Abstract
The human immunodeficiency virus type 1 (HIV-1) Tat protein has been reported to transactivate several cellular genes, including the potent chemotactic factor interleukin-8 (IL-8). Consistent with these in vitro assays, elevated levels of IL-8 protein are found in the serum of HIV-infected individuals. We now extend these observations by demonstrating that Tat induction of IL-8 is linked to the cell cycle. Cells that constitutively express the Tat(1–86) protein (eTat) and control cells (pCEP) were reversibly blocked at the G1/S border with hydroxyurea or thymidine. The cells were subsequently released, and IL-8 expression was monitored by RNase protection assays and enzyme-linked immunosorbent assay (ELISA). RNase protection assays demonstrated that IL-8 mRNA expression is transiently induced, approximately fourfold, as the Tat-expressing cells enter S phase. Consistent with the RNase protection assay, an increase in IL-8 protein was observed in the cell supernatant using an IL-8 ELISA. Similar experiments were performed following a reversible block at the G2/M border with nocodazole and release into G1. Using the RNase protection assay and ELISA, little or no increase in IL-8 expression was observed during G1. Using gel shift as well as an immobilized DNA binding assay, we demonstrate that the increase in IL-8 gene expression correlates with a specific increase in p65 NF-κB binding activity only in the nucleus of the Tat-expressing cells. Moreover, the CREB-binding protein coactivator is present in the complex in the Tat cell line. Finally, we demonstrate that the presence of the proteasome inhibitor MG-132 inhibits the induction of NF-κB binding, as well as IL-8 expression, supporting the role of NF-κB.
Interleukin-8 (IL-8) belongs to the C-X-C chemokine family (29) and is secreted by several different cell types, including monocytes, neutrophils, endothelial cells, fibroblasts, and T lymphocytes. Also known as neutrophil-activating peptide 1, IL-8 is a powerful chemoattractant protein for neutrophils and has been shown to activate neutrophils, causing degranulation, mobilization, and increased CD11/CD18 expression (20). IL-8 is also chemotactic for T lymphocytes, and some reports suggest that IL-8 may play a role in the recruitment of CD4-positive T cells to lymph nodes, sites where the human immunodeficiency virus (HIV) replicates, providing new target cells for virus infection (3). Interestingly, T lymphocytes are 10 times more sensitive to IL-8 than are neutrophils in chemotaxis (14).
In vivo, sera from HIV type 1 (HIV-1)-infected patients carry elevated levels of IL-8 (4, 16). Similarly, after infection of cells in vitro by human T-cell leukemia virus type 1 or HIV-1, levels of IL-8 expression are found to be elevated in the medium (16–18). The retroviral transactivator proteins Tax (17) and Tat (23) are likely to play an important role in the induction of IL-8. In the case of HIV-1 Tat induction, T cells require the presence of a strong cellular activation signal such as that provided by CD3 cross-linking and CD28-mediated costimulation, in addition to Tat, to produce elevated levels of IL-8. This result suggests that the viral protein requires the cooperation of cellular signals to exert its effects (23).
IL-8 production (induced by several stimuli, including IL-1, TNF-α, and phorbol myristate acetate) is primarily regulated at the transcriptional level (10, 21, 22, 27). Nucleotide sequence analysis of the 5′ regulatory region of the IL-8 gene has revealed binding sites for NF-κB, C/EBPβ, and AP-1 (20). Transient transfection analysis of the IL-8 promoter has demonstrated that it is activated by the induction of NF-κB complexes, particularly those containing RelA/p65 (22, 28). C/EBPβ and AP-1 play smaller, but significant, roles in IL-8 expression (11, 12, 19, 22, 28). In this report, we show for the first time that IL-8 gene expression is regulated in a cell cycle-dependent manner in cells constitutively expressing the HIV Tat protein. This induction correlates with the cell cycle-regulated binding of NF-κB to the IL-8 promoter. The cell cycle-regulated binding of NF-κB is accompanied by an increase in binding of the coactivator CREB-binding protein (CBP) to the complex bound to the IL-8 NF-κB binding site. These studies provide the first example of cell cycle-dependent regulation of a cellular gene by the HIV Tat protein.
MATERIALS AND METHODS
Construction of epitope-tagged Tat cell line.
We constructed a cell line that constitutively expressed Tat protein. HeLa cells were stably transfected with either a backbone control plasmid (pCEP4; Invitrogen) or a plasmid expressing Tat(1–86) with a C-terminal epitope tag (pCEP4eTat). HeLa cell lines containing either the control or eTat plasmid were successfully selected by single-cell dilution. Both cell types were selected and maintained under 200 μg of hygromycin per ml. The control pCEP4 HeLa line is designated “pCEP” and the pCEP4eTat HeLa line is designated “eTat” throughout this article (8). Verification of Tat transcriptional activity was achieved by electroporation of reporter plasmids as previously described (8).
