Abstract
The hepatitis C virus (HCV)-encoded protease/helicase NS3 is likely to be involved in viral RNA replication. We have expressed and purified recombinant NS3 (protease and helicase domains) and ΔpNS3 (helicase domain only) and examined their abilities to interact with the 3′-terminal sequence of both positive and negative strands of HCV RNA. These regions of RNA were chosen because initiation of RNA synthesis is likely to occur at or near the 3′ untranslated region (UTR). The results presented here demonstrate that NS3 (and ΔpNS3) interacts efficiently and specifically with the 3′-terminal sequences of both positive- and negative-strand RNA but not with the corresponding complementary 5′-terminal RNA sequences. The interaction of NS3 with the 3′-terminal negative strand [called 3′(−) UTR127] was specific in that only homologous (and not heterologous) RNA competed efficiently in the binding reaction. A predicted stem-loop structure present at the 3′ terminus (nucleotides 5 to 20 from the 3′ end) of the negative-strand RNA appears to be important for NS3 binding to the negative-strand UTR. Deletion of the stem-loop structure almost totally impaired NS3 (and ΔpNS3) binding. Additional mutagenesis showed that three G-C pairs within the stem were critical for helicase-RNA interaction. The data presented here also suggested that both a double-stranded structure and the 3′-proximal guanosine residues in the stem were important determinants of protein binding. In contrast to the relatively stringent requirement for 3′(−) UTR binding, specific interaction of NS3 (or ΔpNS3) with the 3′-terminal sequences of the positive-strand RNA [3′(+) UTR] appears to require the entire 3′(+) UTR of HCV. Deletion of either the 98-nucleotide 3′-terminal conserved region or the 5′ half sequence containing the variable region and the poly(U) and/or poly(UC) stretch significantly impaired RNA-protein interaction. The implication of NS3 binding to the 3′-terminal sequences of viral positive- and negative-strand RNA in viral replication is discussed.
Hepatitis C virus (HCV) is the primary causative agent of parenterally transmitted non-A, non-B hepatitis and affects a significant part of the world's population. HCV infection frequently leads to chronic hepatitis, cirrhosis of the liver, and possibly hepatocellular carcinoma (11, 45). HCV, a member of the family Flaviviridae (44), has been a difficult virus to study due to the lack of an appropriate tissue culture system and an adequate, simple, and low-cost animal model. The RNA genome of HCV has recently been cloned, and the single-stranded, plus-polarity RNA genome of the virus is approximately 9,500 nucleotides long and is flanked by untranslated regions (UTR) at both the 5′ and 3′ ends (22, 28, 48). The 5′ UTR of HCV RNA (341 nucleotides) is highly structured and contains an internal ribosome entry site which extends to nucleotide 370, partially overlapping the structural protein (core) coding sequences (8, 20, 21, 51, 53). The secondary structure of the 5′ UTR appears to be highly conserved among various HCV strains, and structural similarity to members of the pestivirus family has been reported using biochemical approaches (7, 22). The 5′ UTR of the viral RNA is followed by a single large open reading frame that encodes a polyprotein of approximately 3,000 amino acids, which is subsequently proteolytically processed by cellular signal peptidases and two HCV-encoded proteases to produce mature structural and functional proteins (16–18). The 3′ UTR of approximately 200 nucleotides contains three distinct regions—a short region with sequence heterogeneity preceding a poly(U) and/or poly(UC) region of variable length followed by a highly conserved sequence (X region) of approximately 100 nucleotides (6, 24, 30, 48, 55). This X region of conserved sequence forms three stable stem-loop structures, SL-I, SL-II, and SL-III. The major nonstructural proteins include two proteases, NS2 and NS3, followed by NS4A, NS4B, NS5A, and NS5B (reviewed in reference 12). NS4A (a 54-amino-acid polypeptide), however, seems to act as a cofactor for NS3 activity, and the central domain has been implicated as essential for this role of NS4A (3, 4, 15, 33, 34, 49). Almost nothing is known about HCV RNA replication; however, various laboratories have recently demonstrated synthesis of full-length complementary RNA (and dimeric RNA), which can be achieved in vitro by the NS5B protein (38, 40, 60).
The HCV NS3 protein has been the subject of intense study due to its associated protease and helicase activities. The C-terminal 450 amino acids of the NS3 protein constitute a polynucleotide-stimulated NTPase activity (25, 42, 46), a 3′-5′ unwinding activity (19, 25, 47), and a single-stranded-RNA binding activity (19, 47). The N-terminal one-third of NS3 contains a serine protease activity and is responsible for the downstream cleavages in the nonstructural region (31, 50). The three-dimensional structure of NS3 has been solved by X-ray crystallography, and structure-based mutagenesis of NS3 has identified important amino acid residues required for helicase and ATPase activities (9, 29, 37, 59). Recent studies have shown that coexpression of the NS4A protein directs the NS3 protein, which is diffusely distributed in the cytoplasm and nucleus in the absence of NS4A, to the endoplasmic reticulum (54).
Many single-stranded positive-strand RNA viruses encode their own helicases (and NTPases). They are thought to play a role in viral RNA replication. Our previous work has shown that the poliovirus-encoded NTPase (and helicase) 2C specifically interacts with the 3′-terminal sequences of viral negative-strand RNA (2). In addition to its specific RNA binding ability, it is also able to interact with cellular cytoplasmic membranes (5, 10, 13, 14). Since poliovirus RNA synthesis takes place in cytoplasmic membranes, it is believed that the poliovirus 2C protein anchors the negative strand to the cytoplasmic membrane, thus allowing initiation of positive-strand RNA synthesis to occur. Moreover, initiation of positive-strand synthesis is likely to require an unwinding activity to melt the double-stranded structure at the 3′ end of the negative strand formed by initial copying of the input viral plus-strand RNA. The rationale stated above has prompted us to examine the interaction of intact NS3 and the ΔpNS3 (with the helicase domain only) with the 3′-terminal sequence of the negative-strand RNA of HCV. We show that in fact both NS3 and ΔpNS3 interact specifically with the 3′-terminal sequences of HCV negative-strand RNA. This interaction is impaired by deletion of the 3′-terminal sequences of the negative strand. The results presented here suggest that a stem-loop structure within the 3′-terminal sequence is important for interaction with NS3. Unlike the poliovirus-encoded helicase (2C), which interacts with the 3′ UTR of the negative strand but not with the 3′ sequences of the positive strand, NS3 (and ΔpNS3) interacts with both the positive- and negative-strand 3′ UTR sequences. Initial mutagenesis suggests that an intact higher-order structure at the 3′(+) UTR is necessary for the interaction of NS3 with the 3′ terminus of the positive-strand RNA. Thus, our results show that NS3 specifically binds to the 3′ ends of both the positive- and negative-strand RNAs of HCV. The implication of NS3 binding to the 3′-terminal sequences of viral positive- and negative-strand RNA in HCV RNA replication is discussed below.
MATERIALS AND METHODS
Expression and purification of HCV NS3 and ΔpNS3.
The recombinant clones used for expression of the viral wild-type NS3 and the truncated version, ΔpNS3 (with an intact helicase domain but a deleted protease domain) were constructed by PCR amplification using the HCV 1969 cDNA clone (a kind gift from Genevieve Inchauspe, INSERM, Lyon, France) as a template. The coding sequences were ligated into the pET-21b expression vector (Novagen), following standard molecular biology protocols, at the BamHI and HindIII sites using the primer pairs RB-1 and RB-2 (for NS3) and RB-3 and RB-2 (for ΔpNS3) incorporating appropriate enzyme cleavage sites (Table 1).
TABLE 1.
