Abstract
Ankyrin repeat and SOCS box (ASB) family members have a C-terminal SOCS box and an N-terminal ankyrin-related sequence of variable repeats belonging to the SOCS superfamily. While SH2-domain-bearing SOCS proteins are mainly involved in the negative feedback regulation of the protein tyrosine kinase-STAT pathway in response to a variety of cytokines, the roles of ASB family members remain largely unknown. To investigate ASB functions, we screened for ASB3-interacting factors by using antibody array technology and identified tumor necrosis factor receptor II (TNF-R2) as an ASB3 binding target. ASB3 expression and activities are required for (i) TNF-R2 ubiquitination both in vivo and in vitro, (ii) TNF-R2 proteolysis via the proteasome pathway, and (iii) the inhibition of TNF-R2-mediated Jun N-terminal protein kinase (JNK) activation. While the ankyrin repeats of ASB3 interact with the C-terminal 37 amino acids of TNF-R2, the SOCS box of ASB3 is responsible for recruiting the E3 ubiquitin ligase adaptors Elongins-B/C, leading to TNF-R2 ubiquitination on multiple lysine residues within its C-terminal region. Downregulation of ASB3 expression by a small interfering RNA inhibited TNF-R2 degradation and potentiated TNF-R2-mediated cytotoxicity. The data presented here implicate ASB3 as a negative regulator of TNF-R2-mediated cellular responses to TNF-α by direct targeting of TNF-R2 for ubiquitination and proteasome-mediated degradation.
The suppressor of cytokine signaling (SOCS) box has been identified as the functional domain of a diverse family of protein adapters, which include SOCS, ASB, WD-40 repeat and SOCS box (WSB), SPRY domain and SOCS box (SSB), and Ras-related protein Rab. The SOCS box is an approximately 40- to 60-amino-acid motif, and has a predicted helical structure (12). Among the eight SH2-domain-containing SOCS family members, SOCS-1 and SOCS-3 have been most extensively studied and have been shown to act in a classic negative feedback loop to inhibit JAK-STAT signal transduction. In SOCS-1 and SOCS-3, while the SH2 domains are responsible for recognizing phospho-tyrosine motifs of the substrates, as is the case for JAK (24, 29, 34), the SOCS box was found to associate with Elongins-B/C, two proteins that form an E3 ubiquitin ligase complex with Cullin-2 and Rbx-1 (14, 35). This raises the possibility that SOCS proteins may inhibit signaling by functioning as adaptors for an E3 ubiquitin ligase complex, which could mediate the ubiquitination of SOCS binding partners. More definitive evidence of SOCS box recruitment or an E3 ligase has come from studies of the von-Hippel-Lindau (VHL) protein. A structural analysis demonstrated a direct interaction between the VHL protein and the E3 ligase component Elongin-C (32). VHL binds to Elongin-C through the VHLα domain, which is structurally and functionally homologous to the SOCS box (32). Numerous tumors are caused by mutations within the VHLα domain. These mutations prevent Elongin-C recruitment to the VHLα domain, which results in the loss of its activity in inducing proteolysis of its substrate, the transcription factor HIF-1α (15).
The ASB family is the largest among five groups of SOCS box-carrying proteins. Eighteen members of the ASB family have been identified in the National Center for Biotechnology Information human genome database. Recently, the SOCS box of ASB1 has been shown to interact with Elongins-B/C (14), implicating the potential of this group of proteins in mediating protein ubiquitination and degradation. While SOCS family members use the SH2 domain to recruit substrates with phospho-tyrosine motifs, ASB family members are expected to use the ankyrin (ANK) repeats to recruit substrates. The ANK sequence is loosely conserved at the amino acid level but has a tightly conserved secondary structure of approximately 33 amino acids that is found in various copy numbers. Consecutive repeats form V-shaped helix-turn-helix motifs and stack sequentially into bundles (32). As a functional domain specific for mediating protein-protein interactions, the ANK repeat domain has been found in proteins with a wide range of cellular functions. These include transcription factor regulators such as inhibitor of NF-κB (IκB), p53-binding protein 2 (p53BP2), and GABP-β as well as the cyclin-dependent kinase inhibitor family members (30).
To functionally characterize ASB proteins, we cloned ASB1, ASB3, and ASB4. Using an antibody array technology developed in our lab (33), we screened for ASB-interacting factors in cells. We report here that tumor necrosis factor receptor type 2 (TNF-R2) is one of the binding targets of ASB3. By binding to the C-terminal region of TNF-R2, ASB3 mediates TNF-R2 ubiquitination and degradation; ASB3 also inhibits TNF-R2-mediated JNK activation. Furthermore, modulation of asb3 levels by a small interfering RNA (siRNA) affected TNF-R2 protein stability and the extent of TNF-R2-mediated cytotoxicity in a stable cell line, 4E3, expressing TNF-R2. The data presented here therefore implicate ASB3 as a negative regulator of TNF-R2-mediated cellular responses to TNF-α.
MATERIALS AND METHODS
Cell lines and reagents.
The HEK293 (293), HeLa, T47D, MD-468, A431, and DU-145 cell lines were grown in Dulbecco's modified Eagle's medium (Sigma-Aldrich, St. Louis, Mo.) with 10% fetal bovine serum (Invitrogen, Carlsbad, Calif.), penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37°C in 5% CO2 and 95% air. The Jurkat, K562, and 4E3 cell lines were grown in RPMI (Invitrogen) with 10% fetal bovine serum at 37°C in 5% CO2 and 95% air. Recombinant human TNF-α was purchased from R&D Systems (Minneapolis, Minn.).
Plasmids and transient transfection.
ASB3 was cloned from 293 cells by reverse transcription-PCR (RT-PCR). The total RNA was isolated with the Trizol reagent per the manufacturer's protocol and subjected to reverse transcription using Superscript II reverse transcriptase (Invitrogen). ASB3 amplification was performed with the proofreading DNA polymerase PfuTurbo (Stratagene, La Jolla, Calif.) and the primers 5′-CCGGAATTCCATGGATTTTACAGAGGC-3′ and 5′-CCGCTCGAGTTATCCATCTTGAATAGC-3′. ASB3 was cloned in frame with the c-Myc tag into EcoRI and XhoI sites of pcDNA3-c-Myc. c-Myc-ASB3ΔSB was constructed by first excising the insert by EcoRI and XhoI digestion followed by removal of the SOCS box domain of ASB3 by NcoI digestion. The insert was then subcloned in frame with the c-Myc tag into pcDNA3-cMyc by the use of EcoRI and blunt end ligation. TNF-R2 C-terminal-region deletion constructs were generated by PCRs using the proofreading Turbo Taq polymerase and were subcloned into the pcDNA vector. The primers used to generate TNF-R2ΔS424 were 5′-ATGGCGCCCGTCGCCGTCTGG-3′ and 5′-GGGGACCTGCTCGTCCTTCGG-3′; the primers used to generate TNF-R2Δ364 were 5′-ATGGCGCCCGTCGCCGTCTGG-3′ and 5′-GGCCTCCCCGGCCCCACTGGC-3′; and the primers used to generate TNF-R2Δ331 were 5′-ATGGCGCCCGTCGCCGTCTGG-3′ and 5′-CAGGGAGCTGCTGCTGGAGCT-3′. ASB3 ANK deletion mutants were generated by PCRs using the proofreading Turbo Taq polymerase and were ligated into the pcDNA3-Myc vector. The primers used for ANK deletion mutants were as follows: 5′-CCGGAATTCAGGATGGAACTCCTTGCACCAG-3′ and 5′-CCGCTCGAGTTATCCATCTTGAATAGC-3′ for ASB3-ΔANK1-4, 5′-CAGGAATTCTAAAGTAAGCCCTGTTTACTC-3′ and 5′-CCGCTCGAGTTATCCATCTTGAATAGC-3′ for ASB3-ΔANK1-8, and 5′-CAGGAATTCAGTTCCATCCCTGACCCATC-3′ and 5′-CCGCTCGAGTTATCCATCTTGAATAGC-3′ for ASB3-ΔANK12. K→R point mutant TNF-R2 constructs were generated by performing site-directed mutagenesis (Stratagene). All transient transfections were carried out in serum-free medium with the indicated amounts of plasmids by using either Lipofectamine or Lipofectamine 2000 (Invitrogen). Whole extracts from 293 cells (106) after transfection with expression constructs were prepared with RIPA lysis buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 0.1% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol, 1 mM Na3VO4, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM NaF) containing a protease inhibitor cocktail (Roche, Indianapolis, Ind.).
