Abstract
In rotavirus, transcription of the 11 double-stranded RNA genome segments occurs within the structurally intact subviral particle, and nascent transcripts are released through channels penetrating the two capsid layers at the icosahedral vertices. To gain insight into the early molecular events in transcription, we used high-resolution polyacrylamide gel electrophoresis to investigate the length distribution of transcription products at various times following initiation. We observed that, in the subviral particle under normal conditions, transcript initiation and capping are followed by a momentary pause in elongation after the addition of 6 to 7 nucleotides. In the absence of the capping reaction cofactor S-adenosylmethionine, conditions under which the rate of nucleotide incorporation is reduced, we observe a significant decrease in the ratio of paused to full-length transcripts. We propose that this pause site may represent the point at which specific molecular events take place to facilitate processive elongation. Furthermore, our results indicate that the presence of specific ligands on the viral surface, such as VP7 in the mature virion, inhibits polymerase function. From the perspective of the viral replication cycle, this inhibition may serve to ensure that transcription occurs with greatest efficiency only after the virus has entered the cytoplasm and assumed the form of a double-layered particle.
One of the major distinguishing characteristics of viruses that have segmented double-stranded RNA (dsRNA) genomes is that transcription of the genome occurs within the structurally intact core of the virion (reviewed in reference 8). Genome transcription occurs through the concerted action of multiple enzyme activities housed within the viral core, and mature transcripts are released through channels penetrating the viral capsid. In the largest known family of segmented dsRNA viruses, the Reoviridae, whose members all infect higher eukaryotes, the viral core contains all of the enzyme activities needed to synthesize not only the mRNA transcripts themselves but also a properly guanylated and methylated cap structure on the 5′ end, as required by the eukaryotic translation initiation machinery. Rotaviruses, because of their medical importance as the leading cause of life-threatening gastroenteritis in young children (12), have been studied extensively using biochemical and structural techniques and represent a good model system in which to investigate the molecular and chronological events of genome transcription in segmented dsRNA viruses.
Architecturally, the mature rotavirus particle consists of three concentric icosahedrally ordered protein capsid layers which are assembled around a genome of 11 segments of dsRNA (reviewed in reference 22). At the start of the replication cycle, rotavirus attaches to the host cell as a triple-layered particle (TLP). Upon cell entry, the outermost capsid layer, consisting of a shell protein (VP7) and a spike protein (VP4), is removed, and the resulting double-layered particle (DLP) becomes transcriptionally competent. The production of mature mRNA transcripts may be considered to occur in three stages (reviewed in reference 15). Transcription starts with initiation, at which time nucleotidyl transfer begins and capping of the nascent transcript occurs; this is followed by a transition to processive elongation, during which the transcript begins to separate from the genomic template and nucleotidyl transfer continues as the transcription bubble progresses through the template. Elongation at the polymerase active site is followed by translocation of the growing transcript through channels which penetrate the inner VP2 and outer VP6 capsid layers at the icosahedral vertices (13).
The enzymatic steps of transcription, nucleotide incorporation, and cap formation, are performed by two proteins, VP1 and VP3, both of which are housed within the viral core (reviewed in reference (6)). VP1 is the viral RNA-dependent RNA polymerase (28), and VP3 is the enzyme implicated in cap formation (4, 16, 20, 21). Mature rotavirus transcripts are capped with the sequence m7GpppG(m) at the 5′ end (11, 18). The formation of such a cap structure requires the activity of a guanylyltransferase enzyme, which uses GTP as a substrate, as well as a methyltransferase activity, which uses S-adenosylmethionine (SAM) as a substrate (10). Rotavirus VP3 has been shown to possess both guanylyltransferase and methyltransferase activity. Structural studies suggest that the rotavirus core contains 12 copies of VP1 and VP3, which appear to be incorporated as heterodimers anchored to the inner surface of the capsid at each of the icosahedral vertices (24).
Although previous biochemical studies have provided a characterization of the enzymatic reactions which occur during transcription, and structural studies of actively transcribing DLPs have provided a framework in which to understand how mRNA is translocated through the capsid, less is known about how the upstream events of initiation and elongation unfold from a mechanistic perspective. In particular, it is not known what events constitute the point of transition between initiation and processive elongation when this transition takes place following initiation and what structural factors may influence the ability of the endogenous transcription apparatus to make this transition successfully.
To gain insight into the early molecular events in transcription, we used high-resolution denaturing polyacrylamide gel electrophoresis to investigate the length distribution of transcription products at various times following initiation, both in particles which are capable of producing full-length transcription products and in those which are incapable of undergoing the transition from initiation to processive elongation. We observed that, under normal transcription conditions, initiation and capping appear to be followed by a brief pause in elongation after 5 to 6 nucleotides have been transcribed. We propose that this pause site constitutes the point of transition between initiation and a commitment to proceed with full-length elongation. Our results also suggest that the ability of the transcription apparatus to progress successfully from initiation to processive elongation is determined primarily by the extent to which the enzymatic efficiency of the polymerase is modulated by the conformational states of other proteins in the virus.
MATERIALS AND METHODS
Viruses.
Rotavirus strain SA11-4F was propagated in MA-104 cells according to standard procedures and purified as previously described (23). Purified TLPs were stored in TNC buffer (10 mM Tris [pH 7.4], 140 mM NaCl, 10 mM CaCl2), and DLPs were stored in Tris-buffered saline.
In vitro transcription.
In vitro transcription was carried out using a basic transcription reaction buffer containing 100 mM Tris-HCl (pH 8); 10 mM magnesium acetate; 1 mM ATP; a 100 μM concentration (each) of GTP, CTP, and UTP; and 1 μCi of [α-32P]GTP/ml (3,000 Ci/mmol; Amersham Pharmacia Biotech). Reaction mixture also contained 0.5 mM SAM unless otherwise indicated. Reactions requiring radiolabeled SAM contained 0.5 mM S-adenosyl-l-[methyl-3H]methionine (500 mCi/mmol; Amersham-Pharmacia Biotech), and [α-32P]GTP was omitted. Transcription reactions were incubated at 37°C as described in the text. The quantity of virus particles present in each reaction is specified in the text. Because TLP preparations typically also contain a small number of DLPs (usually less than 1%), TLP transcription reactions were supplemented with the anti-VP6 Fab 2A11/E9 at 25 μg/ml when radiolabeled GTP was included in the reaction mixture in order to suppress full-length mRNA production from contaminating DLPs (14).
