Abstract
Paramecium bursaria chlorella virus 1 (PBCV-1) elicits a lytic infection of its unicellular green alga host. The 330-kbp viral genome has been sequenced, yet little is known about how viral mRNAs are synthesized and processed. PBCV-1 encodes its own mRNA guanylyltransferase, which catalyzes the addition of GMP to the 5′ diphosphate end of RNA to form a GpppN cap structure. Here we report that PBCV-1 encodes a separate RNA triphosphatase (RTP) that catalyzes the initial step in cap synthesis: hydrolysis of the γ-phosphate of triphosphate-terminated RNA to generate an RNA diphosphate end. We exploit a yeast-based genetic system to show that Chlorella virus RTP can function as a cap-forming enzyme in vivo. The 193-amino-acid Chlorella virus RTP is the smallest member of a family of metal-dependent phosphohydrolases that includes the RNA triphosphatases of fungi and other large eukaryotic DNA viruses (poxviruses, African swine fever virus, and baculoviruses). Chlorella virus RTP is more similar in structure to the yeast RNA triphosphatases than to the enzymes of metazoan DNA viruses. Indeed, PBCV-1 is unique among DNA viruses in that the triphosphatase and guanylyltransferase steps of cap formation are catalyzed by separate viral enzymes instead of a single viral polypeptide with multiple catalytic domains.
The m7GpppN cap structure of eukaryotic mRNA is formed cotranscriptionally by three enzymatic reactions: (i) the 5′ triphosphate end of the nascent RNA is hydrolyzed to a diphosphate by RNA triphosphatase (RTP), (ii) the diphosphate end is capped with GMP by GTP:RNA guanylyltransferase, and (iii) the GpppN cap is methylated by S-adenosylmethionine:RNA (guanine-N7) methyltransferase (27). DNA viruses have evolved diverse capping strategies. The mRNAs of papovaviruses, parvoviruses, adenoviruses, and herpesviruses are transcribed in the nucleus by RNA polymerase II (Pol II), and their 5′ ends are modified by the host cell's capping and methylating enzymes. However, vaccinia virus and other poxviruses, which replicate in the cytoplasm, encode and encapsidate with the virus particle a multisubunit RNA polymerase and a complete mRNA capping apparatus (26). African swine fever virus (ASFV), which has a cytoplasmic replication phase, also encodes and encapsidates an RNA polymerase and mRNA capping enzymes (24). Baculoviruses, which replicate in the nucleus of insect cells, use Pol II to transcribe early genes, then switch at later times to a virus-encoded transcription system that includes an RNA polymerase and two cap-forming activities— RTP and RNA guanylyltransferase (4, 5, 14). Paramecium bursaria chlorella virus 1 (PBCV-1) encodes an RNA guanylyltransferase (7), but it is not clear whether it encodes an RNA polymerase and additional mRNA-processing activities.
The triphosphatase, guanylyltransferase, and methyltransferase components of the capping apparatus are organized differently in these DNA virus systems. The triphosphatase, guanylyltransferase, and methyltransferase active sites of the vaccinia virus capping enzyme reside in a single 844-amino-acid (844-aa) polypeptide, and the order of the active sites in the primary structure (H2N-triphosphatase/guanylyltransferase/ methyltransferase-COOH) mimics the temporal order of the cap-forming reactions. The triphosphatase and guanylyltransferase active sites of the 464-aa baculovirus capping enzyme are also arrayed in cis in the order H2N-triphosphatase/guanylyltransferase-COOH. The baculovirus capping enzyme is structurally related to the 60-kDa triphosphatase-guanylyltransferase domain of the vaccinia virus capping enzyme. However, baculovirus encodes no discernible homologue of a vaccinia virus RNA (guanine-7)-methyltransferase, and it remains unclear whether a cellular or viral enzyme is responsible for baculovirus cap methylation.
PBCV-1 is the prototype of a family of large icosahedral DNA viruses that replicate in unicellular Chlorella-like green algae (29). The 330-kbp linear PBCV-1 genome encodes 375 polypeptides, which makes PBCV-1 one of the most genetically complex viruses known. Its gene expression strategy is understood only crudely (30). It is believed that the infecting DNA is targeted to the nucleus, where the transcription of early genes ensues within 5 to 10 min postinfection. Transcription of the late class of viral genes commences after the onset of viral DNA replication at 60 to 90 min postinfection. The cis-acting DNA signals that control PBCV-1 transcription and the proteins that comprise the viral transcription machinery are unknown. PBCV-1 encodes polypeptides that resemble the cellular transcription factors TFIIB and TFIIS, but there are no discernible PBCV-1 homologues of cellular or viral RNA polymerase subunits. Thus, PBCV-1 either encodes a novel RNA polymerase or its genome is transcribed by one or more of the RNA polymerases of the algal host cell. Almost nothing is known about Chlorella virus mRNA processing events, except for the fact that PBCV-1 encodes its own mRNA guanylyltransferase, which has been purified and characterized biochemically (7).
Chlorella virus guanylyltransferase is a 330-aa monomeric polypeptide that catalyzes the transfer of GMP from GTP to the 5′ diphosphate end of RNA to form the GpppN cap structure. Its structure and mechanism have been illuminated in atomic detail by X-ray crystallography (6). Chlorella virus guanylyltransferase is monofunctional and has no intrinsic triphosphatase or methyltransferase activities. It is most closely related to the monofunctional yeast RNA guanylyltransferases, more so than to the multifunctional vaccinia virus capping enzyme or the bifunctional triphosphatase-guanylyltransferases of ASFV or baculovirus.
