Abstract
Retroelements (retrotransposons and retroviruses) have two genes in common: gag, which specifies structural proteins that form a virus or virus-like particle, and pol, which specifies catalytic proteins required for replication. For many retroelements, gag and pol are present on separate reading frames. Their expression is highly regulated, and the ratio of Gag to Pol is critical for retroelement replication. The Saccharomyces retrotransposon Ty5 contains a single open reading frame, and we characterized Gag and Pol expression by generating transpositionally active Ty5 elements with epitope tags at the N terminus or C terminus or within the integrase coding region. Immunoblot analysis identified two Gag species (Gag-p27 and Gag-p37), reverse transcriptase (Pol-p59), and integrase (Pol-p80), all of which are largely insoluble in the absence of urea or ionic detergent. These proteins result from proteolytic processing of a polyprotein, because elements with mutations in the presumed active site of Ty5 protease express a single tagged protein (Gag-Pol-p182). Protease mutants are also transpositionally inactive. In a time course experiment, we monitored protein expression, proteolytic processing, and transposition of a Ty5 element with identical epitope tags at its N and C termini. Both transposition and the abundance of Gag-p27 increased over time. In contrast, the levels of Gag-p37 and reverse transcriptase peaked after ∼14 h of induction and then gradually decreased. This may be due to differences in stability of Gag-p27 relative to Gag-p37 and reverse transcriptase. The ratio of Ty5 Gag to Pol averaged 5:1 throughout the time course experiment, suggesting that differential protein stability regulates the amounts of these proteins.
Ty5 is a Ty1/copia group retrotransposon of Saccharomyces (4, 30). Encoded between its long terminal repeats (LTRs) are homologues of retroviral gag and pol genes. For the retroviruses and LTR retrotransposons (collectively referred to as retroelements), gag specifies the structural proteins that form the viral core or the retrotransposon virus-like particle (VLP). Packaged within the particle are retroelement mRNAs and products of the pol gene. These include a protease (PR) that processes the retroelement polyproteins, a reverse transcriptase (RT) and its associated RNase H (RH) that synthesize a cDNA copy of the retroelement from the template mRNA, and an integrase (IN) that inserts the cDNA into a new site in the host chromosome. In addition to gag and pol, retroviruses have an env gene. The env gene product forms the viral envelope and allows the particle to exit the cell as a membrane-bound virion, which fuses with recipient cells during infection.
Assembly of the viral core or virus-like particle requires an excess of “structural” Gag relative to “catalytic” Pol proteins (reviewed in references 9 and 18). For many viruses, the ratio of Gag to Pol approximates 15 to 1, and perturbing this ratio results in a significant loss in replication efficiency. Retroelements use a variety of expression strategies to regulate Gag and Pol expression, including two that are closely tied to protein synthesis—translational suppression and translational frameshifting. Translational suppression is employed by viruses such as murine leukemia virus, for which gag and pol are present in the same reading frame separated by a stop codon (33). Translation of murine leukemia virus mRNA predominantly produces Gag; however, occasionally the gag amber termination codon is decoded by a glutamine tRNA to produce a Gag-Pol fusion protein. Translational suppression is facilitated by downstream sequences that form a pseudoknot, which slows translation and enables the suppressor tRNA to better compete with translation release factors (17, 31).
The second translational mechanism and most common strategy for regulating gag and pol expression is −1 and +1 frameshifting (reviewed in references 9 and 18). For retroelements that use frameshifting, the 3′ coding region of gag typically overlaps the 5′ coding region of pol in either the −1 or the +1 frame. Frameshifting is triggered by secondary structures (stem loops or pseudoknots) or rare codons in the mRNA that cause the ribosome to stall in the overlap region and then to slip forward or backward one nucleotide before continuing translation. Human immunodeficiency virus is an example of a virus that uses −1 ribosomal frameshifting (19). Retroelements that use +1 frameshifting include the yeast retrotransposons Ty1 and Ty3 (3, 10).
The coding regions of some retrotransposons lack stop codons and internal frameshift sites. The ratio of Gag to Pol is nonetheless critical for these elements, and they use alternative strategies to regulate Gag and Pol expression. For example, the copia elements of Drosophila melanogaster produce a single genome-length mRNA that is subject to differential splicing (6, 34). The pol coding sequences are spliced from the majority of transcripts, leading to an abundance of gag mRNA and elevated levels of Gag protein. The Tf1 elements of Schizosaccharomyces pombe use a posttranslational strategy to regulate the stoichiometry of Gag and Pol (1, 25). A single Gag-Pol polyprotein is synthesized, which is processed to yield equimolar amounts of mature Tf1 proteins. As yeast cultures approach stationary phase, Pol is preferentially degraded, resulting in a 26:1 ratio of Gag to Pol. The degradation of Pol triggers Tf1 cDNA synthesis.
Like copia and Tf1, Ty5 encodes a single open reading frame (ORF) and is the only Saccharomyces retrotransposon for which gag and pol are not separated by a +1 frameshift (36). Ty5 and copia are members of the Ty1/copia group of elements (Pseudoviridae), most of which contain a single ORF (4). In previous work, we demonstrated that Ty5 expresses a single mRNA, suggesting that differential mRNA splicing is not used by Ty5 to regulate Gag and Pol levels (36). In this study, we set out to characterize the Ty5 proteins and to assess possible mechanisms that regulate their expression.
MATERIALS AND METHODS
Strains, media, and Ty5 transposition assays.
