Abstract
Major depression (MD) is a serious psychiatric illness afflicting nearly 5% of the world’s population. A large correlational literature suggests that loneliness is a prospective risk factor for MD; observational effects of this nature may be confounded for a variety of reasons. This report uses Mendelian Randomization (MR) to examine potentially causal associations between loneliness and MD. We report on analyses using summary statistics from three large genome wide association studies (GWAS). MR analyses were conducted using three independent sources of GWAS summary statistics. In the first set of analyses, we used available summary statistics from an extant GWAS of loneliness to predict MD risk. We used two sources of outcome data: the Psychiatric Genomics Consortium (PGC) meta-analysis of MD (PGC-MD; N= 142,646) and the Million Veteran Program (MVP-MD; N = 250,215). Finally, we reversed analyses using data from the MVP and PGC samples to identify risk variants for MD and used loneliness outcome data from UK Biobank. We find robust evidence for a bidirectional causal relationship between loneliness and MD, including between loneliness, depression cases status, and a continuous measure of depressive symptoms. The estimates remained significant across several sensitivity analyses, including models that account for horizontal pleiotropy. This paper provides the first genetically-informed evidence that reducing loneliness may play a causal role in decreasing risk for depressive illness, and these findings support efforts to reduce loneliness in order to prevent or ameliorate MD. Discussion focuses on the public health significance of these findings, especially in light of the SARS-CoV-2 pandemic.
Keywords: Loneliness, depression, Mendelian Randomization, causal inference, genetically-informed research
Major depression (MD) is a debilitating psychiatric illness that causes considerable suffering around the globe, afflicting almost 5% of the world’s population at any given time1. The lifetime prevalence of MD is 16.2% in the United States and roughly 11% globally2, and many more people experience subthreshold but clinically significant depressive symptoms3. The impact of depressive illness is high, with recent projections suggesting that MD accounts for the largest share global disease burden, measured by year’s lost to disability4.
Given the high prevalence of MD and the toll it takes on patients, their families, and society at large, the search for modifiable risk factors is of paramount importance5–8. Loneliness—the subjective experience of social isolation—is increasingly recognized a significant public health problem9–12, with new evidence suggesting that rates of loneliness have increased in the last few years13,14, especially in the context of the SARS-CoV-2 pandemic14. Reflective of these facts, in early 2023, the United States Surgeon General issued a formal health advisory on the potentially harmful effects of social disconnection15. Independent of objective social isolation, chronic loneliness is associated with increased risk for a range of mental and physical health morbidities, including MD16. Prior studies and meta-analysis demonstrate that loneliness is highly correlated with MD risk17,18, and that the experience of chronic loneliness is associated with the development of a range of mental disorders14. Moreover, the psychological and epidemiological literatures suggest that loneliness is distinct from depression and may act as a prospective risk factor for depressive symptoms19,20.
What is not yet clear, however, is whether loneliness is causally associated with the risk of MD, or whether MD plays a causal role in the experience of loneliness. Even robust associations emerging from observational research studies are susceptible to confounding or reverse causation, making it difficult to establish causal as opposed to correlational links21. Much of the social integration and health literature is correlational, though it is often discussed as if causal effects are established and well-replicated e.g., see22,23. Relative to traditional observational designs, genetically-informed studies, including co-twin control studies 24 and those using Mendelian Randomization25 (MR) methods, have the potential to inform our understanding of causal influences by accounting for or circumventing a variety of confounding processes and the potential for reverse causation. For example, behavior genetic results the Environmental Risk Longitudinal Twin Study, with over 1,000 same-sex twin pairs, reveal that although there is significant genetic overlap between loneliness and depression, genetic confounding does not account for the entirety of the phenotypic exposure and within-twin pair differences in loneliness are associated with increases in depression, which is consistent with a causal effect26. MR is an analytic method that uses results from genome-wide association studies (GWAS) to examine potentially causal associations between risk exposures and outcomes27,28; the approach is based on an instrumental variable framework that uses genetic variants as proxies for a phenotypic exposure29. New findings from MR analyses suggest that confiding in others—a marker of social support and integration—is associated with decreased risk for MD5 but we are unaware of any MR studies examining the bidirectional association between subjective isolation and MD.
Given the strong genetic basis of loneliness and social integration more broadly, as well as the emergence of large GWAS in this area30–37, MR methods are ideally suited to help answer questions about causal influence. Moreover, the approach is well suited to characterize vertical pleiotropy, when a genetic variant affects an exposure (X), and because of that process, also affects an outcome (Y); to the extent that the phenotypic association between the exposure and outcome is not causal, the genetic variants/instruments associated with the exposure will not be significantly associated with outcome of interest. At the same time, the presence of horizontal pleiotropy—where genetic variants influence the outcome through causal pathways other than through the exposure—can severely bias estimates of causal effects38. This may be especially true in the case of the association between loneliness and MD. For example, recent results from a large GWAS of loneliness reported a genome-wide genetic correlation of rg = 0.64 of loneliness with MD31. A variety of methods are now available for exploring the potential impact of pleiotropy39 and we employ such methods in the current report. Specifically, we conduct bidirectional two-sample MR analyses to examine the potential causal associations (a) from loneliness and MD and (b) from MD to loneliness.
Although our analyses were not preregistered and should therefore be considered largely exploratory, our work was guided by specific predictions. We hypothesized that the association from loneliness to MD risk would be consistent with a causal effect. We also examined the association from MD to the experience of loneliness. To identify the primary genetic instruments for loneliness, we used summary statistics from the recent loneliness meta-analysis, which included 511,280 participants across seven different studies 31. We conducted the MR analyses using two independent sources of outcome data. The first analysis used GWAS summary statistics from the Psychiatric Genomics Consortium (PGC) meta-analysis of MD (PGC-MD; N= 142,646), excluding participants from the UKB and 23andMe sub-samples40. We then sought to replicate our findings using GWAS summary statistics from the Million Veteran Program MVP-MDD; N = 250,21541. Finally, we reversed analyses using data from the MVP and PGC samples to identify risk variants for MD, and harmonized these instruments with loneliness summary statistics from the UK Biobank as the outcome.
METHODS
We used de-identified summary-level data from three sources: A recent GWAS of loneliness that included data from seven different studies31, the Million Veterans Project (MVP) depression GWAS41 (excluding UKB participants), and the Psychiatric Genomics Consortium (PGC) depression GWAS40 (excluding UKB and 23andMe participants). Ethical approval, including informed consent, was obtained in all of the original studies.