Sequences of primers used in the present study
| Name | Sequencea |
|---|---|
| RB-1 | 5′-ATATTAAGGATCCGGCGCCCATCACGG-3′ |
| RB-2 | 3′-ATTAATTAAGCTTCGTGACAACCTCCAGGT-5′ |
| RB-3 | 5′-CGCAATAGGATCCGGTGGACTTTATCCC-3′ |
| RB-4 | 5′-ATATTAGAATTCGCCAGCCCCCTG-3′ |
| RB-5 | 3′-TTATATAAGCTTGGGGGGGTCCTGGAG-5′ |
| RB-6 | 5′-ATCCTCGAATTCTGATGGGGGACA-3′ (mutant I) |
| RB-7 | 5′-GCATTAGAATTCCACCATGAATCA-3′ (mutant II) |
| RB-8 | 5′-AATTAAGAATTCGCCAGgggggTGATGGGGGCGACACTC-3′ (mutant IV) |
| RB-9 | 5′-AATTATGAATTCGCCAGgggggTGATcccccCGACACTC-3′ (mutant V) |
| RB-10 | 5′-TTAAATGAATTCGCCAGtttttTGATaaaaaCGACACTC-3′ (mutant VI) |
| RB-11 | 5′-ATATAAGAATTCGCCAGtttttTGATGGGG-3′ (mutant VIII) |
| RB-12 | 5′-ATATAAGAATTCGCCAGCCCCCTGATaaaaaCGACACTCCA-3′ (mutant IX) |
| RB-13 | 5′-ATATAAGAATTCGCCAGCCCCCTGATcccccCGACACTCCA-3′ (mutant IX-A) |
| RB-14 | 5′-ATATAAGAATTCGCCAGCCCCCcGgcGGGGGCGACACTCCA-3′ (mutant IX-B) |
| RB-15 | 5′-TTAATAGAATTCGCCAGCgCCCTGATGGGcGCGACACTC-3′ (mutant X) |
| RB-16 | 5′-TTAATAGAATTCGCCAGCCgCCTGATGGcGGCGACACTC-3′ (mutant XI) |
| RB-17 | 5′-TTAATAGAATTCGCCAGCCCgCTGATGcGGGCGACACTC-3′ (mutant XII) |
| RB-18 | 5′-ATATAAGAATTCGCCAGcccGGTGATCCgggCGACACTCC-3′ (mutant V-[A]) |
| RB-19 | 5′-ATATAAGAATTCGCCAGGcccGTGATCgggCCGACACTCC-3′ (mutant V-[B]) |
| RB-20 | 5′-ATATAAGAATTCGCCAGGGcccTGATgggCCCGACACTCC-3′ (mutant V-[C]) |
| RB-21 | 5′-TTAATAGAATTCGCCAGGcGGGTGATCCCgCCGACACTCC-3′ (mutant V-[D]) |
| RB-22 | 5′-TTAATAGAATTCGCCAGGccGGTGATCCggCCGACACTCC-3′ (mutant V-[E]) |
| RB-23 | 5′-TTAATAGAATTCGCCAGGcGcGTGATCgCgCCGACACTCC-3′ (mutant V-[F]) |
| RB-24 | 5′-ATTTATGAATTCGCCAGCtCCCTGATGGGaGCGACACTC-3′ (mutant XIX) |
| RB-25 | 5′-GTCTATGAATTCGCCAGCttCCTGATGGaaGCGACACTC-3′ (mutant XX) |
| RB-26 | 5′-GTCTATGAATTCGCCAGCtCtCTGATGaGaGCGACACTC-3′ (mutant XXI) |
| RB-27 | 3′-GCAACGAAGCTTCTCATACTAACGCC-5′ (mutant VII) |
| RB-28 | 5′-TATAATGAATTCCGCCGCCCACAGGAC-3′ |
| RB-29 | 3′-AATTTAAAGCTTTACCTCGAGGTTGCG-5′ |
| RB-30 | 5′-GCATATGAATTCGATGAACGGGGAGC-3′ |
| RB-31 | 3′-ATGGGCAAGCTTACATGATCTGCAGA-5′ |
| RB-32 | 3′-CGCGCCAAGCTTATTAAAGAAGGGAAAAAG-5′ |
| RB-33 | 5′-AGACCCAAGCTTAACGGGG-3′ |
| RB-34 | 3′-ACATGATCTGCAGAGAGGCC-5′ |
| RB-35 | 5′-TAATACGACTCACTATA AGACCCAAGCTTAACGGGGAGC-3′ |
| RB-36 | 3′-GGCTCACGGACCTTTCACAGCTA-5′ |
| RB-37 | 3′-CTAGGGCTAAGATGGAGCCACCA-5′ |
| RB-38 | 5′-CAGCGCAAGCTTTAGTAACCCTACCTC-3′ |
| RB-39 | 3′-CGCGGCGAGCTCCCTCCGAATTAAAGA-5′ |
The lowercase italic letters in the oligonucleotide sequences from RB-8 through RB-26 denote specific altered bases incorporated in the corresponding mutants. The boldface italic letters in forward primer sequence RB-35 represent the T7 polymerase promoter sequence fused to the primer. Restriction enzyme cleavage sites are underlined.
Expression of the target protein in Escherichia coli BL21(DE3)-transformed cells was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside), and the expressed proteins were purified with Co2+ charged resin (Talon; Clonetech) as detailed previously (1). The purities of the isolated protein samples were evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining.
UTR cloning and probe preparation.
The viral 5′ UTR sequence encompassing the first 127 nucleotides was PCR amplified from the HCV 1969 cDNA clone using RB-4 and RB-5 as the forward and reverse primers. These oligonucleotides incorporated the appropriate restriction sites, and the resulting PCR product was gel purified, sequentially digested with HindIII and EcoRI, and ligated into the corresponding sites of the transcription vector pGEM-3 (Promega) using standard protocols. The nucleotide sequences of the recombinant clones used throughout were confirmed by dideoxy sequencing (Amersham). The cloning strategy was chosen so that the positive- or the complementary negative-strand transcripts [5′(+) UTR127 or 3′(−) UTR127, respectively] could be generated following linearization, using either SP6 or T7 RNA polymerase in the presence of [α32P]UTP (3,000 Ci/mmol; Amersham) in an in vitro transcription reaction. The labeled transcripts were purified by denaturing gel electrophoresis before they were used in binding reactions. The 5′ UTR mutants (I to XXI) were obtained using the same vector and were generated with the appropriate forward primers, as indicated in Table 1, and the same reverse primer used for the wild-type 5′ UTR127 (RB-5). For the rescue experiment using the mutants V-[A] to V-[F] (see Fig. 7), the fragment for cloning was derived with mutant V DNA as a template. For mutant VII, the forward primer used was the same as for the wild type (RB-4), and the reverse primer was RB-27. The similar-size RNA from the coding region used for specificity analysis (see Fig. 4B) was obtained from a clone constructed using the primer pairs RB-28 and RB-29.
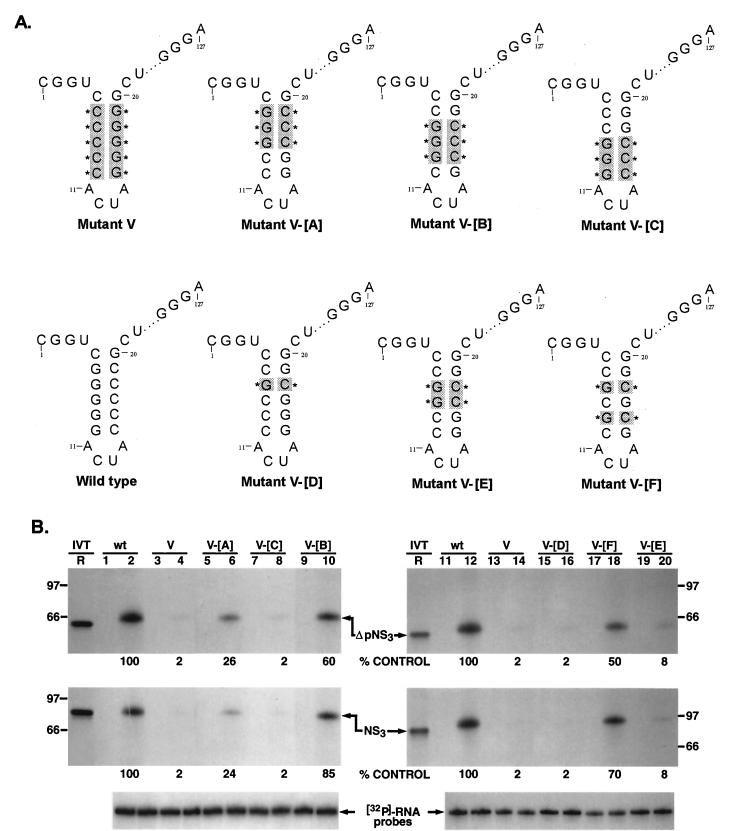
FIG. 7.
Rescue analysis of NS3 and ΔpNS3 binding to mutant V RNA. (A) Schematic illustration of the various mutant probes used for RNA-protein binding analysis. All of the probes contain mutant V RNA as a backbone. The reversion to the wild-type sequence of specific C-G→G-C base pairs in the 3′-proximal stem is marked by shading and an asterisk. The intervening sequences are indicated by dots. (B) Restoration of specific nucleoprotein complex between mutant RNA probes V-[A] to-[E] and NS3 (full length and truncated). Reactions containing either wild-type (wt) or mutant RNA are indicated above the gels. The odd-numbered lanes served as controls without the added proteins, while the even-numbered lanes represent binding reactions containing approximately 100 ng of either purified ΔpNS3 or NS3 protein. Lane R represents the [35S]methionine-labeled in vitro-translated (IVT) ΔpNS3 (top) or NS3 (middle) protein, and the numbers on the left and right indicate the positions of migration of the molecular mass marker proteins in kDa. Only the relevant IVT proteins are shown along with the appropriate binding reactions. The bottom gel shows the results of probe stability analysis with the NS3 protein, the details of which are discussed in Materials and Methods. Quantitation of the RNA-protein complex is indicated as percent control for both NS3 and ΔpNS3 binding.
FIG. 4.
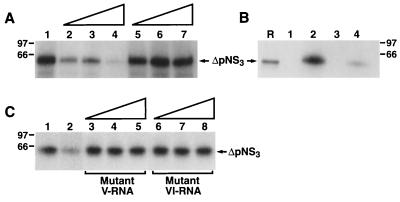
Specificity of ΔpNS3 binding to the negative-strand RNA probe. (A) Cold homologous RNA corresponding to the 3′-terminal 127 bases of the negative strand and a heterologous, similar-size RNA sequence from hepatitis A virus were used in the competition assay as described in Materials and Methods. Reactions were performed without (lane 1) or with 50 (lane 2), 100 (lane 3), or 250 (lane 4) ng of cold homologous RNA or 50 (lane 5), 100 (lane 6), or 250 (lane 7) ng of heterologous RNA. (B) Specific binding to negative-strand RNA. A similar-size RNA obtained from the coding sequence of the HCV negative strand, labeled and processed as described for the UTR127 probe, was added to the binding reaction containing the ΔpNS3 protein. Lane R represents the [35S]methionine-labeled in vitro-translated ΔpNS3 protein. Protein binding to 3′(−) UTR127 probe without (lane 1) and with (lane 2) added ΔpNS3 is shown. Lanes 3 and 4 shows binding to RNA derived from the coding sequence in the absence (lane 3) and presence (lane 4) of ΔpNS3. (C) Competition using inactive mutant V and VI UTR127RNAs. The cold mutant RNAs were added to binding reactions as for panel A. Reactions with no cold competing RNA (control) (lane 1), with homologous RNA (lane 2), and with increasing concentrations (50, 100, and 250 ng/25-μl reaction volume) of mutant V or VI RNA (lanes 3 to 8) are shown. Homologous RNA was used at the same concentration as for lane 3 in panel A.
