°C were quickly thawed and processed by the hot-phenol method (28). Approximately 190 μg of total RNA was isolated from 0.7 × 108 cells. Subsequently, 80 μg of total RNA was mixed with 350 μl RLT buffer and 250 μl of 70% ethanol. The mix was loaded onto an RNeasy Mini-Spin column (QIAGEN), and RNA was eluted in 50 μl of double-distilled water. Total RNA quality was monitored by loading approximately 200 ng onto an RNA Nano 6000 Chip processed with the 2100 Bioanalyzer (Agilent) (see Fig. S1A, top, in the supplemental material). Biotin labeling of RNA was performed as described in the Expression Analysis Technical Manual (Affymetrix) with minor modifications as previously published (51). Approximately 80 μg of labeled cRNA from each reaction was purified using RNeasy Mini-Spin columns, and roughly 300 ng was analyzed on RNA Nano 6000 Chips (see Fig. S1A, bottom, in the supplemental material). A total of 220 μl of the hybridization mixture containing target cRNA at a final concentration of 0.05 μg/μl was transferred onto S98 GeneChips (Affymetrix) and incubated at 45°C on a rotator in a Hybridization Oven 640 (Affymetrix) for 16 h at 60 rpm. The arrays were washed, stained, and scanned as previously described (51). DAT (image), CEL (feature or oligonucleotide probe level), and TXT (gene or probe set level) files of the GeneChips were generated using an Agilent GeneArray scanner (low-intensity settings) and Microarray Analysis Suite 5.0 (Affymetrix).
Microarray data analysis.
Raw data at the feature (oligonucleotide probe) level (CEL files) computed using the algorithm implemented in MAS 5.0 (Affymetrix) were imported into software packages available via the BioConductor project (http://www.bioconductor.org/). Data from CEL files were normalized, analyzed, and clustered using the R language for statistical computing (http://www.r-project.org) and the variance stabilization (VSN) algorithm (19) as implemented in the BioConductor package (http://bioconductor.org), following basic procedures as previously described (51). The Perfect Match values for all probes of a probe set were averaged to yield a single expression value for each gene represented on the array by employing the median polish algorithm as previously published (21). Oligonucleotide probes yielding questionable signals were marked during the image analysis, and the features (probes) at the 3′-most position of each probe set were disregarded during signal computation to decrease the effect of signal artifacts and RNA degradation on data quality (since cRNA synthesis proceeds in a 5′-to-3′ direction). A detailed documentation of all processing and analysis steps (30) is provided as supplemental material on our web portal at http://www.biozentrum.unibas.ch/primig/benomyl/, which provides access to web-specific figures, hyperlinked tables, supporting information, and raw data files. The file provides a code in the R programming language combined with comments in LATEX format. It can be downloaded together with the raw data as a single R package. Within R, the code can be extracted or a PDF document providing an overview of the analysis can be generated. A graphic display of the expression data for each locus is accessible via the GermOnline knowledgebase at http://www.germonline.org (43, 66, 67). A total of 1,189 transcripts that displayed a strong variation of signals between different samples compared to (theoretically) identical replicates were grouped using a hierarchical clustering algorithm (Euclidian distance) and sorted over signal strength during mitotic growth. To search for functional gene ontology (GO) annotations that are correlated with a particular expression pattern in a statistically significant manner, we employed the goCluster tool. Briefly, all loci for which reliable data are available (without prior filtration) are first grouped via their expression patterns using k-means clustering; subsequently, the genes bearing related functional annotation within each cluster are identified using a statistical test (G. Wrobel and M. Primig, submitted for publication). In addition, we directly identified a number of functionally related loci through other GO categories.
MIAME compliance.
The TXT and CEL data files corresponding to cells in rich and presporulation medium (YPD and YPA), sporulation medium (SP4 and SF5), treated samples (SD5, SD8, SB5, and SB8), and cold-shocked samples as well as the appropriate controls (SR4, SR5, SR8, SC5, and SC8) were uploaded to the GEO (National Center for Biotechnology Information) public data repository at http://www.ncbi.nlm.nih.gov/geo/ (12). Files can be retrieved using the accession number GSE1693. CEL and TXT data files of all samples are also available at our web portal.
Other techniques.
Spore viability was analyzed by dissection of tetrads. Flow cytometric analysis of total cellular DNA content and immunoblot analysis was performed as previously described (64). For immunoblotting, α-HA antibody (HA.11; Babco) was used at a 1:500 dilution, α-CLB3 (rabbit; Santa Cruz Biotechnology) was used at a 1:200 dilution, α-Cdc28 antibody was used at a 1:1,000 dilution, and α-Cdc2-Tyr15-P antibody (Cell Signaling Technology) was used at a 1:1,000 dilution. Pairing behavior of green fluorescent protein (GFP)-marked chromosomes was analyzed in vivo with a Zeiss Axioplan 2 microscope.
RESULTS
Benomyl reversibly arrests cells during meiosis.
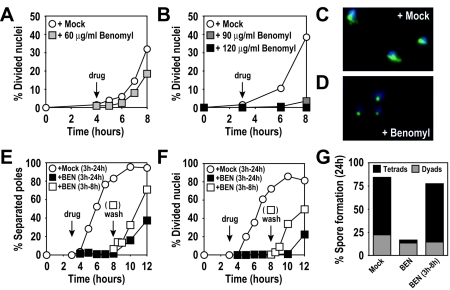
To study the consequences of microtubule perturbation on meiotic cell cycle progression and spore formation, we treated cells with the microtubule-depolymerizing drug benomyl. Previous reports indicated that a benomyl concentration of 60 μg/ml elicits a metaphase I delay in meiotic cells (56). We also observed a small delay in nuclear divisions at this concentration compared to a mock-treated culture (0.4% DMSO) (Fig. 1A). However, at 60 μg/ml, benomyl caused only incomplete microtubule depolymerization, and occasional short spindles were observed (data not shown). To more completely depolymerize microtubules, we incubated cells with 90 μg/ml or 120 μg/ml benomyl. The delay in nuclear division appeared more pronounced as the benomyl concentration was increased (Fig. 1B), with no cells undergoing nuclear divisions at the highest concentration of the spindle toxin, as was observed previously (58). In the presence of benomyl at 120 μg/ml, microtubules disappeared within minutes, and only the spindle pole bodies (SPBs) exhibited weak reactivity with anti-tubulin antibodies (Fig. 1C and D).
FIG. 1.
High levels of benomyl reversibly arrest meiotic cells. (A) Wild-type (A727) cells. At 4 h after meiotic induction at room temperature (black arrow with the label “drug”), cells were resuspended in medium containing 60 μg/ml benomyl (grey squares) or 0.4% DMSO (mock; white circles). (B) Wild-type (A727) cells. At 3 h after meiotic induction at 30°C (black arrow), cells were resuspended in medium containing 90 μg/ml benomyl (dark grey squares), 120 μg/ml benomyl (black squares), or 0.4% DMSO (mock, white circles). The percentages of cells in the results shown in panels A and B that underwent at least one nuclear division were determined by DAPI (4′,6-diamidino-2-phenylindole) staining at the indicated time points. (C and D) Wild-type (A727) cells from cultures treated with 0.4% DMSO (C) or 120 μg/ml benomyl (D) fixed 1 h after treatment. DAPI-stained nuclear masses are shown in blue, and tubulin is shown in green. (E to G) Wild-type (A727) cells. At 3 h after meiotic induction at 30°C (arrow with the label “drug”), cells were resuspended in medium containing 120 μg/ml benomyl (black squares) or 0.4% DMSO (mock; white circles). At 5 h after drug addition (arrow labeled “wash”), the benomyl culture was split, and half the culture was washed and released into fresh sporulation medium containing 0.4% DMSO (white squares). (E) Percentage of cells containing more than one focus of tubulin staining. (F) Percentage of cells having undergone nuclear divisions (DAPI). (G) Spore formation 24 h after induction of meiosis. Asci were classified as containing two spores (dyads) or three or four spores (tetrads) (500 cells were analyzed for each condition).
The benomyl concentration used in this analysis was three to eight times higher than the concentration typically used to arrest mitotically growing cells (35, 50). It was therefore important to determine that such high benomyl concentrations were not toxic to the cells. To investigate this, cells were incubated for 5 h with the compound, washed, and released into sporulation medium. Cells rapidly resumed the meiotic program, initiated meiotic spindle formation as judged by the separation of SPBs (Fig. 1E), and underwent the nuclear divisions with kinetics indistinguishable from mock-treated cells (Fig. 1F). Furthermore, the severe sporulation defect was reversed when benomyl was removed from the medium (Fig. 1G), and spore viability was indistinguishable from that of mock-treated cells (mock treated, 93%; benomyl-treated, 96%; 240 spores analyzed for each condition). Our results indicate that high levels of benomyl effectively inhibit progression through meiosis in a reversible manner without apparent adverse effects on meiotic cell cycle progression and spore formation.