Cell cycle analysis.
eTat or pCEP cells were blocked either with hydroxyurea (Hu) (2 mM) or thymidine (3 mM) for 18 h or by addition of nocodazole (Noco) (50 ng/ml) for 14 h. Following the block, cells were released by being washed twice with phosphate-buffered saline (PBS) and by the addition of complete medium. Samples were collected every 3 h, and nuclear and cytoplasmic cell extracts were made from 3 × 107 cells/time point, as described previously (13) (see below). Supernatants were collected from the same time points and analyzed by an IL-8 ELISA according to the manufacturer's instructions (Biosource International). Samples for 0 h were taken prior to release. At each point, approximately 2 × 106 cells were also processed for cell sorting. Cells were washed with PBS and fixed by addition of 500 μl of 70% ethanol. Cell pellets were washed with PBS and incubated in 1 ml of PBS with 150 μg of RNase A (Sigma) per ml and 20 μg of propidium iodide (Sigma) per ml at 37°C for 30 min. The stained cells were analyzed for red (FL2) fluorescence on a FACScan (Becton Dickinson), and the distribution of cells in the G1, S, and G2/M phases of the cell cycle was calculated from the resulting DNA histogram, using Cell FIT software (Fast Systems, Inc., Gaithersburg, Md.), based on a rectangular S-phase model. When required, nuclear extracts were prepared as previously described (13). The proteasome inhibitor MG-132 (50 μM, final) (Calbiochem) was added to the medium of some cultures at the time of release from cell cycle block.
EMSA.
Nuclear and cytoplasmic extracts were made as previously described (13). Electrophoretic mobility shift assay (EMSA) running conditions were as previously described (13). The IL-8wt, IL-8mκB, and IL-8mC/EBP oligonucleotides were as previously described (28). The HIV NF-κB binding site was created by annealing the oligonucleotides 5′ACAAGGGACTTTCCGCTGGGGA-CTTTCC3′ and 5′GGAAAGTCCCCAGCGGAAAGTCCCTTG3′ and labeling with [γ-32P]ATP.
Biotinylated DNA pull-down experiment.
A double-stranded oligonucleotide corresponding to the NF-κB binding region of the wild-type IL-8 promoter (IL-8wt) or mutant controls (IL-8mκB or IL-8mC/EBP) were labeled with biotin at the 5′ end. Nuclear extracts were prepared, and pull-down experiments were carried out. Briefly, 180 to 240 μg of nuclear proteins was mixed with 3 μl of a 1-mg/ml solution of poly(dI-dC) and 5 μg of biotinylated double-stranded oligonucleotide in gel shift buffer (20 mM HEPES [pH 7.9], 0.1 mM EDTA, 10 mM MgCl2, 20 mM KCl, 1 mM dithiothreitol, and 10% glycerol), and the mixture was incubated 30 min at room temperature in a total volume of 20 μl. The complex was pulled down with streptavidine-agarose beads and then washed in the presence of Sarkosyl (0.03%). The bound proteins were denatured and run on a 4 to 20% Tris-borate-EDTA gel. Western blot analysis was then performed using anti-p65 or anti-CBP antibodies. The sequence of the wild-type IL-8 binding site was 5′AGCTTCATCAGTTGCAAATCGTGGAATTTCCTCTG3′, while the mutant IL-8 NF-κB binding site sequence was 5′AGCTTCATCAGTTCAAATCGTTAACTTTCCTCTG3′. The mutant IL-8 C/EBP binding site sequence was 5′AGCTTCATCAGCTACGAGTCGTGGAATTTCCTCTG3′ (mutations are underlined).
Antibodies.
Commercially available anti-RelA/p65 (sc109X) and CBP antibodies were used for Western blot analyses (Santa Cruz Biotechnology).
RNase protection assay.
Fifteen micrograms of total RNA obtained from blocked and released eTat and pCEP was used. The RiboQuant kit was used according to the manufacturer's instructions (PharMingen) using the Human Cytokine/Chemokine Multi-Probe template hCK-5. Samples were loaded on a 5% acrylamide-urea gel and run at a constant 65 W for 1 h. Gels were subsequently dried and placed on a PhosphorImager cassette for an overnight exposure.
Western blot analysis.
Twenty-five or 50 μg of protein obtained from nuclear and cytoplasmic extracts was run on a 4 to 20% Tris-glycine gel and transferred overnight to Immobilon membranes (Millipore). The membrane was then blocked for 1 h in TNE (10 mM Tris, 50 mM NaCl, 2.5 mM EDTA) containing 0.1% Tween 20 and 5% milk, rinsed, and incubated for 1 h with the appropriate antibody. The blot was washed, incubated with a secondary horseradish peroxidase-conjugated antibody, and developed with the ECL detection system (Amersham).
RESULTS
Expression of IL-8 is cell cycle regulated in eTat cells: induction of IL-8 transcription at S phase.
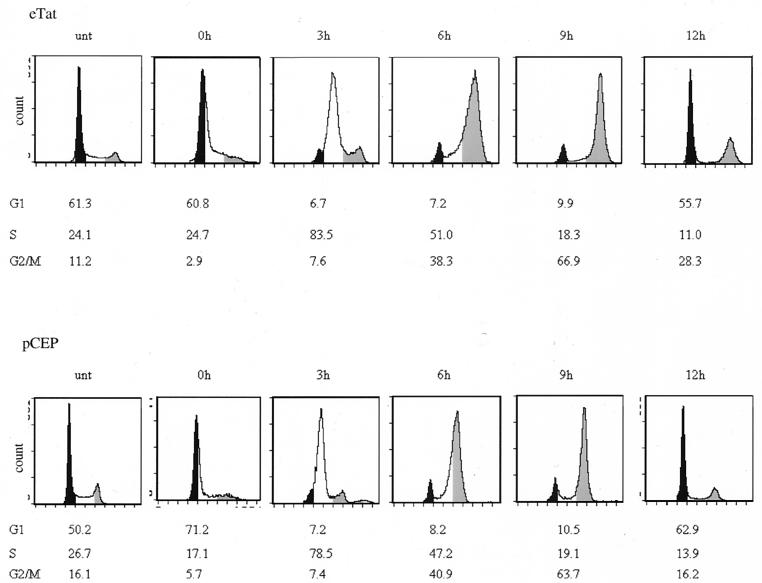
Using a cell line which constitutively expresses an active Tat protein, researchers recently reported that Tat activates transcription from the HIV long terminal repeat in a cell cycle-dependent manner (8). This result prompted us to analyze the potential cell cycle regulation of cellular genes induced by Tat. We arrested the cell cycle of cultured eTat and control pCEP cells using Hu, which arrests cells in early S phase by inhibiting ribonucleotide diphosphate reductase (25). Figure 1 shows a typical fluorescence-activated cell-sorter (FACS) analysis conducted after an 18-h Hu block and subsequent release. After release, eTat and pCEP cells moved through S phase (represented by the 3 h postrelease sample [Fig. 1]) and into the G2/M phase by 6 h postrelease with a peak G2/M population at 9 h postrelease. Cells then entered the G1 phase (12 h postrelease). The cell cycle profiles were similar in both cell lines.
