Abstract
Southampton virus (SHV) is a member of the Norwalk-like viruses (NLVs), one of four genera of the family Caliciviridae. The genome of SHV contains three open reading frames (ORFs). ORF 1 encodes a polyprotein that is autocatalytically processed into six proteins, one of which is p41. p41 shares sequence motifs with protein 2C of picornaviruses and superfamily 3 helicases. We have expressed p41 of SHV in bacteria. Purified p41 exhibited nucleoside triphosphate (NTP)-binding and NTP hydrolysis activities. The NTPase activity was not stimulated by single-stranded nucleic acids. SHV p41 had no detectable helicase activity. Protein sequence comparison between the consensus sequences of NLV p41 and enterovirus protein 2C revealed regions of high similarity. According to secondary structure prediction, the conserved regions were located within a putative central domain of alpha helices and beta strands. This study reveals for the first time an NTPase activity associated with a calicivirus-encoded protein. Based on enzymatic properties and sequence information, a functional relationship between NLV p41 and enterovirus 2C is discussed in regard to the role of 2C-like proteins in virus replication.
Caliciviridae represent a family of small positive-strand RNA viruses. They comprise four genera (22) distinguished by their host range and genome organization (12, 13). Two genera, the Sapporo-like viruses and the Norwalk-like viruses (NLVs), are highly contagious human pathogens that are responsible for outbreaks of epidemic acute gastroenteritis (12). NLVs, also called small round structured viruses, are currently divided into two genogroups based on nucleotide and amino acid sequence diversity (2, 23, 37, 64). The prototype Norwalk virus and Southampton virus (SHV) belong to genogroup 1. At present, there is neither a cell culture nor an animal system available to study the replication of NLVs.
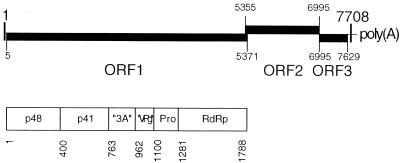
The genome of NLVs is a single copy of single-stranded, positive-sense RNA coding for three open reading frames (ORFs) (26, 28, 36) (Fig. 1). ORF 1 encodes a 200-kDa polyprotein that is autocatalytically processed into nonstructural proteins possibly involved in virus replication (38) (Fig. 1). ORF 2 codes for the major structural protein of 58 kDa, the building block of the viral capsid (17, 27, 65). ORF 3 encodes one minor structural protein of 22 kDa with unknown function (20).
FIG. 1.
Genome organization of SHV. (Top) The positive-strand RNA genome contains three ORFs. ORFs 1 and 3 are in the same frame, whereas ORF 2 is shifted +1 relative to ORFs 1 and 3. ORF 2 overlaps with ORFs 1 and 3 by 17 nt, and 1 nt, respectively. Numbers indicate nucleotide positions in the genome. (Bottom) Primary translation product of ORF 1. The polyprotein is processed by the internal 3C-like proteinase (Pro) at the amino acid positions indicated. The processing products are p48, of unknown function; p41, which shares motifs with picornavirus 2C; two proteins assumed to be equivalents of picornavirus 3A and VPg; the proteinase; and the RdRp. Adapted from references 12, 36, 38, and 39.
Sequence analyses of the ORF 1 of caliciviruses have revealed the presence of motifs in the primary translation product that are associated with distinct functions of nonstructural proteins encoded by picornaviruses and other plus-strand RNA viruses (12). These functions include a trypsin-like cysteine proteinase, an RNA-dependent RNA polymerase (RdRp), and a putative superfamily 3 (SF3) helicase (21). Most attention has been focused on the proteinase, which has been shown to be related to picornavirus 3Cpro based on similarity of sequence and function (12). The RdRp of rabbit hemorrhagic disease virus has been shown to synthesize RNA in a primer- and template-dependent manner (40) as has been demonstrated earlier for picornavirus 3Dpol (19). No function has yet been demonstrated for a calicivirus-encoded putative SF3 helicase. The observation that the translation product of calicivirus ORF 1 shares sequence motifs with picornavirus nonstructural proteins may indicate an evolutionary relationship between the two virus families (23, 24) and similar genome replication strategies.
Picornaviruses have been extensively studied, and among them poliovirus (PV) is one of the best characterized (66). The nonstructural protein 2C contains the motifs A, B, and C related to nucleoside triphosphatase (NTPase) and possibly helicase activity (38). Generally, motifs A and B, first described by Walker et al., appear in a variety of NTP-binding proteins of various functions (63). Motif C consists of an invariant asparagine residue located at a distinct distance downstream of motif B (21). Motif C is distinctive for SF3 helicases encoded by small DNA and RNA viruses (21). Protein 2C of PV and Echovirus 9, both members of the genus Enterovirus (EV), have been demonstrated to exhibit NTPase activities (32, 41, 52, 55). Recently, we have shown that a bacterially expressed fusion protein of glutathione S-transferase (GST) and PV 2C hydrolyzed ATP at least 70-fold more efficiently than other NTPs (52). Mutations in motif A, B, or C abolished ATPase activity. The ATPase activity was strongly inhibited by 1 mM guanidine hydrochloride (52), which at similar concentrations inhibits the RNA replication of PV and other picornaviruses (4, 43, 53, 66). Mutations in PV 2C that confer a guanidine-resistant or -dependent virus phenotype covaried with increased guanidine tolerance of the ATPase (52). On the basis of these data, we refer conveniently to this protein as 2CATPase (52).
SHV was isolated originally from a stool sample of a 2-year-old child during a family outbreak of acute gastroenteritis in Southampton, United Kingdom (36). Lambden et al. have constructed a full-length cDNA of the SHV genome and elucidated the sequence (36). In vitro translation of ORF 1 combined with site-directed mutagenesis and immunodetection of the processing products revealed that the 3C-like proteinase releases a protein of 41 kDa (p41) that contains motifs A to C (38). Using bacterially expressed and purified p41 of SHV we show for the first time an NTP-binding and -hydrolyzing activity associated with a calicivirus protein. The NTPase activity was independent of single-stranded nucleic acids. Comparison of calicivirus-encoded proteins with their picornavirus counterparts offers an additional opportunity to learn more about the molecular biology of human caliciviruses, a group of agents that are serious human pathogens (44).
MATERIALS AND METHODS
Materials.
Chemicals were purchased from Sigma (St. Louis, Mo.) and enzymes were purchased from Roche Biochemicals (Indianapolis, Ind.) unless stated otherwise. Oligodeoxynucleotides were synthesized on an ABI 394 DNA synthesizer (Applied Biosystems, Foster City, Calif.).
Engineering of expression plasmid.