Benomyl treatment prevents Clb-CDK accumulation in meiotic cells.
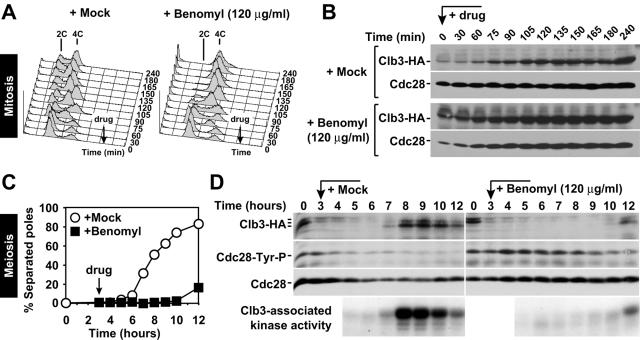
Treatment of mitotically growing cells with 15 μg/ml benomyl causes cells to arrest in metaphase, with unseparated sister chromatids and high levels of mitotic (Clb) CDK activity (32). We obtained similar results when we increased this concentration to 120 μg/ml benomyl (Fig. 2A and B). To determine the effects of high levels of benomyl on meiotic cell cycle progression, we characterized the cell cycle arrest caused by addition of the drug. Exposure to 120 μg/ml benomyl caused cells to arrest with unseparated SPBs (Fig. 1D and 2C), as has been previously observed with mitotically growing cells treated with this drug (22). However, in contrast to mitotically growing cells, meiotic cells treated with benomyl during or after completion of S phase exhibited a dramatic delay in the accumulation of Clb3 protein and associated kinase activity (Fig. 2). Furthermore, Cdc28, the catalytic subunit of CDKs, was phosphorylated on tyrosine 19, which reflects a cell cycle arrest prior to entry into the chromosome segregation phase (41). Thus, whereas mitotic cells treated with high levels of benomyl arrest in metaphase, meiotic cells cultured under similar conditions during S phase and prophase arrested prior to the accumulation of Clb-CDKs that is necessary for entry into meiosis I.
FIG. 2.
Meiotic cells arrest with low levels of Clb3 protein and low Clb3-associated kinase activity. (A and B) Wild-type cells carrying a CLB3-3HA fusion (A4563) were enriched in G1 by acetate starvation and released into YPD medium containing 0.4% DMSO (panel A, left) or 120 μg/ml benomyl (panel A, right). (A) Total cellular DNA content determined by flow cytometry. Black arrows indicate the time of drug addition. (B) Western blot analysis of Clb3 and Cdc28 (loading control). (C and D) Wild-type cells carrying a CLB3-3HA fusion (A4563) at room temperature. At 3 h after meiotic induction (black arrow), cells were resuspended in medium containing 120 μg/ml benomyl (black squares) or 0.4% DMSO (mock; white circles). (C) Percentage of cells containing more than one focus of tubulin staining. (D) Western blot analysis of Clb3 (top panel), Cdc28 tyrosine 19 phosphorylation (second panel), and Cdc28 (loading control, third panel from top), and autoradiogram of H1-kinase activity of Clb3-3HA immunoprecipitated from crude extracts (bottom).
Chromosome pairing is defective in the presence of high levels of benomyl.
To further characterize the arrest induced by high levels of benomyl, we examined chromosome pairing and synapsis. Homologous chromosomes align during prophase, and this process is completed when a multilayered structure, the SC, has formed between homologs (48, 71). We spread meiotic nuclei at various time points before and after benomyl addition and stained them with an antibody against the SC component Zip1 (59). The appearance of the first Zip1 foci was noticeably delayed in benomyl-treated cells compared to mock-treated cells, and SC formation along the entire length of chromosomes was even further delayed (Fig. 3A).
FIG. 3.
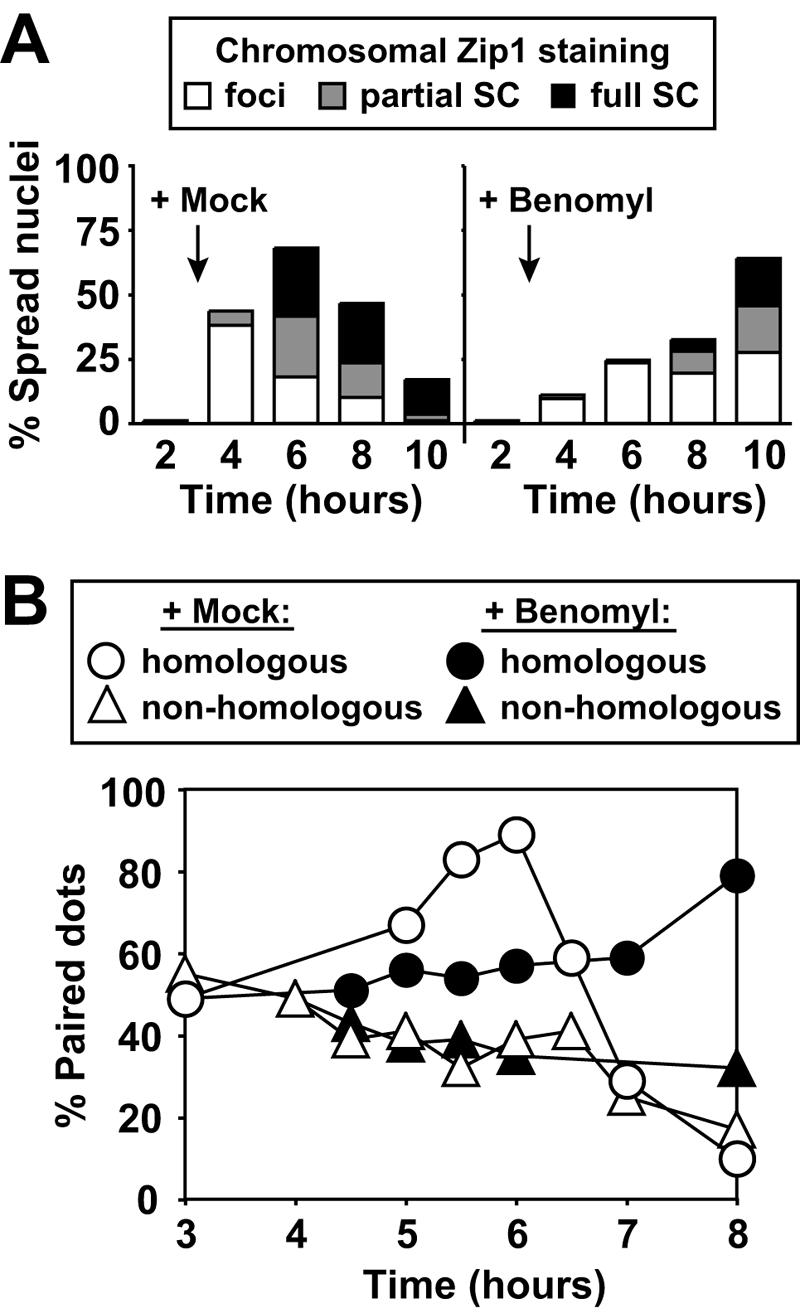
Benomyl exposure causes delayed homolog pairing and SC formation. (A) Cells from the experiment shown in Fig. 2C and D were surface spread and stained for Zip1. The spread nuclei were classified based on the presence of no staining, dispersed Zip1 foci (white), elongated Zip1 foci (partial SC; grey), or Zip1 fully covering chromosomes (full SC; black). Black arrows indicate the time of drug addition. A total of 200 nuclei were counted for each time point. (B) Wild-type cells carrying tetO arrays at homologous (A5009; circles) or nonhomologous (A4967; triangles) chromosomal positions. At 3 h after meiotic induction at room temperature, cells were resuspended in medium containing 120 μg/ml benomyl (black symbols) or 0.4% DMSO (mock; white symbols). Pairing was determined with 100 live cells at the indicated time points. Chromosomes were considered unpaired if two clearly separated GFP dots were visible and paired if the two GFP dots were partially or fully overlapping.
