Abstract
Single-cell transcriptomics has emerged as the preferred tool to define cell identity through the analysis of gene expression signatures. However, there are limited studies that have comprehensively compared the performance of different scRNAseq systems in complex tissues. Here, we present a systematic comparison of two well-established high throughput 3′-scRNAseq platforms: 10× Chromium and BD Rhapsody, using tumours that present high cell diversity. Our experimental design includes both fresh and artificially damaged samples from the same tumours, which also provides a comparable dataset to examine their performance under challenging conditions. The performance metrics used in this study consist of gene sensitivity, mitochondrial content, reproducibility, clustering capabilities, cell type representation and ambient RNA contamination. These analyses showed that BD Rhapsody and 10× Chromium have similar gene sensitivity, while BD Rhapsody has the highest mitochondrial content. Interestingly, we found cell type detection biases between platforms, including a lower proportion of endothelial and myofibroblast cells in BD Rhapsody and lower gene sensitivity in granulocytes for 10× Chromium. Moreover, the source of the ambient noise was different between plate-based and droplet-based platforms. In conclusion, our reported platform differential performance should be considered for the selection of the scRNAseq method during the study experimental designs.
Graphical abstract
1. Introduction
Single-cell RNA sequencing (scRNAseq) enables simultaneous profiling of gene expression of individual cells and is the tool of preference to define cell phenotypes and discover cell subsets and states [1]. Single-cell transcriptomics provides an unprecedented resolution of the composition and functionality of cellular niches in homeostatic tissues and during disease [2,3]. For example, scRNAseq has been used to unravel multicellular dynamic processes during embryogenesis and cell differentiation, tissue regeneration and morphogenesis, disease initiation and progression, and response to stimuli including drug treatments [[4], [5], [6], [7], [8], [9], [10], [11], [12]].
The ability to sequence genomic material combined with barcoding strategies to label each cell and RNA molecule extended the possibility of simultaneous analysis of barcoded cells in the same reaction, dramatically reducing cost and labour [13,14]. The subsequent application of microfluidic devices to encapsulate individual cells in nano-droplet-sized bioreactors, like Drop-seq [6,7], or the development of high-density microwell plates to partition and capture individual cells, as Microwell-Seq [15,16] considerably increased the scRNAseq scale, enabling the analysis of tens-of-thousands of cells in a single reaction. These high throughput scRNAseq methods can be technically challenging and difficult to control to deliver consistent outputs [17]; however, commercial solutions, like 10× Chromium and BD Rhapsody, have streamlined these processes, easing the technical demand and standardising procedures while ensuring consistent reagent quality, [18].
At a first glance, all high throughput scRNAseq methods offer similar data outputs and they can interchangeably be used to interrogate any biological system. However, each method is technically different, with its own intrinsic capabilities, biases and limitations; thus, platform performance could vary with particular cell types and tissues, and specific platforms may be better suited to answer particular biological questions. There is limited information on a systematic comparison of the performance of different high throughput scRNAseq platforms and the available scRNAseq platform comparisons are based on relatively homogenous cell cultures [19,20], or artificial mixed pools [[21], [22], [23]]. Thus, these comparisons are unable to assess each platform's ability to distinguish cellular heterogeneity in a complex tissue scenario, failing to compare performance between cells of different lineages. For example, a recent study comparing five single cells/nucleus RNAseq high-throughput methods concluded that the commercial platform 10× Chromium was the top performing technology compared with the home brew methods [24]. This study, however, did not included other commercial high-throughput platforms, such as the BD Rhapsody; and for the single-cell comparison, it only used cultured cells or PBMC samples which do not require tissue dissociation.
Here, we present a technical and biological comparison of two well-established and widely used 3′-scRNAseq commercial platforms, - 10× Chromium (10× Genomics) and BD Rhapsody (Becton Dickinson); using mammary gland tumours from the MMTV-PyMT mouse model, biologically complex but reproducible samples, [[25], [26], [27], [28]]. 10× Chromium is a droplet-based microfluidic platforms while BD Rhapsody uses microwell-based technology where cells are randomly deposited by gravity into an array of picoliter-size wells. All systems track the cell of origin with a cell barcode and count individual molecules using unique molecular identifiers (UMIs). Although these two platforms are designed to produce similar readouts, essentially a digital count of gene expression in each cell, the device design (microfluidic or microwell), the nature of the capture beads, the molecular design of the barcodes and UMIs, and the essential nature of the molecular workflow for RNA reverse transcription and amplification strongly differs.
In this study, we evaluate how 10× Chromium and BD Rhapsody 3′-scRNAseq platforms manage the challenges of complex tissues to produce meaningful data to discover the strengths and weaknesses of each platform in this context.
2. Material and methods
2.1. Mouse model
The MMTV-Polyoma Middle T antigen (PyMT) was a gift from Dr. William J. Muller (McGill University) and its generation has been previously described [26,27]. At ethical endpoint, (10 % ± 3 % tumour/body weight, which approximately corresponds to 14-week-old animals) the mice were euthanized, and size-matched tumours were harvested and processed for single cell digestions. The tumour location nomenclature used is as follows: Tumour A - right cervical and/or thoracic mammary gland; tumour B - left cervical and/or thoracic mammary gland; tumour C - right abdominal and/or inguinal mammary glands; tumour D - left abdominal and/or inguinal mammary glands. Genotyping was performed at the Garvan Molecular Genetics facility (NATA accredited, ISO 17025) by PCR of DNA extracted from the mouse tail tip using the following primers: CGGCGGAGCGAGGAACTGAGGAGAG and TCAGAA GACTCGGCAGTCTTAGGCG. The touchdown PCR conditions were 94 °C for 10 s of initial denaturation, followed by 10 cycles of 94 °C 10 s, 65-55 °C for 30 s and 72 °C for 1 min and 10 s and then 31 cycles of 94 °C 10 s, 55 °C for 30 s and 72 °C for 1 min and 10 s; the final extension is 72 °C for 3 min. All animals used in this study are heterozygous for the PyMT gene.
All animal experiments were carried out according to guidelines contained within the NSW (Australia) Animal Research Act 1985, the NSW (Australia) Animal Research Regulation 2010 and the Australian code of practice for the care and use of animals for scientific purposes, (8th Edition 2013, National Health and Medical Research Council (Australia)). All experiments involving mice have been approved by the St. Vincent's Campus Animal Research Committee AEC #19/02.
2.2. Tissue digestion and single-cell isolation
PyMT tumours were digested as described in Refs. [29,30]. We aimed to have a minimum of two mice per condition and platform and a minimum of 2 mixed tumours per mice from different locations (Table S1 and S2). Briefly, tumours were manually dissected into 3–5 mm pieces using a surgical scalpel blade and further chopped to 100 μm using a tissue chopper (McIIwain). Samples were enzymatically digested with 15,000 U of collagenase (Sigma Aldrich Cat# C9891) and 5,000 U of hyaluronidase (Sigma Aldrich Cat# H3506) for 30 min at 37 °C. The samples were then further digested by pipetting up and down with 0.25 % trypsin (Gibco Cat# 15090-046), in 1 mM EGTA and 0.1 mg/mL of Polyvinyl alcohol dissolved in Dulbecco's phosphate-buffered saline (DPBS, Gibco Cat# 14190-250) for 1 min at 37 °C in a waterbath. Red blood cells were then lysed with 0.8 % ammonium chloride (Sigma Aldrich Cat# A9434) dissolved in water for 5 min at 37 °C. Single cell suspensions were washed with DPBS containing 2 % of Foetal Bovine Serum (FBS, GE Healthcare Cat# SH30406.02) and spun at 200×g for 5 min at 4 °C between each step. The supernatant was aspirated and 1 mg/mL DNase I (Roche Cat# 10104159001) was mixed with the sample before incubation with each step. Finally, cells were filtered through a 40 μm cell sterile strainer (Corning® Cat# 431750) and resuspended in DPBS with 2 % FBS. For the generation of low-quality-like samples, digested tumours were left overnight for 24 h at 4 °C, and check that viability was reduced by flow cytometry by at least 20 % before processing the sample in autoMACS® Pro (Miltenyi).
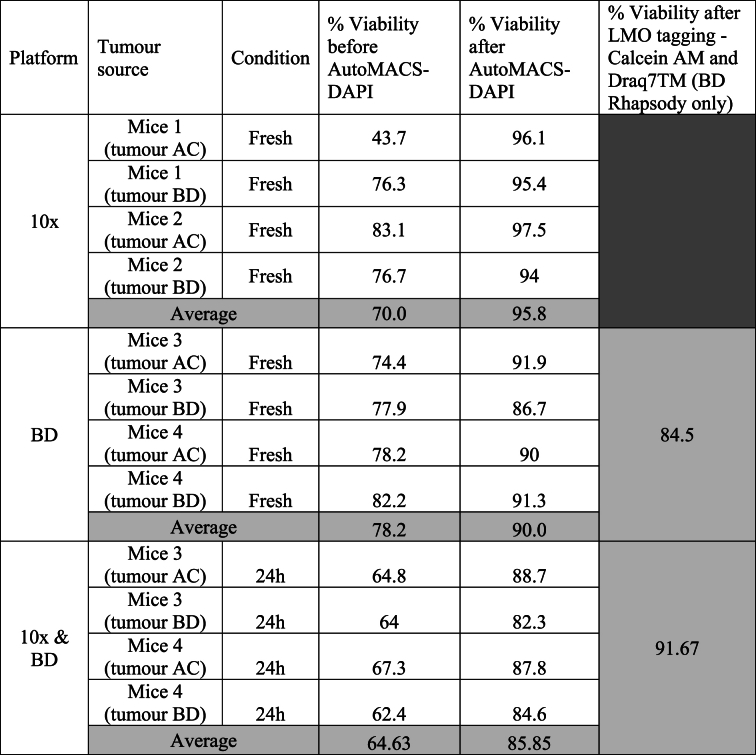
All tumours were labelled with Annexin specific MACS beads using the Dead Cell Removal Kit (Miltenyi Biotec Cat# 130-090-101) following the manufacturers’ instructions and dead cells were removed by passing the labelled cells through the autoMACS® Pro (Miltenyi) (see more details at [17]). All samples showed high percentage of viable cells (≥80 % viability assessed by DAPI in flow cytometry, Table S3). Samples for BD Rhapsody underwent additional steps prior to the cell capture for LMO multiplexing (see below and Fig. S1B). When all samples from both methods were ready for cell capture, high percentage of cell viability was again verified by microscopy with 0.4 % of Trypan blue solution (Sigma-Aldrich, Cat# T8154), to ensure that comparable cell viability was achieved from both single-cell capture methods (≥85 %).
