Abstract
Full osteochondral regeneration remains a major clinical challenge. Among other experimental cartilage regenerative approaches, decellularized cartilage (DCC) is considered a promising material for generating potentially implantable scaffolds useful as cartilage repair strategy. In this work, we focus on screening and comparing different decellularization methods, aiming to generate DCC potentially useful in biomedical context, and therefore, with biological activity and functional properties in terms of induction of differentiation and regeneration. Data indicates that enzymatic and detergents-based decellularization methods differentially affect ECM components, and that it has consequences in further biological behavior. SDS-treated DCC powder is not useful to be further processed in 2D or 3D structures, because these structures tend to rapidly solubilize, or disaggregate, in physiologic media conditions. Conversely, Trypsin-treated DCC powders can be processed to mechanically stable 2D films and 3D solid-foam scaffolds, presumably due to partial digestion of collagens during decellularization, which would ease crosslinking at DCC during solubilization and processing. In vitro cell culture studies indicate that these structures are biocompatible and induce and potentiate chondrogenic differentiation. In vivo implantation of DCC derived 3D porous scaffolds in rabbit osteochondral defects induce subchondral bone regeneration and fibrocartilage tissue formation after implantation. Therefore, this work defines an optimal cartilage tissue decellularization protocol able to generate DCC powders processable to biocompatible and bioactive 2D and 3D structures. These structures are useful for in vitro cartilage research and in vivo subchondral bone regeneration, while hyaline cartilage regeneration with DCC alone as implantable material remains elusive.
Keywords: Cartilage, Decellularization, Extracellular matrix, Collagen
Graphical abstract
Highlights
-
•
Properties, components and further processing ability of decellularized cartilage (DCC) depends on decellularization method.
-
•
Trypsin-treated DCC powders can be solubilized-processed to mechanically stable 2D and 3D porous structures, but not to hydrogels.
-
•
These structures are biocompatible and trigger, both in vitro and in vivo, bone and cartilage differentiation.
-
•
DCC processable to 2D and 3D structures is useful for in vitro cartilage research and in vivo subchondral bone regeneration.
1. Introduction
Hyaline Cartilage is a very specialized tissue with unique properties. Its specific cell-distribution, low vascularization, and matrix composition provide excellent compressive stiffness and load-bearing properties. At present, multiple clinical situations as cartilage injuries, cartilage tissue deterioration and degenerative joint diseases require restorative or replacement surgery. Precisely, some of the repair strategies, including microfracture and autologous chondrocyte implantation, have achieved success in regenerating functional cartilage. However, full osteochondral regeneration remains a major clinical challenge [1].
At experimental level, multiple potentially promising cartilage regenerative approaches have been developed. Focused on cartilage tissue engineering field, potentially implantable, biomimetic or regenerative scaffolds have been fabricated with synthetic, natural and composite materials [2]. Based on the study of the extracellular matrix (ECM) composition, several studies suggest that tissue-specific components trigger tissue-specific differentiation [[3], [4], [5]]. Therefore, decellularized tissues have been postulated as interesting materials to generate implantable and regenerative compounds.
In cartilage field, decellularized cartilage (DCC) is considered a promising material for generating scaffolds useful for cartilage repair [[6], [7], [8], [9], [10], [11]]. Literature is extensive in terms of prospective cartilage tissue decellularization methods and their physical-chemical-mechanical properties testing, while multiple enzymes and detergents have been suggested for cartilage decellularization [[12], [13], [14], [15], [16]]. However, studies performing exhaustive, or conclusive, in vitro cell culture and in vivo implantation assays are scarce [[17], [18], [19]]. DCC alone cannot be processed to hydrogels, and to this purpose pepsin has been thoroughly used to generate soluble and functionalizable DCC, which has been mixed with other polymers to form printable composite hydrogels with tissue regeneration purposes [[20], [21], [22], [23], [24], [25]]. Of note, some clinical trials testing DCC for cartilage regenerative purposes have shown high implant-failure rate [26,27]. Interestingly, decellularization of in vitro cell culture-generated cartilage ECM shows good induction of osteochondral tissue in vivo [28], suggesting that the cartilage ECM may indeed be biologically active when properly processed during decellularization.
