Abstract
Osteosarcoma (OS) has strong invasiveness, early metastasis, high drug resistance, and poor prognosis. At present, OS still lacks reliable biomarkers, which makes early diagnosis of OS more difficult. AOC3 is highly expressed in OS and highly correlated with lung metastasis. qRT-PCR could identify mRNA levels of genes. Immunohistochemistry and Western blot assays could detect protein levels. Immunofluorescence and ELISA assays were applied to evaluate the activation of neutrophils. Additionally, transwell and wound healing assays evaluated cell migration and invasion abilities. Tube formation and sphere-forming assays were applied to detect the angiogenesis. C57BL/6 mice were injected with OS cells to establish a xenograft tumor model to observe the lung metastasis of OS. Flow cytometry is used to evaluate the ability of tumor cells to recruit neutrophils. AOC3 was significantly overexpressed in OS, and down-regulation of AOC3 could inhibit OS migration, invasion, and angiogenesis. AOC3 could increase tumor development and lung metastasis of OS in vivo experiments. The promoting effect of AOC3 on tumor lung metastasis was achieved by recruiting tumor neutrophils. Activated NETs could up-regulate the metastatic ability of OS cells. Tumor neovascularization also played a role in metastasis, and AOC3 supported tumor neovascularization. AOC3 accelerates lung metastasis of OS by recruiting tumor-related neutrophils and utilizing NETs and tumor vascularization formation.
Keywords: Osteosarcoma, AOC3, Neutrophils, NETs, Angiogenesis
Graphical abstract
1. Introduction
OS, osteosarcoma, is the most common primary bone sarcoma and bone malignancy [1,2]. OS is commonly found in the proximal tibia, distal femur, and humerus [3]. OS often causes pathological fractures and severe pain because of its highly invasive ability [4]. It is characterized by early metastasis, increased drug resistance, and poor prognosis [5]. According to statistics, 15%–20 % of OS patients have lung metastasis and about 40 % of OS patients grow metastasis at an advanced stage of the disease [6,7]. At present, the treatment effect of surgery and chemotherapy on metastatic OS is very limited, with a long-term survival rate of less than 30 % [8,9]. Thus, OS still lacks reliable biomarkers, which makes early diagnosis and treatment of OS more difficult.
AOC3, also called VAP-1, is the non-classical endothelial adhesion molecule in leukocyte recycling, can promote leukocyte adhesion [10,11]. Abundant studies have highlighted the important role of AOC3 in inflammation and cancers. For instance, in the inflammatory process, AOC3 participates in the transfer of white blood cells from the blood to inflammatory tissue, where these cells amplify the inflammatory response [[12], [13], [14], [15], [16]]. In OSCC, AOC3 knockdown represses tumor growth and metastasis [17]. In breast cancer, AOC3 is reported to be positively associated with distant metastasis [18].
Neutrophils are polymorphonuclear white blood cells that serve as the innate immune system's 1st line guardians [19]. Under normal circumstances, neutrophils (not activated) move in the peripheral blood circulation [20]. When pathogens invade or endogenous stimuli are released, inflammatory reactions occur and neutrophils are activated [21,22]. Neutrophils, also as a selective molecular promoter, have dual effects in promoting and anti-tumor effects [[23], [24], [25], [26], [27]]. Neutrophils and other mesenchymal factors jointly promote tumor angiogenesis to promote tumor growth in certain types of cancer [24,28]. A previous study has shown that AOC3 acts in renal ischemia injury's pathophysiology by improving the infiltration of neutrophils and generating local H2O2 gradients [29]. Also, AOC3 down-regulation prevents the growth and metastasis of OSCC cells via suppressing neutrophil infiltration [17]. However, whether AOC3 makes an impact on OS related to neutrophils remains unknown.
In this study, we first detected AOC3 expression in OS tissues and cells. Then, we investigated whether AOC3 exerted a role in the lung metastasis of OS and the action mechanism.
2. Materials and methods
2.1. Specimens
The tumor tissues and normal tissues in pairs (n = 20 pairs) were obtained from OS patients (12 patients with lung metastasis and 8 patients without lung metastasis) at Beijing Cancer Hospital from March 2019 to October 2021. Histological diagnosis of patients was conducted blindly by two experienced pathologists. Tissue samples were surgically collected from these patients before the commencement of chemotherapy or radiotherapy. All samples were preserved in liquid nitrogen for long-term storage. This study acquired the Ethics Committee's Ethics approval of Beijing Cancer Hospital (2023KT39) and informed consent was obtained from all the patients. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
2.2. Cell lines
HOS, U2OS, Well5, KHOS, hFOB1.19, and HUVECs were bought and passaged in DMEM media with 10 % FBS and 1 % penicillin/streptomycin in a 5 % (v/v) at 37 °C in CO2 humidified environment.
2.3. Cell transfection
The pcDNA3.1-AOC3, short hairpin RNA (sh)-AOC3, pcDNA3.1-NC, and sh-negative control (NC) were obtained. Incorporation of OS cells (2 × 105 cells) was done with 2 μg pcDNA3.1-AOC3 (OE-AOC3), pcDNA3.1/NC (OE-NC), sh-AOC3, sh-NC according to the guidelines of Lipofectamine 3000. The cells were subjected to other tests following their 2 d-incubation period.
2.4. qRT-PCR
We isolated total RNA from OS cells and hFOB1.19 cells and quantified. Next, RNA was used as a template to form cDNA by means of the reverse transcription kit. The qRT-PCR was done with the SYBR® Green qRT-PCR Kit (Promega, USA). PCR conditions were maintained as follows: 5 min at 95 °C, 95 °C for 10 s at 40 cycles, 60 °C for 30 s, and 72 °C for 15 s. The primers of AOC3, CD31, VEGFA, and GAPDH are listed in Table 1. The AOC3, CD31, and VEGFA mRNA expression levels were estimated with the 2−ΔΔct method. We used GAPDH as the endogenous control.
Table 1.
