Abstract
All eukaryotic cells require a minimal iron threshold to sustain anabolic metabolism. However, the mechanisms by which cells sense iron to regulate anabolic processes are unclear. Here, we report a previously undescribed eukaryotic pathway for iron sensing in which molecular iron is required to sustain active histone demethylation and maintain the expression of critical components of the pro-anabolic mTORC1 pathway. Specifically, we identify the iron-binding histone-demethylase KDM3B as an intrinsic iron sensor that regulates mTORC1 activity by demethylating H3K9me2 at enhancers of a high-affinity leucine transporter, LAT3, and RAPTOR. By directly suppressing leucine availability and RAPTOR levels, iron deficiency (ID) supersedes other nutrient inputs into mTORC1. This process occurs in vivo and is not an indirect effect by canonical iron-utilizing pathways. Because ancestral eukaryotes share homologues of KDMs and mTORC1 core components, this pathway likely predated the emergence of the other kingdom-specific nutrient sensors for mTORC1.
Keywords: Iron, epigenetics, histone demethylase, mTOR, amino acids, metabolism, cancer
Introduction
Mechanisms that integrate diverse environmental cues to modulate energy metabolism are indispensable for both unicellular and multicellular organisms1. Although all cells require iron for survival, it is still unclear how iron levels are sensed to control anabolic processes. The prominent oxygenation of earth’s atmosphere after the rise of the first photosynthetic organisms resulted in a dramatic decrease in iron bioavailability (Fig. 1A)2,3. Iron deficiency (ID), thus, has become common, and robust systems of regulation have evolved across multiple phylogenetic kingdoms to preserve survival in response to ID4,5. Many of these pathways are kingdom specific, such as the POPEYE/Fe-Deficiency Induced Transcript 1 (PYE/FIT) system in Arabidopsis thaliana, Activator of Ferrous Transport 1 (AFT1) in Saccharomyces cerevisiae, and the iron regulatory protein (IRP) and hypoxia inducible factor (HIF) pathways in animals6–9. Because the great oxygenation event preceded the evolution of eukaryotic life on Earth (Fig. 1A), it stands to reason that an additional and evolutionarily conserved mechanism for sensing and responding to iron limitation must be shared among eukaryotes. This pathway would also be expected to inhibit cellular anabolism during ID because the most fundamental anabolic processes, nucleotide biosynthesis and translation, both require iron-utilizing proteins. However, such a pathway has not been characterized to date.
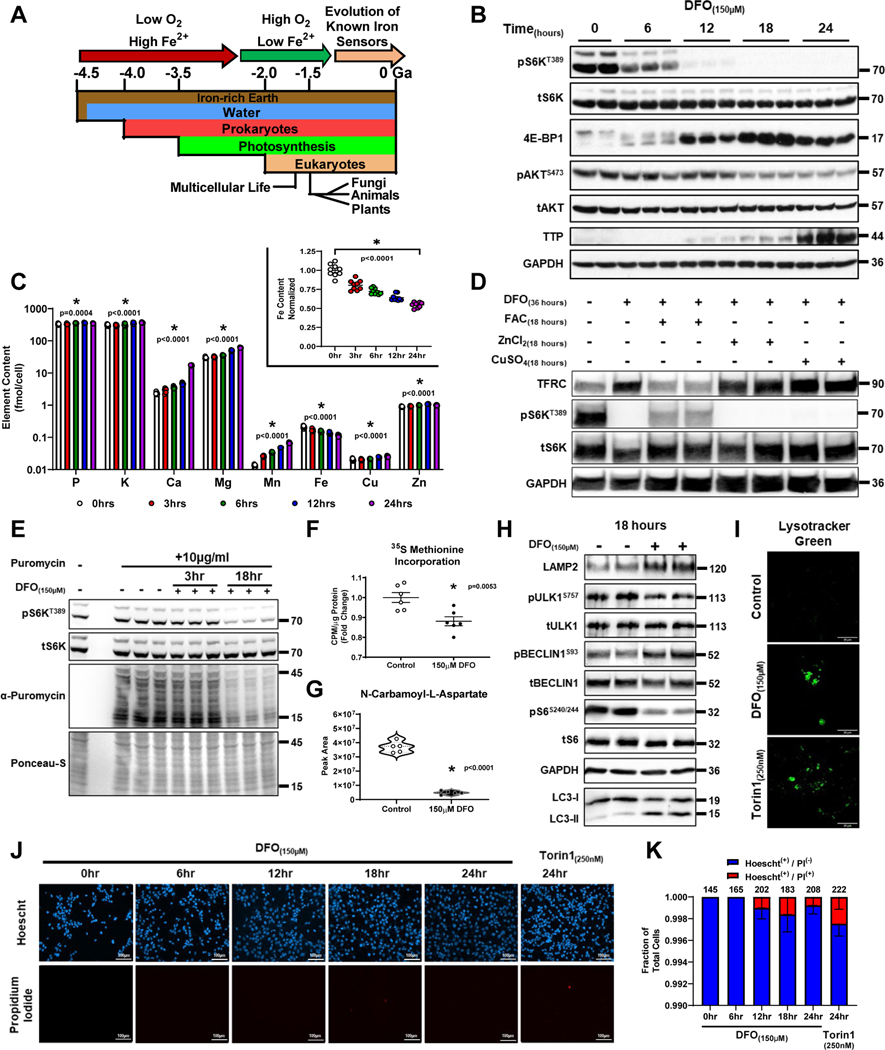
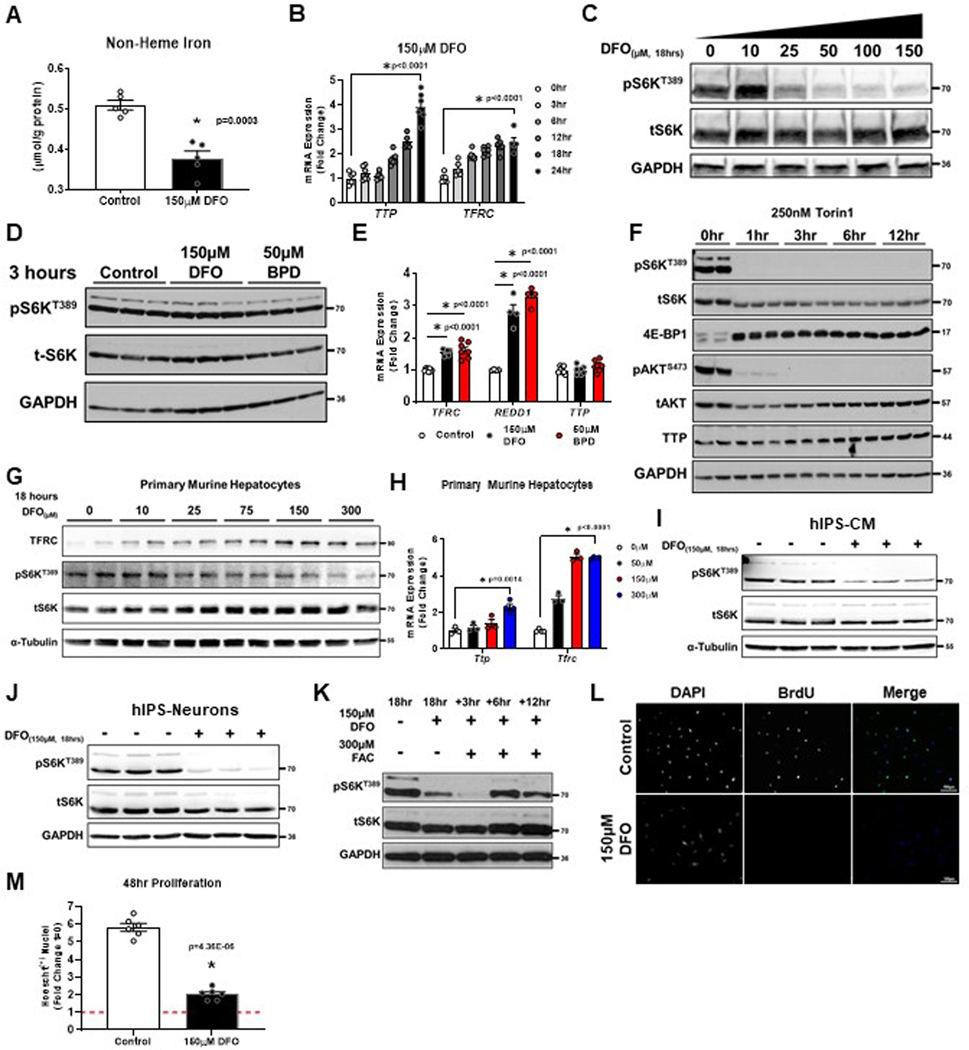
Figure 1. Long-term ID inactivates mTORC1.
(A) Schematic of the evolution of eukaryotic life on earth2,3. (B) Immunoblot of mTORC1 and mTORC2 activity in HEK293T cells treated with 150μM DFO for the indicated times. Representative image of two independent experiments. (C) ICP-MS-based measurement of cellular metal content plotted on Log10-scale in HEK293T cells treated with 150μM DFO for the indicated times. Insert graph depicts normalized changes in Fe content plotted on a linear scale. (n=3 independent samples measured in triplicate, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (D) Immunoblot of mTORC1 activity in HEK293T cells treated with 150μM DFO for 18 hours and then supplemented with equimolar concentrations of either Fe2+, Cu2+ or Zn2+ for an additional 18 hours. Representative image of two independent experiments. (E-F) Incorporation of puromycin (E) and 35S methionine (F) into elongating peptide chains in HEK293T cells after treatment with 150μM DFO for 18 hours. ((F) n=6 replicates per condition, two-tailed unpaired t-test, mean ± SE). (G) Cellular levels of N-carbomyl-L-aspartate after treatment with 150μM DFO for 18 hours measured by HPLC-MS (n=5 replicates per condition, two-tailed unpaired t-test, median ± quartiles). (H) Assessment of autophagy, as measured by ULK1 phosphorylation, LAMP2 levels and BECLIN1S93 phosphorylation, and conversion of LC3I to LC3II in HEK293T cells treated with 150μM DFO for 18 hours. Representative image of two independent experiments. (I) Fluorescent confocal microscopy of lysosomes stained with Lysotracker green in HEK293T cells treated with 150μM DFO for 18 hours or 250nM Torin1 for 6 hours. Representative image of five independent samples. (J) Fluorescent microscopy of cell death using Hoescht and propidium iodide (PI) double staining in HEK293T cells treated with 150μM DFO or 250nM Torin-1 at indicated times. Representative image of five independent samples. (K) Quantification of images in panel J. (n=6 replicates, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). Source numerical data and unprocessed blots are available in source data files. * indicates P value < 0.05 when noted for all panels.
The Jumonji-C domain containing lysine demethylases (Jmj-C KDM) belong to a large family of Fe2+- and 2-oxoglutarate (αKG)-dependent dioxygenases and function as critical regulators of gene transcription through their role in demethylating histones10. Jmj-C KDM proteins have been identified in the plant, animal, and fungi kingdoms, and ancestral Fe2+-dependent dioxygenases can be found in prokaryotes and archae bacteria11,12. Members of this family have varying affinities for iron and therefore likely respond differently to variable degrees of ID. However, whether or not these proteins play an active role in physiologic iron sensing is unknown13,14. Here we present evidence that Mechanistic Target of Rapamycin Complex 1 (mTORC1) activity is inhibited by the Jmj-C KDM family member KDM3B in response to cellular iron deprivation through the transcriptional repression of the amino acid (AA) transporter LAT3 and the obligatory mTORC1 complex member RAPTOR. These data fit a model in which the evolutionary origins of the core mTOR machinery, which predate the emergence of the known amino acid and growth factor sensors, evolved to match anabolic drive with iron availability.
Results
Long-term ID inactivates mTORC1
The effects of iron on mTORC1 activity were assessed by treating HEK293T cells with deferoxamine (DFO), resulting in a reduction in cellular iron and increased expression of TTP and Transferrin Receptor 1 (TFRC) mRNA within 24 hours (Extended Data Fig. 1A, B). Inhibition of mTORC1 activity (as assessed by S6KT389 phosphorylation, 4E-BP1 band-shift, and TTP expression) was first observed at 12 hours (Fig. 1B). This inhibition transitioned sharply above 10μM, with further repression detected up to 50μM (Extended Data Fig. S1C). Late-onset inhibition of mTORC1 activity was not due to ineffective iron chelation, as 3-hour exposure of both DFO and 2,2’-bipyridyl (BPD, another iron chelator with enhanced cell permeability) induced the expression of TFRC, and the hypoxia marker REDD1 without reducing S6KT389 phosphorylation or increasing TTP expression (Extended Data Fig. 1D, E). In contrast, Torin1, a direct mTOR inhibitor, effectively suppressed S6KT389 phosphorylation and induced TTP expression within 1–3 hours (Extended Data Fig. 1F). Non-dividing cells, including primary murine hepatocytes, human induced-pluripotent stem (hIPS) cell derived cardiomyocytes (CM) and hIPS neurons also repressed mTORC1 activity in response to ID (Extended Data Fig. 1G–J). Because other forms of nutrient starvation have rapid effects on mTORC115, but ID causes a delayed effect, we acutely replenished cells with ferric ammonium citrate (FAC) after cells were chelated for 18 hours. S6KT389 phosphorylation returned after 6 hours (Extended Data Fig. 1K), suggesting that iron sensing by the mTORC1 pathway requires protein degradation and/or the translation of new protein.
To determine whether mTORC1-inhibition was attributable to perturbation in the levels of other cationic metals, cellular metal content was assessed over 24 hours of DFO treatment using inductively coupled plasma mass spectrometry (ICP-MS). We observed a significant and selective reduction in the levels of iron that matched the timing of TTP and TFRC expression (Fig. 1C, Extended Data Fig. 1B). We did not observe a decrease in cellular phosphate or potassium levels, indicating cells are able to maintain membrane potential even under prolonged ID (Fig. 1C). Addition of equimolar concentrations of either Fe2+, Cu2+ or Zn2+ after 18 hours of chelation revealed that iron alone is sufficient to rescue mTORC1 activity (Fig. 1D).
We next evaluated the effect of ID on major mTORC1-regulated anabolic and catabolic processes. Treatment with DFO reduced the incorporation of puromycin and 35S methionine into elongating peptide chains (Fig. 1E, F), and decreased the cellular levels of N-carbamoyl-L-aspartate (Fig. 1G), consistent with suppression of protein and pyrimidine synthesis. Additionally, DFO prevented incorporation of BrdU into newly synthesized DNA and significantly reduced the rate of proliferation (Extended Data Fig. 1L, M), consistent with the role of mTORC1 in regulating the G1-S checkpoint16. Treatment of cells with DFO also resulted in activation of autophagy, as evidenced by decreased ULK1 phosphorylation, increased LAMP2 levels and BECLIN1S15 phosphorylation, and conversion of LC3I to LC3II (Fig. 1H). Increased retention of the fluorescent lysosomal dye, Lysotracker Green, in cells treated with either DFO or Torin1 was also observed (Fig 1I). Of note, appreciable cell death among adherent cells treated for 24 hours with DFO or Torin1 was not evident, as determined by propidium iodide (PI) uptake (Fig. 1J, K). Flow cytometry using Annexin-V and PI on cells treated with DFO for 24 hours confirmed that ID did not induce a population of early apoptotic cells (Annexin-V single positive cells) that are impermeable to PI (Supplementary Fig. 1A–C). Thus ID appears to repress mTORC1 activity through a dedicated signaling pathway and not generally due to loss of cell viability.
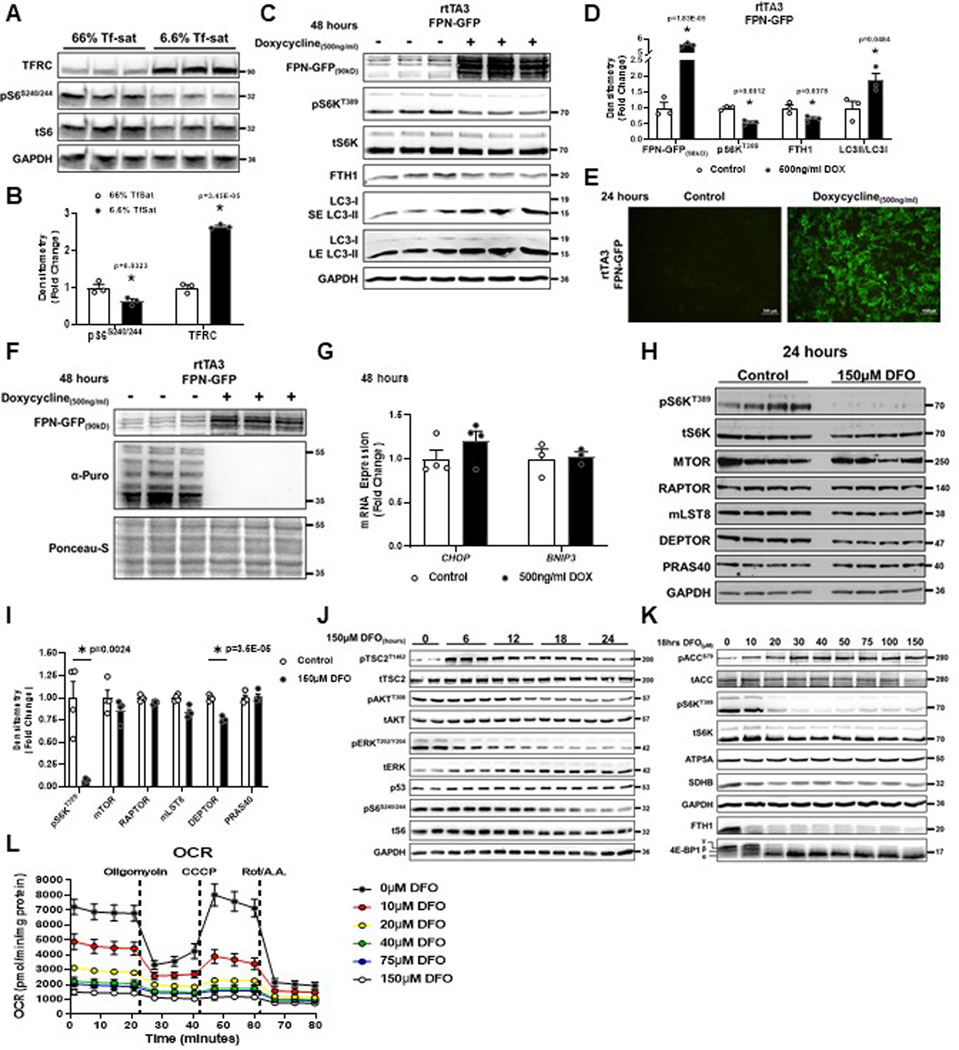
To determine the effects of IDs on mTORC1 under physiological iron deficiency (i.e., without iron chelation), we adjusted the ratio of holo-transferrin (iron-bound) and apo-transferrin (iron-free) in serum-free culture media to match the transferrin saturation (Tf-sat) typically found in healthy individuals (>50%) or patients with ID anemia (<8%). Low Tf-sat conditions increased TFRC expression and reduced pS6S240/244, suggesting that pathologic reduction in cellular iron is sufficient to regulate mTORC1 (Extended Data Fig. 2A, B). Additionally, we utilized a doxycycline inducible system to overexpress the cellular iron exporter SLC40A1 (Ferroportin, FPN) fused to GFP to reduce cellular iron without using a chelator17. Doxycycline treatment resulted in robust GFP expression and reduced levels of ferritin heavy chain (FTH1), effectively achieving cellular ID (Extended Data Fig. 2C–E). We observed concomitant reduction in S6KT389 phosphorylation, increased LC3-II conversion, and repression of translation (Extended Data Fig. 2C–D, F). Doxycycline treatment did not induce markers of ER stress, CHOP and BNIP3, indicating mTORC1 regulation by ID is independent of the general cell stress response (Extended Data Fig. 2G).
ID does not require TSC1/2, HIF, or AMPK signaling to inhibit mTORC1
To determine the mechanism of iron-mediated mTORC1 regulation, we first measured the protein levels of mTORC1 complex components after 24 hours of DFO. We observed minimal to no change in the levels of mTOR, RAPTOR, mLST8, or PRAS40 protein (Extended Data Fig. 2H, I). There was a modest decrease in DEPTOR levels, but as a negative regulator of mTORC118, this could not explain inhibition of mTORC1 activity in ID. Next, we systemically assessed key elements of known nutrient and growth factor pathways for their potential role in ID-mediated mTORC1 inhibition. Iron chelation in the setting of serum deprivation abolished the recovery of mTOR activity after serum repletion (Fig. 2A). ID resulted in the recruitment of TSC2 to the lysosome (Fig. 2B, C), consistent with mTORC1 inhibition. However, TSC2T1462 phosphorylation (associated TSC2 inhibition) increased prior to mTORC1 inhibition and was independent of changes in AKT T308 phosphorylation (Extended Data Fig. 2J). We also observed inactivation of ERK without changes in the levels of p53 (Extended Data Fig. 2J), supporting that prolonged ID promotes cell-cycle arrest but not apoptosis.