To examine the effects of benomyl on homolog pairing, we integrated tandem arrays of the Tet operator sequence (tetO) at the LEU2 locus on chromosome III (LEU2 arrays) or at the URA3 locus on chromosome V (URA3 arrays). The arrays were visualized by expressing a Tet repressor (TetR)-GFP fusion protein, which binds to the tetO arrays (38). To assay pairing, we created diploid cells with LEU2 arrays on both copies of chromosome III, which allowed us to analyze the behavior of a homologous locus. We assessed pairing by determining whether one or two GFP dots were visible within the cell. In this assay, only one GFP dot was visible if the arrays were paired or closely associated. As a control for nonspecific array clustering, we also examined diploid strains with one LEU2 array and one URA3 array, i.e., the two arrays were at nonhomologous positions. Around or shortly after premeiotic S phase (3-h time point), the homologous loci appeared more or less randomly arranged with respect to each other, since colocalization of LEU2/LEU2 (homologous) occurred at similar frequency as colocalization of LEU2/URA3 (nonhomologous) (Fig. 3B). In the mock-treated culture, the homologous LEU2 loci increasingly colocalized during prophase, reaching maximal pairing 6 h after transfer into sporulation medium. Pairing was then lost as cells completed the first meiotic division. The nonhomologous LEU2/URA3 combination, on the other hand, exhibited a slight drop in colocalization, presumably because the ongoing homolog alignment restricted random colocalization of nonhomologous sequences. In the benomyl-treated culture, association of homologous LEU2 loci was very much delayed (Fig. 3B). Thus, this delay in homolog pairing correlated well with the effect of benomyl on SC formation (Fig. 3A). Our results indicate that benomyl interferes with the pairing and synapsis of homologous sequences during meiosis.
Benomyl treatment causes cells to arrest in G1 or G2.
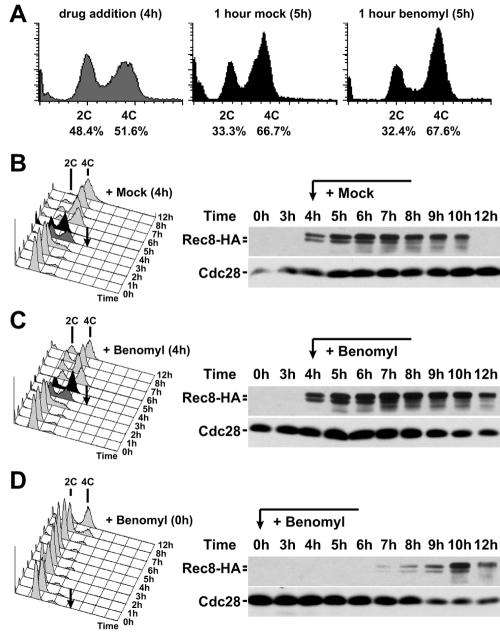
Our results suggest addition of benomyl during or shortly after S phase triggers a cell cycle arrest in G2. We next analyzed in more detail, whether benomyl affected the progression through S phase. To examine the effects of benomyl on premeiotic S phase, we determined the DNA content of benomyl-treated cells. Cells were induced to sporulate, and after 4 h benomyl was added. Progression through premeiotic S phase was not affected by benomyl. The 4C peak continued to increase for at least 1 h after benomyl addition (Fig. 4A to C). Quantification of the extent of DNA replication occurring within an hour of mock or benomyl treatment showed that the extent of DNA replication was similar in the two cultures (Fig. 4A). We did, however, notice that an unusually high fraction of cells with a 2C DNA content persisted in the culture at later time points after benomyl addition (Fig. 4B and C). This finding raised the possibility that the initiation of DNA replication was inhibited by benomyl. Thus, due to the partial asynchrony of sporulating cultures, some cells had not yet started premeiotic S phase at the point of benomyl addition, and these lagging cells were prevented from entering premeiotic S phase by benomyl treatment. To test this possibility, we analyzed the consequences on meiotic progression when benomyl was added to a meiotic culture at the time of meiotic induction (time = 0 h). Analysis of this culture revealed that exposure to benomyl markedly delayed premeiotic DNA synthesis. This delay was due to a defect in entry into the meiotic program because accumulation of an early meiosis-specific protein, Rec8 (27), was significantly delayed (Fig. 4D).
FIG. 4.
Benomyl treatment does not affect premeiotic S phase but prevents entry into meiosis. Wild-type cells carrying a REC8-3HA fusion (A1972) are shown. At 4 h after meiotic induction at room temperature (black arrow), cells were resuspended in medium containing 0.4% DMSO (mock) (B) or 120 μg/ml benomyl (C). Total cellular DNA content was determined by flow cytometry (left). Dark grey cytometry profiles indicate time of drug addition (4 h); black profiles are from 1 h after drug addition (5 h). Western blot samples were used to monitor Rec8 levels during the same time course. (A) Percentages of cells with 2C and 4C DNA content at 4 h and 5 h were determined by quantifying the area beneath the respective peaks ± one-half the respective interpeak distance. (D) Cells were treated as in the experiment described in panel C, except that cells were resuspended directly into sporulation medium containing 120 μg/ml benomyl at the start of the meiotic time course.
The failure of benomyl-treated cells to enter into the meiotic cell cycle was not due to a general toxicity of the drug but was mediated by the absence of microtubules. Cells harboring the benomyl-dependent allele of β-tubulin tub2-150 form overly stable microtubules that need to be destabilized by supplementation of the growth medium with at least 40 μg/ml benomyl (35, 62). Despite this need for benomyl, cells harboring the tub2-150 allele were still sensitive to high doses of benomyl and grew very slowly in medium containing 120 μg/ml benomyl (data not shown). We reasoned, however, that this allele would allow us to shift the sensitivity curve compared to wild-type cells, making the response to benomyl less severe. As described below, cells carrying the tub2-150 allele entered S phase in the presence of the drug (Fig. 5C). Thus, the arrest prior to premeiotic S phase was dependent on microtubule depolymerization. Our results indicate that benomyl causes two cell cycle arrests. It prevents entry into the first meiotic division if the drug is administered during or after premeiotic S phase and entry into the meiotic cell cycle if benomyl is administered during G1.
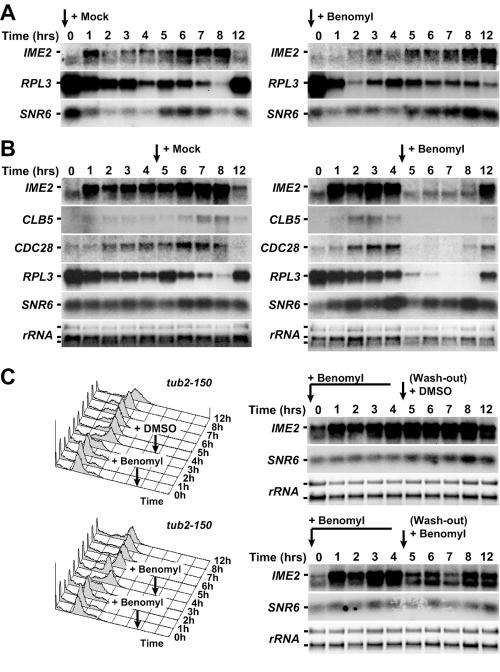
FIG. 5.
Benomyl treatment triggers changes in meiotic gene expression. (A) Northern blot analysis of IME2, RPL3, and SNR6 RNA levels. Wild-type cells (A1972) induced to undergo meiosis at room temperature in medium containing 0.4% DMSO (mock) (left) or 120 μg/ml benomyl (right). (B) The same strain as is shown in panel A was allowed to progress through meiosis for 4 h. After 4 h (black arrow), cells were resuspended in medium containing 0.4% DMSO (mock,) (left) or 120 μg/ml benomyl (right). Northern blot analysis of IME2, CLB5, CDC28, RPL3, and SNR6 RNA levels was carried out; rRNA was used as a loading control. rRNA levels were determined by staining with ethidium bromide; all other RNAs were detected by autoradiography. (C) tub2-150 cells (A5779) were pregrown at 30°C in medium containing 50 μg/ml benomyl and then sporulated at room temperature in medium containing 120 μg/ml benomyl. After 4 h, cells were resuspended in fresh medium containing 0.4% DMSO (mock) (top) or 120 μg/ml benomyl (bottom). Total RNA samples were analyzed by Northern blotting. Mock and benomyl-treated samples were run in the same gel but are separated in these figures for clarity. Total DNA content was analyzed by flow cytometry.
High levels of benomyl interfere with gene expression during meiosis.
The observation that benomyl, added before meiotic induction, caused a significant delay in the production of the meiosis-specific protein, Rec8, raised the possibility that the drug interfered with transcription and/or translation of meiotic factors. To address this possibility, we analyzed RNA levels of the meiosis specific gene IME2 (15). Consistent with the delayed accumulation of Rec8 protein, we also observed delayed accumulation of IME2 mRNA, when benomyl was added at the time of meiotic induction (Fig. 5A). This result suggests that benomyl causes an arrest in G1 by interfering with the accumulation of these two and perhaps other early meiotic cell cycle regulators.