In order to cap its mRNAs, PBCV-1 must either encode its own RTP or rely on the resident enzyme(s) of the host cell to remove the γ-phosphate of the primary transcript so that the diphosphate end can be modified by the PBCV-1 guanylyltransferase. The similarities between Chlorella virus and yeast guanylyltransferases prompted us to search the PBCV-1 proteome for a polypeptide resembling the well-characterized Saccharomyces cerevisiae RTP Cet1p (18). This exercise pinpointed the 193-aa A449R gene product as a candidate RTP. We exploited a yeast-based genetic system for analysis of viral mRNA capping enzymes (12) to show that the Chlorella virus RTP (cvRTP) is functional in vivo in S. cerevisiae in lieu of Cet1p. Purified recombinant cvRTP has intrinsic metal-dependent RTP and nucleoside triphosphatase activities. Mechanistic conservation between cvRTP and yeast RTPs is suggested by mutational analysis of the putative metal-binding site. Our results underscore a close evolutionary connection between the capping apparatus of fungi and Chlorella virus, and they suggest that the capping enzymes of metazoan DNA viruses arose by gene fusion events involving yeast/PBCV-1-like domains.
MATERIALS AND METHODS
Bacterial expression plasmids for cvRTP.
The PBCV-1 A449R gene was amplified by PCR from viral genomic DNA (a gift of James Van Etten). Oligonucleotide primers complementary to the 5′ and 3′ ends of the gene were designed to introduce an NdeI restriction site at the translation initiation codon and a BamHI site 3′ of the translation stop codon. The 0.6-kbp PCR product was digested with NdeI and BamHI and then inserted between the NdeI and BamHI sites of the T7 RNA-polymerase-based vector pET16b so as to fuse the 193-aa A449R polypeptide in frame with an N-terminal 21-aa leader peptide containing 10 tandem histidines. The resulting plasmid, pET-A449R, was sequenced to confirm that the A449R insert was identical to the genomic DNA sequence (GenBank accession number NC 000852). An alanine substitution mutation at position Glu-26 was introduced into the A449R gene by the two-stage overlap extension method (13). The mutated gene was digested with NdeI and BamHI and then inserted into pET16b. The resulting pET-A449R-E26A plasmid insert was sequenced completely to confirm the desired alanine mutation and exclude the acquisition of unwanted changes during amplification or cloning. Plasmids pET-A449R and pET-A449R-E26A then were introduced into Escherichia coli BL21(DE3).
Yeast expression plasmid for cvRTP.
An NdeI-BamHI fragment containing the A449R open reading frame was excised from pET-A449R and inserted between the NdeI and BamHI sites of the yeast shuttle vector pYX1(CEN TRP1). The resulting plasmid pYX-cvRTP encodes the 193-aa polypeptide fused in frame with a 12-aa N-terminal leader peptide (MGSHHHHHHSGH). Expression of the Chlorella virus gene in this plasmid is under the control of the constitutive yeast TPI1 promoter.
Chimeric Chlorella virus-mouse capping enzyme.
A gene encoding cvRTP fused to the guanylyltransferase domain of the mouse capping enzyme [Mce1(211– 597)p] was constructed as follows. The Chlorella virus triphosphatase gene was PCR amplified from pET-A449R using an antisense primer that changed the Val-192 codon to His while introducing an NdeI restriction site at codons 192 to 193. The PCR product was digested with NdeI and then inserted into the NdeI site of pYX1-MCE1(211–597)(CEN TRP1) to yield the fusion gene cvRTP-MCE1(211–597). The mutant fusion gene cvRTP(E26A)-MCE1(211–597) encoding a catalytically defective viral RTP fused to mouse guanylyltransferase was constructed in parallel using the pET-A449R-E28A plasmid as the template for PCR amplification. Expression of the chimeric capping enzyme genes in the yeast plasmids is under the control of the TPI1 promoter. The resulting pYX-cvRTP-MCE1(211–597) and pYX-cvRTP(E26A)-MCE1(211–597) plasmid inserts were sequenced completely to confirm the coding continuity at the fusion junction and exclude the acquisition of unwanted changes during amplification or cloning.
Expression and purification of recombinant cvRTP.
A 1-liter culture of E. coli BL21(DE3)/pET-A449R was grown at 37°C in Luria-Bertani medium containing 0.1 mg of ampicillin per ml until the A600 reached ∼0.5. The culture was placed on ice for 10 min and then adjusted to 0.4 mM isopropyl-1-thio-β-d-galactopyranoside and 2% (vol/vol) ethanol. After further incubation for 17 h at 18°C with constant shaking, the cells were harvested by centrifugation, and the pellet was stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 100 ml of buffer A (50 mM Tris-HCl [pH 7.5]–250 mM NaCl–10% sucrose). Lysozyme was added to a final concentration of 50 μg/ml, and the suspension was incubated on ice for 15 min, then adjusted to 0.1% Triton X-100 and sonicated to reduce the viscosity of the lysate. Insoluble material was removed by centrifugation for 45 min at 17,000 rpm in a Sorvall SS34 rotor. The soluble extract (150 mg of protein) was applied to a 6-ml column of Ni2+-NTA agarose (Qiagen) that had been equilibrated with buffer A containing 0.1% Triton X-100. The column was washed with the same buffer and then eluted stepwise with 11-ml aliquots of buffer B (50 mM Tris-HCl [pH 8.0]–250 mM NaCl–10% glycerol–0.1% Triton X-100) containing 0, 50, 100, 200, and 1,000 mM imidazole. The polypeptide compositions of the column fractions were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The PBCV-1 polypeptide was retained on the column and recovered predominantly in the 200 mM imidazole fraction (3 mg of protein). The E26A mutant protein was purified from a 1-liter culture of E. coli BL21(DE3)/pET-A449R-E26A by the same method. The Ni-agarose enzyme preparations were stored at −80°C. Protein concentrations were determined using Bio-Rad dye reagent with bovine serum albumin as a standard.
