ABSTRACT
The human pathogen Pseudomonas aeruginosa, a leading cause of hospital-acquired infections, inhabits and forms sessile antibiotic-resistant communities called biofilms in a wide range of biotic and abiotic environments. In this study, we examined how two global sensory signaling pathways—the RhlR quorum-sensing system and the CbrA/CbrB nutritional adaptation system—intersect to control biofilm development. Previous work has shown that individually these two systems repress biofilm formation. Here, we used biofilm analyses, RNA-seq, and reporter assays to explore the combined effect of information flow through RhlR and CbrA on biofilm development. We find that the ΔrhlRΔcbrA double mutant exhibits a biofilm morphology and an associated transcriptional response distinct from wildtype and the parent ΔrhlR and ΔcbrA mutants indicating codominance of each signaling pathway. The ΔrhlRΔcbrA mutant gains suppressor mutations that allow biofilm expansion; these mutations map to the crc gene resulting in loss of function of the carbon catabolite repression protein Crc. Furthermore, the combined absence of RhlR and CbrA leads to a drastic reduction in the abundance of the Crc antagonist small RNA CrcZ. Thus, CrcZ acts as the molecular convergence point for quorum- and nutrient-sensing cues. We find that in the absence of antagonism by CrcZ, Crc promotes the expression of biofilm matrix components—Pel exopolysaccharide, and CupB and CupC fimbriae. Therefore, this study uncovers a regulatory link between nutritional adaption and quorum sensing with potential implications for anti-biofilm targeting strategies.
IMPORTANCE
Bacteria often form multicellular communities encased in an extracytoplasmic matrix called biofilms. Biofilm development is controlled by various environmental stimuli that are decoded and converted into appropriate cellular responses. To understand how information from two distinct stimuli is integrated, we used biofilm formation in the human pathogen Pseudomonas aeruginosa as a model and studied the intersection of two global sensory signaling pathways—quorum sensing and nutritional adaptation. Global transcriptomics on biofilm cells and reporter assays suggest parallel regulation of biofilms by each pathway that converges on the abundance of a small RNA antagonist of the carbon catabolite repression protein, Crc. We find a new role of Crc as it modulates the expression of biofilm matrix components in response to the environment. These results expand our understanding of the genetic regulatory strategies that allow P. aeruginosa to successfully develop biofilm communities.
KEYWORDS: quorum sensing, catabolite repression, biofilms, Crc, RhlR, CbrA, nutrient sensing
INTRODUCTION
Bacteria predominantly exist in structured communities called biofilms. Biofilms are defined as aggregates of cells that are embedded in a matrix made of extracellular polymeric substances (EPS) including exopolysaccharides, proteins, lipids, and nucleic acids (1–3). The EPS is crucial for the emergent properties of biofilms such as superior resilience to environmental stresses like antimicrobials and host immune responses. Biofilm formation is a dynamic process that is governed by various intracellular and exogenous stimuli. Often, two-component signaling (TCS) systems integrate and relay the information contained in sensory cues into the control of biofilm formation (4, 5). TCSs are typically composed of sensor histidine kinases (HK) and partner response regulators (RR) that, via phosphorylation cascades, couple stimulus sensing to appropriate changes in behavior (4, 6).
The opportunistic pathogen Pseudomonas aeruginosa forms biofilms in diverse environments, such as soil and water and in host-associated environments, such as burn wounds, lungs of cystic fibrosis (CF) patients, and plant tissues (7–10). Accordingly, P. aeruginosa encodes a large suite of >60 TCS systems that allow it to respond to diverse external cues (11–13). One such TCS is the CbrA/CbrB system that is involved in nutritional adaptation and hierarchical utilization of various carbon sources (14–17). CbrA represents a non-canonical sensor HK that also functions as a histidine transporter (17). CbrA has two putative sensor domains—SLC5 and PAS, yet the sensory stimulus activating its kinase is unknown. CbrA kinase phosphorylates its cognate RR CbrB that activates the expression of the small RNA CrcZ (17, 18; Fig. 1A). CrcZ sequesters complexes of the catabolite repression protein Crc with Hfq to antagonize Crc function in the carbon catabolite repression (CCR) process termed “reverse diauxie” or reverse CCR (rCCR; 19, 20). In the presence of preferred carbon sources, CrcZ-mediated antagonism is relieved and Crc/Hfq post-transcriptionally inhibits the expression of enzymes involved in the catabolism of non-preferred carbon sources. In addition to the metabolic regulation of carbon and nitrogen utilization in P. aeruginosa, Cbr TCS plays an important role in various virulence-associated processes including biofilm formation and antibiotic resistance (15).
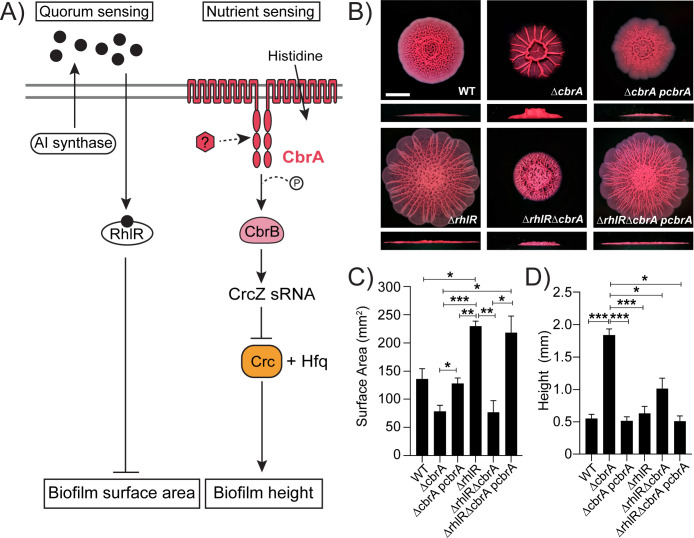
Fig 1.
P. aeruginosa ΔcbrA and ΔrhlR mutants have distinct hyper-rugose biofilm phenotypes. (A) Schematic of the RhlR quorum sensing and CbrA nutrient sensing pathways. The two gray horizontal lines represent the cytoplasmic membrane, black circles represent autoinducer sensed by RhlR, red hexagon represents the unknown signal that activates CbrA sensor kinase. CbrA also functions as a histidine transporter, but its kinase function appears to be independent of histidine transport (17). (B) Colony biofilm phenotypes of WT PA14 and the designated mutants on Congo red agar medium after 120 h of growth. Scale bar, 5 mm. (C) Colony biofilm surface area quantitation for the indicated strains after 120 h of growth. Error bars represent standard deviation of three independent experiments. (D) Colony biofilm height quantitation for the indicated strains after 120 h of growth. Error bars represent standard deviation of three independent experiments. (C and D) Only pairwise comparisons that had P value < 0.05 are denoted. Statistical significance was determined using Welch’s ANOVA with Dunnett’s T3 multiple comparisons test in GraphPad Prism software. ***P < 0.001, **P < 0.01, *P < 0.05.
Another sensory cue commonly detected by bacteria is their population density via the chemical communication process called quorum sensing (21). Quorum sensing relies on the production of extracellular signaling molecules called autoinducers and their subsequent detection by cognate receptors (21, 22) and thereby allows groups of bacteria to coordinate their gene expression patterns in response to changes in population density to exhibit collective behaviors. Quorum sensing controls biofilm development and virulence in P. aeruginosa (23–25). The P. aeruginosa quorum-sensing circuit consists of two canonical LuxI/R pairs: LasI/R and RhlI/R (26–28). LasI produces and LasR responds to the autoinducer 3OC12-homoserine lactone (3OC12-HSL), while RhlR binds to the autoinducer C4-HSL, the product of RhlI. While LasR promotes biofilm formation (23, 29), RhlR represses biofilms (25; Fig. 1A).
Bacteria encounter multiple sensory cues simultaneously and must integrate information from each cue into a response. In this study, we examine how the CbrA/CbrB signaling pathway intersects with the RhlR-dependent quorum sensing system to control biofilm development. We find that the combined absence of quorum sensing via RhlR and nutrient sensing via CbrA results in (i) a biofilm morphology and an associated transcriptional response that indicates codominance of the RhlR and CbrA/CbrB pathways and (ii) a significant reduction in the levels of the small RNA CrcZ. We further find that the ΔrhlRΔcbrA mutant gives rise to spontaneous suppressor mutations in the crc gene that allows biofilm spreading. Transcriptomic profiling of biofilms of wildtype (WT) and mutant P. aeruginosa strains and reporter assays demonstrate that Crc promotes the expression of Pel exopolysaccharide and Cup fimbriae components of the biofilm matrix. Thus, our work uncovers a new role for Crc as a master regulator of biofilm formation that allows the integration of quorum and nutritional cues.
RESULTS
CbrA-mediated nutrient-sensing and RhlR-mediated quorum-sensing codominantly control biofilm development
To study the role of the CbrA/CbrB TCS in P. aeruginosa biofilm formation, we first generated in-frame marker-less deletion of the cbrA gene in the WT P. aeruginosa UCBPP-PA14 (hereafter called PA14) background. PA14 exhibits a characteristic rugose-center/smooth-periphery colony biofilm phenotype on Congo red biofilm medium, while the ΔcbrA mutant exhibits a distinct colony biofilm phenotype with decreased surface area coverage and increased height, defined as the maximum vertical rise from the base of the colony, when compared to WT (Fig. 1B through D; Fig. S1). To uncouple the two functions of CbrA—HK and histidine transport (30–32)—and determine whether biofilm regulation is a consequence of information flow from CbrA HK to CbrB RR, we generated a ΔcbrB mutant. The ΔcbrB mutant produces hyper-rugose biofilms identical to the ΔcbrA mutant (Fig. S2). In addition, we built a cbrAH766A mutant where the histidine residue that undergoes autophosphorylation is changed to alanine to abolish kinase function of CbrA. Similar to the ΔcbrB strain, the cbrAH766A mutant has a biofilm phenotype that resembles the ΔcbrA mutant (Fig. S2). Introduction of a plasmid expressing cbrA under its native promoter to the ΔcbrA and cbrAH766A mutants restored biofilm formation to WT levels (Fig. 1B through D; Fig. S2). These analyses indicate that CbrA kinase activity is required for the repression of biofilm development in P. aeruginosa.