The primer sequences for qRT-PCR in this study.
| Genes | Sequences (5’∼3′) | |
|---|---|---|
| AOC3 | Forward | CGGTGCTGGCGAGAAGTTTG |
| Reverse | TCTGCCCAGGCCCAGTTCTTC | |
| CD31 | Forward | CACAACAAACAAGCTAGCAAGA |
| Reverse | TTTGGCTGCAACTATTAAGGTG | |
| VEGFA | Forward | ACCATGAACTTTCTGCTGTCTTGGGTGCAT |
| Reverse | TCACCGCCTCGGCTTGTCACATCTGCAAGT | |
| GAPDH | Forward | ACCACAGTCCATGCCATCAC |
| Reverse | TCCACCACCCTGTTGCTGTA |
2.5. Mouse tumorigenesis assay
C57BL/6 mice (wild-type, 8-wks-old, male) were acquired and housed for one week to make them well-adapted to the environment. To mimic tumor metastasis, after transfected with sh-AOC3/sh-NC, we injected KHOS cells (5 × 106 cells in 200 μL of PBS, labeled by LUC) into the tail vein of each mouse (n = 3/group).
To construct an orthotopic transplantation tumor model, we suspended KHOS cells (5 × 106) transfected with OE-AOC3/OE-NC in PBS (200 μL) and injected into the tibial bone marrow cavity using a microliter syringe. In one group of mice (n = 3/group), anti-Ly6G (BE0075-1, BioXCel, USA) was injected via intraperitoneal injection to consume neutrophils. Mice were given two initial intraperitoneal injections (200 μg/mouse) of anti-Ly6G every 3 days before injecting tumor cells and then maintenance injection twice a week.
After a period of four weeks, the IVIS Lumina Series III (PerkinElmer, USA) system was used to observe OS metastasis of mice to their lungs. According to the manufacturer's recommendation, Cyanine 5.5 (Lumiprobe, Hunt Valley, MD, USA) labeled the anti-AOC3 was injected into mice via tail vein injection. After 1 d, mice were imaged to analyze the lung metastasis of OS. Finally, mice were euthanized with an overdose of pentobarbital. The Ethics Committee of Beijing Viewsolid Biotechnology Co. Ltd. (VS2126A00157) approved the animal experiments. All methods are reported in accordance with ARRIVE guidelines.
2.6. Immunohistochemistry (IHC) staining
We fixed, embedded, and sliced tumor tissues into segments (4-μm-thick). Afterward, these segments were deparaffinized and rehydrated. Antigen restoring was conducted using citrate buffer (high temperature; pH 6.0). These sections were incubated with primary antibodies: anti-VAP1 (ab42885, dilution: 1 μg/mL), anti-Ki67 (ab15580, 0.1 μg/mL), anti-Myeloperoxidase (MPO) (ab9535, dilution: 1/25) and anti-CD31 (ab28364, dilution: 1/50) at refrigeration temperature. On the next day, we incubated these segments with the secondary antibody Goat F(ab')2 Anti-Rabbit IgG F(ab')2 (HRP) (ab6112, dilution: 1: 500) for 30 min. Segments were stained with diaminobenzidine and hematoxylin and examined the results by means of a microscope.
2.7. H&E staining
We fixed mice lung tissues in neutral formalin (10 %) and embedded them in paraffin. We cut tissues into 4 μm segments and stained them with H&E. The findings were detected under a microscope.
2.8. Neutrophil isolation from peripheral blood of healthy individuals
We isolated peripheral blood's neutrophils by density centrifugation employing a Pancoll™ gradient. In brief, we diluted 10 mL blood containing EDTA in 10 mL PBS and layered on pancoll™ (10 mL). Following centrifugation for 30 min, we separated neutrophils via dextran sedimentation from the erythrocyte-rich pellet and eliminated residual erythrocytes by hypotonic lysis and re-suspended neutrophils in DMEM with low endotoxin bovine serum albumin (0.5 %). We assessed neutrophil purity by forward and side scatter flow cytometric analysis.
To explore whether AOC3 mediated the release of neutrophil extracellular traps (NETs), isolated neutrophils were treated with ASP8232 (AOC3 inhibitor) for 1 d before exposure to KHOS cells.
For investigating the effect of NETs on OS cells, isolated neutrophils were treated with hFOB1.19 or KHOS cells for 12 h in the absence and presence of DNase I (25 U/mL; Sigma-Aldrich, Shanghai, China).
2.9. Chemotaxis assay
We utilized a transwell chamber for evaluating the neutrophils' chemotactic activity. Neutrophils were placed in the upper chamber from the peripheral blood of individuals (healthy status; activity and purity above 95 %). The supernatant of sh-AOC3/sh-NC transfected KHOS cells was assembled and placed in the bottom chamber (200 μL). The neutrophils were cultivated at 37 °C for 2 d. Next, we removed the upper chamber's cells with cotton swabs. We exposed the bottom chamber's cells to crystal violet (0.2 %) for staining for 5 min. Then, we utilized a microscope (inverted) to count the neutrophil number.
2.10. Transwell assay for detection of KHOS cell invasion and migration
To evaluate the invasion and migration capabilities of KHOS cells, KHOS cells (2 × 104 cells) in the medium were inoculated in the top chamber (with or without a 2 mg/mL Matrigel-coated membrane). About 500 μL complete medium was appended to the bottom compartment. After 3 d incubation, we removed the cells using a cotton swab and mixed the cells moving to the bottom of the membrane employing 4 % formaldehyde for 10 min. Finally, we stained the cells with crystal violet (0.5 %) for 15 min.
2.11. Flow cytometry
Peripheral blood was collected from mice, and 1 mL of 1 × RBC buffer was used to dissolve the red blood cells and incubate them at room temperature until the sample became semi-transparent. The sample was neutralized with PBS and centrifuge at 4 °C for 5 min. We cultured the sample with APC-labeled anti-CD11b antibody (ab8878, Abcam, dilution: 0.1 μg for 106 cells) and PerCP-labeled anti-Ly6G antibody (ab238132, Abcam, dilution: 1/600) in Brilliant stain buffer on ice for 1/2 h. We washed the sample with PBS and then centrifuged it at 4 °C, 400×g for 5 min, and discarded its supernatant. We filtered the debris with a mesh and analyzed the percolated cells using an Attune flow cytometer (Applied Biosystems, USA).
