Abstract
Understanding of newborn immune ontogeny in the first week of life will enable age-appropriate strategies for safeguarding vulnerable newborns against infectious diseases. Here we conducted an observational study exploring the immunological profile of infants longitudinally throughout their first week of life. Our Expanded Program on Immunization - Human Immunology Project Consortium (EPIC-HIPC) studies the epigenetic regulation of systemic immunity using small volumes of peripheral blood samples collected from West African neonates on days of life (DOL) 0, 1, 3, and 7. Genome-wide DNA methylation and single nucleotide polymorphism markers are examined alongside matched transcriptomic and flow cytometric data. Integrative analysis reveals that a core network of transcription factors mediates dynamic shifts in neutrophil-to-lymphocyte ratios (NLR), which are underpinned by cell-type specific methylation patterns in the two cell types. Genetic variants are associated with lower NLRs at birth, and healthy newborns with lower NLRs at birth are more likely to subsequently develop sepsis. These findings provide valuable insights into the early-life determinants of immune system development.
Subject terms: DNA methylation, Neonatal sepsis, Epigenetics in immune cells, Immunogenetics
Evaluating the immune status of newborns helps recognition of those who are at higher risk for serious infectious diseases. Here authors identify lower, epigenetically inferred,
neutrophil-to-lymphocyte ratios during the first week of life as risk factor for sepsis and provide insight into the underpinning epigenetic and transcriptional patterns.
Introduction
Newborn mortality is a global public health concern worldwide1,2. Infections, including sepsis, are a leading cause of preventable deaths3. Although substantial progress has been made in reducing mortality in older children, there has been little progress in preventing neonatal deaths occurring in the first week of life4. Successful vaccine interventions offer limited immune protection during the first week of life when immense developmental transitions are occurring5. To devise age-appropriate strategies to reduce mortality during this period of heightened vulnerability, understanding newborn immunity in the first week is essential6.
The current understanding of immune ontogeny in the first week of life is limited because biosampling neonates is challenging7. Studies have focused on umbilical cord blood because of its accessibility or have investigated immune ontogeny over monthly time intervals beyond the neonatal period8,9. Innovations in neonatal biosampling have enabled understanding of the molecular10, cellular, and proteomic11 networks mediating early immune ontogeny from small volumes of peripheral blood, allowing longitudinal assessments. Previously, in a pilot study (EPIC-001), we described a developmental trajectory of newborn ontogeny characterized by molecular changes in interferon, complement, and neutrophil signaling within the first seven days of life10. Others have found changes in peripheral neutrophil and lymphocyte populations within days of birth, reflecting a stereotypical ontogenesis program in preterm and term infants11,12, aligning with the concept of a highly conserved layered immune system13, in which the transition from the fetal to early infant immune profile occurs through consecutive periods of developmental programming14. However, the genomic contributors remain unclear.
Epigenetic regulation is crucial in hematopoiesis and mediates dynamic shifts in ontogeny during postnatal life15. Epigenetic regulation includes post-translational modifications to DNA, histone, chromatin remodeling factors, and non-coding RNA signaling that regulates genome and cellular functions16. DNA methylation remodeling via DNA methyltransferases (DNMT) and ten-eleven translocation (TET) enzymes potentiates gene expression during hematopoiesis, facilitating the age-dependent plasticity of the immune system15. DNA methylation remodeling at methylation-sensitive transcription factor-binding motifs shapes the hematopoietic landscape by modulating the sensitivity of progenitor cells to lineage-defining transcription factors16. This mechanism may underlie the dynamic skewing of hematopoiesis in newborns11 and age-dependent changes within distinct leukocyte populations17. We hypothesize that epigenetic modifications, particularly DNA methylation, play a crucial role in shaping the immune system during the first week of life. Specifically, we aim to investigate whether dynamic changes in DNA methylation patterns are associated with shifts in immune cell populations and gene expression profiles during this critical period of development using whole blood samples from two newborn cohorts (EPIC-002, main cohort; EPIC-003, Gambian Ontogeny cohort; hereafter referred to as the validation cohort in this report).
We present a genome-wide analysis of dynamic epigenetic changes, contributing to a deeper understanding of the immune landscape in newborns. This research expands our knowledge of neonatal immunity and may inform the development of targeted interventions to improve newborn health.
Results
Whole blood methylation dynamics in the first week of life
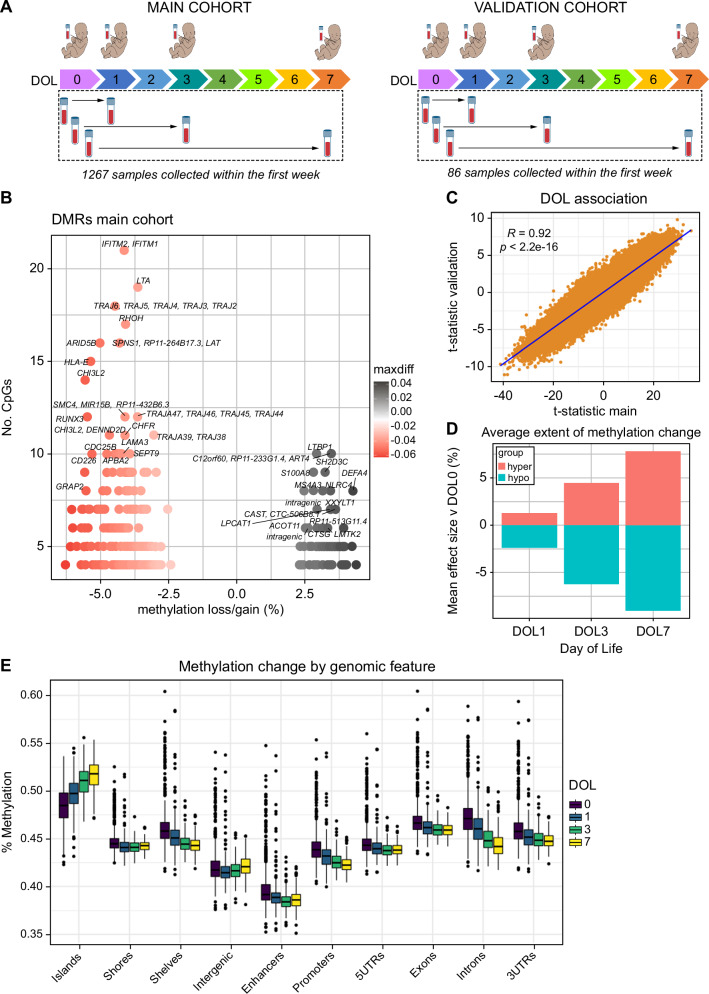
Genome-wide DNA methylation and single-nucleotide polymorphism (SNP) microarrays were performed at multiple time points. In EPIC-002, n = 648 participants had available DNA methylation data at Visit 1 and 619 at Visit 2 (total n = 1,267, 88% of the cohort). In the EPIC-003 validation cohort, 43/45 newborns (96% of participants) had available methylation data at both Visit 1 and Visit 2 (total n = 86 samples) (Fig. 1A). We conducted a longitudinal analysis of DNA methylation levels in 1267 newborn samples from EPIC-002, using days of life (DOL) as a categorical outcome variable. Overall, 333 genomic regions comprising 22,836 individual CpGs underwent dynamic remodeling over the first week of life in the main cohort (FDR < 0.05 & beta ± 2%, Fig. 1B). These associations were also observed in EPIC-003 (22,676/22,836 FDR < 0.05), suggesting that methylation remodeling is a robust process (Fig. 1C and Supplementary Fig. 1). The effect sizes increased continuously with each DOL in the cross-sectional analysis (Fig. 1D). Methylation loss as a function of DOL was observed more frequently than methylation gain (260 hypomethylated versus 73 hypermethylated regions). Most genomic features lost methylation with an increasing number of days of life, except for CpG islands, which exclusively gained methylation (Fig. 1E) in the EPIC-002 and EPIC-003 cohorts (Supplementary Fig. 2). Since CpG islands and gene promoters have overlapping features, we stratified this analysis into island- and non-island-associated promoters, observing similar gains in methylation across categories, suggesting CpG islands are important reservoirs of methylation gain during early immune ontogeny (Supplementary Fig. 2).
Fig. 1. Genome-wide analysis of DNA methylation in peripheral blood samples shows rapid changes in the first week of life.
