Abstract
Background/aim
Breast cancer is the most prevalent cancer in women, emphasizing need for noninvasive blood biomarkers to aid in treatment selection. Previous studies have demonstrated elevated levels of plasma circulating cell-free DNA (ccfDNA) in breast cancer patients. Both ccfDNA and mitochondrial DNA (mtDNA) are fragments released into the bloodstream. In this study, we investigated effectiveness of ccfDNA and mtDNA as indicators of treatment response and explored their potential as monitoring biomarkers. Additionally, we compared these markers with circulating tumor cell (CTC) data and assessed their relationship with epithelial-mesenchymal transition (EMT).
Materials and methods
Thirty-six female breast cancer patients and 21 healthy females were included in the study. Quantitative polymerase chain reaction (qPCR) was performed on plasma samples to measure levels of ND1, ND4, ALU115, ALU247, and GAPDH, and DNA integrity was determined by calculating ratios of ALU247/ALU115 and ND4/ND1.
Results
After treatment, patients had a significant decrease in ccfDNA levels and a significant increase in mtDNA copy number (mtDNAcn). However, there was no significant change in ccfDNA and mtDNA integrity. When comparing all groups, patients exhibited higher levels of ALU115 and ALU247 compared to controls. Moreover, patients demonstrated significantly lower ccfDNA integrity than controls.
Conclusion
This study represents the first comprehensive investigation of plasma ccfDNA levels, mtDNAcn, and integrities collectively. Furthermore, it is the first study to explore the relationship between these markers and CTCs, cancer stem cell markers, treatment response, and metastatic status. Our findings suggest that plasma ccfDNA and mtDNA may serve as potential biomarkers for assessing chemotherapy response and can be employed alone or in combination with other biomarkers to monitor treatment efficacy in breast cancer patients.
Keywords: Breast cancer, ccfDNA, mtDNA, neoadjuvant therapy, EMT
1. Introduction
Breast cancer is the most frequent cancer worldwide and ranks as the second leading cause of cancer-related death in women [1]. Multiple molecular changes that lead to uncontrolled self-renewal, proliferation, transformation, and metastasis of normal cells cause cancer [2,3]. Early detection of cancer and accurate identification of metastases have considerably improved the survival rates of women with breast cancer by enhancing treatment options. Monitoring treatment response is crucial to prevent the continuation of ineffective treatments, minimize unnecessary side effects, and assess the effectiveness of new therapeutics [1,2,4,5]. Accumulating findings over the last couple of years have emphasized the potential use of circulating nucleic acids in peripheral blood, such as DNA, mRNA, and microRNA, in the breast cancer diagnosis, prognosis, and monitoring of response to anticancer therapy [6]. While the use of tumor tissue specimens remains important, the utility of biopsy samples is limited because they may not capture tumor heterogeneity, and recurrent biopsies are not practical. A considerable alternative method is “liquid biopsy”, which allows for sensitive and targeted serial sampling during therapy [5].
Changes in the levels of circulating tumor cells (CTCs), extracellular free DNAs (cfDNAs), and mitochondrial DNA (mtDNA), known as liquid biopsies, have received great attention as cancer biomarkers in plasma and serum. In addition to plasma and serum, liquid biopsies are a minimally invasive tool for detecting molecular biomarkers in body fluids such as peripheral blood, urine, saliva, cerebral spinal fluid, and breast milk [7–13]. This approach provides a perspective for real-time monitoring of tumor dynamics in an individual cancer patient. While tissue biopsy from a cancerous location remains the mainstay of diagnosis, liquid biopsy presents an alternative to the restricted solid biopsy approaches due to several advantages, including the ability to perform sequential sampling for monitoring tumor progression, treatment response, metastasis, and disease recurrence [4,9,11].
In 1948, two French researchers, Mandel and Métais, discovered the presence of cell-free DNA in the blood of both healthy and diseased humans [7,10,11,14–19]. While cfDNA might have seemed insignificant when it was first discovered in the human circulatory system, its clinical significance was recognized when researchers observed differences between the properties of cfDNA in healthy individuals and cancer patients [16]. The vast majority of cfDNA is released through apoptosis or necrosis of tumor cells in oncological patients [20,21]. Besides screening healthy and at-risk patient groups for early detection and cancer treatment, cfDNA serves as a biomarker for multiple indications in oncology, including staging and prognosis, tumor localization, initial therapy stratification, monitoring of local or systemic treatment response, identification of acquired resistance mechanisms, monitoring of recurrence [8,16,22].
The cfDNA profile found in a single blood sample contains a mixture of both “wild-type” and genetically and epigenetically modified DNA fragments released by diverse cells from various processes, tissues, and organs under environmental factors [23]. All cells seem to have the ability to continuously release cell-specific DNA into the extracellular environment. An important point here is that the concentration of cfDNA and the concentration of tumor-derived DNA in tumor microenvironment, as well as in other healthy cells, can vary significantly between individuals [16].
cfDNA presents as ALU (Arthrobacter luteus) sequences. More than 50%–65% of the human genome consists of repetitive DNA [18,24]. ALU families belonging to the class of retroelements called short interspersed nuclear elements (SINEs) in the more than 10% of the human genome, with a copy number of approximately 1.4 million, are the most abundant ALU [18,25]. They are typically about 300 nucleotides in length [14,26–28]. While the source of cfDNA in healthy individuals is merely by apoptosis, producing evenly sized shorter DNA fragments (ALU 115); on the other hand, in cancers, necrosis contributes uneven longer DNA fragments (ALU 247) to the shorter fragments from apoptosis [14,29–31]. Analysis of ccfDNA integrity is a factor independent of the genetic or epigenetic status of cfDNA and is theoretically representative of all tumors. ccfDNA integrity is calculated as the ratio of the concentration of longer DNA fragments to shorter fragments in plasma or serum [32,33].
Mitochondria are eukaryotic cell organelles that play a central role in energy production, cell proliferation, and apoptosis. It is the main source and target of intracellular reactive oxygen species (ROS), which plays an important role in breast carcinogenesis [34]. Recent advances aimed at increasing the diagnostic and prognostic value for cancer patients have also targeted the circulating mitochondrial genome due to its unique and distinctive properties. Circulating mitochondrial DNA is known to have short length, relatively simple molecular structure, and high copy number. These properties make it an easily accessible, noninvasive biomarker for diagnosing various types of solid tumors, complementing the function of liquid biopsy [35]. In addition to the inconsistent association between peripheral blood mtDNA copy number and breast cancer risk, breast cancer may alter the observed mtDNA levels in peripheral blood, emphasizing the need for designing forward-looking studies [36].