2.3. Flow cytometry
The viability and cellular content of the main cell compartments of the tumours was assessed by flow cytometry. Digested tissue samples were washed with DPBS (Gibco Cat#14190136) supplemented with 2 % FBS (GE Healthcare Cat# SH30406.02) and centrifuged at 200×g for 5 min at 4°C. The pellet was then resuspended with 2 % FBS in PBS for use in flow cytometry analysis. For flow cytometry analysis, single cell suspensions were incubated with antibodies for anti-mouse EpCAM (BioLegend Cat#118205, RRID: AB_1134176) and anti-mouse CD45 (Clone 30-F11, BioLegend, Cat# 103114 RRID: AB_312979) on ice in the dark for 30 min before they were washed, centrifuged, and resuspended with 2 % FBS in DPBS for analysis using the BD FACSymphony™ Cell Analyser. To check viability, cells were stained with DAPI in a 0.5 μg/mL concentration (Invitrogen, D1306) at 4 °C for 3 min immediately before running the samples in the flow cytometer. Flow cytometry data were analysed using the software package FlowJo (version 10.4.2).
2.4. 10× Chromium
We aimed to capture 8,000 cells with >85 % cell viability from each condition. Libraries were prepared using the Chromium Next GEM Single Cell 3ʹ Kit from 10× Genomics (Cat# PN-1000269) following the manufacturer's instructions. Sequencing was performed on an Illumina NovaSeq™ 6000 System (Illumina, Cat# 20012850) using the NovaSeq 6000 S4 Reagent Kit (200 cycles) (Illumina, Cat # 20028313) using the following configuration: 28bp for Read 1, 91bp for Read 2 and 8bp for Index, to an estimated depth of 20,000–30,000 reads per cell. Cell Ranger pipeline v3.0.1 was used for Fastq file generation, alignment to the mm10 (Release M19 (GRCm38.p6) transcriptome reference and UMI counting. Barcodes corresponding to empty droplets were excluded using cell calling algorithm from Cell Ranger based on EmptyDrops [31]. We used “subset” function from Seurat v5.0.1., for random cell subsampling.
2.5. BD Rhapsody
We adopted the MULTI-seq protocol [32] based on lipid-modified oligos (LMOs) for multiplexing different samples in the same BD Rhapsody cartridge (Fig. S1B). Single-cell suspensions from four different tumours were washed twice with DPBS (Gibco Cat#14190136) and incubated in a 200 nM solution containing equal amounts of anchor LMO (generously gifted by Prof. Gartner's laboratory) and sample barcode oligonucleotides (©Integrated DNA Technologies, IDT, Inc) on ice for 5min. After LMO-barcode labelling, we incubated the co-anchor LMO (gifted by Prof. Gartner's laboratory, final concentration of 200 nM) in each sample for 5min on ice. 1 mL of 1 % BSA (Sigma-Aldrich Cat#A9418) was added in each sample to quench the LMO binding and washed once with 1 % BSA. The tumour samples were pooled in equal proportions, washed twice with 1 % BSA and resuspended in the cold Sample Buffer (BD Rhapsody Cat. No. 650000062). Altogether, the multiplexing step added 30 min extra prior to the cell capture, which represents 11 % of the total time for sample processing (typically 4 h for 4 single-plex samples [29]). Two extra QC steps were performed to ensure that the multiplexing workflow did not compromise cell viability: 1) 0.4 % Trypan blue cell staining (Sigma-Aldrich, T8154) to make sure cell viability was comparable to the 10× samples (see above in Tissue digestion and single-cell isolation section), and then, 2) immediately before cells were loaded into a BD Rhapsody cartridge, a high percentage of cell viability was corroborated in the BD Rhapsody™ Scanner using Calcein AM (Thermo Fisher Scientific, Cat# C1430) and Draq7TM (Cat# 564904) in the pooled multiplexed samples (Table S3). Cell capture, cDNA synthesis and exonuclease I treatment were performed using the standard protocols from BD Rhapsody Express Single-Cell Analysis System Instrument User guide (Doc ID 214062).cDNA and LMO libraries were prepared using a custom protocol combining BD Rhapsody's mRNA whole transcriptome analysis (WTA) library preparation protocol (Doc ID 23-21711-00 Rev 7/2019) and the MULTI-seq protocol [32]. First, we performed 2 sequential random priming and extension (RPE) reactions of the Exonuclease I-treated beads containing the whole cellular transcriptome and LMO cDNA to increase assay sensitivity using half of the reagents for each round from the BD Rhapsody™ WTA Amplification Kit (Cat#633801). After this step we performed two parallel workflows, one for the WTA libraries, using the RPE product from the supernatant and the other one for the LMO libraries from the post-RPE beads. The RPE supernatant underwent WTA library preparation following the manufacturers' instructions from the BD Rhapsody™ System mRNA Whole Transcriptome Analysis (WTA) Library Preparation Protocol (Doc ID 23-21711-00 Rev 7/2019) that included the steps of purifying RPE product, performing RPE PCR, purifying the RPE PCR amplification product, performing WTA Index PCR and purifying the WTA index PCR product. The LMO libraries were obtained from the post-RPE beads, which were resuspended and washed in the cold Bead Resuspension Buffer (BD Rhapsody Cat. No. 650000062)and after three washes, the beads were resuspended in 80 μL of the MULTI-seq cDNA amplification mix containing 1 μL of 2.5 μM Multi-seq primer 5′-CTTGGCACCCGAGAATTCC-3′, 69 μL PCR Master Mix (Part# 91–118 from BD Rhapsody™ WTA Amplification Kit, Cat# 633801), 10 μL Universal Oligo (Part# 650000074 from BD Rhapsody™ WTA Amplification Kit, Cat# 633801) and the PCR was performed as follows: 95 °C, 3 min; 95 °C, 30 s, 60 °C, 1 min and 72 °C, 1 min for 14 cycles; 72 °C, 2 min; 4 °C hold. Next, we purified the PCR products of the LMOs using a 0.6× ratio of the SPRI beads (Beckman Coulter Cat# B23317) and keeping the supernatant, followed by a left side selection using 1.8× ratio. The LMO library was eluted in 69 μL of Elution Buffer (Part# 91–1084 from BD Rhapsody™ WTA Amplification Kit, Cat# 633801) and was quantified using Qubit (dsDNA HS Assay, Invitrogen™ Cat# Q32851) and the expected DNA size (∼95–115 bp) was confirmed with the Agilent 4200 TapeStation system (Agilent High Sensitivity D5000 ScreenTape Assay, Agilent Cat# 5067–5593 and 5067–5592). Next, we used 3.5 ng of LMO cDNA purified product to perform a LMO Index PCR, the PCR mix had 26.25 μl of 2× KAPA HiFi HotStart Ready mix (KAPABiosystems Cat# KK2601), 2.5 μl of 10 μM Library Forward Primer (Part# 91–1085 from BD Rhapsody™ WTA Amplification Kit, Cat# 633801), 2.5 μl of 10 μM Library Reverse Primer (reagent part from BD Rhapsody™ WTA Amplification Kit, Cat# 633801, a different primer was chosen for each sample for sequencing multiplexing); and the PCR conditions were 95 °C, 5 min; 98 °C, 15 min; 60 °C, 30 min; 72 °C, 30 min; 8 cycles; 72 °C, 1 min; 4 °C hold A final cleanup of the LMO libraries using a 1.6× SPRI bead ratio was performed before sequencing.
WTA libraries and LMO libraries were pooled and sequenced on the NovaSeq™ 6000 System (Illumina, Cat# 20012850) using the NovaSeq 6000 S1 Reagent Kit (100 cycles) (Illumina, Cat # 20028319) using the following configuration: 100bp for Read 1, 100bp for Read 2 and 8bp for Index to an estimated depth of 25,000 reads per cell for the WTA library and 2,500 reads per cell for the LMOs.
Fastq files from BD Rhapsody samples were processed using BD Rhapsody™ WTA Analysis Pipeline, v1.9.1 in Seven Bridges Genomics cloud platform. WTA reads were aligned to the mm10 reference genome (Release M19 (GRCm38.p6) and LMO sequences (8bp) were included as sample tags, allowing a maximum of 1 mismatch. The sample multiplexing option of the WTA Analysis pipeline was used to determine the sample of origin of each cell and singlets were used for downstream analysis. The downsampling to reduce the number of reads was performed using the “Donwsampling tool” in the Seven Bridges Genomics pipeline.
2.6. Single-cell data integration, clustering, and annotation
Single-cell clustering and annotation were performed using Seurat v5.0.1. A Seurat object of each sample was created using CreateSeuratObject() function without a minimum gene or counts filtering. First, samples from the same platform were merged and normalized and variable genes were detected using the SCTransform method [33]. Cell clustering was determined by shared nearest neighbour (SNN) modularity using 15 dimensions and a resolution of 0.3 and dimensional reduction was performed using RunUMAP() using 15 dimensions in all three platforms. Secondly, data from both platforms were integrated utilizing either reciprocal PCA (RPCA), canonical correlation analysis (CCA), Harmony [34] and Join PCS to identify anchors. RPCA integrated object with 5 anchors and normalized using SCTransform were used for further analysis. Cell identities were annotated using SingleR signatures based on the mouse cell type reference generated by the Immunologic Genome Project [35] and based on single cell reference mapping using the TransferData() function and the previously characterized PyMT tumour dataset as reference [30]. Silhouette score per cell at each resolution was calculated using silhouette() function from the cluster R package and represented as a boxplot per platform using ggplot2. Bar plots for comparing cluster abundance across platforms were generated using dittoSeq package and dotplots and boxplots using ggplot2. Gene markers of specific cell types or clusters were identified using FindMarkers() or FindAllMarkers() functions from Seurat using logfc.threshold = 1, min.pct = 0.3 and logfc.threshold = 0.25, min.pct = 0.25, respectively. To determine gene sets enriched in the upregulated genes of a specific cluster we used the enrichGO() function from the clusterProfiler package, p-value was adjusted using Benjamini-Hochberg formula and the q value cutoff set at 0.2. Gene expression correlation between different samples was calculated using the DESeq2 package. Cell communication analysis was performed using the CellChat R package [36].
2.7. Public data processing and analysis
10× Chromium and BD Rhapsody from bone marrow and whole blood gene expression and antibody matrices (BioProject ID: PRJNA734283) were downloaded and re-processed using Seurat v4.2.1. Filtered 10× cells based on the default settings of Cell Ranger or based on a custom threshold of Protein UMI counts per cell (>10) as Qi et al. [37] were first demultiplexed using the Hashtag oligo (HTO) libraries and the HTODemux() function from Seurat. Barcodes with more than one hashtag were discarded. Cells from the bone marrow and whole blood in each platform were merged, clustered, and visualised as explained above.
2.8. Ambient noise detection
The single-cell Ambient Remover (scAR) model was used to determine the gene expression profile of the empty droplets or wells representing the ambient noise, the ratio of ambient noise in each cell and to create a denoised matrix [38]. For the mouse datasets, for the setup_anndata() function, the whole list of barcodes selected were used as the unfiltered matrix and the putative cells based on either Cell Ranger of the BD Rhapsody pipeline for the filtered matrix, using a probability threshold of 0.995. Empty barcodes were used to calculate the ambient profile and train the model. For the human dataset from BD Rhapsody the noise ratio was calculated the same way than the mouse dataset for both the Gene expression and protein matrices. For the human datasets generated in 10× Chromium, the putative cells used in the setup_anndata() function were based on the manually filtering the UMI Protein counts (>10). DecontX was also used to detect ambient noise, named contamination in this package [39] and decontX function was run per sample without using the empty barcodes as background.