A Overview of study design and sample collection from each day of life (DOL). B Volcano plot of differentially methylated regions significantly associated with DOL. The X-axis represents the average methylation change from DOL0-7, and the y-axis shows the number of CpGs within each region, which is a function of region size. C Correlation of t-test statistics for DOL regression model between main and validation cohorts. Each point is an individual observation from n = 308,655 data points measured in both cohorts. The solid line indicates the linear regression fit for DOL. A two-sided P-value for Pearson’s correlation coefficient (R) is shown. D Cross-sectional analyses of mean effect size (% methylation) at each time point, stratified by hypermethylation (gain) or hypomethylation (loss). E Methylation ratios expressed as percentage (y-axis) for DOL associated CpGs stratified by genomic feature (x-axis). Boxplots show the median and interquartile range (IQR, 25th-75th percentile). Whiskers extend to the most extreme data points within 1.5 times the IQR from the box hinges (the 25th and 75th percentiles). Points outside this range are considered outliers and are plotted individually). UTR = untranslated region.Total n = 1267 samples.
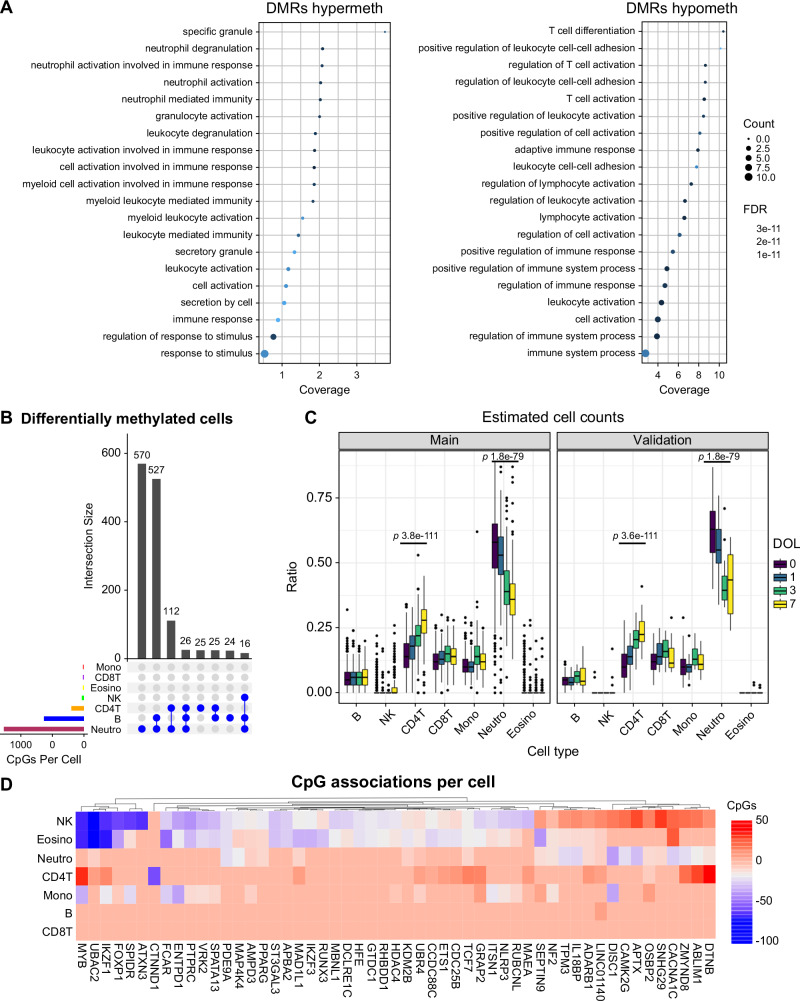
Widespread hypomethylation during the first week of life was observed on chromosome 14, which encodes germline T cell receptor gene segments (TRAJ2-TRAJ6; TRAJ42-TRAJ47; Fig. 1B), reflecting changes in preferential gene use and junctional diversity. The AT-rich interacting domain 5 B (ARID5B) gene, an epigenetic regulator essential for the development of hematopoietic cells18, was among the largest regions of methylation loss. Notably, various interferon (IFN)-induced antiviral proteins (IFITM1, IFITM2), lymphotoxin alpha (LTA) inflammatory cytokine19, Ras superfamily protein (RHOH) cell survival and growth regulator20, and human leukocyte antigen B (HLA-B), lost methylation. Hypermethylation was observed in regions encoding genes such as DEFA4, which encodes the alpha-defensin-4 peptide, an antimicrobial component of the neutrophil primary granule response21. The calcium-binding alarmin protein S100A8, a marker of septic shock22, NLRC4 inflammasome innate response protein23, a regulator of transforming growth factor beta (LTBP1)24 and ART4 glycoprotein, which carries Dombrock blood group antigens25, were genes that gained methylation. To characterize the biology associated with these methylation dynamics, we conducted a gene ontology enrichment analysis of hypomethylated and hypermethylated regions associated with DOL. Hypermethylated regions were strongly enriched in neutrophil- and myeloid-related pathways, whereas hypomethylated regions were enriched in lymphocyte processes (Fig. 2A).
Fig. 2. Differentially methylated cell type analysis reveals enrichment in neutrophils and lymphocyte populations.
A Dotchart of gene ontology enrichment analysis of the DOL ontogeny signature highlighting significantly enriched pathways. B Upset plot showing counts of shared and unique differentially methylated CpGs among blood cell populations from DMCT analysis. C Deconvoluted whole blood cellular ratios inferred from DNA methylation profiles. Boxplots show the median and interquartile range (IQR, 25th-75th percentile). Whiskers extend to the most extreme data points within 1.5 times the IQR from the box hinges (the 25th and 75th percentiles). Points outside this range are considered outliers and are plotted individually). P-values are one-way repeated measures ANOVA. D Heatmap visualization of the number of CpG associations per cell for the top 50 ranked genes. Positive associations are shown in red, and negative associations in blue. *** = P < 0.001, repeated measures ANOVA. Total n = 1267 samples.
Differentially methylated cell type analysis identifies distinct neutrophil and lymphocyte enrichment of ontogeny signature
To determine the cell-type specificity of this ontogeny signature, we deconvoluted whole blood methylation profiles from each cohort into inferred immune cell ratios and performed differentially methylated cell type (DMCT) analysis using the same linear model as before, incorporating the interaction between DOL and immune cell ratios. The DOL ontogeny methylation signature was almost exclusively of neutrophils, B, and CD4 + T cells (Fig. 2B), consistent with the gene ontology analysis. A significant trend for reduction in neutrophil counts and increase in T-lymphocytes with each DOL was observed in both cohorts (Fig. 2C) by repeated measures ANOVA (P < 0.05), and confirmed by flow cytometry (Supplementary Fig. 3). In longitudinal models of DOL adjusted for deconvoluted blood cell counts as covariates, only 53 of 22,836 associations were attenuated, indicating that methylation remodeling was not a consequence of cell composition changes. Neutrophils harbored the greatest degree of cell type-specific differential methylation (570 unique CpGs) and shared many associations with B cells (527 shared CpGs), and to a lesser degree with CD4 + T cells (112 shared CpGs) (Fig. 2B). Methylation changes shared between cell types often exhibited pleiotropy. Essential transcription factors with chromatin remodeling functions that regulate hematopoiesis (FOXP1, MYB, and IKZF1) were positively associated with CD4 + T-cell counts and negatively associated with other cell types such as eosinophils and NK cells (Fig. 2D).