RESULTS
Identification of Chlorella virus protein A449R as a candidate RTP.
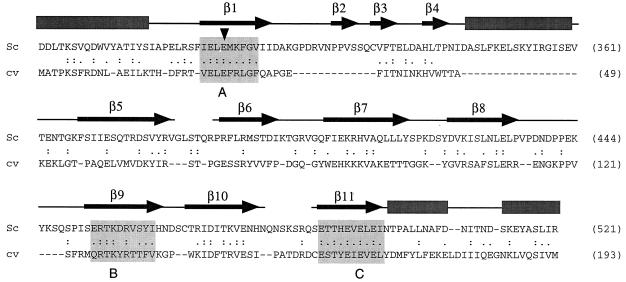
The budding yeast S. cerevisiae encodes a capping apparatus that consists of separate triphosphatase (Cet1p; 549-aa), guanylyltransferase (Ceg1p; 459-aa), and methyltransferase (Abd1p; 436-aa) gene products (27). The yeast RTP Cet1p exemplifies a growing family of metal-dependent phosphohydrolases that includes the RTPs encoded by other fungi (Candida albicans and Schizosaccharomyces pombe) and by several groups of eukaryotic DNA viruses (poxviruses, ASFV, and baculoviruses) (4, 8, 14, 23; Y. Pei, B. Schwer, S. Hausmann, and S. Shuman, submitted for publication). The yeast/viral triphosphatase family is defined by two glutamate-rich peptide motifs (motifs A and C), which are essential for catalytic activity and comprise the metal-binding site, and by a basic peptide motif (motif B), which is implicated in binding the 5′ triphosphate moiety of the substrate (Fig. 1). The crystal structure of S. cerevisiae RTP reveals that the active site is located within the hydrophilic core of a topologically closed eight-stranded β barrel—the so-called triphosphate tunnel (18). The β strands comprising the tunnel (β1 and β5 to β11) are displayed over the Cet1p amino acid sequence shown in Fig. 1.
FIG. 1.
Structural similarity between S. cerevisiae RTP and the Chlorella virus A449R gene product. The amino acid sequence of S. cerevisiae (Sc) RTP Cet1p from residues 279 to 521 is aligned to that of the predicted Chlorella virus (cv) A449R polypeptide. Gaps in the alignment are indicated by dashes. Numbers of amino acids in sequences are given in parentheses on the right-hand side of the figure. The secondary structure of Cet1p is displayed above the amino acid sequence. Conserved motifs A (β1), B (β9), and C (β11) that define the metal-dependent RTP family are highlighted in the shaded boxes. The conserved glutamate in motif A that was subjected to mutational analysis is indicated by an arrowhead.
We used the software PSI-BLAST (1) to search the NCBI database for proteins related to the biologically active C-terminal domain Cet1(241–539)p. The initial search highlighted the extensive sequence similarity between Cet1p and other fungal triphosphatases but revealed no viral homologues. The first iteration of the search identified a short segment of the Chlorella virus A449R gene product (a 193-aa polypeptide) with weak similarity to Cet1p (BLAST score 35). The similarity of A449R to Cet1p and other fungal triphosphatases was rated as statistically significant in the second iteration of the search (BLAST score 90).
The small region of similarity between A449R and yeast RTP identified by the computer-based sequence search extended from strands β9 to β11 in Cet1p and embraced conserved motif B (RTKXR) in β9 and motif C (EVELE) in β11 (Fig. 1). The A449R and Cet1p sequences were then aligned manually using the tertiary structure of Cet1p and the available structure-activity relationships for fungal RNA triphosphatases as a guide (18, 22, 23). We readily identified in A449R a counterpart of conserved motif A (β1) and putative counterparts of β strands 5, 6, 7, 8, and 10 that comprise the walls of the triphosphate tunnel in Cet1p (Fig. 1). The sequence similarity extended for the entire length of the 193-aa A449R polypeptide and included 52 positions of side-chain identity plus 38 positions of side-chain similarity (Fig. 1). Most of the hydrophilic amino acids that comprise the active site of yeast RTPs and are important for its catalytic activity are also present in the A449R protein, leading to the prediction that this gene product is a component of the Chlorella virus capping apparatus. In the experiments presented below, we tested genetically and biochemically whether the Chlorella virus A449R gene product has the requisite activities of a cap-forming enzyme in vivo and in vitro.
Probing the function of cvRTP using a yeast-based genetic system.
There are no methods available to manipulate the PBCV-1 genome in vivo in a controlled fashion. Thus, it is not possible to test directly whether the A449R gene is essential for PBCV-1 replication or to probe its role in mRNA processing. To circumvent the latter problem, we exploited a yeast-based system for genetic analysis of virus-encoded capping enzymes (12). The system provides the capacity to answer the following questions concerning the putative cvRTP. (i) Can the viral protein function in the cap-synthetic pathway and sustain the growth of yeast cells that lack the endogenous RTP Cet1p? (ii) If so, does complementation of the yeast cet1Δ mutation by the viral protein depend on its catalytic activity in cap formation?