The 3′ UTR sequence was PCR amplified from the plasmid pCV-H77C (a kind gift from Jens Bukh, National Institutes of Health [NIH]). The amplified product was cloned into pGEM-3 between the HindIII and EcoRI sites using the primer set RB-30 and RB-31. The clone with the 98-nucleotide conserved X sequence deleted (mutant A [see Fig. 10A]) was made using RB-30 as the forward primer and RB-32 as the reverse primer.
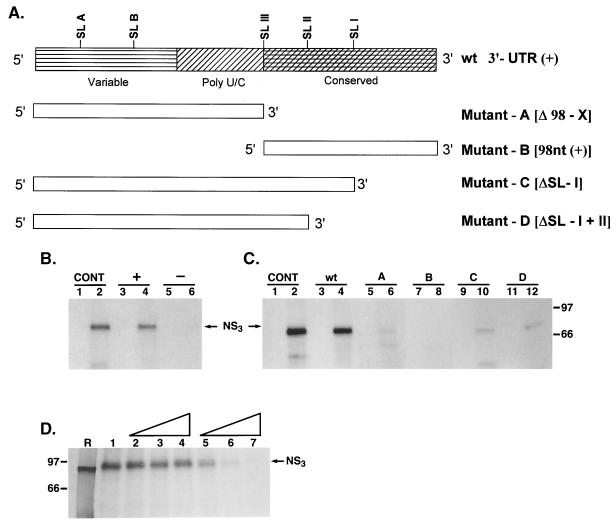
FIG. 10.
Analysis of NS3 binding to the 3′ UTR of the positive strand. (A) Schematic representation of the viral 3′ UTR RNA and the derived mutants used in analysis. The mutants with deleted RNA sequences (mutants A to D) are shown. (B) UV-cross-linking analysis of NS3 binding to 3′ UTR RNA of the positive strand (lanes 3 and 4) and the corresponding complementary region of the negative-strand 5′ UTR probe (lanes 5 and 6). The specific interaction of NS3 with the negative-strand 3′ UTR127 probe was included as a control (CONT) (lanes 1 and 2) (see Materials and Methods). (C) Binding analysis using mutant RNA probes (A, B, C, and D). NS3 binding to the mutant RNA probes (lanes 5 to 12) is compared to its binding to the wild-type 3′(+) UTR probe (lanes 1 and 2). Reactions with mutant RNA probes are indicated above the gel. The reactions in lanes 1 and 2 are similar to those in panel B. The odd-numbered lanes in each set contain control reactions with no added protein, while the even-numbered lanes represent binding reactions containing approximately 100 ng of purified NS3 protein. The positions of UV-cross-linked complexes are indicated. (D) Specificity of NS3 interaction with 3′ UTR probe RNA. Cold homologous RNA corresponding to the 3′ UTR and a heterologous viral RNA with poly(U) sequence were added to reactions as detailed in Materials and Methods. Reaction mixtures contained no competitor RNA (control reaction [lane 1]); 50 (lane 2), 250 (lane 3), or 500 (lane 4) ng of cold heterologous RNA; or similar amounts of homologous RNA (lanes 5 to 7). Lane R is the [35S]methionine-labeled in vitro-translated NS3 protein. The positions of the proteins are marked.
The additional HCV 3′ UTR clone with 13 uridine residues, pHCV-3′(+), and the plasmid containing the 98-nucleotide fragment of the 3′ UTR conserved region (mutant B [see Fig. 10A]) were kind gifts from M. Lai, University of Southern California. Mutants C and D were obtained by using pHCV-3′(+) DNA as the template following two cycles of PCR amplification with RB-33 and -34 as the forward and reverse primers, respectively, in the first cycle and RB-35 and -36 (for mutant C) and RB-35 and -37 (mutant D) in the second cycle as described earlier (24, 40). The probe added in the control reactions in one experiment (see Fig. 10) was generated using T7 RNA polymerase from the plasmid pGEM-4Z with the 5′(+) UTR127 sequence ligated between the HindIII and EcoRI sites. This manipulation was essential in order to use T7 RNA polymerase for 3′(−) UTR127 probe synthesis.
The heterologous cold RNA added during competition in the specificity reactions (see Fig. 10D) was derived from a clone containing the intact poliovirus 3′ UTR. Briefly, the UTR was amplified from an infectious cDNA clone using the oligonucleotides RB-38 (sense) and RB-39 (antisense), incorporating either HindIII or SacI enzyme sites. The gel-purified product was ligated to pSP-64 (A) vector (Promega) at the corresponding sites. The recombinant plasmid obtained was digested sequentially once again with HindIII and EcoRI, and the required fragment with poly(A) sequence was isolated and recloned into pGEM-3. From this clone, UTR RNA with poly(U) residues was derived using T7 polymerase and was added to the binding reactions.
RNA binding, UV-cross-linking analysis, and probe stability analysis.
The binding reaction (25 μl) contained binding buffer III, 20 mM dithiothreitol, 2 mM ATP, 15 μg of yeast tRNA, RNasin (30 U), and approximately 100 ng of purified protein. The binding reaction was preincubated with purified NS3 or ΔpNS3 in a 30°C water bath for 5 min, following which the 32P-labeled RNA probe (200,000 cpm/25-μl sample) was added. The competitor RNAs were preincubated with the protein for 10 min prior to the addition of the probe. The incubation was continued for an additional 15 min to facilitate the binding of the probe RNA to the protein. At termination, samples were cross-linked by UV irradiation as detailed in references 1 and 2, and samples were analyzed by SDS–14% PAGE. The signal intensity from each lane was quantitated with a laser densitometer scanner (Molecular Dynamics), and the data were analyzed with the Image-QuaNT software program. Each reaction was performed two or three times for reproducibility.
The various binding buffers used were as follows: binding buffer I, 5 mM HEPES (pH 7.9), 0.5 mM MgCl2, 25 mM KCl, and 0.5% glycerol; binding buffer II, 5 mM HEPES (pH 7.9), 2 mM MgCl2, 25 mM KCl, 10 mM NaCl, and 0.5% glycerol; and binding buffer III, 5 mM HEPES (pH 7.9), 2 mM MgCl2, 25 mM KCl, and 0.5% glycerol. Binding buffer III was used throughout this study.
The stabilities of the 32P-labeled wild-type or mutant RNA probes in the presence or absence of the viral proteins were examined following incubation under standard binding conditions. The samples were deproteinized twice, followed by alcohol precipitation of the RNA, which was subsequently analyzed on a denaturing sequencing gel (8% acrylamide–8 M urea).
In vitro transcription and translation.
The standard protocol for in vitro transcription was followed. Briefly, the expression clone, pET-NS3 or pET-ΔpNS3, was linearized using HindIII, purified, and used as a template for the synthesis of capped mRNA. The mRNAs obtained were used in translation reactions with rabbit reticulocyte lysate (Promega) according to the manufacturer's instructions. In vitro-synthesized proteins were labeled with 40 μCi of [35S]methionine (specific activity, >100 Ci/mmol; Amersham) and resolved along with UV-cross-linked protein samples on SDS-PAGE gels. In selected experiments, the coupled transcription-translation system (Promega) was also used.
Immunoprecipitation.
The in vitro-translated protein or the protein-nucleotidyl complexes were immunoprecipitated with the same concentrations of monoclonal antibody to NS3 (AUSTRAL Biologicals, San Ramon, Calif.) or a nonspecific antibody targeted toward CREB. Three separate UV-cross-linked samples were pooled for immunoprecipitations. The reaction mixtures were incubated at 4°C for 3 h with 1× radioimmunoprecipitation assay buffer (20 mM Tris-HCl [pH 7.5], 0.5% deoxycholate, 1% NP-40, and 150 mM NaCl), 0.5 mM phenylmethyl sulfonyl fluoride, and 5 mg of bovine serum albumin. Immune complexes were adsorbed on protein A-Sepharose beads (5 mg/reaction; Pharmacia) for 1 h at 4°C, following which the beads were washed five times with radioimmunoprecipitation assay buffer to reduce background binding and washed once with phosphate-buffered saline. The proteins were finally eluted with Laemmli sample buffer and resolved on an SDS–14% PAGE gel, followed by fixing and autoradiography.
RESULTS
HCV protease/helicase NS3 interacts specifically with the 3′ terminus of HCV negative-strand RNA.
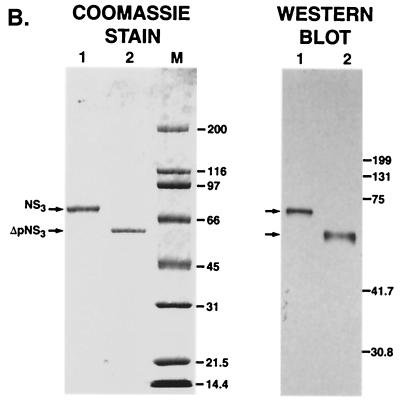
In order to examine the interaction of NS3 with the 3′-terminal sequence(s) of HCV negative-strand RNA, both NS3 and the truncated protein (ΔpNS3) were expressed in E. coli BL21(DE3) cells. Both proteins contained a T7 tag at the N terminus and six histidine residues in tandem at the C terminus. The additional amino acids contributed by the epitopes increase the mass of the expressed proteins by approximately 2 kDa. The bacterially expressed proteins were purified by Co2+ immobilized affinity chromatography, and the purified polypeptides were analyzed by SDS-PAGE followed by Coomassie blue staining. As shown in Fig. 1B, both NS3 and ΔpNS3, with estimated molecular masses of 75.6 and 55.6 kDa, respectively, were purified to near homogeneity (approximately 98%). To reconfirm that the proteins seen in the Coomassie blue-stained gel were indeed NS3 and ΔpNS3, Western blot analysis was performed with available antibodies against the N-terminal T7 tag. The signals obtained in the immunoblot analysis corresponded correctly with the expected migrations of the polypeptides.