2.12. Immunofluorescence (IF) assay
We performed an IF assay for detecting MPO and Cit-H3 expression levels associated with tumor-associated neutrophils (TANs) infiltration and the NET generation. On the glass slide, We cultured KHOS cells or neutrophils to 70 % confluence. After washing two times with 1 × PBS, the cells were fixed with paraformaldehyde (4 %) for 1 h. Next, cells were permeabilized with 0.2 % Triton X-100 (#X100) in 1 × PBS for 15 min at 37 °C and closed by normal goat serum (10 %) at 25 °C for 1 h. We incubated cells with an Anti-MPO antibody (ab45977, dilution: 5 μg/mL) and Anti-Histone H3 (citrulline R8) (ab219406, Abcam, dilution: 1/1000) at 1 d at 4 °C. The cells were incubated with the goat anti-mouse IgG Alexa 488 conjugated fluorescence secondary antibody (R37120, dilution 1/1000) for 1 h at 25 °C following washing PBS. Ultimately, the cells were stained with DAPI and studied by means of a microscope.
2.13. Western blot
Protein extraction was conducted based on instructions from a radioimmunoprecipitation assay kit (Beyotime, China). We separated extracted proteins through SDS-PAGE and then transferred them to the PVDF membrane. After membranes were blocked for 2 h, they were incubated with primary antibodies like Anti-VAP1 (ab42885, Abcam, dilution: 1 μg/ml), Anti-Histone H3 (citrulline R8) (ab219406, dilution: 1/1000) and anti-GAPDH (ab9485, dilution 1/2500) at 4oC for 1 d. Next, we added a peroxidase-labeled secondary antibody (anti-rabbit IgG, ab6721, dilution 1:2000) to incubate for 2 h. Finally, we observed protein bands with the chemiluminescence system (ECL) and evaluated them via Image J Software.
2.14. ELISA
Neutrophils (1 × 105) re-suspended with complete medium (300 μL) were placed in 96-well plates. The supernatant was collected 3 days later. We performed ELISA to identify the MPO-DNA complex according to the instructions of a Cell Death ELISA kit (Roche Diagnostics, Germany). We recorded the absorbance value when the value of 450 nm.
2.15. CCK-8 assay
The proliferative capability of HUVECs was assessed by means of a Cell Counting Kit-8 (Beyotime, Shanghai, China). HUVECs (1 × 103/well) treated with KHOS cell culture medium were seeded in a 96-well plate for incubating 1 d, 2 d, and 3 d. Afterward, the Cell Counting Kit solution (10 μL) was added into each well for incubating for another at 37 °C for 2 h. A microplate reader (Molecular Devices, USA) could determine the optical density (OD) value at 450 nm.
2.16. Wound healing assay
HUVECs (3 × 105 cells/well) were plated in 6-well plates, which were cultured with a complete medium that included extracellular matrix molecule (10 μg/mL). After that, a cell monolayer formed. To generate a wound, a pipette tip (200 μL) was used to scratch a straight line on the plate bottom, and the wound distance at 0 h was recorded. Next, HUVECs were cultured with a serum-free medium at 37 °C for 1 d. Finally, we observed the wound distance at 1 d and photographed it via an inverted microscope (Carl Zeiss, Oberkochen, Germany). The formula for calculating the cell migration rate is as follows: (1–24 h wound distance/0 h wound distance) × 100.
2.17. Transwell assay for evaluation of HUVEC invasion
HUVECs in serum-free medium were inoculated in the top chamber with a Matrigel-coated membrane (2 mg/mL). Meantime, 500 μL complete medium was added to the bottom chamber. We removed HUVECs from the top of the filters using a cotton swab and HUVECs moved to the bottom of the membrane and fixed with 4 % formaldehyde for 10 min after incubation for 2 d. Finally, HUVECs were dyed with crystal violet (0.5 %) for 15 min.
2.18. Tube formation assay
We inoculated HUVECs (2 × 104 cells/well) into the 96-well plate that was coated with Matrigel. The KHOS cells' supernatant was added. After 8 h culture, the HUVEC's tubular structure was observed under an inverted microscope.
2.19. Statistical analysis
Data were displayed mean ± standard deviation of at least three replications. GraphPad Prism 7.0 (GraphPad Software, USA) was utilized for data analysis. We analyzed multiple group differences using a one-way ANOVA. We performed Single group comparisons by means of a student's t-test. P < 0.05 indicated statistical significance.
3. Results
3.1. AOC3 is up-regulated in OS tissues, especially in lung metastasis tissues
The data were sourced from the TARGET and GEO accession number: GSE14359. The TARGET database identified 191 DEGs in metastatic and non-metastatic OS, of which 65 were highly expressed and 126 were lowly expressed. GSE14359 (OS dataset in GEO database) included a total of 20 samples, including 10 samples from OS, 8 samples from patients with lung metastasis of OS, and 2 samples from the normal control group. There were 164 co-up-regulated and 34 co-down-regulated genes. Only AOC3 and vWF genes were up-regulated in lung metastasis compared with primary OS and normal bone tissue (Fig. 1A). Prognostic analysis has suggested that high AOC3 expression level is closely associated with poor prognosis (vWF prognosis is not significant and not presented) (Fig. 1B). The result of qRT-PCR showed that AOC3 expression was evidently boosted in non-metastatic OS tissues when comparing with the normal tissues, and AOC3 expression was significantly boosted in metastatic OS tissues when comparing with non-metastatic OS tissues (Fig. 1C). IHC staining also revealed that AOC3 expression was higher in the metastatic group when compared with the non-metastatic group (Fig. 1D). We then detected the expression level of AOC3 in hFOB1.19, Well5, KHOS, HOS, and U2OS. AOC3 had a higher expression level in Well5, KHOS, HOS, and U2OS cells compared to hFOB1.19 cells (Fig. 1E).
Fig. 1.
AOC3 was up-regulated in OS tissues, especially in lung metastasis tissues (A) TARGET and GSE14359 databases screened differentially expressed genes in OS. T-Tumor, M-Metastasis, C-Control. (B) Analyzing the relationship between high expression of AOC3 and poor prognosis by TARGET database. (C) qRT-PCR was used to detect the mRNA levels of AOC3 in normal tissues, OS tissues, and metastatic OS tissues. (D) Immunohistochemical assay was used to detect the protein levels of AOC3 in normal tissues, OS tissues, and metastatic OS tissues. (E) qRT-PCR was used to detect the mRNA expression of AOC3 in osteoblast lines (hFOB1.19), metastatic cell lines (Well5, KHOS), and non-metastatic cell lines (HOS, U2OS). *P < 0.05, **P < 0.01, ***P < 0.001. Each experiment was executed in triplicate.