A plasmid containing the PstI-NsiI fragment (nucleotides [nt] 953 to 2663) of SHV cDNA (gift from P. Lambden) was used as the template in a PCR with Pfu polymerase (Stratagene, La Jolla, Calif.) and the oligodeoxynucleotides 5′-GGAATTCTAGAAGCGCTGTTTCAGGGACCTGAAGAC and 5′-GCATCGATGCATGCTATTACTGTAGCTGGAACTCATCC. The amplified DNA product was phosphorylated with T4 kinase and subsequently digested with EcoRI. Plasmid pGEX-KG (25) was linearized with HindIII. The single-stranded overhangs were digested using mung bean nuclease (New England Biolabs, Beverly, Mass). Subsequently, the DNA was digested with EcoRI and dephosphorylated with calf intestine phosphatase (New England Biolabs). After gel purification, the PCR-derived and pGEX-KG-derived fragments were ligated by T4 ligase. Competent Escherichia coli DH5α cells were transformed with the ligated products. Individual clones were picked, and the p41-coding sequence of the purified plasmid clone, designated pGEX-p41, was checked for correct orientation and sequence.
To create a plasmid for the expression of mutant p41 harboring a Q at position 168 instead of the wild-type (wt) K in motif A (-p41 K168Q), we used megaprimer PCR-mediated site-directed mutagenesis (16). The antisense primer 5′-GCCTTGGTCTGGCCAATCCCAGG changed G to C and A to C at positions 1702 and 1703, respectively (positions in the SHV genome). The HindIII-DraIII fragment was excised from the final PCR product and ligated between the corresponding sites of pGEX-p41.
Expression and purification of GST-p41.
The wt and mutant GST-p41 were expressed in E. coli BL21(DE3) transformed with the corresponding plasmids. Protein expression was induced by 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at a cell optical density at 600 nm of 1 in 2×YT medium (58). Expression was allowed to proceed at 25°C for 3.5 h. Protein purification was performed as previously described for PV 2C fused to GST (52).
ATP-binding assay.
The ATP-binding assay was performed according to the method described by Clertant and Cuzin (14), with some modifications. Radiolabeled oxidized ATP was prepared by combining 8 μl of [α-33P]ATP (10 μCi/μl, 3,000 Ci/mmol; NEN, Boston, Mass.), 2 μl of 24 mM HCl, and 2 μl of 24 mM sodium m-periodate (NaIO4). The reaction mixture was incubated at room temperature in the dark for 30 min. The reaction was stopped by the addition of 3 μl of 50% glycerol. The binding reaction was prepared on ice and contained 5 μg of protein, 1 mM magnesium acetate, 5 mM dithiothreitol, 100 mM HEPES-KOH (pH 7.0), 1.7 μl of H2O, and 1 μl of radiolabeled oxidized ATP. After gentle mixing, 2 μl of 0.1 M sodium cyanoborohydride (NaCNBH3; Fluka, St. Louis, Mo.) was added immediately, and the reaction mixtures were kept on ice for 17 h. Subsequently, 5 μl of fivefold-concentrated sodium dodecyl sulfate (SDS) loading buffer was added, and the samples were boiled for 2 min and separated by SDS-polyacrylamide gel electrophoresis (PAGE) (35). A prestained protein marker (Bio-Rad, Hercules, Calif.) was run in the same gel. The gel was fixed in 30% methanol–7.5% acetic acid, enhanced using Enhance (NEN), vacuum dried, and exposed to BioMax MS film (Kodak, Rochester, N.Y.).
ATPase assay.
The standard ATPase assay mixture contained 1 μg of protein, 0.5 mM ATP, 5 mM manganese chloride, 0.01% Triton X-100, 5 mM dithiothreitol, and 20 mM HEPES-KOH (pH 7.5) in a total volume of 60 μl. The amount of inorganic phosphate was measured as a blue molybdate complex in a spectrophotometer as described elsewhere (45, 52).
Helicase assay.
A radiolabeled, partially double stranded DNA-RNA heteroduplex was generated to serve as the substrate in the helicase assay. Fifty picomoles of the dephosphorylated oligodeoxynucleotide 5′-TAGGGGGGGGGGGGCCCCCCCCTA was phosphorylated with polynucleotide kinase in the presence of 5 μl of [γ-32P]ATP (10 μCi/μl, 6,000 Ci/mmol; NEN) in a 20-μl reaction volume at 37°C for 1 h. To the reaction mixture, 1 μl of poly(C) (6 nmol/μl of CMP; Pharmacia Biotech, Piscataway, N.J.), 29 μl of H2O, and 50 μl of 2× hybridization buffer (40 mM HEPES-NaOH [pH 7.6], 1 M NaCl, 2 mM EDTA, 0.2% SDS) were added. The mixture was heated to 95°C and cooled to 25°C at a cooling rate of −0.5°C/min. Unincorporated [γ-32P]ATP was removed by gel filtration through a Nick Spin column (Pharmacia Biotech) as specified in the manufacturer's manual. The substrate was purified further by Tris-borate-EDTA PAGE according to the method described by Kim et al. (30). The DNA-RNA substrate was suspended in 80 μl of H2O. A typical unwinding reaction mixture contained 1 μl of substrate, 0.5 μg of protein, 20 mM HEPES-KOH (pH 7.2), 3 mM ATP, 8 mM MnCl2, 5 mM dithiothreitol, and 0.01% Triton X-100. The reaction was carried out at room temperature for 30 min and subsequently processed for autoradiography as described elsewhere (30).
Sequences.
The amino acid sequences of SHV p41 (accession number Q04544 [36]) and PV type 1, strain Mahoney 2C (accession number P03299 [31]), were retrieved from the SwissProt database accessed through the ExPASy proteomics server.
Sequence alignments.
Using SHV p41 and PV 2C as query sequences, related sequences were retrieved from the SwissProt and TrEMBL databases, using the FASTA program (48, 49) accessed through the EBI server. Six sequences of NLVs and 44 sequences of EVs were aligned separately. Two consensus sequences were generated, one for p41 of NLVs and one for 2C of EVs, using the default settings of the Multalin program (15) at the INRA server (France). The output of the consensus sequences was modified by replacing non-amino-acid acronyms with the most frequent residue in a particular position. Because of the limited amount of NLV sequences available, a consensus could not be found for each residue. In those cases, although rare, the residue present in the SHV p41 was selected as the consensus. Finally, the two consensus sequences were aligned to each other using the Align program at the Genestream Network server.
Secondary protein structure prediction.
