Abstract
Background and Aims:
In the classical form of α1-antitrypsin deficiency, a misfolded variant α1-antitrypsin Z accumulates in the endoplasmic reticulum of liver cells and causes liver cell injury by gain-of-function proteotoxicity in a sub-group of affected homozygotes but relatively little is known about putative modifiers. Here, we carried out genomic sequencing in a uniquely affected family with an index case of liver failure and 2 homozygous siblings with minimal or no liver disease. Their sequences were compared to sequences in well-characterized cohorts of homozygotes with or without liver disease, and then candidate sequence variants were tested for changes in the kinetics of α1-antitrypsin variant Z degradation in iPS-derived hepatocyte-like cells derived from the affected siblings themselves.
Approach and Results:
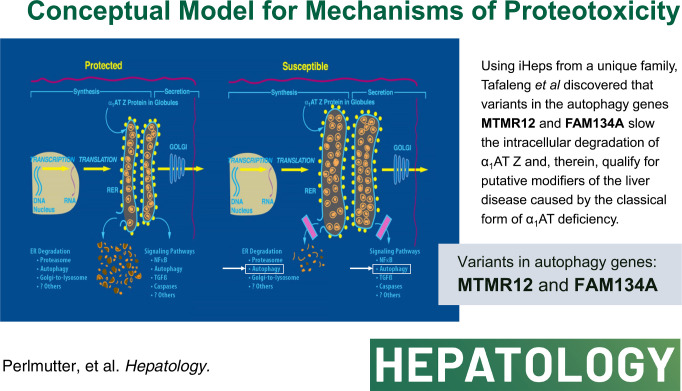
Specific variants in autophagy genes MTMR12 and FAM134A could each accelerate the degradation of α1-antitrypsin variant Z in cells from the index patient, but both MTMR12 and FAM134A variants were needed to slow the degradation of α1-antitrypsin variant Z in cells from a protected sib, indicating that inheritance of both variants is needed to mediate the pathogenic effects of hepatic proteotoxicity at the cellular level. Analysis of homozygote cohorts showed that multiple patient-specific variants in proteostasis genes are likely to explain liver disease susceptibility at the population level.
Conclusions:
These results validate the concept that genetic variation in autophagy function can determine susceptibility to liver disease in α1-antitrypsin deficiency and provide evidence that polygenic mechanisms and multiple patient-specific variants are likely needed for proteotoxic pathology.
INTRODUCTION
The classical form of α1-antitrypsin deficiency (ATD) is a rare, autosomal co-dominant disease caused by a point mutation that impairs the folding of the secretory glycoprotein, α1-antitrypsin (AT). AT is predominantly synthesized in the liver and constitutes one of the most abundant glycoproteins in the circulating bloodstream.1 A sub-population of affected homozygotes develop chronic obstructive pulmonary disease, exclusively adult in onset, by a loss-of-function mechanism in which loss of the protease inhibitory function of AT allows neutrophil proteases to damage the lung connective tissue matrix.2
A smaller sub-population of affected homozygotes develop chronic liver disease, and work in transgenic mice has proven that this sequelae is determined by a gain-of-function proteotoxic mechanism. Accumulation of the misfolded and aggregation-prone α1-antitrypsin Z variant (ATZ) in the endoplasmic reticulum (ER) initiates a series of reactions that result in liver cell dysfunction, fibrosis, and carcinogenic liver cell hyperproliferation.1 The disorder can cause liver disease in infancy and childhood that is severe enough to require liver transplantation, but recent analysis has shown that most of the cases requiring liver transplantation occur in homozygotes at 50–65 years of age and may be exacerbated by other cause of liver disease in that age group.3 There is still not complete certainty on the incidence and severity of liver disease among ZZ homozygotes, but it is clear that most completely escape clinically significant liver disease.1 An important study using nationwide screening of newborns in Sweden indicated that only 8%–10% of affected homozygotes have clinical liver disease up to the fourth decade of life.4,5
In numerous studies in mammalian cell line and mouse models we have shown that ATZ accumulation in the ER is mitigated by proteostasis mechanisms, including, most notably, endoplasmic reticulum-associated degradation (ERAD) and autophagy.1 In most of those studies, the capacity of ERAD appears to be limited, and autophagy plays a dominant role. Recent studies have shown that ATZ degradation may also involve vesicular pathways that are called ER-phagy pathways, and several of them have subtle differences from canonical macro-autophagy, including a pathway that has been called ER-to-lysosome-associated degradation and another that involves the action of p62 with the E3-ubiquitin ligase tripartite-motif containing 13.6 We have hypothesized that genomic variants in the molecular components of these pathways would be ideal candidates for genetic modifiers of the liver disease phenotype in ATD homozygotes.1
There is still relatively limited knowledge about genetic and environmental modifiers of liver disease susceptibility in ATD homozygotes. In 1 study, a single nucleotide polymorphism in the MAN1B1 gene was found to be over-represented statistically in a cohort of infants with end-stage liver disease.7 This variant was shown to reduce intracellular (IC) levels of the mannosidase. Subsequent experiments showed that mannosidase alpha class 1B member 1 (Man1B1) is localized to the Golgi and plays a role in how the Golgi may participate in ERAD,8 but this Man1B1 variant has not been tested on the kinetics of ATZ degradation in a human model system and publicly available reference databases of genetic variations from global populations that have become available more recently, such as genome aggregation database and NHLBI’s Trans-Omics for Precision Medicine (TOPMed) database,9 show that this variant is very common in the general population. A single nucleotide polymorphism in the upstream flanking region of the AT gene has also been implicated in susceptibility to liver disease in ATD homozygotes,10 but statistical validation of the variant could not withstand a reasonable re-classification of patient sub-populations.
Because the incidence of this disorder has been estimated at 1 in 3000–5000 and a much smaller number of affected individuals have severe liver disease, it has been challenging to approach the modifier issue using classical population genetic strategies. In the following study, we carried out genomic sequencing of a unique family to identify potential genetic modifiers of the liver disease phenotype. The index subject, homozygous for the ATZ allele, underwent liver transplantation for liver failure at 18 months of age. His older brother was also homozygous and had elevated liver enzymes in the first few years of life, but these were normalized by early childhood. The younger, also homozygous, brother never developed abnormalities of liver enzymes or clinical signs of liver disease. The studies were done when the brothers had reached 19–22 years of age with induced pluripotent stem cell-derived hepatocyte-like cells (iHeps) derived from peripheral blood mononuclear cells obtained at that time and also iHeps derived from skin fibroblast samples that had been obtained from the index subject and his older brother when they were infants. We reasoned that the sibling relationships would allow us to optimize the signal-to-noise ratio for selecting putative modifiers. We focused on genes with known function in proteostasis mechanisms and variants predicted to damage protein function. The strategy also assumed that pathogenic variants would impact the cell biological mechanism of ATZ accumulation. Further, because we predicted that such variants would have relatively subtle effects on the fate of the ATZ variant, we used classical pulse-chase radiolabeling in iHeps from the brothers to detect changes in the kinetics of ATZ secretion and degradation.
