Abstract
Fowlpox virus (FPV), a pathogen of poultry, can persist in desiccated scabs shed from infected hosts. Although the mechanisms which ensure virus survival are unknown, it is likely that some type of remedial action against environmentally induced damage is required. In this regard, we have identified an open reading frame (ORF) coding for a putative class II cyclobutane pyrimidine dimer (CPD)-photolyase in the genome of FPV. This enzyme repairs the UV light-induced formation of CPDs in DNA by using blue light as an energy source and thus could enhance the viability of FPV during its exposure to sunlight. Based on transcriptional analyses, the photolyase gene was found to be expressed late during the FPV replicative cycle. That the resultant protein retained DNA repair activity was demonstrated by the ability of the corresponding FPV ORF to complement functionally a photolyase-deficient Escherichia coli strain. Interestingly, insertional inactivation of the FPV photolyase gene did not impair the replication of such a genetically altered virus in cultured cells. However, greater sensitivity of this mutant than of the parental virus to UV light irradiation was evident when both were subsequently photoreactivated in the absence of host participation. Therefore, FPV appears to incorporate its photolyase into mature virions where the enzyme can promote their survival in the environment. Although expression of a homologous protein has been predicted for some chordopoxviruses, this report is the first to demonstrate that a poxvirus can utilize light to repair damage to its genome.
Among the environmental hazards encountered by living organisms is exposure to UV light, which can damage DNA. This induced formation of potentially lethal cyclobutane pyrimidine dimers (CPDs) and (6-4) photoproducts can be reversed by a photoreactivation process (17) dependent on the presence of blue light (300 to 500 nm) (23, 38). Such repairs are performed by enzymes (photolyases) which bind to the dimers, capture the energy from photons via associated chromophores, and then use electron transfer to split the dimers and restore the DNA to its original form (39). Photolyases maintain specificity for only one type of potential substrates and are designated as such (39, 47). The CPD-photolyases can be further separated into class I and class II on the basis of divergent primary structures, the former being more closely related to the (6-4) photolyases (24). Since representatives of all three groups of repair enzymes have been found in each of the three domains of life (24), there is no correlation between evolutionary status and type of photolyase.
For most DNA-containing viruses, the absence of associated photolyase genes precludes their ability to utilize sunlight for repairing UV light-induced genomic damage. Therefore, any DNA repair dependent on photoreactivation is conditional on successful infection of a host which can provide the necessary enzyme. Indeed, it is well documented that through this process the viability of UV-irradiated viruses can be restored (19). Moreover, the photoreactive ability of marine bacteria has been implicated as the reason for high concentrations of infectious phage in surface water despite the unmitigated exposure to solar radiation (50). Although some viruses such as bacteriophage T4 (34) and possibly the Chlorella virus PBCV-1 (30) encode enzymes which are involved in the excision repair of pyrimidine dimers resulting from UV light irradiation, these proteins would be active only when the virus is internalized in a bacterium and an alga, respectively. In contrast, at least some poxviruses may have circumvented the direct requirement for host intervention. Determination of the nucleotide sequences of the complete genomes of the Melanoplus sanguinipes entomopox virus (2), the leporipoxviruses myxoma virus (11) and Shope fibroma virus (52), and the avipoxvirus fowlpox virus (3) has identified a putative class II CPD-photolyase gene in the DNA of each. Since the encoded enzyme utilizes photons in lieu of physiologically renewed compounds as an energy source and does not require a supply of nucleotides for restoration of damaged DNA, the potential for autonomous genomic repair by extracellular pox virions exists.
In view of the complexity of poxvirus genomes, it is not surprising that these highly successful pathogens (10) have acquired the ability to protect themselves from the perils of their immediate surroundings. Clearly, their production of proteins which mimic host immunoregulators can be construed as being directed toward the provision of a habitat favorable for replication (28). Likewise, their mass accumulation into inclusion bodies may offer some resiliency to environmental insults (8). Moreover, the capture and retention of a gene encoding a light-activated DNA repair enzyme could be indicative of genetic elasticity which would enable poxviruses to adjust to an otherwise hostile environment.
The stability of desiccated poxviruses has proven invaluable for purposes such as the mass vaccination against smallpox of people in underdeveloped countries. However, this property can also be problematic when the virus is present in naturally formed scabs. For example, the avian virus FPV can persist in the dander shed from infected poultry and may later infect naive birds, especially those in multiple-aged flocks, to produce an outbreak of fowlpox (48). Since the released virus is exposed to a variety of wavelengths of light, an FPV-encoded photolyase could contribute to the observed persistence. To address this possibility, we characterized a class II CPD-photolyase open reading frame (ORF) discovered during sequencing of the genome of an FPV field strain. In addition to demonstrating that the gene product was indeed an active enzyme, we also created a photolyase-deficient FPV and found that this mutant, unlike the parental virus, was unresponsive to photoreactivation.
MATERIALS AND METHODS
Virus and cells.
An FPV isolate originating from a fowlpox outbreak in poultry at the University of Illinois (48) was used in this study. Initially this virus was propagated in the chorioallantoic membranes of developing chicken embryos obtained from a specific-pathogen-free flock. For virus purification, pock lesions from infected membranes were collected, ground, and clarified by low-speed centrifugation (450 × g for 10 min). After additional grinding and clarification, virus was pelleted from the supernatants (53,000 × g for 30 min). The resuspended pellet was layered onto a discontinuous gradient consisting of layers of 20, 25, 30, and 40% sucrose in Tris-buffered saline (50 mM Tris [pH 8.0], 150 mM NaCl, 5 mM EDTA) and then centrifuged in a Beckman SW28 rotor at 4°C and 29,800 × g for 70 min. Virus bands at the 25 to 30% and 30 to 40% sucrose interfaces were diluted in Tris-buffered saline and pelleted at 4°C and 53,000 × g for 60 min. The virus pellet was resuspended in Tris-buffered saline and stored at −80°C.
Later, the FPV isolate was propagated in monolayers of the quail fibroblast cell line QT-35 as previously described (42). Whereas the purified virus served as a source of DNA for cloning, the tissue culture-grown virus was used in all subsequent biological assays.