FIG. 1.
Cell cycle analysis of eTat and control pCEP cell lines following Hu block. Log-phase growing cells were blocked with Hu (2 mM) for 18 h and released by removing the inhibitor and adding fresh media. Samples were collected every 3 h, and the cells were processed for FACS analysis. Cells were removed from media at each time point, washed with PBS without Mg2+ or Ca2+, fixed with 70% ethanol, and stained with propidium iodide (Fast Systems, Inc.), followed by cell sorting analysis on a Coulter EPICS cell analyzer. The proportion of cells in G1, S, or G2/M are plotted as the percentage of total cells at each time point. The black sections represent the G1-phase population, the white sections are the S-phase population, and the gray sections represent the G2/M population. The shading on the figures was added manually. The results are representative of three different experiments.
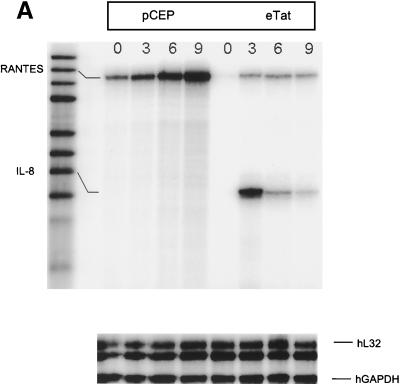
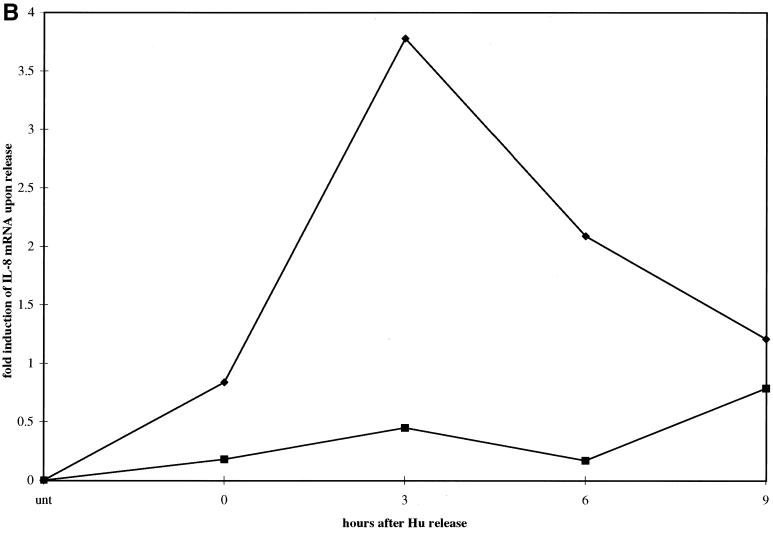
To analyze cell cycle-dependent gene expression, pCEP and eTat cells were reversibly blocked with Hu for 18 h and then released. Cells were collected every 3 h for 9 h, and RNA was extracted. Fifteen micrograms of total RNA from each sample was then used in an RNase protection assay using the hCK-5 cytokine probe set (PharMingen). This allowed us to study the expression of a total of seven different cytokine and chemokine genes, including the genes for lymphotoxin, RANTES, IP-10, MIP-1β, MIP-1α, MCP-1, IL-8, and I-309, along with two housekeeping genes, the L32 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes, as controls (Fig. 2A). The results of this experiment demonstrated that the expression of one of the cytokine genes, IL-8, was regulated as the cells progressed through S phase. Strikingly, the IL-8 mRNA levels increased from 0 h (G1/S) to 3 h (S), then decreased at 6 h (G2/M) and further at 9 h (G2/M) in the eTat cells. In contrast, the levels of IL-8 mRNA were low and roughly constant in the pCEP cells. Densitometric analysis of the RNA levels (Fig. 2B) showed a 3.5- to 4-fold induction of IL-8 expression at 3 h in eTat cells. The level of RNA expression and the sensitivity of the RNase protection technique did not allow us to detect fluctuation in several of the other genes analyzed in this assay. We did reproducibly observe, however, that RANTES was expressed at higher levels in pCEP cells and that its transcription peaked at the G2/M phase (Agbottah et al., unpublished data). The levels of L32 and GAPDH mRNAs were constant in pCEP and eTat throughout the cell cycle.
FIG. 2.