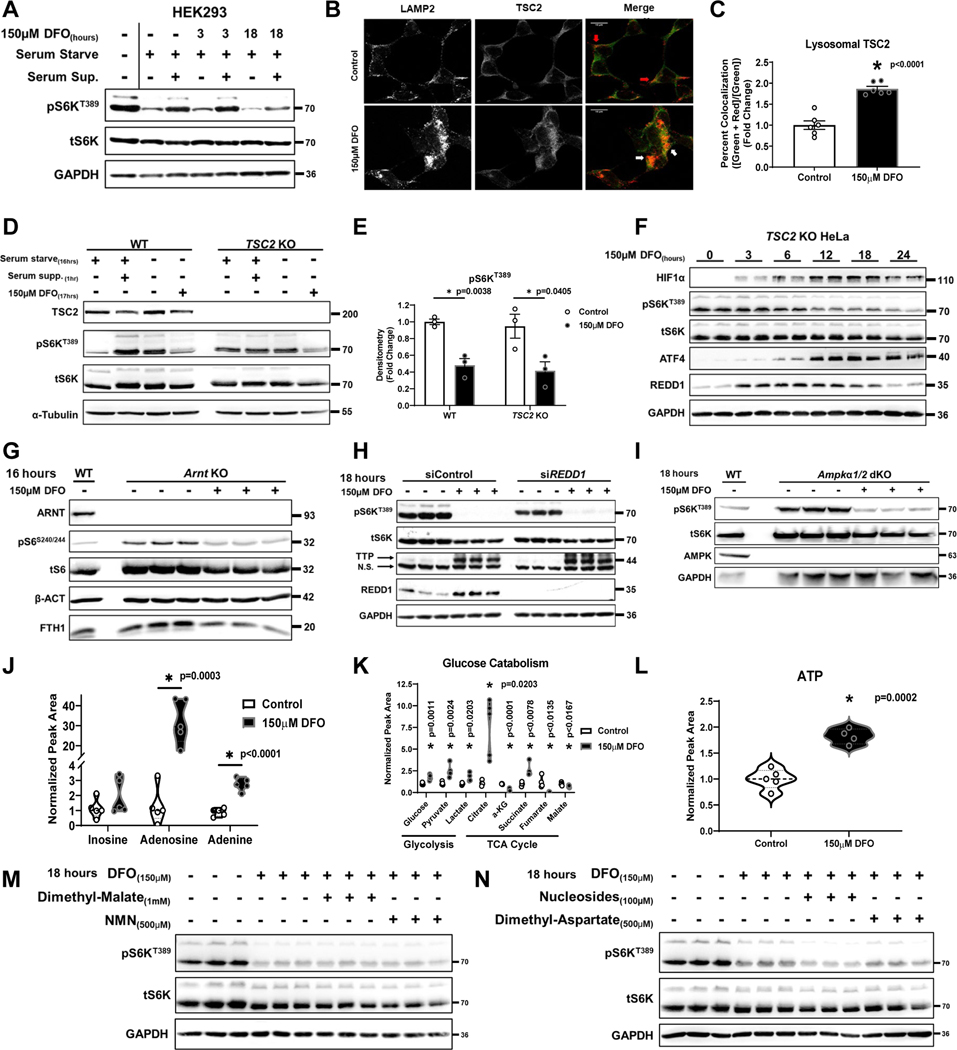
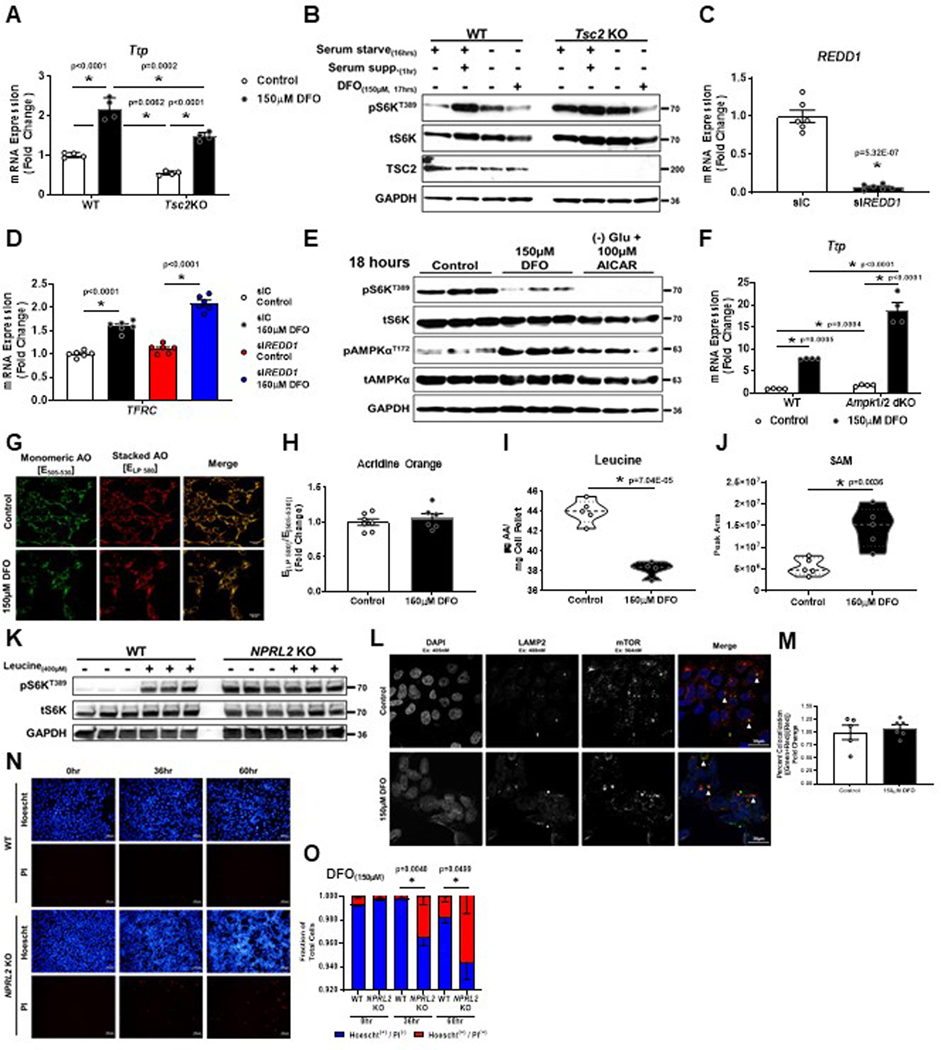
Figure 2. ID does not require TSC1/2 or AMPK signaling to inhibit mTORC1.
(A) Immunoblot of mTORC1 activity in HEK293 cells that were serum starved overnight followed by the addition of serum for 1 hour in the presence or absence of 150μM DFO for the indicated times. Representative image of three independent experiments. (B) Recruitment of TSC2 to the lysosome in HEK293 cells treated with DFO for 24 hours. Representative image of five independent samples. (C) Summary of the results in panel C (n=6 replicates per condition, two-tailed unpaired t-test, mean ± SE). (D) Immunoblot of mTORC1 activity in WT and TSC2 KO HeLa cells that were serum starved overnight followed by the addition of serum for 1 hour in the presence or absence of 150μM DFO for 17 hours. Representative image of two independent experiments. (E) Summary graph of immunoblot in panel D (n=3 samples per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (F) Immunoblot of mTORC1 activity and hypoxia regulated factors in TSC2 KO HeLa cells treated with 150μM DFO for the indicated times. Representative image of two independent experiments. (G) Immunoblot of mTORC1 activity in Arnt KO MEFs treated with 150μM DFO for 16 hours. Representative image of two independent experiments. (H) Immunoblot of mTORC1 activity in HEK293T cells treated with REDD1 siRNA in the presence and absence of 150μM DFO for 18 hours. Representative image of one experiment with 3 independent samples. (I) Immunoblot of mTORC1 activity in WT and Ampkα 1/2 dKO MEFs treated with 150μM DFO for 18 hours. Representative image of two independent experiments. (J) Cellular levels of the purines inosine, adenosine, and adenine in HEK293T cells after treatment with 150μM DFO for 18 hours measured by HPLC-MS (n=5 replicates per condition, two-tailed unpaired t-test, median ± quartiles). (K) Targeted metabolomics in HEK293T cells treated with 150μM DFO for 18 hours (n=4 replicates per condition, two-tailed unpaired t-test, median ± quartiles). (L) Total cellular ATP pools in HEK293T cells after 18 hours of treatment with 150μM DFO (n=4 replicates per condition, two-tailed unpaired t-test, median ± quartiles). (M) Immunoblot of mTORC1 activity in HEK293T cells in the presence and absence of 150μM DFO for 18 hours, supplemented with 1mM dimethyl malate or 500μM NMN. Representative image of one experiment with 3 independent samples. (N) Immunoblot of mTORC1 activity in HEK293T cells in the presence and absence of 150μM DFO for 18 hours, supplemented with 100μM nucleoside cocktail (Adenosine, Guanosine, Thymidine and Cytidine), or 500μM dimethyl aspartate. Representative image of one experiment with three independent samples. Source numerical data and unprocessed blots are available in source data files. * indicates P value < 0.05 when noted for all panels.
It is possible that ID induces mitochondrial dysfunction and serves as an upstream signal to repress mTORC1. Consistent with previous studies17, treating cells with low dose DFO (10μM) was sufficient to induce ID (evidenced by a reduction in FTH1 and SDHB levels), increase ACCS79 phosphorylation (a marker of AMPK activation and mitochondrial stress) (Extended Data Fig. 2K) and suppress mitochondrial oxygen consumption (Extended Data Fig. 2L). Despite these markers of mitochondrial dysfunction, there was no change in mTORC1 activity with low dose DFO treatment (Extended Data Fig. 2K). To determine whether iron deprivation requires a component of the growth factor signaling pathway to regulate mTORC1, we utilized TSC2 knockout (KO) HeLa cells and Tsc2 KO mouse embryonic fibroblasts (MEFs), which maintain mTORC1 activity in the absence of growth factors19–21. Tsc2 KO MEFs displayed loss of Ttp mRNA expression at baseline, consistent with mTORC1 hyper-activation (Extended Data Fig. 3A). However, both TSC2 KO HeLa cells and Tsc2 KO MEFs show reduced S6KT389 phosphorylation and increased Ttp expression after iron chelation, despite being completely resistant to serum starvation (Fig. 2D, E, and Extended Data Fig. 3B).
ID can activate hypoxia response pathways through stabilization of the HIF1/2α proteins22,23 and transcription of REDD1, which inhibits mTORC1 through the TSC1/2 pathway24,25. However, mTORC1 activity still decreased after 12 hours of DFO treatment in TSC2 KO HeLa cells (Fig. 2F). The expression of HIF1α, ATF4 and REDD1 preceded the reduction in S6KT389 phosphorylation and was partially reversed at 24 hours, diverging from mTORC1 activity. Additionally, we demonstrated repression of S6S240/244 phosphorylation in Arnt KO MEFs treated with DFO (Fig. 2G), confirming that HIF signaling is not required for mTORC1 inhibition. Equivalent reduction in S6KT389 phosphorylation and TTP induction in DFO treated REDD1 KD cells was also observed. (Fig. 2H, Extended Data Fig. 3C, D).
ID activates AMP-activated protein kinase (AMPK), a negative regulator of mTORC126 (Extended Data Fig 2K, 3E). However, iron chelation still caused mTORC1 inhibition and increased Ttp expression in Ampkα 1/2 double KO MEFs (Fig. 2I, Extended Data Fig. 3F). A reduction in purines and their precursors have also been shown to have an inhibitory effect on mTORC1 activity27, however, ID increased cellular inosine, adenosine, and adenine levels (Fig. 2J). Thus, inhibition of mTORC1 by ID is not sensed through growth factors, HIF, REDD1, AMPK, or purine levels.
Targeted metabolomics on DFO-treated HEK293T cells were performed to assess the effects of perturbations in metabolic pathways on mTORC1 activity. We observed increased levels of glucose, pyruvate, lactate, citrate, and succinate with concomitant decreases in α-ketoglutarate (αKG), fumarate, and malate (Fig. 2K). These findings are consistent with TCA cycle arrest at points of iron-catalyzed reactions (aconitase and succinate dehydrogenase) and subsequent transition to anaerobic glycolytic metabolism. Total cellular ATP levels were not depleted after prolonged iron chelation, suggesting that ID cells maintain sufficient energy pools via glycolysis (Fig. 2L). We next supplemented DFO-treated cells with either dimethyl malate (to replace the lost TCA cycle carbons), NMN (to replenish cytosolic NAD+ levels due to ETC blockade), dimethyl aspartate (to supply the precursor for nucleotide biosynthesis and support the malate-aspartate shuttle), or a nucleoside cocktail (to replace loss of de novo nucleotide biogenesis) (Fig. 2M, N). None of these metabolites were able to rescue mTORC1 activity.
ID leads to mTORC1 inhibition through leucine sensing
Activation of mTORC1 by nutrients requires its recruitment to the lysosomal surface in an AA-dependent manner28. AA repletion after AA deprivation caused full reactivation of mTORC1 in cells treated with DFO for 3 hours, but not 18 hours (Fig. 3A). Furthermore, we observed dissociation of mTOR from lysosomes in cells chelated for 18 hours (Fig. 3B, C), that was not due to lysosomal deacidification (Extended Data Fig. 3G, H). These data suggest that ID actively regulates the AA sensing branch of mTORC1.
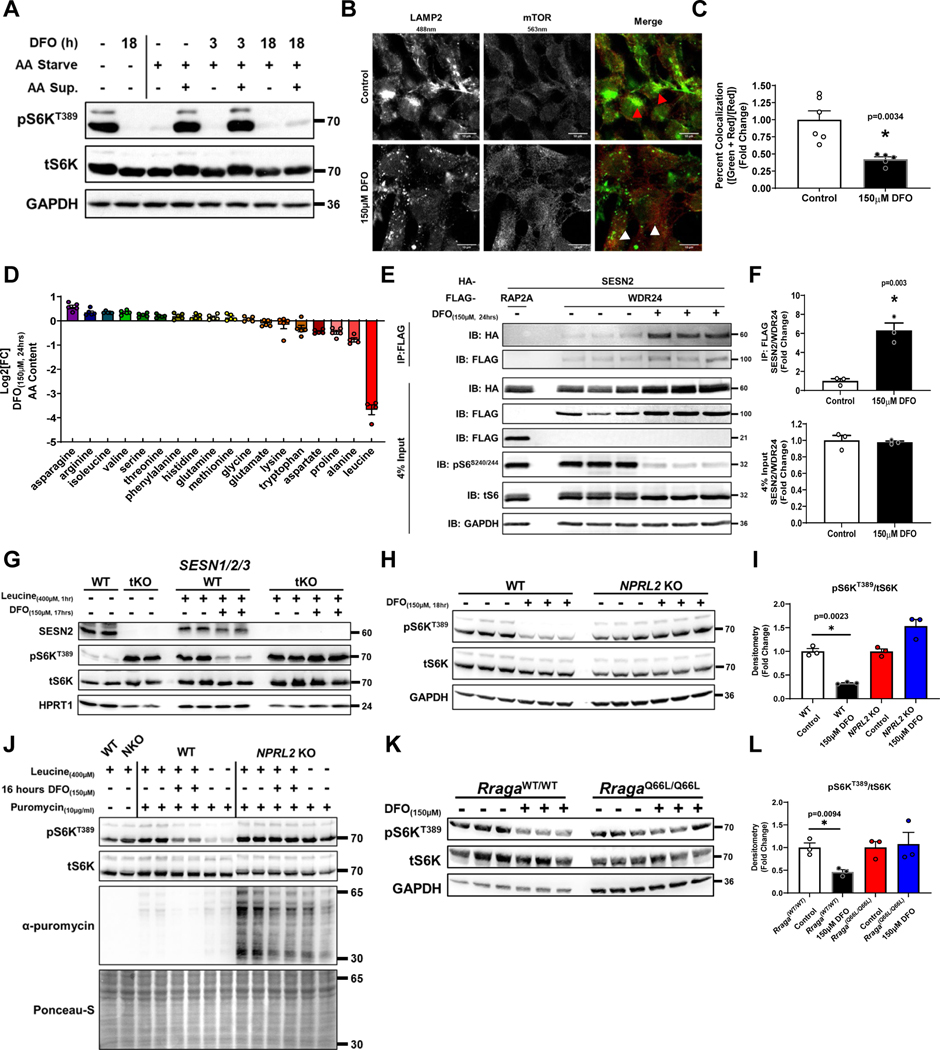
Figure 3. ID causes mTORC1 inhibition through leucine sensing.
(A) Immunoblot of mTORC1 activity in HEK293T cells starved of AA for 3 hours followed by restimulation for 1 hour in the presence or absence of 150μM DFO for the indicated times. Representative image of three independent experiments. (B) Fluorescent confocal microscopy showing dissociation of mTOR from the lysosome in HEK293T cells treated with 150μM DFO for 18 hours. Representative image of five independent samples. (C) Summary graph of the images in panel B (n=6 replicates control; n=5 replicates 150μM DFO, two-tailed unpaired t-test, mean ± SE). (D) HPLC-MS based measurement of 16 amino acids in MEFs treated with 150μM DFO for 24 hours. (n=5 replicates, mean ± SE). (E) Co-IP of HA-SESTRIN2 and FLAG-WDR24 (a member of the GATOR2 complex) in HEK293T cells treated with 150μM DFO for 24 hours. Cell lysates were subjected to immunoprecipitation with anti-FLAG antibody and analyzed by immunoblotting. RAP2A = negative control. Representative image of three independent experiments. (F) Summary of co-IP studies in panel E (n=3 replicates per condition, unpaired t-test, mean ± SE). (G) Immunoblot of mTORC1 activity in WT and SESN1/2/3 tKO HEK293T cells starved overnight of leucine followed by the addition of 400μM leucine for 1 hour in the presence or absence of 150μM DFO. Representative image of two independent experiments. (H) Immunoblot of mTORC1 activity in WT and NPRL2 KO HEK293T in the presence or absences of 150μM DFO for 18 hours. Representative image of two independent experiments. (I) Summary graph of immunoblot in panel H (n=3 samples per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (J) Incorporation of puromycin into elongating peptide chains in WT and NPRL2 KO 293T cells in the presence and absence of 150μM DFO and 400μM leucine for 16 hours. Representative image of two independent experiments. (K) Immunoblot of mTORC1 activity in Rraga+/+ (WT) and RragaQ66L/Q66L (KI) MEFs treated with 150μM DFO for 18 hours. Representative image of two independent experiments. (L) Summary graph of immunoblot in panel K. (n=3 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). Source numerical data and unprocessed blots are available in source data files. * indicates P value < 0.05 when noted for all panels.
To determine if intracellular AA levels are specifically regulated by ID, we profiled the concentrations of 16 AAs using HPLC-MS in DFO-treated MEFs and observed a marked decreased in the levels of leucine, with minimal changes in the levels of methionine and arginine (Fig. 3D). Similar results were observed in HEK293T cells (Extended Data Fig. 3I). We observed a significant increase in SAM levels, indicating ID does not regulate mTORC1 through SAM (Extended Data Fig. 3J). To confirm the decrease in cellular leucine levels by ID is sufficient to affect mTORC1 signaling, we assessed the interaction of SESTRIN2 with WDR24 of the GATOR2 complex, which occurs in the absence of leucine29. ID increased SESTRIN2 and WDR24 interaction (Fig. 3E, F). Additionally, SESTRIN1/2/3 triple KO (SESN tKO) and NPRL2 KO 293T cells, which are not responsive to changes in leucine levels29,30, maintain full mTORC1 activity despite ID (Fig. 3G–I, Extended Data Fig. 3K). Consistent with these findings, mTOR remained localized to lysosomes despite DFO treatment in NPRL2 KO cells (Extended Data Fig. 3L, M). Moreover, NPRL2 KO cells were resistant to ID induced suppression of translation, and puromycin incorporation in NPRL2 KO cells subjected to ID was equivalent to cells starved of leucine (Fig. 3J). ID also did not reduce S6KT389 phosphorylation in RragaQ66L knock-in (KI) MEFs, which are similarly resistant to leucine deprivation31 (Fig. 3K–M). However, sustained mTORC1 activation and translation under prolonged ID in NPRL2 KO 293T cells resulted in significant cell death, but not WT counterparts (Extended Data Fig. 3N, O). Thus, the regulation of mTORC1 by iron is mediated upstream of SESTRIN, GATOR1/2, and the RAG proteins, and ID-induced translational repression is likely a downstream consequence of mTORC1 inactivation.
ID prevents leucine uptake
Leucine is an essential AA and must be imported by cells from the extracellular environment. However, DFO-treated cells remained resistant to leucine mediated activation of mTORC1 even at supra-physiologic levels (Fig. 4A). Cellular uptake of 14[C]-leucine in HEK293T cells and 3[H]-leucine in MEFs was significantly inhibited by ID (Fig. 4B, C). In contrast, mTORC1 inhibition by rapamycin caused a compensatory increase in leucine uptake (Fig. 4D), indicating that reduced leucine uptake is a primary effect of ID. Furthermore, repression of leucine uptake is not a consequence of using a pharmacologic chelator. Overexpression of FPN using doxycycline resulted in similar reduction in leucine uptake (Fig. 4E).
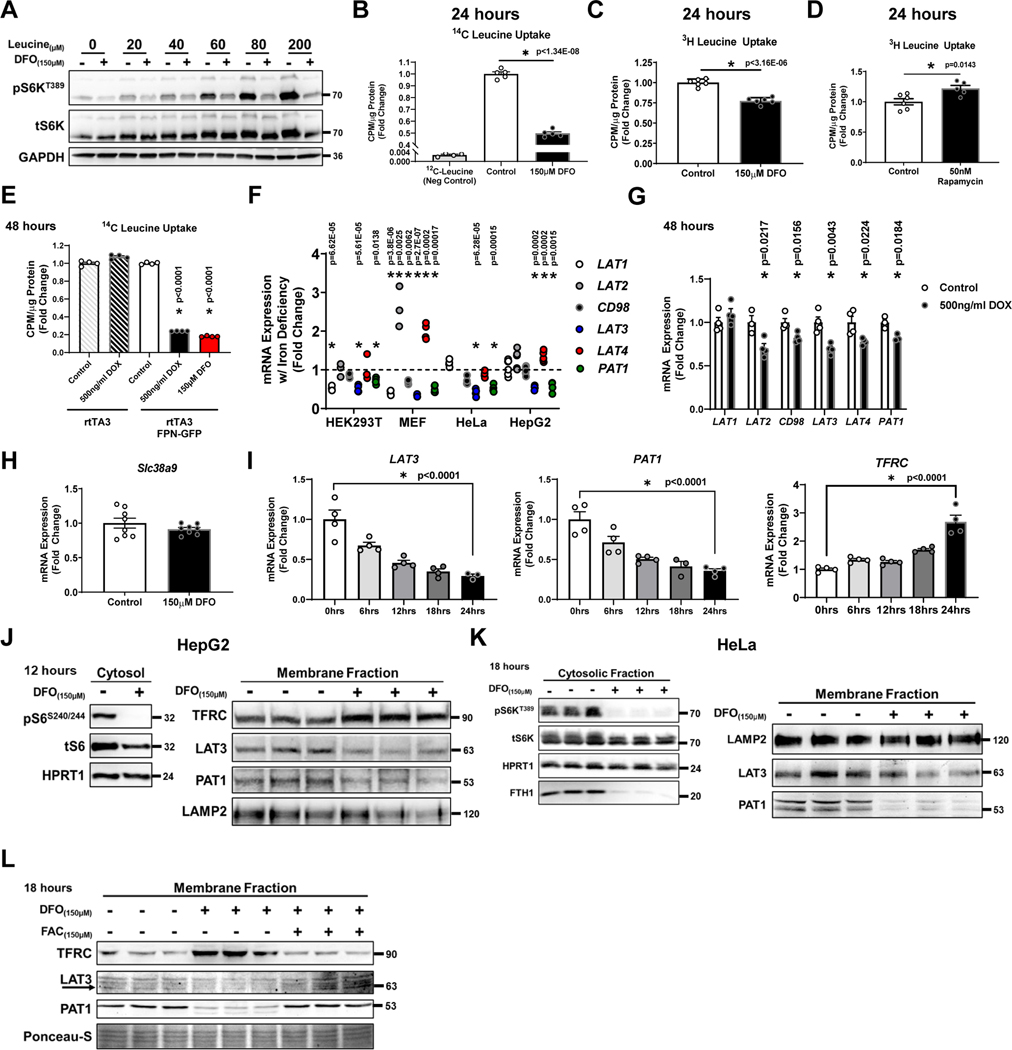
Figure 4. ID prevents leucine uptake.