There are hotspot locations for deletions along the mtDNA circle, 90% of which are deletions of the nicotinamide adenine dinucleotide dehydrogenase 4 (ND4) sequence, reflecting a population of viable mitochondria but with poor mtDNA integrity [37]. ND4 subunits are often missing in complex I and are a common indicator of mtDNA damage [38]. In contrast, the loss of ND1 subunits has a much more detrimental effect on complex I and the mitochondria itself, making ND1 deletions rare in viable mitochondria. Therefore, the rarely deleted ND1 copy number is a suitable marker for the total mtDNA copy number and the ND4/ND1 ratio can be used to assess the proportion of intact mtDNA [37–40].
Breast cancer treatment typically incorporates a multimodality strategy that includes surgery, radiation, and systemic therapy. Neoadjuvant chemotherapy (NACT) has become a standard treatment for patients with advanced breast cancer. If breast-conserving surgery is not possible, neoadjuvant chemotherapy can be utilized [41]. It has become a standard-of-care for patients with locally advanced breast cancer. NACT provides a unique opportunity for real-time monitoring of tumor response and evaluation of drug efficacy. Secondly, it can reduce the stage of tumors and thus promote the chances of breast-conserving surgery [42–44].
Currently, the need for diagnostic and prognostic biomarkers continues. However, using a combination of blood biomarkers with a noninvasive method is critical for treatment selection. According to the studies reviewed, ccfDNA and mtDNA play crucial roles in human cancers, particularly in their significant involvement in metastasis. Given the lack of research on the relationship between multiple markers, circulating tumor cells, and epithelial-mesenchymal transition, the present study aims to contribute to clinical research in disease diagnosis, prognosis, and treatment.
In this study, we investigated the changes in plasma cfDNA and mtDNA copy number and integrity before and after NACT in breast cancer patients. By correlating these findings with CTC molecular analysis data obtained from the same patients in our previous study, we aimed to examine the relationship of these biomarkers with epithelial-mesenchymal transformation (EMT). This approach aims to predict the likelihood of metastasis development and consequently forecast the clinical course of these patients.
2. Materials and methods
2.1. Study subjects and ethical approval
Breast cancer patients receiving neoadjuvant chemotherapy (n = 36) and healthy individuals as control (n = 21) were included in this retrospective study. Plasma retrieved from specimens stored at −80 °C. The present study was approved by the Marmara University School of Medicine Ethics Committee (approval ID 09.2022.246). Informed consent was obtained from all the recruited subjects.
2.2. Isolation of DNA
Blood samples were collected in EDTA tubes. To obtain plasma, centrifugation was performed at 2000 × g for 10 min at 4 °C. DNA was extracted from plasma samples using the QIAmp DNA mini kit (Qiagen, Hilden, Germany, cat. number: 51304) according to the manufacturer’s instructions and stored at −20 °C until use.
2.3. RT-PCR of ALU elements and mtDNA copy number
One microliter of genomic DNA was used as a template for each real-time polymerase chain reaction (RT PCR) using BlasTaq 2X SYBR Green Master Mix (abm Good) on a real-time PCR device (LightCycler 480, Roche).
The quantitative values obtained from the 115 bp (shorter fragments) primers represent the total level of ccfDNA (ng/μL), while the quantitative values from the 247 bp (longer fragments) primers were used to calculate the integrity of ccfDNA. The sequences of the ALU 115 and ALU 247 primers were as follows: ALU 115 forward 5′-CCTGAGGTCAGGAGTTCGAG-3′ and reverse 5′-CCCGAGTAGCTGGGATTACA-3′; ALU 247 forward 5′-GTGGCTCACGCCTGTAATC-3′ and reverse 5′-CAGGCTGGAGTGCAGTGG-3′ [45]. The absolute amount of ccfDNA in each sample was determined by a standard curve using 10-fold dilutions (10, 1, 0.1, 0.01, 0.001) of genomic DNA obtained from peripheral blood of a healthy donor volunteer. A negative control (without template) was run in each reaction plate.
We analyzed the levels of ND1, ND4, and GAPDH to determine mtDNA copy numbers in plasma samples. ND1 copy number serves as a convenient marker for the total amount of mtDNA. The sequences of the ND1 and ND4 were obtained from NCBI. The ND1 sequences were as follows: forward 5′-ATGGCCAACCTCCTACTCCT-3′ and reverse 5′-GGGCCTTTGCGTAGTTGTAT-3′. The ND4 sequences are: forward 5′-GATGAGGCAACCAGCCAGAA-3′ and reverse 5′-GTAGGGGAAGGGAGCCTACT-3′. After obtaining the Ct values, the mtDNA copy number was calculated using the following formula: 2−DCt.
Real-time PCR amplification was performed with the following cycles: Initial holding at 95 °C for 3 min, followed by 50 cycles of 95 °C for 15 s and 60 °C for 60 s. Negative template control was run in each plate. All reactions were conducted in triplicate in 96-well plates.
2.4. DNA integrity determination
DNA integrity was calculated as the ratio of ALU 247 to ALU 115 for ccfDNA and the ratio of ND4 to ND1 for ccf-mtDNA. In this study, mtDNA integrity was calculated by proportioning the amounts obtained as a result of the 2−DCt formula of the ND1 and ND4 primer sets while the ratio of longer to shorter fragments (ALU 247 (ng/μL) / ALU 115 (ng/μL)) demonstrates the integrity of ccfDNA in each sample [14,38,45].
2.5. Statistical analysis
In this study, analysis was conducted on patients who received neoadjuvant chemotherapy before treatment, after treatment, and a control group. The SPSS 17.0 program was used to examine levels of ccfDNA, ccfDNA integrity, mtDNA copy number, and mtDNA integrity. The results were assessed individually using the Wilcoxon signed-rank test and collectively using the Mann–Whitney U test. A significance level of p < 0.05 was considered unless otherwise specified for statistical analyses.
3. Results
In this study, data were obtained from 34 women before treatment and 30 women after treatment with locally advanced breast cancer. As a control group, 21 age-matched healthy volunteers were included in the study. The mean ages of the participants were 50.35 (32–80) years. The age range of the control group was less than 50 years. The clinicopathologic characteristics of the patients before and after NACT were given in our previous study and Table 1 [41].
Table 1.