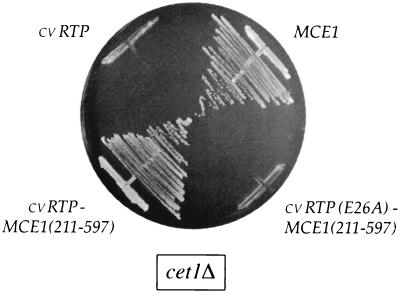
To express the putative cvRTP in yeast, we cloned the A449R gene (henceforth referred to as cvRTP) into a yeast CEN plasmid and placed it under the transcriptional control of the strong constitutive TPI1 promoter. The capacity of the viral protein to replace the essential yeast RTP was tested by plasmid shuffle in yeast cet1Δ cells that contain CET1 on a URA3 plasmid. The cet1Δ strain is unable to form colonies on medium containing 5-fluoroorotic acid (5-FOA), a drug that selects against the URA3 plasmid, unless it is transformed with a second plasmid bearing CET1 or a functional homologue from another source. For example, transformation with a TRP1 plasmid bearing the MCE1 gene, which encodes the 597-aa mammalian triphosphatase-guanylyltransferase, allows growth of cet1Δ cells on 5-FOA (Fig. 2). In contrast, we found that expression of cvRTP did not complement the cet1Δ mutation (Fig. 2).
FIG. 2.
cvRTP activity in vivo in yeast. The yeast cetlΔ strain YBS20 (MATa trp1 his3 ura3 leu2 ade2 can1 cet1::LEU2 p360-CET1) was transformed with CEN TRP1 plasmids containing either MCE1, cvRTP, a cvRTP-MCE1(211–597) chimera, or a mutated fusion gene, cvRTP(E26A)-MCE1(211–597). Single Trp+ transformants were patched to agar plates lacking tryptophan and then streaked on agar medium containing 0.75 mg of 5-FOA per ml. The plates were photographed after incubation for 3 days at 30°C.
The likely explanation for why cvRTP could not replace Cet1p is that the cvRTP failed to localize to the intranuclear sites of pre-mRNA synthesis. Targeting of cap formation to nascent pre-mRNAs is achieved via the binding of the guanylyltransferase component of the cellular capping apparatus to the phosphorylated carboxyl-terminal domain (CTD) of elongating Pol II (2, 3, 9, 10, 15, 20, 25, 32). The CTD, consisting of tandem repeats of a heptapeptide of the consensus sequence YSPTSPS, is extensively phosphorylated in the context of the transcription elongation complex. In yeast, the guanylyltransferase Ceg1p binds to CTD-PO4, whereas the triphosphatase Cet1p does not. Formation of a Cet1p-Ceg1p complex in trans allows the yeast guanylyltransferase to chaperone Cet1p to the transcription complex. The guanylyltransferase-binding site of S. cerevisiae Cet1p is contained within a 21-aa peptide segment (residues 239 to 259) that flanks the catalytic domain (11, 17). Because cvRTP contains no counterpart for this domain, we would not expect it to form the requisite complex with yeast guanylyltransferase.
The in vivo requirement for a Ceg1p-binding site on RTP can be bypassed by linking the Cet1p triphosphatase catalytic domain (minus the Ceg1p-binding domain) in cis to the guanylyltransferase domain of the mammalian capping enzyme, Mce1(211–597)p, which by itself binds avidly to the phosphorylated CTD (9, 10, 17). Mammalian guanylyltransferase can target Cth1p, an S. cerevisiae RTP not normally involved in capping, and thereby convert it into a cap-forming enzyme in vivo (22). The mammalian guanylyltransferase can even act as chaperone for vaccinia virus capping enzyme when the two are fused in cis, thereby allowing vaccinia virus RTP to complement the cet1Δ mutation (12). These results suggested that Mce1(211–597)p can be used as a vehicle to deliver other viral mRNA processing enzymes to the yeast transcription elongation complex in vivo.
We tested whether fusion of the putative cvRTP to Mce1(211–597)p might correctly target cvRTP and thereby result in a gain of function in vivo. A chimeric gene, cvRTP-MCE1(211–597), was cloned into a CEN TRP1 vector such that its expression was under the control of the yeast TPI1 promoter. The instructive finding was that cet1Δ cells transformed with the fusion gene grew on 5-FOA (Fig. 2). The cvRTP-MCE1(211–597) cells grew well on rich medium at either 25, 30, or 37°C (data not shown). We conclude that the Chlorella virus A449R protein can function as an RTP in mRNA cap formation in yeast cells when it is targeted appropriately to the transcription complex.
RNA triphosphatase activity of cvRTP.
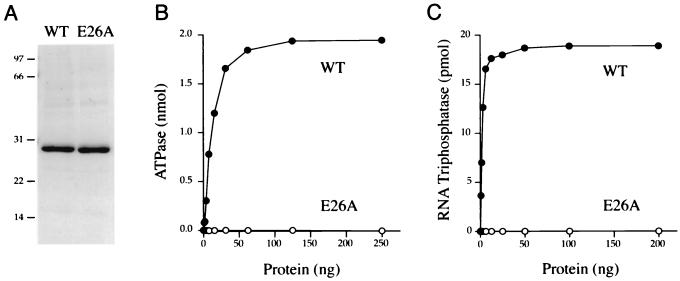
The cvRTP gene was cloned into a T7 RNA polymerase-based pET vector so as to place the open reading frame in frame with an N-terminal leader encoding a 21-aa peptide with 10 tandem histidines. The expression plasmid was introduced into E. coli BL21(DE3), a strain that contains the T7 RNA polymerase gene under the control of the lac promoter. The His-tagged cvRTP protein was purified from a soluble extract of isopropyl-β-d-thiogalactopyranoside-induced bacteria by adsorption to Ni2+-agarose and elution with 200 mM imidazole. The preparation was highly enriched with respect to the 29-kDa cvRTP polypeptide, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 3A).
