Abstract
Some forms of Sjögren’s syndrome (SS) follow a clinical course accompanied by systemic symptoms caused by lymphocyte infiltration and proliferation in the liver, kidneys, and other organs. To better understand the clinical outcomes of SS, here we used minor salivary gland tissues from patients and examine their molecular, biological, and pathological characteristics. A retrospective study was performed, combining clinical data and formalin-fixed paraffin-embedded (FFPE) samples from female patients over 60 years of age who underwent biopsies at Okayama University Hospital. We employed direct digital RNA counting with nCounter® and multiplex immunofluorescence analysis with a PhenoCycler™ on the labial gland biopsies. We compared FFPE samples from SS patients who presented with other connective tissue diseases (secondary SS) with those from stable SS patients with symptoms restricted to the exocrine glands (primary SS). Secondary SS tissues showed enhanced epithelial damage and lymphocytic infiltration accompanied by elevated expression of autophagy marker genes in the immune cells of the labial glands. The close intercellular distance between helper T cells and B cells positive for autophagy-associated molecules suggests accelerated autophagy in these lymphocytes and potential B cell activation by helper T cells. These findings indicate that examination of FFPE samples from labial gland biopsies can be an effective tool for evaluating molecular histological differences between secondary and primary SS through multiplexed analysis of gene expression and tissue imaging.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12903-024-04869-4.
Keywords: Autoimmune disease, Xerostomia, Multiplex immunostaining, Spatial analysis, Autophagy
Introduction
Sjögren’s syndrome (SS) is a chronic autoimmune disease that primarily affects the exocrine glands, including the salivary and lacrimal glands, leading to dryness of the mouth and eyes [1]. However, symptoms appear not only in the exocrine glands but also in systemic organs. SS is frequently associated with other connective tissue diseases and extraglandular lesions such as interstitial pneumonia. A differential diagnosis between SS and other autoimmune diseases is necessary in cases where SS is suspected. It is also important to distinguish SS from IgG4-related diseases associated with dry eyes and dry mouth [2]. SS is known to increase the risk of developing other fatal diseases. Malignant lymphoma is a predictable complication that should be considered as its incidence in patients with SS has increased in recent years [3]. Patients with primary SS are at a 10-44 times higher risk of lymphoma than healthy individuals, which is more serious than the risk reported for systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) [4]. Although immune complexes in the serum are known to be important for the lesion expansion of SS, no specific marker that reflects the variety of clinical manifestations of SS is available [5]. Better diagnostic techniques are required to provide appropriate treatment.
Autophagy is an intracellular cleansing and recycling system in all eukaryotes, including yeast, plants, and animals [6], and essential for the development and homeostasis of the immune system. Abnormalities in autophagy are known to be involved in development of the autoimmune diseases [7]. Autophagy-related proteins include beclin 1, microtubule associated protein 1 light chain 3 alpha and beta (LC3A and LC3B), sequestosome 1 (SQSTM1, known as P62), and sirtuin 1 (SIRT1). Among them, LC3B and SQSTM1 are generally used as markers for monitoring autophagy [8]. A genome-wide association study carried out to identify the disease susceptibility genes reported autophagy-related factors as candidates for systemic autoimmune diseases. Autophagy related 5 (ATG5) has been identified as one of the disease susceptibility genes in SLE, a disease closely related to SS [9]. It would be worthwhile to investigate the involvement of these genes during SS progression.
Technological advances in human immunological analysis have made it possible to obtain vast amounts of information from even small samples [10]. New biomarkers and pathways involved in immune responses have been identified using systems biology approaches with human tissues or serum [11, 12]. Here, we performed multiplexed immunohistochemical staining with nucleic acid-barcoded antibodies in conjunction with gene expression profiling using digitally counted RNAs to analyze small amounts of labial gland samples. We subsequently analyzed correlation of spatial protein information at the single-cell level with matched global gene expression data to computationally identify differences in labial gland tissue organization between primary SS and secondary SS.
Methods
Patients with SS
A retrospective study design was employed for the analysis of patients with suspected SS who underwent biopsy between January 2010 and January 2021 at Okayama University Hospital. A total of 514 patients were grouped and analyzed according to their medical records. Primary SS is defined as SS with stable symptoms localized to the exocrine glands, whereas secondary SS is defined as SS complicated by systemic connective tissue disease [13]. The inclusion criteria were women over 60 years of age who were SS-A-positive, had biopsy histopathology showing more than one focus per 4 mm² (more than 50 lymphocytes per duct), and exhibited clinical exocrine gland symptoms. The exclusion criteria were patients with primary SS who had symptoms other than exocrine gland symptoms and who had not been followed up for more than one year. After applying the exclusion criteria, 14 patients with primary SS and 19 patients with secondary SS were selected for clinical data analysis. The top four samples with the highest concentration and quality of total RNA extracted from FFPE samples were used for gene expression profiling analysis and spatial analysis. The concentration and quality of total RNA extracted from FFPE samples were measured by 4200 Tape Station (Aglent, CA, USA) and Qubit 4 Fluorometer (Invitrogen, MA, USA). In terms of quality, samples containing more than 45% RNA fragments of approximately 200 nucleotides or less were excluded. Samples with a measured concentration of less than 10.0 ng/µl were also excluded to reduce analysis error. Since all patients presented with clinical symptoms and met the inclusion criteria, all patients were diagnosed with SS based on the revised classification standards and the American College of Rheumatology (ACR) or European League Against Rheumatism (EULAR) in 2016 and the Japanese criteria for diagnosing SS in 1999 [14, 15]. A flowchart of the patient selection process is shown in Figure S1A. None of the patients included in the present analysis were on any steroid or salivary medications at the time of sampling.
Patients with mucoceles
A total of 214 patients who underwent mucocele biopsy between January 2016 and January 2021 at Okayama University Hospital were analyzed as controls for patients with SS. Labial gland samples from female patients over 60 years of age with mucocele were included. Samples containing no minor salivary gland tissue were excluded, as only labial gland tissues with mucocele were collected and examined. A flowchart of the patient selection process is shown in Figure S1B.
Clinical data analysis
Salivary and lacrimal gland tests were performed to examine the exocrine gland dryness symptoms of patients. Cases were included if they met the following criteria: Gum test ≤ 10 mL/10 min, Saxon test ≤ 2.0 g/2 min, and Schirmer test ≤ 5.0 mm. Because blood data was collected multiple times for a single patient, data from the date of biopsy was used for the analysis.
Hematoxylin and eosin (H&E) staining
Labial glands were fixed in a 10% formalin solution at room temperature. Paraffin-embedded sections of the labial glands were prepared according to standard methods and stained with H&E. The areas of lymphocyte infiltration in the tissue were analyzed using ImageJ software (https://imagej.net/ij/index.html).
