Abstract
Transforming growth factor β (TGF-β) inhibits proliferation and promotes cell migration. In TGF-β-treated MCF10A mammary epithelial cells overexpressing HER2 and by chromatin immunoprecipitation, we identified novel Smad targets including protein tyrosine phosphatase receptor type kappa (PTPRK). TGF-β up-regulated PTPRK mRNA and RPTPκ (receptor type protein tyrosine phosphatase kappa, the protein product encoded by the PTPRK gene) protein in tumor and nontumor mammary cells; HER2 overexpression down-regulated its expression. RNA interference (RNAi) of PTPRK accelerated cell cycle progression, enhanced response to epidermal growth factor (EGF), and abrogated TGF-β-mediated antimitogenesis. Endogenous RPTPκ associated with EGF receptor and HER2, resulting in suppression of basal and ErbB ligand-induced proliferation and receptor phosphorylation. In MCF10A/HER2 cells, TGF-β enhanced cell motility, FAK phosphorylation, F-actin assembly, and focal adhesion formation and inhibited RhoA activity. These responses were abolished when RPTPκ was eliminated by RNA interference (RNAi). In cells expressing RPTPκ RNAi, phosphorylation of Src at Tyr527 was increased and (activating) phosphorylation of Src at Tyr416 was reduced. These data suggest that (i) RPTPκ positively regulates Src; (ii) HER2 signaling and TGF-β-induced RPTPκ converge at Src, providing an adequate input for activation of FAK and increased cell motility and adhesion; and (iii) RPTPκ is required for both the antiproliferative and the promigratory effects of TGF-β.
The transforming growth factor β (TGF-β) ligands play an important role in cell proliferation, lineage determination, extracellular matrix production, cell motility, apoptosis, and modulation of immune function (24). TGF-β was originally reported to induce anchorage-independent growth of mouse fibroblasts (28). Subsequent studies indicated that TGF-β is a potent inhibitor of cell proliferation and a tumor suppressor (37, 46). Consistent with its tumor suppressor role, many cancers lose or attenuate TGF-β-mediated antimitogenic action by mutational inactivation of TGF-β receptors or their signal transducers or by less known mechanisms (16, 17, 19, 23, 49, 51). On the other hand, there is increasing evidence that excess production and/or activation of TGF-β in tumors can accelerate cancer progression by a combination of autocrine and paracrine mechanisms which include enhancement of tumor cell motility and survival; increase in tumor angiogenesis, extracellular matrix production, and peritumoral proteases; and inhibition of immune effectors in tumor hosts (reviewed in references 11 and 48).
More recent data suggest that TGF-β can cooperate with oncogenes in tumor progression. Overexpression of active TGF-β1 or an activated type I TGF-β receptor (TβRI) in the mammary gland of transgenic mice results in acceleration of metastases derived from Neu-induced mammary tumors (31, 43). In transgenic mice bearing polyomavirus middle T antigen-expressing mammary tumors, inhibition of TGF-β with the soluble fusion protein TβRII:Fc results in increased apoptosis of tumor cells and a reduction in both circulating tumor cells and lung metastases (30). In the same transgenic model, conditional induction of active TGF-β1 in mice bearing established mammary cancers, increased lung metastases >10-fold without a detectable effect on mammary tumor proliferation or size (32). Mice expressing soluble TβRII under the regulation of the mouse mammary tumor virus (MMTV) long terminal repeat promoter exhibit high levels of the TGF-β antagonist in the circulation which suppress metastases from Neu-induced mammary tumors as well as metastases resulting from injected B16 melanoma cells (57). Finally, exogenous as well as transduced TGF-β has been shown to confer motility and invasiveness to MCF10A human mammary nontransformed epithelial cells that overexpress the HER2 (ErbB2) tyrosine kinase (41, 47). Taken together, these data suggest that oncogenic signals are permissive for TGF-β-induced signals associated with tumor cell motility and potentially metastatic progression.
The cooperation between TGF-β and receptor tyrosine kinases (RTKs) such as HER2 may occur through at least four possible mechanisms: (i) transcriptional modulation that targets the same downstream genes through TGF-β-induced transcription factor Smads, (ii) activation of the Smad-independent signaling pathway, (iii) inhibition of TGF-β-induced antiproliferative effects through the up-regulation of the inhibitory Smad7, and (iv) autocrine induction of TGF-β and ligands that activate RTKs. Upon ligand binding, activated TGF-β receptors phosphorylate Smad2/3, which assemble with Smad4, followed by the translocation of Smad2/3/4 complexes into the nucleus (12, 25). Smad2/3/4 complexes interact with other transcription factors to selectively bind to and transactivate TGF-β target gene promoters. Therefore, to identify TGF-β-regulated genes that can cooperate with HER2 signaling, we performed chromatin immunoprecipitation (ChIP) in TGF-β-treated MCF10A/HER2 cells. Chromatin Smad targets (ChSTs) were precipitated by Smad antibodies and cloned. The ChSTs identified herein included the regulatory regions of protein tyrosine phosphatase receptor-type kappa (PTPRK), serine/threonine kinase 24 (STK24), integrin-α9 (ITGA9), and vimentin-similar genes.
In this study, we found that PTPRK expression is up-regulated by TGF-β-Smad signaling in tumor and nontumor human mammary epithelial cells. The overall levels of RPTPκ (receptor type protein tyrosine phosphatase kappa, the protein product encoded by the PTPRK gene) are relatively suppressed in breast cancer cells compared to nontumor cells. RPTPκ associated with the epidermal growth factor (EGF) receptor (ErbB1) and HER2 (ErbB2) and inhibited ligand-induced ErbB receptor phosphorylation, suggesting a tumor-suppressive role for this receptor phosphatase. HER2 overexpression suppressed RPTPκ expression, suggesting a novel mechanism for HER2-mediated tumorigenesis. We examined the role of RPTPκ on TGF-β function in both normal and breast cancer cells using RNA interference (RNAi). The results from these studies indicated that RPTPκ is required for both TGF-β-induced antiproliferation and cell motility. These data suggest that, similar to TGF-β, RPTPκ exhibits a dual tumor suppressor and tumor-promoting role in mammary epithelial cells.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
All cells were from the American Type Culture Collection. MDA231, MCF7, and Phoenix-Ampho cells were grown in Dulbecco's modified Eagle's medium (DMEM; Cambrex) containing 10% fetal bovine serum (FBS; HyClone) in a humidified 5% CO2 incubator at 37°C. A431 cells were maintained in improved modified Eagle's medium zinc option (Invitrogen) containing 10% FBS. MCF10A, MCF10A/VEC, and MCF10A/HER2 cells were generated and grown as described previously (47). The following reagents were used: human recombinant TGF-β1 (R&D Systems), human EGF and heregulin (Calbiochem), trastuzumab (Herceptin; Vanderbilt University Medical Center Pharmacy), ZD1839 (gefitinib; provided by Alan Wakeling, AstraZeneca Pharmaceuticals), U0126 (Promega, Madison, WI), SB202190 and puromycin (Calbiochem), and G418 (Research Products International Corp.).
The RPTPκ expression plasmid contains a reverse transcription (RT)-PCR-generated full-length 4.3-kb human PTPRK cDNA coding region, controlled by the cytomegalovirus promoter. The RPTPκ expression plasmid or its empty vector control were transfected into cells using FuGENE 6 (Roche) followed by selection in G418 (1 mg/ml). Smad2, Smad3, Smad4, and Smad7 adenoviruses were kindly provided by Kohei Miyazono (Japanese Foundation for Cancer Research, Tokyo, Japan) (15). To generate retroviruses expressing small interfering RNA (siRNA) against human PTPRK, two complementary oligonucleotides, 5′-phos-GATCCCCGATCATTTGATCCTGCAGTTTCAAGAGAACTGCAGGATCAAATGATCTTTTTGGAAT and 5′-phos-AGCTATTCCAAAAAGATCATTTGATCCTGCAGTTCTCTTGAAACTGCAGGATCAAATGATCGGG were annealed and inserted into BglII/HindIII sites of the pSUPER.retro vector (8). As controls, we used siRNA against mouse PTPRK. The oligo-nucleotides 5′-phos-GATCCCCGATCATTTGAcCCTGCtGTTTCAAGAGAACaGCAGGgTCAAATGATCTTTTTGGAAT and 5′-phos-AGCTATTCCAAAAAGATCATTTGAcCCTGCtGTTCTCTTGAAACaGCAGGgTCAAATGATCGGG (lowercase indicates the difference between human and mouse PTPRK genes) were annealed and inserted into the same vector. The resulting plasmids were transfected into Phoenix-Ampho cells (34) to produce human and mouse small interfering PTPRK (siPTPRK) virions, which were harvested at 48 h after transfection. Human PTPRK siRNA (siPTPRK) or mouse PTPRK siRNA (control siRNA) was used to infect cells. Stably transduced colonies were selected in puromycin (1 ng/ml). RPTPκ was examined by immunoblot analysis.
ChIP assay and cloning of novel Smad targets.