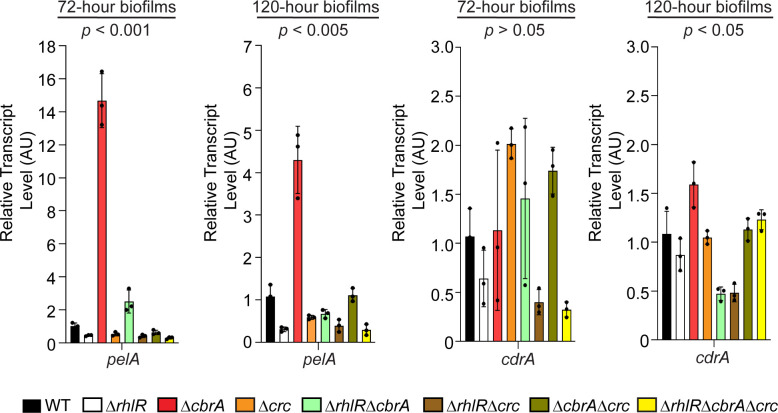
The production of EPS is a defining feature of biofilms, and the matrix composition can vary depending on the bacterial species. P. aeruginosa biofilm matrix has been reported to be composed of three different exopolysaccharides—Pel, Psl, and alginate—depending on the strain (reviewed in reference 33). In PA14, Pel is the major exopolysaccharide under commonly used laboratory growth conditions (34). Pel biosynthetic enzymes are encoded by the pelABCDEFG operon (pel). In addition, the matrix protein CdrA interacts with Pel exopolysaccharide and promotes structural stability in biofilm aggregates (35). Here, we assessed the expression of pelA and cdrA in the biofilms of WT and ΔcbrA mutant using quantitative RT-PCR. We find that pelA transcript levels are increased by 15- and 5-fold in the ΔcbrA mutant compared to WT at 72 h and 120 h timepoints, respectively (Fig. 2). In contrast, cdrA transcript levels do not exhibit any significant differences between WT and the ΔcbrA mutant (Fig. 2). Next, we assessed the contribution of Pel to the ΔcbrA mutant biofilms by comparing the ΔpelA and ΔcbrAΔpelA double mutants in biofilm assays. The absence of PelA abolished the formation of colony biofilms and pellicles (Fig. S3 and S4). Both the ΔpelA and ΔcbrAΔpelA mutants had completely smooth colony morphologies and lower biofilm height compared to WT (Fig. S4A through C). Furthermore, to test the generality of these observations, we employed a solid surface-associated (SSA) biofilm assay using polystyrene multi-well plates and found that the ΔpelA and ΔcbrAΔpelA mutants showed significantly less crystal violet staining compared to the ΔcbrA mutant (Fig. S4D). Thus, CbrA represses pel expression, and Pel exopolysaccharide is required for the ΔcbrA mutant to exhibit its characteristic hyper-rugose biofilm phenotype.
Fig 2.
P. aeruginosa ΔcbrA mutant exhibits increased pel expression in biofilms. Relative expression of pelA and cdrA genes normalized to 16S RNA, ostA, and rpsO transcript levels in WT PA14 and indicated mutants after 72 and 120 h of colony biofilm growth. AU denotes arbitrary units. Error bars represent standard deviation of three biological replicates. Statistical significance was determined using Welch’s ANOVA in GraphPad Prism software. For pelA expression at 72 h, the following comparisons were statistically significant (P < 0.001): WT vs ΔrhlR, and ΔcbrA vs all other strains. For pelA expression at 120 h, the following comparisons were statistically significant (P < 0.005): WT vs ΔrhlR, and ΔcbrA vs all other strains. For cdrA expression at 120 h, the following comparisons were statistically significant (P < 0.05): ΔcbrA vs ΔrhlRΔcbrA and ΔrhlRΔcrc mutants.
We have previously reported that the P. aeruginosa quorum-sensing receptor RhlR represses colony biofilm development and the hyper-rugosity conferred by the absence of RhlR requires Pel exopolysaccharide (Fig. 1; 25). Our quantitative RT-PCR analyses on biofilm samples demonstrate that RhlR promotes the expression of pelA as the ΔrhlR mutant showed fourfold reduction in pelA transcript levels compared to WT in mature biofilms (Fig. 2). Intriguingly, while both ΔrhlR and ΔcbrA mutants appear to be hyper-rugose compared to WT, the ΔcbrA mutant exhibits increased biofilm height and higher levels of pel expression as opposed to the increased surface area coverage and decreased pel transcript levels of the ΔrhlR mutant (Fig. 1B through D and 2; Fig. S1). To explore the combined effect of Rhl and Cbr pathways on biofilm formation, we deleted rhlR and cbrA genes together. The ΔrhlRΔcbrA double mutant showed a growth profile indistinguishable from WT and its parent ΔrhlR and ΔcbrA mutants but formed colony biofilms that were markedly distinct from those of the WT and single ΔrhlR and ΔcbrA mutants, i.e., had significantly lower surface coverage than ΔrhlR mutant and lower height than ΔcbrA mutant (Fig. 1B through D; Fig. S1). Furthermore, the ΔrhlRΔcbrA double mutant showed decreased pel expression compared to the ΔcbrA mutant (Fig. 2). Introduction of a plasmid expressing cbrA under its native promoter to the ΔrhlRΔcbrA double mutant resulted in a hyper-rugose biofilm that resembles the ΔrhlR mutant in terms of biofilm morphology, surface area, and height (Fig. 1B through D). We conclude that RhlR and CbrA control different properties, surface area coverage and vertical rise, respectively, of a growing biofilm.
Loss-of-function mutations in crc affects biofilm development
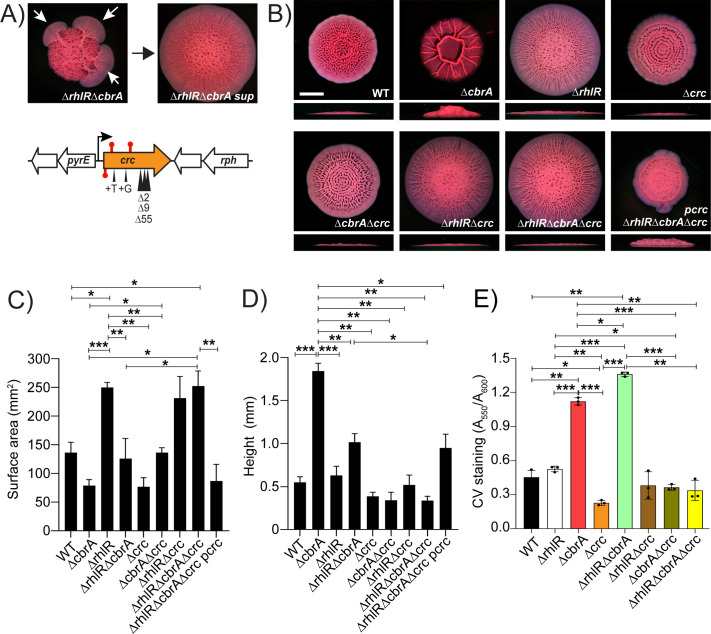
The ΔrhlRΔcbrA mutant gave rise to suppressor flares after 72 h of biofilm growth that allowed biofilm expansion similar to the ΔrhlR mutant (Fig. 3A; Fig. S5). We isolated 12 spontaneously arising suppressor mutants from ΔrhlRΔcbrA colony biofilms; 10 suppressors contained deletions, insertions, or missense mutations in the crc gene, while the remaining 2 suppressors harbored mutations in the rplP and rplC genes that encode for 50S ribosomal protein L16 and 50S ribosomal protein L3, respectively (Fig. 3A; Table S1). We generated Δcrc single, ΔrhlRΔcrc and ΔcbrAΔcrc double, and ΔrhlRΔcbrAΔcrc mutants and analyzed their biofilm-forming behaviors. The absence of Crc showed little effect in WT and ΔrhlR backgrounds but substantially altered the biofilms of the ΔcbrA and ΔrhlRΔcbrA mutants (Fig. 3B through D). Specifically, the deletion of crc abolished the vertical rise of the ΔcbrA mutant colony biofilm (Fig. 3B through D) and led to increased surface area coverage of ΔrhlRΔcbrA double mutant colony biofilms (Fig. 3B through D; Fig. S1). Introduction of a plasmid expressing crc under its native promoter to the ΔrhlRΔcbrAΔcrc triple mutant resulted in colony biofilm phenotype identical to the ΔrhlRΔcbrA double mutant validating our deletion analyses (Fig. 3B through D). We conclude that Crc promotes colony biofilm height when CbrA is inactive and restricts colony biofilm expansion when signal transduction via both RhlR and CbrA are absent.
Fig 3.
P. aeruginosa ΔrhlRΔcbrA mutant biofilms generate spontaneous mutations that allow biofilm expansion. (A) (Top) Shown is a representative isolation of a suppressor mutation of the ΔrhlRΔcbrA biofilm phenotype. The white arrows in the left panel indicate flares radiating from the biofilm diagnostic of the emergence of suppressor mutations. The right panel shows the biofilm phenotype of a mutant following isolation. (Bottom) Chromosomal arrangement of the crc (orange) gene. Large white arrows represent open reading frames (lengths not to scale), black bent arrow indicates promoter, red stem-loops indicate STOP mutations, and black triangles indicate the locations of insertion and deletion suppressor mutations. (B) Colony biofilm phenotypes of WT PA14 and the designated mutants on Congo red agar medium after 120 h of growth. Scale bar, 5 mm. (C) Colony biofilm surface area quantitation for the indicated strains after 120 h of growth. Error bars represent the standard deviation of three independent experiments. (D) Colony biofilm height quantitation for the indicated strains after 120 h of growth. Error bars represent the standard deviation of three independent experiments. (E) Biofilm development assays followed by crystal violet staining for WT and indicated mutant strains. Error bars represent the standard deviation of three biological replicates. (C–E) Only pairwise comparisons that had P value < 0.05 are denoted. Statistical significance was determined using Welch’s ANOVA with Dunnett’s T3 multiple comparisons test in GraphPad Prism software. ***P < 0.001, **P < 0.01, *P < 0.05.
To explore the extent of biofilm regulation by Crc, we analyzed the ability of WT and the Δcrc single and combination mutants to form SSA biofilms on polystyrene. Crystal violet staining of these SSA biofilms showed a significant increase in mature biofilm formation in the ΔcbrA and ΔrhlRΔcbrA mutants compared to WT (Fig. 3E). We note that there was no significant difference between WT and ΔrhlR mutants in this assay. Nonetheless, the absence of Crc significantly decreased SSA biofilms in WT, as well as ΔcbrA and ΔrhlRΔcbrA mutant backgrounds (Fig. 3E). Taken together, our data suggest a new role for Crc beyond reverse diauxie, i.e., Crc promotes biofilm development.