Clinicopathologic features of patients.
| Parameters | Patients | Parameters | Patients |
|---|---|---|---|
|
| |||
| Age | Node | ||
| >50 | 17 | Positive | 28 |
| <50 | 16 | Negative | 5 |
|
| |||
| Menopause | ER | ||
| Pre- | 17 | ER (+) | 23 |
| Post- | 16 | ER (−) | 10 |
|
| |||
| Grade | |||
| G0 | 22 | PR | |
| G1 | 2 | PR (+) | 13 |
| G2 | 6 | PR (−) | 20 |
| G3 | 3 | ||
|
| |||
| Molecular subtype | |||
| Luminal A | 7 | HER2 | |
| Luminal B | 16 | HER2 (+) | 12 |
| TNBC | 5 | HER2 (−) | 21 |
| HER2 enriched | 5 | ||
|
| |||
| Histologic subtype | |||
| Invasive ductal carcinoma | 26 | Lymphatic invasion | |
| İnvasive lobular carcinoma | 2 | Positive | |
| Invasive breast carcinoma | 2 | Negative | 13 |
| Invasive mucinous carcinoma | 1 | 6 | |
| Medullary carcinoma | 2 | ||
|
| |||
| Tumor size | |||
| T1 | 4 | Masculine invasion | |
| T2 | 19 | Positive | |
| T3 | 5 | Negative | 1 |
| T4 | 4 | 18 | |
3.1. ccfDNA amount and integrity index at pre- and posttreatment
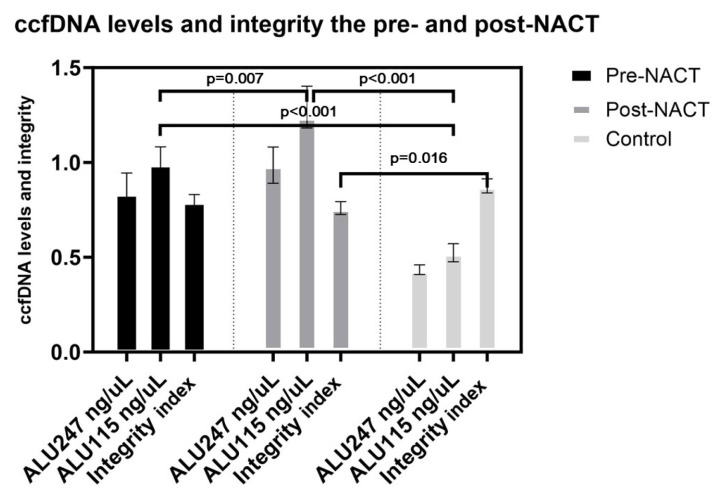
While the mean ALU values increased, the ccfDNA integrity index decreased posttreatment. The ALU values of the healthy controls were lower than the pre- and posttreatment values (p < 0.01) (Figure 1).
Figure 1.
ccfDNA levels and integrity the pre- and post-NACT.
3.2. mtDNA amount and integrity index at pre- and posttreatment
Analysis of the pretreatment and post-treatment groups revealed a significant reduction in mtDNA copy number in patients prior to treatment compared to posttreatment group (mean: 6956.58 to 1395.16, p = 0.014) (Figure 2). Following treatment, an average decrease in mtDNA integrity from 0.58 to 0.50 was observed, although statistical significance was not reached (p = 0.135) (Figure 3).
Figure 2.
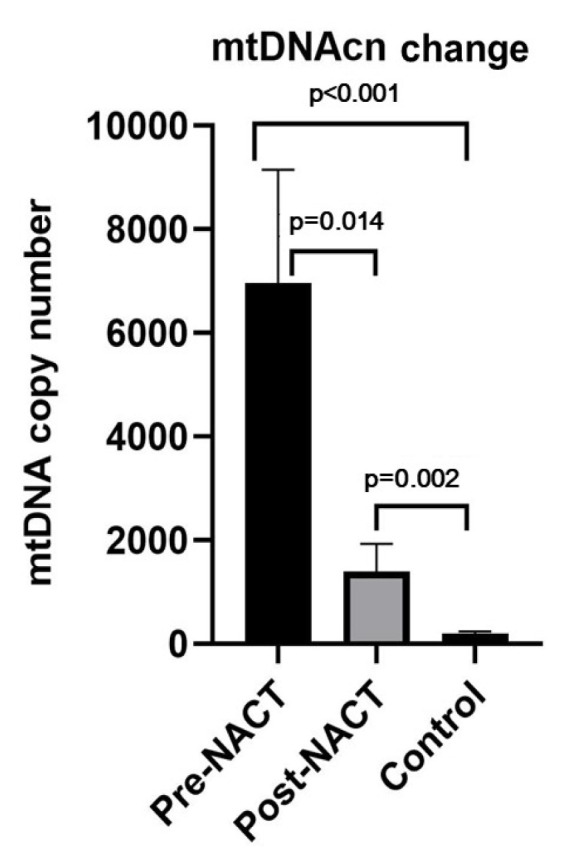
mtDNAcn the pre- and post-NACT.
Figure 3.
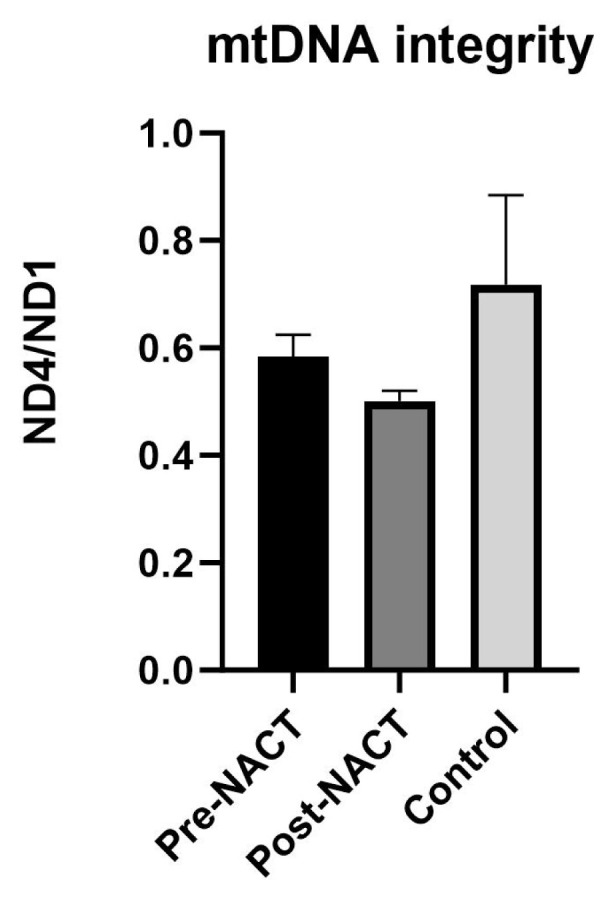
mtDNA integrity the pre- and post-NACT
The mtDNA copy number pretreatment was found to be higher than in the control group, which was statistically significant (p < 0.001) (Figure 2). Analysis of the mtDNA integrity index revealed a value of 0.58 in breast cancer patient’s pretreatment. In contrast, the control group demonstrated a mean integrity index of 0.72. However, statistical analysis showed no significant difference between the two groups (p = 0.690) (Figure 3).
When we examine the difference between the posttreatment and control groups; the mtDNA copy number posttreatment was approximately 7.34 times higher than the that in the control group (p = 0.002) (Figure 2). However, the mean mtDNA integrity index was 0.72 in the control group, while it was 0.50 in the posttreatment group, but this difference was not statistically significant (p = 0.528) (Figure 3).
