Abstract
Simian virus 40 large T antigen is a multifunctional oncoprotein that is required for numerous viral functions and the induction of cellular transformation. T antigen contains a J domain that is required for many of its activities including viral DNA replication, transformation, and virion assembly. J-domain-containing proteins interact with Hsc70 (a cellular chaperone) to perform multiple biological activities, usually involving a change in the conformation of target substrates. It is thought that Hsc70 associates with T antigen to assist in performing its numerous activities. However, it is not clear if T antigen binds to Hsc70 directly or induces the binding of Hsc70 to other T-antigen binding proteins such as pRb or p53. In this report, we show that T antigen binds Hsc70 directly with a stoichiometry of 1:1 (dissociation constant = 310 nM Hsc70). Furthermore, the T-antigen–Hsc70 complex formation is dependent upon ATP hydrolysis at the active site of Hsc70 (ATP dissociation constant = 0.16 μM), but T-antigen–Hsc70 complex formation does not require nucleotide hydrolysis at the T-antigen ATP binding site. N136, a J domain-containing fragment of T antigen, does not stably associate with Hsc70 but can form a transient complex as assayed by centrifugation analysis. Finally, T antigen does not associate stably with either of two yeast Hsc70 homologues or an amino-terminal fragment of Hsc70 containing the ATPase domain. These results provide direct evidence that the T-antigen–Hsc70 interaction is specific and that this association requires multiple domains of both T antigen and Hsc70. This is the first demonstration of a nucleotide requirement for the association of T antigen and Hsc70 and lays the foundation for future reconstitution studies of chaperone-dependent tumorigenesis induced by T antigen.
Simian virus 40 large T antigen is a multifunctional, multidomain protein that is the focus of intense study as an effector for neoplastic transformation, DNA replication and other molecular processes (for reviews see references 3, 9, and 32). The mechanism by which a single protein can perform so many functions is enigmatic. Recently it was demonstrated that T antigen is a molecular chaperone protein that contains a functional J domain (5, 22, 37). The J domain is essential for multiple functions of T antigen, including transformation (37), induction of increased cellular division (38), inhibition of apoptosis (36), release of Rb/E2F family complexes (39, 44), and viral DNA replication (30). Thus, it has been proposed that the action of the J domain, combined with the ability to dock various substrates, including Rb family members (pRb, p107, p130) and p53, can account at least in part for the multiple activities of T antigen (3, 37).
J-domain-containing proteins (J proteins) bind to partner DnaK-homologue chaperones (DnaKs), stimulating the ATPase activity of the DnaKs. When DnaK hydrolyzes ATP, it can change the conformation of its bound substrate to perform a number of functions, including protein folding and unfolding (17), protein import and export across the endoplasmic reticulum and mitochondrial membranes (2), and the disruption of multiprotein complexes (1, 39, 45). The prototypic mammalian DnaK is Hsp70, which is induced during cellular stress and prevents illicit protein aggregation, as well as assisting in the refolding of denatured proteins (17). Hsc70 consists of an amino-terminal ATPase domain, as well as a carboxyl-terminal peptide binding domain. Hsc70 is highly similar to Hsp70 at the amino acid level and is thought to be the constitutively expressed, functional equivalent of Hsp70 (17). In Saccharomyces cerevisiae, 14 different DnaK homologous proteins are known to exist, and it is thought that there are as many or more in mammalian cells (33).
Nuclear magnetic resonance and biochemical analysis has demonstrated that J proteins interact with the ATPase domain of DnaK proteins through a conserved HPD loop (4), and structure-function analysis has demonstrated that mutation of any of these three residues abolishes the functional interaction between J proteins and DnaKs (21, 41). Consistent with these observations, mutation of the HPD loop in T antigen renders it nonfunctional for multiple activities (3, 5, 37, 39, 44). It should be noted that whereas the amino-terminal ATPase domain of Hsc70 is required for association with J-domain-containing proteins, at least in some circumstances, regions of Hsc70 outside the ATPase domain (including its carboxyl-terminal EEVD motif of the peptide binding domain) are also important for interaction with J proteins (8, 12).
While the essential nature of the J-domain chaperone function of T antigen is clearly established for altering some cellular growth control mechanisms and viral functions, it remains undetermined which DnaK homologue(s) T antigen associates with to perform these functions. Several studies have demonstrated that in the context of a cellular lysate, the T-antigen–Hsc70 complex can be isolated and the association of the components of this complex requires a functional J domain (5, 34, 35, 40). However, it has also been well documented that at least two T-antigen cellular binding targets, pRb and p53, also bind to Hsc70 (11, 18, 29). Therefore, another plausible hypothesis is that T antigen does not directly associate with Hsc70, but rather the T-antigen–Hsc70 association is indirect and mediated by pRb, p53, or other T-antigen binding proteins.
This study seeks to understand better the interaction of T antigen with Hsc70. The results show that T antigen associates with Hsc70 directly, and the stoichiometry of binding is 1:1. This association requires ATP binding and ATP hydrolysis by Hsc70, but not by T antigen. Two yeast DnaK homologues fail to efficiently form the stable ATP-dependent complex, suggesting that the T-antigen–Hsc70 association is specific for the mammalian Hsc70 chaperones. The T-antigen–Hsc70 interaction is J-domain dependent, but surprisingly N136, an amino-terminal fragment containing the J domain, is not sufficient for complex formation, implying that regions of T antigen more carboxyl terminal to the J domain are also required for the stable association of T antigen and Hsc70. Sedimentation velocity centrifugation analysis demonstrates that N136 can, however, transiently associate with Hsc70. Finally, it is shown that the ATPase domain of Hsc70 is not sufficient for T-antigen–Hsc70 complex formation. These data imply that domains carboxy terminal to the ATPase domain are required for the stable association of Hsc70 and T antigen. The results presented here reveal an elaborate molecular machine that is fueled by ATP turnover to drive the in vivo functions of T antigen as an effector for neoplastic transformation, DNA replication, and virion assembly.
MATERIALS AND METHODS
Buffers.
Buffer I contains 20 mM HEPES at pH 7.8 with NaOH, 6 mM MgCl2, 40 mM KCl, and 0.1% NP-40. PBS at pH 7.3 contains 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl.
Protein purification.
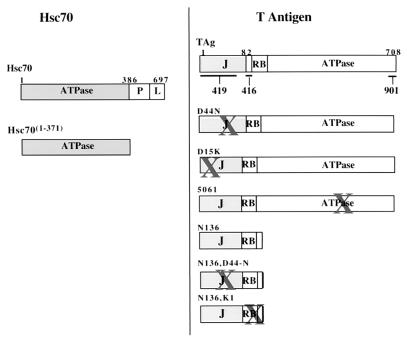
Expression of T antigen and mutants of T antigen (Fig. 1) have been described previously (30, 37). 5061 contains an insertional mutation at Gly431 with Ala-Leu-Glu and has been described (7, 10, 31). Mutant D44N (5110) harbors a single amino acid substitution in the conserved HPD loop of the J domain, and N136 encodes a truncated amino-terminal fragment made by inserting a stop codon after amino acid (aa) 136 of wild-type T antigen (37). A recombinant baculovirus that expresses the wild-type T antigen (Autographa californica nuclear polyhedrosis virus 941T) and a baculovirus transfer plasmid containing the wild-type T-antigen gene (pVL941T) were kindly provided by Robert Lanford (Southwest Foundation for Biomedical Research) and have been described previously (25). Baculovirus transfer plasmids pVL941-N136 and -5061 were generated as described for pVL941T. Baculoviruses were constructed as previously described (7).
