Summary
Cyclic-phospholipids-based vesicles can play a role in facilitating the chemical evolution of protocells from the structurally simple to the functionally more complex form. Here, we present a protocol for preparing decanoic acid-derived cyclic phospholipid and glyceryl-diester phosphate-containing vesicles. We describe steps for sample preparation, equilibration, and image acquisition using confocal microscopy. This protocol has the potential for preparing a wide variety of these phospholipid-based artificial cell constructs.
For complete details on the use and execution of this protocol, please refer to Pulletikurti et al.1
Subject areas: Biophysics, Microscopy, Chemistry
Graphical abstract
Highlights
-
•
Steps for preparing cyclic-phospholipid-based vesicles
-
•
Instructions for acquisition of images and analysis of vesicles from the samples
-
•
Troubleshooting steps and guidance on sample preparation and equilibration
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Cyclic-phospholipids-based vesicles can play a role in facilitating the chemical evolution of protocells from the structurally simple to the functionally more complex form. Here, we present a protocol for preparing decanoic acid-derived cyclic phospholipid and glyceryl-diester phosphate-containing vesicles. We describe steps for sample preparation, equilibration, and image acquisition using confocal microscopy. This protocol has the potential for preparing a wide variety of these phospholipid-based artificial cell constructs.
Before you begin
Lipid synthesis
Timing: 3 days (for step 1)
Timing: 2 days from GMD (for step 2)
Timing: 4 days (for step 3)
Timing: 6 days (for step 4)
The lipids required for the vesicle preparation can be synthesized following the step-by-step protocols as described in Ortuno et al.2 and Pulletikurti et al.1
-
1.
Glyceryl monodecanoate (GMD) can be synthesized by following the detailed step-by-step synthetic protocols described in Ortuno et al.2
-
2.
Cyclic phospholipid decanoate can be synthesized by following the detailed step-by-step synthetic protocols described in Ortuno et al.2
-
3.
Glyceryl 2,3-didecanoate 1-phosphate (GDDP1) can be synthesized by following the detailed step-by-step synthetic protocols described in Pulletikurti et al.1
-
4.
Glyceryl 1,3-didecanoate 2-phosphate (GDDP2) can be synthesized by following the detailed step-by-step synthetic protocols described in Pulletikurti et al.1
Note: Store all the synthesized lipids in 4°C refrigerator and ensure the purity of the lipids by NMR analysis. (For the NMRs please see the Supporting Information in Ortuno et al. Figure S31-S32 for GMD; Figure S55-S57 for cPC10. And see the Supporting Information in Pulletikurti et al; Figure S75-S77 for GDDP1; Figure S84-S86 for GDDP2)
Buffer and dye (rhodamine 6G, R6G) preparation
Timing: 30 min
-
5.
Prepare 0.2 M Bis-Tris buffer of pH 6.3 or pH 6.6 and 2.6 mM Rhodamine 6G (R6G) according to the recipe in the materials and equipment section.
Note: Cyanine5 carboxylic acid (Cy5) or Nile red dyes can be used as a substitute for R6G.
General lab preparation
Timing: 30 min
-
6.
Ensure the working condition of the hot plate by checking the sensor and temperature stability.
-
7.
Check vortex functioning and keep at the full speed touch mode.
-
8.
Check the pH meter functioning and calibrate with pH standard solutions if required.
-
9.
Check the functioning of the hot air gun.
Note: This section deals with hazardous and corrosive materials. Ensure to follow lab safety measures, wearing lab coat, goggles and nitrile gloves and wash your hands once you finish with your experiments.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Decanoic acid (DA) | TCI | Cas #334-48-5 |
| Bis-Tris | RPI Research Products | Cas #6976-37-0 |
| HEPES | Oakwood Chemical | Cas #7365-45-9 |
| Hydrochloric acid (HCl) | Fisher Chemical | Cas #7647-01-0 |
| Rhodamine 6G (R6G) | Combi-Blocks | Cas # 989-38-8 |
| Software and algorithms | ||
| Fiji ImageJ 1.53t | Cardona et al.3 | https://imagej.net/ij/download.html |
| BioRender | Scripps Research Institute | Scientific Image and Illustration Software | BioRender |
| Other | ||
| pH meter | Fisherbrand, Accumet | AB250 |
| Hot air gun | Master ProHeat | PH 1100 |
| Heating plate | IKA | RCT BS 1 |
| Sonicator | Branson | 1510 |
| Vortex | Corning | LSE vortex mixer |
| Shaker | IKA | KS 260 |
| Microscopy | Zeiss Microscope | LSM710/LSM880 |
| 96-well plate | Corning | Part #15100170 |
| Chambered cover glass 8 well | Thermo Scientific Nunc Lab-Tek | Part #12565338 |
Note: The above reagents or chemicals can be obtained from any other company, such as Sigma-Aldrich, VWR and Thermo Fisher scientific etc.