RT-PCR.
Cells (106) growing in six-well clusters were washed with ice-cold phosphate-buffered saline and lysed with Trizol (Invitrogen). Total RNAs were isolated according to the manufacturer's protocol and subjected to reverse transcription with Superscript II reverse transcriptase (Invitrogen). The primers for ASB3 amplification were the same as those used for ASB3 cloning. The primers used for β-actin amplification were 5′-TGCGTGACATTAAGGAGAAG-3′ and 5′-GCTCGTAGCTCTTCTCCA-3′. Semiquantitative RT-PCR primers used for determinations of siRNA knockout efficiencies were as follows: for ASB3 amplification, 5′-CCAGATAAATGAACTTCATTTGGC-3′ and 5′CCGCTCGAGTTATCCATCTTGAATAGC-3′; and for β-actin amplification, 5′-TGCGTGACATTAAGGAGAAG-3′ and 5′-GCTCGTAGCTCTTCTCCA-3′.
Antibody array, immunoprecipitation, and immunoblot analysis.
Antibody arrays were prepared by spotting 300 commercial antibodies from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.) onto a polyvinylidene difluoride (PVDF) membrane according to a previously published protocol (14). Antibody arrays were preincubated for 4 h at room temperature with 5% milk in Tris-buffered saline with Tween 20 (TBST) followed by incubation with whole-cell extracts (1.5 mg) overnight at 4°C. Unbound proteins were removed by washing with TBST at room temperature for three times. The array was then blotted with a horseradish peroxidase (HRP)-conjugated c-Myc antibody (Santa Cruz Biotechnology Inc.) for 4 h at room temperature followed by TBST washes at room temperature. Signal detection was performed by enhanced chemiluminescence (ECL) detection (Amersham Biosciences, Piscataway, N.J.). For immunoprecipitations, cells were washed twice with ice-cold phosphate-buffered saline and lysed in RIPA lysis buffer. Cleared cell extracts were immunoprecipitated with the indicated antibodies, followed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis and immunoblotting coupled with fluorescent signal detection with an Odyssey fluorescence scanner (LI-COR Biosciences, Lincoln, Nebr.). The following antibodies were used for this study. Anti-c-Myc (9E10), anti-TNF-R2 (N-18 and L-20), anti-ubiquitin (PD41), and anti-IκBα (C-20) were purchased from Santa Cruz Biotechnology Inc. Anti-hemagglutinin (anti-HA) (12CA5), anti-mouse-IRDye800, and anti-rabbit-Alexa fluor 680 were purchased from Roche, Rockland Immunochemicals (Gilbertsville, Pa.), and Molecular Probes (Eugene, Oreg.), respectively.
In vitro kinase assay.
Anti-HA precipitates were prepared from 293 cells that were transiently transfected with HA-JNK1. The HA precipitates were washed extensively in kinase assay buffer (20 mM Tris [pH 7.6], 10 mM MgCl2, 3 mM EDTA, 1 mM vanadate, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM NaF, and protease inhibitor cocktail) and then incubated in kinase assay buffer containing 0.2 μCi/μl [γ-32P]ATP (Amersham Biosciences) for 30 min at 30°C. Purified glutathione S-transferase (GST)-c-Jun was added as an exogenous substrate, and 1 μM ATP was added to initiate the reaction. The reactions were stopped by the addition of an equal volume of 2× SDS-PAGE sample buffer and then analyzed by SDS-PAGE followed by autoradiography.
In vitro ubiquitination assay.
Whole extracts prepared from 293 cells transfected with c-Myc-ASB3 or TNF-R2 were incubated with agarose beads conjugated with anti-c-Myc or anti-TNF-R2 for immunoprecipitation. After being washed extensively with lysis buffer, the antibody-conjugated beads were resuspended in 50 μl of reaction buffer containing 20 mM HEPES (pH 7.3), 10 mM MgCl2, 1 mM dithiothreitol, 2 mM ATP, 5 μg/ml ubiquitin (Ub) or methyl-Ub, 50 μM ubiquitin-activating enzyme (E1), and 1 μM ubiquitin-conjugating enzyme (E2-UbcH5a) (Boston Biochem, Cambridge, Mass.). The reactions were incubated for 1.5 h at 37°C and were terminated by the addition of 2× SDS loading buffer. The reactions were then separated by SDS-PAGE and transferred to a PVDF membrane followed by anti-ubiquitin immunoblot analysis.
Luciferase reporter and cell death quantitation assays.
293 cells were cotransfected with 5× Gal4-luc, c-Jun-Gal4, a Renilla luciferase vector, and other constructs as indicated (1.5 μg of DNA in total). After 24 h, cells were harvested and luciferase activities were determined by use of a dual-luciferase assay kit from Promega (Madison, Wis.) according to the manufacturer's protocol and a Lumat luminometer from Berthold Technologies (Oak Ridge, Tenn.). The results shown are representative of three independent experiments performed in duplicate. Cell death was determined by the release of Renilla luciferase activity as described previously (17). 4E3 cells (2 × 106) were transfected with the pRL-TK Renilla reporter construct (1.5 μg) with either a control siRNA (200 nM) or the ASB3 siRNA (200 nM) by the use of Lipofectamine 2000. After 24 h of transfection, the cells were split into two wells and were either left untreated or treated with TNF-α (10 ng/ml). The cells were lysed at the indicated times post-TNF-α treatment, and the Renilla activity was measured by the addition of the Stop & Glow substrate (Promega, Madison, Wis.) with a Lumat luminometer. Cell death was calculated with the following formula: percent specific cytotoxicity = [100 − (experimental Renilla activity/control Renilla activity)] × 100.
RESULTS
ASB3 is ubiquitously expressed and recruited by TNF-R2.
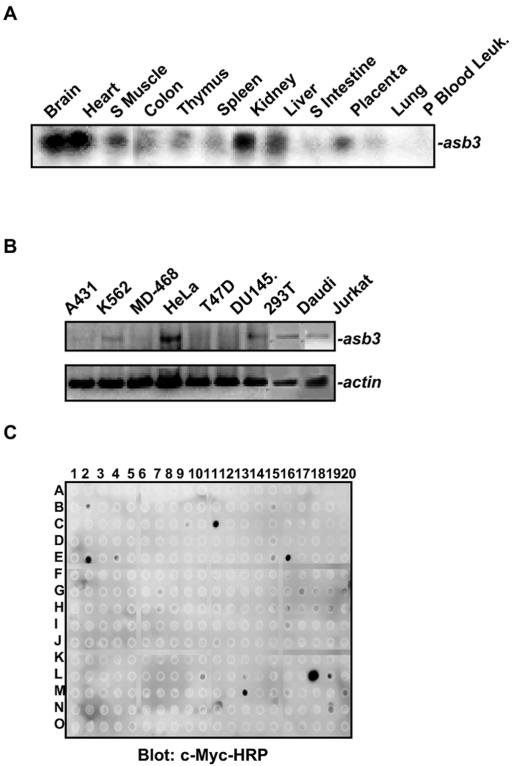
We cloned the ASB3 cDNA from the human embryonic kidney cell line HEK293. Although asb3 was widely expressed, the highest asb3 mRNA levels were detected in the brain, heart, skeletal muscle, and kidney (Fig. 1A). Among the human cancer cell lines tested, HeLa cells showed the highest expression level of asb3, whereas the breast cancer cell lines MD-468 and T47D and the prostate cancer cell line DU-145 lacked detectable asb3 mRNA, as shown by RT-PCR analysis (Fig. 1B). To identify ASB3 binding targets, we performed antibody array screening (33). Whole extracts prepared from 293 cells transiently transfected with c-Myc-tagged ASB3 were incubated with an antibody array containing 300 immobilized antibodies. After extensive washes, the array was immunoblotted with an HRP-conjugated c-Myc antibody followed by ECL detection. Positive signals (cMyc-HRP) were detected at spots printed with antibodies against factors such as TNF-R2, Toso, and survivin (Fig. 1C). In a similar approach, exogenous GST-ASB3 proteins were added to whole extracts prepared from 293 cells and incubated with an antibody array, followed by anti-GST-HRP and ECL detection analysis. Positive (GST-HRP) signals were detected at the spots immobilized with antibodies against TNF-R1, TNF-R2, p105, and SMAD4 (data not shown). Therefore, these antibody array analyses strongly indicated that ASB3 interacts with TNF-α signaling factors in cells.
