Abstract
Penciclovir (PCV), an antiherpesvirus agent in the same class as acyclovir (ACV), is phosphorylated in herpes simplex virus (HSV)-infected cells by the viral thymidine kinase (TK). Resistance to ACV has been mapped to mutations within either the TK or the DNA polymerase gene. An identical activation pathway, the similarity in mode of action, and the invariant cross-resistance of TK-negative mutants argue that the mechanisms of resistance to PCV and ACV are likely to be analogous. A total of 48 HSV type 1 (HSV-1) and HSV-2 isolates were selected after passage in the presence of increasing concentrations of PCV or ACV in MRC-5 cells. Phenotypic analysis suggested these isolates were deficient in TK activity. Moreover, sequencing of the TK genes from ACV-selected mutants identified two homopolymeric G-C nucleotide stretches as putative hot spots, thereby confirming previous reports examining Acvr clinical isolates. Surprisingly, mutations identified in PCV-selected mutants were generally not in these regions but distributed throughout the TK gene and at similar frequencies of occurrence within A-T or G-C nucleotides, regardless of virus type. Furthermore, HSV-1 isolates selected in the presence of ACV commonly included frameshift mutations, while PCV-selected HSV-1 mutants contained mostly nonconservative amino acid changes. Data from this panel of laboratory isolates show that Pcvr mutants share cross-resistance and only limited sequence similarity with HSV mutants identified following ACV selection. Subtle differences between PCV and ACV in the interaction with viral TK or polymerase may account for the different spectra of genotypes observed for the two sets of mutants.
The introduction of penciclovir [PCV;9-(4-hydroxy-3-hydroxymethylbut-1-yl)guanine] and its prodrug, famciclovir, (FCV), resulted in the use of antivirals alternative to acyclovir (ACV) for treatment of herpes simplex virus (HSV) infections. Biochemical studies have indicated that PCV, like ACV, is phosphorylated by the viral thymidine kinase (TK) to a monophosphate and subsequently converted by cellular enzymes to a triphosphate, which inhibits the HSV DNA polymerase (Pol) (44). Although PCV and ACV have identical activation pathways and similar modes of action (14, 44), and the frequencies with which resistance in HSV arises to PCV and ACV in cell culture are identical (36), the affinities and therefore the fine molecular interactions of PCV, ACV, and their triphosphates with TK and Pol differ (14). The last point raises the possibility that drug-resistant mutants selected by these antiviral agents may differ.
Resistance to acyclovir typically arises by a single mutation in either the TK or Pol gene (11, 23, 29). The viral TK, unlike DNA polymerase, is not essential for virus replication in cell culture (13), although in vivo analyses implicate it in HSV virulence, pathogenicity, and reactivation from latency (9, 15, 20, 41). Mutations in HSV TK are the most common causes of clinical resistance to ACV (7, 34), and the majority of mutants completely lack TK activity (TK−). TK− variants are invariably cross resistant to PCV and ACV because these antivirals share a dependence upon the viral TK for phosphorylation (3, 4). Missense point mutations and single-base deletions or insertions which shift the translational reading frame of the protein generally confer this phenotype (11, 23, 29).
HSV isolates, whether from patients or cell culture, are heterogeneous populations and thus contain preexisting drug-resistant TK variants (six to eight mutants per 104 plaque-forming viruses) (12, 31, 36). The infidelity of the HSV DNA replication process is directly responsible for this naturally occurring variation (17, 24, 25, 28), with errors in the viral DNA introduced spontaneously during DNA replication and not requiring the presence of drug. However, exposure to a nucleoside analog may provide selective pressure leading to the enrichment of such preexisting drug-resistant viruses. Since ACV-selected mutants derived in cell culture have been partially predictive of those which have emerged from the clinic, an in vitro examination of PCV-selected HSV should further understanding of the selection process for clinically PCV-resistant HSV.
To address whether viruses selected for resistance to PCV and ACV are similar, a series of HSV type 1 (HSV-1) and HSV-2 mutants from a single virus preparation (either HSV-1 SC16 or HSV-2 SB5) were selected in vitro with PCV or ACV in MRC-5 cells. Classically, ACV-resistant HSV mutants have been selected by serial passage in the presence of increasing concentrations of antiviral (11), and this approach was used in the present study. A comparison of the phenotypes, genotypes, and biochemical properties of mutants selected in vitro for PCV or ACV resistance is presented in this report.
MATERIALS AND METHODS
Cell lines and virus strains.
Vero (American Type Culture Collection [ATCC]), an African green monkey kidney cell line; MRC-5 (ATCC), a diploid human embryonic lung cell line; and 143 TK− (ATCC), a human osteosarcoma cell line, were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS) and incubated at 37°C and 5% CO2. HSV-1 SC16, SC16-S1, and DM21 were generously provided by S. Safrin (Gilead Sciences, Foster City, Calif.). HSV-2 SB5 (ATCC VR-2546) is a plaque-purified derivative of HSV-2 strain 333. D21 cells, a line derived from BUHK-TK cells which constitutively expresses an HSV TK gene, were a kind gift from H. Field (University of Cambridge, Cambridge, United Kingdom). These transformed cells were maintained in DMEM with 10% FCS and HAT supplement (hypoxanthine, aminopterin, and thymidine).
Compounds.
PCV (BRL 39123) was synthesized at SmithKline Beecham Pharmaceuticals. ACV, vidarabine (Ara-A), bromovinyl-deoxyuridine (BVDU), iodo-deoxyuridine (IdU), and foscarnet (FOS) were obtained from Sigma Chemical Co. (St. Louis, Mo.). Cidofovir (CDU; GS-504) was generously provided by J. Smith (Gilead Sciences). For cell culture assays, 5- to 40-mg/ml stock solutions were prepared in dimethyl sulfoxide or sterile water and stored at −20°C.
Selection of resistant isolates.
Confluent MRC-5 cell monolayers (seeded with 7 × 105 cells/well) prepared in six-well dishes were infected with 0.1 PFU of SC16 or SB5/cell in 0.5 ml of Hanks balanced salt solution for 1 h at 37°C. The inoculum was then removed, and 2 ml of medium containing 1 μg of PCV or ACV per ml was added per well. When a culture demonstrated complete cytopathic effect, typically after 3 days, it was freeze-thawed three times (pass 1). Next, 0.5 ml of the resulting virus pool was passaged in the presence of a threefold-higher concentration of antiviral (pass 2). The pass 2 virus samples were then passaged in the presence of an approximately threefold-higher concentration of antiviral (10 μg/ml) to produce the final pass 3 samples. The pass 3 virus pools were titrated, and single plaque isolates were purified by limiting dilution from each pool. Virus from a single plaque was amplified in Vero cells. Following three rounds of plaque purification to ensure homogeneity, virus stocks were prepared in Vero cells which were infected at 0.01 PFU/cell. The stocks were titrated in Vero cells, and 50% inhibitory concentrations (IC50s) were determined by plaque reduction assay (PRA) in MRC-5 cells as described below.
PRA.
PRAs were performed according to the method detailed previously (36). Briefly, fourfold dilutions of ACV, PCV, CDV, and IdU were tested, ranging from 0.09 to 100 μg/ml for known resistant control strains (SC16-S1) or from 0.02 to 25 μg/ml for wild-type sensitive strains (SC16 and SB5). For all viruses, four-fold dilutions of FOS and Ara-A were tested, ranging from 1.56 to 400 μg/ml, and for BVDU, concentrations ranged either from 0.05 to 50.0 μg/ml (HSV-2) or from 0.01 to 10.0 μg/ml (HSV-1). The cultures were incubated for 48 h, fixed with 10% formaldehyde, and finally stained with crystal violet (0.5% [wt/vol] in 70% methanol). The plaques were counted, and IC50s were calculated by the Kärber method (21). The criterion used to define an HSV-1 SC16 or HSV-2 SB5 variant as resistant was an IC50 greater than 10-fold above that for the parental wild-type virus tested in the same assay (36). The mean IC50s from duplicate tests are presented in the tables, and the relative resistances of an isolate in different tests remained similar.