Data Sources and Instruments
Loneliness.
When loneliness was the exposure, we drew on summary data from the largest GWAS conducted on this topic to date, which included 511,280 participants from the UKB, 23andMe, Netherlands Twin Register, Health & Retirement Study, Rotterdam Study, Sweden-SALTY and Sweden TwinGene studies31. The average age of the participants across the different studies was 57.5 years-old. Information on the genotyping, imputation, and quality control of each sample is included the original meta-analysis; only participants from European descent were included in the original study.
The genetic risk statistic from the meta-analysis are used in the current report and represent the standardized sample-sized weighted effect size, derived across the seven samples, many of which measured loneliness differently. (As reported in Abdellaoui et al.\31 Supplementary Table 1, the loneliness phenotype assessments ranged from a single binary question “Do you often feel lonely?” (UKB) to a single “Did you feel lonely during the past week?” (4-point scale) in the Rotterdam Study, Sweden-SALTY and Sweden-TwinGene Studies and the use of a 9-point questionnaire (4-point scale) in the 23andMe sample.) In order to conduct independent, two-sample MR analyses, when loneliness was the outcome, we relied on the available outcome summary statistics for the raw binary loneliness item (“Do you feel lonely?” yes/no) in the UKB-only data42. The available summary statistics include data from 100,988 who endorsed feeling lonely and 463,062 who responded negatively to the item.
Depression.
As noted above, we included two data sources assessing MD. The first set of summary data comes from the PGC meta-analysis, excluding UKB and 23andMe participants (see Supplementary Table 1 in the Wray at el. report40). The GWAS summary statistics used here included PGC data from 45,396 adult MD cases and 97,250 adult controls of European ancestry. Information on the diagnostic assessment of case status is available in the original reports; as reported by Wray et al.40, all diagnosed cases met international consensus criteria for MDD, and the assessment methodologies were cross-checked by expert reviewers. The second set of summary data is from the MVP, which is a large observational cohort study of mostly male (92%); participants were an average of 63.7 years-old (SD = 13.39 years); in the MVP data, 12% of participants diagnosed with depression were female43. GWAS summary statistics included MVP data from 83,810 MD cases (diagnosed using International Classification of Diseases (ICD) codes for MDD derived from the Electronic Health Record) and 166,405 controls of European ancestry41. The MVP data also includes the 2-item Patient Health Questionnaire PHQ;44 which is a continuous measure of depressive symptoms. The widely used PHQ-2 asks people to report on an item assessing loss of interest of pleasure in doing things and on an item assessing feeling down, depressed, or hopeless; to assess the presence of these symptoms, the PHQ-2 uses a Likert-type scale ranging from 0 (Not at all) to 3 (nearly every day).
Statistical Analysis.
We conducted two-sample Mendelian randomization (MR) analyses in the R computing environment using the “TwoSampleMR” package (R Project for Statistical Computing). This package harmonizes exposure and outcome data according to a common allele for each SNP, along with information on effect size, standard error, and p-value information about the association between specific gene variants and the exposure and outcome phenotype, respectively. Genetic variants missing in outcome GWAS were replaced with proxy instruments from the 1,000 Genomes European reference panel. The MR approach relies on three key underlying assumptions about the exposure-outcome association45, including that (I) the genetic markers are strongly associated with the exposure; (II) the genetic markers are not associated with confounders of the risk-outcome association—often referred to as independence assumption46; and, (III) the genetic markers are not associated with the outcome through any other path than through the exposure. The third assumption can be a challenge in practice because GWASs provide increasing evidence that pleiotropy is more of a rule than an exception47. We used the inverse variance-weighted (IVW) method as our primary estimator48; the IVW statistic is a weighted average of the regression estimates (for the individual SNP instruments), and the significance the average estimate can be interpreted as evidence of a causal association. Thus, the overall IVW estimate can be used to determine if the average exposure-outcome association is reliably different from zero and consistent with a causal effect. The other MR methods are used to test the consistency and replicability of the effects under different model assumptions. To account for potential violation of the MR assumptions and pleiotropic effects, we conducted parallel sensitivity analyses including MR Egger49, weighted median50, weighted mode, MR Robust Adjusted Profile Score RAPS51, and MR Pleiotropy Residual Sum and Outlier (MR-PRESSO) methods46. We performed these tests to evaluate the stability of causal associations relative to the IVW estimate. Briefly, MR Egger assumes a non-zero intercept to account for pleiotropy, which provides a consistent causal estimate when the Instrument Strength Independent of Direct Effect (InSIDE) assumption is met. We evaluated the reliability of the Egger regression estimate using the I2GX statistic52. The weighted mode- and median-based estimates are more robust to outliers by fitting a regression with greater weight for genetic variants with more precise ratio estimates. MR-RAPS uses a profile-likelihood function to estimate the variance of the pleiotropic effect distribution, downweighting outliers in causal estimation. MR-PRESSO detects global pleiotropy by comparing the residual sum of squares against the expected distance under the null hypothesis of no pleiotropic effect and assesses the impact of removing outliers46.
As a group, these analyses help us identify potential pleiotropic effects and minimize the potential of a false positive finding, wherein genetic variants influence the outcome through causal pathways other than the exposure29. We also calculated the mean F-statistic to test instrument strength (F>10 being sufficiently strong) and Cochran’s Q to estimate heterogeneity between the SNP effects which might suggest pleiotropy. Steiger filtering was used to reduce risk of potential reverse causation by excluding variants explaining greater variance in the outcome than in the exposure53. Finally, we applied the Causal Analyses Using Summary Effect estimates (CAUSE) method, which is useful for assessing correlated pleiotropy, a form of horizontal pleiotropy in which a variant affects an exposure and outcome through a shared heritable factor39. CAUSE modeling uses a genome-wide approach to identify risk variants for the exposure, then creates two nested models: A sharing model that allows horizontal pleiotropy (where the causal effect is fixed to zero) and a causal model in which a causal effect is freely estimated. The model fit is then compared within a Bayesian framework by comparing the two nested models and using a pointwise posterior density comparison approach to determine the extent to which the posterior distributions would predict a new set of data. A rejection of the “sharing model” indicates that the estimated posterior distribution are better fit under the “causal model.” Thus, rejecting the sharing model means that the data are consistent with a causal effect 39.