FIG. 1.
ASB3 expression and protein-binding patterns. (A) A human adult tissue Northern blot (BD Biosciences Clontech, Palo Alto, Calif.) was probed with a 32P-labeled probe generated from ASB3 cDNA. (B) With different cell lines, as indicated, RT-PCR was performed to examine asb3 (top) and β-actin (bottom) mRNA expression. (C) Whole extracts (1.5 mg) prepared from 293 cells transfected with c-Myc-ASB3 were incubated with an antibody array (300 antibodies immobilized on a PVDF membrane) overnight at 4°C. After being washed, the array was blotted with anti-c-Myc-HRP for 4 h followed by ECL detection. Anti-c-Myc-HRP-positive signals were detected at the spots printed with antibodies against c-Myc (C11), FAST (E2), HEF-1 (E16), Survivin (L10), TNF-R2 (L18), Toso (L19), and VHR (M13).
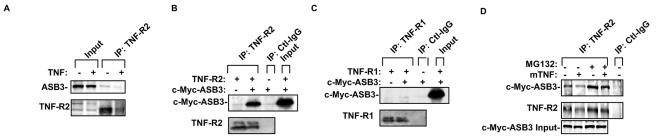
To confirm the protein-protein interactions detected with the antibody arrays, we performed coimmunoprecipitation assays. This approach confirmed the interaction between transfected ASB3 and endogenous TNF-R2 (Fig. 2A). The interaction between TNF-R2 and ASB3 was markedly enhanced when both ASB3 and TNF-R2 were transfected into 293 cells (Fig. 2B). When ASB3 was cotransfected with TNF-R1 into 293 cells, however, no protein interaction was observed (Fig. 2C). Moreover, under similar conditions, ASB1 and ASB4 were not recovered from TNF-R2 immunoprecipitations (data not shown). It is worthwhile to note that while TNF-R2 and ASB3 seem to associate in the absence of a ligand, a reduction in the TNF-R2 protein level in response to TNF-α treatment was repeatedly detected for cells expressing ASB3 (Fig. 2A). Membrane-associated tumor necrosis factor alpha (mTNF-α) has been reported to preferentially bind to TNF-R2 (8, 20). Transient transfection of mTNF-α reduced the TNF-R2 protein level and subsequent TNF-R2-ASB3 complex formation in 293 cells (Fig. 2D). Treatment of the cells with the 26S proteasome inhibitor MG132 reversed the negative effect of mTNF-α on TNF-R2 and stabilized TNF-R2-ASB3 complex formation (Fig. 2D). These results strongly indicate that ASB3 can associate with TNF-R2 and affects TNF-R2's stability in a TNF-α stimulation-dependent manner.
FIG. 2.
TNF-R2 recruits ASB3 in cells. (A) c-Myc-ASB3 was transiently transfected into 293 cells, followed by treatment with or without TNF-α for 30 min. Whole extracts prepared from these cells were immunoprecipitated with anti-TNF-R2 followed by Western blot analysis with anti-c-Myc or anti-TNF-R2. (B) 293 cells were transfected with TNF-R2 alone (1 μg pcDNA3 empty vector and 1 μg TNF-R2) or with TNF-R2 and c-Myc-ASB3 (1 μg TNF-R2 and 1 μg c-Myc-ASB3). Forty-eight hours after transfection, cell extracts were prepared for the same immunoprecipitation/Western blot analysis as that described for panel A. As an immunoprecipitation control, immunoglobulin G (normal mouse IgG) was incubated with the extracts prepared from 293 cells expressing TNF-R2 and c-Myc-ASB3. (C) 293 cells were transfected with TNF-R1 alone or with TNF-R1 and c-Myc-ASB3 under similar conditions to those used for panel B. Anti-TNF-R1 precipitates were analyzed for ASB3 binding under similar conditions to those used for panel B. (D) 293 cells were transfected with TNF-R2 (0.1 μg), c-Myc-ASB3 (1 μg), and either empty vector or mTNF-α (0.5 μg). After 48 h, the cells were either left untreated or treated with 10 μM MG132, followed by protein extraction and immunoprecipitation as described for panel A.
The ANK repeats of ASB3 interact with the C terminus of TNF-R2 while the SOCS box binds to Elongin-C.
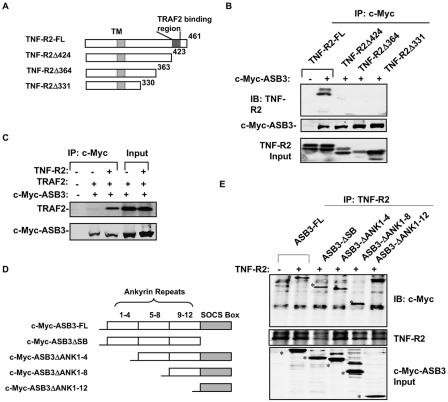
To determine the ASB-3 docking region, we created a series of TNF-R2 C-terminal deletion constructs (Fig. 3A) and cotransfected them with ASB3 into 293 cells. Deletion of the C-terminal 37 residues of TNF-R2 (S424 to S461) abrogated its interaction with ASB3, as shown by coimmunoprecipitation analysis (Fig. 3B), establishing this C-terminal region as the binding site for ASB3. Since TRAF2 has also been shown to bind this region of TNF-R2 (25), we examined the binding interactions between ASB3, TRAF2, and TNF-R2. When ASB3 and TRAF2 were cotransfected into 293 cells, ASB3 immunoprecipitation could not efficiently pull down TRAF2 (Fig. 3C). However, the interaction between ASB-3 and TRAF2 was dramatically enhanced upon TNF-R2 coexpression (Fig. 3C), whereas increasing levels of TRAF2 expression did not alter ASB3-TNF-R2 complex formation (data not shown). These findings indicate that ASB3 and TRAF2 do not interact with each other directly and that both of them can be recruited by TNF-R2 independently.
FIG. 3.
ANK repeats of ASB3 and the C terminus of TNF-R2 are responsible for their interaction. (A) Schematic illustration of TNF-R2 deletion mutants. TM, transmembrane domain. (B) ASB3 (1 μg) was transfected into 293 cells with full-length (FL) TNF-R2 (1 μg) or a deletion mutant, as indicated. After transfection for 48 h, whole-cell extracts were prepared and immunoprecipitated with anti-c-Myc, followed by anti-TNF-R2 or anti-c-Myc blotting as indicated. TNF-R2 expression levels are shown in the bottom panel. (C) 293 cells were cotransfected with ASB3 (1 μg) and TRAF2 (1 μg), with or without TNF-R2, as indicated. Whole-cell extracts were prepared and immunoprecipitated with anti-c-Myc, followed by Western blot analysis with anti-TRAF2 or anti-c-Myc. (D) Schematic illustration of c-Myc-ASB3 with ANK deletion mutations. (E) 293 cells were cotransfected with full-length ASB3 (1 μg) or an ANK deletion mutant and with TNF-R2 (1 μg) or an empty vector (1 μg). Forty-eight hours after transfection, whole-cell extracts were prepared for immunoprecipitation with anti-TNF-R2, followed by Western blot analysis with anti-c-Myc or anti-TNF-R2. The ASB3 expression input (anti-c-Myc) is shown in the bottom panel. Asterisks mark anti-c-Myc signals.