3.3. The results of Wilcoxon signed-rank test at pre- and posttreatment
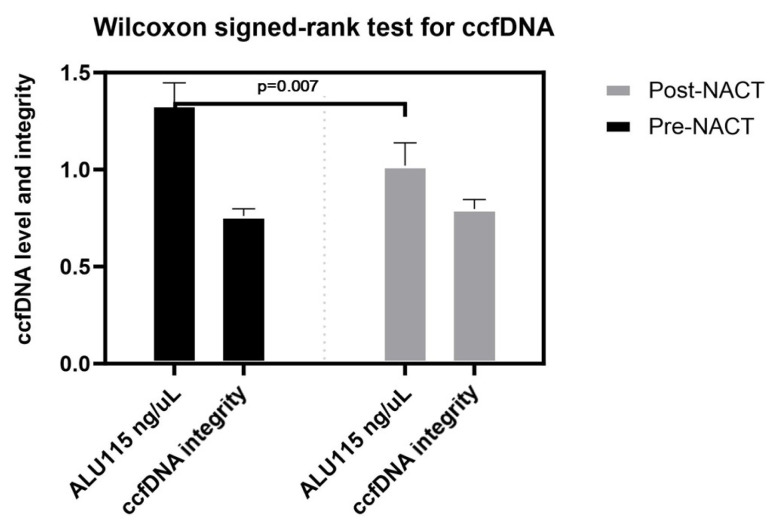
We have pre- and posttreatment data for only 28 of 34 patients. When examine the pre- and posttreatment results of this patient group, the posttreatment ccfDNA levels were lower than those of the pretreatment group, and the difference was statistically significant (p = 0.007). The ccfDNA integrity index of the posttreatment group was higher than that of the pretreatment group, but the difference was not statistically significant (p = 0.665) (Figure 4).
Figure 4.
Wilcoxon signed-rank test for ccfDNA.
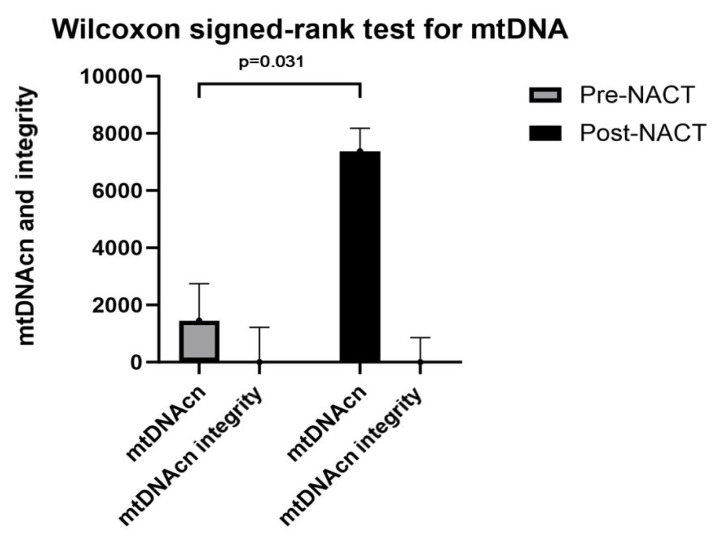
The mtDNA copy number was significantly higher in the posttreatment group than in the pretreatment group (p = 0.031). The mtDNA integrity index in the posttreatment group was higher than that in the pretreatment group, while this difference was not statistically significant (p = 0.820) (Figure 5).
Figure 5.
Wilcoxon signed-rank test for mtDNA.
3.4. ccfDNA, mtDNA, CTC, EMT, and ALDH1 at pre- and posttreatment
We aimed to examine the relationship between changes in plasma cfDNA and mtDNA copy numbers and integrity with CTC, EMT, and ALDH1 data obtained in a previous study by our team [41]. Detailed results of CTC, EMT, and ALDH1 pre- and post- NACT were presented in our previous study [41]. All cfDNA and mtDNA results, along with other biomarker findings, were given in Table 2. There was no correlation found between ccfDNA levels and mtDNA copy number with CTC, EMT, and stem cell markers pre- and post-NACT. The results were not statistically significant (p > 0.05).
Table 2.
Pre- and post-NACT ccfDNA, mtDNAcn, CTC, EMT, ALDH1, and pathologic and clinical response.
| ccfDNA | mtDNA cn |
CTC | EMT | ALDH1 | Therapy response | Metastasis during | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. | Molecular subgroup | Neoadjuvant chemotherapy regime | Pre-NACT | Post-NACT | Pre-NACT | Post-NACT | Pre-NACT | Post-NACT | Pre-NACT | Post-NACT | Pathologic | Clinical | |
| P1 | Lum A | 4′ EC + 12′ Paclitaxel | - | - | N | N | N | PR | CR | N | |||
| P2 | Lum B | - | - | - | P | N | N | P | P | N | PR | PR | N |
| P3 | Lum B, HER2+ | - | - | - | N | P | N | N | N | N | PR | CR | N |
| P4 | Lum B, HER2+ | 4′ EC + 12′ Paclitaxel + Herceptin | ↑ | ↑ | N | N | N | N | N | N | PR | PR | N |
| P5 | Lum B, HER2+ | 12′ Paclitaxel + 4′ Herceptin + 4′ EC | ↑ | ↑ | N | N | N | N | N | N | CR | PR | N |
| P6 | Lum B, HER2+ | - | - | - | N | N | P | CR | PR | N | |||
| P7 | Lum B | 4′ EC + 12′ Paclitaxel | ↑ | ↓ | N | N | N | P | P | N | CR | CR | N |
| P8 | Lum B | 4′ EC + 12′ Paclitaxel | ↑ | ↓ | N | N | N | N | N | N | PR | PR | N |
| P9 | Lum B | 4′ EC + 12′ Paclitaxell | ↑ | ↑ | N | N | N | N | N | N | CR | CR | N |
| P10 | Lum A | 4′ EC + 12′ Paclitaxel | ↓ | ↓ | N | N | N | N | P | N | PR | PR | N |
| P11 | TN | 4′ EC + 9′ Paclitaxel | ↓ | ↓ | N | N | N | P | N | N | PR | PR | N |
| P12 | Lum A | - | ↑ | ↓ | P | N | P | P | P | N | PR | PD | P |
| P13 | Lum A | Anastrozole | ↑ | ↑ | N | N | N | N | N | N | PR | PR | N |
| P14 | HER2 enriched | - | - | N | N | P | N | N | N | PR | PD | N | |
| P15 | Lum B, HER2+ | 4′ EC + 12′ Paclitaxel + 4′ Herceptin | ↑ | ↓ | N | N | N | N | N | N | PR | CR | N |
| P16 | Lum B, HER2+ | 4′ EC + 12′ Paclitaxel + 4′ Herceptin | ↑ | ↓ | N | N | N | N | N | N | PR | PR | N |
| P17 | Lum A | 12′ Paclitaxel + 4′ EC | ↑ | ↓ | P | N | P | N | P | N | PR | PR | P |
| P18 | Lum B | 12′ Paclitaxel + 4′ AC | ↑ | ↓ | N | N | N | N | N | N | PR | PR | N |
| P19 | TN | 4′ AC + 10′ Paclitaxel | ↓ | ↓ | N | N | N | N | N | N | PR | PD | P |
| P20 | Lum B, HER2+ | - | - | - | N | N | N | N | N | P | PR | PR | N |
| P21 | Lum A | 4′ EC + 12′ Paclitaxel | ↑ | ↓ | N | N | N | N | N | N | PR | PR | N |
| P22 | Lum B | 4′ EC + 9′ Paclitaxel | ↑ | ↓ | N | N | N | N | N | N | CR | CR | N |
| P23 | TN | 4′ EC + 12′ Paclitaxel | ↓ | ↑ | N | N | N | N | N | N | CR | CR | N |
| P24 | Lum B | 4′ EC | ↓ | ↓ | N | N | N | P | N | P | PR | PR | N |
| P25 | Lum B | 4′ EC + 11′ Paclitaxel | ↑ | ↓ | N | N | N | N | N | N | PR | PR | N |
| P26 | HER2 enriched | 12′ Paclitaxel + 4′ Herceptin + 4′ AC | ↑ | ↓ | N | N | N | N | P | N | CR | CR | N |
| P27 | Lum B | 4′ AC | ↑ | ↑ | N | P | N | N | P | N | PR | PR | N |
| P28 | TN | 4′ EC + 12′ Paclitaxel | ↑ | ↓ | P | P | N | N | N | N | PR | PR | N |
| P29 | HER2 enriched | 12′ Paclitaxel + 6′ Herceptin + 4′ AC | - | - | N | N | N | N | N | P | CR | CR | P |
| P30 | Lum B | 4′ EC + 12′ Paclitaxel | ↑ | ↓ | P | N | N | N | P | N | PR | PR | P |
| P31 | Lum B, HER2+ | 12′ Paclitaxel + 4′ Herceptin + 4′ AC | ↑ | ↓ | N | P | N | N | N | N | PR | PR | N |