Sequence analysis of the FPV genome.
DNA obtained by phenol-chloroform extraction of gradient-purified FPV was digested with HindIII (Life Technologies, Gaithersburg, Md.), and the resulting fragments were randomly inserted into the corresponding site of pUC19. Recombinant plasmids were differentiated based on the relative sizes of and, when necessary, the presence of unique restriction endonuclease sites within the inserted DNA. The nucleotide sequences of the terminal regions of selected FPV genomic fragments were determined by using universal forward and reverse primers. For completing and verifying the sequence of the photolyase gene, both the universal and internal region-specific primers were used in conjunction with various pieces of cloned FPV DNA. The obtained nucleotide and predicted amino acid sequences were analyzed using Genetics Computer Group software (16) and compared to published sequences using the BLAST and BLASTP functions provided by the National Center for Biotechnology Information (4). The FPV photolyase protein was screened for motifs by using the Prosite database. The amino acid sequences of various photolyases were aligned by using the Clustal W program (45). The alignment was manually adjusted, and a phylogenetic tree was generated by the maximum-likelihood method (1). Analyses were performed using software available at Biology Workbench (http://workbench.sdsc.edu), the expert protein analysis system proteomics server available at the Swiss Institute of Bioinformatics website (http://www.expasy.ch), or BioNavigator (http://www.ebioinformatics.com.).
Southern hybridization.
Intracellular FPV DNA was isolated from infected QT-35 cells, digested with various combinations of restriction endonucleases, and electrophoresed in a 0.8% agarose gel as previously described (42). Conditions for Southern hybridization and preparation of the radioactively labeled probe have been described previously (49). A 725-bp photolyase-specific probe was obtained by HindIII and BamHI digestion of a 16-kb HindIII fragment of the FPV genome.
Cloning of the FPV photolyase gene.
A 3.5-kb EcoRI-PvuII fragment of the viral genome found to contain the entire photolyase gene was ligated with EcoRI-SmaI-digested pUC19. The resulting plasmid, pPHEP1, was maintained in Escherichia coli DH5α (Life Technologies).
Primers.
Based on the determined sequence of the FPV photolyase gene and flanking regions, forward (PFE; CGGAATTCACAATGATTAGCCCCCAATC) and reverse (PRE; CGGAATTCTATTATCTTAATTGTATTTCTG) primers were designed for the amplification of an intact ORF having unique EcoRI recognition sites located at each terminus. For detecting the photolyase gene transcript by reverse transcription (RT)-PCR, a forward primer (132.F; TGCAGATGGTGAGAGAAGG) representing an internal region of this FPV gene was used in conjunction with primer PRE. To detect read-through transcripts that would originate from other FPV ORFs and then extend into the photolyase gene, forward (PT.F; TACATTTACGGAAAATAGCTGG) and reverse (PT.R; AAGAAATCCGTTATGATTACTCC) primers were designed for the amplification of a region extending from 88 nucleotides upstream to 404 nucleotides downstream of the initiation codon of the photolyase gene. For specific detection of the FPV thymidine kinase gene RNA, forward (TK1; TCCGGTAAAACATCGGAG) and reverse (TK2; AACGAAGCGTCGCAATAG) primers corresponding to and complementary with internal portions of this gene were designed based on the published sequence (GenBank accession no. M16617) (9). Likewise, similarly positioned forward (129.F2; GAACAGGTCATGGATGGTC) and reverse (129.R2; GCATTGCTCTATCGACAT) primers to be used for detecting A-type inclusion body protein gene (FPV ORF 191 [(3)]) transcripts were designed based on our determination of this gene's nucleotide sequence (W. M. Schnitzlein, V. Srinivasan, and D. N. Tripathy, unpublished data).
Transcriptional analysis of the FPV photolyase gene.
Monolayers of QT-35 cells were infected with FPV at a multiplicity of infection of 100. To prevent virus DNA synthesis, cells were continuously exposed to cytosine arabinoside (40 μg/ml; Sigma, St. Louis, Mo.) starting at 1 h prior to virus infection. At 8 h postinfection, total RNA was isolated from virus-infected cells in the presence and absence of cytosine arabinoside by using Trizol (Life Technologies) according to the manufacturer's protocol.
RNA preparations were screened for the presence of FPV photolyase gene-specific transcripts by RT-PCR. Briefly, 1 μg of RNA and 0.05 μM reverse primer PRE were denatured at 70°C for 10 min and then placed on ice. First-strand cDNA synthesis using Superscript II reverse transcriptase (Life Technologies) then proceeded for 1 h at 42°C as instructed by the manufacturer and was terminated by incubation at 70°C for 15 min. PCR was performed in a 50-μl volume containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 0.2 μM primers 132.F and PRE, 0.25 U of Taq polymerase, and 5 μl of completed RT reaction mixture. Conditions for this PCR consisted of one cycle of denaturation at 94°C for 2 min, 35 cycles of denaturation at 92°C for 30 s, annealing at 45°C for 30 s, and extension at 72°C for 2 min, and a final cycle of elongation at 72°C for 10 min.
Overall, FPV thymidine kinase and A-type inclusion body protein gene RNAs were reversed transcribed as described above except that primers TK2 and 129.R2, respectively, replaced primer PRE. Likewise, thymidine kinase or A-type inclusion body protein gene transcript-specific primers, TK1 and TK2 or 129.F2 and 129.R2, respectively, were used in PCR. Although the composition of the PCR mixture remained constant, the conditions were altered to one cycle of denaturation at 94°C for 2 min, 35 cycles of denaturation at 92°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s, and a final cycle of elongation at 72°C for 5 min.
To evaluate amplification of transcripts reading through to the photolyase gene, RT was performed as described above except that (i) random hexamers (0.05 μM; Life Technologies) were used as primers and (ii) the reaction conditions were altered to include an initial primer annealing step at 25°C for 10 min. PCR using primers PT.F and PT.R was performed as described for amplification of the thymidine kinase and A-type inclusion body protein gene cDNAs.
Phenotypic complementation of photolyase-deficient E. coli.