FIG. 1.
Summary of T antigen and Hsc70 proteins and sites of epitopes for anti-T antigen antibodies. The left panel corresponds to the Hsc70 constructs used in this study and the right panel corresponds to the T antigen constructs used. X indicates the area of targeted mutation. The position of the epitopes for anti-T antigen antibodies, PAb419, PAb416, and 901, are indicated on the wild-type T antigen. J denotes the position of the J domain, while RB indicates the position of the LXCXE Rb family binding motif. The carboxyl-terminal T-antigen ATPase and amino-terminal Hsc70 ATPase domains are indicated.
Wild-type and mutant T antigens were purified essentially as described (6) except that the gel filtration step was eliminated. Purification of Ssalp, BiP, and BiP(1–386) has been described (28). Purified bovine brain and recombinant Hsc70, as well as Hsc70(1–386) were purchased from StressGen Biotechnologies, Victoria, British Columbia, Canada.
All proteins used in the experiments were >95% pure as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue R-250. Protein concentration was determined by the Bradford method (Bio-Rad, Hercules, Calif.) with bovine serum albumin as the standard.
Antibodies.
The T-antigen-specific polyclonal antibodies (PAbs) PAb416 (which recognizes an epitope between aa 91 and 95), and PAb419 (which recognizes an epitope between aa 1 and 82) have been described previously (28). Anti-T-antigen antibody 901 recognizes an epitope in the last carboxyl-terminal 30 aa of T antigen (aa 684 to 698) and was kindly provided by Judith Tevethia (The Pennsylvania State University Medical School, Hershey). Antibodies were expressed in hybridoma cells, and the media were collected. The media were passed over a protein G column and eluted using a glycine solution at pH 2.5 into a Tris neutralization buffer of pH 9.6. Fractions most enriched for antibody were pooled and dialyzed into PBS.
Immunoprecipitation of T-antigen–Hsc70 complex.
Unless otherwise noted, 1 μg of T antigen or mutants of T antigen were incubated with Hsc70 (3 μg) in the presence or absence of MgATP with an ATP regeneration system (50 μM GDP manose, 40 μM creatine phosphate, 0.2 mg of phosphocreatine kinase per ml) in a volume of 100 μl of buffer I. The reaction mixtures were then incubated with 2 to 3 μg of appropriate antibody for 30 min at 22°C in the presence of 50 μl of a 50% slurry of protein A-Sepharose CL-4B beads (Pharmacia Biotech Inc., Uppsala, Sweden) in buffer I to immunoprecipitate T antigen. The reaction mixtures were captured via a 20-s spin in a microcentrifuge at 16,000 × g and washed three times with 1 ml of PBS. The final pellets were resuspended in 2× Laemmli sample buffer containing the reducing agents dithiothreitol and β-mercaptoethanol and heated at 100°C for 5 min. These samples were then loaded onto an 8% acrylamide SDS gel and electrophoresed. The Coomassie blue-stained gels were analyzed using an Astra 600S digital scanner (Umax Technologies Inc., Fremont, Wash.) and quantified using NIH Image software (version 2.1). T antigen and Hsc70 showed a linear increase in Coomassie blue staining intensity as a function of protein concentration.
Sedimentation velocity centrifugation.
The interaction of wild-type or mutant T antigen (600 nM) and bovine brain Hsc70 (500 nM) was evaluated by incubating these proteins for 30 min in the presence of 1 mM MgATP at 22°C in buffer I. The reaction solution (60 μl) was then layered onto a 5 to 15% linear glycerol gradient in buffer I and centrifuged at 137,000 × g for 60 min at 10°C on a Beckman TLS-55 rotor. The resulting gradients were fractionated (12-μl aliquots) from top to bottom (least to most dense), which was followed by SDS-PAGE analysis. The Coomassie blue-stained gels were scanned, quantified using NIH Image software (version 2.1), and graphed based on the percent total protein collected in each fraction.
Data analysis.
The T-antigen–Hsc70 equilibrium binding data were fitted to one of the following equations by a nonlinear least-squares method using KaleidaGraph software (Synergy Software, Reading, Pa.). The binding stoichiometry of T antigen and Hsc70 (Fig. 2A) was determined by plotting the data as Hsc70 concentration that immunoprecipitated with T antigen, using anti-T-antigen antibodies (PAb416) as a function of total Hsc70 concentration. The analysis assumes a single binding site on Hsc70 for each T-antigen molecule, and Hsc70 concentrations were as low as the T-antigen concentration used in the experiment. Therefore, the data were fitted to the following quadratic equation:
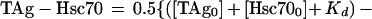 |
1 |
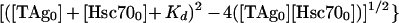 |
where TAg-Hsc70 is the concentration of Hsc70 bound to T antigen; TAg0 is the total T-antigen concentration, Hsc700 is the total Hsc70 concentration; and Kd is the dissociation constant. Figure 2B shows the ATP concentration dependence of TAg-Hsc70 complex formation, and these data were fitted to the following quadratic equation:
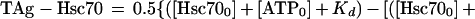 |
2 |
 |
The data in Fig. 2C were plotted as the fraction of Hsc70 partitioning as the TAg-Hsc70 complex as a function of ATPγS concentration. The concentration of MgATP used in the experiment, 50 μM, is significantly higher than the Kd of ATP (Kd,ATP), 0.16 μM (Fig. 2B), and suficiently high to lead to maximal TAg-Hsc70 complex formation. Furthermore, 50 μM ATP is significantly higher than the concentrations of Hsc70 (0.3 μM) and T antigen (0.3 μM) used in the experiment. Therefore, these data were fitted to a hyperbola to obtain the K1/2,ATPγS in the presence of 50 μM MgATP. The apparent Kd,ATPγS was obtained from the following equation:
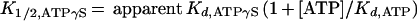 |
3 |
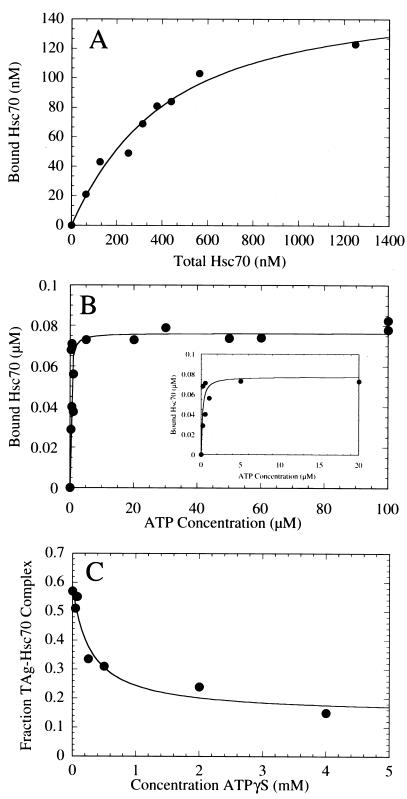
FIG. 2.