3.2. AOC3 promotes lung metastasis of OS
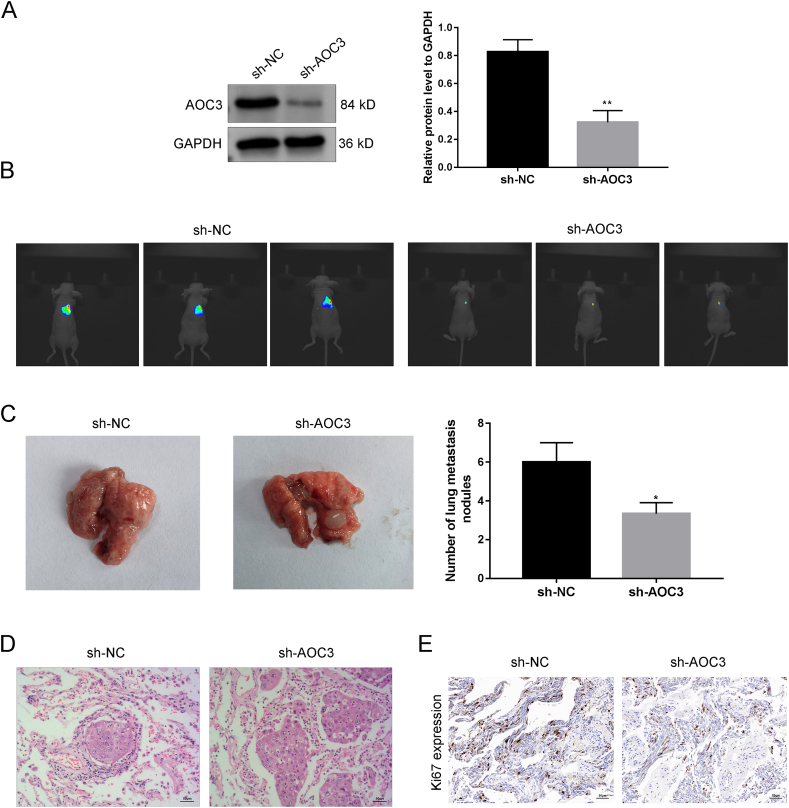
Western blotting assay could verify the transfection efficiency of sh-AOC3 in KHOS cells, and the AOC3 level in the sh-NC group was significantly higher than compared with the sh-AOC3 group (Fig. 2A). IVIS showed the metastasis of OS to the lungs, indicating a decrease in lung cancer cell signaling in the sh-AOC3 group (Fig. 2B). Quantification of lung metastasis in mice OS revealed a decrease in lung metastasis after AOC3 knockdown (Fig. 2C). According to HE staining analysis of lung tissues, compared with the sh-NC group, the sh-AOC3 group showed decreasing lung inflammatory infiltration in mice (Fig. 2D). The immunohistochemical results confirmed a decrease in the Ki67 level in the sh-AOC3 group, indicating that AOC3 down-regulation inhibited tumor metastasis (Fig. 2E).
Fig. 2.
AOC3 promotes lung metastasis of OS (A) The transfection efficiency of sh-AOC3 was verified by Western blotting. (B) KHOS cells were injected into the tail vein to construct an OS mice model, and the lung metastasis of OS was observed by IVIS.(C) The number of pulmonary nodules in mice was recorded. (D) HE analysis of mouse lung tissue. (E) Immunohistochemical experiments were used to detect Ki67 expression in lung tissue of mice. **P < 0.01. Each experiment was executed in triplicate.
3.3. AOC3's promoting effect on lung metastasis by neutrophils
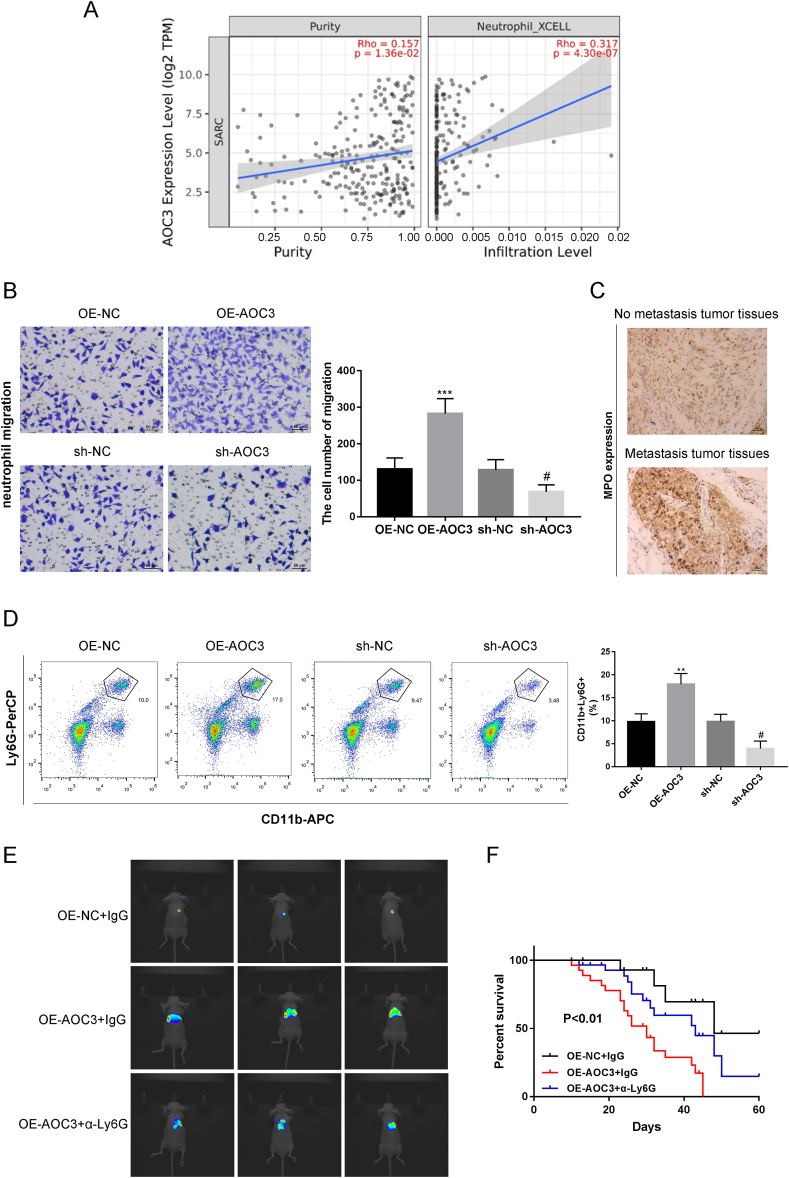
Timer 2.0 database was used to predict a significant correlation between AOC3 expression and neutrophil infiltration in sarcoma (Fig. 3A). The number of neutrophils infiltrating the bottom chamber in the sh-AOC3 group was reduced compared with the sh-NC group. The OE-AOC3 group significantly increased compared with the OE-NC group, suggesting that AOC3 significantly influenced the chemotactic activity of neutrophils (Fig. 3B). IHC staining showed that the MPO (a marker of neutrophils) in the metastatic group was higher than the non-metastatic group, indicating that metastatic tumors recruited more neutrophil infiltration than primary tumors (Fig. 3C). Flow cytometry analysis proved that overexpression of AOC3 increased the percentage of neutrophils with CD11b+ and Ly6G+ in mice bearing orthotopic transplanted tumors, while knocking