FFPE sample preparation and RNA profiling
For RNA profiling of FFPE samples, 30 μm thick serial sections from chilled paraffin blocks were placed on film-coated slides (Leica, Wetzlar, Germany), and dried on a hot plate at 40 °C for 2 h. Toluidine blue staining was performed for laser microdissection (LMD), and only the glandular tissue portion was collected using an LMD7000 microscope (Leica). RNA was extracted from FFPE samples using a NucleoSpin® total RNA FFPE XS kit (Takara, Shiga, Japan). FFPE samples with foil were deparaffinized and lysed with Proteinase K. The samples were then treated with DNase and washed and after which RNA was eluted through the column. The amount and quality of the RNA samples was measured using a Qubit® 2.0 Fluorometer (Invitrogen) and a 4150 TapeStation automated electrophoresis platform (Agilent, CA, USA), respectively. The top four samples with concentrations higher than 10 ng/µL, and those with more than 30% of the RNA longer than 200 nucleotides were selected for digital RNA analysis.
Direct digital RNA counting analysis
nCounter® (NanoString Technologies, WA, USA) was used for comprehensive gene expression analysis by direct digital counting of mRNA [16]. For the nCounter® hybridization assay for oligonucleotide counting, nCounter® Low RNA Input Kit (NanoString Technologies) was used to assess the data reproducibility. The Human Immunology V2 Panel, which comprises 447 genes related to human immunology, was used for the analysis. Hybridization of the cleaved indexing oligonucleotides with fluorescent barcodes was performed using nCounter® TagSets Reagents (NanoString Technologies). For experiments in which the indexing oligonucleotide was double-stranded, the aspirates were denatured at 95 °C for 5 min and then placed on ice for 2 min. A master mix was prepared by adding hybridization buffer (NanoString Technologies) to the Hyb Code. To read the RNA detection probes, 9.6 nM in situ capture probe was added, and 7 µL aliquots of the master mix were added to each well. Depending on the experiment, 10 µL aliquots of aspirate were added to each tube, and each hybridization was adjusted to a final volume of 15 µL with diethyl pyrocarbonate-treated water. The hybridizations were performed at 65 °C overnight in a LightCycler® PCR system. After hybridization, the samples were processed using the nCounter® Prep Station and Digital Analyzer according to the manufacturer’s instructions. Gene expression comparison and Gene Ontology (GO) enrichment analysis was performed using nSolver™ analysis software (NanoString Technologies) and DAVID Bioinformatics resources (https://david.ncifcrf.gov/).
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
Each pooled RNA sample was reverse transcribed to cDNA. Each cDNA was used for RT-qPCR, which was performed on a LightCycler® Nano System (Roche, Basel, Switzerland) using FastStart™ Essential DNA Green Master Mix (06402712001; Roche). The RT-qPCR results for each sample were normalized to that of the reference gene Glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The results are expressed as normalized ratios, and each measurement was repeated thrice. The synthetic primers used are as follows: ATG5: 5’-CACAAGCAACTCTGGATGGGATTG-3′ and 5′-GCAGCCACAGGACGAAACAG-3′, LC3B: 5′-GAGAAGCAGCTTCCTGTTCTGG-3′ and 5′-GTGTCCGTTCACCAACAGGAAG-3′, SQSTM1: 5′-TAGGAACCCGCTACAAGTGC-3′ and 5′-GAGAAGCCCTCAGACAGGTG-3′ and GAPDH:5′-CCACTCCTCCACCTTTGACG-3 ′and 5′- CCACCACCCTGTTGCTGTAG-3′.
Multiplexed immunofluorescence staining
Prior to multiplex immunostaining, FFPE samples of labial gland tissues and pharyngeal tonsils were used to validate the staining characteristics of each antibody. Commercial sources provided purified, carrier-free, monoclonal, and polyclonal anti-human antibodies for multiplexed immunofluorescence staining. Oligonucleotide-barcoded antibody staining of tissue sections mounted on coverslips was performed using a commercially available PhenoCycler™ staining kit (Akoya Biosciences, MA, USA), following the manufacturer’s instructions for FFPE tissue [17, 18]. Coverslip slides (Akoya Biosciences) for mounting the samples were prepared by immersing the samples in poly-L-lysine solution (Sigma-Aldrich, MO, USA) for 12 h. The samples were cut into 6.0 μm sections using a microtome and mounted. The coverslips were baked at 60 °C, cooled, deparaffinized, and then rehydrated. Antigen retrieval was performed using citrate buffer (pH 6.0) in a pressure cooker. The antibodies were conjugated to maleimide-modified short DNA oligonucleotides at a weight/weight ratio of 2:1 of the oligonucleotide to antibody, with a minimum of 100 µg of antibody per reaction. The samples were incubated with barcoded antibodies (Table S4) in blocking buffer for 3 h in a humidified chamber at room temperature. After washing with phosphate-buffered saline, the tissue sections were sequentially fixed with 1.6% paraformaldehyde, methanol (on ice), and fixative reagent. Stained sample coverslips were stored in storage buffer at 4 °C for up to two weeks. Barcoded fluorescent reporters corresponding to the barcoded primary antibodies were added to 96-well plates containing Reporter Stock Solution, and nuclear staining was performed for each multiplex cycle. PhenoCycler™ imaging was performed acquired in six cycles of three colors (Alexa Fluor 488, Atto 550, and Cy5), with DAPI. Images were acquired at 20-fold magnification using a BZX-810 microscope (Keyence, Osaka, Japan) with a metal halide light source and Plan Apo λNA 0.75 20x air objective (Nikon, Tokyo, Japan).
Spatial profiling of labial glands by multiplex imaging
Spatial profiling of labial glands was performed by multiplex imaging. The spatial distributions and marker intensities of the target cells were analyzed using the QuPath spatial analysis toolbox (https://qupath.github.io) [19, 20]. Cell segmentation using the StarDist Extension was first performed to identify single cells based on DAPI staining. QuPath performs machine learning for classification based on various types of epithelial, stroma, and immune cells. The phenotypes of the cells were determined by confirming the classification through machine learning, specifying the positive and negative results for each of the 30 cells per antibody. Cell categorization data were calculated by analyzing whole glandular tissue. Immune marker positive cells were defined as cells that were positive for CD3e, CD4, CD8, CD20, CD11b or CD11c. Cells positive for E-cadherin (E-cad), aquaporin5 (AQP5) or cytokeratin 7 (KRT7) was based on location information from QuPath. In the neighborhood analysis, the area was determined by the constituent cells located in the vicinity of the cell in a 30-µm area centered on the cell.
Statistics
The data are presented as the mean values with standard deviations. Bonferroni correction was used for multiple comparisons among mucoceles, primary SS, and secondary SS. Paired t-test was used to compare the means of the between primary SS and secondary SS. Spearman’s rank correlation coefficients were used to determine the relationship between LC3B and SQSTM1 protein expression levels. Values of p < 0.05 were considered significant. Data analysis was performed using RStudio (https://posit.co/) with R version 4.2.2.