IL-8 mRNA is induced in a cell cycle-dependent manner in Tat-expressing cells. For the RNase protection assay, 15 μg of total RNA was obtained from 3 × 107 eTat and pCEP cells blocked with Hu. (A) Samples were collected at 0, 3, 6, and 9 hrs, processed, and hybridized with Human Cytokine/Chemokine Multi-Probe template hCK-5 according to the manufacturer's instructions. Samples were then loaded on a 5% acrylamide-urea gel and run at a constant 65 W for 1 h. Gels were subsequently dried and placed on a PhosphorImager cassette for an overnight exposure and analyzed. (B) Quantification of the PhosphorImager signal from panel A, normalizing the IL-8 signal to the signal obtained from L32 and GAPDH internal standard signals. The results are representative of three independent experiments.
A similar experiment was conducted with Noco, a drug which reversibly blocks the cells at the G2/M boundary (5). In contrast to the results obtained as the cells transitioned through S phase, the level of IL-8 mRNA expression in G2/M and G1 was not significantly above background (data not shown) (see below). These results demonstrate that IL-8 transcription is specifically induced during the S phase of the cell cycle in cells expressing the Tat protein.
IL-8 protein is overexpressed in eTat cells following Hu block and release.
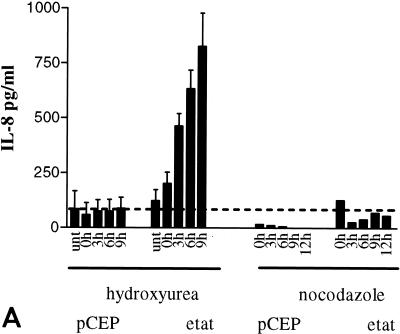
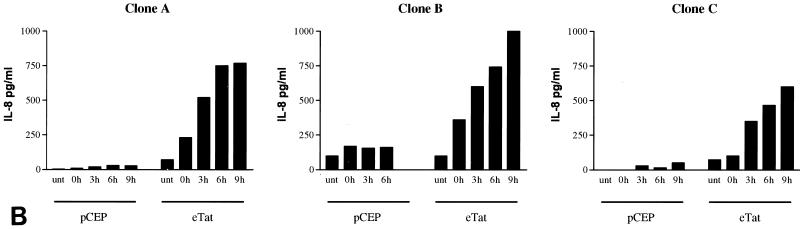
We next investigated whether overexpression of IL-8 mRNA was reflected in levels of secreted IL-8 protein that were higher in the supernatant of eTat cells than in that of pCEP cells. Cell culture supernatants were collected for 9 h following Hu or Noco block and release and assayed by ELISA (Fig. 3A). The levels of IL-8 increased dramatically starting at 3 h postrelease (approximately 450 pg/ml) in the eTat supernatant, reaching as high as 828.0 ± 151.0 pg/ml at 9 h post-Hu release. In contrast, the levels of IL-8 were low and fairly constant in the supernatant obtained from pCEP cells (170 to 188 pg/ml) (Fig. 3A). To address the possibility that IL-8 expression might be due to clonal variation in cells, and not to Tat expression, we tested several other clones of pCEP and eTat cells for IL-8 expression (Fig. 3B). The results of this assay clearly demonstrate that IL-8 expression is induced in each Tat-expressing eTat cell clone during S phase. These results argue strongly that IL-8 induction is due to Tat and is not an artifact of clonal variation.
FIG. 3.
IL-8 is secreted from Tat-expressing cells in a cell cycle-dependent manner. (A) ELISA of IL-8 from supernatant of eTat or control cells. Control pCEP and eTat cells were blocked with Hu (2 mM) or Noco (50 ng/ml) for 15 to 18 h, washed, and released by incubation in fresh medium. Cell supernatants were collected for either 9 h (Hu) or 12 h (Noco) and analyzed for IL-8 protein using an ELISA kit (Biosource International). Fifty microliters of supernatant per collection time point was used for ELISA. The results presented are the means of three independent experiments ± standard deviations. (B) ELISA of IL-8 from supernatants of three independent clones of pCEP or eTat cells after Hu block and release.
Consistent with the RNA analysis, after a Noco (G2/M) block, IL-8 protein levels did not significantly change in either cell line following release into G1 (Fig. 3A). Taken together, these results confirm the mRNA data showing that IL-8 production is an S-phase phenomenon that is independent of the blocking agent used and is not due to cell cycle arrest per se.
To demonstrate that the induction of IL-8 was specific for the cell cycle and not due to non-cell cycle-related activity of Hu, we also used a thymidine block to synchronize the cells at the G1/S border. Following release, we collected the supernatant and analyzed IL-8 secretion by ELISA. The cells were also analyzed by FACS to assess the cell cycle progression. As observed after the Hu block, we detected a significant increase of IL-8 protein in the supernatant of eTat cells after release as the cells progressed through S phase, whereas the levels of IL-8 were constant and low in pCEP (data not shown).
Induction of NF-κB binding to the IL-8 promoter at the S phase of the cell cycle in eTat cells.