down AOC3 reduced its percentage (Fig. 3D). We validated whether AOC3 participated in lung metastasis of OS through the recruitment of granulocytes. Constructing an orthotopic transplant tumor model, specific depletion of neutrophils in mice with anti-Ly6G can reverse AOC3-induced lung metastasis, leading to a decrease in lung metastasis in mice and prolonging survival (Fig. 3E and F).
Fig. 3.
Promoting effect of AOC3 in lung metastasis is mediated by neutrophil (A) The TIMER2.0 database was used to predict the correlation between AOC3 expression and neutrophil infiltration. (B) Transwell assay for detecting neutrophil chemotactic activity. (C) Immunohistochemical evaluation of MPO (neutrophil marker) levels in both metastatic and non-metastatic groups. (D) Flow cytometry analysis of the percentage of CD11b+ and Ly6G+ neutrophils. A microliter syringe was used to inject KHOS cells into the tibial bone marrow cavity of mice to construct an orthotopic transplant tumor model, and specifically consume neutrophils in mice using anti Ly6G antibodies. (E) The lung metastasis of OS was observed by IVIS. (F) Survival analysis was conducted on mice in the OE-AOC3+IgG, OE-AOC3+α-Ly6G, and OE-NC + IgG groups. ***P < 0.001, #P < 0.05. Each experiment was executed in triplicate.
3.4. Tumoral AOC3 mediates the release of NETs
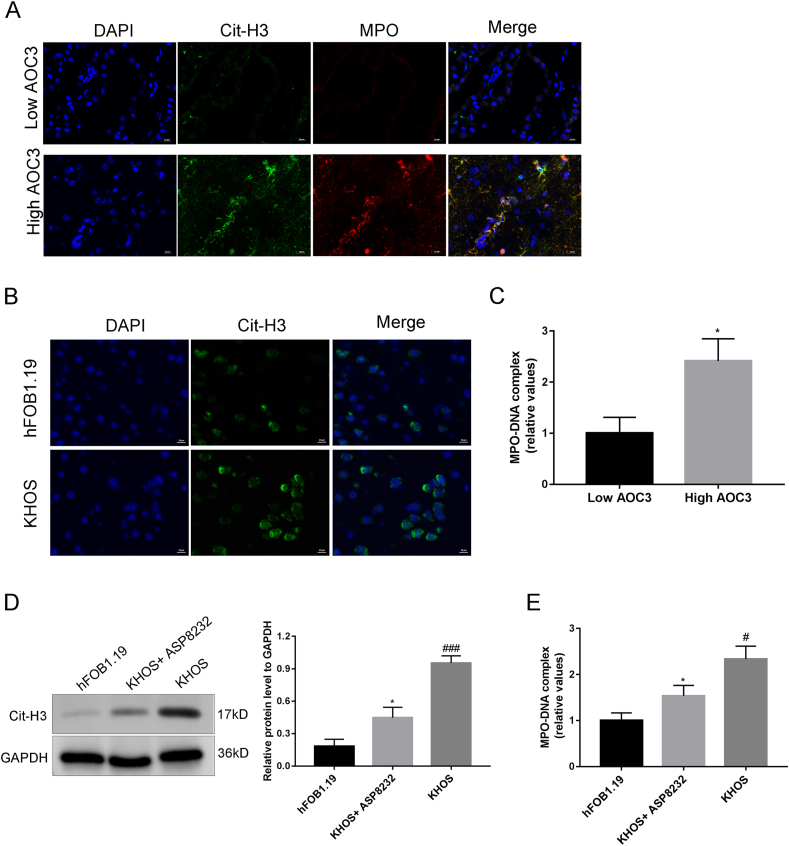
Immunofluorescence assay could detect MPO and Cit-H3 expression levels (markers of NETs) in OS samples, and the high AOC3 expression level in tumor cells was associated with the infiltration of tumor neutrophils and the increase in NETs production (Fig. 4A). Neutrophils were treated with the supernatant of KHOS or hFOB1.19 cells for 12 h from peripheral blood of healthy individuals. Immunofluorescence and ELISA results showed that in comparison with the neutrophils in the hFOB1.19 group, the neutrophils in the KHOS group exhibited phenotypes similar to those derived from their tumor tissues, increased Cit-H3 expression, and increased NETs release (Fig. 4B and C). In addition, neutrophils were treated with ASP8232 (AOC3 inhibitor) before exposure to KHOS. Compared with the KHOS group, their tumor-induced MPO and Cit-H3 in the KHOS + ASP8232 group were up-regulated and DNA release was markedly antagonized (Fig. 4D and E).
Fig. 4.