Unless otherwise indicated, reaction mixtures were prepared for electrophoresis using the following procedure. First, the reaction mixture was passed through a Micro Bio Spin P-30 column equilibrated in 10 mM Tris buffer (Bio-Rad) to remove salts and unincorporated nucleotides as well as short oligonucleotide transcription products (length, less than ∼20 bp) which may have been released prematurely from the particles. The gel filtration matrix in these columns has a size exclusion limit equivalent to a 20-bp double-stranded DNA fragment or a 40-kDa globular protein. The eluate was lyophilized and resuspended in 10 μl of denaturing TBE (Tris-borate-EDTA) sample buffer (containing 7 M urea and 0.1% SDS) and then boiled for 5 min to ensure complete disruption of the particles. For reactions in which the purpose was to examine the total yield of initiated transcripts (including both particle-bound and prematurely released oligonucleotides), reaction mixtures were simply diluted fourfold with denaturing TBE sample buffer and boiled for 5 min prior to electrophoresis.
High-resolution denaturing polyacrylamide gel electrophoresis (PAGE).
The products of in vitro transcription were resolved by electrophoresis on 20% polyacrylamide gels (0.4 mm thickness) containing 7 M urea. Electrophoresis was typically carried out at 400 V until the bromophenol blue dye front migrated 20 cm (between 22 and 24 h). Following electrophoresis, both the gel and one supporting glass plate were wrapped in a thin plastic film and autoradiographed for an appropriate period of time as needed to visualize the transcription products of interest.
Quantitation of 32P-labeled transcription products.
To quantitate the amount of radioactivity associated with specific transcription products resolved by denaturing PAGE, regions of the gel containing the product of interest were placed in contact with a Molecular Dynamics Phosphor Storage Screen. Following exposure, the signal stored in the phosphor screen was quantitated on a Molecular Dynamics Storm 860 PhosphorImager operated at a 200-μm scanning interval, and image analysis was carried out using the ImageQuant software package (Molecular Dynamics). Quantitation was performed on the desired bands by drawing a rectangular mask around the edge of each band and noting the total number of counts present within the mask. Background correction was performed by subtracting the counts obtained within similar-sized masks drawn around corresponding areas from a negative control reaction.
Quantitation of 3H-labeled transcription products.
For reactions in which the 5′ cap was labeled using S-adenosyl-l-[methyl-3H]methionine, regions of the gel containing the transcription products of interest were excised, crushed, and mixed with ∼3 volumes of a standard nucleotide elution buffer containing 0.5 M ammonium acetate and 10 mM magnesium acetate (pH7). The gel slices were incubated with the elution buffer for 36 h at 40°C with rapid agitation. Following elution of the nucleic acid, the supernatant (∼200 μl) was withdrawn and mixed with 5 ml of CytoScint ES scintillation fluid (ICN Biomedicals). Radioactivity was quantitated using a Beckman 1010LS liquid scintillation counter operating within the tritium counting window.
Pulse-chase analysis of transcription in TLPs and structurally converted DLPs.
TLPs were added to a standard transcription reaction buffer containing SAM and supplemented with [α-32P]GTP. The reaction was incubated at 37°C for 5 min. After this initial pulse incubation, both the TLP reaction, as well as a negative control reaction containing [α-32P]GTP but no particles, were passed successively through two Micro Bio Spin P-30 columns equilibrated in 10 mM Tris-HCl buffer (pH 8.6) and 10 mM magnesium acetate to remove unincorporated radiolabeled nucleotides. Particles eluting in the void volume were divided into three equal portions. One portion was set aside as a sample for gel electrophoretic analysis. The second portion was added to a standard transcription reaction mixture without radiolabeled nucleotides and incubated for 10 min at 37°C. A third portion was incubated for 5 min at room temperature with 5 mM BAPTA [1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid] (Sigma) to remove the outermost capsid layer (VP7 capsid plus VP4 spikes) without substantially affecting the Mg2+ concentration in the buffer. The resulting DLPs were then added to a standard transcription reaction mixture without radionucleotides and incubated for 10 min at 37 °C. Following incubation, these two chase reactions were used directly as samples for gel electrophoresis.
RESULTS
Short oligonucleotides are produced during transcription.
As with most transcription systems, the production of mature, full-length mRNA transcripts in rotavirus is believed to occur in several stages, beginning with initiation and followed by a transition to processive elongation and, finally, translocation out of the particle interior. To visualize the manner in which growing transcripts progress from initiation to elongation, we examined the length distribution of transcription products as a function of time in both native DLPs, the viral form in which transcription ordinarily occurs, and mature TLPs, a viral form in which full-length transcripts cannot be made. Virus particles were incubated in a standard in vitro transcription reaction mixture, and, at various time points, aliquots of the reaction mixture were removed and analyzed by high-resolution denaturing PAGE.
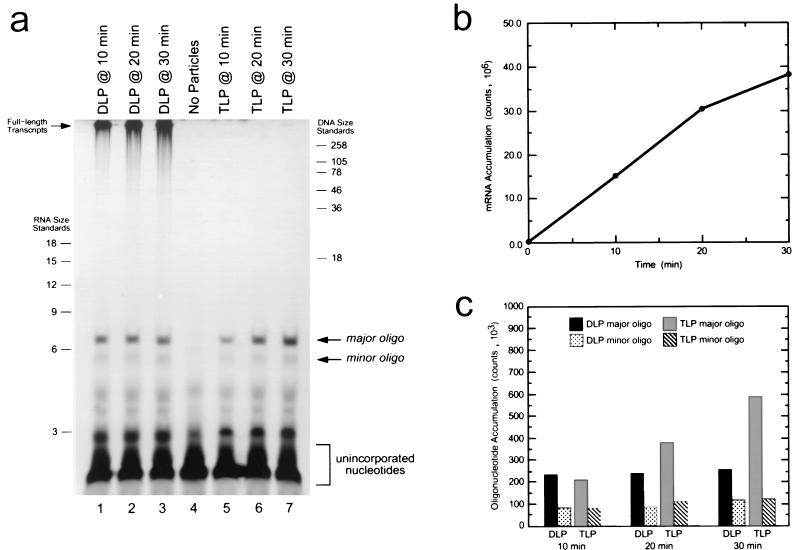
In the DLP reaction (Fig. 1a, lanes 1 to 3), two distinct size classes of transcription products were observed. As expected, the majority of the transcription products were full-length mRNAs. These remained at the top of the gel and accumulated steadily throughout the course of the reaction (Fig. 1b). In the lower portion of the gel, where transcription products shorter than 20 nucleotides could be resolved, a series of short oligonucleotide species having various discrete lengths were also observed. Of these, most were also present to some degree in a control reaction that did not contain any particles (lane 4), indicating that they did not likely represent specific products of transcription. However, two oligonucleotide species migrating above and below the 6-nucleotide single-stranded RNA (ssRNA) size marker were present specifically in the DLP reaction; these were designated the major oligo and minor oligo. Measurement of the radioactivity present in the major oligonucleotide product indicated that, unlike the full-length transcripts, the quantity of the major oligonucleotide species remained essentially constant throughout the course of the reaction (Fig. 1c). This observation indicates that, under conditions which permit full-length elongation, these oligonucleotides do not accumulate appreciably as the reaction progresses, suggesting that they exist only transiently during the transcription process.