To determine whether the benomyl-induced G2 arrest was accompanied by transcriptional down-regulation, we analyzed the consequences of adding benomyl 4 h after transfer into meiosis-inducing conditions. mRNA levels of IME2 and the cell cycle regulator CLB5 dropped sharply upon benomyl addition (Fig. 5B). Surprisingly, mRNA levels of two constitutively expressed genes, CDC28 and RPL3, showed a similar response, while expression of the large rRNA and the small nuclear RNA SNR6 remained unchanged upon benomyl addition (Fig. 5B). Our results suggest that benomyl treatment during G1 or S phase/G2 leads to changes in meiotic gene expression, with a number of important cell cycle genes being drastically down-regulated.
The down-regulation of meiotic transcripts depends at least in part on microtubule depolymerization.
To determine whether the decline in mRNA levels brought about by benomyl was due to the microtubule-depolymerizing effect of the drug, we examined the consequences of benomyl treatment on IME2 mRNA levels in tub2-150 cells (35, 62). Cells carrying the tub2-150 allele were grown in the presence of 50 μg/ml benomyl and upon transfer into meiosis-inducing medium were incubated with 120 μg/ml benomyl. tub2-150 cells entered the meiotic cell cycle normally, and IME2 mRNA accumulated to normal levels (Fig. 5C). When benomyl was removed after 4 h and cells were transferred into medium lacking the drug, IME2 mRNA levels did not increase further, indicating that transcription was fully induced in the presence of high levels of benomyl (Fig. 5C). When benomyl was added again at a concentration of 120 μg/ml (during or after completion of DNA replication; time = 4 h), IME2 expression decreased somewhat, although not nearly as dramatically as in wild-type cells (compare Fig. 5B and C). These results suggest that the decline in IME2 RNA levels (and presumably that of other transcripts) brought about by benomyl is at least in part due to the microtubule-depolymerizing effect of the drug.
Benomyl causes a global change in meiotic gene expression.
A large number of genes, including loci that are also involved in mitotic growth, are differentially regulated as cells progress through meiosis (52). Previous work has identified at least seven broad meiotic expression profiles, six of which involve transient up-regulation of transcription during the process (7, 44). To determine how general the effects of benomyl on gene expression were, we examined the effect of benomyl on meiotic gene expression at a genome-wide level as cells progressed through meiosis in the presence or absence of benomyl. The experimental protocol is outlined in Fig. 6A. Duplicate samples were harvested from cultures at several stages in premeiotic and meiotic development: after saturation in rich medium (YPD), after acetate starvation (YPA), and 4 h after induction of meiosis (SP4). At this point, cells were filtered and resuspended in normal SPO medium or in medium containing DMSO or benomyl. Duplicate samples were then harvested 1 h later from the SPO culture (SF5), DMSO culture (SD5), or benomyl culture (SB5) and 4 h later from the DMSO culture (SD8) or benomyl culture (SB8).
FIG. 6.
Benomyl treatment triggers a transcriptional response from genes involved in detoxification, cell cycle control, and the meiotic program. (A) Flow chart of the experimental protocol. Boxes indicate the samples analyzed in the results shown in Fig. 6 (see also Fig. S2 in the supplemental material). Box labels correspond to column labels in panels B and C. The flow chart also indicates the total time cultures spent in sporulation (SPO) medium. The time spent in SPO medium corresponds to the x axis of the graphs shown in panels D and E. (B and C) Loci involved in drug response and mitotic or meiotic cell cycles, as well as sporulation. The complete data set was analyzed using goCluster and split into 100 subgroups with similar expression using partitioning around medoids (26). The GO categories associated with the genes in each of the resulting groups were analyzed for statistically significant overrepresentation by comparing the frequencies of occurrence of the same GO term within the group and all transcripts represented on the microarray, respectively. The hypergeometric distribution was employed to determine the resulting P values (18). To correct for the multiple testing procedure, we used data from 100 randomized data sets to determine a P value cutoff that would result in a false discovery rate of 10%. A total of 586 different GO terms were identified as being enriched in one or several of the 100 clusters. Blue and red indicate low and high expression values, respectively. (D and E) Expression patterns of manually selected genes involved in drug response, cell cycle control and the regulation of mitotic and meiotic gene expression. Signals are given as log2 transformed values on the y axis and are graphed versus the total time the cultures spent in SPO medium. Black arrows indicate the time of drug treatment or shift to low temperature. Note that low abundance mRNAs that are also cell cycle regulated (e.g., SWI4) are often not or only barely detectable in nonsynchronized heterogeneously growing cell populations.
Total RNA samples from these time points were hybridized to Affimetrix S98 GeneChips covering approximately 6,400 yeast transcripts. A total of 1,189 transcripts displayed a strong variation of signals between different samples. These loci were grouped using a hierarchical clustering algorithm and sorted by signal strength during mitotic growth, such that genes strongly expressed in rich medium and presporulation medium were preferentially placed on top of the cluster (see Fig. S2 in the supplemental material). To get an overview of the gene functions dominating the different subparts of the hierarchical clustering, we used the goCluster tool to identify nonoverlapping branches of the hierarchical tree that showed the strongest enrichment of genes annotated with a common GO term (15a). The effects of benomyl were most obvious 1 h after drug addition, comparing DMSO-treated (SD5) with benomyl-treated (SB5) cells (Fig. 6; see also Fig. S2 in the supplemental material). A substantial number of genes were up-regulated by benomyl treatment in a highly coordinated manner, notably, genes encoding factors involved in drug response and transport (GO:0015893, drug transport; GO:0050896, response to stimulus), as well as subunits of the proteasome and genes required for ubiquitin-dependent proteolysis (GO:0006511, ubiquitin-dependent protein catabolism). Among the genes whose expression decreased in the presence of benomyl, we identified a large number of genes involved in protein translation (GO:0006412, protein biosynthesis), and factors implicated in meiotic development (GO:0030437, sporulation [sensu fungi]; GO:0030476, spore wall assembly [sensu fungi]; and GO:030154, cell differentiation). This suggests that benomyl treatment up-regulates genes involved in protein turnover and the response to stress, while negatively affecting many factors involved in cell growth and the cell cycle. Analysis of the transcriptional effects of benomyl on mitosis also showed an up-regulation of stress response genes and transporters; but in contrast to the meiotic cell cycle, no transcripts were down-regulated (S. Biggins, personal communication).
We then analyzed the transcriptional effects of benomyl on individual genes within those functional groups. In addition to causing an up-regulation of genes involved in protein turnover, the presence of benomyl led to a detoxification response. In particular, we observed increased expression of genes involved in transport (SNQ2 and YOR1), multidrug and chemical stress resistance (CIN5, PDR5, PDR16, and SNG1) (Fig. 6B), and stress response, including FLR1, a gene encoding a benomyl-inducible multidrug resistance permease (Fig. 7). At the same time, benomyl treatment caused a widespread down-regulation of meiosis-specific genes and general cell cycle factors (Fig. 6C). These include genes essential for meiotic transcriptional control (IME2, IME4, and NDT80) and early meiotic functions, such as recombination (EXO1, MSH4, MSH5, REC102, REC107, REC114, SAE3, and SPO11), SC formation (HOP1, HOP2, MND1, MER1, RED1, ZIP1, and ZIP2), sister chromatid cohesion and chromosome segregation (CSM2, IRR1/SCC3, REC8, SCC2, and SGO1), spindle pole body formation (SPO1), the recombination checkpoint (PCH2, MEK1, and DDC1), and control of M phase (CDC5, MND2, and SPO13). As a consequence of the decreased expression of early meiotic genes, factors involved in postmeiotic functions like spore wall formation and maturation (e.g., ADY4, DON1, SMA1, SMK1, SPO74, SPR3, SPR6, and SSP2) were also expressed later and at a lower level. Among factors important for both mitotic and meiotic cell cycle progression, we observed coordinated transcriptional down-regulation of the protein kinase CDC28, five of its six B-type cyclin regulatory subunits (CLB1, CLB3, CLB4, CLB5, and CLB6; note that CLB2 is not expressed during the meiotic cell cycle) (Fig. 6C and D), and components of the APC/C (APC4, APC5, APC11, CDC20, CDC23, and CDC26).
FIG. 7.