Tumoral AOC3 mediates the release of neutrophil extracellular traps (NETs) (A) Immunofluorescence evaluation of MPO (red) and Cit-H3 (green) levels in OS samples. (B) Immunofluorescence assay was used to detect the expression intensity of Cit-H3 in control neutrophils exposed to hFOB1.19 and neutrophils exposed to cancer cells. (C) The concentration of MPO was evaluated by ELISA. (D) The Western blot assay detected the expression level of Cit-H3 in neutrophils that exposed to KHOS and treated with ASP8232 (AOC3 inhibitor). (E) ELISA detected the MPO level in neutrophils that exposed to KHOS and treated with ASP823. *P < 0.05, **P < 0.01, #P < 0.05,##P < 0.01, ###P < 0.001. Each experiment was executed in triplicate. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. NETs can enhance the metastatic ability of OS cells
Immunofluorescence assay showed that DNase I could effectively disrupt NET release exposed to KHOS (Fig. 5A). Transwell assay showed that compared to the KHOS group, neutrophils exposed to KHOS had stronger migration activity, while DNase I partially reversed the promoting effect of KHOS (Fig. 5B).
Fig. 5.
NETs can upregulate the metastatic ability of OS cells Analysis of clinical pathological characteristics data of patients. (A) Immunofluorescence evaluation of the inhibitory effect of DNase I on the release of NETs by neutrophils. (B) Migration ability of KHOS cells was detected by Transwell assay. ***P < 0.001, ##P < 0.01. Each experiment was executed in triplicate.
3.6. AOC3 promotes tumor neovascularization
HUVECs were incubated with a KHOS culture medium. CCK-8 results showed that AOC3 silencing significantly reduced the number of live cells in HUVECs (Fig. 6A). The transwell assay and wound healing assay confirmed that the invasion and migration capabilities of HUVECs were significantly inhibited after knocking down AOC3 (Fig. 6B and C). In addition, we determined that the low level of AOC3 prevented angiogenesis partly, presenting as a reduction of tube formation and the down-regulation of VEGFA (endothelial growth factor) expression in HUVECs (Fig. 6D and E). Immunohistochemical and PCR assays were conducted on the tumor tissue of the mice, which displayed that the expression of CD31 (a marker of vascular endothelial cells) and VEGFA in the sh-AOC3 group was reduced when compared with the sh-NC group (Fig. 6F and G).
Fig. 6.
AOC3 promotes tumor neovascularization HUVECs were incubated with KHOS cell culture medium. (A) CCK-8 was used to evaluate the cell viability of HUVECs. (B) Evaluation of HUVECs migration through wound healing assay. (C) Evaluation of HUVECs invasion through Transwell assay. (D) Tube formation assay was used to detect the angiogenesis. (E) VEGFA concentration was detected by ELISA. (F) Detecting the expression of AOC3 and CD31 in orthotopic transplanted tumors of mice through immunohistochemistry staining. (G) The expression of CD13 and VEGFA in tumor tissue of mice was detected by qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001. Each experiment was executed in triplicate.
4. Discussion
OS is characterized by strong invasion, early metastasis, high drug resistance, and poor prognosis [1,3,4,[30], [31], [32]]. AOC3 is abnormally expressed in various cancers. For example, in colorectal cancer, low AOC3 expression level is related to poor prognosis [33]. In breast cancer, AOC3 is highly expressed and positively correlated with lymphatic invasion and distal metastasis [18]. In particular, AOC3 is highly expressed in tumor tissues of glioma patients, and its expression is related to the poor prognosis of glioma patients [34]. Consistent with this report, our results have displayed that AOC3 is highly expressed in OS tissues and cells, especially in metastatic OS, and high expression of AOC3 was closely related to the poor prognosis of OS patients. Subsequently, we silenced AOC3 in KHOS cells and established an OS mice model. We found that AOC3 down-regulation significantly weakened the spontaneous metastasis of OS to the lungs. The experimental results we obtained were similar to the following studies. Silencing of AOC3 inhibits OSCC cell proliferation and tumor metastasis in vivo [17]. Inhibiting AOC3 and inhibiting endothelial cell activation can reduce tumor cell survival and metastasis, and AOC3 recruits myeloid cells to facilitate lung metastasis of breast cancer and colorectal cancer in mice [35]. AOC3 can exacerbate tumor malignancy by enhancing leukocyte recruitment and is associated with OS immune infiltration [36,37]. In this study, we predicted using TIMER2.0 and found a significant correlation between AOC3 expression and neutrophil infiltration in sarcoma. Overexpression of AOC3 enhanced the chemotactic activity of neutrophils toward tumor cells, especially toward metastatic tumor cells, while knocking down AOC3 reduced this chemotactic activity. Tumor cells recruit TANs and play a key role in the metastasis and incidence of many cancers [38]. Therefore, we validated whether AOC3 participated in lung metastasis of OS through neutrophil recruitment. When we specifically consumed neutrophils in mice using anti-Ly6G, we found that AOC3-induced lung metastasis was reversed, leading to a decrease in lung metastasis and prolonging survival. These findings suggested that the silencing of AOC3 prevented lung metastasis via reducing neutrophil recruitment in OS. Our findings were similar to a previous study, which indicates that AOC3 down-regulation inhibits the growth and metastasis of OSCC cells by suppressing neutrophil infiltration [17].
When neutrophils become strongly activated, these cells can form NETs [29]. More importantly, AOC3 overexpression is reported to increase the extravasation of neutrophils to inflammatory sites [10,12]. Here, we detected the expression of MPO (as a maker of neutrophils) and Cit-H3 (as a maker of NETs) in OS samples, indicating that overexpression of AOC3 in tumor cells is associated with increased neutrophil infiltration and NETs production. However, when neutrophils were treated with ASP8232 (an inhibitor of AOC3) when compared with the group treated with KHOS alone, the expression of Cit-H3 was up-regulated and the release of NETs was significantly antagonized. The above experimental results confirm that AOC3 mediates the process of tumor neutrophils releasing NETs. According to previous studies, NETs can promote tumor metastasis by promoting cancer cell invasion, capturing circulating tumor cells, and awakening dormant cancer cells [27,[39], [40], [41]]. Similarly, we found that DNase I could block the release of NETs from neutrophils exposed to KHOS cells and then prevent the migration of KHOS cells.