T-antigen–Hsc70 complex formation. (A) Saturation binding of Hsc70 to T antigen. This graph presents the quantification of the Hsc70 concentration dependence seen in the Coomassie blue-stained gel shown in Fig. 3C. T antigen at 120 nM was incubated with Hsc70 (0 to 1,250 nM) in the presence of 1 mM MgATP plus an ATP regeneration system as described in Materials and Methods. The concentration of Hsc70 partitioning with T antigen was plotted as a function of the total Hsc70 concentration. The fit of the data to the quadratic equation (equation 1) yields the apparent Kd,Hsc70 310 ± 60 nM, with maximum binding at 159 ± 13.0 nM Hsc70. (B) ATP-dependent association of Hsc70 with T antigen. The gel in Fig. 3A was quantified, and the association of Hsc70 (300 nM) with T antigen (120 nM) was plotted as a function of MgATP concentration. The data were fitted to the quadratic equation (equation 2) which provides the apparent Kd, ATP, 0.16 ± 0.07 μM, with maximum binding at 76 ± 4.5 nM Hsc70. Only the data from 0.2 to 100 μM MgATP are shown, and the inset presents the data from 0 to 20 μM MgATP. (C) Inhibition by ATPγS of the ATP-induced T-antigen–Hsc70 association. T-antigen–Hsc70 complex formation (300 nM T antigen, 300 nM Hsc70) was initiated in the presence of 50 μM MgATP plus increasing concentrations of MgATPγS (0 to 4,000 μM). The T-antigen–Hsc70 complexes were immunoprecipitated with anti-T-antigen antibody PAb416 and quantified. The fraction of T-antigen–Hsc70 complex was plotted as a function of ATPγS concentration, and the fit of the data to a hyperbola provides the K0.5,ATPγS, 286 ± 132 μM ATPγS. The apparent Kd, ATPγS P is 0.91 μM (equation 3).
RESULTS
To investigate the association of T antigen with Hsc70, purified Hsc70 and T-antigen wild-type and mutant proteins were obtained as described in Materials and Methods (Fig. 1). The purification of these proteins has been reported previously (28, 30, 37), and all proteins were purified to >95% homogeneity as determined by Coomassie blue staining (data not shown). Of the potential T-antigen partner DnaK homologues, Hsc70 was chosen for the following reasons: (i) T antigen can bind Hsc70 in the context of a cellular lysate (5, 34, 35, 40), (ii) T antigen stimulates ATP hydrolysis by Hsc70 (37), and (iii) Hsc70 enhances T-antigen-mediated disruption of Rb/E2F family complexes in vitro (39).
T antigen binds directly to Hsc70.
T antigen was incubated with Hsc70 in the presence of 1 mM MgATP and an ATP regeneration system or in the absence of nucleotide to evaluate the ATP requirement for T-antigen–Hsc70 association. These reactions were then immunoprecipitated using anti-T-antigen antibodies, and the bound immunocomplexes were visualized by SDS-PAGE with Coomassie blue staining (Fig. 3). We hypothesized that if Hsc70 were in fact a DnaK binding partner of T antigen, then their association should be ATP dependent as reported for other J-DnaK protein interactions (19). In the absence of nucleotide, T antigen was efficiently immunoprecipitated as expected, but no Hsc70 was detected (Fig. 3A, lanes 2 and 3). If, however, 1 mM MgATP and an ATP regeneration system were included in the reaction mixture, then both T antigen and Hsc70 were coprecipitated (Fig. 3A, lane 4). As a negative control, an irrelevant antibody (PAb240 anti-p53) was incubated in the reaction, and association of neither Hsc70 nor T antigen was detected in the immunoprecipitate (data not shown).
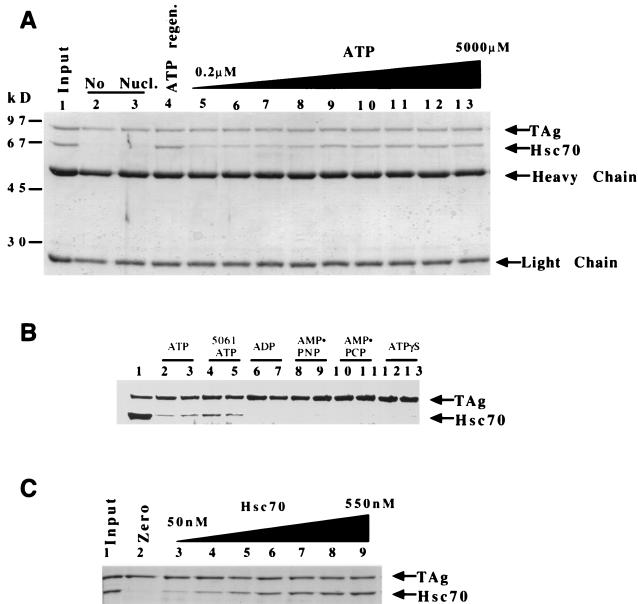
FIG. 3.
Hsc70 binding to T antigen requires ATP hydrolysis and is concentration dependent. (A) T antigen (120 nM) and Hsc70 (300 nM) were incubated and then immunoprecipitated for T antigen. A Coomassie blue-stained gel shows associated proteins. Results for 100% input of anti-T-antigen PAb416 and 50% input of T antigen and Hsc70 are shown (lane 1). Reactions containing no ATP (lanes 2 and 3), an ATP regeneration system (1 mM) (lane 4), or increasing amounts of ATP (0.2 to 5,000 μM) are shown in order (lanes 5 to 13) 0.2, 0.5, 1.0, 5.0, 50, 100, 500, 1,000, 5,000 (μM), respectively. Heavy chain and light chain refer to the antibody that coprecipitates in the immunoprecipitation reaction. (B) Two concentrations (1 or 4 mM) of ATP (lanes 2 and 3), ADP (lanes 6 and 7), AMP-PNP (lanes 8 and 9), AMP-PCP (lanes 10 and 11), and ATPγS (lanes 12 and 13) were incubated with T antigen and Hsc70, and the reactions were immunoprecipitated for T antigen with PAb416 as described in Materials and Methods. Lanes 4 and 5 show the reaction with 5061, a T-antigen ATP binding mutant, incubated with ATP (1 and 4 mM, respectively). (C) Hsc70 concentration dependence. T antigen (120 nM) was incubated with Hsc70 (0 to 550 nM). Lane 1, 50% input of T antigen and 1 μg of Hsc70 protein; lanes 2 to 9; Hsc70 at 0, 50, 110, 230, 290, 340, 400, and 550 nM, respectively.
To explore further the ATP dependence of this association, we incubated T antigen and Hsc70 in the presence of increasing ATP concentrations. An increase in Hsc70 association with T antigen was observed as a function of the ATP concentration used in the reactions (Fig. 3A lanes 5 to 13). We conclude that the association of T antigen with Hsc70 is specific and ATP dependent.
In order to determine if nucleotide binding is sufficient to stimulate the association of Hsc70 with T antigen, or whether ATP hydrolysis is required, the experiment was repeated with the addition of ADP or one of three different slowly hydrolyzable or nonhydrolyzable ATP analogs (MgADP, MgAMP-PNP, MgAMP-PCP, MgATPγS) at two different concentrations (1 or 4 mM). Each failed to induce Hsc70 association with T antigen (Fig. 3B, lanes 6 to 13), yet wild-type T antigen was shown to efficiently associate with Hsc70 in the presence of MgATP (Fig. 2B, lanes 2 and 3). These results indicate that ATP hydrolysis is required for T-antigen–Hsc70 complex formation.