Materials and equipment
Bis-Tris buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Bis-Tris | 0.2 M | 1.67 g |
| Deionized water | NA | 35 mL |
| Total | NA | 40 mL |
Note: Adjust the pH to 6.3 ± 0.05 or pH 6.6 ± 0.05 by adding 6 N HCl and make up the total volume to 40 mL. Store at 23°C–25°C (room temperature) for 6 months.
HEPES buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| HEPES | 0.2 M | 1.67 g |
| Deionized water | NA | 8 mL |
| Total | NA | 10 mL |
Note: Adjust the pH to 7.4 ± 0.05 and make up the total volume to 10 mL. Store at 23°C–25°C (room temperature) for 6 months.
Rhodamine 6G (R6G)
| Reagent | Final concentration | Amount |
|---|---|---|
| Rhodamine 6G (R6G) | 2.6 mM | 6.2 mg |
| 0.2 M HEPES buffer | NA | 4.97 mL |
| Total | NA | 4.97 mL |
Note: Store at −20°C for 2 years and protect from light. R6G stock solution can also be prepared either in MilliQ water or in Bis-Tris buffer.
Step-by-step method details
Protocol for the preparation of 40 mM of 2:1 DA:cPC10
Timing: 30 min
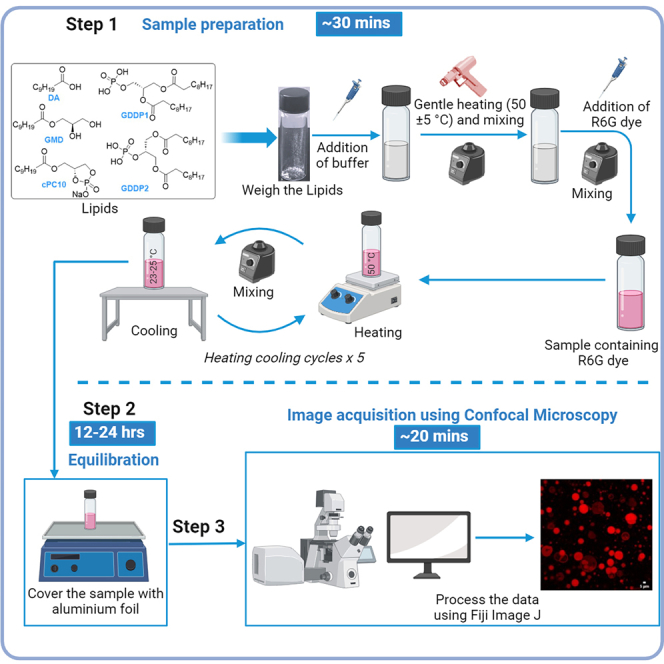
This step enables the preparation of vesicles from a 2:1 DA: cPC10-based system using five heating-cooling cycle procedure.
-
1.
Weigh 4.6 mg of decanoic acid (DA, Mol. Wt. 172.27) and 4.4 mg of cPC10 (Mol. Wt. 330.29) as solids in a 4.0 mL scintillation glass vial.
-
2.
Add 1.0 mL of 0.2 M Bis-Tris buffer, pH 6.6 to the glass vial containing the above mixture (for 40 mM of total concentration of the mixture).
-
3.
Gently heat the above sample using a hot air gun for 20 s at 50 ± 5°C and vortex for 10 s to get a uniform suspension.
Note: Ambient temperature mode was used on the hot air gun for gentle heating.
-
4.
Add 3.84 μL of 2.6 mM of rhodamine 6G dye solution to the above lipid sample for 0.01 mM of final dye concentration and vortex for 10 s.
-
5.
Heat the above lipid sample using the IKA stirrer-heater with Aluminum heat block at 50°C for 2 min (as showed in the image) and remove the sample from the hot plate and vortex for 10 s.
-
6.Allow the sample to cool down to room temperature (23°C–25°C) for 1 min by placing the sample at room temperature (23°C–25°C).