FIG. 1.
Analysis of transcription products in DLP and TLP. (a) Visualization of transcription products by 20% denaturing PAGE. Lanes 1 to 3, DLP transcription products sampled after 10, 20, and 30 min of incubation at 37 °C (each reaction aliquot contained 1-μg particles); lane 4, negative-control reaction containing no particles, sampled after 30 min of incubation; lanes 5 to 7, TLP transcription products sampled after 10, 20, and 30 min of incubation. Each reaction aliquot contained 2-μg particles. RNA and DNA size standards are indicated (length in nucleotides). (b) Quantitation of full-length mRNA yield in the DLP transcription reaction. (c) Quantitation of the major and minor oligoncleotide species in the DLP and TLP reactions at each of the indicated time points. Quantitation was performed as described in the Materials and Methods.
In the TLP reaction (Fig. 1a, lanes 5 to 7), the products of transcription consisted entirely of short oligonucleotide species migrating in the lower portion of the gel. Interestingly, the length distribution of oligonucleotides in this region was identical to that observed in the DLP reaction, and likewise, the two oligonucleotide species migrating near the 6-nucleotide ssRNA size marker were present in the TLP reaction and absent in the control reaction, suggesting that they were specific products of transcription. Unlike in the DLP reaction, however, the quantity of the major oligonucleotide species increased steadily over time (Fig. 1c). This observation suggests that, under conditions where initiation can occur but full-length elongation is prevented, these oligonucleotides represent aborted transcripts which gradually accumulate as a result of repeatedly occurring cycles of initiation followed by limited elongation.
Short oligonucleotides produced in DLPs during transcription are particle associated.
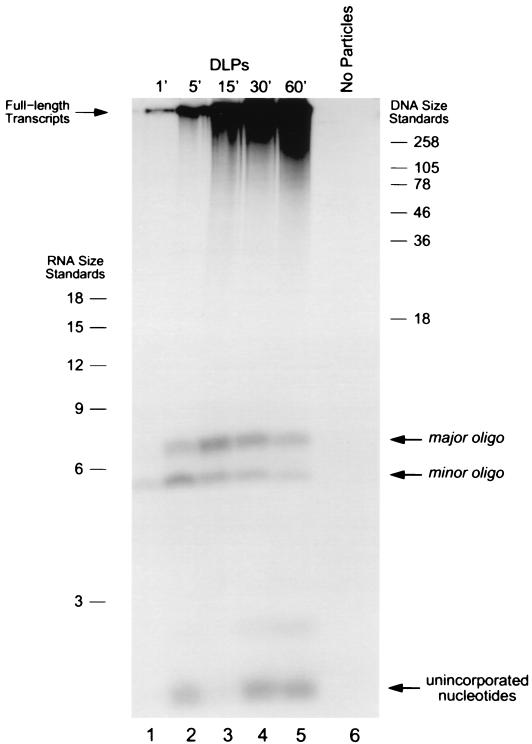
To determine whether these oligonucleotide transcription products are particle associated, as would be expected if they represent early intermediates in the elongation process, we used gel filtration to separate actively transcribing DLPs and elongated transcripts from free nucleotides and oligonucleotide species shorter than ∼20 bases. Aliquots taken from a standard transcription reaction at various time points were passed through a centrifugal gel filtration column, and material eluting in the void volume was then analyzed by high-resolution denaturing PAGE (Fig. 2).
FIG. 2.
Analysis of particle-bound oligonucleotide transcription products in DLP. Lanes 1 to 5 contain DLP transcription products sampled after incubation at 37°C for the indicated period of time. Prior to electrophoresis, each reaction aliquot was passed through a size exclusion column to remove free nucleotides and unbound oligonucleotides shorter than 20 bases. Material eluting in the void volume was resolved on the gel. Each reaction aliquot contained 6-μg particles. Close inspection of the intensity of major and minor oligonucleotide bands in the gel indicates that the ratio of the two products varied slightly over the time course of the reaction and, additionally, was different from that observed in Fig. 1. Variations in the ratio of the major to minor oligonucleotide products were routinely observed throughout our studies and are likely to reflect subtle variations in reaction conditions. In all of our experiments, however, the quantity of the minor oligonucleotide species was consistently observed to be less than that of the major oligonucleotide species. Lane 6 contains a negative control reaction with no particles, sampled after 15 min of incubation. The intensity of the band corresponding to unincorporated nucleotides is not the same in each reaction aliquot, perhaps because of the varying efficiency with which the size exclusion columns removed free radiolabeled nucleotides after transcription. RNA and DNA size standards are indicated (length in nucleotides).
As previously observed, two classes of transcription products were produced: full-length mRNAs and two species of short oligonucleotides migrating near the 6-nucleotide ssRNA size marker. Although the full-length mRNAs are significantly larger than the size exclusion limit of the gel filtration matrix and thus would be expected to pass through without being retained in the column, the short oligonucleotides, which are much smaller than the exclusion limit, would not be expected to elute in the void volume unless they were incorporated within a much larger molecular complex. The fact that these short oligonucleotides were not retained on the gel filtration column suggests that they were particle associated. This observation further indicates that these short oligonucleotide transcription products are transient intermediates during either the initiation or elongation processes. Similar experiments indicated that the short oligonucleotides produced under transcription conditions in TLPs are initially particle associated as well (data not shown).
Both major and minor oligonucleotide transcription products are capped.
The observation that specific, particle-associated oligonucleotide transcripts having discrete lengths and transient lifetimes can be observed at a very early point in the transcription process suggests that one or more of the initial steps in mRNA production occurs more slowly than the other steps, giving rise to a population of transcripts which are temporarily stalled at a specific point. To determine whether this delay in transcription occurs before or after the events involved in cap formation, we investigated whether or not a methylated cap can be detected on the 5′ end of either of these oligonucleotide transcripts. Methylation is recognized to be the final step in cap formation and involves the transfer of a methyl group from S-adenosylmethionine (SAM) to the penultimate Gppp moiety attached by the guanylyltransferase (4, 11, 26). The rotavirus methyltransferase has also been observed to attach a methyl group to the first sequence-derived G base uniformly conserved in all rotavirus mRNA transcripts, although with less efficiency and only following the attachment and methylation of a G base on the 5′ end (11).