METHODS
Exome sequencing
We sequenced DNA samples from the unique family, including the 3 brothers and parents, 30 ATD homozygous Z allele individuals with liver disease (cases), and 50 ATD homozygous Z allele carriers with no liver disease (controls) using established procedures at the McDonnell Genome Institute at Washington University in St. Louis. A detailed description of the sequencing technique, sequence alignment, variant calling, quality control, annotation, gender confirmation, population estimation, and relatedness checks is provided in the Supplemental Materials section, http://links.lww.com/HEP/I396.
Variant filtering and prioritization
We undertook this discovery process in several stages. At each stage, evidence suggested slightly different candidate genes and variants.
Initially, we had data from a trio of 3 siblings, each of which was homozygous for the Z allele, but only 1 of whom had accompanying liver disease. Following exome sequencing and annotation, we examined filtered variants based on allele frequency in the ExAC database using an allele frequency < 1%, then further filtered variants to keep only those exonic protein-coding variants (nonsense, splice, missense, and frameshift) predicted to be damaging by Polyphen 2 and SIFT. For the variants meeting these criteria, we then assessed 3 potential inheritance models to identify candidate genetic variants underlying liver disease: homozygous, heterozygous, and compound heterozygous using Mendelscan (http://gmt.genome.wustl.edu/2014/03/10/mendelscan_v1.2.1_released.html). In the heterozygous model, a variant needed to be present in a single copy in the affected brother but absent in both unaffected siblings. For the homozygous model, the affected sibling needed to be a homozygous carrier for the alternate allele, while the unaffected siblings both were either homozygous for the reference allele or heterozygous. For the compound het model, we identified genes in which the affected brother contained at least 2 deleterious variants with evidence the variants are carried on opposite haplotypes, while the unaffected siblings do not. After initial identification, we examined each candidate variant for genotype quality across each individual.
The heterozygous model generated 17 potential candidate variants in 16 genes, the heterozygous 5 variants in 3 genes, and the compound het model identified a further 2 genes (4 variants). In silicovisualization was performed to further remove false positive variant calls using the Integrative Genomics Viewer.11
After incorporating an additional 30 similarly affected ATD cases with accompanying liver disease, we again attempted to filter variants absent in the 2 unaffected siblings for functional consequence (CADD score > 18, PolyPhen>0.15 or SIFT < 0.05) and global frequency in ExAC populations (minor allele frequency [MAF] < 0.01). Stratifying these individuals by age of onset (infants, juveniles, adolescents, and adults), no qualifying putatively functional variants were common to all age groups. Within each age group, we identified several genes containing either variants with multiple carriers and/or multiple variants. We saw 2 genes with variants only in infants, 17 genes with variants in juveniles, 6 genes with variants only in adolescents, and 2 genes with variants only in adulthood.
With the addition of 50 control individuals homozygous for the Z allele but without liver disease, we relaxed some of the stringent assumptions of global allele frequency and took a more objective approach to variant discovery by performing 2 main analyses: (1) single variant case/control analysis and (2) identifying variants uniquely carried only by cases or only by controls. We then tested genetic variants for association between cases with ATD and liver disease and controls with ATD but no liver disease. Due to the relatedness identified among our cases and controls, we ran association testing both using an unrelated subset of individuals, prioritizing cases and otherwise randomly selecting 1 individual from each family, and also including all individuals using a linear mixed model approach, which uses a relationship matrix created from the genetic data (genetic relationship matrix) to adjust for both known and cryptic relatedness within our study sample.
In the analysis of unrelated individuals, we use Firth-bias corrected logistic regression,12 which corrects for low counts for rare variants, to test for association between cases with liver disease and controls with no liver disease. This analysis consisted of 28 cases and 46 controls. For analysis of all individuals taking the relationship into account, we used a linear mixed model approach, EMMAX, that treats case/control values on a linear scale, which provides reliable p-value estimates for the association, though beta estimates are not appropriately interpretable. Both Firth logistic regression and EMMAX13 are implemented in the EPACTS software (https://genome.sph.umich.edu/wiki/EPACTS).
Following the identification of genes in each of the methods proposed above, we further prioritized candidates for additional follow-up experiments based on the overlap of genes implicated in autophagy pathways. We used 3 sources of identified autophagy genes: a proteomics selection experiment to define the human autophagy system,14 genes in the human autophagy database, and proteins differentially expressed in mouse models of ATD.15
For 3 candidate variants, we sought further validation of genotype results through an orthogonal technology. We validated each of these variants, rs141977664 (chr1:36074927 G/A, p.Ala123Val in PSMB2), rs140836761 (chr5:32230027 G/A, p.Arg701Cys in MTMR12), and rs150609294 (chr11:121384931 A/C, p.Asn371Thr in SORL1), with Sanger sequencing at GENEWIZ followed by manual inspection of both forward and reverse sequence traces. Except for genotypes not called through exome sequencing, we validated all genotypes, and no additional initially uncalled samples were carriers for these rare variants.
Mammalian cell models and immunoblot analysis
We used the HTO/Z cell line15 with inducible expression of ATZ and then engineered for KI of variants in specific autophagy genes using the Genome Engineering & Stem Cell Center (GESC@MGI) at Washington University in St. Louis. Steady-state levels of ATZ were analyzed by immunoblot and densitometric quantification as described.15
Generation of iHeps
Fibroblasts and blood mononuclear cells were reprogrammed using lentiviral-mediated gene transfer and nucleofection as described.16 Directed differentiation of iPScs into iHeps was carried out by plating cells on growth factor reduced Matrigel and using 4 steps of endoderm induction, hepatic specification, hepatic induction, and hepatic maturation with various concentrations of activin A, bone morphogenic protein 4, FGF2, HGF, and oncostatin M as described.16 Each of the lines was evaluated after differentiation by expression of albumin, asialoglycoprotein receptor, alpha-feto protein, ultrastructural features characteristic of primary hepatocytes, and relative secretion of albumin in pulse-chase experiments. Despite the different origins of the lines (skin fibroblasts versus peripheral blood mononuclear cells) and the manipulations that were part of the CRISPR-mediated editing, the differentiated state of the lines was similar in terms of each of these parameters.
HTO/Z cells and iHeps that underwent CRISPR-mediated editing were subcultured to generate single-cell clones, and the selected clones were shown to have the appropriate mutation in the target site region by NGS in each case. Other details on CRISPR-mediated editing of HTO/Z cells and iHeps are described in the Supplemental Methods section, http://links.lww.com/HEP/I396.
Pulse-chase radiolabeling of iHeps
Methods for biosynthetic labeling with 35S methionine, immunoprecipitation, sodium dodecyl sulfate polyacrylamide gel electrophoresis/fluorography, and densitometric analysis have been published.17 The pulse period was 1 hour and the chase for various time intervals up to 4 hours. All fluorograms were subjected to densitometric analysis using ImageJ software with the relative densitometric value of T0 set at 100% and the remainder of the data set expressed as % of this value.