The Saccharomyces cerevisiae strain YPH499 (MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1) was used in this study (28). The XL1-Blue Escherichia coli strain was used for recombinant DNA manipulations (Stratagene). Bacterial and yeast strains were grown and transformed using standard methods (2). Synthetic complete medium lacking uracil (SC−U) was used to select yeast cells with Ty5 plasmids, and rich medium (YPD) was used for nonselective growth. Ty5 transcription is regulated by the GAL1-10 upstream activation sequence (36); therefore, changing the carbon source from glucose to galactose induces transcription. Before addition of galactose, cells were grown for 10 to 26 h on raffinose as the carbon source. This insured that glucose was metabolized completely before induction. Liquid cultures also contained 2% (wt/vol) Casamino Acids. Quantitative Ty5 transposition was carried out as previously described (36).
Cloning and plasmids.
All plasmids were derived from pSZ152, which contains a Ty5 element from Saccharomyces paradoxus (36). Epitope-tagged Ty5 elements were constructed using PCR site-directed mutagenesis (16) with primers that encoded the epitope.
(i) The plasmid with a His-tagged RT (pIP19) was constructed using overlapping primers DVO447 (5′-ATC-TGT-TAG-TGA-TGG-TGA-TGG-TGA-TGC-GAT-CCT-CTC-ATT-TTT-GCA-GTT-TCT-GGT-TCC-CTC-3′) and DVO448 (5′-CTG-CAA-AAA-TGA-GAG-GAT-CGC-ATC-ACC-ATC-ACC-ATC- ACT-AAC-AGA-TCG-AGG-TCG-ACG-GTA-3′). These primers were designed to place the His tag (MRGSH6; Qiagen) directly in front of the Ty5 stop codon.
(ii) The T7 epitope (MASMTGGQQMG; Novagen) was inserted at the C terminus of RT by first amplifying Ty5 with DVO465 (5′-GCT-CTA-ACT-GCT-ATT-ATC-3′), which is upstream of a PflMI site, and the mutagenic primer DVO560 (5′-CCA-TCG-ATT-AAC-CCA-TTT-GCT-GTC-CAC-CAG-TCA- TGC-TAG-CCA-TTT-TTG-CAG-TTT-CTG-GTT-C-3′), which carries the T7 tag and a ClaI restriction site. The amplified product and pSZ152 were digested with PflMI and ClaI and ligated. Because the his3AI marker cassette was removed by ClaI digestion, it was then reinserted to create pXW182.
(iii) The plasmid with an N-terminal His tag, pNK520, was constructed using the overlapping primers DVO557 (5′-ATG-AGA-GGA-TCG-CAT-CAC-CAT-CAC-CAT-CAC-ACA-TAT-AAG-CTA-GAT-CG-3′) and DVO558 (5′-GTG- ATG-GTG-ATG-GTG-ATG-CGA-TCC-TCT-CAT-AAT-GTT-GTA-AGT- TTA-TTG-G-3′). The first 10 amino acid residues of the resulting Ty5 element were MRGSH6.
(iv) It was previously found that a portion of the C terminus of integrase could be modified without compromising transposition (X. Gai and D. Voytas, unpublished data). One integrase derivative (pXW29) contained a BglII site at nucleotide position 2845. This construct was used as a template for amplification with a Ty5-specific primer and the mutagenic primer DVO543 (5′-CGG-AAT-AGA- TCT-GTG-ATG-GTG-ATG-GTG-ATG-CGA-TCC-TCT-TTG-TAT-CCT- TGT-TGG-TGG-3′). The amplification product carried the RGSH6 epitope tag as well as a BglII site. This product was reinserted into pXW29, giving rise to pWW32. The wild-type residues IQHS were changed to IQRGSH6RS.
(v) Protease was mutated by PCR with primer DVO246 (5′-CAT-GTG-TGA-AGT-AAT-CGC-GAT-GAT-ACA-AAT-CCA-ATC-TGA-3′) and a Ty5-specific primer. This inserted a NruI restriction site into the presumed active site of Ty5's aspartic protease and changed the residues DTGC to IIAI. The resulting PCR product was used to replace the corresponding wild-type sequence of pSZ152, thus creating pNK259. The protease mutation was moved into the epitope-tagged constructs described above, giving rise to pIP20, pNK526, pIP38, and pNK527 (Fig. 1).
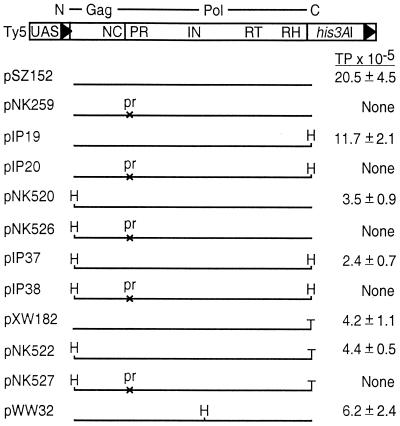
FIG. 1.
Epitope-tagged Ty5 elements and their respective transposition frequencies. The genomic organization of Ty5 is shown at the top. Boxes with filled arrowheads represent LTRs; UAS identifies the GAL1 upstream activating sequence that drives Ty5 transcription. The Ty5 ORF is indicated by a single line above the element. The location of regions encoding conserved amino acid sequence domains within gag and pol are indicated. Replication by reverse transcription is monitored by the his3AI marker gene (8, 36). Below the map of Ty5 are the various constructs used in this study. Construct names are on the left, and transposition frequencies (TP) are on the right. “None” indicates a transposition frequency of ≤0.04 × 10−5. pr, protease mutation. Epitope tags were placed at various positions: T, T7 tag (MASMTGGQQMG); H, His tag (RGSH6).
(vi) Several plasmids were created by swapping restriction fragments from the various constructs. A C-terminal His tag (pIP19) was combined with either the N-terminal His tag (pNK520) or the N-terminal His tag protease mutant (pNK526) to produce pIP37 and pIP38, respectively. The C-terminal T7 tag (pXW182) was combined with the N-terminal His tag (pNK520) to produce pNK522.