Results
Overview of the selected patients
This study aimed to determine histological differences between primary SS and secondary SS using a retrospective analysis of comprehensive gene expression profiles obtained from FFPE samples combined with patient clinical data (Figures S1 and S2). We focused on women over 60 years of age to exclude the involvement of sex hormones in autoimmune diseases [21]. The mean age at menopause in Asian women has been reported to be 49 years, and one SD (70% included) amounts to approximately 53 years [22]. Males were excluded because salivary glands are known to be morphologically sexually dimorphic organs [23]. Of the 514 biopsied SS samples, 404 patients were women (78.6%), 218 of whom were over 60 years of age. It indicates that half of the patients biopsied for suspected SS were women over 60 years of age. And, among the 250 biopsied samples of mucocele, 133 patients were women (53.2%), 11 of whom were over 60 years of age. The patient backgrounds are summarized in Table S1. In addition, the blood test was performed to compare the two groups of SS. Most blood test data were not significantly different, although the hematocrit levels were lower in the secondary SS group, but within normal levels.
Comparison of immune-related gene expressions and identification of differential markers between mucocele, primary SS and secondary SS with digital RNA counting
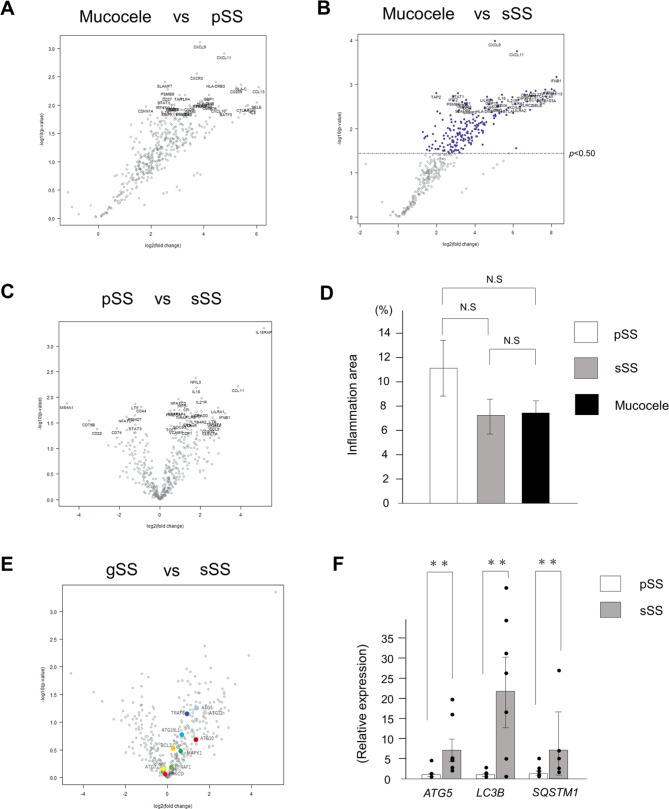
Digital RNA count analysis was performed using nCounter® to profile the labial gland biopsies. We used the Human Immunology V2 panel consisting of 447 genes because it contains many genes in common with the top genes expressed in oral gland tissues [24]. We compared mucocele samples with the respective SS samples, because normal labial tissues were not available in this retrospective study. We found that most genes were upregulated in the SS samples, as shown in the volcano plots (Fig. 1A, B). Although the total RNA in this analysis was from damaged FFPE tissues, we confirmed experimental reproducibility based on consistent results in all samples. Most genes in primary SS and secondary SS did not exhibit any predominant differences; however, some genes, such as IL18RAP, exhibited predominant differences (Fig. 1C and Table S2). The percentages of lymphocytic foci in the labial gland tissues were determined by H&E staining, and no significant differences were observed in the amount of lymphocyte infiltration into the epithelial tissues among the three groups (Fig. 1D and Table S3). Next, we performed GO analysis by using nSolver™ software to determine the immune-related genes involved in each disease group. Based on the volcano plot results, we narrowed down the signaling pathways involved and their associated genes (Figure S3). Through this GO analysis and comparison of various gene groups, no significant GO terms were found among the 32 gene groups related to the immunity panel that consisted of commercially available probe sets. However, we found that ten types of autophagy-related genes were upregulated in secondary SS (Fig. 1E, Figure S3 and Table S4). To substantiate nCounter® results, the top 100 upregulated genes in secondary SS were extracted for another GO analysis by using DAVID Bioinformatics resources. Seven genes, including ATG12, were extracted as autophagy-related genes. In the biological process category, secondary SS showed significantly increased autophagy-related genes compared with primary SS (p = 0.0096). To confirm the results of the digital RNA analysis, RT-qPCR analysis was performed with representative autophagy markers LC3B and SQSTM1 in addition to ATG5, which was included in the nCounter® gene set. Secondary SS exhibited significantly increased expression of ATG5, LC3B and SQSTM1 compared to primary SS (Fig. 1F). Comparison of ATG5 expression between the two groups in terms of values obtained by nCounter® showed no significant differences, but there was a trend in which secondary SS values were higher than primary SS values, identical to the results of RT-qPCR analysis. Although conventional histological analysis showed no difference between the primary SS and secondary SS groups, digital RNA counting and RT-qPCR analysis revealed a significant increase in the expression of autophagy-related genes in secondary SS compared to that in primary SS.
Fig. 1.