RESULTS
From Loneliness to Depression.
This set of analyses investigated the putatively causal role of loneliness (exposure) on MD (outcome). We conducted this first set of analyses with the PGC-MD data, then sought to replicate findings using the MVP-MD data (see Methods). Using a conventional threshold of P < 5 × 10−8 from the loneliness GWAS31, 17 instruments were available that, when clumped and harmonized with the genome-wide PGC outcome data, yielded 15 instruments for the analyses. All F-statistics suggested instruments were sufficiently associated with the loneliness exposure (F >10, see Supplementary Table 1; the genetic variants are reported in Supplementary Table 7).
Figure 1 displays the beta estimates using five MR regression methods in parallel. Using the PGC-MD outcome data, we identified a significant positive association in the IVW analysis, suggesting a causal influence from loneliness to MD (IVW = .78, 95% CI = 0.36, 1.21). The other four MR effect estimates also identified significant positive associations. We identified significant evidence of heterogeneity as measured by the I2GX statistic and Cochran’s Q statistic (Supplementary Tables 2 and 3). Steiger filtering identified two variants that explained greater variance in MD than in loneliness; sensitivity analyses removing these variants attenuated effect estimates, however, all results remained significant (Supplemental Table 4). We also applied the Causal Analyses Using Summary Effect Estimates (CAUSE) method to assess for correlated pleiotropy (see Methods). Using the PGC-MD outcome data, CAUSE estimates, shown in Table 1, suggest loneliness is causally associated with MD (p = .004). Overall, analyses with the PGC support a causal effect of loneliness on major depression.
Figure 1.
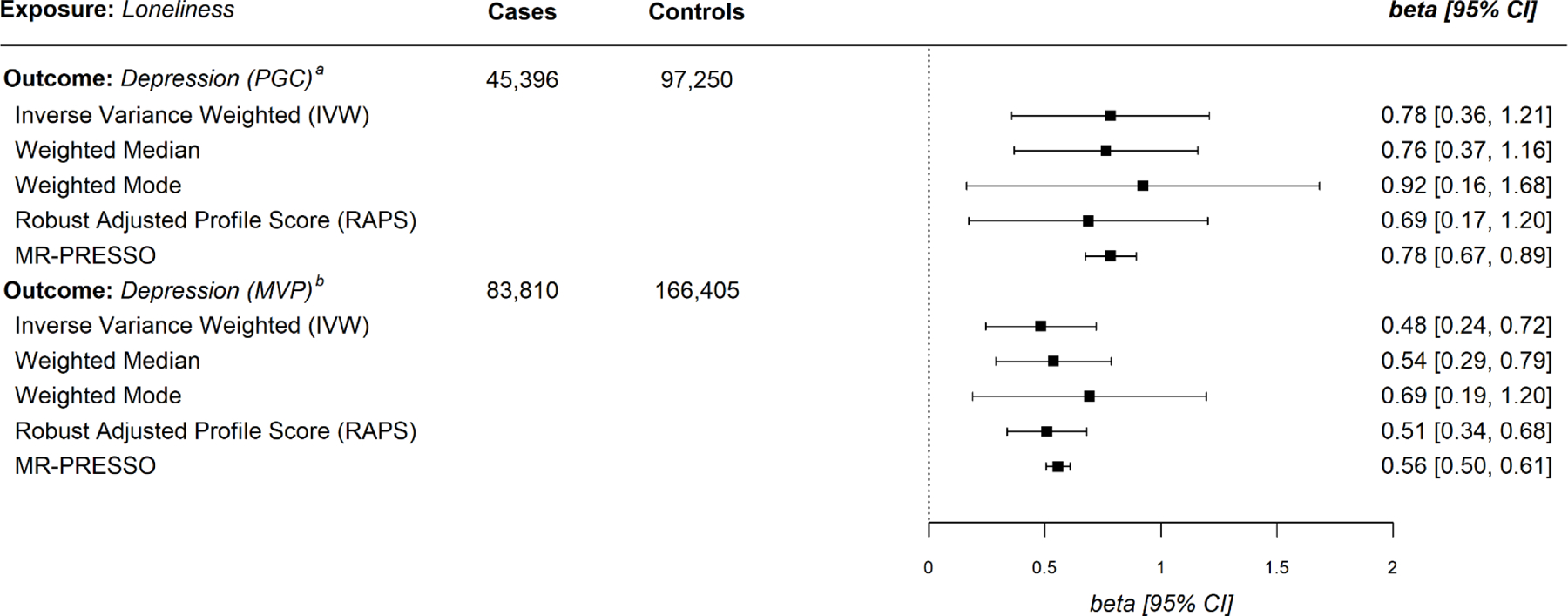
Mendelian Randomization Estimates for Loneliness Exposure with Major Depressive Disorder as the Outcome. a Psychiatric Genomics Consortium; b Million Veteran Program
Table 1.
Causal Analysis Using Summary Effect Estimates (CAUSE) Parameter Estimates
| Exposure * | Outcome | Model † | γ | η | q | p |
|---|---|---|---|---|---|---|
| Loneliness | MD: PGC | Sharing | -- | 3.36 (2, 5.25) | 0.39 (0.17, 0.63) | 0.004 |
| Loneliness | MD: PGC | Causal | 2.01 (1.26, 2.75) | −0.13 (−7.21, 6.16) | 0.04 (0, 0.26) | |
| Loneliness | MD: MVP | Sharing | -- | 1.63 (0.64, 3.39) | 0.25 (0.04, 0.52) | 0.008 |
| Loneliness | MD: MVP | Causal | 0.74 (0.38, 1.1) | −0.01 (−4.24, 4.1) | 0.04 (0, 0.25) | |
| MD: PGC | Loneliness | Sharing | -- | 0.03 (−0.02, 0.09) | 0.12 (0, 0.41) | 0.053 |
| MD: PGC | Loneliness | Causal | 0.01 (0.01, 0.02) | 0.00 (−0.08, 0.09) | 0.04 (0, 0.26) | |
| MD: MVP | Loneliness | Sharing | -- | 0.05 (−0.04, 0.11) | 0.16 (0.01, 0.47) | 0.014 |
| MD: MVP | Loneliness | Causal | 0.03 (0.01, 0.04) | 0.00 (−0.12, 0.11) | 0.04 (0.00, 0.26) | |
| Loneliness | MD (PHQ-2) | Sharing | -- | 1.82 (0.93, 3.38) | 0.29 (0.09, 0.54) | 0.007 |
| Loneliness | MD (PHQ-2) | Causal | 0.82 (0.45, 1.19) | −0.01 (−3.07, 3.00) | 0.05 (0.00, 0.27) |
Note. All loneliness summary statistics were identified from Elsworth et al. GWAS;
CAUSE calculates the proportion of variants exhibiting correlated pleiotropy by producing two mixture models; a sharing model which fixes the exposure effect at 0, and a causal model which sets the exposure effect as a free parameter. If the posterior distribution is better fit under the causal model the exposure is presumed to have an independent effect on the outcome, as statistically evaluated by the p-value; γ The causal effect of the exposure on the outcome; in the sharing model γ is fixed at 0, in the causal model γ is a free parameter; η Effect of unobserved heritable shared factor on the outcome; q Effect of uncorrelated factor on the outcome.; p = One-sided p-value testing whether posteriors estimated under the causal model are better fit than the sharing model.