A mutant of T antigen (5061) defective for ATP hydrolysis (7) was examined to determine whether the ATP requirement for complex formation is due to the catalytic activity of T antigen or Hsc70. When incubated with Hsc70 in the presence of 1 mM MgATP and the ATP regeneration system (Fig. 3B, lanes 4 and 5), Hsc70 was immunoprecipitated with the T-antigen 5061 mutant. Note that the association of Hsc70 with 5061 is as robust as its association with wild-type T antigen under the same experimental conditions (Fig. 3B, lanes 2 and 3), yet no ATP hydrolysis can occur at the T-antigen nucleotide-binding site. These results indicate that the ATPase requirement for T-antigen–Hsc70 complex formation is associated with Hsc70 rather than T antigen.
To determine the stoichiometry of the T-antigen–Hsc70 complex, T antigen (120 nM) was incubated with increasing concentrations of Hsc70 (Fig. 3C) in the presence of 1 mM MgATP and the ATP regeneration system. As the concentration of Hsc70 was increased, an increasing concentration of Hsc70 coprecipitated with T antigen (Fig. 3C, lanes 3 to 9). The concentration of both Hsc70 and T antigen immunoprecipitated by T-antigen antibodies was quantified from the gel shown in Fig. 3C, as well as two other independent experiments. These data are presented in Fig. 2A. The fit of the data to equation 1 provides a Kd,Hsc70 of 310 nM, with maximum binding at 160 nM Hsc70. These results indicate that the stoichiometry of binding is 1:1, because the predicted maximum binding concentration of Hsc70 is close to the input concentration of T antigen (120 nM) that was included in the reaction mixture.
The ATP-dependent association of T antigen and Hsc70 was quantified from the gel presented in Fig. 3A and from another independent experiment. The concentration of Hsc70 that immunoprecipitated with T antigen increased as a function of ATP concentration (Fig. 2B and 3A), and the Kd, ATP, 0.16 μM, implies a relatively tight affinity with nucleotide. Furthermore, we observe the maximal Hsc70 partitioning as the T-antigen–Hsc70 complex at 76 nM (Fig. 2B; equation 2), which is consistent with the Kd,Hsc70 at 310 nM (Fig. 2A). Because the concentration of Hsc70 (300 nM) is similar to the Kd,Hsc70, (310 nM) only 76 nM T antigen of the 120 nM T antigen (∼60%) is complexed with Hsc70.
To explore further the nucleotide dependence of T-antigen–Hsc70 association, we incubated the T-antigen–Hsc70 complex (120 nM T antigen, 300 nM Hsc70) with increasing amounts of the slowly hydrolyzable ATP analog, ATPγS, in the presence of 50 μM MgATP (Fig. 2C). The results show that the fraction of T-antigen–Hsc70 complex immunoprecipitated by T-antigen antibodies decreased as a function of ATPγS concentration, indicating that ATPγS binds the active site of Hsc70 and competes with ATP. The apparent Kd,ATPγS in the presence of 50 μM ATP is 0.91 μM ATPγS (Fig. 2C; equation 3). Furthermore, the observation that T-antigen–Hsc70 complex formation decreases as a function of ATPγS implies that ATP hydrolysis in addition to nucleotide binding is required to trap the T-antigen–Hsc70 complex.
J domains mediate direct contact with Hsc70 in multiple biological systems (4, 17). A J-domain mutant of T antigen was assayed for the ability to associate with Hsc70 to test if the direct association between Hsc70 and T antigen is dependent on a functional J domain. D44N is a point mutant in the highly conserved HPD loop of the J domain that is essential for contact with Hsc70 in bacterial and mammalian J-protein–DnaK interactions (3). D44N fails to associate with Hsc70 even when one- to threefold higher concentrations of D44N relative to wild-type protein are included in the association assay (Fig. 4A, lanes 6 to 8). D15K, another point mutant in the amino terminus that is functional for J domain activity, associates with Hsc70 as well as wild-type T antigen (data not shown). We conclude that an intact J domain is required for the direct association of Hsc70 with T antigen.
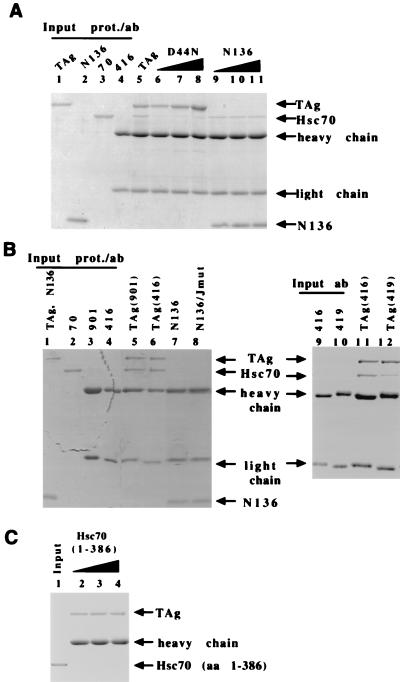
FIG. 4.
Direct T-antigen–Hsc70 association requires multiple domains of both T antigen and Hsc70. (A) The J domain point mutant D44N and truncated T-antigen N136 fail to bind to Hsc70. Increasing amounts of D44N (120, 240, and 360 nM [lanes 6 to 8, respectively]) or N136 (600, 1,200, and 1,800 nM [lanes 9 to 11, respectively]) were incubated with Hsc70 and immunoprecipitated with anti-T-antigen antibody PAb416. The Coomassie blue-stained SDS-PAGE gel is shown. A positive-control reaction with wild-type T antigen (120 nM [lane 5]) is shown. Results for 50% input of Hsc70 (lane 3) or 100% input of PAb416 (lane 4) are shown. One microgram of T antigen (lane 1) and N136 (lane 2) are shown for migration markers. (B) T antigen and Hsc70 were incubated and immunoprecipitated with anti-T antigen PAb416 (lanes 7 and 11), anti-T antigen 901 (lane 5), or anti-T antigen PAb419 (lane 12). Mutants N136 (lane 7) and N136/D44N (lane 8) were incubated with Hsc70 and immunoprecipitated with PAb416. All reactions were conducted in the presence of an ATP regeneration system. Results for 50% input of T antigen and N136 (lane 1) and Hsc70 (lane 2) are shown. Results for 100% input of antibodies 901 (lane 3), PAb416 (lanes 4 and 9), and PAb419 (lane 12) are also shown. (C) Hsc70(1–386) at 300, 600, and 1,300 nM (lanes 2 to 4, respectively) was incubated with T antigen in the presence of 1 mM MgATP. Hsc70(1–386) (300 nM) is shown as a migration marker (lane 1). Abbreviations: prot., protein; ab, antibody; TAg, T antigen. Heavy chain and light chain refer to the antibody that coprecipitates in the immunoprecipitation reaction.
T-antigen J domain is not sufficient to stably bind Hsc70.
Because the J domain is required for T-antigen–Hsc70 complex formation, we next evaluated whether the J domain is sufficient for complex formation. N136 is an amino-terminal fragment of T antigen that contains the J domain and Rb binding motif (37). N136 retains some of the biological activities of the full-length molecule, including the abilities to associate with pRb and to stimulate the ATPase activity of Hsc70 (37). When incubated with Hsc70 in the presence of the ATP regeneration system, N136 is defective for association with Hsc70 (Fig. 4B, lane 7). Multiple preparations of N136 and N136 mutants defective for Rb binding and J-domain activity are also completely defective for association with Hsc70 (Fig. 4B, lane 8; Table 1). Even when N136 is incubated at a molar excess of 10-fold (Fig. 4A, lane 10) more than the wild-type T-antigen positive-control reaction (Fig. 4A, lane 5), N136 binds to only 23% of the amount of Hsc70 that binds to wild-type T antigen (shown is the conservative gel of multiple experiments; at molar concentrations of N136 greater than 1500 nM the amount of antibody becomes limiting [Fig. 4A, lane 11]). Taken together, the above data suggest that the J domain is required but not sufficient for T-antigen–Hsc70 complex formation.