FIG. 3.
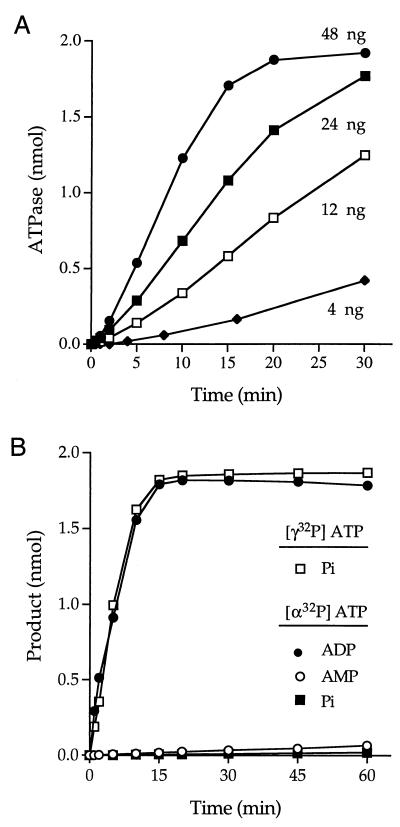
RTP and ATPase activities of cvRTP. (A) Protein purification. Aliquots (3 μg) of the Ni2+-agarose preparations of wild-type (WT) cvRTP and the E26A mutant protein were analyzed by electrophoresis through a 15% polyacrylamide gel containing 0.1% sodium dodecyl sulfate. Polypeptides were visualized by staining with Coomassie blue dye. The positions and sizes (in kilodaltons) of marker proteins are indicated on the left-hand side. (B) ATPase activity. Reaction mixtures (10 μl) containing 50 mM Tris-HCl [pH 7.5], 5 mM dithiothreitol (DTT), 1 mM MnCl2, 0.2 mM [γ-32P]ATP, and either WT or E26A proteins as specified were incubated for 15 min at 37°C. The reactions were quenched by adding 2.5 μl of 5 M formic acid. Aliquots of the mixtures were applied to a polyethyleneimine-cellulose thin-layer chromatography (TLC) plate, which was developed with 1 M formic acid–0.5 M LiCl. 32Pi release was quantitated by scanning the chromatogram with a FUJIX phosphorimager and is plotted as a function of input protein. (C) RTP activity. Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 10 mM MgCl2, 20 pmol (of triphosphate termini) of γ-32P-poly(A), and either WT or E26A proteins as specified were incubated for 15 min at 37°C. The reactions were quenched with formic acid and the products were analyzed by TLC. 32Pi release is plotted as a function of input protein.
We found that recombinant cvRTP is indeed an RNA triphosphatase. Activity was assayed by the liberation of 32Pi from 2 μM γ-32P-labeled triphosphate-terminated poly(A) in the presence of 10 mM magnesium chloride. The extent of γ-phosphate hydrolysis during a 15-min incubation at 30°C was proportional to the amount of input protein (Fig. 3C). In the linear range of enzyme dependence, 120 fmol of 32Pi was released per fmol of cvRTP. This value corresponds to a turnover number of ∼0.1 s−1, which is lower than the values reported for the hydrolysis of γ-32P-poly(A) by S. cerevisiae Cet1p (1 s−1), C. albicans CaCet1p (1.4 s−1), baculovirus LEF4 (1 s−1), and vaccinia virus D1 (0.8 s−1) but similar to the turnover number of S. pombe Pct1p (0.2 s−1) (4, 8, 21, 23; Pei et al., submitted for publication).
Metal-dependent nucleoside triphosphatase activity of cvRTP.
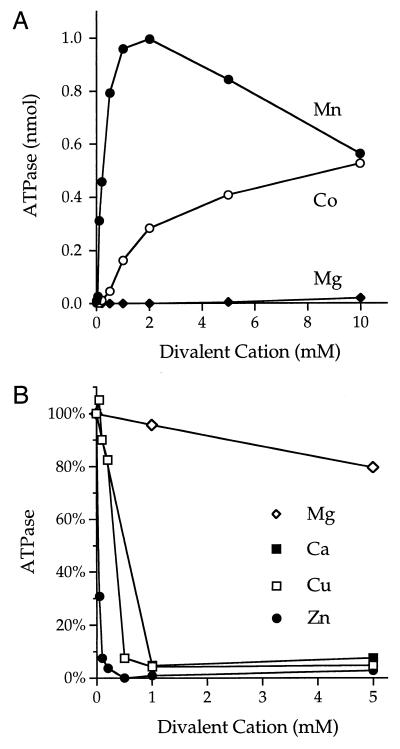
The signature biochemical feature of the fungal/viral triphosphatase family is its ability to hydrolyze nucleoside triphosphates to nucleoside diphosphates in the presence of manganese or cobalt (8). The divalent cation specificity of the nucleoside triphosphatase is distinct from the RTP function, which is optimal in magnesium. We found that recombinant cvRTP catalyzed the release of 32Pi from [γ-32P]ATP in the presence of manganese and that the extent of ATP hydrolysis increased as a function of input enzyme (Fig. 3B).