Our hypothesis was that a proper cartilage tissue decellularization method should lead to ECM with biological activity and functional properties in terms of induction of differentiation and regeneration. Therefore, the aim of this manuscript was to test and optimize multiple cartilage decellularization methods, characterize the resulting DCC, and more importantly, define their bioactive properties at in vitro cell culture and in vivo osteochondral regenerative applications.
According to the observed results, decellularization with enzymes and detergents differentially affect ECM components. SDS treated samples retain better cell-adhesion properties, which may be useful in in vitro cartilage studies. However, they are not useful to be processed in 2D or 3D structures, because these structures tend to rapidly solubilize in physiologic media. Interestingly, Trypsin treated DCC can be processed to mechanically stable 2D film and 3D porous scaffolds, which makes them suitable for in vitro cell culture testing and in vivo implantation procedures. In vitro, these structures are biocompatible and induce chondrogenic differentiation, while in vivo, the 3D porous scaffolds induce subchondral bone regeneration in osteochondral defects.
2. Results and discussion
2.1. Different decellularization protocols yield DCC with different ECM composition
The first step in a tissue decellularization approach is selecting the decellularization agents. In this case, our initial target was producing DCC useful for generating implantable and bioactive structures. According to the literature, enzymes as Trypsin and DNase, and detergents as SDS and TritonX-100 were previously assayed for cartilage decellularization, but data is limited regarding comparative studies among them. Therefore, the first objective of this research was testing the feasibility of different decellularization methods and compare the outcome in terms of ECM composition.
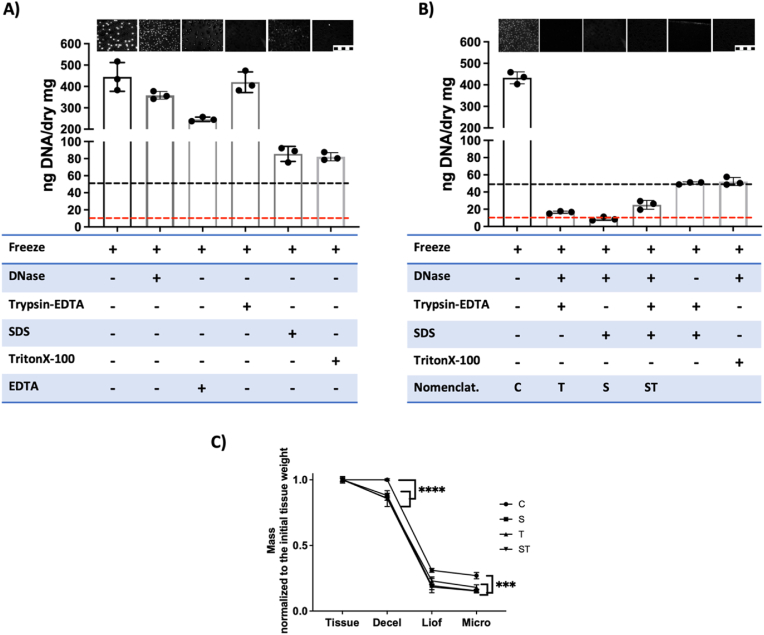
Selected enzymes and detergents were tested and decellularization was assessed by testing the DNA content. When decellularization was tested using individual treatments (Fig. 1A), none of the treatments reached the decellularization threshold of less than 50 ng of DNA/mg of dry weight, as defined by the ASTM Standard Guide F3354 for Evaluating Extracellular Matrix Decellularization Processes [29]. Note that DNA staining images show no nuclei staining at Trypsin treatment, while the DNA quantification indicates presence of DNA in these samples, suggesting that DNA fragments were surely retained at ECM (Fig. 1A). Then, assays were performed using different combinations of treatments, as shown in Fig. 1B. It was observed that the combination of DNase with SDS or Trypsin treatments was enough to reach the decellularization threshold, but not with TritonX-100 (Fig. 1B). We hypothesize that the DNase treatment helped to clean the DNA released by Trypsin or SDS treatments, yielding a reduction of DNA in samples to the range of 10 ng of DNA/mg of dry weight in 3 of the decellularization treatments. Therefore, these combinatorial treatments were defined as S and T (Fig. 1B) and they were selected for further studies. The combination of detergents and enzymes in a single treatment is also common in literature, and therefore, the SDS-Trypsin treatment was also selected for further studies (defined as ST in Fig. 1B).