2.9. Statistical analysis
scRNAseq data was generated from 16 different tumours across 4 different mice. Specific statistical details for sample size and the threshold for statistical significance can be found either in the figure legend or in the method section. The data in bar graphs are presented as mean ± SEM, and the statistical analysis employed an unpaired t-test. Box plots display data by depicting the minimum and maximum values as whiskers, the median of the lower half (quartile 1), and the median of the upper half (quartile 3) as a box. The graphs and statistical analyses for scRNAseq were performed using R and for flow cytometry using GraphPad Prism.
3. Results
3.1. Experimental design and data processing
Our comparison is achieved by using mammary gland tumours from the widely used Polyoma Middle T antigen (PyMT) transgenic mouse model [26]. These tumours present multiple lineages and high cell diversity while retaining reproducibility due to the limited variability from a genetically controlled congenic mouse strain (FVBn) [30,40,41]. In addition, tumours are biologically challenging samples, as they are not homeostatic tissues, and present tissue damage, hypoxia or cell death; features that will put to the test any scRNAseq method. Tumour fragments from the 4 different mice (4 tumours per mouse, Table S1) were randomized, digested and enriched for viable tumour cells using Magnetic Associated Cell Separation (MACS) as previously described [29]. Single cell suspensions containing viable cells (≥85 %, Table S3) were loaded into a 10× Chromium, or multiplexed using lipid-modified oligos (LMOs) and then loaded into BD Rhapsody chip (Tables S1 and S3 and Figs. S1A and B) and run following the corresponding molecular workflows (see Methods section, Figs. S1A and S1B).
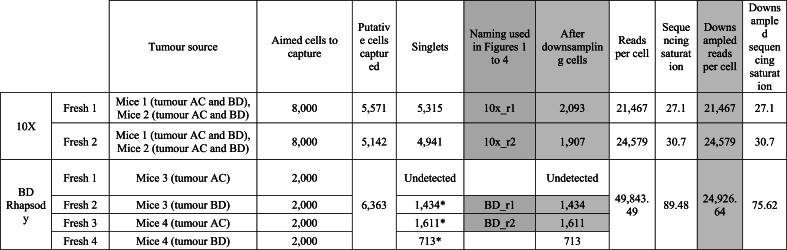
Each platform has a different read structure for barcodes and UMIs and each system has a different processing bioinformatic pipeline. Therefore, to replicate the standard user experience, we have used the intended pipeline for each platform to generate a Digital Gene Expression (DGE) matrix per cell: cellranger for 10× Chromium and the whole transcriptome analysis (WTA) pipeline in Seven Bridges Genomics cloud platform for BD Rhapsody. In all cases, data was subsequently integrated and visualised using the Seurat R package v5.0.1 [42]. A critical step for single-cell data processing is to determine how many real cell barcodes were detected. 10× Chromium's pipeline, Cell Ranger (>v3.0) and BD's pipeline in the Seven Bridges Genomics platform, now include a step that detects putative cells. Therefore, we filtered out empty barcodes based on the default parameters of each platform. Doublets were excluded using DoubletFinder [43] in the case of 10× Chromium, while for BD Rhapsody we used the information from the sample tags used to demultiplex tumour samples. After removing the empty droplets and doublets, we obtained 10,713 cells in 10× Chromium, and 6,363 for BD Rhapsody (Table S1). For BD Rhapsody we had 4 sample tags, but unfortunately one of the LMOs was not detected and thus that sample had to be excluded from the study (Fresh 1 BD Rhapsody, see details in Table S1) as we could not distinguish the cells from this sample from the untagged cells from the other samples. From the 3 remaining BD Rhapsody LMO-tagged samples and after demultiplexing the number of singlets detected was 1,434 and 1,611 cells, while the third one had 713 cells. A total of 5,315 cells and 4,941 singlets were identified in 10× Chromium from the two replicates. To keep a consistent sequencing depth per cell in our downstream technical and cell type comparisons (Fig. 1, Fig. 2, Fig. 3, Fig. 4), we selected the two replicates with the highest number of cells from BD Rhapsody (Fresh 2, 1,434 cells and Fresh 3, 1,611 cells) and the two existing replicates from 10× Chromium (Fresh 1, 5,315 and Fresh 2, 4,941 cells) and subsampled the later to obtain around 4,000 cells as well as downsampled the sequencing depth of BD Rhapsody to reach around similar sequencing depth per cell (∼24,000 reads). This resulted in 2,093 and 1,907 cells from 10× Chromium that were compared with 1,434 and 1,611 cells from BD Rhapsody at similar sequencing depth per cell (Table S1).
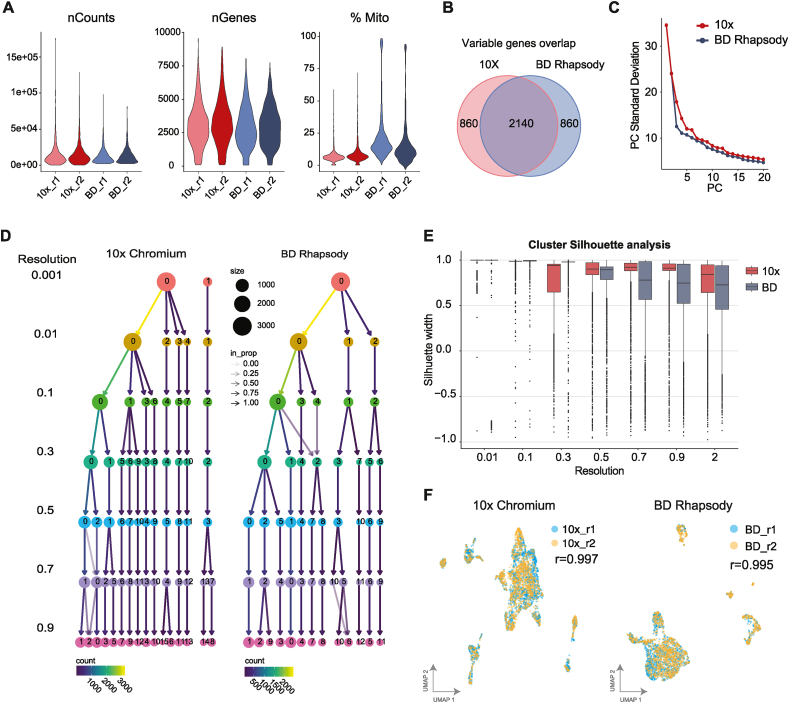
Fig. 1.
Measurements of the technical quality, sensitivity and reproducibility among scRNAseq platforms. A. Violin plots showing the number of UMIs (nCounts), and genes (nGenes) detected per cell, as well as the percentage of mitochondrial content (%Mito) per cell in 10× Chromium (red) and BD Rhapsody (blue) each using two biological replicates (r1, r2). B. Overlap between the top 3000 most variable genes in each platform. C. Principal component (PC) plot showing the total variability identified in each system measured as the standard deviation at increasing PCs. D. Cluster tree showing the phylogenetic relationship of clusters in each platform at increasing resolutions. E. Box plot of the Silhouette scores (y axis) between the 10× Chromium (red) and BD Rhapsody (blue) samples at different resolutions (x axis). The silhouette ranges from −1 to +1, where a high value indicates that the object is well matched to its own cluster and poorly matched to neighbouring clusters. F. UMAP plots showing the overlap between two replicates in each platform, the “r” corresponds to the Pearson correlation coefficient of gene expression between replicates.
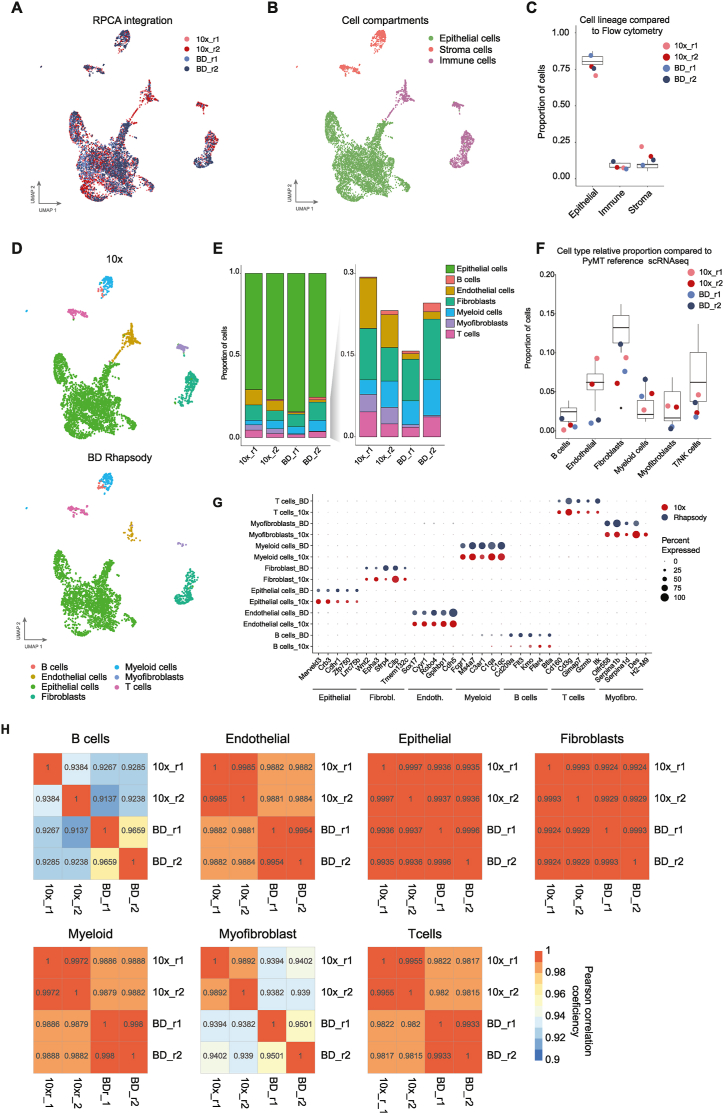
Fig. 2.
Identification of the main cell lineages in tumours by each scRNAseq platform. A UMAP plot of all integrated samples using the Reciprocal Principal Component Analysis (RPCA) method with 5 anchors. B. UMAP plot showing the three main cell compartments: epithelial, immune and stroma cells identified in all cells from all replicates and platforms after integration. C. Proportion of cells from each main cell lineage. The box plot represents the relative proportion of each cell lineage identified by flow cytometry, EpCAM+ (epithelial cells), CD45+ (leukocytes), and EpCAM−/CD45− (stroma), and the colour dots represent each of our samples from the single cell RNAseq dataset. D. UMAP plots showing the cell types assigned using label transfer analysis of the PyMT reference scRNAseq previously published [30], and split based on the platform. E. Bar plot of the proportion of cells from each cell type per sample (left) and magnified and excluding the epithelial cells (right). F. Cell type relative proportions of each cell type per sample. The box plot represents the proportions found in the PyMT reference scRNAseq [30] and the colour dots represent each of our samples from our single cell dataset. G. Dot plot showing the gene expression comparison of the top cell lineage marker genes between platforms. H. Heatmaps of the Pearson Correlation coefficient between 10× Chromium and BD Rhapsody samples (r1 and r2) in the different annotated cell types.