MCF10A/HER2 cells were grown to 60% confluence on a 150-mm dish and treated with TGF-β1 (2 ng/ml). Cells were then washed with phosphate-buffered saline (PBS) and cross-linked with 1% formaldehyde in PBS at 37°C for 10 min. Cross-linking was stopped by adding 1 M glycine to a final concentration of 125 mM and incubating the mixture at room temperature for 5 min. Cells were washed with ice-cold PBS twice and scraped into 1 ml of ice-cold PBS containing leupeptin and aprotinin (2 μg/ml each). After collecting the cells by centrifugation, the cell pellet was resuspended in 300 μl of lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], 200 μM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, plus leupeptin and aprotinin) and incubated on ice for 10 min followed by sonication (7 W, 20 s [three times]) to shear the chromatin to 300- to 1,000-bp fragments. After centrifugation, 20 μl of the supernatant was set aside as the chromatin fraction input. The remaining supernatant was diluted (1:5) in dilution buffer (20 mM Tris-HCl [pH 8.1], 150 mM NaCl, 2 mM EDTA, 0.01% SDS, 1% Triton X-100, plus protease and phosphatase inhibitors). To preclear the sample, 2 μg sonicated salmon sperm DNA, 2 μg bovine serum albumin (BSA), 5 μg normal mouse immunoglobulin G (IgG), and 50 μl protein A/G-Sepharose (a 50% slurry of 1:1 in dilution buffer) were added and rotated for 2 h at 4°C. Precleared chromatin was divided into 2 aliquots; 5 μg Smads monoclonal antibodies (2.5 μg Smad2/3 plus 2.5 μg Smad4) or 5 μg actin monoclonal antibody was added to each aliquot and rotated overnight at 4°C followed by addition of 50 μl protein A/G-Sepharose, 2 μg of salmon sperm DNA, and 2 μg BSA and rotation for 1 h at 4°C. Sepharose beads were harvested by centrifugation and washed sequentially in 1 ml (each) of low-salt wash buffer (20 mM Tris-HCl [pH 8.1], 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), high-salt wash buffer (20 mM Tris-HCl [pH 8.1], 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), LiCl wash buffer (0.25 M LiCl, 10 mM Tris-HCl [pH 8.1], 1 mM EDTA, 1% NP-40, 1% deoxycholate), and TE buffer (twice) (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) on a rotating platform at 10-min intervals per wash. Precipitated Smads were confirmed by SDS-polyacrylamide gel electrophoresis and immunoblotting using 5 μl beads. Protein-chromatin complexes were eluted from the beads with 100 μl elution buffer (0.1 M NaHCO3, 1% SDS) by rotating at room temperature for 15 min. To reverse the formaldehyde cross-links, 10 μg RNase and 6 μl of 5 M NaCl were added and samples were incubated at 65°C overnight. Twenty microliters of 1 M Tris-HCl [pH 6.5], 10 μl of 0.5 M EDTA, and 20 μg proteinase K were added to each sample followed by an incubation at 45°C for 2 h. DNA was obtained by phenol-chloroform extraction followed by ethanol precipitation and resuspension in 20 μl TE buffer [pH 8.0]. PCR (30 cycles) was performed using 2-μl ChIP samples as templates and gene-specific primers.
For cloning of novel Smad targets, about 108 cells were used in the ChIP assay except that salmon sperm DNA was omitted in both the preclearing and immunoprecipitation steps. The purified Smad-binding DNA fragments were treated with T4 DNA polymerase to create blunt ends. A DNA linker containing an XhoI site was created by annealing two complementary oligonucleotides: 5′-GATCGGTAAGATCTCGAGGATTCGGTAGGCTGAACG and 5′-phos-CGTTCAGCCTACCGAATCCTCGAGATCTTACC. The linker was added to the blunt-ended DNA fragments at a 100:1 molar ratio followed by an overnight ligation at 15°C using T4 DNA ligase. After digestion with XhoI, DNA fragments were purified using the QIAGEN PCR purification kit and cloned into the pBluescript II KS(−) vector (Stratagene) precut with SalI. Escherichia coli transformants containing inserts longer than 250 bp were sequenced for further analysis.
DNA affinity immunoblotting (DAI) assay.
The Smad-binding regions were released from the corresponding ChST clones by restriction enzyme digestion followed by 3′-end biotinylation using a biotin 3′-end DNA-labeling kit (Pierce) according to the manufacturer's protocol. A 250-bp β-actin cDNA region was biotinylated and used as a control. The labeled probes were purified using the nucleotide removal kit (QIAGEN). MCF10A/HER2 cells were cotransduced with Smad2 (multiplicity of infection [MOI], 3), Smad3 (MOI, 3), and Smad4 (MOI, 15) adenoviruses and lysed in NP-40 lysis buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 1% Nonidet P-40, 0.1 mM EDTA, plus protease and phosphatase inhibitors) 20 h posttransduction. For each DNA-protein binding reaction, 200 μg of protein extract was incubated with 0.5 pmol of biotinylated DNA probes, 2 μg poly(dI-dC), and 2 μg BSA, either with or without 5 pmol of unlabeled competitor (CAGA)12 double-stranded oligonucleotides in a 500-μl reaction mixture diluted in DNA binding buffer (20 mM Tris-Cl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 5% glycerol, 0.1% Triton X-100, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride) for 1 h at 4°C. Streptavidin magnetic beads (0.1 mg; Promega) were added for 1 h immediately after. The beads were washed with DNA binding buffer five times and then boiled in 20 μl of 2× SDS-gel loading buffer before SDS-polyacrylamide gel electrophoresis and immunoblotting.
Extraction of total RNA, RT-PCR, and Northern hybridization.
Total RNA was extracted using the RNeasy mini kit (QIAGEN). RT-PCR was carried out using the Titanium one-step RT-PCR kit (BD Biosciences, San Jose, CA). For each RT-PCR, 100 ng RNA was added to a 50-μl reaction system according to the manufacturer's protocol (50°C for 1 h followed by 30 cycles of PCR amplification) using gene-specific primers. The PCR products were analyzed in 1.2% agarose gels. Primers used for the RT-PCR of human PTPRK gene are 5′-GATGGCTACCAGAGACCAAGTCATTACAT and 5′-TATAATTTGCGGCCGCCTAAGATGATTCCAGGTACTCCAA, which generate a 1.5-kb PCR product. Primers that generate a 250-bp β-actin cDNA PCR product, 5′-AGCCATGTACGTTGCTATCC and 5′-CTTAATGTCACGCACGATTT, were used as internal controls for the RT-PCR. For Northern hybridization, 20-μg RNA samples were used. The PTPRK and β-actin cDNA fragments were labeled using the Prime-It II random primer labeling kit (Stratagene) in a 50-μl reaction mixture containing 25 ng of denatured DNA template, 10 μl random oligonucleotides primers, 1× dCTP primer buffer, 333 nM [α-32P]dCTP (3,000 Ci/mmol), and 5 U of Exo(−) Klenow DNA polymerase at 37°C for 10 min. Probes were purified using the nucleotide removal kit (QIAGEN), denatured, and incubated with the prehybridized membrane in 5 ml ExpressHyb hybridization solution (BD Biosciences). Hybridization was carried out overnight at 50°C.
Immunoprecipitation, Rho/Rac activity assay, and immunoblotting.
Cells were washed twice with ice-cold PBS and lysed in NP-40 lysis buffer. After sonication for 10 s and centrifugation (14,000 rpm), the protein concentration in the supernatants was measured using the bicinchoninic acid protein assay reagent (Pierce). Immunoprecipitation and immunoblotting were performed as described previously (13, 47). Horseradish peroxidase-conjugated secondary antibodies (Promega) were used for all immunoblots. Primary antibodies included: RPTPκ antiserum no. 116 (provided by Axel Ullrich, Max-Planck-Institut für Biochemie, Martinsried, Germany) (14); RhoA, Smad2/3, Smad4, c-Jun, P-Tyr, p38, FAK, Rb, cyclin D1, cyclin E, p27, and c-Src (Santa Cruz Biotechnology); Rac1, P-Smad2, and E-cadherin (BD Biosciences); P-p38, P-Src (Tyr527), and P-Src (Tyr416) (Cell Signaling, Beverly, MA.); EGF receptor (EGFR) and HER2/ErbB2 (NeoMarkers); actin and vinculin (Sigma); and total mitogen-activated protein kinase (MAPK) and phospho-MAPK (P-MAPK; Promega). To pull down GTP-bound (active) Rho and Rac, a glutathione S-transferase (GST) fusion protein of rhotekin-Rho binding domain or GST-Pak binding domain fusion protein precoupled to agarose-glutathione beads (Cytoskeleton, Inc., Denver, CO) were utilized as described previously (6). Eluted RhoA and Rac1 were detected by immunoblotting.
Flow cytometric analysis of cell cycle distribution.
Cells were harvested by trypsinization and labeled with 50 μg/ml propidium iodide (Sigma) containing 125 units/ml protease-free RNase (Calbiochem). A total of 15,000 stained nuclei were analyzed in a FACSCalibur flow cytometer (BD Biosciences) as described previously (47).
Cell adhesion and motility.
Cells were trypsinized and resuspended in DMEM-Ham's medium (1:1) containing 5% horse serum ± 2 ng/ml TGF-β1; 2 × 105 cells/well were added to uncoated six-well dishes and allowed to attach at 37°C. After 1, 2, or 4 h, cells that had not attached were removed and the attached cells were trypsinized and counted. Cell adhesion was calculated as the percentage of attached cells over the total cell input per well. For the wound closure assay, cells in complete medium were allowed to reach confluence on a six-well plate and then incubated in serum-free medium for 30 h. The monolayers were scraped with a plastic pipette tip as described previously (13) and replenished with fresh serum-free medium ± 2 ng/ml TGF-β1. Phase-contrast images were photographed at 0, 12, and 24 h after wounding.
IFA.
Cell indirect immunofluorescence assays (IFA) were performed as described previously (6). Fluorescent images were captured using a Princeton Instruments cooled charge-coupled device digital camera from a Zeiss Axiophot upright microscope. The fluorescent antibodies are Oregon Green anti-mouse IgG, Texas Red anti-rabbit IgG, and Texas Red-phalloidin (Molecular Probes, Eugene, Oregon). For IFA in tumor tissues, primary mammary tumors and metastatic lung tumors were harvested from MMTV-Neu × MMTV-HA-Alk5T204D bigenic and MMTV-Neu transgenic mice. Paraffin-embedded tumor samples were sectioned (3 μm), rehydrated, and stained with Mayer's hematoxylin (Sigma) before double-staining with an RPTPκ antiserum and a hemagglutinin (HA) monoclonal antibody.
Transcription reporter assays.