TABLE 1.
Mutant T antigen association with Hsc70a
| T antigen protein | Binds Hsc70 + ATP | Binds Hsc70 − ATP | Binds Hsc70(1–371) + ATP |
|---|---|---|---|
| TAg | + | − | − |
| D44N | − | − | NDb |
| D15K | + | − | ND |
| 5061 | + | − | ND |
| N136 | − | − | − |
| N136, D44N | − | − | ND |
| N136, K1 | − | − | ND |
Purified T antigen or mutants of T antigen were incubated in the presence (+) or absence (−) of MgATP with Hsc70 or the ATPase fragment of Hsc70 (Hsc70(1–371)). These reactions were then immunoprecipitated with antibodies to T antigen, and the ability to coimmunoprecipitate Hsc70 was determined.
ND, not determined.
Three different anti-T-antigen antibodies were used in separate reactions of the T-antigen–Hsc70 immunoprecipitation binding assay to discard the possibility that the antibody was modulating the efficiency of this reaction. Antibody PAb416 recognizes an amino-terminal epitope between amino acids 91 and 95 (16, 27), PAb419 recognizes an amino-terminal epitope in the region of the J domain between amino acids 1 and 82 (16, 27), and antibody 901 recognizes a carboxyl-terminal epitope in the last 30 aa between amino acids 684 and 698 (Materials and Methods). Antibody PAb419 has also been shown to alter the conformation of the carboxyl terminus of T antigen (23). When PAb416 or antibody 901 was incubated in the immunoprecipitation reaction mixture in the presence of an ATP regeneration system, similar amounts of both T antigen and Hsc70 are coimmunoprecipitated (Fig. 4B, lanes 5 and 6). If, however, PAb419 is used in the immunoprecipitation reaction, the amount of T antigen immunoprecipitated is unchanged, but a marked reduction in the amount of Hsc70 that coprecipitates is observed. This experiment was performed five times, and each time PAb419 immunoprecipitated as much or more T antigen than PAb416. The amount of Hsc70 that was coprecipitated with PAb419 ranged from 19 to 45% of the amount that precipitated with PAb416, with the average being 28%. Shown is a representative gel (Fig. 4B [compare lanes 11 and 12]). We conclude that two unique antibodies that recognize epitopes of T antigen in either the amino-terminal or carboxyl-terminal portions of the protein successfully precipitate a T-antigen–Hsc70 complex. However, PAb419 inhibits the ability of T antigen to associate with Hsc70.
Hsc70 ATPase domain is not sufficient to bind T antigen.
Several studies have shown that even though the amino-terminal ATPase domain of Hsc70 is essential for association with J proteins, the carboxyl-terminal domains of Hsc70 may contribute to the interaction (8, 12, 20). To determine if the ATPase domain is sufficient to bind to T antigen, increasing amounts of a 386-aa fragment (Hsc701–386) that consists of the amino-terminal ATPase domain of Hsc70 was incubated with T antigen. No association is observed between Hsc70(1–386) and T antigen (Fig. 4C, lanes 2 to 4), even when a fourfold-higher molar concentration of Hsc70(1–386) over the concentration at which wild-type Hsc70 readily associates with T antigen was used.
Because Hsc70(1–386) is a recombinant Hsc70 expressed in Escherichia coli, and all of the other experiments thus far have used Hsc70 purified from bovine brain, we tested a full-length recombinant Hsc70 that was expressed in E. coli and purified in the same manner as Hsc70(1–386). No difference between the recombinant and the endogenous Hsc70s was detected (data not shown). Therefore, we conclude that the ATPase domain of Hsc70 is not sufficient to promote T-antigen–Hsc70 complex formation.
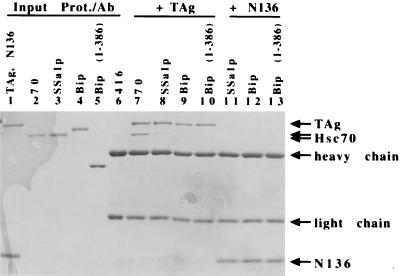
Different yeast DnaK homologues were incubated with T antigen or N136 in the presence of an ATP regeneration system to test the specificity of the stable interaction between Hsc70 and T antigen. Ssalp (a yeast cytosolic Hsc70 homologue), BiP (an endoplasmic reticulum Hsc70 homologue), and the ATPase domain of BiP (BiP1–386) were tested. Even though approximately equal molar amounts of mammalian or yeast Hsc70 proteins were included in the assay to measure association, mammalian Hsc70 bound to T antigen with the greatest affinity (Fig. 5 compare lane 7 with lanes 8 to 10). Ssalp association with T antigen was detected, but at <5% of mammalian Hsc70 (Fig. 5, compare lanes 7 and 8). Very little (approximately 1%) association between BiP or BiP(1–386) and T antigen was detected (Fig. 5, lanes 9 and 10). Furthermore, consistent with our previous results, no association between N136 and any of the yeast Hsc70 constructs was detected (Fig. 5, lanes 11 to 13). These data suggest that there is specificity to the stable interaction of T antigen with mammalian Hsc70 versus other DnaK homologues.
FIG. 5.
T-antigen binding to Hsc70 is specific. Anti-T-antigen antibody (PAb416) was used to immunoprecipitate reactions including T antigen (TAg) (1 μg) and 3 μg of Hsc70 (lane 7), Ssa1p (lane 8), BiP (lane 9), or BiP(1–386) (lane 10). An amino-terminal fragment of T antigen, N136 (1 μg), was incubated with 3 μg of Ssa1p (lane 11), BiP (lane 12), or BiP(1–386) (lane 13). Results for 50% input for T antigen and N136 (lane 1), Hsc70 (lane 2), Ssa1p (lane 3), BiP (lane 4), and BiP(1–386) (lane 5) and 100% input of PAb416 (lane 6) are shown. Heavy chain and light chain refer to the antibody that coprecipitates in the immunoprecipitation reaction.