-
a.Again, place the sample in the same heat block at 50°C.
-
b.Heat the sample for 1 min.
-
c.Remove the sample from the heat block and vortex for 10 s.
-
d.Allow the sample to cool down to room temperature (23°C–25°C) for 1 min by keeping at room temperature (23°C–25°C).
-
a.
-
7.
Repeat the above step (step 6) three more times and finally vortex for 10 s.
-
8.
Cover the sample with aluminum foil to protect it from the light.
-
9.
Place the thus prepared sample on a low-speed shaker at 150 rpm at room temperature (23°C–25°C) for 12–24 h to equilibrate the sample.
-
10.
Analyze the sample by confocal microscopy (LSM 710 or LSM 880 Zeiss microscope) at different time intervals. See “steps for imaging the sample”. See Figure 1.
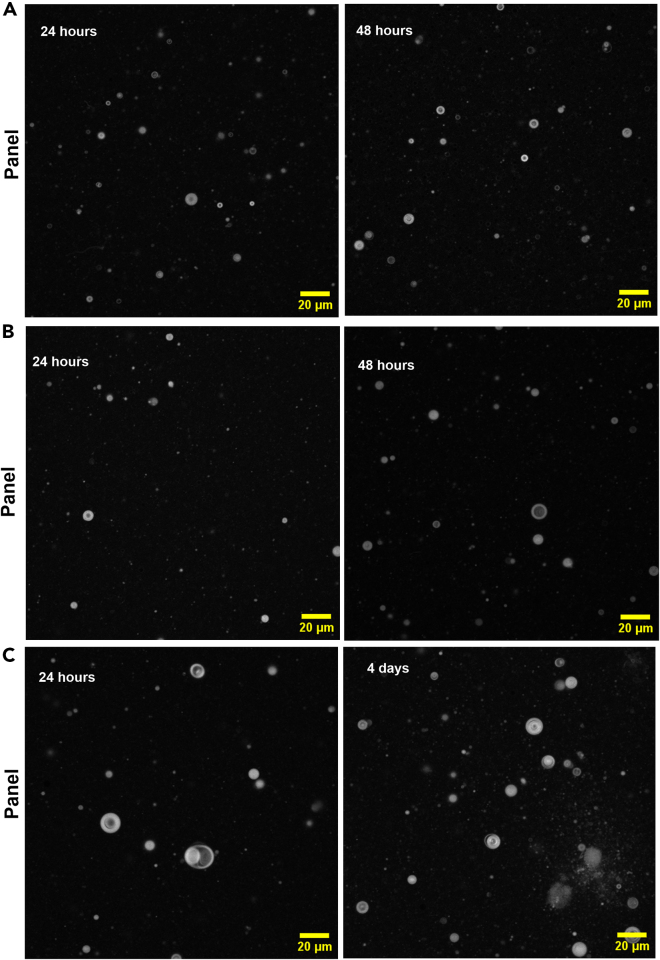
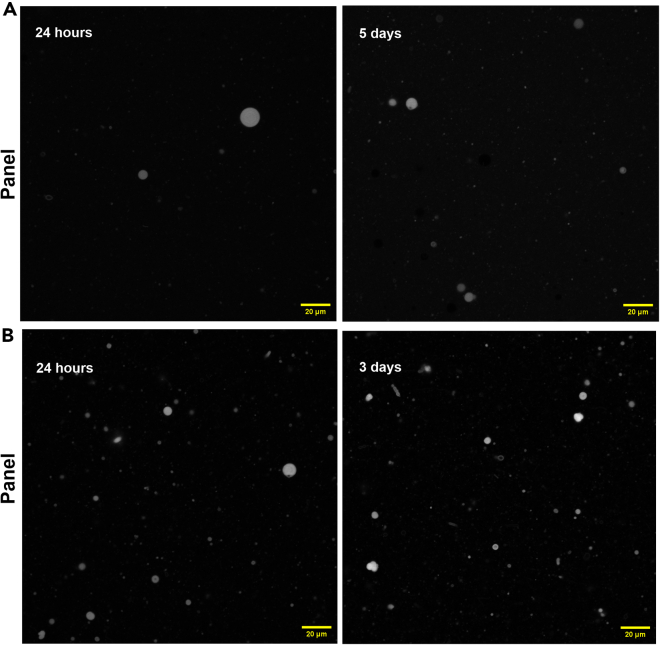
Figure 1.