To generate an intact FPV photolyase gene flanked by EcoRI recognition sites, plasmid pPHEP1 was used as the template in a PCR together with primers PFE and PRE. Amplification conditions were similar to those using the photolyase gene cDNA as the template except that the length of the initiation denaturation was reduced to 1 min and the number of repetitive cycles was decreased to 30. The EcoRI-digested amplicon was ligated into the corresponding site of plasmid pBAD 24 (21). In this plasmid there is a Shine-Dalgarno bacterial ribosome binding site positioned immediately upstream of its EcoRI recognition site. Further upstream is the E. coli arabinose promoter, which regulates expression of an inserted gene and whose activity is repressed in the presence of glucose and induced in the presence of arabinose.
Recombinant plasmid pPHE1, containing the correctly oriented photolyase gene, was used to transform the photolyase-deficient E. coli strain CSR 603 (recA1 uvrA6 phr-1; provided by the Escherichia coli Genetic Stock Center, Yale University). UV sensitivity of and capability of photoreactivation by the transformed bacteria were assayed in the following manner. Individual colonies of both the parental bacterium and one harboring plasmid pPHR1 were separately grown to mid-log phase in Luria broth (LB) supplemented with 0.2% glucose. At this point, aliquots of the bacterial cultures were pelleted at 450 × g and 4°C for 10 min and resuspended to their original volume in LB containing either 0.2% arabinose or glucose. After incubation for 3 h at 37°C, each culture was diluted 100-fold in LB and then exposed to UV light (0.0, 0.1, 0.2, 0.3, and 0.4 J/m2) at 254 nm in a Spectroline EF-16 shortwave UV lamp (Spectronics Corp., Westbury, N.Y.). Cultures were then diluted an additional 100-fold in LB, and equal aliquots were kept either in darkness or exposed to Plexiglas-filtered white light for 1 h at ambient conditions. The number of survivors in each situation was based on the CFU obtained after incubation of diluted samples on LB (with 100 μg of ampicillin/ml) agar plates in darkness for 16 h at 37°C.
Generation of photolyase-deficient FPV.
The insertion vector pPHL1 was generated by ligating the vaccinia virus P11 promoter-lacZ gene fusion, released from pVBX5 (41) by XbaI digestion, with XbaI-digested pPHEP1. In pPHL1, the lacZ gene is transcribed in the same direction as the photolyase gene and nearly bisects this FPV ORF. Plasmid pPHL1 was maintained in E. coli DH5α and purified on an anion-exchange resin column (Qiagen, Valencia, Calif.) prior to use in transfection with FPV.
To produce recombinant virus, approximately 106 QT-35 cells in one well of a six-well cluster plate (Costar, Cambridge, Mass.) were infected with FPV at a multiplicity of infection of 5. At 6 h postinfection, the monolayer was overlaid with 2.5 ml of growth medium (42) lacking serum but containing 2 μg of pPHL1 and 12 μl of Lipofectamine reagent (Life Technologies). After an additional 18 h, an equivalent amount of medium supplemented with 4% serum was added and the infection was allowed to proceed for 48 h more. At this time, cytopathic effect was nearly complete and the cells were harvested and stored at −80°C. Recombinant viruses were then identified and plaque purified six times on the basis of the ability to produce β-galactosidase (lacZ gene product) and thus hydrolyze the substrate Bluo-gal (Life Technologies) as previously described (41, 49). Purity and genotype of the recombinant virus was verified by using primers PRF and PRE to amplify the region flanking the lacZ gene insertion site in conjunction with Elongase enzyme (Life Technologies).
Growth kinetics of photolyase-deficient FPV.
Monolayers of QT-35 cells were infected with either parental or recombinant FPV at a multiplicity of infection of 0.1 for 1 h at ambient conditions and then washed twice with medium to remove unattached virus. At 0, 8, 18, 36, and 42 h postinfection, monolayers were stored at −80°C. After two rounds of freezing and thawing, the amount of infectious virus in each sample was determined by plaque assays in QT-35 cell monolayers (42).
UV light inactivation of photolyase-deficient FPV.
QT-35 cells infected with either parental or recombinant FPV were frozen and thawed twice in the presence of the overlaying medium. Intact cells and cellular debris were then removed by centrifugation at 450 × g and 4°C for 10 min. The supernatants were titrated for the presence of infectious virus and stored at −80°C. The amounts of cell-free virus in the supernatants were normalized to 4 × 107 PFU/ml, and 0.5 ml of each virus solution was irradiated with UV light at 0.0, 0.1, 0.2, 0.3, or 0.4 J/m2. Samples were then divided equally and either kept in darkness or exposed to Plexiglas-filtered white light for 1 h at ambient conditions. The number of survivors in each situation was determined by titration in QT-35 monolayers.
Nucleotide and protein sequence databases.
Accession numbers presented are from the GenBank, DJB, PIR PRF, or SwissPro database.
Nucleotide sequence accession number.
The FPV photolyase gene promoter and ORF sequence has been deposited in GenBank under accession no. AF246697.
RESULTS
Identification of a photolyase gene in the FPV genome.
Based on sequence analysis of one of the terminal regions of an approximately 16-kb HindIII fragment of the genome of an FPV field strain isolated in the early 1970s (48), an incomplete ORF was detected. Comparison of its predicted amino acid sequence to the PIR database indicated that it represented the carboxyl half of a class II CPD-photolyase. In an attempt to obtain the intact gene, a radioactive probe representing the known portion was used in Southern hybridization with restriction endonuclease-digested FPV DNA. Partial sequencing (1.52 kb) of an identified 3.5-kb EcoRI-PvuII fragment enabled the remainder of the photolyase gene to be determined. The entire ORF plus its putative promoter was later found to be identical to that recently reported for an FPV (3) derived from a vaccine manufactured in the early 1960s (fowlpox challenge virus disclosure; Animal and Plant Health Inspection Service Center for Veterinary Biologics, Ames, Iowa). Interestingly, comparison to a cited nucleotide sequence derived from the genome of an FPV of unknown origin (36) revealed one discontinuous and two adjacent nucleotide mismatches. These alterations resulted in the nonconservative replacements of a glutamic acid and alanine moiety in the photolyase of the FPV field isolate by an alanine and valine, respectively.