In the next series of analyses, we sought to replicate these findings in the larger MVP-MD outcome data set. As shown in the lower panel of Figure 1, in the MVP-MD outcome data, we again identified a significant positive association in the IVW analysis, suggesting a causal influence from loneliness to MD (IVW = .48, 95% CI = 0.24, 0.72). The other four regression-based estimates, including RAPS and MR-PRESSO analyses, yielded effects similar to the IVW. These findings are consistent with those observed using the PGC-MD outcome data. With the MVP-MD data, both Cochran’s Q and I2GX statistic revealed significant heterogeneity (Supplementary Tables 2 and 3). The I2GX analysis yielded no values > .6, suggesting the MR Egger estimate may not be reliable; we therefore removed MR Egger from subsequent analyses. Steiger filtering did not identify instruments explaining greater variance in MD than in loneliness. Sensitivity analysis using CAUSE are shown in Table 1 and again illustrate that loneliness was causally associated with MD here also (p = .008). Overall, the MVP data provide consistent evidence that loneliness plays a causal role in the risk for MD.
The MVP data also provided an opportunity to examine the causal association between loneliness on depressive symptoms using the PHQ-2 measure (see Methods). Using a conventional threshold of P < 5 × 10−8 from the loneliness GWAS 31, we identified 14 genetic instruments for a harmonized analysis with the MVP-PHQ-2 outcome data. As shown in Supplemental Table 31, four of the MR effect estimates were reliably different from zero, suggesting a causal effect from loneliness to depressive symptoms on the PHQ-2 (IVW = .70, 95% CI = 0.42, 0.98; sensitivity analyses presented in Supplemental Tables 32-34). As with the MD diagnostic analysis in the MVP data, CAUSE modeling rejected the sharing parameter (p = .007; Table 1). Taken together, our findings using MVP data provide robust evidence that loneliness is associated with both depression case status and symptoms in a manner that is consistent with a causal effect.
From Depression to Loneliness.
In the next set of analyses, we reversed the direction of MR, treating MD status as the exposure and loneliness as the outcome. (As noted in the Methods, when loneliness was used as the outcome variable, we relied on data from a UKB-only sample; in this case, the outcome is a binary report of current loneliness—see Figure 2.) We conducted the analyses with each of the PGC- and MVP-MD exposure data. Within the PGC-MD data, at the conventional P < 5 × 10−8 threshold, only one genetic instrument was harmonized with loneliness as the outcome; we therefore used a reduced P < 5 × 10−7 threshold and identified 11 instruments after clumping and harmonization with the extant UKB summary data for loneliness. We identified significant positive effects in the five main MR analyses (IVW = .021, 95% CI = 0.006, 0.036, see Figure 2). All F-statistics suggested instruments were sufficiently associated with the loneliness exposure (F >10, see Supplementary Table 1; the genetic variants are reported in Supplementary Table 7). Both Cochran’s Q and I2GX revealed significant heterogeneity (Supplementary Tables 2 and 3). Steiger filtering did not identify instruments explaining greater variance in loneliness than MD. However, CAUSE sensitivity analysis using the PGC-MD exposure on loneliness did not suggest a causal effect; therefore, we were unable to formally rule-out correlated pleiotropy (p = .053; Table 1). Given the limited number of genetic variants identified in the PGC-MD, we conducted a final sensitivity analysis using the variants first identified in the Wray et al. GWAS40; after clumping and harmonization with the extant UKB summary data for loneliness, this analysis included 35 genetic instruments.1 Using this expanded set of genetic instruments and repeating our analyses with the UKB outcome data, the MR effect estimates were directionally consistent with those reported above (see Supplemental Table 27).
Figure 2.