Confocal microscopy imaging of the 40 mM of the 2:1 DA:cPC10 system with R6G (0.01 mM) at pH 6.6
Each panel (A–C) corresponds to three different sets of experiments. Vesicle sizes range from 1–20 μm in diameter. Scale bars represent 20 μm for all panels.
Protocol for the preparation of 40 mM of 5:1 GMD:cPC10
Timing: 30 min
This step enables the formation of vesicles from a 5:1 GMD:cPC10-based system using five heating-cooling cycle procedure (Figure 2).
-
11.
Weigh 4.1 mg GMD (Mol. Wt. 246.35) and 1.1 mg cPC10 (Mol. Wt. 330.29) in a 4 mL scintillation glass vial.
-
12.
Add 500 μL of 0.2 M Bis-Tris buffer, pH 6.3 (for 40 mM total concentration).
-
13.
Gently heat the sample using a hot air gun for 60 s at 50 ± 5°C and vortex for 10 s.
Note: Ambient temperature mode was used on the hot air gun for gentle heating.
-
14.
Again, heat the sample with a hot air gun for 10 s and vortex for 10 s to make a complete homogenous solution.
-
15.
Add 1.92 μL of Rhodamine 6G dye from a 2.6 mM stock solution to the above sample to make final concentration of 0.04 mM R6G and vortex for 10 s.
-
16.Place the sample in a hot plate at 50°C.
-
a.Heat the sample for 2 min.
-
b.Remove the sample from the hot plate and vortex for 10 s.
-
c.Allow the sample to cool down for 1 min by placing the sample at room temperature (23°C–25°C).
-
a.
-
17.Again, place the sample in the same hot plate at 50°C.
-
a.Heat the sample for 1 min.
-
b.Remove the sample from the hot plate and vortex for 10 s.
-
c.Allow the sample to cool down for 1 min by keeping at room temperature (23°C–25°C).
-
a.
-
18.
Repeat the above step (step 17) three more times.
-
19.
Cover the sample using aluminum foil to protect the sample from light.
-
20.
Place the sample in a low-speed shaker at 150 rpm at room temperature (23°C–25°C) for 12–24 h.
-
21.
Analyze the sample by confocal microscopy (LSM 710 Zeiss microscope) at different time intervals). See “Steps for Imaging the Sample”. See Figure 3.
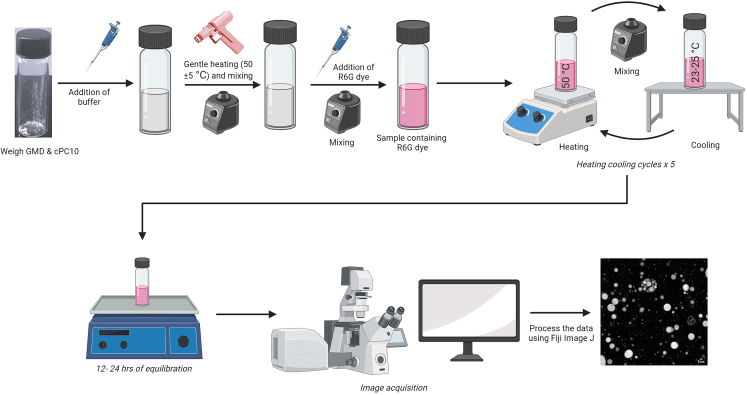
Figure 2.