The FPV-encoded photolyase is predicted to be 464 amino acids in length and to have a molecular mass of approximately 54.5 kDa, physical attributes similar to those of other photolyases. The primary structure of this virus protein is most similar (55% identity, 72% similarity) to that of the South American opossum photolyase (53) but is also well conserved (53 to 54% identity, 68 to 70% similarity) in other poxvirus-encoded photolyases. Regions of homology between these proteins are interspersed throughout the molecules and are especially prevalent in their carboxyl halves, where the class II DNA photolyase Prosite sequences (PS01083 and PS01084) are located (Fig. 1). As previously noted for the FPV enzyme (3), there is a conservative replacement of glutamic acid by aspartic acid at the third position of the first Prosite sequence. This substitution has also occurred in both leporipoxvirus photolyases. The only other deviation from the first Prosite sequence is at the ninth position of the insectpox virus protein, where there is a conservative replacement of arginine by lysine. As for the second consensus motif, the only divergence is at the last position, where a nonconservative and a semiconservative replacement of asparagine by lysine (leporipoxviruses) and serine (insectpox virus), respectively, is evident.
FIG. 1.
Multiple amino aid sequence alignments of poxvirus and opossum photolyases. The deduced amino acid sequences of photolyases of FPV (accession no. AF246697) myxoma virus (MXV; accession no. AAF15015), Shope fibroma virus (SFV; accession no. AAF18004), M. sanguinipes entomopox virus (MSV; accession no. AAC97743), and short-tailed opossum (Monodelphis domestica) (STP; accession no. S50083) were aligned with the Clustal W program and analyzed for the presence of specific motifs in the Prosite database. Regions of conserved amino acids are shaded, and locations of the class II DNA photolyase signature sequences (PS01083 and PS01084) are underlined.
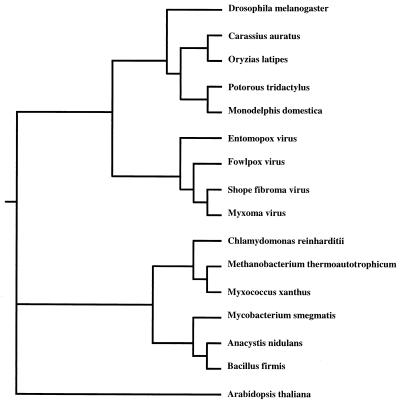
Since photolyase is apparently not produced by orthopoxviruses due to the apparent absence of the corresponding gene (20, 32, 44), it was of interest to determine the evolutionary relationship of the known poxvirus photolyases to each other and representatives from the three domains of life. A phylogenetic comparison of class II photolyases suggested that the FPV enzyme is most closely related to those of the leporipoxviruses (Shope fibroma virus and myxoma virus) (Fig. 2). Moreover, the inclusion of the insectpox virus (M. sanguinipes) counterpart within this clade is indicative of a common progenitor. Furthermore, the poxvirus photolyases appear to share a common ancestry with those of eukaryotes, and this group as a whole seems to have diverged from the eubacteria and the archaebacteria as suggested by Kanai et al. (24) and also from plants.
FIG. 2.
Phylogenetic tree of class II DNA photolyases. The amino acid sequences of vertebrate and invertebrate photolyases were aligned by using the Clustal W program. The alignment was manually edited, and a phylogenetic tree was generated by the maximum-likelihood method. Sources of enzymes: fruit fly (Drosophila melanogaster), accession no. S52047; goldfish (Carassius auratus), accession no. P34205; Japanese medaka (Oryzias latipes), accession no. BAA05043; long-nosed potoroo (Potorous tridactylus), accession no. BAA05041; short-tailed opossum (Monodelphis domestica), accession no. S50083; entomopox virus, accession no. AAC97743; fowlpox virus, accession no. AF246697; Shope fibroma virus, accession no. AAF18004; myxoma virus, accession no. AAF15015; Chlamydomonas reinharditii, accession no. AAD39433; Methanobacterium thermoautotrophicum, accession no. P12769; Myxococcus xanthus, accession no. AAC43723; Mycobacterium smegmatis, accession no. AAF04135; cyanobacterium (Anacystis nidulans), accession no. P05327; Bacillus firmis, accession no. 2017201A; thale cress (Arabidopsis thaliana), accession no. BAA74701.
Temporal expression of the FPV photolyase.
For predicting the temporal expression of poxvirus genes, consensus sequences representing portions of natural and/or mutated vaccinia virus early, intermediate, and late promoters have been described (5, 14, 15, 22, 37). Unfortunately, the region immediately upstream of and including the initiation codon of the FPV photolyase gene (ATATTGAATCTATATTGTTTTTTAGTTATATAAAAACATG) does not conform to any of these. There is a somewhat A-rich stretch reminiscent of the early promoter consensus sequence (AAAAAATGAAAAAAA/TA), but its location would require transcription to initiate within the promoter instead of at least the usual 10 to 20 nucleotides further downstream. Moreover, the only proximal early transcriptional termination signal (TTTTTNT) (54) is located within the FPV photolyase gene at a site 454 nucleotides upstream from its termination codon. Thus, it would appear that early production of photolyase is precluded, although a T5NT motif is present at unlikely sites within some apparently early and intermediate Shope fibroma virus genes (52). Since the consensus intermediate promoter AAANAAN11–13TAAA and late promoter TAAAT(G) sequences are also absent, it is likely that the FPV photolyase is not strongly expressed.