FIG. 1.
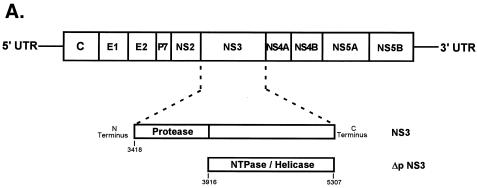
HCV-encoded NS3 protein expression. (A) Schematic representation of HCV genome organization marking the relative position of NS3 protein. The numbers correspond to nucleotide positions in the HCV 1969 cDNA clone. (B) On the left is a Coomassie blue-stained gel of purified NS3 and ΔpNS3. The positions of the NS3 (lane 1) and ΔpNS3 (lane 2) proteins are indicated, along with the protein marker lane (M). On the right is a Western blot analysis of the purified proteins resolved on a 14% gel; lanes 1 and 2 correspond to the same protein samples run on the Coomassie blue-stained gel. The numbers on the right correspond to the migrations of the molecular mass marker proteins and are marked in kDa.
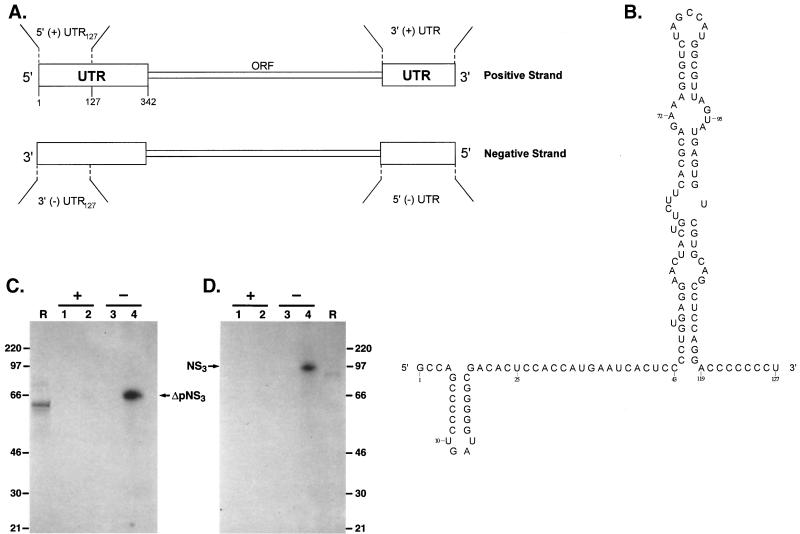
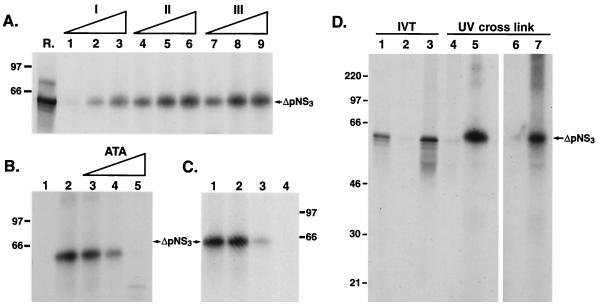
To determine whether NS3 and ΔpNS3 interact with the terminal sequence of the HCV RNA, 32P-labeled RNA probes representing the first 127 nucleotides from the 5′ terminus of positive-strand RNA or the corresponding sequence from the 3′ terminus of negative-strand RNA were prepared (Fig. 2A). Initial experiments with the full-length 3′(−) UTR showed interaction with NS3. Later, however, it was found that the majority of the NS3 binding was localized within the first 127 nucleotides of the 3′(−) UTR (data not shown). The predicted secondary structure of the 5′(+) UTR127 is shown in Fig. 2B. The structure of the 5′(+) UTR127, but not that of the 3′(−) UTR127, has been confirmed by chemical and enzymatic analysis (7, 20). Our M-fold analysis of the structure of 3′(−) UTR127 revealed multiple forms having very similar ΔG values(−50 to −48 kcal/mol [data not shown]). We have used a predicted structure of 3′(−) UTR127 which resembles that of the 5′(+) UTR127 until the actual secondary structure of the 3′(−) UTR127 is determined experimentally. The 32P-labeled RNA probes were incubated with purified NS3 and ΔpNS3, and the UV-cross-linked RNA-protein complexes were visualized by SDS-PAGE followed by autoradiography. The results clearly show that both NS3 and ΔpNS3 are capable of interacting with the 3′(−) UTR127, but no detectable complex was seen with the corresponding complementary sequence of the 5′(+) UTR127 of HCV RNA (Fig. 2C and D, lanes 3 and 4 versus lanes 1 and 2). No complexes were observed when the NS3 or ΔpNS3 protein was excluded from the complete reaction. The protein-nucleotide complexes migrated slightly more slowly than the [35S] methionine-labeled in vitro-translated protein (Fig. 2C and D, compare lanes R with lanes 4). We have previously shown that the poliovirus-encoded 2C protein (also with helicase and ATPase activities) migrates more slowly than the protein itself when cross-linked to one or more nucleotides (2). This is presumably due to the negative charges contributed by the nucleotides covalently linked to the protein. The interaction of ΔpNS3 with the 3′(−) UTR127 was linear with increasing concentration of the purified protein, and significant binding was obtained when the reaction buffer contained 5 mM HEPES, 2 mM MgCl2, 25 mM KCl, 10 mM NaCl, and 0.5% glycerol (Fig. 3A, lanes 4 to 6). Reducing MgCl2 to 0.5 mM (lanes 1 to 3) resulted in significant loss of binding. Omitting NaCl from the reaction mixture did not have a significant effect on binding of ΔpNS3 to the 3′(−) UTR127 (lanes 7 to 9). The formation of the ΔpNS3-RNA complex was sensitive to aurine tricarboxylic acid (ATA), a well-known inhibitor of protein-nucleic acid interaction. Almost total inhibition of binding was observed at 10 μM ATA (Fig. 3B, lane 5). Both SDS and proteinase K also inhibited complex formation (data not shown). The extents of nucleoprotein complex formation were similar when ΔpNS3 was preincubated at 30 and 40°C before being added to the binding reaction mixture (Fig. 3C, lanes 1 and 2). However, binding was significantly inhibited when preincubation was performed at 65 and 90°C (Fig. 3C, lanes 3 and 4), indicating the heat-labile nature of the protein.
FIG. 2.
HCV-encoded NS3-RNA interaction. (A) Schematic representation of the viral positive-strand UTR used in the present study. (B) Predicted secondary structure of the first 127 nucleotides of the sequence of the positive strand, 5′(+) UTR127 (adapted from reference 20). (C and D) Analysis of purified ΔpNS3 and NS3 binding to the 5′ positive- or 3′ negative-strand UTR127 RNA probe. The binding reaction mixture contained either 5′(+) UTR127 (lanes 1 and 2) or 3′(−) UTR127 (lanes 3 and 4) probe. Lanes 1 and 3 are controls with no protein added while in lanes 2 and 4 approximately 100 ng of ΔpNS3 (C) or NS3 (D) was added. The migration of the [35S]methionine-labeled in vitro-translated wild-type or truncated NS3 is shown in lanes R, and the numbers correspond to the migrations of rainbow molecular mass markers (Amersham) in kDa. The relative position of the UV-cross-linked RNA-protein complex is indicated.
FIG. 3.
Characterization of the interaction between ΔpNS3 and 3′ UTR127 RNA. (A) Effect of binding buffer composition on nucleoprotein complex formation. Reaction mixtures containing ΔpNS3 were incubated with RNA probes under various binding buffers as detailed in Materials and Methods. The protein was analyzed for each binding buffer condition at three concentrations (approximately 50, 100, and 150 ng/25-μl reaction volume), and UV-cross-linked complexes were resolved by SDS–14% PAGE. Lane R represents in vitro-translated [35S]methionine-labeled ΔpNS3 protein, and the numbers on the left indicate the positions of the molecular mass marker proteins in kDa. (B) Effect of nucleoprotein inhibitor ATA on complex formation. Reactions with (lane 2) or without (lane 1) ΔpNS3 and reactions in the presence of increasing concentrations (2.5, 5.0, and 10 μM) of ATA and ΔpNS3 (lanes 3 to 5) are shown. (C) Effect of heat denaturation of ΔpNS3 on RNA interaction. The ΔpNS3 protein was preincubated for 10 min at 30, 40, 65, and 90°C (lanes 1 to 4) prior to its addition to the binding reaction and then was processed as described above. (D) Immunoprecipitation analysis of the UV-cross-linked RNA-protein complex. Purified ΔpNS3 UV cross-linked to 3′(−) UTR127 was immunoprecipitated as detailed in Materials and Methods. [35S]methionine-labeled in vitro-translated (IVT) ΔpNS3 protein was analyzed directly (lane 1) or following immunoprecipitation with anti-NS3 (lane 3) or a nonspecific antibody (lane 2). Lanes 4 to 7 contain UV-cross-linked RNA-protein complex either loaded directly (lanes 4 and 5) or following immunoprecipitation with the control (lane 6) or anti-NS3 (lane 7) antibody. The reaction mixtures in lanes 5 to 7 contained ΔpNS3, but lane 4 had no added ΔpNS3. The portion of the gel containing lanes 6 and 7 was overexposed to visualize the ΔpNS3 band.