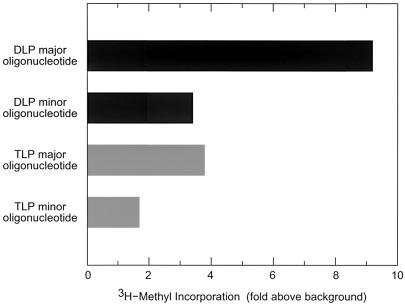
Transcription was carried out in the presence of tritium-labeled SAM, and the resulting particle-associated oligonucleotides were isolated and examined for the incorporation of tritium (Fig. 3). The results indicate that both the major and minor oligonucleotide species in the DLP and the TLP became labeled with tritium. Because capped and uncapped oligonucleotides migrate quite differently in high-resolution denaturing polyacrylamide gels (30, 31), the detection of a tritium label in both the major and minor bands implies that these species consisted entirely of capped oligonucleotides (i.e., contain a Gppp moiety), as uncapped oligonucleotides, were they to arise, would not be found at the same locations in the gel. Taken together, these observations suggest that the momentary pause in transcription, which gave rise to the observed oligonucleotides, occurred after cap formation. The observation that transcripts are capped shortly after initiation suggests that the polymerase (VP1) and capping enzyme (VP3) are positioned in close proximity to one another in the viral core, in agreement with structural studies (24).
FIG. 3.
Analysis of 3H-methyl incorporation in oligonucleotide transcription products. The quantity of tritium radioactivity present in each of the indicated RNA species is expressed with respect to background. To estimate the level of background radioactivity, an average value of 55.4 cpm (range, 49 to 63 cpm) was obtained from five independent measurements taken from regions of the polyacrylamide gel which were adjacent to those from which oligonucleotide transcription products were isolated. A series of parallel reactions run with both 3H and 32P labeling (data not shown) suggested that the variation in tritium signal strength between the major and minor oligonucleotides of DLPs and TLPs was due primarily to differences in the quantity of the oligonucleotide species present in the gel, as indicated by 32P incorporation.
Oligonucleotides are not observed during transcript elongation in DLPs when the efficiency of the polymerase is reduced.
To examine whether a decrease in the rate of nucleotide incorporation leads to a reduction in the number of initiated transcripts which appear to be stalled early in the elongation process, as would be characteristic of a transcription pause site, we carried out transcription in both the presence and absence of the capping reaction cofactor SAM and compared the ratio of particle-bound oligonucleotides to full-length transcripts in the two reactions. Although the precise mechanism is unknown, biochemical studies have shown that the enzymatic efficiency of the polymerase is improved (as reflected by a reduction in the apparent Km value for each of the nucleotide triphosphates and an increase in the overall Vmax for the transcription reaction) when SAM is continuously supplied in the reaction mixture (26). A similar phenomenon has also been observed in the segmented dsRNA virus cypovirus (9).
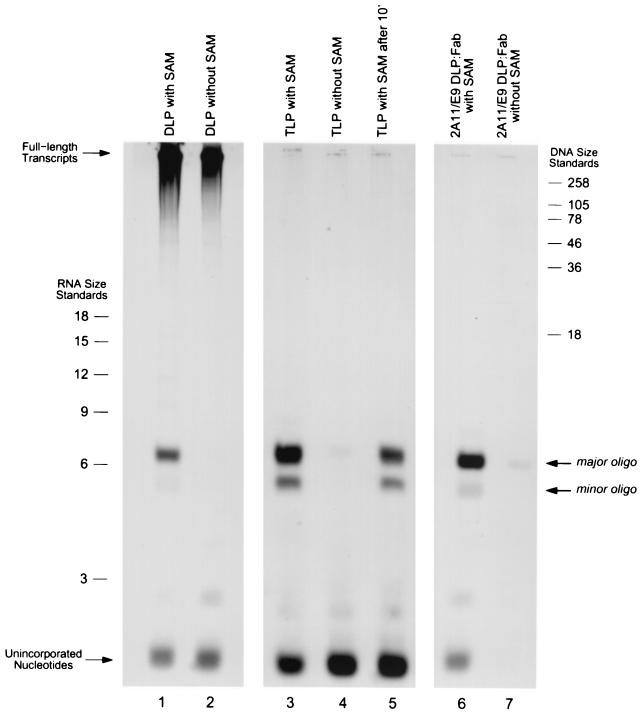
As expected, in the presence of SAM (Fig. 4, lane 1), both full-length transcripts and short oligonucleotides were observed, as this represents the condition under which transcription ordinarily occurs. However, in the absence of SAM (lane 2), the overall yield of full-length mRNA transcripts was reduced by approximately 50%, consistent with previous reports (26), and no short oligonucleotides in the vicinity of the 6-nucleotide ssRNA size marker were observed. To determine whether, in the absence of SAM, oligonucleotides were produced but failed to remain particle associated, we also examined reaction mixtures which had not been subjected to gel filtration prior to electrophoresis and observed essentially no accumulation of short oligonucleotides in the vicinity of the 6-nucleotide ssRNA size marker (data not shown). These results suggest that, under conditions in which the rate of nucleotide incorporation is reduced, the event that ordinarily causes transcript elongation to stall momentarily at a point downstream of cap formation is no longer appreciably slower than other events in the elongation process, and therefore, transcripts are able to progress from the point of initiation to full-length without pausing. This result further suggests that the short particle-associated oligonucleotide transcription products observed in DLPs are paused elongation intermediates and not prematurely terminated transcripts.
FIG. 4.
Length distribution of transcription products in the presence and absence of the capping reaction cofactor SAM. Lanes 1 and 2 contain DLP transcription products in the presence and absence of SAM. Each reaction mixture contained 5-μg particles and was incubated at 37°C for a total of 20 min. Following transcription, reaction mixtures were passed over size exclusion columns to remove unincorporated nucleotides and any free oligonucleotides shorter than 20 bases. Though identical amounts of radiolabeled GTP were included in each reaction mixture, the reason for which the intensity of the band corresponding to unincorporated nucleotides is not the same in each reaction aliquot may be due to the varying efficiency with which the size exclusion columns removed free radiolabeled nucleotides after transcription; additionally, this band may also include nucleotides which are specifically or nonspecifically bound inside the particle during gel filtration. RNA and DNA size standards are indicated (length in nucleotides).