An illustration demonstrating the overall procedure
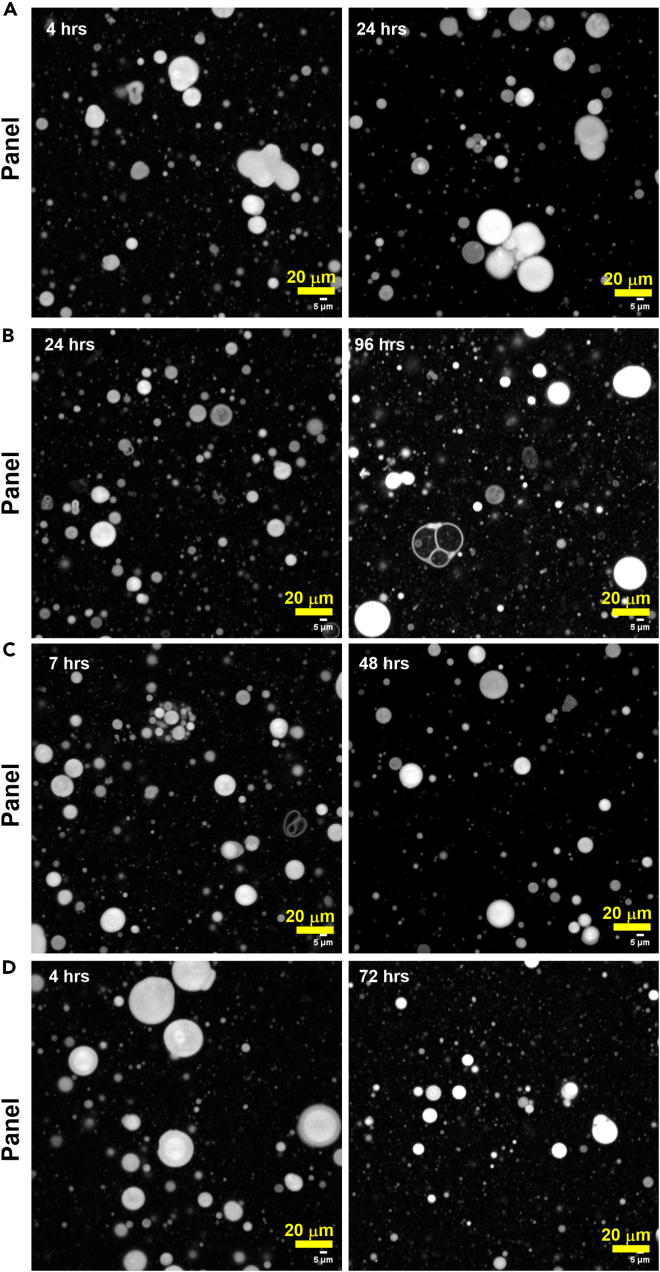
Figure 3.
Confocal Images of 5:1 GMD:cPC10 system labeled with R6G (0.04 mM) in 0.2 M Bis-Tris buffer, pH 6.3
Each panel (A–D) corresponds to four different sets of experiments. Vesicle sizes range from 1–25 μm in diameter. Scale bars represent 20 μm for all panels.
Protocol for the preparation of 20 mM of 5:1 GMD:GDDP1
Timing: 40 min
This step enables the preparation of vesicles from a 5:1 GMD: GDDP1-based system using five heating-cooling cycle procedure.
-
22.
Weigh 2.5 mg GMD (Mol. Wt. 246.35) and 4.8 mg GDDP1 (Mol. Wt. 480.58) separately in two different 4 mL scintillation glass vials.
-
23.
Add 500 μL of 0.2 M Bis-Tris buffer, pH 6.3 to each glass vial for a 20 mM individual concentration.
-
24.
Gently heat each sample using a hot air gun for 60 s at 50 ± 5°C and vortex for 10 s.
Note: Ambient temperature mode was used on the hot air gun for gentle heating.
-
25.
Again, heat each sample with hot air gun for 10 s and vortex for 10 s to make a complete homogenous solution.
-
26.
Sonicate the vial containing GDDP1 for 60 s and mix well by vortexing it for 10 s.
-
27.
Add 3.82 μL of R6G dye from a 2.6 mM stock solution to each of above sample (GMD containing glass vial and GDDP1 containing glass vial) to make final concentration of 0.02 mM R6G and vortex for 10 s.
-
28.
Add 500 μL of the GMD sample to a new glass vial followed by 100 μL of GDDP1 (resulting in a ratio of 5:1 GMD: GDDP1), and vortex the sample for 10 s.
-
29.Place the sample in a hot plate at 50°C.
-
a.Heat the above prepared sample to 50°C for 2 min.
-
b.Remove the sample from the hot plate and vortex for 10 s.
-
c.Allow the sample to cool down for 1 min by placing the sample at room temperature (23°C–25°C).
-
a.
-
30.Again, place the sample in the same hot plate at 50°C.
-
a.Heat the sample for 1 min.
-
b.Remove the sample from the hot plate and vortex for 10 s.
-
c.Allow the sample to cool down for 1 min by keeping at room temperature (23°C–25°C).
-
a.
-
31.
Repeat step 30 three more times.
-
32.
Cover the sample using aluminum foil to protect the sample from light.
-
33.
Place the sample in a low-speed shaker at 150 rpm at room temperature (23°C–25°C) for12–24 h.