Viral DNA isolation and subcloning.
Confluent Vero cell monolayers (seeded with 106 cells/100-mm-diameter dish) were infected with HSV mutants at 5 PFU/cell. Approximately 20 h after infection, the cells were washed gently with phosphate-buffered saline and then treated with 4.0 ml of lysis buffer (1% sodium dodecyl sulfate–1% Sarkosyl in 10 mM Tris [pH 8.0], 2 mM EDTA). The cells were scraped into the lysis buffer, and 200 μl of RNaseA (10-mg/ml stock) was added. After 1 h at 37°C, 200 μl of proteinase K (2-mg/ml stock) was added, and the mixture was incubated for an additional 2 h at 42°C. Two phenol-chloroform extractions were performed prior to DNA precipitation. The infected cell DNA (20 μg) was digested with BamHI (SC16 derivatives) or HindIII/EcoRI (SB5 derivatives) to allow subcloning of a fragment of viral DNA containing the TK open reading frame into a modified derivative of pSG5 (Stratagene). For some samples, the TK gene was amplified by PCR using Pfu DNA polymerase (Stratagene), and the coding region was directly sequenced in both directions.
Sequencing.
HSV-1 SC16 and HSV-2 SB5 subclones containing a 3.6- or 3.5-kb fragment, respectively, were evaluated by PCR to verify that they contained the TK open reading frame. Positive clones were subjected to terminator cycle sequencing using an automated model 377 DNA sequencer (Perkin-Elmer Applied Biosystems). The primers utilized for HSV-1 SC16 derivatives were as follows: SBA9188, GGCATAAGGCATGCCCATTG; SBA9189, CAATCGCGAACATCTACACC; and SBA9190, GCTTGACCTGGCTATGCTG. The primers utilized for HSV-2 SB5 derivatives were as follows: SBB1165, GCGGTGGTAATGACCAGCGC; SBB1166, CCAACACGGTGCGGTACCTG; SBB1167, CAGGGAGGCGATAGGGTGCC; SBB1168, GTCATGCTTCCCATGAGGTACC. The overlap between sequencing runs was evaluated to allow for the identification of mutations within the primer sequence.
Antibodies and Western analysis.
Rabbit polyclonal antiserum raised to a glutathione S-transferase–HSV TK fusion protein was generously provided by S. Albelda (University of Pennsylvania, Philadelphia). This antiserum cross-reacts with both HSV-1 and HSV-2 TKs. Confluent MRC-5 cell monolayers in 12-well plates were infected at 5 PFU/cell in 500 μl of Hanks balanced salt solution at 37°C and 5% CO2 for 1 h. Following adsorption, the inoculum was removed and replaced with 1.0 ml of medium (DMEM plus 5% FCS). Eight hours postinfection, the medium was removed, the cell monolayers were rinsed with phosphate buffered saline, and the viral proteins were harvested in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. Equal volumes of protein were loaded in all wells and membranes were treated according to the manufacturer's recommendations (ECL; Amersham Life Science).
TK assay.
Viral TK activity was determined by a modification of the method described by Coen et al. (9) performed as reported previously (36).
Plaque autoradiography.
Plaque autoradiography was performed according to the method described by Tenser et al. (42). The radiolabeled cells were fixed with 10% formaldehyde, stained with crystal violet, air dried, and placed in contact with X-ray film for 5 days at room temperature.
RESULTS
Selection of antiviral-resistant HSV.
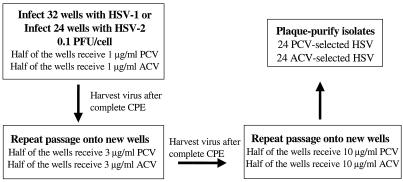
Drug-resistant mutants were randomly selected from a parental virus preparation of two wild-type laboratory HSV strains, HSV-1 SC16 and HSV-2 SB5, by three serial passages of the virus in MRC-5 cells treated with escalating concentrations of PCV or ACV as outlined in Fig. 1. From the resulting 56 virus pools, a total of 48 viruses were plaque purified three times: 13 SC16 and 11 SB5 mutants were selected in the presence of increasing concentrations of PCV, and 15 SC16 and 9 SB5 mutants were selected with increasing concentrations of ACV. The naming convention used for plaque isolates is based on their derivation, that is, virus type, drug selection, and plaque number (e.g., 1P1 is an HSV-1 isolate selected for resistance to PCV, plaque isolate number 1; 2A1 is an HSV-2 isolate selected for resistance to ACV, plaque isolate number 1). In three instances, two plaques were purified from the same well, and these viruses are designated with the letters A and B (e.g., 1P3-A and 1P3-B represent two plaque isolates of HSV-1 selected for resistance to PCV from the same experimental well).
FIG. 1.
Flow diagram illustrating methodology for selecting drug-resistant HSV-1. A stock of wild-type HSV-1 (containing approximately 5,000 total mutants [36]) was used to initiate 32 individual infections. The infection represents an input of approximately 40 mutant viruses per experimental well or 1,280 total variants within the 32 infections to select drug-resistant virus. The same methodology was followed using a stock of wild-type HSV-2 (containing approximately 2.1 × 106 total mutants [36]) to initiate 24 individual infections, which represents an input of approximately 490 mutant viruses per experimental well or 11,760 total variants within the 24 infections to select drug-resistant virus. CPE, cytopathic effect. Eleven PCV-selected and 9 ACV-selected HSV-2 isolates were plaque purified.
Antiviral susceptibility of drug-selected HSV-1 isolates.
The susceptibilities of the 28 drug-selected HSV-1 isolates and the parental HSV-1 strain (SC16) to PCV and ACV were determined by PRA in MRC-5 cells (Tables 1 and 2). PCV, ACV, and BVDU all rely on viral TK expression for initial phosphorylation, although BVDU diphosphorylation is also directed by the viral TK (3, 16, 44). Generally, TK− strains are resistant to these compounds yet remain sensitive to FOS, Ara-A, and CDV, which inhibit HSV replication independent of the viral TK. The susceptibilities of the drug-selected HSV-1 isolates to non-TK-dependent antivirals were determined to identify potential DNA polymerase mutants or TK-polymerase double mutants.
TABLE 1.
Susceptiblities of PCV-selected HSV-1 in MRC-5 cells
| SC16 isolateb | IC50 (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|
| ACV (0.2) | PCV (0.3) | BVDU (<0.01) | FOS (29.1) | Ara-A (1.5) | CDV (0.3) | |
| 1P1 | >100 | >100 | 12.6 | 31.8 | 8.3 | 0.6 |
| 1P2 | >100 | >100 | >50.0 | 20.7 | 8.1 | 0.7 |
| 1P3-A | 62.1 | 55.4 | >10.0 | 45.0 | 1.6 | 0.4 |
| 1P3-B | 48.4 | 54.4 | >50.0 | 81.3 | 1.9 | 0.1 |
| 1P4 | >100 | 39.2 | 10.9 | 70.0 | 3.3 | 0.1 |
| 1P5 | 20.0 | >100 | 8.4 | 10.0 | 0.5 | 0.1 |
| 1P6 | >100 | >100 | 16.7 | 8.3 | 4.3 | 0.4 |
| 1P7 | >100 | >100 | 33.2 | 11.6 | 4.6 | 0.5 |
| 1P8 | >100 | 62.1 | 27.5 | 84.0 | 2.1 | 0.5 |
| 1P9 | 88.4 | 58.8 | 23.0 | 43.1 | 1.6 | 0.7 |
| 1P10 | >100 | >100 | 25.5 | 52.1 | 1.9 | 0.4 |
| 1P11 | 0.11 | 0.4 | <0.01 | 38.8 | 1.2 | 0.4 |
| 1P12 | >100 | >100 | 28.0 | 25.2 | 4.6 | 0.5 |
Susceptibility data for wild-type HSV-1 SC16 are from the initial virus preparation used for these studies and are listed in parentheses.