Cells were seeded in six-well plates and transfected with 1 μg of the Smad reporter construct p(CAGA)12-Luciferase (provided by J.-M. Gauthier, Laboratoire GlaxoSmithKline, Les Ulis Cedex, France) along with 0.02 μg of pCMV-Renilla using FuGENE 6 according to the manufacturer's protocol. Firefly and Renilla reniformis luciferase activities were measured using the dual-luciferase assay system (Promega), and the data were normalized utilizing the ratio of firefly to R. reniformis luciferase as described previously (47).
RESULTS
Identification of ChST.
After ChIP, over 50 different ChST regions were cloned and sequenced; 22 of them were located within the proximal 1-kb promoter or intron regions adjacent to transcription start sites of known genes (Table 1). These potential Smad target genes included kinases, phosphatases, and integrins. We initially focused on 4 genes: PTPRK, STK24, ITGA9, and vimentin-similar gene.
TABLE 1.
ChSTs cloned from MCF10A/HER2 cells
| Genomic locus | Target gene description |
|---|---|
| 6q22.2-23.1 | PTPRK |
| 6q22.33 | Vimentin-similar gene (LOC377430) |
| 13q31.2-32.3 | STK24 (yeast STE20 homolog) |
| 3p21.3 | ITGA9 |
| 19p13.2 | Syntrophin-associated serine/threonine kinase |
| 1p34.1 | Erythroblast membrane-associated protein |
| 11q13.4 | Odd Oz/len-m homolog 4 |
| 3p22.3 | Oxysterol binding protein-like 10 |
| 17p12 | DKFZP566O084 protein |
| 11q13.3 | Suppressor of potassium transport defect 3 |
| 8p23.3 | F-box only protein 25 |
| 14q24 | RAD51-like 1; RecA-like protein |
| 10p12.33-12.32 | DnaJ (Hsp40) homolog, subfamily C, member 1 |
| 2q37 | UDP glycosyltransferase 1 family, polypeptide A cluster |
| 22q13 | Thyroid autoantigen 70kDa (Ku antigen) |
| 4q21-23 | unc-5 netrin receptor homolog C |
| 10q21.3 | Modulator recognition factor 2 |
| 1q12 | Phosphodiesterase 4D interacting protein |
| 16q22 | Hypothetical protein FLJ12688 |
| 8q24.13 | Hypothetical protein FLJ32440 |
| 5q33.1 | Hypothetical protein FLJ36748 |
| 8p21.2 | Hypothetical protein MGC22776, 99% similar to insulin-like growth factor-binding protein 6 precursor |
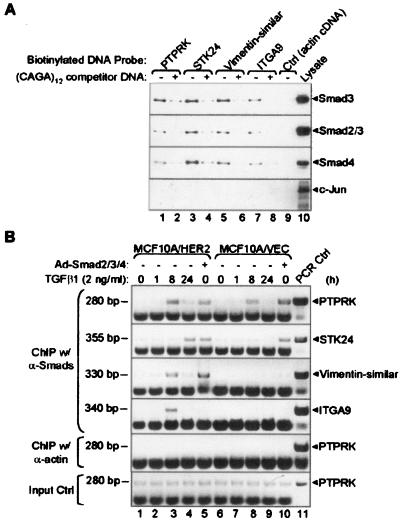
We confirmed the interaction between Smads and the target DNA fragments with DNA affinity immunoblotting. Cell lysates from MCF10A/HER2 cells that had been transduced with the Smad2/3/4 adenoviruses were incubated with different biotinylated ChST regions. After pulling down biotin-DNA-protein complexes, Smad proteins were found in all 4 ChST regions but not in a biotinylated actin cDNA control (Fig. 1A, lanes 1, 3, 5, 7, 9). Immunoblotting with a (control) c-Jun antibody did not reveal c-Jun bound to any of the ChST regions. In DNA competition experiments, unlabeled (CAGA)12 double-stranded oligonucleotides, which specifically bind Smads, were added. The interactions between Smads and the ChST regions were blocked by the addition of (CAGA)12, as fewer Smads were detected bound to the labeled DNA probes (Fig. 1A, lanes 2, 4, 6, 8). Thus, the DAI assay confirmed the accuracy of the Smad target DNA pool we had established by ChIP and suggested that the target genes controlled by these ChST regions are transcriptionally regulated by Smads.
FIG. 1.
TGF-β induces binding of Smads to ChST DNAs. (A) DAI assay. Biotin-labeled ChST regions found on the indicated target genes were incubated with lysates of MCF10A/HER2 cells expressing Smad2/3/4 adenoviruses in the presence (+) (lanes 2, 4, 6, 8) or absence (−) (lanes 1, 3, 5, 7) of a 10-fold excess of unlabeled (CAGA)12 competitor DNA. A biotinylated 250-bp β-actin cDNA was used as a control (lane 9). A whole-cell lysate was loaded as a positive control (lane 10). The presence of Smads on ChST DNAs was detected using antibodies against Smad2/3 and Smad4; a c-Jun antibody was used as a negative control. (B) Time course ChIP assay. MCF10A/HER2 or MCF10A/VEC cells were treated with TGF-β1 (2 ng/ml) for 0, 1, 8, or 24 h or infected by adenoviruses encoding Smad2/3/4 for 24 h prior to the ChIP assay. After precipitation with a combination of Smad2/3/4 antibodies, primers encompassing ChST regions in the indicated genes (right) were used for PCR amplification. An actin antibody was used as a negative control. To control for equal chromatin input, cell lysates before immunoprecipitation were used as templates for PCR (bottom panel). Plasmids containing corresponding ChST regions were used as templates to control for size of the PCR products (lane 11). The sizes of the PCR products are indicated on the left of each panel. α-Smads, anti-Smads; α-actin, anti-actin.
We next determined the in vivo association of Smads with ChST DNAs in TGF-β-treated cells by using ChIP with Smad antibodies followed by gene-specific PCR. In MCF10A/HER2 cells, TGF-β induced binding of Smads to PTPRK, STK24, vimentin-similar, and ITGA9 ChST regions at as early at 8 to 24 h (Fig. 1B). In MCF10A/VEC cells, a similar result was obtained with PTPRK but not with the other 3 genes, thus suggesting that cofactors regulated by HER2 overexpression are required for Smad binding to STK24, vimentin-similar, and ITGA9 gene promoters in response to TGF-β. Overexpressed (adenovirus-encoded) Smads were able to interact with PTPRK, STK24, and vimentin-similar ChST regions in MCF10A/HER2 cells, whereas in vector control cells, this interaction was only detected for PTPRK and STK24 (Fig. 1B, lanes 5 and 10). In neither cell line were the ectopic Smads able to interact with the ITGA9 ChST region, implying the possibility that a non-Smad TGF-β-induced pathway might be required for the production/activation of cofactor(s) necessary for Smad binding to the ITGA9 promoter in MCF10A/HER2 cells (Fig. 1B, lane 3, ITGA9 panel)
TGF0β up-regulates PTPRK expression.
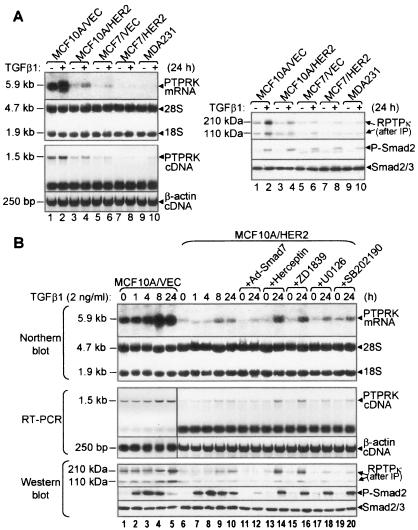
PTPRK expression was determined by Northern blotting, RT-PCR, and immunoblotting. Treatment with TGF-β (24 h) induced both PTPRK mRNA and RPTPκ protein in nontumor MCF10A/VEC and MCF10A/HER2 cells as well MCF7/VEC, MCF7/HER2 (stably transfected with HER2), and MDA231 human breast cancer cells (Fig. 2A). Interestingly, both basal and TGF-β-induced RPTPκ levels were significantly lower in cancer cell lines compared to MCF10A/VEC cells. In MCF10A/HER2 and vector control cells, TGF-β maximally induced PTPRK mRNA at 8 h and RPTPκ protein at 24 h (Fig. 2B, lanes 1 to 10). Transduction with a Smad7 adenovirus suppressed Smad2 phosphorylation and ligand-induced PTPRK expression (Fig. 2B, lanes 11 to 12). In MCF10A/HER2 cells, preincubation with the HER2 blocking antibody Herceptin (54) or with the EGF receptor tyrosine kinase inhibitor ZD1839 (gefitinib, “Iressa”) enhanced ligand-induced PTPRK expression (Fig. 2B, lanes 13 to 16). Consistent with previous reports (29), ZD1839 blocked HER2 phosphorylation in the HER2-overexpressing cells (data not shown). Finally, the MEK1/2 inhibitor U1026 and the p38 inhibitor SB202190 did not affect ligand-induced RPTPκ levels, suggesting that transducers other than Erk and p38 downstream of HER2 repress PTPRK expression (Fig. 2B, lanes 17 to 20).
FIG. 2.
Expression of PTPRK is transcriptionally up-regulated by TGF-β. (A) Subconfluent monolayers of the indicated cell lines were treated with TGF-β1 for 24 h and then subjected to Northern hybridization and RT-PCR (left panel) and immunoblot analysis (right panel). A 5.9-kb PTPRK mRNA and a 1.5-kb PTPRK cDNA band were detected by Northern blotting and RT-PCR, respectively. The 28S and 18S rRNA bands were used as loading controls for the Northern blot. A 250-bp β-actin cDNA region was amplified as a control for RT-PCR. To detect RPTPκ proteins, 500 μg of total protein from the indicated cell lysates was precipitated with an RPTPκ antiserum. Immune complexes were next subjected to an RPTPκ immunoblot using the same antiserum (right). P-Smad2 and Smad2/3 immunoblots of cell lysates (30 μg of protein/lane) were used to confirm response to TGF-β in the same experiment. (B) Time course of TGF-β-induced PTPRK expression and effect of HER2 overexpression. Subconfluent MCF10A/VEC or MCF10A/HER2 cells were serum starved for 24 h before being treated with TGF-β1 for the indicated time (lanes 1 to 10). Lanes 11 and 12, cells were infected with adenovirus encoding Smad7 (MOI, 15) 24 h before adding TGF-β1; lanes 13 and 14, Herceptin (10 μg/ml) was added 8 h before addition of TGF-β1; lanes 15 to 20, ZD1839 (1 μM), U0126 (10 μM), or SB202190 (10 μM) was added 1 h before TGF-β1. After treatment, samples were prepared for Northern blotting, RT-PCR, and immunoblot for PTPRK gene expression as described for panel A.