Protein preparation.
Yeast strains growing on plates were used to inoculate a small volume of SC−U–glucose medium, and cultures were grown to saturation (30°C). Cells were harvested by centrifugation (1,500 × g, 4°C, 5 min), resuspended in SC−U–raffinose medium (starting optical density at 600 nm [OD600], 0.4 to 0.6), and then grown at 30°C for an additional 10 to 26 h. The raffinose culture was centrifuged (1,500 × g, 4°C, 5 min), resuspended in SC−U–galactose medium (starting OD600, 0.4 to 0.6), and grown at room temperature (22 to 25°C) until the OD600 reached 2.5 to 3.5. Harvested cells were disrupted using the glass bead method (2). The cell lysate was centrifuged (20,000 × g, 60 min), and the supernatant was collected. The remaining pellet was extracted with disruption buffer containing 8 M urea; a volume equivalent to that of the supernatant was used to enable comparisons between the soluble and insoluble fractions. A Dounce homogenizer aided in extracting proteins from the pellet. The extract was centrifuged (20,000 × g, 15 min, 4°C) and the supernatant was collected. For the time course study, proteins were extracted from an equivalent number of cells (based on OD600) and harvested at various time points after induction on galactose media. Transposition was also measured at each time point using our standard quantitative assay (36).
To evaluate Gag solubility, proteins were prepared as described above from strains with pNK522. Aliquots of the cell lysate (100 μl) were centrifuged (20,000 × g, 5 min, 4°C), and the resulting pellet was extracted with 100 μl of Tris-buffered saline (TBS) containing one of the following: 8 M urea; 0.1, 1.0, or 2.0% sodium dodecyl sulfate (SDS); or 3.0 or 6.0% Triton X-100. The extracted sample was centrifuged (20,000 × g, 5 min, 4°C) and the supernatant was collected (soluble proteins). The remaining pellet was resuspended in 100 μl of TBS (insoluble protein suspension). Equal volumes (20 μl) of soluble proteins and the insoluble protein suspension were mixed with SDS-polyacrylamide gel electrophoresis loading buffer and analyzed by immunoblotting.
To characterize the protease mutant, proteins were prepared as described above from yeast cells carrying pIP20. After centrifugation of the cell lysate, the soluble His-tagged proteins were purified using Qiagen's nickel-nitrilotriacetic acid agarose. A 250-μl aliquot of the soluble protein was diluted with 750 μl of a modified Qiagen binding buffer (100 mM NaH2PO4, 10 mM Tris-Cl [pH 8.0], 10 mM imidazole, 5% glycerol). The diluted sample was incubated with 125 μl of nickel-nitrilotriacetic acid agarose for 1 h at room temperature. The slurry was loaded into a gravity flow column, and 1 ml of unbound sample was collected. The column was washed once with 600 μl of binding buffer containing 10 mM imidazole and once with 600 μl of binding buffer containing 20 mM imidazole and then eluted with 600 μl of binding buffer containing 250 mM imidazole. The column was stripped with 600 μl of binding buffer containing 20 mM EDTA. The collected fractions (60 μl) were used for immunoblot analysis.
Immunoblot analysis.
Epitope-tagged Ty5 proteins were subjected to SDS-polyacrylamide gel electrophoresis and then electrophoretically transferred to a nitrocellulose membrane (NitroBind; Micron Separations Inc.) in a modified transfer buffer (25 mM Tris, 0.2 M glycine, 0.1% SDS, 10% methanol) (2). The membrane was blocked with 3% (wt/vol) bovine serum albumin in TBS-Tween-Triton (TBSTT) (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20, 0.2% Triton X-100). The membrane was incubated for 1 h with either RGS-His monoclonal antibody (Qiagen) or the T7 tag monoclonal antibody (Novagen) that had been diluted 1:1,000 in blocking buffer. The membrane was then incubated for 1 h with a horseradish peroxidase-conjugated sheep anti-mouse antibody that had been diluted 1:4,000 in 10% nonfat dry milk-TBSTT. Detection was accomplished using enhanced chemiluminescent detection reagents (ECL; Amersham). Finally, the membrane was exposed to Fuji medical X-ray film. Densitometry was performed on scanned films using the NIH Image (version 1.62) program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/).
RESULTS
Characterizing the Ty5 proteins.
Ty5 has a single ORF whose product has amino acid similarity to the structural (Gag) and enzymatic (Pol) proteins required for replication of other retrotransposons and retroviruses. To characterize Ty5 protein expression, we modified the coding region by the addition of epitope tags. Two short epitope tags were chosen to minimize effects on protein function: (i) a His tag that allows purification under native or denaturing conditions through metal-chelate chromatography and (ii) a T7 tag that allows for protein purification using resin-conjugated antibodies. In both cases, monoclonal antibodies were available for detecting tagged proteins by immunoblot analysis. We positioned epitope tags at the very N and C termini of the Ty5 polyprotein, and elements encoding these tags transposed at levels two- to sixfold lower than those of the wild type (Fig. 1). Addition of both tags reduced transposition by at most ninefold. We previously identified a nonessential region in the presumed C terminus of IN (Gai and Voytas, unpublished). This region was also chosen as a site for His tag insertion (Fig. 1, pWW32), and the modified element transposed at levels approximately threefold lower than those of the wild type. Several of the tagged constructs were further altered to carry mutations in a region of the Ty5 polyprotein that shows similarity to aspartic proteases encoded by other retrotransposons and retroviruses (the DTG motif). For Ty1 and Ty3, mutations in this motif abolish polyprotein processing (23, 35). All Ty5 constructs with a mutation in the DTG motif were transposition defective (Fig. 1).