Each plaque isolate was purified from a unique starting well, except 1P3-A and 1P3-B, which are separate plaque isolates purified from the same well.
TABLE 2.
Susceptiblities of ACV-selected HSV-1 in MRC-5 cells
| SC16 isolateb | IC50 (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|
| ACV (0.2) | PCV (0.3) | BVDU (<0.01) | FOS (29.1) | Ara-A (1.5) | CDV (0.3) | |
| 1A1 | >100 | >100 | >10.0 | 29.1 | 2.3 | 0.5 |
| 1A2 | >100 | >100 | >10.0 | 36.5 | 4.0 | 0.6 |
| 1A3 | 56.0 | 55.6 | 17.4 | 7.8 | 1.4 | 0.4 |
| 1A4 | 36.0 | >100 | >10.0 | 8.0 | 4.1 | 0.6 |
| 1A5 | >100 | >100 | 26.9 | 30.7 | 2.9 | 0.2 |
| 1A6 | 26.3 | >100 | >10.0 | 7.6 | 2.4 | 0.4 |
| 1A7 | 22.4 | 29.1 | >10.0 | 50.6 | 3.9 | 1.0 |
| 1A8 | >100 | 23.5 | >10.0 | 30.8 | 3.5 | 0.6 |
| 1A9 | 52.5 | 37.8 | >10.0 | 6.9 | 2.3 | 0.6 |
| 1A10 | 0.2 | 0.4 | <0.01 | 21.0 | 1.1 | 0.3 |
| 1A11-A | 20.4 | 8.5 | >10.0 | 35.8 | 2.5 | 0.5 |
| 1A11-B | 17.3 | 24.0 | 12.2 | 10.0 | 2.6 | 0.2 |
| 1A12 | 25.8 | >100 | >10.0 | 14.8 | 4.5 | 0.4 |
| 1A13 | 60.4 | 37.5 | >10.0 | 23.0 | 6.7 | 0.4 |
| 1A14 | 19.4 | 47.8 | >10.0 | 36.4 | 2.8 | 0.8 |
Susceptibility data for wild-type HSV-1 SC16 are from the initial virus preparation used for these studies and are listed in parentheses.
Each plaque isolate was purified from a unique starting well, except 1A11-A and 1A11-B, which are separate plaque isolates purified from the same well.
A single HSV-1 isolate purified after passage in the presence of increasing PCV remained sensitive to PCV by PRA (Table 1, isolate 1P11), whereas resistance to PCV was confirmed for the remaining 12 isolates. Furthermore, one HSV-1 isolate (Table 2, isolate 1A10) selected after passage with increasing concentrations of ACV remained susceptible to ACV, although the other 14 isolates were ACV resistant. The criterion set to classify an isolate as resistant when using PCV or ACV was an IC50 for it greater than 10-fold above that for the parental wild-type virus tested in the same assay (hence, a PCV IC50 of ≥3.0 μg/ml or an ACV IC50 of ≥2.0 μg/ml). The IC50s for the two susceptible HSV-1 isolates (1P11 [PCV IC50 = 0.40 and ACV IC50 = 0.11 μg/ml] and 1A10 [PCV IC50 = 0.40 and ACV IC50 = 0.20 μg/ml]) were well below this criterion, whereas the IC50s for the authentic PCV- or ACV-resistant viruses were greater than 8 μg of PCV/ml or 17 μg of ACV/ml (Tables 1 and 2, respectively).
All confirmed resistant HSV-1 isolates selected in the presence of either PCV or ACV were cross resistant to both PCV and ACV. Although cross-resistance occurred regardless of the antiviral used in the selection process, 0.5- to 5-fold differences in susceptibility were evident. For example, the IC50 of PCV against isolate 1P5 was fivefold higher than the IC50 of ACV; nonetheless, isolate 1P5 is clearly resistant to both agents (Table 1). Additionally, these isolates were examined for susceptibility to BVDU. BVDU is a potent and selective inhibitor of HSV-1, although wild-type HSV-2 strains are markedly less susceptible to it (16). All PCV- or ACV-selected isolates which demonstrated resistance to those agents were also cross resistant to BVDU (all BVDU IC50s for HSV-1 were >8.0 μg/ml).
No DNA polymerase mutants or TK-Pol double mutants were identified among the panel of resistant HSV-1 isolates, since all were susceptible to the non-TK-dependent antivirals FOS, CDV, and Ara-A by PRA (Tables 1 and 2). However, since screening with these compounds alone is not sufficient to guarantee the identification of such mutants, a second test was used. As described below, D21 cells, a cell line expressing the HSV TK, were used to facilitate the identification of Pol mutants.
Antiviral susceptibility of drug-selected HSV-2 isolates.
The susceptibilities of the 20 drug-selected HSV-2 isolates as well as the parental HSV-2 strain (SB5) to PCV and ACV were determined by PRA in MRC-5 cells (Tables 3 and 4). Although two HSV-2 isolates purified after three serial passages in the presence of increasing PCV retained sensitivity (Table 3, isolates 2P3-A and 2P3-B), resistance to PCV was confirmed for the remaining 10 isolates. Additionally, a single HSV-2 isolate (Table 4, isolate 2A1) selected after three serial passages in MRC-5 cells remained susceptible to ACV, although the other eight isolates were confirmed to be ACV resistant. The IC50s for the three drug-susceptible HSV-2 isolates, 2P3-A, 2P3-B, and 2A1, were well below the criterion used to define resistance (10 times the IC50 for the parental wild-type virus, i.e., a PCV IC50 of ≥7.5 μg/ml or an ACV IC50 of ≥5.0 μg/ml). The IC50s for all other HSV-2 isolates were greater than 45 (PCV) or 26 (ACV) μg/ml. Isolate 2A6 remained a potential mixture of two virus populations (the wild-type and a mutant virus, expressing a full-length and truncated TK protein, respectively) even after three rounds of plaque purification and was not further characterized (Fig. 2).
TABLE 3.
Susceptiblities of PCV-selected HSV-2 in MRC-5 cells
| SB5 isolateb | IC50 (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|
| ACV (0.5) | PCV (0.75) | BVDU (8.1) | FOS (35.0) | Ara-A (1.8) | CDU (0.7) | |
| 2P1 | 80.6 | >100 | 21.2 | 14.4 | 6.5 | 0.5 |
| 2P2 | >100 | >100 | >50 | 40.5 | 2.0 | 0.9 |
| 2P3-A | 0.11 | 0.4 | 4.0 | 25.0 | 2.0 | 0.7 |
| 2P3-B | 0.11 | 0.4 | 4.5 | 18.5 | 2.1 | 0.5 |
| 2P4 | 67.0 | >100 | 26.1 | 21.2 | 0.9 | 1.8 |
| 2P5 | 26.0 | 63.0 | 18.2 | 24.5 | 2.6 | 2.9 |
| 2P6 | 60.0 | 45.0 | >10.0 | 40.5 | 2.5 | 0.5 |
| 2P7 | 50.0 | >100 | >10.0 | 10 | 1.0 | 0.3 |
| 2P8 | >100 | 68.4 | >50 | 48.2 | 4.3 | 2.2 |
| 2P9 | 29.8 | >100 | 45.0 | 32.3 | 2.8 | 3.0 |
| 2P10 | >100 | 60.7 | >10.0 | >400 | 7.8 | 0.5 |
Susceptibility data for wild-type HSV-2 SB5 are from the initial virus preparation used for these studies and are listed in parentheses.
Each plaque isolate was purified from a unique starting well, except 2P3-A and 2P3-B, which are separate plaque isolates purified from the same well.
TABLE 4.