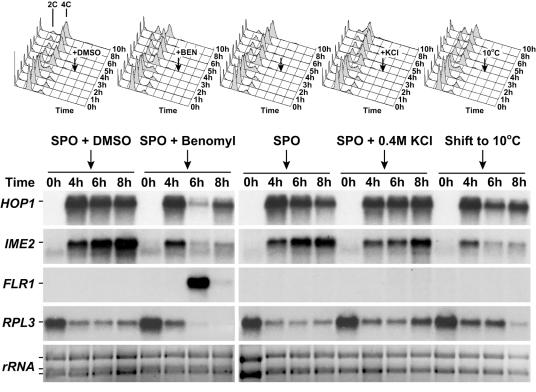
Benomyl-induced changes in meiotic gene expression are not a general stress response but resemble the transcriptional response to low temperatures. At 4 h after induction of meiosis at room temperature (black arrows), wild-type cells (A1972) were either cultured at room temperature in SPO medium containing various additives (0.4% DMSO, 0.4% DMSO plus 120 μg/ml benomyl, no additive, or 0.4 M potassium chloride) or cultured at 10°C. Cellular DNA content was determined by flow cytometry (top panels). Total RNA samples were analyzed by Northern blotting (bottom panels). All samples were run in the same gel but are separated in this figure for clarity.
Microtubule depolymerization may affect the expression of this large group of genes by influencing the expression of a smaller set of transcription factors that regulate the former. To test this, we directly examined a number of transcription factors involved in stress response, cell cycle regulation, and meiotic expression. Indeed, we found that mRNA levels of several transcription factors, SWI4, NDD1 (G1- and G2/M-specific induction), ABF1 (general regulator of mitotic and meiotic genes), as well as IME1, UME6, and NDT80 (required for meiotic activation and rerepression) were down-regulated at the transcriptional level in the presence of benomyl (Fig. 6E). This was not a general effect, since other transcription factors involved in stress response (Gcn4) and cell cycle control (Swi6, Mpb1, Mcm1, and Fkh1) did not show a significant decrease in their expression levels (see the S. cerevisiae section of GermOnline at http://www.germonline.org). These results suggest that the widespread transcriptional changes that occur after benomyl-induced microtubule destabilization could be correlated with the down-regulation of a set of transcription factors that regulate these genes.
Cold shock causes effects similar to those of benomyl.
Low temperatures are also known to destabilize microtubules (14). To determine whether cold temperatures affected transcription in a manner similar to that of benomyl, we analyzed the transcriptional changes in sporulating cultures which, instead of being exposed to benomyl, were shifted to low temperatures (10°C) for 1 h (SC5) or 4 h (SC8). Low-temperature stress did not lead to induction of genes involved in detoxification and drug transport but did cause, among other effects, a general shutdown of the meiotic transcriptional program very similar to that observed with benomyl-treated cells (Fig. 6B and C). One notable exception is the transcription factor IME1, whose expression, while down-regulated by benomyl, does not appear to be markedly affected by low-temperature stress (Fig. 6E). Taken together, these results suggest that a wide array of genes involved in cell proliferation and meiotic progression, including the three major early meiotic kinases IME2, CDC28, and CDC5 are down-regulated to halt cell cycle progression in the presence of damaged microtubules.
The down-regulation of meiotic genes by low temperatures or benomyl treatment was not caused by a general meiotic stress response pathway, because cells lacking the major stress kinase HOG1 still arrested in G2 after benomyl treatment (data not shown), and exposure of meiotic cells to hypertonic stress (0.4 M potassium chloride) did not elicit a change in gene expression comparable to that induced by benomyl or cold treatment. mRNA levels of IME2, HOP1, and RPL3 declined after treatment with benomyl or exposure to cold but remained at normal levels after exposure to hypertonic stress (Fig. 7). Furthermore, cell cycle arrest occurred in either G1 or G2 after exposure to benomyl or low temperatures but not if cells were exposed to hypertonic stress (Fig. 7, top). On the other hand, expression analysis suggested that the detoxification response was benomyl specific. We confirmed this by analyzing expression of the multidrug resistance permease gene, FLR1, by Northern blot analysis (Fig. 7). Thus, microtubule perturbation (chemically or by low temperature) but not other stress causes a specific transcriptional response that leads to meiotic cell cycle arrest in G1 or G2.
G2 arrest caused by benomyl treatment is independent of known meiotic checkpoints.
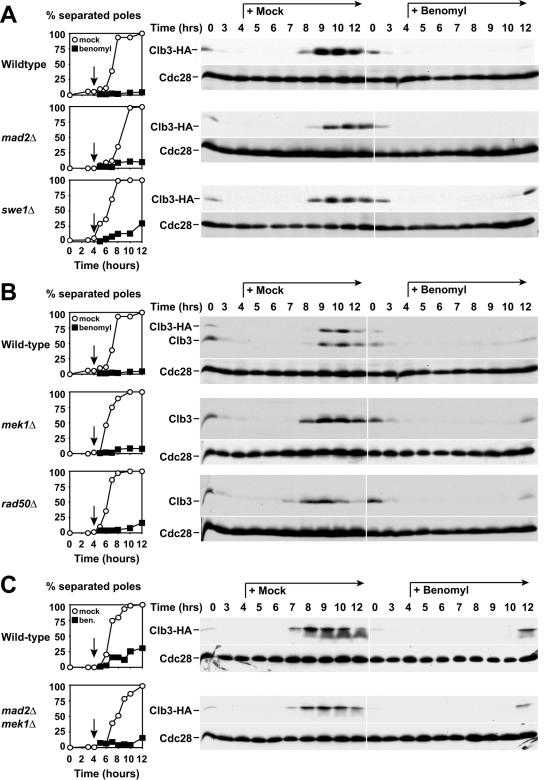
Several checkpoints have been characterized to date that cause a cell cycle arrest prior to meiotic chromosome segregation. We therefore determined whether any of the known meiotic checkpoints were responsible for the G2 arrest brought about by benomyl treatment. Since microtubule destabilization appeared to be the cause for the arrest, we first tested whether the G2 arrest we observed was dependent on the spindle assembly checkpoint. Deletion of the spindle assembly checkpoint component MAD2 did not bypass the G2 arrest. mad2Δ cells treated with benomyl exhibited the same delay in Clb3 accumulation as wild-type cells (Fig. 8A). Thus, the G2 arrest caused by benomyl treatment is not due to activation of the spindle checkpoint.
FIG. 8.
The benomyl-induced G2 arrest does not require the spindle checkpoint or the recombination checkpoint. Wild-type (4563), mad2Δ (A4843), swe1Δ (A4704), mek1Δ (A4838), rad50Δ (A1771), and mad2Δ mek1Δ (A12927) cells were studied. At 4 h after induction of meiosis at room temperature (black arrow), cells were resuspended in medium containing 0.4% DMSO (mock; white circles and left panels) or 120 μg/ml benomyl (black squares and right panels). Graphs on the left indicate percentages of cells containing more than one focus of tubulin staining. Western blot analysis was used to monitor Clb3 and Cdc28 (loading control) levels during the same time course. (A and C) A4563, A4843, A4704, and A12927 carry a CLB3-3HA fusion. Polyclonal α-Clb3 antibody was used to determine Clb3 levels shown in panel B.
The recombination checkpoint (pachytene checkpoint) arrests cells in G2 by down-regulating Clb-CDK activity and by inhibiting the transcription factor Ndt80 responsible for the transcriptional activation of genes necessary for entry into meiosis I and spore formation (49). Clb-CDK activity is kept low by at least two mechanisms. The protein kinase Swe1 phosphorylates Cdc28 on tyrosine 19, thereby inhibiting its activity (31). At the same time, CLB cyclin transcription is repressed through the inhibition of Ndt80 (63). To determine whether the recombination checkpoint is required for the G2 arrest caused by benomyl treatment, we analyzed the response of mek1Δ and swe1Δ mutants to benomyl. Mek1 is the meiotic homolog of the mitotic DNA-damage checkpoint kinase Rad53 and a central player in the recombination checkpoint (3, 69). Cells lacking MEK1 arrested with low levels of Clb3 protein after exposure to benomyl (Fig. 8B). Similar results were obtained when Swe1 was inactivated (Fig. 8A). We furthermore excluded the possibility that the recombination checkpoint and the spindle assembly checkpoint acted together to cause the arrest by examining the response of mek1Δ mad2Δ double mutants to benomyl treatment (Fig. 8C). Our results indicate that inactivation of both the recombination checkpoint and the spindle checkpoint does not allow benomyl-treated cells to enter meiosis I.
Finally, we tested whether the presence of unprocessed meiotic double-strand breaks contributed to the G2 arrest caused by benomyl treatment. Cells lacking RAD50 do not form meiotic double-strand breaks (6), yet they still arrested in G2 with low levels of Clb3 protein upon exposure to benomyl (Fig. 8B). We conclude that the benomyl-triggered G2 arrest is independent of double-strand break formation and is not due to activation of either the recombination or the spindle checkpoint.