RESULTS
Potential modifiers of liver disease phenotype from sequence analysis of unique family
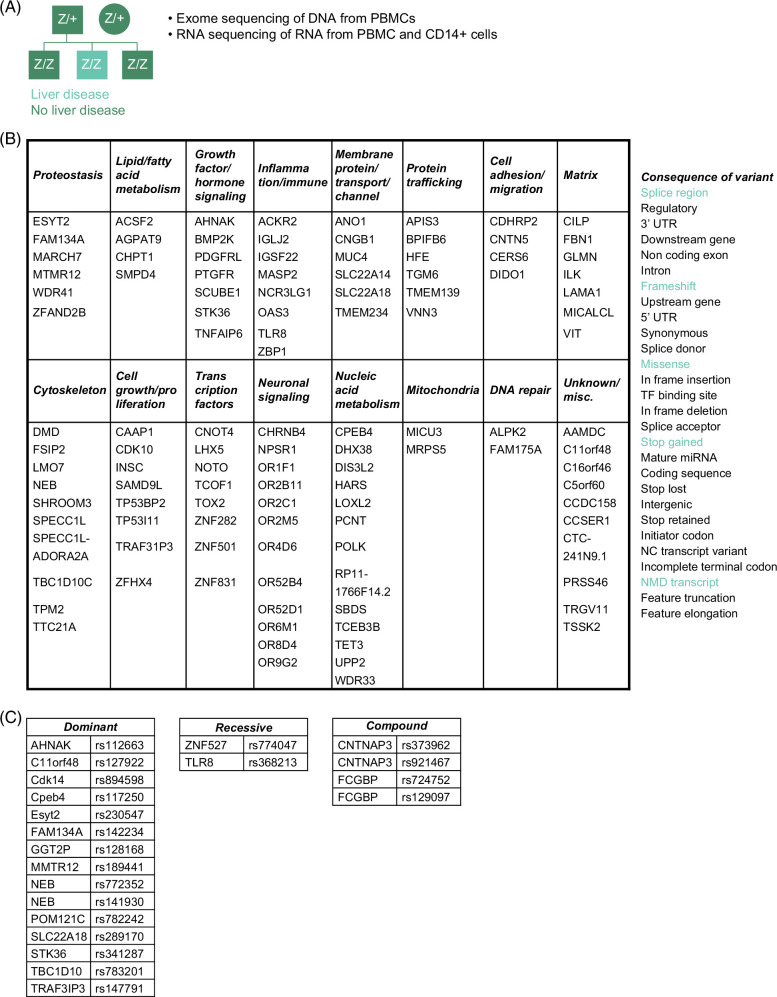
We analyzed whole-exome sequence data from the 3 siblings and their parents to identify variants that were found differentially in the siblings (Figure 1A). Filtering was carried out to identify variants based on allele frequency and likely to be damaging to protein function (loss-of-function), and this narrowed the number of variants to 115 in 113 genes (Figure 1B). From this list, we narrowed further to genes that were known to have some function in proteostasis and variants that were expressed heterozygously, that is, present as heterozygote in the index case and absent in the protected sibs. Second, we used a control cohort to determine the MAF of variants in the candidate proteostasis genes. The control cohort consisted of 101 ATZ homozygous subjects over the age of 30 years with no lung disease (by spirometry) and no signs or symptoms of liver disease (by questionnaire) from the AAT Genetic Modifier Study.18 Exome chip analysis for selected variants was carried out on all of these subjects, and whole-exome sequencing on a sub-group of 50 subjects. Single nucleotide variants in 2 genes were prioritized for further analysis----MTMR12rs140836761 (Chr5:32229921 G>A); FAM134Ars142234261 (Chr2:219182235A>G) (Figure 1C and Supplemental Tables 1-3, http://links.lww.com/HEP/I397). In each case, the variant was heterozygous in the index subject and completely absent in the protected siblings. First, myotubularin-related protein 12 (MTMR12) functions in autophagosome-lysosome fusion, and a sequence variant in that gene (distinct from the variant identified here) has been implicated in a degenerative myopathy.19,20 It has an MAF of 0.003 in the control cohort. Second, FAM134A has recently been shown to play a role in ER-phagy, both working in concert with, and independently of, FAM134B,21 and its MAF in the control cohort was 0.
FIGURE 1.
Unique family (A), potentially damaging variants (B), and those prioritized for further analysis (C) are described in the text. The variants in fibrillin-1 (FBN) and homeostatic iron regulator (HFE) are described as “ conflicting interpretation of pathogenicity” by the ClinVar database. Abbreviaton: PBMC, peripheral blood mononuclear cells.
Interestingly, the MAN1B1 single nucleotide variant rs4567that had been identified as a modifier of the liver disease phenotype7,8 was found to be homozygous for the minor allele (A/A) in all 3 siblings (Supplemental Fig. S1, http://links.lww.com/HEP/I398) and so could not be implicated as a putative modifier in this experimental paradigm.
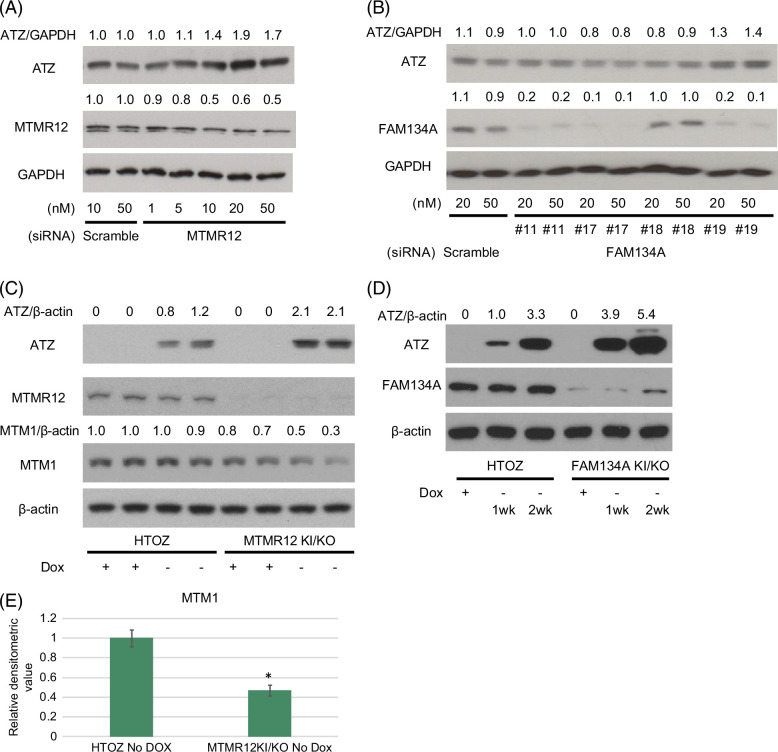
To determine if variants in MTMR12 and FAM134A could be potential modifiers of the liver disease phenotype, we first investigated the effects of knocking down their expression with small interfering RNA on steady-state levels of ATZ in the HTO/Z cell line, a HeLa-based cell line engineered for inducible expression of ATZ that has been extensively characterized in previous studies by our team (Figure 2). The results show dose-dependent increases in ATZ levels in HTO/Z cells treated with that small interfering RNA to MTMR12 (Figure 2A) and FAM134A (Figure 2B) but no effect for small interfering RNA to a negative control protein extended synaptotagmin 2, known to play a role in tethering of vesicles from the ER to the plasma membrane (Supplemental Fig. S2, http://links.lww.com/HEP/I398).