Secondary protein structure predictions were performed using Profile network prediction HeiDelberg (PHD) (56, 57) accessed through the CUBIC server, using SHV p41 and PV 2C as query sequences. The PHD program created a consensus from a family of sequences related to the query sequence. Based on the consensus sequence, a one-dimensional protein structure was predicted. The predicted structure was graphically displayed using Canvas program (Deneba Systems, Miami, Fla.).
RESULTS
Purification and enzymatic characterization of SHV p41.
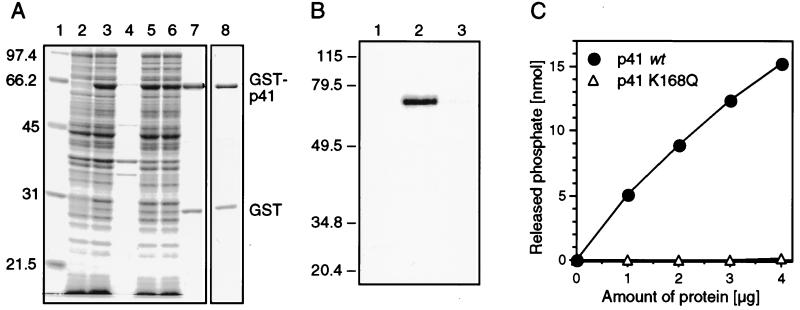
The nonstructural protein p41 of SHV is a processing product of the polyprotein encoded by ORF 1. The amino acids at either end of SHV p41 have been elucidated (38), enabling us to express authentic SHV p41 in E. coli. The GST fusion protein expression system was chosen because of its versatility and ease of protein purification. The purification steps were monitored by SDS-PAGE (Fig. 2A). High expression of GST-p41 was achieved in the presence of 0.1 mM IPTG at room temperature (lanes 2 and 3). GST-p41 migrated with an apparent molecular mass of 64 kDa, slightly faster than with the calculated molecular mass of 67.4 kDa. It may not be accidental that PV 2CATPase also migrates faster than its molecular weight would predict, for which reason it originally escaped detection (34). After cell lysis and solubilization, GST-p41 remained in the supernatant during centrifugation at 12,000 × g (lanes 4 and 5). Binding of GST-p41 to glutathione-Sepharose appeared to be inefficient, since a considerable amount of GST-p41 failed to bind to the resin (lane 6). Nevertheless, after extensive washing of the resin, a sufficiently pure GST-p41 preparation was eluted (lane 7). The amount of protein obtained was typically between 3.6 and 4 mg per liter of bacterial culture. The purity was estimated to be 75%. A major contaminant was GST which was the result of either degradation of the p41 moiety or premature termination of translation. We observed this phenomenon, which may be intrinsic to this particular GST expression system, in an earlier study (52). Using the same expression and purification strategy, we also produced the mutant p41 K168Q, a protein harboring a mutation in motif A (Fig. 2A, lane 8; see also Fig. 6).
FIG. 2.
Purification and characterization of SHV p41. (A) Coomassie blue-stained SDS-polyacrylamide gel. Lanes: 1, molecular weight marker (sizes in kilodactons are indicated on the left); 2 and 3, bacterial lysates before (lane 2) and after (lane 3) induction of GST-p41 expression; 4 and 5, insoluble (lane 4) and soluble (lane 5) fractions of lysed bacteria expressing GST-p41; 6, soluble fraction after absorption to glutathione-Sepharose; 7 and 8, eluates containing GST-p41 (lane 7) and GST-p41 K168Q (lane 8). (B) Chemical cross-linking of oxidized [α-33P]ATP to GST (lane 1), GST-p41 (lane 2), and GST-p41 K168Q (lane 3). (C) Release of inorganic phosphate from ATP in the presence of increasing amounts of GST-p41 or GST-p41 K168Q, as indicated.
FIG. 6.
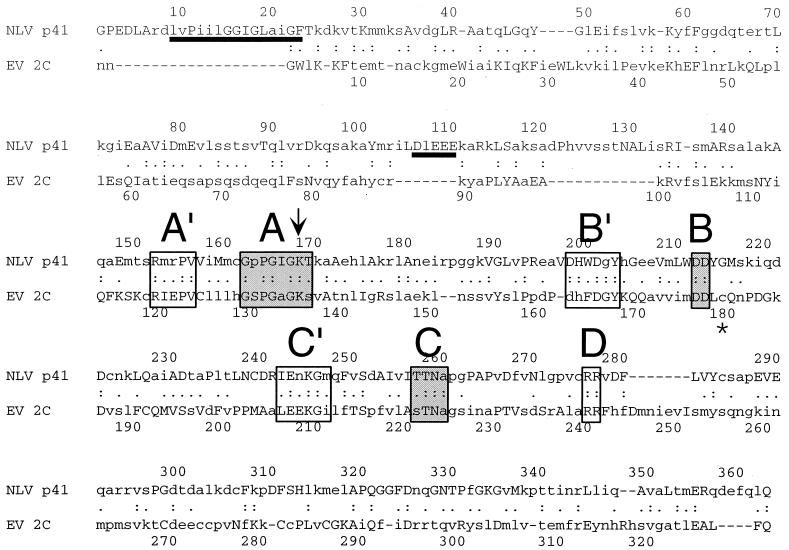
Alignment of the consensus sequence of NLV p41 with that of EV 2C. Capital letters indicate residues that are invariant in all sequences of a genus. The classical motifs of SF3 helicases are depicted in shaded boxes and labeled A to C. Regions of high similarity (A′ to C′) preceding motifs A to C and two highly conserved arginine residues (motif D) are framed. A hydrophobic and an acidic region appearing in the consensus sequence of p41, but not in that of 2C, are underlined. The arrow indicates the position of the K-to-Q mutation in GST-p41 K168Q. A glycine residue at position 179 in PV 2CATPase (asterisk) renders the ATPase activity resistant to 10 mM guanidine hydrochloride (52).
Since the presence of motifs A and B in p41 suggests binding of NTP, we tested GST-p41 for its ability to bind oxidized [33P]ATP. Chemical cross-linking of oxidized ATP with GST-p41 resulted in a covalent complex that migrated with an apparent molecular mass of approximately 64 kDa, similar to that of GST-p41 (Fig. 2B, lane 2). In contrast, oxidized ATP could not be cross-linked to GST alone (lane 1) or to GST-p41 K168Q, which lacks a functional phosphate-binding loop (lane 3).