Susceptiblities of ACV-selected HSV-2 in MRC-5 cells
| SB5 isolateb | IC50 (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|
| ACV (0.5) | PCV (0.75) | BVDU (8.1) | FOS (35.0) | Ara-A (1.8) | CDV (0.7) | |
| 2A1 | 0.8 | 1.2 | 5.9 | 26.6 | 1.4 | 1.1 |
| 2A2 | >100 | >100 | 12.4 | 28.7 | 6.2 | 0.7 |
| 2A3 | >100 | >100 | 12.6 | 119.3 | 2.4 | 2.0 |
| 2A4 | >100 | >100 | >10.0 | 50.0 | 2.0 | 1.0 |
| 2A5 | >100 | >100 | 16.4 | 18.9 | 3.6 | 1.1 |
| 2A6 | >100 | >100 | NTc | NT | NT | NT |
| 2A7 | >100 | >100 | >10.0 | 42.0 | 3.3 | 1.5 |
| 2A8 | 65.2 | >100 | >50.0 | 10.0 | 2.1 | 1.7 |
| 2A9 | 75.0 | 86.1 | 25.0 | 23 | 1.9 | 1.2 |
Susceptibility data for wild type HSV-2 SB5 are from the initial virus preparation used for these studies and are listed in parentheses.
Each plaque isolate was purified from a unique starting well.
NT, not tested.
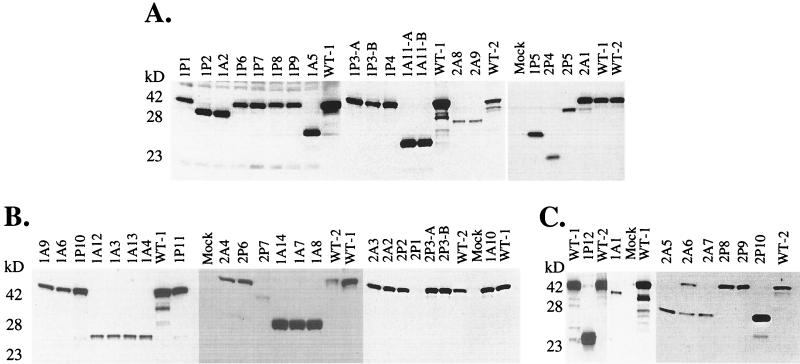
FIG. 2.
Western blot analysis of TK protein products from HSV plaque isolates. The full-length TK protein product is approximately 43 kDa. Individual blots of HSV plaque isolates (A to C) are shown. The name of the plaque isolate (described in Materials and Methods) is indicated at the top of each lane. WT-1, wild-type HSV-1 SC16; WT-2, wild-type HSV-2 SB5; Mock, mock infected.
All drug-resistant HSV-2 viruses selected with either PCV or ACV were also cross resistant to both agents. Although cross-resistance occurred regardless of the antiviral used in the selection process, differences among the IC50s were less marked (two- to threefold) than those among the IC50s for HSV-1 isolates. The panel of HSV-2 isolates selected for resistance to ACV (Table 4) were cross resistant to PCV without marked differences among the IC50s (all were >100 μg of PCV or ACV/ml). Resistance to BVDU was apparent, although less clearly defined than with the HSV-1 isolates, and was consistent with previous observations (16). Furthermore, screening the panel of HSV-2 isolates against DNA polymerase inhibitors in a PRA resulted in the identification of an HSV mutant which may contain a DNA polymerase mutation. HSV-2 isolate 2P10 was resistant to FOS, with an IC50 greater than 400 μg/ml (Table 3), suggesting this isolate may be a double mutant in both the TK and DNA polymerase genes. Notably, this virus retained susceptibility to the other DNA polymerase inhibitors, Ara-A and CDV.
Biochemical characterization of drug-selected HSV isolates.
The panel of resistant HSV-1 SC16 and HSV-2 SB5 progeny was examined by Western blotting for viral TK gene expression. Total cell lysates following HSV infection were probed with antiserum specific for the viral TK polypeptide (Fig. 2A through C). SC16 and SB5, the wild-type parental viruses, produced a full-length 43-kDa protein which reacted with TK antiserum. Plaque isolates containing frameshift mutations would be expected to express truncated TK or extended products, although PCV and ACV resistance do not demand the synthesis of such TK proteins.
The majority of PCV-selected drug-resistant HSV-1 isolates (9 of 12) generated full-length TK polypeptides (Fig. 2). However, the ACV-selected HSV-1 SC16 isolates in this assay generally expressed a truncated TK polypeptide (13 to 14). Conversely, among the HSV-2 SB5 isolates, PCV selection and ACV selection both resulted in a more even distribution of isolates expressing either full-length or truncated TK gene products. The exception was HSV-2 isolate 2P1, which did not express a detectable TK product with the polyclonal TK antiserum (Fig. 2B), although a comparable amount of the viral DNA polymerase protein relative to the wild-type parental control virus was evident (data not shown). The variation in the amount of TK product detected among samples (for example, Fig. 2B, isolates 2P6, 2P7, and 1A14) cannot be attributed to differences in TK expression. Parallel blots using antiserum against the viral DNA Pol illustrated similar differences across isolates, suggesting that the apparent variation in TK protein between isolates is the result of differences in total protein loaded or a variation in infection (data not shown).
Since most Acvr mutants characterized to date are TK− the TK activities of the PCV- and ACV-selected isolates were evaluated. Mock-infected TK− human osteosarcoma 143 cells or extracts of cells infected with the known TK− deletion mutant, HSV-1 DM21 (15), were below the limit of detection of the TK assay (<0.3% TK activity). Extracts of cells infected with the parental wild-type HSV-1 SC16 or HSV-2 SB5 exhibited high TK activity, defined as 100%. All drug-resistant HSV plaque isolates produced little or no TK enzymatic activity in this assay, with values ranging from <0.3 to 7%. Isolate HSV-1 SC16S1 (10), an Acvr Pcvr strain of SC16 which expresses an altered TK, produced 13 to 20% TK activity in this assay. Hence, all plaque isolates examined in this report are most likely TK−, consistent with the majority of Acvr viruses reported to date. However, the important distinction between TK− and TK-partial remains difficult to distinguish from in vitro TK assays alone.
Lastly, the TK polypeptide expressed from mutant 2P10 was truncated (∼28 kDa) relative to the full-length product. This multi-drug-resistant virus therefore most likely accumulated independent genetic lesions within both the TK coding sequence and the DNA polymerase coding sequence to confer resistance to both FOS and the TK-dependent antiviral agents.
Plaque autoradiography of drug-selected HSV isolates.
Vero cell monolayers infected with the panel of HSV isolates were exposed to [125I]iododeoxycytidine in order to evaluate the phosphorylation of this substrate and its subsequent incorporation into viral DNA. Autoradiographs of wild-type virus (SC16 and SB5) yielded a black-rimmed plaque due to strong incorporation of radiolabel into the replicating HSV DNA, whereas the entire panel of drug-resistant viruses failed to incorporate the substrate. Furthermore, all isolates were also severely impaired relative to the parental virus in the ability to phosphorylate IdU, as judged by the antiviral activity of IdU in a further PRA (data not shown), since all IdU IC50s were greater than 30.0 μg/ml (IdU IC50s for HSV-1 SC16 and HSV-2 SB5 were 1.9 and 3.4 μg/ml, respectively). These results further support the notion that all of the mutants studied are TK−.
DNA sequence analysis of drug-selected HSV isolates.
The HSV TK coding region is 1,128 nucleotides in length and encodes a protein of 376 amino acids. The proposed ATP and nucleoside binding sites are defined by amino acids 49 to 66 and 162 to 178, respectively (22). The TK genes of the panel of antiviral-resistant HSV isolates were sequenced, and comparisons of the nucleotide and amino acid changes, as well as the predicted polypeptide sizes, are shown in Tables 5 through 8. The sequencing data are in complete agreement with results from Western analysis.
TABLE 5.