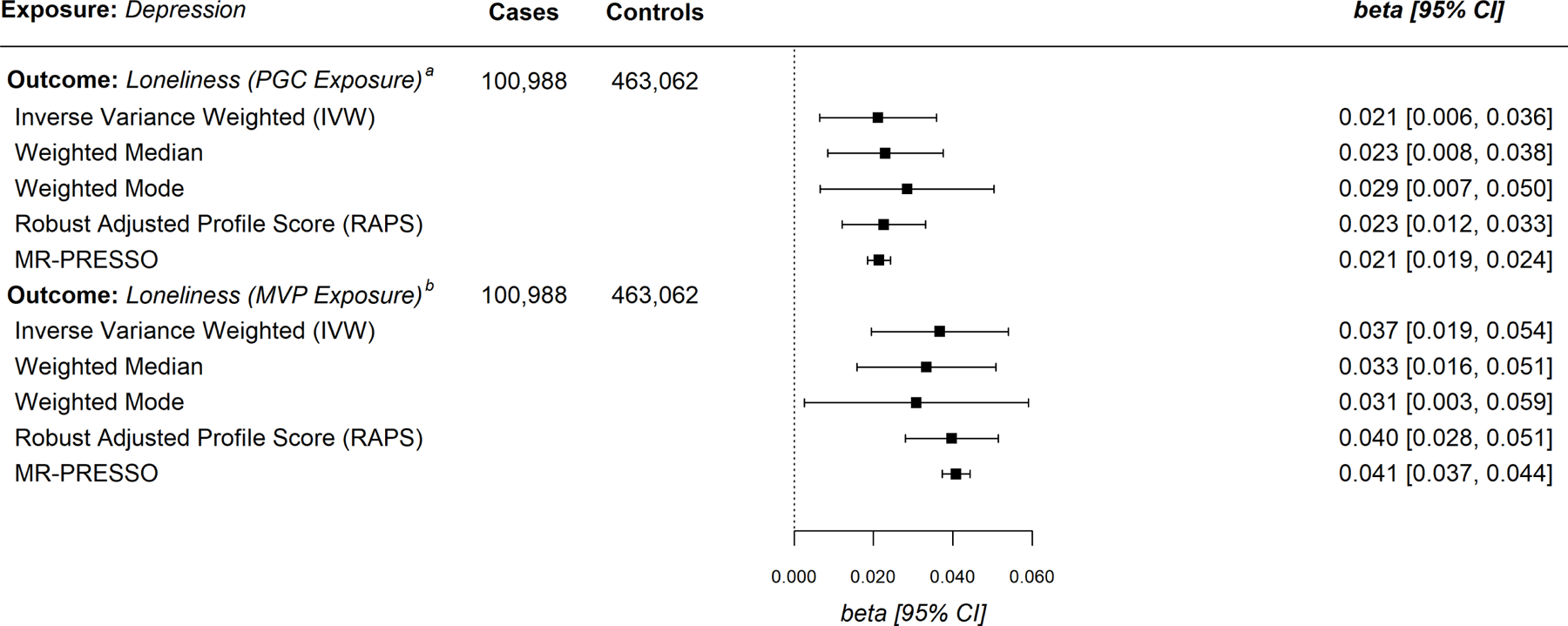
Mendelian Randomization Estimates for Major Depressive Disorder Exposure with Loneliness as the Outcome. a Psychiatric Genomics Consortium; b Million Veteran Program
We next sought to replicate these effects using the MVP data. Using a conventional P < 5 × 10−8 threshold, we identified only 8 genetic instruments; we therefore used a relaxed P < 5 × 10−7 threshold and following harmonization with the UKB-only outcome data, we identified 19 independent genetic instruments with F-statistics >20, providing no evidence of weak instrument bias in the MVP data (Supplementary Table 1). As shown in the lower panel of Figure 2, we find evidence of a causal effect from MD to loneliness— all five of the main MR methods revealed significant positive effects 2 (IVW = .037, 95% CI = 0.019, 0.054). Both Cochran’s Q and I2GX statistics revealed significant heterogeneity (Supplementary Tables 2 and 3). Steiger filtering identified no variants more strongly associated with loneliness than with MD. The CAUSE modeling rejected the sharing parameter (p = .007; Table 1), indicating that estimating a causal effect in the model predicting loneliness to MD would improve the model fit; thus, using the MVP exposure data, results of the CAUSE modeling are consistent with a causal effect from MD to loneliness. Similar to our approach with the PGC data, we conducted an additional sensitivity analysis using the 223 genetic variants identified by Levey et al.41 in their GWAS, which yielded 137 instruments after clumping and harmonization with the UKB-only outcome data. All five of the MR estimates were consistent with those reported in the lower panel of Figure 2 (IVW = .075, 95% CI = 0.065, 0.085; see Supplemental Table 17).
DISCUSSION
This report used two-sample MR to examine the potentially causal association between loneliness—the subjective experience of social isolation—and MD. The observational research literature, including multiple longitudinal studies, suggests a strong positive association between loneliness and MD risk10,19,54,55, but the question of whether loneliness is a causal risk for MD remains largely unknown. Studying this issue is complicated by the fact that the genome-wide genetic correlation between loneliness and MD is high (rg = .64 see30,31) and the constructs studied here share significant (phenotypic and genetic) overlap with neuroticism31,33,56; these facts raise questions of whether loneliness is an independent causal risk factor for MD, or perhaps part-and-parcel of the genetic architecture of MD itself21 or the broader construct of negative affectivity33. To address this issue, we used a variety of MR methods to examine horizontal pleiotropy and conducted a series of sensitivity analyses to test the robustness of the observed associations. Using outcome data from two large-scale GWAS studies, we found robust evidence across different MR techniques and using multiple, independent data sets in support of a causal association from loneliness to MD. In addition, using the summary statistics from the MVP, we identified a potential causal effect from loneliness to depressive symptoms, as assessed by the PHQ-2. We also identified evidence suggesting a causal effect in the reverse direction, from MD to loneliness.
The clearest case for a causal effect from loneliness to depression would be a RCT showing that risk for depressive symptoms or cases of MD are reduced following reductions in loneliness. Although this evidence does not yet exist, there are widespread efforts to reduce population-wide loneliness9,12, and social disconnection is increasingly recognized as a public health priority. The U.S. Surgeon General’s recent Advisory on the Healing Effects of Social Connection and Community15 outlines a national strategy to mitigate the negative effects of social disconnection and loneliness, and these efforts include strengthening the social infrastructures of communities, enacting pro-connection social policies, mobilizing the health sector (including direct interventions to reduce loneliness among individuals), reforming digital environments and social media, deepening knowledge through funding and a national research agenda, and building a culture of connection in schools, workplaces, and communities.
Results from the current study provide broad support for reducing loneliness in service of treating depression and reducing depressive symptoms (and vice versa), although the findings do not speak to which of public health strategies outlined above might be the most effective way to achieve this goal. Moreover, the exact mapping of effect sizes from this report to a clinical intervention study are a challenge as well. For example, the IVW effect size in the PGC analyses from loneliness to depression (b = .78) suggests that a one standard deviation reduction in loneliness would be associated with a 54% reduction in MD case status, but this does not translate to a direct intervention effect. Rather, these results are consistent with recent guidelines for interpreting results from MR investigations: When the aim of MR is to test a causal hypothesis, as it is here, establishing the existence of a causal effect using strong genetic instruments is of greater importance than estimating the exact size of the causal effect 38. The findings reported here are consistent with a causal effect from loneliness to MD and depressive symptoms, and this work provides a robust basis for intervention studies that seek to target and reduce loneliness in service of reducing depressive risk, but our findings do not speak to the specific magnitude of the likely intervention effect. Overall, the current paper contributes to the growing body of MR research on loneliness as a causal risk for a range of morbidities31,35,36,57 and the current work may be especially timely in light of the SARS-CoV-2 pandemic, which has engendered lifestyle changes and economic burdens around the globe—including lockdowns, social isolation, and job loss— and hastened concerns about mental illness and emotional distress58,59.