Transcript initiation is significantly diminished in TLPs when the efficiency of the polymerase is reduced.
Under normal transcription conditions, native DLPs and mature TLPs both appear to be equally capable of performing the enzymatic steps required for the early stages of transcription, including transcript initiation, cap formation, and limited elongation up to the point which, in DLPs, is the site of momentary pausing prior to processive full-length elongation. To investigate whether the limited transcription observed in the TLP is dependent upon polymerase efficiency, we carried out transcription in the presence and absence of SAM and examined the length distribution of the resulting particle-bound transcription products.
As expected, when SAM was included in the reaction mixture (Fig. 4, lane 3), two species of particle-associated oligonucleotide transcription products migrating near the 6-nucleotide ssRNA size marker were observed, just as in the DLP. No full-length transcripts were produced. However, in the absence of SAM, virtually no transcription products whatsoever were observed (Fig. 4, lane 4). Likewise, as with the DLP, reaction mixtures not subjected to gel filtration prior to electrophoresis also contained virtually no oligonucleotides within the vicinity of the 6-nucleotide ssRNA size marker (data not shown). These results suggest that, when the efficiency of the polymerase is reduced in the architectural context of the TLP, transcription initiation becomes inhibited. This surprising result contrasts with that observed in the DLP, where a reduction in polymerase efficiency merely resulted in a lower overall yield of full-length transcripts.
To ensure that incubation of TLPs in a transcription reaction mixture lacking SAM did not somehow irreversibly compromise polymerase function, we also carried out transcription under conditions in which SAM was added to the reaction mixture after 10 min of incubation in an otherwise complete transcription reaction mixture (Fig. 4, lane 5). We observed that, upon the addition of SAM, the limited transcriptional competence of the TLP was restored. Taken together, these results suggest that the presence of the outer capsid layer in the mature TLP has a detrimental effect on the enzymatic activity of the polymerase in the core and that this inhibition can be partially overcome when SAM is provided to enhance the rate of nucleotide incorporation.
The precise mechanism by which the attachment of the outermost capsid layer in the TLP reduces the enzymatic efficiency of the polymerase in the core is not understood, nor is it known what other factors may also play a role in hindering transcript elongation. Structural studies have provided evidence that a subtle conformational change, transmitted from the capsid surface into the viral interior upon VP7 binding to specific sites on VP6, may be involved (14). To determine whether enzyme efficiency is also reduced in other forms of the virus in which ligand attachment has altered the transcriptional properties of the particle, we carried out a similar analysis using a DLP complexed with an antibody fragment (2A11/E9) shown previously to bind to the VP6 surface in a manner similar to that of the native outer capsid protein VP7 and to inhibit full-length mRNA production (14).
When SAM was included in the reaction mixture, the 2A11/E9 DLP-Fab complex produced short oligonucleotides migrating near the 6-nucleotide ssRNA size marker and no full-length transcripts (Fig. 4, lane 6), consistent with previous observations (14). However, when SAM was omitted from the transcription reaction mixture, virtually no transcription products of any kind were produced (Fig. 4, lane 7). These results suggest that the presence of the Fab on the DLP surface affected the enzymatic efficiency of the polymerase to the point that transcript initiation could not occur in the absence of SAM, just as was seen in the TLP.
Taken together, the results shown in Fig. 4 indicate that, in addition to the stimulatory effect of SAM on the rate of nucleotide incorporation, the architectural configuration of the viral capsid also influences the enzymatic efficiency of the polymerase. This influence is especially apparent from a comparison of the reactions shown in lanes 2 and 7. Architecturally, these two particle forms differed only by the presence or absence of 2A11/E9 Fabs on the capsid surface. However, in the absence of SAM, native DLPs (Fig. 4, lane 2) were still able to produce full-length transcripts, while Fab-bound DLPs (Fig. 4, lane 7) were essentially rendered incapable of transcript initiation.
The oligonucleotide transcription products are precursors of full-length mRNA transcripts.
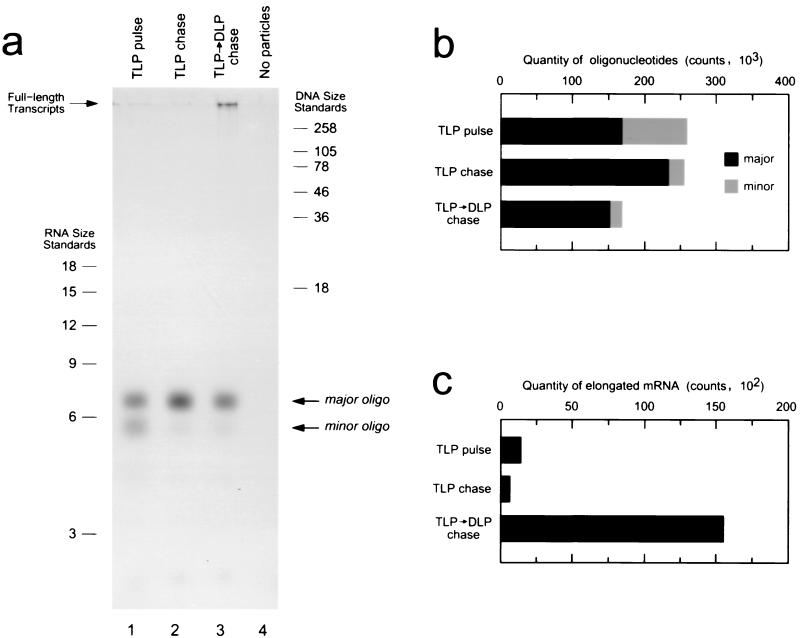
The observation that the short oligonucleotide transcription products in the DLP reaction are capped, have a transient lifetime, and are particle-associated suggests that they are elongation intermediates, temporarily stalled while in the process of becoming full-length transcripts. To establish that these oligonucleotides are indeed precursors of full-length transcripts, we employed a pulse-chase strategy. TLPs were first allowed to begin transcription using a reaction mixture supplemented with radiolabeled GTP; following a brief incubation at 37°C, radiolabeled nucleotides were removed from the reaction mixture using two successive centrifugal gel filtration columns. The TLPs were then converted to DLPs with the addition of the calcium-specific chelator BAPTA, and finally, transcription was resumed by supplying unlabeled nucleotide triphosphates. The length distribution of transcription products before and after structural conversion was compared using high-resolution denaturing PAGE (Fig. 5).
FIG. 5.