Integrative analysis identifies transcription factor changes mediating neutrophil to lymphocyte ratios
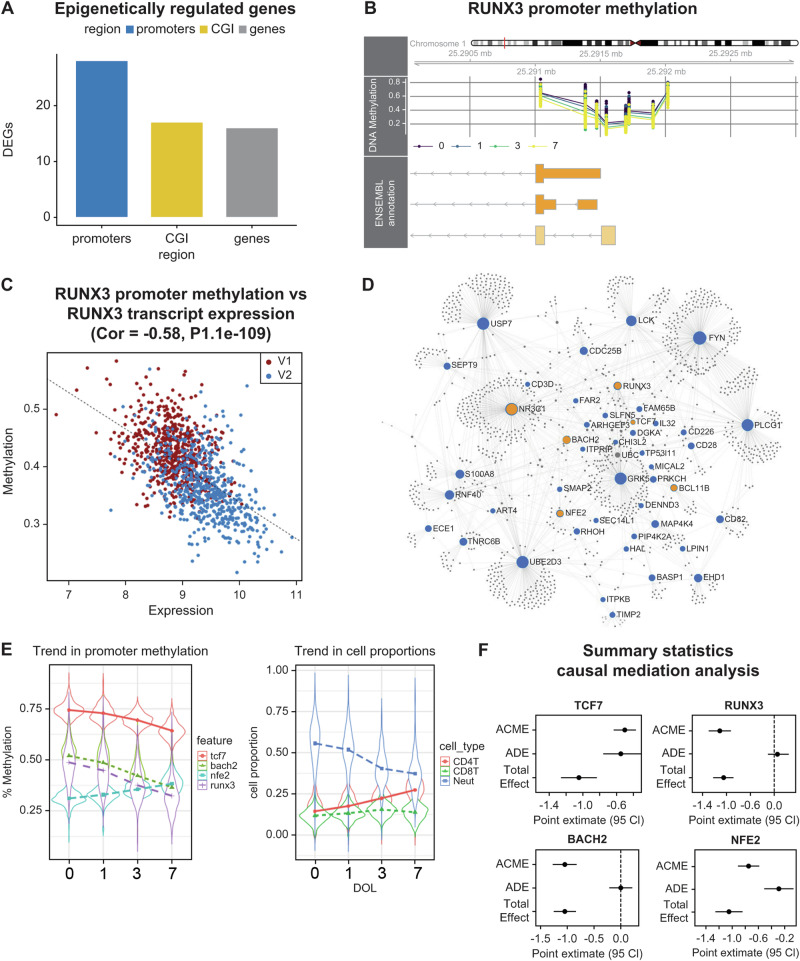
We performed an integrative analysis of blood methylomes with transcriptomic datasets from 606 participants from the main EPIC-002 cohort who had matched RNA-seq data available (n = 1212). The DOL-associated regions correlated with the expression of 61 target genes (Fig. 3A–C, Table 1, complete list in Supplementary Data 1). Methylation changes in gene promoters and CpG islands were associated with gene silencing, whereas changes in gene bodies were positively correlated with gene expression, consistent with prior evidence that gene body methylation modulates transcriptional isoform usage26. The 61 epigenetically regulated genes encoded hematopoietic transcription factors (RUNX3, TCF7, NR3C1, NFE2, RNF10, and BACH2), protein kinases (PRKCH, GRK5, LCK, FYN, and MAP4K4), and cell differentiation markers (CD3G, CD28, CD226, CD82, and ART4). From these target genes, we inferred a complex regulatory protein interaction network (PPI enrichment P = 1.33 × 10−15) significantly enriched for pathways involved in immune system regulation, signal transduction, cell cycle, and apoptosis (Fig. 3D, Table 2; complete list in Supplementary Data 2). The transcription factor dynamics influencing blood cell compositional changes were interesting based on the data of time-dependent trends in transcription factor promoter methylation and the corresponding changes in blood cell proportions (Fig. 3E). To further explore whether the methylation of transcription factors mediates changes in blood composition, we employed a causal mediation framework constructing individual regression models addressing the effect of age (in days) at time of sample collection on neutrophil to lymphocyte ratios (NLR), including promoter methylation levels as mediator variables. Both RUNX3 and BACH2 completely mediated the effect of age on blood cell ratios, whereas significant partial mediation was observed for other transcription factors (Fig. 3F, statistics in Supplementary Data 3), suggesting that these transcription factors play specialized roles in mediating the effect of age on blood cell ratios.
Fig. 3. Integrative analysis of newborn DNA methylation and transcriptomic changes in the main cohort.
A Number of differentially expressed genes (DEG) correlated with day of life associated with differentially methylated regions in each genomic context. B Visualization of RUNX3 promoter methylation levels measured on the main cohort. Average methylation ratios are shown on the y-axis colored by day of life (0, 1, 3, and 7). Each point on the plot is a CpG dinucleotide. Their relative position to RUNX3 promoter isoforms is shown in the orange boxes. C Pearson’s correlation between RUNX3 promoter methylation and gene expression. Data points are colored according to visit number (V1 = day of life 0; V2 = day of life 1, or 3, or 7). D First-order protein-protein interaction network from InnateDB. Key transcription factors are colored in yellow, blue nodes are seed nodes from the list of epigenetically regulated genes, and gray nodes are interactors. Interactions between nodes are denoted by edge lines. E Violin plots showing average changes in transcription factor promoter methylation and cell proportions as a function of day of life (DOL). Plots show the median (white dot) and interquartile range (IQR), and the thin line extends to the most extreme data points within 1.5x IQR. Points outside this are considered outliers and plotted individually. The violin shape represents the probability density of the data. F Summary statistics from causal mediation analysis. Point estimates from mediation models are shown with 95% confidence intervals (CI). ACME = average causal mediation effect, ADE = average direct effect. Total n = 1265.
Table 1.
Differentially methylated and expressed genes associated with ontogeny
| Symbol | RegionType | Gene | Correlation | P-Value | adjPValue |
|---|---|---|---|---|---|
| RHOH | promoters | ENSG00000168421 | − 0.76 | 2.31E-228 | 1.41E-226 |
| DENND2D | promoters | ENSG00000162777 | − 0.75 | 1.01E-221 | 2.05E-220 |
| ART4 | promoters | ENSG00000111339 | − 0.75 | 7.80E-217 | 1.19E-215 |
| CD3D | promoters | ENSG00000160654 | − 0.70 | 2.81E-176 | 2.14E-175 |
| LCK | promoters | ENSG00000182866 | − 0.69 | 1.17E-170 | 6.47E-170 |
| DGKA | promoters | ENSG00000065357 | − 0.68 | 9.51E-167 | 4.83E-166 |
| CHI3L2 | promoters | ENSG00000064886 | − 0.68 | 7.88E-164 | 3.43E-163 |
| CD28 | promoters | ENSG00000178562 | − 0.67 | 2.87E-161 | 1.10E-160 |
| PCED1B | promoters | ENSG00000179715 | − 0.67 | 1.22E-159 | 4.38E-159 |
| IL32 | promoters | ENSG00000008517 | − 0.65 | 3.37E-148 | 1.03E-147 |
| BACH2 | promoters | ENSG00000112182 | − 0.65 | 5.14E-148 | 1.49E-147 |
| TTC39C | promoters | ENSG00000168234 | − 0.64 | 7.26E-141 | 1.93E-140 |
| FYN | promoters | ENSG00000010810 | − 0.63 | 7.19E-135 | 1.76E-134 |
| CLEC2D | promoters | ENSG00000069493 | − 0.60 | 8.38E-121 | 1.83E-120 |
| S100A8 | promoters | ENSG00000143546 | − 0.60 | 4.02E-118 | 8.17E-118 |
| BASP1 | promoters | ENSG00000176788 | − 0.59 | 1.69E-116 | 3.22E-116 |
| RCAN3 | promoters | ENSG00000117602 | − 0.59 | 1.98E-115 | 3.67E-115 |
| HAL | promoters | ENSG00000084110 | − 0.59 | 1.49E-114 | 2.53E-114 |
| RUNX3 | promoters | ENSG00000020633 | − 0.58 | 1.06E-109 | 1.66E-109 |
| CDC25B | promoters | ENSG00000101224 | − 0.57 | 2.02E-104 | 2.93E-104 |
| NFE2 | promoters | ENSG00000123405 | − 0.55 | 6.87E-97 | 9.53E-97 |
| SCML4 | promoters | ENSG00000146285 | − 0.55 | 1.02E-96 | 1.39E-96 |
| FAM69A | promoters | ENSG00000154511 | − 0.55 | 2.92E-96 | 3.88E-96 |
| FAM134B | promoters | ENSG00000154153 | − 0.54 | 1.30E-91 | 1.58E-91 |
| TP53I11 | promoters | ENSG00000175274 | − 0.52 | 1.43E-85 | 1.61E-85 |
| FAR2 | promoters | ENSG00000064763 | − 0.52 | 1.59E-83 | 1.71E-83 |
| CD226 | promoters | ENSG00000150637 | − 0.51 | 2.77E-82 | 2.86E-82 |
| CD82 | promoters | ENSG00000085117 | − 0.51 | 7.45E-80 | 7.57E-80 |
Correlation: Pearson’s correlation coefficient between lead CpG in DMR (most significant) and transcript expression for genes within 1.5 kb from the DMR. P-values are shown from the Pearson’s coefficient test. P-values adjusted by False Discovery Rate.
An Abbreviated list of gene promoter associations is shown here, a complete set of regions in Supplementary Data 1.
Table 2.