Fig. 1.
Enzymatic and detergent treatments provide proper decellularization. A-B) Decellularization yields using different enzymatic and detergent treatment protocols. Inserted images indicate the visual assessment of the presence of DNA by DAPI staining (Scale 10 μm). Table indicate the treatment performed (+). Graphics provide quantitative DNA amount (n = 3); Dotted lines indicate the acceptable DNA amount threshold (Black, 50 ng DNA/mg of dry tissue; red, 10 ng DNA/mg of dry tissue). A) Individual treatments, B) Combinations of treatments, and the nomenclature used in the further assays (C, control; T, DNase-Trypsin-EDTA; S, DNase-SDS; ST, DNase-SDS-Trypsin-EDTA decellularization treatments). C) Mass loss at different steps of the process (n = 3) (Decel, decellularization; Liof, liofilization; Micro, Micronization). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We recorded the weight of samples in each one of the sample processing steps, aiming to define the mass loss in each step (Fig. 1C). Compared to the non-decellularized material processed as a control, the 3 decellularization protocols induce mild mass loss. Thereafter, sample lyophilization induced a massive reduction in weight, in accordance with cartilage tissue properties, in which water contributes to up to 80 % of its wet weight [30].
We characterized the effect of the different decellularization methods on ECM by histology and quantitative techniques (Fig. 2, Fig. 3). Cartilage ECM is composed by a collagen network interlocked by a mesh of proteins and proteoglycans. Therefore, they were the target of the study.
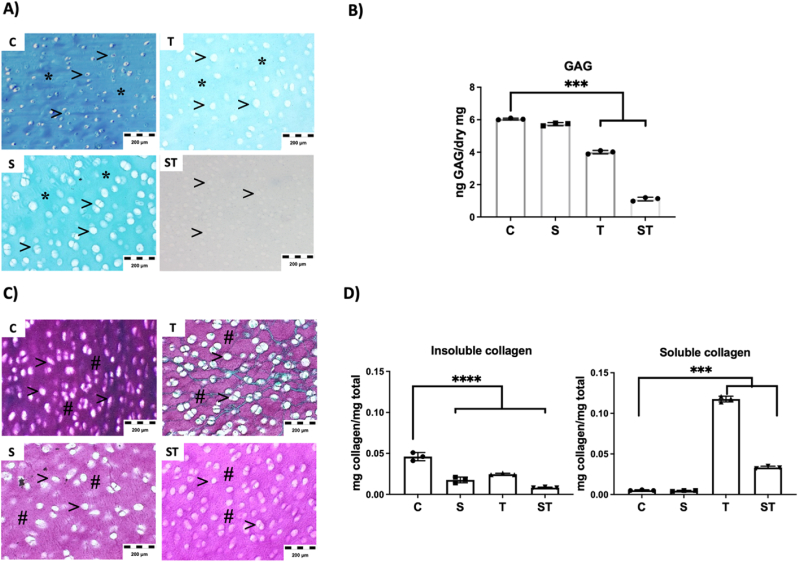
Fig. 2.