(A) Immunoblot of mTORC1 activity in cells treated with 150μM DFO and cultured in leucine-free media for 15 hours. At t=15 hours, cells were supplemented with increasing concentrations of leucine for 3 hours. Representative image of two independent experiments (B) 14C-Leucine uptake in HEK293T cells with and without 150μM DFO treatment for 18 hours (n=5 replicates per condition, two-tailed unpaired t-test, mean ± SE). (C) 3H-leucine uptake into MEFs treated with 150μM DFO for 24 hours (n=6 replicates per, two-tailed unpaired t-test, mean ± SE). (D) 3H-Leucine uptake into MEF cells treated with 50nM rapamycin for 24 hours (n=6 replicates control; n=5 replicates 50nM Rapamycin, two-tailed unpaired t-test, mean ± SE). (E) 14C-Leucine uptake into HEK293T cells transfected with rtTA3/FPN-GFP or rtTA3/eGFP control and in the presence and absence of 500ng/ml doxycycline treatment for 48 hours or 150μM of DFO for 18 hours (n=4 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (F) mRNA levels of the cell surface leucine transporters LAT1–4 and the lysosomal leucine regulator PAT1 in indicated cell types and tissues. Internal controls: POLR2A (HEK, HeLa), 18S (HepG2), Snrk (MEF) (n=4 replicates HEK293T LAT1; n=3 HEK293T LAT2; n=3 HEK293T CD98; n=4 HEK293T LAT3; n=3 HEK293T LAT4; n=4 HEK293T PAT1; n=4 MEF LAT1; n=4 MEF LAT2; n=4 MEF CD98; n=4 MEF LAT3; n=4 MEF LAT4; n=5 MEF PAT1; n=4 HeLa LAT1; n=4 HeLa CD98; n=4 HeLa LAT3; n=4 HeLa LAT4; n=4 HeLa PAT1; n=6 HepG2 LAT1; n=6 HepG2 LAT2; n=6 HepG2 CD98; n=6 HepG2 LAT3; n=6 HepG2 LAT4; n=5 HepG2 PAT1, two-tailed unpaired t-test, mean ± SE). (G) mRNA of various leucine transporters in HepG2 cells transfected with rtTA3/FPN-GFP plasmids in and treated with 500ng/ml doxycycline for 48 hours. Internal control: 18S (n=4 replicates per condition for all conditions except; n=3 LAT2 control; n=3 PAT1 control; n=3 PAT1 500ng/ml DOX, unpaired t-test, mean ± SE). (H) Slc38a9 mRNA levels in MEFs treated with DFO for 24 hours. Internal control: Polr2a (n=8 replicates per condition, two-tailed unpaired t-test, mean ± SE). (I) mRNA levels of LAT3, PAT1 and TFRC1 at indicated time points in HeLa cells treated with150μM DFO: Internal control: POLR2A (n=4 replicates per time point except; n=3 LAT3 24hrs; n=3 PAT1 18hrs, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (J) Immunoblotting of cytosolic and membrane fractions isolated from HepG2 cells treated with 150μM DFO for 12 hours. Immunoblot of mTORC1 activity demonstrating the effectiveness of DFO in the cytoplasmic fraction is shown on the left. Representative image of two independent experiments. (K) Immunoblot of mTORC1 activity in cytosolic fraction and LAT3 and PAT1 in the membrane fraction of HeLa cells treated with 150μM DFO for 18 hours. Representative image of two independent experiments. (L) Immunoblot of LAT3 and PAT1 in iron deficient-HEK293T cells supplemented with FAC. Representative image from one experiment with three independent samples. Source numerical data and unprocessed blots are available in source data files. * indicates P value < 0.05 when noted for all panels.
To determine the mechanism for the reduced leucine uptake with ID, we measured the mRNA levels of the membrane leucine transporters LAT1–4 and the lysosomal leucine regulator PAT1. LAT3 and PAT1 were the only transporters consistently downregulated in various cell lines subjected to ID by DFO treatment or overexpression of FPN (Fig 4F, G). Leucine can also be effluxed from the lysosome through an arginine-dependent mechanism involving SLC38A932,33. However, expression of Slc38A9 in MEFs did not change with iron chelation (Fig. 4H). The timing of LAT3 and PAT1 repression coincided with the reduction in S6KT389 phosphorylation and induction of TFRC mRNA (Fig. 1B, 4I). We correspondingly observed reduction in LAT3 and PAT1 protein levels in cellular membrane fractions isolated from HepG2 and HeLa cells treated with DFO (Fig. 4J, K,). Furthermore, levels of LAT3 and PAT1 were restored in iron deficient HEK293T cells supplemented with FAC (Fig. 4L).
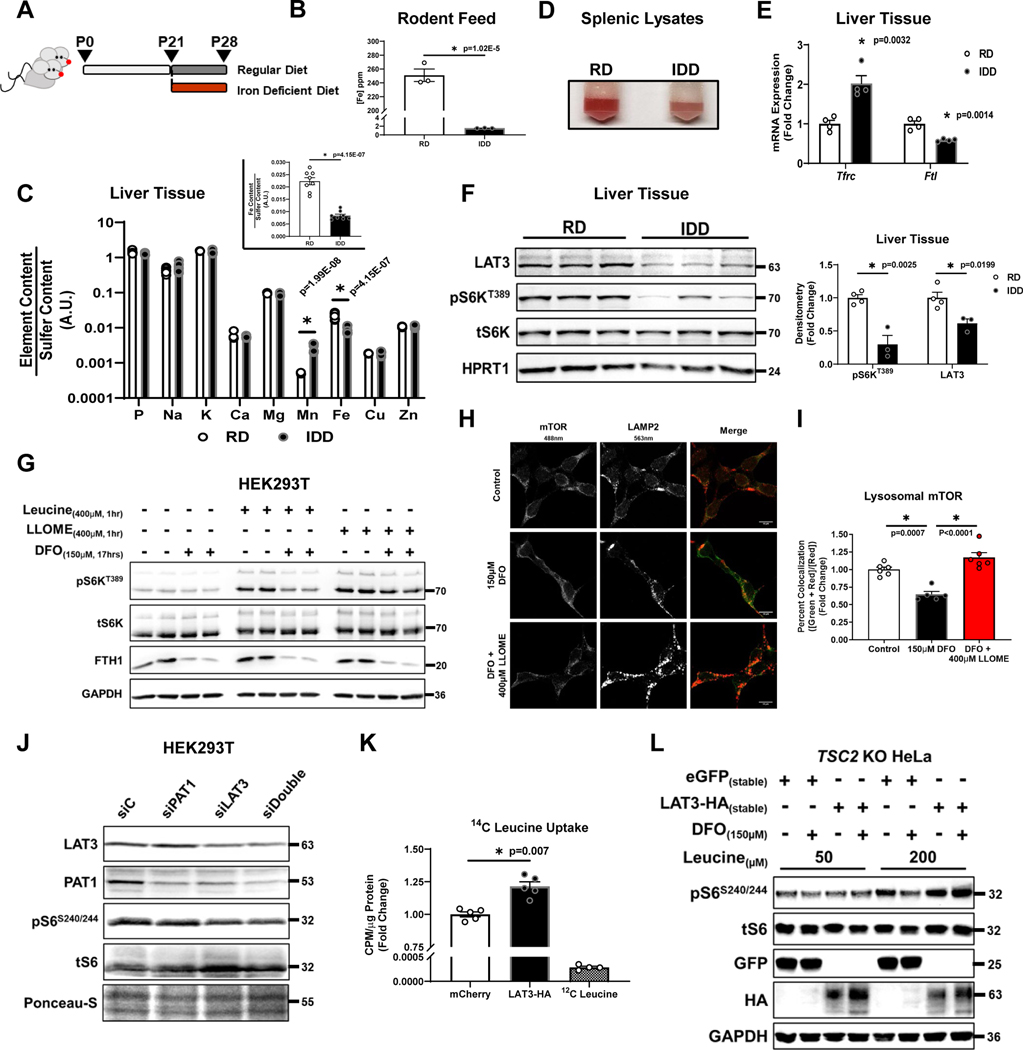
We utilized an in vivo model of acute ID where weaning-age pups (P21) were randomized to regular diet (RD; 250 ppm Fe) or iron deficient diet (IDD; 2 ppm Fe) for 1 week (Fig. 5A). Iron content of the two diets were confirmed by ICP-MS (Fig. 5B). We used this model to evaluate mTORC1 activity in the liver for three reasons: 1) to focus on acute signaling processes upstream of mTORC1; 2) the liver is disproportionately affected by acute ID due to its role in iron mobilization; and 3) LAT3 is the dominant leucine transporter in the liver and is highly expressed (Supplementary Fig. 2A, B). Seven days of IDD resulted in 62% reduction in hepatic iron content as quantified by ICP-MS (Fig. 5C). The pallor of splenic lysates (a marker of iron deficiency anemia), and characteristic changes in hepatic expression of Tfrc and Ftl mRNA further confirmed ID (Fig. 5D, E). Consistent with in vitro results, livers from mice fed IDD displayed reduced LAT3 expression and S6KT389 phosphorylation (Fig. 5F). Together, these data support that physiologic reduction in iron without pharmacologic chelation can regulate mTORC1 activity in vivo.
Figure 5. ID inhibits mTORC1 through LAT3.
(A) Schematic depicting our in vivo ID protocol. (B) ICP-MS-based measurement of iron content in ppm from regular and iron deficient rodent diets. (n=3 replicates per group, two-tailed unpaired t-test, mean ± SE). (C) ICP-MS-based measurement of cellular metal content plotted on Log10-scale in livers from mice fed 7 days of IDD. Insert graph depicts normalized changes in Fe content plotted on a linear scale. (n=8 replicates per group, two-tailed unpaired t-test, mean ± SE). (D) Image of splenic lysates from mice treated with regular and ID diet. Representative image from five independent samples. (E) mRNA levels of Tfrc1 and Ftl in mice treated with regular and ID diet. Internal control: Polr2a (n=4 mice per group, two-tailed unpaired t-test, mean ± SE). (F) Immunoblot of mTORC1 activity and LAT3 in the liver of mice treated with iron deficient diet. Summary graph of immunoblot is shown to the right (n=4 samples RD; n=3 IDD, two-tailed unpaired t-test, mean ± SE). Representative image of two independent experiments. (G) Immunoblot of mTORC1 activity in cells treated with 150μM DFO and cultured in leucine-free media for 16 hours. At t=16 hours, cells were supplemented with 400μM L-leucine or Leucyl-Leucine-O-Methyl-Ester (LLOME) for 1 hour. Representative image of three independent experiments. (H) Recruitment of mTORC1 to the lysosome in HEK293T cells treated with 150μM DFO for 18 hours and supplemented with 400μM LLMOE for 1 hour. Representative image of six independent samples. (I) Summary of the results in panel E (n=6 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (J) Immunoblot of mTORC1 activity and LAT3 and PAT1 levels in HEK293 cells treated with Lat3 or Pat1 siRNA or with both siRNAs. Representative image of one experiment (K) 14C-Leucine uptake into HEK293T cells transfected with mCherry control or LAT3-HA for 48 hours (n=5 replicates per condition, two-tailed unpaired t-test, mean ± SE). (J) Immunoblot of mTORC1 activity in TSC2 KO HeLa cells stably expressing eGFP or HA-tagged LAT3 and treated with DFO for 18 hours. Representative image of one experiment. Source numerical data and unprocessed blots are available in source data files. * indicates P value < 0.05 when noted for all panels.
To confirm that ID regulates mTORC1 by preventing leucine uptake, we utilized a cell-permeable form of leucine (Leucyl-Leucine-O-Methyl-Ester (LLOME)). Addition of LLOME, but not L-leucine, was sufficient to fully rescue mTORC1 activity under ID, as evidenced by recovery of S6KT389 phosphorylation and re-recruitment of mTOR to the lysosome (Fig. 5G–I). To test whether LAT3 and/or PAT1 are sufficient for the ID-mediated repression of leucine transport, we knocked down LAT3 and PAT1 individually and together. Knockdown (KD) of LAT3, but not PAT1, was sufficient to repress mTORC1 activity in HEK293T cells (Fig. 5J). Interestingly, KD of LAT3 alone reduced the levels of PAT1, suggesting coregulation between the two transporters. To test whether reduction in LAT3 is required for ID-mediated mTORC1 inhibition, we generated a C-terminal HA-tagged LAT3 overexpression construct (LAT3-HA) containing the codon-optimized human LAT3 coding sequence together with the 5’- and 3’-UTRs. Overexpression of this construct led to increased 14[C]-leucine uptake, confirming its functionality (Fig. 5K). Stable overexpression of LAT3-HA, but not eGFP, in TSC2 KO HeLa cells rescued mTORC1 activity in ID cells. (Fig. 5L), and this process was dependent on sufficient extracellular leucine available for uptake. Together, these data demonstrate that repression of LAT3 is both necessary and sufficient for mTORC1 inhibition by ID.
ID increases global histone methylation
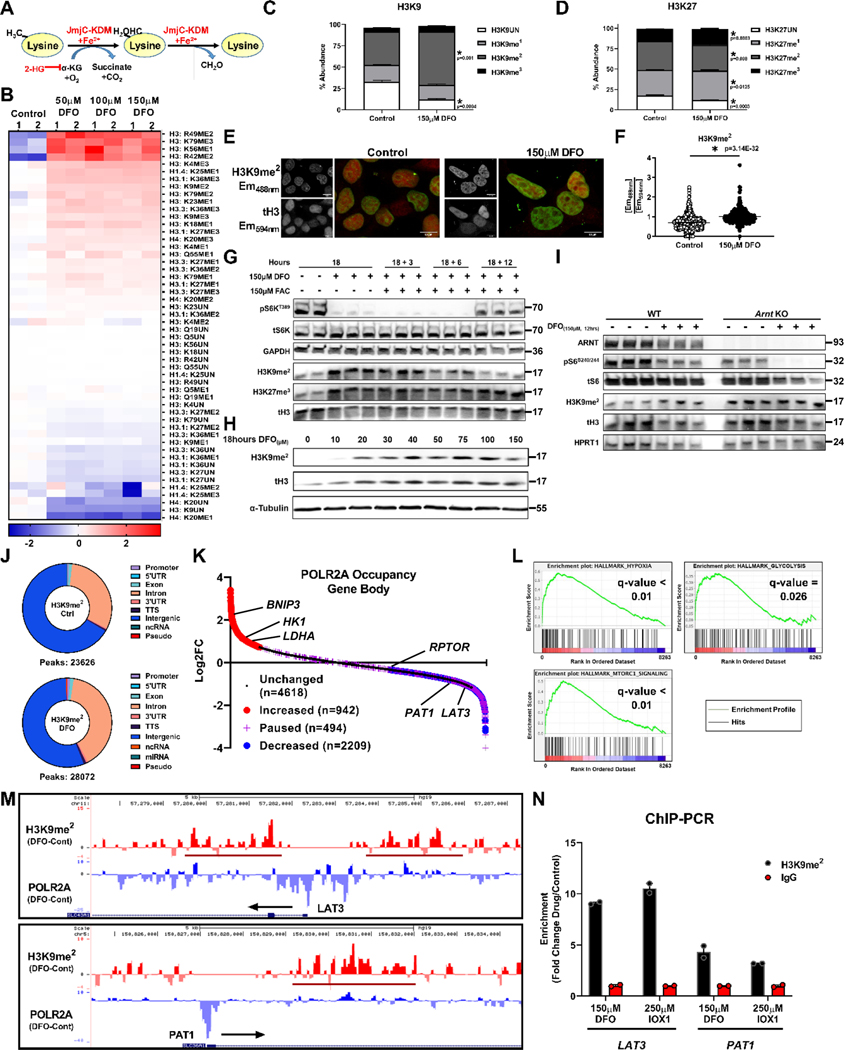
The delayed response by mTORC1 to ID through a reduction in LAT3 mRNA indicates that iron regulates transcription. Among factors that regulate epigenetics and transcription, Jmj-C histone demethylases utilize Fe2+ and αKG for catalysis11 (Fig. 6A). We performed histone-mass spectrometry (histone-MS) to unbiasedly measure global changes in histone methylation in ID. Iron chelation resulted in significant increases in histone lysine methylation across multiple residues, including H3K9, H3K27, H3K4, and H3K36 (Fig. 6B). Among the histone marks associated with transcriptional repression, we observed a marked increase in H3K9me2 and H3K27me3 (Fig. 6C, D). H3K9me2 had the greatest absolute increase in percent abundance of all histone marks (from 38% to 60% of all H3K9 peptides) and was the second-most abundant mark in the DFO treated samples (Supplementary Fig. 3A, B). This data was validated via immunofluorescence (IF) using antibodies specific for H3K9me2 and total histone H3 (tH3), which showed increased H3K9me2 fluorescence in ID (Fig. 6E, F).
Figure 6. ID increases global histone methylation.
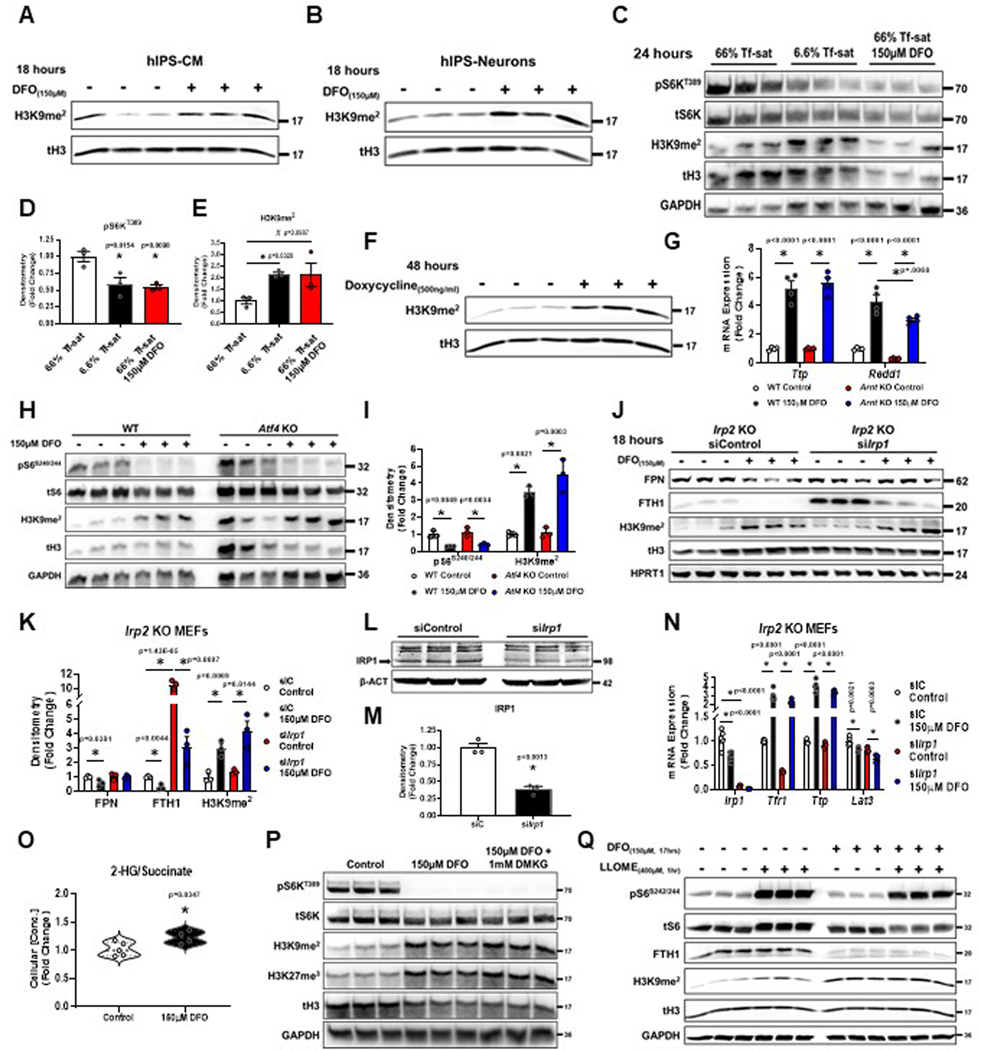
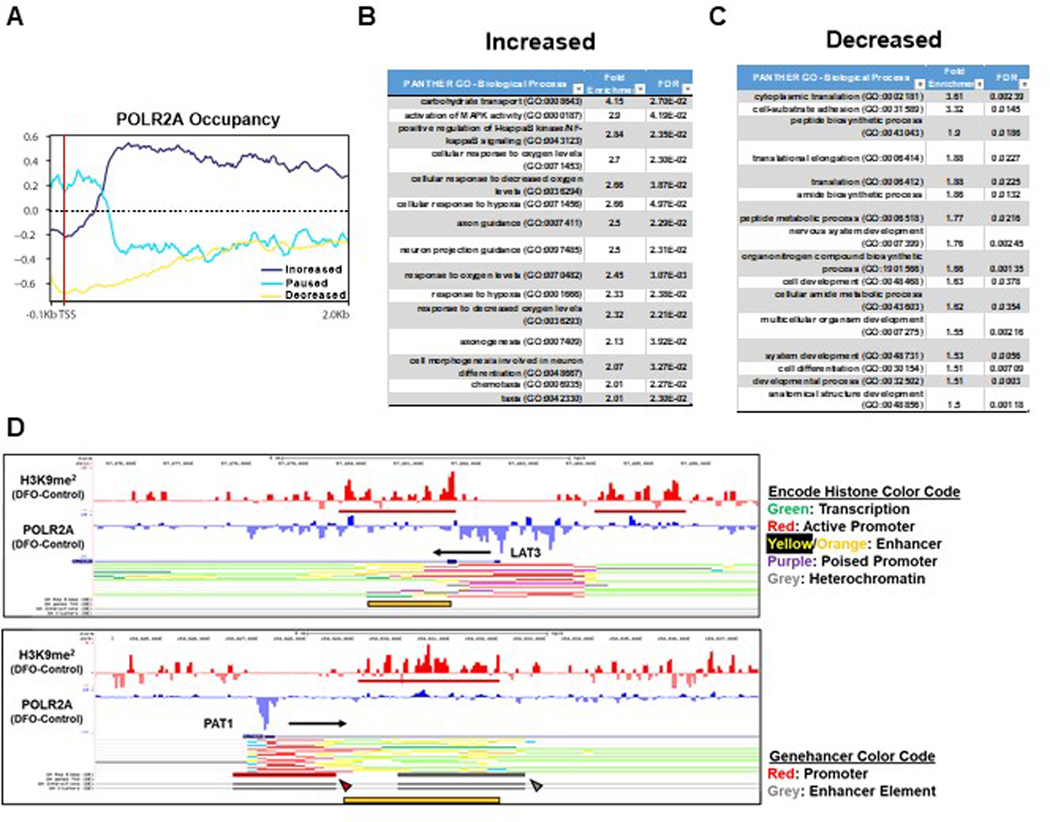
(A) Schematic of lysine demethylation catalyzed by Jmj-C KDMs in the presence of Fe2+, O2, and αKG. (B) Heatmap of results from histone-mass spectrometry (histone-MS) in HEK293T cells treated with DFO at indicated concentrations for 12 hours. (Two representative samples of n=3 independent samples measured in triplicate.) (C, D) Percentage of H3K9 (C) and H3K27 (D) methylation in HEK293T cells in the presence of 150μM DFO from histone-MS experiment in panel B. (n=3 independent samples measured in triplicate, two-tailed unpaired t-test, mean ± SE). (E) Fluorescent confocal microscopy of H3K9me2 immunostaining in HEK293T cells treated with 150μM DFO for 12 hours. Representative image from 5 independent samples. (F) Summary graph of the images shown in panel E (n=667 (control) and n=298 (150μM DFO) cells per group, two-tailed unpaired t-test, mean ± SE). (G) Immunoblot of mTORC1 activity and H3 methylation in HEK293T cells treated with 150μM DFO for 18 hours and supplemented with Fe2+ (delivered as FAC) for the indicated times. Representative image of two independent experiments. (H) Immunoblot of H3K9me2 levels in HepG2 cells treated with DFO for 18 hours at the indicated doses. Data paired with Extended Data Fig. 2K. Representative image of two independent experiments. (I) Immunoblot of mTORC1 activity and H3K9me2 in WT and Arnt KO MEFs treated with 150μM DFO for 12 hours. Representative image of two independent experiments. (J) Donut charts depicting distribution of called H3K9me2 peaks from ChIP-seq performed on HEK293T cells treated with 150μM DFO for 12 hours. (K) Ranking of POLR2A occupancy within gene bodies based on log2FC after treatment with 150μM DFO. (L) Enrichment plots from Gene Set Enrichment Analysis (GSEA) performed on the ranked POLR2A list from panel K. FDR(q) values below 0.25 were considered statistically significant. (M) UCSC genome browser tracks for the LAT3 (top) and PAT1 (bottom) gene loci. POLR2A and H3K9me2 tracks from ChIP-seq analysis were loaded and represented as the difference in normalized reads between the DFO and control groups. Regions of H3K9me2 enrichment in the DFO group are underlined in red. Direction of transcription is indicated by a black arrow. (N) ChIP-PCR of LAT3 and PAT1 in HEK293T cells. Cells treated with 150μM DFO or 250μM IOX1 for 12 hours and vehicle controls were followed by IP of lysates using an antibody against H3K9me2. IgG was used as a negative control for the IP (n=2 replicates per condition). Source numerical data and unprocessed blots are available in source data files. * indicates P value < 0.05 when noted for all panels.