To test for potential ATPase activity of GST-p41, we incubated GST-p41 or GST-p41 K168Q in the presence of ATP and manganese ions. Inorganic phosphate was produced in reactions containing GST-p41 (Fig. 2C). The amount of inorganic phosphate increased with increasing amounts of GST-p41. In contrast, no significant increase in inorganic phosphate concentration was observed when the reactions contained GST-p41 K168Q instead of wt GST-p41. Therefore, hydrolysis of ATP required wt p41 and was not the result of a contaminating bacterial ATPase. Taken together, these experiments demonstrated that p41 of a calicivirus is able to bind and hydrolyze ATP.
Properties of the NTPase activity of SHV p41.
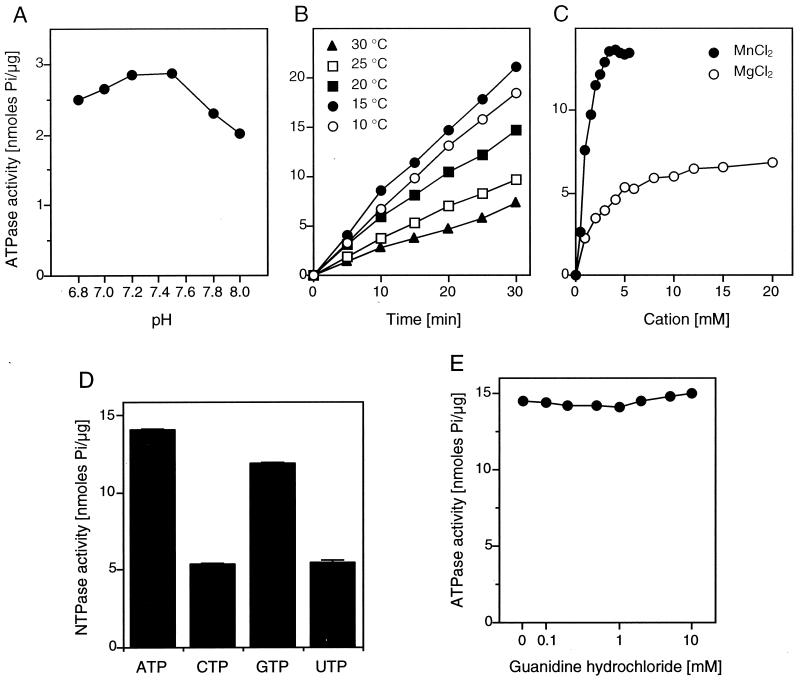
To characterize the ATPase activity of p41 in more detail, we have established the reaction conditions at which ATP is hydrolyzed very efficiently by SHV p41. The addition of bovine serum albumin or glycerol, both substances thought to enhance enzyme stability, did not increase the ATPase activity of p41 (not shown); 0.01% Triton X-100, a mild nonionic detergent, increased the ATPase activity by 50%, but higher concentrations of the detergent eliminated its stimulatory effect (not shown). The ATPase activity had a broad pH curve with optimal activity between pH 7.2 and 7.5 (Fig. 3A). Time course experiments at various temperatures indicated that ATP is hydrolyzed most efficiently at 15°C (Fig. 3B), in contrast to the optimal reaction temperature of 37°C for PV 2CATPase (52). Surprisingly, hydrolysis by p41 was linear at all temperatures tested over a period of 30 min. This may indicate that the lower activity at higher temperatures was not primarily due to thermal denaturation of the enzyme. Indeed, preincubation of p41 at 30°C only marginally reduced its activity at 15°C (not shown). As expected, ATP hydrolysis by p41 was dependent on divalent cations as cofactors. Manganese was the preferred cofactor (Fig. 3C); 3.5 to 5 mM manganese chloride appeared to be optimal at an ATP concentration of 0.5 mM. From Fig. 3C, the velocity of ATP hydrolysis was estimated to be 1.4 s−1. A 50% reduction in velocity was obtained when manganese chloride was replaced by 20 mM magnesium chloride (Fig. 3C). Zinc and calcium ions did not serve as cofactors in the ATPase reaction, as found for many other ATPases including PV 2CATPase (not shown). In the presence of manganese, 10 mM magnesium chloride or 10 mM calcium chloride inhibited the ATPase activity by 10 or 20%, respectively. Zinc chloride at a concentration of 0.3 mM inhibited ATP hydrolysis completely (not shown). The ATPase activity of p41 was moderately sensitive to sodium chloride. A concentration of 100 mM inhibited ATP hydrolysis by 30% (not shown). Based on these observations, we have defined our standard reaction conditions (see Materials and Methods).
FIG. 3.
NTPase activity of GST-p41. (A) Effect of pH. Reactions were carried out at 37°C for 1 h, and the amount of released phosphate was plotted. (B) Time course of ATP hydrolysis at various temperatures. (C) ATP hydrolysis in the presence of either manganese chloride or magnesium chloride. Reactions were performed at 15°C for 15 min. (D) Release of inorganic phosphate from various substrates (averages of three independent reactions with standard deviations). (E) ATPase activity of p41 in the presence of various concentrations of guanidine hydrochloride.
To find out whether NTPs other than ATP serve as substrates for p41, we replaced ATP by CTP, GTP, or UTP under standard conditions. It appeared that all NTPs were hydrolyzed (Fig. 3D). Therefore, p41 is an NTPase (and hence is referred to below as p41NTPase). Hydrolysis was most efficient with ATP, closely followed by GTP. CTP and UTP were hydrolyzed with an efficiency of approximately 35% of that of ATP.
To test whether p41NTPase is a target for guanidine inhibition as well, we performed ATPase reactions in the presence of various concentrations of guanidine hydrochloride. In contrast to poliovirus protein 2CATPase (52), p41NTPase did not lose its ATPase activity in the presence of up to 10 mM guanidine hydrochloride (Fig. 3E). It may be significant that amino acid residue (aa) 218 of SHV p41 is a glycine. A glycine residue at the corresponding position in PV 2CATPase renders the ATPase activity resistant to up to 10 mM guanidine (52) and confers a guanidine-resistant phenotype to PV (53).
Inhibitory effect of RNA on the activity of SHV p41NTPase.