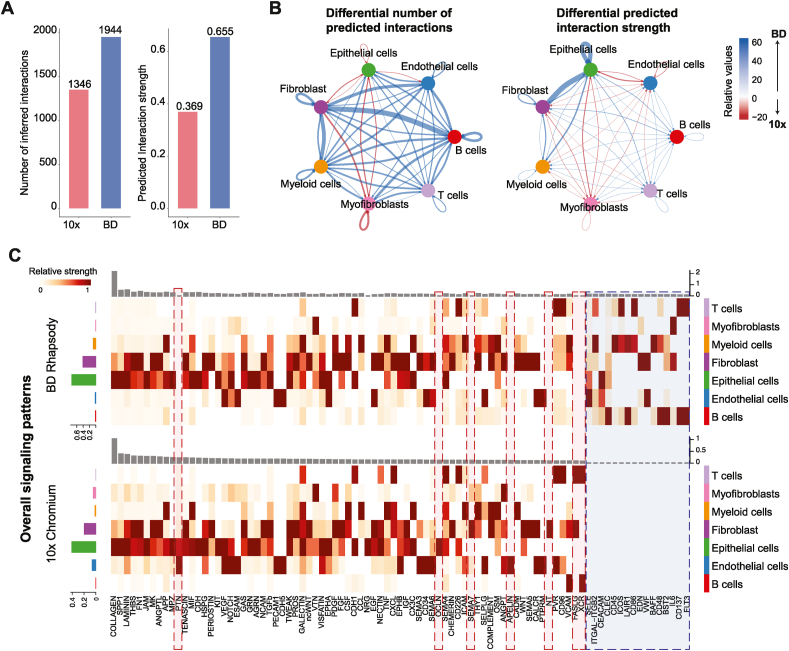
Fig. 3.
Comparison of cell-cell communication prediction in each platform. A. Bar plot showing the overall number of putative interactions (left) and predicted interaction strength in each platform (right). B. Circle plot visualizing the differential cell–cell interaction networks predicted by each platform. In the left panel, the thickness of the line represents the number of differential interactions predicted between the connecting cell types, while in the right panel, the width of the line corresponds to the differential strength of the interaction by assessing the level of expression of the ligand-receptor pairs after correcting the differences in cell type abundance. Blue lines represent higher number or stronger predicted interactions in BD Rhapsody while red lines correspond to predicted interactions enriched in 10×. C. Heatmaps representing the relative signalling strength of the signalling pathways across platforms. The top grey bar plots show the total signalling strength of each signalling pathway combining all cell types, and the coloured bar plots on the left show the total signalling strength of each cell type combining all pathways.
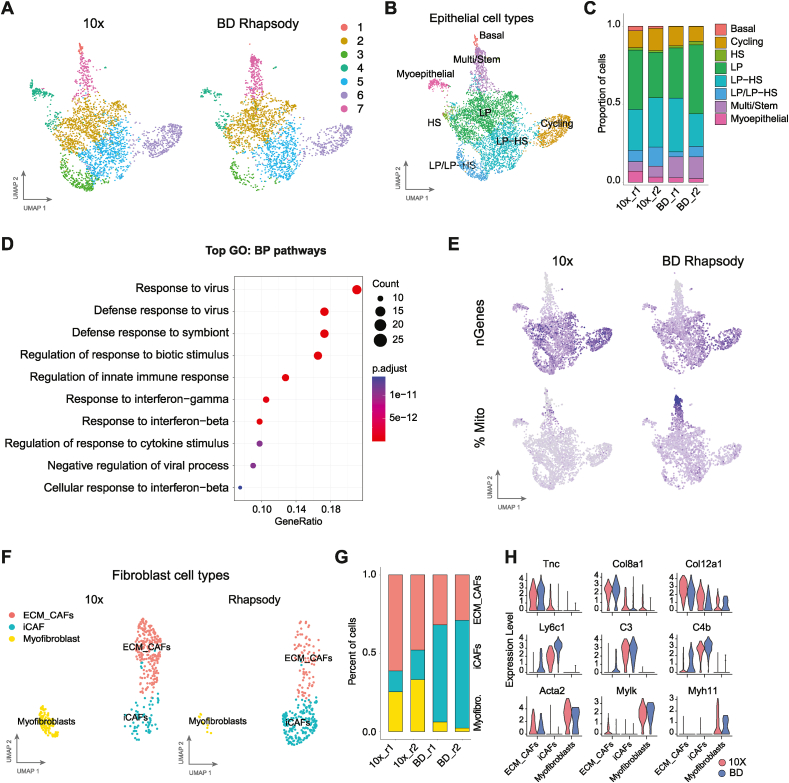
Fig. 4.
Cancer-epithelial and fibroblast subtype detection across platforms. A. UMAP projections of epithelial cells showing the unsupervised KNN clusters split by platform. B UMAP plot showing the epithelial cell types based on the annotated PyMT reference [30]. (LP: luminal progenitor; HS: Hormone sensing) C. Bar plot of the proportion of cells from each epithelial cell type per sample. D. Gene Set Enrichment Analysis of the marker genes from cluster 3 (A) versus the rest of the luminal progenitor cells (GO: Gene Ontology. BP: Biological Processes). E. UMAP visualization of the number of genes (n_Genes) detected and the percentages of mitochondrial genes (% Mito) per cell in each platform. F. UMAP projections showing the fibroblast subtypes divided by platform. (iCAF: immune cancer-associated fibroblasts; ECM_CAFs: extracellular-matrix synthesis cancer-associated fibroblasts). G. Bar plot representation of the proportion of each fibroblast subtype per sample. H. Violin plots for the expression of marker genes for ECM CAFs (top), iCAFs (middle) and myofibroblasts (bottom) comparing each fibroblast subtypes between 10× (red) and BD Rhapsody (blue) platforms.
3.2. Technical comparison: assessing sensitivity and reproducibility
An important quality metric for scRNA-seq is the sensitivity of gene and UMI detection per cell. This sensitivity metric was different between the platforms (Fig. 1A); we detected a median of 2,995.5 and 2,791 genes, and 10,513.5 and 9,880 UMIs per cell in 10× Chromium and BD Rhapsody, respectively. Based on this analysis 10× Chromium and BD Rhapsody had a similar UMI and genes counts per cell at an equivalent number of reads per cell (Table S1). The proportion of mitochondrial genes is another metric commonly used to assess cell quality. Here we found that cells processed with 10× Chromium have significantly less mitochondrial content than BD Rhapsody samples, where 10× had a median of 6.5 % of mitochondrial transcripts and BD Rhapsody 15.3 % (Fig. 1A, right plot).
An essential application for scRNAseq is to define single-cell phenotypes by their gene expression signature through dimensional reduction, principal component analysis (PCA), and clustering algorithms [44]. We used the top 3,000 variable genes for dimensional reduction analysis in both platforms, noting that the variable genes detected in each platform were slightly different, with 2,140 (71.3 %) common genes (Fig. 1B). The total variability identified in the system, measured as the standard deviation at increasing principal components (PCs) had similar standard deviations per PC between platforms (Fig. 1C). When we examined the clustering capabilities at increasing resolutions, we found that at the lowest resolution, only 10× Chromium was able to split the cells into two clusters suggesting that their detected transcriptome was more distinct; moreover, at increasing resolutions (>0.1), we found that 10× had a greater number of clusters and more consistent cluster composition than BD Rhapsody (Fig. 1D). Statistical analysis using Silhouette confirmed that at resolutions higher than 0.3, Chromium 10× data had slightly higher silhouette score, suggesting that cells were more cohesive with their own cluster and more different to the cells from other clusters in 10× (Fig. 1E). In conclusion, BD Rhapsody and 10× Chromium had similar gene variability but 10× Chromium outperformed BD Rhapsody at consistently assigning cells to cluster at increasing resolutions.
Finally, to assess the reproducibility of each platform, we looked at the correlation of gene expression between replicates (Fig. 1F). We found that both 10× Chromium and BD Rhapsody had over 0.99 correlation values confirming high reproducibility for both platforms.
3.3. Cell type identification and lineage biases between platforms
To directly compare the performance of each platform to resolve tumour heterogeneity, we merged (Fig. S1C) and integrated (Fig. 2A and S1C) the data from the two platforms into one Seurat object. For the integration, we tested canonical Reciprocal PCA integration (RPCA) (Fig. 2A), correlation analysis (CCA), Join Principal Components Space (PCS) and Harmony Integration [34] (Fig. S1D). We found a similar level of integration using all four methods. RPCA integration has been reported to be less prone to overcorrection [45], therefore, we used this approach for the downstream analyses.
For cell type identification, first, we annotated the main cell lineage compartments in PyMT tumours (epithelial, immune and stroma) in the integrated object (Fig. 2B) and we compared their relative proportion in each platform to the measured proportion of these main cell types for PYMT tumours by flow cytometry, as it is the current gold standard method for cell type annotation and validation (Fig. 2C). Overall, we found similar proportions between scRNAseq and flow cytometry, but the samples analysed through 10× Chromium had a slightly higher number, but consistent between replicates, of stroma cells and fewer epithelial cells. Next, we assigned each cell cluster to the main cell types based on SingleR [35] (Figs. S2A and B) and further refined based on label transfer of the cell types from our previously published PyMT tumour reference [30] (Figs. S2C–S2E). We found that all major cell types: B cells, T cells, myeloid cells, endothelial cells, epithelial cells, fibroblasts and myofibroblasts were detected in both platforms (Fig. 2D), with similar proportions of cell types between replicates and platforms, except for endothelial cells and myofibroblasts (Fig. 2E). BD Rhapsody identified a lower percentage of endothelial cells, from 1 % of the total cells in BD Rhapsody compared to 9-6% in 10× Chromium, and myofibroblasts, 10 times less detected in BD Rhapsody compared to 10× Chromium. Consistently, the relative proportion of endothelial cells and myofibroblasts in BD Rhapsody deviated from the expected inter-tumour variability in the PyMT tumour scRNAseq reference atlas across eight tumours [30], suggesting that these differences were not a consequence of tumour heterogeneity (Fig. 2F). Of note, the proportion of fibroblasts was very variable between 10×/BD Rhapsody and the PyMT tumour reference atlas this could be due to a technical bias of the Drop-seq platform used in this atlas [30]. We further confirmed the consistency of cell annotation across platforms by looking at the top markers of each cell type and confirmed that these markers were similarly expressed in all cells captured in both platforms (Fig. 2G) We also confirmed a high correlation between platforms when we compared the gene expression differences of the same cell types (Pearson correlation coefficient >0.9, Fig. 2H).