The evolutionary theory of loneliness suggests that acute experiences of subjective social isolation are universal and provides a critical signal about one’s social standing10. In these situations, loneliness is believed to provide motivation to connect or reconnect with other people60. In some cases, however, loneliness becomes enduring, possibly due to negative social cognition, behavioral conformation, or social withdrawal9,61. When experienced chronically, loneliness is associated with a state of vigilance in which a person monitors their belonging and ruminates on their perceived lack of social connection62. Belongingness is a fundamental human need and when thwarted, the disconnection between social needs, desires, and perceived availability of social resources can lead to depression63. The data available for this report focuses on the point-prevalence of both loneliness and the lifetime prevalence of depression, making it difficult to disentangle unique associations between acute or chronic loneliness on the causal risk for depressive illness or depressive symptoms. Although disentangling the predictive utility of these dimensions of loneliness is needed in future research, the findings reported here indicate that treating the experience of loneliness— whether acute or chronic— as a modifiable risk factor for depression is scientifically justified 5. Furthermore, loneliness itself is an aversive psychological state, and the findings reported here also suggest that reducing depressive symptoms should have a concomitant effect on reducing perceptions of social isolation. Finally, although our paper has addressed loneliness as a key risk for MD, it will be important for future MR studies to also examine a number of related social connection constructs, including social isolation, which is uniquely and independently associated with a range of poor health outcomes16. In the U.S.-based All of Us Research Program (https://allofus.nih.gov/) has an extensive measurement of social determinant variables and a relatively large cohort with genetic data is available to study social influences on health.
The analyses reported here include several strengths that increase confidence in the causal conclusions. First, we used separate exposure and outcome datasets to facilitate a two-sample MR design, which avoids risk of bias toward observational estimates. Not only are the GWAS discovery samples included here large, but we replicated each of our main regression-based MR analyses in a second GWAS dataset, which increases confidence these effects are robust across European-ancestry populations. In addition, we found evidence that loneliness appears to exert a causal effect on MD case status as well as with depressive symptoms, as assessed by the PHQ-2 in the MVP sample. The overall consistency across our primary and sensitivity analyses increases confidence in our findings. Second, in situations where the genome-wide correlation between constructs is high, as it is here between loneliness and MDD (see31), the application of CAUSE modeling is a strength and can help determine whether correlated horizontal pleiotropy fully explains this genetic correlation. CAUSE uses a Bayesian model fitting strategy to determine if the absence of a causal parameter degrades model fit; in this case (i.e., when the models reject the sharing parameter alone), the posterior distributions for fitting new data would be improved with the inclusion of a causal effect. Applying CAUSE to the current data, we rejected the sharing model using the MVP data and found evidence for bidirectional causal effects. In our MD to loneliness analyses, although the CAUSE modeling was consistent with a causal effect in the MVP data, we could not rule-out correlated horizontal pleiotropy in the PGC sample (p =.053). In our view, the preponderance of the evidence reported here is suggestive of a causal effect, but we recognize that the CAUSE analyses with the PGC data leave open the possibility that horizontal pleiotropy biases these results. Finally, both the MVP and PGC data sets provide GWAS summary data on MD caseness, which allow us to make specific statements about clinical risk for MD status. With the MVP data, we also showed that loneliness appears to be a causal risk marker for depressive symptoms (as measured by the two-item PHQ that assesses continuous symptoms of anhedonia and low mood).
Despite these strengths, this study should be interpreted in the context of its limitations. First, our measurement of loneliness was relatively crude, and we are unable to disentangle whether the observed causal effects vary as a function of acute versus chronic loneliness. Although the loneliness → MD analyses relied on a standardized metric of loneliness derived from a GWAS sample of over 500,000 people 31, unit differences in this standardized score are difficult to interpret; moreover, when we reversed the direction for the MD → loneliness, we relied on the binary loneliness outcome from the UKB-only sample. Although this latter measure is nonspecific, it is not an unreasonable metric—e.g., an abbreviated 3-item UCLA loneliness scale has strong psychometric properties 64; however, in the future, it would be ideal for researchers to assess the degree to which participants perceive their loneliness to be relatively acute and transient versus relatively longstanding and chronic. In addition, it is quite possible the effects of loneliness on depression vary by age; the current analyses cannot stratify the MR effects by age, but it is possible that causal effects may operate differentially for younger and older adults. Moreover, the analyses reported here only speak to adult samples. Second, the measure of continuous depressive symptoms in the MVP is relatively crude and focuses solely on anhedonia and low mood. In future research, it will be ideal to relate loneliness to specific depressive symptoms (e.g., guilt or suicidality); doing so requires large-scale individual GWASs for the specific symptoms. Third, the genetic analyses are limited to participants of European ancestry, which limits the generalizability of the findings and leaves unaddressed the impact of loneliness on individuals of non-European ancestry who may be at increased risk for MD. This issue is gaining increased attention in the literature 41 and future studies are needed to fully evaluate the generalizability of these results. Fourth, due to the unavailability of sex-stratified GWAS for loneliness, we were unable to assess for causal differences by sex. We note that depression measured in the MVP predominantly consisted of men, whereas the PGC cohort consisted of a mixed-sex population. The observed MR estimates may differ in stratified samples. Fifth, in revealing evidence for a bidirectional causal effect in the association between loneliness and depression, results from this paper cannot speak to the specific temporal ordering of loneliness and depression. Loneliness and depressive illness (or symptoms) are intertwined causal risk factors, but we find no evidence a specific directional effect. Finally, although we have taken care in trying to address horizontal pleiotropy, we note that other methods exist for doing so and that these tools may be useful to this area of study in the future. For example, Latent Heritability Confounder MR (LHC-MR;65) uses genome-wide summary data to model potential bidirectional causal influences while also accounting for unmeasured and shared confounding via genomic structural equation modeling. A challenge for future research is interrogating questions of unmeasured confounding when two constructs share a high genetic correlation.
Overall, the present results suggest that reducing loneliness would impact risk for MD, but additional work is needed to further substantiate these causal assertions. New research integrating MR and classic twin direction of causality models—the MR-DoC specification 66— illustrates novel ways to examine potential causal effects in the context of high genetic overlap 67. Because population-wide epidemiological studies often do not assess psychological constructs at this depth, the integration of polygenic risk scores in large twin studies via the MR-DoC model may be especially informative in developing deeper insights into the specific psychological features of loneliness that convey its risk. Applying these models to the association between loneliness and MD or specific depressive symptoms would be an excellent next step in the field.