FIGURE 2.
Immunoblot analysis of the effect of knocking down and/or knocking in the MTMR12 and FAM134A variants in the HTO/Z cell line with inducible expression of ATZ. (A) Knocking down MTMR12 with siRNA (relative densitometric values for ATZ levels compared to GAPDH levels are shown at the top and relative densitometric values for MTMR12 levels are shown above the MTMR12 blot); (B) Knocking down FAM134A with siRNA (relative densitometric values for ATZ levels compared to GAPDH levels are shown at the top and relative densitometric values for FAM134A levels are shown above the FAM134A blot); (C) ATZ, MTMR12, MTM1 and β-actin levels in HTO/Z cell line with MTMR12 variant knock-in/knockout background (relative densitometric values for ATZ levels compared to β-actin levels are shown at the top and relative densitometric values for MTM1 levels compared to β-actin levels are shown above the MTM1 blot); (D) ATZ, FAM134A and β-actin levels in HTO/Z cell line with FAM134A variant knock-in/knockout background (relative densitometric values for ATZ levels compared to β-actin are shown at the top); (E) MTM1 levels in HTO/Z cell line after removal of dox for 3 weeks, parent on the left and with MTMR12 variant knock-in/knockout background on the right (relative densitometric values on the vertical axis; n = 4 separate experiments for each bar; *p < 0.001 for students t-test). Abbreviations: ATZ, α1-antitrypsin variant Z; FAM134A, family with sequence similarity 134 member A; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HTO, HeLa tet-off cell line; MTMR12, myotubularin-related protein 12; siRNA, small interfering RNA.
Next we examined the possibility of knocking in the specific variants for MTMR12 and FAM134A in the HTO/Z line to determine the effect on ATZ levels. We were only able to establish clones with the KI on a knockout background. Immunoblot analysis (Figure 2C, D) in these KI/knockout lines shows decreased levels of either MTMR12 or FAM134A in the designated line and increased accumulation of ATZ in each case. These results were consistent with the possibility that the MTMR12 and FAM134A variants could accentuate ATZ accumulation and potentially qualify as liver disease susceptibility modifiers.
MTMR12 is a member of the myotubularin-related protein family of conserved lipid phosphatases. It is catalytically inactive but has been shown to stabilize the myotubularin myotubularin protein 1 (MTM1), which is catalytically active as a lipid phosphatase and known to function in autophagosome-lysosome fusion.19,20 A variant in MTMR12 associated with centronuclear myopathy, distinct from the variant we found in the index patient here, has been shown to destabilize MTM1 in a zebrafish model.19 To determine whether the same phenomenon occurs with the variant of MTMR12 identified in our index patient, we investigated steady-state levels of MTM1 in the HTO/Z cell line with KI/knockout of MTMR12. The results show a significant reduction in levels of MTM1 when the MTMR12 variant is engineered (Figure 2C and E), validating the concept that de-stabilization of MTM1 is the mechanism by which this variant impacts the autophagy pathway in the index patient.
We also investigated autophagic function in the HTO/Z cell lines with the editing of the MTMR12 and FAM134A genes (Supplemental Fig S3, http://links.lww.com/HEP/I398). The results show that the putative damaging variants reduce the LC3II/LC3I ratio in the absence and presence of bafilomycin, indicating that autophagic flux is suppressed by the damaging effect of these variants on the function of MTMR12 and FAM134A in autophagy.
Analysis of variants in MTMR12 and FAM134A in iHeps from the unique family
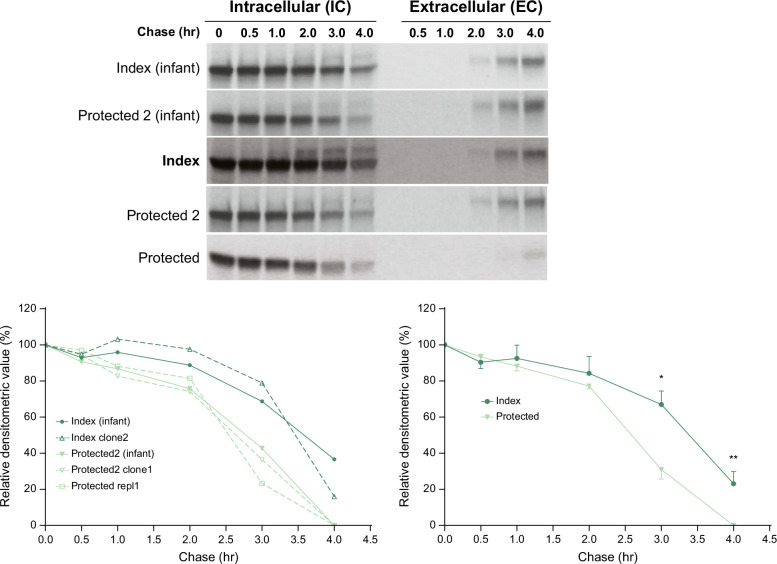
Next, we examined the possibility that the impact of the MTMR12 and FAM134A variants could be evaluated in iHeps from the unique family. Our study had shown that iHeps from ZZ homozygotes with severe clinical liver disease had reduced kinetics of IC degradation of ATZ compared to ZZ homozygotes with no liver disease.16 We reasoned that it would be ideal for determining the impact of the putative modifiers in the cells derived from the 3 siblings. iHeps were derived from skin fibroblasts of the index patient and his older sibling obtained when they were infants. iHeps were derived from peripheral blood mononuclear cell samples from all 3 sibling 20 years later. The iHeps were subjected to pulse-chase radiolabeling and immune capture for the ATZ polypeptide (Figure 3). The results show slower kinetics of disappearance for ATZ in the IC contents from the index case as compared to his siblings, and this was apparent at the 2 different ages for the index case and older sibling. The separation in kinetics was apparent at chase time points of 3 and 4 hours. When multiple replicates were compared, the differences were significant at both of these chase time points (composite results, lower right of Figure 3). The time of appearance of ATZ in the extracellular (EC) fluid was not significantly different except for a delay from 2 to 4 hours in the iHeps from the younger “protected” sibling, which could only be explained by more robust IC degradation in that case. Taken together, these results provide strong evidence for the conclusion that liver disease susceptibility in this family correlates with impairment in IC degradative mechanism(s). The fact that the difference in the rate of disappearance from the IC compartment of the index case as compared to his older “protected” sibling is seen in samples from early infancy and adulthood is most consistent with a mechanism that is likely to be genetic or epigenetic.