Top 10 enriched Reactome pathways in the ontogeny protein-protein interaction network
| Pathway | P.Value | FDR |
|---|---|---|
| Immune System | 2.11E-60 | 2.88E-57 |
| Adaptive Immune System | 4.25E-53 | 2.90E-50 |
| Antigen processing: Ubiquitination & Proteasome degradation | 6.17E-29 | 2.80E-26 |
| Class I MHC-mediated antigen processing & presentation | 2.25E-27 | 7.68E-25 |
| Signaling by the B Cell Receptor (BCR) | 4.66E-27 | 1.27E-24 |
| Signaling by ERBB4 | 2.30E-25 | 5.21E-23 |
| Signaling by Interleukins | 1.74E-24 | 3.40E-22 |
| Cytokine Signaling in the Immune System | 1.65E-23 | 2.81E-21 |
| Innate Immune System | 1.39E-22 | 2.11E-20 |
| Signaling by SCF-KIT | 2.20E-22 | 2.99E-20 |
| Downstream Signaling Events Of B Cell Receptor (BCR) | 2.53E-22 | 3.14E-20 |
| Signaling by EGFR in Cancer | 4.85E-21 | 5.51E-19 |
| Signaling by NGF | 5.58E-20 | 5.85E-18 |
| Signaling by EGFR | 6.40E-20 | 6.23E-18 |
| Downstream signal transduction | 4.34E-18 | 3.95E-16 |
| Signaling by ERBB2 | 6.10E-18 | 4.89E-16 |
| DAP12 signaling | 6.10E-18 | 4.89E-16 |
| TCR signaling | 1.14E-17 | 8.63E-16 |
| DAP12 interactions | 1.91E-17 | 1.37E-15 |
| Interleukin-3, 5 and GM-CSF signaling | 9.04E-17 | 6.16E-15 |
P-Values for the hypergeometric test are shown. FDR = False Discovery Rate. Complete the table of statistics in Supplementary Data 2.
Reduced NLRs at birth were a pre-morbid risk factor for subsequent early-onset sepsis
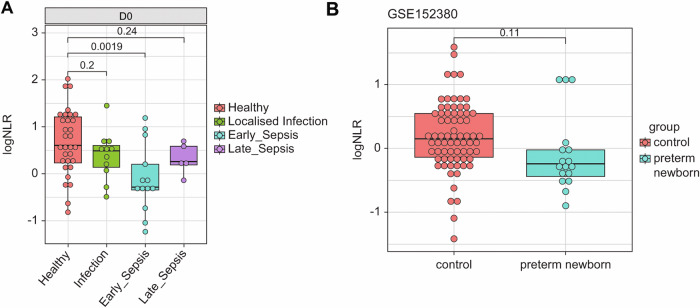
We explored whether variations in baseline NLR at DOL0 were related to clinical inflammatory outcomes, such as infections, including sepsis, occurring in the first week. In the EPIC-002 main cohort, twelve newborns developed acute localized infections, and 21 developed sepsis within the first week of life and were matched to 33 healthy controls from the same cohort based on sex, vaccination status, DOL, and time of blood collection after birth. We examined the correlation between the epigenetically inferred NLR and flow cytometry-derived NLR and found strong significant positive correlations (R = 0.72, P < 2.2−16, Supplementary Fig. 4). Healthy controls had a significantly higher epigenetic NLR at birth than the neonates who subsequently developed early onset sepsis (Fig. 4A). In linear mixed-effect models adjusted for sex, birth weight z-scores, gestational age, and subject ID, the baseline epigenetic NLR was significantly associated with ‘any sepsis’ outcome diagnosed between DOL1-7 (estimate, − 1.2; SD, 0.55; P = 0.03). This was statistically corroborated when we used the flow cytometry-derived NLR in the same models (estimate − 0.5, std.error 0.21, P = 0.02). We also explored whether epigenetic NLRs were influenced by preterm birth since preterm neonates are at a higher risk of sepsis. As there were no preterm births in this cohort, we accessed publicly available umbilical cord blood whole-blood methylation profiles from 72 term and 18 preterm newborns27 and calculated the epigenetically inferred NLR. The NLR scores were lower in preterm newborns than those in term controls (Fig. 4B) due to significantly lower neutrophil counts and elevated B-lymphocyte and NK cell counts (Supplementary Fig. S5). We assessed the possible prenatal characteristics explaining the variation in the baseline epigenetic NLR using linear regression. Birth weight, gestational age, the season of birth, APGAR scores, and maternal age were not significant predictors of baseline NLR (Table 3). Since the epigenetic NLR at birth had a high degree of inter-individual variation, we hypothesized potential gene-environment influence.
Fig. 4. Lower Neutrophil to lymphocyte ratios at birth in term newborns who later developed sepsis and in preterm newborns.
A Median difference in epigenetic NLR at birth (D0) between infants that developed localized infection or clinical sepsis within the first seven days in the EPIC-002 cohort. Exact p-values by t-test. Total n = 54. B Difference in median epigenetic NLR from preterm and term (control) umbilical cord blood samples. Exact p-values by t test. Total n = 90. Boxplots show the median and interquartile range (IQR, 25th-75th percentile). Whiskers extend to the most extreme data points within 1.5 times the IQR from the box hinges (the 25th and 75th percentiles). Points outside this range are considered outliers and are plotted individually).
Table 3.
Summary statistics of the general linear model
| Estimate | Std.Error | P-value | |
|---|---|---|---|
| Intercept | 2.10 | 1.08 | 0.05 |
| Sex | 0.40 | 0.32 | 0.21 |
| Apgar9* | 0.80 | 0.74 | 0.29 |
| Apgar101 | 0.19 | 0.71 | 0.80 |
| Birthweight z-score | 0.30 | 0.17 | 0.08 |
| Birth Season | − 0.34 | 0.35 | 0.32 |
| Maternal Age | − 0.01 | 0.03 | 0.80 |
| Maternal Antibody2 | − 0.50 | 0.32 | 0.12 |
| Breastfeeding | 0.39 | 0.50 | 0.43 |
| Gestational age | 0.09 | 0.16 | 0.59 |
Std.Error = Standard error.
1= Apgar score parameter estimates compared to reference group APGAR 8.0.
2Maternal anti-HepB (HBsAg) serological status.
Genome-wide association study of SNP markers and baseline NLR identifies novel genetic variants
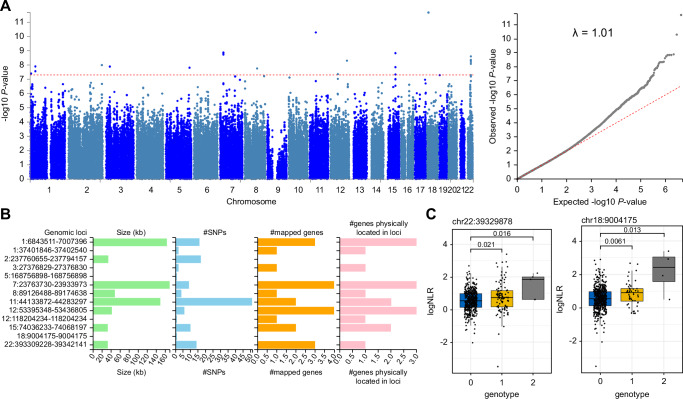
To explore the genetic contribution to NLR at birth, we conducted a genome-wide association study (GWAS) of ~ 8.5 million genotypes from 557/720 individuals from the EPIC-002 cohort regressed on birth NLRs as a continuous variable. Principal component analysis showed that the study population clustered with the African super-population reference data from the 1000 genomes project (Supplementary Fig. 6), suggesting no major population stratification. We detected 13 associations at genome-wide significance (Table 4) and used the model statistics as input to the FUMA GWAS tool to identify genomic risk loci and perform expression quantitative trait loci (eQTL) analysis. The strongest significant genome-wide genetic signals were detected at 13 genomic risk loci (Fig. 5A, B, Supplementary Datas 4, 5). The top SNP associations indicated an allelic variation in the baseline NLRs (Fig. 5C). The lead SNP on chromosome 7 tagged a known variant (rs73080951) previously identified as a protein quantitative trait locus for type I and II interferon receptor expression28. The variants on chromosome 22 were whole blood expression QTLs (GTExv8 catalog) for the CBX6 polycomb transcriptional repressor protein, which is required to balance pluripotency and differentiation in mammals (Table 5)29. We estimated the broad-sense heritability of NLR at birth using GWAS statistics and linkage disequilibrium (LD) scores from the UK Biobank African population. The heritability estimate was high at 0.87 (std.dev = 1.3), suggesting that a substantial portion of the variation in the baseline NLR was due to variation in genotypes, although we expect this estimate to be inflated due to the small sample size. Nevertheless, our data provide cogent evidence that host genetic factors partly influence baseline NLR.
Table 4.