TK mutations identified in PCV-selected HSV-1 SC16
| Clonal virus | Mutationa
|
Predicted length (aa)b | |
|---|---|---|---|
| Nucleotide | Amino acid | ||
| 1P1 | C424T; C860T | H142Y; T287M | 376 |
| 1P2 | T37C; ΔC553 | F13L; FS 185 | 346 |
| 1P3-A | G16T | G6C | 376 |
| 1P3-B | G559A; T614C | G200D; L205S | 376 |
| 1P4 | G16T | G6C | 376 |
| 1P5 | +C554 | FS 185 | 227 |
| 1P6 | C860T | T287M | 376 |
| 1P7 | +C700 | FS 234 | 375 |
| 1P8 | G559A; T800G | G200D; V267G | 376 |
| 1P9 | G559A | G200D | 376 |
| 1P10 | +T929 | FS 310 | 377 |
| 1P12 | +T364 | FS 122 | 227 |
The designation C424T represents a change from the wild-type nucleotide C to a T at base 424; the others follow this pattern. Δ, base deletion; +, base insertion; FS, frameshift in the coding sequence.
aa, amino acids.
TABLE 8.
TK mutations identified in Acvr HSV-2 SB5 plaque isolates
| Clonal virus | Mutationa
|
Predicted length (aa)b | |
|---|---|---|---|
| Nucleotide | Amino acid | ||
| 2A2 | +G775 | FS 259 | 390 |
| 2A3 | T206C; T514C | L69P; C172R | 376 |
| 2A4 | +T9; ΔC25; C863T | FS 3, 9; T288M | 376 |
| 2A5 | +C556 | FS 186 | 228 |
| 2A7 | +C556 | FS 186 | 228 |
| 2A8 | +C467; +C556 | FS 156, 186 | 263 |
| 2A9 | ΔC43; ΔG137; +C556 | FS 15, 46, 156 | 262 |
The designation T206C represents a change from the wild-type nucleotide T to a C at base 206. The others follow this pattern. Δ, base deletion; +, base insertion; FS, frameshift in the coding sequence.
aa, amino acids.
Most HSV-1 isolates selected with ACV contained frameshift mutations, thereby altering the coding sequence and termination site of the TK product. The majority of PCV-selected HSV-1 isolates contained single or double point mutations, resulting in an amino acid change (Table 5). Although TK sequence differences were mostly unique to individual isolates, for viruses selected with PCV, two changes appeared more than once (nucleotide 559, G to A; nucleotide 860, C to T). Additionally, a large number of mutations were apparent more than once for HSV-1 isolates selected with ACV (nucleotide 13, C to G; nucleotide 16, G to T; nucleotide 437, plus G; nucleotide 860, C to T). Two identical changes were observed in both PCV- and ACV-selected isolates: nucleotide 16, G to T, and nucleotide 860, C to T.
For PCV-selected HSV-2 isolates, the transition at nucleotide 863 (C to T) was present in three isolates, and the change from G to A at nucleotide 116 was present in two isolates. The ACV-selected HSV-2 isolates often (four out of seven) contained a frameshift mutation at nucleotide 556 (plus C), and a single isolate was also observed to contain the change at nucleotide 863 (C to T).
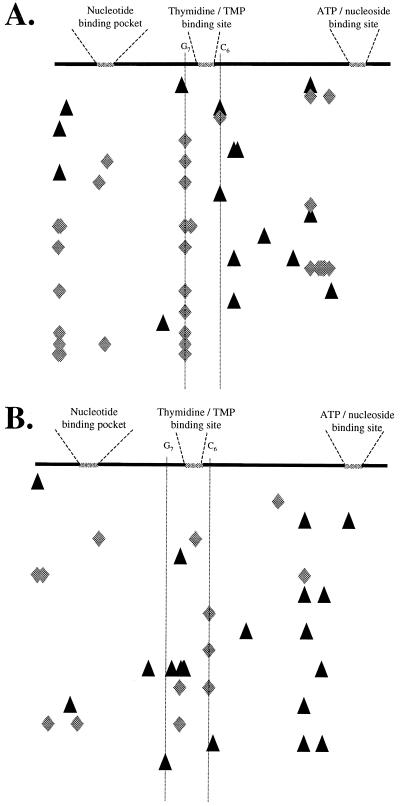
Alignment of the mutations in the TK coding sequence for HSV-1 and HSV-2 isolates illustrates that genetic lesions found in ACV-selected isolates generally accumulate in two distinct homopolymeric nucleotide stretches within the TK gene (Fig. 3, G7 and C6 stretches). Surprisingly, mutations identified among isolates selected with PCV were generally not in these homopolymeric regions. Mutations within PCV-selected isolates encoding nonfunctional TK proteins were randomly distributed throughout the TK gene and generally equally associated with A-T nucleotides or G-C nucleotides (15 mutations at A-T nucleotides out of 37 total [A-T and G-C] changes), whereas ACV-selected isolates contained 7 mutations at A-T nucleotides out of 46 total changes. Lastly, although PCV-selected mutations are not concentrated in the homopolymeric Acvr hot spots, PCV-selected variants across virus types may be preferentially located within the predicted alpha-helical region immediately upstream of the ATP-nucleoside binding site (Fig. 3).
FIG. 3.
Alignment of mutations within TK coding sequence. The schematic representations depict the HSV-1 (A) and HSV-2 (B) TK polypeptides and three conserved domains, the nucleotide binding pocket, the thymidine binding site, and the ATP binding site. The homopolymeric hot-spot regions are indicated (G7 and C6). Below the cartoon, the locations of base changes for Pcvr (triangles) or Acvr (diamonds) HSV-1 or HSV-2 are indicated. Each row represents a unique HSV drug-resistant clone.
The HSV-2 mutant 2P10 carries a single-nucleotide deletion within the TK gene, which results in expression of a truncated TK polypeptide. This incomplete TK gene product most likely is responsible for conferring resistance to PCV and ACV, although a mutation within the TK coding region is not likely to account for FOS resistance. To address this, the sequence of the 2P10 Pol coding region will need to be determined and recombinant viruses will have to be generated to assess whether the change(s) compared to wild-type HSV-2 SB5 further contributes to PCV-ACV and/or FOS resistance. However, a PRA in cells constitutively expressing the HSV TK polypeptide can provide an immediate indication as to whether the Pol contains a mutation(s) that directly impacts TK-dependent nucleoside analog resistance.
Antiviral susceptibility of PCV-selected HSV isolates in TK-transformed cells.
The entire panel of HSV mutants described here has been tested in D21 cells, which constitutively express the HSV TK product, for susceptibility to PCV. All PCV IC50s, excluding that for HSV mutant 2P10, were below 0.11 μg/ml, inconsistent with the isolates having DNA polymerase mutations which confer PCV resistance and consistent with rescue of their TK− phenotype. Although the PCV IC50 for HSV-2 2P10 in D21 cells was only 0.5 μg/ml (compared to a PCV IC50 of 60.7 μg/ml in MRC-5 cells), the PCV IC50 remained significantly higher than the values attained for the parental HSV-2 SB5 and the remaining HSV-2 mutant isolates. It remains to be determined whether a single or multiple nucleotide changes were responsible for the Fosr phenotype of 2P10.
DISCUSSION
Studies of drug resistance in laboratory herpesviruses have furthered understanding of antiviral mechanisms, viral enzyme structure-function, and clinical drug resistance (8, 26). This report represents the largest laboratory collection of PCV- and ACV-resistant HSV characterized to date, a total of 48 isolates. Resistance to ACV has been previously mapped to mutations within the TK and DNA polymerase genes for both clinical and laboratory HSV isolates (11, 23, 29). The mechanisms of HSV resistance to PCV and ACV are expected to be analogous, given the similarity between the two agents (4, 44), and indeed, all of the 48 plaque isolates described here are deficient in TK activity.
Darby et al. (11) and Nugier et al. (29) noted that mutations in the nucleoside binding site (amino acids 161 to 192) are frequently observed in response to selective pressure of ACV, and nine mutations in this study were found in that region. Other mutations identified within our drug-selected mutants were clustered near the nucleotide binding site (amino acids 50 to 63), as well as upstream of the C-terminal ATP-nucleoside binding region (Fig. 3). The highly conserved (2) glutamic acid at residue 226, which the TK crystal structure implicates in the formation of the nucleoside binding pocket (5), was found to be mutated in plaque isolate 2P6. It is unclear whether the change at residue 226 or the additional frameshift at residue 290 was initially responsible for the resistance phenotype of 2P6. The majority of mutants examined in this study contained more than one lesion within the TK gene, with few instances of identical mutations shared between independent isolates. Examples are isolates 1A4, 1A5, and 1A13 (selected by ACV), which all contained a frameshift at amino acid 146 and other, nonidentical changes adjacent to the nucleoside binding site. Given that these plaque isolates were selected by sequential passage in the presence of ACV, the exact mutation initially relevant to the establishment of the TK− phenotype cannot be identified with certainty.