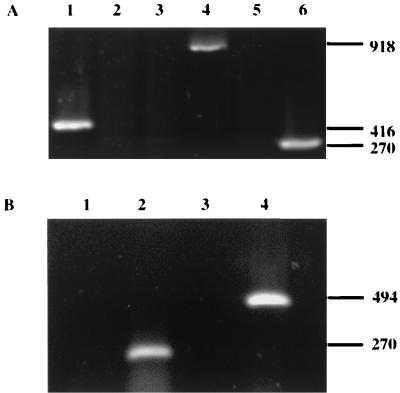
To determine whether and presumably when the FPV photolyase gene is transcribed, RNA isolated from QT-35 cells infected with FPV in the presence and absence of cytosine arabinoside was subjected to RT-PCR. When primers capable of directing the specific amplification of a photolyase transcript were used, products were generated only with RNA produced after the onset of viral DNA replication (Fig. 3A). Similar results were obtained using primers specific for an A-type inclusion body protein gene transcript predicted to be produced late based on the conservation of the TAAATG motif within its promoter. In contrast, the early FPV thymidine kinase transcript (9) was detected only in RNA from cells treated with cytosine arabinoside. No amplicons were observed when the RT step was omitted or the source of RNA was an uninfected monolayer (data not shown). Thus, the FPV photolyase gene appeared to be transcriptionally regulated by an intermediate or late promoter. However, since the mRNAs produced after the onset of DNA synthesis fail to terminate accurately (13), the amplicons generated in the presence of the photolyase gene-specific primers could conceivably be derived from read-through transcripts. To address this possibility, PCR was performed on nonspecifically reversed-transcribed FPV-infected cell RNA using primers directing the specific amplification of cDNA corresponding to a region flanking the initiation codon of the photolyase gene and located upstream of that portion of the gene previously amplified. Although the predicted 494-bp product was readily obtained when FPV DNA was used as the template, a similar-sized amplicon was not observed when cDNA was substituted for the virus genome (Fig. 3B). This inadequacy could not be attributed to template preparation, as evidenced by successful amplification in the presence of the photolyase mRNA-specific primers. Therefore, authentic transcription of the FPV photolyase gene was being demonstrated.
FIG. 3.
Requirement of DNA replication for expression of the FPV photolyase gene. (A) RNAs extracted from FPV-infected QT-35 cells in the presence (lanes 1, 3, and 5) or absence of (lanes 2, 4, and 6) of cytosine arabinoside were subjected to RT-PCR using primers specific for the FPV thymidine kinase (lanes 1 and 2), A-type inclusion body protein (lanes 3 and 4), or photolyase (lanes 5 and 6) gene transcript. (B) RNAs extracted from FPV-infected QT-35 cells were reverse transcribed in the presence of random hexamer primers and then subjected to PCR using primers specific for either the FPV photolyase gene transcript (lane 2) or potential read-through transcripts encompassing a region including the photolyase gene promoter and 29% of the downstream coding sequence (lane 3). The extracted RNAs as well as virion DNA were also directly subjected to PCR using either the photolyase gene (lane 1) or read-through (lane 4) transcript-specific primer. The amplicons were electrophoresed in a 1.0% agarose gel, and their sizes (in base pairs) are indicated at the right.
FPV photolyase gene encodes a functionally active enzyme.
To examine whether the FPV photolyase is functional, its activity in the photolyase-deficient E. coli strain CSR-603 (40) was indirectly assessed. For this purpose, the corresponding viral gene was placed under the transcriptional regulation of the bacterial arabinose promoter located in plasmid pBAD24 (21), and the resulting recombinant plasmid pPHE1 was introduced into the E. coli mutant. Expression of the foreign gene was induced or repressed by growth of the transformed bacterium in the presence of arabinose or glucose, respectively (21). The relative photoreactive capability of repairing UV light-induced lethal damage to the bacterial genome was then determined by comparing the viability curves of the transformed bacteria exposed to various doses of UV light and then either kept in darkness or placed in filtered white light (photoreactivated). A similar inactivation of the transformed bacterium grown in the presence of glucose was observed regardless of subsequent treatment and was comparable to that of the induced but not photoreactivated strain (Fig. 4). Likewise, placement of the nontransformed parent strain in white light failed to reverse the lethal effect of UV light exposure. In contrast, the induced and photoreactivated transformed bacterium exhibited an increased (up to approximately 1,000-fold) resistance to UV light inactivation. Thus, it appeared that the FPV enzyme could phenotypically complement the photolyase-negative E. coli.
FIG. 4.
Phenotypic complementation of photolyase-deficient E. coli. Plasmid pPHR1 was used to transform E. coli strain CSR 603. Exponentially growing cultures of the resultant bacterium (Phr) were kept for 3 h in the presence of arabinose (Ara) or glucose (Glu) to induce or repress, respectively, expression of the introduced FPV photolyase gene. Diluted cultures were then exposed to the indicated amounts of UV light and subsequently photoreactivated (PR) or kept in darkness (dark) for 1 h at ambient conditions. Survivability was based on CFU of the treated bacterial suspensions relative to CFU of the original stocks. For comparison, a UV inactivation curve of the parental, untransformed bacterium which had been placed in arabinose-containing medium and photoreactivated (CSR 603) is shown. The data represent the average of three independent determinations; standard deviations are indicated as error bars.
FPV photolyase is nonessential for virus replication in tissue culture.
To resolve the essentiality of virus-encoded photolyase for virus survival, elimination of this enzymatic activity was attempted. Rather than deleting the photolyase gene from the FPV genome, the intention was to render this ORF nonfunctional by insertion inactivation. In this procedure, a plasmid (pPHL1) containing a transcriptional marker unit consisting of the vaccinia virus P11 promoter fused to the E. coli lacZ gene and flanked by portions of the FPV photolyase gene was generated and transfected into FPV-infected cells. As a result of recombination between the homologous nucleotide sequences in the plasmid and replicating virus genome, transcription of the photolyase gene is disrupted due to insertion of the foreign DNA. Progeny from such a transfection can then be screened for the presence of recombinants based on their ability to express the inserted gene.
Using the above-mentioned protocol, putative photolyase-deficient FPV were identified as blue plaques due to hydrolysis of the supplied Bluo-gal substrate by the lacZ gene product β-galactosidase. One isolate was plaque purified to homogeneity (only blue plaques were obtained in two consecutive rounds of purification), and its recombinant status was determined by using PCR to amplify the region flanking the insertion site. As anticipated, only an amplicon whose size was indicative of the insertion event was obtained (data not shown). The generation of a pure population of FPV having an inactivated photolyase gene indicated that its protein product was not required for virus replication in tissue culture. Furthermore, comparison of the growth kinetics of the recombinant and parental viruses (Fig. 5) demonstrated that the mutagenic event did not alter the rate of virus replication in an established quail cell line.