Decellularization treatment affects ECM components, assessed by histology and quantitative assays. A-B) Assessment of the presence of cartilage GAGs at decellularized samples, A) Alcian blue histology staining of decellularized tissues, (*, Blue stained ECM indicate presence of GAGs at sample. >, White areas correspond the area filled by cells. They are empty and enhanced at decellularized samples). B) Quantitative data of GAG amount in decellularized powders (n = 3). C-D) Assessment of the presence of collagens at decellularized samples, C) Sirius red histology staining of decellularized tissues, (#, Purple stained ECM indicate collagens at sample. >, White areas corresponding to cell spaces). D) Quantitative data of insoluble and soluble collagens present in decellularized powders (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
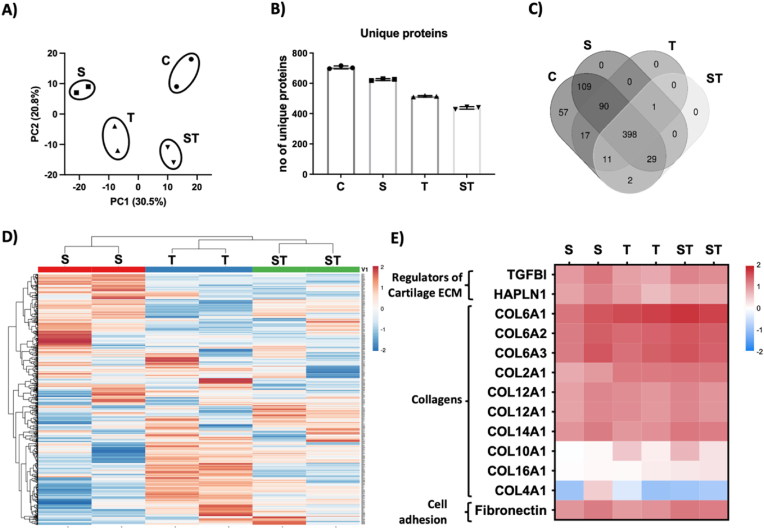
Fig. 3.
Decellularization treatment affects ECM components, assessed by histology proteomics assays. A) Principal component analysis of proteomic data. The analysis groups separately each treatment (n = 2 per treatment), B) Total number of unique proteins observed in each treatment, C) Venn diagram of common proteins observed among different decellularization treatments and compared to control initial samples, D-E) Heat map representation of, D) All detected proteins and, E) selected proteins including collagens and cell adhesion molecules.
In all histology images the tissue is observed as a ECM specifically stained from GAGs or collagen. There are also white spots, which correspond to cell area, and they seem empty and increased in size at decellularized samples. This is in accordance with the activity of the decellularization reagents and procedures, which not only remove the cells, but also affect the ECM in close-contact with the removed cells. Glycosaminoglycans (GAGs) measurement is relevant to define the integrity of ECM components after decellularization, because they can become soluble and be removed from ECM [31]. Data indicate a reduction of GAGs due to Trypsin (T) treatment and almost total depletion of GAGs in the doble SDS-Trypsin (ST) treatment (Fig. 2A and B). Previous reports already described the diffusion of GAGs out from cartilage tissue due to Trypsin cleavage, without apparent affectation of the collagen network and organization at histology [32,33]. Our histology studies of collagen confirmed similar results (Fig. 2C), with similar histology staining at control and Trypsin treated samples (T), while SDS treated samples (S and ST) showed slightly reduced collagen staining (Fig. 2C).
However, quantification data of insoluble and soluble collagens showed a different scenario (Fig. 2D). Mature collagen fibrils are formed by a triple helix and N-terminal and C-terminal telopeptides, which are involved in the formation of cross-links between different collagen fibrils. These structures are insoluble, while soluble collagens lack of cross-links and telopeptides. Note that an intact triple helix is resistant to digestion by Trypsin, while a triple helix with disruptions, partially folded structures or denaturalized are sensitive to Trypsin digestion. In these cases, Trypsin cleaves the collagen telopeptide domains and therefore fibril cross-links are liberated, transforming these collagens to their soluble form [34,35]. In our data a reduction of insoluble collagens is observed at Trypsin treated samples (T), and an increase of soluble collagens as a consequence of their digestion (Fig. 2D).
On the other hand, SDS detergent denaturalizes collagen by unravelling the triple helix of collagen, which explains the reduction of insoluble collagen observed at SDS treated samples (S) without increasing soluble collagens (Fig. 2D). According to the effects of SDS and Trypsin in collagens, we could expect an even higher increase in soluble collagens in the SDS-Trypsin treatment (ST), but data show only limited increase in solubilized collagens (Fig. 2D). It has been described that SDS interacts with Trypsin forming spontaneous Trypsin-SDS complex by hydrogen bonds and van der Waals forces. This molecular docking is dynamic, but located at Trypsin active center, affecting and reducing the hydrolytic activity of the Trypsin [36]. In any case, further proteomic studies revealed that, irrespectively of being in soluble or insoluble form, indeed collagens were present at all DCC samples (Fig. 3E).