The absence of phenazines contributes to the colony biofilm spreading phenotype of the ΔrhlR mutant (25) and one known regulatory target of Crc is phzM that encodes for a key enzyme involved in the biosynthesis of the phenazine pyocyanin (36). Therefore, we assessed the contribution of phenazines to the ΔrhlRΔcbrA double-mutant phenotype. We deleted the two phenazine biosynthesis operons phzA1-G1 and phzA2-G2 (Δphz) in the WT, the ΔcbrA single-, and the ΔrhlRΔcbrA double-mutant backgrounds and assayed pyocyanin production and biofilm development. As reported previously, the Δphz mutant did not produce pyocyanin and its biofilm spread radially outward and covered more surface area than WT (Fig. S6A through C; 37). Mutation of the phenazine biosynthesis operons, however, failed to increase biofilm spreading in cells lacking CbrA (Fig. S6B and C). Likewise, there was no detectable change in biofilm morphology of the ΔrhlRΔcbrAΔphz mutant when compared with its parent ΔrhlRΔcbrA strain (Fig. S6B and C). We infer that the absence of CbrA is epistatic to the absence of phenazines during colony biofilm development.
CrcZ small RNA serves as the point of convergence for quorum and nutrient-sensing cues
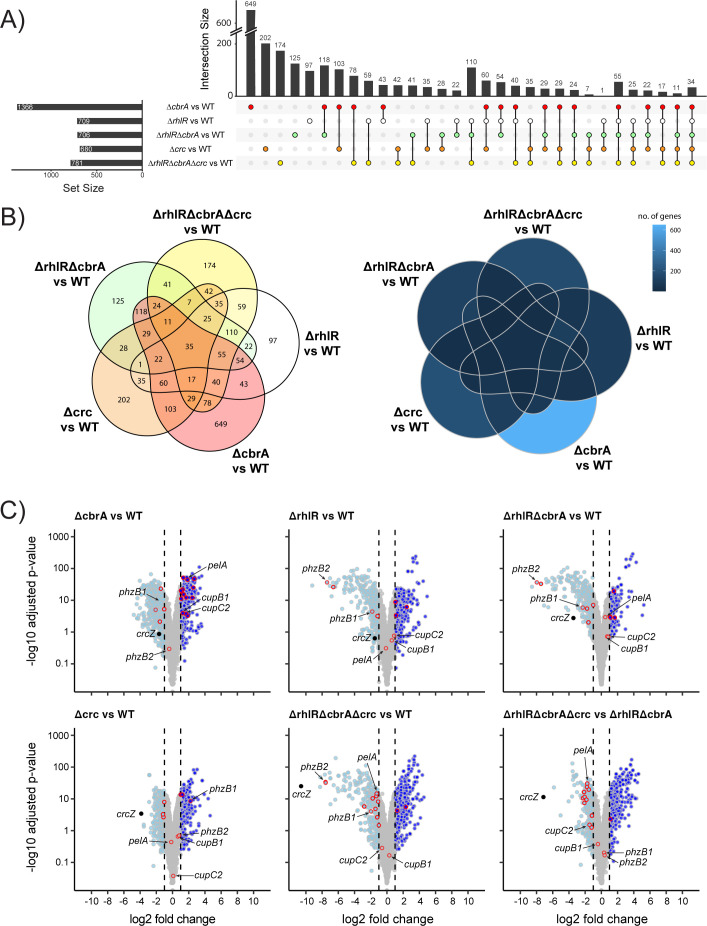
To define the molecular basis underpinning the different ΔrhlR and ΔcbrA biofilm phenotypes, we used RNA-seq to compare the global transcriptional profiles of the biofilms of WT PA14 and the ΔrhlR, ΔcbrA, and ΔrhlRΔcbrA double mutants grown on Congo red agar medium for 72 h. Principal component analysis (PCA) of normalized read counts for 5,978 genes showed that biofilm samples of each mutant clustered separately from WT (Fig. S7). Comparative transcriptomic analysis revealed a total of 1,366 differentially expressed genes (DEGs; genes with expression fold changes ≤−2 and ≥2, and the P values (Padj), adjusted using the Benjamini-Hochberg procedure, <0.05; see Table S3 in the supplemental material) in the ΔcbrA mutant (Fig. 4A and B). In the ΔrhlR mutant, we find a total of 709 DEGs, of which 326 transcripts overlapped with the CbrA regulon. Comparing the ΔrhlRΔcbrA double to the ΔcbrA single mutant showed a 51.6% reduction in the number of DEGs in the double mutant (Fig. 4A and B). Furthermore, 125 genes were uniquely regulated in the ΔrhlRΔcbrA mutant (Fig. 4A and B). We conclude that the combined absence of each regulator—CbrA and RhlR—gives rise to a transcriptomic response distinct from mere addition of their individual regulons.
Fig 4.
RNA-seq analysis of biofilms of PA14 and mutants. (A) Upset plot showing overlaps in genes that are differentially regulated in ΔrhlR, ΔcbrA, Δcrc, ΔrhlRΔcbrA double, and ΔrhlRΔcbrAΔcrc triple mutants compared to WT. Numbers on top of each vertical bar indicate number of genes differentially regulated in each intersection. Set size indicates the total number of genes that are significantly differentially regulated in a particular mutant when compared to WT. (B) Venn diagrams showing overlaps in genes that are differentially regulated in ΔrhlR, ΔcbrA, Δcrc, ΔrhlRΔcbrA double, and ΔrhlRΔcbrAΔcrc triple mutants compared to WT. Numbers (left) or shade of blue (right) indicate number of genes differentially regulated in each intersection. (C) Volcano plots of RNA-seq data for ΔrhlR, ΔcbrA, Δcrc, ΔrhlRΔcbrA, and ΔrhlRΔcbrAΔcrc compared to WT, and ΔrhlRΔcbrAΔcrc compared to ΔrhlRΔcbrA. Light blue solid circles with gray outlines represent genes with expression fold-changes ≤−2 and the P values (Padj), adjusted using the Benjamini-Hochberg procedure, <0.05; dark blue solid circles with gray outlines represent genes with expression fold-changes ≥ 2 and the P values (Padj), adjusted using the Benjamini-Hochberg procedure, <0.05. Gray solid circles represent genes with gene expression fold changes ≥−2 or ≤2 or Padj ≥ 0.05. Genes reported to be associated with biofilms are outlined in red.
Next, we determined the transcriptomic response to the absence of Crc. The Crc regulon consists of 680 genes, of which ~48% are also regulated by CbrA and ~36% coregulated by RhlR (Fig. 4A and B; Table S3). We note that, similar to the ΔrhlRΔcbrA biofilm samples, a subset of genes was uniquely regulated in the ΔrhlRΔcbrAΔcrc triple mutant, i.e., these 174 genes were not differentially expressed in the ΔcbrA, ΔrhlR, and Δcrc single mutant strains compared to WT (Fig. 4A and B). Furthermore, binning the DEGs according to their function defined by their PseudoCAP categories (38) suggests that RhlR functions downstream of CbrA mediated regulation of genes in categories such as “Biosynthesis of cofactors, prosthetic groups and carrier” and “Antibiotic resistance and susceptibility” (Fig. S8). Notably, in biofilms, Crc appears to both positively and negatively regulate several categories unrelated to carbon catabolite repression such as “Cell wall/LPS/Capsule,” “Protein secretion/export apparatus,” and “Adaptation, Protection” (Fig. S8). Taken together, the combined absence of RhlR, CbrA, and Crc results in a distinct regulon that does not overlap with the single mutants. We infer that CbrA, RhlR, and Crc regulons exhibit complex non-linear interactions.
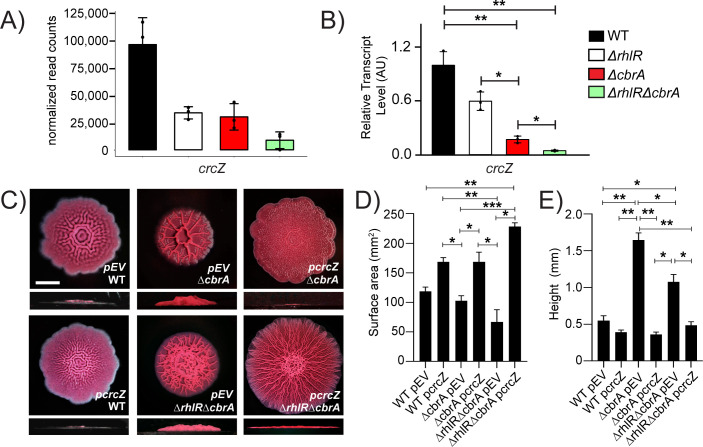
The expression of the small RNA CrcZ that antagonizes Crc to allow coordinated utilization of carbon sources is activated by CbrA/CbrB (Fig. 1A; 14). Thus, CrcZ levels are downregulated in the ΔcbrA mutant compared to WT (Fig. 4C). However, our RNA-seq data revealed a more severe reduction in CrcZ abundance in the ΔrhlRΔcbrA double mutant compared to WT than in the ΔrhlR and ΔcbrA single mutants (Fig. 4C and 5A). Indeed, quantitative RT-PCR shows that crcZ transcript levels are ~6-fold lower in the ΔcbrA mutant than WT but 23-fold lower in the ΔrhlRΔcbrA mutant compared to WT (Fig. 5B). Furthermore, consistent with previous report that CrcZ small RNA is stabilized by Crc in Pseudomonas putida (39), we find a significant reduction in CrcZ abundance in the Δcrc and ΔrhlRΔcbrAΔcrc mutants compared to WT PA14 (Fig. 4C; Table S3). Thus, our data suggest that low levels of CrcZ promote biofilm morphology with reduced surface coverage and increased height.
Fig 5.