-
34.
Analyze the sample by the confocal microscopy (LSM 710 Zeiss microscope) at different time intervals. See “Steps for Imaging the Sample”. See Figure 4.
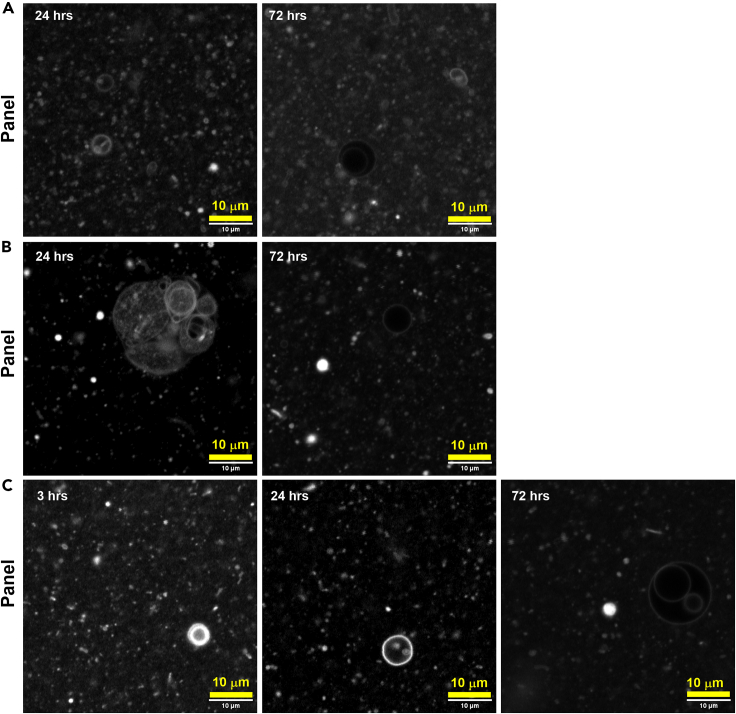
Figure 4.
Confocal Microscope image of 5:1 GMD:GDDP1 system
20 mM (Panel A) and 10 mM (Panel B, C) total concentration of the lipid labeled with 0.02 mM of R6G in 0.2 M Bis-Tris buffer, pH 6.3. Each panel corresponds to three different sets of experiments. Vesicle sizes range from 1–20 μm in diameter. Scale bars represent 10 μm for all panels.
Protocol for the preparation of 40 mM of 5:1 GMD:GDDP2
Timing: 30 min
This step enables the preparation of vesicles from a 5:1 GMD: GDDP2-based system using five heating-cooling cycle procedure.
-
35.
Weigh 8.2 mg of GMD (Mol. Wt. 246.35) and 3.2 mg of GDDP2 (Mol. Wt. 480.58) in a 4 mL scintillation glass vial.
-
36.
Add 1000 μL of 0.2 M Bis-Tris buffer, pH 6.6 to the above mixture (for 40 mM total concentration).
-
37.
Gently heat the above sample using a hot air gun for 20 s at 50 ± 5°C and vortex for 10 s.
Note: Ambient temperature mode was used on the hot air gun for gentle heating.
-
38.
Add 3.84 μL of 2.6 mM of rhodamine 6G dye solution for the final dye concentration of 0.01 mM and vortex for 10 s.
-
39.Place the sample in a hot plate at 50°C.
-
a.Heat the sample at 50°C for 2 min.
-
b.Remove the sample from the hot plate and vortex for 10 s.
-
c.Allow the sample to cool down for 1 min by placing the sample at room temperature (23°C–25°C).
-
a.
-
40.Again, place the sample in the same heat block at 50°C.
-
a.Heat the sample for 1 min.
-
b.Remove the sample from the heat block and vortex for 10 s.
-
c.Allow the sample to cool down for 1 min by keeping at room temperature (23°C–25°C).
-
a.
-
41.
Repeat the above step (step 40) three more times and finally vortex for 10 s.
-
42.
Cover the sample with aluminum foil to protect from the light.
-
43.
Place the above sample on a low-speed shaker at 150 rpm at room temperature (23°C–25°C) for 12–24 h to equilibrate the sample.
-
44.
Analyze the sample by confocal microscopy (LSM 710 or LSM 880 Zeiss microscope) at different time intervals. See “Steps for Imaging the Sample”. See Figure 5.
Figure 5.