Pulse-chase analysis of transcription products in TLPs and structurally converted DLPs. (a) Visualization of radiolabeled transcription products using high-resolution denaturing PAGE. Each lane shows transcription products arising from 1-μg particles. Lane 1, oligonucleotide transcripts synthesized in TLP following 5 min of incubation at 37°C in a reaction mixture containing radiolabeled GTP (pulse reaction); lane 2, oligonucleotide transcripts in TLP following a 5-min pulse incubation and subsequent 10-min chase incubation at 37°C; lane 3, transcription products arising from a 5-min pulse incubation in TLP followed by structural conversion to DLP and subsequent 10-min chase incubation at 37°C; lane 4, negative-control reaction containing radiolabeled GTP but no particles, incubated at 37°C for 5 min. The absence of a detectable signal from unincorporated nucleotide precursors indicates that the use of two successive size exclusion columns was sufficient to remove the radiolabeled GTP from the reaction following the pulse. RNA and DNA size standards are indicated (length in nucleotides). (b) Quantitation of the radioactivity present in the two oligonucleotide species in each reaction. (c) Quantitation of the radioactivity present in the region of the gel corresponding to elongated transcripts.
When TLPs were incubated briefly with a transcription reaction mixture containing radiolabeled GTP (the TLP pulse incubation), the only transcription products observed were the major and minor particle-associated oligonucleotide species migrating near the 6-nucleotide ssRNA size marker (Fig. 5a, lane 1), consistent with previous observations. Comparison of the relative yield of these two reaction products indicates that, following the pulse incubation, approximately 65% of the radioactivity was contained in the major oligonucleotide species and 35% was contained in the minor oligonucleotide species (Fig. 5b).
When TLPs were allowed to continue transcribing using unlabeled nucleotide triphosphates (the TLP chase incubation), no additional transcription products were observed (Fig. 5a, lane 2). Interestingly, following the TLP chase, more than 90% of the radioactivity was contained in the major oligonucleotide species and less than 10% was contained in the minor oligonucleotide species, while the overall yield of mRNA was essentially unchanged (Fig. 5b). This result suggests that the minor oligonucleotide species is a precursor of the major oligonucleotide transcription product.
When the TLPs were structurally converted to DLPs and then incubated with unlabeled nucleotide triphosphates (the TLP-to-DLP chase incubation), radiolabeled full-length transcripts were produced (Fig. 5a, lane 3). As expected, the appearance of full-length mRNA was marked by a drop in the overall quantity of short oligonucleotides (Fig. 5b) and an increase in the quantity of radiolabeled mRNA migrating in the region of full-length transcripts (Fig. 5c). Because radiolabeled mRNA precursors were provided only during the TLP pulse incubation, the radiolabel present in these full-length transcripts must have been incorporated while the particles were still in the form of TLPs. These results clearly demonstrate that transcripts initiated in the structural context of the TLP can be successfully elongated once the particle is converted to the DLP form and the inhibitory effect of the outermost capsid layer is eliminated. Extending these observations to the early events of the transcription process in the DLP, it is likely that the short oligonucleotide transcription products seen in the DLP are also precursors of full-length transcripts.
Close inspection of the length distribution of transcription products in this reaction indicates that not all of the oligonucleotides could be elongated into full-length transcripts upon structural conversion, as a fraction of both the major and minor oligonucleotide species remained after the TLP-to-DLP chase incubation. Similar results were obtained in several independent experiments. The short oligonucleotides which could not be elongated after structural conversion may represent initiated transcripts which were somehow disrupted as a consequence of the treatments required either to remove the unincorporated radiolabeled nucleotides from the reaction mixture following the pulse or to convert the particles from TLP to DLP form. It is conceivable that such treatments could potentially cause the transcript or template to become misaligned in the polymerase active site or could otherwise adversely affect the integrity of the transcription bubble. Once disrupted, such oligonucleotides perhaps cannot undergo successful elongation and are then aborted. Alternatively, these oligonucleotides may represent transcripts which were prematurely aborted within the TLP during the pulse phase and then released from the particle upon conversion to DLP form.
DISCUSSION
In order to gain an increased understanding of the molecular events occurring early in the transcription process in rotavirus, we investigated the length distribution of transcription products arising under various conditions of polymerase efficiency in particles which are capable of producing full-length transcripts (DLPs) as well as in particles which are not (TLPs). We observed that, under normal conditions, DLPs produce both full-length mRNA transcripts and two short oligonucleotide species. These oligonucleotide species, which migrate in a polyacrylamide gel with an apparent length between 5 and 7 bases, are particle associated, contain methylated cap structures, and have a transient lifetime, suggesting them to be precursors of full-length transcripts.
The production of oligonucleotides during transcription has been well documented in many diverse transcription systems. Biochemical studies of genome transcription in the related viruses mammalian orthoreovirus and insect cypovirus have reported that transcription of full-length mRNA is accompanied by the production of a very large molar excess of short oligonucleotides two to three bases in length, some with caps but most without (1, 7, 9, 30–32). These oligonucleotides were identical in sequence with the conserved 5′ ends of mature mRNAs and were believed to be aborted transcripts resulting from failed initiation attempts. Indeed, abortive reinitiation has also been observed in prokaryotic and eukaryotic transcription systems as well (3, 17).
Because of the widespread nature of reiterative initiation in transcription systems, it is likely that some form of this phenomenon accompanies transcription in rotavirus as well. However, for several reasons, the oligonucleotide transcription products we describe in the current study appear to be distinct from oligonucleotides arising through abortive reinitiation. First, the major and minor oligonucleotide species appear to be longer than the largely uncapped di- and trinucleotide species ordinarily associated with the phenomenon of abortive reinitiation. Second, while the products of abortive reinitiation typically accumulate in vast molar excess of successfully elongated transcripts, we observe that the major and minor oligonucleotide species produced in the DLP do not accumulate appreciably during the reaction and indeed appear to have a finite lifetime, as would be characteristic of stalled elongation intermediates as opposed to aborted transcripts arising through reiterative initiation. Longer oligonucleotides having these characteristics have not, to our knowledge, been observed in orthoreovirus or cypovirus and indeed may arise because of fundamental architectural differences between “turreted” reoviruses, which include orthoreovirus and cypovirus, and “nonturreted” reoviruses, which include rotavirus and orbivirus. Such architectural differences are especially prominent in the vicinity of endogenous transcription apparatus and indeed may impose significant constraints on the mechanism of mRNA production and capping (15).