To determine how the different cell population ratios, gene marker dropout or ambient noise can affect downstream analysis, we performed cell-to-cell communication predictions using the CellChat software on these two platforms independently. We found that overall BD Rhapsody detected a larger number of predicted interactions and higher predicted interaction strength (Fig. 3A). In line with the number of cells detected, BD Rhapsody predicted fewer and weaker interactions between myofibroblasts with epithelial cells or fibroblasts and between endothelial and epithelial cells (Fig. 3B, red lines), however, it detected more predicted interactions between most of the cell line pairs and it had stronger putative interactions within the epithelial compartment, between the fibroblast and the epithelial cells and between myeloid cells and epithelial or fibroblast cells (Fig. 3B, blue lines). CellChat analyses also revealed that some signalling genes were missing in BD Rhapsody such as PTN and OCLN in the epithelial cells, NT and SMA7 in fibroblasts and APELIN in endothelial cells, while 10× did not detect 16 genes related to cell-to cell communication pathways including ITGAL, CD45 and BAFF from myeloid cells, CLEC, CD48 and CD137 from B cells or ICOS and CD86 from B cells (Fig. 3C). Overall BD Rhapsody seems to predict more cell-cell interactions and as expected, changes in cell type proportions had an impact on the predicted interaction strength detected with those cell types.
In summary, 10× Chromium detected more endothelial cells and myofibroblasts, which resulted in different levels of cell-cell interactions predicted with those cell types in each platform, however overall, the genes identify in BD Rhapsody per cell subtype were better on predicting cell to cell interactions, as a higher number and stronger putative interactions were detected.
3.4. Cell subtype proportions across platforms
To further evaluate how each platform can distinguish among cellular subtypes within the main cell types in tumours, we performed subclustering of some of these main cell types. First, we selected the most abundant cell type in PyMT tumours, the epithelial compartment; unsupervised clustering revealed visual differential distribution between platforms in some cell clusters including clusters 1 and 3 out of the 7 clusters (Fig. 4A and S3A). To infer the identity of each cell subcluster, we again used the scRNAseq PyMT tumour atlas [30] of the epithelial cell subtypes in the integrated epithelial cluster (Figs. S3B and S3C), which resulted in the annotation of 8 epithelial subtypes manually (Fig. 4B). As expected in this transgenic mouse model [[26], [27], [28]], most cells were classified as luminal progenitors (LP), including a subset of hormone-sensitive luminal progenitor (LP-HS) cells where no major differences were observed between platforms (Fig. 4C). Interestingly, the cell clusters where we visually observed differences (Fig. 4A) were classified as basal (cluster 1) and luminal progenitor/luminal hormone-sensing (LP/LP-HS) (cluster 3) (Fig. 4B and C). Cluster 3, or LP/LP-HS cells, was missing from the PyMT reference (previously done using Drop-seq) and was heavily comprised of cells from the 10× Chromium platform. Gene set enrichment analysis identified that this 10x-specific luminal progenitor cluster was enriched with genes involved in immune responses, including antiviral response and interferon (Fig. 4D). This suggests that the 10× Chromium platform detects more epithelial cells that are actively interacting with the immune system. Interestingly, basal cells (cluster 1) were almost undetected in BD Rhapsody (3 cells detected), while in the 10× platform comprised around 2 % of the epithelial compartment. We also identify differential proportion of cells between platforms in the Multi/Stem cluster (Fig. 4C) with the highest fraction found in the BD Rhapsody replicates. Even though, the gene expression profile of the cluster 7 correlated with the multipotent/stem cell type (Figs. S3B and C), we found that these clusters also had the highest percentage of mitochondrial gene content, and the lowest number of genes detected in both platforms (Fig. 4E), potentially suggesting damaged cells. Considering cells with high mitochondrial content but low number of genes detected as the definition of damaged cells, the 10× Chromium platform had the lowest ratio of damaged cells (Fig. 1A), which may explain the absence of these cells and suggest that these are damaged cells.
Next, we did the same analysis in a less abundant population, fibroblasts. We again assigned cell types based on the fibroblast subsets in scRNAseq PyMT tumour reference [30] (Fig. 4F) and found that within the fibroblast partitions, there were three clusters that correlated with either myofibroblasts or secretory cancer-associated fibroblasts (CAFs) which were subdivided into extracellular-matrix synthesis CAFs (ECM-CAFs) and inflammatory CAFs (iCAFs) (Fig. 4F). The 10× Chromium platform detected a lower percentage of inflammatory CAFs, while the BD Rhapsody platform detected fewer ECM-CAFs and myofibroblasts (Fig. 4G). In this context, the expression of key markers of the inflammatory CAFs, like Ly6c1 and C4b, was higher in BD Rhapsody, while the myoepithelial markers Acta2, Mylk and Myh11 were higher in the 10× data (Fig. 4H).
Together these data suggest that, even though all major cell types were represented in both platforms, there are still cell type detection biases intrinsic to each platform which should be considered when analysing the tissue heterogeneity.
3.5. Ambient noise comparison between 10× chromium and BD Rhapsody in high and low-quality samples
One of the limitations of single-cell data is the technical noise caused by the ambient RNA contamination [39], amplification bias during library preparation and index swapping during sequencing [46]. The nature of the cell capture method (microwell or oil droplet) and the differences in the molecular workflows for RNA amplification in each platform (Fig. S1A) could result in a differential origin of the technical and/or ambient noise. Ambient RNA is defined as the mRNA molecules that have been released from dead or stressed cells and are part of the cell suspension. The ambient noise is especially present in samples that require tissue digestion and in low-quality samples, such as tissues stored for an extended period before processing, or tissues with active cell dead or hypoxia regions like tumour tissues.
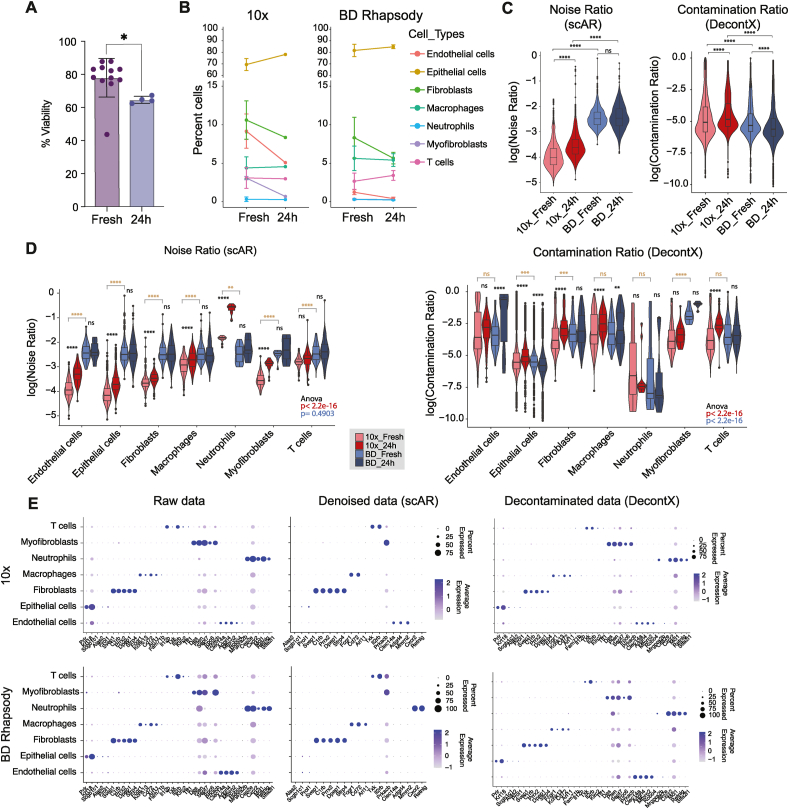
Our data suggest that noise might be handled differently by the commercial platforms (Fig. 1, Fig. 4E). Thus, we studied how these two commercial platforms, performed with the technical noise from challenging samples. To recreate this “low quality sample”, we incubated PyMT digested tumours for 24 h at 4 °C. After 24 h, cell viability dropped 20 % compared with the freshly digested sample as measured by DAPI positive cells in flow cytometry, indicating a substantial cell decay (Fig. 5A, S4 and Table S3). As done before with fresh PyMT tumour samples, we removed dead cells using autoMACS® Pro and processed the samples for scRNAseq using the 10× Chromium or BD Rhapsody platforms. Cell viability was checked before and after autoMACS® Pro, and even damaged cells had a viability of ≥85 % before running the single cell RNAseq experiments, reflecting a real-life scenario.
Fig. 5.
Analysis of the ambient noise in challenging samples in 10× Chromium and BD Rhapsody. A. Bar plot showing the percentage of cell viability measured by flow cytometry as the percentage of DAPI negative cells in digested fresh tumours (Fresh) and simulated low-quality tumour samples (24 h). B. Line plot showing the changes in percentage of cells for each cell type between fresh tumours and low-quality tumours (24 h) measured from the scRNAseq data. C. Violin plots showing noise ratio by scAR (left) and contamination ratio by DecontX per cell in each condition. Statistical analysis of the comparisons between fresh and low-quality (24 h) samples (black) or between fresh 10× and fresh BD (brown) was performed using paired t-test adjusted with the Bonferroni method, ****adjusted p-value<0.001; ns = not significant. D. Violin plots split by cell type of the noise ratio by scAR (left) and the contamination ratio by DecontX (right) detected in each cell. Statistical analysis of the noise ratio comparisons per cell type between fresh and low quality (24 h) samples in each platform (black) or between fresh 10× and fresh BD (brown) was performed using paired t-test adjusted with the Bonferroni method, **** adjusted p-value <0.001; ns = not significant. One-way ANOVA was also performed to compare the noise ratio distribution across all cell types in the fresh samples in 10× Chromium (p-value = 0.4903), or BD Rhapsody (p-value = 2.2e-16). E. Dot plot showing average expression of the top gene markers of each cell type using the integrated raw data or denoised (scAR) or decontaminated (DecontX) gene expression matrix.
First, we compared the tissue heterogeneity of the damaged samples to the fresh cells. As for this analysis, we increased the number of cells to ∼4000 cells per condition (Table S2), which allowed us to further split the myeloid cell compartment into neutrophils and macrophages. We found that endothelial cells, fibroblasts and myofibroblasts were consistently lost on the damaged sample regardless of the platform (Fig. 5B).