To further document that TGF-β signaling up-regulates PTPRK expression in vivo, we examined endogenous phosphatase expression in mammary tumors from MMTV-Neu × MMTV-HA-Alk5T204D bigenic mice. These mice overexpress Neu, the rat homolog of HER2, and a constitutively active mutant of TβRI (Alk5) (53), both under the control of the MMTV promoter (R. S. Muraoka-Cook and C. L. Arteaga, unpublished data). Immunofluorescent staining of MMTV-Neu × MMTV-HA-Alk5T204D mammary tumor sections depicted high RPTPκ protein levels in both nontumor epithelial cells and a subset of cancer cells (see Fig. S1a in the supplemental material). When double stained using an HA antibody, tumor cells that expressed higher levels of HA-Alk5T204D consistently overexpressed RPTPκ (see Fig. S1a to e in the supplemental material). Similarly, in lung metastases from bigenic mice, RPTPκ was highly expressed in normal epithelial cells and a subset of tumor cells that expressed a higher Alk5T204D level (see Fig. S1f to h in the supplemental material) than (HA-negative) primary and metastatic tumors from MMTV-Neu mice (see Fig. S1k to r in the supplemental material). These data suggest that PTPRK expression is up-regulated by TGF-β signaling in vivo.
RPTPκ modulates cell proliferation and ErbB receptor phosphorylation.
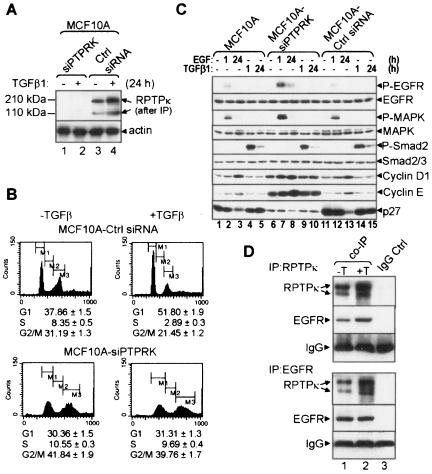
To determine the function of RPTPκ, we constructed MCF10A cells that stably express siRNA against human PTPRK (designated “siPTPRK”). At passage 2 after selection in puromycin, expression of siPTPRK inhibited basal and TGF-β-induced RPTPκ protein levels by >90% (Fig. 3A). MCF10A-siPTPRK cells exhibited a lower G1 and higher S and G2/M fractions than MCF10A cells expressing control siRNA. Further, TGF-β induced a cell cycle delay in MCF10A-control siRNA cells but not in MCF10A-siPTPRK cells (Fig. 3B). Compared to parental and control siRNA cells, MCF10A-siPTPRK cells showed higher EGF-induced P-EGFR and P-MAPK and contained increased levels of basal and EGF-induced cyclin D1 and cyclin E and decreased levels of the Cdk inhibitor p27Kip1 (Fig. 3C). TGF-β-induced Smad2 phosphorylation was similar in all 3 lines, suggesting that RPTPκ knockdown did not affect serine/threonine phosphorylation. At passage 3 after selection in puromycin, the majority of MCF10A-siPTPRK cells showed altered morphology, multiple (3 to 4) nuclei, and increased cell size (see Fig. S2A in the supplemental material). This phenotype was temporally associated with increased apoptosis as indicated by a higher subgenomic DNA fraction and a striking increase in the percentage of cells unable to exclude trypan blue (see Fig. S2B in the supplemental material). TGF-β-induced Smad transcriptional activity was not affected by the expression of siPTPRK, as determined with a transiently transfected p(CAGA)12-Luciferase reporter in knockdown cells and controls (see Fig. S3 in the supplemental material). These results suggested that RPTPκ inhibits cell proliferation by associating with and modulating EGFR signaling. To examine an RPTPκ/EGFR association, we precipitated lysates of MCF10A cells treated or not with TGF-β with RPTPκ or EGFR antibodies. EGFR and RPTPκ coprecipitated with RPTPκ or EGFR antibodies, respectively, suggesting a protein-protein association which was enhanced by treatment with TGF-β (Fig. 3D).
FIG. 3.
RPTPκ associates with EGFR and modulates EGFR signaling in MCF10A cells. (A) Passage 2 subconfluent MCF10A cells stably expressing siRNA against PTPRK or control siRNA were treated with TGF-β1 for 24 h (+) or not (−), lysed, and tested in an RPTPκ immunoblot procedure after immunoprecipitation as indicated in Fig. 2A. An immunoblot for actin was used as a control. (B) Same-passage cells were grown in medium containing 5% horse serum with or without TGF-β1 (2 ng/ml). After 24 h, nuclei were labeled with propidium iodide and cell cycle distribution was analyzed by flow cytometry as indicated in Materials and Methods. The percentages of gated cells in the G1, S, and G2/M phases of the cell cycle are shown. (C) The indicated cells were serum-starved for 8 h before treatment with EGF (10 ng/ml) or TGF-β1 (2 ng/ml) for 1 and 24 h, respectively. Cell lysates were prepared and subjected to immunoblot analysis with the antibodies indicated at the right of the panels. (D) Five hundred micrograms of MCF10A cell lysates was precipitated using RPTPκ or EGFR antibodies (lane 1). Normal IgG was used as a control (lane 2). The precipitates were subjected to immunoblot analyses using RPTPκ and EGFR antibodies.
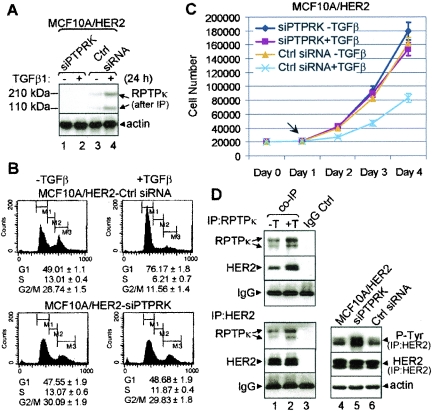
Stable expression of the same siPTPRK clone in MCF10A/HER2 cells abolished basal and TGF-β-induced RPTPκ levels (Fig. 4A). MCF10A/HER2-siPTPRK cells showed a cell cycle distribution identical to that of control siRNA cells. However, similar to MCF10A cells, siPTPRK expression in HER2-overexpressing cells abrogated the antimitogenic effect of TGF-β, as determined by flow cytometry and monolayer growth (Fig. 4B and C). The effects of siPTPRK on MCF10A/HER2 cells suggested an interaction between the phosphatase and HER2. To test this, we precipitated MCF10A/HER2 cell lysates with RPTPκ or HER2 antibodies. Both the endogenous RPTPκ and HER2 were present in the HER2 and RPTPκ precipitates, respectively, supporting a receptor-phosphatase association. This association was increased by treatment with TGF-β (Fig. 4D, lanes 1 to 3). Further, MCF10A/HER2-siPTPRK cells contained higher levels of tyrosine-phosphorylated HER2 than controls (Fig. 4D, lanes 4 to 6). TGF-β-induced transcriptional activity measured with a p(CAGA)12-Lucifrease reporter showed that MCF10A/HER2-siPTPRK cells respond to TGF-β as well as control cells (see Fig. S3 in the supplemental material).
FIG. 4.
RPTPκ modulates HER2 phosphorylation and cell cycle progression. (A) Subconfluent passage 2 MCF10A/HER2 cells stably expressing siPTPRK or control siRNA were treated (+) or not (−) with TGF-β1 for 24 h, lysed, and tested in a RPTPκ immunoblot procedure after precipitation, as indicated in the legend to Fig. 2A. An immunoblot for actin was used as a control. (B) Cell cycle distribution of passage 2 MCF10A/HER2-siPTPRK and control cells with (+) or without (−) TGF-β1 (24 h) measured by flow cytometry as described in the legend to Fig. 3B. (C) MCF10A/HER2-siPTPRK or control siRNA cells were seeded on 12-well plates in complete growth medium at a density of 2 × 104 cells/well. TGF-β1 (2 ng/ml) was added the next day (arrow); cells were trypsinized and counted (Coulter) every 24 h. Each data point represents the mean ± standard deviation of the results from 4 wells. (D) Lanes 1 and 2, 500 μg of MCF10A/HER2 cell lysates was precipitated with RPTPκ antiserum, HER2 antibody, or normal IgG (control). RPTPκ and HER2 were detected in the antibody pull downs by immunoblot analysis. Lanes 3 to 5, cell lysates were precipitated with an HER2 antibody followed by P-Tyr or HER2 immunoblots. An immunoblot for actin was used as a control. IP, immunoprecipitation.