SNPs associated with epigenetically inferred NLR
| CHR | ID | A1 | UNADJ | GC | BONF | HOLM | SIDAK_SS | FDR_BH |
|---|---|---|---|---|---|---|---|---|
| 18 | 18:9004175:C:T | T | 2.10E-12 | 2.76E-12 | 2.3E-05 | 2.3E-05 | 2.3E-05 | 2.3E-05 |
| 11 | 11:44282120:A:G | A | 5.20E-11 | 6.60E-11 | 5.6E-04 | 5.6E-04 | 5.6E-04 | 2.8E-04 |
| 7 | 7:23763730:A:T | T | 1.33E-09 | 1.63E-09 | 1.4E-02 | 1.4E-02 | 1.4E-02 | 2.6E-03 |
| 7 | 7:23796946:C:T | T | 1.47E-09 | 1.80E-09 | 1.5E-02 | 1.5E-02 | 1.5E-02 | 2.6E-03 |
| 7 | 7:23793021:C:A | A | 1.47E-09 | 1.80E-09 | 1.5E-02 | 1.5E-02 | 1.5E-02 | 2.6E-03 |
| 15 | 15:74047924:T:C | C | 1.48E-09 | 1.81E-09 | 1.5E-02 | 1.5E-02 | 1.5E-02 | 2.6E-03 |
| 7 | 7:23773195:A:G | G | 1.86E-09 | 2.27E-09 | 1.9E-02 | 1.9E-02 | 1.9E-02 | 2.7E-03 |
| 22 | 22:39329878:C:T | T | 2.50E-09 | 3.05E-09 | 2.6E-02 | 2.6E-02 | 2.5E-02 | 3.2E-03 |
| 22 | 22:39324344:C:A | A | 4.36E-09 | 5.28E-09 | 4.5E-02 | 4.5E-02 | 4.4E-02 | 3.7E-03 |
| 22 | 22:39333937:G:T | T | 4.71E-09 | 5.71E-09 | 4.8E-02 | 4.8E-02 | 4.7E-02 | 3.7E-03 |
| 22 | 22:39329780:C:T | T | 4.71E-09 | 5.71E-09 | 4.8E-02 | 4.8E-02 | 4.7E-02 | 3.7E-03 |
| 22 | 22:39329593:G:A | A | 4.71E-09 | 5.71E-09 | 4.8E-02 | 4.8E-02 | 4.7E-02 | 3.7E-03 |
| 22 | 22:39332920:C:T | T | 4.71E-09 | 5.71E-09 | 4.8E-02 | 4.8E-02 | 4.7E-02 | 3.7E-03 |
CHR: Chromosome.
A1 = Effect Allele.
UNADJ = unadjusted P-value, linear regression under an additive model.
GC = Genomic Control.
BONF = Bonferroni P-value.
HOLM = Holm corrected P-value.
SIDAK_SS = Sidak correction single step.
FDR_BH = False discovery rate correction Benjamini Hochberg method.
Fig. 5. Genome-wide association analysis of baseline NLR trait in African newborns from the main cohort.
A GWAS summary statistics for linear regression under the additive genetic model with genome-wide significance threshold is shown in red (left panel). The quantile-quantile plot of P-values with genomic inflation score (right panel). B Thirteen genomic-risk loci were identified from GWAS summary statistics using the FUMA GWAS tool. Genomic location is shown on the y-axis with region size, number of SNPs and mapped genes shown for each region. C Median difference in log NLR shown as a function of genotype for two top-ranked SNPs on Chromosome 22 (left panel) and Chromosome 18 (right panel). Exact P-values are shown using the Wilcox test. Boxplots show the median and interquartile range (IQR, 25th-75th percentile). Whiskers extend to the most extreme data points within 1.5 times the IQR from the box hinges (the 25th and 75th percentiles). Points outside this range are considered outliers and are plotted individually). Genotype codes indicate the presence of a minor allele of the SNP variant; 0 = homozygous major allele, 1 = hemizygous, 2 = homozygous minor allele. Total n = 557.
Table 5.
Expression quantitative trait loci mapping results
| CHR | ID | A1 | UNADJ | STAT | FDR | ENS | SYMBOL | TISSUE |
|---|---|---|---|---|---|---|---|---|
| 22 | 22:39314326:C:T | T | 2.37E-05 | − 0.196894 | 7.44E-20 | ENSG00000183741 | CBX6 | Whole Blood |
| 22 | 22:39321323:A:G | A | 5.78E-05 | 0.188351 | 7.44E-20 | ENSG00000183741 | CBX6 | Whole Blood |
| 22 | 22:39324344:A:C | A | 9.69E-05 | 0.189103 | 7.44E-20 | ENSG00000183741 | CBX6 | Whole Blood |
CHR: Chromosome.
ID = Unique SNP identifier consists of chr:pos:Allele1:Allele2.
UNADJ = unadjusted P-value for linar regression test.
STAT = Signed test statistic.
FDR = False Discovery Rate P-value of eQTLs.
ENS = Ensembl Gene Identifier.
SYMBOL = Official Gene Symbol.
TISSUE = Tissue source from GTEx/v8 data source.
Discussion
Our study has identified genetic and epigenetic factors that contribute to newborn immune ontogeny and correlate with the risk for neonatal sepsis in the first week of life, a time of immense developmental transition and susceptibility to infectious diseases6. We observed dynamic changes in promoter methylation and gene expression in four hematopoietic transcription factors, NFE2, BACH2, RUNX3, and TCF7, and a broader program of epigenetic change at 333 genomic regions, and 61 expressed genes that were associated with time-dependent changes in NLR. These ontogenetic patterns were independent of gestational age or sex. Similar trends in changing blood composition were described in Scandinavian and American newborn cohorts11,12, reflecting a stereotypic program of ontogeny across diverse populations. The changes in methylated DNA we observed in lymphocytes and neutrophils likely reflect the changing contributions of hematopoietic niches (thymus and bone marrow), which impart distinct DNA methylation profiles to progenitor cells, layering the systemic compartment with modified cellular phenotypes30. While we cannot establish the causality of these associations, statistical analysis suggests methylated alleles in these gene promoters were mediators of dynamic shifts in NLR. Functional perturbation studies using in vitro fate mapping and single-cell analysis are planned to decipher the causal role of transcription factor changes in hematopoietic lineage decisions31. Together, these findings expand the current understanding of the molecular determinants of newborn immune ontogeny.
NLR measured by full blood count is a clinically useful prognostic marker of inflammation32,33, infection, and sepsis33, and we report it also as a useful marker of early ontogeny. Although many previous studies have described the clinical utility of NLR for sepsis diagnosis or prognosis among hospitalized infants33, we found that low NLR at birth among healthy newborns was a pre-morbid risk factor for subsequent sepsis. It is possible lower NLRs could indicate an underlying predisposition, but it is difficult to draw causal inferences in this observational study. Of note, we inferred NLR from the ratio of methylated alleles in blood samples34. This molecular NLR may be a promising marker for severe disease risk stratification in newborns, as we found that it correlated strongly with flow cytometric measures of NLR, but exhibited stronger associations with early onset sepsis than NLR derived by flow. Analyzing publicly available cord blood data, we also observed a lower NLR in preterm newborns known to be at higher risk of sepsis, which should encourage future studies of NLR in preterm infants.
Considering the in-utero factors that determine the NLR set-point at birth, our GWAS implicates host genetics, but other environmental factors, such as the microbiota, were not explored and could also contribute. Consistent with previous reports35, inter-individual variability in NLR at birth was high. A few GWAS of adult NLRs have been conducted in European populations36,37. A total of 151 genetic associations have been described, with broad-sense heritability estimated at 36%37. Our heritability estimate of 87% from this African cohort is high, explained by our inclusion of non-European LD scores from the UK Biobank to calculate heritability, which can produce noisy estimates in small sample sizes. Nevertheless, these data confirm a role for genetic factors driving NLR. Given population differences in LD structure, meta-analysis across European and African cohorts for NLR genetic associations will provide further insights into the genetic architecture of blood cell traits. Together, our findings suggest genetic and epigenetic factors play a role in the development of sepsis and suggest that molecular markers may be useful for developing new diagnostic tools to identify those at highest risk for and possibly prevent sepsis. Future studies could develop novel molecular predictors of sepsis risk and strategies to boost NLRs in those most vulnerable.