The observation that capped, particle-associated oligonucleotide transcripts having transient lifetimes arise at a specific point early in the transcription process suggests that the movement of the dsRNA template through the polymerase during elongation is not a continuous process but rather is temporarily interrupted shortly after initiation and cap formation. This brief pause in nucleotide incorporation appears to divide the elongation phase of transcription into two parts. Prior to stalling, the polymerase is able to elongate the initiated transcript to approximately 5 to 7 nucleotides in length; following the pause, the polymerase is able to elongate the transcript processively all the way to the end of the dsRNA template, generating full-length mRNA. This pause site may represent the point at which the transcription apparatus must carry out specific steps to enable full-length elongation to take place.
It is not known which molecular event early in the transcription process causes the polymerase to pause shortly after initiation and cap formation. Evidently, this event occurs at a rate which is lower than the basic rate of nucleotide incorporation under ordinary conditions. Under conditions in which the rate of nucleotide incorporation is reduced, as is the case when SAM is omitted from the reaction mixture (26), the transcription apparatus no longer stalls while carrying out this step, indicating that pausing at this point in elongation occurs because of a difference in the rate at which various steps involved in elongation may proceed. From a consideration of the apparent length of the paused transcripts, the circumstances under which they arise, and the events that likely must occur to facilitate processive elongation, several possibilities emerge as to the reason why the transcription apparatus stalls momentarily before proceeding with full-length elongation.
One possibility is that the brief pause in elongation may be necessary in order to allow the viral helicase to become properly engaged on the template ahead of the polymerase. In all transcription systems operating on a double-stranded template, a helicase is required to open the transcription bubble downstream of the polymerase. Such a helicase has, in fact, been positively identified in the closely related viruses bluetongue virus (26) and mammalian orthoreovirus (2, 19). It is probable that, at the start of transcription, the polymerase is already engaged on a partially unwound template and, as such, can incorporate several nucleotides before requiring the activity of the viral helicase to begin further unwinding. Following the proper engagement of the helicase, nucleotide incorporation may then proceed unhindered, giving rise to full-length transcripts without additional delays.
Another early event possibly requiring a brief pause in elongation involves the initial separation of the nascent transcript from the transcription bubble. Studies in other transcription systems have suggested that, during elongation, the template strand and nascent transcript are base paired for 8 to 10 nucleotides as a hybrid duplex within the transcription bubble (25, 29), and the polymerase itself is believed to take an active role in separating the growing transcript from the template (5). Although little is known about how this may occur during rotavirus transcription, it is conceivable that, following the incorporation of the first several nucleotides, a specific event involving the polymerase, or perhaps a domain of the inner capsid protein VP2, is required to promote the initial separation of the nascent transcript from the template and the subsequent closure of the transcription bubble before processive elongation may occur.
A third transcriptional event possibly requiring a brief pause early in transcript elongation involves the export of the nascent transcript from the core. During rotavirus transcription, nascent transcripts are translocated across the intact double-layered capsid through channels at the icosahedral vertices (13). At the outset of transcription, the transcripts emerging from the transcription bubbles must be threaded through the exit channels in the inner capsid layer. Though these channels are in close proximity to the transcription enzyme complexes in the viral core (24), it is possible that the threading of the transcripts into the channels may generally require a temporary pause in elongation several nucleotides downstream of initiation in order to allow sufficient time for this to occur properly.
The transcriptional behaviour of TLPs, which differ from DLPs in that they contain an additional external capsid layer, provides further insight into the nature of the events accompanying transcriptional pausing during elongation. Under ordinary transcription conditions, TLPs produce only the major and minor oligonucleotide species and no full-length transcripts. These oligonucleotides contain methylated caps and appear to be particle associated initially but are eventually released. This behavior contrasts with that observed for mature orthoreovirus, where predominantly uncapped di- and trinucleotide aborted transcription products accumulate to high levels within the particle interior and are released only when the outermost capsid layer is removed (7).
The observation that cap formation and limited nucleotide incorporation can occur in the TLP suggests that the TLP is not strictly transcriptionally incompetent; both the polymerase and the capping enzymes are functional, and capped oligonucleotide transcripts having an apparent length of 5 to 7 bases are made. However, in the presence of the outermost capsid layer, the transcription apparatus in the core nonetheless appears to be unable to perform at least one of the critical events required early in the elongation phase to facilitate the transcription of full-length mRNA. These observations also imply that the events accompanying the momentary pause in elongation are somehow susceptible to disruption from changes in the architecture of the capsid, as has been suggested from structural studies in rotavirus (14). The conformation of the viral capsid has also been suggested to play a role in inhibiting transcript elongation in mature orthoreoviruses (7).
Interestingly, in addition to preventing the transcription apparatus from carrying out the events needed to facilitate full-length elongation, the attachment of specific ligands on the viral surface also appears to alter the function of the polymerase. Although TLPs retain the ability to perform limited transcript elongation under ordinary conditions, omission of SAM appears to render the transcription apparatus incapable of initiation (see Fig. 4). The ability of SAM, a substrate for the capping enzyme, to affect polymerase function indicates that these two enzymes are likely in close contact with one another in the viral core, as has been suggested from other studies (24, 26). Were cap formation to be the only component of the transcription process affected by the omission of SAM, then TLPs should remain capable of transcript initiation even in the absence of SAM, as is seen in DLPs. However, such a result is clearly not observed, implying that the polymerase in the TLP may have an altered structure because of the presence of an additional capsid layer on the viral surface, leading to a reduction in enzymatic efficiency. In the presence of SAM, this reduction in polymerase efficiency may be partially compensated for to the extent that initiation and limited elongation may proceed. Although the precise reason for which TLPs cannot elongate beyond the pause site remains unknown, it is possible that the efficiency of the polymerase is still not adequate for the transcription apparatus to carry out the specific downstream events needed for the transition to processive elongation.
The contrasting behavior of the transcription apparatus under various conditions illustrates the degree to which polymerase function is sensitive to the architectural environment in which it is operating. The presence of SAM tends to boost polymerase function, while the presence of specific ligands on the viral surface tends to inhibit polymerase function. From the perspective of the viral replication cycle, these two opposing influences, both of which likely affect the transcription apparatus through structural interactions, serve to ensure that transcription occurs with greatest efficiency only after the virus has entered the cytoplasm and assumed the form of a DLP. Likewise, the inhibition of transcript elongation in the TLP may benefit the virus by ensuring that transcription does not begin to occur prematurely, under circumstances in which genome transcription would not be productive.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI-36040 (B.V.V.P.) and DK-30144 (M.K.E.).
We thank R. F. Ramig for critical reading of the manuscript.