Comparison of immune-related gene expressions between mucocele and SS by digital RNA counting and RT-qPCR. (A) Volcano plot comparing gene expression between the mucocele and primary SS by nCounter® system analysis. The left side shows the group of genes for which expression was upregulated in mucoceles. The right side shows the group of genes for which expression was upregulated in primary SS. (B) Volcano plot comparing gene expression between the mucocele and secondary SS. The left side shows the group of genes for which expression was upregulated in mucocele. The right side shows the group of genes for which expression was upregulated in secondary SS. Genes with P-value < 0.5 are highlighted in dark blue. (C) Volcano plot comparing gene expression between primary SS and secondary SS. The left side shows the group of genes for which expression was upregulated in primary SS. The right side shows the group of genes for which expression was upregulated in secondary SS. (D) The percentages of lymphocytic foci areas in the labial gland tissue determined by hematoxylin and eosin (H&E) staining. The Bonferroni method was employed for comparison between the three groups. (E) Volcano plot comparing gene expression between primary SS and secondary SS with highlighting autophagy-related factors. The expression of autophagy-related ten genes (colored) was upregulated in secondary SS. (F) Comparison of Autophagy related 5 (ATG5), LC3B and sequestosome 1 (SQSTM1) expression between primary SS and secondary SS using qRT-PCR. Bars indicate mean values and dots indicate their respective values. ** p < 0.01
Cell profiling by multiplex immunostaining using a PhenoCycler™
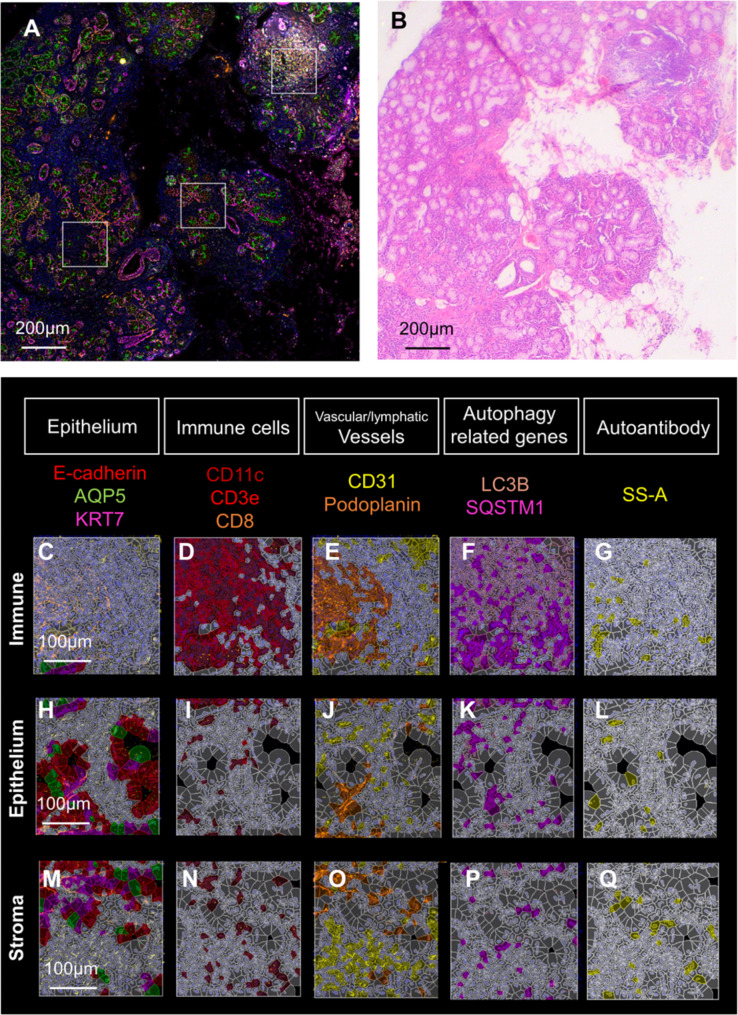
Multiplex immunostaining was performed using a PhenoCycler™ to profile gene expression in each cell and compare the differences between groups using the spatial information of each cell. In this study, simultaneous staining was performed on 18 target molecules using three fluorochromes: Alexa Fluor 488, Atto 550, and Cy5, repeated for six cycles, as well as DAPI to label nuclei (Table S4). SS samples showed SS-A-positive cells, although these cells were not found in any mucocele samples (Figures S4 and S5). The open-source software QuPath was used for automated quantitative and semiquantitative analyses of protein expression levels in complete slide images. The algorithms employed by QuPath enable spatial expression analysis on a per-cell basis. QuPath analysis can extract positive cells via machine learning according to cell location data and protein expression levels. To perform machine learning, analysis of cell labeling with epithelial, stromal, and immune cell-specific markers was first carried out (Fig. 2). Then, machine learning was performed via supervised learning where the correct answers were given to the training data from three images. Positive cell determination was performed after supervised learning on all genes. For example, the localization of three proteins, CD11c, CD3e, and CD8, represented immune cells (Figure S2). Per-cell citation analysis demonstrated accumulation of immune cells in immune areas (Fig. 2D). However, CD11c, a dendritic cell marker, was also distributed in the stroma of epithelial tissues. CD11b-positive cells (macrophages) were especially abundant around the ducts, in agreement with published data [26] (Fig. 2D, I, N). The distribution of CD3e (T cell) was significantly different between mucocele and SS (Figure S6). Immune areas were prominent around ducts in SS samples (Figure S6E-L). Vessels (vascular and lymphatic) were mainly present in the immune areas (Fig. 2E, J, O). LC3B and SQSTM1 were expressed throughout but predominantly in the immune areas (Fig. 2F). Moreover, SS-A was localized to immune cells, as well as to epithelial and stroma tissues (Fig. 2G, L, Q).
Fig. 2.
Cell profiling by multiplex immunostaining. (A) Multiplex immunostaining and the corresponding histological (H&E) images are presented. Multiplex immunostaining is 19-color superimposed image by PhenoCycler™ analysis. The three white squares delineate, from left to right, the stroma area, denoted as stroma, the epithelial area, denoted as epithelium, and the lymphocytic foci, denoted as “Immune”, which is the same as immune area in the text. Since epithelial and stromal tissues were intermingled, the panels for “Epithelium” are images rich in epithelial tissues and those for “Stroma” are rich in connective tissue. Scale bars representing 200 μm. (B) H&E image, accompanied by scale bar: 200 μm. (C–Q) Single-cell profiles based on multiplex immunostaining. The top sections show areas of lymphocytic foci (C–G), the middle sections show the epithelium (H–L). and the bottoms sections show the stroma (M–Q). (C, H, M) Localization of E-cadherin (E-cad), aquaporin5 (AQP5), and cytokeratin7 (KRT7) in epithelial tissue. Similarly, the followings are shown: immune cells defined by CD11c, CD3e, and CD8 (D, I, N); vascular lymphatics defined by podoplanin and CD31 (E, J, O); autophagy-related protein markers defined by light chain 3 alpha (LC3B) and SQSTM1 (F, K, P); and the area reactive to the autoantibody SS-A (G, L, R)
Difference in tissue damage between primary SS and secondary SS
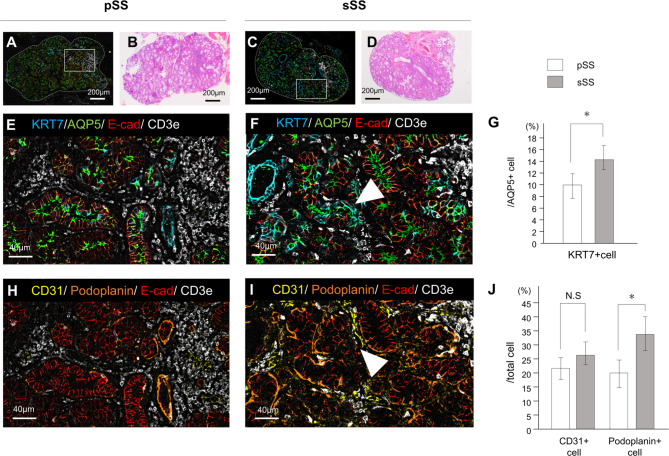
Using cell profiles, we first estimated the percentages of tissue categories and cells specifically found in SS labial glands. Each SS sample was performed with n = 4, and mucocele was performed with n = 2. The three tissue categories were set as follows: epithelium, immune system, and stroma. As shown in the pie chart in Figure S7A, no differences were observed in the percentage of these three categorized tissues between the mucocele, primary SS and secondary SS groups. Additionally, a detailed cell-by-cell analysis of the three areas showed that the percentage of immune cells in the immune area remained unchanged (Figure S7B). However, differences were observed between the epithelium and stroma areas (Figure S7C, D). In salivary gland tissue, KRT7 labels duct cells and AQP5 labels acinar cells. However, after salivary duct ligation or irradiation, acinar cells also express KRT7, indicating that the acinar cells regenerate duct cells when damaged. The percentage of AQP5-positive cells co-expressing KRT7 was significantly higher in secondary SS [25], indicating more substantial damage to the epithelial tissues (Fig. 3A-G). We also investigated the percentage of vascular and lymphatic tissues expressing CD31 and podoplanin. CD31 is expressed in all vascular endothelial cells, whereas podoplanin is a lymphatic endothelial cell marker. Podoplanin-positive cells were significantly more abundant in secondary SS than in primary SS, whereas CD31-positive cells tended to be more abundant in secondary SS, although the difference was not significant (Fig. 3H-J). The ratio of lymphocytic foci per total area did not differ between the secondary SS and primary SS samples. Although routine histological analyses of the biopsies suggest similar levels of damage, a more detailed analysis of SS using multiplex immunohistochemistry revealed differences in tissue damage levels. Additionally, tissue damage to the exocrine glands was more advanced in secondary SS than in localized stable SS, based on the analysis of epithelial and vascular/lymphatic tissues.