| P32 | Lum A | 4′ AC + 9′ Paclitaxel | ↓ | ↓ | N | N | N | N | N | N | PR | PD | N |
| P33 | HER2 enriched | 10′ Paclitaxel + 7′ Herceptin + 4′ AC | ↑ | ↑ | N | P | N | N | N | N | CR | PR | N |
| P34 | TN | 4′ AC + 12′ Paclitaxel | ↑ | ↑ | N | P | N | N | N | N | PR | PR | P |
| P35 | HER2 enriched | 4′ AC + 12′ Paclitaxel + 4′ Herceptin | ↑ | - | N | N | N | N | N | N | CR | CR | N |
| P36 | Lum B, HER2+ | 12′ Paclitaxel + 4′ Herceptin + 4′ AC | - | - | P | N | N | - | - | N | |||
Abbreviations: ALDH1: tumor stem cells marker; CR: complete response; CTC: circulating tumor cell; EMT: epithelial-mesenchymal transition; HER2: Human epidermal growth factor receptor 2; Lum A: luminal A; Lum B; luminal B; NACT: neoadjuvant chemotherapy; N: negative; P: positive; PD: progressive response; PR: partial response; TN: triple negative.
3.5. Relationship of ccfDNA levels and mtDNAcn with breast cancer type, therapy response, and metastasis
When we compare the same patient group with previous study data, only 1 out of 6 patients who tested negative for metastasis and exhibited complete pathological and clinical responses showed negative CTC, EMT, and ALDH1 markers both before and after NACT. However, mtDNA copy number decreased, while ccfDNA level increased after treatment, but the difference was not statistically significant (p > 0.05). Patients’ ccfDNA and mtDNA levels were analyzed according to breast cancer type. No significant results were found between them (p > 0.05) (Table 2).
4. Discussion
Breast cancer stands as the leading form of cancer affecting women globally, presenting numerous challenges for effective treatment [1]. Traditional methods such as tissue biopsy, although widely employed, have limitations in terms of comprehensively detecting the disease and monitoring treatment response. Additionally, the site of metastases can pose obstacles to biopsy procedures [9,16]. Therefore, there is a critical clinical need for noninvasive biomarkers that can aid in the diagnosis and follow-up of breast cancer [46].
Breast cancer exhibits a complex molecular landscape, necessitating innovative approaches for its detection and monitoring. The advent of liquid biopsy, which involves the analysis of circulating tumor cells (CTCs), cell-free DNAs (cfDNAs), and mitochondrial DNA (mtDNA) in plasma and serum, has emerged as a promising noninvasive tool in cancer diagnostics [9,16,47]. Cancer cells can enter the bloodstream early in the disease process, even before the detection of a tumor, and can disseminate throughout the body. By capturing and analyzing CTCs, cfDNAs, and mtDNA released through apoptosis, necrosis, or active release during tumor growth, liquid biopsy enables the comprehensive assessment of disease dynamics [47].
Plasma ccfDNA and mtDNA were evaluated as blood biomarkers to assess neoadjuvant chemotherapy response in breast cancer patients. Changes in ccfDNA levels, mtDNA copy numbers, and integrity were detected. The relationship between CTC molecular analysis and EMT was explored, aiming to predict metastasis development and estimate patient outcomes.
Consistent with previous findings, breast cancer patients exhibited higher plasma levels of circulating cell-free DNA (ccfDNA) compared to controls, alongside elevated levels of ALU 115 and ALU 247. This is likely due to increased release of fragmented DNA from apoptotic and necrotic cells in breast cancer patients. Similarly, a study involving breast and prostate cancer patients reported a higher ccfDNA integrity index in prostate cancer patients compared to controls, while breast cancer patients had a lower index. The amount of cfDNA released into circulation is influenced by factors such as cancer stage, tumor mutation load, and DNA clearance rate. The ALU DNA integrity index has been suggested as a more advantageous marker than absolute ccfDNA levels, as it correlates with tumor cell death. However, variations in DNA integrity index among different cancer types and individual cases highlight its heterogeneity and complexity. When we examined all the patients we have, ccfDNA levels increased significantly posttreatment, while the ccfDNA integrity index decreased and was not statistically significant. Severe destruction of cells and different number of pre- and posttreatment data may affect this situation. In studies conducted with various cancer patients differences in the integrity index were observed while ccfDNA levels increased posttreatment [43,45,48,49]. These findings highlight the importance of ALU 115 and ALU 247 levels, as well as ccfDNA integrity, as potential biomarkers for breast cancer.