CrcZ small RNA allows integration of Rhl- and Cbr-signaling pathways and modulates biofilm development. (A) Normalized read counts obtained via Median Ratio Normalization (MRN) analysis for crcZ gene from RNA-seq run on biofilm samples of WT and indicated mutants. (B) Relative expression of crcZ gene normalized to 16S RNA, ostA, and rpsO transcript levels measured by qRT-PCR in WT PA14 and indicated mutants after 120 h of colony biofilm growth. AU denotes arbitrary units. Error bars represent the standard deviation of three biological replicates. (C) Colony biofilm phenotypes of WT PA14 and the designated mutants on Congo red agar medium after 120 h of growth. Scale bar, 5 mm. (D) Colony biofilm surface area quantitation for the indicated strains after 120 h of growth. Error bars represent the standard deviation of three independent experiments. (E) Colony biofilm height quantitation for the indicated strains after 120 h of growth. Error bars represent the standard deviation of three independent experiments. (B, D, E) Only pairwise comparisons that had P value < 0.05 are denoted. Statistical significance was determined using Welch’s ANOVA with Dunnett’s T3 multiple comparisons test in GraphPad Prism software. ***P < 0.001, **P < 0.01, *P < 0.05.
To assess the contribution of CrcZ small RNA to biofilm development, we introduced a plasmid expressing crcZ from Plac promoter to WT PA14, the ΔcbrA single, and ΔrhlRΔcbrA double mutants. Overexpression of CrcZ small RNA led to increased surface area coverage and a severe reduction in biofilm height in the ΔcbrA mutant compared to the empty vector control (Fig. 5C through E). Furthermore, overexpression of CrcZ small RNA in the ΔrhlRΔcbrA double mutant resulted in a hyper-rugose biofilm that resembled the ΔrhlR mutant in terms of biofilm morphology, surface area, and height (Fig. 5C through E). We conclude that the abundance of CrcZ small RNA is a key determinant of biofilm development in response to signals transduced via CbrA and RhlR. We further conclude that drastic reduction in crcZ expression in the ΔrhlRΔcbrA double mutant leads to uncontrolled Crc activity which, in turn, restricts surface area coverage and promotes height, likely explaining why crc suppressors arose in the ΔrhlRΔcbrA mutant to allow biofilm expansion.
Crc activates the expression of biofilm matrix components
Closer inspection of the transcriptomics data revealed downregulation of Pel biosynthesis genes such as pelA in the ΔrhlRΔcbrAΔcrc triple mutant compared to WT and ΔrhlRΔcbrA double mutant (Fig. 4C; Table S3), suggesting that Crc can be a positive regulator of Pel biosynthesis. Accordingly, we performed qRT-PCR for pelA from colony biofilm samples in the WT and a series of crc mutants—Δcrc, ΔrhlRΔcrc, ΔcbrAΔcrc, and ΔrhlRΔcbrAΔcrc. We find that there is a twofold reduction in the relative abundance of pelA transcript in the Δcrc mutant compared to WT at 72 h and 120 h timepoints, respectively, and 27- and 8-fold reduction compared to the ΔcbrA strain at 72 h and 120 h timepoints, respectively (Fig. 2). Furthermore, the absence of Crc is epistatic to ΔcbrA as pelA transcript levels are drastically reduced in the ΔcbrAΔcrc and ΔrhlRΔcbrAΔcrc mutants (Fig. 2). Thus, we conclude that Crc activates pel expression and the high expression of pel in the ΔcbrA and ΔrhlRΔcbrA mutants is due to the unchecked activity of Crc in the absence of CrcZ small RNA.
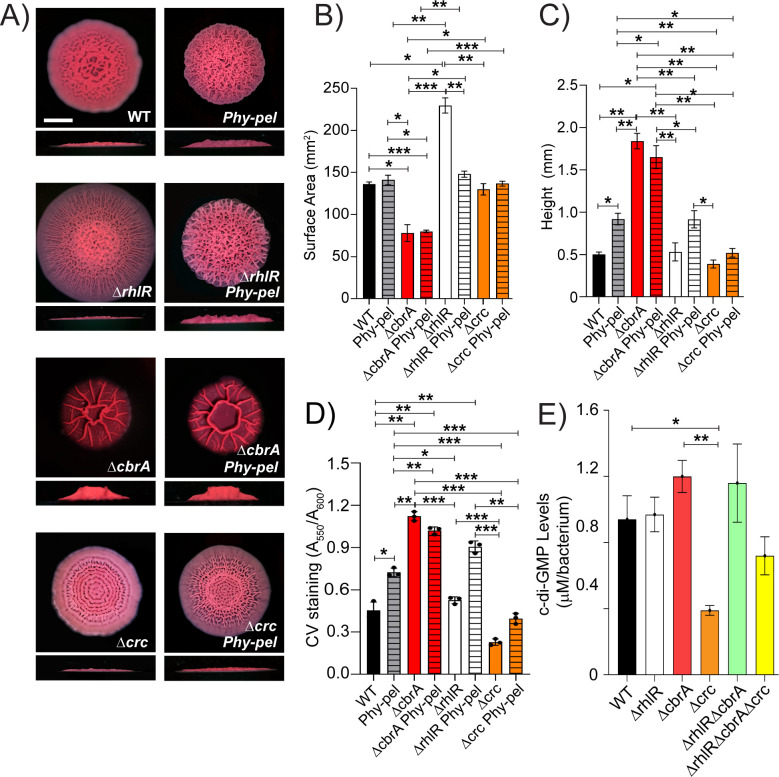
To determine whether different expression level of pel is involved in the distinct surface area and height of the ΔrhlR and ΔcbrA mutants, we generated a Pel overexpression strain where we replaced the native promoter of the pelABCDEFG operon (PpelA) with a constitutively expressed artificial promoter region comprised of transcription terminators T1 and T2 from the Escherichia coli rrnB gene followed by the Bacillus subtilis Physpank promoter (we call this strain Phy-pel). Compared to WT, relative transcript abundance of pelA is increased about 18-fold in the Phy-pel strain (Fig. S9). Accordingly, the Phy-pel strain exhibits a hyper-wrinkled biofilm phenotype that is distinct from WT in the colony biofilm assay (Fig. 6A through C) and increased attachment in the SSA biofilm assay (Fig. 6D). Consistent with the result that RhlR activates the expression of pel (Fig. 2), overexpression of pel is epistatic to ΔrhlR as the ΔrhlR Phy-pel strain shows reduced biofilm spreading and increased attachment to polystyrene surface compared to the parent ΔrhlR mutant (Fig. 6). Although the abundance of pelA transcript is similar between ΔcbrA and Phy-pel mutants, the Phy-pel strain produced a hyper-rugose biofilm morphology that is markedly distinct from the ΔcbrA mutant biofilm and showed lower crystal violet staining (Fig. 6; Fig. S9). Furthermore, the absence of cbrA is epistatic to overexpression of pel as the ΔcbrA Phy-pel double mutant formed biofilms identical to the ΔcbrA mutant (Fig. 6). Notably, the Δcrc Phy-pel double mutant mirrors the Δcrc mutant biofilm phenotypes in the colony biofilm and solid-surface attachment assays (Fig. 6). Thus, we conclude that the absence of crc is epistatic to transcriptional activation of the pelABCDEFG operon.
Fig 6.
Crc-dependent increase in pel expression is post-transcriptional. (A) Colony biofilm phenotypes of WT PA14 and the designated mutants on Congo red agar medium after 120 h of growth. Scale bar, 5 mm. (B) Colony biofilm surface area quantitation for the indicated strains after 120 h of growth. Error bars represent the standard deviation of three independent experiments. (C) Colony biofilm height quantitation for the indicated strains after 120 h of growth. Error bars represent the standard deviation of three independent experiments. (D) Biofilm crystal violet staining assays for WT and indicated mutant strains. Error bars represent the standard deviation of three biological replicates. (E) c-di-GMP levels of the indicated strains. Error bars represent the standard deviation of three biological replicates. (B–E) Only pairwise comparisons that had P value < 0.05 are denoted. Statistical significance was determined using Welch’s ANOVA with Dunnett’s T3 multiple comparisons test in GraphPad Prism software. ***P < 0.001, **P < 0.01, *P < 0.05.
Because Crc functions as a post-transcriptional regulator in reverse diauxie, we hypothesized that Crc mediates an increase in pel expression via a post-transcriptional mechanism. One well-known post-transcriptional mode of control of Pel biosynthesis is the second messenger cyclic di-guanosine monophosphate (c-di-GMP) such that high c-di-GMP levels correlate with biofilm formation (reviewed in reference 40). Furthermore, Hfq, and thereby carbon catabolite repression, has been linked to c-di-GMP concentration in cells (reviewed in reference 41). This prompted us to determine intracellular c-di-GMP concentration of the mutants in this study. Figure 6E shows that while c-di-GMP concentration is lowered in the Δcrc mutant compared to WT, none of the ΔrhlR, ΔcbrA, ΔrhlRΔcbrA, and ΔrhlRΔcbrAΔcrc mutants displayed any discernable changes from WT. We conclude that c-di-GMP levels do not contribute to the difference in biofilm development of the ΔrhlR, ΔcbrA, and ΔrhlRΔcbrA mutants.