Tumor neovascularization acts in tumor invasion and metastasis [42,43]. We further investigated whether AOC3 regulated angiogenesis in OS and found that AOC3 down-regulation caused a significant decrease in live cell number. The abilities of migration and invasion, and the number of angiogenesis in HUVECs. These outcomes indicate that AOC3 supports tumor neovascularization, providing an outlook for developing new approaches to improve the efficacy of antiangiogenic therapy.
There are some limitations in our study. First, upstream regulatory mechanisms of AOC3 expression are not investigated in this study. Second, the mechanisms by which AOC3 overexpression affects tumor-associated neutrophil recruitment, neutrophil extracellular trap formation, and tumor vascularization are not explored. Thus, further studies on these aspects are still required.
5. Conclusions
AOC3 was highly expressed in OS and affected lung metastasis by mediating the NETs release from neutrophils and influencing the formation of tumor neovascularization. Our research provides new targets for early OS diagnosis and treatment.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81802689).
Ethics declarations
This study acquired the Ethics Committee's Ethics approval of Beijing Cancer Hospital (2023KT39).
The Ethics Committee of Beijing Viewsolid Biotechnology Co. LTD (VS2126A00157) approved the animal experiments.
Data availability
All data generated or analyzed are included in this published article.
CRediT authorship contribution statement
Luxia Qi: Writing – review & editing, Writing – original draft, Supervision, Conceptualization. Tian Gao: Writing – review & editing, Data curation. Chujie Bai: Writing – review & editing, Data curation. Zhanfei Guo: Writing – review & editing, Data curation. Linjing Zhou: Writing – review & editing, Formal analysis. Xiaodong Yang: Writing – review & editing, Formal analysis. Zhengfu Fan: Writing – review & editing, Methodology, Formal analysis, Conceptualization. Guifang Zhang: Writing – review & editing, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37070.
Contributor Information
Zhengfu Fan, Email: zhengfufan@126.com.
Guifang Zhang, Email: xxchzhangguifang@126.com.
List of abbreviations
- OS
Osteosarcoma
- AOC3
Amine oxidase copper containing 3
- NETs
neutrophil extracellular traps
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Ritter J., Bielack S.S. Osteosarcoma. Ann. Oncol. 2010;21(Suppl 7):vii320–325. doi: 10.1093/annonc/mdq276. [DOI] [PubMed] [Google Scholar]
- 3.Sarhadi V.K., Daddali R., Seppanen-Kaijansinkko R. Mesenchymal Stem cells and extracellular vesicles in osteosarcoma pathogenesis and therapy. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222011035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franchi A. Epidemiology and classification of bone tumors. Clin Cases Miner Bone Metab. 2012;9:92–95. [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson E., Brown H.L. Understanding osteosarcomas. JAAPA. 2018;31:15–19. doi: 10.1097/01.JAA.0000541477.24116.8d. [DOI] [PubMed] [Google Scholar]
- 6.Taran S.J., Taran R., Malipatil N.B. Pediatric osteosarcoma: an updated review. Indian J. Med. Paediatr. Oncol. 2017;38:33–43. doi: 10.4103/0971-5851.203513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean D.C., Shen S., Hornicek F.J., Duan Z. From genomics to metabolomics: emerging metastatic biomarkers in osteosarcoma. Cancer Metastasis Rev. 2018;37:719–731. doi: 10.1007/s10555-018-9763-8. [DOI] [PubMed] [Google Scholar]
- 8.Jafari F., Javdansirat S., Sanaie S., Naseri A., et al. Osteosarcoma: a comprehensive review of management and treatment strategies. Ann. Diagn. Pathol. 2020;49 doi: 10.1016/j.anndiagpath.2020.151654. [DOI] [PubMed] [Google Scholar]
- 9.Tsukamoto S., Errani C., Angelini A., Mavrogenis A.F. Current treatment considerations for osteosarcoma metastatic at presentation. Orthopedics. 2020;43:e345–e358. doi: 10.3928/01477447-20200721-05. [DOI] [PubMed] [Google Scholar]
- 10.Stolen C.M., Marttila-Ichihara F., Koskinen K., Yegutkin G.G., et al. Absence of the endothelial oxidase AOC3 leads to abnormal leukocyte traffic in vivo. Immunity. 2005;22:105–115. doi: 10.1016/j.immuni.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Smith D.J., Salmi M., Bono P., Hellman J., et al. Cloning of vascular adhesion protein 1 reveals a novel multifunctional adhesion molecule. J. Exp. Med. 1998;188:17–27. doi: 10.1084/jem.188.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tohka S., Laukkanen M., Jalkanen S., Salmi M. Vascular adhesion protein 1 (VAP-1) functions as a molecular brake during granulocyte rolling and mediates recruitment in vivo. Faseb. J. 2001;15:373–382. doi: 10.1096/fj.00-0240com. [DOI] [PubMed] [Google Scholar]
- 13.Salmi M., Kalimo K., Jalkanen S. Induction and function of vascular adhesion protein-1 at sites of inflammation. J. Exp. Med. 1993;178:2255–2260. doi: 10.1084/jem.178.6.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salmi M., Tohka S., Jalkanen S. Human vascular adhesion protein-1 (VAP-1) plays a critical role in lymphocyte-endothelial cell adhesion cascade under shear. Circ. Res. 2000;86:1245–1251. doi: 10.1161/01.res.86.12.1245. [DOI] [PubMed] [Google Scholar]
- 15.Boomsma F., Bhaggoe U.M., van der Houwen A.M., van den Meiracker A.H. Plasma semicarbazide-sensitive amine oxidase in human (patho)physiology. Biochim. Biophys. Acta. 2003;1647:48–54. doi: 10.1016/s1570-9639(03)00047-5. [DOI] [PubMed] [Google Scholar]
- 16.Yraola F., Albericio F., Royo M. Inhibition of VAP1: quickly gaining ground as an anti-inflammatory therapy. ChemMedChem. 2007;2:173–174. doi: 10.1002/cmdc.200600245. [DOI] [PubMed] [Google Scholar]
- 17.Xu Q., Chen X., Yu T., Tang Q., et al. Downregulation of VAP-1 in OSCC suppresses tumor growth and metastasis via NF-kappaB/IL-8 signaling and reduces neutrophil infiltration. J. Oral Pathol. Med. 2022;51:332–341. doi: 10.1111/jop.13285. [DOI] [PubMed] [Google Scholar]