To determine which domain of ASB3 is responsible for TNF-R2 binding, we engineered a series of ASB3 constructs with deletions of N-terminal ANK repeats or the C-terminal SOCS box (Fig. 3D). Deletion of the ANK repeats from ANK1 to ANK4 (ASB3ΔANK1-4) and ANK1 to ANK8 (ASB3ΔANK1-8) did not produce any detectable effect on ASB3-TNF-R2 complex formation (Fig. 3E). In contrast, further deletion of the remaining four ANK repeats (i.e., ASB3ΔANK1-12) completely disrupted the interaction between ASB3 and TNF-R2, suggesting that a partial ANK repeat domain is necessary and sufficient for ASB3 and TNF-R2 interactions. Furthermore, deletion of the SOCS box did not affect ASB3-TNF-R2 complex formation (Fig. 3E).
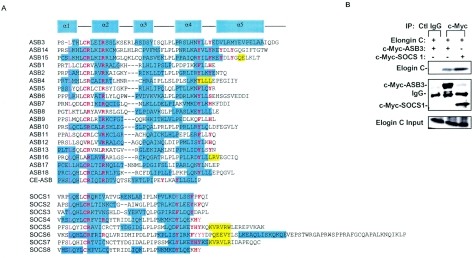
Primary and secondary structural analyses indicated that the ASB protein family and the SOCS family share important structural features within their SOCS box domains, such as conserved leucine and cysteine residues which are required for Elongin-B/C binding within the SOCS box α1-α2-helix (Fig. 4A) (14). After profiling ubiquitinated proteins by using our antibody array technology, we found that multiple proteins, including TNF-R2, are ubiquitinated in 293 cells that have been transiently transfected with ASB3 and HA-ubiquitin (13), suggesting that ASB3 may function as a ubiquitin ligase. The SOCS box in SOCS family members has been proposed to be responsible for recruiting Elongins-BC/CUL-2/Rbx-1 for the formation of a ubiquitin E3 ligase complex (14). The interaction between ASB3 and Elongin-C was clearly detected in 293 cells (Fig. 4B), suggesting that ASB family members may also assemble ubiquitin E3 ligase complexes through their interactions with Elongin-C, thereby mediating the ubiquitination of their substrates (14, 18, 35).
FIG. 4.
ASB3 recruits Elongin-C. (A) Alignment of SOCS boxes of ASB family members and SOCS family members from the human genome database. CE-ASB is a putative ASB identified from the Caenorhabilitis elegans genome database (accession no. U39995). The α-helices (cyan shading) and β-strands (yellow shading) were predicted with the 3D-PSSM web tool, version 2.5.6 (http://www.sbg.bio.ic.ac.uk/∼3dpssm/). Tyrosine residues of α4-helix, the leucine and cysteine residues responsible for the Elongin-C (14) interaction, and the two conserved arginine residues of the α2-helix are shown with red letters. (B) 293 cells were cotransfected with ASB3 or SOCS-1 and Elongin-C and Elongin-B. Whole-cell extracts were prepared for immunoprecipitation with control IgG or anti-c-Myc, followed by Western blot analysis with anti-Elongin-C (top) or anti-c-Myc (middle). The bottom panel shows the expression of Elongin-C.
The SOCS box domain of ASB3 is required for TNF-R2 ubiquitination.
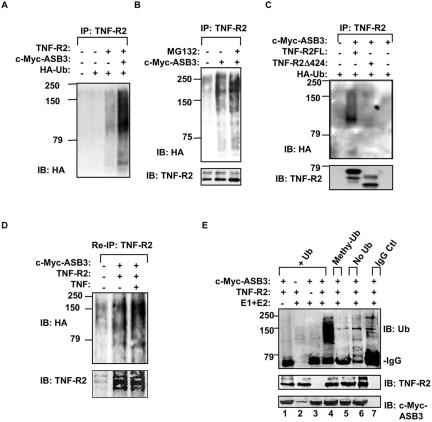
In 293 cells, TNF-R2 ubiquitination was strongly induced when ASB3 was transfected with TNF-R2 (Fig. 5A). Neither ASB3ΔSB (Fig. 6B) nor ASB3ΔANK1-12 was able to induce TNF-R2 ubiquitination (data not shown). Likewise, TNF-R2Δ424, which lacked the ASB3 binding site, was not ubiquitinated in the presence of ASB3 (Fig. 5C). Additionally, polyubiquitinated TNF-R2 accumulated when cells were pretreated with a proteasome inhibitor for 30 min, suggesting that ubiquitinated TNF-R2 may be targeted for proteasome-mediated degradation (Fig. 5B). To determine whether TNF-R2 or TNF-R2-associated proteins were ubiquitinated, we performed a two-step immunoprecipitation assay by boiling anti-TNF-R2 immunoprecipitates for 5 min, followed by a second round of immunoprecipitation with anti-TNF-R2 (17). After the removal of TNF-R2-associated proteins by boiling, the polyubiquitination pattern of TNF-R2 precipitates after reimmunoprecipitation was apparently not perturbed. In addition, the TNF-R2 ubiquitination intensity was enhanced by a TNF-α treatment (Fig. 5D). We repeatedly observed TNF-α-dependent TNF-R2 ubiquitination following reimmunoprecipitation, presumably due to an enhanced sensitivity of detection of ubiquitinated TNF-R2 with this assay. Together, these data suggest that TNF-R2 is indeed ubiquitinated and that this modification is dependent on TNF-α.
FIG. 5.
ASB3 promotes TNF-R2 ubiquitination. (A) 293 cells were transfected with an empty vector (1 μg), TNF-R2 (1 μg), c-Myc-ASB3 (1 μg), and HA-ubiquitin (0.5 μg) as indicated. Twenty-four hours after transfection, whole-cell extracts were prepared and subjected to immunoprecipitation with anti-TNF-R2, followed by Western blot analysis with anti-HA. (B) 293 cells were cotransfected with full-length TNF-R2 and with an empty vector or ASB3 and HA-ubiquitin, as indicated. TNF-R2 ubiquitination was analyzed as described for panel A except that cells were pretreated with MG132 (25 μM) for 30 min prior to cell harvest. TNF-R2 levels are shown in the bottom panel. (C) 293 cells were cotransfected with full-length (FL) TNF-R2 or TNF-R2Δ424 and with ASB3 and HA-ubiquitin, as indicated. TNF-R2 ubiquitination was analyzed as described for panel A. TNF-R2 levels are shown in the bottom panel. (D) 293 cells were transfected with TNF-R2 as described for panel A and then left untreated or treated with TNF-α for 30 min. The cells were then lysed for immunoprecipitation with anti-TNF-R2. The immunoprecipitates were eluted from the beads by boiling in the presence of 0.5% SDS for 5 min, followed by reimmunoprecipitation (re-IP) with anti-TNF-R2. After separation by SDS-PAGE, the proteins were transferred to a PVDF membrane, followed by blotting with anti-HA (top) and anti-TNF-R2 (bottom). (E) In vitro ubiquitination assay. Immunoprecipitated TNF-R2 and/or immunoprecipitated c-Myc-ASB3 was incubated in a control reaction mixture with only Ub and ATP (lane 1). For lanes 2, 3, and 4, immunoprecipitated TNF-R2 and/or immunoprecipitated c-Myc-ASB3 was incubated in a complete reaction mixture containing E1 (50 μM), E2 (1 μM), Ub, and ATP. For lane 5, Ub was replaced with an equal amount of methyl-Ub, whereas for lane 6, no Ub was added. For lane 7, normal mouse IgG plus goat IgG precipitates were incubated in a complete reaction mixture. The reactions were separated by SDS-PAGE, transferred to a PVDF membrane, and blotted with anti-ubiquitin (top). The same membrane was reblotted with anti-TNF-R2 (middle) or anti-c-Myc (bottom).
FIG. 6.