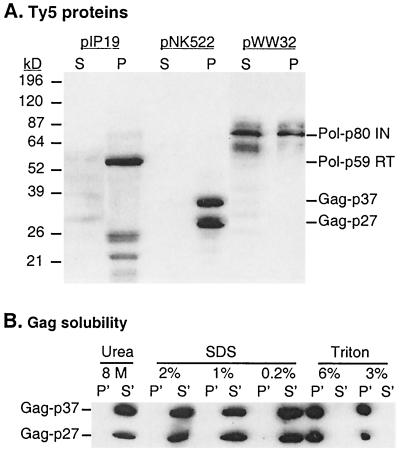
Yeast cells with tagged Ty5 elements were grown in liquid galactose media to induce transcription and transposition. Cells were harvested in late log phase and then lysed by the glass bead method (see Materials and Methods). Lysates were centrifuged, and proteins from the supernatant (soluble fraction) and pellets (insoluble fraction) were analyzed by immunoblotting using monoclonal antibodies specific for the epitope (Fig. 2A). An insoluble 59-kDa protein was the most abundant species expressed by the element with a C-terminal His tag (pIP19). Because the C terminus of the Ty5 protein shows similarity to reverse transcriptase and RNase H, we designated this protein RT (Pol-p59). For the element carrying an N-terminal His tag (pNK522), two Gag species of 27 and 37 kDa were observed (Gag-p27 and Gag-p37). These proteins were of approximately equal abundance and, like RT, were found only in the insoluble fraction. For the element with a His tag near the presumed catalytic domain of integrase (pWW32), comparable levels of an ∼80-kDa protein were observed in both the soluble and insoluble fractions, suggesting that this species is Ty5 IN (Pol-p80). To characterize the protein insolubility, we extracted insoluble proteins expressed by an N-terminally tagged Ty5 element (pNK522) with several concentrations of two detergents and 8 M urea (Fig. 2B). Up to 6% Triton X-100 was unable to solubilize the His-tagged Gag protein. In contrast, concentrations of SDS greater than 0.2% or 8 M urea were very effective in releasing the pellet-bound proteins.
FIG. 2.
Immunoblot analysis of His-tagged Ty5 proteins. (A) Ty5 transcription was induced in yeast cells carrying various epitope-tagged Ty5 elements. These include His tags located at the C terminus of RT (pIP19), at the N terminus of Gag (pNK522), or within IN (pWW32) (Fig. 1). Cells were harvested, lysed, and centrifuged to obtain total soluble yeast protein (S) (see Materials and Methods). The remaining pellet (P) was extracted with a volume of 8 M urea equivalent to that of the supernatant. The exception was pIP19 (His-tagged RT), for which the pellet was extracted with one-fifth the supernatant volume. Equal volumes (20 μl) of all fractions were loaded onto SDS gels and transferred to nitrocellulose. (B) In order to further evaluate Ty5 Gag solubility, pellets were extracted with buffer containing one of the following: 8 M urea; 2, 1, or 0.2% SDS; or 6 or 3% Triton X-100. The extracted pellets were centrifuged and the supernatants (S′) were recovered. The remaining pellets (P′) were resuspended in a volume of SDS loading buffer equal to that of the supernatant to enable direct comparisons with the soluble proteins.
Ty5 is translated into a single polyprotein.
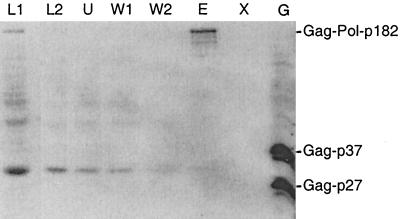
One likely explanation for how the different Ty5 proteins are generated is that a single polyprotein is synthesized and subsequently processed by a Ty5-encoded protease. To test this, we analyzed expression of a Ty5 protein with a His-tagged RT and a mutation in the presumed active site of protease (pIP20). If Ty5 protease is responsible for cleaving the polyprotein, then we should detect in the mutant an approximately 182-kDa protein corresponding to the primary translation product. Proteins were prepared from a strain expressing the Ty5 protease mutant, and immunoblot analysis revealed a large protein in the predicted size range as well as other nonspecific proteins with lower molecular weights (Fig. 3). The high-molecular-weight species was not previously observed in wild-type strains (e.g., Fig. 2A). To confirm that the putative polyprotein was encoded by Ty5, we subjected the protein extract to metal chelate chromatography to enrich for His-tagged proteins. The high-molecular-weight protein specifically bound the resin (Fig. 3, lane E) in contrast to the lower-molecular-weight, cross-reacting proteins (Fig. 3, lanes U, W1, and W2). This indicates that the large protein is encoded by Ty5, and we designate it Gag-Pol-p182 based on its mobility and its molecular weight extrapolated from the Ty5 amino acid sequence. The absence of lower-molecular-weight His-tagged proteins in the mutant indicates that Ty5's aspartic protease is required for cleaving the polyprotein.
FIG. 3.
Ty5 polyprotein identification and purification. After 32 h of growth on galactose media, protein was prepared from yeast cells with a Ty5 element encoding a C-terminal His tag and a protease mutation. The protein sample was electrophoresed on an SDS gel, blotted to nitrocellulose, and probed with the RGSH6 antibody (L1). In contrast to results in Fig. 2A, the p59 RT signal was absent and a large protein at approximately 182 kDa was observed. To determine if the large protein carried a His tag, the protein sample was diluted with binding buffer (L2) and subjected to nickel chelate chromatography. Additional lanes represent unbound flowthrough (U), 10 mM imidazole wash (W1), 20 mM imidazole wash (W2), 250 mM imidazole eluate (E), 20 mM EDTA eluate (X), and a Gag-positive control (G).