In 28 patients with both pre- and posttreatment data, a significant decrease in ccfDNA levels and a nonsignificant increase in ccfDNA integrity were observed posttreatment. These findings align with a study by Adusei et al., who also reported a decline in serum ALU 247 and ALU 115 levels and an increase in ccfDNA integrity after three cycles of chemotherapy in breast cancer patients [14]. These findings emphasize the dynamic nature of plasma ccfDNA levels in response to treatment in breast cancer patients. The significant decrease in ccfDNA levels following treatment indicates its potential as a monitoring tool for treatment response. Further investigations are warranted to explore the underlying mechanisms driving the alterations in ccfDNA levels and integrity, and to evaluate their clinical implications in breast cancer management.
Chemotherapy treatment leads to the destruction of cancer cells, resulting in the release of cellular DNA into the bloodstream and subsequently increased levels of circulating cell-free DNA (ccfDNA) in the blood [48]. However, posttreatment measurements reveal lower ccfDNA levels, indicating a reduction in cancer cell population and activity, as well as an efficient clearance system for cfDNA. The extensive destruction of cancer cells during treatment may contribute to the release of longer DNA fragments, potentially affecting DNA integrity [14]. Studies in breast and colorectal cancer patients have demonstrated a decrease in ALU 115 levels and an increase in integrity following treatment [31,50,51]. These observations highlight the dynamic nature of ccfDNA and its potential as a valuable biomarker in monitoring chemotherapy response.
Our findings revealed a higher mitochondrial DNA (mtDNA) copy number in breast cancer patients compared to controls, with the highest levels observed pretreatment. These results were statistically significant. In contrast, mtDNA integrity was higher in the control group, although this difference was not statistically significant. Studies have reported conflicting results regarding mtDNA copy number, with some showing higher levels in the patient group and others demonstrating lower levels [52–58]. Consistent with our results, a study in breast cancer patients found the highest mtDNA copy number in late-stage cancer patients, while healthy individuals and early-stage cancer patients exhibited lower levels [59]. Additionally, elevated ND1 levels have been observed in thyroid and colorectal cancer patients compared to normal individuals, potentially indicating increased replication-induced mtDNA damage and the need for compensatory mtDNA molecules in tumor tissues [38,40,57]. Despite the mtDNA integrity index not reaching statistical significance and being lower in patients due to fragmented DNA and increased copy number, further research could explore its potential as a marker for treatment follow-up in breast cancer.
In our current study, we also aimed to examine the relevance of the previous findings to our research. However, we did not observe any statistically significant associations between ccfDNA levels, mtDNA copy numbers (mtDNAcn), and CTCs, EMT, ALDH1, treatment response, or metastasis. To gain a comprehensive understanding of these biomarkers’ clinical significance, further investigations involving larger patient cohorts are warranted. Serial monitoring and characterization of these biomarkers at specific time points during treatment are essential to elucidate their potential as clinically meaningful indicators. This study is the first to comprehensively investigate plasma ccfDNA levels, mtDNA copy number, and their integrities simultaneously. It is also the first to explore the relationship between these biomarkers and CTCs, cancer stem cell markers, treatment response, and metastatic status. Differences in biomarker levels observed in our study may stem from variations in factors such as blood collection periods, sample preprocessing, storage, DNA isolation procedures, as well as clinical characteristics including tumor stage, tumor size, and patient age. The findings highlight the potential of ccfDNA and mtDNA as biomarkers for monitoring chemotherapy response in breast cancer. Noninvasive methods for cancer detection and monitoring have gained significant attention in recent years. These approaches offer the potential for improved patient comfort, reduced invasiveness, and enhanced accessibility. The development of reliable biomarkers that can be detected through noninvasive means is crucial to address these clinical needs. However, due to the limited dataset and lack of pre- and posttreatment results, further confirmation in larger patient cohorts is necessary to validate our findings.
Acknowledgments
This study was retrospectively registered with TÜBİTAK 1001 project (115S272) (M.Ü. Faculty of Medicine Clinical Research Ethics Committee protocol code: 09.2019.283). It received a specific grant from Marmara University Research Fund (BAPKO) under project TYL-2022-10552.
Funding Statement
It received a specific grant from Marmara University Research Fund (BAPKO) under project TYL-2022-10552.
Footnotes
Ethical approval: The ethical suitability of this study was reviewed and unanimously approved by the Marmara University Faculty of Medicine Clinical Research Ethics Committee (Ethics committee protocol code: 09.2022.246).
References
- 1. Al-Bashaireh AM, Khraisat O, Alnazly EK, Aldiqs M. Inflammatory markers, metabolic profile, and psychoneurological symptoms in women with breast cancer: a literature review. Cureus. 2021;13(11) doi: 10.7759/cureus.19953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Theil G, Fornara P, Bialek J. Position of circulating tumor cells in the clinical routine in prostate cancer and breast cancer patients. Cancers. 2020;12(12):1–21. doi: 10.3390/cancers12123782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tetikoğlu S. MSc. Karadeniz Technical University; Trabzon, Türkiye: 2020. Investigation of the long-term cytotoxic effect of bee venom on liver and metastatic breast cancer cells. (in Turkish with an abstract in English) [Google Scholar]
- 4. Peled M, Agassi R, Czeiger D, Ariad S, Riff R, et al. Cell-free DNA concentration in patients with clinical or mammographic suspicion of breast cancer. Scientific Reports. 2020;10(14601):1–9. doi: 10.1038/s41598-020-71357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dawson SJ, Tsui DWY, Murtaza M, Biggs H, Rueda O, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. The New England Journal of Medicine. 2013;368(13):1199–1209. doi: 10.1056/nejmoa1213261. [DOI] [PubMed] [Google Scholar]