There was no detectable ATP hydrolysis in the absence of a divalent cation. Hydrolysis of 0.2 mM ATP was optimal at 1 to 2 mM MnCl2 and declined slightly at 5 to 10 mM MnCl2 (Fig. 4A). ATP hydrolysis with cobalt as the cofactor was optimal at 10 mM CoCl2 (Fig. 4A). Magnesium did not support ATP hydrolysis (Fig. 4A) nor did calcium, copper, or zinc (data not shown). A mixing experiment showed that addition of up to 5 mM magnesium had little effect on ATP hydrolysis promoted by 1 mM manganese (Fig. 4B). In contrast, the ATPase activity was virtually abolished by the inclusion of 1 mM calcium, copper, or zinc (Fig. 4B). Zinc was the most potent of the inhibitory metals; 50 and 100 μM zinc reduced ATP hydrolysis by 70% and 90%, respectively.
FIG. 4.
Divalent cation dependency and specificity. (A) Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 7.5), 0.2 mM [γ-32P]ATP, 30 ng of cvRTP, and MgCl2, MnCl2, or CoCl2 as indicated were incubated for 15 min at 37°C. 32Pi release is plotted as a function of divalent cation concentration. (B) Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 7.5), 0.2 mM [γ-32P]ATP, 30 ng of cvRTP, 1 mM MnCl2, and either MgCl2, CaCl2, CuSO4, or ZnSO4 at the concentration specified were incubated for 15 min at 37°C. The extents of ATP hydrolysis in the presence of manganese plus magnesium, calcium, copper, or zinc were normalized to the control activity in the presence of manganese alone (defined as 1.0).
The manganese-dependent ATPase activity of cvRTP in 50 mM Tris-HCl buffer was optimal between pH 7 and 8; the extent of ATP hydrolysis at pH 9 was 2̃0% of the value at pH 7.5 (data not shown).
Kinetic analysis of ATP hydrolysis.
The rate of 32Pi release from [γ-32P]ATP varied linearly with the amount of input cvRTP (Fig. 5A). From a plot of reaction rate versus enzyme we calculated a turnover number of 1 s−1. The quantitative conversion of [α-32P]ATP to [α-32P]ADP was catalyzed by cvRTP. The rate of [α-32P]ADP formation was essentially identical to the rate of 32Pi release from [γ-32P]ATP assayed in a parallel reaction mixture (Fig. 5B). We detected little formation of [α-32P]AMP from [α-32P]ATP, even after 60 min of incubation, by which time all of the nucleotide had been converted to ADP. We conclude that cvRTP catalyzes the hydrolysis of ATP to ADP plus Pi and is unable to further hydrolyze the ADP reaction product. The cvRTP also catalyzed manganese-dependent hydrolysis of [α-32P]GTP to [α-32P]GDP, [α-32P]dATP to [α-32P]dADP, [α-32P]CTP to [α-32P]CDP, [α-32P]dCTP to [α-32P]dCDP, and [α-32P]UTP to [α-32P]UDP (data not shown).
FIG. 5.
Kinetics of ATP hydrolysis. (A) Reaction mixtures containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 1 mM MnCl2, 0.2 mM [γ-32P]ATP, and cvRTP as specified (in nanograms per 10 μl) were incubated at 37°C. Aliquots (10 μl) were withdrawn at times indicated and quenched immediately with formic acid. (B) Reaction mixtures (110 μl) containing 50 mM Tris-HCl (pH 7.5), 1 mM MnCl2, 5 mM DTT, 0.2 mM [γ-32P]ATP or [α-32P]ATP as specified, and 1.1 μg of cvRTP were incubated at 37°C. Aliquots (10 μl) were withdrawn at times indicated and quenched immediately with formic acid. The reaction products were analyzed by TLC. The extent of 32Pi, [α32P]ADP, or [α32P]AMP formation (from 2 nmol of input ATP per sample) is plotted as a function of time.
Kinetic parameters were determined by measuring ATPase activity as a function of input [γ-32P]ATP concentration. From a double-reciprocal plot of the data, we calculated a Km of 7 μM ATP and a Vmax of 1.5 s−1 (data not shown). The turnover number of cvRTP in ATP hydrolysis is lower than the values reported for Cet1p (25 s−1), CaCet1p (17 s−1), Pct1p (67 s−1), vaccinia virus D1 (10 s−1), and baculovirus LEF4 (30 s−1), but it is similar to the turnover number of S. cerevisiae Cth1p (2 s−1) (4, 8, 21–23; Pei et al., submitted). The Km of cvRTP for ATP (7 μM) falls in the lower range of values reported for other family members: Cet1p (3 μM), CaCet1p (9 μM), Pct1p (19 μM), LEF4 (43 μM), Cth1p (75 μM), and D1 (800 μM).
cvRTP activity is abolished by replacement of motif A Glu-26 with alanine.
Glu-26 in motif A of cvRTP is strictly conserved in the RTPs encoded by S. cerevisiae, C. albicans, S. pombe, poxviruses, ASFV, and baculoviruses. The equivalent glutamate of Cet1p (Glu-307 [highlighted by the vertical arrowhead in Fig. 1]) directly coordinates manganese in the active site and is essential for catalysis by Cet1p in vitro and for Cet1p function in vivo (8, 18). The same motif A glutamate is also essential for catalysis by CaCet1p, Cth1p, Pct1p, vaccinia virus D1, and baculovirus LEF4 (14, 22, 23, 31; Pei et al., submitted). If cvRTP is a true member of the yeast/viral triphosphatase enzyme family with a similar mechanism of metal-assisted catalysis, then removal of the Glu-26 carboxylate of cvRTP should elicit a significant loss of function. We replaced Glu-26 with alanine by site-directed mutagenesis of the cvRTP gene, produced recombinant His-tagged E26A protein in bacteria, and then purified it from a soluble extract by Ni2+-agarose chromatography (Fig. 3A). The E26A mutant was unable to hydrolyze triphosphatase-terminated RNA or ATP at levels of input protein well in excess of the amount sufficient for maximal release of 32Pi by wild-type cvRTP (Fig. 3B and C). From these data, we calculated that the specific RTP and ATPase activities of E26A were <0.1% of the activity of wild-type enzyme. We conclude that the invariant glutamate of motif A is essential for the phosphohydrolase activity of cvRTP in vitro. The in vivo RNA cap-forming activity of the cvRTP-Mce1(211–597) fusion protein was also abolished by introducing an alanine in lieu of Glu-26 in motif A (Fig. 2). Thus, complementation of cet1Δ by cvRTP was contingent on its ability to hydrolyze the 5′ β-γ phosphoanhydride bond of RNA.