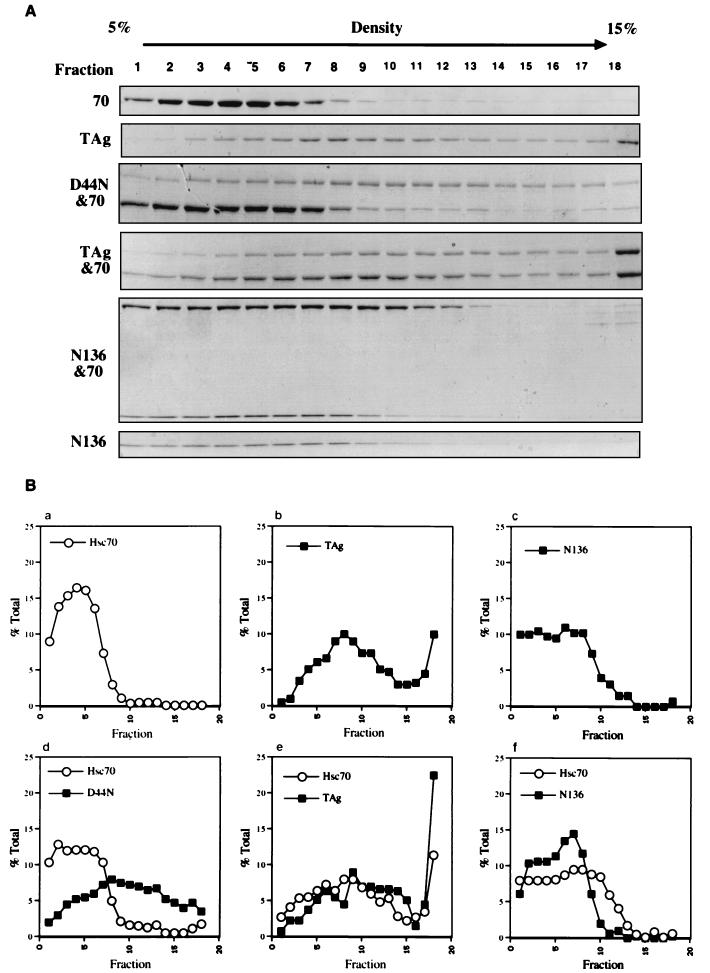
Thus far, all the binding data presented have been determined using immunoprecipitation assays. Even though irrelevant antibodies fail to immunoprecipitate any Hsc70 when treated with T antigen and Hsc70 and two different antibodies that recognize independent epitopes of T antigen show identical results, the formal possibility exists that the immunoprecipitation assay could detect nonphysiological interactions between T antigen and Hsc70 induced by the antibodies. Therefore, an independent assay was employed to test for the direct association of T antigen and Hsc70. T antigen or mutants of T antigen were incubated with Hsc70 in the presence of ATP (4 mM) and in the absence of antibody. The reactions were then layered onto a 5 to 15% glycerol gradient. Because T antigen forms large homo-oligomers in the presence of ATP (7), the control reaction of T antigen alone demonstrates that T antigen is distributed throughout the gradient (Fig. 6A, second panel from top; Fig. 6Bb). In contrast, a majority of the Hsc70 (90%) from the Hsc70-alone reaction remains concentrated in the least dense nine fractions of the gradient (Fig. 6A, top panel; Fig. 6Ba). However, inclusion of T antigen induces a redistribution of Hsc70 with only ∼50% found in fractions 1 to 9 (Fig. 6A, fourth panel from top; Fig. 6Be). As expected, the negative control D44N and Hsc70 reaction did not change the distribution of Hsc70. Note that 90% of the Hsc70 is found in the least dense nine fractions (Fig. 6A, third panel from top; Fig. 6Bd). Therefore, T antigen alters the migration profile of Hsc70 in a J-domain-dependent manner.
FIG. 6.
Sedimentation velocity centrifugation. Wild-type and mutant T antigens were incubated in the presence or absence of Hsc70 plus 4 mM MgATP and then layered onto a 5 to 15% glycerol gradient. The reaction mixtures were centrifuged, and fractions were collected. (A) Fractions were analyzed via SDS-PAGE and Coomassie blue staining. (B) The relative percent of the total Hsc70 or T antigen in each fraction was graphed.
N136 was incubated with Hsc70, and the reaction was layered onto the gradient. A reproducible slight shift in the density of Hsc70 is observed when N136 is incubated with Hsc70 since only 77% of Hsc70 is in the least dense nine fractions (Fig. 6A, fifth panel from top; Fig. 6Bf). The migration of N136 is not affected by the presence of Hsc70 in the reaction mixture (Fig. 6A, bottom panel versus panel second from bottom; Fig. 6Bc versus f). We conclude that N136, when incubated with Hsc70, induces a slight change in the distribution of Hsc70. The centrifugation data provide an independent confirmation that Hsc70 and T antigen directly interact, consistent with the immunoprecipitation data.
DISCUSSION
The stable association of T antigen with Hsc70 is direct.
Previous studies have shown that T antigen can bind to Hsc70 in the context of a cellular lysate, and this association is dependent upon an intact J domain (5, 34, 35, 40). However, it is not known if Hsc70 is an endogenous DnaK-like chaperone partner of T antigen, since it is possible that Hsc70 could indirectly associate with T antigen through other T-antigen binding proteins. For example, both p53 and pRb form stable complexes with Hsc70 as well as T antigen. In this report we use immunoprecipitation and sedimentation velocity centrifugation to show that T antigen forms a direct complex with mammalian Hsc70. In contrast, T antigen does not bind to either of two different yeast DnaK homologues efficiently (Fig. 4), arguing that the interaction with mammalian Hsc70 is specific. Furthermore, inclusion of an equimolar amount of purified p53 in the T-antigen–Hsc70 association assay does not increase the affinity of Hsc70 for the complex (data not shown). The above data, combined with the fact that T antigen can stimulate the ATPase activity of mammalian Hsc70 in vitro (37), demonstrates that Hsc70 is indeed a chaperone partner of T antigen.
The association of Hsc70 with T antigen is concentration dependent, with the stoichiometry of binding at 1:1 (120 nM T antigen, 1 mM ATP, Kd = 310 nM Hsc70). This Kd is within a twofold range of the published dissociation constant of 600 nM for the yeast J protein auxilin and Hsc70 (19). Thus, the association between J proteins and Hsc70s of different species occurs with a similar affinity. However this does not imply that the chaperone components of different species are necessarily interchangeable, as we have previously demonstrated the inability of T-antigen chimeras containing an E. coli or yeast J domain to function for simian virus 40 activities in vivo (40).
Stable association between T antigen and Hsc70 is dependent upon ATP hydrolysis by Hsc70.
Stable complex formation between T antigen and Hsc70 requires ATP and is ATP concentration dependent (Fig. 2B, 3A). However, nucleotide binding is not sufficient to promote complex formation because ADP, AMP-PNP, AMP-PCP, and ATPγS all fail to induce complex formation (Fig. 3B). Furthermore, ATPγS competes with ATP for binding to Hsc70 and promotes a decrease in the fraction of T-antigen–Hsc70 complex that immunoprecipitates with T-antigen antibodies (Fig. 2C). Therefore, complex formation between Hsc70 and T antigen requires ATP hydrolysis and not simply ATP binding.
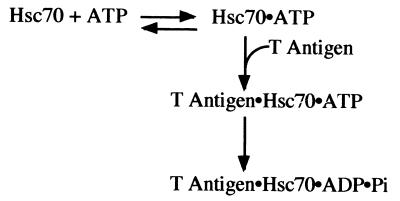
A T-antigen mutant that fails to bind ATP associates with Hsc70 as well as wild-type T antigen does (Fig. 3B); therefore, these data indicate that ATP hydrolysis at the T-antigen nucleotide binding site is not required for T-antigen–Hsc70 complex formation. The Kd, ATP for the T-antigen–Hsc70 complex is 0.16 μM, and this dissociation constant is similar to other published values for the Kd, ATP of mammalian Hsc70 (15, 42, 43). The results presented in Fig. 2 and 3 demonstrate that Hsc70 must bind and hydrolyze ATP to trap the stable T-antigen–Hsc70 intermediate. The observation that T antigen stimulates the ATPase activity of Hsc70 (37) as well as the results presented here indicates that T antigen and Hsc70 are two components of a molecular machine whose conformational changes are driven by ATP turnover (depicted in Fig. 7). T antigen liberates E2F4 from p130 in an ATP-dependent manner, and this is thought to contribute to tumorigenesis (39). The results presented here lay the foundation for understanding how T antigen, in conjunction with Hsc70, acts as a molecular machine to disrupt tumor suppressor complexes and induce transformation.
FIG. 7.