Proteolytic processing and transposition: a time course study.
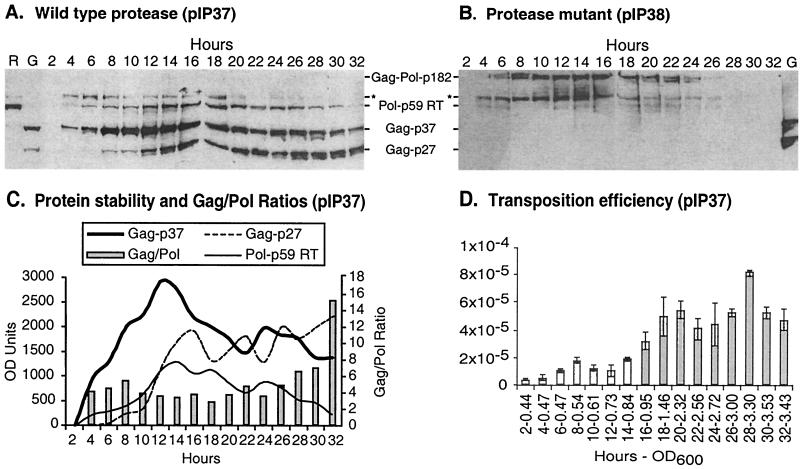
We sought to understand the kinetics of Ty5 protein processing and the relationship of processing to transposition. A yeast strain with pIP37 (His-Gag, His-RT) was induced for transposition by growth on galactose. At 2-h intervals after induction, the culture's OD600 was measured, and samples were harvested. Proteins were extracted from equivalent numbers of cells and subjected to immunoblot analysis using antibodies that recognize the His tag. Because Gag and RT contain identical epitopes, this allowed direct comparisons of protein abundance. As observed in previous experiments, the Ty5 proteins were insoluble (Fig. 4A); none were detected in immunoblots performed with soluble fractions (data not shown). Therefore, the amount of insoluble protein represents the total Ty5 protein expressed. We also stained all gels after electrophoretic transfer to membranes to ensure that no protein remained in the gels. As a control, a study with a parallel time course was conducted with a dually His-tagged protease mutant. In addition to Gag-Pol-p182, a second high-molecular-weight species of comparable size was observed. Because this protein is specific to extracts from the protease mutant, it may represent a polyprotein breakdown product. A protein somewhat larger than RT was apparent in proteins prepared from both the wild type and the protease mutant (Fig. 4A and B). This protein was observed only with C-terminal His tags (Fig. 4A). We do not know the origin of this protein, but because it is found in both wild-type and protease mutant extracts, we do not believe it represents a processed form of Ty5 protein. Rather, it may also represent a polyprotein breakdown product.
FIG. 4.
Ty5 protein expression and transposition: a time course analysis. (A) A yeast strain carrying a Ty5 element with His tags at its N and C termini (pIP37) was induced for transposition by growth in liquid galactose media. Culture samples were taken every 2 h and analyzed for protein expression by immunoblotting. Time points are indicated above the blot. Independent preparations of both His-tagged RT (R) and His-tagged Gag (G) were run as protein references. (B) An immunoblot was prepared as for panel A except that a dually tagged element with a protease mutation was used (pIP38). (C) Densitometry analysis of the blot in panel A. The Gag/Pol ratios were calculated by summing the OD units for Gag-p27 and Gag-p37 and then dividing by the OD units for Pol-p59. (D) Transposition efficiency for the time course in panel A. Transposition frequencies are the averages for three culture samples taken from each time point.
Shortly after induction, Gag-p37 began to accumulate (Fig. 4A and C) and increased in amount until h 12 to 14, after which its abundance slowly declined. By 8 h after induction, Gag-p27 began to appear and slowly increased throughout the remainder of the time course experiment. The relatively late appearance of Gag-p27 suggests that the 37-kDa Gag species is initially released from the polyprotein and then subsequently processed at its C terminus to generate the 27-kDa product. The accumulation of RT (Pol-p59) paralleled that of Gag-p37; however, it had significantly lower abundance (Fig. 4A and C). Despite fluctuations in protein amounts, the ratio of Gag (p27 plus p37) to Pol (p59) was relatively constant throughout the time course experiment, with the average being 5:1 (Fig. 4C). The only striking change occurred when the culture reached saturation (h 32), at which point the ratio shifted to 15:1.
The frequency of transposition was calculated at each of the time points. The dually His-tagged Ty5 element carries a his3AI marker, which enables selection of transposition events by histidine prototrophy conferred when Ty5 cDNA enters the genome. This can occur by integration of the cDNA, mediated by the Ty5-encoded integrase, or through homologous recombination between the cDNA and donor Ty5 element (22). Transposition (which in this study refers to both integration and cDNA recombination) was observed as early as 2 h after induction, before any Ty5 proteins were evident. Transposition frequencies formed two plateaus (Fig. 4D): from h 2 to 14 the frequency was less than 2 × 10−5; h 16 was an inflection point, and then the frequency increased to somewhat more than 4 × 10−5. The range of transposition was 26-fold, from 0.31 × 10−5 at h 2 to 8.2 × 10−5 at h 28. There was no apparent correlation between transposition and the kinetics of protein processing or abundance.
DISCUSSION
The Ty5 proteins.