Fig. 3.
Difference in tissue damage between primary SS and secondary SS. (A, B, E, H) Histological analyses of primary SS and (C, D, F, I) secondary SS samples were performed. Tissue samples were imaged at low magnification (A–D) along with the corresponding H&E images. Scale bar: 200 μm (A–D), 40 μm (E, F, H, I). (E, F) Four-color multiplex immunostaining showing epithelial tissue damage: KRT7, a duct marker; AQP5, an acinar marker; and E-cad, an adhesion factor in epithelial tissues. Localization of CD3e indicates immune cells in the vicinity. White arrowhead in (F) indicates one of the representative cells double positive for KRT7 and AQP5. (G) Difference in the percentage of KRT7-positive cells among AQP5-positive cells. *p < 0.05. (H, I) Four-color multiplex immunostaining of the vascular lymphoid tissue is shown: CD31, a vascular endothelial cell marker; podoplanin, a lymphatic endothelial cell marker; and E-cad, an adhesion factor for epithelial tissue. Localization of CD3e indicates immune cells in the vicinity. White arrowhead in (I) indicates one of the representative CD31-positive cells surrounding the acinar cells. (J) Differences in the percentages of CD31-positive and podoplanin-positive cells relative to the total cell count are shown. *p < 0.05
Autophagy marker expression in the area of lymphocytic foci
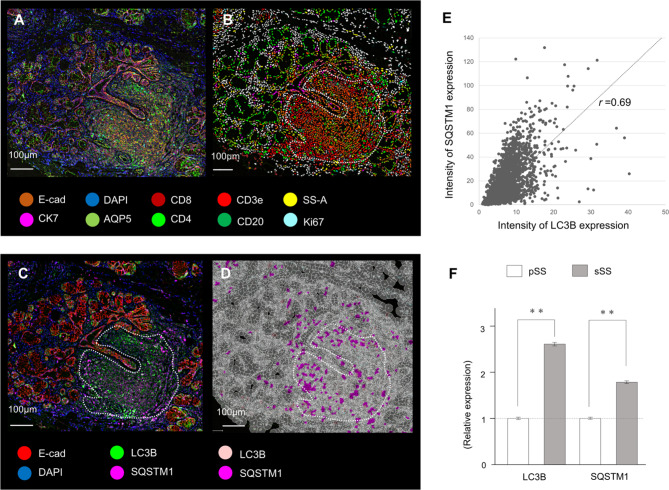
It was reported that higher levels of autophagy marker expression were detected in CD4-positive T cells from patients with SLE [26]. This prompted us to examine the expression of autophagy markers on the PhenoCyclerTM platform. To identify the presence of immune responses, only the regions with lymphocyte infiltration were extracted and subjected to cellular analysis with autophagy markers LC3B and SQSTM1, as shown in Fig. 4A-D. It is widely accepted that the expression levels of LC3B and SQSTM1, which are key autophagy markers, are not only determined by their presence or absence but also by their intensity [27]. Therefore, we compared the expression levels of LC3B and SQSTM1 and found a correlation between the two (r = 0.69) (Fig. 4E). The average expression level of LC3B and SQSTM1 in the immune area was higher for secondary SS than for primary SS (Fig. 4F). These findings are consistent with those obtained from digital RNA counting analysis. Our results indicate that autophagy may be upregulated in immune areas infiltrated by lymphocytes in the salivary gland tissue, with secondary SS exhibiting higher expression of autophagy markers.
Fig. 4.
Autophagy marker expression in the area of lymphocytic foci. (A) Multiplex immunostaining image of a ten-color overlay of primary SS samples. (B) Corresponding cell-type map from the multiplex immunostaining image using the QuPath spatial analysis toolbox. The lymphocyte infiltration area is circled with a dotted line. (C) Multiplex immunostaining image of LC3B and SQSTM1. The lymphocyte infiltration area is circled with a dotted line. Scale bar: 100 μm. (D) Corresponding cell-type map of the autophagy markers LC3B and SQSTM1. The thin white line indicates the boundary between cells. (E) Correlation between the intensity of LC3B and SQSTM1 protein expression in lymphocyte infiltration areas. (F) Comparison of the expression levels of autophagy markers in primary SS and secondary SS, presented as a relative comparison with primary SS. **p < 0.01
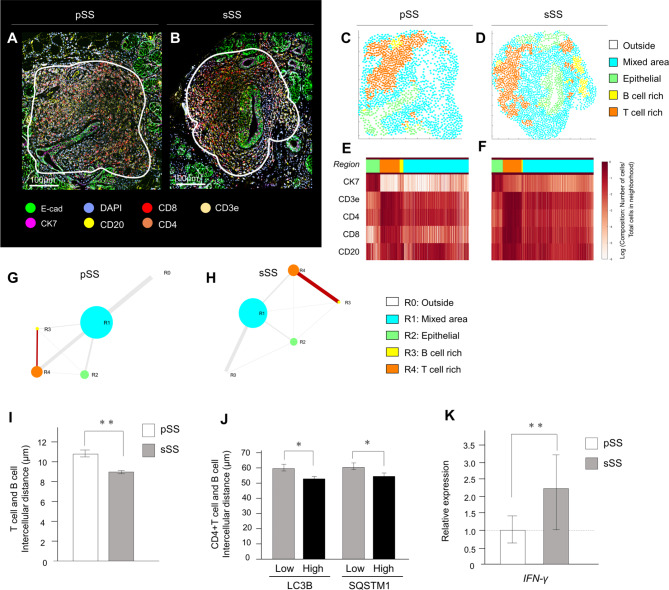
Comparison of the distance between T cells and B cells by cellular neighborhood analysis
To better understand the significance of autophagy marker-positive cells in the tissues, we performed neighborhood analysis, as shown in Fig. 5A and B. Based on the spatial arrangement of cell types within tissues, we predicted cell-cell interaction networks, where each cell in the dataset and its four nearest neighbors were identified using a distance map of cells. Neighborhood analysis [17] was used to determine the local composition of cells within a circular area of the tissue, with the size of the circle indicating the area occupied, and the thickness of the line indicating the distance in contact (Fig. 5C-F). A comparison of the distance between the T cell and the B cell-rich area between the primary SS and secondary SS revealed that the two areas were closer in proximity in secondary SS, with a greater distance between the two area boundaries. These results suggest closer proximity between the areas of T cell and B cell areas in secondary SS than in primary SS (Fig. 5G, H). Because the distance between the T- and B-cell regions was found to be close within the cell population, the intercellular distance was checked on a per-cell basis. The intercellular distance between T cells and B cells was 10.58 ± 0.38 μm in primary SS and 0.88 ± 0.21 μm in secondary SS, indicating that the distance between T and B cells in secondary SS was closer on a per-cell basis (Fig. 5I). Additionally, the distance between helper T cells and B cells in LC3B- and SQSTM1-positive areas significantly increased, as shown in Fig. 5J. To examine the relationship between helper T cells and B cells, we examined the expression of IFN-γ, which is secreted by helper T cells when activating B cells. Gene expression analysis (Fig. 5K) indicated that the upregulation of IFN-γ in the secondary SS might be a result of the activation of B cells by autophagy marker-positive helper T cells, thereby exacerbating tissue inflammation.