Next, to assess the technical noise found in each platform, we used the single-cell Ambient Remover (scAR) method, a universal model to detect noise across single-cell platforms based on probabilistic deep learning of the ambient signal [38]. Interestingly, around 5 % of cells in both platforms and in any condition (fresh or damaged) were considered empty cells as their transcriptome was indistinguishable from the ambient signal of the empty barcodes (Fig. S5A). Those assigned “empty cells” also had a high percentage of mitochondrial content and low gene content (Fig. S5B) and therefore confirming that these assigned “cells” were instead damaged cells that needed to be removed from downstream analysis. When we compared the noise ratio of the remaining cells, we found that BD Rhapsody had significantly more ambient noise per cell than 10× Chromium (Fig. 5C, left plot Fresh samples), however, the ambient contamination (noise ratio) was significantly increased in the damaged samples (24 h) in the 10× Chromium but not in BD Rhapsody (Fig. 5C, left plot). To further explore the origin of ambient RNA, we used an additional decontamination tool, DecontX, a Bayesian method to estimate and remove contamination in individual cells [39] without modelling the noise from the empty droplets, but from each putative cell. Fig. 5C right plot, shows that the differences in the contamination ratio were less pronounced between platforms than with scAR. Altogether the ambient RNA has different origins, empty droplets and from ambient RNA from putative cells and each platform handles this noise differently, with BD Rhapsody having more noise in empty droplets (noise) while the ambient RNA in the putative cells (contamination) is comparable between platforms.
We also confirmed our previous analyses of a higher percentage of mitochondrial content in Rhapsody (Fig. 1A), and as expected, in the damaged samples the mitochondrial content increased similarly in both platforms (Fig. S5C, left plot). The number of genes found per cell decreased in the damaged samples compared to their fresh counterpart; however, the decrease on the number of genes was more pronounced with BD Rhapsody (Fig. S5C, right plot). We explored how a standard filtering based on mitochondrial content and number of genes would perform to denoise the data. As the overall mitochondria content per cell was different per platform (Fig. 1A and S5C), we used the threshold based on the percentiles of each platform, cells with more than the 25 percentile of mitochondria content and less than the 10 % percentile of number of genes per cells were labelled as low-quality control (lowQC) (Figs. S5D and S5E). Even though, this stringent threshold removed the majority of “empty cells” (Fig. S5F) this type of filtering does not clean the ambient RNA contamination per cell type (Fig. S5G vs Fig. 5G). In fact, the noise ratio does not correlate well with either the percentage of mitochondria genes (r = 0.33), or the number of genes (r = −0.12) (Fig. S5H).
Next, to evaluate which cell lineages are most affected by the technical noise in each platform, we compared the noise ratio per cell type and platform in each sample condition, using both scAR and DecontX (Fig. 5D and E). The analysis with scAR, showed that in BD Rhapsody all cell types had a similar level of noise, regardless of their damaged condition, while 10× Chromium's noise was notably different between cell types and there were significant differences between fresh and damaged conditions (Fig. 5D, left plot). Particularly, in 10× Chromium, neutrophils had the highest percentage of noise which increased to an even higher ratio in the damaged samples. As expected, neutrophils also had the lowest number of genes detected especially in Chromium 10× (Fig. S6A), independently of sequencing depth (Fig. S6B). To understand which cell types and genes are driving the ambient signal, we analysed the expression across cell types of the top genes contributing to the ambient pool based on scAR analysis (Fig. S6C). Remarkably, in 10× Chromium, the ambient expression of the S100a9 neutrophil marker was >25-fold times higher than any other cell type marker and was even higher in the damaged samples, despite the low number of neutrophils identified in the sample; while in BD Rhapsody the ambient gene expression was more uniformly distributed and did not show major differences between fresh and damaged sample (Fig. S6C). To confirm the neutrophil biases from 10× Chromium in the ambient profile, we correlated the average transcriptome of the single cells with the ambient gene expression and observed a higher presence of neutrophil marker genes in ambient RNA than in the single cells in 10× Chromium, but not in BD Rhapsody (Fig. S6D). Interestingly, the same cell type analysis for the distribution of the ambient RNA calculated using only putative cells with DecontX did not show major cell type biases either in 10× Chromium or BD Rhapsody, and less pronounced differences between platforms with myofibroblasts showing the highest differences in contamination between platforms, where BD Rhapsody had higher contamination (Fig. 5D, right plot).
The scAR and DecountX algorithms can denoise the count matrix based on the ambient composition. When we used the denoised data using scAR to resolve cell type clusters, we found that clusters are more distinct using denoised data and there is less background expression of non-specific markers compared to the raw data (Fig. 5E) or the data processed with standard QC filtering (Fig. S5G). As expected, the expression of the neutrophil markers was completely lost in the 10× Chromium data in this cell subtype (Fig. 5E) The decontaminated data using DecontX on the other hand maintained the neutrophil marker however had a comparable non-specific background than the raw data or QC filtering (Fig. 5E and S5G). This dichotomy may be explained by the different approaches used by the two ambient RNA removal tools. For scAR, we modelled ambient RNA based on the empty barcodes and putative cells, while for DecontX we measured ambient RNA using only putative cells (“in cell”) and based on the expression of key gene markers in other cells populations. Therefore, this suggests that the signature of neutrophils mainly comes from droplets that have captured neutrophils but due to its in-drop degradation the amount of RNA detected in the sequencing is very low and thus considered an empty droplet. This explanation also supports the fact that when the empty droplets are not considered for ambient RNA calculations, DecontX, all cell types have a similar contribution to the noise between platforms (Fig. 5C right plot).
In conclusion, the differential molecular design, the microfluidic versus microwell format and the sample quality are sources of the ambient noise. In BD Rhapsody there is a generalised level of noise that is independent of the cell of origin and sample quality, however there is some bias towards myofibroblast specially in low-quality samples; while in 10× Chromium, ambient noise is cell type-specific and overrepresented by the neutrophil population specially in empty droplets suggesting in droplet neutrophil-specific RNA degradation.
3.6. Cell type bias noise validation on public datasets
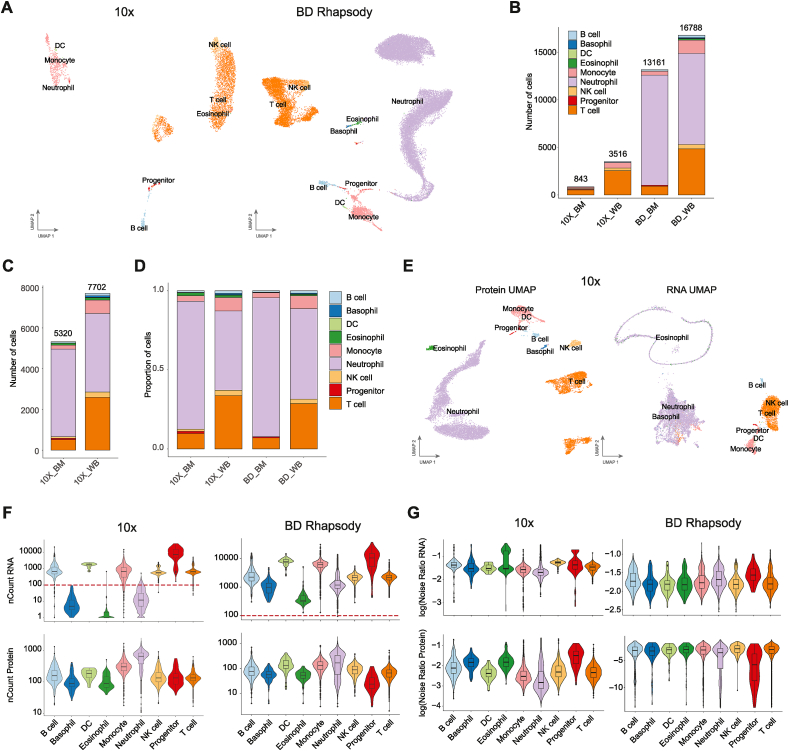
To determine if the cell type biases and technical noise differences between 10× Chromium and BD Rhapsody were universal, we assessed another two public data sets, human whole blood and bone marrow, where more neutrophils are expected, and both platforms were used [37]. This data was processed using CITEseq (10× Chromium) and Ab-Seq + whole transcriptome analysis (BD Rhapsody) and therefore it also included 30 antibody-derived barcodes in both datasets. Additionally, hashtag oligos (HTO) were used in 10× Chromium platform to demultiplex samples.
We first compared the abundance of each cell type detected based on the antibody markers in both platforms using the default parameters for filtering established by Cell Ranger and the Rhapsody pipelines (Fig. 6A). Interestingly, the whole blood and the bone marrow samples processed with 10× had virtually no granulocytes (neutrophils, basophils, or eosinophils) (Fig. 6B). This is in line with previous reports of a low number of neutrophils in human datasets detected using this platform [47,48]. Interestingly, if we filtered the 10× Chromium cell barcodes using the custom threshold based on the Protein UMI counts per cell as Qi et al. [37], we recovered 5,320 and 7,702 cells, in the bone marrow and whole blood samples, respectively, from which the majority were neutrophils (Fig. 6C). In addition, using this custom filtering for all samples, resulted in very similar relative proportions of each cell type in the bone marrow and whole blood between the 10× and BD Rhapsody platforms (Fig. 6D). However, even though the UMAP using the protein matrix was able to distinguish all major cell types in 10× Chromium (Fig. 6E), the low gene sensitivity in the granulocyte population, with a median of fewer than 100 counts per cell (Fig. 6F), did not manage to further resolve the granulocyte clusters at the transcriptome level (Fig. 6F and 6E right panel). In fact, when we run the scAR algorithm using the threshold for filtering cells based on Protein UMI counts, we found that in 10× Chromium some granulocytes, especially eosinophils, still had a higher RNA noise ratio, although not as pronounced as in our mouse dataset in comparison with other cell types (Fig. 6, Fig. 5D). Overall, the noise ratio from both the RNA and the protein data was more similar across cell types in BD Rhapsody than in 10× Chromium, confirming that in BD Rhapsody the ambient noise is not cell type specific (Fig. 6G). In summary, 10× Chromium was unable to detect granulocytes in human samples based exclusively on their RNA content, and their low gene detection hinders the denoising algorithms to distinguish empty droplets from real cells with low UMI counts.
Fig. 6.
Performance evaluation for the detection of human granulocytes between 10× Chromium and BD Rhapsody. A. UMAPs of cell types found in human whole blood and bone marrow samples processed with 10× Chromium or BD Rhapsody using the transcriptome for dimension reduction. Cell barcodes were filtered using either Cell Ranger or BD Rhapsody pipelines. B. Bar plot showing the number of cells found in each cell type per condition. C. Bar plot showing the number of cells found in each cell type in 10× Chromium after custom filtering based on a minimum of 10 Protein UMI counts. D. Bar plot of the relative proportion of cells from each cell type per condition using the same custom threshold than panel C for 10× Chromium. E. UMAP plots of the human datasets processed with 10× Chromium and filtered based on custom threshold where the dimensional reduction was done using either the protein data (left) or the RNA data (right) F. Violin plots of the number of RNA and Protein UMI counts in each cell split by cell type. The red dotted line highlights a 100 counts threshold. The y axis is shown in logarithmic scale. G. Noise ratio in each cell using either from the RNA (top) or the protein (bottom) data matrix and split by cell type. (BM: bone marrow; WB: whole blood).