Most retroelements carry gag and pol on separate reading frames separated by a stop codon or frameshift, and this genetic organization is important for the differential expression of these two genes. Retroelement mRNAs typically direct the synthesis of Gag, which is encoded at the 5′ end of the element. Occasionally, a Gag-Pol fusion protein is synthesized that results from translational suppression of the gag stop codon or ribosomal frameshifting that occurs within an overlap region between gag and pol (reviewed in references 9 and 18). These expression strategies produce an excess of Gag relative to Pol, which is required for assembly of a functional virus or virus-like particle. The single ORF present in Ty5 is unusual among characterized retroelements and suggests that equal amounts of Gag and Pol are expressed. To investigate Ty5 protein expression, we created a series of Ty5 elements with epitope tags at the N terminus, at the C terminus, or near the conserved catalytic domain of integrase. All epitope-tagged elements were transpositionally competent and transposed at levels not more than ninefold lower than that of the wild type.
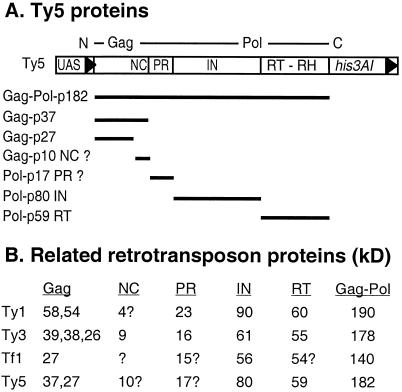
Immunoblot analysis of proteins expressed from an N-terminally tagged Ty5 element revealed two primary protein species, which we believe represent the major forms of Ty5 Gag (Gag-p37 and Gag-p27). Because of the N-terminal location of the tag, the 10-kDa difference between the two proteins must represent a C-terminal processing event. Ty5 Gag has a zinc finger motif (36), and by extrapolating molecular weights from the Ty5 amino acid sequence, we predict that the finger motif resides within the presumed 10-kDa protein. Finger motifs are characteristic of retroviral nucleocapsid proteins (NC), which bind template mRNA, help assemble the particle shell, and have sizes (60 to 90 amino acids) similar to that of the predicted Ty5 protein (29). Confirming the existence of the 10-kDa protein and determining its possible role in transposition will require additional experimentation. Other yeast retrotransposons express multiple Gag species (Fig. 5): Ty1 encodes a 58-kDa Gag protein that is proteolytically cleaved at its C terminus into a 54-kDa product, the major protein found in Ty1 virus-like particles (13). The 4-kDa C terminus of Ty1 Gag that is released during maturation may carry out an NC role, despite the fact that it lacks a zinc finger motif (27). The phylogenetically distant Ty3 retrotransposons produce multiple forms of Gag, which are encoded by GAG3. Among these are a 39- or 38-kDa protein and its two processed products, a 26-kDa capsid (the outer protein shell of a virus particle) and a 9-kDa nucleocapsid (14). Tf1 of Schizosaccharomyces pombe is similar to Ty5 in that it encodes a single ORF. The Tf1 polyprotein is processed to generate a 27-kDa Gag species (1). Like Ty1, Tf1 does not encode a zinc finger motif characteristic of nucleocapsid proteins (26).
FIG. 5.
Summary of Ty5 proteins. (A) Estimated sizes of the processed Ty5 proteins are based on their mobility on SDS-polyacrylamide gel electrophoresis gels. The sizes of nucleocapsid and protease were estimated based on the position of conserved amino acid sequence domains and the sizes of adjacent Ty5 proteins. (B) Comparison of protein sizes, in kilodaltons, between Ty5 and other retrotransposons. Question marks indicate protein sizes that were extrapolated from amino acid sequence data. Relevant references for the other retrotransposons are found in the text.
Immunoblots prepared from cells expressing a C-terminally tagged element identified a predominant protein species of 59 kDa that we designate RT (Pol-p59). Based on molecular weight extrapolations from the Ty5 amino acid sequence, Pol-p59 is large enough to encompass all of the amino acid sequence domains that characterize RT and RH. In comparison, Ty1 and Ty3 RT are 60 and 55 kDa, respectively (13, 14) (Fig. 5B). Immunoblots prepared from cells expressing a centrally located His tag revealed an 80-kDa protein, which we believe is IN (Pol-p80). The His tag resides within a poorly conserved region downstream of the catalytic domain of IN. Our lab has previously described a role in targeting integration for sequences further downstream from the His tag (amino acid 1094) (11). When a His tag is inserted at position 1094, the resulting protein is identical in size to the IN species observed in this study (W. Xie and D. Voytas, unpublished data). This places the protease cleavage site downstream of residue 1094, which is consistent with the observed size of RT. Comparatively, Ty1, Ty3, and Tf1 IN are 90, 61, and 56 kDa, respectively (13, 15, 25). We provide evidence that Ty5 also encodes a protease (see below), but because it was not epitope tagged, we did not detect it in our experiments. Based on the protein species observed and the derived amino acid sequence of Ty5, we predict that protease approximates 17 kDa. Proteases encoded by Ty1 and Ty3 are 23 and 16 kDa, respectively (13, 23).
Protease is required for Ty5 protein processing.