Our findings should be interpreted within the context of study limitations. We were restricted to small volumes of whole blood tissue, limiting our ability to detect cell-specific effects or conduct functional studies. Our study focused on an African cohort, prioritizing the detection of moderate to large effect sizes. Despite this, our sample size, modest for GWAS, limited the identification of the full spectrum of genetic associations. We did not consider the differential effect of participant sex on immune ontogeny, as our models were adjusted for sex to identify stereotypic ontogenetic patterns. However, our results are meaningful, identifying target molecules for follow-up studies. While our study provides valuable insights into the dynamic epigenetic landscape during early life in an African cohort, it is important to acknowledge that the generalizability of these findings to other populations remains unclear. The specific genetic and environmental factors influencing neonatal immune development may vary across different populations, potentially leading to distinct epigenetic signatures and immune responses. Future studies in diverse populations are needed to validate our findings and determine the extent to which the observed patterns are universal or population-specific. In summary, by integrating analysis of epigenetic, transcriptomic, and flow cytometric data in a large cohort using longitudinal sampling, this study extends insight into the molecular basis of newborn immune ontogeny and identifies promising candidates for clinical translation.
Methods
Sample collection
The EPIC-002 main cohort of 720 term newborns was an unselected sample of healthy newborns enrolled in a larger clinical trial at the Medical Research Council Unit, The Gambia. The sample size for EPIC-002 was optimized for multi-omic analysis as described previously38. The protocol was approved by the local Ethics Committees and by the Institutional Review Board at Boston Children’s Hospital (IRB-P00024239) and is registered at clinical trials. gov: https://clinicaltrials.gov/ct2/show/NCT03246230. Informed consent was obtained from the parents or guardians of participants. No specific inclusion or exclusion criteria were applied beyond those of the parent trial, which were healthy term newborns (gestational age > 36 weeks), born vaginally (as is the vast majority (> 90%) of births in our study population), 5-min Apgar scores > 8, and birth weights > 2.5 kg. Subjects were screened for HIV-I, HIV-II, and hepatitis B and excluded if positive. Venous blood samples (2 mL) were obtained from all neonates within the first 24 hours of life (DOL0, Visit 1). Participants were randomized to a second blood draw (Visit 2) at either DOL1, DOL3, or DOL7. Blood was collected in heparinized collection tubes (Becton Dickinson) for multi-omic analyses as described previously10. An Ontogeny cohort (EPIC-003) comprising 45 unselected newborns was recruited from the same study site for validation purposes, following the same study protocol, which is used in this report for validating statistical models. Neonatal sepsis observed in EPIC-002 was diagnosed by a senior clinician for neonates hospitalized over the course of the study. Neonatal sepsis was defined as (a) blood culture-proven bacterial sepsis with a clinically significant pathogen (culture-confirmed sepsis) or (b) without a positive blood culture, fever syndrome, excessive crying, poor feeding, vomiting, mottled skin, high-pitched cry, convulsions, and laboratory parameters such as neutrophilia (clinically diagnosed sepsis). Sepsis was classified as early onset (< 72 h) or late onset (> 72 h) depending on time of occurrence. Neonates requiring hospital admission were classified as the ‘localized infection’ comparator group if they had no systemic signs, negative blood cultures, and alternative diagnoses, including a case of pneumonia associated with congenital heart disease, bronchiolitis, gastroenteritis, pustular skin infection, ophthalmia neonatorum, and jaundice. Due to the smaller size of EPIC-003, no hospitalizations occurred.
Epigenetics
Whole blood samples were randomized by DOL, sex, and vaccination status in 96-well plates prior to genomic DNA extraction using the Chemagic DNA 400 kit H96 (cat # CMG-1491). DNA samples were quantified using the Qant-iT HS kit (cat# Q33120) and sent to the Australian Genome Research Facility in Melbourne, Western Australia, for bisulfite conversion and genotyping using Illumina Infinium MethylationEPIC Beadchip v1 arrays. Raw.iDAT files were preprocessed using the Minfi package39 from the Bioconductor Project (http://www.bioconductor.org) in R software (http://cran.r-project.org/). Sample quality was assessed using control probes on the array and concordance between the estimated and reported sex and genotype based on MDS clustering of the SNP genotyping control probes. Samples discordant for sex, genotype, or failed quality control were removed prior to analysis. Between-array normalization was performed using the stratified quantile method to correct for type 1 and 2 probe biases. Probes exhibiting a P-detection call rate of > 0.01 in one or more samples, containing SNPs at the single-base extension site or CpG assay site, probes measuring non-CpG loci, and reported to have off-target effects by McCartney et al. 40 were removed. After sample and probe filtering, the final EPIC-002 dataset consisted of 1267 samples and 747,905 CpG probes, and the EPIC-003 dataset consisted of 90 samples and 771,309 probes (Supplementary Data 6). Methylation ratios were derived as β values = with log 2 transformation to M values for statistical analysis.
Genotyping and imputation
Genomic DNA samples were genotyped at the Australian Genome Research Facility using an Illumina Global Screening Array v3 with a multi-disease drop-in. Genotype calling was performed with the gencall algorithm within GenomeStudio (Illumina). Quality control was performed using the plinkQC package (v 0.3.4)41 to remove samples with > 5% missing data and high relatedness (PI_HAT > 0.2) in identity-by-descent analysis for all pairs of samples or with mismatched ancestry estimates based on principal component analysis of merged data with the 1000 genomes phase 3 data set. We excluded SNPs characterized by > 5% missing values, a Hardy-Weinberg equilibrium p-value of < 0.001, and a minor allele frequency of < 5%. Quality-controlled data were then imputed with the Haplotype Reference Consortium hg19 r1.1 reference panels using Beagle 5.4 on the Michigan imputation server. Imputed genotypes were filtered to remove SNPs with a minor allele frequency of < 5% and Hardy–Weinberg equilibrium p-value < 0.001, with an r2 value greater than 0.3, leaving 8,461,000 variants for analysis.
RNASeq
Total RNA was extracted from each sample using a Paxgene Blood RNA kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. RNA quantification and quality assessment were performed using an Agilent 2100 Bioanalyzer (Santa Clara, CA). The NEBNextPoly(A) mRNA Magnetic Isolation Module (NEB; Ipswich, MA, USA) was used to capture polyadenylated RNA. Strand-specific cDNA libraries were generated using the KAPA RNAHyperPrep Kit (Roche, Basel, Switzerland) on a Freedom EVO 100 (Tecan, Männedorf, Switzerland). cDNA libraries were prepared across 33 separate batches, and sequencing was completed over 19 sequencing runs. Thirteen batches of samples were sequenced on a HiSeq2500 (Illumina; San Diego, CA, USA) using high-output single-read runs of 100bp-long sequence reads (including adapter/index sequences). Six batches of samples were sequenced on HiSeqX (Illumina, San Diego, CA, USA) to generate paired-end reads of 150 bp (the second read pair was discarded). Seven samples were sequenced on both platforms to evaluate variations between the sequencing platforms. FastQC v0.11.9 and MultiQC v1.8 1 were used to assess the sequence quality, and short sequence reads were aligned to the hg38 human reference genome (Ensembl GRCh38v98) using STAR v2.7.3a42. Counts of the number of reads mapped per gene were generated using htSeq count (HTSeq v0.11.2 3)43. High-read-count globin genes (< 10 counts in 53 or more samples or the smallest number of biological replicates within each treatment group) were removed from the count matrix. Counts were then normalized using vst-transformation and adjusted for the cDNA library preparation batch using ComBat-seq44 (a covariate matrix was supplied that included sex, DOL, and vaccine group to preserve any effects associated with major covariates).
Flow cytometry
Following venipuncture, 200 µL of whole blood cells were stained for viability and stored in Smart tube solutions prior to red blood cell lysis and cell fixation (Smart tube, CA, USA), and frozen at − 80 °C. The samples were thawed in a water bath at 10 °C and centrifuged at 600 × g for 10 min. Cell pellets were washed by centrifugation in phosphate-buffered saline (PBS) and stained according to the manufacturer’s recommendations. Each sample was stained with antibodies specific to the anchor markers in the two flow panels. Each panel contained 13 surface markers and was designed to identify B cell subsets and the main myeloid cell populations, including neutrophils, dendritic cells (DCs), natural killer (NK) cells, monocytes, and T cell subsets (Supplementary Table 1)10. After staining, the blood cells were washed in PBS, and flow cytometry data were acquired using an LSRII flow cytometer (BD Biosciences). The samples were run in four different batches, with up to 200 samples analyzed daily in 96 well plates. Initial quality control was performed by inspecting the compensated cytometry data using FlowJo software (version 9.9, Becton, Dickinson, and Company) to ensure optimal experimental conditions and population gating according to predefined gating strategies. Batch control was achieved by processing the same whole blood internal control samples, and the batch data were monitored to assess instrument performance and perform routine checks on the cytometer45. The cell counts of all gated populations were normalized to the counting beads to derive the cell counts per microliter of blood. Unbiased automated gating tools were used to generate cell counts of different cells circulating in the blood at a given time point and are described in the Supplementary Methods.