4. Discussion
Single-cell RNA-seq allows the classification of different cell types based on the gene expression profile of individual cells. The rise of commercial instruments and kits has enabled the use of this methodology by the broad scientific community. However, there are still technical challenges during sample preparation, cell capture and library preparation [49], that can affect the bioinformatic readout and consequently, the biological interpretation driven from the data. Therefore, it is crucial to understand the advantages and limitations of each platform to best tailor the selection of a platform to the sample type and the biological question that wants to be answered, in addition to the costs and expertise level of the user. Here, we have compared the most popular commercial platforms, the microfluidic-based system, 10× Chromium, and the microwell plate-based system, BD Rhapsody, using mammary gland tumours from the PyMT breast cancer mouse model [[25], [26], [27], [28]]. Analysing samples that require tissue digestion prior to the single-cell experiments is more challenging as the damaged cells can introduce additional ambient noise and different cell-type vulnerabilities can change the observed tissue heterogeneity. An advantage of our design is the comparison of tumour samples from a transgenic mouse model digested using the same protocol and in the same laboratory, which allows us to discard sample preparation as the source of variability. In addition, we have created low-quality samples using tumours from the same mouse model with reduced viability to assess how the quality of the sample affects the readout from 10× Chromium and BD Rhapsody. Frequently, the samples processed for scRNAseq have high levels of damaged cells, for example, tumour samples have necrotic regions or increased cell death due to their high cell turnover or exposure to treatments, such as radiotherapy or chemotherapy. Transportation of the biological material to the laboratory where cell capture is performed may also increase the cellular stress of the sample. In this study, we have used a biologically relevant tissue and replicated the standard user experience to perform a realistic assessment of the different technologies in challenging samples.
Our comprehensive data analysis identified, first, at the technical level, that 10× Chromium and BD Rhapsody had comparable overall gene sensitivity and reproducibility at similar sequencing depths. Next, we measured tissue heterogeneity and found a lower percentage of endothelial and myofibroblast cells in BD Rhapsody compared to 10× Chromium. However, comparing their ratios with the flow cytometry data, we found that 10× Chromium tends to detect more cells from the stroma than BD Rhapsody and flow cytometry. This suggests that 10× Chromium may enrich stroma cells which could be beneficial if you are interested in those populations. Interestingly, endothelial, and myofibroblast cells are reduced in low-quality samples in both platforms. We also found differences within the epithelial compartment, including an immune-responsive cluster that were enriched in the 10× Chromium samples. Orthogonal validation of this cell population is needed to understand its origin.
In the immune compartment, the most notable differences were found within the myeloid cells, especially in granulocytes, which 10× Chromium was not able to detect granulocytes neither in the PyMT mammary tumours nor in the human whole blood or bone marrow datasets, due to the low gene detection, which misclassifies them as empty droplets. It is known that granules in neutrophils, eosinophils and basophils have high levels of nucleases including RNases [50]. This has been suggested to be the reason why granulocytes have lower genes and counts detected than other cell types [37,51]. We have also confirmed in our PyMT dataset and human blood and bone marrow public datasets that granulocytes, including neutrophils, have the lowest number of genes detected. Interestingly, BD Rhapsody's neutrophils had an average of 1,240 or 602 genes while in 10× Chromium neutrophils only had 551 or 10 in the mouse and human datasets, respectively. We hypothesise that there are two factors that may contribute to this phenomenon; firstly, neutrophils may be more sensitive to the pressure from the microfluidic devices which could cause their breakage and loss of RNA to the ambient pool in the 10× Chromium system; secondly, we hypothesise that the 10× platform is more sensitive to the presence of RNases released by neutrophils than BD Rhapsody. Supporting this theory, Qi et al. found that cell doublets of neutrophils and T cells from 10× Chromium had less number of UMIs detected than T cell singlets, suggesting that the RNases from the neutrophils degraded the RNA of the T cells [37]. The exact recipes of the lysis buffers used in 10× Chromium and BD Rhapsody are not disclosed, but it is possible that BD Rhapsody has stronger detergents or RNase inhibitors that degrade RNases. BD Rhapsody workflow also includes washes of the beads before cDNA generation which could remove any remaining RNases or strong detergent from the lysis buffer to not interfere with the reverse transcription reaction. Moreover, a recent report describing TAS-seq, a BD Rhapsody-based technique with improved cDNA amplification step, resulted in better cellular composition fidelity, especially in neutrophils, using their system compared to 10× Chromium and Smart-seq [52]. Together, this demonstrates that BD Rhapsody is better suited for granulocytes research, and new strategies for cDNA amplification that increase the number of genes detected per cell will further improve the transcriptional resolution of this challenging cell population.
Mitochondrial content has been commonly used as a sign of damaged cells, where their cytoplasmatic RNA has been lost but the RNA contained in the mitochondria remains [53]. In fact, we see an increase in the mitochondrial fraction in the damaged samples. Surprisingly, all cells processed with the BD Rhapsody have a higher percentage of mitochondrial genes, even though they are not considered damaged. This phenomenon has also been recently reported by others [54,55]. [56]Our experimental design included additional steps for multiplexing only in the BD Rhapsody samples prior to the cell capture (Figs. S1A and B) that may have contributed to a higher mitochondrial content. However, this is highly unlikely as the additional time required for the overall 4-h process was 30 min and cell viability did not substantially change after this step (Table S3). Nevertheless, to further explore this, we have analysed the mitochondrial content from publicly available whole blood and bone marrow samples (Fig. 6), where multiplexing was performed in 10× Chromium but not in BD Rhapsody samples. We found that mitochondrial content was consistently higher in BD Rhapsody when we compared two cell types that have equivalent RNA quantity, T cells and monocytes (Fig. S7) [37]. Furthermore, Gao et al. showed that the mitochondrial content in BD Rhapsody was higher comparing the demo PBMCs dataset from both platforms where multiplexing was not performed [55]. A possible explanation for this mitochondrial disparity could be that the lysis buffer of BD Rhapsody is more effective at digesting the organelles including mitochondria and therefore releasing the mitochondrial RNA into the cytosol to bind to the beads. For this reason, different thresholds of mitochondrial content should be used to filter out damaged cells in BD Rhapsody or 10× Chromium.
Ambient RNA released during the sample preparation and cell capture introduces noise to the data affecting the downstream analysis. Here, we have compared the level of ambient noise using two different computational frameworks, scAR [38] and DecontX [39] to identify and define vulnerabilities across platforms. We run these two methods using different strategies to calculate the ambient RNA. For scAR, the noise was measured for each barcode corresponding to putative cells and the gene expression of the ambient RNA from empty barcoded droplets; while for DecontX, we analysed the data without using the empty barcodes as background, and thus we only calculated the ambient RNA coming from droplets containing putative cells. We found that using scAR, 10× Chromium ambient pool was biased towards neutrophils, however this was not seen when the ambient RNA was only measured in putative cells (DecontX). This confirms that the RNA from captured neutrophils is likely degraded by the RNases from these cells reducing the number of UMIs making it hard to distinguish between real or empty barcodes, as previously discussed. This confounding factor explains why scAR detected less ambient RNA per cell overall in 10× Chromium compared to BD Rhapsody, as the ambient expression in 10× was mostly made of neutrophil marker genes that are not likely detected in putative cells from other cell types while BD ambient RNA pool was comprise by RNA from all cell types. In fact, DecontX found similar levels of ambient RNA per cell across platforms. This analysis highlights the importance of understanding the origin of ambient RNA calculated by denoising tools before using them to clean up the data. This issue has been in fact considered by 10× Genomics and they recommend including additional steps on their workflows to optimise single cell assays for neutrophils/granulocytes, these include: immediate sample processing within 2 h after sample collection, supplementing with RNAse inhibitors in the wash and resuspension buffers, avoid long incubations on ice, increasing the PCR cycles during cDNA amplification, sorting neutrophils to enrich this population and to change the filtering parameters in Cell Ranger using the “force cell” parameter to recover cells with little RNA (neutrophils).
In this study, we also adapted the LMO sample multiplexing approach from MULTI-seq [32] for our BD Rhapsody samples (Fig. S1B, see Material and Methods) using 4 LMO sample tags with a lower-than-expected capture efficiency of 59 % likely due to a drop out of one LMO (Table S1). The MULTI-seq method uses a highly efficient approach for DNA sample tagging in live cells based on the affinity of lipids to the cell membranes which ensures a full capture of these LMO-oligos in the cells as well as in the bead capture [32]. The unexpected lower capture efficiency of our adapted MULTI-seq method could be due to the addition of an incorrect amount of one of the sample barcode oligonucleotides that resulted in a non-efficient hybridization to the anchor LMO. This is further supported by the fact that the rest of the LMO-tagged samples had an average capture efficiency of 79 %, over the expected amount of captured cells/sample (1,591 singlets/sample, Table S1). These LMOs for sample tagging are now commercially available through Sigma-Aldrich (Cat# LMO001) and could be used for sample multiplexing in both platforms, as previously described in the case of 10× Chromium [32] or following the method described in this study for BD Rhapsody. Alternatively, BD offers a multiplexing kit (BD® Single-Cell Multiplexing Kit) using polyadenylated DNA barcodes conjugated to a universal antibody.
A limitation of our comparison analysis is the fact that we have used mouse mammary gland tumours, human whole blood and bone marrow only. Other tissues where different cell types are found may show other cell type biases, especially for lowly represented cell types. However, this study highlights the importance of using different technologies when assessing the population ratios of a sample and avoiding comparing cell type ratios across biological conditions performed in different platforms. Based on our analysis, we also found that the expected quality of the sample and origin of the tissue should also be considered when choosing a platform to perform single-cell RNAseq, as damaged tissues or tissues with high levels of RNases, such as bone marrow or spleen [51], would be more susceptible to RNA loss in 10× Chromium.
Complementary strategies, such as performing multi-omic studies where two or more molecular layers of information are investigated per cell [57], or spatial transcriptomics [58], as well as alternative methods for tissue preservation such as the ALTEN system [11] or parafilm fixation in combination with fixed RNA profiling [51,59], may also help dissect the true heterogeneity of complex tissues and overcome the current limitations of single-cell RNAseq.
Ethics declarations
This study was reviewed and approved by the St. Vincent's Campus Animal Research Committee with the approval number: 19/02, dated March 01, 2019.
Data availability
Data generated in this paper is available through Gene Expression Omnibus (GEO): GSE229765. PyMT mouse data processed with Dropseq is available in GSE158677. Human data sets were downloaded from PRJNA73428.
CRediT authorship contribution statement
Yolanda Colino-Sanguino: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Laura Rodriguez de la Fuente: Validation, Resources, Methodology, Data curation. Brian Gloss: Methodology, Data curation. Andrew M.K. Law: Resources, Methodology. Kristina Handler: Writing – review & editing, Methodology. Marina Pajic: Resources. Robert Salomon: Resources. David Gallego-Ortega: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Fatima Valdes-Mora: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by Cancer Institute NSW Fellowship (2019/CDF002-CDF181218) to FVM and Cancer Institute NSW Fellowship (DG00625) and Cancer Council NSW project grant (RG18-03), NHMRC (2012941), NBCF Elaine Henry Fellowship (IIRS-21-096) to DGO; YCS is supported by Cure Brain Cancer Foundation (20210917).