Fig. 5.
Comparison of distances between T cells and B cells via cellular neighborhood analysis. (A, B) Multiplex immunostaining images of ten-color overlay images, with the white-circled area in the lymphocytic foci area for analysis. Scale bar: 100 μm. (C, D) Four distinct cellular neighborhoods (CNs) were identified in SS samples: epithelial, B cell-rich, T cell-rich, and mixed. (E, F) The heatmap for marker expression in each area. (G, H) Schematic diagrams comparing the distances between the areas, with the distance between the four areas representing the length of each CN edge distance tangential to each other. The intercellular distance between the T and B cells was measured as the distance between the nuclei. (I) Interaction of T cells and B cells in the secondary SS was significantly closer than that in the primary SS. **p < 0.01. (J) The distances between CD4 + T cells and CD20 + B cells after dividing the intensity of autophagy marker expression between the two groups. *p < 0.05. (K) Difference in INF-γ expression by digital RNA analysis. **p < 0.01
Discussion
We conducted a retrospective analysis of FFPE samples and clinical symptoms in patients with SS, suggesting a deeper involvement of autophagy in secondary SS than in pSS tissues. Furthermore, in secondary SS samples, elevated expression of autophagy markers was predominantly found in immune cells. Helper T cells, which are particularly autophagic, were found in close proximity to B cells and appeared involved in their interaction, possibly resulting in increased cytokine expression that influenced the progression of SS. Regarding this, Alessandri et al. found that autophagy levels in the CD4-naive T cells of primary SS were already enhanced compared to healthy controls, as shown with LC3B (specifically LC3B-II) expression levels [28]. It has been known that the phosphatidylethanolamine-conjugated isoform of LC3B, LC3B-II, is localized to autophagosomes. Thus, the ratio of LC3B (LC3B-I)/LC3B-II was reported to be significantly higher in SS [28]. Immunofluorescence studies also indicated colocalization of autophagosome components ATG5 and LC3B-II (ATG8) in CD4-positive T cells from primary SS salivary glands [28, 29]. Histological analysis with transmission electron microscopy and autophagic flux analysis are needed to prove autophagy activity [30]. In the case of SLE patients, it was previously reported that the number of autophagosomes, examined by the amount of LC3B-II, significantly increased in CD4-positive T cells from patients, while T cells from the SLE patients were resistant to autophagic induction [26]. These findings strongly support the involvement of autophagy in the etiology of autoimmune diseases and support the results of the present study.
Another study showed the correlation between SS and autophagy, demonstrating that LAMP3 inhibits autophagy after autophagosome formation to induce cell death in a subset of patients with SS [31]. Our digital count RNA analysis demonstrated that secondary SS exhibited higher LAMP3 levels than primary SS (data not shown). It is therefore possible that lysosomal LC3B is mobilized in the cells of secondary SS patient tissues by inducing a lysosomal osmotic imbalance in the lysosomal membranes. A positive correlation between the EULAR Sjogren’s Syndrome Disease Activity Index and LC3B expression was also reported, which is consistent with the finding that LC3B expression is higher in secondary SS (Fig. 1F). Additionally, positive correlation of the immune cell infiltration area score with LC3B and ATG5 has been reported [26]. This study showed that LC3B and SQSTM1 were positive in areas of lymphocyte infiltration, suggesting that autophagic activity increases as tissue inflammation progresses.
Previous studies have reported that 45–50% of salivary gland-infiltrating cells are CD4-positive T cells, 20% are CD8-positive T cells, and 20% are B cells, indicating that T cells are important in early SS pathogenesis [32]. No difference in the percentage of immune cells was found between patients with SS with local symptoms and those with connective tissue disease-related complications. However, the present study demonstrated that B cells were activated by helper T cells, as judged from the close intercellular distance between helper T cells and B cells. Activation of the B cell response is considered a hallmark of SS, and may play a critical role in autoimmune response and lymphoproliferative processes in SS. B cell activation is accompanied by the production of anti-SS-A antibodies and an increased risk of lymphoma development through chronic activation of B cells [33, 34]. Antibody-producing B cells are known to circulate from tissues to the peripheral blood as short-lived plasma blasts [26]. This study analyzed only labial salivary gland samples, and therefore further blood examinations are necessary to understand the involvement of this mechanism in SS and other rheumatic diseases.
Although we focus on autophagy in this study, other cellular processes may also be involved in the progression of SS. For example, IL18RAP is an inflammatory response effector that has been reported to be involved in SS and other autoimmune diseases [35, 36]. Several reports have indicated that this gene is a novel target for therapeutic intervention in type 1 diabetes [37] and is dependent on reactive oxygen species (ROS) production [38]. Many reports have suggested the involvement of ROS in autoimmune diseases, and elevated expression of oxidative stress markers has been observed in the SS patients and animal models [39]. ROS act as a mediator between immune cells, thereby affecting antigen presentation, which is involved in T cell activation [40, 41]. Since there are also studies linking ROS and autophagy [27, 35], it is possible that autophagy is activated by IL18RAP and ROS. It is highly possible that each of these genes is related to the prognosis of patients with SS, but this requires further investigation.