Proteomic study helped us to define the effect of each decellularization treatment at cartilage tissue (Fig. 3). The principal component analysis revealed that the proteome was different in each case, and also different from control initial tissue (Fig. 3A). The study identified approximately 700 unique proteins at control cartilage tissue, which were reduced due to decellularization protocols, being Trypsin treated samples the ones showing less number of proteins (Fig. 3B). Almost 400 of the proteins were detected in control and all DCC, suggesting all decellularization treatments preserved these proteins (Fig. 3C). However, the heat map showing the relative amount of each protein indicates that the amount of these proteins was different between treatments, and similar between replicas (Fig. 3D). Interestingly, the main representatives of cartilage ECM, the collagens, showed similar amounts in all DCC, indicating the preservation of collagens in all cases (Fig. 3E). Moreover, other ECM proteins relevant for cartilage homeostasis also seem quite stable after decellularization. In example, TGFBI is an ECM protein relevant for cartilage homeostasis [37], while CRTL1, or cartilage link protein, is key in stabilizing cartilage ECM components as aggrecan and hyaluronic acid (HA) [38].
In all DCC, type IV collagen was underrepresented (Fig. 3E). In normal cartilage tissue, Laminin and type IV collagen are present at pericellular matrix area [39]. Laminin is a cell-adhesive glycoprotein relevant in tissue morphogenesis and homeostasis [40], while in combination with type IV collagen, they form a network relevant to maintain hypoxic and antiangiogenic environments in cartilage tissue context [41,42]. According to our results, the Trypsin treatment reduces the presence of type IV collagen, suggesting that the pericellular matrix is altered due to Trypsin decellularization process. On the other hand, Fibronectin is also a cell-adhesive glycoprotein, but it is ubiquitous and binds covalently to multiple collagens [43], thereby playing roles not only in cell-adhesion, but also in the organization of the cartilage extracellular matrix [44]. It is known that Fibronectin sensitiveness to Trypsin cleavage is dependent on factors such as glycosylation [45]. According to our data, it seems that fibronectin in cartilage ECM is resistant to the Trypsin decellularization treatment.
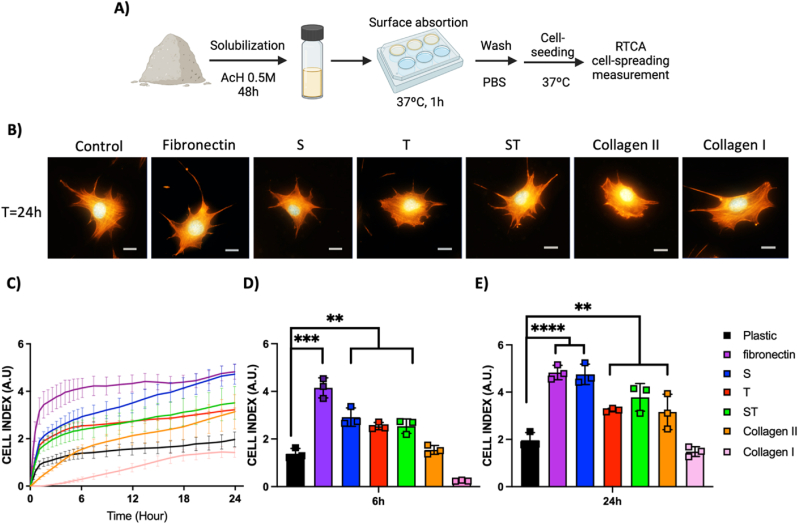
Aiming to prove the biological activity of the cell-adhesion molecules present at DCC, we solubilized the material in acetic acid and deposited it at plastic surface to allow surface coating by adsorption. For comparative purposes, other cell-adhesion molecules were adsorpted on control wells. Then, surfaces were washed and further on chondrocyte cell adhesion was tested on them (Fig. 4A). For these assays, ATDC5 chondrogenic cell line was selected as a well-stablished in vitro cell model useful to study factors that influence cell behaviors during chondrogenesis [46]. Cells seeded on different treated wells were fixed at 24h and cytoskeleton was stained aiming to observe cell morphology and spreading by microscopy (Fig. 4B). This technique allowed us to assess cell adhesion and spreading in all conditions, but we observed that microscopy is not an accurate way to define these parameters due to the variability at cell size and spreading within each well. The Real Time Cell Analysis (RTCA) technique based on impedance and microsensor electrodes allows label-free, real-time, and continuous monitoring of cell adhesion because the measurement is related to the surface area covered by cells in the whole well. Therefore, RTCA technique was selected as an accurate sensitive detection method to measure cell adhesion and spreading in a kinetic study (Fig. 4C). As it was already described [47,48], fibronectin coating induced the best initial chondrocyte cell spreading and adhesion among all the control adhesion molecules tested (Fig. 4D). Thereafter at 24 h, the S treatment-derived coating exhibited a cell behavior similar to fibronectin, while T and ST treatment-derived coatings show similar cell spreading than collagen 2 treated surfaces (Fig. 4 E). In all these cases, the cell spreading was higher than control non-coated plastic surface and collagen I control coating. All it suggests that, after decellularization protocols, biologically active adhesion molecules were still present at DCC samples and these molecules adsorpted on cell culture plastic surface helped cell adhesion and spreading.