We thank the Garvan-Weizmann Centre for Cellular Genomics, the Molecular Genetics Facility, the Australian BioResources Pty Ltd facility and the KCCG Sequencing Laboratory from the Garvan Institute. We would also like to thank Dr Antoine de Weck for his guidance on the use of scAR and Prof Zev J. Gartner and Dr. Christopher S. McGinnis for gifting us the MULTI-seq kit reagents.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37185.
Contributor Information
David Gallego-Ortega, Email: David.GallegoOrtega@uts.edu.au.
Fatima Valdes-Mora, Email: FValdesMora@ccia.org.au.
Appendix A.
Table S1.
Number of cells and reads after filtering and downsampling for platform comparison analysis.
* For BD Rhapsody we used the sample tag information to determine the number of real cells singlets associated to each sample tag, multiplets or undetermined cells are excluded. *Only samples Fresh 2 and Fresh 3 were used for Figures 1, 2,3 and 4. Renamed as BD_r1and BD_r2 in the figures to facilitate the reader*For BD Rhapsody, Fresh 2, 3 and 4 samples were used for Figure 5 as also shown in Table 2Tumour location nomenclature: Tumour A - right cervical and/or thoracic mammary gland; tumour B - left cervical and/or thoracic mammary gland; tumour C - right abdominal and/or inguinal mammary glands; tumour D - left abdominal and/or inguinal mammary glandsDownsampled cells and reads (grey columns) were used for Figures 1-4.
Table S2.
Number of cells and reads per sample used for fresh versus damaged comparison analysis.
| Tumour source | Naming used in Fig. 5 | N Cell used for analysis | Reads per cell | ||
|---|---|---|---|---|---|
| 10× | Fresh 1 | Mice 1 (tumour AC and BD), Mice 2 (tumour AC and BD) |
10×_Fresh | 4,000 | 23,023 |
| 24 h | Mice 3 (tumour BD), Mice 4 (tumour AC) |
10×_24 h | 4,000 | 31,365 | |
| BD Rhapsody | Fresh 2, 3 and 4 | Mice 3 (tumour BD), Mice 4 (tumour AC) |
BD_Fresh | 3,758 | 49,843 |
| 24 h | Mice 3 (tumour BD), Mice 4 (tumour AC) |
BD_24 h | 4,186 | 40,675 | |
* Tumour location nomenclature: Tumour A - right cervical and/or thoracic mammary gland; tumour B - left cervical and/or thoracic mammary gland; tumour C - right abdominal and/or inguinal mammary glands; tumour D - left abdominal and/or inguinal mammary glands.
Table S3.
Cell viability percentage measured by flow cytometry.
* Tumour location nomenclature: Tumour A - right cervical and/or thoracic mammary gland; tumour B - left cervical and/or thoracic mammary gland; tumour C - right abdominal and/or inguinal mammary glands; tumour D - left abdominal and/or inguinal mammary glands.
Appendix B. Supplementary data
The following is the supplementary data to this article:
References
- 1.Tanay A., Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331. doi: 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shalek A.K., et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wills Q.F., et al. Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat. Biotechnol. 2013;31(8):748–752. doi: 10.1038/nbt.2642. [DOI] [PubMed] [Google Scholar]
- 4.Liu S., Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Res. 2016;5 doi: 10.12688/f1000research.7223.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pal B., et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat. Commun. 2017;8(1):1627. doi: 10.1038/s41467-017-01560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macosko E.Z., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein A.M., et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tirosh I., et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539(7628):309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farbehi N., et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife. 2019;8 doi: 10.7554/eLife.43882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herring C.A., et al. Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution. Cell. 2022;185(23):4428–4447 e28. doi: 10.1016/j.cell.2022.09.039. [DOI] [PubMed] [Google Scholar]
- 11.Law A.M.K., et al. ALTEN: a high-fidelity primary tissue-engineering platform to assess cellular responses ex vivo. Adv. Sci. 2022;9(21) doi: 10.1002/advs.202103332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Twigger A.J., et al. Transcriptional changes in the mammary gland during lactation revealed by single cell sequencing of cells from human milk. Nat. Commun. 2022;13(1):562. doi: 10.1038/s41467-021-27895-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimshony T., et al. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2(3):666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Jaitin D.A., et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343(6172):776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X., et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172(5):1091–1107 e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Chen H., et al. High-throughput Microwell-seq 2.0 profiles massively multiplexed chemical perturbation. Cell Discov. 2021;7(1):107. doi: 10.1038/s41421-021-00333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomon R., et al. Droplet-based single cell RNAseq tools: a practical guide. Lab Chip. 2019;19(10):1706–1727. doi: 10.1039/c8lc01239c. [DOI] [PubMed] [Google Scholar]
- 18.Svensson V., Vento-Tormo R., Teichmann S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018;13(4):599–604. doi: 10.1038/nprot.2017.149. [DOI] [PubMed] [Google Scholar]
- 19.Ziegenhain C., et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017;65(4):631–643 e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., et al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol Cell. 2019;73(1):130–142 e5. doi: 10.1016/j.molcel.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Dueck H.R., et al. Assessing characteristics of RNA amplification methods for single cell RNA sequencing. BMC Genom. 2016;17(1):966. doi: 10.1186/s12864-016-3300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svensson V., et al. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods. 2017;14(4):381–387. doi: 10.1038/nmeth.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian L., et al. Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat. Methods. 2019;16(6):479–487. doi: 10.1038/s41592-019-0425-8. [DOI] [PubMed] [Google Scholar]
- 24.Ding J., et al. Author Correction: systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 2020;38(6):756. doi: 10.1038/s41587-020-0534-z. [DOI] [PubMed] [Google Scholar]
- 25.Gallego-Ortega D., et al. ELF5 drives lung metastasis in luminal breast cancer through recruitment of Gr1+ CD11b+ myeloid-derived suppressor cells. PLoS Biol. 2015;13(12) doi: 10.1371/journal.pbio.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guy C.T., Cardiff R.D., Muller W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell Biol. 1992;12(3):954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin E.Y., et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003;163(5):2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maglione J.E., et al. Transgenic Polyoma middle-T mice model premalignant mammary disease. Cancer Res. 2001;61(22):8298–8305. [PubMed] [Google Scholar]
- 29.Rodriguez de la Fuente L., et al. Tumor dissociation of highly viable cell suspensions for single-cell omic analyses in mouse models of breast cancer. STAR Protoc. 2021;2(4) doi: 10.1016/j.xpro.2021.100841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valdes-Mora F., et al. Single-cell transcriptomics reveals involution mimicry during the specification of the basal breast cancer subtype. Cell Rep. 2021;35(2) doi: 10.1016/j.celrep.2021.108945. [DOI] [PubMed] [Google Scholar]
- 31.Lun A.T.L., et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20(1):63. doi: 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGinnis C.S., et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods. 2019;16(7):619–626. doi: 10.1038/s41592-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20(1):296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korsunsky I., et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aran D., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20(2):163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin S., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi J., et al. Multimodal single-cell characterization of the human granulocyte lineage. bioRxiv. 2021:2021. 06.12.448210. [Google Scholar]
- 38.Sheng C., et al. Probabilistic modeling of ambient noise in single-cell omics data. bioRxiv. 2022 2022.01.14.476312. [Google Scholar]
- 39.Yang S., et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol. 2020;21(1):57. doi: 10.1186/s13059-020-1950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Araujo A.M., et al. Stromal oncostatin M cytokine promotes breast cancer progression by reprogramming the tumor microenvironment. J. Clin. Invest. 2022;132(7) doi: 10.1172/JCI148667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papanicolaou M., et al. Temporal profiling of the breast tumour microenvironment reveals collagen XII as a driver of metastasis. Nat. Commun. 2022;13(1):4587. doi: 10.1038/s41467-022-32255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stuart T., et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902 e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329–337 e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pezzotti N., et al. Approximated and user steerable tSNE for progressive visual analytics. IEEE Trans. Vis. Comput. Graph. 2017;23(7):1739–1752. doi: 10.1109/TVCG.2016.2570755. [DOI] [PubMed] [Google Scholar]
- 45.Hao Y., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587 e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costello M., et al. Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms. BMC Genom. 2018;19(1):332. doi: 10.1186/s12864-018-4703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Travaglini K.J., et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 2020;587(7835):619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin J.C., et al. Single-cell analysis of Crohn's disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178(6):1493–1508 e20. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ke M., et al. Single cell RNA-sequencing: a powerful yet still challenging technology to study cellular heterogeneity. Bioessays. 2022;44(11) doi: 10.1002/bies.202200084. [DOI] [PubMed] [Google Scholar]
- 50.Lu L., et al. Immune modulation by human secreted RNases at the extracellular Space. Front. Immunol. 2018;9:1012. doi: 10.3389/fimmu.2018.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monaco G., et al. RNA-seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019;26(6):1627–1640 e7. doi: 10.1016/j.celrep.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shichino S., et al. TAS-Seq is a robust and sensitive amplification method for bead-based scRNA-seq. Commun. Biol. 2022;5(1):602. doi: 10.1038/s42003-022-03536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ilicic T., et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. doi: 10.1186/s13059-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salcher S., et al. Comparative analysis of 10X Chromium vs. BD Rhapsody whole transcriptome single-cell sequencing technologies in complex human tissues. Heliyon. 2024;10(7) doi: 10.1016/j.heliyon.2024.e28358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao C., Zhang M., Chen L. The comparison of two single-cell sequencing platforms: BD Rhapsody and 10x genomics Chromium. Curr. Genom. 2020;21(8):602–609. doi: 10.2174/1389202921999200625220812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salcher S., et al. Comparative analysis of whole transcriptome single-cell sequencing technologies in complex tissues. bioRxiv. 2023 doi: 10.1016/j.heliyon.2024.e28358. 2023.07.03.547464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J., Hyeon D.Y., Hwang D. Single-cell multiomics: technologies and data analysis methods. Exp. Mol. Med. 2020;52(9):1428–1442. doi: 10.1038/s12276-020-0420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moses L., Pachter L. Museum of spatial transcriptomics. Nat. Methods. 2022;19(5):534–546. doi: 10.1038/s41592-022-01409-2. [DOI] [PubMed] [Google Scholar]
- 59.Janesick A., et al. High resolution mapping of the breast cancer tumor microenvironment using integrated single cell, spatial and in situ analysis of FFPE tissue. bioRxiv. 2022 doi: 10.1038/s41467-023-43458-x. 2022.10.06.510405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this paper is available through Gene Expression Omnibus (GEO): GSE229765. PyMT mouse data processed with Dropseq is available in GSE158677. Human data sets were downloaded from PRJNA73428.