Few studies have been carried out for elderly pSS patients, as the lower frequency of autoantibodies in elderly patients compared to younger pSS patients makes it difficult to diagnose SS by blood tests [42, 43]. On the contrary, this study was conducted in SS patients over 60 years of age to exclude the influence of sex hormones, and because most patients with xerostomia are elderly [44]. It has been reported that the early onset of SS is associated with more severe systemic disease [45]. The ability to predict the progression from SS to lymphoma is a particularly important area of ongoing research. In addition to Sjögren’s syndrome, minor salivary glands are known to be potential sites for tumorigenesis [46]. In recent years, spatial analysis has made great strides in oncology research. Techniques such as spatial transcriptomics are often utilized to understand the tumor microenvironment, the spatial distribution of cancer cells, and the interactions between different cell types. The methodologies employed in their study have been widely utilized across various fields of research.
Conclusion
The pathogenic mechanisms underlying SS may not be unique to this disease and rather be common in other autoimmune diseases. Patients with secondary SS were found to have more advanced tissue damage in minor salivary glands than those with primary SS. Notably, the activation of autophagy was found to be particularly relevant in CD4 + T cells, with a significant correlation between the expressions levels of autophagy-related genes and the histological severity of the disease. Since the level of autophagy in peripheral blood T lymphocytes positively correlated with patient disease activity and damage indices [47], we suggest the potential of autophagy-related molecules as useful biomarkers for monitoring disease progression and therapeutic responses.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are deeply grateful to Dr. Daisuke Ennishi, Dr. Tomida Shuta, and Dr. Tomohiro Urata (Okayama University Hospital Center for Comprehensive Genomic Medicine, Okayama, Japan) for insightful comments and suggestions regarding nCounter® analysis. We would like to express our gratitude to Dr. Natalie Atyeo (National Institute of Dental and Craniofacial Research, National Institutes of Health, MD, USA) for her invaluable assistance in proofreading the English content of this manuscript. We would like to offer special thanks to Mr. Naoya Hosono (BioStream Co., Ltd, Tokyo, Japan) for their support with the PhenoCycler™ analysis.
Abbreviations
- ACR
The American College of Rheumatology
- AQP5
Aquaporin5
- ATG5
Autophagy related 5
- E-cad
E-cadherin
- EULAR
European League Against Rheumatism
- FFPE
Formalin-fixed paraffin-embedded
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GO
Gene Ontology
- KRT7
Cytokeratin 7
- LC3A
Microtubule associated protein 1 light chain 3 alpha
- LC3B
Microtubule associated protein 1 light chain 3 beta
- LMD
Laser microdissection
- pSS
Primary SS
- RA
Rheumatoid arthritis
- ROS
Reactive oxygen species
- SIRT1
Sirtuin 1
- SLE
Systemic lupus erythematosus
- SQSTM1
Sequestosome 1
- SS
Sjögren’s syndrome
- sSS
Secondary SS
Author contributions
H.O.M. and T.N. wrote the main manuscript text. K.T., T.T., T.K., J.W., S. Ibaragi, S. Iida, T.Y., and H.N. provided resources. H.O. and T.S. supervised the work. All authors reviewed the manuscript.
Funding
This work was supported by a research fellowship from Japan Society for the Promotion of Science (https://www.jsps.go.jp), KAKENHI (grant number JP21K10093 to H.O.M., JP19KK0230 to H.O.M., JP 22KJ230305 to H.O.M.). The funders play no roles in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Institutional review board statement
The Institutional Ethical Review Board of the Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences and Okayama University Hospital approved the study protocol (approval number: No. 2105-035). All methods were performed in accordance with the relevant guidelines and regulations.
Informed consent
Informed consent was obtained from all subjects involved in the study. Written consent was not obtained because the data were analyzed anonymously.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hitomi Ono-Minagi, Email: hitomiono1216@gmail.com.
Hideyo Ohuchi, Email: ohuchi-hideyo@okayama-u.ac.jp.
References
- 1.Verstappen GM, Pringle S, Bootsma H, Kroese FGM. Epithelial-immune cell interplay in primary Sjogren syndrome salivary gland pathogenesis. Nat Rev Rheumatol. 2021;17(6):333–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umehara H, Okazaki K, Kawa S, Takahashi H, Goto H, Matsui S, et al. The 2020 revised comprehensive diagnostic (RCD) criteria for IgG4-RD. Mod Rheumatol. 2021;31(3):529–33. [DOI] [PubMed] [Google Scholar]
- 3.Retamozo S, Brito-Zeron P, Ramos-Casals M. Prognostic markers of lymphoma development in primary Sjogren syndrome. Lupus. 2019;28(8):923–36. [DOI] [PubMed] [Google Scholar]
- 4.Yadlapati S, Efthimiou P. Autoimmune/Inflammatory arthritis Associated Lymphomas: who is at risk? Biomed Res Int. 2016;2016:8631061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischbach M, Char D, Christensen M, Daniels T, Whaley K, Alspaugh M, Talal N. Immune complexes in Sjogren’s syndrome. Arthritis Rheum. 1980;23(7):791–5. [DOI] [PubMed] [Google Scholar]
- 6.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1–2):169–74. [DOI] [PubMed] [Google Scholar]
- 7.Yin H, Wu H, Chen Y, Zhang J, Zheng M, Chen G, et al. The therapeutic and pathogenic role of Autophagy in Autoimmune diseases. Front Immunol. 2018;9:1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian HR, Yang Y. Functional role of autophagy in gastric cancer. Oncotarget. 2016;7(14):17641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–7. [DOI] [PubMed] [Google Scholar]
- 10.Schultze JL, Buttner M, Becker M. Swarm immunology: harnessing blockchain technology and artificial intelligence in human immunology. Nat Rev Immunol. 2022;22(7):401–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arunachalam PS, Wimmers F, Mok CKP, Perera R, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyawaki K, Kato K, Sugio T, Sasaki K, Miyoshi H, Semba Y, et al. A germinal center-associated microenvironmental signature reflects malignant phenotype and outcome of DLBCL. Blood Adv. 2022;6(7):2388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alani H, Henty JR, Thompson NL, Jury E, Ciurtin C. Systematic review and meta-analysis of the epidemiology of polyautoimmunity in Sjogren’s syndrome (secondary Sjogren’s syndrome) focusing on autoimmune rheumatic diseases. Scand J Rheumatol. 2018;47(2):141–54. [DOI] [PubMed] [Google Scholar]
- 14.Fujibayashi T, Sugai S, Miyasaka N, Hayashi Y, Tsubota K. Revised Japanese criteria for Sjogren’s syndrome (1999): availability and validity. Mod Rheumatol. 2004;14(6):425–34. [DOI] [PubMed] [Google Scholar]