Conclusion
Major depressive disorder (MD) is a serious psychiatric illness that afflicts many people around the globe and is associated with considerable emotional pain and disability. In the face of the SARS-CoV-2 pandemic, rates of MD have increased dramatically, making the search for modifiable causal risk factors an important and timely endeavor5. This report used a suite of two-sample Mendelian Randomization (MR)27 methods to examine the potentially causal, bidirectional associations between loneliness—the subjective experience of social isolation—and MD. Across two independent samples using a variety of sensitivity analyses, we find evidence for a bidirectional causal effect between loneliness and depression. The current findings advance our understanding of the relationship between loneliness and MD and may lay the basis for improved prevention and treatment. Results from this study may be useful for informing public health interventions that seek to reduce or alter loneliness in service of reducing risk for depressive illness, and we would expect to see decreases in loneliness as depression abates as well12.
Supplementary Material
Funding:
DAS was partially supported by a grant from the National Institute of Aging (R01AG078361–01). KWC was partially supported by a Kaplen Fellowship on Depression from the Harvard Medical School, a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation, and funding from the National Institute of Mental Health (K08MH127413). DFL was supported by a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation. REW is supported by a postdoctoral fellowship from the South-Eastern Regional Health Authority (2020024).
Footnotes
Conflict of Interest Disclosures: MBS in the past 3 years has received consulting income from Actelion, Acadia Pharmaceuticals, Aptinyx, Bionomics, BioXcel Therapeutics, Clexio, EmpowerPharm, GW Pharmaceuticals, Janssen, Jazz Pharmaceuticals, and Roche/Genentech. He has also received research support from NIH, Department of Veterans Affairs, and the Department of Defense. He is on the scientific advisory board for Brain and Behavior Research Foundation and the Anxiety and Depression Association of America. Dr. Stein has stock options in Oxeia Biopharmaceuticals and Epivario.
The Wray et al. GWAS included UKB participants, and therefore the genetic instruments were first identified in a sample that included UKB participants. For the current sensitivity analysis, in an effort to maintain independent samples for the exposure and outcome data, we used the instruments identified by Wray et al. in the PGC-MDD exposure sample that does not include UKB participants.
Data Availability Statement Data Sharing:
The data used in this study comes from several sources. Genetic summary statistics for the loneliness instrument are available through the UK Biobank and by request to the authors. The PGC MD summary statistic data is publicly available: https://www.med.unc.edu/pgc/download-results/mdd/. The MVP MD summary statistic data is available on dbGaP via Study Accession: phs001672.v6.p1. MR source code is published on the Open Scientific Framework: https://osf.io/sc6we/
REFERENCES
- 1.Ferrari A et al. Global variation in the prevalence and incidence of major depressive disorder: a systematic review of the epidemiological literature. Psychological medicine 43, 471 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Lim GY et al. Prevalence of depression in the community from 30 countries between 1994 and 2014. Scientific reports 8, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kessler RC et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). Jama 289, 3095–3105 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Smith K Mental health: a world of depression. Nature News 515, 180 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Choi KW et al. An exposure-wide and Mendelian randomization approach to identifying modifiable factors for the prevention of depression. American Journal of Psychiatry 177, 944–954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whisman MA, Sbarra DA & Beach SR Intimate Relationships and Depression: Searching for Causation in the Sea of Association. Annual Review of Clinical Psychology 17(2021). [DOI] [PubMed] [Google Scholar]
- 7.Choi KW et al. Assessment of bidirectional relationships between physical activity and depression among adults: a 2-sample mendelian randomization study. JAMA psychiatry 76, 399–408 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrman H et al. Time for united action on depression: a Lancet–World Psychiatric Association Commission. The Lancet 399, 957–1022 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Cacioppo S, Grippo AJ, London S, Goossens L & Cacioppo JT Loneliness Clinical Import and Interventions. Perspectives on Psychological Science 10, 238–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cacioppo JT & Patrick W Loneliness: Human nature and the need for social connection, (WW Norton & Company, 2008). [Google Scholar]
- 11.Beutel ME et al. Loneliness in the general population: prevalence, determinants and relations to mental health. BMC Psychiatry 17, 97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt-Lunstad J, Robles TF & Sbarra DA Advancing social connection as a public health priority in the United States. American Psychologist 72, 517–530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Killgore WD, Cloonan SA, Taylor EC & Dailey NS Loneliness: A signature mental health concern in the era of COVID-19. Psychiatry Research 290, 113117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bu F, Steptoe A & Fancourt D Loneliness during a strict lockdown: Trajectories and predictors during the COVID-19 pandemic in 38,217 United Kingdom adults. Social Science & Medicine 265, 113521 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Office of the Surgeon General. Our Epidemic of Loneliness and Isolation: The U.S. Surgeon General’s Advisory on the Health Efffects of Social Connection and Community. (2023). [PubMed] [Google Scholar]
- 16.Holt-Lunstad J & Perissinotto C Social isolation and loneliness as medical issues. New England Journal of Medicine 388, 193–195 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Erzen E & Çikrikci Ö The effect of loneliness on depression: A meta-analysis. International Journal of Social Psychiatry 64, 427–435 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Van den Brink R et al. Prognostic significance of social network, social support and loneliness for course of major depressive disorder in adulthood and old age. Epidemiology and Psychiatric Sciences 27, 266–277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich LM & Gullone E The clinical significance of loneliness: A literature review. Clinical Psychology Review 26, 695–718 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Cacioppo JT, Hawkley LC & Thisted RA Perceived social isolation makes me sad: 5-year cross-lagged analyses of loneliness and depressive symptomatology in the Chicago Health, Aging, and Social Relations Study. Psychology and Aging 25, 453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews T et al. Social isolation, loneliness and depression in young adulthood: a behavioural genetic analysis. Social Psychiatry and Psychiatric Epidemiology 51, 339–348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YC et al. Social relationships and physiological determinants of longevity across the human life span. Proceedings of the National Academy of Sciences 113, 578–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murthy VH & Murthy VH Together, (Harper Collins Publishers, 2020). [Google Scholar]
- 24.Turkheimer E & Harden K Behavior genetic research methods: Testing quasi-causal hypotheses using multivariate twin data. in Handbook of Research Methods in Personality and Social Psychology (eds. Reis HT & Judd CM) 159–187 (Cambridge University Press, New York, 2014). [Google Scholar]