FIGURE 3.
Kinetics of ATZ fate in index versus protected subjects. Analysis of pulse-chase radiolabeling in iHeps from index subjects and protected siblings. Fluorograms are shown at the top and densitometric analysis at the bottom. The densitometric results for each line are shown at the left bottom, and the composite for index versus protected at the right bottom. The lines were derived from skin fibroblasts collected during infancy for index and protected 2 subjects and separate PBMCs collected from the index and both protected subjects as adults. The younger sibling is referred to as “protected” and the older sib as “protected 2”. Multiple replicates are shown in the graph at the lower left. The composite of replicates (n = 3 for index; n = 4 for protected) is shown in the graph at the lower right. Bars represent SEM. The difference between the index and protected subjects for the composite of replicates is significant at 3 hours (*p = 0.0092) and 4 hours (**p = 0.0096). Abbreviations: ATZ, α1-antitrypsin variant Z; EC, extracellular; iHeps, induced hepatocyte-like cells; IC, intracellular; PBMC, peripheral blood mononuclear cells.
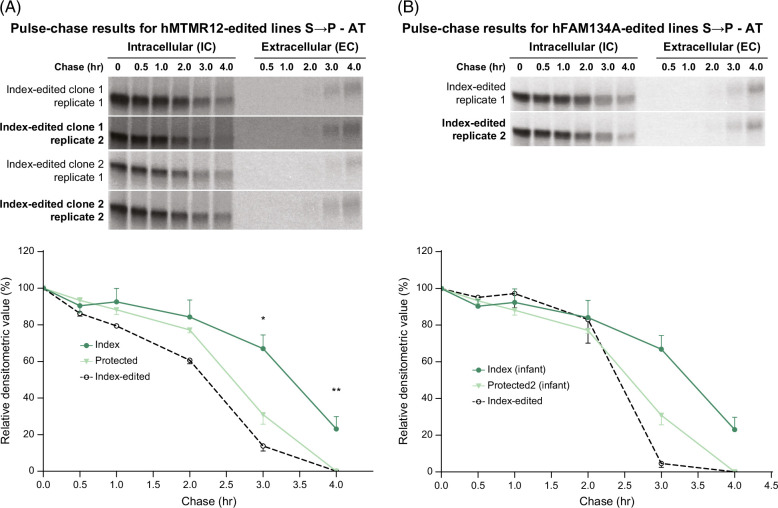
With this data, we used iHeps from the index case to investigate the effect of correcting the putative modifier variants on the kinetics of degradation of ATZ. This was done by genome editing of the iPSCs from the index case such that they were converted to the MTMR12 allele that characterized the protected siblings. The results in this edited line showed an acceleration of the degradation of ATZ (Figure 4A). The rate of disappearance of ATZ in the iHeps from the index case with correction of the MTMR12 variant was similar to that in the iHeps from the youngest “protected” sib. The effect of the correction was demonstrated in 2 independent clones from the gene editing, and statistical analysis of the n = 4 replicates showed significant differences from the index control at both 3 and 4 hours of the chase period. Analysis of the timing of the appearance of ATZ in the EC fluid showed a delay in 1 of the edited replicates but, if anything, this is consistent with the conclusion that an increased rate of IC degradation is occurring in the edited clones. There was no difference in the rate of disappearance from IC and appearance in EC for albumin in the independent clones of iHeps from the index patient with or without the correction of the MTMR12 variant and as compared to his protected sib (Supplemental Fig. S4A, http://links.lww.com/HEP/I398), also providing evidence that the relative differentiation of the iHeps was similar for each of the comparisons.
FIGURE 4.
Effect of correcting MTMR12 (A) and FAM134A (B) variants in the index subject on the kinetics of ATZ fate. Fluorograms for the edited clones are shown at the top, and densitometric analysis compared to unedited iHeps from the index and protected subjects is shown in the graphs at the bottom. Bars represent SEM. In (A), the difference between the index control (n = 3) and index-edited cells (n = 4) was significant at 3 hours (*p = 0.0006) and at 4 hours (**p = 0.0096). In (B), the difference between index control (n = 3) and the total (n = 6) of index-edited together with protected control was significant at 3 hours (p < 0.01) and 4 hours (p < 0.01). Abbreviations: AT, α1-antitrypsin; ATZ, α1-antitrypsin variant Z; EC, extracellular; FAM134A, family with sequence similarity 134 member A; iHeps, induced hepatocyte-like cells; IC, intracellular; MTMR12, myotubularin-related protein 12.
The results were similar when we investigated the effect of correcting the FAM134A variant in iPSCs from the index case (Figure 4B). This was done by genome editing that converted the FAM134A allele to that of the protected siblings, and then the cells were differentiated into iHeps. The results show that degradation of ATZ was accelerated and reverted almost identically to the kinetic profile of iHeps from the protected sibling with distinct differences at the 3 and 4-hour time points. The kinetics of albumin were identical in each of these clones (Supplemental Fig. S4B, http://links.lww.com/HEP/I398). Taken together, these results were consistent with a conclusion that variants in MTMR12 and FAM134A could each represent modifiers of the liver disease phenotype.