HSV isolates selected in the presence of ACV (19 of 21) and HSV-2 isolates selected for PCV resistance (8 of 9) commonly included frameshift mutations which disrupted both the thymidine and ATP binding pockets of TK. However, the PCV-selected HSV-1 mutants contained mostly nonconservative amino acid changes (7 of 12) rather than frameshifts. Sequences from the panel of ACV-resistant viruses identified two homopolymeric nucleotide stretches (G7 and C6) as putative hot spots within the HSV TK gene, confirming previous reports on Acvr isolates (18, 38). Although we identified HSV-1 and HSV-2 mutations in both G7 and C6, the in vitro-selected HSV-1 isolates generally contained mutations within the G7 string, whereas alterations within the downstream C6 tract were typically associated with HSV-2. This virus type-specific distribution may be attributed to chance, although type-specific or strain-specific responses to selective pressure cannot be ruled out. It unclear whether any type-specific differences are related to the higher incidence of spontaneous mutations in HSV-2 than in HSV-1 (36).
Surprisingly, resistance mutations in the TK coding sequence after selection with PCV were generally not in the G7 or C6 hot spots associated with ACV resistance. Those PCV mutations which did occur at a G or C nucleotide were generally at single G or C bases evenly distributed throughout the TK gene. Moreover, while ACV mutations were primarily within G or C nucleotides, PCV mutations were present equally at A-T and G-C nucleotides.
The G7 and C6 elements represent the two longest homopolymer stretches within the TK coding region of HSV and directly flank the thymidine-TMP binding site. Regions containing homopolymeric nucleotides in other genomes, such as T4 bacteriophage, polyomavirus, and the mouse immunoglobulin heavy chain locus, have also been shown to be especially susceptibile to mutations, and the frequency of mutation may be proportional to the number of reiterated nucleotides (30, 39, 45). The mechanism which gives rise to the hot-spot nature of these nucleotide stretches most likely involves a localized mispairing within the homopolymer (39). Additionally, polymerases may preferentially slip or stutter within such mispaired regions, contributing further errors. The exact location of a homopolymeric element is also known to have an impact on the recombination, deletion, or mutation frequency within a given region of DNA (37). Since we observed clustering of base changes within the C6 block as opposed to the G7 string for ACV-selected HSV-2, local topology may also influence the selection or mutation process involved in resistance to PCV and ACV.
While a recent analysis of the Acvr Kawaguchi strain of varicella-zoster virus (VZV) revealed TK-deficient variants similar to the classical Acvr HSV mutants, with deletions which result in frameshifts and premature termination (19), the VZV TK gene lacks a G7 element, and the location of the single C6 tract is not conserved relative to its HSV-1 and HSV-2 homologs. Most ACV-resistant VZV mutants reportedly lack frameshift mutations and are more likely to express full-length TK than Acvr HSV mutants (40). Furthermore, Acvr VZV mutations are located throughout the coding region of TK and are not highly localized to G-C-rich regions, such as the C6 string. Thus, the locations of Acvr VZV mutations are generally similar to those we identified for Pcvr HSV-1 (40) and support the concept that the context and relative proximity of identical homopolymeric regions can influence whether they are mutational hot spots.
It was anticipated that many of the mutants resulting from selection with PCV or ACV would carry identical mutations in TK, since all isolates were selected in parallel, presumably from preexisting mutants contained in the wild-type parental mixture. In fact, statistical analyses of the differences in the distribution of mutations between the PCV- and ACV-selected plaque isolates suggest that significant drug-related differences exist (P = 0.028 for HSV-1 and P = 0.082 for HSV-2). An alternative hypothesis to drug-specific selection of different preexisting mutants is that PCV and ACV or their triphosphates, rather than preferentially selecting a subset of preexisting mutants, may play a more active role in the introduction of specific errors into the HSV genome during replication. Although unlikely, given their proven safety profiles in normal cells, it is possible that in HSV-infected cells specific HSV polymerase replication errors might be enhanced.
Sasadeusz et al. proposed that ACV triphosphate (ACV-TP) may be capable of modifying the enzymatic or proofreading properties of the HSV DNA Pol to enhance polymerase stuttering in G-C-rich regions (38). However, it is difficult to rationalize how ACV-TP, an obligate chain terminator, could influence the viral DNA Pol without being incorporated and terminating the newly synthesized DNA chain. Nevertheless, the possibility remains open that ACV-TP, which unlike PCV-TP lacks a 3′ hydroxyl group, could promote an enhanced local misalignment within homopolymer stretches and thereby provide multiple sites for misaligned, although complementary, base pairing. While the Ki of PCV-TP with the Pol is significantly higher than that of ACV-TP (PCV Ki, 8.5 μM; ACV Ki, 0.07 μM), PCV-TP is actually a more efficient and complete chain terminator under physiological conditions (with competing natural nucleotides present) than is ACV-TP (14, 35). The more complete chain termination activity of PCV-TP, the weaker interaction with the Pol compared to ACV-TP, and the lack of hot-spot mutations in PCV-selected HSV-1 are all consistent with such a mechanism.
The observation that TK mutations in PCV-selected mutants were very often different from those in ACV-selected mutants suggests that subtle differences in the mechanisms of action of these two antiviral agents may be pivotal in the genotypes of mutants that arise under PCV or ACV pressure. Interestingly, antiviral studies of the nucleoside analogs lamivudine (3TC) and PCV, two potent inhibitors of hepatitis B virus, (HBV), also suggest that both the fine molecular interaction of these inhibitors with viral polymerases and their base-pairing interactions may directly influence the type of drug-resistant variants selected. A substantial difference in the Ki for inhibition of recombinant HBV polymerase by PCV-TP and 3TC-TP is apparent (4.8 and 0.25 μM, respectively), similar to the differences in the PCV-TP and ACV-TP affinities for inhibition of the HSV polymerase (46, 47). Although mutations within the catalytic site of the HBV reverse transcriptase have been found to occur following treatment of chronic HBV infection with either 3TC or FCV, variants containing a point mutation in the YMDD motif are most readily found after 3TC treatment (27, 43). Other mutations within the catalytic domain, aside from the YMDD motif, were identified following FCV treatment (1). Certainly, the identification of a Pcvr HBV mutant which retains sensitivity to 3TC reinforces the idea that the interaction of each of these nucleoside analogs with the polymerase is unique (33). Hence, distinct structural differences between PCV and 3TC, leading to differences in the kinetic interaction of these agents with the polymerase, may directly influence the selection or generation of particular mutants. Previous reports indicate that there are Acvr TK and DNA Pol mutants which remain sensitive to PCV. Such variants reinforce the biochemical data indicating that the interactions between TK and PCV or ACV, and likewise between the HSV polymerase and the triphosphates of PCV or ACV, are distinct and may have biological consequences (3, 6, 32).
In summary, we have confirmed earlier reports that HSV TK mutations selected by ACV preferentially occur at the homopolymeric G7 and C6 stretches, or hot spots. Conversely, HSV TK mutations selected by PCV generally were more uniformly distributed throughout the TK gene. Although the differential distribution of mutant sequences selected by PCV and ACV could result from chance, our data suggest that there are subtle, yet distinct, selection or mutation differences for PCV and ACV. A formal possibility remains that the observed selection differential could be influenced by strain differences. Further experiments using virus strains other than HSV-1 SC16 and HSV-2 SB5 could help address these issues. Nevertheless, the overall phenotypic effect of both ACV- and PCV-selected mutations was the loss of TK activity.