To rule out the possibility that the RNA-protein complex detected by UV-cross-linking analysis is due to the presence of one or more contaminating E. coli proteins, the ΔpNS3-3′(−) UTR complex was immunoprecipitated with an antiserum specific to NS3. As expected, the ΔpNS3-RNA complex was specifically immunoprecipitated by anti-NS3 but not by an unrelated antibody (Fig. 3D, lanes 6 and 7). These results confirmed that the protein-nucleotidyl complex contains ΔpNS3. The same experiment was repeated with purified recombinant NS3, and the results were similar to those shown in Fig. 3D. The possibility that the T7 and/or His tags fused to the NS3 (or ΔpNS3) protein contributed to RNA binding was ruled out by the demonstration that a mutant NS3 protein that had internal deletions but still retained the tag was unable to interact with the UTR sequence (data not shown).
Specificity of RNA-protein interaction.
To determine if the binding of ΔpNS3 to the 3′(−) UTR127 RNA was specific, competition binding assays were performed in which unlabeled homologous and heterologous RNAs were added to the binding reaction during formation of the 32P-labeled UTR-ΔpNS3 complex. While the addition of 20- and 50-fold molar excesses of unlabeled 3′(−) UTR127 resulted in approximately 60 to 70% reduction in binding, the intensity of the labeled complex was reduced to almost 10% by the addition of a 100-fold molar excess of unlabeled homologous RNA (Fig. 4A, lanes 2 to 4). No significant reduction in the intensity of the RNA-protein complex was noted in the presence of 20-, 50-, and 100-fold molar excesses of cold, heterologous, similar-size RNA from hepatitis A virus (Fig. 4A, lanes 5 to 7). Additionally, when an unrelated similar-size labeled RNA fragment from the HCV negative strand was used in the binding reaction, only approximately 4% as much RNA-ΔpNS3 complex formation was observed as with the wild type 3′(−) UTR127 probe (Fig. 4B, compare lane 2 with lane 4). Finally, two mutant 3′(−) UTR127 RNAs that were defective in binding NS3 and ΔNS3 (mutants V and VI; see below) were unable to compete with 3′(−) UTR127 probe in the binding assay over a range of concentrations (Fig. 4C). Taken together, these results suggest that the interaction of ΔpNS3 with the 3′(−) UTR127 sequence is specific.
3′-Terminal sequences of the negative-strand UTR127 RNA are important for protein binding.
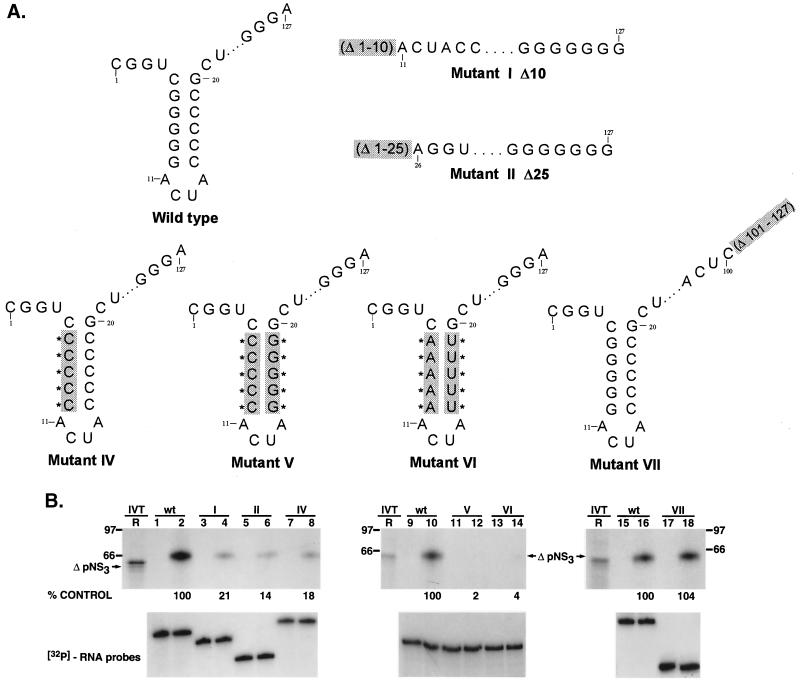
To determine sequences within the 3′ UTR RNA (127 nucleotides) of HCV required for NS3 binding, we initially deleted 10 and 25 nucleotides from the 3′ terminus. These mutants were termed Δ10 (mutant I) and Δ25 (mutant II). Compared to the wild-type 3′(−) UTR127 Fig. 5B, lane 2, both mutants I and II were defective in RNA binding (lanes 4 and 6). While deletion of the first 10 nucleotides from the 3′ terminus of the negative-strand (−10; mutant I) (the minus signs preceding the nucleotide numbers denote the position in the negative-strand RNA starting from the 3′ end) resulted in almost 80% decrease in RNA binding (lane 4), removal of the first 25 nucleotides (−25; mutant II) reduced binding by approximately 85% (lane 6). In both mutants I and II, a stem-loop structure within the first 20 nucleotides from the 3′ end was deleted (Fig. 5A). Therefore, the initial results suggested that the predicted stem-loop spanning nucleotides −5 through −20 may be important for ΔpNS3-RNA interaction. To determine if the stem-loop was somehow involved in NS3 binding, the stem was destabilized by replacing the five guanosine residues with cytosines (Fig. 5A, mutant IV). As expected, like mutants I and II, mutant IV was also defective in ΔpNS3 binding (18% binding remaining [Fig. 5B, lane 8]). It therefore appeared that the stem within the stem-loop structure adjacent to the 3′ terminus of the negative-strand UTR127 RNA was important for NS3 binding. Deletion of the last 27 nucleotides from the UTR RNA (nucleotides −101 through −127; mutant VII) did not significantly alter protein binding (Fig. 5B, compare lanes 16 and 18 [mutant VII]). Similar results were obtained when ΔpNS3 was replaced by NS3 in the binding reaction with the above-mentioned mutants (data not shown).
FIG. 5.
Analysis of HCV-encoded ΔpNS3 interaction with negative-strand 3′ UTR127 mutant RNA probes. (A) Schematic illustration of the mutant RNAs used in the binding assay. Mutants with deleted (mutants I, II, and VII) or altered (mutants IV, V, and VI) bases compared to the wild-type RNA are indicated by shading and Δ or ∗, respectively. The intervening sequences are indicated by dots. (B) UV-cross-linking analysis of ΔpNS3 protein binding to mutant RNAs. Lanes 1, 2, 9, 10, 15, and 16 represent binding to wild-type (wt) RNA and the remaining lanes show binding to the mutant RNAs as indicated above each set of lanes. The odd-numbered lanes served as control reactions with no added protein, while the even-numbered lanes represent binding reactions containing approximately 100 ng of purified ΔpNS3. Lane R is the [35S]methionine-labeled in vitro-translated ΔpNS3, and the numbers on the left and right indicate the positions of the rainbow molecular mass marker proteins in kDa. The lower gels show the stability of the RNA probes in the absence and presence of ΔpNS3 protein, the details of which are discussed in Materials and Methods. The relative binding of ΔpNS3 protein to mutant RNAs compared to the control wild-type RNA is shown as percent control.
To rule out the possibility that these mutant RNAs were unstable under the binding assay conditions, parallel reactions were examined for RNA stability. After incubation of various mutant 32P-labeled RNA probes with (or without) protein, the labeled RNAs were isolated by phenol-chloroform extraction and examined by gel analysis. As shown in the lower gels of Fig. 5B, all of the mutant RNAs tested were just as stable as the wild-type 3′(−) UTR127 RNA probe.
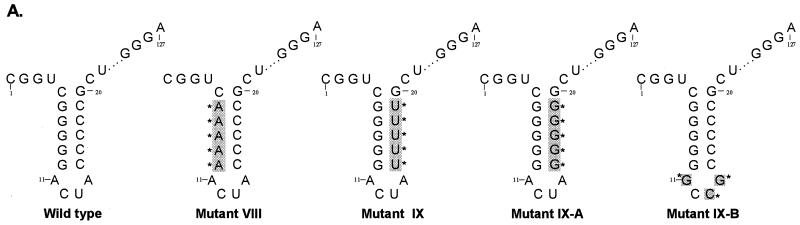
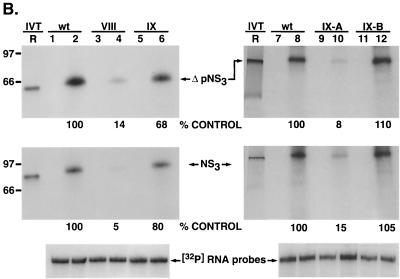
To address the question of whether the stem-loop structure alone is important for binding or whether the identities of the bases within this structure are also relevant, we replaced the five guanosine and cytosine pairs by the alternative purine and pyrimidine bases, adenosine and uridine (mutant VI). In this case, no significant binding of ΔpNS3 (or NS3 [data not shown]) was observed (Fig. 5B, lanes 13 and 14). This suggested that the presence of guanosine-cytosine pairs in the stem rather than just a double-stranded structure was important for interaction with NS3. Also, when the positions of the five consecutive guanosine-cytosine pairs in wild-type RNA were flipped (nucleotides −6 through −10; mutant V), no protein-RNA complex could be detected (Fig. 5B, lanes 11 and 12). Again, as shown at the bottom of Fig. 5B, this was not due to reduced stability of the mutant RNAs. The relative importance of the five guanosine residues from positions −6 through −10 for protein interaction was also demonstrated by replacing either the five guanosines or the five cytosines in the stem with adenosines and uridines, respectively (Fig. 6A). As shown, in Fig. 6B, while G-to-A substitutions reduced ΔpNS3 binding by 86% over the control (mutant VIII; lanes 3 and 4), C-to-U substitutions reduced binding by only 32% (mutant IX; lanes 5 and 6). When NS3 was used in the binding assay, the results were more pronounced; while G-to-A substitution affected binding by 95%, C-to-U changes inhibited binding by only 20% of the control level. To test the possibility that an intact double-stranded structure of the stem is important for RNA-protein interaction, the stem was destabilized by replacing all cytosine residues with guanosine residues (mutant IX-A) (Fig. 6A). This mutant affected ΔpNS3 and NS3 binding by 92 and 85%, respectively, compared to the wild-type control, suggesting the importance of an intact helical structure of the stem (Fig. 6B, lanes 8 and 10). To test the contribution of the ACUA loop in protein binding, these nucleotides were replaced by GCCG (mutant IX-B) (Fig. 6A). These changes in the loop did not have any significant effect on NS3 (or ΔpNS3) binding (Fig. 6B, lanes 8 and 12). These results indicated that in addition to the double-stranded structure, the presence of the five guanosines in the 3′-proximal arm of the stem was important for interaction with NS3.