Confocal Images of 5:1 GMD:GDDP2 system labeled with R6G (0.01 mM) in 0.2 M Bis-Tris buffer, pH 6.6
Panel A: 40 mM of the lipid sample; Panel B: 10 mM of the lipid sample. Vesicle sizes range from 1–10 μm. Scale bars represent 20 μm for all panels.
Step for imaging the sample
Timing: 15 min (for step 45)
Timing: 5 min (for step 46)
This step enables acquisition of images and analysis of vesicles from the samples prepared as described above.
-
45.Preparing the microscope instrument and sample for imaging.
-
a.After switching on the instrument, touch on Microscope on the screen and choose 40x oil immersion objective (DIC M27). Place a drop of oil on the center of the objective and fix the chambered cover glass 8-well or the 96-well plate.
-
b.Place 50–100 μL of the above prepared sample on the glass chamber.
-
c.From the desktop open the ZEN application > click on open system.
-
d.Click on bright field and later adjust the focus to locate the sample (clear vision).
-
e.Go to acquisition and click on smart setup to select the dye (R6G). For R6G channel, the laser excitation was at 514 nm, laser power of 1.5%–2% was used; the detection range was set from 521 to 699 nm; the detector gain range was 400–600 and digital offset was 0.
-
f.Click on live to visualize the sample and take a snapshot to capture the images. (Frame size - 512 or 1024), Pixel size (0.21 μm)
-
a.
-
46.
Steps for Processing an image using Fiji Image J.
This step enables the processing of the images obtained from confocal microscopy using the Fiji Image J app.-
a.Open Fiji Image J application.
-
b.Choose the image file from the respective folder.
-
c.Go to “Analyze-> Tools-> scale bar”.
-
d.If required, adjust the brightness by Image >Adjust > Brightness/Contrast.
-
e.Save the file by clicking “File-> Save as-> .Gif” format.Note: The images were taken at different locations of the same sample and the representative images were provided here.If a different microscope system is used, appropriate instrument parameters should be optimized for needed image acquisition/analysis.
-
a.
Expected outcomes
Cyclic phospholipids are a single chain amphiphiles (fatty acid derived phospholipids) that give rise to a heterogeneous library of vesicles with diverse morphologies and tolerance to a range of metal ions, temperature, and pH.3,4 Furthermore, cyclic-phospholipid-based vesicles have been shown to retain encapsulated nucleic acids during growth and division, acquire nucleotides from their surroundings, and be compatible with the nonenzymatic extension of an RNA oligonucleotide.5
The method described in this paper provides a simple and effective way to prepare protocells containing decanoic acid derived cyclophospholipid (cPC10) amphiphile mixed with decanoic acid or glyceryl monodecanoate. Additionally, we delineate methods for generating vesicles using a mixture of phosphorylated glyceryl didecanoate and glyceryl monodecanoate. The protocol consists of five heating and cooling steps followed by an equilibration process. The resulting vesicles can be analyzed using either the LSM 710 or LSM 880 Zeiss microscope, and the captured images can be processed using the Fiji Image J application.
Limitations
This protocol offers a robust method for preparing vesicles. However, the specific type of vesicles that may form using this method cannot be predicted. It will always result in a heterogeneous mixture of vesicles, including multilamellar vesicles (MLV); multivesicular vesicles (MVV), unilamellar vesicles (ULV) and giant unilamellar vesicles (GUV).
Troubleshooting
Problem 1
Weighing small quantities of DA is tedious. (Step 1).
Potential solution
DA has a low melting point and can melt to liquid by gently keeping the DA sample (in an Eppendorf or in a glass vial) in hot water bath (bath set temperature 45°C–50°C). The DA melt can be easily weighed by using a pipette.
Problem 2
Sometimes dissolving the lipid compounds can be slow depending on the sample amount.
Potential solution
Vortex the sample thoroughly until homogeneous solution is formed. Heat gently using hot air gun (less than 1 min) when required.
Problem 3
For DA-based systems, vesicle formation is poor at pH > 7.0 due to the pKa of DA (6.4),.6,7 (Step 2).
Potential solution
Ensure the pH of the buffer and sample is maintained at the required pH (between pH 4.0 to 6.6). If pH jumps above pH 7.0 or below pH 4.0 by any case, vesicles can be reformed (turbid solution) by adjusting the pH to required pH (between pH 4.0 to pH 6.6) with aq. 6 N HCl or 4 N NaOH solution.