DISCUSSION
Our analyses reveal a profound effect of microtubule destabilization on meiotic progression in budding yeast. In the presence of the microtubule-depolymerizing drug benomyl, cells fail to enter the meiotic program and arrest in G1. When benomyl is added during or after premeiotic S phase, it causes cells to arrest in G2 with low levels of Clb-CDK activity and incompletely paired chromosomes. Upon microtubule depolymerization, cells respond with a complex change in the pattern of meiotic gene expression that affects both meiosis-specific genes, as well as loci important for both mitotic and meiotic cell cycle progression. In particular, treatment with benomyl or low-temperature stress causes a shutdown of the meiotic transcriptional cascade. Our data further indicate that this transcriptional change is not a general stress response but specific to perturbation of the microtubule cytoskeleton and is likely to be responsible for the cell cycle and developmental arrest. Finally, our results show that the effects of benomyl on meiotic cell cycle progression are not mediated by any known checkpoint pathways pointing to the existence of a novel mechanism responsible for monitoring microtubule integrity and responding to perturbations.
Novel response to microtubule perturbation.
It has been reported that treatment of cells with only 60 μg/ml benomyl causes a delay in metaphase I (56). However, at this concentration, cells were still able to segregate their chromosomes, indicating that a transient meiotic spindle could still form (56). When benomyl is added at a concentration of 120 μg/ml, microtubules completely depolymerize. We find that treatment of meiotic cells with such a high dose of benomyl causes a G1 arrest when it is added during induction of meiosis, or a G2 arrest when cells are treated during S phase or G2. Furthermore, the G2 arrest is accompanied by a dramatic drop in mRNA levels of meiosis-specific genes and meiotically expressed cell cycle regulators. A key question is whether these events are the result of a general stress response or of a specific response mediated by microtubule perturbations. Several lines of evidence indicate that the latter is the case. First, benomyl is a well-characterized microtubule-depolymerizing agent, and the concentration of benomyl used in this study elicits the characteristic metaphase arrest during the mitotic divisions. Second, at least the G2 arrest caused by benomyl is fully reversible. When the drug is removed, cells progress through meiosis and form viable spores with normal efficiency. Third, cells carrying the tub2-150 allele, in which microtubules are stabilized and less vulnerable to microtubule-depolymerizing agents (35, 62), are able to enter meiosis and progress through premeiotic S phase in the presence of 120 μg/ml benomyl, indicating that the G1 arrest caused by benomyl is mediated by the drug's microtubule-depolymerizing function. The transcriptional response observed when benomyl is added 4 h after induction of meiosis is also at least in part mediated by benomyl-induced microtubule perturbations. In tub2-150 cells, renewed exposure to benomyl 4 h after transfer into meiosis-inducing conditions did cause a drop in RNA levels, but this drop was significantly less dramatic than that observed when wild-type cells are treated in this way. Given that tub2-150 microtubules still respond to benomyl, some microtubule depolymerization is likely to occur when the cell are reexposed to the drug, which may explain the partial drop in RNA levels that is observed.
A fourth line of evidence indicating that the cell cycle response to benomyl is not a general stress response is that hypertonic stress, a condition not known to affect microtubule structures, fails to cause a cell cycle arrest in G2 or to down-regulate mRNAs. In contrast, low temperature (14) affects cell cycle progression and genome-wide transcript levels in a manner qualitatively very similar to that of benomyl. We were not able to examine the effects of other fungal microtubule drugs such as thiabendazole, carbendazim, or nocodazole on meiotic cell cycle progression because these drugs are not sufficiently soluble in sporulation medium. However, it is interesting that microtubule-depolymerizing drugs such as colchicine and vinblastine that are structurally quite different from benomyl (11) perturb meiotic prophase in other organisms, such as mice, ciliates, and plants (1, 23, 33, 55, 70). Taken together, these findings indicate that the response of meiotic cells to high levels of benomyl or low temperatures is specific and due to microtubule perturbations.
Benomyl treatment causes a cell cycle arrest in G1 and G2.
The response of cells to benomyl during entry into the meiotic cell cycle (G1) and the response during S phase/G2 appear similar. The G2 arrest is accompanied by changes in expression levels of several hundred genes. Likewise, transcripts that are affected by benomyl during S phase/G2, such as IME2 and RPL3, are also down-regulated by benomyl treatment during G1. We do not know how benomyl causes a cell cycle arrest in G1 or G2, but our data indicate that the arrest is a consequence of the fact that it causes microtubules to depolymerize. Cells carrying the tub2-150 allele enter and progress through premeiotic S phase efficiently even when benomyl is added to the medium. In this context, it is interesting that disruption of microtubule dynamics by inactivating the microtubule motor KAR3 or its associated factor CIK1 also causes defects in meiotic entry and a meiotic cell cycle arrest in G2 (4, 53, 54). It is possible that a similar transcriptional response underlies the meiotic defects of these mutants.
The nature of the cell cycle arrest is also unclear. The G2 arrest elicited by benomyl is triggered by neither the activation of the spindle assembly nor the recombination checkpoint nor their combined activation. The arrest is also independent of the stress kinase Hog1, since hog1Δ cells still arrest in G2 after benomyl treatment. This suggests that a novel, as-yet-uncharacterized pathway mediates cell cycle arrest in G2 in response to microtubule perturbations caused by benomyl and low-temperature stress. We do not know much about this response, the factors involved, or the nature of the signal triggering it—possible candidates for signals would be unattached kinetochores or the level of free tubulin dimers. It is clear, however, that one of the consequences of this response is a dramatic change in gene expression. How are these changes in transcription mediated? Obvious targets of the response mechanism to microtubule depolymerization would be transcription factors involved in stress response or mitotic and meiotic gene expression. Meiosis is controlled by a complex transcriptional cascade (25). Induction of early meiotic genes is necessary for the correct expression of the subsequent middle and mid-late meiotic genes. Thus, observed delays in induction of later meiotic genes are likely a consequence of a failure early in the expression cascade. However, the transcriptional decrease also coordinately affects general cell cycle factors including most components of the APC/C and other genes involved in chromosome segregation and cell cycle progression. We therefore favor the idea that the expression and/or activity of a number of mitotic and meiotic transcriptional regulators might be coordinately decreased in response to microtubule depolymerization. Indeed, the meiotic transcription factors IME1, UME6, ABF1, and NDT80, as well as the cell cycle regulators SWI4 and NDD1, were expressed at lower levels in response to benomyl, while their transcriptional levels were reestablished when cells began to escape from the arrest. Note that the IME1 promoter does not appear to respond to temperature stress during early meiosis (Fig. 8D), so the inactivation of the meiotic cascade by cold shock may involve, if Ime1 is affected at all, a posttranslational mechanism. A possible explanation for this orchestrated response of transcription factors to microtubule instability could be auto- and cross-regulation of the factors, which renders them interdependent. It has been suggested that Abf1 and Ndt80 are involved in their own regulation (16, 25, 40). Moreover, Ume6 is required for correct IME1 and NDT80 expression (68). Finally, Abf1 may be required for normal IME1 and UME6 expression (45). Irrespective of the mechanism eliciting this response, it is clear that that the cell cycle arrest caused by benomyl is a consequence of the transcriptional response, because mRNA levels of factors essential for entry into meiosis I, such as B-type cyclins and the CDK Cdc28, are down-regulated.
Why does microtubule perturbation cause a G1 or G2 arrest during the meiotic cell cycle?
In mitotically dividing cells, the sole arrest elicited by microtubule depolymerization is a metaphase arrest (22) with no known effects on gene expression (S. Biggins, personal communication). In cells undergoing the meiotic cell cycle, complete microtubule depolymerization causes cells to arrest in G1 or G2. Why is the response to benomyl different between these two types of cell cycles? The decision not to enter the meiotic cell cycle when microtubules become unstable may be related to the fact that the meiotic cell cycle occurs under conditions where nutrients are limited. Delays in cell cycle progression caused by perturbations of the microtubule cytoskeleton or any other stresses could thus lead to cell death. Microtubule depolymerization and other stress-sensing mechanisms may therefore be in place during G1 that prevent entry into the cell cycle when conditions are not favorable for completion of this cell cycle. In this regard, it is interesting that treatment of cells with the DNA replication inhibitor hydroxyurea also inhibits entry into the meiotic cell cycle (10).