- 6. Hamam R, Hamam D, Alsaleh KA, Kassem M, Zaher W, et al. Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death and Disease. 2017;8(9):e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barbany G, Arthur C, Lied A, Nordenskj M, Rosenquist R, et al. Cell-free tumour DNA testing for early detection of cancer – a potential future tool. Journal of Internal Medicine. 2019;286:118–136. doi: 10.1111/joim.12897. [DOI] [PubMed] [Google Scholar]
- 8. Huang CC, Du M, Wang L. Bioinformatics analysis for circulating cell-free DNA in cancer. Cancers (Basel) 2019 Jun 11;11(6):805. doi: 10.3390/cancers11060805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Junqueira-Neto S, Batista IA, Costa JL, Melo SA. Liquid biopsy beyond circulating tumor cells and cell-free DNA. Acta Cytologica. 2019;63:479–488. doi: 10.1159/000493969. [DOI] [PubMed] [Google Scholar]
- 10. Panagopoulou M, Esteller M, Chatzaki E. Circulating cell-free DNA in breast cancer: searching for hidden information towards precision medicine. Cancers (Basel) 2021;13(728):1–25. doi: 10.3390/cancers13040728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ponti G, Manfredini M, Tomasi A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Critical Reviews in Oncology/Hematology. 2019;141(2019):36–42. doi: 10.1016/j.critrevonc.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 12. Stewart CM, Tsui DWY. Circulating cell-free DNA for non-invasive cancer management. Cancer Genetics. 2018;228–229:169–179. doi: 10.1016/j.cancergen.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delmonico L, Alves G, Bines J. Cell free DNA biology and its involvement in breast carcinogenesis. Advances in Clinical Chemistry. 2020;97:171–223. doi: 10.1016/bs.acc.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 14. Adusei E, Ahenkorah J, Adu-aryee NA, Adutwum-Ofosu KK, Tagoe EA, et al. Reduced serum circulation of cell-free DNA following chemotherapy in breast cancer patients. Medical Sciences. 2021;9(2):37. doi: 10.3390/medsci9020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Cambridge Philosophical Society. 2018;93(3):1649–1683. doi: 10.1111/brv.12413. [DOI] [PubMed] [Google Scholar]
- 16. Bronkhorst AJ, Ungerer V, Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomolecular Detection and Quantification. 2019;17(November 2018):100087. doi: 10.1016/j.bdq.2019.100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duvvuri B, Lood C. Cell-Free DNA as a Biomarker in Autoimmune Rheumatic Diseases. Frontiers in Immunology. 2019;10(502):1–21. doi: 10.3389/fimmu.2019.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gezer U, Bronkhorst AJ, Holdenrieder S. The utility of repetitive cell-free DNA in cancer liquid biopsies. Diagnostics. 2022;12:1363. doi: 10.3390/DIAGNOSTICS12061363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meddeb R, Pisareva E, Thierry AR. Guidelines for the preanalytical conditions for analyzing circulating cell-free DNA. Clinical Chemistry. 2019;65(5):623–633. doi: 10.1373/clinchem.2018.298323. [DOI] [PubMed] [Google Scholar]
- 20. Pös O, Biró O, Szemes T, Nagy B. Circulating cell-free nucleic acids: Characteristics and applications. European Journal of Human Genetics. 2018;26(7):937–945. doi: 10.1038/s41431-018-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szilágyi M, Pös O, Márton É, Buglyó G, Soltész B, et al. Circulating cell-free nucleic acids: main characteristics and clinical application. International Journal of Molecular Sciences. 2020;21(18):6827. doi: 10.3390/ijms21186827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang P, Bahreini A, Gyanchandani R, Lucas P, Hartmaier R, et al. Sensitive detection of mono- and polyclonal ESR1 mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clinical Cancer Research. 2016;22(5):1130–1137. doi: 10.1158/1078-0432.CCR-15-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Oliveira IBD, Hirata RDC. Circulating cell-free DNA as a biomarker in the diagnosis and prognosis of colorectal cancer. Brazilian Journal of Pharmaceutical Sciences. 2018;54(1):e17368. doi: 10.1590/s2175-97902018000117368. [DOI] [Google Scholar]
- 24. Jiang Y, Zong W, Ju S, Jing R, Cui M. Promising member of the short interspersed nuclear elements (Alu elements): mechanisms and clinical applications in human cancers. Journal of Medical Genetics. 2019;56:639–645. doi: 10.1136/jmedgenet-2018-105761. [DOI] [PubMed] [Google Scholar]
- 25. Park MK, Lee JC, Lee JW, Hwang SJ. Alu cell-free DNA concentration, Alu index, and LINE-1 hypomethylation as a cancer predictor. Clinical Biochemistry. 2021;94(February):67–73. doi: 10.1016/j.clinbiochem.2021.04.021. [DOI] [PubMed] [Google Scholar]
- 26. Hussein NA, Mohamed SN, Ahmed MA. Plasma ALU-247, ALU-115, and cfDNA integrity as diagnostic and prognostic biomarkers for breast cancer. Applied Biochemistry and Biotechnology. 2019;187(3):1028–1045. doi: 10.1007/s12010-018-2858-4. [DOI] [PubMed] [Google Scholar]
- 27. Umetani N, Kim J, Hiramatsu S, Reber H, Hines O, et al. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clinical Chemistry. 2006;52(6):1062–1069. doi: 10.1373/clinchem.2006.068577. [DOI] [PubMed] [Google Scholar]
- 28. Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. Journal of Clinical Oncology. 2006;24(26):4270–4276. doi: 10.1200/JCO.2006.05.9493. [DOI] [PubMed] [Google Scholar]
- 29. Arko-Boham B, Aryee NA, Blay RM, Owusu EDA, Tagoe EA, et al. Circulating cell-free DNA integrity as a diagnostic and prognostic marker for breast and prostate cancers. Cancer Genetics. 2019;235–236:65–71. doi: 10.1016/j.cancergen.2019.04.062. [DOI] [PubMed] [Google Scholar]
- 30. Lehner J, Stötzer OJ, Fersching D, Nagel D, Holdenrieder S. Circulating plasma DNA and DNA integrity in breast cancer patients undergoing neoadjuvant chemotherapy. Clinica Chimica Acta. 2013;425:206–211. doi: 10.1016/J.CCA.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 31. Lehner J, Stötzer OJ, Fersching DMI, Nagel D, Holdenrieder S. Plasma DNA integrity indicates response to neoadjuvant chemotherapy in patients with locally confined breast cancer. International Journal of Clinical Pharmacology and Therapeutics. 2013;51(1):59–62. doi: 10.5414/CPP51059. [DOI] [PubMed] [Google Scholar]