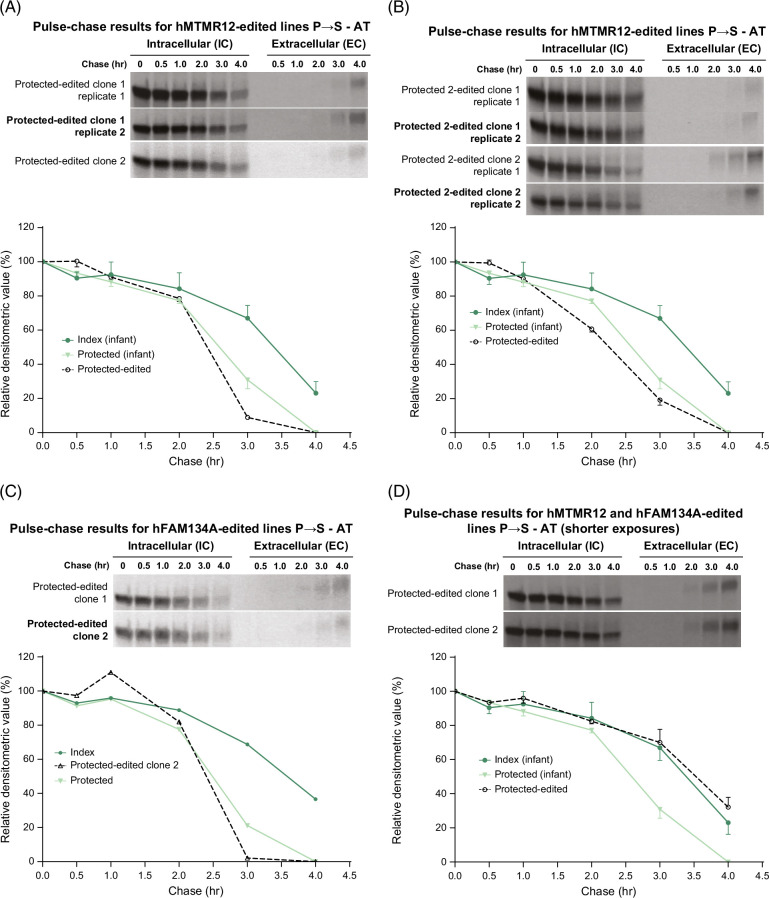
Next we investigated the effects of introducing the putative pathogenic variants in MTMR12 and FAM134A, characteristic of the index subject, into the iHeps of the protected sibs. We first used genome editing for the MTMR12 variant into the cells from the younger sibling (Figure 5A). Interestingly, this editing had no effect on the kinetics of ATZ degradation. Indeed the kinetic profile was identical to that in the unedited cells from the younger sibling (Figure 5A). To determine if this surprising result was peculiar to this iHep line, we also evaluated iHeps from the older sibling (labeled “protected 2”). After editing the MTMR12 variant of the index subject, the kinetics of ATZ degradation were identical to those of the unedited cells from the older sib (Figure 5B). Furthermore, using the same approach, the FAM134A variant had no effect on the kinetics of ATZ degradation in the iHeps of the younger protected sibling (Figure 5C). There were some variations in the timing of the appearance of ATZ in the EC fluid among the edited clones, but these could not account for the differences in the rate of IC degradation from the unedited clones. Kinetics of albumin secretion were not affected in these lines (Supplemental Fig. S5, http://links.lww.com/HEP/I397).
FIGURE 5.
Effect of introducing MTMR12 and FAM134A variants into iHeps from protected subjects on the kinetics of ATZ fate. Fluorograms from the edited clones are shown at the top and densitometric analysis compared to unedited index and protected subjects is shown in the graphs below in each case. The MTMR12 variant was introduced into iHeps from the younger sibling (“protected”) in (A) and from the older sibling (“protected 2”) in (B) and the FAM134A variant was introduced into iHeps from ‘protected’ sib in (C). In (D) both of the variants have been introduced together in the “protected” subject. Bars represent SEM. In (A), (B), and (C), the difference between index control (n = 3) and the total (n = 6) of protected-edited and protected control was significant at 3 and 4 hours (p < 0.01). In (D), the difference between protected control (n = 4) and the total (n = 5) of protected-edited and index control was significant at 3 and 4 hours (p < 0.01). Abbreviations: AT, α1-antitrypsin; ATZ, α1-antitrypsin variant Z; EC, extracellular; FAM134A, family with sequence similarity 134 member A; iHeps, induced hepatocyte-like cells; IC, intracellular; MTMR12, myotubularin-related protein 12.
To examine the possibility that the modifier effect might require both variants, we next introduced the FAM134A variant from the index subject into the iHeps of the protected younger sibling into which we had previously engineered the index MTMR12 variant (Figure 5D). These results were striking. In 2 separate clones, the kinetics of ATZ disappearance reverted to the susceptible pattern with a significant delay at the 3 and 4-hour time points, almost identical to the rate in the iHeps from the index case. Kinetics of albumin secretion were not affected (Supplemental Fig. S6, http://links.lww.com/HEP/I397). These results provide robust evidence that variants in MTMR12 and FAM134A together can slow the degradation of ATZ and conceptually model accentuation of the proteotoxic consequences of homozygosity for the ATZ variant.
We also investigated autophagic flux in the iHep model cells (Supplemental Fig S7, http://links.lww.com/HEP/I397). In each case, the LC3II to LC3I ratio increased with bafilomycin treatment (Supplemental Fig S7A, http://links.lww.com/HEP/I397). When comparing the ratio in iHeps from the index case with and without editing, there is a marked increase when the MTMR12 variant is corrected and a lesser, but still considerable, increase when the FAM134A variant is corrected (Supplemental Fig S7B, http://links.lww.com/HEP/I397). The ratio was increased in the iHeps from the protected compared to the index subject but the introduction of both MTMR12 and FAM134A variants in the iHeps from the protected subject did not lead to a reduction in the ratio. This may mean that the introduction of these variants is sufficient to slow the selected degradation of ATZ but not sufficient to slow bulk autophagic flux.
Whole-exome sequencing for the MTMR12 and FAM134A variants in a cohort of ATD subjects with severe liver disease
Next, we carried out whole-exome sequencing analysis on a cohort of 30 well-characterized ATD ZZ homozygotes who had undergone liver transplantation. This liver disease cohort includes subjects ranging in age from 6.7 months to 55 years of age (3 less than 1 y of age; 10 at 1–5 y of age; 8 at 5–10 y of age; 5 at 10–20 y of age; 4 at 20–55 y). We detected the identical MTMR12 variant in a single male child who underwent liver transplantation at 5.8 years of age. This subject had damaging variants in 2 other proteostasis genes (PSMB2, DNAJC18) but not in FAM134A. We did not detect the FAM134A variant in any of the other subjects of the liver disease cohort. We analyzed the DNA sequences from the liver disease cohort for other damaging variants, including protein-coding variants (nonsense, splice, and missense) predicted as deleterious by both SIFT and Polyphen, in proteostasis genes unique to the index subject but not in his siblings—none were discovered. We also analyzed the DNA sequences from the liver disease cohort for other proteostasis genes with variants predicted to be damaging and which had allele frequencies below 0.01 in the control cohort. There were 31 variants that fit these criteria and at least 1 in each of the 30 subjects. One of these variants was present in 4 subjects in the liver disease cohort, HOOK2, but its MAF in the control cohort was 0.14. Three of the variants, PSMB2, SORL1, and CPVL, were each present in 2 of the patients but only PSMB2 and SORL1 had a MAF less than 0.01 in the control cohort. Twelve other variants were present in 2 subjects from the liver disease cohort, but none of these had known proteostasis functions.
Taken together with the above studies of MTMR12 and FAM134A, these results suggest genetic modifiers of the liver disease phenotype in ATD, which have known proteostasis function, are likely to be due to numerous different variants akin to locus heterogeneity.