TNF-R2 C terminus bears multiple lysine sites for ubiquitination. (A) Sequence alignment of TNF-R2 C termini of human, bovine, mouse, and rat proteins. (B) 293 cells were cotransfected with wild-type TNF-R2 or various TNF-R2 mutants with Lys→Arg substitutions and with HA-ubiquitin. Twenty-four hours after transfection, whole-cell extracts were prepared for immunoprecipitation with anti-TNF-R2, followed by Western blot analysis with anti-HA (top panel). Anti-TNF-R2 immunoprecipitates were probed with anti-c-Myc (middle panel), and immunoprecipitated TNF-R2 levels are shown in the bottom panel.
ASB3-dependent TNF-R2 ubiquitination was further confirmed in an in vitro ubiquitination assay. In the presence of E1, E2, and ubiquitin, immunopurified ASB3 was able to catalyze the polyubiquitination of TNF-R2 purified from 293 cells (Fig. 5E). Under the same conditions, ASB3 was unable to induce TNF-R2 ubiquitination when ubiquitin was replaced with methyl-ubiquitin, a non-chain-elongating form of ubiquitin (Fig. 5E). Neither ASB3 nor TNF-R2 alone induced ubiquitination under complete reaction conditions (Fig. 5E), suggesting that neither of these proteins has the ability to self-ubiquitinate. Together, these findings strongly indicate that TNF-R2 undergoes ubiquitination modification by recruiting ASB3, which in turn can recruit the Elongin-BC/CUL-2/Rbx-1 E3 ligase complex.
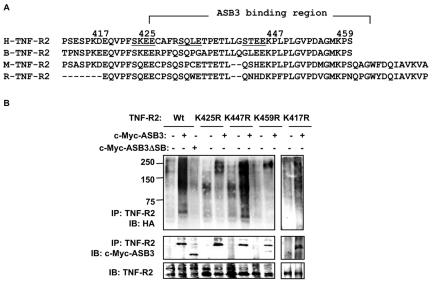
The TNF-R2 C-terminal region has multiple lysine residues for ubiquitination and degradation.
Human TNF-R2 has four lysine residues (Lys417, Lys425, Lys447, and Lys459) in its C-terminal region, where ASB3 docks (Fig. 6A). To determine whether these lysine residues are responsible for TNF-R2 ubiquitination, we made TNF-R2 constructs with K→R mutations and evaluated their ubiquitination profiles in 293 cells. Immunoblotting confirmed that all lysine point mutants were expressed at a level similar to that of wild-type TNF-R2 (Fig. 6B). TNF-R2 polyubiquitination, however, was markedly impaired for both TNF-R2K425R and TNF-R2K459R, whereas the ubiquitination levels of TNF-R2K417R and TNF-R2K447R appeared to be unaffected (Fig. 6B, top panel). A Lys→Arg substitution in all four single point mutants (TNF-R2K417R, TNF-R2K425R, TNF-R2K447R, and TNF-R2K459R) did not affect ASB3 docking within the C terminus of TNF-R2 (Fig. 6B, middle panel), indicating that the reduction in polyubiquitination was not due to a loss of the interaction between TNF-R2 mutants and ASB3 or to gross conformational changes in the mutant proteins. These data suggest that residues K425 and K459 are both specific ubiquitin acceptor sites.
Polyubiquitin modification via K48-linked ubiquitin chains serves as the recognition signal for the 26S proteasome, targeting ubiquitin-modified proteins for proteasome-mediated degradation (9, 10). The TNF-R2 protein was steadily degraded over the course of a 6-h cycloheximide (CHX) chase in 293 cells when TNF-R2 was cotransfected with ASB3 but not with ASB3ΔSB (Fig. 7A), indicating the requirement of the SOCS box for TNF-R2 degradation. Under the same conditions, the stability of TNF-R2-associated proteins such as TRAF2 (TNF receptor-associated factor 2) was unaffected by ASB3, suggesting that ASB3 specifically targets TNF-R2 for degradation (Fig. 7A, bottom panel). ASB3-mediated TNF-R2 degradation proceeds through a proteasome-mediated pathway, as its degradation was blocked by treatment of the cells with MG132, a reversible inhibitor of the 26S proteasome, but not by treatment with NH4Cl, a lysosomal inhibitor used for CHX chase experiments (Fig. 7B). Unlike wild-type TNF-R2, the TNF-R2 K425→R and K459→R mutants both remained relatively stable in the CHX chase experiment, corroborating our previous findings that both the TNF-R2K425R and TNF-R2K459R mutants are resistant to ubiquitination. In contrast, using TNF-R2K447R as an example of a mutant with a wild-type ubiquitination profile, we observed that this K→R substitution did not alter the TNF-R2 degradation kinetics (Fig. 7C), consistent with our previous results showing that K447 is not a ubiquitination site (Fig. 6B).
FIG. 7.
ASB3 induces TNF-R2 proteasome-dependent degradation. (A) 293 cells were cotransfected with TNF-R2 (1 μg) and 1.5 μg c-Myc-ASB3 or c-Myc-ASB3ΔSB for 48 h, followed by a CHX (25 μg/ml) chase for the indicated times. Whole-cell extracts were immunoblotted with anti-TNF-R2 (top panel), anti-c-Myc (middle panel), and anti-TRAF2 (low panel). (B) 293 cells were cotransfected with TNF-R2 and the empty pcDNA vector (EV), c-Myc-ASB3, or c-Myc-ASB3ΔSB, followed by a CHX chase in the presence or absence of MG132 (10 μM) or NH4Cl (15 mM). TNF-R2 and ASB3 proteins were detected as described for panel A. (C) 293 cells were cotransfected with wild-type TNF-R2 or TNF-R2 lysine mutants and with either an empty vector or ASB3 and ubiquitin for 48 h, followed by a CHX chase (25 μg/ml) for an additional 5 h. Cells were either left untreated or treated with TNF-α (10 ng/ml) for 30 min. TNF-R2 and ASB3 protein levels were immunoblotted with anti-TNF-R2 or anti-c-Myc as described above. The right panel shows the TNF-R2 protein levels prior to the CHX chase. (D) Inhibition of ASB3 and Elongin-C expression by ASB3 and Elongin-C siRNAs. 293 cells were transfected with a siRNA directed against green fluorescent protein (GFP; control siRNA), ASB3, Elongin-C, or both ASB3 and Elongin-C. The levels of TNF-R2 proteins (top) and β-tubulin (bottom) were analyzed by immunoblotting with anti-TNF-R2 and anti-β-tubulin, respectively. Elongin-C, asb3, and β-actin mRNA levels were measured by RT-PCR.
The transfection of a siRNA directed against endogenous ASB3 and the ubiquitin E3 ligase adapter Elongin-C noticeably down-regulated the expression of these genes in 293 cells. Consequently, TNF-R2 protein levels in these cells were stabilized (Fig. 7D). These data provide strong genetic evidence that ASB3 and its SOCS box-mediated recruitment of the Elongin-C E3 ligase complex (14) are responsible for TNF-R2 ubiquitination and degradation.
ASB3 inhibits TNF-R2-mediated JNK activation and apoptosis induction.