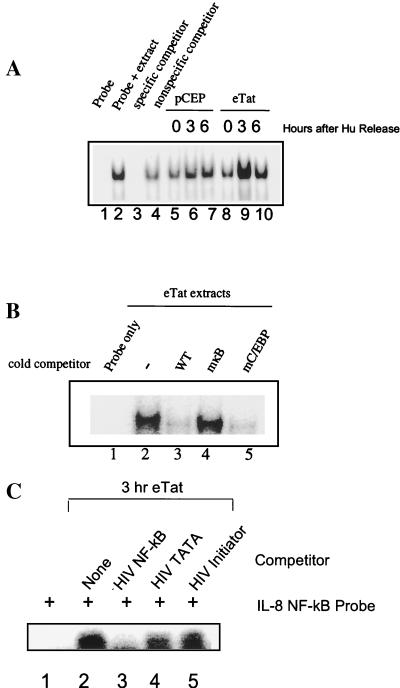
The IL-8 promoter was previously shown to be regulated by NF-κB. We conducted a series of EMSAs to measure NF-κB binding activity during the cell cycle. pCEP and eTat cells were blocked with Hu, released, and collected at 0, 3, and 6 h postrelease. Cells were subsequently fractionated into nuclear and cytoplasmic extracts. We examined the levels of nuclear NF-κB by mobility shift assay with a probe corresponding to the IL-8wt sequence (Fig. 4A). Three hours following release from the Hu block, an increase in NF-κB binding in eTat but not in pCEP cells was noted (Fig. 4A, lanes 6 and 9). NF-κB binding decreased at 6 h postrelease (S to G2/M transition) (lanes 7 and 10).
FIG. 4.
NF-κB is induced during S phase in eTat cell nuclear extracts. (A) EMSA using 5 μg of nuclear extracts obtained from control pCEP and eTat cells after Hu block (2 mM, 15 h) and release. Lane 1, IL-8wt probe alone; lane 2, eTat untreated extracts; lanes 3 and 4, same as lane 2, with competition using a 60-fold excess of unlabeled IL-8wt and IL-8mκB, respectively; lanes 5 through 7, pCEP extracts at 0, 3, and 6 h postrelease; lanes 8 through 10, eTat extracts at 0, 3, and 6 h postrelease. (B) EMSA using 5 μg of nuclear extracts obtained from eTat cells 3 h post-Hu block (2 mM, 15 h). Lane 1, IL-8wt probe alone; lane 2, eTat at 3 h post-Hu release; lanes 3 through 5, same as lane 2, with competition using a 60-fold excess of unlabeled IL-8wt, IL-8mκB, and IL-8mC/EBP probes, respectively. The results presented in panels A and B are representative of two and three independent experiments, respectively. (C) EMSA using 5 μg of nuclear extract from eTat cells after Hu block (2 mM, 15 h) and release. Lane 1, IL-8wt probe; lane 2, IL-8wt probe plus eTat nuclear extract; lane 3, IL-8wt probe plus eTat extract plus 25-fold excess HIV NF-κB competitor; lane 4, IL-8wt probe plus eTat extract plus 25-fold excess HIV TATA competitor; lane 5, IL-8wt probe plus eTat extract plus 25-fold excess HIV initiator competitor.
To determine the specificity of the DNA binding activity, competitor oligonucleotides were synthesized with base substitutions in the NF-κB (IL-8mκB) or neighboring C/EBP (IL-8mC/EBP) site (Fig. 4B). The results of competition experiments demonstrate that the gel shift complex is due to NF-κB binding. An excess of unlabeled wild-type oligonucleotide strongly diminished the signal (Fig. 4B, lane 3). In contrast, IL-8mκB had little effect (lane 4). The IL-8mC/EBP oligonucleotide, which possesses only the NF-κB binding sequence, competed for binding as efficiently as the wild type (lane 5). We also used oligonucleotides containing the NF-κB binding site from the HIV promoter as competitors. The results presented in Fig. 4C demonstrate that the HIV NF-κB oligonucleotides, but not the HIV TATA or initiator oligonucleotides, compete for binding of protein to the IL-8 NF-κB site. We also analyzed the gel shift activity of transcription factor Sp1 in the eTat and pCEP cell extracts. No significant change in Sp1 binding activity was observed in the 0-, 3-, 6-, and 9-h samples from eTat and pCEP cells (data not shown).
NF-κB complexes bound to the IL-8 probe are more stable in eTat cell extracts.
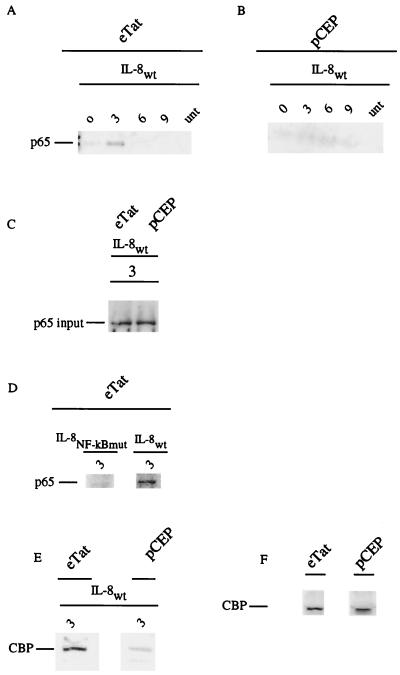
We next analyzed the composition of the NF-κB complex bound to biotinylated oligonucleotides containing either the wild-type or mutant IL-8 NF-κB binding site. Nuclear extracts obtained from pCEP or eTat cells, blocked with Hu and released, were incubated with the probes. Following pull-down, the complexes were subjected to Western blot analysis, using antibodies specific to the p65 NF-κB subunit and to CBP. As seen in Fig. 5A and B, and consistent with the gel shift analysis, bound p65 was most abundant in nuclear extracts prepared from the eTat cells at 3 h after release from Hu (Fig. 5A, lane 2, versus Fig. 5B, lane 2). Similar to the RNA induction curve and gel shift binding, NF-κB binding was transient. The level of NF-κB binding activity returned to baseline by 6 h after release. The lack of p65 binding in pCEP nuclear extracts was not due to the absence of p65 in the nucleus of these cells, as demonstrated by straight Western blotting (Fig. 5C). To demonstrate the specificity of NF-κB binding, parallel assays were run with an oligonucleotide containing a mutant IL-8 NF-κB binding site. Consistent with the results presented in Fig. 5A, a significant level of NF-κB p65 was observed 3 h after release when the IL-8wt probe was used (Fig. 5D). In contrast, no binding of NF-κB p65 was observed when the IL-8 NF-κB mutant probe was used (Fig. 5D).