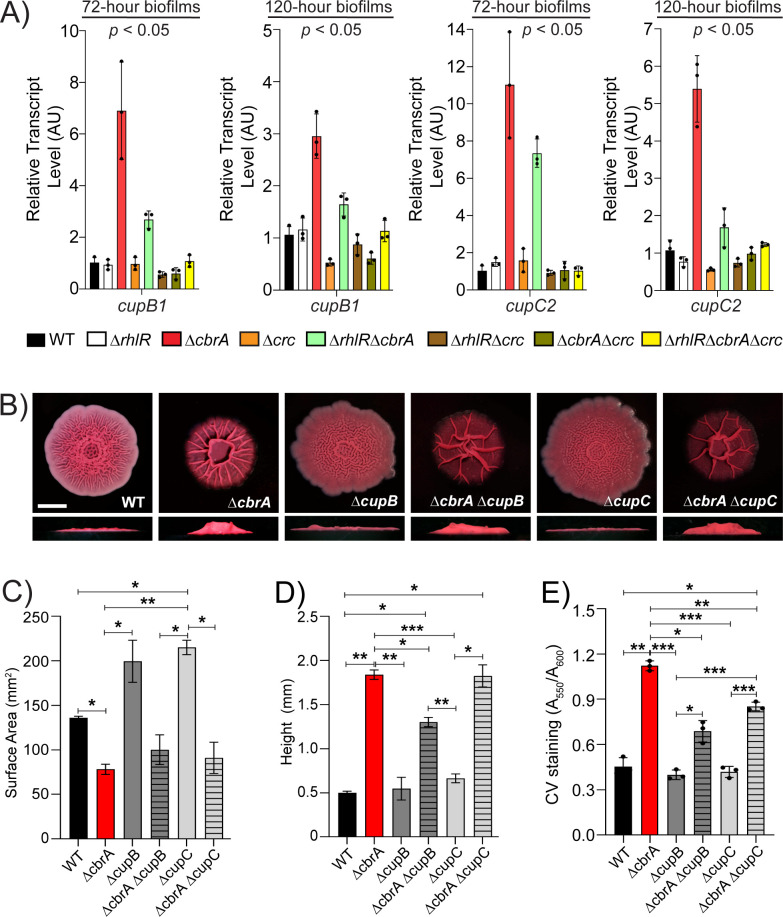
In addition to Pel and CdrA, the biofilm matrix involves a large arsenal of cell surface-associated structures, including type IV pili, Fap fibrils, and the cup fimbriae (42–47). P. aeruginosa can produce five types of cup fimbriae—CupA, CupB, CupC, CupD, and CupE, products of cupA, cupB, cupC, cupD, and cupE gene clusters each of which encodes an usher, a chaperone, and at least one fimbrial subunit. Our RNA-seq data revealed that cupB and cupC biosynthetic operons are upregulated in the ΔcbrA and ΔrhlRΔcbrA mutants (Fig. 4C; Table S3). To assess the coordinated regulation of the Cup components of the biofilm matrix by RhlR, CbrA, and Crc over time, we performed quantitative RT-PCR for cupA1, cupB1, cupC2, cupD4, and cupE1 from colony biofilm samples after 72 and 120 h of growth, respectively, in the WT and the ΔrhlR, ΔcbrA, Δcrc, ΔrhlRΔcbrA, ΔrhlRΔcrc, ΔcbrAΔcrc, and ΔrhlRΔcbrAΔcrc mutants (Fig. 7A; Fig. S9). Our data show that the relative mRNA levels of the cupB1 and cupC2 genes increased greater than fivefold at 72 h and more than threefold at 120 h in the ΔcbrA single and ΔrhlRΔcbrA double mutants, but these increases in transcript levels were abolished in the ΔcbrAΔcrc double and ΔrhlRΔcbrAΔcrc triple mutants (Fig. 7A; Fig. S9). This suggests that, in addition to Pel, Crc promotes the expression of CupB and CupC matrix components.
Fig 7.
The cupB and cupC fimbrial operons are overexpressed in the absence of CbrA. (A) Relative expression of cupB1 and cupC2 genes normalized to 16S RNA, ostA, and rpsO transcript levels in WT PA14 and indicated mutants after 72 and 120 h of colony biofilm growth. AU denotes arbitrary units. Error bars represent the standard deviation of three biological replicates. Statistical significance was determined using Welch’s ANOVA in GraphPad Prism software. For cupB1 expression at 72 and 120 h, the following comparisons were statistically significant (P < 0.05): ΔcbrA vs all other strains. For cupC2 expression at 72 h, the following comparisons were statistically significant (P < 0.05): ΔcbrA vs all other strains except ΔrhlRΔcbrA, and ΔrhlRΔcbrA vs all other strains except ΔcbrA. For cupC2 expression at 120 h, the following comparisons were statistically significant (P < 0.05): ΔcbrA vs all other strains except ΔrhlRΔcbrA mutant. (B) Colony biofilm phenotypes of WT PA14 and the designated mutants on Congo red agar medium after 120 h of growth. Scale bar, 5 mm. (C) Colony biofilm surface area quantitation for the indicated strains after 120 h of growth. Error bars represent the standard deviation of three independent experiments. (D) Colony biofilm height quantitation for the indicated strains after 120 h of growth. Error bars represent the standard deviation of three independent experiments. (E) Biofilm crystal violet staining assays for WT and indicated mutant strains. Error bars represent the standard deviation of three biological replicates. (C–E) Only pairwise comparisons that had P value < 0.05 are denoted. Statistical significance was determined using Welch’s ANOVA with Dunnett’s T3 multiple comparisons test in GraphPad Prism software. ***P < 0.001, **P < 0.01, *P < 0.05.
To determine the role of CupB and CupC fimbriae in biofilms of the ΔcbrA mutant, we generated deletion mutants where we removed entire cupB (cupB1-B5) and cupC (cupC1-C3) operons in WT and ΔcbrA backgrounds. The ΔcbrAΔcupB mutant exhibited significant, albeit minor, decrease in biofilm height compared to the ΔcbrA mutant while for the ΔcbrAΔcupC mutant, surface area coverage and height parameters show values similar to the ΔcbrA mutant (Fig. 7B through D). Notably, the absence of CupB and CupC reduced SSA biofilms in the ΔcbrA mutant (Fig. 7E). We conclude that Crc promotes the expression of CupB and CupC fimbriae that, in turn, contribute to the formation of hyper-rugose biofilms.
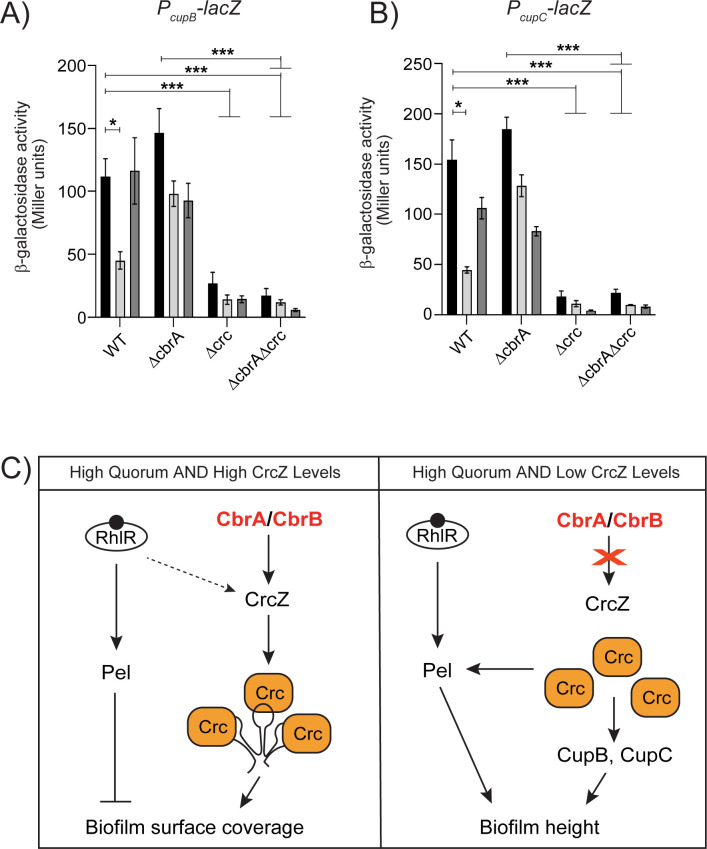
The levels of CrcZ small RNA have been reported to vary according to the carbon source in the growth medium which, in turn, affects catabolite repression (14). Therefore, we probed the expression of biofilm matrix genes in response to carbon sources by generating transcriptional reporter fusions to the cupB and cupC promoters (PcupB-lacZ and PcupC-lacZ). We incorporated the reporter fusion into an intergenic region on the chromosomes of WT P. aeruginosa and the ΔcbrA, Δcrc and ΔcbrAΔcrc mutants. The PcupB-lacZ and PcupC-lacZ reporters exhibited approximately four- and eightfold lower expression in the Δcrc and ΔcbrAΔcrc mutants than the WT in tryptone broth (Fig. 8A and B). These results show that Crc is absolutely required for the expression of cupB and cupC operons in both WT and ΔcbrA backgrounds. In P. aeruginosa, “reverse” carbon catabolite repression is known to be triggered by organic acids such as pyruvate (48). Furthermore, pyruvate activates the expression of crcZ in both P. aeruginosa and P. putida (49). Therefore, we tested pyruvate as preferred carbon source and glucose as non-preferred sugar. The expression of the reporters in the WT was reproducibly and significantly lowered in the presence of pyruvate but not glucose (Fig. 8A and B). Taken together, our data lead to a regulatory model in which environmental cues that reduce the levels of CrcZ small RNA allow for Crc-dependent activation of biofilm matrix gene expression in P. aeruginosa (Fig. 8C).
Fig 8.
Crc promotes the transcription of cupB and cupC fimbrial operons. (A and B) β-Galactosidase assays of PcupB-lacZ transcriptional (A) or PcupC-lacZ transcriptional (B) fusions for background genotypes indicated on the x-axis and grown in tryptone broth with no addition (black), 50 mM pyruvate (light gray), or 50 mM glucose (dark gray). Error bars represent SEM of three biological replicates. Only pairwise comparisons that had P value < 0.05 are denoted. Statistical significance was determined using Welch’s ANOVA with Dunnett’s T3 multiple comparisons test in GraphPad Prism software. ***P < 0.001, *P < 0.05. (C) Model for Crc-dependent activation of biofilm matrix components. At high population density, RhlR-mediated quorum sensing activates the expression of Pel exopolysaccharide while Crc, depending on the abundance of CrcZ small RNA, promotes the expression of Pel, CupB, and CupC fimbriae components of the biofilm matrix. The arrow indicates activation and T-bar indicates inhibition.