The timing of ID-induced H3K9me2 methylation corresponded closely with mTORC1 inhibition and was reversed upon the re-addition of iron (Fig. 5G). The levels of H3K27me3, however, were not reversed, suggesting that ID does not regulate mTORC1 activity through H3K27me3 (Fig. 5G). We also observed a dramatic increase in H3K9me2 levels between cells treated with 10μM and 20μM DFO (Fig. 5H). This transition in H3K9me2 methylation matches the repression in mTORC1 activity beginning at 20μM DFO (Extended Data Fig. 1C, and 2K). DFO also increased the levels of H3K9me2 in terminally differentiated hIPS-CM and hIPS-neurons, suggesting DNA replication and cell division are not required for ID to regulate histone methylation (Extended Data Fig. 4A, B). ID induced by low Tf-sat (6.6%) culture media or overexpression of doxycycline-inducible FPN both resulted in increased H3K9me2 levels (Extended Data Fig. 4C–F). Thus, physiologic ID can alter H3K9me2 levels and mTORC1 activity, independent of proliferation or pharmacologic chelation.
We next determined whether regulation of H3K9me2 levels during ID was indirectly mediated by HIF1/2 activation. Although baseline H3K9me2 levels were higher in Arnt KO MEFs, ID still elicited robust inhibition of S6S240/244 phosphorylation and an increase in H3K9me2 levels (Fig. 6I). Ttp expression was equally induced in WT and Arnt KO MEFs, whereas basal and post-chelation Redd1 mRNA levels were reduced in Arnt KO MEFs, consistent with defective HIF signaling (Extended Data Fig. 4G). Cytosolic heme levels regulate the translation of the transcription factor ATF4 through Heme-Regulated Inhibitor (HRI) and eIF2α34, and ATF4 can cooperate with KDM4C to activate the transcription of multiple amino acid transporters35. However, Atf4 KO MEFs treated with DFO displayed equivalent reduction in S6S240/244 phosphorylation and increased H3K9me2 levels compared to WT MEFs (Extended Data Fig. 4H, I). We next tested whether regulation of H3K9me2 by ID required an intact IRP system. Irp1 KD in Irp2 KO MEFs significantly reduced IRP1 mRNA and protein levels, reduced baseline Tfrc mRNA expression, increased baseline FTH1 protein levels and failed to repress FPN levels after iron chelation (Extended Data Fig. 4J–N). Despite loss of the IRP system, ID increased H3K9me2 and Ttp mRNA levels, and decreased the mRNA levels of Lat3 (Extended Data Fig. 4J–N). Our data suggest a model in which an intrinsically Fe2+-dependent Jmj-C KDM protein regulates H3K9me2 levels and mTORC1 activity independently of the HIF, ATF4, and IRP pathways.
Jmj-C KDMs can be inhibited by the metabolite 2-hydroxyglutarate (2-HG), which competes with αKG at the active-site36 (Fig. 6A). Although iron chelation resulted in a slight increase in the ratio of 2-HG/succinate (Extended Data Fig. 4O), supplying cells with excess cell permeable dimethyl-αKG (DMKG) did not prevent histone hypermethylation or restore mTORC1 activity (Extended Data Fig. 4P). Finally, we tested whether alterations in histone methylation by ID were upstream of its effects on leucine transport and mTORC1 activity. Although addition of LLOME for 1 hour to DFO treated cells fully rescued mTORC1 activity, it had no effect on H3K9me2 levels (Extended Data Fig. 4Q).
We next performed chromatin immunoprecipitation followed by sequencing (ChIP-Seq) targeting H3K9me2 and POLR2A to identify genes regulated by ID. We observed an increase in the total number of H3K9me2 peaks, mostly within introns, in DFO-treated cells (Fig. 6J). We next focused on transcriptional changes that occurred during ID as inferred by changes in POLR2A occupancy. We utilized a scoring method that quantifies the fold change in POLR2A occupancy within predefined regions of the promoter and gene body37 to categorize genes defined by increased POLR2A binding, POLR2A loss, promoter pausing, or no change (Fig. 6K, Extended Data Fig. 5A). About half of the genes with a POLR2A peak had no change in occupancy and more than twice the number of genes lost POLR2A compared to genes that increased recruitment of POLR2A (Fig. 6K). Gene set enrichment analysis (GSEA)38,39 revealed signatures consistent with the hallmarks of hypoxia, glycolysis and mTORC1 signaling (Fig. 6L). Additionally, gene-ontology analysis of genes with increased POLR2A occupancy revealed overrepresentation of genes related to hypoxia signaling and glycolysis (Extended Data Fig. 5B). Translation and transcription related processes were enriched among genes which had lost POLR2A occupancy (Extended Data Fig. 5C). Both LAT3 and PAT1 had enriched H3K9me2 signals in enhancer regions located within 5 Kb of their transcription start sites (TSS) and reduced POLR2A occupancy (Fig. 6M). The increase in H3K9me2 signal in the introns of LAT3 and PAT1 corresponded to annotated enhancer regions for these genes, suggesting that these peaks are functionally relevant (Extended Data Fig. 5D). To validate our ChIP-seq data, we performed ChIP-PCR using primers that flank the regions of increased H3K9me2 signal in the promoters and introns of LAT3 and PAT1. We observed enrichment of these regions in cells treated with either DFO or the Jmj-C-specific inhibitor IOX1. Thus, inactivity of one or more KDM family members is likely responsible for the regulation of H3K9me2 levels at the LAT3 and PAT1 loci (Fig. 6N).
ID leads to repression of core mTORC1 genes in Arabidopsis thaliana and Saccharomyces cerevisiae
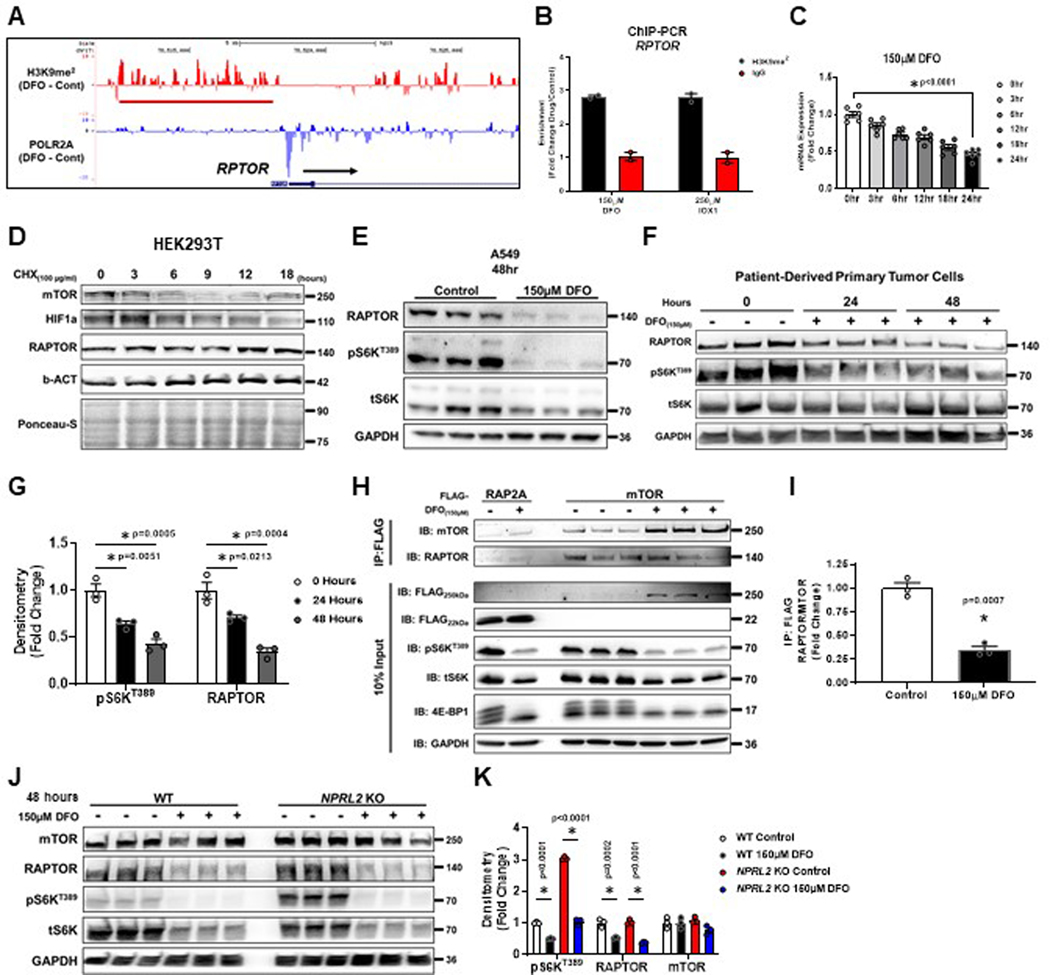
An unexpected result of our ChIP-Seq analysis was the observation that H3K9me2 signal was enriched in the promoter for RPTOR, and this coincided with loss of POL2RA occupancy (Extended Data Fig. 6A). This was confirmed using ChIP-PCR in cells treated with DFO and IOX1 (Extended Data Fig. 6B). We measured the levels of RPTOR mRNA over 24 hours of ID and observed a continuous decrease, similar to LAT3 and PAT1 (Extended Data Fig. 6C). However, RAPTOR protein levels did not change in that time period (Extended Data Fig. 2H, I). RAPTOR protein has been reported to be highly stable40, and unlike mTOR itself, RAPTOR protein levels remained constant over an 18-hour period in HEK293 cells treated with cycloheximide (Extended Data Fig. 6D). Considering its inherent stability, we extended iron chelation to 48 hours, and observed nearly total loss of RAPTOR protein and S6KT389 phosphorylation in both an established cell line and patient-derived primary tumor cell cultures (Extended Data Fig. 6E–G). We also observed a corresponding decrease in mTORC1 complex assembly (Extended Data Fig. 6H, I). Reduction in the core mTORC1 complex after 48 hours of ID meant that mTORC1 activity was repressed in NPRL2 KO HEK293T cells despite being resistant to leucine deprivation (Extended Data Fig. 6J, K). Together, these data suggest that eukaryotic cells retain a mechanism to suppress mTORC1 mediated anabolic processes after extended periods of ID, independent of the status of other nutrient sources.
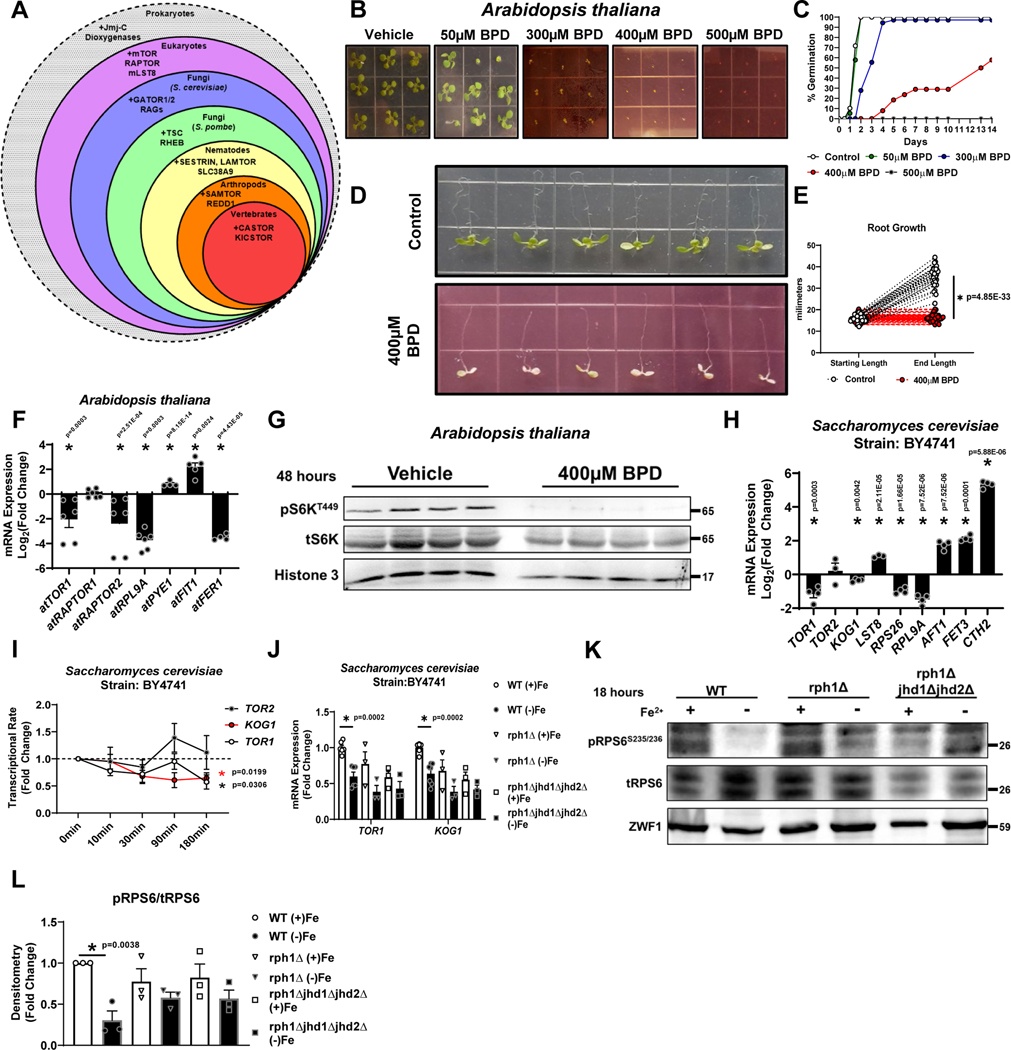
Animals, fungi and plants share homologues for mTOR, RPTOR, and mLST8, but other mTORC1 regulatory machineries are not conserved across kingdoms41. Homologues to Jmj-C KDMs are also present within all three kingdoms (Fig. 7A). To study whether mTORC1 regulation by ID and the role of Jmj-C KDMs in this process is evolutionarily conserved, we used Saccharomyces cerevisiae and Arabidopsis thaliana as model organisms. Iron chelation prevented germination and post-germination growth of stratified seeds, and root growth in 5-day old seedlings when grown on MS medium containing BPD, consistent with anabolic arrest (abiotic stress) in A. thaliana (Fig. 7B–E). Iron chelation also led to repression of atTOR, atRAPTOR2, atRPL9A (transcriptional marker of TOR activity) and the iron storage marker ferritin (atFER1), while the levels of the iron response genes atPYE1 and atFIT1 were upregulated (Fig. 7F). We also observed marked inhibition of A. thaliana S6K phosphorylation, demonstrating that TOR activity in A. thaliana is indeed regulated by iron (Fig. 7G).
Figure 7. ID leads to epigenetic repression of core mTORC1 genes in Arabidopsis thaliana and Saccharomyces cerevisiae.
(A) Schematic of conservation of mTOR components and its regulatory proteins among various species. (B) Representative images of post-germination growth of A. thaliana seeds germinating on MS medium containing indicated concentrations of BPD (14 days after stratification) from three independent experiments. (C) Summary graph of seed germination rates in panel B. Representative data from three independent experiments. (D) A. thaliana root growth on MS medium containing 0 (Control) or 400μM BPD for 4 days. Seedlings were transferred 5 days after stratification on BPD-free MS medium. (E) Summary bar graph of root length in panel (D) (n=30 replicates control; n=24 BPD, two-tailed unpaired t-test, mean ± SE). (F) mRNA expression of indicated TOR components and markers of ID and TOR activity in A. thaliana seedlings after treatment with 400μM BPD. Internal control: At ACTIN2 (n=6 replicates for all groups except n=4 for atPYE1 and atFER1, unpaired t-test, mean ± SE). A. thaliana seedlings were transferred at 5 days after stratification on MS medium to MS medium supplemented with or without BPD. Samples were collected 4 days post-transfer. (G) Immunoblot of A. thaliana TOR activity in seedlings treated with 400μM BPD for 48 hours. Representative image of one experiment. (H) mRNA expression of indicated TOR components and markers of ID in S. cerevisiae cells cultured in Fe dropout media containing 100μM BPS for 18 hours. Internal control: ScACT1 (n=4 replicates for all groups except n=3 for TOR2 and LST8, two-tailed unpaired t-test, mean ± SE). (I) Transcriptional rate of TOR1, TOR2 and KOG1 within 180 minutes of iron chelation in S. cerevisiae (n=3 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE, (black *) = P < 0.05 for TOR1, (red *) = P < 0.05 for KOG1). (J) mRNA expression of TOR1 and KOG1 in WT, ΔRph1 and ΔRph1/ΔJhd1/ΔJhd2 S. cerevisiae cells cultured in Fe dropout media containing 100μM BPS for 18 hours. Internal control: Sc ACT1 (n=6 replicates per condition for WT; n=3 ΔRph1 and n=3 ΔRph1/ΔJhd1/ΔJhd2, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (J) Immunoblot of TOR activity in WT, Δrph1, and Δrph1Δjhd1Δjhd2 cells cultured in Fe dropout media containing 100μM BPS for 18 hours. Representative image of three independent experiments. (L) Summary bar graph of image in panel K (n=3 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). Source numerical data and unprocessed blots are available in source data files. * indicates P value < 0.05 when noted for all panels.
In S. cerevisiae, ID resulted in decreased mRNA levels of TOR1, KOG1 (homologue of RPTOR), and RPS26 and RPL9A (transcriptional targets of TOR activity42), while markers of iron deficiency AFT1, FET3, and CTH2 were upregulated, confirming effective chelation and decreased TOR activity (Fig. 7H). To assess whether decreases in TOR1 and KOG1 mRNA were due to transcriptional repression, the transcriptional rate (TR) of TORC1 complex genes were determined using genomic run-on experiments under ID conditions43. Consistent with our steady-state RNA data, the TR for KOG1 and TOR1 decreased within 180 minutes of iron chelation (Fig. 7I). S. cerevisiae possess only three genes that encode functional Jmj-C KDMs (RPH1, JHD1, and JHD2) with conservation of the Fe2+ binding site motif [HXD/E…H]44. Although S. cerevisiae histones do not contain appreciable H3K9 methylation, the Jmj-C KDM Rph1 is thought to retain the ability to demethylate H3K9 methyl-marks45. Therefore, we tested the ability for rph1Δ and rph1Δjhd1Δjhd2Δ yeast mutants to repress TOR1 and KOG1 mRNA levels in response to ID. Yeast rph1Δ cells displayed partial repression of TOR1 and KOG1 at baseline, whereas rph1Δjhd1Δjhd2Δ cells demonstrated full repression of TOR1 and KOG1 mRNA at baseline and no response to ID (Fig. 7J). These findings were consistent with the level of phosphorylation of the TORC1 target Rps6, with rph1Δ and rph1Δjhd1Δjhd2Δ yeast demonstrating insensitivity to ID (Fig. 7K, L). Thus, the three iron-binding Jmj-C KDMs serve as major players in iron sensing in yeast.
Regulation of mTORC1 activity by ID is mediated through KDM3B
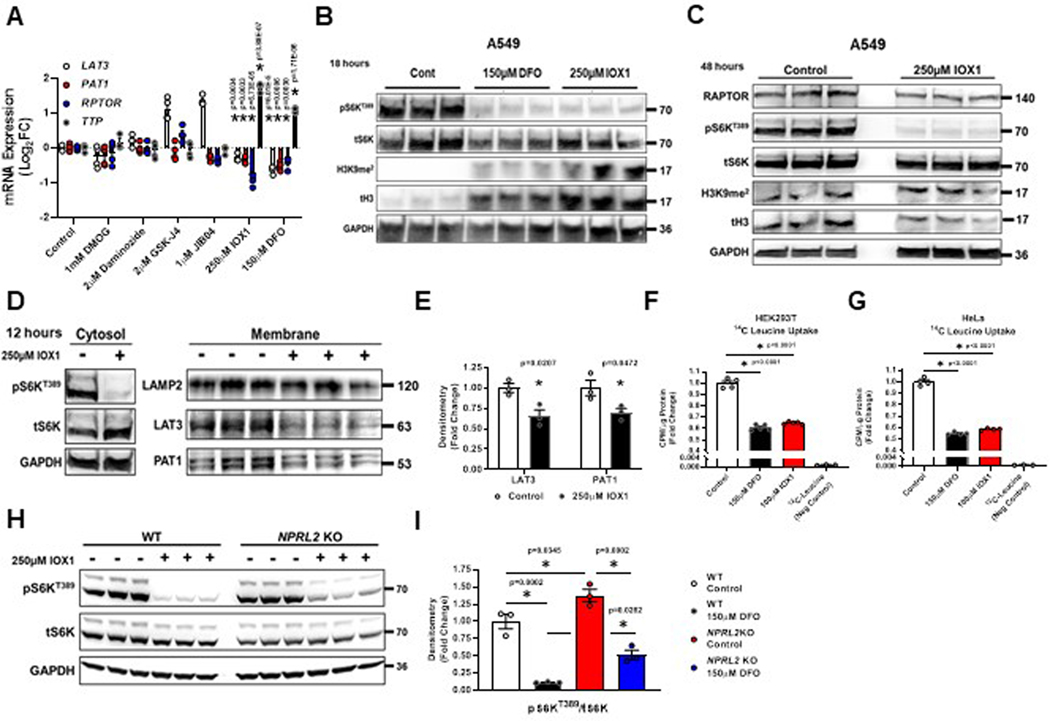
To identify which Jmj-C domain containing KDM family member(s) is responsible for mediating iron sensing upstream of mTORC1 in mammalian cells, we used pharmacologic inhibitors of various Jmj-C domain containing proteins. Administration of DMOG (EGLN), daminozide (KDM2/7), GSK-J4 (KDM6), and JIB-04 (KDM4/5/6) failed to mimic the effect of DFO on LAT3, PAT1, RPTOR and TTP mRNA expression (Figure S17A). Only the pan-JmjC inhibitor IOX1, which inhibits the KDM3 family and disrupts iron binding to KDM proteins46,47, resulted in simultaneous repression of LAT3, PAT1 and RPTOR mRNA (Extended Data Fig. 7A). Like DFO, IOX1 also increased the levels of H3K9me2 and repressed S6KT389 phosphorylation at both 18 and 48 hours (Extended Data Fig. 7B, C). Additionally, IOX1 reduced the levels of LAT3, PAT1 and RAPTOR protein (Extended Data Fig. 7C–E). IOX1 treatment also suppressed 14[C]-leucine uptake in HEK293T and HeLa cells to an equivalent extent as DFO (Extended Data Fig. 7F, G). Finally, the reduction in S6KT389 phosphorylation after IOX1 treatment was partially abrogated in NPRL2 KO 293T cells (Extended Data Fig. 7H, I).