FIG. 5.
Growth kinetics of parental and photolyase-deficient recombinant FPV. QT-35 cell monolayers were infected at 0.1 PFU/cell with parental (Phr+) or recombinant (Phr−) FPV. At the indicated times, the cells and the medium were frozen and titered for the amount of infectious virus. The data represent the average of two independent determinations; standard deviations are indicated as error bars.
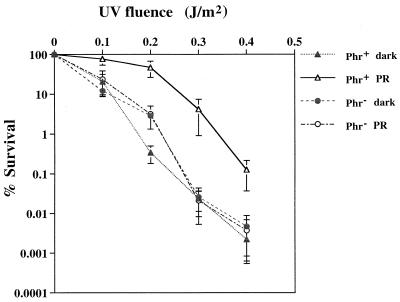
FPV contains a virus-encoded photolyase.
Although no phenotypic differences between the recombinant and parental FPV were detected with regard to interactions with the cell host, the two viruses did differ in the ability to use white light to reverse the lethal effects of previous exposure to UV light (Fig. 6). Whereas the UV light inactivation curve of either type of virus in the absence of photoreactivation was similar to that of the photoreactivated recombinant FPV, subsequent exposure of the unaltered, parental virus to white light resulted in an approximately 100-fold increase in survivability. Presumably, the observed enhanced resistance of photoreactivated, parental virus to UV light inactivation can be attributed to an active, virus-encoded photolyase. Since the infectivity of cell-free virus was assayed, it is likely that this enzyme is an integral part of mature virions.
FIG. 6.
UV light inactivation of parental and photolyase-deficient recombinant FPV. Extracellular parental (Phr+) and recombinant (Phr−) virions were exposed to the indicated doses of UV light and then either photoreactivated (PR) or kept in darkness (dark) for 1 h at ambient conditions. Survivability was based on titers of the treated virus suspensions relative to titers of the original stocks. The results represent the average of three independent determinations; standard deviations are indicated as error bars.
DISCUSSION
Although poxviruses are known to incorporate enzymes such as DNA-dependent RNA polymerase (7, 44), poly(A) polymerase (33), and mRNA guanyltransferase and mRNA methyltransferase (6, 31) into virions, similar retention of a DNA repair enzyme had not been previously shown. The requirement by extracellular FPV for white light to overcome the otherwise lethal effects of exposure to UV light clearly indicates that this avipoxvirus contains an active photolyase. Moreover, based on the required expression of the FPV photolyase gene for phenotypic rescue of transformed photolyase-deficient bacteria and the increased UV light sensitivity of virions whose genomes contained an insertionally inactivated photolyase gene, this enzyme appears to be virus encoded. Although the presence of a host-provided photolyase in mature virions cannot be excluded, its protective contribution would be minimal, as evidenced by the similarity of the UV light inactivation curves of photoreactivated recombinant and nonphotoreactivated parental virus. However, by analogy to better-characterized class II photolyases (26, 27, 53), the complete FPV enzyme should have at least flavine adenine dinucleotide (FAD) as an active-site cofactor. Since FPV cannot independently synthesize this chromophore, it and possibly another used as a photoantenna (24) would be of host origin. In any case, the ability of photolyase to utilize photons instead of a renewable source of energy such as phosphorylated compounds for the cleavage of CPD dimers makes it ideal for functioning in an otherwise inert virion.
Temporally, production of photolyase appears to occur late during the FPV infectious cycle, at a time relegated primarily to the synthesis of structural proteins. However, the region located immediately upstream of the FPV photolyase gene lacks the canonical TAAAT(G) sequence characteristic of poxvirus late promoters (15). In reconciling this anomaly, it should be realized that this motif has been associated with strong regulatory elements (22, 37). Natural alterations such as replacement of the T at the +5 position with an A do not appear to eliminate functionality, as evidenced by the vaccinia virus A2L gene promoter (51). Likewise, the same deliberate manipulation of this nucleotide in a strong promoter reduced transcriptional activity only by 48% (15). Since this substitution in the vaccinia virus 28-kDa gene promoter rendered it inactive, other features such as the composition of the region upstream of the promoter influence the level of its activity (15). In this regard, the FPV photolyase gene promoter has a stretch of six T residues near the beginning of an 85% AT-rich 20-nucleotide sequence which is separated by a 6-bp spacer region from its presumed TAAAA transcriptional start site. Despite such favorable physical conditions (15), this FPV promoter is apparently very weak, as evidenced by its relative strength being approximately 1,000-fold less than that of the strong vaccinia virus 11-kDa gene promoter (V. Srinivasan and D. N. Tripathy, unpublished data). Conceptually, this finding is not surprising. A low level of photolyase synthesis would be expected since only a few copies of an enzyme, compared to the multitude of a structural protein, would be required per virion.
Interestingly, the poxvirus late promoter signature sequence is also absent at the corresponding site in the photolyase gene promoters of an entomopox virus and two leporipoxviruses. For one of the leporipoxviruses, myxoma virus, photolyase was predicted to be expressed early on the basis of some similarity between its gene promoter and the consensus poxvirus early promoter (11). No such homology with early or even intermediate transcriptional regulatory regions was observed for the FPV counterpart. Since the photolyase gene is probably conserved in all poxviruses infecting invertebrates and lower vertebrates, it is likely that its relative time of expression and the mode of action of its product have also remained unchanged. Therefore, one would expect photolyase to be produced during the morphogenesis of poxvirus particles into which this enzyme is incorporated.