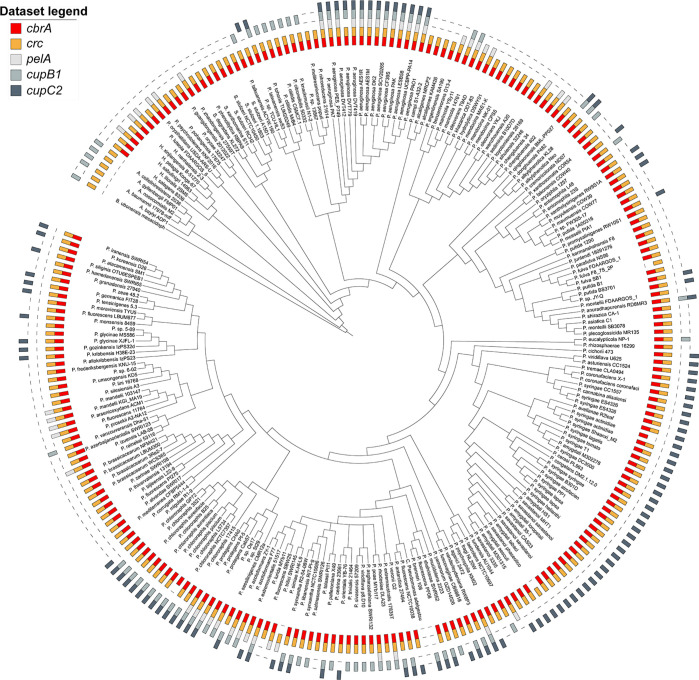
Bacteria of the genus Pseudomonas are metabolically versatile, can be isolated from a wide range of niches, and preferentially catabolize organic acids as they lack a functional glycolytic pathway (reviewed in reference 50). This “reverse” carbon catabolite repression is thought to provide adaptive advantage in different niche colonization. As such, the CbrA/CbrB/CrcZ/Crc signaling cascade is conserved in most pseudomonads, and components of this pathway have been found to be exchangeable between P. aeruginosa and P. putida (49). We explored whether Crc in other species of Pseudomonas might also be involved in controlling biofilm formation via phylogenetic analysis of the co-occurrence of cbrA, crc, pelA, cupB1, and cupC2 in the genomes of diverse pseudomonads (Fig. 9). Consistent with previous reports, majority of the pseudomonads encode for CbrA and Crc orthologs. PelA and CupB1 are found to co-occur in a subset of the genomes that include several CF isolates of P. aeruginosa, while CupC2 is present in ~45% of the genomes analyzed here. For example, pseudomonads that colonize the rhizosphere of plants such as P. stutzeri, P. chlororaphis, P. syringae, and P. protegens encode for genes required to assemble CupC fimbriae. These bacteria are exposed to glucose as well as organic acids like succinate and pyruvate that are commonly found in root exudates and likely derive ecological advantage by integrating nutritional cues to control adhesion to root surfaces. Thus, it will be interesting to know whether Crc has a conserved function in the regulation of CupC fimbriae which might also contribute to biofilm development in the environmental pseudomonads.
Fig 9.
The CbrA and Crc proteins and their regulatory targets Pel, CupB, and CupC are conserved in diverse pseudomonads. Co-occurrences of CbrA and Crc are depicted in red and orange, respectively, while that of their regulatory target genes obtained from this study—pelA, cupB1, and cupC2—are depicted in shades of gray as indicated in the data set legend.
DISCUSSION
Combinatorial gene regulation by multiple two-component systems is highly utilized in bacteria in responding to environmental cues to allow distinct gene expression patterns. P. aeruginosa is a versatile human pathogen that causes both acute and chronic infections such as in the lungs of cystic fibrosis (CF) patients and in patients suffering from burn and diabetic wounds that are often associated with worse disease outcomes (51–54). In this study, we sought to define the interaction of Rhl quorum sensing with another global regulatory system, i.e., CbrA/CbrB pathway. As the sensory signal that activates CbrA kinase is unknown (17), we studied biofilm development and biofilm-associated transcriptomic changes in the absence of RhlR, CbrA, and both. Our RNA-seq analysis sheds light on the transcriptomic landscape of WT P. aeruginosa biofilms and provides a map of the regulatory network orchestrated by RhlR, CbrA, and Crc. Furthermore, our data highlight the complexity of sensory regulation mediated by these two signaling pathways in P. aeruginosa biofilms and reveals a previously unexplored facet of Crc as it promotes the expression of specific biofilm matrix components—Pel exopolysaccharide and CupB/CupC fimbriae.
P. aeruginosa, one of the major causes of morbidity and mortality of CF patients, colonize the lung mucus which is a complex substrate that provides carbon, nitrogen, and other energy sources during infection (55). Recent studies have highlighted the importance of carbon catabolite repression in virulence-associated processes such as quorum sensing, phenazine production, and antibiotic resistance (56, 57). Although glucose is a less preferred carbon source for P. aeruginosa, it is present at high concentration in the mucus of both CF and non-CF patients, with CF mucus typically having higher glucose content than those of other pulmonary diseases (55, 56, 58). Our work suggests that glucose (or another less preferred carbon source) could function as an inducer of biofilm formation via the CbrA/CbrB/CrcZ/Crc pathway. Additional work is required to define the molecular mechanism by which Crc activates the expression of pel, cupB, or cupC genes. We speculate that one possible mechanism for increased pel expression might be at the post-transcriptional level as the absence of Crc decreases biofilm formation even after exchanging the PpelA promoter with an artificial constitutive promoter. Nonetheless, given that both carbon catabolite repression and Pel biosynthetic genes are widespread in bacteria (59), we expect future studies defining the molecular details underlying the link between reverse diauxie and pel expression would be informative.
The material properties of the matrix contribute to biofilm growth and architecture. For instance, in Vibrio cholerae and Bacillus subtilis, matrix production allows biofilm-dwelling cells to establish an osmotic pressure difference between the biofilm interior and the external environment that promotes biofilm expansion (60, 61). Specifically, V. cholerae colony biofilms were found to expand more in the absence of the matrix protein RbmA (60). Furthermore, biofilm matrix components contribute to processes such as signaling, migration, and genetic exchange over the course of biofilm development (reviewed in reference 62). B. subtilis biofilm matrix protein BslA and exopolysaccharide EPS are essential for sliding motility that allows colony expansion via surface spreading (63). Our work suggests that in PA14, lower Pel polysaccharide levels allow greater surface area coverage as observed in the ΔrhlR mutant. Conversely, increased Pel polysaccharide levels limits biofilm expansion as is the case with ΔcbrA and ΔrhlR ΔcbrA mutants. Increased matrix combined with reduced levels of CrcZ in the ΔrhlR ΔcbrA mutant leads to spontaneous mutations in Crc that, in turn, decreases Pel levels to allow biofilm growth and expansion.
The cup genes in P. aeruginosa are poorly expressed in standard laboratory conditions making it difficult to study their regulation, and therefore, few regulators such as the H-NS-like protein MvaT that represses cupA gene expression and the two-component systems named Roc1 and Roc2 that induce the expression of cupB and cupC genes have been described (46, 64). Nonetheless, the Cup fimbrial structures have been reported to facilitate bacterial attachment to host tissue and promote biofilm formation and pathogenesis (43, 65). We uncover the significant contribution of CupB and CupC fimbriae to the biofilms made by the ΔcbrA mutant (Fig. 7A) suggesting the CbrA/CbrB system downregulates Cup fimbriae expression under laboratory settings. Furthermore, our finding that organic acids such as pyruvate can repress the expression of cupB and cupC fimbrial genes hints at a change in adhesion depending on nutritional cues that might contribute to biofilms within hosts.
Transcriptome analysis may reveal both direct and indirect targets of Crc. Crc/Hfq binding to transcripts is thought to involve at least one CA motif (AAnAAnAA) in the neighborhood of the translational start site in target mRNAs (16, 18). Accordingly, we scanned the 5′ untranslated region of Crc-regulated genes in two RNA-seq data sets—Δcrc vs WT and ΔrhlRΔcbrA vs ΔrhlRΔcbrAΔcrc mutants. We found 32 and 73 Crc-regulated transcripts having a putative CA motif, respectively, but none of these transcripts belong to biofilm-matrix biosynthesis genes (Table S4). One caveat is that the Crc/Hfq recognition motif for biofilm-matrix biosynthesis genes might be distinct from the aforementioned CA motif. Another possibility is that Crc indirectly activates the expression of biofilm matrix components. This study provides the basis for future biochemical characterization of Crc to define how it functions as a positive regulator of biofilm development. Taken together, by characterizing the convergence of global signaling pathways on biofilm development, we have expanded our understanding of the molecular interplay between key regulators—CbrA and RhlR and identified Crc as a master regulator of biofilm matrix gene expression in response to environmental stimuli.
MATERIALS AND METHODS
Strains and growth conditions
P. aeruginosa UCBPP-PA14 strain was grown in lysogeny broth (LB) (10 g tryptone, 5 g yeast extract, 5 g NaCl per L), in 1% tryptone broth (TB) (10 g tryptone per L), and on LB plates fortified with 1.5% Bacto agar at 37˚C. When appropriate, antimicrobials were included at the following concentrations: 400 µg/mL carbenicillin, 50 µg/mL gentamycin, 100 µg/mL irgasan.
Strain construction
Strains and plasmids were constructed as described previously (25). To construct marker-less in-frame chromosomal deletions in P. aeruginosa, DNA fragments flanking the gene of interest were amplified, assembled by the Gibson method, and cloned into pEXG2 (66). The resulting plasmids were used to transform Escherichia coli SM10λpir and, subsequently, mobilized into P. aeruginosa PA14 via biparental mating. Exconjugants were selected on LB containing gentamicin and irgasan, followed by recovery of deletion mutants on LB medium containing 5% sucrose. Candidate mutants were confirmed by PCR. The cbrA complementation plasmid was constructed by inserting DNA containing the promoter of dksA and the entire cbrA open-reading frame using HindIII and XbaI, followed by cloning into similarly digested pUCP18. The crc complementation plasmid was constructed by inserting DNA containing the promoter and entire crc open-reading frame using HindIII and XbaI, followed by cloning into similarly digested pUCP18. The crcZ overexpression plasmid was constructed by inserting DNA containing the entire crcZ gene using HindIII and XbaI, followed by cloning into similarly digested pUCP18.
To construct the PcupB-lacZ and PcupC-lacZ transcriptional reporter fusions, 500 bp of DNA upstream of the cupB1 and cupC1 genes and the DNA encoding the lacZ open-reading frame were amplified using P. aeruginosa PA14 genomic DNA and the plasmid pIT2 as templates, respectively. Next, two DNA fragments of ~730 bp, one corresponding to the intergenic region ~700 bp downstream of the P. aeruginosa PA14_20500 gene and the other corresponding to ~1,000 bp upstream of the P. aeruginosa PA14_20510 gene, were amplified from P. aeruginosa PA14 genomic DNA. The four DNA fragments were assembled by the Gibson method and cloned into pEXG2. The resulting plasmid was used to transform E. coli SM10λpir and subsequently mobilized into P. aeruginosa PA14 WT and the ΔrhlR and ΔrhlI mutants via biparental mating as described above.
Colony biofilm assay
One microliter of overnight P. aeruginosa cultures grown at 37°C in 1% tryptone broth was spotted onto 60 × 15 mm petri plates containing 10 mL 1% tryptone medium fortified with 40 mg per L Congo red and 20 mg per L Coomassie brilliant blue dyes and solidified with 1% agar. Colonies were grown at 25°C under dark conditions, and images were acquired using a Zeiss AxioZoom v16 microscope.