- 18.Lai Y.C., Chang S.J., Kostoro J., Kwan A.L., et al. Vascular adhesion protein-1 as indicator of breast cancer tumor aggressiveness and invasiveness. APMIS. 2018;126:755–761. doi: 10.1111/apm.12885. [DOI] [PubMed] [Google Scholar]
- 19.Chan L., Karimi N., Morovati S., Alizadeh K., et al. The roles of neutrophils in cytokine storms. Viruses. 2021;13 doi: 10.3390/v13112318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H., Chen S., Xie M., Zhou C., et al. The complex roles of neutrophils in APAP-induced liver injury. Cell Prolif. 2021;54 doi: 10.1111/cpr.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliveira C., Navarro-Xavier R.A., Anjos-Vallota E.A., Martins J.O., et al. Effect of plant neutrophil elastase inhibitor on leucocyte migration, adhesion and cytokine release in inflammatory conditions. Br. J. Pharmacol. 2010;161:899–910. doi: 10.1111/j.1476-5381.2010.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantovani A., Cassatella M.A., Costantini C., Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 23.Veglia F., Tyurin V.A., Blasi M., De Leo A., et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature. 2019;569:73–78. doi: 10.1038/s41586-019-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Guoqiang L., Sun M., Lu X. Targeting and exploitation of tumor-associated neutrophils to enhance immunotherapy and drug delivery for cancer treatment. Cancer Biol Med. 2020;17:32–43. doi: 10.20892/j.issn.2095-3941.2019.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szczerba B.M., Castro-Giner F., Vetter M., Krol I., et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566:553–557. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 26.Snoderly H.T., Boone B.A., Bennewitz M.F. Neutrophil extracellular traps in breast cancer and beyond: current perspectives on NET stimuli, thrombosis and metastasis, and clinical utility for diagnosis and treatment. Breast Cancer Res. 2019;21:145. doi: 10.1186/s13058-019-1237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albrengues J., Shields M.A., Ng D., Park C.G., et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. 2018;361 doi: 10.1126/science.aao4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granot Z. Neutrophils as a therapeutic target in cancer. Front. Immunol. 2019;10:1710. doi: 10.3389/fimmu.2019.01710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C., Yalavarthi S., Tambralli A., Zeng L., et al. Inhibition of neutrophil extracellular trap formation alleviates vascular dysfunction in type 1 diabetic mice. Sci. Adv. 2023;9 doi: 10.1126/sciadv.adj1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrena A., Wang J., Zhang R., Karadal-Ferrena B., et al. SKP2 knockout in Rb1/p53 deficient mouse models of osteosarcoma induces immune infiltration and drives a transcriptional program with a favorable prognosis. Mol. Cancer Therapeut. 2023 doi: 10.1158/1535-7163.MCT-23-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain R.K. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell. 2014;26:605–622. doi: 10.1016/j.ccell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong Z., Liao Z., He Y., Wu C., et al. Advances in the biological functions and mechanisms of miRNAs in the development of osteosarcoma. Technol. Cancer Res. Treat. 2022;21 doi: 10.1177/15330338221117386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward S.T., Weston C.J., Shepherd E.L., Hejmadi R., et al. Evaluation of serum and tissue levels of VAP-1 in colorectal cancer. BMC Cancer. 2016;16:154. doi: 10.1186/s12885-016-2183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang S.J., Tu H.P., Lai Y.C., Luo C.W., et al. Increased vascular adhesion protein 1 (VAP-1) levels are associated with alternative M2 Macrophage activation and poor prognosis for human gliomas. Diagnostics. 2020;10 doi: 10.3390/diagnostics10050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferjancic S., Gil-Bernabe A.M., Hill S.A., Allen P.D., et al. VCAM-1 and VAP-1 recruit myeloid cells that promote pulmonary metastasis in mice. Blood. 2013;121:3289–3297. doi: 10.1182/blood-2012-08-449819. [DOI] [PubMed] [Google Scholar]
- 36.Marttila-Ichihara F., Castermans K., Auvinen K., Oude Egbrink M.G., et al. Small-molecule inhibitors of vascular adhesion protein-1 reduce the accumulation of myeloid cells into tumors and attenuate tumor growth in mice. J. Immunol. 2010;184:3164–3173. doi: 10.4049/jimmunol.0901794. [DOI] [PubMed] [Google Scholar]
- 37.Pan R., Pan F., Zeng Z., Lei S., et al. A novel immune cell signature for predicting osteosarcoma prognosis and guiding therapy. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1017120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing X., Bai Y., Song J. The Heterogeneity of neutrophil recruitment in the tumor Microenvironment and the formation of Premetastatic Niches. J Immunol Res. 2021;2021 doi: 10.1155/2021/6687474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J., Wysocki R.W., Amoozgar Z., Maiorino L., et al. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci. Transl. Med. 2016;8:361ra138. doi: 10.1126/scitranslmed.aag1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cools-Lartigue J., Spicer J., McDonald B., Gowing S., et al. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Invest. 2013;123:3446–3458. doi: 10.1172/JCI67484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cedervall J., Zhang Y., Olsson A.K. Tumor-induced NETosis as a risk factor for metastasis and organ failure. Cancer Res. 2016;76:4311–4315. doi: 10.1158/0008-5472.CAN-15-3051. [DOI] [PubMed] [Google Scholar]
- 42.Jiang W., Shi X., Sun L., Zhang Y., et al. Exosomal miR-30a-5p promoted intrahepatic cholangiocarcinoma progression by increasing angiogenesis and vascular permeability in PDCD10 dependent manner. Int. J. Biol. Sci. 2023;19:4571–4587. doi: 10.7150/ijbs.83170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang M., Lin Y., Wang C., Deng L., et al. New insights into antiangiogenic therapy resistance in cancer: mechanisms and therapeutic aspects. Drug Resist. Updates. 2022;64 doi: 10.1016/j.drup.2022.100849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed are included in this published article.