The reason why meiotic cells whose microtubules have disassembled arrest in G2 rather than metaphase I may lie in the way in which the meiotic cell cycle is organized. Once cells have entered meiosis I, cell cycle events are no longer coupled with the developmental program. For example, the monopolin complex, a kinetochore-bound protein complex that promotes the attachment of sister chromatids to microtubules emanating from the same spindle pole (46), dissociates from kinetochores in a manner uncoordinated with other cell cycle events. When cells are arrested in metaphase I through the inactivation of the APC/C activator Cdc20, the developmental program continues. Cdc20-depleted cells initiate spore formation despite arresting with metaphase I spindles (29). Furthermore, the monopolin complex dissociates from kinetochores at the time when it dissociates from wild-type cells despite being arrested in metaphase I (29). Thus, if microtubule depolymerization were to cause cell cycle arrest in metaphase, as it does during the mitotic divisions, the results for meiotic development would be disastrous. During the metaphase I arrest, the monopolin complex would dissociate from kinetochores and upon repolymerization of microtubules, attachment of microtubules to sister kinetochores in a cooriented manner would not occur; hence, meiosis I segregation would fail. In contrast, halting of cells in a cell cycle stage in which the cell cycle is still coordinated with the developmental program would allow cells to resume the meiotic cell cycle upon repolymerization of microtubules and therefore permit the successful completion of the meiotic cell cycle. This is achieved by the arrest in G2, prior to the commitment to meiosis I. At this stage, both the developmental program and the cell cycle program still rely on the same transcription factor, Ndt80, for their initiation.
Transcriptional changes in response to microtubule depolymerization are not a phenomenon restricted to budding yeast. Changes in RNA levels upon colchicine treatment have been observed in a variety of tissue culture cells (9) and a microtubule-associated transcription factor that activates transcription following microtubule instability has been reported (72). Furthermore, as we observed with budding yeast, several widely divergent organisms, including mouse, Allium, and lily, exhibit a meiotic prophase arrest or delay upon exposure to microtubule poisons (33, 55, 60). Indeed, the existence of a prophase “colchicine checkpoint” in mouse spermatocytes has been previously suggested (60, 61). Interestingly, mammalian tissue culture cells show an arrest response to the microtubule drugs colchicine and nocodazole even in mitotic prophase (39, 47). It will be interesting to determine whether these arrests are caused by a transcriptional response similar to that observed in budding yeast.
Supplementary Material
Acknowledgments
We are grateful to N. Kleckner and F. Solomon for reagents and advice and to S. Biggins for sharing unpublished results. We thank F. Solomon and members of the Amon lab for their critical reading of the manuscript. We thank L. Hermida for administration of the GermOnline database.
P.D. and G.W. are supported by the Department of Clinical and Biological Sciences and the Biozentrum, respectively. M.C. and C.N.-W. are supported by the Swiss Institute of Bioinformatics. This research was supported by a National Institutes of Health grant (GM62207) to A.A. A.A. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allen, J. W., J. B. Gibson, P. A. Poorman, L. C. Backer, and M. J. Moses. 1988. Synaptonemal complex damage induced by clastogenic and anti-mitotic chemicals: implications for non-disjunction and aneuploidy. Mutat. Res. 201:313-324. [DOI] [PubMed] [Google Scholar]
- 2.Amon, A., M. Tyers, B. Futcher, and K. Nasmyth. 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74:993-1007. [DOI] [PubMed] [Google Scholar]
- 3.Bailis, J. M., and G. S. Roeder. 2000. Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell 101:211-221. [DOI] [PubMed] [Google Scholar]
- 4.Bascom-Slack, C. A., and D. S. Dawson. 1997. The yeast motor protein, Kar3p, is essential for meiosis I. J. Cell Biol. 139:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buonomo, S. B., K. P. Rabitsch, J. Fuchs, S. Gruber, M. Sullivan, F. Uhlmann, M. Petronczki, A. Toth, and K. Nasmyth. 2003. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev. Cell 4:727-739. [DOI] [PubMed] [Google Scholar]
- 6.Cao, L., E. Alani, and N. Kleckner. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61:1089-1101. [DOI] [PubMed] [Google Scholar]
- 7.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 8.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 9.Cleveland, D. W., M. A. Lopata, P. Sherline, and M. W. Kirschner. 1981. Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell 25:537-546. [DOI] [PubMed] [Google Scholar]
- 10.Davis, L., M. Barbera, A. McDonnell, K. McIntyre, R. Sternglanz, Q. Jin, J. Loidl, and J. Engebrecht. 2001. The Saccharomyces cerevisiae MUM2 gene interacts with the DNA replication machinery and is required for meiotic levels of double strand breaks. Genetics 157:1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downing, K. H. 2000. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu. Rev. Cell Dev. Biol. 16:89-111. [DOI] [PubMed] [Google Scholar]
- 12.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, M. L., Jr., C. J. Bode, C. A. Dougherty, R. T. Marquez, and R. H. Himes. 2001. Mutagenesis of beta-tubulin cysteine residues in Saccharomyces cerevisiae: mutation of cysteine 354 results in cold-stable microtubules. Cell Motil. Cytoskeleton 49:67-77. [DOI] [PubMed] [Google Scholar]
- 15.Guttmann-Raviv, N., S. Martin, and Y. Kassir. 2002. Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol. Cell. Biol. 22:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Harris, M. A., J. Clark, A. Ireland, J. Lomax, M. Ashburner, R. Foulger, K. Eilbeck, S. Lewis, B. Marshall, C. Mungall, J. Richter, G. M. Rubin, J. A. Blake, C. Bult, M. Dolan, H. Drabkin, J. T. Eppig, D. P. Hill, L. Ni, M. Ringwald, R. Balakrishnan, J. M. Cherry, K. R. Christie, M. C. Costanzo, S. S. Dwight, S. Engel, D. G. Fisk, J. E. Hirschman, E. L. Hong, R. S. Nash, A. Sethuraman, C. L. Theesfeld, D. Botstein, K. Dolinski, B. Feierbach, T. Berardini, S. Mundodi, S. Y. Rhee, R. Apweiler, D. Barrell, E. Camon, E. Dimmer, V. Lee, R. Chisholm, P. Gaudet, W. Kibbe, R. Kishore, E. M. Schwarz, P. Sternberg, M. Gwinn, L. Hannick, J. Wortman, M. Berriman, V. Wood, N. de la Cruz, P. Tonellato, P. Jaiswal, T. Seigfried, and R. White. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32:D258-D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honigberg, S. M., and K. Purnapatre. 2003. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116:2137-2147. [DOI] [PubMed] [Google Scholar]
- 18.Hosack, D. A., G. Dennis, Jr., B. T. Sherman, H. C. Lane, and R. A. Lempicki. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber, W., A. von Heydebreck, H. Sultmann, A. Poustka, and M. Vingron. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl. 1):S96-S104. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, L. H., L. F. Lau, D. L. Smith, C. A. Mistrot, K. G. Hardwick, E. S. Hwang, A. Amon, and A. W. Murray. 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science 279:1041-1044. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry, R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs, C. W., A. E. Adams, P. J. Szaniszlo, and J. R. Pringle. 1988. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 107:1409-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaczanowski, A., J. Gaertig, and J. Kubiak. 1985. Effect of the antitubulin drug nocodazole on meiosis and postmeiotic development in Tetrahymena thermophila. Induction of achiasmatic meiosis. Exp. Cell Res. 158:244-256. [DOI] [PubMed] [Google Scholar]
- 24.Kane, S. M., and R. Roth. 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kassir, Y., N. Adir, E. Boger-Nadjar, N. G. Raviv, I. Rubin-Bejerano, S. Sagee, and G. Shenhar. 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224:111-171. [DOI] [PubMed] [Google Scholar]
- 26.Kaufman, L., and P. J. Rousseeuw. 1987. Clustering by means of mediods, p. 405-416. In Y. Dodge (ed.), Statistical data analysis based on the L1-norm and related methods. North-Holland/Elsevier, Amsterdam, The Netherlands.