FIG. 5.
The NF-κB species p65 binds strongly to the IL-8 promoter sequence in Tat-containing cells. eTat and pCEP cells were blocked with Hu (2 mM) and released. (A, B, and D) Nuclear extracts taken at different points were incubated with either the IL-8wt or the mutant IL-8mκB probe in gel shift buffer at room temperature as previously described. The complexes were washed with 0.03% Sarkosyl and analyzed by Western blot analysis with anti-p65 antibody. (C) p65 Western blot using nuclear extracts taken at 3 h postrelease from pCEP and eTat cells. (E) The wild-type IL-8 probe was incubated with the 3-h eTat or pCEP extract and washed, and Western blot analysis was done with an anti-CBP antibody. (F) Western blot analysis of CBP input from eTat and pCEP extracts. The results presented are representative of two independent experiments.
It has recently been reported that the association of the coactivator CBP with p65 is important for the induction of transcription activity (26). To determine whether CBP was present in the IL-8 NF-κB binding complex, the 3-h nuclear extract from eTat and pCEP was incubated with the biotinylated oligonucleotide, centrifuged, washed, and analyzed by Western blotting. The results presented in Fig. 5E demonstrate that CBP was present in the complex of proteins bound to the IL-8 NF-κB site. Consistent with the results of the p65 Western blot, the level of CBP bound to the IL-8 NF-κB probe was significantly higher in the NF-κB pull-down from eTat extracts. The CBP input Western blot (Fig. 5F) did not reveal a significant difference between CBP levels in eTat and pCEP cells.
MG-132 treatment abolishes NF-κB binding to the IL-8 promoter sequence as well as IL-8 protein synthesis.
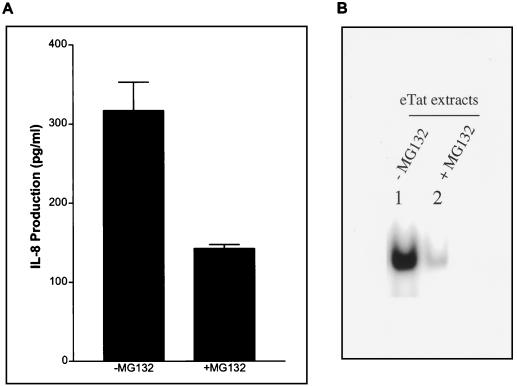
Since we demonstrated that the IL-8 promoter sequence is regulated by NF-κB through the cell cycle, it was of interest to determine what effect the proteasome inhibitor MG-132 would have on IL-8 expression. MG-132 inhibits proteasome function and thus inhibits IκBα degradation, interfering with NF-κB activation and translocation to the nucleus. eTat cells were treated with MG-132 immediately after release from the Hu block, and samples were harvested as described above. An IL-8 ELISA was conducted on the culture supernatants. The results of this experiment demonstrate that treatment with MG-132, which inhibits NF-κB translocation to the nucleus, significantly reduced the levels of IL-8 protein (Fig. 6A). These observations are consistent with the enhancement of IL-8 expression by a transient activation of NF-κB during S phase.
FIG. 6.
A proteasome inhibitor blocks NF-κB signaling and IL-8 production during S phase. (A) ELISA of IL-8 from supernatant of eTat cells. eTat cells were blocked with Hu (2 mM) and released. MG-132 (50 μM) was added at the time of release. The supernatant was collected at 3 h postrelease and analyzed for IL-8 protein using ELISA. The result shown is representative of three independent experiments ± standard deviation. (B) EMSA using the IL-8wt probe and 5 μg of nuclear extracts obtained from eTat after Hu block (2 mM, 15 h) and release. MG-132 (50 μM) was added at the time of release. Lane 1, eTat at 3 h postrelease without MG-132 treatment; lane 2, eTat at 3 h postrelease with MG-132 (50 μM) treatment.
eTat cells were treated with MG-132 immediately after release, and time course nuclear extracts were also prepared as described above. As seen on the gel shift (Fig. 6B), MG-132 treatment abolished NF-κB binding on the IL-8 NF-κB promoter sequence. This observation explains the inhibition of IL-8 production (Fig. 6A).
Finally, the cytoplasmic fractions from control and MG-132-treated cells were run on a sodium dodecyl sulfate gel and analyzed by Western blotting using an antibody which recognizes IκBα (data not shown). In the MG-132-treated cells, a slower-migrating species of IκBα, corresponding to the phosphorylated form, was detected at 3 h postrelease (data not shown). This result suggests that NF-κB induction occurs at 3 h post-Hu release (S phase) in eTat cells by phosphorylation of IκBα and its subsequent degradation by the proteasome.
DISCUSSION
The data presented here demonstrate that the IL-8 gene is expressed in a cell cycle-dependent manner in cells that express the HIV-1 Tat protein. There is a novel induction of stable NF-κB binding to the IL-8 promoter during the S phase of the cell cycle only in Tat-expressing cells. Consistent with the timing of NF-κB induction, the IL-8 mRNA level rises during mid-S phase, and the level of IL-8 protein in the medium reaches its maximum shortly after this. Blocking IκBα degradation with the proteasome inhibitor MG-132 abolishes both the appearance of NF-κB in the nucleus and the increase in IL-8.