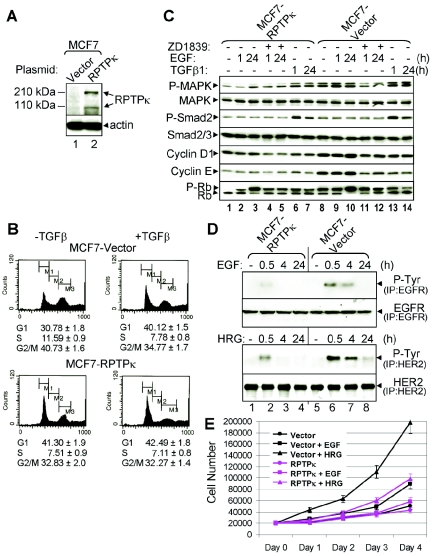
To complement these results, we stably overexpressed a full-length human PTPRK cDNA in MCF7 cells, which exhibit low endogenous PTPRK levels compared to the high-expressing MCF10A cells (Fig. 2A). In transfected cells, immunoblotting with an RPTPκ antiserum recognized the ectopic protein as 210-kDa (precleavage) and 110-kDa (postcleavage) forms (Fig. 5A) (14, 39). Consistent with the cell cycle permissive effect of siPTPRK (Fig. 3 and 4), overexpression of RPTPκ resulted in the opposite: inhibition of cell cycle progression as indicated by higher G1 and lower S phase fractions than controls, thus mimicking the effects of TGF-β (Fig. 5B). In line with this result, MCF7-RPTPκ cells expressed lower basal and EGF-induced levels of P-MAPK, cyclin D1, cyclin E, and P-Rb than vector control cells (Fig. 5C). In RPTPκ-overexpressing cells, TGF-β still induced MAPK phosphorylation, suggesting that the tyrosine phosphatase did not affect TGF receptor serine-threonine kinases (Fig. 5C, lanes 1, 7, 8). In both cell types, TGF-β-induced phosphorylation of Smad2 was similar. We next examined the effect of RPTPκ overexpression on ligand-induced ErbB phosphorylation. EGF and heregulin induced phosphorylation of EGFR and HER2, respectively, but in MCF7-RPTPκ, the duration and magnitude of ligand-induced EGFR and HER2 phosphorylation were reduced compared to that of MCF7 vector cells (Fig. 5D). Consistent with these data, EGF- and heregulin-stimulated growth in monolayers was reduced in RPTPκ-transfected cells compared to controls (Fig. 5E). These results further suggest that RPTPκ associates with ErbB tyrosine kinases and down-regulates ErbB receptor signaling output.
FIG. 5.
RPTPκ modulates ErbB receptor phosphorylation in MCF-7 cells. (A) MCF7 cells stably expressing a PTPRK cDNA or control vector were lysed and subjected to immunoblot procedures with RPTPκ and actin antibodies. (B) DNA histograms of propidium iodide-labeled nuclei from MCF7-vector and MCF7-RPTPκ cells treated (+) or not (−) with TGF-β1 (24 h). (C) MCF7-RPTPκ and vector control cells were serum starved for 8 h before treatment with EGF (10 ng/ml) or TGF-β1 (2 ng/ml). ZD1839 (1 μM) was added 1 h prior to EGF where indicated. Total proteins from cell lysates were tested in immunoblot procedures utilizing the antibodies indicated at the left of each panel. (D) MCF7-RPTPκ and vector control cells were serum starved for 8 h before treatment with EGF (10 ng/ml) or heregulin (HRG; 10 nM) for the indicated time. Cell lysates were precipitated with EGFR or HER2 antibodies followed by P-Tyr, EGFR, or HER2 immunoblot analysis as indicated in Materials and Methods. IP, immunoprecipitation. (E) MCF7-RPTPκ and vector control cells were seeded on 12-well plates in low-serum DMEM (1% FBS) containing EGF (10 ng/ml) or heregulin (HRG; 10 nM) at a density of 2 × 104 cells/well. Cells were trypsinized and counted every 24 h. Each data point represents the mean ± standard deviation of the results from 4 wells.
To determine the antiproliferative function of RPTPκ in nonmammary cells, we stably transfected EGFR-overexpressing A431 human squamous cancer cells with the RPTPκ expression vector (see Fig. S4A in the supplemental material). Overexpression of RPTPκ in A431 cells also inhibited their proliferation (see Fig. S4B and C in the supplemental material). Tyrosine-phosphorylated EGFR was minimally detectable in A431-RPTPκ cells, whereas it remained high in parental and vector control cells (see Fig. S4D in the supplemental material). An important mechanism to regulate the function of protein tyrosine phosphatases (PTPs) is alteration of their substrate specificity and signaling output via restriction of their subcellular localization (26). Therefore, to study the localization of RPTPκ, we performed double-labeled IFA in parental A431 cells. EGFR was observed predominantly at cell membranes in both intercellular junctions (59) and leading edges. Endogenous RPTPκ was also detected on cell membranes and colocalized with EGFR, especially at the leading edges of cells (see Fig. S4E in the supplemental material).
RPTPκ is required for TGF-β-enhanced cell adhesion and migration.
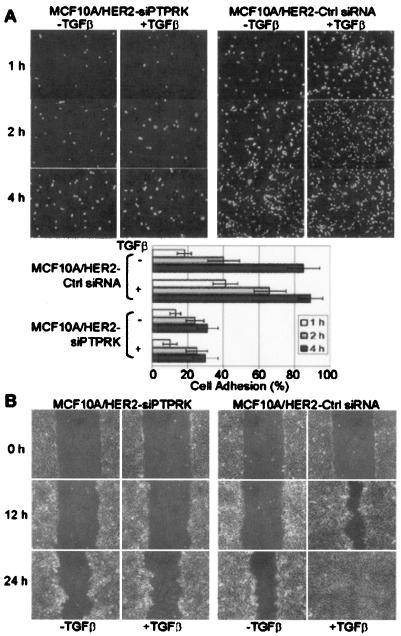
TGF-β enhances motility of HER2-overexpressing MCF10A cells (41, 47) where PTPRK was identified as a Smad target (Fig. 1). Thus, we next examined whether RPTPκ was required for this response to TGF-β. MCF10A/HER2-siPTPRK cells showed delayed adhesion compared to cells expressing control siRNA. TGF-β accelerated cell adhesion in control cells but not in cells lacking RPTPκ (Fig. 6A). In addition, TGF-β accelerated wound closure of MCF10A/HER2 cell monolayers but not MCF10A/HER2-siPTPRK cell monolayers (Fig. 6B), implying that RPTPκ is required for TGF-β-induced cell adhesion and migration.
FIG. 6.
RPTPκ is required for TGF-β-induced cell adhesion and migration. (A) Adherent MCF10A/HER2-siPTPRK or control cells were incubated in DMEM-Ham's medium (1:1) containing 5% horse serum with (+) or without (−) TGF-β1 (2 ng/ml) for 8 h. After trypsinization and resuspension in the same medium with or without TGF-β1, cells were seeded on uncoated six-well tissue culture plates and allowed to attach at 37°C. After 1, 2, or 4 h, the monolayers were washed and the cells that had attached were trypsinized and counted. Each bar represents the mean ± standard deviation of results from six wells from three independent experiments. (B) Close-to-confluent monolayers of MCF10A/HER2-siPTPRK and control cells were serum starved for 30 h before wounding. Serum-free medium with or without TGF-β1 (2 ng/ml) was replenished, and wound closure was monitored until 24 h as indicated in Materials and Methods.
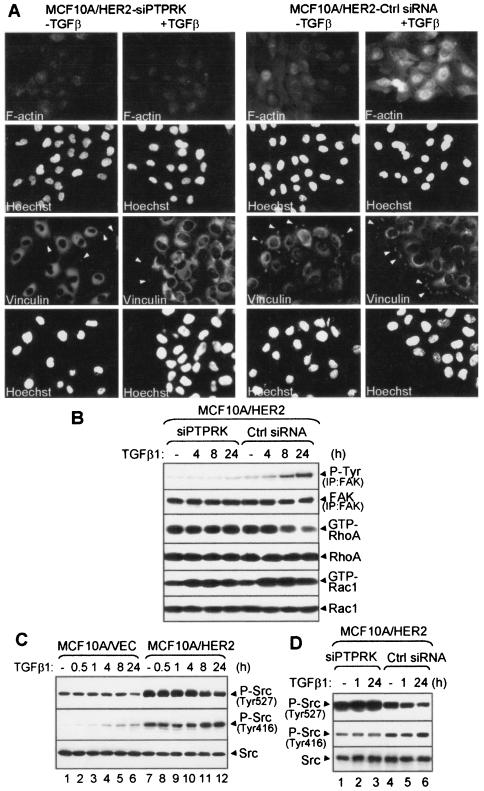
The effects on motility and adhesion suggested a role for RPTPκ on cytoskeletal organization and focal adhesions. IFA staining of actin filaments indicated that TGF-β induced F-actin assembly and accumulation of vinculin at focal adhesions and leading edges in MCF10A/HER2 control cells but not in RPTPκ knockdown cells (Fig. 7A). Formation of focal adhesions in response to TGF-β was impaired in MCF10A-siPTPRK cells, but vinculin content in both cell types, as measured by immunoblotting, was not altered (data not shown). In a double-labeled IFA using RPTPκ and vinculin antibodies, RPTPκ colocalized with vinculin-containing focal adhesions at the leading edges of TGF-β-treated MCF10A/HER2 cells (see Fig. S5 in the supplemental material), suggesting a possible requirement of RPTPκ in focal adhesions and F-actin assembly and/or adhesion-induced cell signaling.
FIG. 7.
RPTPκ mediates rearrangement of cytoskeletal elements and modulates Src and FAK phosphorylation. (A) Filamentous actin stress fiber assembly and focal adhesion formation in MCF10A/HER2-siPTPRK and control cells. Prior to IFA, cells were grown in medium with (+) or without (−) TGF-β1 (2 ng/ml) for 24 h. IFA shows F-actin staining and vinculin-containing focal adhesions. Hoechst, nuclear staining. Arrows indicate focal adhesion formation at cell leading edges. (B) MCF10A/HER2-siPTPRK and control cells were serum starved for 8 h before treatment with TGF-β1 for the indicated times. Cell lysates were precipitated with a FAK antibody followed by P-Tyr or FAK immunoblot analysis. The GTP-bound form of RhoA or Rac1 were pulled down using GST-rhotekin or GST-Pak binding domain beads, respectively. Eluates from the beads as well as total cell lysates (30 μg/lane) were immunoblotted with RhoA or Rac1 antibodies. (C) MCF10A/VEC and MCF10A/HER2 cellswere serum starved for 8 h before treatment with TGF-β for the indicated times. Cell lysates were subjected to immunoblot analysis using P-Src (Tyr527), P-Src (Tyr416), and total Src antibodies. At 24 h, there was a 40% reduction in P-Src (Tyr527) in TGF-β-treated MCF10A/HER2 cells compared to untreated controls. (D) MCF10A/HER2-siPTPRK and control cells were serum starved for 8 h before treatment with TGF-β1 for the indicated times. Cell lysates were subjected to immunoblot analysis as described for panel C.