FIG. 6.
Analysis of NS3 and ΔpNS3 binding to the 3′ negative-strand UTR127 mutant RNAs. (A) Schematic illustration of the mutant RNAs used in binding analysis. The altered bases in the mutant RNAs compared to the wild-type RNA are shaded and marked by asterisks. The intervening sequences are indicated by dots. (B) Specific RNA binding activity of NS3 and ΔpNS3 proteins using the mutant RNA probes. Reactions showing binding to either wild-type (wt) or mutant (mutant VIII, IX, IX-A, and IX-B) RNA probes are indicated above the gel. The odd-numbered lanes contain control reactions with no added proteins, while the even-numbered lanes represent binding reactions containing either the ΔpNS3 or NS3 protein. Lanes R represent [35S]methionine-labeled in vitro-translated (IVT) ΔpNS3 or NS3, respectively, and the numbers on the left indicate the positions of the molecular mass marker proteins in kDa. Only the relevant IVT proteins are shown along with the appropriate binding reactions. The bottom gel shows the result of probe stability analysis with NS3 protein, the details of which are discussed in Materials and Methods. Quantitation of RNA-protein complexes is indicated as percent control.
G-C pairs at positions −7, −8, and −9 in the 3′ UTR127 RNA-proximal stem are important for protein binding.
To determine if all five G-C pairs present in the stem were important for NS3 binding, we introduced mutations into the binding-inactive backbone of mutant V to restore NS3 binding. It is important to note that in mutant V RNA, the positions of the five Gs and Cs in the stem were flipped so that the Cs were 3′ proximal in the mutant instead of the Gs being 3′ proximal, as in the wild-type 3′(−) UTR127. Replacing three C-G pairs at positions −6, −7, and −8 in the mutant V backbone with the wild-type G-C sequence (mutant V-[A]) restored only 26% binding compared to the wild-type (Fig. 7B, lanes 5 and 6) while having three G-C pairs at positions −8, −9, and −10 (mutant V-[C]) did not bring back any significant level of ΔpNS3 binding (lanes 7 and 8). Similar results were obtained when ΔpNS3 was replaced with NS3 in the binding reaction (Fig. 7B, middle). Restoring three G-C base pairs at positions −7, −8, and −9 (mutant V-[B]), however, brought back 60 to 85% of the binding activity compared to the wild type (lanes 9 and 10). Mutant V-[D], in which only the −7 position had a G-C reversion, retained only 2% of the binding activity, and introducing two G-C base pairs at positions −7 and −8 brought the binding activity to only 8% of the control (lanes 19 and 20; mutant V-[E]). Interestingly, replacing two G-C pairs at positions −7 and −9, however, restored 50 to 70% of the binding activity compared to that of the wild-type UTR127 RNA (lanes 17 and 18; mutant V-[F]). These results suggested that the G-C base pairs at positions −7, −8, and −9 in the negative strand may be important for protein binding.
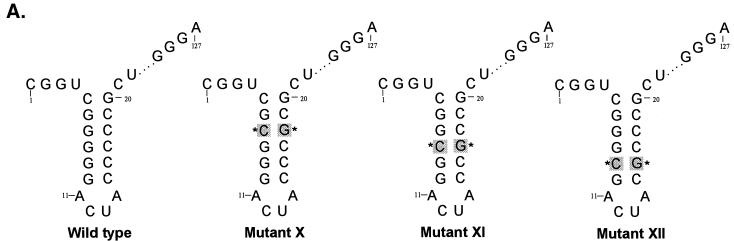
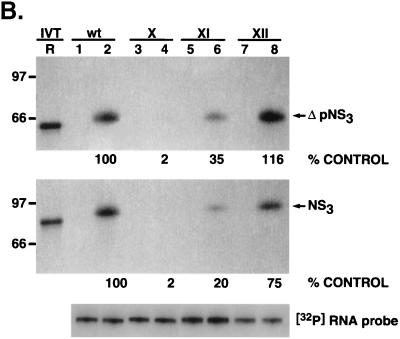
To confirm the above results, specific point mutations confined within these G-C base pairs were introduced into the wild-type UTR127 RNA backbone. Changing the G-C pair at position −7 to the flipped sequence C-G (mutant X) (Fig. 8, lanes 3 and 4) severely impaired both ΔpNS3 and NS3 binding compared to the wild-type (2% of the binding remained). Mutant XI, with the G-C at position −8 changed to C-G, retained approximately 20 to 35% of the binding (lanes 5 and 6), while changing the G-C base pair at position −9 to C-G in mutant XII had only a marginal effect, if any (lanes 7 and 8). These results suggested that the G-C pairs in the stem at the −7 and −8 positions are critical for interaction of 3′(−) UTR127 with NS3.
FIG. 8.
Analysis of NS3 and ΔpNS3 protein binding to 3′ negative-strand UTR127 RNA mutants with a single G-C→C-G change. (A) Schematic illustration of the mutant RNA probes used in binding analysis. The base pair sequences in the mutant RNAs that are altered compared to the wild-type sequence are marked by shading and asterisks. The intervening sequences are indicated by dots. (B) Specific RNA binding activity of NS3 and ΔpNS3 proteins to mutant RNA probes. Reaction lanes showing binding to either wild-type (wt) or mutant (X, XI, and XII) RNA probes are indicated above the gels. The odd-numbered lanes contain control reactions with no added proteins, while the even-numbered lanes represent binding reactions containing approximately 100 ng of either purified ΔpNS3 or NS3 protein. Lane R represents the [35S]methionine-labeled in vitro-translated (IVT) ΔpNS3 or NS3 protein, and the numbers on the left indicate the positions of migration of the molecular mass marker proteins in kDa. Only the relevant IVT proteins are shown along with the appropriate binding reactions. The bottom gel shows the result of probe stability analysis with NS3 protein, the details of which are discussed in Materials and Methods. Quantitation of the RNA-protein complex is indicated as percent control for both NS3 and ΔpNS3 binding.
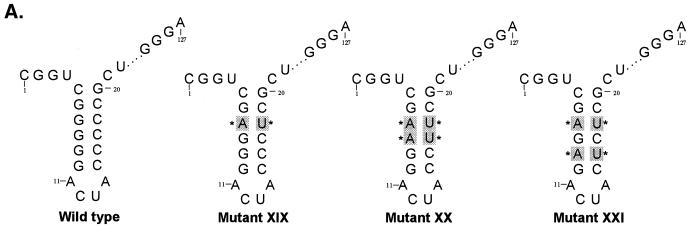
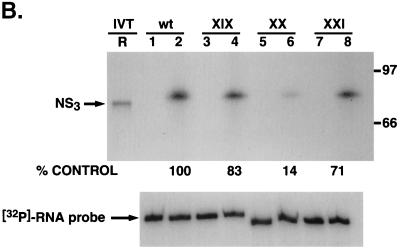
Consistent with the results obtained with NS3 and mutant V and VI UTR protein interaction, we replaced the G-C base pair with the purine-pyrimidine pair, A-U, either singly or in double pairs at the −7, −8, and −9 positions in the wild-type RNA backbone (Fig. 9A). As anticipated, significant loss of binding (85% compared to the wild-type control RNA) was apparent when the base pairs at positions −7 and −8 (Fig. 9B, lanes 5 and 6; mutant XX) were replaced. However, by contrast, 71 to 83% of the binding (compared to the wild-type RNA) remained when either the base pair at position −7 (mutant XIX) or the base pairs at positions −7 and −9 (mutant XXI) were changed to A-U. It is worth pointing out that while replacement of the G-C pair at the −7 position in the wild-type backbone with a C-G pair decreased protein-RNA interaction drastically (Fig. 8B, compare lane 4 with lane 2), the replacement of the same G-C pair with A-U did not significantly alter NS3-RNA interaction (Fig 9B, compare lane 4 with lane 2). The precise reason for this discrepancy is not clear, but it could be due to the fact that the presence of a purine base at position −7 of the 3′-proximal stem may be crucial for specific binding.
FIG. 9.