Within the Chordopoxvirinae subfamily (poxviruses infecting vertebrates) of the Poxviridae family, the presence of a gene predicted to encode a photolyase has been detected only in the DNAs of members of the genera Avipoxvirus (3) and Leporipoxvirus (11, 52). Such an ORF was not found in the sequenced genomes of the human-specific poxviruses variola virus (32, 44) and molluscum contagiosum virus (43). Thus, based on the absence of this DNA repair mechanism in placental mammals (12, 25, 29), photoreactivity has become vestigial, initially for the host and then for the pathogen. Since the responsible enzyme is active in the internal organs of opossums and chickens (12) and presumably other vertebrates, its evolutionary loss may be the inconsequential result of its replacement by other, possibly more efficient DNA repair enzymes which function independently of light activation. In this regard, the E. coli photolyase seems to be bifunctional in that it can stimulate the nucleotide excision repair system in the absence of light (35). Likewise, yeast photolyase also binds to DNA damaged by agents besides UV light but may inhibit repair by rendering the lesions inaccessible to the excision enzymes (18). In the case of FPV, as evidenced by the generation of recombinants unable to produce photolyase, any secondary function(s) of photolyase either is not necessary for virus replication or is provided by another virus protein or a host substitute. In view of the apparent requirement of direct transmission of the human poxviruses compared to the possibility of indirect routes for other poxviruses, the primary purpose of photolyase is most likely stabilization of virions during exposure to vectors and the environment. In that case, the ability of leporipoxviruses to infect rabbits, a mammal apparently lacking a photolyase gene (25), would not be compromised.
Clearly, the pathogenicity and persistence of the photolyase-deficient virus in its natural host, the chicken, needs to be evaluated. Although attenuated FPV is currently used in vaccination programs, its stability in the scabs and dander of immunized poultry, and possibly insect vectors, could pose a threat to immunologically naive chickens. Perhaps continual exposure of photolyase-deficient FPV to either natural sunlight or artificial incandescent light would cause its inactivation at a greater rate than occurs for vaccine strains. Such a property could be of value in designing a new generation of FPV to be used for immunization of poultry and perhaps mammalian species.
ACKNOWLEDGMENTS
We sincerely thank Jeffrey F. Gardner, University of Illinois at Urbana-Champaign, for providing the pBAD 24 expression vector and Mary Berlyn, curator, E. coli Genetic Stock Center, Yale University, for providing the photolyase-deficient E. coli CSR 603.
This work was supported by a grant to D.N.T. from the U.S. Department of Agriculture (RRF, NC-228).
REFERENCES
- 1.Adachi J, Hasegawa M. Molphy version 2.3 programs for molecular phylogenetics based on maximum likelihood. Computer Science Monographs no. 28. Tokyo, Japan: Institute of Statistical Methods; 1996. [Google Scholar]
- 2.Afonso C L, Tulman E R, Lu Z, Oma E, Kutish G F, Rock D L. The genome of Melanoplus sanguinipesentomopox virus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afonso C L, Tulman E R, Lu Z, Zsak L, Kutish G F, Rock D L. The genome of fowlpox virus. J Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldick Jr C J, Keck J G, Moss B. Mutational analysis of the core, spacer, and initiator regions of vaccinia virus intermediate-class promoters. J Virol. 1992;66:4710–4719. doi: 10.1128/jvi.66.8.4710-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbosa E, Moss B. mRNA (nucleoside-2′-)-methyltransferase from vaccinia virus. Characteristics and substrate specificity. J Biol Chem. 1978;253:7698–7702. [PubMed] [Google Scholar]
- 7.Baroudy B M, Moss B. Purification and characterization of DNA-dependent RNA polymerase from vaccinia virions. J Biol Chem. 1980;225:4372–4380. [PubMed] [Google Scholar]
- 8.Bergion A, Dales S. Comparative observations on poxviruses of invertebrates and vertebrates. In: Maramorsch K, Kurstad E, editors. Comparative virology. New York, N.Y: Academic Press; 1971. pp. 171–205. [Google Scholar]
- 9.Boyle D B, Coupar B E H, Gibbs A J, Seigman L J, Both G W. Fowlpox virus thymidine kinase: nucleotide sequence and relationships to other thymidine kinases. Virology. 1987;156:355–365. doi: 10.1016/0042-6822(87)90415-6. [DOI] [PubMed] [Google Scholar]
- 10.Buller R M L, Palumbo G J. Poxvirus pathogenesis. Microbiol Rev. 1991;55:80–122. doi: 10.1128/mr.55.1.80-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron C, Mitchell S H, Chen L, Barrett J, Cao J, Macaulay C, Willer D, Evans D, McFadden G. The complete DNA sequence of myxoma virus. Virology. 1999;264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 12.Cook J S. Photoreactivation in animal cells. Photophysiology. 1970;5:191–233. [PubMed] [Google Scholar]
- 13.Cooper J A, Witteck R, Moss B. Extension of the translational map of the left end of the vaccinia virus genoma to 21 kilobase pairs. J Virol. 1981;39:733–745. doi: 10.1128/jvi.39.3.733-745.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davison A J, Moss B. Structure of vaccinia virus early promoters. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 15.Davison A J, Moss B. Structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dulbecco R. Reactivation of ultraviolet-inactivated bacteriophage by visible light. Nature. 1949;163:949–950. doi: 10.1038/163949b0. [DOI] [PubMed] [Google Scholar]
- 18.Fox M E, Feldman B J, Chu G. A novel role for DNA photolyase: binding to DNA damaged by drugs is associated with enhanced cytotoxicity in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:8071–8077. doi: 10.1128/mcb.14.12.8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 20.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of the vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. , 517–563. [DOI] [PubMed] [Google Scholar]