- 15.Shiboski CH, Shiboski SC, Seror R, Criswell LA, Labetoulle M, Lietman TM, et al. 2016 American College of Rheumatology/European League against Rheumatism Classification Criteria for Primary Sjogren’s syndrome: a Consensus and Data-Driven Methodology Involving Three International Patient cohorts. Arthritis Rheumatol. 2017;69(1):35–45. [DOI] [PMC free article] [PubMed]
- 16.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–25. [DOI] [PubMed] [Google Scholar]
- 17.Kuswanto W, Nolan G, Lu G. Highly multiplexed spatial profiling with CODEX: bioinformatic analysis and application in human disease. Semin Immunopathol. 2023;45(1):145–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black S, Phillips D, Hickey JW, Kennedy-Darling J, Venkataraaman VG, Samusik N, et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc. 2021;16(8):3802–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37(2):364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339(6123):1084–8. [DOI] [PubMed] [Google Scholar]
- 22.Shiozawa S, Tanaka Y, Shiozawa K. Single-blinded controlled trial of low-dose oral IFN-alpha for the treatment of xerostomia in patients with Sjogren’s syndrome. J Interferon Cytokine Res. 1998;18(4):255–62. [DOI] [PubMed] [Google Scholar]
- 23.Ono Minagi H, Sarper SE, Kurosaka H, Kuremoto KI, Taniuchi I, Sakai T, Yamashiro T. Runx1 mediates the development of the granular convoluted tubules in the submandibular glands. PLoS ONE. 2017;12(9):e0184395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oyelakin A, Horeth E, Song EC, Min S, Che M, Marzullo B, et al. Transcriptomic and Network Analysis of minor salivary glands of patients with primary Sjogren’s syndrome. Front Immunol. 2020;11:606268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ono Minagi H, Usami Y, Sakai M, Sakai T. Morphological differences between regenerating salivary glands after salivary gland duct ligation and embryonic salivary glands. Ann Anat. 2020;229:151482. [DOI] [PubMed] [Google Scholar]
- 26.Alessandri C, Barbati C, Vacirca D, Piscopo P, Confaloni A, Sanchez M, et al. T lymphocytes from patients with systemic lupus erythematosus are resistant to induction of autophagy. FASEB J. 2012;26(11):4722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colafrancesco S, Vomero M, Iannizzotto V, Minniti A, Barbati C, Arienzo F, et al. Autophagy occurs in lymphocytes infiltrating Sjogren’s syndrome minor salivary glands and correlates with histological severity of salivary gland lesions. Arthritis Res Ther. 2020;22(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alessandri C, Ciccia F, Priori R, Astorri E, Guggino G, Alessandro R, et al. CD4 T lymphocyte autophagy is upregulated in the salivary glands of primary Sjogren’s syndrome patients and correlates with focus score and disease activity. Arthritis Res Ther. 2017;19(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto H, Zhang S, Mizushima N. Autophagy genes in biology and disease. Nat Rev Genet. 2023;24(6):382–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshii SR, Mizushima N. Monitoring and measuring autophagy. Int J Mol Sci. 2017;18(9). [DOI] [PMC free article] [PubMed]
- 31.Tanaka T, Warner BM, Michael DG, Nakamura H, Odani T, Yin H, et al. LAMP3 inhibits autophagy and contributes to cell death by lysosomal membrane permeabilization. Autophagy. 2022;18(7):1629–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroese FG, Abdulahad WH, Haacke E, Bos NA, Vissink A, Bootsma H. B-cell hyperactivity in primary Sjogren’s syndrome. Expert Rev Clin Immunol. 2014;10(4):483–99. [DOI] [PubMed] [Google Scholar]
- 33.Ibrahem HM. B cell dysregulation in primary Sjogren’s syndrome: a review. Jpn Dent Sci Rev. 2019;55(1):139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nocturne G, Mariette X. B cells in the pathogenesis of primary Sjogren syndrome. Nat Rev Rheumatol. 2018;14(3):133–45. [DOI] [PubMed] [Google Scholar]
- 35.Myhr CB, Hulme MA, Wasserfall CH, Hong PJ, Lakshmi PS, Schatz DA, et al. The autoimmune disease-associated SNP rs917997 of IL18RAP controls IFNgamma production by PBMC. J Autoimmun. 2013;44:8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F. Interleukin–18 binding protein: Biological properties and roles in human and animal immune regulation (review). Biomed Rep. 2024;20(6):87. 10.3892/br.2024.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma J, Lam IKY, Lau CS, Chan VSF. Elevated Interleukin-18 receptor accessory protein mediates enhancement in reactive oxygen species production in neutrophils of systemic Lupus Erythematosus patients. Cells. 2021;10(5). [DOI] [PMC free article] [PubMed]
- 38.Chang KC, Liu PF, Chang CH, Lin YC, Chen YJ, Shu CW. The interplay of autophagy and oxidative stress in the pathogenesis and therapy of retinal degenerative diseases. Cell Biosci. 2022;12(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redza-Dutordoir M, Averill-Bates DA. Interactions between reactive oxygen species and autophagy: special issue: death mechanisms in cellular homeostasis. Biochim Biophys Acta Mol Cell Res. 2021;1868(8):119041. [DOI] [PubMed] [Google Scholar]
- 40.Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22(3):377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondo M, Kumagai S, Nishikawa H. Metabolic advantages of regulatory T cells dictated by cancer cells. Int Immunol. 2024;36(2):75–86. [DOI] [PubMed] [Google Scholar]
- 42.Tishler M, Yaron I, Shirazi I, Yaron M. Clinical and immunological characteristics of elderly onset Sjogren’s syndrome: a comparison with younger onset disease. J Rheumatol. 2001;28(4):795–7. [PubMed] [Google Scholar]
- 43.Goules AV, Argyropoulou OD, Pezoulas VC, Chatzis L, Critselis E, Gandolfo S, et al. Primary Sjogren’s syndrome of early and late onset: distinct clinical phenotypes and Lymphoma Development. Front Immunol. 2020;11:594096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ono Minagi H, Yamanaka Y, Sakai T. Evaluation of the Saxon test for patients with hyposalivation without Sjogren’s syndrome. J Oral Rehabil. 2020;47(12):1550–6. [DOI] [PubMed] [Google Scholar]
- 45.Anquetil C, Hachulla E, Machuron F, Mariette X, Le Guern V, Vittecoq O, et al. Is early-onset primary Sjogren’s syndrome a worse prognosis form of the disease? Rheumatology (Oxford). 2019;58(7):1163–7. [DOI] [PubMed] [Google Scholar]
- 46.Aldelaimi AA, Enezei HH, Aldelaimi TN, Mohammed KA, Al-Ani RM. Salivary gland diseases: a retrospective clinicopathological study of 159 cases. Cureus. 2022;14(9):e29589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan M, Zhang QB, Liu TH, Yang YY, Zheng JX, Zhou WJ, et al. Autophagy dysfunction may be involved in the pathogenesis of ankylosing spondylitis. Exp Ther Med. 2020;20(4):3578–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.