This role is similar to the functions of another receptor type PTP, PTPα. PTPα−/− cells show defects in migration and delayed F-actin stress fiber assembly as well as focal adhesion formation (60), possibly due to a loss of PTPα-mediated, integrin-stimulated phosphorylation of focal adhesion kinase (FAK). FAK has been reported to suppress Rho activity to promote focal adhesion turnover (36). FAK can be activated by cell adhesion, G-proteins, and growth factors such as TGF-β (45, 50, 61). In MCF10A/HER2 cells, TGF-β induced FAK phosphorylation and suppressed RhoA activity and these effects were abolished in RPTPκ knockdown cells (Fig. 7B). TGF-β treatment did not alter P-FAK or RhoA in MCF10A cells (data not shown). Basal levels of P-FAK were also lower in RPTPκ knockout cells than in control cells. Basal and TGF-β-stimulated Rac1 activity, as measured by the binding of GTP-bound Rac1 to a GST-Pak binding domain fusion protein, was similar in both cell types. This result suggests that RPTPκ is required for TGF-β-induced FAK activation and RhoA suppression.
A major regulatory pathway of FAK is the Src tyrosine kinase. The fact that HER2 is a potent activator of Src (58) suggested that the induction of P-FAK by TGF-β in MCF10A-HER2 cells but not in controls (Fig. 7B) might be due to a higher level of active Src, which provides enough of a threshold for TGF-β-induced P-FAK. Src is inactive when phosphorylated at Tyr-527; dephosphorylation of this residue results in a conformational change that facilitates Src autophosphorylation at Tyr-416 and activation of its catalytic activity (35). By immunoblot analyses using phosphospecific antibodies, (active) P-Src (Tyr416) was high in MCF10A/HER2 cells but undetectable in MCF10A vector cells. Conversely, levels of P-Src (Tyr527) were lower in vector cells. Treatment with TGF-β for 24 h modestly increased P-Src (Tyr416) and reduced P-Src (Tyr527) in both cell lines (Fig. 7C). Interestingly, MCF10A/HER2-siPTPRK cells exhibited increased P-Src (Tyr527) and decreased P-Src (Tyr416) compared to control cells (Fig. 7D), implying that RPTPκ may regulate Src. Indeed, treatment with TGF-β for 24 h modestly reduced P-Src (Tyr527) in control but not in cells lacking RPTPκ (Fig. 7D). Since this response correlates temporally with TGF-β-induced PTPRK mRNA and protein expression (Fig. 2B), this result suggests a causal link between the receptor phosphatase with ligand-induced reduction in P-Src (Tyr527) and activation of Src and FAK. Consistent with this result, MCF7-RPTPκ cells (shown in Fig. 5) contained decreased Src phosphorylation at Tyr527 and increased the phosphorylation at Tyr416 (see Fig. S6 in the supplemental material).
DISCUSSION
We have examined novel mechanisms by which TGF-β cooperates with HER2 in the progression of mammary tumor cells. We established a Smad-bound DNA pool from a ChIP assay in MCF10A/HER2 cells and identified several novel Smad target genes including PTPRK, STK24, ITGA9, and vimentin-similar gene. In the ChIP assay, a stronger affinity between Smads and certain DNA regions results in a higher abundance of this DNA in the precipitated pool. To obtain enough DNA fragments for cloning and sequencing, we PCR amplified the Smad target DNA pool originally obtained by ChIP. Since PCR amplifies template DNA logarithmically, the differences in abundance between high-affinity and low-affinity DNA regions were further amplified by PCR. As a result, the amplified Smad target DNA pool should contain more copies for high-affinity than low-affinity DNA regions and, therefore, may represent physiologically important TGF-β-regulated genes. This may also explain why other more known TGF-β-induced Smad-target DNA regions were not found in the DNA pool. In addition, genetic and epigenetic factors, such as HER2 overexpression, can alter TGF-β-mediated gene expression. Thus, the Smad target DNA pool may represent TGF-β-Smad target genes in mammary epithelial cells that overexpress HER2 but not in cells with low HER2 levels. Indeed, TGF-β only induced Smads to bind to and regulate STK24, ITGA9, and vimentin-similar gene expression in HER2-overexpressing cells (Fig. 1B), suggesting that cofactors regulated by HER2 signaling modulate Smad-mediated transcription.
We focused on the PTPRK gene, whose expression was up-regulated by TGF-β in both tumor and nontumor mammary cells. PTPRK expression has been found in the spleen, prostate, ovary, kidney, and liver and in keratinocytes and epidermal cell lines (20, 56) and is up-regulated by TGF-β in keratinocytes (55). PTPRK mRNA and protein levels were lower in breast cancer cells than in MCF10A cells. Overexpression of HER2 markedly decreased RPTPκ, and blockade of HER2 function with Herceptin or gefitinib partially restored RPTPκ expression (Fig. 2B). Furthermore, down-regulation of RPTPκ by RNAi accelerated cell cycle progression, enhanced the response to EGF, and abrogated TGF-β-mediated antimitogenic action. Finally, overexpression of RPTPκ inhibited basal and ligand (EGF and heregulin)-induced proliferation and ErbB receptor signaling in cancer cells. These results may explain the reported ability of TGF-β to reversibly inhibit the proliferative responses to EGF receptor ligands (9, 38). They also suggest, as for TGF-β, a tumor suppressor role for RPTPκ. Consistent with this notion, studies have shown that deletions in chromosome 6q22-23, the genomic locus for the PTPRK gene, are one of the most common alterations in high-grade non-Hodgkin's lymphomas (33, 62). However, knockdown of RPTPκ in MCF10A cells was not sufficient to maintain dysregulated proliferation but, instead, resulted in multiple nuclei, abnormal cell morphology, and apoptosis (Fig. 3; see Fig. S2 in the supplemental material). This result implies that besides the accelerated cell proliferation caused by the suppression of RPTPκ, other oncogenic signals are also required to synchronize nuclear division and cytokinesis and therefore sustain prolonged dysregulated cell division.
PTPs dynamically modulate the initiation, duration, and termination of signal transduction mediated by tyrosine kinases, thus playing an important role in cell behavior (27), integrin signaling (40), cell-cell (1), and cell-ECM assembly (4). Many PTPs recognize RTKs or RTK adaptor proteins (i.e., PTP1B binds the platelet-derived growth factor receptor, PTP SHP1 binds c-Kit, and PTPα binds Grb2) and negatively regulate RTK-induced cellular transformation (21, 22, 42, 44). Other receptor type transmembrane PTPs, such as PTPα and PTPɛ, can support the transformed phenotype by activating Src/Yes/Fyn kinases (7, 18). In this study, RPTPκ induced growth inhibition at least in part through suppression of ErbB receptor phosphorylation. PTPα expression is also associated with breast cancer cell growth inhibition (5). In addition, PTPα has been reported to mediate integrin-stimulated FAK phosphorylation; PTPα−/− cells show defects in migration and delayed F-actin stress fiber assembly as well as focal adhesion formation (60). Our data indicate that RNAi of RPTPκ increases the inactivating phosphorylation of Src at Tyr527 and disrupts focal adhesion formation, F-actin assembly, TGF-β-induced FAK phosphorylation, and cell migration, further suggesting that RPTPκ and PTPα share common functions.
PTPα and RPTPκ also exhibit different functional characteristics. RPTPκ, as well as PTPs μ, ρ, and λ, contain an extracellular domain consisting of MAM (meprin/A5-protein/PTPμ), IgG, and fibronectin type III motifs. This complex extracellular domain is likely to have adhesive functions in cell-cell interactions (2). Indeed, RPTPκ has been reported to mediate homophilic intercellular interactions (39) and colocalize with β- and γ-catenin at adherens junctions (14). PTPα and ɛ do not have the extracellular domain found in RPTPκ but are heavily glycosylated (3). Overexpression of PTPα, but not RPTPκ, inhibits insulin-stimulated glucose transport (10). RPTPκ associates with armadillo family members (14), but the same association has not been found with PTPα. The mechanisms regulating expression of both PTPs also diverge. HER2 overexpression induces PTPα expression in human ovarian cancer cells (52) but suppresses RPTPκ expression in breast cancer cells (Fig. 2). RPTPκ expression is also induced by high cell density (14), suggesting an involvement in contact inhibition of cell growth. Although RPTPκ interacts with and dephosphorylates β-catenin in vitro (14), the cellular substrates of the phosphatase's catalytic activity remain to be identified. RPTPκ is proteolytically cleaved and the cleavage products remain associated (20); the biological significance of this proteolytic processing is unclear.