Statistical analysis
All statistical analyses were performed using R software developed by CRAN (v4.1.2). Blood cell proportions were deconvoluted from methylation Beta values using the EpiDISH method under ‘RPC’ mode46. For the primary epigenetic analysis, we included all participants from EPIC-002 and EPIC-003 with available DNA methylation data regardless of whether they had complete follow-up samples. Longitudinal linear regression modeling of M-value methylation ratios on the sample plate and sex was initially performed using the Limma framework47, which handles missing data through pairwise comparisons between available time points for each individual, utilizing all available data. Residuals from the model fit were extracted and used in the matrix decomposition for repeated measures in the mixOmics package to decompose individual variation48. To analyze differentially methylated cell types, a regression model was fitted to the decomposed dataset with DOL as a numeric predictor and adjusted for Sentrix slide ID as a batch variable. The estimated cell counts were used for multiple interaction testing, as implemented in EpiDISH49. To identify differentially methylated regions, a regression model was fitted to the decomposed dataset with DOL as the main numeric predictor for differentially methylated cell type CpGs using limma, adjusted for covariates sex and sample plate ID as a batch variable. Limma statistics were then used as inputs to the DMRcate package50 for the de novo identification of DMRs using a bandwidth smoothing window of 1 kb, a scaling factor of 2, a minimum of four CpGs, and a minimum effect size of 2% to define DMRs. Only DMRs with a minimum smoothed FDR of P < 0.05 were reported. For all epigenetic analyses, genome-wide significance was declared at a false discovery rate adjusted P-value of ≤ 0.05. To evaluate reproducibility in the validation cohort, we fitted the same regression model of DOL on methylation M values adjusted for sex and sample plate to both cohorts and performed Pearson’s correlation analysis on t-statistics from both datasets. Ontology testing of DMRs was performed using the MissMethyl package51. The integration of DNA methylation and RNA-seq data was achieved using the mCSEA package52. Both RNAseq and methylation ratios were subset to those with matching data before ranking probes based on the association with visit number (V1 or V2) and correlation of the lead CpG with RNAseq counts under the default parameters. To derive epigenetic NLR scores, we used epiDISH cell counts to calculate NLR= . The natural log10 of the NLR was used to compare the ratio data. General linear mixed-effect models were used to determine whether the NLR at DOL0 was a significant predictor of sepsis status. Linear regression using the DOL NLR as the independent variable was used to identify prenatal predictors. For causal mediation analysis, we used lead CpG for each tested gene promoter, neutrophil counts, and DOL, in regression models to estimate the total effect and the effect of lead CpG as a mediator variable. Non-parametric bootstrapping with 1000 simulations was used to derive the estimates and confidence intervals using the mediate package in R (v4.5.0). For GWAS, linear regression of variants using NLR scores at DOL0 as a predictor was conducted under an additive genetic model adjusted for sex and the top five principal components from genetic ancestry analysis using a standardized covariance structure and adjusted for genomic control implemented in the PLINK2 software53. SNPs were genome-wide significant at a threshold of P ≤ 5 × 10−8. We used the FUMA GWAS tool (v1.5.4) using a position mapping window size of 10 kb (AFR reference population 1KG/Phase3) for functional annotation of SNP associations. Heritability estimation was performed on the GWAS summary statistics using the LD score regression implemented in the LDSC package54, incorporating the UK BioBank African population LD scores. DNA methylation analysis was performed using the following Bioconductor packages: EpiDISH (v2.10.0) for cell-type deconvolution, Limma (v3.50.3) for differential methylation analysis, mixOmics (v6.18.1) for within-subject variance estimation, DMRcate (v2.8.5) for identification of differentially methylated regions, MissMethyl (v1.28.0) for imputation of missing methylation values, mCSEA (v1.14) for methylation enrichment analysis, and methylclock (v1.0.1) for estimating epigenetic age acceleration. The tidyverse suite of packages (v2.0.0) was employed for data manipulation and visualization. Genetic association analyses were conducted using FUMA GWAS (v1.5.4) for functional mapping and annotation of GWAS results, PLINK2 (v2.00) for data management and statistical analysis, and LDSC (v1.0.1) for estimating genetic heritability. Preprocessing of Illumina methylation array data was performed using Minfi (v1.40.0). Mediation analyses were performed using the mediation package (v4.5.0), and linear mixed-effects models were fitted using lmerTest (v3.1-3).
Data management and quality assurance
Data Management, governance, and quality assurance were managed by a dedicated Data Management Core (DMC) team at Boston Children’s Hospital. DMC oversaw the curation of raw datasets generated by core assay labs and the production of their canonical forms within the EPIC-HIPC network. A typical assay core-generated raw dataset was comprised of [1] a “Features” matrix of samples (as rows) and assay features (as columns) values and [2] a “Metadata” matrix of the sample identifiers and assay-specific processing and quality control features for each sample in the “Features’ matrix. Each EPIC-HIPC sample had a unique global sample identifier associated with the subject and biosample extraction time. Sample identifiers were checked for concordance between “Features,” “Metadata,” and the central EPIC-HIPC database of sample identifiers, clinical, and assay core processing parameters. Second, the sample-wise distributions of the feature values were qualitatively investigated for technical anomalies. For genetic association testing, the sex and kinship of the participants derived from their sample feature variables were compared with their clinical records.
SAGER guidelines
Subject sex was recorded in clinical notes and determined by genetic inference from whole blood samples. In cases where recorded sex differed from genetically inferred sex, subjects were excluded from analysis (QC details in Supplementary Data 6). Regression modeling was adjusted for sex such that findings are generalizable to both sexes.
Inclusion and Ethics
This research was conducted in partnership with local researchers who were involved in the design, data collection, analysis, and reporting of the study. The research is locally relevant and the study protocol was acceptable to parents and local IRB. The design of this study was considerate and intentionally minimized invasive sampling or any changes to routine care.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We wish to thank the Australian Genome Research Facility microarray team for their incredible service. Biostatistical support was kindly provided by Dr. Matt Cooper of the Telethon Kids Biometrics team. We would like to thank Editage [http://www.editage.com] for editing and reviewing this manuscript for the English language. Our thanks go to all parents in The Gambia for allowing us to enroll their healthy newborns and to the dedicated field team at the MRC Unit in The Gambia. OL thanks the leadership of Boston Children’s Hospital, including Drs. Wendy Chung, Nancy Andrews, Kevin Churchwell, and Mr. Gus Cervini, for their support of the PVP. This study was funded in part by the NIH NIAID Human Immunology Project Consortium U19AI118608 and the Immune Development in Early Life (IDEAL) U19AI168643 (Principal Investigator O.L). Dr. A.A. was supported by an NIH/NIAID Mentored Clinical Scientist Research Career Development Award (No.1K08AI168487). Dr D.M was supported by the Western Australian Future Health Research and Innovation Fund (IG2021/3). Dr O.O. was supported by an NIH Loan Repayment Program award through the National Institute on Minority Health and Health Disparities (NIMHD).
Author contributions
Conception and design of the work (D.M., J.D.A., A.O., P.R., S.T., B.K., O.L., R.E.W.H., and T.K.). Data collection (D.M., N.K., N.A., R.B., B.C., M.L., O.I., O.O., R.F., T.M.B., A.A., C.P.S., S.M., B.K.D., K.S., R.B., K.M., A.A., and A.H.Y.L.) Drafting the article (D.M.). Critical revisions (all authors). Final approvals (O.L., and T.K.).
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Methylation data files generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE272800. The SNP array data files are available under restricted access for participant confidentiality at the following link (https://www.immport.org/shared/study/SDY1538), and access requests should be directed to the corresponding author. Source data are provided in this paper.
Code availability
Analysis scripts have been deposited at Gihub: (https://github.com/pvpdmac/epichipc/tree/main/Epigenetics) and are publicly available as of the date of publication.
Competing interests
OL is the inventor of patents helped by Boston Children’s Hospital relating to vaccine adjuvants (EP3709998A1) and human in vitro systems that model responses to immunomodulators and vaccines (e.g., US20150152385A1). Hi is an advisor to GlaxoSmithKline (GSK) and Hillevax and a cofounder of and advisor to Ovax Inc. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
A list of members and their affiliations appears in the Supplementary Information.