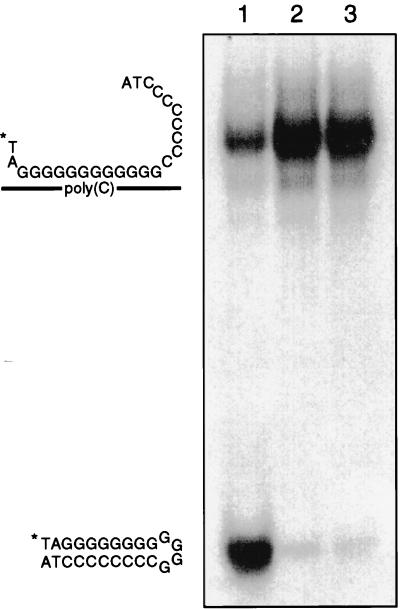
Since the presence of motif C identifies p41NTPase as a member of SF3 helicases, we tested p41NTPase for potential helicase activity. A heteroduplex consisting of a 32P-labeled oligodeoxynucleotide hybridized to poly(C) was used as the substrate in unwinding assays (Fig. 4). The substrate contained single-stranded 3′ and 5′ overhangs, which are required for duplex-unwinding activities of most RNA helicases (29). A preparation of purified hepatitis C virus (HCV) NS3 helicase (generous gift from D. W. Kim) was used as a positive control for substrate unwinding. The substrate was efficiently unwound by NS3, as evidenced by the fast migration of the labeled oligonucleotide in gel electrophoresis (Fig. 4, lane 1). As expected, the NTPase-deficient mutant p41 K168Q was not able to separate the labeled oligonucleotide from the slow-migrating duplex (lane 3). More importantly, wt p41NTPase was also inactive in this unwinding assay (lane 2), a result suggesting, but not proving, that p41NTPase does not possess helicase activity.
FIG. 4.
Helicase assay using a DNA-RNA heteroduplex as the substrate. (Left) Graphic presentation showing locations of substrate and product. ∗, radiolabeled 5′ end of oligonucleotide. (Right) Autoradiogram of the helicase reaction products separated on a nondenaturing acrylamide gel. Lanes: 1, NS3 helicase of HCV; lane 2, GST-p41; 3, GST-p41 K168Q.
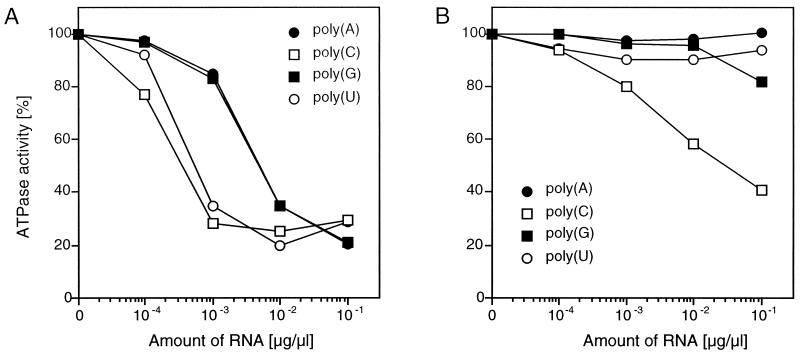
The NTPase activity of most RNA helicases is stimulated in the presence of single-stranded RNA (29). Thus, we tested the effect of ribonucleoside homopolymers on the ATPase activity of p41NTPase. Poly(C) and poly(U) at a concentration of 1 ng/μl inhibited ATP hydrolysis by 70% (Fig. 5A). It appeared that poly(A) and poly(G) were also potent inhibitors of the NTPase, although to a lesser extent than poly(C) and poly(U). In the presence of 20 mM magnesium, which may reflect a biologically more relevant cofactor for ATP hydrolysis than manganese, the ATPase activity of p41NTPase was not stimulated by homopolymeric RNAs (Fig. 5B). Significant inhibition, however, was observed only with poly(C). Taken together, the results indicate that RNA had no stimulatory effect on the NTPase activity of p41NTPase. Moreover, ATP hydrolysis was inhibited by homopolymeric RNA under otherwise optimal conditions for ATP hydrolysis. We conclude from these results that p41NTPase is distinct from typical RNA helicases.
FIG. 5.
Effect of homopolymeric RNA on the ATPase activity of GST-p41 in the presence of 5 mM manganese chloride (A) or 20 mM magnesium chloride (B). The amount of inorganic phosphate released in the absence of RNA was taken as 100%.
Amino acid sequence comparison between NLV p41 and EV 2C.
When the first sequences of ORF 1 of Norwalk virus and SHV were deciphered in 1993, it became apparent that ORF 1 encodes a protein that shares motifs found in protein 2C of picornaviruses (28, 36). Using FASTA, six deduced protein sequences spanning the 2C-like motifs of NLVs were retrieved from SwissProt and TrEMBL databases. Based on these six sequences, a consensus sequence (NLV p41) was calculated and aligned with the consensus sequence of 44 EV 2C sequences (Fig. 6). The two consensus sequences showed an overall identity of 20.3%. A hydrophobic region between residues 9 and 23 and an acidic region between residues 106 and 110 present in the NLV p41 consensus did not align with EV 2C. These regions contribute to the larger size of p41 (363 aa) than of 2C (329 aa). Additional gaps in the alignment of the two consensus sequences appear in regions of low sequence identity (lowercase letters in Fig. 6) located near the amino and carboxy termini of both consensus sequences. However, a high degree of similarity was found in the area of motifs A to C. Mutational analyses in PV have revealed that these motifs are involved in RNA replication (42, 61) and required for ATP hydrolysis (41, 52, 55). Each of these motifs appeared to be preceded by conserved regions, designated A′, B′, and C′ according to their locations upstream of motifs A, B, and C, respectively (Fig. 6). A′ to C′ partially overlap with and extend the conserved regions identified previously by Koonin and Dolja (33). Region A′ has the sequence RxxPV and is separated from motif A by five mostly hydrophobic residues. Region B′ consists of the sequence DHxDGY, of which DxY is invariant in all EV and NLV sequences. Region B′ and motif B are separated by eight residues. Finally, region C′ consists of the sequence ExKG flanked by one hydrophobic residue at both sides. The nine residues between C' and motif C are mostly hydrophobic with the exception of an invariant serine residue. We also identified two invariant arginine residues, termed motif D, 17 and 18 aa downstream of the conserved asparagine residue in motif C. Motif D is conserved among all picornavirus and calicivirus sequences (28) (data not shown), which indicates a critical function of motif D in the life cycle of these viruses.
Comparison of domain organizations of SHV p41 and PV 2C.