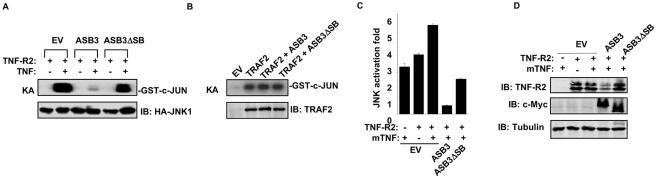
TNF-R2, in response to TNF-α, activates the JNK pathway via the recruitment of TRAF2 (26). We examined the effect of ASB3 on JNK activation by performing in vitro JNK kinase assays. In a cell-free system, TNF-R2 mediated JNK activation in response to TNF-α treatment (Fig. 8A). In addition, the coexpression of ASB-3 but not a mutant with the SOCS box deleted (ASB3ΔSB) dramatically inhibited TNF-R2-dependent JNK activation (Fig. 8A). Consistent with our observations (Fig. 8B), the overexpression of TRAF2 alone has been shown to be sufficient for the induction of JNK activation, independent of a receptor association (24). In vitro JNK protein kinase activation by TRAF2 was not affected by ASB3 overexpression in 293 cells (Fig. 8B), indicating that ASB-3 likely functions upstream of TRAF2. We then evaluated the effect of ASB3 on JNK signaling as a function of c-Jun transcriptional activity measured by a c-Jun-Gal4- and Gal4-responsive luciferase reporter assay. While the expression of mTNF-α or TNF-R2 alone could activate JNK, TNF-R2 and mTNF-α together further enhanced JNK activation (Fig. 8C). The expression of ASB3, however, dramatically inhibited JNK activation by mTNF-α in 293 cells (Fig. 8C). Interestingly, ASB3ΔSB still partially inhibited JNK activation in both an in vitro kinase assay (Fig. 8A) and an in vivo reporter assay (Fig. 8B), even though ASB3ΔSB had no detectable effect on TNF-R2 stability (Fig. 8D). Furthermore, analyses of TNF-R2 protein levels in the same cells revealed that TNF-R2 was down-regulated in the presence of ASB3 (Fig. 8D). Together, these findings provide further evidence that ASB3 is a TNF-R2-specific negative regulator which inhibits TNF-R2-JNK signal transduction, mainly by altering the TNF-R2 protein's stability.
FIG. 8.
ASB3 inhibits JNK activation by TNF-R2. (A) 293 cells were cotransfected with TNF-R2 and HA-JNK1 and with pcDNA3, ASB3, or ASB3ΔSB. Forty-eight hours after transfection, whole-cell extracts were prepared and immunoprecipitated with anti-HA (JNK1). Immunoprecipitated HA-JNK1 was analyzed in an in vitro kinase assay with GST-c-Jun as the substrate. The bottom panel shows immunoprecipitated HA-JNK1 blotted with anti-HA. (B) 293 cells were cotransfected with HA-JNK1 and with TRAF-2 and ASB3 (or ASB3ΔSB). The immunoprecipitated HA-JNK1 was analyzed in an in vitro kinase assay with GST-c-Jun as the substrate. The expression levels of TRAF2 in the cell extracts (anti-TRAF2 blotting) are shown in the bottom panel. (C and D) 293 cells were cotransfected with c-Jun-Gal4- and Gal4-responsive luciferase reporters, Renilla luciferase reporter constructs, and pcDNA, mTNF-α, TNF-R2, c-Myc-ASB3, or c-Myc-ASB3ΔSB, as indicated. After 24 h, whole-cell extracts were prepared and split into half for immunoblotting with the indicated antibodies (D) and for a dual-luciferase assay (C).
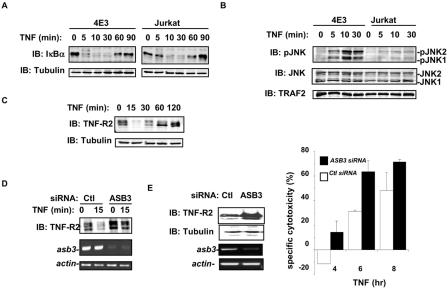
To further explore the regulatory effect of ASB3 on TNF-R2 signal transduction and TNF-R2-mediated cellular effects, we examined Jurkat cells and a derivative Jurkat cell line (4E3) which stably expresses TNF-R2 (3). A quick and transient down-regulation of TNF-R2 protein levels in 4E3 cells was observed in response to TNF-α treatment (Fig. 9C). A knockdown of ASB3 in 4E3 cells attenuated TNF-R2 down-regulation in both untreated cells and cells treated with TNF-α for 15 min and 4 h (Fig. 9D and E). JNK activation following TNF-α treatment was markedly enhanced in 4E3 cells compared to that in Jurkat cells (Fig. 9B), consistent with previous findings that the overexpression of TNF-R2 can preferentially trigger JNK pathway activation (16, 20). In contrast, IκBα degradation by TNF-α was moderately enhanced in 4E3 cells compared to that in parental Jurkat cells (Fig. 9A).
FIG. 9.
ASB3 knockout potentiates TNF-α-induced apoptosis in 4E3 cells. (A) 4E3 and Jurkat cells were treated with TNF-α (10 ng/ml) for the indicated times. Whole-cell extracts were subjected to Western blot analysis with anti-IκBα (top) or anti-β-tubulin (bottom). (B) 4E3 and Jurkat cells were treated as described for panel A. Whole-cell extracts were examined by Western blot analysis with specific antibodies against phospho-JNK (Cell Signaling, Beverly, Mass.), JNK (Pharmingen, San Diego, Calif.), and TRAF2 (Santa Cruz Biotechnology Inc.). (C) Whole-cell extracts were prepared from 4E3 cells following TNF-α treatment for the indicated times. The protein levels of TNF-R2 (top panel) and β-tubulin (bottom panel) were analyzed by Western blot analysis with anti-TNF-R2 and anti-β-tubulin. (D) 4E3 cells, transfected with either control (ctl) siRNA (GFP sequence) or ASB3 siRNA for 48 h, were treated with TNF-α for 15 min. Whole extracts prepared from these cells were subjected to Western blot analysis with anti-TNF-R2. RT-PCRs were performed with the RNAs prepared from these cells to evaluate asb3 expression or β-actin expression. (E) 4E3 cells were transfected with either GFP siRNA (ctl) or ASB3 siRNA for 48 h. Whole-cell extracts prepared from these cells were subjected to Western blot analysis with anti-TNF-R2 and anti-β-tubulin. RT-PCRs were performed with RNAs prepared from these cells to evaluate asb3 expression or β-actin expression. 4E3 cells were transfected with a Renilla luciferase reporter construct (pRLTK; Promega) and either control siRNA or ASB3 siRNA for 48 h. Each group was split in two, with one group maintained in medium and the other treated with TNF-α. At the indicated times, the percentage of specific cytotoxicity was determined by measuring the loss of Renilla luciferase activity. The means and standard deviations of three independent experiments are shown.
Since TNF-α-activated TNF-R2 has been shown to induce T-cell apoptosis via JNK activation (3, 5), we examined whether ASB3 can regulate TNF-R2-dependent apoptosis in 4E3 cells. In 4E3 cells, apparent cell death, as measured by a loss of Renilla luciferase activity (Fig. 9E), was induced by a TNF-α treatment at 6 h, agreeing with previous reports that the overexpression of TNF-R2 sensitizes 4E3 cells to apoptosis in response to TNF-α (3, 17). A knockdown of the asb3 mRNA in 4E3 cells resulted in stabilization of the TNF-R2 protein level (Fig. 9E) and further potentiated apoptosis induction in response to TNF-α treatment (Fig. 9E). Collectively, these studies indicate that ASB3 can affect T-cell signaling via the polyubiquitination and degradation of TNF-R2 and can thereby regulate downstream signaling events in response to TNF-α.
DISCUSSION
In contrast to the rapid progress that has been made in elucidating the functions of SOCS proteins, little is known about ASB protein functions. We demonstrated here that ASB3 is a component of the TNF-R2 signaling complex and possesses a role in providing negative regulation for JNK activation, JNK-mediated gene regulation, and apoptosis induction in response to TNF-α. Our findings also delineate the putative mechanism underlying this regulation, including the recruitment of the ASB3/Elongin-BC/Cullin/Rbx-1 E3 ligase complex and the ubiquitination of TNF-R2 followed by its degradation. While several factors have been found to inhibit TNF-R2 signaling by targeting downstream TNF-R2 signaling molecules (17, 34), ASB3 does so by directly targeting TNF-R2.