REFERENCES
- 1.Bellamy A R, Nichols J L, Joklik W K. Nucleotide sequences of reovirus oligonucleotides: evidence for abortive RNA synthesis during virus maturation. Nat New Biol. 1972;238:49–51. doi: 10.1038/newbio238049a0. [DOI] [PubMed] [Google Scholar]
- 2.Bisaillon M, Bergeron J, Lemay G. Characterization of the nucleoside triphosphates phosphohydrolase and helicase activities of the reovirus λ1 protein. J Biol Chem. 1997;272:18298–18303. doi: 10.1074/jbc.272.29.18298. [DOI] [PubMed] [Google Scholar]
- 3.Carpousis A J, Gralla J D. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 4.Chen D, Luongo C L, Nibert M L, Patton J T. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology. 1999;265:120–130. doi: 10.1006/viro.1999.0029. [DOI] [PubMed] [Google Scholar]
- 5.Daube S S, von Hippel P H. RNA displacement pathways during transcription from synthetic RNA-DNA bubble duplexes. Biochemistry. 1994;33:340–347. doi: 10.1021/bi00167a044. [DOI] [PubMed] [Google Scholar]
- 6.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Chanock R M, Melnick J L, Roizman B, Hope R E, editors. Virology. New York, N.Y: Raven Press; 1996. pp. 1625–1655. [Google Scholar]
- 7.Farsetta D L, Chandran K, Nibert M L. Transcriptional activities of reovirus RNA polymerase in recoated cores: initiation and elongation are regulated by separate mechanisms. J Biol Chem. 2000;275:39693–39701. doi: 10.1074/jbc.M004562200. [DOI] [PubMed] [Google Scholar]
- 8.Fields B N. Reoviridae. In: Fields B N, Knipe D M, Chanock R M, Melnick J L, Roizman B, Hope R E, editors. Virology. New York, N.Y: Raven Press; 1996. pp. 1553–1555. [Google Scholar]
- 9.Furuichi Y. Allosteric stimulatory effect of S-adenosylmethionine on the RNA polymerase in cytoplasmic polyhedrosis virus. J Biol Chem. 1981;256:483–493. [PubMed] [Google Scholar]
- 10.Furuichi Y, Muthukrishnan S, Tomasz J, Shatkin A J. Mechanism of formation of reovirus mRNA 5′-terminal blocked and methylated sequence, m7GpppGmpC. J Biol Chem. 1976;251:5043–5053. [PubMed] [Google Scholar]
- 11.Imai M, Akatani K, Ikegami N, Furuichi Y. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J Virol. 1983;47:125–136. doi: 10.1128/jvi.47.1.125-136.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, Knipe D M, Chanock R M, Melnick J L, Roizman B, Hope R E, editors. Virology. New York, N.Y: Raven Press; 1996. pp. 1657–1708. [Google Scholar]
- 13.Lawton J A, Estes M K, Prasad B V V. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 14.Lawton J A, Estes M K, Prasad B V V. Comparative structural analysis of transcriptionally competent and incompetent rotavirus-antibody complexes. Proc Natl Acad Sci USA. 1999;96:5428–5433. doi: 10.1073/pnas.96.10.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawton J A, Estes M K, Prasad B V V. Mechanism of genome transcription in segmented dsRNA viruses. Adv Virus Res. 2000;57:185–230. doi: 10.1016/S0065-3527(00)55004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Mattion N M, Estes M K. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology. 1992;188:77–84. doi: 10.1016/0042-6822(92)90736-9. [DOI] [PubMed] [Google Scholar]
- 17.Luse D S, Jacob G A. Abortive initiation by RNA polymerase II in vitro at the adenovirus 2 major late promoter. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 18.McCrae M A, McCorquodale J G. Molecular biology of rotaviruses: V. Terminal structure of viral RNA species. Virology. 1983;126:204–212. doi: 10.1016/0042-6822(83)90472-5. [DOI] [PubMed] [Google Scholar]
- 19.Noble S, Nibert M L. Characterization of an ATPase activity in reovirus cores and its genetic association with core-shell protein λ1. J Virol. 1997;71:1282–1291. doi: 10.1128/jvi.71.3.2182-2191.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patton J T, Chen D. RNA-binding and capping activities of proteins in rotavirus open cores. J Virol. 1999;73:1382–1391. doi: 10.1128/jvi.73.2.1382-1391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizarro J M, Pizarro J L, Fernández J, Spencer E. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. J Gen Virol. 1991;72:325–332. doi: 10.1099/0022-1317-72-2-325. [DOI] [PubMed] [Google Scholar]
- 22.Prasad B V V, Estes M K. Molecular basis of rotavirus replication: structure-function correlations. In: Chiu W, Burnett R, Garcia R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 239–268. [Google Scholar]
- 23.Prasad B V V, Wang G J, Clerx J P, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 24.Prasad B V V, Rothnagel R, Zeng C Q, Jakana J, Lawton J A, Chiu W, Estes M K. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 25.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 26.Spencer E, García B I. Effect of S-adenosylmethionine on human rotavirus RNA synthesis. J Virol. 1984;52:188–197. doi: 10.1128/jvi.52.1.188-197.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stäuber N, Martinez-Costas J, Sutton G, Monastyrskaya K, Roy P. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J Virol. 1997;71:7220–7226. doi: 10.1128/jvi.71.10.7220-7226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valenzuela S, Pizarro J, Sandino A M, Vasquez M, Fernandez J, Hernandez O, Patton J, Spencer E. Photoaffinity labeling of rotavirus VP1 with 8-azido-ATP: identification of the viral RNA polymerase. J Virol. 1991;65:3964–3967. doi: 10.1128/jvi.65.7.3964-3967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson K S, Conant C R, von Hippel P H. Determinants of the stability of transcription elongation complexes: interactions of the nascent RNA with the DNA template and the RNA polymerase. J Mol Biol. 1999;289:1179–1194. doi: 10.1006/jmbi.1999.2814. [DOI] [PubMed] [Google Scholar]
- 30.Yamakawa M, Furuichi Y, Shatkin A J. Reovirus transcriptase and capping enzymes are active in intact virions. Virology. 1982;118:157–168. doi: 10.1016/0042-6822(82)90329-4. [DOI] [PubMed] [Google Scholar]
- 31.Yamakawa M, Furuichi Y, Nakashima K, LaFiandra A J, Shatkin A J. Excess synthesis of viral mRNA 5′-terminal oligonucleotides by reovirus transcriptase. J Biol Chem. 1981;256:6507–6514. [PubMed] [Google Scholar]
- 32.Zarbl H, Hastings K E M, Millward S. Reovirus core particles synthesize capped oligonucleotides as a result of abortive transcription. Arch Biochem Biophys. 1980;202:348–360. doi: 10.1016/0003-9861(80)90437-3. [DOI] [PubMed] [Google Scholar]