ATP-promoted association of T-antigen–Hsc70 complex.
Stable association requires multiple domains of both T antigen and Hsc70.
Stable complex formation between T antigen and Hsc70 requires a functional J domain, because no association between D44N and Hsc70 was detected in either the immunoprecipitation assays or the sedimentation velocity centrifugation assay. D44N is a prototypic J-domain mutant used in a number of studies (5, 35, 36, 38, 39). Even though D44N retains other J-domain-independent activities of T antigen, including the ability to associate with pRb and p130 (37, 39), it is defective for the induction of viral DNA replication, the inhibition of apoptosis (36), and the disruption of Rb/E2F family complexes (38, 39). Thus, the direct and stable association of Hsc70 may be an important J-domain activity of T antigen that is required for the successful completion of multiple viral functions.
The J domain of T antigen alone is insufficient to associate with Hsc70 in a stable manner. N136, an amino-terminal fragment of T antigen containing the J domain, failed to complex with Hsc70, even when a molar excess of N136 (10-fold more than the positive control wild-type T-antigen reaction) was included in the reaction (Fig. 2A). Interestingly, N136 retains partial biological activities of T antigen, including the ability to bind pRb and transform C3H10T1/2 cells, albeit less efficiently than full-length T antigen (37). These results suggest that for at least some transforming activities, T antigen does not require a stable interaction with Hsc70.
We also observed that N136 induced a redistribution of Hsc70 to denser glycerol fractions during the sedimentation velocity centrifugation assay. This may be explained by a transient interaction between N136 and Hsc70, which could account for the ability of N136 to stimulate the ATPase activity of Hsc70 (37). However, the fact that N136 does not cofractionate with the Hsc70 in these fractions is inconsistent with a stable complex between N136 and Hsc70, as also confirmed by the immunoprecipitation data. An alternative possibility is that N136 induces the self-association of Hsc70 into homo-oligomers. For example, it has been shown that other J-domain-containing proteins can induce the oligomerization of Hsc70 (13, 19, 24). The gradient data show that the ability to transiently associate with Hsc70 requires only the J domain while the immunoprecipitation data show that the J domain alone is not sufficient to effectively complex with Hsc70, suggesting that additional domains of T antigen, carboxyl terminal to the J domain, are required for the stable association between Hsc70 and T antigen. It is possible that the J domain of T antigen is sufficient to induce homo-oligomerization of Hsc70, but stable T-antigen–Hsc70 complex formation requires the carboxyl-terminal portion of both the T antigen and Hsc70 polypeptides. Alternatively, it is possible that the J-domain-containing fragment induces a conformational change in Hsc70 without changing its oligomeric state.
There are at least two possible reasons why the carboxyl-terminal regions of T antigen are required to bind to Hsc70. First, it is possible that a specific Hsc70-binding sequence exists between amino acids 137 and 708 of T antigen that contacts Hsc70. For example, there is a weak homology to the DnaK binding protein GrpE at aa 501 to 520 of T antigen, and the cocrystal structure of GrpE and DnaK reveals that this region of GrpE directly binds to DnaK. Second, it is possible that a nonspecific sequence of amino acids is required to associate with the peptide binding region of Hsc70. It has been reported that very small amounts of J proteins are needed to stimulate the ATPase activity of Hsc70, and incubating larger amounts of protein can drive binding of Hsc70 to J proteins through the peptide binding domain of Hsc70 (26). We cannot rule out a similar mechanism in our studies. However, the yeast Hsc70 homologues fail to bind T antigen in a stable manner. This argues that our in vitro conditions detect a specific interaction between Hsc70 and T antigen, because related Hsc70 homologues fail to associate with T antigen.
Two different anti-T-antigen antibodies, one which recognizes an amino-terminal epitope (PAb416) and one which recognizes a carboxyl-terminal epitope (901), demonstrate the ability of T antigen to bind to Hsc70 in the immunoprecipitation assay. Interestingly, when PAb419 is used in the reaction, less Hsc70 is found complexed with T antigen (Fig. 4B, lanes 11 and 12). This results is observed whether we incubate T antigen with PAb419 before or after Hsc70 (data not shown). We speculate that this could be for one of two reasons. First, it is possible that since PAb419 recognizes an epitope within the J domain of T antigen, it may block efficient contact between the J domain and Hsc70, thereby preventing or disrupting association. Alternatively, since PAb419 has been shown to alter the conformation of the carboxyl terminus of T antigen (23), it is possible that PAb419 alters the conformation of a carboxyl-terminal Hsc70 binding site of T antigen. Nuclear magnetic resonance perturbation studies reveal that J proteins contact the ATPase domain of Hsc70 through their J domains (14). However, the carboxyl-terminal peptide binding and EEVD domains of Hsc70 are also important for the association of Hsc70 with J proteins (8, 12). Consistent with these data, we have shown that the ATPase domain of Hsc70 does not form a stable complex with T antigen. These results suggest that the carboxyl-terminal region of Hsc70, including either the peptide binding domain or EEVD motif, is required for stable association with T antigen. Future mapping experiments are necessary to determine which carboxyl-terminal regions of Hsc70 and T antigen are required for their stable association.
Our findings demonstrate that Hsc70 binds to T antigen with the affinity, stoichiometry, and ATP dependence similar to those of other J protein-DnaK complexes, such as auxilin and Hsc70 (19). This observation suggests a functional conservation between diverse species for the mechanism of how substrates are presented to Hsc70 and how Hsc70 homologues interact with J proteins. However, this hypothesis does not imply that the components of these interactions are interchangeable. On the contrary, there are species-specific elements within the T-antigen J domain that are required for transformation (40). This fact, combined with our finding that two yeast Hsc70s fail to efficiently bind T antigen (Fig. 4), argues that there is specificity to T-antigen–DnaK–protein interactions. It remains to be determined if Hsc70 alone or other DnaK-like proteins are involved in J-domain-mediated viral functions.
ACKNOWLEDGMENTS
We thank J. L. Brodsky and members of his laboratory for use of reagents, experimental advice, and critical reading of the manuscript. We also acknowledge P. Cantalupo for technical assistance and T. Harper and A. Mackey for help with the figures.
This work was supported by grants from the NIH (CA40586 to J.M.P. and GM-54141 to S.P.G.). S.P.G. was supported in part by an ACS Junior Faculty Research Award (JFRA-618).