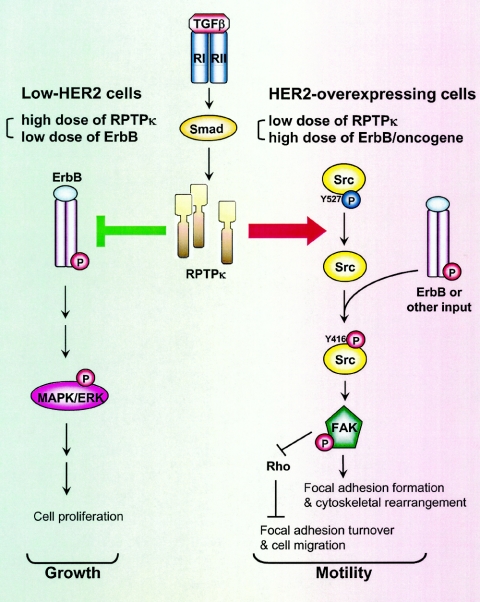
The results presented herein support dual functions of RPTPκ in cell transformation (Fig. 8). RPTPκ expression is up-regulated by TGFβ and down-regulated by HER2 signaling. RPTPκ can associate with ErbB receptors and suppress signal transduction, thus delaying cell proliferation. On the other hand, RPTPκ is involved in F-actin stress fiber assembly, focal adhesion formation, Src activation, FAK phosphorylation, and downregulation of RhoA. We speculate that by inhibiting RhoA to induce turnover of focal adhesions (36) while leaving Rac1 function unchanged (Fig. 7B), RPTPκ contributes to TGF-β-induced cell motility. In nontumor mammary epithelial cells, basal and TGF-β-induced levels of RPTPκ are high and the antiproliferative effects of the phosphatase are dominant. In HER2-overexpressing cells, however, TGF-β and HER2 have antagonistic effects on RPTPκ expression but TGF-β can still induce RPTPκ levels to a threshold that allows ligand-induced enhancement of cell migration and adhesion. In addition, the high levels of HER2-activated Src provide an adequate threshold of Src activity for TGF-β-enhanced adhesion and migration. The loss of these TGF-β responses in siPTPRK cells (Fig. 6 and 7) implies that RPTPκ is required for these processes. This may be a function of the subcellular localization of RPTPκ and the association of RPTPκ with HER2 and other proteins in TGF-β-treated cells. In summary, the data presented suggest that RPTPκ phenocopies the dual role of TGF-β in epithelial cells: high expression is associated with tumor suppression, whereas limited expression is associated with cell behaviors reflective of increased transformation.
FIG. 8.
Diagram of RPTPκ-mediated functions. RPTPκ protein levels are higher in nontransformed cells than in tumor cells. The expression of RPTPκ is up-regulated by TGF-β and down-regulated by HER2. RPTPκ associates with ErbB receptors and reduces their phosphorylation and signaling output, thus inhibiting both basal and ErbB ligand-induced cell proliferation. RPTPκ is also involved in the activation of the Src tyrosine kinase, perhaps by inhibiting the inactivating phosphorylation at Src-Tyr527. In HER2-overexpressing tumor cells but not in nontumor cells, Src is activated, providing an adequate threshold of activity that permits or facilitates TGF-β-induced phosphorylation/activation of FAK, focal adhesion formation, F-actin stress fiber assembly, and suppression of RhoA, leading to enhanced cell adhesion and migration. RNA interference of PTPRK abrogates these TGF-β-mediated processes.
Supplementary Material
Acknowledgments
This work was supported in part by grant R01 CA62212 (to C.L.A.), Breast Cancer Specialized Program of Research Excellence (SPORE) grant P50 CA98131, and Vanderbilt-Ingram Comprehensive Cancer Center Support grant CA68485.
We thank Axel Ullrich for providing the RPTPκ antiserum; Joe Zhao, Jae Youn Yi, and Yasuhiro Koh for helpful comments; and Teresa Dugger for technical and administrative support.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aberle, H., H. Schwartz, and R. Kemler. 1996. Cadherin-catenin complex: protein interactions and their implications for cadherin function. J. Cell. Biochem. 61:514-523. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, A., J. Sasin, N. Bottini, I. Friedberg, A. Osterman, A. Godzik, T. Hunter, J. Dixon, and T. Mustelin. 2004. Protein tyrosine phosphatases in the human genome. Cell 117:699-711. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, J. N., O. H. Mortensen, G. H. Peters, P. G. Drake, L. F. Iversen, O. H. Olsen, P. G. Jansen, H. S. Andersen, N. K. Tonks, and N. P. Moller. 2001. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 21:7117-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angers-Loustau, A., J. F. Cote, and M. L. Tremblay. 1999. Roles of protein tyrosine phosphatases in cell migration and adhesion. Biochem. Cell Biol. 77:493-505. [PubMed] [Google Scholar]
- 5.Ardini, E., R. Agresti, E. Tagliabue, M. Greco, P. Aiello, L. T. Yang, S. Menard, and J. Sap. 2000. Expression of protein tyrosine phosphatase alpha (RPTPalpha) in human breast cancer correlates with low tumor grade, and inhibits tumor cell growth in vitro and in vivo. Oncogene 19:4979-4987. [DOI] [PubMed] [Google Scholar]
- 6.Bakin, A. V., C. Rinehart, A. K. Tomlinson, and C. L. Arteaga. 2002. p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J. Cell Sci. 115:3193-3206. [DOI] [PubMed] [Google Scholar]
- 7.Brandt, D. T., A. Goerke, M. Heuer, M. Gimona, M. Leitges, E. Kremmer, R. Lammers, H. Haller, and H. Mischak. 2003. Protein kinase C delta induces Src kinase activity via activation of the protein tyrosine phosphatase PTP alpha. J. Biol. Chem. 278:34073-34078. [DOI] [PubMed] [Google Scholar]
- 8.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 9.Coffey, R. J., Jr., N. J. Sipes, C. C. Bascom, R. Graves-Deal, C. Y. Pennington, B. E. Weissman, and H. L. Moses. 1988. Growth modulation of mouse keratinocytes by transforming growth factors. Cancer Res. 48:1596-1602. [PubMed] [Google Scholar]
- 10.Cong, L. N., H. Chen, Y. Li, C. H. Lin, J. Sap, and M. J. Quon. 1999. Overexpression of protein tyrosine phosphatase-alpha (PTP-alpha) but not PTP-kappa inhibits translocation of GLUT4 in rat adipose cells. Biochem. Biophys. Res. Commun. 255:200-207. [DOI] [PubMed] [Google Scholar]
- 11.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 29:117-129. [DOI] [PubMed] [Google Scholar]
- 12.Derynck, R., and Y. E. Zhang. 2003. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425:577-584. [DOI] [PubMed] [Google Scholar]
- 13.Dumont, N., A. V. Bakin, and C. L. Arteaga. 2003. Autocrine transforming growth factor-beta signaling mediates Smad-independent motility in human cancer cells. J. Biol. Chem. 278:3275-3285. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, M., T. Muller, M. M. Lerch, and A. Ullrich. 1996. Association of human protein-tyrosine phosphatase kappa with members of the armadillo family. J. Biol. Chem. 271:16712-16719. [DOI] [PubMed] [Google Scholar]
- 15.Fujii, M., K. Takeda, T. Imamura, H. Aoki, T. K. Sampath, S. Enomoto, M. Kawabata, M. Kato, H. Ichijo, and K. Miyazono. 1999. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol. Biol. Cell 10:3801-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gobbi, H., W. D. Dupont, J. F. Simpson, W. D. Plummer, Jr., P. A. Schuyler, S. J. Olson, C. L. Arteaga, and D. L. Page. 1999. Transforming growth factor-beta and breast cancer risk in women with mammary epithelial hyperplasia. J. Natl. Cancer Inst. 91:2096-2101. [DOI] [PubMed] [Google Scholar]
- 17.Goggins, M., M. Shekher, K. Turnacioglu, C. J. Yeo, R. H. Hruban, and S. E. Kern. 1998. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 58:5329-5332. [PubMed] [Google Scholar]
- 18.Granot-Attas, S., and A. Elson. 2004. Protein tyrosine phosphatase epsilon activates Yes and Fyn in Neu-induced mammary tumor cells. Exp. Cell Res. 294:236-243. [DOI] [PubMed] [Google Scholar]
- 19.Hahn, S. A., M. Schutte, A. T. Hoque, C. A. Moskaluk, L. T. da Costa, E. Rozenblum, C. L. Weinstein, A. Fischer, C. J. Yeo, R. H. Hruban, and S. E. Kern. 1996. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271:350-353. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, Y. P., H. Wang, P. D'Eustachio, J. M. Musacchio, J. Schlessinger, and J. Sap. 1993. Cloning and characterization of R-PTP-kappa, a new member of the receptor protein tyrosine phosphatase family with a proteolytically cleaved cellular adhesion molecule-like extracellular region. Mol. Cell. Biol. 13:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, F., M. A. Sells, and J. Chernoff. 1998. Transformation suppression by protein tyrosine phosphatase 1B requires a functional SH3 ligand. Mol. Cell. Biol. 18:250-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenz, U., A. D. Bergemann, H. N. Steinberg, J. G. Flanagan, X. Li, S. J. Galli, and B. G. Neel. 1996. Genetic analysis reveals cell type-specific regulation of receptor tyrosine kinase c-Kit by the protein tyrosine phosphatase SHP1. J. Exp. Med. 184:1111-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markowitz, S., J. Wang, L. Myeroff, R. Parsons, L. Sun, J. Lutterbaugh, R. S. Fan, E. Zborowska, K. W. Kinzler, B. Vogelstein, et al. 1995. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science 268:1336-1338. [DOI] [PubMed] [Google Scholar]
- 24.Massague, J. 1998. TGF-beta signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 25.Massague, J., and Y. G. Chen. 2000. Controlling TGF-beta signaling. Genes Dev. 14:627-644. [PubMed] [Google Scholar]
- 26.Mauro, L. J., and J. E. Dixon. 1994. ‘Zip codes’ direct intracellular protein tyrosine phosphatases to the correct cellular ‘address’. Trends Biochem. Sci. 19:151-155. [DOI] [PubMed] [Google Scholar]
- 27.Moghal, N., and P. W. Sternberg. 1999. Multiple positive and negative regulators of signaling by the EGF-receptor. Curr. Opin. Cell Biol. 11:190-196. [DOI] [PubMed] [Google Scholar]