- 25.Davey Smith G & Ebrahim S ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? International Journal of Epidemiology 32, 1–22 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Matthews T et al. Social isolation, loneliness and depression in young adulthood: a behavioural genetic analysis. Social Psychiatry and Psychiatric Epidemiology 51, 339–348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davey Smith G & Hemani G Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Human Molecular Genetics 23, R89–R98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanderson E et al. Mendelian randomization. Nature Reviews Methods Primers 2, 6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawlor DA, Harbord RM, Sterne JA, Timpson N & Davey Smith G Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Statistics in Medicine 27, 1133–1163 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Rødevand L et al. Polygenic overlap and shared genetic loci between loneliness, severe mental disorders, and cardiovascular disease risk factors suggest shared molecular mechanisms. Translational Psychiatry 11, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdellaoui A et al. Phenome-wide investigation of health outcomes associated with genetic predisposition to loneliness. Human Molecular Genetics 28, 3853–3865 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sbarra D Social Integration and Sleep Disturbance: A Gene-Environment Interaction Study. Collabra: Psychology 2(2016). [Google Scholar]
- 33.Abdellaoui A et al. Associations between loneliness and personality are mostly driven by a genetic association with neuroticism. Journal of Personality 87, 386–397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boomsma DI, Cacioppo JT, Muthén B, Asparouhov T & Clark S Longitudinal genetic analysis for loneliness in Dutch twins. Twin Research and Human Genetics 10, 267–273 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Wootton RE et al. Bidirectional effects between loneliness, smoking and alcohol use: evidence from a Mendelian randomization study. Addiction 116, 400–406 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Day FR, Ong KK & Perry JR Elucidating the genetic basis of social interaction and isolation. Nature Communications 9, 1–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goossens L et al. The Genetics of Loneliness Linking Evolutionary Theory to Genome-Wide Genetics, Epigenetics, and Social Science. Perspectives on Psychological Science 10, 213–226 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Burgess S et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Research 4(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrison J, Knoblauch N, Marcus JH, Stephens M & He X Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nature Genetics 52, 740–747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wray NR et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics 50, 668–681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levey DF et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in> 1.2 million individuals highlight new therapeutic directions. Nature Neuroscience, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elsworth BL et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv (2020). [Google Scholar]
- 43.Gaziano JM et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. Journal of Clinical Epidemiology 70, 214–223 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Löwe B, Kroenke K & Gräfe K Detecting and monitoring depression with a two-item questionnaire (PHQ-2). Journal of Psychosomatic Research 58, 163–171 (2005). [DOI] [PubMed] [Google Scholar]
- 45.VanderWeele TJ, Tchetgen EJT, Cornelis M & Kraft P Methodological challenges in mendelian randomization. Epidemiology (Cambridge, Mass.) 25, 427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verbanck M, Chen C. y., Neale B & Do R Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nature Genetics 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burgess S & Thompson SG Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. American Journal of Epidemiology 181, 251–260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowden J, Davey Smith G & Burgess S Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. International Journal of Epidemiology 44, 512–525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgess S & Thompson SG Interpreting findings from Mendelian randomization using the MR-Egger method. European Journal of Epidemiology 32, 377–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowden J, Davey Smith G, Haycock PC & Burgess S Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genetic Epidemiology 40, 304–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Q, Wang J, Hemani G, Bowden J & Small DS Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Annals of Statistics 48, 1742–1769 (2020). [Google Scholar]
- 52.Bowden J et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I 2 statistic. International Journal of Epidemiology 45, 1961–1974 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hemani G, Bowden J & Davey Smith G Evaluating the potential role of pleiotropy in Mendelian randomization studies. Human Molecular Genetics 27, R195–R208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cacioppo JT, Hughes ME, Waite LJ, Hawkley LC & Thisted RA Loneliness as a specific risk factor for depressive symptoms: cross-sectional and longitudinal analyses. Psychology and Aging 21, 140 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Kraav S-L et al. Depression and loneliness may have a direct connection without mediating factors. Nordic Journal of Psychiatry, 1–5 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Lo M-T et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nature Genetics 49, 152–156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andreu-Bernabeu A et al. Polygenic contribution to the relationship of loneliness and social isolation with schizophrenia. Nature Communications 13(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mencacci C & Salvi V Expected effects of COVID-19 outbreak on depression incidence in Italy. Journal of Affective Disorders 278, 66 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bueno-Notivol J et al. Prevalence of depression during the COVID-19 outbreak: A meta-analysis of community-based studies. International Journal of Clinical and Health Psychology 21, 100196 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qualter P et al. Loneliness across the life span. Perspectives on Psychological Science 10, 250–264 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Cacioppo JT & Hawkley LC Perceived social isolation and cognition. Trends in Cognitive Sciences 13, 447–454 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bangee M, Harris RA, Bridges N, Rotenberg KJ & Qualter P Loneliness and attention to social threat in young adults: Findings from an eye tracker study. Personality and Individual Differences 63, 16–23 (2014). [Google Scholar]
- 63.Allen K-A, Kern ML, Rozek CS, McInerney DM & Slavich GM Belonging: a review of conceptual issues, an integrative framework, and directions for future research. Australian Journal of Psychology, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hughes ME, Waite LJ, Hawkley LC & Cacioppo JT A short scale for measuring loneliness in large surveys. Research on Aging 26, 655–672 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Darrous L, Mounier N & Kutalik Z Simultaneous estimation of bi-directional causal effects and heritable confounding from GWAS summary statistics. Nature Communications 12, 7274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minică CC, Dolan CV, Boomsma DI, de Geus E & Neale MC Extending causality tests with genetic instruments: An integration of Mendelian randomization with the classical twin design. Behavior Genetics 48, 337–349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Vries LP et al. Genetic evidence for a large overlap and potential bidirectional causal effects between resilience and well-being. Neurobiology of Stress, 100315 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study comes from several sources. Genetic summary statistics for the loneliness instrument are available through the UK Biobank and by request to the authors. The PGC MD summary statistic data is publicly available: https://www.med.unc.edu/pgc/download-results/mdd/. The MVP MD summary statistic data is available on dbGaP via Study Accession: phs001672.v6.p1. MR source code is published on the Open Scientific Framework: https://osf.io/sc6we/