The first described SOCS box protein, SOCS-1, was demonstrated to inhibit both interleukin-6-induced receptor phosphorylation and STAT activation (6, 23, 31) by targeting JAK (7). For SOCS family members, the C-terminal helical SOCS box has been shown to be responsible for E3 ligase complex formation by recruiting Elongins-B/C and Cul2 (35). The α-helixes found in the C termini of ASB family members correspond to a typical SOCS box domain (ASB3 residues 460 to 518). A detailed sequence analysis indicated that the SOCS box regions of the SOCS and ASB families share conserved residues responsible for binding to Elongin-C (L462 and C466 in ASB3) (14). In addition, ASB3 has two arginine residues (R467 and R471) that are conserved among the 18 known family members and that may also contribute to the interaction with Elongin-C (Fig. 4A). The region within the SOCS box that has been demonstrated to associate with the ubiquitin ligase complex adapters Elongins-B/C (termed the BC box) has the consensus sequence A(P,S,T)LXXXCXXXA(I,L,V) (14). This consensus sequence was originally identified in Elongin-A and the VHL tumor suppressor protein (15) and is also present in ASB3 (S461LTHLCRLEI) and SOCS-1, both of which were shown here to bind to Elongin-C. TNF-R2 polyubiquitination is evident by both in vitro and in vivo methods and is dependent on ASB3 binding. The SOCS box domain of ASB3 is critical for this process, as demonstrated by the loss of TNF-R2 polyubiquitination when TNF-R2 was coexpressed with a mutant with a SOCS box deletion (ASB3ΔSB). SOCS-1 has been shown to regulate the half-lives of the signaling molecules JAK and VAV by bridging them to the E3 ubiquitin ligase complex (5, 7). It appears reasonable that ASB3 acts similarly as the substrate recognition component of the E3 ubiquitin ligase complex through its interaction with Elongin-C in mediating the degradation of TNF-R2.
The amino acid sequence of ASB3 predicts a 518-amino-acid peptide with a short N-terminal sequence followed by 12 ANK repeats. ANK proteins, such as cyclin-dependent kinase (cdk) inhibitor family members (e.g., p16INK4a) and inhibitors of NF-κB signal transduction (i.e., IκB family members), have a wide range of biological activities (30). ANK repeats form helix-turn-helix motifs that are linked together by loops (30). Studies have shown that some loops serve as sites of protein-protein interactions, while the ANK repeats provide a stabilizing platform (1). Although ASB3 has 12 ANK repeats, a partial ANK repeat region (i.e., ANK9-12) was sufficient for its interaction with TNF-R2, suggesting that ASB3 may require only a partial recognition of its substrate for interaction. Similarly, both p16INK4a and IκBβ use partial ANK repeats for interaction with their substrates (1, 19). Additional ANK repeats may therefore simply function to further stabilize substrate binding or may be involved simultaneously in interactions with other substrates (30). Like the ANK repeats of p16INK4a and IκBβ (1, 19), the loop sequences of the ASB3 ANK repeats contain charged residues, which may be responsible for mediating ionic interactions with residues in the substrate. Although a conserved ANK repeat interaction motif has not yet been identified, Arg47 within the ANK repeats of p16INK4a has been shown to interact with Glu7 of an SXXE motif in the N terminus of its substrate, cdk4 (1). This interaction is primarily mediated by interactions between charged residues (1). The C terminus of TNF-R2 bears three such SXXE motifs, while TRAF2 has been shown to dock on one of these SXXE motifs (25). Whether SXXE motifs provide a modular binding motif for ANK repeats therefore becomes a very intriguing question.
The TNF-R2 C terminus also provides a docking site for TRAF2. The major known signaling factor downstream of TNF-R2 is TRAF2, which is a RING finger domain-containing protein (27). Reimmunoprecipitation experiments with TNF-R2 demonstrated that ASB3 mediates the ubiquitination of TNF-R2 itself (Fig. 5D). This excludes the possibility of misinterpreting the detection of polyubiquitinated TNF-R2 for that of TRAF2 or other TNF-R2-associated proteins. Additionally, our in vitro ubiquitination results indicated that TNF-R2 does not self-ubiquitinate and, in all likelihood, is not ubiquitinated by TRAF2. This is evidenced by the in vitro ubiquitination data shown in Fig. 5E (lane 1), which indicate that TRAF2 coimmunopurified with TNF-R2 from transfected 293 cells (data not shown) was unable to mediate a ubiquitination reaction. Only the addition of ASB3 to the in vitro ubiquitin reactions catalyzed TNF-R2 polyubiquitination.
Although TNF-R2 couples to the prosurvival TRAF2/NF-κB signaling pathway, TNF-R2 activation has also been shown to induce apoptosis in T cells and several tumor cells (3, 4, 11, 21, 28, 36). In these cells, signals from TNF-R2 dramatically potentiate the magnitude and kinetics of caspase activation and apoptosis induction from TNF-R1, which presumably overcomes the induced NF-κB antiapoptotic response (3). It was previously demonstrated that the overexpression of TNF-R2 in Jurkat cells overcomes the cells' innate resistance to TNF-α-induced apoptosis (3). This suggests that the cellular response to TNF-α can be affected by the level of plasma-membrane-bound TNF-R2. Consistent with these findings, we observed that the overexpression of TNF-R2 in Jurkat cells rendered these cells susceptible to TNF-α-induced cell death. Additionally, a knockdown of endogenous asb3 in the 4E3 cell line stabilized TNF-R2 and potentiated TNF-R2-mediated cytotoxicity in response to TNF-α. ASB3 modification of the surface availability of TNF-R2 may therefore be another cellular mechanism for altering cellular responses to TNF-α stimulation.
TNF-R2 plays a crucial role in several inflammatory responses, and the up-regulation of this receptor has been linked to several diseases, including rheumatoid arthritis (2). Etanercept or Enbrel is a recombinant form of TNF-R2 used to treat rheumatoid arthritis via antagonizing TNF-α actions (2, 22). Our data demonstrate that ASB3 attenuates TNF-R2 signaling in response to TNF-α by mediating TNF-R2 proteolysis. These findings imply that targeting ASB3 may provide a novel means for the optimization of therapeutic efforts aimed at neutralizing the inflammatory effects of TNF-α.
Acknowledgments
We thank J. Singer and W. Min for discussions and for reading the manuscript. We thank F. K. Chan for the 4E3 cell line, Z. Q. Pan for HA-ubiquitin, J. W. Conaway for Elongin-C, and D. V. Goeddel for TRAF2 and TNF-R2 expression constructs.
This work was supported by an RO1 grant (CA-82549) from the National Institutes of Health (NIH) (to Y.E.C.) and by a COBRE grant from the NIH to Brown University. A.S.C. was supported by an NIH predoctoral training grant (GM-07601).