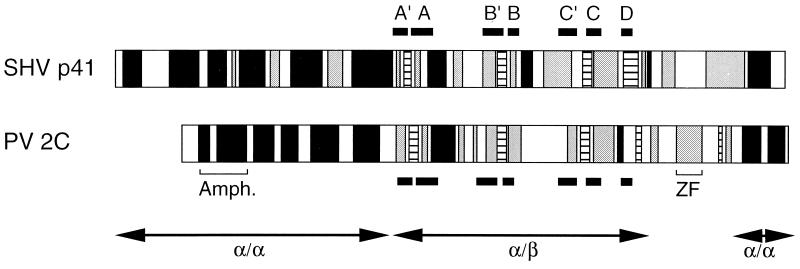
At present, a crystal structure encompassing motifs A to C of a member of the SF3 helicases is not available. To test whether the similarity in amino acids sequence between p41 and 2C is also reflected in protein structure and domain organization, we created individual one-dimensional structure predictions for p41 and 2C. Using the PHD program, we found patterns of α helices and β strands that appeared to be remarkably similar (Fig. 7). Both predictions contained a large domain dominated by α helices in the amino-terminal third of the protein. The central domain with motifs A to D contained four β strands that are well aligned between the two predictions. A small helical region appeared at the carboxy termini of both predictions. It thus appears that p41 and 2C have similar domain organizations consisting of two α/α domains that flank a central α/β domain. The α/β domain contains all of the highly conserved motifs found by sequence alignment (Fig. 6) and is thus likely to perform similar catalytic core functions in the two proteins.
FIG. 7.
Putative secondary protein structures of SHV p41 and PV 2CATPase, predicted by the PHD method (56, 57). Black, gray, and dashed regions are predicted to fold into α helices, turns, and β strands, respectively. White regions indicate no prediction. Regions of high sequence similarity (boxed or framed in Fig. 2) are labeled and indicated by black bars. The locations of known structural features of PV 2C (Amph., amphipathic helix [46]; ZF, zinc finger [51]) are indicated. The putative domain organization into α/α and α/β folds is shown by double-headed arrows.
DISCUSSION
NLVs are human enteric caliciviruses for which no cell culture or small-animal system is available. Consequently, the tools for the study of virus replication and mechanisms of viral pathogenicity are restricted to in vitro assays. These assays have mostly addressed the proteolytic activity of the virally encoded trypsin-like cysteine proteinase (12), an enzyme responsible for the cleavage of the ORF 1-encoded polyprotein. One of the cleavage products is p41, a protein with sequence motifs that suggest a relationship to SF3 helicases. In this study, we have addressed NTP binding, NTP hydrolysis, and helicase activities of SHV p41. By sequence comparison, we found regions of high similarity between NLV p41 and EV 2C.
Using a bacterially expressed GST-p41 fusion protein, we demonstrated that p41 bound ATP in a manner that required an intact motif A. P41 was able to hydrolyze ATP at a rate of up to 1.4 s−1, which is slightly higher than that reported for PV 2CATPase (0.9 s−1) (52). In contrast to PV 2CATPase, which prefers strongly ATP as substrate (52), p41 was found to be a promiscuous NTPase with a marginal preference for ATP over GTP and then CTP/UTP. To our surprise, the ATPase activity of p41 was most efficient at 15°C and in the presence of relatively high (≥3.5 mM) concentration of manganese ions. Optimal enzyme activity under rather nonbiological conditions may be explained by missing co factors that possibly play a role in the infected cell.
The NTPase activity of SHV p41NTPase was neither dependent on nor stimulated by homopolymeric RNA. A stimulation of NTPase activity by RNA is a common property of helicases (29). Moreover, whereas an RNA-DNA heteroduplex was readily unwound by HCV NS3, an RNA helicase (30), this heteroduplex was not unwound by p41NTPase. It is noteworthy that numerous attempts in several laboratories (52, 55; H. Shimizu, personal communication) have also failed to identify a helicase activity for picornavirus protein 2C. Thus, although 2C-like proteins like p41 of caliciviruses and 2C of picornaviruses have adopted the NTP-binding and hydrolysis motifs of SF3 helicases, they seem to use their ability to hydrolyze NTPs for a function distinct from nucleic acid unwinding. Under optimal assay conditions, the ATPase activity of p41NTPase was sensitive to the presence of homopolymeric RNA, an observation arguing against helicase activity and suggesting that the NTPase activity may need to be down regulated at some point during viral replication.
The NTPase activity of p41NTPase is the third enzymatic function identified for a calicivirus-encoded protein. In contrast to the proteolytic and RNA-synthesizing activities, the significance of the NTPase, however, is not obvious. In a reconstituted in vitro system, PV RNA polymerase (3Dpol) is able to initiate RNA synthesis in the absence of 2CATPase (47). However, in an in vitro translation-transcription system primed with PV RNA, initiation of RNA synthesis required the guanidine-sensitive function (4, 43), which has been identified to be the ATPase activity of 2CATPase (52). 3Dpol has also been demonstrated to possess an unwinding activity (10), allowing the polymerase to melt RNA duplexes of more than 1,000 bp in length while elongating a nascent RNA chain. However, mutations in motifs characteristic for SF3 helicases in 2CATPase debilitate PV RNA replication in the infected cell (42, 61). Thus, it seems that functions of 2C-like proteins are indispensable in the infected cell but dispensable in some biochemical reactions involving highly purified components such as initiation of RNA synthesis (47) and RNA duplex unwinding (10).
A large body of genetic and biochemical information has suggested that protein 2C of picornaviruses is involved in many processes during virus replication, yet its precise mechanism of function has not been determined. Inhibitors such as guanidine hydrochloride, benzimidazole derivatives, and derivatives of hydantoin target functions of 2C required for RNA replication (4, 43, 66) and encapsidation (62). Expression of picornavirus protein 2C in mammalian cells results in the formation of vesicles (1, 11, 59, 60). In infected cells, these virus-induced vesicles are tightly associated with the viral replication complexes (7, 9), the sites of RNA replication and virus assembly (6, 50). Protein 2C, as well as the processing precursor 2BC, are believed to spatially organize the replication complex (6, 8). Such a role of protein 2C is supported by its membrane (18, 60) and RNA-binding (3, 54) properties.
The morphology of the novel membranous structures that appear in eukaryotic cells expressing protein 2C of PV or hepatitis A virus has been studied in detail by electron microscopy (1, 11, 59, 60). A mutation in motif A of PV 2CATPase resulted in membrane structures distinct from those induced by wt 2C (11). The mutation corresponds to our mutation K168Q in p41NTPase, which affected ATP binding (Fig. 2B). Thus, NTP-bound 2C and p41 may have a structural function that is regulated by the intrinsic NTPase activity. One may speculate that 2C-like proteins are involved in RNA replication and encapsidation by creating a dynamic structural scaffold that responds to the changing requirements during virus replication. Experiments that address the biochemical functions of SHV p41NTPase and their effects on eukaryotic cells might help to clarify the mechanism by which caliciviruses and picornaviruses replicate and cause disease.