Problem 4
GDDP1 and GDDP2 have the potential to undergo hydrolysis, which could impact the vesicle formation. (Steps 22 and 35).
Potential solution
Before preparing the vesicles, ensure their purity by checking the 1H-and 31P-NMR of the samples. Store the compounds in the refrigerator at 4°C. Refer to Pulletikurti et al.2 for NMR spectra.
Problem 5
GDDP1 compound as a solid has a sticky (waxy) nature that can result in improper mixing and affect the quality of vesicles. (Steps 22–26).
Potential solution
Always ensure that a homogenous stock solution is prepared by thorough vortexing.
Problem 6
Sometimes, there can be an inconsistent population of vesicles that needs to be addressed.
Potential solution
For optimal vesicle density, it is essential to carry out 5 heating and cooling cycles and double-check the heating temperature (50°C).
Problem 7
This protocol will always result in a heterogeneous mixture of vesicles.
Potential solution
If there is the desire to obtain uniform-sized vesicles, then the sample can be can be subjected to an extrusion procedure using a polycarbonate membrane as described in Hunter et al.8
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ramanarayanan Krishnamurthy (rkrishna@scripps.edu).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Kollery, S. Veena (vks@scripps.edu) and Pulletikurti, S (spulletikurti@scripps.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The published article by (Pulletikurti et al.)1 includes all datasets generated or analyzed during this study.
Acknowledgments
We gratefully acknowledge the support from the NASA Astrobiology-Exobiology grant to A.A.D. and R.K. (80NSSC20K0625) and the Simons Foundation grant to R.K. (327124FY19). S.P. thanks the NASA NPP program for support. We thank the Scripps Research Core Microscopy Facility for assistance with confocal microscopy measurements.
Author contributions
R.K. conceived the project, and A.A.D. and R.K. supervised the research. S.P., K.S.V., A.A.D., and R.K. designed the experiments. S.P. and K.S.V. conducted the experiments and generated the data.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Ashok A. Deniz, Email: deniz@scripps.edu.
Ramanarayanan Krishnamurthy, Email: rkrishna@scripps.edu.
References
- 1.Pulletikurti S., Veena K.S., Yadav M., Deniz A.A., Krishnamurthy R. Experimentally modeling the emergence of prebiotically plausible phospholipid vesicles. Chem. 2024;10:1839–1867. doi: 10.1016/j.chempr.2024.02.007. [DOI] [Google Scholar]
- 2.Ortuno V.E., Pulletikurti S., Veena K.S., Krishnamurthy R. Synthesis and hydrolytic stability of cyclic phosphatidic acids: implications for synthetic- and proto-cell studies. Chem. Commun. 2022;58:6231–6234. doi: 10.1039/D2CC00292B. [DOI] [PubMed] [Google Scholar]
- 3.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toparlak Ö.D., Karki M., Egas Ortuno V., Krishnamurthy R., Mansy S.S. Cyclophospholipids Increase Protocellular Stability to Metal Ions. Small. 2020;16 doi: 10.1002/smll.201903381. [DOI] [PubMed] [Google Scholar]
- 5.Toparlak Ö.D., Sebastianelli L., Egas Ortuno V., Karki M., Xing Y., Szostak J.W., Krishnamurthy R., Mansy S.S. Cyclophospholipids Enable a Protocellular Life Cycle. ACS Nano. 2023;17:23772–23783. doi: 10.1021/acsnano.3c07706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanicky J.R., Shah D.O. Effect of Premicellar Aggregation on the pKa of Fatty Acid Soap Solutions. Langmuir. 2003;19:2034–2038. doi: 10.1021/la020672y. [DOI] [Google Scholar]
- 7.Wellen B.A., Lach E.A., Allen H.C. Surface pKa of octanoic, nonanoic, and decanoic fatty acids at the air–water interface: applications to atmospheric aerosol chemistry. Phys. Chem. Chem. Phys. 2017;19:26551–26558. doi: 10.1039/C7CP04527A. [DOI] [PubMed] [Google Scholar]
- 8.Hunter D.G., Frisken B.J. Effect of Extrusion Pressure and Lipid Properties on the Size and Polydispersity of Lipid Vesicles. Biophys. J. 1998;74:2996–3002. doi: 10.1016/S0006-3495(98)78006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article by (Pulletikurti et al.)1 includes all datasets generated or analyzed during this study.