The single ORF present in the Ty5 element suggests that it specifies a large polyprotein that is processed to give rise to the various products described above. Typically, a dimeric aspartic protease cleaves retroelement-encoded polyproteins (20). Evidence for a Ty5-encoded protease was first suggested by amino acid sequence comparisons with closely related elements such as copia, Ta1, and Ty1, all of whose products have a motif conserved among protease active sites (D[S/T]G) (20, 36). We mutated Ty5's DTG motif in several of the epitope-tagged elements and found that this abolished transposition. Elements with the mutation expressed an approximately 182-kDa protein, which we purified by nickel affinity chromatography, confirming that it carried a His tag. These results indicate that the DTG residues are important for transposition and protein processing, and they likely define the active site of Ty5 protease. It is unlikely that Ty5 expresses proteins from spliced mRNAs, like the single-ORF copia elements of D. melanogaster (6, 34). We have previously shown that Ty5 expresses a single, genome-length mRNA (36), and elements with a protease mutation and epitope tags at both the N and C termini accumulate primarily the 182-kDa protein. This latter observation also suggests that frameshifting events do not occur that introduce stop codons during translation to generate truncated protein products. Nonetheless, we do observe some bands that cross-react with the epitope-specific antibodies, which we believe are polyprotein breakdown products. Until these cross-reacting proteins are better characterized, we cannot completely exclude the possibility that Ty5 employs a nonconventional expression mechanism.
Ty5 protein insolubility.
Virus-like particles produced by Ty1 and Ty3 are soluble and can be purified on sucrose density gradients (12, 14). Tf1 VLPs, however, are less soluble and are readily pelleted from cell lysates by low-speed centrifugation (1). A characteristic feature of Ty5 proteins is their high insolubility, and denaturing agents such as ionic detergents or urea are required to bring them into solution. Insolubility may be an inherent feature of Ty5 proteins, or it may be due to a defect in particle assembly. The latter is consistent with the observation that some Ty1 gag mutants express proteins that are highly insoluble (5). Immunolocalization studies also indicate that mutant forms of Ty1 Gag and Ty5 proteins form aggregates within the cytoplasm (reference 7 and unpublished data). Ty5 VLP assembly may be hindered because Ty5 originates from a heterologous host (S. paradoxus) or because the Ty5 element used in these studies carries naturally occurring mutations. Two independent Ty5 gag mutations have recently been isolated which, when combined, increase Ty5 transposition more than 10-fold (X. Gao, D. Rowley, and D. Voytas, unpublished data). These mutant elements are being tested to determine whether they form VLPs and whether they confer changes in protein solubility.
Ty5 protein processing and the onset of transposition.
The ratio of Gag to Pol is critical for retroelement replication. Altering Ty1 frameshifting, for example, changes the relative abundance of these two proteins and has deleterious consequences for transposition (3, 32). Our data indicate that mature Ty5 Gag and Pol proteins are generated from processing of a single polyprotein, suggesting that they are produced in equimolar amounts. To test this, we measured the relative abundance of Gag and Pol (RT) using a Ty5 element with identical His tags at its N and C termini. Expression was monitored over a time course after induction of transcription so that protein abundance could be correlated with processing and transposition. To ensure that all Ty5 proteins were accounted for, gels were stained after immunoblotting to confirm that electrophoretic transfer was complete, and immunoblots were performed with both soluble and insoluble protein extracts from each of the time points.
Gag-p37 appeared shortly after induction (4 h). This protein peaked at approximately h 14 and then slowly declined in abundance. In contrast, Gag-p27 appeared 4 h after Gag-p37 and increased throughout the time course. The expression profiles for these two proteins suggest an order for Ty5 Gag processing events: the protease cleavage site between Gag and PR that generates Gag-p37 is likely among the first sites cleaved. This is also the first site cleaved in the Ty1 polyprotein (27). Once Gag-p37 is released, a subsequent cleavage near its C terminus gives rise to Gag-p27. Other sites within the polyprotein must be cleaved efficiently, as high-molecular-weight processing intermediates were rarely observed. Throughout the time course experiment, the RT profile paralleled that of Gag-p37; however, RT was of significantly lower abundance. Despite changes in the relative amounts of individual proteins, the ratio of Gag (p27 plus p37) to Pol (p59) was relatively constant and averaged 5:1. This is somewhat lower than that observed for retroviruses (e.g., ∼15 to 1) (reviewed in reference 18) and other retrotransposons (Ty1, 33:1 [21]; Tf1, 26:1 [1]). Because Ty5 proteins were measured from whole-cell extracts, the Gag to Pol ratio could be skewed by the accumulation of nonproductive protein aggregates. The relative amounts of Gag and Pol may differ in VLPs and may more closely approximate those of other retrotransposons.
How is it that levels of Ty5 Pol are significantly less than those of Gag when both are derived from the same polyprotein? One explanation is that Ty5 Gag and Pol are turned over at different rates, similar to the proteins expressed by the Tf1 elements of S. pombe. For Tf1, entry into stationary phase triggers changes in protein abundance (1). In log-phase cultures, the amounts of Tf1 Gag and Pol are equivalent. As the medium is depleted of nutrients, Pol is degraded over an approximately 6-h time period, resulting in a Gag/Pol ratio of 26:1. Tf1 cDNA levels increase when the culture is saturated, indicating that the bulk of reverse transcription occurs when there is a molar excess of Tf1 Gag. The abrupt shift in the amounts of Tf1 proteins and cDNA differs from what we observed with Ty5. Ty5 proteins gradually changed in abundance throughout the growth of the culture, and the Ty5 Gag/Pol ratio remained relatively constant. Transposition was detected at the earliest time point and increased 26-fold over the course of the experiment. There was no clear correlation between Ty5 transposition and the Gag/Pol ratio. Nevertheless, an increase in this ratio (from 5:1 to 15:1) was observed after the culture reached saturation. Ty5 protein abundance, therefore, may be sensitive to culture conditions, but culture conditions do not appear to be the primary signal that triggers changes in protein stability and transposition.