REFERENCES
- 1.Byeon, I. J., J. Li, K. Ericson, T. L. Selby, A. Tevelev, H. J. Kim, P. O'Maille, and M. D. Tsai. 1998. Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol. Cell 1:421-431. [DOI] [PubMed] [Google Scholar]
- 2.Carpentier, I., B. Coornaert, and R. Beyaert. 2004. Function and regulation of tumor necrosis factor type 2. Curr. Med. Chem. 11:2205-2212. [DOI] [PubMed] [Google Scholar]
- 3.Chan, F. K., and M. J. Lenardo. 2000. A crucial role for p80 TNF-R2 in amplifying p60 TNF-R1 apoptosis signals in T lymphocytes. Eur. J. Immunol. 30:652-660. [DOI] [PubMed] [Google Scholar]
- 4.Chen, G., and D. V. Goeddel. 2002. TNF-R1 signaling: a beautiful pathway. Science 296:1634-1635. [DOI] [PubMed] [Google Scholar]
- 5.Deng, Y., X. Ren, L. Yang, Y. Lin, and X. Wu. 2003. A JNK-dependent pathway is required for TNFalpha-induced apoptosis. Cell 115:61-70. [DOI] [PubMed] [Google Scholar]
- 6.Endo, T. A., M. Masuhara, M. Yokouchi, R. Suzuki, H. Sakamoto, K. Mitsui, A. Matsumoto, S. Tanimura, M. Ohtsubo, H. Misawa, T. Miyazaki, N. Leonor, T. Taniguchi, T. Fujita, Y. Kanakura, S. Komiya, and A. Yoshimura. 1997. A new protein containing an SH2 domain that inhibits JAK kinases. Nature 387:921-924. [DOI] [PubMed] [Google Scholar]
- 7.Frantsve, J., J. Schwaller, D. W. Sternberg, J. Kutok, and D. G. Gilliland. 2001. Socs-1 inhibits TEL-JAK2-mediated transformation of hematopoietic cells through inhibition of JAK2 kinase activity and induction of proteasome-mediated degradation. Mol. Cell. Biol. 21:3547-3557.11313480 [Google Scholar]
- 8.Grell, M., E. Douni, H. Wajant, M. Lohden, M. Clauss, B. Maxeiner, S. Georgopoulos, W. Lesslauer, G. Kollias, K. Pfizenmaier, et al. 1995. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 83:793-802. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann-Petersen, R., and C. Gordon. 2004. Proteins interacting with the 26S proteasome. Cell. Mol. Life Sci. 61:1589-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann-Petersen, R., M. Seeger, and C. Gordon. 2003. Transferring substrates to the 26S proteasome. Trends Biochem. Sci. 28:26-31. [DOI] [PubMed] [Google Scholar]
- 11.Heller, R. A., K. Song, N. Fan, and D. J. Chang. 1992. The p70 tumor necrosis factor receptor mediates cytotoxicity. Cell 70:47-56. [DOI] [PubMed] [Google Scholar]
- 12.Hilton, D. J., R. T. Richardson, W. S. Alexander, E. M. Viney, T. A. Willson, N. S. Sprigg, R. Starr, S. E. Nicholson, D. Metcalf, and N. A. Nicola. 1998. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc. Natl. Acad. Sci. USA 95:114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov, S. S., A. S. Chung, Z. L. Yuan, Y. J. Guan, K. V. Sachs, J. S. Reichner, and Y. E. Chin. 2004. Antibodies immobilized as arrays to profile protein post-translational modifications in mammalian cells. Mol. Cell. Proteomics 3:788-795. [DOI] [PubMed] [Google Scholar]
- 14.Kamura, T., S. Sato, D. Haque, L. Liu, W. G. Kaelin, Jr., R. C. Conaway, and J. W. Conaway. 1998. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, Ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kibel, A., O. Iliopoulos, J. A. DeCaprio, and W. G. Kaelin, Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269:1444-1446. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S. Y., A. Reichlin, A. Santana, K. A. Sokol, M. C. Nussenzweig, and Y. Choi. 1997. TRAF2 is essential for JNK but not NF-kappaB activation and regulates lymphocyte proliferation and survival. Immunity 7:703-713. [DOI] [PubMed] [Google Scholar]
- 17.Li, X., Y. Yang, and J. D. Ashwell. 2002. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 416:345-347. [DOI] [PubMed] [Google Scholar]
- 18.Liu, Y., J. Li, F. Zhang, W. Qin, G. Yao, X. He, P. Xue, C. Ge, D. Wan, and J. Gu. 2003. Molecular cloning and characterization of the human ASB-8 gene encoding a novel member of ankyrin repeat and SOCS box containing protein family. Biochem. Biophys. Res. Commun. 300:972-979. [DOI] [PubMed] [Google Scholar]
- 19.Malek, S., D. B. Huang, T. Huxford, S. Ghosh, and G. Ghosh. 2003. X-ray crystal structure of an IkappaBbeta × NF-kappaB p65 homodimer complex. J. Biol. Chem. 278:23094-23100. [DOI] [PubMed] [Google Scholar]
- 20.McFarlane, S. M., G. Pashmi, M. C. Connell, A. F. Littlejohn, S. J. Tucker, P. Vandenabeele, and D. J. MacEwan. 2002. Differential activation of nuclear factor-kappaB by tumour necrosis factor receptor subtypes. TNFR1 predominates whereas TNFR2 activates transcription poorly. FEBS Lett. 515:119-126. [DOI] [PubMed] [Google Scholar]
- 21.Medvedev, A. E., A. Sundan, and T. Espevik. 1994. Involvement of the tumor necrosis factor receptor p75 in mediating cytotoxicity and gene regulating activities. Eur. J. Immunol. 24:2842-2849. [DOI] [PubMed] [Google Scholar]
- 22.Moreland, L. W., S. W. Baumgartner, M. H. Schiff, E. A. Tindall, R. M. Fleischmann, A. L. Weaver, R. E. Ettlinger, S. Cohen, W. J. Koopman, K. Mohler, M. B. Widmer, and C. M. Blosch. 1997. Treatment of rheumatoid arthritis with a recombinant human tumor necrosis factor receptor (p75)-Fc fusion protein. N. Engl. J. Med. 337:141-147. [DOI] [PubMed] [Google Scholar]
- 23.Naka, T., M. Narazaki, M. Hirata, T. Matsumoto, S. Minamoto, A. Aono, N. Nishimoto, T. Kajita, T. Taga, K. Yoshizaki, S. Akira, and T. Kishimoto. 1997. Structure and function of a new STAT-induced STAT inhibitor. Nature 387:924-929. [DOI] [PubMed] [Google Scholar]
- 24.Narazaki, M., M. Fujimoto, T. Matsumoto, Y. Morita, H. Saito, T. Kajita, K. Yoshizaki, T. Naka, and T. Kishimoto. 1998. Three distinct domains of SSI-1/SOCS-1/JAB protein are required for its suppression of interleukin 6 signaling. Proc. Natl. Acad. Sci. USA 95:13130-13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, Y. C., V. Burkitt, A. R. Villa, L. Tong, and H. Wu. 1999. Structural basis for self-association and receptor recognition of human TRAF2. Nature 398:533-538. [DOI] [PubMed] [Google Scholar]
- 26.Reinhard, C., B. Shamoon, V. Shyamala, and L. T. Williams. 1997. Tumor necrosis factor alpha-induced activation of c-Jun N-terminal kinase is mediated by TRAF2. EMBO J. 16:1080-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothe, M., V. Sarma, V. M. Dixit, and D. V. Goeddel. 1995. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science 269:1424-1427. [DOI] [PubMed] [Google Scholar]
- 28.Sarin, A., M. Conan-Cibotti, and P. A. Henkart. 1995. Cytotoxic effect of TNF and lymphotoxin on T lymphoblasts. J. Immunol. 155:3716-3718. [PubMed] [Google Scholar]
- 29.Sasaki, A., H. Yasukawa, A. Suzuki, S. Kamizono, T. Syoda, I. Kinjyo, M. Sasaki, J. A. Johnston, and A. Yoshimura. 1999. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells 4:339-351. [DOI] [PubMed] [Google Scholar]
- 30.Sedgwick, S. G., and S. J. Smerdon. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24:311-316. [DOI] [PubMed] [Google Scholar]
- 31.Starr, R., T. A. Willson, E. M. Viney, L. J. Murray, J. R. Rayner, B. J. Jenkins, T. J. Gonda, W. S. Alexander, D. Metcalf, N. A. Nicola, and D. J. Hilton. 1997. A family of cytokine-inducible inhibitors of signalling. Nature 387:917-921. [DOI] [PubMed] [Google Scholar]
- 32.Stebbins, C. E., W. G. Kaelin, Jr., and N. P. Pavletich. 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284:455-461. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Y., T. R. Wu, S. Cai, T. Welte, and Y. E. Chin. 2000. Stat1 as a component of tumor necrosis factor alpha receptor 1-TRADD signaling complex to inhibit NF-kappaB activation. Mol. Cell. Biol. 20:4505-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasukawa, H., H. Misawa, H. Sakamoto, M. Masuhara, A. Sasaki, T. Wakioka, S. Ohtsuka, T. Imaizumi, T. Matsuda, J. N. Ihle, and A. Yoshimura. 1999. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. 18:1309-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, J. G., A. Farley, S. E. Nicholson, T. A. Willson, L. M. Zugaro, R. J. Simpson, R. L. Moritz, D. Cary, R. Richardson, G. Hausmann, B. J. Kile, S. B. Kent, W. S. Alexander, D. Metcalf, D. J. Hilton, N. A. Nicola, and M. Baca. 1999. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. USA 96:2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng, L., G. Fisher, R. E. Miller, J. Peschon, D. H. Lynch, and M. J. Lenardo. 1995. Induction of apoptosis in mature T cells by tumour necrosis factor. Nature 377:348-351. [DOI] [PubMed] [Google Scholar]