Caliciviridae and Picornaviridae are evolutionary only distantly related (5, 24). Both families include numerous pathogens that infect a wide array of different tissues in a variety of hosts. It is thus not surprising that 2C-like proteins have diverged to accommodate different protein-protein and/or protein-RNA interactions occurring during replication in a tissue-and host-specific environment (24). To keep sequence diversity minimal, we restricted our sequence comparison to the 2C-like proteins of NLVs and EVs. Both genera include predominantly human pathogens, and both contain viruses that infect the gastrointestinal tract. Comparing the consensus sequences obtained from each genus, regions of high similarity have been identified. These regions precede the known motifs A to C by distinct spacing and were thus termed A′ to C′. A′ to C′ overlap with the more extensive sequence homologies in these regions found in alignments of picornavirus sequences (67). In addition, two invariant arginine residues, called motif D, were found at a constant distance downstream of motif C. Motif D has been found in other picorna- and caliciviruses (28, 67) but is not present in the SF3 helicases for which helicase activity has been demonstrated, such as simian virus 40 large T antigen and E1 of papillomaviruses (not shown). It will be interesting to see whether motifs A′, B′, C′, and D are involved in unique functions of 2C-like members of SF3 helicases.
Individual secondary structure predictions for NLV p41 and EV 2C revealed that the seven conserved motifs map to a central domain consisting of almost identical patterns of α helices and β sheets. The same result was obtained when 2C-like protein sequences of picornaviruses, caliciviruses, and picorna-like plant viruses were all aligned and used as input for a structure prediction (60). Both approaches combined with sequence comparisons support a model in which the central α/β domain carries out the core function of 2C-like proteins (60). Taken together, our results suggest that SHV p41 and most likely the corresponding polypeptides of all caliciviruses are NTPases that are functionally related to protein 2C of picornaviruses.
ACKNOWLEDGMENTS
We are grateful to Paul Lambden for helpful discussions and for the generous gift of a plasmid containing a partial SHV cDNA. We thank Dong Wook Kim for the preparation of HCV NS3 helicase. We thank A. Gorbalenya for invaluable advice. We are indebted to Aniko Paul and Elizabeth Rieder for critically reading the manuscript.
This work was supported by grants R37-AI15122 and R01-AI32100 from the National Institutes of Health.
REFERENCES
- 1.Aldabe R, Carrasco L. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem Biophys Res Commun. 1995;206:64–76. doi: 10.1006/bbrc.1995.1010. [DOI] [PubMed] [Google Scholar]
- 2.Ando T, Mulders M N, Lewis D C, Estes M K, Monroe S S, Glass R I. Comparison of the polymerase region of small round structured virus strains previously classified in three antigenic types by solid-phase immune electron microscopy. Arch Virol. 1994;135:217–226. doi: 10.1007/BF01309781. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee R, Echeverri A, Dasgupta A. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J Virol. 1997;71:9570–9578. doi: 10.1128/jvi.71.12.9570-9578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton D J, Flanegan J B. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol. 1997;71:8482–8489. doi: 10.1128/jvi.71.11.8482-8489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berke T, Matson D O. Reclassification of the Caliciviridae into distinct genera and exclusion of hepatitis E virus from the family on the basis of comparative phylogenetic analysis. Arch Virol. 2000;145:1421–1436. doi: 10.1007/s007050070099. [DOI] [PubMed] [Google Scholar]
- 6.Bienz K, Egger D, Pfister T. Characteristics of the poliovirus replication complex. Arch Virol Suppl. 1994;9:147–157. doi: 10.1007/978-3-7091-9326-6_15. [DOI] [PubMed] [Google Scholar]
- 7.Bienz K, Egger D, Rasser Y, Bossart W. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology. 1980;100:390–399. doi: 10.1016/0042-6822(80)90530-9. [DOI] [PubMed] [Google Scholar]
- 8.Bienz K, Egger D, Troxler M, Pasamontes I. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J Virol. 1990;64:1156–1163. doi: 10.1128/jvi.64.3.1156-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caliguiri L, Tamm I. Membranous structures associated with translation and transcription of poliovirus RNA. Science. 1969;166:885–886. doi: 10.1126/science.166.3907.885. [DOI] [PubMed] [Google Scholar]
- 10.Cho M W, Richards O C, Dmitrieva T M, Agol A, Ehrenfeld E. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J Virol. 1993;67:3010–3018. doi: 10.1128/jvi.67.6.3010-3018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 12.Clarke I N, Lambden P R. The molecular biology of caliciviruses. J Gen Virol. 1997;78:291–301. doi: 10.1099/0022-1317-78-2-291. [DOI] [PubMed] [Google Scholar]
- 13.Clarke I N, Lambden P R. Organization and expression of calicivirus genes. J Infect Dis. 2000;181(Suppl. 2):S309–S316. doi: 10.1086/315575. [DOI] [PubMed] [Google Scholar]
- 14.Clertant P, Cuzin F. Covalent affinity labelling by periodate-oxidized [α-32P]ATP of the large-T proteins of polyoma and SV40 viruses. J Biol Chem. 1982;257:6300–6305. [PubMed] [Google Scholar]
- 15.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta A K. Efficient amplification using “megaprimer” by asymmetric polymerase chain reaction. Nucleic Acids Res. 1995;23:4530–4531. doi: 10.1093/nar/23.21.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dingle K E, Lambden P R, Caul E O, Clarke I N. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from genetic group II small round structured virus. J Gen Virol. 1995;76:2349–2355. doi: 10.1099/0022-1317-76-9-2349. [DOI] [PubMed] [Google Scholar]
- 18.Echeverri A C, Dasgupta A. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology. 1995;208:540–553. doi: 10.1006/viro.1995.1185. [DOI] [PubMed] [Google Scholar]
- 19.Flanegan J B, Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A) Proc Natl Acad Sci USA. 1977;74:3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass P J, White L J, Ball J M, Leparc-Goffard I, Hardy M E, Estes M K. Norwalk virus open reading frame encodes a minor structural protein. J Virol. 2000;74:6581–6591. doi: 10.1128/jvi.74.14.6581-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 22.Green K Y, Ando T, Balayan M S, Berke T, Clarke I N, Estes M K, Matson D O, Nakata S, Neill J D, Studdert M J, Thiel H-J. Taxonomy of the caliciviruses. J Infect Dis. 2000;181(Suppl. 2):S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- 23.Green S M, Dingle K E, Lambden P R, Caul E O, Ashley C R, Clarke I N. Human enteric Caliciviridae: a new and prevalent small round structured virus group defined by RNA-dependent RNA polymerase and capsid diversity. J Gen Virol. 1994;75:1883–1888. doi: 10.1099/0022-1317-75-8-1883. [DOI] [PubMed] [Google Scholar]