- 27.Klein, F., P. Mahr, M. Galova, S. B. C. Buonomo, C. Michaelis, K. Nairz, and K. Nasmyth. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements and recombination during yeast meiosis. Cell 98:91-103. [DOI] [PubMed] [Google Scholar]
- 28.Kohrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 29.Lee, B. H., and A. Amon. 2003. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300:482-486. [DOI] [PubMed] [Google Scholar]
- 30.Leisch, F. 2002. Sweave: dynamic generation of statistical reports using literate data analysis, p. 575-580. In W. Hardle and B. Ronz (ed.), Compstat 2002—proceedings in computational statistics. Physika Verlag, Heidelberg, Germany.2002
- 31.Leu, J. Y., and G. S. Roeder. 1999. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell 4:805-814. [DOI] [PubMed] [Google Scholar]
- 32.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37:251-282. [DOI] [PubMed] [Google Scholar]
- 33.Loidl, J. 1988. The effect of colchicine on synaptonemal complex formation in Allium ursinum. Exp. Cell Res. 178:93-97. [DOI] [PubMed] [Google Scholar]
- 34.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 35.Machin, N. A., J. M. Lee, and G. Barnes. 1995. Microtubule stability in budding yeast: characterization and dosage suppression of a benomyl-dependent tubulin mutant. Mol. Biol. Cell 6:1241-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marston, A. L., B. H. Lee, and A. Amon. 2003. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev. Cell 4:711-726. [DOI] [PubMed] [Google Scholar]
- 37.Marston, A. L., W. H. Tham, H. Shah, and A. Amon. 2004. A genome-wide screen identifies genes required for centromeric cohesion. Science 303:1367-1370. [DOI] [PubMed] [Google Scholar]
- 38.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 39.Mikhailov, A., and C. L. Rieder. 2002. Cell cycle: stressed out of mitosis. Curr. Biol. 12:R331-R333. [DOI] [PubMed] [Google Scholar]
- 40.Miyake, T., J. Reese, C. M. Loch, D. T. Auble, and R. Li. 2004. Genome-wide analysis of ARS (autonomously replicating sequence) binding factor 1 (Abf1p)-mediated transcriptional regulation in Saccharomyces cerevisiae. J. Biol. Chem. 279:34865-34872. [DOI] [PubMed] [Google Scholar]
- 41.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 42.Nairz, K., and F. Klein. 1997. mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 11:2272-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Primig, M., C. Wiederkehr, R. Basavaraj, C. Sarrauste de Menthiere, L. Hermida, R. Koch, U. Schlecht, H. G. Dickinson, M. Fellous, J. A. Grootegoed, R. S. Hawley, B. Jegou, B. Maro, A. Nicolas, T. Orr-Weaver, T. Schedl, A. Villeneuve, D. J. Wolgemuth, M. Yamamoto, D. Zickler, N. Lamb, and R. E. Esposito. 2003. GermOnline, a new cross-species community annotation database on germ-line development and gametogenesis. Nat. Genet. 35:291-292. [DOI] [PubMed] [Google Scholar]
- 44.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 45.Prinz, S., F. Klein, H. Auer, D. Schweizer, and M. Primig. 1995. A DNA binding factor (UBF) interacts with a positive regulatory element in the promoters of genes expressed during meiosis and vegetative growth in yeast. Nucleic Acids Res. 23:3449-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabitsch, K. P., M. Petronczki, J. P. Javerzat, S. Genier, B. Chwalla, A. Schleiffer, T. U. Tanaka, and K. Nasmyth. 2003. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell 4:535-548. [DOI] [PubMed] [Google Scholar]
- 47.Rieder, C. L., and R. Cole. 2000. Microtubule disassembly delays the G2-M transition in vertebrates. Curr. Biol. 10:1067-1070. [DOI] [PubMed] [Google Scholar]
- 48.Roeder, G. S. 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11:2600-2621. [DOI] [PubMed] [Google Scholar]
- 49.Roeder, G. S., and J. M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395-403. [DOI] [PubMed] [Google Scholar]
- 50.Saunders, W., D. Hornack, V. Lengyel, and C. Deng. 1997. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J. Cell Biol. 137:417-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlecht, U., P. Demougin, R. Koch, L. Hermida, C. Wiederkehr, P. Descombes, C. Pineau, B. Jegou, and M. Primig. 2004. Expression profiling of mammalian male meiosis and gametogenesis identifies novel candidate genes for roles in the regulation of fertility. Mol. Biol. Cell 15:1031-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlecht, U., and M. Primig. 2003. Mining meiosis and gametogenesis with DNA microarrays. Reproduction 125:447-456. [DOI] [PubMed] [Google Scholar]
- 53.Shanks, R. M., C. Bascom-Slack, and D. S. Dawson. 2004. Analysis of the kar3 meiotic arrest in Saccharomyces cerevisiae. Cell Cycle 3:363-371. [PubMed] [Google Scholar]
- 54.Shanks, R. M., R. J. Kamieniecki, and D. S. Dawson. 2001. The Kar3-interacting protein Cik1p plays a critical role in passage through meiosis I in Saccharomyces cerevisiae. Genetics 159:939-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shepard, J., E. R. Boothroyd, and H. Stern. 1974. The effect of colchicine on synapsis and chiasma formation in microsporocytes of Lilium. Chromosoma 44:423-437. [Google Scholar]
- 56.Shonn, M. A., R. McCarroll, and A. W. Murray. 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289:300-303. [DOI] [PubMed] [Google Scholar]
- 57.Shonn, M. A., A. L. Murray, and A. W. Murray. 2003. Spindle checkpoint component Mad2 contributes to biorientation of homologous chromosomes. Curr. Biol. 13:1979-1984. [DOI] [PubMed] [Google Scholar]
- 58.Sora, S., P. Melchioretto, P. Primignani, and M. L. Agostoni Carbone. 1988. Caffeine interactions with methyl methanesulphonate, hycanthone, benlate, and cadmium chloride in chromosomal meiotic segregation of Saccharomyces cerevisiae. Mutat. Res. 201:9-16. [DOI] [PubMed] [Google Scholar]
- 59.Sym, M., J. A. Engebrecht, and G. S. Roeder. 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72:365-378. [DOI] [PubMed] [Google Scholar]
- 60.Tepperberg, J. H., M. J. Moses, and J. Nath. 1997. Colchicine effects on meiosis in the male mouse. I. Meiotic prophase: synaptic arrest, univalents, loss of damaged spermatocytes and a possible checkpoint at pachytene. Chromosoma 106:183-192. [DOI] [PubMed] [Google Scholar]
- 61.Tepperberg, J. H., M. J. Moses, and J. Nath. 1999. Colchicine effects on meiosis in the male mouse. II. Inhibition of synapsis and induction of nondisjunction. Mutat. Res. 429:93-105. [DOI] [PubMed] [Google Scholar]
- 62.Thomas, J. H., N. F. Neff, and D. Botstein. 1985. Isolation and characterization of mutations in the beta-tubulin gene of Saccharomyces cerevisiae. Genetics 111:715-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tung, K. S., E. J. Hong, and G. S. Roeder. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. USA 97:12187-12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers, and A. Amon. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2:709-718. [DOI] [PubMed] [Google Scholar]
- 65.Visintin, R., E. S. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818-823. [DOI] [PubMed] [Google Scholar]
- 66.Wiederkehr, C., R. Basavaraj, C. Sarrauste de Menthiere, L. Hermida, R. Koch, U. Schlecht, A. Amon, S. Brachat, M. Breitenbach, P. Briza, S. Caburet, M. Cherry, R. Davis, A. Deutschbauer, H. G. Dickinson, T. Dumitrescu, M. Fellous, A. Goldman, J. A. Grootegoed, R. Hawley, R. Ishii, B. Jegou, R. J. Kaufman, F. Klein, N. Lamb, B. Maro, K. Nasmyth, A. Nicolas, T. Orr-Weaver, P. Philippsen, C. Pineau, K. P. Rabitsch, V. Reinke, H. Roest, W. Saunders, M. Schroder, T. Schedl, M. Siep, A. Villeneuve, D. J. Wolgemuth, M. Yamamoto, D. Zickler, R. E. Esposito, and M. Primig. 2004. GermOnline, a cross-species community knowledgebase on germ cell differentiation. Nucleic Acids Res. 32:D560-D567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiederkehr, C., R. Basavaraj, C. Sarrauste de Menthiere, R. Koch, U. Schlecht, L. Hermida, B. Masdoua, R. Ishii, V. Cassen, M. Yamamoto, C. Lane, M. Cherry, N. Lamb, and M. Primig. 2004. Database model and specification of GermOnline Release 2.0, a cross-species community annotation knowledgebase on germ cell differentiation. Bioinformatics 20:808-811. [DOI] [PubMed] [Google Scholar]
- 68.Williams, R. M., M. Primig, B. K. Washburn, E. A. Winzeler, M. Bellis, C. Sarrauste de Menthiere, R. W. Davis, and R. E. Esposito. 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA 99:13431-13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu, L., B. M. Weiner, and N. Kleckner. 1997. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 11:106-118. [DOI] [PubMed] [Google Scholar]
- 70.Zickler, D., and N. Kleckner. 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32:619-697. [DOI] [PubMed] [Google Scholar]
- 71.Zickler, D., and N. Kleckner. 1999. Meiotic Chromosomes: integrating structure and function. Annu. Rev. Genet. 33:603-754. [DOI] [PubMed] [Google Scholar]
- 72.Ziegelbauer, J., B. Shan, D. Yager, C. Larabell, B. Hoffmann, and R. Tjian. 2001. Transcription factor MIZ-1 is regulated via microtubule association. Mol. Cell 8:339-349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.