Crystal violet staining of SSA biofilms
P. aeruginosa was cultured overnight in 200 µL LB in a BioTek Synergy Neo2 microplate reader at 37°C under shaking conditions. The OD600 of the culture was taken by the plate reader to calculate the amount of culture needed to inoculate 200 µL 1% tryptone broth so that the final OD600 is 0.005. Biofilms were developed for 72 h in stationary 96-well polystyrene plate at 25°C in dark conditions. After 72 h, OD600 measurement was taken, and the supernatant was discarded. The cultures were washed vigorously with tap water and were left to dry for an hour. The biofilms were then stained 250 µL 0.1% crystal violet solution for a half an hour. The excess solution was poured out, and the samples were washed thrice with tap water and let to dry overnight. For elution, 250 µL of 33% glacial acetic acid solution was used in each well, and samples were left to rest for an hour. The crystal violet stain was quantified at OD550 using the microplate reader.
RNA-seq
P. aeruginosa colony biofilms were harvested with a sterile plastic inoculation loop. Half of each biofilm was picked up and transferred into 600 µL of Tri-reagent (Zymo Research) and disrupted by pipetting with a 1 mL tip. Total RNA was extracted with the Zymo Direct-zol RNA kit following the manufacturer’s instructions. Samples were subjected to DNase treatment using Ambion DNase I (RNase-free) kit, followed by rRNA depletion and library preparation with Illumina Stranded Total RNA Prep with Ribo-Zero kit. Libraries were sequenced on Illumina’s NextSeq 2000, with 51 bp paired-end reads and 10 bp long index reads (51-10-10-51).
RNA-seq analysis
RNA-seq reads were processed using the nf-core RNAseq pipeline version 3.4. Briefly, low-quality bases and contaminant adapters were trimmed using Cutadapt version 3.4 and Trimgalore version 0.6.7. RNA-seq reads passing quality filters were mapped against the reference genome of P. aeruginosa UCBPP_PA14 strain (GenBank assembly accession number CP000438.1) using HISAT2 version 2.2.0 (with parameters --rna-strandness RF, --no-mixed, --no-discordant, --no-spliced-alignment [67]). Next, read mappings for each annotated gene were counted using the featureCounts program within the Subread package version 2.0.1 (with parameters -B -C and -s 2 [68]). Analysis of differentially expressed genes (DEGs) was performed using the DESeq2 package 1.28.0 (69). Genes were considered significantly differentially expressed when the P value (Padj), adjusted using the Benjamini-Hochberg procedure, was <0.05. RNA-seq analysis pipeline is available on GitHub at https://github.com/trestle-biosciences/rnaseq-cbra-rhlr-crc-mukherjee. Assessment of functional classification enrichment was performed by assigning DEGs to at least one of 27 manually defined and curated PseuodCAP functional classifications (38). The percentage of genes in each category that exhibited downregulation or upregulation was then calculated.
qRT-PCR
P. aeruginosa colony biofilms were harvested with a sterile plastic inoculation loop. Half of each biofilm was picked up and transferred into 600 µL of Tri-reagent (Zymo Research) and disrupted by pipetting with a 1 mL tip. Total RNA was extracted with the Zymo Direct-zol RNA kit following the manufacturer’s instructions and quantified with a BioTek Synergy Neo2 microplate reader. 0.5 µg of total RNA was used for reverse-transcription using the TaKaRa PrimeScript RT Reagent Kit with gDNA Eraser. The resulting cDNA was used for real-time PCR using the Applied biosystems PowerTrack SYBR Green Master Mix on a Bio-Rad C1000 Touch Thermal Cycler. The results were exported into RDML v1.1 format and analyzed with web-based LinRegPCR (https://www.gear-genomics.com/rdml-tools/). Relative fold change was calculated with the qBase method (70) using 16 s, clpX, ostA, and rpsO as reference genes.
β-Galactosidase assay
Briefly, bacterial cultures were grown to OD600 ~1.0. Pellets of 1 mL cultures were collected and redissolved in 1 mL Z-buffer with the addition of 200 µg lysozyme for permeabilization. Samples were then incubated in 30°C for 15 min and diluted as needed in a total volume of 500 µL. To start the reaction, 100 µL of 4 mg/mL of ONPG solution was added to the samples and time was recorded. The samples were incubating in 30°C until sufficient color change was observed. The reaction was quenched by 250 µL 1M Na2CO3. Standard activity was calculated in Miller units.
Intracellular c-di-GMP measurements
For intracellular c-di-GMP determination, cells were grown in tryptone broth and an aliquot was taken for OD measurement. One milliliter of cells was harvested, resuspended in 100 µL nucleotide extraction solution (40% acetonitrile, 40% methanol, 0.1% formic acid, and 19.9% water), and incubated at −20°C for 20 min. The samples were centrifuged at 15,000 rpm for 5 min, and the supernatant was transferred to a fresh tube. The samples were dried under vacuum, resuspended in 100 µL HPLC grade H2O, and analyzed by HPLC-MS/MS as described previously (71). Intracellular c-di-GMP levels were determined by fitting the peak intensity to a standard curve with the known concentrations of c-di-GMP and by normalizing the levels to the total number of cells.
Phylogenetic tree construction
Genomes and associated proteomes GFF annotation files of strains from the Pseudomonas (238), Acinetobacter (5), and Burkholderia (1) genera were downloaded from the NCBI data sets database. To determine orthologous relationships between protein-coding genes, we used OrthoFinder version 2.5.4. The analysis was performed on an AWS EC2 instance type (c6a.48xlarge) with default settings. OrthoFinder computed hierarchical orthologous groups (HOGs) for each internal node in the species tree. To improve HOG prediction accuracy, an outgroup proteome (Burkholderia) was used to root the resulting species tree. HOGs are sets of proteins descended from a single gene in the ancestral species corresponding to the respective internal node. In this study, we focused on analyzing HOGs associated with the species tree node representing the last common ancestor of all A. Pseudomonas, Acinetobacter, and Burkholderia. Specifically, we examined HOGs containing cbrA, crc, and members of the pel, cupB, and cupC operons. For visualizing and annotating phylogenetic trees, custom Python scripts were employed to generate the data sets for annotation in the Interactive Tree of Life (iTol) tool (https://itol.embl.de/). Jupyter notebooks for downloading genomes from NCBI, processing OrthoFinder results, and creating figures are available on GitHub at https://github.com/JonWinkelman/dash_app_pseudomonas.
ACKNOWLEDGMENTS
We thank Eric Littman and the Microbiome Metagenomics Facility at the Duchossois Family Institute, University of Chicago, for help with whole genome sequencing. RNA sequencing was performed by Microbial Genome Sequencing Center (MiGS). Bioinformatics analysis was carried out by Trestle LLC. We thank Jonathan Winkelman for help with generating the phylogenetic tree and Dan Kearns for comments on the manuscript.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under grant R35GM139537 to C.M.W. and grants R35GM150803 and R00GM129424 and the Searle Scholars Program Grant SSP-2022-104 to S.M. The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Sampriti Mukherjee, Email: sampriti@uchicago.edu.
Anupama Khare, National Cancer Institute, Bethesda, Maryland, USA.
DATA AVAILABILITY
All relevant data are provided within the article and the supplemental information. Whole genome sequencing and RNA-seq data sets generated in this paper have been deposited to SRA with accession number PRJNA1109357.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00372-24.
Figures S1 to S9; Tables S1 and S2; captions for Tables S3 and S4.
RNA-seq data sets.