Contributor Information
David Martino, Email: david.martino@thekids.org.au.
the EPIC-HIPC consortium:
David Martino, Nelly Amenyogbe, Rym Ben-Othman, Bing Cai, Olubukola Idoko, Oludare A. Odumade, Reza Falsafi, Travis M. Blimkie, Andy An, Casey P. Shannon, Sebastiano Montante, Bhavjinder K. Dhillon, Joann Diray-Arce, Al Ozonoff, Kinga K. Smolen, Ryan R. Brinkman, Kerry McEnaney, Asimenia Angelidou, Peter Richmond, Scott J. Tebbutt, Beate Kampmann, Ofer Levy, Robert E. W. Hancock, Amy H. Y. Lee, and Tobias R. Kollmann
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52283-9.
References
- 1.Collaborators, G. B. D. C. O. D. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: a systematic analysis for the Global Burden of disease study 2017. Lancet392, 1736–1788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oza, S., Cousens, S. N. & Lawn, J. E. Estimation of daily risk of neonatal death, including the day of birth, in 186 countries in 2013: a vital-registration and modelling-based study. Lancet Glob. Health2, e635–e644 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann-Struzek, C. et al. The global burden of paediatric and neonatal sepsis: a systematic review. Lancet Respir. Med.6, 223–230 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Collins, A., Weitkamp, J.-H. & Wynn, J. L. Why are preterm newborns at increased risk of infection? Arch. Dis. Child. Fetal Neonatal Ed.103, F391–F394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kollmann, T. R., Marchant, A. & Way, S. S. Vaccination strategies to enhance immunity in neonates. Science368, 612–615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kollmann, T. R., Kampmann, B., Mazmanian, S. K., Marchant, A. & Levy, O. Protecting the newborn and young Infant from infectious diseases: Lessons from immune Ontogeny. Immunity46, 350–363 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Coppini, R., Simons, S. H. P., Mugelli, A. & Allegaert, K. Clinical research in neonates and infants: Challenges and perspectives. Pharm. Res.108, 80–87 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Brook, B., Harbeson, D., Ben-Othman, R., Viemann, D. & Kollmann, T. R. Newborn susceptibility to infection vs. disease depends on complex in vivo interactions of host and pathogen. Semin. Immunopathol.39, 615–625 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Harbeson, D., Ben-Othman, R., Amenyogbe, N. & Kollmann, T. R. Outgrowing the immaturity Myth: The cost of defending from neonatal infectious disease. Front. Immunol.9, 1077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, A. H. et al. Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat. Commun.10, 1092 (2019). [DOI] [PMC free article] [PubMed]
- 11.Olin, A. et al. Stereotypic immune system development in newborn children. Cell174, 1277–1292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson, L. S. et al. Single-cell analysis of the neonatal immune system across the gestational age continuum. Front. Immunol.12, 714090 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsaid, R. et al. Hematopoiesis: A layered organization across chordate species. Front. Cell Dev. Biol.8, 606642 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornef, M. W. & Torow, N. ‘Layered immunity’ and the ‘neonatal window of opportunity’ – timed succession of non‐redundant phases to establish mucosal host–microbial homeostasis after birth. Immunology159, 15–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bermick, J. & Schaller, M. Epigenetic regulation of pediatric and neonatal immune responses. Pediatr. Res.91, 297–327 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Izzo, F. et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat. Genet.52, 378–387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martino, D., Maksimovic, J., Joo, J. H., Prescott, S. L. & Saffery, R. Genome-scale profiling reveals a subset of genes regulated by DNA methylation that program somatic T-cell phenotypes in humans. Genes Immun.13, 388–398 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Wang, P. et al. The role of ARID5B in acute lymphoblastic Leukemia and beyond. Front. Genet.11, 598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke, R. et al. Lymphotoxin-alpha gene and risk of myocardial infarction in 6,928 cases and 2,712 controls in the ISIS case-control study. PLoS Genet.2, e107 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tetlow, A. L. & Tamanoi, F. The Ras superfamily G-proteins. Enzymes33 Pt A, 1–14 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Basingab, F. et al. Alterations in immune-related defensin alpha 4 (DEFA4) gene expression in health and disease. Int. J. Inflam.2022, 9099136 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, S. et al. S100A8/A9 in Inflammation. Front. Immunol.9, 1298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu, Y. et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature490, 539–542 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Miyazono, K., Olofsson, A., Colosetti, P. & Heldin, C. H. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J.10, 1091–1101 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durousseau de Coulgeans, C., Chiaroni, J., Bailly, P. & Chapel-Fernandes, S. Sequencing of the ART4 gene in sub-Saharan cohorts reveals ethnic differences and two new DO alleles: DO*B-Ile5Thr and DO*B-Trp266Arg. Transfusion55, 2376–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Maunakea, A. K. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature466, 253–257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spada, E. et al. Epigenome wide association and stochastic epigenetic mutation analysis on cord blood of preterm birth. Int. J. Mol. Sci.21, 5044 (2020). [DOI] [PMC free article] [PubMed]
- 28.Lundtoft, C. et al. Function of multiple sclerosis-protective HLA class I alleles revealed by genome-wide protein-quantitative trait loci mapping of interferon signalling. PLoS Genet.16, e1009199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santanach, A. et al. The Polycomb group protein CBX6 is an essential regulator of embryonic stem cell identity. Nat. Commun.8, 1235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lessard, S., Beaudoin, M., Benkirane, K. & Lettre, G. Comparison of DNA methylation profiles in human fetal and adult red blood cell progenitors. Genome Med.7, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobsen, S. E. W. & Nerlov, C. Haematopoiesis in the era of advanced single-cell technologies. Nat. Cell Biol.21, 2–8 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Song, M., Graubard, B. I., Rabkin, C. S. & Engels, E. A. Neutrophil-to-lymphocyte ratio and mortality in the United States general population. Sci. Rep. UK11, 464 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zahorec, R. Neutrophil-to-lymphocyte ratio, past, present and future perspectives. Bratisl. Med. J.122, 474–488 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Koestler, D. C. et al. DNA Methylation-derived neutrophil-to-lymphocyte ratio: An epigenetic tool to explore cancer inflammation and outcomes. Cancer Epidemiol. Biomark. Prev.26, 328–338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moosmann, J. et al. Age- and sex-specific pediatric reference intervals for neutrophil-to-lymphocyte ratio, lymphocyte-to-monocyte ratio, and platelet-to-lymphocyte ratio. Int J. Lab Hematol.44, 296–301 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Kachuri, L. et al. Genetic determinants of blood-cell traits influence susceptibility to childhood acute lymphoblastic leukemia. Am. J. Hum. Genet.108, 1823–1835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, B. D. et al. 2SNP heritability and effects of genetic variants for neutrophil-to-lymphocyte and platelet-to-lymphocyte ratio. J. Hum. Genet.62, 979–988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Idoko, O. T. et al. Clinical Protocol for a Longitudinal Cohort Study Employing Systems Biology to Identify Markers of Vaccine Immunogenicity in Newborn Infants in The Gambia and Papua New Guinea. Front. Pediatr.8, 197 (2020). [DOI] [PMC free article] [PubMed]
- 39.Aryee, M. J. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics30, 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCartney, D. L. et al. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genom. Data9, 22–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.plinkQC: Genotype quality control in genetic association studies (2020).
- 42.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y., Parmigiani, G. & Johnson, W. E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform.2, lqaa078 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montante, S. et al. Breastfeeding and neonatal age influence neutrophil-driven ontogeny of blood cell populations in the first week of human life. J. Immunol. Res.2024, 1117796 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teschendorff, A. E., Breeze, C. E., Zheng, S. C. & Beck, S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinforma.18, 105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohart, F., Gautier, B., Singh, A. & Le Cao, K. A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol.13, e1005752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, S. C., Breeze, C. E., Beck, S. & Teschendorff, A. E. Identification of differentially methylated cell types in epigenome-wide association studies. Nat. Methods15, 1059–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters, T. J. et al. De novo identification of differentially methylated regions in the human genome. Epigenet. Chromatin8, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phipson, B., Maksimovic, J. & Oshlack, A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics32, 286–288 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Martorell-Marugan, J., Gonzalez-Rumayor, V. & Carmona-Saez, P. mCSEA: detecting subtle differentially methylated regions. Bioinformatics35, 3257–3262 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet.47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Methylation data files generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE272800. The SNP array data files are available under restricted access for participant confidentiality at the following link (https://www.immport.org/shared/study/SDY1538), and access requests should be directed to the corresponding author. Source data are provided in this paper.
Analysis scripts have been deposited at Gihub: (https://github.com/pvpdmac/epichipc/tree/main/Epigenetics) and are publicly available as of the date of publication.