It remains to be seen whether the genotypic changes identified for HSV mutants selected in cell culture with PCV will also be characteristic of resistant isolates from patients treated with PCV or the oral prodrug FCV. An examination of several clinical isolates resistant to PCV during or after treatment with FCV or PCV will be required to verify that the genotypes selected in vitro are characteristic of resistance selection in treated populations.
TABLE 6.
TK mutations identified in ACV-selected HSV-1 SC16
| Clonal virus | Mutationa
|
Predicted length (aa)b | |
|---|---|---|---|
| Nucleotide | Amino acid | ||
| 1A1 | C860T; ΔG919 | T287M; FS 307 | 346 |
| 1A2 | ΔC553 | FS 185 | 346 |
| 1A3 | +G437 | FS 146 | 227 |
| 1A4 | G176C; +G437 | G59A; FS 146 | 227 |
| 1A5 | C148G; +G437 | L50V; FS 146 | 227 |
| 1A6 | C860T | T287M | 376 |
| 1A7 | C13G; T21A; +2G437; ΔC462 | P5A; H7Q; FS 146; FS 153 | 227 |
| 1A8 | C13G; +G437 | P5A; FS 146 | 227 |
| 1A9 | C860T; +C886; +T893; +GCC896; +A901; +G905; +G919 | T287M; FS 296, 298, 301, 302, 307 | 349 |
| 1A11-A | G16T; +G437 | G6C; FS 146 | 227 |
| 1A11-B | +G437 | FS 146 | 227 |
| 1A12 | G16T; +G437 | G6C; FS 146 | 227 |
| 1A13 | G16T; G167T; +G437 | G6C; G56V; FS 146 | 227 |
| 1A14 | C13G; T21A; +G437 | P5A; H7Q; FS 146 | 227 |
The designation C148G represents a change from the wild-type nucleotide C to a G at base 148; the others follow this pattern. Δ, base deletion; +, base insertion; FS, frameshift in the coding sequence.
aa, amino acids.
TABLE 7.
TK mutations identified in Pcvr HSV-2 SB5 plaque isolates
| Clonal virus | Mutationa
|
Predicted length (aa)b | |
|---|---|---|---|
| Nucleotide | Amino acid | ||
| 2P1 | +T7 | FS 3 | 31 |
| 2P2 | C863T; +C1003 | G39E; T288M; FS 335 | 390 |
| 2P4 | ΔC467 | FS 156 | 182 |
| 2P5 | C863T; ΔT927 | T288M; FS 309 | |
| 2P6 | G676A; ΔAC862–863 | E226K; FS 290 | 347 |
| 2P7 | +A366; ΔG439; ΔC471; ΔC476; ΔT482; +T917 | FS 122, 147, 157, 159, 161, 306 | 347 |
| 2P8 | G116A; C863T | G39E; T288M | 376 |
| 2P9 | C573A; C863T; +A923 | F191L; T288M; FS 308 | 390 |
| 2P10 | ΔT419 | FS 140 | 182 |
The designation C863T represents a change from the wild-type nucleotide C to a T at base 863; the others follow this pattern. Δ, base deletion; +, base insertion; FS, frameshift in the coding sequence.
aa, amino acid.
ACKNOWLEDGMENTS
We thank S. Albelda, A. Awan, H. Field, J. Smith, S. Safrin, and P. Shaffer for generous gifts of reagents; A. Hager for technical assistance with plaque purification; J. Mao for primer synthesis; B. Gagnon for statistical analyses; R. Boon for support from SmithKline Beecham Consumer Healthcare; and S. Dillon, F. Del Vecchio, D. Earnshaw, and K. Esser for scientific advice and critical review of the manuscript.
REFERENCES
- 1.Aye T T, Bartholomew M M, Shaw T, Bowden D S, Breschkin A M, McMillan J S, Angus P. Hepatitis B virus polymerase mutations during antiviral therapy in a patient following liver transplantation. Hepatology. 1997;26:1148–1153. doi: 10.1016/s0168-8278(97)80125-0. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramaniam N K, Veerisetty V, Gentry G A. Herpesviral deoxythymidine kinases contain a site analogous to the phosphoryl-binding arginine-rich region of porcine adenylate kinase; comparison of secondary structure predictions and conservation. J Gen Virol. 1991;71:2979–2987. doi: 10.1099/0022-1317-71-12-2979. [DOI] [PubMed] [Google Scholar]
- 3.Boyd M R, Bacon T H, Sutton D, Cole M. Antiherpesvirus activity of 9-(4-hydroxy-3-hydroxymethylbut-1-yl) guanine (BRL 39123) in cell culture. Antimicrob Agents Chemother. 1987;31:1238–1242. doi: 10.1128/aac.31.8.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd M R, Safrin S, Kern E R. Penciclovir—a review of its spectrum of activity, selectivity, and cross-resistance pattern. Antivir Chem Chemother. 1993;4:3–11. [Google Scholar]
- 5.Brown D G, Visse R, Sandhu G, Davies A, Rizkallah P J, Melitz C, Summers W C, Sanderson M R. Crystal structure of the thymidine kinase from herpes simplex virus type 1 in complex with deoxythymidine and ganciclovir. Nat Struct Biol. 1995;2:876–881. doi: 10.1038/nsb1095-876. [DOI] [PubMed] [Google Scholar]
- 6.Chiou H C, Kumura K, Hu A, Kerns K M, Coen D M. Penciclovir-resistance mutations in the herpes simplex virus DNA polymerase gene. Antivir Chem Chemother. 1995;6:281–288. [Google Scholar]
- 7.Christophers J, Clayton J, Craske J, Ward R, Collins P, Trowbridge M, Darby G. Survey of resistance of herpes simplex virus to acyclovir in northwest England. Antimicrob Agents Chemother. 1998;42:868–872. doi: 10.1128/aac.42.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coen D M. The implications of resistance to antiviral agents for herpesvirus drug targets and drug therapy. Antivir Res. 1991;15:287–300. doi: 10.1016/0166-3542(91)90010-o. [DOI] [PubMed] [Google Scholar]
- 9.Coen D M, Kosz-Vnenchak M, Jacobson J G, Leib D A, Bogard C L, Schaffer P A, Tyler K L, Knipe D M. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc Natl Acad Sci USA. 1989;86:4736–4740. doi: 10.1073/pnas.86.12.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darby G, Field H J. Altered substrate specificity of herpes simplex virus thymidine kinase confers acyclovir resistance. Nature. 1981;289:81–83. doi: 10.1038/289081a0. [DOI] [PubMed] [Google Scholar]
- 11.Darby G, Lawrence G, Inglis M M. Evidence that the ‘active centre’ of the herpes simplex virus thymidine kinase involves an interaction between three distinct regions of the polypeptide. J Gen Virol. 1986;67:753–758. doi: 10.1099/0022-1317-67-4-753. [DOI] [PubMed] [Google Scholar]
- 12.Dasgupta U B, Summers W C. Ultraviolet reactivation of herpes simplex virus is mutagenic and inducible in mammalian cells. Proc Natl Acad Sci USA. 1978;75:2378–2381. doi: 10.1073/pnas.75.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubbs D R, Kit S. Mutant strains of herpes simplex deficient in thymidine kinase-inducing activity. Virology. 1964;22:493–502. doi: 10.1016/0042-6822(64)90070-4. [DOI] [PubMed] [Google Scholar]
- 14.Earnshaw D L, Bacon T H, Darlison S J, Edmonds K, Perkins R M, Vere H R. Mode of antiviral action of penciclovir in MRC-5 cells infected with herpes simplex virus type 1 (HSV-1), HSV-2, and varicella-zoster virus. Antimicrob Agents Chemother. 1992;36:2747–2757. doi: 10.1128/aac.36.12.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efstathiou S, Kemp S, Darby G, Minson A C. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J Gen Virol. 1989;70:869–879. doi: 10.1099/0022-1317-70-4-869. [DOI] [PubMed] [Google Scholar]
- 16.Fyfe J A. Differential phosphorylation of (E)-5-(2-bromovinyl)-2′-deoxyuridine monophosphate by thymidylate kinases from herpes simplex viruses types 1 and 2 and varicella zoster virus. Mol Pharmacol. 1982;21:432–437. [PubMed] [Google Scholar]