Analysis of NS3 binding to additional 3′UTR127 RNA mutants with specific G-C→A-U changes. (A) Schematic illustration of the mutant RNA probes used in the binding analysis. The base pair sequence in the mutant RNAs that have been altered compared to the wild-type sequence are marked by shading and asterisks. The intervening sequences are indicated by dots. (B) Specific RNA binding activity of NS3 protein to mutant RNA probes. Reaction lanes with binding to either wild-type (wt) or mutant (XIX, XX, and XXI) RNA probes are indicated above the gels. The odd-numbered lanes contain control reactions with no added protein while the even-numbered lanes represent binding reactions containing 100 ng of purified NS3 protein. Lane R represents the [35S]methionine-labeled in vitro-translated NS3 protein, and the numbers on the right indicate the migrations of the molecular mass marker proteins in kDa. The lower gel represents the result of probe stability analysis with and without NS3 protein in the binding reaction, as in the experiments discussed above. The relative binding of NS3 protein to mutant RNAs compared to the wild-type RNA is shown as percent control.
NS3 also interacts with the 3′-terminal sequences of the positive-strand RNA.
Since initiation of negative-strand synthesis is likely to occur at or near the 3′ terminus of the positive-strand RNA, we examined whether NS3 is capable of interacting with the 3′-terminal sequences of the positive-strand RNA. The 3′ UTR sequence of genomic RNA was cloned into the transcription vector, and RNA was transcribed from the clone, similar to the procedure with the negative-strand UTR127 probe. The 32P-labeled 3′(+) UTR RNA was incubated with purified recombinant NS3, and the resulting RNA-protein complexes were analyzed by UV cross-linking. As shown in Fig. 10B, NS3 readily formed complexes with the 3′(+) UTR of HCV (lanes 3 and 4). The intensity of the protein-nucleotidyl complex was 70% of that with 3′(−) UTR127 RNA (Fig. 10B, compare lane 4 with lane 2). The 5′-terminal sequences of the negative strand [5′(−) UTR] that are complementary to the 3′(+) UTR sequence showed very little binding to NS3 [approximately 3% compared to the 3′(+) UTR binding (Fig. 10B, lanes 5 and 6)]. These results suggested that NS3 is capable of interacting with the 3′-terminal sequences of both positive- and negative-strand RNA.
The 3′(+) UTR of HCV RNA contains a variable sequence followed by a stretch of poly(U) and/or poly(UC), which varies in size in various HCV strains, and a 98-nucleotide 3′-terminal X region with a fairly conserved sequence (Fig. 10A). To determine if these regions play a role in NS3 binding, UV cross-linking studies were carried out with mutant 32P-labeled 3′(+) UTR probes in which either the 98-nucleotide X region (mutant A) (Fig. 10A) or the preceding variable sequence and the poly(U) and/or poly(UC) stretch were deleted (mutant B) (Fig. 10A). As shown in Fig. 10C, the RNA containing the variable sequence along with the poly(U) and/or poly(UC) stretch showed very little NS3 binding in the absence of the 98-nucleotide X region (lanes 5 and 6; mutant A). Similarly, the 98-nucleotide conserved region without the variable and poly(U) and/or poly(UC) sequences was totally defective in NS3 binding (Fig. 10C, lanes 7 and 8; mutant B). When the 3′-terminal SL-I or both (SL-I and -II were deleted from the entire 3′UTR (mutant C and D; ΔSL-I and ΔSL-I and II), NS3 binding to the 3′(+) UTR was drastically reduced (Fig. 10C, lanes 9 to 12). While these data suggest that SL-I is important, mutant B (with the 98-nucleotide X region only), which still contains SL-I, is also defective in NS3 binding. These results suggest that the presence of the intact 3′-terminal sequences, including the variable region, the poly(U) and/or poly(UC) stretch, and the 98-nucleotide X region, is required for interaction with NS3. The interaction between NS3 and the 3′(+) UTR appears to be specific, since an excess of unlabeled homologous RNA was able to compete out RNA-protein complex formation, whereas a similar-size heterologous RNA was totally inactive in the competition assay (Fig. 10D). These results suggest that the overall structure (or sequence) of the 3′(+) UTR RNA is required for specific binding of NS3. Additional mutagenesis will be required to clarify the role of the various sequence and/or secondary structural elements within the 3′(+) UTR.
DISCUSSION
We have shown in the present study that the HCV-encoded protease/helicase protein NS3 interacts with the 3′-terminal sequences of the viral positive-and negative-strand RNA. Specifically, the helicase portion of NS3 is responsible for RNA-protein interaction, as indicated by the similar binding profiles of the full-length NS3 protein and the truncated protein lacking the protease domain (ΔpNS3). The interactions of NS3 with the 3′ UTR127 of the negative-strand RNA and the 3′ UTR of the positive-strand RNA appear to be specific, since the formation of the nucleoprotein complexes is successfully competed by the unlabeled homologous RNA but not by similar-size RNAs derived from hepatitis A virus or poliovirus. The specificity of NS3-RNA interaction is also validated by the fact that only the 3′(−) UTR127 and 3′(+) UTR sequences, but not the corresponding complementary sequences from the 5′(+) and 5′(−) termini, interact with NS3. The interaction of NS3 with the 3′-terminal sequences of the negative-strand UTR RNA appears to depend on a stem-loop structure near the 3′ terminus of the RNA. Additional mutagenesis confined to the stem-loop structure spanning nucleotides −5 through −20 suggests that an intact double-stranded structure of the stem and the orientation of three G-C pairs (at positions −7, −8, and −9) within this stem are important elements for specific interaction of NS3 with the negative-strand RNA.
In the past, various investigators have reported on the RNA binding activity of the helicase portion of NS3. The majority of the RNA binding studies reported to date have used partially single-stranded or single-stranded RNA, including homopolymeric RNAs. Kanai et al. demonstrated binding of NS3 to poly(U) Sepharose resin with an apparent dissociation constant of 2 × 10−7 M (26). Other investigators have used filter binding (35), gel retardation (19), and fluorescence quenching (43) assays to assess the RNA binding activity of NS3. Except for some preference for poly(U), none of these studies detected any specific interaction of NS3 with HCV RNA. The preference for poly(U) can be explained by the fact that poly(U) stretches are present at the 3′ UTR terminus in HCV positive-strand RNA. Our binding results, along with those of the competition analysis, clearly demonstrate that the requirements of NS3 interaction with the 3′(+) UTR are much more complex than the mere presence of the poly(U) stretch. In fact, the 5′ half of the 3′(+) UTR, which contains the poly(U) stretch, is unable to interact with NS3 (or ΔpNS3) in the absence of the 3′ half of the UTR under our assay conditions (Fig. 10C). In contrast to the relatively stringent requirement for 3′(−) UTR127 binding, the interaction of NS3 with the 3′(+) UTR appears to require an intact structure of the entire 3′(+) UTR RNA under the assay conditions used in this study. To our knowledge, the results presented in this paper represent the first demonstration of a specific interaction of HCV NS3 protein with the 3′-terminal sequences of the HCV RNA. The importance of the 5′ and 3′ UTR sequences in viral infectivity have recently been confirmed in the chimpanzee model (32, 56–58). It should be pointed out that the 3′-terminal sequences of the negative- and positive-strand UTR contain some extra nucleotides that are contributed by the multiple cloning site. These sequences most likely do not play any role in NS3 (or ΔpNS3) recognition, as several of the internal mutations within the viral UTR sequences result in total blockage of RNA-protein interaction even though the mutant RNAs still contain the extra 3′-terminal nucleotides.
All pestiviruses and flaviviruses contain conserved helicase motifs in their NS3 proteins, suggesting an important role of the helicase in the life cycles of these viruses. Other positive-strand RNA viruses, such as poliovirus, rhinovirus, and coxsackie virus, also encode a distinct but homologous protein (called 2C) with RNA binding and NTPase activities and putative helicase activity (39, 41). We have shown previously that the poliovirus-encoded 2C protein specifically interacts with the 3′ UTR sequences of the viral negative-strand RNA (2). Both the precursor, 2BC, and the mature protein, 2C, are also capable of interacting with the cellular membranes (5, 10, 13, 14). It has been postulated that 2C may anchor viral negative-strand RNA to the cytoplasmic membrane so that initiation of positive-strand RNA synthesis can occur at the 3′ end of negative-strand RNA (2). Given the functional similarity between poliovirus 2C and the HCV helicase (NS3 and ΔpNS3), it is tempting to speculate that NS3 might have a similar role in HCV RNA synthesis. Recent results have suggested that interaction of NS3 with NS4A directs NS3 to the endoplasmic reticulum (54). Moreover, since NS3 appears to interact with the 3′ UTR of the positive- as well as the negative-strand RNA, a likely scenario might be that the NS3-NS4A complex bound to either the positive- or negative-strand 3′ UTR may anchor RNA-protein complexes to the cytoplasmic membrane. The polymerase NS5B (and possibly NS5A) can then join this complex by protein-protein and/or protein-RNA interaction and initiate positive- or negative-strand RNA synthesis. In fact, direct interaction of HCV NS5B with NS3 and NS4A has been reported (23). Future experiments will be directed to see if NS3-UTR interaction is influenced by NS4A either alone or when present as the NS3-NS4A fusion polypeptide. Also, in the closely related dengue virus, another member of the family Flaviviridae, both NS3 and NS5 have been shown to interact in vivo in CV-1 and HeLa cells (27). It is also possible that one or more host cell proteins could play a role in the membrane association of the viral RNA replication complex through participation of a membrane-associated protein, hVAP-33 (52). This SNARE-like protein containing a membrane-spanning domain has been shown to interact with both NS5A and NS5B. As reliable systems to study HCV RNA replication become available, it will be possible to address mechanistic questions to assess the roles of various viral proteins in RNA replication.