- 28.Moses, H. L., E. L. Branum, J. A. Proper, and R. A. Robinson. 1981. Transforming growth factor production by chemically transformed cells. Cancer Res. 41:2842-2848. [PubMed] [Google Scholar]
- 29.Moulder, S. L., F. M. Yakes, S. K. Muthuswamy, R. Bianco, J. F. Simpson, and C. L. Arteaga. 2001. Epidermal growth factor receptor (HER1) tyrosine kinase inhibitor ZD1839 (Iressa) inhibits HER2/neu (erbB2)-overexpressing breast cancer cells in vitro and in vivo. Cancer Res. 61:8887-8895. [PubMed] [Google Scholar]
- 30.Muraoka, R. S., N. Dumont, C. A. Ritter, T. C. Dugger, D. M. Brantley, J. Chen, E. Easterly, L. R. Roebuck, S. Ryan, P. J. Gotwals, V. Koteliansky, and C. L. Arteaga. 2002. Blockade of TGF-beta inhibits mammary tumor cell viability, migration, and metastases. J. Clin. Investig. 109:1551-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muraoka, R. S., Y. Koh, L. R. Roebuck, M. E. Sanders, D. Brantley-Sieders, A. E. Gorska, H. L. Moses, and C. L. Arteaga. 2003. Increased malignancy of Neu-induced mammary tumors overexpressing active transforming growth factor β1. Mol. Cell. Biol. 23:8691-8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muraoka-Cook, R. S., H. Kurokawa, Y. Koh, J. T. Forbes, L. R. Roebuck, M. H. Barcellos-Hoff, S. E. Moody, L. A. Chodosh, and C. L. Arteaga. 2004. Conditional overexpression of active transforming growth factor beta1 in vivo accelerates metastases of transgenic mammary tumors. Cancer Res. 64:9002-9011. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura, M., M. Kishi, T. Sakaki, H. Hashimoto, H. Nakase, K. Shimada, E. Ishida, and N. Konishi. 2003. Novel tumor suppressor loci on 6q22-23 in primary central nervous system lymphomas. Cancer Res. 63:737-741. [PubMed] [Google Scholar]
- 34.Oliff, A., M. D. McKinney, and O. Agranovsky. 1985. Contribution of the gag and pol sequences to the leukemogenicity of Friend murine leukemia virus. J. Virol. 54:864-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsons, S. J., and J. T. Parsons. 2004. Src family kinases, key regulators of signal transduction. Oncogene 23:7906-7909. [DOI] [PubMed] [Google Scholar]
- 36.Ren, X. D., W. B. Kiosses, D. J. Sieg, C. A. Otey, D. D. Schlaepfer, and M. A. Schwartz. 2000. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J. Cell Sci. 113:3673-3678. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, A. B., M. A. Anzano, L. M. Wakefield, N. S. Roche, D. F. Stern, and M. B. Sporn. 1985. Type beta transforming growth factor: a bifunctional regulator of cellular growth. Proc. Natl. Acad. Sci. USA 82:119-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell, W. E., R. J. Coffey, Jr., A. J. Ouellette, and H. L. Moses. 1988. Type beta transforming growth factor reversibly inhibits the early proliferative response to partial hepatectomy in the rat. Proc. Natl. Acad. Sci. USA 85:5126-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sap, J., Y. P. Jiang, D. Friedlander, M. Grumet, and J. Schlessinger. 1994. Receptor tyrosine phosphatase R-PTP-kappa mediates homophilic binding. Mol. Cell. Biol. 14:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoenwaelder, S. M., and K. Burridge. 1999. Bidirectional signaling between the cytoskeleton and integrins. Curr. Opin. Cell Biol. 11:274-286. [DOI] [PubMed] [Google Scholar]
- 41.Seton-Rogers, S. E., Y. Lu, L. M. Hines, M. Koundinya, J. LaBaer, S. K. Muthuswamy, and J. S. Brugge. 2004. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc. Natl. Acad. Sci. USA 101:1257-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin, D. Y., T. Ishibashi, T. S. Choi, E. Chung, I. Y. Chung, S. A. Aaronson, and D. P. Bottaro. 1997. A novel human ERK phosphatase regulates H-ras and v-raf signal transduction. Oncogene 14:2633-2639. [DOI] [PubMed] [Google Scholar]
- 43.Siegel, P. M., W. Shu, R. D. Cardiff, W. J. Muller, and J. Massague. 2003. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc. Natl. Acad. Sci. USA 100:8430-8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su, J., A. Batzer, and J. Sap. 1994. Receptor tyrosine phosphatase R-PTP-alpha is tyrosine-phosphorylated and associated with the adaptor protein Grb2. J. Biol. Chem. 269:18731-18734. [PubMed] [Google Scholar]
- 45.Thannickal, V. J., D. Y. Lee, E. S. White, Z. Cui, J. M. Larios, R. Chacon, J. C. Horowitz, R. M. Day, and P. E. Thomas. 2003. Myofibroblast differentiation by transforming growth factor-beta1 is dependent on cell adhesion and integrin signaling via focal adhesion kinase. J. Biol. Chem. 278:12384-12389. [DOI] [PubMed] [Google Scholar]
- 46.Tucker, R. F., G. D. Shipley, H. L. Moses, and R. W. Holley. 1984. Growth inhibitor from BSC-1 cells closely related to platelet type beta transforming growth factor. Science 226:705-707. [DOI] [PubMed] [Google Scholar]
- 47.Ueda, Y., S. Wang, N. Dumont, J. Y. Yi, Y. Koh, and C. L. Arteaga. 2004. Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks TGF-induced cell motility. J. Biol. Chem. 279:24505-24513. [DOI] [PubMed] [Google Scholar]
- 48.Wakefield, L. M., and A. B. Roberts. 2002. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 12:22-29. [DOI] [PubMed] [Google Scholar]
- 49.Wang, D., T. Kanuma, H. Mizunuma, F. Takama, Y. Ibuki, N. Wake, A. Mogi, Y. Shitara, and S. Takenoshita. 2000. Analysis of specific gene mutations in the transforming growth factor-beta signal transduction pathway in human ovarian cancer. Cancer Res. 60:4507-4512. [PubMed] [Google Scholar]
- 50.Wang, H., V. Radjendirane, K. K. Wary, and S. Chakrabarty. 2004. Transforming growth factor beta regulates cell-cell adhesion through extracellular matrix remodeling and activation of focal adhesion kinase in human colon carcinoma Moser cells. Oncogene 23:5558-5561. [DOI] [PubMed] [Google Scholar]
- 51.Wang, J., L. Sun, L. Myeroff, X. Wang, L. E. Gentry, J. Yang, J. Liang, E. Zborowska, S. Markowitz, J. K. Willson, et al. 1995. Demonstration that mutation of the type II transforming growth factor beta receptor inactivates its tumor suppressor activity in replication error-positive colon carcinoma cells. J. Biol. Chem. 270:22044-22049. [DOI] [PubMed] [Google Scholar]
- 52.Wiener, J. R., S. K. Kassim, Y. Yu, G. B. Mills, and R. C. Bast, Jr. 1996. Transfection of human ovarian cancer cells with the HER-2/neu receptor tyrosine kinase induces a selective increase in PTP-H1, PTP-1B, PTP-alpha expression. Gynecol. Oncol. 61:233-240. [DOI] [PubMed] [Google Scholar]
- 53.Wieser, R., J. L. Wrana, and J. Massague. 1995. GS domain mutations that constitutively activate T beta R-I, the downstream signaling component in the TGF-beta receptor complex. EMBO J. 14:2199-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yakes, F. M., W. Chinratanalab, C. A. Ritter, W. King, S. Seelig, and C. L. Arteaga. 2002. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer Res. 62:4132-4141. [PubMed] [Google Scholar]
- 55.Yang, Y., M. Gil, S. M. Byun, I. Choi, K. H. Pyun, and H. Ha. 1996. Transforming growth factor-beta1 inhibits human keratinocyte proliferation by upregulation of a receptor-type tyrosine phosphatase R-PTP-kappa gene expression. Biochem. Biophys. Res. Commun. 228:807-812. [DOI] [PubMed] [Google Scholar]
- 56.Yang, Y., M. C. Gil, E. Y. Choi, S. H. Park, K. H. Pyun, and H. Ha. 1997. Molecular cloning and chromosomal localization of a human gene homologous to the murine R-PTP-kappa, a receptor-type protein tyrosine phosphatase. Gene 186:77-82. [DOI] [PubMed] [Google Scholar]
- 57.Yang, Y. A., O. Dukhanina, B. Tang, M. Mamura, J. J. Letterio, J. MacGregor, S. C. Patel, S. Khozin, Z. Y. Liu, J. Green, M. R. Anver, G. Merlino, and L. M. Wakefield. 2002. Lifetime exposure to a soluble TGF-beta antagonist protects mice against metastasis without adverse side effects. J. Clin. Investig. 109:1607-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2:127-137. [DOI] [PubMed] [Google Scholar]
- 59.Yu, X., S. Miyamoto, and E. Mekada. 2000. Integrin alpha 2 beta 1-dependent EGF receptor activation at cell-cell contact sites. J. Cell Sci. 113:2139-2147. [DOI] [PubMed] [Google Scholar]
- 60.Zeng, L., X. Si, W. P. Yu, H. T. Le, K. P. Ng, R. M. Teng, K. Ryan, D. Z. Wang, S. Ponniah, and C. J. Pallen. 2003. PTP alpha regulates integrin-stimulated FAK autophosphorylation and cytoskeletal rearrangement in cell spreading and migration. J. Cell Biol. 160:137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhai, J., H. Lin, Z. Nie, J. Wu, R. Canete-Soler, W. W. Schlaepfer, and D. D. Schlaepfer. 2003. Direct interaction of focal adhesion kinase with p190RhoGEF. J. Biol. Chem. 278:24865-24873. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., R. Siebert, P. Matthiesen, Y. Yang, H. Ha, and B. Schlegelberger. 1998. Cytogenetical assignment and physical mapping of the human R-PTP-kappa gene (PTPRK) to the putative tumor suppressor gene region 6q22.2-q22.3. Genomics 51:309-311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.