Evidence that synergy between NF-κB and other transcriptional activators involves the assembly of multicomponent complexes that use the transcriptional coactivators p300 and CBP has been presented (9, 26, 30). In our experiments, we observed an increase of p65 and CBP, stably binding to the IL-8 NF-κB site in extracts made from Tat-expressing cells at 3 h post-Hu release, coincident with the appearance of IL-8 mRNA. It will be of interest to determine if Tat transiently facilitates the association of p65 and CBP during S phase. Along these lines, Tat has been shown to interact both physically and functionally with p300 and CBP in activation of the HIV long terminal repeat (1, 6, 15). Perkins et al. (24) have suggested that NF-κB activity is controlled by cyclin-dependent kinases associated with the p300 transcriptional integrator. In this model, cycE-CDK2 bound to p300 with RelA exerts some kinase-dependent inhibitory influence over RelA bound in the same complex. Since Tat interacts with p300 (1, 6, 7, 15), it would be of interest to determine if Tat modifies the p300-CDK2-cycE-RelA complex, decreasing CDK2's ability to inhibit the transcriptional activity of RelA. Alternatively, Tat may simply enhance the assembly of a stable NF-κB complex containing p65 and CBP. It is of interest that the differences in NF-κB binding between eTat and pCEP cells were best observed when the binding reaction mixtures contained 0.03% Sarkosyl. This result, in fact, suggests that the binding of NF-κB from eTat cells is more stable.
Elevated levels of circulating IL-8, a potent chemotactic factor for T lymphocytes and granulocytes, are found in HIV-infected individuals. Ott et al. (23) recently reported that the HIV-1 transactivator protein Tat increased IL-8 secretion in T-cell lines following CD3- and CD28-mediated costimulation. Full-length Tat (Tat101) enhanced IL-8 transcription through increased transcription factor binding to the CD28-responsive element (CD28RE) in the IL-8 promoter. Expression of Tat72 also enhanced IL-8 production following T-cell stimulation via a different mechanism, most likely posttranscriptional. CD28RE in the IL-8 promoter was characterized as a low-affinity NF-κB binding site recognized by a complex including p50, p65, and c-Rel. Interestingly, transcription factor binding to “classical” NF-κB sites such as those present in the HIV-1, human IL-2, and lymphotoxin promoters, also recognized by p50 and p65 following CD3-plus-CD28-mediated costimulation, was unaffected by Tat101. These experiments identify CD28RE in the IL-8 promoter as an NF-κB recognition site and a Tat-responsive element. Moreover, these results suggest that Tat has the capacity to differentially induce binding of NF-κB to different responsive promoters. It will be of interest to determine the correlation between NF-κB binding to CD28RE and the cell cycle. Interestingly, stimulation of T cells with CD3 and CD28 has been shown to induce a strong increase in the number of cells in S phase (2).
IL-8 may play an important role in virus production and infection. We have observed that the presence of IL-8 in a cell culture medium, at physiologically relevant concentrations, is sufficient to protect infected cells against cell death induced by HIV expression (data not shown). Therefore, one could argue that the effect of a Tat, S-phase-driven expression of IL-8 could be to protect the cell from death, allowing more virus production. In addition, the secreted IL-8 in the medium could serve as a chemoattractant, providing new target cells for the virus. Further study of IL-8 and other cytokines regulated by Tat will provide insights into the mechanisms by which HIV can modify the extracellular environment to the advantage of virus replication.
ACKNOWLEDGMENTS
R.M. and P.F.L. contributed equally to this work.
We thank Kaneko Osamu and Wilfrid Mahieux for technical help, Nancy Rice for the generous gift of IκBα antibodies, and Janet Duvall for preparation of the manuscript.
M.A.H. was a participant in the HHMI/NIH Research Scholars Program.
REFERENCES
- 1.Benkirane M, Chun R F, Xiao H, Ogryzko V V, Howard B H, Nakatani Y, Jeang K T. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273:24898–24905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- 2.Boonen G, van Dijk A, Verdonck L, van Lier R, Rijksen G, Medema R. CD28 induces cell cycle progression by IL-2-independent down-regulation of p27(kip1) expression in human peripheral T lymphocytes. Eur J Immunol. 2000;29:789–798. doi: 10.1002/(SICI)1521-4141(199903)29:03<789::AID-IMMU789>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Cheynier R, Henrichwark S, Hadida F, Pelletier E, Oksenhendler E, Autran B, Wain-Hobson S. HIV and T cell expansion in splenic white pulps is accompanied by infiltration of HIV-specific cytotoxic T lymphocytes. Cell. 1994;78:373–387. doi: 10.1016/0092-8674(94)90417-0. [DOI] [PubMed] [Google Scholar]
- 4.Denis M, Ghadirian E. Dysregulation of interleukin 8, interleukin 10, and interleukin 12 release by alveolar macrophages from HIV type 1-infected subjects. AIDS Res Hum Retrovir. 1994;10:1619–1627. doi: 10.1089/aid.1994.10.1619. [DOI] [PubMed] [Google Scholar]
- 5.Hoebeke J, Van Nijen G, De Brabander M. Interaction of oncodazole (R 17934), a new antitumoral drug, with rat brain tubulin. Biochem Biophys Res Commun. 1976;69:319–324. doi: 10.1016/0006-291x(76)90524-6. [DOI] [PubMed] [Google Scholar]