Fig. 4.
Decellularized ECM possesses biologically active cell-adhesion related proteins. A) Schematic of ECM protein absorption on cell-culture plastic surfaces and further ATDC5 cell line seeding, showing initial and 24 h cell-morphology and spreading, B) Confocal images of cells at different surfaces stained for actin cytoskeleton (orange) and Nuclei (DAPI, Blue), C-E) RTCA data, C) quantitative Kinetic cell adhesion on multiple protein-adsorption treated surfaces (The color code indicates the protein adsorbed in each case) (n = 3), D-E) cell adhesion at selected time points, D) at 6 h, E) at 24 h after initial cell-seeding (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In sum, DCC characterization data indicates that each decellularization agent affects the ECM in different way. Both SDS and Trypsin treatments reduce the insoluble collagen amounts, while the enzymatic activity of Trypsin treatment seems also to alter some collagens, transforming them to their soluble form. Compared to SDS detergent treatment, the Trypsin enzymatic treatment removes more proteins and GAGs, while collagens and bioactive molecules related to cell adhesion remain in all DCC samples.
2.2. Biocompatible 2D and 3D structures can be generated from Trypsin treated DCC powders
Once the decellularization outcomes were assessed, the further aim was using DCC samples to generate approaches with added value or with properties useful in biomedical context. Therefore, we applied multiple postprocessing techniques to generate 2D and 3D structures and, more importantly, we tested their biological activity, functional properties and potential specific applications.
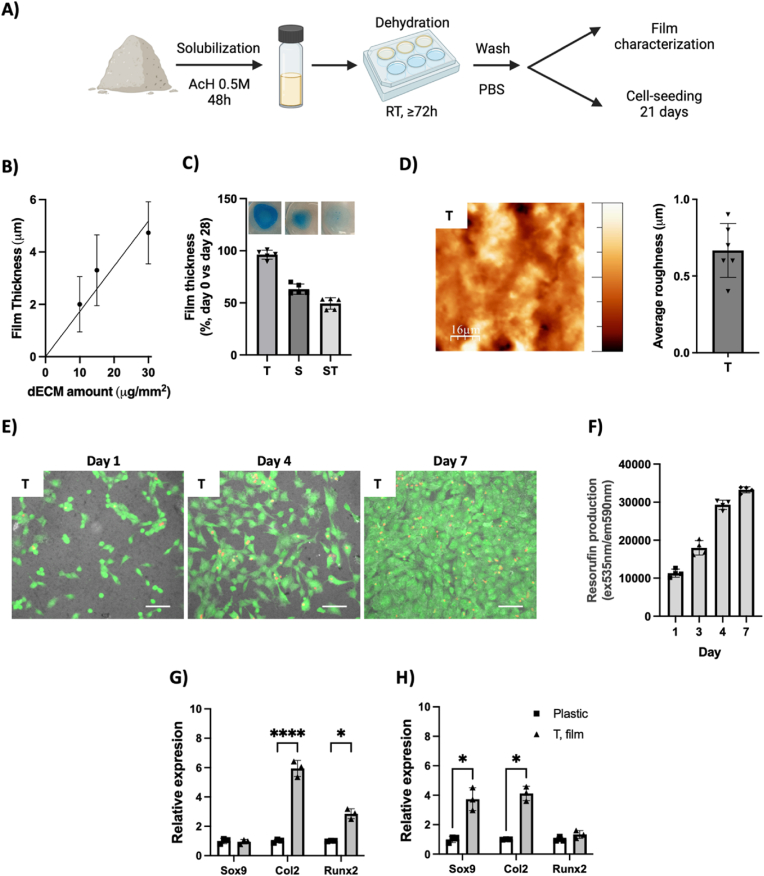
First, we tested the ability of the different DCC to create surface coating materials, or films, potentially useful as in vitro cell culture cartilage differentiation testing platform. In this case, a DCC powder suspension was created using acetic acid, then it was deposited on cell culture wells and finally films were generated by dehydration (Fig. 5A). Film characterization studies revealed that film thickness was dependent on the amount of DCC deposited (Fig. 5B). DPBS was added on the DCC films and samples were incubated at 37 °C for 28 days (Fig. 5C). Data indicates that DCC films from Trypsin decellularization (T) were stable in these conditions, with presence of GAGs at them (see blue color at inserted images). These samples also show a surface roughness measured as 0.66 μm (SD 0.17 μm) (Fig. 5D). However, the films from SDS treatments (S and ST) were not stable and they partially dissolve during the assay, which leads us to discard them for further studies. Therefore, ATDC5 cells were seeded on T films and tested. Fig. 5E shows attached viable (Green) cells at material surface and further proliferation of them until cell confluence, which was reached at day 7 in the tested culture conditions and is in accordance with