- 17.Hall J D, Almy R E. Evidence for control of herpes simplex virus mutagenesis by the viral DNA polymerase. Virology. 1982;116:535–543. doi: 10.1016/0042-6822(82)90146-5. [DOI] [PubMed] [Google Scholar]
- 18.Hwang C C, Chen H H. An altered spectrum of herpes simplex virus mutations mediated by an antimutator DNA polymerase. Gene. 1995;152:191–193. doi: 10.1016/0378-1119(94)00712-2. [DOI] [PubMed] [Google Scholar]
- 19.Ida M, Kageyama S, Sato H, Kamiyama T, Yamamura J, Kurokawa M, Morohashi M, Shiraki K. Emergence of resistance to acyclovir and penciclovir in varicella-zoster virus and genetic analysis of acyclovir-resistant variants. Antivir Res. 1999;40:155–166. doi: 10.1016/s0166-3542(98)00057-6. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson J G, Ruffner K L, Kosz-Vnenchak M, Hwang C B C, Knipe D M, Coen D M. Herpes simplex virus thymidine kinase and specific stages of latency in murine trigeminal ganglia. J Virol. 1993;67:6903–6908. doi: 10.1128/jvi.67.11.6903-6908.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karber G. Bertrag zur kollektiven behandlung pharmakologischer reihenversuche. Arch Exp Pathol Pharmakol. 1931;162:480–483. [Google Scholar]
- 22.Kit S. Thymidine kinase. Microb Sci. 1985;2:369–375. [PubMed] [Google Scholar]
- 23.Kit S, Sheppard M, Ichimura H, Nusinoff-Lehrman S, Ellis N M, Fyfe J A, Otsuka H. Nucleotide sequence changes in the thymidine kinase gene of herpes simplex virus type 2 clones from a patient treated with acyclovir. Antimicrob Agents Chemother. 1987;31:1483–1490. doi: 10.1128/aac.31.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knopf C W. Properties of herpes simplex virus DNA polymerase and characterization of its associated exonuclease activity. Eur J Biochem. 1979;98:231–244. doi: 10.1111/j.1432-1033.1979.tb13181.x. [DOI] [PubMed] [Google Scholar]
- 25.Knopf C W, Weisshart K. Comparison of exonucleolytic activities of herpes simplex virus type-1 DNA polymerase and DNase. Eur J Biochem. 1990;191:263–273. doi: 10.1111/j.1432-1033.1990.tb19119.x. [DOI] [PubMed] [Google Scholar]
- 26.Larder B A, Darby G. Virus drug resistance: mechanisms and consequences. Antivir Res. 1984;4:1–42. doi: 10.1016/0166-3542(84)90023-8. [DOI] [PubMed] [Google Scholar]
- 27.Ling R D, Mutimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplanted recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 28.Marcy A I, Olivo P D, Challberg M D, Coen D M. Enzymatic activities of overexpressed herpes simplex virus DNA polymerase purified from recombinant baculovirus-infected insect cells. Nucleic Acids Res. 1990;18:1207–1215. doi: 10.1093/nar/18.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nugier F, Collins P, Larder B A, Langlois M, Aymard M, Darby G. Herpes simplex virus isolates from an immunocompromised patient who failed to respond to acyclovir treatment express thymidine kinase with altered substrate specificity. Antivir Chem Chemother. 1991;2:295–302. [Google Scholar]
- 30.Okada Y, Streisinger G, Owen J, Newton J, Tsugita A, Inouye M. Molecular basis of a mutational hot spot in the lysozyme gene of bacteriophage T4. Nature. 1972;236:338–341. doi: 10.1038/236338a0. [DOI] [PubMed] [Google Scholar]
- 31.Parris D S, Harrington J E. Herpes simplex virus variants resistant to high concentrations of acyclovir exist in clinical isolates. Antimicrob Agents Chemother. 1982;22:71–77. doi: 10.1128/aac.22.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelosi E, Mulamba G B, Coen D M. Penciclovir and pathogenesis phenotypes of drug-resistant herpes simplex virus mutants. Antivir Res. 1998;37:17–28. doi: 10.1016/s0166-3542(97)00054-5. [DOI] [PubMed] [Google Scholar]
- 33.Pichoud C, Seigneres B, Wang Z, Trepo C, Zoulin F. Transient selection of a hepatitis B virus polymerase gene mutant associated with a decreased replication capacity and famciclovir resistance. Hepatology. 1999;29:230–237. doi: 10.1002/hep.510290119. [DOI] [PubMed] [Google Scholar]
- 34.Pottage J C, Kessler H A. Herpes simplex virus resistance to acyclovir: clinical relevance. Infect Agents Dis. 1995;4:115–124. [PubMed] [Google Scholar]
- 35.Reardon J E, Spector T. Herpes simplex virus type 1 DNA polymerase. Mechanism of inhibition by acyclovir triphosphate. J Biol Chem. 1989;264:7405–7411. [PubMed] [Google Scholar]
- 36.Sarisky R T, Nguyen T T, Duffy K E, Wittrock R J, Leary J J. Difference in incidence of spontaneous mutations distinct between herpes simplex virus types 1 and 2. Antivir Agents Chemother. 2000;44:1524–1529. doi: 10.1128/aac.44.6.1524-1529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarisky R T, Weber P C. Role of anisomorphic DNA conformations in the negative regulation of a herpes simplex virus type 1 promoter. Virology. 1994;204:569–579. doi: 10.1006/viro.1994.1571. [DOI] [PubMed] [Google Scholar]
- 38.Sasadeusz J J, Tufaro F, Safrin S, Schubert K, Hubinette M M, Cheung P K, Sacks S L. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J Virol. 1997;71:3872–3878. doi: 10.1128/jvi.71.5.3872-3878.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Streisinger G, Owen J. Mechanism of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985;109:633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talarico C L, Phelps W C, Biron K K. Analysis of the thymidine kinase genes from acyclovir-resistant mutants of varicella-zoster virus isolated from patients with AIDS. J Virol. 1993;67:1024–1033. doi: 10.1128/jvi.67.2.1024-1033.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenser R B, Edris W A. Trigeminal ganglion infection by thymidine kinase-negative mutants of herpes simplex virus after in vivo complementation. J Virol. 1987;61:2171–2174. doi: 10.1128/jvi.61.7.2171-2174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenser R B, Jones C J, Ressel S J, Fralish F A. Thymidine plaque autoradiography of thymidine kinase-positive and thymidine kinase-negative herpesviruses. J Clin Microbiol. 1983;17:122. doi: 10.1128/jcm.17.1.122-127.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell D L J. Mutation in the HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 44.Vere Hodge R A, Cheng Y C. The mode of action of penciclovir. Antivir Chem Chemother. 1993;4:13–24. [Google Scholar]
- 45.Wilson J B, Hayday A, Courtneidge S, Fried M. A frameshift at a mutational hot spot in the polyoma virus early region generates two new proteins that define T-antigen functional domains. Cell. 1986;44:477–487. doi: 10.1016/0092-8674(86)90469-1. [DOI] [PubMed] [Google Scholar]
- 46.Xiong X, Flores C Y H, Toole J J, Gibbs C S. Mutations in hepatitis B DNA polymerase associated with resistance to lamivudine do not confer resistance to adefovir in vitro. Hepatology. 1998;28:1669–1673. doi: 10.1002/hep.510280629. [DOI] [PubMed] [Google Scholar]
- 47.Xiong X, Yang H, Westland C E, Zou R, Gibbs C S. In vitro evaluation of hepatitis B virus polymerase mutations associated with famciclovir resistance. Hepatology. 2000;31:219–224. doi: 10.1002/hep.510310132. [DOI] [PubMed] [Google Scholar]