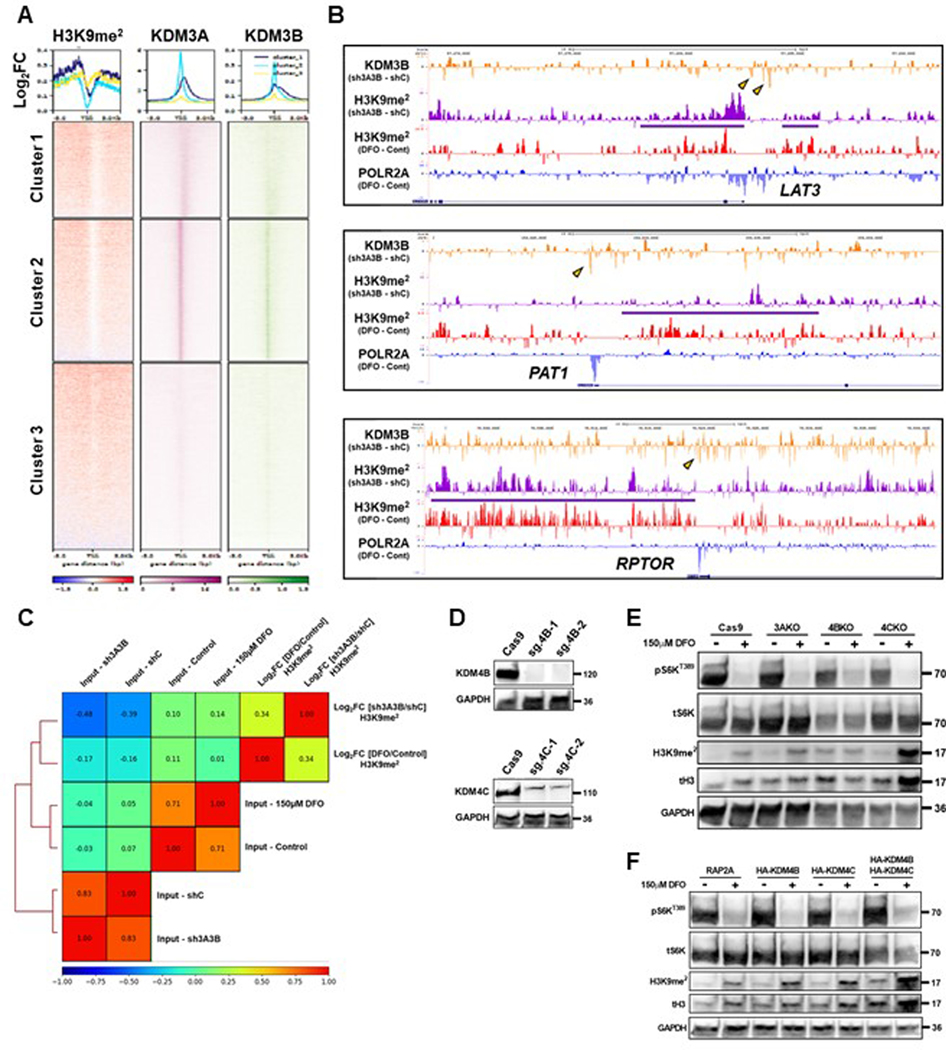
Of the Jmj-C demethylases, the KDM3 family members possess the most prominent activity towards H3K9me2 and H3K9me1 to maintain euchromatin (Figure 8A). We generated heatmaps of the log2FC in H3K9me2 signal within 5 Kb of TSSs between DFO treatment and control among genes with called POLR2A peaks and plotted them against published KDM3A and KDM3B ChIP-seq data48,49. Genes with the greatest fold change in H3K9me2 levels after ID corresponded with basal occupancy of KDM3A and KDM3B (Extended Data Fig. 8A). Analysis of double knockdown of KDM3A and KDM3B in HCT116 cells from published ChIP-seq data49 revealed loss of KDM3B occupancy and increased H3K9me2 signal at the loci of LAT3, PAT1, and RPTOR (Extended Data Fig. 8B). We next tested whether the increased H3K9me2 signal upon KDM3A/3B double knockdown correlated with ID-mediated changes in H3K9me2 signal generally across POL2RA occupied genes. Hierarchical clustering and correlation analysis revealed a higher positive Pearson correlation in Log2FC H3K9me2 signal between KDM3A/3B double knockdown and DFO treatment than that of the input tracks (Extended Data Fig. 8C). Together, these data suggest that KDM3A or KDM3B directly mediate iron-dependent changes in H3K9me2 levels as part of a general chromatin remodeling process, in which regulators of mTORC1 are specific targets.
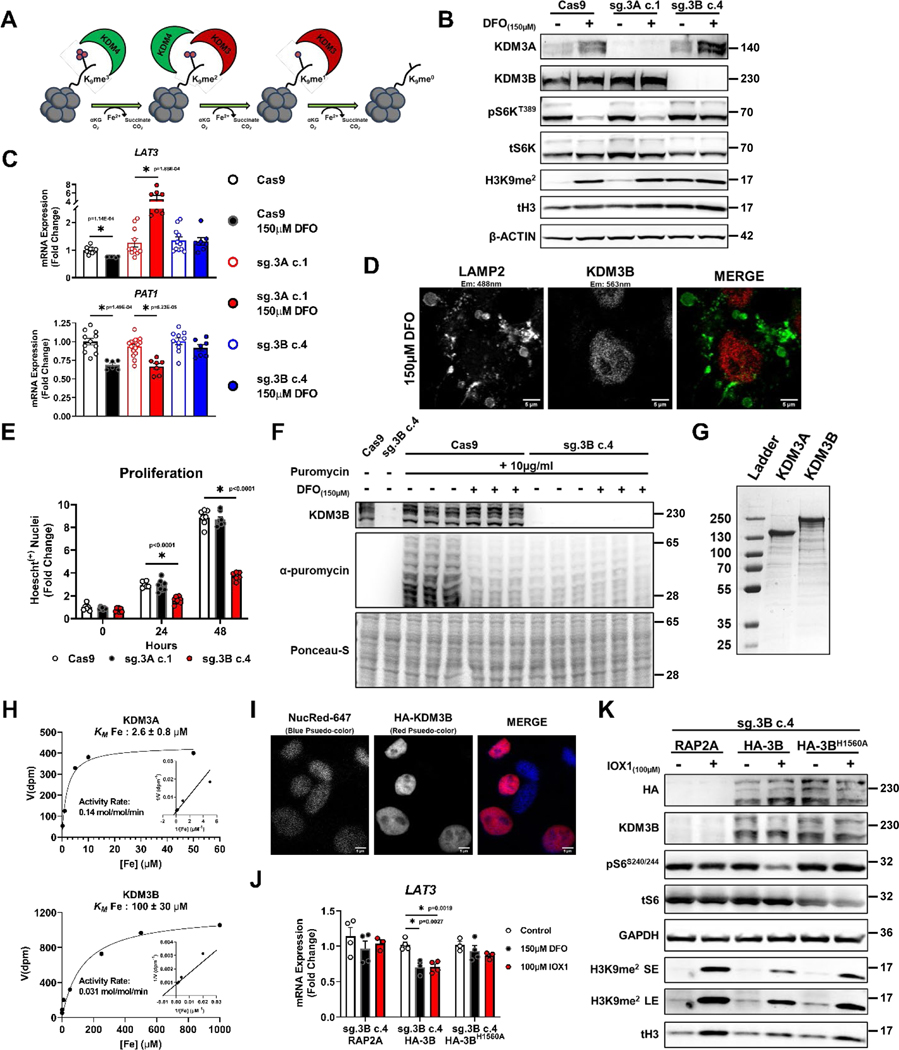
Figure 8. Regulation of mTORC1 activity by ID is mediated through KDM3B.
(A) Schematic presentation of the role of KDM3 and KDM4 family proteins in demethylating histone H3. (B) Immunoblot depicting successful editing of KDM3A and KDM3B in HEK293T cells transfected with CRISPR/Cas9 and indicated sgRNA followed by clonal selection. The experiments were performed in the presence and absence of 150μM DFO for 18 hours. Representative image of two independent experiments. (C) mRNA levels of LAT3 and PAT1 in HEK293 cell controls (Cas9), with KDM3A deletion (sg.3A c.1) and KDM3B deletion (sg.3B c.4) in the presence and absence of 150μM DFO for 18 hours. Internal control: 18S (LAT3: n=11 Cas9 control; n=6 Cas9 150μM DFO; n=11 sg.3A c.1 control; n=7 sg.3A c.1 150μM DFO; n=11 sg.3B c.4 control; n=8 sg.3B c.4 150μM DFO, PAT1: n=11 Cas9 control; n=6 Cas9 150μM DFO; n=16 sg.3A c.1 control; n=7 sg.3A c.1 150μM DFO; n=10 sg.3B c.4 control; n=7 sg.3B c.4 150μM DFO replicates, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (D) HEK293T cells stained with antibodies against LAMP2 and KDM3B in the presence of 150μM DFO for 18 hours, demonstrating the nuclear localization of KDM3B in ID. Representative image of five independent samples. (E) Quantification of viable WT, KDM3A KO, and KDM3B KO 293T cells over a 48-hour period, (n=7 replicates WT 0hr; n=8 KDM3A KO 0hr; n=8 KDM3B KO 0hr; n=6 WT 24hrs; n=8 KDM3A KO 24hrs; n=8 KDM3B KO 24hrs; n=7 replicates WT 48hrs; n=8 KDM3A KO 48hrs; n=6 KDM3B KO 48hrs;, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (F) Incorporation of puromycin into elongating peptide chains in HEK293T cells with and without KDM3B deletion and after treatment with 150μM DFO for 18 hours. Representative image of one experiment (G) SDS-PAGE gel of purified N-terminal FLAG-tagged murine KDM3A and human full-length KDM3B overexpressed using a baculoviral overexpression system. Representative image of three independent experiments (H) Enzyme kinetics of KDM3A and KDM3B in the presence of increasing concentrations of iron. (I) Nuclei staining of HEK293T cells with overexpression of HA-KDM3B to demonstrate proper nuclear localization of the protein. Representative image of five independent samples. (J) mRNA levels of LAT3 in response to 150μM DFO and 100μM IOX1 in KDM3B KO HEK293T cells with overexpression of RAP2A, WT KDM3B, and an iron-binding deficient mutant of KDM3B (KDM3BH1560A) Internal control: SNRK (n=4 replicates control RAP2A; n=4 150μM DFO RAP2A; n=3 100μM IOX1 RAP2A; n=4 replicates control HA-3B; n=3 150μM DFO HA-3B; n=4 100μM IOX1 HA-3B; n=3 replicates control HA-3BH1560A; n=4 150μM DFO HA-3BH1560A; n=3 100μM IOX1 HA-3BH1560A, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (K) Immunoblot of mTORC1 activity in KDM3B KO HEK293T cells with overexpression of WT KDM3B and KDM3BH1560A treated with 100μM IOX1 for 18 hours. Representative image of one experiment. Source numerical data and unprocessed blots are available in source data files. * indicates P value < 0.05 when noted for all panels.
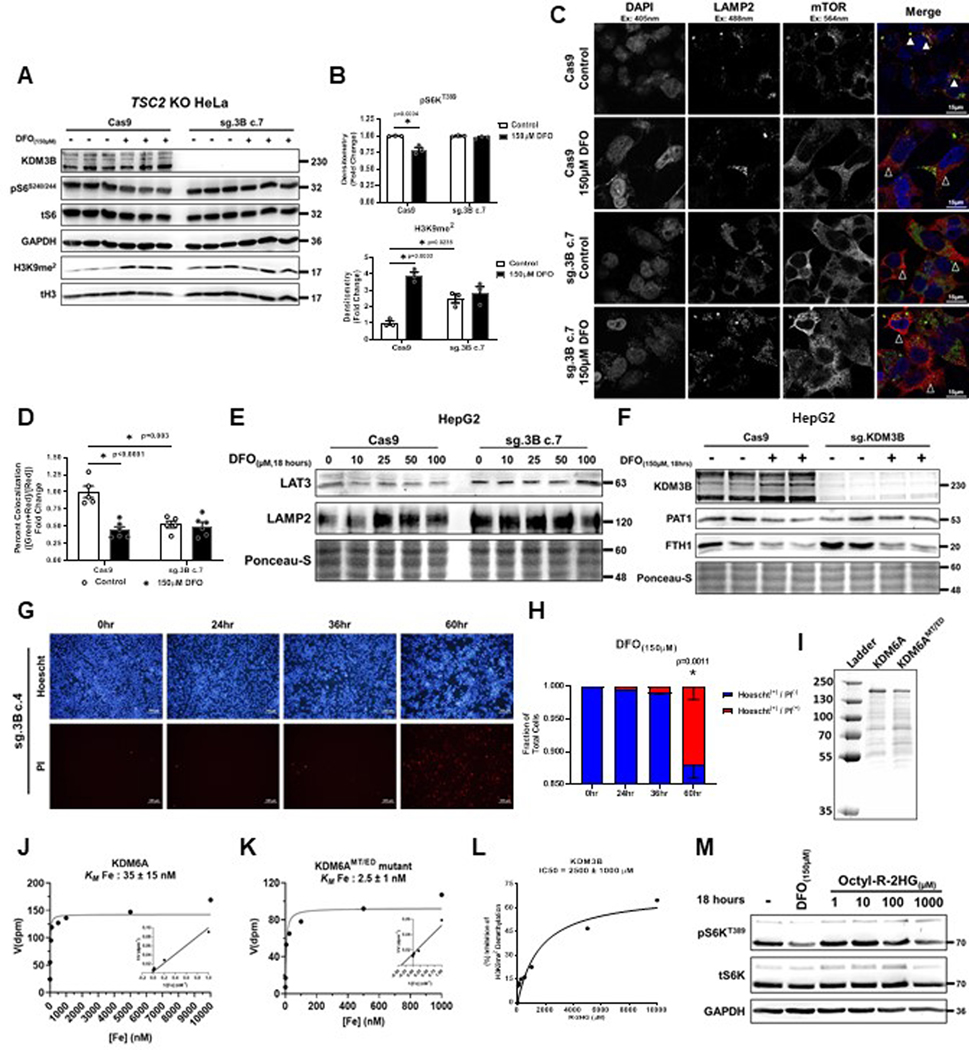
To identify which KDM3 member is responsible for sensing cellular iron levels and regulating mTORC1 activity, CRISPR-Cas9 was utilized to delete KDM3A and KDM3B in HEK293T cells (Fig. 8B). Given the ability for the KDM4 family to demethylate both H3K9me3 and H3K9me2 marks (Fig. 8A), we also generated KDM4B and KDM4C KO lines (Extended Data Fig. 8D). KDM4A was excluded since it was previously identified as a negative regulator of mTORC1 through its direct interaction with DEPTOR in the cytosol50. Additionally, the H3K9me2 demethylase PHF8 (KDM7B) was excluded based on the inability of its inhibitor daminozide to replicate the effects of DFO (Extended Data Fig. 7A). Consistent with our JIB-04 data, KDM4B and KDM4C KO cells did not demonstrate any difference in mTORC1 activity in response to DFO (Extended Data Fig. 8E). Overexpression of KDM4B and KDM4C, individually and more so in combination, increased basal H3K9me2 levels, consistent with their predominant role in catalyzing demethylation of H3K9me3 to H3K9me2 (Extended Data Fig. 8F). However, this had no effect on the increase in H3K9me2 levels or repression of mTORC1 activity in response to ID (Extended Data Fig. 8F).
Only KDM3B KO cells showed significantly increased H3K9me2 levels at baseline and failed to inhibit S6KT389 phosphorylation in response to ID, suggesting an inability for these cells to sense changes in iron levels (Fig. 8B). We confirmed these findings in an independent KDM3B KO line generated in TSC2 KO HeLa cells (Extended Data Fig. 9A, B). We also observed chronic dislocation of mTOR from the lysosome that was unresponsive to DFO treatment in KDM3B KO HepG2 cells (Extended Data Fig. 9C, D). The inability of mTORC1 to respond to ID in KDM3B KO cells correlated with a lack of LAT3 and PAT1 mRNA repression (Fig. 8C). Similarly, LAT3 and PAT1 protein expression was not repressed in KDM3B KO cells treated with DFO (Extended Data Fig. 9E, F). KDM3B remained within the nucleus and did not associate with lysosomal structures in ID cells (Fig. 8D), suggesting that KDM3B acts as a cellular iron sensor through its role in histone demethylation and not through an independent moonlighting function. KDM3B KO cells displayed significantly reduced proliferation and translation rates at baseline and ID-mediated repression of protein translation was abrogated in KDM3B KO cells (Fig. 8E, F). Similar to NPRL2 KO cells, KDM3B KO 293T cells demonstrated significantly increased cell death after 60 hours of ID (Extended Data Fig. 9G, H). These data are consistent with the inability of KDM3B KO cells to appropriately coordinate changes in mTORC1 activity with iron levels.
We determined the Michaelis constant for iron (Km[Fe]) of both KDM3A and KDM3B using affinity-purified, recombinant protein (Fig. 8G). The Km[Fe] for KDM3B was 100 ± 30μM (40-fold higher than KDM3A), suggesting that KDM3B is likely to respond to decreases in cellular iron before KDM3A (Fig. 8H). Additionally, the rate of catalysis for KDM3B was approximately 4.5-fold slower than KDM3A (apparent kcat for KDM3A: 0.14 mol/mol/min and KDM3B: 0.031 mol/mol/min) (Fig. 8H). The relative slowness by which KDM3B demethylates H3K9me2 parallels the delayed effect of ID on H3K9me2 levels and mTORC1 activity observed in living cells. Because the transcriptional response to ID and hypoxia share many features, we assessed whether the oxygen sensor KDM6A, which has a high Km[O2], has a similarly high Km[Fe]. Both WT KDM6A and KDM6AMT/ED mutant (which binds oxygen more tightly) had approximately 3×103 fold lower Km[Fe] than KDM3B, indicating that KDM6A does not function as a physiologic iron sensor and oxygen sensing by Jmj-C KDMs is not necessarily linked to iron sensing (Extended Data Fig. 9I–K).
We next tested whether KDM3B was sensitive to levels of the onco-metabolite R-2HG. Competitive inhibition studies performed on purified KDM3B protein determined that the IC50[R-2HG] was approximately 2.456 ± 1.04mM (Extended Data Fig. 9L), suggesting that KDM3B is a poor sensor of R-2HG levels. Treatment of HEK293T cells with increasing concentrations of octyl-R-2HG for 18 hours had no effect on mTORC1 activity at doses up to 1mM (the maximum potential dose given the limit of solubility of octyl-R-2HG) (Extended Data Fig. 9M).
Finally, we tested if expression of WT KDM3B in KDM3B KO cells could restore sensitivity to ID. We first confirmed that our overexpression construct properly localized to the nucleus (Fig. 8I). Expression of WT KDM3B, but not an iron-binding deficient mutant of KDM3B (KDM3BH1560A), restored LAT3 mRNA repression in response to DFO and IOX1 (Fig. 8J) as well as the ability of cells to repress mTORC1 activity in response to IOX1, demonstrating that the ability for KDM3B to bind iron is requisite to function as a regulator of mTORC1 activity (Fig. 8K).
Discussion
Iron signaling has traditionally been thought to be handled by the dedicated IRP system, which is primarily involved in iron acquisition from the environment8. Here we show that iron is necessary for active histone demethylation and that cells dynamically regulate chromatin in response to the availability of iron in the environment. We also find that the anti-anabolic effect of ID is mediated t mTORC1 and by KDM3B-directed changes in H3K9me2 levels in enhancer regions of genes necessary to facilitate mTORC1 activity. KDM3B requires iron to demethylate lysine residues on histones, and our studies demonstrate that cellular iron levels get sufficiently low to cause inhibition of this protein both in vitro and in vivo. The observation that this system can be activated in mice with systemic ID and in patient-derived primary tumor cells exposed to iron chelators highlight the physiological significance of our findings. Overall, our data demonstrate the presence of a sophisticated and evolutionarily conserved iron sensing mechanism that is engaged to shut down anabolic processes in cases of prolonged ID. This pathway has profound implications for proliferative diseases, which rely heavily on iron and mTORC1-mediated anabolism.
The observation that the inhibition of mTORC1 activity in response to ID is conserved in other eukaryotic organisms, including yeast and plants, suggests that this pathway evolved prior to the introduction of more recent iron sensors such as the IRP, AFT and FIT proteins, which are restricted to different phylogenetic kingdoms. Additionally, by demonstrating that iron can exert transcriptional control over RAPTOR in yeast, plants and mammals, and TOR in plants and yeast, we propose a paradigm in which the core mTORC1 complex was initially developed as a pro-anabolic mediator of iron sufficiency, and later evolved to integrate signals from additional nutrients and growth factors.
We found no evidence that cells chelated with 150μM DFO enter fulminant “metabolic collapse”, a state characterized by ATP depletion, p53 activation, and irreversible progression to cell death51. To the contrary, our data indicate DFO treated cells undergo a normal response to metabolic stress, enabling them to maintain cellular ATP levels, mitigate p53 activation, and preserve their viability. Additionally, supplementation of essential metabolites whose synthesis relies on iron-dependent proteins did not reactivate mTORC1, suggesting that loss of these metabolites in ID does not contribute to mTORC1 inhibition in this setting. However, addition of LLOME in ID was sufficient to rapidly relocate mTORC1 to the lysosome and rescue mTORC1 activity, which suggests that mTORC1 inactivation by ID is due to a dedicated signaling pathway and not generalized metabolic collapse. We attribute the delayed effect of ID on mTORC1 activity to the time it takes to turnover LAT3 and RAPTOR proteins while transcription of these genes is repressed.