The organization of gag and pol on separate reading frames is typical of only the small subset of retroelements that have been characterized to date. We have already discussed two exceptions, namely, copia of D. melanogaster and Tf1 of S. pombe. Many other examples are found in plants, where most retrotransposons carry gag and pol on a single ORF, and this includes both Ty1/copia and Ty3/gypsy elements (reviewed in reference 24). Among the single-ORF retrotransposons, Tf1 and now Ty5 appear to regulate levels of Gag and Pol by differential protein stability. Posttranslational mechanisms, therefore, may be widely used to meet the apparently universal requirement among retroelements for an excess of Gag relative to Pol. Additional studies that test whether changes in Ty5 protein stability affect transposition will be required to substantiate this hypothesis.
ACKNOWLEDGMENTS
We thank Ning Ke, Weiwu Xie, and Xiaowu Gai for constructing several of the epitope-tagged Ty5 elements.
This work was supported by NIH grant GM51425 and by Hatch Act and State of Iowa funds.
Footnotes
Journal paper J-19002 of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa, project no. 3383.
REFERENCES
- 1.Atwood A, Lin J H, Levin H L. The retrotransposon Tf1 assembles virus-like particles that contain excess Gag relative to integrase because of a regulated degradation process. Mol Cell Biol. 1996;16:338–346. doi: 10.1128/mcb.16.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene/Wiley Interscience; 1987. [Google Scholar]
- 3.Belcourt F M, Farabaugh P J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke J D, Eickbush T, Sandmeyer S B, Voytas D F. Pseudoviridae. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carsten E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. New York, N.Y: Academic Press; 2000. pp. 349–351. [Google Scholar]
- 5.Brachmann C B, Boeke J D. Mapping the multimerization domains of the Gag protein of yeast retrotransposon Ty1. J Virol. 1997;71:812–817. doi: 10.1128/jvi.71.1.812-817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brierley C, Flavell A J. The retrotransposon copia controls the relative levels of its gene products post-transcriptionally by differential expression from its two major mRNAs. Nucleic Acids Res. 1990;18:2947–2951. doi: 10.1093/nar/18.10.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brookman J L, Stott A J, Cheeseman P J, Adamson C S, Holmes D, Cole J, Burns N R. Analysis of TYA protein regions necessary for formation of the Ty1 virus-like particle structure. Virology. 1995;212:69–76. doi: 10.1006/viro.1995.1454. [DOI] [PubMed] [Google Scholar]
- 8.Curcio J M, Garfinkel D J. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farabaugh P J. Programmed translational frameshifting. Annu Rev Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 10.Farabaugh P J, Zhao H, Vimaladithan A. A novel programmed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. . (Erratum, 75:826.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gai X, Voytas D F. A single amino acid change in the yeast retrotransposon Ty5 abolishes targeting to silent chromatin. Mol Cell. 1998;1:1051–1055. doi: 10.1016/s1097-2765(00)80105-7. [DOI] [PubMed] [Google Scholar]
- 12.Garfinkel D J, Boeke J D, Fink G R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42:507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 13.Garfinkel D J, Hedge A M, Youngren S D, Copeland T D. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65:4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen L J, Chalker D L, Orlinsky K J, Sandmeyer S B. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J Virol. 1992;66:1414–1424. doi: 10.1128/jvi.66.3.1414-1424.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen L J, Sandmeyer S B. Characterization of a transpositionally active Ty3 element and identification of the Ty3 integrase protein. J Virol. 1990;64:2599–2607. doi: 10.1128/jvi.64.6.2599-2607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honigman A, Wolf D, Yaish S, Falk H, Panet A. cis acting RNA sequences control the gag-pol translation readthrough in murine leukemia virus. Virology. 1991;183:313–319. doi: 10.1016/0042-6822(91)90144-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- 19.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 20.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami K, Pande S, Faiola B, Moore D P, Boeke J D, Farabaugh P J, Strathern J N, Nakamura Y, Garfinkel D J. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993;135:309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ke N, Voytas D F. High frequency cDNA recombination of the Saccharomyces retrotransposon Ty5: the LTR mediates formation of tandem elements. Genetics. 1997;147:545–556. doi: 10.1093/genetics/147.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchner J, Sandmeyer S. Proteolytic processing of Ty3 proteins is required for transposition. J Virol. 1993;67:19–28. doi: 10.1128/jvi.67.1.19-28.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, Bennetzen J L. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 25.Levin H L, Weaver D C, Boeke J D. Novel gene expression mechanism in a fission yeast retroelement: Tf1 proteins are derived from a single primary translation product. EMBO J. 1993;12:4885–4895. doi: 10.1002/j.1460-2075.1993.tb06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin H L, Weaver D C, Boeke J D. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol Cell Biol. 1990;10:6791–6798. doi: 10.1128/mcb.10.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merkulov G V, Swiderek K M, Brachmann C B, Boeke J D. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J Virol. 1996;70:5548–5556. doi: 10.1128/jvi.70.8.5548-5556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–70. [PubMed] [Google Scholar]
- 30.Voytas D F, Boeke J D. Yeast retrotransposon revealed. Nature. 1992;358:717. doi: 10.1038/358717a0. [DOI] [PubMed] [Google Scholar]
- 31.Wills N M, Gesteland R F, Atkins J F. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H, Boeke J D. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc Natl Acad Sci USA. 1990;87:8360–8364. doi: 10.1073/pnas.87.21.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshinaka Y, Katoh I, Copeland T D, Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci USA. 1985;82:1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshioka K, Honma H, Zushi M, Kondo S, Togashi S, Miyake T, Shiba T. Virus-like particle formation of Drosophila copia through autocatalytic processing. EMBO J. 1990;9:535–541. doi: 10.1002/j.1460-2075.1990.tb08140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youngren S D, Boeke J D, Sanders N J, Garfinkel D J. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol Cell Biol. 1988;8:1421–1431. doi: 10.1128/mcb.8.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou S, Ke N, Kim J M, Voytas D F. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]