The results presented here do not exclude the possibility that NS3 also interacts with other regions of viral positive- or negative-strand RNA. However, since initiation of RNA synthesis is likely to start at the 3′ terminus of positive- and negative-strand RNA, it is likely that the interaction presented here is important for viral RNA replication. It is also important to note that specific binding of a protein to an RNA sequence and/or structure does not necessarily provide functional relevance to the RNA-protein interaction. Additional studies must be conducted to assess the importance of the mutations in both the 3′(−) and 3′(+) UTR that alter NS3 binding. Such studies may now be possible with the advent of the HCV replicon system that has recently been reported (36). Future work will be directed towards evaluation of the 3′(+) and 3′(−) UTR mutations in viral RNA synthesis.
In summary, we have demonstrated specific interaction of the HCV NS3 (helicase domain) with the 3′(−) and 3′(+) UTR sequences. The initial mutagenesis studies reported here have confirmed the requirement for the specific structure and sequence in NS3 binding. We suggest that the binding of NS3 to the 3′-terminal sequences of positive- and negative-strand RNA plays an important role in HCV RNA replication.
ACKNOWLEDGMENTS
This work was supported by the NIH grants AI-45733 and AI-27451.
We are grateful to Genevieve Inchauspe (INSERM, Lyon, France), Michael Lai (University of Southern California, Los Angeles) and Jens Bukh (NIH) for providing the various reagents used in this study.
We thank Weimin Tsai for the illustrations and his expert technical assistance, Winnie Kim for her help during cloning procedures, Akemi Yamane for critical reading of the manuscript, and Raquel Izumi, Kathy Weidman, and Arun Venkateshan for helpful suggestions and constructive comments throughout this study.
REFERENCES
- 1.Banerjee R, Igo M, Izumi R, Datta U, Dasgupta A. In vitro replication of RNA viruses. In: Cann A J, editor. RNA viruses. Oxford, United Kingdom: Oxford University Press; 2000. pp. 141–178. [Google Scholar]
- 2.Banerjee R, Echeverri A, Dasgupta A. Polio virus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J Virol. 1997;71:9570–9578. doi: 10.1128/jvi.71.12.9570-9578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Ahlborn-Laake L, Yasargil K, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager R L, Lohmann V, Wilkinson T, Koch J O. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol. 1995;69:7519–7528. doi: 10.1128/jvi.69.12.7519-7528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienz K, Egger D, Troxler M, Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic regions. J Virol. 1990;64:1156–1163. doi: 10.1128/jvi.64.3.1156-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight K J, Rice C M. Secondary-structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:5041–5045. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown E A, Zhang H, Ping L H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukh J, Purcell R H, Miller R H. Sequence analysis of the 5′ coding region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H S, Ha N C, Kang L W, Chung K M, Back S H, Jang S K, Oh B H. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J Biol Chem. 1998;273:15045–15052. doi: 10.1074/jbc.273.24.15045. [DOI] [PubMed] [Google Scholar]
- 10.Cho M W, Tetrina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 11.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 12.Clark B. Molecular virology of hepatitis C virus. J Gen Virol. 1997;78:2397–2410. doi: 10.1099/0022-1317-78-10-2397. [DOI] [PubMed] [Google Scholar]
- 13.Echeverri A, Banerjee R, Dasgupta A. Amino terminal region of poliovirus 2C is sufficient for membrane binding. Virus Res. 1998;54:217–223. doi: 10.1016/s0168-1702(98)00016-1. [DOI] [PubMed] [Google Scholar]
- 14.Echeverri A, Dasgupta A. Amino terminal region of poliovirus 2C mediates membrane binding. Virology. 1995;208:540–553. doi: 10.1006/viro.1995.1185. [DOI] [PubMed] [Google Scholar]
- 15.Failla C, Tomei L, De Francesco R. Both NS3 and NS4 are required for processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grakoui A, Mc Court D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of the proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gwack Y, Kim D W, Han J H, Choe J. Characterization of RNA binding and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem Biophys Res Commun. 1996;225:654–659. doi: 10.1006/bbrc.1996.1225. [DOI] [PubMed] [Google Scholar]
- 20.Honda M, Beard M R, Ping L H, Lemon S M. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda M, Brown E A, Lemon S M. Stability of a stem loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 22.Inchauspe G, Zebedee S, Lee D H, Sugitani M, Nasoff M, Prince A M. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc Natl Acad Sci USA. 1991;88:10292–10296. doi: 10.1073/pnas.88.22.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishido M, Fujita T, Hotta H. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem Biophys Res Commun. 1998;244:35–40. doi: 10.1006/bbrc.1998.8202. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Lai M M C. Determination of the secondary structure of and cellular proteins binding to the 3′ untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin L, Peterson D L. Expression, isolation and characterization of the hepatitis C virus ATPase-RNA helicase. Arch Biochem Biophys. 1995;323:47–53. doi: 10.1006/abbi.1995.0008. [DOI] [PubMed] [Google Scholar]
- 26.Kanai A, Tanabe K, Kohara M. Poly(U) binding activity of hepatitis C virus NS3 protein, a putative RNA helicase. FEBS Lett. 1995;376:221–224. doi: 10.1016/0014-5793(95)01283-x. [DOI] [PubMed] [Google Scholar]
- 27.Kapoor M, Zhang K, Ramachandra M, Kusukawa J, Ebner K E, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 28.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the hepatitis C virus genome from Japanese patients with non A, non B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J K, Morgenstern K A, Lin C, Fox T, Dwyer M D, Landro J A, Chambers S P, Markland W, Lepre C A, O'Malley E T, Harbeson S L, Rice C M, Murcko M A, Caron P R, Thompson J A. Crystal structure of the hepatitis C virus protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 30.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolykhalov A A, Agapov E V, Rice C M. Specificity of the hepatitis C NS3 serine protease: effects of substitution at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J Virol. 1994;68:7525–7533. doi: 10.1128/jvi.68.11.7525-7533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 33.Lin C, Thompson J A, Rice C M. A central region in the hepatitis C virus NS4A protein allows formation of active NS3-NS4A serine proteinase complex in vivo and in vitro. J Virol. 1995;69:4373–4380. doi: 10.1128/jvi.69.7.4373-4380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin C, Pragai B M, Grakoui A, Xu J, Rice C M. Hepatitis C virus NS3 serine proteinase: trans cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin C, Kim J L. Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J Virol. 1999;73:8798–8807. doi: 10.1128/jvi.73.10.8798-8807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohmann V, Korner F, Koch J-O, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNA in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 37.Love R A, Parge A E, Wickersham J A, Hostomsky Z, Habuka N, Moomaw E W, Adachi T, Hostomska Z. The crystal structure of hepatitis C virus NS3 proteins reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 38.Luo G, Hamatake R K, Mathis D M, Racela J, Rigat K L, Lemm J, Colonno R J. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol. 2000;74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirzayan C, Wimmer E. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology. 1994;199:176–187. doi: 10.1006/viro.1994.1110. [DOI] [PubMed] [Google Scholar]
- 40.Oh J-W, Ito T, Lai M M C. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J Virol. 1999;73:7694–7702. doi: 10.1128/jvi.73.9.7694-7702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfister T, Wimmer E. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J Biol Chem. 1999;274:6992–7001. doi: 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- 42.Porter D J. A kinetic analysis of the oligonucleotide modulated ATPase activity of the helicase domain of the NS3 protein from hepatitis C virus. The first cycle of interaction of ATP with the enzyme is unique. J Biol Chem. 1998;273:14247–14253. doi: 10.1074/jbc.273.23.14247. [DOI] [PubMed] [Google Scholar]
- 43.Preugschat F, Averett D R, Clark B E, Porter D J T. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J Biol Chem. 1996;271:24449–24457. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- 44.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 931–960. [Google Scholar]
- 45.Saito I T, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Choo Q-L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzich J A, Tamura J K, Palmer-Hill F, Warrener P, Grakoui A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;70:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tai C L, Chi W K, Chen D S, Hwang L H. The helicase activity associated with hepatitis C virus nonstructural protein (NS3) J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka T, Kato N, Cho M-J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno S. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomei L, Failla C, Santolini E, Francesco R D, Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu H, Gao L, Shi S T, Taylor D R, Yang T, Mircheff A K, Wen Y, Gorbalenya A E, Hwang S B, Lai M M. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology. 1999;263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- 53.Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolk B, Sansonno D, Krausslich H-G, Dammacco F, Rice C M, Blum H E, Moradpour D. Subcellular localization, stability, and trans cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J Virol. 2000;74:2293–2304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada N, Tanihara K, Takada A, Yorihuzi T, Tsutsumi M, Shimomura H, Tsuji T, Date T. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology. 1996;223:255–261. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]
- 56.Yanagi M, St. Claire M, Emerson S U, Purcell R H, Bukh J. In vivo analysis of the 3′ UTR of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci USA. 1999;96:2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanagi M, Purcell R H, Emerson S U, Bukh J. Transcripts from a single full length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yanagi M, St. Claire M, Shapiro M, Emerson S U, Purcell R H, Bukh J. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 59.Yao N, Hesson T, Cable M, Hong Z, Kwong A D, Lee H V, Weber P C. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 60.Zhong W, Uss A S, Ferrari E, Lau J Y, Hong Z. De novo initiation of RNA synthesis by hepatis C virus nonstructural protein 5B polymerase. J Virol. 2000;74:17–22. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]