The human genome encodes more than sixty members of the 2-oxoglutarate (αKG)-dependent dioxygenases family52. Because all members of this family require iron, oxygen and αKG for catalytic activity, a major question has focused on the differential effects of a reduction in cellular iron (and oxygen) levels on these proteins. Our data, along with those reported on oxygen sensing by KDM6A53, indicate that only certain members of this family have a Km for these substrates that is high enough to sense their physiological levels inside the cell. We propose that, among dioxygenases, the EGLN family of prolyl-hydroxylases and KDM3B are likely the first to respond to cellular ID, leading to HIF activation and chromatin remodeling. The combined effect of these two processes promotes a metabolic switch to anaerobic glucose catabolism and repression of anabolism, which are both necessary to survive prolonged ID.
Our study has some limitations, and a number of unanswered questions remain. Whether ID specifically disrupts leucine import at the cell membrane or prevents the loading of leucine into the lysosome and its subsequent efflux therefrom, a process termed “inside-out” signaling33,54–56, is not clear. Since leucine sensing by mTORC1 evolved subsequent to the Jmj-C KDMs, it is possible that KDM3B regulates, the expression of yet unidentified proteins necessary to facilitate the appropriate subcellular trafficking of leucine. Additionally, expression of the various Jmj-C proteins can vary across different tissues and the Km[Fe] for some KDMs are still unknown57. There are likely additional KDMs capable of regulating H3K9 and other histone methyl-marks in response to ID. We have observed that sequential passaging of KDM3B KO cells resulted in the return of H3K9me2 to basal levels and coincided with mTORC1 activity regaining sensitivity to ID, despite the continued absence of KDM3B. This underscores the essentiality of cellular iron sensing through the demethylation of H3K9 and suggests a layer of redundancy through potential compensation by other KDMs.
Continued challenges in determining whether a KDM member is a physiologic iron sensor include the lack of methods to accurately measure the free iron concentration in the nucleus, making predictions of percent metalation of given KDMs challenging. Finally, a crucial question when defining a nutrient sensor is whether the candidate sensor functions as a generalist, regulating multiple pathways in tandem, or are its effects limited to a specific function. Known nitrogen sensors for mTORC1 (i.e., SESN, SAMTOR, and CASTOR) act as dedicated regulators of GATOR1 and GATOR2 through direct protein-protein interactions. In contrast, the cellular oxygen sensors HIF1/2α, KDM5A, and KDM6A, operate more broadly, and exert their regulatory effects through genome-wide changes in histone acetylation and methylation, respectively. In the case of iron sensing through KDM3B, our data suggest a model in which the regulation of mTORC1 is one part of a global adaptive response to ID and suppression of anabolic processes is a specific effect mediated through the repression of mTORC1 at the epigenetic level.
Methods
Our research complies with all relevant ethical regulations and guidelines. All animal studies were approved by the Institutional Animal Care and Use Committee at Northwestern University and were performed in accordance with guidelines from the National Institutes of Health. Human tumor sample was collected according to Northwestern Institutional Review Board (IRB) approved protocols.
Cell lines and culture
HEK293 (CRL-1573), HEK293T (CRL-3216), and HepG2 (HB-8065) cells were obtained from ATCC. Ampkα1/2 dKO MEFs and A549 cells were generous gifts from Dr. Navdeep Chandel. Arnt KO, Tsc2 KO, and Irp2 KO MEFs were described previously58. SESN1/2/3 triple KO and NPRL2 KO HEK293T cells were generous gifts from Dr. David Sabatini. RragaQ66L and corresponding WT MEFs were generous gifts from Dr. Efeyan Alejo. HEK293T rtTA3/FPN-GFP stable cells were a gift from Dr. Yatrik Shah. All cells were cultured in Dulbecco’s Modified Eagle’s Medium with high glucose, 1mM sodium pyruvate, and 2mM glutamine (Hyclone) supplemented with 10% FBS (Atlanta Biologicals) unless otherwise indicated. Atf4 KO MEFs were grown in the aforementioned media supplemented with 10μM 2-mercaptoethanol (Sigma) and 10μM L-cystine (Sigma). Experiments on modified transferrin saturation were performed in serum free DMEM supplemented with 1% BSA (Goldbio),10μg/ml insulin (Sigma), 10ng/ml hEGF (Thermo), 10μM 2-mercaptoethanol, 0.04μM sodium selenite (Sigma), 1:1000 chemically defined lipid concentrate (Thermo), and indicated ratios of holo- and apo-Transferrin (Sigma) totaling 2.25μM. For experiments involving FPN overexpression, 24 hours after transfection, cells were washed once with PBS and switched to DMEM + 5% FBS with doxycycline or vehicle for the indicated times. For experiments involving serum starvation, cells were washed once with PBS and switched to DMEM without FBS for the indicated times. For experiments involving total AA deprivation, cells were washed once with PBS and switched to HBSS (Fisher Scientific) + 10% FBS + 25mM glucose (Sigma) + 1mM sodium pyruvate (Fisher Scientific) for the indicated times. For experiments involving leucine deprivation, cells were washed once with PBS and switched to leucine-free RPMI-1640 (United States Biologicals) with 10% FBS for the indicated times.
Mouse lines and iron deficient diet
C57BL/6 mice were housed in the barrier facility at Northwestern University with 12 h light and 12 h dark cycle, in ventilated cages with bedding and standard enrichment (temperature: 68–72ºF, humidity: 30–60%). Mice received either normal chow (TD 2916, Harlan-Teklad) or iron deficient diet containing 2–6 ppm of iron (TD 80394, Harlan-Teklad), which was fed to weaned P21 pups and continued until P28. All animal studies were approved by the Institutional Animal Care and Use Committee at Northwestern University and were performed in accordance with guidelines from the National Institutes of Health. IACUC protocols approved under the corresponding author used in this study include IS00019227, IS00015124, IS00013689, IS00006808, IS00002409, IS00001404, IS00000777, IS00000763.
Isolation and culturing of primary hepatocytes
After anesthetization with Avertin (created from freshly prepared tribromoethanol; Sigma), the portal vein of each mouse was cannulated with a 24 G × 3/4-inch i.v. catheter (Terumo), and the liver was perfused with Ca2+- and Mg2+-free HBSS (Fisher Scientific) for 5 minutes, followed by perfusion DMEM containing 0.05% type IV collagenase (Worthington) for 5 minutes. The perfusate was drained by an incision of the femoral artery. After perfusion, the liver was removed and placed in ice-cold DMEM (with 4.5 g/l glucose and L-glutamine with no sodium pyruvate, Cytiva) and cells were gently released. The cell suspension was filtered through a 100-μm nylon strainer (BD Falcon, BD Biosciences) and centrifuged at 50 g for 1 minute. The cell pellets were washed 3 times with ice-cold DMEM and then resuspended in DMEM containing 5% FBS (Atlanta Biologicals). Cell viability (~80%) was determined by trypan blue exclusion. Cells were plated on type I collagen–coated dishes at a density of 5 × 104 cells/cm2 and incubated at 37°C under 5% CO2. After 1.5 hours of attachment, the media was aspirated, cells were washed with PBS and incubated in complete media (DMEM containing 10% FBS and 1X antibiotic-antimycotic). Experiments involving cultured primary hepatocytes were completed within 48 hours of isolation.
Generation and culture of hIPS-Neurons
iPSCs were differentiated into cortical glutamatergic neurons using a modified version of a protocol based on Ngn2 overexpression59–61. This protocol exploits the combinatorial effects of small molecules that inhibit the TGF-β/SMAD pathways (to efficiently neuralize iPSCs) 62, and the exogenous expression of Ngn2 gene under Tet-inducible promoter. They give rise to homogenous populations represented by >85% vGLUT2/MAP2-positive neurons. Stem cells were dissociated as single cells using Accutase, re-suspended in mTeSR1 with 10 μM ROCK inhibitor (Y-27632, DNSK International, #129830–38-2), then incubated with lentiviruses (FUW-M2rtTA, TetO-Ngn2-Puro, TetO-FUW-EGFP) in suspension for 5 min before plating (95,000 cells/cm2). After 24 h (day 1), lentivirus was removed and replaced with knockout serum replacement medium (KOSR) consisting of KnockOut DMEM supplemented with Knockout replacement serum KSR, nonessential amino acids (NEAA), Glutamax (Life Technologies), 55 μM β-mercaptoethanol (Gibco, Cat# 21985023), 10 μM SB431542 (DNSK International), 100 nM LDN-193189 (DNSK International), 2 μM XAV939 (DNSK International) and 3 μg/ml of doxycycline (Sigma). On the following day (Day 2), media was replaced with a 1:1 ratio of KOSR to neural induction media (NIM) composed of DMEM: F12 supplemented with NEAA, Glutamax, N2 (Gibco, Life Technologies), 0.16% D-glucose (Sigma) and 2 μg/ml heparin sulfate (Sigma). Doxycycline (2 μg/ml) and puromycin (2 μg/ml; Sigma) were added to this NIM media. On Day 3, the media was replaced with NIM containing doxycycline (3 μg/ml) and puromycin (2 μg/ml). All neurons were frozen in 10% DMSO/Hyclone FBS (VWR) on Day 4. For all experimental analysis, iPSC-derived neurons were plated on primary CD1 mouse cortical glia, derived as previously described63. Glial cells were first plated on PDL/laminin-coated plates or coverslips in glia media composed of MEM (Life Technologies) supplemented with Glutamax (0.6%), D-glucose, and 10% horse serum (Life Technologies). After 5–7 days, neurons were thawed (Day 5 post-induction) and plated, at a density of 20,000/cm2, directly onto the monolayer of mouse glia in Neurobasal medium (NBM), supplemented with NEAA, Glutamax, N2 and B27 (Life Technologies) containing BDNF (10 ng/mL, R&D systems), 2% Hyclone FBS, doxycycline (3 μg/ml) and ROCK inhibitor. Half of the media was replaced the next day and then every other day thereafter with NBM supplemented with NEAA, Glutamax, N2 and B27 containing BDNF (10 ng/mL), 2% Hyclone FBS and doxycycline (2 μg/ml).
Generation and culture of hIPS-CM
Pluripotent and reprogramming cells were generated as previously described64. In summary, cells were maintained at 37 ºC in Heracell VIOS 160i humidified incubators (Thermo Scientific) with 5% CO2 and 5% O2. During differentiation cells were maintained at 5% CO2 and atmospheric O2. All cultures (pluripotent and differentiation) were maintained in 2 mL medium per 9.6 cm2 of surface area or equivalent. All cultures were routinely tested for mycoplasma using a MycoAlert PLUS Kit (Lonza) and a Varioskan LUX (Thermo Scientific) plate reader. The hIPSC line 19c3 was previously derived from peripheral blood mononuclear cells of a healthy male using Sendai virus (Invitrogen) using B8. Protocols were approved by the Northwestern University Institutional Review Board. This hIPSC line was modified to express an exogenous TNNT2 promoter-driven Zeocin resistance cassette for cardiomyocyte purification. hIPSCs were passaged at a ratio of 1:15 every 4 days using 0.5 mM EDTA for 6 min at RT, achieving 80% confluence. Cells were routinely maintained in B8 medium on 1:800 growth factor reduced Matrigel (Corning) diluted in DMEM (Corning), except for the first 24 h after passage when B8 was supplemented with 2 μM thiazovivin (LC Labs, T-9753), hereby referred to as B8T medium. Differentiation into cardiomyocytes was performed according to previously described protocol with slight modifications65,66. Briefly, hIPSCs were split at a 1:15 ratio using 0.5 mM EDTA and grown in B8 medium for 4 days reaching ~75% confluence. At the start of differentiation (day 0), B8 medium was changed to R6C, consisting of RPMI 1640 (Corning, 10–040-CM), supplemented with 6 μM of glycogen synthase kinase 3-β inhibitor CHIR99021 (LC Labs, C-6556). On day 1, medium was changed to RPMI 1640 basal medium alone, and on day 2 medium was changed to RBA-C59, consisting of RPMI 1640 supplemented with 2 mg/mL fatty acid-free bovine serum albumin (GenDEPOT, A0100), 200 μg/mL l-ascorbic acid 2-phosphate (Wako, 321–44823) and 0.5 μM Wnt-C59 (Biorbyt, orb181132). Medium was then changed on day 4 and then every other day with RBAI consisting of RPMI 1640 supplemented with 500 μg/mL fatty acid-free bovine serum albumin, 200 μg/mL l-ascorbic acid 2-phosphate, and 1 μg/mL E. coli-derived recombinant human insulin (Gibco, A11382IJ). Contracting cells were noted from day 7, differentiated cardiomyocytes were treated with 25 μg/mL of Zeocin from day 10 to day 14. On day 20 of differentiation, cardiomyocytes were dissociated using DPBS for 20 min at 37 °C followed by 1:200 Liberase TH (Roche, 5401151001) diluted in DPBS for 20 min at 37 °C, centrifuged at 300 × g for 5 min, and filtered through a 100 μm cell strainer (Falcon). Cells were then plated in RBAI+10% Cosmic Calf Serum (Hyclone) for 2 days on 1:800 Matrigel-coated plates for each assay, media was then switched back to RBAI which was changed every 2–3 days and cells were assayed on d30.
Patient-derived tumor cells (PDC)
Primary cells were established from a resected cholangiocarcinoma (T3N2, + perineural invasion). Patient had no history of neo-adjuvant chemotherapy. Tumor sample was collected according to Northwestern Institutional Review Board (#STU00007180) using an opt-in consent process. No financial compensation was provided to the donor for participation in this study. To establishment a primary tumor cell culture, tumor tissue was dissected into small pieces < 1mm3 and digested in a mixture of collagenase 2 (Worthington) and trypsin (cytiva). Cell culture media (DMEM supplemented with 10% FBS and 1X antibiotic-antimycotic (Corning) was added 1:1, and undigested debris was pelleted by centrifugation. The supernatant containing primary tumor cells was passed through a 45μM filter to achieve a single cell suspension. Cells were washed 2x with fresh media, counted and plated into flasks. The cells were passaged once and expanded to generate frozen stocks. For experiments, cells were thawed, subcultured and used within 5 passages.
Organ harvesting
At the time of tissue harvest, mice were anesthetized with 250 mg/kg dose of freshly prepared Tribromoethanol (Avertin; Sigma) and tissue was excised and rinsed in phosphate buffered saline to remove excess blood. The tissue to be processed for biochemical assays was then freshly frozen in liquid nitrogen and stored at −80 °C until the assay.
Drug treatments
All chemicals used in for experiments were dissolved in either H2O, DMSO, or 100% EtOH according to their specification sheets. A complete list of chemical compounds can be found in Supplementary Data Table 1. All treatments began when cells reached approximately 60% confluency unless otherwise noted.
Gene downregulation, overexpression, and generation of knockout cell lines using Crispr-Cas9
siRNAs against mouse Irp1, human REDD1, LAT3, and PAT1 (Dharmacon) were transfected using Dharmafect I Transfection Reagent (Dharmacon) according to the manufacturer’s protocols. siRNA concentrations of 25nM were used to achieve knockdown. Transfection into MEFs and HeLa cells were performed using Lipofectamine 3000 according to manufacturer’s protocols (Thermofisher). All other transfections were performed in HEK293T cells using either polyethylenimine (Polysciences) or the CaPO4 method. A list of all plasmids can be found in Supplementary Data Table 1. Transient transfections were performed 48 hours prior to initiation of experiments. In cases where plasmids conferred resistance to puromycin, transfected, cells were selected 24 hours after transfection using 4μg/ml puromycin (Sigma) for 24 hours. To knockout KDM3A, KDM3B, KDM4B, and KDM4C in HEK293T and KDM3B in TSC2 KO HeLa and HepG2 cells, small guide RNAs (sgRNA) were designed using CRISPOR67 (http://crispor.tefor.net/crispor.py) and cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene). A complete list of guide sequences and the exons they target can be found in Supplementary Data Table 1. After cloning, sgRNA-Cas9 plasmids were transfected into cells and selected with 4μg/ml puromycin for 48 hours. Individual cells were then seeded into 96-well plates and expanded as clones. Clonal cells were screened via immunoblotting to confirm knockout status.
RNA isolation, reverse transcription and quantitative real time (RT)-PCR
RNA was isolated from cells or tissues using RNA-STAT60 (Teltest). Reverse transcription was carried out using qScript cDNA Synthesis Kit (Quanta Bio). The resulting cDNA was amplified quantitatively using PerfeCTa SYBR Green Mix (Quanta Bio) on a 7500 Fast Real-time PCR System (Applied Biosystems). The relative gene expression was determined using differences in Ct values between gene of interest and control genes. A complete list of primers is included in Supplementary Data Table 1.
Immunoblotting and co-immunoprecipitation
Cells and tissue were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with 1X Protease Arrest (Fisher Scientific), and phosphatase inhibitors sodium fluoride (10mM, Sigma) and sodium pyrophosphate (15mM, Sigma). Protein concentration in samples was determined using the BCA Protein Quantification Kit (Pierce). Equal amounts of protein were loaded on a tris-glycine polyacrylamide gel (Life Technologies) and transferred to nitrocellulose membrane. After blocking with tris-buffered saline containing 0.05% Tween 20 (Fisher) and 5% BSA, the membrane was incubated O/N at 4°C in primary antibody against indicated proteins. The following day, membranes were incubated with HRP-conjugated anti-mouse or anti-rabbit secondary antibodies (Jackson Immunoresearch) for 2 hours at RT or O/N at 4°C. Immunoblots were developed using chemiluminescent substrate (Pierce). A complete list of primary antibodies is included in Supplementary Data Table 1. For co-IP experiments, transfected cells were lysed in IP-buffer (0.3% CHAPS, 40mM HEPES, 2mM EDTA, 1X Protease Arrest and phosphatase inhibitors). Protein content was quantified and 400μg −1mg of protein was loaded onto 40μl of precleared packed protein G agarose beads (Sigma) with anti-FLAG M2 antibody (Sigma) and incubated O/N at 4°C. The following day, the beads were washed 4x in wash buffer (lysis buffer + 150mM NaCl) followed by boiling the samples in 50μl of 1x Laemmli sample buffer (Boston BioProducts) for 15min at 95°C. CoIP samples were then loaded onto tris-glycine polyacrylamide gels and processed as described above.
Cell proliferation and viability experiments
For IF experiments, cells were seeded in 12-well plates coated in 0.02% gelatin + 5μg/ml fibronectin. At the time of the assay, cells were washed once with PBS and incubated with 10μg/ml Hoescht 33342 and 5μg/ml propidium iodide diluted in 1x HBSS for 15 minutes. After incubation, excess dye was washed away with 1x HBSS, and images were taken using a Zeiss Axio Observer.Z1 fluorescence microscope. Viable cells were quantified using ImageJ as the fraction of Hoescht(+) and PI(−) over total Hoescht(+) cells. For flow cytometry experiments, cells were seeded in TC-treated 10cm dishes at 2.5×106 cells/dish. At the time of the assay, the supernatant containing dead, floating cells were collected into 50ml conical tubes. Adherent cells were trypsinized and then added to the same conical tubes. Cells were pelleted, washed 1x in phenol-free DMEM containing 10% serum, and suspended in the same media to a concentration of 1×106 cells/ml. 100μl aliquots containing 1×105 cells were prepared and cells were stained by adding 5μL Annexin-V AlexaFluor 488 conjugate (Thermo A13120) to each tube. After 15 minutes, 400μL of phenol-free DMEM + 10% serum was added together with propidium iodide at a final concentration of 1μg/mL. No-stain and single stain control samples were also prepared. Samples were then transferred to ice and flowed on a BD FACSAria SORP system and analyzed using BD FACSDiva 8.0.1 analysis software.
Quantification of cellular metal content by ICP-MS
Cells (1.7 × 104 cells/cm2) were seeded in T225 flasks (Cat# 431082, Corning, Inc, Corning, NY), and two T225 flasks were used for quantification of intracellular inorganic elements at each time point. After 24 h incubation in a 95% CO2 5% O2 incubator at 37 °C, cells were treated with DFO (0.15 mM) for 0, 3, 6, 12, and 24 h, respectively. They were harvested at appropriate time points immediately following a short (<2 min) trypsin-EDTA (Cat# 25200056, Thermo Fisher Scientific, Waltham, MA) treatment at room temperature. ICP-MS samples were prepared using the Wash-Free Gd ICP-MS Assay (unpublished manuscript). Cells were pelleted, weighed, and digested in concentrated nitric acid together with a reference element Gd-DOTA (Cat# M-147, Macrocyclics, Plano, TX). Each element concentration was measured by ICP-MS that was performed on a computer-controlled (QTEGRA software) Thermo iCapQ ICP-MS (Thermo Fisher Scientific, Waltham, MA) operating in KED mode and equipped with an ESI SC-2DX PrepFAST autosampler (Omaha, NE). A set of element calibration standards (0.01, 0.05, 0.1, 0.5, 1, 3, 5, 10, 15, 20, 40, 100, 200, and 400 ppb) were prepared by diluting the multiple element ICP standard solution (Cat# NWU-15, Inorganic Venture, VA) with 3 wt % nitric acid in ultrapure deionized water. Sample element concentrations were determined in Kinetic Energy Discrimination (KED) mode with 10% H2/He gas to minimize polyatomic interferences. Ultrapure deionized water (18.2 MΩ cm resistivity) was obtained from a Milli-Q Biocel system with Q-Gard® 2 Purification Cartridge (Millipore Inc., Billerica, MA).
Non-heme iron quantification
Cells were lysed in RIPA buffer, spun at 16,000xg for 15 minutes and the supernatant collected. Equal amounts of protein were mixed with protein precipitation solution (1:1 1 N HCl (Sigma) and 10% trichloroacetic acid (Sigma)) and heated to 95 OC for 1 hour to release iron. Precipitated protein was removed by centrifugation at 4 OC at 16,000xg for 10 min, the supernatant was mixed with the equal volume of chromogen solution (0.5 mM ferrozine (Sigma), 1.5 M sodium acetate (Sigma), 0.1% [v/v] thioglycolic acid (Sigma)), and the absorbance was measured on a Spectra Max Plus microplate reader at 562nm. Absolute iron values were determined was based on a standard curve generated using ferric ammonium citrate (Sigma).
Total cellular amino acid measurements
Total cellular amino acid levels (free + peptide) were determined using a Beckman 121 MB amino acid analyzer. Cell pellets were hydrolyzed with 6M HCl for 24 h at 110 °C in sealed tubes after replacing oxygen with nitrogen. The solution obtained was filtered through a 0.25μm membrane filter and then analyzed for amino acids.
Seahorse Assay
The day before the assay, the Seahorse cartridge was placed in ddH2O overnight at 37°C in a non-CO2 incubator and one hour prior to the assay switched to the XF calibrant. 48 hours prior to the assay, cells were seeded into the Seahorse 96-well plate at 15,000 cells/100μl per well and cultured in normal culture media. 24 hours later, DFO was added to wells at the indicated concentrations. Just prior to the assay, the plates were washed and incubated for one hour in serum-free DMEM without bicarbonate or phenol-red supplemented with 11mM glucose, 1mM Na-pyruvate, and 2mM L-glutamine. Oligomycin (1.5μM final), CCCP (1μM final), and Rotenone/Antimycin A (2μM each final) were diluted in DMEM and 30μL were added to ports A, B, and C. Drugs were plunged sequentially into all wells at the indicated times. After the run was complete, the assay buffer was removed and 50μL RIPA buffer was added, followed by measurement of protein concentration for normalization.