- 24.Gromeier M, Wimmer E, Gorbalenya A E. Genetics, pathogenesis, and evolution of picornaviruses. In: Domingo E, Webster R G, Holland J J, editors. Origin and evolution of viruses. New York, N.Y: Academic Press; 1999. pp. 287–343. [Google Scholar]
- 25.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X, Graham D Y, Wang K, Estes M K. Norwalk virus genome cloning and characterization. Science. 1990;250:1580–1583. doi: 10.1126/science.2177224. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X, Wang M, Graham D Y, Estes M K. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X, Wang M, Wang K, Estes M K. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 29.Kadaré G, Haenni A-L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 31.Kitamura N, Semler B L, Rothberg P G, Larsen G R, Adler C J, Dorner A J, Emini E A, Hanecak R, Lee J, van der Werf S, Anderson C W, Wimmer E. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- 32.Klein M, Eggers H J, Nelsen-Salz B. Echovirus 9 strain Barty non-structural protein 2C has NTPase activity. Virus Res. 1999;65:155–160. doi: 10.1016/s0168-1702(99)00110-0. [DOI] [PubMed] [Google Scholar]
- 33.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 34.Korant B D. Cleavage of poliovirus-specific polypeptides aggregates. J Virol. 1973;12:556–563. doi: 10.1128/jvi.12.3.556-563.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Lambden P R, Caul E O, Ashley C R, Clarke I N. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259:516–519. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- 37.Lew J F, Kapikian A Z, Valdesuso J, Green K Y. Molecular characterization of Hawaii virus and other Norwalk-like viruses: evidence for genetic polymorphism among human caliciviruses. J Infect Dis. 1994;170:535–542. doi: 10.1093/infdis/170.3.535. [DOI] [PubMed] [Google Scholar]
- 38.Liu B, Clarke I N, Lambden P R. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J Virol. 1996;70:2605–2610. doi: 10.1128/jvi.70.4.2605-2610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B L, Viljoen G J, Clarke I N, Lambden P R. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J Gen Virol. 1999;80:291–296. doi: 10.1099/0022-1317-80-2-291. [DOI] [PubMed] [Google Scholar]
- 40.López Vázquez A, Martín Alonso J M, Casais R, Boga J A, Parra F. Expression of enzymatically active rabbit hemorrhagic disease virus RNA-dependent RNA polymerase in Escherichia coli. J Virol. 1998;72:2999–3004. doi: 10.1128/jvi.72.4.2999-3004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirzayan C M, Wimmer E. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology. 1994;199:176–187. doi: 10.1006/viro.1994.1110. [DOI] [PubMed] [Google Scholar]
- 42.Mirzayan C M, Wimmer E. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology. 1992;189:547–555. doi: 10.1016/0042-6822(92)90578-d. [DOI] [PubMed] [Google Scholar]
- 43.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 44.Monroe S S, Ando T, Glass R I. Introduction: human enteric Caliciviruses—an emerging pathogen whose time has come. J Infect Dis. 2000;181(Suppl. 2):S249–S251. doi: 10.1086/315594. [DOI] [PubMed] [Google Scholar]
- 45.Noble S, Nibert M L. Characterization of an ATPase activity in reovirus cores and its genetic association with core-shell protein λ1. J Virol. 1997;71:2182–2191. doi: 10.1128/jvi.71.3.2182-2191.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul A V, Molla A, Wimmer E. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology. 1994;199:188–199. doi: 10.1006/viro.1994.1111. [DOI] [PubMed] [Google Scholar]
- 47.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 48.Pearson W R. Rapid and sensitive comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 49.Pearson W R, Lipman D J. Improved tool for biological sequence analysis. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfister T, Egger D, Bienz K. Poliovirus subviral particles associated with progeny RNA in the replication complex. J Gen Virol. 1995;76:63–71. doi: 10.1099/0022-1317-76-1-63. [DOI] [PubMed] [Google Scholar]
- 51.Pfister T, Jones K W, Wimmer E. A cysteine-rich motif in poliovirus protein 2C ATPase is involved in RNA replication and binds zinc in vitro. J Virol. 2000;74:334–343. doi: 10.1128/jvi.74.1.334-343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfister T, Wimmer E. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J Biol Chem. 1999;274:6992–7001. doi: 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- 53.Pincus S E, Wimmer E. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J Virol. 1986;60:793–796. doi: 10.1128/jvi.60.2.793-796.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodríguez P L, Carrasco L. Poliovirus protein 2C contains two regions involved in RNA binding activity. J Biol Chem. 1995;270:10105–10112. doi: 10.1074/jbc.270.17.10105. [DOI] [PubMed] [Google Scholar]
- 55.Rodríguez P L, Carrasco L. Poliovirus protein 2C has ATPase and GTPase activities. J Biol Chem. 1993;268:8105–8110. [PubMed] [Google Scholar]
- 56.Rost B. PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 1996;183:63–98. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 57.Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins. 1994;20:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 59.Teterina N L, Bienz K, Egger D, Gorbalenya A E, Ehrenfeld E. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology. 1997;237:66–77. doi: 10.1006/viro.1997.8775. [DOI] [PubMed] [Google Scholar]
- 60.Teterina N L, Gorbalenya A E, Egger D, Bienz K, Ehrenfeld E. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J Virol. 1997;71:8962–8972. doi: 10.1128/jvi.71.12.8962-8972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teterina N L, Kean K M, Gorbalenya A E, Agol V I, Girard M. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J Gen Virol. 1992;73:1977–1986. doi: 10.1099/0022-1317-73-8-1977. [DOI] [PubMed] [Google Scholar]
- 62.Vance L M, Moscufo N, Chow M, Heinz B A. Poliovirus 2C region functions during encapsidation of viral RNA. J Virol. 1997;71:8759–8765. doi: 10.1128/jvi.71.11.8759-8765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the a- and b-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Jiang X, Madore H P, Gray J, Desselberger U, Ando T, Seto Y, Oishi I, Lew J F, Green K Y, Estes M K. Sequence diversity of small, round-structured viruses in the Norwalk virus group. J Virol. 1994;68:5982–5900. doi: 10.1128/jvi.68.9.5982-5990.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White L J, Hardy M E, Estes M K. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J Virol. 1997;71:8066–8072. doi: 10.1128/jvi.71.10.8066-8072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wimmer E, Hellen C U, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 67.Zimmern D. Evolution of RNA viruses. In: Holland J J, Domingo E, Alquist P, editors. RNA genetics. Vol. 2. Boca Raton, Fla: CRC Press; 1988. pp. 211–240. [Google Scholar]