DISCUSSION
Our biochemical and genetic studies of cvRTP provide new insights into the evolution of the mRNA capping apparatus. The extensive sequence similarity between cvRTP and the catalytic domains of the S. cerevisiae, C. albicans, and S. pombe RTPs (especially the β strands that comprise the triphosphate tunnel), the similar catalytic repertoires of these enzymes in metal-dependent hydrolysis of triphosphate-terminated RNA and free nucleoside triphosphates, and the inactivation of all four proteins by alanine substitutions for the metal-binding glutamate of motif A suggest to us that the active site folds of the Chlorella virus and fungal RTPs are conserved as β barrels. The 193-aa cvRTP is significantly smaller than the smallest known fungal RTP—S. pombe Pct1p (303 aa)—and it is likely to represent the minimal functional unit of the yeastlike RTP family.
How is this minimization achieved? cvRTP lacks all of the structural elements flanking the catalytic domain of yeast Cet1p (including the Ceg1p-binding site and the Cet1p-Cet1p dimerization interface), and it is also missing one of the α helices found within the catalytic domain of Cet1p (see Fig. 1). The missing α-helix is located on a lateral surface of Cet1p (18), and its deletion in cvRTP would not pose a major problem in maintaining connectivity of the β4 and β5 strands in the tertiary structure modeled according to the sequence alignment shown in Fig. 1. The other three α-helices of Cet1p that are apparently conserved in cvRTP comprise the hydrophobic core that supports the “floor” of the triphosphate tunnel (18). Determination of a crystal structure for cvRTP, together with additional comparative mutational analyses, will provide important clues to how the unique tunnel architecture of the yeastlike RTPs evolved and whether the minimal cvRTP (like the minimal Chlorella virus guanylyltransferase) is a precursor of the larger capping enzymes of fungi.
The yeast and algal virus triphosphatases are clearly related to the RTP domains of the capping enzymes of metazoan DNA viruses. Together they comprise a family of metal-dependent phosphohydrolases with the signature ability to hydrolyze NTPs in the presence of manganese and cobalt. Results from mutational analysis of the vaccinia and baculovirus triphosphatases argue that motifs A and C are components of the metal-binding site (12, 14, 31), just as they are in yeast Cet1p. There are as yet no crystal structures for poxvirus or baculovirus capping enzymes—and our attempts at structure-based sequence alignments of the vaccinia virus or baculovirus capping enzymes to Cet1p failed to highlight conserved counterparts to β5, 6, 7, 8, and 10 strands of the Cet1p tunnel. Thus, it is possible that the active sites of the viral triphosphatases have a more open tertiary structure than do the fungal and Chlorella virus enzymes.
It is remarkable that cvRTP is more similar in its structure to the yeast RTPs than it is to the triphosphatase domains of the capping enzymes of the other large eukaryotic DNA viruses— poxviruses, ASFV, and baculoviruses. Indeed, PBCV-1 is unique among the DNA viruses in that the triphosphatase and guanylyltransferase reactions are catalyzed by separately encoded viral proteins rather than a single viral protein composed of multiple functional domains. Again, Chlorella virus is more akin to yeasts in its genetic separation of the triphosphatase and guanylyltransferase functions. The close relationship between the capping systems of yeasts and Chlorella virus may simply reflect the fact that the host alga for PBCV-1 is a unicellular eukaryote with a cell wall and is thus nearer in the evolutionary scheme to budding and fission yeasts than to the metazoan hosts for poxviruses, ASFV, and baculoviruses.
The triphosphatase and guanylyltransferase activities are linked in cis within a single polypeptide in the vaccinia virus, ASFV, and baculovirus capping enzymes. How might this have occurred? We envision a gene rearrangement event early in virus evolution, perhaps even prior to the emergence of metazoa (16), that fused an ancestral yeastlike metal-dependent triphosphatase to a guanylyltransferase to create the polyfunctional cap-forming proteins that we see today in metazoan DNA viruses. It is extremely unlikely that the poxvirus, ASFV, or baculovirus capping enzymes are derived from the capping apparatus of their metazoan host cells. Although all metazoan organisms examined to date do encode a bifunctional capping enzyme with an N-terminal RTP domain linked in cis to a C-terminal guanylyltransferase domain (9, 20, 28, 32), the metazoan RTPs are members of the cysteine phosphatase superfamily of metal-independent phosphohydrolases and are completely divergent in both structure and mechanism from the fungal/viral family of metal-dependent triphosphatases (18, 19, 30). Moreover, there are no discernible homologues of the fungal/viral RTPs in available metazoan proteomes. Because the only yeastlike RTPs extant in metazoans are those encoded by large DNA viruses such as poxviruses, ASFV, and baculoviruses, we surmise that the viral proteins are derived from ancestral capping enzymes predating the evolution of the present metazoan capping enzymes.