Metabolomics
Metabolomics services were performed by the Metabolomics Core Facility at Robert H. Lurie Comprehensive Cancer Center of Northwestern University. Samples were analyzed by High-Performance Liquid Chromatography and High-Resolution Mass Spectrometry and Tandem Mass Spectrometry (HPLC-MS/MS). Specifically, system consisted of a Thermo Q-Exactive in line with an electrospray source and an Ultimate3000 (Thermo) series HPLC consisting of a binary pump, degasser, and auto-sampler outfitted with an Xbridge Amide column (Waters; dimensions of 4.6 mm × 100 mm and a 3.5μm particle size). The mobile phase A contained 95% (vol/vol) water, 5% (vol/vol) acetonitrile, 20mM ammonium hydroxide, 20mM ammonium acetate, pH = 9.0; B was 100% Acetonitrile. The gradient was as following: 0 min, 15% A; 2.5 min, 30% A; 7 min, 43% A; 16 min, 62% A; 16.1–18 min, 75% A; 18–25 min, 15% A with a flow rate of 400 μL/min. The capillary of the ESI source was set to 275 °C, with sheath gas at 45 arbitrary units, auxiliary gas at 5 arbitrary units and the spray voltage at 4.0 kV. In positive/negative polarity switching mode, an m/z scan range from 70 to 850 was chosen and MS1 data was collected at a resolution of 70,000. The automatic gain control (AGC) target was set at 1 × 106 and the maximum injection time was 200ms. The top 5 precursor ions were subsequently fragmented, in a data-dependent manner, using the higher energy collisional dissociation (HCD) cell set to 30% normalized collision energy in MS2 at a resolution power of 17,500. The sample volumes of 10μl were injected. Data acquisition and analysis were carried out by Xcalibur 4.0 software and Tracefinder 2.1 software, respectively (both from Thermo Fisher Scientific).
3H-leucine and 14C-leucine uptake
After cells were subjected to the indicated treatment in complete media for the indicated times, cells were washed with once with PBS and then incubated with 800nM 3H-leucine or 14C-leucine (Perkin Elmer) diluted in leucine-free RPMI-1640 w/ 10% FBS for 5–60 minutes. Then cells were washed two times with cold PBS to remove residual extracellular radioactive leucine, and immediately lysed in RIPA buffer. Protein content of each sample was determined by BCA assay. Radioactivity of the lysate was determined by scintillation counting and normalized to the protein concentration of each sample.
35S-methionine based measurement of protein synthesis
HEK293T cells were preloaded with 35S-methionine (Perkin Elmer) for two passages and then plated in 10cm dishes at 60% confluency. At the start of the experiment, cells were washed 3x with PBS and chased for 24 hours in normal growth media containing either iron chelator or vehicle control. Cells were washed 1x with PBS and immediately lysed in RIPA buffer. The protein fraction was precipitated with 10% trichloroacetic acid. Pellets were washed once with acetone and resolubilized with 1N NaOH with 0.1% SDS. Radioactivity of the resolubilized protein fraction was determined by scintillation counting and normalized to the protein concentration of each sample as determined by BCA assay.
Confocal imaging
For immunocytochemistry, cells were plated on 18×18mm coverglass coated with 5 μg/ml fibronectin (Sigma) in 0.2% gelatin (BD) and treated as described. At the end of the experiment, cells were fixed using 4% paraformaldehyde for 10 min and subsequently washed 3x in 1X PBS. Cells were permeabilized in 1X permeabilization solution (PBS + 0.25% triton x-100 (Sigma)) for 1 hour and then incubated with designated primary antibodies overnight at 4O C in PBS-T+ (PBS + 0.05% Triton x-100 + 5% donkey serum (Sigma)). Coverslips were then washed 3x in 1X PBS-T and stained with secondary antibodies (Jackson immunochem) in PBS-T+ for 2 hours, washed 3x in 1X PBS, and then mounted on slides using Prolong Diamond mounting media (Life Technologies). A list of all primary and secondary antibodies is provided in Table S2. For imaging of lysosomal acidification, cells were incubated with acridine orange (Invitrogen) at 2μg/ml for 15minutes and washed 1x with HBSS (Fisher) before imaging in 1X HBSS supplemented with 5mM HEPES (HBSS-H; Sigma). For imaging of lysosomal content, cells were stained with 500nM lysotracker green (Life Technologies) in 1X HBSS-H and directly imaged. All images were acquired on a Zeiss LSM 510 Meta confocal microscope. Images were quantified using ImageJ 1.53t.
Epiproteomic Histone Modification Panel
LC-MS/MS studies on histones were performed by the Proteomics Core Facility at Robert H. Lurie Comprehensive Cancer Center of Northwestern University. Briefly, 1×106 cells per sample were treated with DFO or vehicle for 12 hours. Cells were then collected, lysed, nuclei were isolated, and histones were extracted as previously described68,69. Histones were then digested with trypsin and separated by nano-liquid chromatography (75 μm i.d., 15 cm long, packed with MagicC18aq media, particle size 3 μm) coupled to a TSQ Quantum Ultra mass spectrometer. Data were analyzed with Skyline 70 and relative quantification was performed by peak area.
ChIP-seq/PCR assays and analysis
ChIP assays were performed as previously described71 using antibodies against IgG (Sigma) and H3K9me2 (CST 4658S) and POLR2A (CST 14958S). For ChIP-seq analysis, H3K9me2 and POLR2A peaks were called with SICER72 and MACS v1.4.273 respectively using default parameters and were annotated with HOMER74. Heatmaps were generated using deepTools75. Using BEDTools (https://academic.oup.com/bioinformatics/article/26/6/841/244688) and in-house perl scripts, raw counts at each peak were converted to RPKM values with total library counts, and Log2 fold change values between conditions were then computed with these normalized values. The source code of Ceto pipeline and custom scripts used for analyzing the NGS data from this study is available at the Github sites: https://github.com/ebartom/NGSbartom and https://github.com/ebartom/Shapiroetal2023.
Plant materials and growth conditions
Arabidopsis thaliana wild type (Col-0) was obtained from Arabidopsis Biological Resource Centre (ABRC). Plants were grown on Murashige and Skoog (MS) basal medium76; Sigma-Aldrich, full-strength MS, with Ferrous sulfate heptahydrate 27.80 mg/L), with 1% (w/v) sugar (Sigma-Aldrich), 0.05% (w/v) MES (Fisher), 0.8% (w/v) Agar (PhytoTechnology Laboratories, Plant TC), pH 5.7, in the square petri dishes (Fisher), under a 12h/12h light/dark cycle in the growth chamber (SANYO versatile environment test chamber, light level: 5), at 22 ºC, relative humidity 50%.
Plant treatment with BPD and phenotypic analysis
To assay germination and post-germination growth of Arabidopsis thaliana in response to BPD, the MS basal medium (Sigma-Aldrich; full-strength MS, with Ferrous sulfate heptahydrate 27.80 mg/L), with 1% (w/v) sugar (Sigma-Aldrich), 0.05% (w/v) MES (Fisher), 0.8% (w/v) Agar (PhytoTechnology Laboratories, Plant TC), pH 5.7, was supplemented DFO (Vehicle) or indicated concentrations of BPD (Sigma). The seeds were sown and stratified in the MS medium containing indicated concentrations of BPD at 4 °C for 3 days, and then they were placed at 22 ºC under the light condition described above. The seed germination (emergence of radicals) was scored at the indicated times after stratification. To assay Arabidopsis thaliana seedling growth in response to BPD, seeds were germinated after stratification on common MS medium for 5 days, and then transferred to MS medium supplemented with indicated concentrations of BPD. Seedling growth was investigated, and then seedling samples were collected at the indicated times after the transfer. Root length was analyzed by ImageJ.
Yeast culture and experiments
Saccharomyces cerevisiae yeast strains used in this work include wild-type BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), rph1Δ (BY4741 rph1::KanMX4, from Research Genetics) and rph1Δjhd1Δjhd2Δ (BY4741 rph1::hphMX4 jhd1::KanMX4 jhd2::HISMX6, constructed for this study). Yeast cells were cultivated at 30ºC in synthetic complete media lacking or containing 100μM of the Fe2+-specific chelator bathophenanthroline disulfonic acid (BPS, Sigma) for the indicated times. Total yeast RNA and proteins were extracted and analyzed by quantitative RT-PCR and Western blot, respectively, as described previously43. Rps6 and Rps6-Ser235-Ser236 antibodies from Cell Signaling Technology were used to detect total Rps6 (tRPS6) and phosphorylated Rps6 (pRPS6), respectively.
Baculovirus generation, protein production and purification
The N-terminal FLAG tagged murine KDM3A (AA 115–1323) and human full-length KDM3B (AA 1–1761) were generated by PCR and subcloned into pFastBac1 plasmid, and the sequences were confirmed by DNA sequencing. KDM3A and KDM3B bacmids were generated using E.coli DH10Bac competent cells with the standard Bac-to-Bac protocol (Invitrogen) and the corresponding baculoviruses were generated by transducing the bacmid DNA into Sf9 insect cells using flashBACTM System (Oxford Expression Technologies). Recombinant proteins were produced by infecting Sf9 insect cells with the corresponding baculoviruses for 72 h at 27 ˚C. The cells were homogenized in a buffer containing 10mM Tris, 150mM NaCl, 100mM glycine, 0.1% (v/v) Triton X-100, pH 7.8 and a protease inhibitor cocktail tablet without EDTA. The cell lysates were centrifuged at 21,000 g for 30 min and the soluble fractions containing the FLAG-tagged proteins were affinity purified using the anti-FLAG M2 affinity gel (Sigma). The gel beads were washed with TBS buffer (50mM Tris, 150mM NaCl, pH 7.4, protease inhibitor cocktail tablet without EDTA) and the proteins were eluted with TBS buffer containing additionally 150μg/mL FLAG-peptide. The protein concentration was measured with Nanodrop and the protein aliquots were stored at −70˚C until further use. Human full-length WT KDM6A and KDM6AMT/ED mutant variant were recombinantly produced using Sf9 insect cells and purified as previously described53.
Enzyme Kinetic assays
The KDM3A and KDM3B enzymes were affinity purified using buffers without iron. These purified enzymes were pre-incubated with 50 μM DFO at room temperature for 20 min and purified using PD MiniTrap G-25 column (GE Healthcare) prior to the kinetic assays. The kinetic assays were done as previously described with slight modifications77. Briefly, The 50 μL reaction system consisted of 50mM Tris-HCl, pH 7.8, 2mg/mL BSA (Roche), 60μg/mL catalase (Sigma), 0.1mM DTT, 2mM sodium ascorbate, 10% v/v DMSO, 0.4 −0.8μM of KDM3A or KDM3B enzyme and 200μM 2-oxo [1-14C] glutarate (Perkin-Elmer). The histone peptide substrate H3K9me2 (Innovagen) containing an additional glycine and a biotinylated lysine at its C-terminus was used at saturating concentrations (1mM). FeSO4 was added to the reaction mix at six to seven different concentrations from 1nM to 50 μM (for KDM3A) and from 1μM to 1mM (for KDM3B). The reactions were carried out at 37˚C for 10 min and 180 min for KDM3A and KDM3B, respectively. The reactions were stopped by adding 100 μL of 1M KCl, pH 5 and the amount of 14C-labeled CO2 generated during reaction was counted using Tri-carb 2900TR scintillation device (Perkin-Elmer). The Fe2+ KM values were calculated from Michaelis-Menten saturation curves and Lineweaver-Burk plots using Graphpad Prism. The activity rates of the enzymes (mol product / mol enzyme / min) were calculated using Vmax values obtained from Michaelis-Menten curves. Similarly, iron was chelated out from the affinity purified KDM6A and the KDM6AMT/ED mutant by incubating the enzyme with 5 μM DFO for 10 min on ice followed by purification using the PD MiniTrap G-25 column. The kinetic assays were done in the same way as for KDM3A and KDM3B, but using histone peptide H3K27me3, FeSO4 concentration from 1nM to 10μM and 20 min reaction time at 37˚C. The Fe2+ KM values for KDM6A WT and the KDM6AMT/ED mutant were calculated the same way as above
Statistical and Reproducibility
Unpaired two-tailed Student’s t-tests and one-way ANOVA were used to determine statistical significance when appropriate. P < 0.05 was considered to be statistically significant, as indicated by an asterisk ‘*’. Significant one-way ANOVA were followed by Tukey’s post-hoc analysis. Analysis was performed using Graphpad Prism 9. All error bars were calculated and displayed as standard error of measurement (SEM) unless otherwise noted in the figure legends. The Investigators were not blinded to allocation during experiments and outcome assessment. No statistical methods were used to predetermine sample size. No data were excluded from analyses. For in vivo experiments, animals were assigned to experimental groups using simple randomization, without investigator blinding. Both male and female age-matched litter-mate mice were included. Chromatin immunoprecipitation and sequencing was performed without replicates. ChIP-PCR experiments were performed in duplicate.
Data Availability:
NSG data generated for this study are available at the Gene Expression Omnibus (GEO) under accession number GSE214019.
The histone mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD043952.
All source data have been provided in Source Data Files. All other reagents and material supporting the findings of this study are available from the corresponding author on reasonable request.
Code Availability:
The source code of Ceto pipeline and custom scripts used for analyzing the NGS data from this study is available at the Github site: https://github.com/ebartom/NGSbartom and https://github.com/ebartom/Shapiroetal2023.
Extended Data
Extended Data Figure 1. Long-term ID inactivates mTORC1 in multiple cell types.
(A) Non-heme iron levels in HEK293T cells treated with 150μM DFO. (n=5 replicates, two-way unpaired t-test, mean ± SE). (B) mRNA levels of TTP and TFRC at indicated time points in HEK293T cells treated with 150μM DFO. Internal control: POLR2A (n=6 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (C) Immunoblot of mTORC1 activity in HEK293T cells treated with indicated concentrations of DFO. Representative image of two independent experiments (D) Immunoblot of mTORC1 activity in HEK293T cells treated with 150μM DFO or 50μM BPD for 3 hours. Representative image of two independent experiments. (E) mRNA levels of indicated genes 3 hours after addition of 150μM DFO or 50μM BPD. Internal control: POLR2A (n=8 TFRC Control; n=7 TFRC 150μM DFO and 50μM BPD; n=4 REDD1; n=8 TTP replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (F) Immunoblot of mTORC1 activity in response to 250nM Torin1 at indicated time points. Representative image of two independent experiments. (G) Immunoblot of mTORC1 activity in primary murine hepatocytes treated with increasing concentrations of DFO. Representative image of two independent experiments (H) mRNA of Ttp and Tfrc in primary murine hepatocytes treated with indicated concentrations of DFO. Internal control: Polr2a (n=3 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (I) Immunoblot of mTORC1 activity in hPS-CM treated with 150μM DFO. Representative image of one experiment (J) Immunoblot of mTORC1 activity in hiPS-neurons treated with 150μM DFO. Representative image of one experiment. (K) Immunoblot of HEK293T cells chelated for 18 hours followed by addition of ferric ammonium citrate (FAC) for indicated times. Representative image of two independent experiments. (L) BrdU incorporation in HEK293T cells treated with 150μM DFO. Representative image of six independent samples. (M) Quantification of cellular proliferation by Hoescht staining 48 hours after treatment with DFO (n=6 replicates, two-way unpaired t-test, mean ± SE). * indicates P value < 0.05 when noted for all panels. Source numerical data and unprocessed blots are available in source data files.
Extended Data Figure 2. Non-pharmacologic means of inducing ID inactivates mTORC1.
(A) Immunoblot of mTORC1 activity in HEK293T cells treated with high Tf-sat (66%) or low Tf-sat (6.6%) media for 18 hours. Representative image of two independent experiments. (B) Summary of immunoblot in panel A (n=3 replicates, two-way unpaired t-test, mean ± SE). (C) Immunoblot of mTORC1 activity in HepG2 cells transfected with the FPN-GFP fusion protein and TET inducible rtTA3 plasmids in the presence and absence of 500ng/ml doxycycline for 48 hours. Representative image of two independent experiments. (D) Summary of immunoblot in panel C (n=3 replicates, two-way unpaired t-test, mean ± SE). (E) Fluorescent microscopy of cells transfected with FPN-GFP construct and treated with 500ng/ml doxycycline for 24 hours demonstrating appropriate expression and localization of the FPN-GFP fusion protein. Representative image of three independent samples. (F) Immunoblot of puromycin incorporation in rtTA3/FPN-GFP stable HEK293T cells in the presence and absence of 500ng/ml doxycycline for 48 hours. Representative image of one experiment. (G) mRNA expression of ER stress makers CHOP and BNIP3 in HepG2 cells transfected with rtTA3/FPN-GFP plasmids and treated with 500ng/ml doxycycline for 48 hours. Internal control: 18S (n=4 replicates, two-way unpaired t-test, mean ± SE). (H) Immunoblot of protein levels of the key components of the mTORC1 complex after 24 hours of 150μM DFO in HEK293T cells. Representative image of two independent experiments. (I) Summary of results shown in panel A (n=4 replicates, two-way unpaired t-test, mean ± SE). (J) Immunoblot of total and phosphorylated TSC2, AKT, ERK, GSK3β, and S6 proteins and total P53 at different time points after treatment with 150μM of DFO in HEK293T cells. Representative image of one experiment. (K) Immunoblot of mTORC1 activity and mitochondrial function in HepG2 cells treated with DFO for 18 hours at the indicated doses. Representative image of one experiment. (L) Oxygen Consumption Rate (OCR) measured by the Seahorse Assay in HepG2 cells treated for 24 hours of DFO at the indicated doses (n=10 replicates per group, mean ± SE). * indicates P value < 0.05 when noted for all panels.
Extended Data Figure 3. Leucine sensing is required for mTORC1 inactivation by ID.
(A) Ttp mRNA in WT and Tsc2 KO MEFs treated with 150μM DFO. Internal control: Snrk (n=4 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (B) Immunoblot of mTORC1 activity in WT and Tsc2 KO MEFs with indicated treatments. Representative image of three independent experiments. (C) REDD1 mRNA in HEK293T cells treated with siREDD1. Internal control: POL2RA (n=6 replicates per condition, two-way unpaired t-test, mean ± SE). (D) TFRC mRNA in HEK293T cells treated with siREDD1 and 150μM DFO. Internal control: POL2RA (n=6 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (E) Immunoblot of mTORC1 and AMPK activity with indicated treatments. Representative image of one experiment (F) Ttp mRNA in WT and Ampkα1/2 dKO cells treated with 150μM DFO. Internal control: Polr2a (n=4 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (G) Acridine orange staining in HEK293T cells treated with 150μM DFO. Representative image of six independent samples. (H) Summary of the results in Panel G (n=7 replicates control; n=6 150μM DFO, two-way unpaired t-test, mean ± SE). (I) Leucine levels in HEK293T cells treated with 150μM DFO (n=5 replicates control; n=4 150μM DFO two-way unpaired t-test, median ± quartiles). (J) SAM levels in HEK293T cells treated with 150μM DFO (n=5 replicates control; n=4 150μM DFO, two-way unpaired t-test, median ± quartiles). (K) Immunoblot of mTORC1 activity in WT and NPRL2 KO HEK293T cells with indicated treatments. Representative image of one experiment. (L) mTORC1 localization to lysosomes in NPRL2 KO cells treated with 150μM DFO. Representative image of six independent samples. (M) Quantification of images in panel L. (n=5 replicates control; n=6 150μM DFO, two-way unpaired t-test, mean ± SE) (N) Cell death using Hoescht and propidium iodide (PI) in WT and NPRL2 KO HEK293T cells treated with 150μM DFO for indicated times. Representative image of six independent samples. (O) Quantification of images in panel N. (n=6 replicates, two-way unpaired t-test, mean ± SE). * indicates P value < 0.05 when noted for all panels.
Extended Data Figure 4. ID increases H3K9 di-methylation independent of ATF4, the IRP system, and 2-HG.
(A) Immunoblot of H3K9me2 in hiPS-CM treated with 150μM DFO. Representative image of one experiment. (B) Immunoblot of H3K9me2 in hiPS-Neurons treated with 150μM DFO. Representative image of one experiment. (C) Immunoblot of H3K9me2 levels in MEFs with indicated treatments. Representative image of two independent experiments. (D, E) Summary of immunoblot in panel C (n=3 replicates, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (F) Immunoblot of H3K9me2 in HepG2 cells expressing rtTA3/GFP-FPN with indicated treatments. Representative image of two independent experiments. (G) mRNA of indicated genes in WT and Arnt KO MEFs treated with 150μM DFO. Internal control: 18S (n=4 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (H) Immunoblot of H3K9me2 levels in WT and Atf4 KO MEFs with indicated treatments. Representative image of one experiment. (I) Summary of immunoblot in panel H (n=3 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (J) Immunoblot of H3K9me2 levels in WT and Irp1/2 KD/KO MEFs with indicated treatments. Representative image of two independent experiments. (K) Summary of immunoblot in panel J (n=3 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (L) Immunoblot of IRP1 levels in Irp2 KO MEFs treated with indicated siRNA. Representative image of one experiment. (M) Summary of immunoblot in panel L (n=3 replicates per condition, two-way unpaired t-test, mean ± SE). (N) mRNA of indicated genes in WT and Irp1/2 KD/KO MEFs treated with 150μM DFO. Internal control: Snrk (n=6 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (O) Ratio of 2-HG/succinate levels in HEK293T cells treated 150μM DFO (n=5 replicates control; n=4 150μM DFO, two-way unpaired t-test, median ± quartiles). (P) Immunoblot of H3K9me2 in HEK293T cells with indicated treatments. Representative image of one experiment. (Q) Immunoblot of H3K9me2 in HEK293T cells with indicated treatments. Representative image of one experiment. * indicates P value < 0.05 when noted for all panels.
Extended Data Figure 5. ID alters occupancy of POLR2A at the promoters of genes involved in metabolic pathways.
(A) Fold change in POLR2A occupancy within predefined regions of the promoter and gene body to categorize genes defined by increased POLR2A binding, POLR2A loss, and promoter-pausing. (B) Gene-ontology (GO) terms of genes enriched in the increased POLR2A group after ID. (C) GO terms of genes that had decreased POLR2A after ID. (D) UCSC genome browser tracks for the LAT3 (top) and PAT1 (bottom) gene loci. POLR2A and H3K9me2 tracks from ChIP-seq analysis were loaded and represented as the difference in normalized reads between the DFO and control groups. Regions of H3K9me2 enrichment in the DFO group are underlined in red. Direction of transcription is indicated by a black arrow. Encode Histone (LAT3 and PAT1) and Genehancer (PAT1) browser tracks are displayed beneath and represent predicted enhancer regions which align with regions of increased H3K9me2 signal in response to DFO. Yellow bar, red and grey arrows indicate enhancer regions designated by Encode Histone and Genehancer browser tracks.