- 32. Condappa A, McGrowder D, Aiken W, McLaughlin W, Gossell-Williams M. Evaluation of plasma circulating cell free DNA concentration and integrity in patients with prostate cancer in Jamaica: a preliminary study. Diseases. 2020;8:34. doi: 10.3390/diseases8030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eskander NS, Mansour L, Abdelaal A, Saad E, Mohamed D. Circulating cell free DNA integrity index as a biomarker for response to chemotherapy in patients with metastatic colorectal carcinoma. Asian Pacific Journal of Cancer Prevention. 2022;23(1):339–348. doi: 10.31557/APJCP.2022.23.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thyagarajan B, Wang R, Nelson H, Barcelo H, Koh WP, et al. Mitochondrial DNA copy number is associated with breast cancer risk. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0065968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Afrifa J, Zhao T, Yu J. Circulating mitochondria DNA, a non-invasive cancer diagnostic biomarker candidate. Mitochondrion. 2019;47:238–243. doi: 10.1016/j.mito.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 36. Yu M. Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sciences. 2011;89:65–71. doi: 10.1016/j.lfs.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 37. Campa D, Barrdahl M, Santoro A, Severi G, Baglietto L, et al. Mitochondrial DNA copy number variation, leukocyte telomere length, and breast cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Breast Cancer Research. 2018:20–29. doi: 10.1186/s13058-018-0955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jiang Z, Bahr T, Zhou C, Jin T, Chen H, et al. Diagnostic value of circulating cell-free mtDNA in patients with suspected thyroid cancer: ND4/ND1 ratio as a new potential plasma marker. Mitochondrion. 2020;55:145–153. doi: 10.1016/j.mito.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rygiel KA, Grady JP, Taylor RW, Tuppen HAL, Turnbull DM. Triplex real-time PCR-an improved method to detect a wide spectrum of mitochondrial DNA deletions in single cells. Scientific Reports. 2015;5(July):1–12. doi: 10.1038/srep09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Y, Zhou J, Yuan Q, Su J, Li Q, et al. Quantitative detection of circulating MT-ND1 as a potential biomarker for colorectal cancer. Bosnian Journal of Basic Medical Sciences. 2021;21(5):577–586. doi: 10.17305/bjbms.2021.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akkiprik M, Koca S, Uğurlu MÜ, Ekren R, Peker Eyüboğlu İ, et al. Response assessment with molecular characterization of circulating tumor cells and plasma MicroRNA profiling in patients with locally advanced breast cancer during neoadjuvant chemotherapy. Clinical Breast Cancer. 2020;20(4):332–343e3. doi: 10.1016/j.clbc.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 42. Magbanua MJM, Swigart LB, Wu HT, Hirst G, Yau C, et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Annals of Oncology. 2021;32(2):229–239. doi: 10.1016/j.annonc.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ma G, Wang J, Huang H, Han X, Xu J, et al. Identification of the plasma total cfDNA level before and after chemotherapy as an indicator of the neoadjuvant chemotherapy response in locally advanced breast cancer. Cancer Medicine. 2020;9(7):2271–2282. doi: 10.1002/cam4.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chopra S, Davies EL. Breast cancer key points. Medicine. 2019;11(9):1–6. doi: 10.1016/j.mpmed.2019.11.009. [DOI] [Google Scholar]
- 45. Katopodis P, Anikin V, Kishore U, Carter T, Hall M, et al. Circulating tumour cells and circulating cell-free DNA in patients with lung cancer: a comparison between thoracotomy and video-assisted thoracoscopic surgery. BMJ Open Respiratory Research. 2021;8(1):1–10. doi: 10.1136/bmjresp-2021-000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, et al. Breast cancer. Nature Reviews Disease Primers. 2019 Sep 23;5(1):66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 47. Banys-Paluchowski M, Fehm TN, Grimm-Glang D, Rody A, Krawczyk N. Liquid biopsy in metastatic breast cancer: current role of circulating tumor cells and circulating tumor DNA. Oncology Research and Treatment. 2022;(45):4–10. doi: 10.1159/000520561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Swystun LL, Mukherjee S, Liaw PC. Breast cancer chemotherapy induces the release of cell-free DNA, a novel procoagulant stimulus. Journal of Thrombosis and Haemostasis. 2011;9:2313–2321. doi: 10.1111/j.1538-7836.2011.04465.x. [DOI] [PubMed] [Google Scholar]
- 49. Salem R, Ahmed R, Shaheen K, Abdalmegeed M, Hassan H. DNA integrity index as a potential molecular biomarker in colorectal cancer. Egyptian Journal of Medical Human Genetics. 2020;21(38) doi: 10.1186/s43042-020-00082-4. [DOI] [Google Scholar]
- 50. Zhu F, Ma J, Ru D, Wu N, Zhang Y, et al. Plasma DNA integrity as a prognostic biomarker for colorectal cancer chemotherapy. Journal of Oncology. 2021;2021 doi: 10.1155/2021/5569783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheng J, Holland T, Markus L, Surowy H, Cuk K, et al. Circulating free DNA integrity and concentration as independent prognostic markers in metastatic breast cancer CI Confidence interval. Breast Cancer Research and Treatment. 2018;169(1):69–82. doi: 10.1007/s10549-018-4666-5. [DOI] [PubMed] [Google Scholar]
- 52. Mahmoud EH, Fawzy A, Ahmad OK, Ali AM. Plasma circulating cell-free nuclear and mitochondrial DNA as potential biomarkers in the peripheral blood of breast cancer patients. Asian Pacific Journal of Cancer Prevention. 2016;16(18):8299–8305. doi: 10.7314/APJCP.2015.16.18.8299. [DOI] [PubMed] [Google Scholar]
- 53. Pasha HA, Rezk NA, Riad MA. Circulating cell free nuclear DNA, mitochondrial DNA and global DNA methylation: potential noninvasive biomarkers for breast cancer diagnosis. Cancer Investigation. 2019;37(9):432–439. doi: 10.1080/07357907.2019.1663864. [DOI] [PubMed] [Google Scholar]
- 54. Safi M, Najib AR. Evaluation of circulating cell free nuclear and mitochondrial DNA levels in Syrian patients with breast tumor. Experimental and Therapeutic Medicine. 2020;21(1):1–7. doi: 10.3892/etm.2020.9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Toh YL, Wong E, Chae JW, Yap NY, Yeo AHL, et al. Association of mitochondrial DNA content and displacement loop region sequence variations with cancer-related fatigue in breast cancer survivors receiving chemotherapy. Mitochondrion. 2020;54(July):65–71. doi: 10.1016/j.mito.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 56. Rai NK, Panjwani G, Ghosh AK, Haque R, Sharma LK. Analysis of mitochondrial DNA copy number variation in blood and tissue samples of metastatic breast cancer patients (A pilot study) Biochemistry and Biophysics Reports. 2021;26(January):100931. doi: 10.1016/j.bbrep.2021.100931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weerts MJA, Sieuwerts AM, Smid M, Look M, Foekens J, et al. Mitochondrial DNA content in breast cancer: impact on in vitro and in vivo phenotype and patient prognosis. Oncotarget. 2016;7(20):29166–29176. doi: 10.18632/oncotarget.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tuchalska-Czuroń J, Lenart J, Augustyniak J, Durlik M. Is mitochondrial DNA copy number a good prognostic marker in resectable pancreatic cancer. Pancreatology. 2019;19(1):73–79. doi: 10.1016/j.pan.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 59. Keserű JS, Soltész B, Lukács J, Márton É, Szilágyi-Bónizs M, et al. Detection of cell-free, exosomal and whole blood mitochondrial DNA copy number in plasma or whole blood of patients with serous epithelial ovarian cancer. Journal of Biotechnology. 2019;298:76–81. doi: 10.1016/j.jbiotec.2019.04.015. [DOI] [PubMed] [Google Scholar]