DISCUSSION
In this study, we use exomic sequencing analysis of a unique family to identify potentially damaging variants in proteostasis genes that could increase susceptibility to liver disease in homozygotes for the ATZ variant in the classical form of ATD. The analysis used a uniquely rigorous approach to determining the pathogenicity of candidate variants by testing them in the iHeps derived from the affected subjects themselves and furthermore by testing them for kinetic differences. Only 1 of 3 brothers who were ATZ homozygotes had severe liver disease requiring liver transplantation at 18 months of age. Analysis of genome sequences from this family provided a favorable signal-to-noise background for prioritizing putative variants, and kinetic studies of iHeps from the 3 siblings showed that there was a specific and “hard-wired” difference in rate of degradation that correlated with the liver disease phenotype. Variants in 2 proteostasis genes that were identified in the index case but not in his siblings met the criteria for being genetic modifiers of liver disease susceptibility using a functional genomics analytical strategy. The most stringent of these criteria was accelerating the degradation of ATZ by correcting each of these variants in iHeps from the index case himself and by slowing the degradation of ATZ when introducing both variants in iHeps from one of the siblings. These results suggest that a combination of at least 2 variants in proteostasis genes may be necessary for predisposing the host to liver disease and, further, suggest that the genetic modifier mechanism is likely “polygenic”. One of the variants was detected in another child with severe liver disease from homozygous ZZ ATD who required liver transplantation at age 5. Variants of other proteostasis genes that could potentially meet criteria for liver disease modifiers varied widely among a well-characterized cohort of 30 ATZ homozygotes who underwent liver disease, with most in individual patients and a few in 2 patients at most, providing evidence that genetic modifiers that can account for conversion to the liver disease phenotype by exacerbating cellular accumulation of ATZ are likely to be heterogeneous.
The results of this study provide the most powerful validation to date of 2 concepts that have been proposed to explain the marked variation in liver disease phenotype among ATZ homozygotes. In addition to the rather obvious concept that genetic and environmental modifiers determine liver disease susceptibility, we have hypothesized that variation in proteostatic mechanisms, and more specifically in autophagy genes, would influence the relative degree of IC ATZ accumulation or the cellular response to ATZ accumulation and lead to more or less proteotoxicity.
It is not surprising that the first 2 variants that have the capability to modify ATZ accumulation and liver disease by gain-of-function proteotoxicity are involved in autophagic function. We have shown that autophagy is the dominant mechanism for disposal of ATZ in a variety of model systems and that drugs that enhance autophagy reduce the accumulation and toxicity of ATZ in vivo in pre-clinical animal models.17 In unique systems with inducible expression of ATZ, we have shown that the contribution of the other major disposal pathway, ERAD, is limited,6 and it remains highly unlikely that the proteasome can accommodate large polymers and/or aggregates of ATZ. The variant in MTMR12 was selected for further study because it was predicted to lead to functional damage, and other studies have shown that loss of MTMR12 function leads to instability in myotubularin MTM1 and, in turn, reduced lipid phosphatase activity for autophagy function.19,20 Likewise, FAM134A was selected because it is known to be a receptor for autophagic regulation of the ER,21 also called the ER-phagy pathway, and would therefore be likely to contribute to the autophagic disposal of ATZ from the ER. Linkage to autophagy also provides an explanation for why there is a peak in the incidence of liver failure from ATD at ages 50–65 years,3 a timing consistent with a physiological decline in autophagic function.
There are several limitations of this study that need to be considered in its conclusions. For 1 thing, we only analyzed variants that were in coding regions and were predicted to damage protein function and not mutations that act by gain-of-function or arise from non-coding regions, including regulatory elements. Second, our study design is only applicable to variants that affect the primary pathology of hepatocellular proteotoxicity or what could be called ATD-specific variants. There are likely to be genetic modifiers that impact the response of the liver to ATZ accumulation in ways that are not ATD-specific, including, for example, variants that affect the cholestatic, inflammatory and/or fibrotic responses. Third, our conclusions are also limited by the criteria used in the study in which we are comparing the absence of the liver disease phenotype to the most severe form of liver injury, in which liver transplantation is required. This means that there are likely to be other variants that account for the wide variation in the severity of liver disease that is known to occur in patients with ATD. In other words, our approach could not assess for common variants, including those that lead to variation in the severity of liver disease in non-ATD disorders. Recent approaches using polygenic risk scores, coupled with a large, well-phenotyped cohort, could provide valuable insight into these types of other considerations for genetic effects underlying this disease.
One of the surprising results of the study is that correcting the MTMR12 and FAM134A variants each accelerated the degradation of ATZ in the iHeps from the index subject, but both variants were required to slow the degradation of ATZ in the iHeps from the protected subject. The most likely explanation for this lies in the overall capacity, multiplicity, and temporally adaptive properties within the different autophagolysosomal mechanisms that participate in the degradation of ATZ6 such that correcting 1 gene is sufficient to accelerate the process but impaired functioning of 2 genes is necessary to slow it down. Indeed, the fact that most ATD homozygotes escape severe liver disease is consistent with the prediction that multiple variants would be needed to predispose to hepatic proteotoxic consequences.
In summary, using a stringent functional genomics strategy, we identified a combination of variants in proteostasis genes that could modify the primary pathology and liver disease susceptibility in a unique family affected by the classical form of ATD. These determinations provide powerful proof-in-principle for a 2-hit conceptual model for liver disease progression in ATD and, at a more general level, for the role of proteostasis mechanisms in the clinical manifestations of a proteotoxic genetic disease. Further studies of many more patients with ATD with liver failure will be needed to determine if these 2 variants affect other families with this genetic disease, but the current results provide a basis for further considering the autophagy pathway as a therapeutic target using the personalized medicine paradigm.
Supplementary Material
AUTHOR CONTRIBUTIONS
Conceptualization: David H. Perlmutter, Ira J. Fox, Gary A. Silverman, and Stephen Pak. Data curation: Edgar N. Tafaleng, Jie Li, Yan Wang, Adam E. Locke, Thomas J. Nicholas, Yung-Chun Wang, Michael H. Cho, Edwin K Silverman, and Sheng Chih Jin, Formal analysis: Edgar N. Tafaleng, Jie Li, Yan Wang, Tunda Hidvegi, Alex Soto-Gutierrez, Adam E. Locke, Thomas J. Nicholas, Yung-Chun Wang, Michael H. Cho, Edwin K Silverman, Sheng Chih Jin, Ira J. Fox, Gary A. Silverman, Stephen Pak, and David H. Perlmutter. Funding acquisition, Investigation, Methodology, Project administration, Resources: Ira J. Fox, Gary A. Silverman, and David H. Perlmutter. Software: Tunda Hidvegi, Adam E. Locke, Thomas J. Nicholas, Yung-Chun Wang, Michael H. Cho, Edwin K Silverman, and Sheng Chih Jin. Supervision: Edwin K Silverman, Gary A. Silverman, Sheng Chih Jin, Ira J. Fox, and David H. Perlmutter. Validation: Tunda Hidvegi, Adam E. Locke, Thomas J. Nicholas, Yung-Chun Wang, Edwin K Silverman, Sheng Chih Jin, Ira J. Fox, and David H. Perlmutter. Visualization: Edgar N. Tafaleng, Jie Li, Yan Wang, Tunda Hidvegi, Adam E. Locke, Yung-Chun Wang, and Sheng Chih Jin. Writing-original draft: David H. Perlmutter. Writing-review and editing: David H. Perlmutter, Ira J. Fox Gary A. Silverman, Michael H. Cho, and Edwin K Silverman.