Extended Data Figure 6. ID increases H3K9 di-methylation within the promoter of RPTOR and correlates with decreased RPTOR expression.
(A) H3K9me2 signal and POL2RA occupancy in the promoter for RPTOR from ChIP-Seq analysis. (B) ChIP-PCR of RPTOR in HEK293T cells. Cells treated with 150μM DFO or 250μM IOX1 for 12 hours and vehicle controls were followed by IP of lysates using an antibody against H3K9me2. IgG was used as a negative control for the IP (n=2 replicates). (C) RPTOR mRNA levels at indicated time points after the addition of 150μM DFO. Samples matched with Figure 4I. Internal control: POLR2A (n=4 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (D) Immunoblot of RPTOR, mTOR and HIF1-α levels in HEK293T cells treated with 100 μg/ml cycloheximide (CHX) for indicated time points. HIF1-α, which is rapidly turned over under normoxic conditions via the actions of the EGLN and Von-Hippel Lindau (VHL) proteins, was used as a positive control. Representative image of one experiment. (E) Immunoblot of mTORC1 activity and complex member RAPTOR in A549 cells after treatment with 150μM DFO for 48 hours. Representative image of two independent experiments. (F) Immunoblot of mTORC1 activity and complex member RAPTOR in patient-derived primary tumor cell cultures treated with 150μM DFO. Representative image of one experiment. (G) Quantification of immunoblot in panel (F) (n=3 replicates, one-way ANOVA and Tukey’s post-hoc test, mean ± SE.) (H) Immunoblot of lysates from cells transfected with FLAG-RAP2A or FLAG-mTOR and treated with or without 150μM DFO for 48 hours. Immunoprecipitation was performed with anti-FLAG antibody. RAP2A = negative control. (I) Summary of IP studies from panel (H) (n=3 replicates, two-way unpaired t-test, mean ± SE). Representative image of two independent experiments (J) Immunoblot of mTORC1 activity in WT and NPRL2 KO HEK293T treated with and without 150μM DFO for 48 hours. Representative image of one experiment. (K) Quantification of immunoblot in panel (J) (n=3 replicates, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). * indicates P value < 0.05 when noted for all panels.
Extended Data Figure 7. The Jumonji-C KDM family inhibitor IOX1 mimics the actions of ID on mTORC1 activity.
(A) RT-PCR of AA transporters, RPTOR and TTP in HEK293T cells treated with various Jmj-C domain inhibitors for 12 hours. Internal control: POLR2A (n=4 replicates per condition; except n=3 TTP 1mM DMOG, two-way unpaired t-test, mean ± SE). (B) Immunoblot of mTORC1 activity and H3K9me2 levels in A549 cells treated with 150μM DFO or 250μM IOX1 for 18hrs. Representative image of two independent experiments. (C) Immunoblot of indicated proteins in A549 cells treated with 250μM IOX1 for 48 hours. Representative image of two independent experiments. (D) Immunoblot of cytosol and membrane fractions from HepG2 cells treated with 250μM IOX1 for 12 hours. Representative image of one experiment. (E) Densitometry summary of data in Panel D (n=3 replicates per condition, two-way unpaired t-test, mean ± SE). (F) 14[C]-leucine uptake into HEK293T cells treated with 150μM DFO or 100μM IOX1 for 18 hours (n=5 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (G) 14[C]-leucine uptake into HeLa cells treated with 150μM DFO or 100μM IOX1 for 18 hours (n=4 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (H) Immunoblot of mTORC1 activity WT and NPRL2 KO HEK293T cells treated with 250μM IOX1 for 18 hours. Representative image of two independent experiments (I) Quantification of immunoblot in panel H (n=3 replicates per group, one-way ANOVA with Tukey’s post-hoc test. mean ± SE). * indicates P value < 0.05 when noted for all panels.
Extended Data Figure 8. H3K9 di-methyl ChIP-seq signal in iron chelated samples correlate with changes after loss of KDM3A and KDM3B.
(A) Heatmap of the log2FC in H3K9me2 signal between DFO treatment and control plotted against KDM3A and KDM3B occupancy. K-means cluster =3. (B) Browser tracks with KDM3B and H3K9me2 track data from HCT116 cells treated with control shRNA (shC) or shRNA targeting KDM3A and KDM3B (sh3A3B). Track data for KDM3A was downloaded from the GEO database accession GSE127624. Track data for KDM3B and H3K9me2 from HCT116 cells were downloaded from the GEO database accession GSE71885. (C) Hierarchical clustering and correlation analysis between indicated samples using deepTools plotCorrelation. Values indicate the Pearson correlation between the sample listed in the corresponding row and column. (D) Immunoblot showing deletion of KDM4A and 4B, using CRISPR-Cas technology in HEK293T cells. Representative image of one experiment. (E) Immunoblot of mTORC1 activity and H3K9me2 in Cas9 (WT) and KDM3A, KDM4B or KDM4C KO HEK293T cells in the presence and absence of 150μM DFO for 18 hours. Representative image of one experiment. (F) Immunoblot of mTORC1 activity and H3K9me2 in HEK293T cells with overexpression of HA-tagged KDM4A, KDM4B or combination of the two proteins treated with 150μM DFO for 18 hours. Representative image of one experiment.
Extended Data Figure 9. KDM3B KO cells do not repress LAT3/PAT1 expression or mTORC1 activity, and have increased cell death during ID.
(A) Immunoblot of mTORC1 activity and H3K9me2 in Cas9 (WT) and sg.3B c.7 (KDM3B KO) TSC2 KO HeLa cells in the presence and absence of 150μM DFO for 18 hours. Representative image of one experiment. (B) Summary of immunoblot in panel A (n=3 replicates per group, one-way ANOVA with Tukey’s post-hoc test. Mean ± SE). (C) Assessment of mTORC1 on the lysosome in KDM3B KO HepG2 cells treated with 150μM DFO for 18 hours. Representative image of five independent samples. (D) Quantification of images in panel C. (n=5 replicates per condition, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (E) Immunoblot of LAT3 protein in KDM3B KO HepG2 cells treated with DFO for 18 hours. Representative image of one experiment. (F) Immunoblot of PAT1 protein in KDM3B KO HepG2 cells treated with DFO for 18 hours. Representative image of one experiment. (G) Fluorescent microscopy of cell death using Hoescht and propidium iodide (PI) double staining in KDM3B KO HEK293T cells treated with 150μM DFO for 0, 24, 36 and 60 hours. Representative image of six independent samples. (H) Quantification of images in panel G. (n=6 replicates, one-way ANOVA and Tukey’s post-hoc test, mean ± SE). (I) SDS-PAGE gel of purified N-terminal FLAG-tagged WT KDM6A and KDM6AMT/ED mutant expressed using a baculoviral overexpression system. Representative image of three independent experiments. (J, K) Enzyme kinetics of WT KDM6A (J) and KDM6AMT/ED (K) in the presence of increasing concentrations of iron. (L) Enzyme kinetics of KDM3B in the presence of increasing concentrations of R-2HG. (M) Immunoblot of mTORC1 activity in HEK293T cells after treatment with 150μM DFO or increasing concentrations of octyl-R-2HG. Representative image of one experiment. * indicates P value < 0.05 when noted for all panels.
Supplementary Material
Supplemental Data Figure 1. Iron chelation does not cause appreciable apoptosis among adherent cells. (A) Gating strategy for flow cytometry using Annexin-V and PI on HEK293T cells treated with DFO for 24 hours. Adherent cells were trypsinized and flowed together with floating cells. (B) Representative flow plots for Control and DFO samples with summary percentages ± SEM for each quadrant. (C) Quantification of images in panel D. (n=3 replicates, two-way unpaired t-test, mean ± SE). * indicates P value < 0.05 when noted for all panels.
Supplemental Data Figure 2. The leucine transporter SLC43A1 is highly expressed in liver tissue. (A) Normalized expression (NX) of SLC43A1 across multiple human tissues (https://www.proteinatlas.org/ENSG00000149150-SLC43A1/tissue). (B) Table listing the normalized expression of leucine transporters in the liver. Data for panels A and B was acquired from The Human Protein Atlas version 20.1 and Ensembl version 92.38, available from http://www.proteinatlas.org “Uhlén M et al., Tissue-based map of the human proteome. Science (2015) PubMed: 25613900 DOI: 10.1126/science.1260419”.
Supplementary Data Figure 3. ID causes global hyper-methylation of histones (A-B) Unbiased measure of global changes in histone methylation measured by histone-MS in response to ID. Graphs are presented as rank ordered lists by change in %abundance with 150μM DFO - control (A) and total %abundance in the 150μM DFO group (B). Data matched with Figure 6B. (n=3 independent samples measured in triplicate, mean ± SE).
Acknowledgements:
We would like to thank Hailey Harris, Meng Shang, Chunlei Chen, Yihan Chen, Xinghang Jiang, and Kai Xu for their technical help, managing our mouse colony and genotyping the mice used in this study. We also thank Dr. Christopher J. Payne for his sound advice and providing initial aliquots of histone methylation antibodies to run preliminary experiments. We thank Dr. Yatrik Shah for providing us with reagents to perform the FPN overexpression studies. We thank Dr. Evangelos Kiskinis and Mennat Gharib for generously providing us with hIPS-Neurons. We thank Dr. Paul Burridge for providing us with hIPS cells for differentiation into cardiomyocytes. We thank Dr. Nathan A. Lack for generously providing us the KDM3B plasmids. Metal analysis was performed at the Northwestern University Quantitative Bio-element Imaging Center generously supported by the National Research Resource for Quantitative Mapping in the Life Sciences (QE-Map) Award Number P41GM135018. Metabolomics services were performed by the Metabolomics Core Facility at Robert H. Lurie Comprehensive Cancer Center of Northwestern University. Proteomics services were performed by Dr. Young Ah Goo, in conjunction with Dr. Neil L. Kelleher at the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center, instrumentation award (S10OD025194) from NIH Office of Director, and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. We thank Dr. Matthew J. Schipma, and Brian Wray at the Northwestern University Sequencing Core and Patrick A. Ozark for their contribution to the ChIP-seq analyses. This work was supported through core services and support from the Northwestern University George M. O’Brien Kidney Research Core Center (NU GoKidney), an NIH/NIDDK funded program (P30 DK114857). This work was supported by the Northwestern University – Flow Cytometry Core Facility supported by Cancer Center Support Grant (NCI CA060553). Flow Cytometry Cell Sorting was performed on a BD FACSAria SORP system and BD FACSymphony S6 SORP system, purchased through the support of NIH 1S10OD011996–01 and 1S10OD026814–01. Additional funding for this work was provided through NRSA 5F30DK109608–02, and T32-DK007169 awarded to J.S.S., NHLBI HL127646, HL140973, and HL138982 awarded to H.A., U54CA193419 and R01GM038784 awarded to T.V.O., and the Spanish Ministry of Science, Innovation and Universities (MICINN) grant BIO2017–87828-C2–1-P and FEDER funds to SP. Z.S.W. was supported in part by the National Institutes of Health Training Grant T32GM008449 through Northwestern University’s Biotechnology Training Program.
Footnotes
Competing Interests:
The authors declare no competing interests.
Inclusion and Ethics
We have complied with all relevant ethical regulations and have received prior approval from relevant regulatory institutions and review boards.
References
- 1.Efeyan A, Comb WC & Sabatini DM Nutrient-sensing mechanisms and pathways. Nature 517, 302–310, doi: 10.1038/nature14190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyons TW, Reinhard CT & Planavsky NJ The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315, doi: 10.1038/nature13068 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Hedges SB, Blair JE, Venturi ML & Shoe JL A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol 4, 2, doi: 10.1186/1471-2148-4-2 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hentze MW, Muckenthaler MU, Galy B & Camaschella C Two to tango: regulation of Mammalian iron metabolism. Cell 142, 24–38, doi: 10.1016/j.cell.2010.06.028 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Coffey R & Ganz T Iron homeostasis: An anthropocentric perspective. J Biol Chem 292, 12727–12734, doi: 10.1074/jbc.R117.781823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hindt MN & Guerinot ML Getting a sense for signals: regulation of the plant iron deficiency response. Biochim Biophys Acta 1823, 1521–1530, doi: 10.1016/j.bbamcr.2012.03.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramos-Alonso L, Romero AM, Martinez-Pastor MT & Puig S Iron Regulatory Mechanisms in Saccharomyces cerevisiae. Front Microbiol 11, 582830, doi: 10.3389/fmicb.2020.582830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairo G & Recalcati S Iron-regulatory proteins: molecular biology and pathophysiological implications. Expert Rev Mol Med 9, 1–13, doi: 10.1017/S1462399407000531 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirota K An intimate crosstalk between iron homeostasis and oxygen metabolism regulated by the hypoxia-inducible factors (HIFs). Free Radic Biol Med 133, 118–129, doi: 10.1016/j.freeradbiomed.2018.07.018 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Islam MS, Leissing TM, Chowdhury R, Hopkinson RJ & Schofield CJ 2-Oxoglutarate-Dependent Oxygenases. Annu Rev Biochem 87, 585–620, doi: 10.1146/annurev-biochem-061516-044724 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Klose RJ, Kallin EM & Zhang Y JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7, 715–727, doi: 10.1038/nrg1945 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hausinger RP FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol 39, 21–68, doi: 10.1080/10409230490440541 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Tsukada Y et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816, doi: 10.1038/nature04433 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Cloos PA et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442, 307–311, doi: 10.1038/nature04837 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Sancak Y et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501, doi: 10.1126/science.1157535 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fingar DC et al. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol 24, 200–216, doi: 10.1128/mcb.24.1.200-216.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz AJ et al. Hepcidin sequesters iron to sustain nucleotide metabolism and mitochondrial function in colorectal cancer epithelial cells. Nat Metab 3, 969–982, doi: 10.1038/s42255-021-00406-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson TR et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137, 873–886, doi: 10.1016/j.cell.2009.03.046 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoki K, Li Y, Zhu T, Wu J & Guan KL TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4, 648–657, doi: 10.1038/ncb839 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Bilanges B et al. Tuberous sclerosis complex proteins 1 and 2 control serum-dependent translation in a TOP-dependent and -independent manner. Mol Cell Biol 27, 5746–5764, doi: 10.1128/MCB.02136-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H et al. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest 112, 1223–1233, doi: 10.1172/JCI17222 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang GL, Jiang BH, Rue EA & Semenza GL Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 92, 5510–5514, doi: 10.1073/pnas.92.12.5510 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaakkola P et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472, doi: 10.1126/science.1059796 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Brugarolas J et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 18, 2893–2904, doi: 10.1101/gad.1256804 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sofer A, Lei K, Johannessen CM & Ellisen LW Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol 25, 5834–5845, doi: 10.1128/MCB.25.14.5834-5845.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishan S, Richardson DR & Sahni S The Anticancer Agent, Di-2-Pyridylketone 4,4-Dimethyl-3-Thiosemicarbazone (Dp44mT), Up-Regulates the AMPK-Dependent Energy Homeostasis Pathway in Cancer Cells. Biochim Biophys Acta 1863, 2916–2933, doi: 10.1016/j.bbamcr.2016.09.011 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Hoxhaj G et al. The mTORC1 Signaling Network Senses Changes in Cellular Purine Nucleotide Levels. Cell Rep 21, 1331–1346, doi: 10.1016/j.celrep.2017.10.029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancak Y et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303, doi: 10.1016/j.cell.2010.02.024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfson RL et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48, doi: 10.1126/science.aab2674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bar-Peled L et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106, doi: 10.1126/science.1232044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efeyan A et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683, doi: 10.1038/nature11745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194, doi: 10.1126/science.1257132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyant GA et al. mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell 171, 642–654 e612, doi: 10.1016/j.cell.2017.09.046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S et al. HRI coordinates translation necessary for protein homeostasis and mitochondrial function in erythropoiesis. Elife 8, doi: 10.7554/eLife.46976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao E et al. KDM4C and ATF4 Cooperate in Transcriptional Control of Amino Acid Metabolism. Cell Rep 14, 506–519, doi: 10.1016/j.celrep.2015.12.053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu C et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478, doi: 10.1038/nature10860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen FX et al. PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell 162, 1003–1015, doi: 10.1016/j.cell.2015.07.042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mootha VK et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Kim K et al. mTORC1-independent Raptor prevents hepatic steatosis by stabilizing PHLPP2. Nat Commun 7, 10255, doi: 10.1038/ncomms10255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxton RA & Sabatini DM mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976, doi: 10.1016/j.cell.2017.02.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ & Heitman J The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev 13, 3271–3279, doi: 10.1101/gad.13.24.3271 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romero AM et al. A genome-wide transcriptional study reveals that iron deficiency inhibits the yeast TORC1 pathway. Biochim Biophys Acta Gene Regul Mech 1862, 194414, doi: 10.1016/j.bbagrm.2019.194414 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Tu S et al. Identification of histone demethylases in Saccharomyces cerevisiae. J Biol Chem 282, 14262–14271, doi: 10.1074/jbc.M609900200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klose RJ et al. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Mol Cell Biol 27, 3951–3961, doi: 10.1128/MCB.02180-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King ON et al. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors. PLoS One 5, e15535, doi: 10.1371/journal.pone.0015535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hopkinson RJ et al. 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-Oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation. Chem Sci 4, 3110–3117, doi: 10.1039/C3SC51122G (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, doi: 10.1038/nature11247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J et al. KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/beta-catenin signalling. Nat Commun 8, 15146, doi: 10.1038/ncomms15146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carbonneau M et al. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nat Commun 7, 12700, doi: 10.1038/ncomms12700 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altman BJ & Rathmell JC Metabolic stress in autophagy and cell death pathways. Cold Spring Harb Perspect Biol 4, a008763, doi: 10.1101/cshperspect.a008763 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose NR, McDonough MA, King ON, Kawamura A & Schofield CJ Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev 40, 4364–4397, doi: 10.1039/c0cs00203h (2011). [DOI] [PubMed] [Google Scholar]
- 53.Chakraborty AA et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363, 1217–1222, doi: 10.1126/science.aaw1026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zoncu R et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678–683, doi: 10.1126/science.1207056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milkereit R et al. LAPTM4b recruits the LAT1–4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nat Commun 6, 7250, doi: 10.1038/ncomms8250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida S, Pacitto R, Yao Y, Inoki K & Swanson JA Growth factor signaling to mTORC1 by amino acid-laden macropinosomes. J Cell Biol 211, 159–172, doi: 10.1083/jcb.201504097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Losman JA, Koivunen P & Kaelin WG Jr. 2-Oxoglutarate-dependent dioxygenases in cancer. Nat Rev Cancer 20, 710–726, doi: 10.1038/s41568-020-00303-3 (2020). [DOI] [PubMed] [Google Scholar]
Methods-Only References
- 58.Bayeva M et al. mTOR regulates cellular iron homeostasis through tristetraprolin. Cell Metab 16, 645–657, doi: 10.1016/j.cmet.2012.10.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798, doi: 10.1016/j.neuron.2013.05.029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nehme R et al. Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Rep 23, 2509–2523, doi: 10.1016/j.celrep.2018.04.066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simkin D et al. Dyshomeostatic modulation of Ca(2+)-activated K(+) channels in a human neuronal model of KCNQ2 encephalopathy. Elife 10, e64434, doi: 10.7554/eLife.64434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chambers SM et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27, 275–280, doi: 10.1038/nbt.1529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Giorgio FP, Boulting GL, Bobrowicz S & Eggan KC Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell 3, 637–648, doi: 10.1016/j.stem.2008.09.017 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Kuo HH et al. Negligible-Cost and Weekend-Free Chemically Defined Human iPSC Culture. Stem Cell Reports 14, 256–270, doi: 10.1016/j.stemcr.2019.12.007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burridge PW, Holmstrom A & Wu JC Chemically Defined Culture and Cardiomyocyte Differentiation of Human Pluripotent Stem Cells. Curr Protoc Hum Genet 87, 21 23 21–21 23 15, doi: 10.1002/0471142905.hg2103s87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burridge PW et al. Chemically defined generation of human cardiomyocytes. Nat Methods 11, 855–860, doi: 10.1038/nmeth.2999 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Concordet JP & Haeussler M CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46, W242–W245, doi: 10.1093/nar/gky354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcia BA et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc 2, 933–938, doi: 10.1038/nprot.2007.106 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng Y et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc Natl Acad Sci U S A 109, 13549–13554, doi: 10.1073/pnas.1205707109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.MacLean B et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968, doi: 10.1093/bioinformatics/btq054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szczepanski AP et al. ASXL3 bridges BRD4 to BAP1 complex and governs enhancer activity in small cell lung cancer. Genome Med 12, 63, doi: 10.1186/s13073-020-00760-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zang C et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958, doi: 10.1093/bioinformatics/btp340 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589, doi: 10.1016/j.molcel.2010.05.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramirez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165, doi: 10.1093/nar/gkw257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murashige T & Skoog F A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol Plantarum 15, 473–497, doi:DOI 10.1111/j.1399-3054.1962.tb08052.x (1962). [DOI] [Google Scholar]
- 77.Laukka T, Myllykoski M, Looper RE & Koivunen P Cancer-associated 2-oxoglutarate analogues modify histone methylation by inhibiting histone lysine demethylases. J Mol Biol 430, 3081–3092, doi: 10.1016/j.jmb.2018.06.048 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data Figure 1. Iron chelation does not cause appreciable apoptosis among adherent cells. (A) Gating strategy for flow cytometry using Annexin-V and PI on HEK293T cells treated with DFO for 24 hours. Adherent cells were trypsinized and flowed together with floating cells. (B) Representative flow plots for Control and DFO samples with summary percentages ± SEM for each quadrant. (C) Quantification of images in panel D. (n=3 replicates, two-way unpaired t-test, mean ± SE). * indicates P value < 0.05 when noted for all panels.
Supplemental Data Figure 2. The leucine transporter SLC43A1 is highly expressed in liver tissue. (A) Normalized expression (NX) of SLC43A1 across multiple human tissues (https://www.proteinatlas.org/ENSG00000149150-SLC43A1/tissue). (B) Table listing the normalized expression of leucine transporters in the liver. Data for panels A and B was acquired from The Human Protein Atlas version 20.1 and Ensembl version 92.38, available from http://www.proteinatlas.org “Uhlén M et al., Tissue-based map of the human proteome. Science (2015) PubMed: 25613900 DOI: 10.1126/science.1260419”.
Supplementary Data Figure 3. ID causes global hyper-methylation of histones (A-B) Unbiased measure of global changes in histone methylation measured by histone-MS in response to ID. Graphs are presented as rank ordered lists by change in %abundance with 150μM DFO - control (A) and total %abundance in the 150μM DFO group (B). Data matched with Figure 6B. (n=3 independent samples measured in triplicate, mean ± SE).
Data Availability Statement
NSG data generated for this study are available at the Gene Expression Omnibus (GEO) under accession number GSE214019.
The histone mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD043952.
All source data have been provided in Source Data Files. All other reagents and material supporting the findings of this study are available from the corresponding author on reasonable request.