- 21.Guzman L, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanggi M, Bannwarth W, Stunnenberg H G. Conserved TAAAT motif in vaccinia virus late promoters: overlapping TATA box and site of transcriptional initiation. EMBO J. 1986;5:1071–1076. doi: 10.1002/j.1460-2075.1986.tb04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hearst J E. The structure of photolyase: using photon energy for DNA repair. Science. 1995;268:1858–1859. doi: 10.1126/science.7604259. [DOI] [PubMed] [Google Scholar]
- 24.Kanai S, Kikuno R, Toh H, Ryo H, Todo T. Molecular evolution of the photolyase-blue light photoreceptor family. J Mol Evol. 1997;45:535–548. doi: 10.1007/pl00006258. [DOI] [PubMed] [Google Scholar]
- 25.Kato T, Jr, Todo T, Ayaki H, Ishizaki K, Morita T, Mitra S, Ikenaga M. Cloning of a marsupial DNA photolyase gene and the lack of related nucleotide sequences in placental animals. Nucleic Acids Res. 1994;22:4119–4124. doi: 10.1093/nar/22.20.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S T, Malhotra K, Ryo H, Sancar A, Todo T. Purification and characterization of Drosphilia melanogasterDNA photolyase. Mutat Res. 1996;363:97–104. doi: 10.1016/0921-8777(96)00003-1. [DOI] [PubMed] [Google Scholar]
- 27.Kleiner O, Butenandt J, Carell T, Batschauer A. Class II DNA photolyase from Arabidopsis thalianacontains FAD as a cofactor. Eur J Biochem. 1999;264:161–167. doi: 10.1046/j.1432-1327.1999.00590.x. [DOI] [PubMed] [Google Scholar]
- 28.Kotwal G J. Poxviral mimicry of complement and chemokine system components: what's the end game? Immunol Today. 2000;21:242–248. doi: 10.1016/s0167-5699(00)01606-6. [DOI] [PubMed] [Google Scholar]
- 29.Li Y F, Kim S T, Sancar A. Evidence for lack of DNA photoreactivating enzyme in humans. Proc Natl Acad Sci USA. 1993;90:4389–4393. doi: 10.1073/pnas.90.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Z, Li Y, Zhang Y, Kutish G F, Rock D L, van Etten J L. Analysis of 45 kb of DNA located at the left end of the Chorella virus PBCV-1 genome. Virology. 1995;206:339–352. doi: 10.1016/s0042-6822(95)80049-2. [DOI] [PubMed] [Google Scholar]
- 31.Martin S A, Paoletti E, Moss B. Purification of mRNA guanylytransferase and mRNA (guanine-7-)-methyltransferase from vaccinia virions. J Biol Chem. 1975;250:9322–9329. [PubMed] [Google Scholar]
- 32.Massung R F, Lu L-I, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 33.Moss B, Rosenblum E W, Gershowitz A. Characterization of a polyadenylate polymerase from vaccinia virions. J Biol Chem. 1975;250:4722–4729. [PubMed] [Google Scholar]
- 34.Nakabeppu Y, Sekiguchi M. Physical association of pyrimidine dimer DNA glycosylase and apurinin/apyrimidinic DNA endonuclease essential for repair of ultraviolet-damaged DNA. Proc Natl Acad Sci USA. 1981;78:2742–2746. doi: 10.1073/pnas.78.5.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozer Z, Reardon J T, Hsu D S, Malhotra K, Sancar A. The other function of DNA photolyase: stimulation of excision repair of chemical damage to DNA. Biochemistry. 1995;34:15886–15889. doi: 10.1021/bi00049a002. [DOI] [PubMed] [Google Scholar]
- 36.Paoletti, E., J. Tartaglia, and J. Taylor. December 1998. Poxvirus-canine distemper virus (CDV) recombinants and compositions and methods employing recombinants. U.S. patent 5,756,102. Sequence 48.
- 37.Rosel J L, Earl P L, Weir J P, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII fragment. J Virol. 1986;60:436–439. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sancar A. Structure and function of DNA photolyase. Biochemistry. 1994;33:2–9. doi: 10.1021/bi00167a001. [DOI] [PubMed] [Google Scholar]
- 39.Sancar A. No end of history for photolyases. Science. 1996;272:48–49. doi: 10.1126/science.272.5258.48. [DOI] [PubMed] [Google Scholar]
- 40.Sancar A, Rupert C S. Determination of plasmid molecular weights from ultraviolet sensitivities. Nature. 1978;272:471–472. doi: 10.1038/272471a0. [DOI] [PubMed] [Google Scholar]
- 41.Schnitzlein W M, Tripathy D N. Utilization of vaccinia virus promoters by fowlpox virus recombinants. Anim Biotechnol. 1990;1:161–174. [Google Scholar]
- 42.Schnitzlein W M, Ghildyal N, Tripathy D N. Genomic and antigenic characterization of avipoxviruses. Virus Res. 1988;10:65–76. doi: 10.1016/0168-1702(88)90058-5. [DOI] [PubMed] [Google Scholar]
- 43.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 44.Shchelkunov S N, Marennikova S S, Blinov V M, Resenchuk S M, Totmenin A V, Chizhikov V E, Guturov V V, Safronov P F, Kurmanov R K, Sandaknchiev L S. Entire coding sequence of the variola virus. Dokl Akad Nauk. 1993;328:629–632. [PubMed] [Google Scholar]
- 45.Spencer E, Shuman S, Hurwitz J. Purification and properties of vaccinia virus DNA dependent RNA polymerase. J Biol Chem. 1980;255:5388–5395. [PubMed] [Google Scholar]
- 46.Thompson J D, Higgins D C, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Todo T, Ryo H, Yamamoto K, Toh H, Inui T, Ayaki H, Nomura T, Ikenaga M. Similarity among the Drosphilia(6-4) photolyase, a human photolyase homologue, and the DNA photolyase-blue-light receptor family. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- 48.Tripathy D N, Hanson L E, Killinger A H. Atypical fowlpox in a poultry farm in Illinois. Avian Dis. 1974;18:84–90. [PubMed] [Google Scholar]
- 49.Tripathy D N, Schnitzlein W M. Expression of avian influenza virus hemagglutinin by recombinant fowlpox virus. Avian Dis. 1991;35:186–191. [PubMed] [Google Scholar]
- 50.Weinbauer M G, Wilhelm S W, Suttle A, Garza D R. Photoreactivation compensates for UV damage and restores infectivity to natural marine viral communities. Appl Environ Microbiol. 1997;63:2200–2205. doi: 10.1128/aem.63.6.2200-2205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinrich S L, Hruby D E. A tandemly-oriented late gene cluster within the vaccinia virus genome. Nucleic Acids Res. 1986;14:3003–3016. doi: 10.1093/nar/14.7.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willer D O, McFadden G, Evans D H. The complete genome sequence of Shope (rabbit) fibroma virus. Virology. 1999;264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 53.Yasui A, Eker A P M, Yasuhira S, Yajima H, Kobayashi T, Takao M, Oikawa A. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J. 1994;13:6143–6151. doi: 10.1002/j.1460-2075.1994.tb06961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuen L, Moss B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci USA. 1987;84:6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]