ACKNOWLEDGMENTS
The authors thank Dr Rick Sifers for providing DNA samples and Drs Leon Murphy, Beat Nyfeler, Dan Koboldt, and Rick Wilson for assisting in sequence technology.
FUNDING INFORMATION
These studies were supported by the US Public Health Service grant P01-DK096990.
CONFLICTS OF INTEREST
Alex Soto-Gutierrez owns stock in Pittsburgh ReLiver and Von Baer Wolff. Adam E. Locke is employed by and owns stock in Regeneron. Michael H. Cho received grants from Bayer. Edwin K Silverman received grants from Bayer and Northpond Laboratories. Ira J. Fox consults for Miromatrix. He owns stock in Pittsburgh ReLiver and Von Baer Wolff. The remaining authors have no conflicts to report.
Footnotes
Abbreviations: AT, α1-antitrypsin; ATD, α1-antitrypsin deficiency; ATZ, α1-antitrypsin variant Z; CPVL, carboxypeptidase vitellogenic like; DNAJC12, DNAJ heat shock protein family (Hsp40) member C12; EC, extracellular; ER, endoplasmic reticulum; ERAD, endoplasmic reticulum-associated degradation; FAM134A, family with sequence similarity 134 member A; HOOK2, Hook microtubule tethering protein 2; HTO, HeLa tet-off cell line; IC, intracellular; iHeps, induced hepatocyte-like cells; KI, knock-in; MAF, minor allele frequency; MAN1B1, mannosidase alpha class 1B member 1; MTM1, myotubularin protein 1; MTMR12, myotubularin-related protein 12; PSMB2, proteasome 20S subunit beta 2; SORL1, sortilin related receptor 1.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.hepjournal.com.
Contributor Information
Edgar N. Tafaleng, Email: ent8@pitt.edu.
Jie Li, Email: jieli@wustl.edu.
Yan Wang, Email: wangyan8518@gmail.com.
Tunda Hidvegi, Email: tunde@wustl.edu.
Alex Soto-Gutierrez, Email: alexsotoguti@gmail.com.
Adam E. Locke, Email: adam.locke@regeneron.com.
Thomas J. Nicholas, Email: thomas.nicholas@utah.edu.
Yung-Chun Wang, Email: yung-chun@wustl.edu.
Stephen Pak, Email: stephen.pak@wustl.edu.
Michael H. Cho, Email: remhc@channing.harvard.edu.
Edwin K. Silverman, Email: reeks@channing.harvard.edu.
Gary A. Silverman, Email: gsilverman@wustl.edu.
Sheng Chih Jin, Email: jin810@wustl.edu.
Ira J. Fox, Email: foxi@upmc.edu.
David H. Perlmutter, Email: perlmutterd@wustl.edu.
REFERENCES
- 1.Perlmutter DH. α1-antitrypsin deficiency: A misfolded protein variant with unique effects on the endoplasmic reticulum. Endoplasmic Reticulum Stress Dis. 2016;3:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strnad P, McElvaney NG, Lomas DA. Alpha1-antitrypsin deficiency. N Engl J Med. 2020;382:1443–1455. [DOI] [PubMed] [Google Scholar]
- 3.Chu AS, Chopra KB, Perlmutter DH. Is severe progressive liver disease caused by alpha-1-antitrypsin deficiency more common in children or adults? Liver Transpl. 2016;22:886–894. [DOI] [PubMed] [Google Scholar]
- 4.Sveger T. Liver disease in alpha1-antitrypsin deficiency detected by screening of 200,000 infants. N Engl J Med. 1976;294:1316–1321. [DOI] [PubMed] [Google Scholar]
- 5.Mostafavi B, Diaz S, Tanash HA, Piitulainen E. Liver function in alpha-1-antitrypsin deficient individuals at 37 to 40 years of age. Medicine. 2017;96:e6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Moretti F, Hidvegi T, Sviben S, Fitzpatrick JAJ, Sundaramoorthi H, et al. Mulitple genes core to ERAD, macro-autophagy and lysosomal degradation pathways participate in the proteostasis response in α1-antitrypsin deficiency. Cell Mol Gastroenterol Hepatol. 2024;17:1007–1024. (online ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan S, Huang L, McPherson J, Muzny D, Rouhani F, Brantly M, et al. Single nucleotide polymorphism-mediated translational suppression of endoplasmic reticulum mannosidase I modifies the onset of end-stage liver disease in alpha1-antitrypsin deficiency. Hepatology. 2009;50:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun AH, Collette JR, Sifers RN. The cytoplasmic tail of human mannosidase Man1b1 contributes to catalysis-independent quality control of misfolded alpha1-antitrypsin. Proc Natl Acad Sci USA. 2020;117:24825–24836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 58,831 diverse genomes from the NHLBI TOPMed program. Nature. 2021;590:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chappell S, Hazdic N, Stockley R, Guetta-Baranes T, Morgan K, Kalsheker N. A polymorphism of the alpha1-antitrypsin gene represents a risk factor for liver disease. Hepatology. 2008;47:127–132. [DOI] [PubMed] [Google Scholar]
- 11.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotech. 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 13.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S, Freimer NB, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidvegi T, Schmidt BZ, Hale P, Perlmutter DH. Accumulation of mutant α1-antitrypsin Z in the ER activates caspases-4 and -12, NFκB and BAP31 but not the unfolded protein response. J Biol Chem. 2005;280:39002–39015. [DOI] [PubMed] [Google Scholar]
- 16.Tafaleng EN, Chakraborty S, Han B, Hale P, Wu W, Soto‐Gutierrez A, et al. Induced pluripotent stem cells model personalized variations in liver disease due to α1-antitrypsin deficiency. Hepatology. 2015;62:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant α1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. [DOI] [PubMed] [Google Scholar]
- 18.DeMeo DL, Sandhaus RA, Barker AF, Brantly ML, Eden E, McElvaney NG, et al. Determinants of airflow obstruction in severe alpha-1-antitrypsin deficiency. Thorax. 2007;62:806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta VA, Hnia K, Smith LL, Gundry SR, McIntire JE, Shimazu J, et al. Loss of catalytically inactive lipid phosphatase myotubularin-related protein 12 impairs myotubularin stability and promotes centronuclear myopathy in zebrafish. PloS Genetics. 2013;9:e1003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fetalvero KM, Yu Y, Goetschkes M, Liang G, Valdez RA, Gould T, et al. Defective autophagy and mTORC1 signaling in myotubularin null mice. Molecular Cell Biol. 2013;33:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reggio A, Buonomo V, Berkane R, Bhaskara RM, Tellechea M, Peluso I, et al. Role of FAM134A paralogues in endoplasmic reticulum remodeling, ER-phagy and collagen quality control. EMBO Reports. 2021;22:e52289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.