Does Chlorella virus encode the full ensemble of three cap-forming enzymes or does it rely on a host enzyme to catalyze cap methylation? The present study establishes that genetic complementation of yeast capping mutants by viral polypeptides fused to a mammalian delivery vehicle can be used to identify new viral cap-forming enzymes in advance of biochemical studies. We now anticipate applying the yeast complementation approach, guided where possible by phylogenetic insights, to search for the elusive methyltransferase component of a Chlorella virus capping apparatus.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho E J, Rodriguez C R, Takagi T, Buratowski S. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:3482–3487. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross C H, Shuman S. RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein. J Virol. 1998;72:10020–10028. doi: 10.1128/jvi.72.12.10020-10028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guarino L A, Jin J, Dong W. Guanylyltransferase activity of the LEF-4 subunit of baculovirus RNA polymerase. J Virol. 1998;72:10003–10010. doi: 10.1128/jvi.72.12.10003-10010.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Håkansson K, Doherty A J, Shuman S, Wigley D B. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 7.Ho C K, Van Etten J L, Shuman S. Expression and characterization of an RNA capping enzyme encoded by Chlorella virus PBCV-1. J Virol. 1996;70:6658–6664. doi: 10.1128/jvi.70.10.6658-6664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho C K, Pei Y, Shuman S. Yeast and viral RNA 5′ triphosphatases comprise a new nucleoside triphosphatase family. J Biol Chem. 1998;273:34151–34156. doi: 10.1074/jbc.273.51.34151. [DOI] [PubMed] [Google Scholar]
- 9.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 10.Ho C K, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 11.Ho C K, Lehman K, Shuman S. An essential surface motif (WAQKW) of yeast RNA triphosphatase mediates formation of the mRNA capping enzyme complex with RNA guanylyltransferase. Nucleic Acids Res. 1999;27:4671–4678. doi: 10.1093/nar/27.24.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho C K, Martins A, Shuman S. A yeast-based genetic system for functional analysis of viral mRNA capping enzymes. J Virol. 2000;74:5486–5494. doi: 10.1128/jvi.74.12.5486-5494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 14.Jin J, Dong W, Guarino L A. The LEF-4 subunit of baculovirus RNA polymerase has RNA 5′-triphosphatase and ATPase activities. J Virol. 1998;72:10011–10019. doi: 10.1128/jvi.72.12.10011-10019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komarnitsky P, Cho E, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen M, Gunge N, Meinhardt F. Kluyveromyces lactis killer plasmid pGKL2: evidence for a viral-like capping enzyme encoded by ORF 3. Plasmid. 1998;40:243–246. doi: 10.1006/plas.1998.1367. [DOI] [PubMed] [Google Scholar]
- 17.Lehman K, Schwer B, Ho C K, Rouzankina I, Shuman S. A conserved domain of yeast RNA triphosphatase flanking the catalytic core regulates self-association and interaction with the guanylyltransferase component of the mRNA capping apparatus. J Biol Chem. 1999;274:22668–22678. doi: 10.1074/jbc.274.32.22668. [DOI] [PubMed] [Google Scholar]
- 18.Lima C D, Wang L K, Shuman S. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- 19.Martins A, Shuman S. Mechanism of phosphoanhydride cleavage by baculovirus phosphatase. J Biol Chem. 2000;275:35070–35076. doi: 10.1074/jbc.M005748200. [DOI] [PubMed] [Google Scholar]
- 20.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myette J R, Niles E G. Domain structure of the vaccinia virus mRNA capping enzyme: expression in Escherichia coli of a subdomain possessing the RNA 5′-triphosphatase and guanylyltransferase activities and a kinetic comparison to the full-size enzyme. J Biol Chem. 1996;271:11936–11944. doi: 10.1074/jbc.271.20.11936. [DOI] [PubMed] [Google Scholar]
- 22.Pei Y, Ho C K, Schwer B, Shuman S. Mutational analyses of yeast RNA triphosphatases highlight a common mechanism of metal-dependent NTP hydrolysis and a means of targeting enzymes to pre-mRNAs in vivo by fusion to the guanylyltransferase component of the capping apparatus. J Biol Chem. 1999;274:28865–28874. doi: 10.1074/jbc.274.41.28865. [DOI] [PubMed] [Google Scholar]
- 23.Pei Y, Lehman K, Tian L, Shuman S. Characterization of Candida albicans RNA triphosphatase and mutational analysis of its active site. Nucleic Acids Res. 2000;28:1885–1892. doi: 10.1093/nar/28.9.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pena L, Yanez R J, Revilla Y, Vinuela E, Salas M L. African swine fever virus guanylyltransferase. Virology. 1993;193:319–328. doi: 10.1006/viro.1993.1128. [DOI] [PubMed] [Google Scholar]
- 25.Schroeder S C, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 27.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2000;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 28.Takagi T, Moore C R, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 29.Van Etten J L, Meints R H. Giant viruses infecting algae. Annu Rev Microbiol. 1999;53:447–494. doi: 10.1146/annurev.micro.53.1.447. [DOI] [PubMed] [Google Scholar]
- 30.Wen Y, Yue Z, Shatkin A J. Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism. Proc Natl Acad Sci USA. 1998;95:12226–12231. doi: 10.1073/pnas.95.21.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu L, Martins A, Deng L, Shuman S. Structure-function analysis of the triphosphatase component of vaccinia virus mRNA capping enzyme. J Virol. 1997;71:9837–9843. doi: 10.1128/jvi.71.12.9837-9843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]