Crc binding motif analysis.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 2. Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. doi: 10.1038/nrmicro.2016.94 [DOI] [PubMed] [Google Scholar]
- 3. Karygianni L, Ren Z, Koo H, Thurnheer T. 2020. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol 28:668–681. doi: 10.1016/j.tim.2020.03.016 [DOI] [PubMed] [Google Scholar]
- 4. Liu C, Sun D, Zhu J, Liu W. 2019. Two-component signal transduction systems: a major strategy for connecting input stimuli to biofilm formation. Front Microbiol 9:3279. doi: 10.3389/fmicb.2018.03279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prüß BM. 2017. Involvement of two-component signaling on bacterial motility and biofilm development. J Bacteriol 199:1–12. doi: 10.1128/JB.00259-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capra EJ, Laub MT. 2012. Evolution of two-component signal transduction systems. Annu Rev Microbiol 66:325–347. doi: 10.1146/annurev-micro-092611-150039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. doi: 10.1586/eri.13.12 [DOI] [PubMed] [Google Scholar]
- 8. Markou P, Apidianakis Y. 2014. Pathogenesis of intestinal Pseudomonas aeruginosa infection in patients with cancer. Front Cell Infect Microbiol 3:115. doi: 10.3389/fcimb.2013.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manfredi R, Nanetti A, Ferri M, Chiodo F. 2000. Pseudomonas spp. complications in patients with HIV disease: an eight-year clinical and microbiological survey. Eur J Epidemiol 16:111–118. doi: 10.1023/a:1007626410724 [DOI] [PubMed] [Google Scholar]
- 10. Kim M, Christley S, Khodarev NN, Fleming I, Huang Y, Chang E, Zaborina O, Alverdy JC. 2015. Pseudomonas aeruginosa wound infection involves activation of its iron acquisition system in response to fascial contact. J Trauma Acute Care Surg 78:823–829. doi: 10.1097/TA.0000000000000574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol 62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x [DOI] [PubMed] [Google Scholar]
- 12. Francis VI, Stevenson EC, Porter SL. 2017. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol Lett 364:104. doi: 10.1093/femsle/fnx104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang BX, Cady KC, Oyarce GC, Ribbeck K, Laub MT. 2021. Two-component signaling systems regulate diverse virulence-associated traits in Pseudomonas aeruginosa. Appl Environ Microbiol 87:e03089-20. doi: 10.1128/AEM.03089-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sonnleitner E, Abdou L, Haas D. 2009. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 106:21866–21871. doi: 10.1073/pnas.0910308106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yeung ATY, Bains M, Hancock REW. 2011. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J Bacteriol 193:918–931. doi: 10.1128/JB.00911-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sonnleitner E, Wulf A, Campagne S, Pei X-Y, Wolfinger MT, Forlani G, Prindl K, Abdou L, Resch A, Allain FH-T, Luisi BF, Urlaub H, Bläsi U. 2018. Interplay between the catabolite repression control protein CRC, Hfq and RNA in Hfq-dependent translational regulation in Pseudomonas aeruginosa. Nucleic Acids Res 46:1470–1485. doi: 10.1093/nar/gkx1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Monteagudo-Cascales E, Santero E, Canosa I. 2022. The regulatory hierarchy following signal integration by the CbrAB two-component system: diversity of responses and functions. Genes (Basel) 13:375. doi: 10.3390/genes13020375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kambara TK, Ramsey KM, Dove SL. 2018. Pervasive targeting of nascent transcripts by Hfq. Cell Rep 23:1543–1552. doi: 10.1016/j.celrep.2018.03.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park H, McGill SL, Arnold AD, Carlson RP. 2020. Pseudomonad reverse carbon catabolite repression, interspecies metabolite exchange, and consortial division of labor. Cell Mol Life Sci 77:395–413. doi: 10.1007/s00018-019-03377-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McGill SL, Yung Y, Hunt KA, Henson MA, Hanley L, Carlson RP. 2021. Pseudomonas aeruginosa reverse diauxie is a multidimensional, optimized, resource utilization strategy. Sci Rep 11:1457. doi: 10.1038/s41598-020-80522-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. doi: 10.1146/annurev.micro.55.1.165 [DOI] [PubMed] [Google Scholar]
- 22. Mukherjee S, Bassler BL. 2019. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol 17:371–382. doi: 10.1038/s41579-019-0186-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298. doi: 10.1126/science.280.5361.295 [DOI] [PubMed] [Google Scholar]
- 24. Rumbaugh KP, Griswold JA, Hamood AN. 2000. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect 2:1721–1731. doi: 10.1016/s1286-4579(00)01327-7 [DOI] [PubMed] [Google Scholar]
- 25. Mukherjee S, Moustafa D, Smith CD, Goldberg JB, Bassler BL. 2017. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog 13:e1006504. doi: 10.1371/journal.ppat.1006504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brint JM, Ohman DE. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive luxr-luxi family. J Bacteriol 177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pearson JP, Passador L, Iglewski BH, Greenberg EP. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 92:1490–1494. doi: 10.1073/pnas.92.5.1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seed PC, Passador L, Iglewski BH. 1995. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J Bacteriol 177:654–659. doi: 10.1128/jb.177.3.654-659.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sakuragi Y, Kolter R. 2007. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J Bacteriol 189:5383–5386. doi: 10.1128/JB.00137-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang X-X, Gauntlett JC, Oldenburg DG, Cook GM, Rainey PB. 2015. Role of the transporter-like sensor kinase CbrA in histidine uptake and signal transduction. J Bacteriol 197:2867–2878. doi: 10.1128/JB.00361-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Monteagudo-Cascales E, García-Mauriño SM, Santero E, Canosa I. 2019. Unraveling the role of the CbrA histidine kinase in the signal transduction of the CbrAB two-component system in Pseudomonas putida. Sci Rep 9:9110. doi: 10.1038/s41598-019-45554-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wirtz L, Eder M, Schipper K, Rohrer S, Jung H. 2020. Transport and kinase activities of CbrA of Pseudomonas putida KT2440. Sci Rep 10:5400. doi: 10.1038/s41598-020-62337-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang Z, Nero T, Mukherjee S, Olson R, Yan J. 2021. Searching for the secret of stickiness: how biofilms adhere to surfaces. Front Microbiol 12:686793. doi: 10.3389/fmicb.2021.686793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR. 2015. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A 112:11353–11358. doi: 10.1073/pnas.1503058112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reichhardt C, Jacobs HM, Matwichuk M, Wong C, Wozniak DJ, Parsek MR. 2020. The versatile Pseudomonas aeruginosa biofilm matrix protein CdrA promotes aggregation through different extracellular exopolysaccharide interactions. J Bacteriol 202:e00216-20. doi: 10.1128/JB.00216-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang J, Sonnleitner E, Ren B, Xu Y, Haas D. 2012. Catabolite repression control of pyocyanin biosynthesis at an intersection of primary and secondary metabolism in Pseudomonas aeruginosa. Appl Environ Microbiol 78:5016–5020. doi: 10.1128/AEM.00026-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dietrich LEP, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. 2013. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol 195:1371–1380. doi: 10.1128/JB.02273-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hernández-Arranz S, Sánchez-Hevia D, Rojo F, Moreno R. 2016. Effect of Crc and Hfq proteins on the transcription, processing, and stability of the Pseudomonas putida CrcZ sRNA. RNA 22:1902–1917. doi: 10.1261/rna.058313.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ha D-G, O’Toole GA. 2015. c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa. Microbiol Spectr 3:MB-0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fu Y, Yu Z, Zhu L, Li Z, Yin W, Shang X, Chou SH, Tan Q, He J. 2021. The multiple regulatory relationship between RNA-chaperone Hfq and the second messenger c-di-GMP. Front Microbiol 12:689619. doi: 10.3389/fmicb.2021.689619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O’Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x [DOI] [PubMed] [Google Scholar]
- 43. Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci U S A 98:6911–6916. doi: 10.1073/pnas.111551898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x [DOI] [PubMed] [Google Scholar]
- 45. de Bentzmann S, Aurouze M, Ball G, Filloux A. 2006. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVB Pili. J Bacteriol 188:4851–4860. doi: 10.1128/JB.00345-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruer S, Stender S, Filloux A, de Bentzmann S. 2007. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J Bacteriol 189:3547–3555. doi: 10.1128/JB.00093-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dueholm MS, Petersen SV, Sønderkær M, Larsen P, Christiansen G, Hein KL, Enghild JJ, Nielsen JL, Nielsen KL, Nielsen PH, Otzen DE. 2010. Functional amyloid in Pseudomonas. Mol Microbiol 77:1009–1020. doi: 10.1111/j.1365-2958.2010.07269.x [DOI] [PubMed] [Google Scholar]
- 48. Wolff JA, MacGregor CH, Eisenberg RC, Phibbs PV. 1991. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J Bacteriol 173:4700–4706. doi: 10.1128/jb.173.15.4700-4706.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Valentini M, García-Mauriño SM, Pérez-Martínez I, Santero E, Canosa I, Lapouge K. 2014. Hierarchical management of carbon sources is regulated similarly by the CbrA/B systems in Pseudomonas aeruginosa and Pseudomonas putida. Microbiology (Reading) 160:2243–2252. doi: 10.1099/mic.0.078873-0 [DOI] [PubMed] [Google Scholar]
- 50. Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x [DOI] [PubMed] [Google Scholar]
- 51. Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. 1999. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun 67:5854–5862. doi: 10.1128/IAI.67.11.5854-5862.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hammond JH, Dolben EF, Smith TJ, Bhuju S, Hogan DA. 2015. Links between Anr and quorum sensing in Pseudomonas aeruginosa biofilms. J Bacteriol 197:2810–2820. doi: 10.1128/JB.00182-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, Miller SI. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros 8:66–70. doi: 10.1016/j.jcf.2008.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Feltner JB, Wolter DJ, Pope CE, Groleau MC, Smalley NE, Greenberg EP, Mayer-Hamblett N, Burns J, Déziel E, Hoffman LR, Dandekar AA. 2016. LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa . mBio 7:e01513-16. doi: 10.1128/mBio.01513-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189:8079–8087. doi: 10.1128/JB.01138-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mould DL, Finger CE, Conaway A, Botelho N, Stuut SE, Hogan DA. 2024. Citrate cross-feeding by Pseudomonas aeruginosa supports lasR mutant fitness. mBio 15:e0127823. doi: 10.1128/mbio.01278-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gil-Gil T, Valverde JR, Martínez JL, Corona F. 2023. In vivo genetic analysis of Pseudomonas aeruginosa carbon catabolic repression through the study of CrcZ pseudo-revertants shows that CRC-mediated metabolic robustness is needed for proficient bacterial virulence and antibiotic resistance. Microbiol Spectr 11:e0235023. doi: 10.1128/spectrum.02350-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. La Rosa R, Johansen HK, Molin S. 2019. Adapting to the airways: metabolic requirements of Pseudomonas aeruginosa during the infection of cystic fibrosis patients. Metabolites 9:234. doi: 10.3390/metabo9100234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Whitfield GB, Howell PL. 2021. The matrix revisited: opening night for the Pel polysaccharide across eubacterial kingdoms. Microbiol Insights 14:1178636120988588. doi: 10.1177/1178636120988588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yan J, Nadell CD, Stone HA, Wingreen NS, Bassler BL. 2017. Extracellular-matrix-mediated osmotic pressure drives Vibrio cholerae biofilm expansion and cheater exclusion. Nat Commun 8:327. doi: 10.1038/s41467-017-00401-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Seminara A, Angelini TE, Wilking JN, Vlamakis H, Ebrahim S, Kolter R, Weitz DA, Brenner MP. 2012. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc Natl Acad Sci U S A 109:1116–1121. doi: 10.1073/pnas.1109261108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dragoš A, Kovács ÁT. 2017. The peculiar functions of the bacterial extracellular matrix. Trends Microbiol 25:257–266. doi: 10.1016/j.tim.2016.12.010 [DOI] [PubMed] [Google Scholar]
- 63. Grau RR, de Oña P, Kunert M, Leñini C, Gallegos-Monterrosa R, Mhatre E, Vileta D, Donato V, Hölscher T, Boland W, Kuipers OP, Kovács ÁT. 2015. A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. mBio 6:e00581. doi: 10.1128/mBio.00581-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. 2005. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 102:11082–11087. doi: 10.1073/pnas.0502663102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Busch A, Waksman G. 2012. Chaperone-usher pathways: diversity and pilus assembly mechanism. Philos Trans R Soc Lond B Biol Sci 367:1112–1122. doi: 10.1098/rstb.2011.0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. doi: 10.1038/nprot.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. doi: 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 69. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19. doi: 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Massie JP, Reynolds EL, Koestler BJ, Cong JP, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci U S A 109:12746–12751. doi: 10.1073/pnas.1115663109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S9; Tables S1 and S2; captions for Tables S3 and S4.
RNA-seq data sets.
Crc binding motif analysis.
Data Availability Statement
All relevant data are provided within the article and the supplemental information. Whole genome sequencing and RNA-seq data sets generated in this paper have been deposited to SRA with accession number PRJNA1109357.