REFERENCES
- 1.Alfano C, McMacken R. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage lambda DNA replication. J Biol Chem. 1989;264:10709–10718. [PubMed] [Google Scholar]
- 2.Brodsky J L. Post-translational protein translocation: not all hsc70s are created equal. Trends Biochem Sci. 1996;21:122–126. [PubMed] [Google Scholar]
- 3.Brodsky J L, Pipas J M. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J Virol. 1998;72:5329–5334. doi: 10.1128/jvi.72.7.5329-5334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 5.Campbell K S, Mullane K P, Aksoy I A, Stubdal H, Zalvide J, Pipas J M, Silver P A, Roberts T M, Schaffhausen B S, DeCaprio J A. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 6.Cantalupo P, Saenz-Robles M T, Pipas J M. Expression of SV40 large T antigen in baculovirus systems and purification by immunoaffinity chromatography. Methods Enzymol. 1999;306:297–307. doi: 10.1016/s0076-6879(99)06019-x. [DOI] [PubMed] [Google Scholar]
- 7.Castellino A M, Cantalupo P, Marks I M, Vartikar J V, Peden K W, Pipas J M. trans-dominant and non-trans-dominant mutant simian virus 40 large T antigens show distinct responses to ATP. J Virol. 1997;71:7549–7559. doi: 10.1128/jvi.71.10.7549-7559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheetham M E, Caplan A J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fanning E. Simian virus 40 large T antigen: the puzzle, the pieces, and the emerging picture. J Virol. 1992;66:1289–1293. doi: 10.1128/jvi.66.3.1289-1293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farber J M, Peden K W, Nathans D. trans-dominant defective mutants of simian virus 40 T antigen. J Virol. 1987;61:436–445. doi: 10.1128/jvi.61.2.436-445.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fourie A M, Hupp T R, Lane D P, Sang B C, Barbosa M S, Sambrook J F, Gething M J. HSP70 binding sites in the tumor suppressor protein p53. J Biol Chem. 1997;272:19471–19479. doi: 10.1074/jbc.272.31.19471. [DOI] [PubMed] [Google Scholar]
- 12.Freeman B C, Myers M P, Schumacher R, Morimoto R I. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao B, Eisenberg E, Greene L. Effect of constitutive 70-kDa heat shock protein polymerization on its interaction with protein substrate. J Biol Chem. 1996;271:16792–16797. doi: 10.1074/jbc.271.28.16792. [DOI] [PubMed] [Google Scholar]
- 14.Greene M K, Maskos K, Landry S J. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha J H, McKay D B. Kinetics of nucleotide-induced changes in the tryptophan fluorescence of the molecular chaperone Hsc70 and its subfragments suggest the ATP-induced conformational change follows initial ATP binding. Biochemistry. 1995;34:11635–11644. doi: 10.1021/bi00036a040. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 18.Inoue A, Torigoe T, Sogahata K, Kamiguchi K, Takahashi S, Sawada Y, Saijo M, Taya Y, Ishii S, Sato N, et al. 70-kDa heat shock cognate protein interacts directly with the N-terminal region of the retinoblastoma gene product pRb. Identification of a novel region of pRb-mediating protein interaction. J Biol Chem. 1995;270:22571–22576. doi: 10.1074/jbc.270.38.22571. [DOI] [PubMed] [Google Scholar]
- 19.Jiang R F, Greener T, Barouch W, Greene L, Eisenberg E. Interaction of auxilin with the molecular chaperone, Hsc70. J Biol Chem. 1997;272:6141–6145. doi: 10.1074/jbc.272.10.6141. [DOI] [PubMed] [Google Scholar]
- 20.Karzai A W, McMacken R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- 21.Kelley W L. Molecular chaperones: how J domains turn on Hsp70s. Curr Biol. 1999;9:R305–308. doi: 10.1016/s0960-9822(99)80185-7. [DOI] [PubMed] [Google Scholar]
- 22.Kelley W L, Georgopoulos C. The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc Natl Acad Sci USA. 1997;94:3679–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kernohan N M, Hupp T R, Lane D P. Modification of an N-terminal regulatory domain of T antigen restores p53-T antigen complex formation in the absence of an essential metal ion cofactor. J Biol Chem. 1996;271:4954–4960. doi: 10.1074/jbc.271.9.4954. [DOI] [PubMed] [Google Scholar]
- 24.King C, Eisenberg E, Greene L E. Interaction between Hsc70 and DnaJ homologues: relationship between Hsc70 polymerization and ATPase activity. Biochemistry. 1999;38:12452–12459. doi: 10.1021/bi9902788. [DOI] [PubMed] [Google Scholar]
- 25.Lanford R E. Expression of simian virus 40 T antigen in insect cells using a baculovirus expression vector. Virology. 1988;167:72–81. doi: 10.1016/0042-6822(88)90055-4. [DOI] [PubMed] [Google Scholar]
- 26.Laufen T, Mayer M P, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindner K, Mole S E, Lane D P, Kenny M K. Epitope mapping of antibodies recognising the N-terminal domain of simian virus large tumour antigen. Intervirology. 1998;41:10–16. doi: 10.1159/000024910. [DOI] [PubMed] [Google Scholar]
- 28.McClellan A J, Endres J B, Vogel J P, Palazzi D, Rose M D, Brodsky J L. Specific molecular chaperone interactions and an ATP-dependent conformational change are required during posttranslational protein translocation into the yeast ER. Mol Biol Cell. 1998;9:3533–3545. doi: 10.1091/mbc.9.12.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrick B A, He C, Witcher L L, Patterson R M, Reid J J, Pence-Pawlowski P M, Selkirk J K. HSP binding and mitochondrial localization of p53 protein in human HT1080 and mouse C3H10T1/2 cell lines. Biochim Biophys Acta. 1996;1297:57–68. doi: 10.1016/0167-4838(96)00089-1. [DOI] [PubMed] [Google Scholar]
- 30.Peden K W, Pipas J M. Simian virus 40 mutants with amino-acid substitutions near the amino terminus of large T antigen. Virus Genes. 1992;6:107–118. doi: 10.1007/BF01703060. [DOI] [PubMed] [Google Scholar]
- 31.Peden K W, Srinivasan A, Vartikar J V, Pipas J M. Effects of mutations within the SV40 large T antigen ATPase/p53 binding domain on viral replication and transformation. Virus Genes. 1998;16:153–165. doi: 10.1023/a:1007941622680. [DOI] [PubMed] [Google Scholar]
- 32.Prives C. The replication functions of SV40 T antigen are regulated by phosphorylation. Cell. 1990;61:735–738. doi: 10.1016/0092-8674(90)90179-i. [DOI] [PubMed] [Google Scholar]
- 33.Rassow J, von Ahesen O, Bomer U, Pfanner N. Molecular chaperones: towards a characterization of the heat-shock protein 70 family. Trends Cell Biol. 1997;7:129–133. doi: 10.1016/S0962-8924(96)10056-8. [DOI] [PubMed] [Google Scholar]
- 34.Sawai E T, Butel J S. Association of a cellular heat shock protein with simian virus 40 large T antigen in transformed cells. J Virol. 1989;63:3961–3973. doi: 10.1128/jvi.63.9.3961-3973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slinskey A, Barnes D, Pipas J M. Simian virus 40 large T antigen J domain and Rb-binding motif are sufficient to block apoptosis induced by growth factor withdrawal in a neural stem cell line. J Virol. 1999;73:6791–6799. doi: 10.1128/jvi.73.8.6791-6799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan C S, Cantalupo P, Pipas J M. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol Cell Biol. 2000;20:6233–6243. doi: 10.1128/mcb.20.17.6233-6243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan C S, Tremblay J D, Fewell S W, Lewis J A, Brodsky J L, Pipas J M. Species-specific elements in the large T-antigen J domain are required for cellular transformation and DNA replication by simian virus 40. Mol Cell Biol. 2000;20:5749–5757. doi: 10.1128/mcb.20.15.5749-5757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai J, Douglas M G. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 42.Wang C, Lee M R. High-level expression of soluble rat hsc70 in Escherichia coli: purification and characterization of the cloned enzyme. Biochem J. 1993;294:69–77. doi: 10.1042/bj2940069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei J, Hendershot L M. Characterization of the nucleotide binding properties and ATPase activity of recombinant hamster BiP purified from bacteria. J Biol Chem. 1995;270:26670–26676. doi: 10.1074/jbc.270.44.26670. [DOI] [PubMed] [Google Scholar]
- 44.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zylicz M, Ang D, Liberek K, Georgopoulos C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]