- 6.Hottiger M O, Felzien L K, Nabel G J. Modulation of cytokine-induced HIV gene expression by competitive binding of transcription factors to the coactivator p300. EMBO J. 1998;17:3124–3134. doi: 10.1093/emboj/17.11.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hottiger M O, Nabel G J. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–8256. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashanchi F, Agbottah E T, Pise-Masison C A, Mahieux R, Duvall J, Kumar A, Brady J N. Cell cycle-regulated transcription by the human immunodeficiency virus type 1 Tat transactivator. J Virol. 2000;74:652–660. doi: 10.1128/jvi.74.2.652-660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 10.Kowalski J, Denhardt D T. Regulation of the mRNA for monocyte-derived neutrophil-activating peptide in differentiating HL60 promyelocytes. Mol Cell Biol. 1989;9:1946–1957. doi: 10.1128/mcb.9.5.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunsch C, Lang R K, Rosen C A, Shannon M F. Synergistic transcriptional activation of the IL-8 gene by NF-kappa B p65 (RelA) and NF-IL-6. J Immunol. 1994;153:153–164. [PubMed] [Google Scholar]
- 12.Kunsch C, Rosen C A. NF-κB subunit-specific regulation of the interleukin-8 promoter. Mol Cell Biol. 1993;13:6137–6146. doi: 10.1128/mcb.13.10.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert P F, Ludford-Menting M J, Deacon N J, Kola I, Doherty R R. The nfkb1 promoter is controlled by proteins of the Ets family. Mol Biol Cell. 1997;8:313–323. doi: 10.1091/mbc.8.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen C G, Anderson A O, Appella E, Oppenheim J J, Matsushima K. The neutrophil-activating protein (NAP-1) is also chemotactic for T lymphocytes. Science. 1989;243:1464–1466. doi: 10.1126/science.2648569. [DOI] [PubMed] [Google Scholar]
- 15.Marzio G, Tyagi M, Gutierrez M I, Giacca M. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci USA. 1998;95:13519–13524. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto T, Miike T, Nelson R P, Trudeau W L, Lockey R F, Yodoi J. Elevated serum levels of IL-8 in patients with HIV infection. Clin Exp Immunol. 1993;93:149–151. doi: 10.1111/j.1365-2249.1993.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori N, Mukaida N, Ballard D W, Matsushima K, Yamamoto N. Human T-cell leukemia virus type I Tax transactivates human interleukin 8 gene through acting concurrently on AP-1 and nuclear factor-kappaB-like sites. Cancer Res. 1998;58:3993–4000. [PubMed] [Google Scholar]
- 18.Mori N, Murakami S, Oda S, Prager D, Eto S. Production of interleukin 8 in adult T-cell leukemia cells: possible transactivation of the interleukin 8 gene by human T-cell leukemia virus type I tax. Cancer Res. 1995;55:3592–3597. [PubMed] [Google Scholar]
- 19.Mukaida N, Mahe Y, Matsushima K. Cooperative interaction of nuclear factor-kappa B- and cis-regulatory enhancer binding protein-like factor binding elements in activating the interleukin-8 gene by pro-inflammatory cytokines. J Biol Chem. 1990;265:21128–21133. [PubMed] [Google Scholar]
- 20.Mukaida N, Shiroo M, Matsushima K. Genomic structure of the human monocyte-derived neutrophil chemotactic factor IL-8. J Immunol. 1989;143:1366–1371. [PubMed] [Google Scholar]
- 21.Oeth P A, Parry G C, Kunsch C, Nantermet P, Rosen C A, Mackman N. Lipopolysaccharide induction of tissue factor gene expression in monocytic cells is mediated by binding of c-Rel/p65 heterodimers to a κB-like site. Mol Cell Biol. 1994;14:3772–3781. doi: 10.1128/mcb.14.6.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto S, Mukaida N, Yasumoto K, Horiguchi H, Matsushima K. Molecular mechanism of interleukin-8 gene expression. Adv Exp Med Biol. 1993;351:87–97. doi: 10.1007/978-1-4615-2952-1_10. [DOI] [PubMed] [Google Scholar]
- 23.Ott M, Lovett J L, Mueller L, Verdin E. Superinduction of IL-8 in T cells by HIV-1 Tat protein is mediated through NF-kappaB factors. J Immunol. 1998;160:2872–2880. [PubMed] [Google Scholar]
- 24.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 25.Schlegel R, Pardee A B. Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science. 1986;232:1264–1266. doi: 10.1126/science.2422760. [DOI] [PubMed] [Google Scholar]
- 26.Sheppard K-A, Rose D W, Haque Z K, Kurokawa R, McInerney E, Westin S, Thanos D, Rosenfeld M G, Glass C K, Collins T. Transcriptional activation by NF-κB requires multiple coactivators. Mol Cell Biol. 1999;19:6367–6378. doi: 10.1128/mcb.19.9.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonoda Y, Kasahara T, Yamaguchi Y, Kuno K, Matsushima K, Mukaida N. Stimulation of interleukin-8 production by okadaic acid and vanadate in a human promyelocyte cell line, an HL-60 subline. Possible role of mitogen-activated protein kinase on the okadaic acid-induced NF-kappaB activation. J Biol Chem. 1997;272:15366–15372. doi: 10.1074/jbc.272.24.15366. [DOI] [PubMed] [Google Scholar]
- 28.Stein B, Baldwin A S., Jr Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-κB. Mol Cell Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taub D D. Chemokine-leukocyte interactions. The voodoo that they do so well. Cytokine Growth Factor Rev. 1996;7:355–376. doi: 10.1016/s1359-6101(97)89237-4. [DOI] [PubMed] [Google Scholar]
- 30.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]