viable cell metabolism quantification study (Fig. 5F). Some red nuclei staining indicate the presence of some dead cells in all timepoints, which is the standard in this cell line. For long term cell culture and gene expression studies, cells were seeded on control plastic surfaces or DCC T films (Fig. 5G and H) and cultured in control media (Fig. 5G) or chondrogenic induction media (Fig. 5H). After 21 days in control media, and compared to control surface, ATDC5 cells grown on T films show increased expression of Runx2 transcription factor and Collagen 2 extracellular matrix component (Fig. 5G). Therefore, our data suggests that DCC components at T films can induce chondrogenic differentiation by stimulation of the expression of Runx2 transcription factor. It has been already reported that multiple ECM-derived signaling may converge to stimulate Runx2, while at ATDC5 cells, Runx2 induces chondrogenesis [49]. On the other hand, when cells were cultured with chondrogenic differentiation media (Fig. 5H), T films induce in cells higher expression of Sox9 transcription factor compared to the control surface. Precisely, Sox9 is related to chondrocyte maturation and induces the degradation of Runx2 [50,51].
Fig. 5.
Surface coating by films formation is feasible with Trypsin-treated DCC and it promotes in vitro cartilage differentiation. A) Schematic of surface coating procedure and further testing. B) Relationship between used DCC amount and formed film thickness (n = 3), C) Quantification of film thickness after 28 days incubation at 37 °C and 5%CO2 (data normalized to film thickness at day 0) (n = 5), and inserted images of films stained with Alcian Blue as representative of film stability or dilution. D) Representative AFM image showing the surface roughness of T films and their average roughness measurement (n = 6). E-H) cell culture studies on Trypsin-obtained DCC films (T). E-F) Time-course studies of, E) Cell morphology and viability observed by CalceinAM, a viable cell-permeant green-fluorescent dye. Dead cells are observed by EtBr red-fluorescent nuclear staining. Surface roughness is provided by superposition of the transmitted-light image. F) Quantification of cell metabolic activity at different time point by (Resazurin-Resorufin fluorescence assay). G-H) Gene expression study of selected cartilage markers at cells cultured 21 days on control plastic surface or Trypsin-obtained DCC films (Data normalized to plastic grown cells) (n = 3), E) Cells cultured at control media, F) Cells cultured at cartilage differentiation media. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In sum, film formation study suggests that Trypsin decellularization yields a DCC able to be dissolved to form surface coatings by dehydration, which are stable at cell culture conditions. These surface coatings are biocompatible in terms of cell adhesion and proliferation, and they induce chondrogenic differentiation.
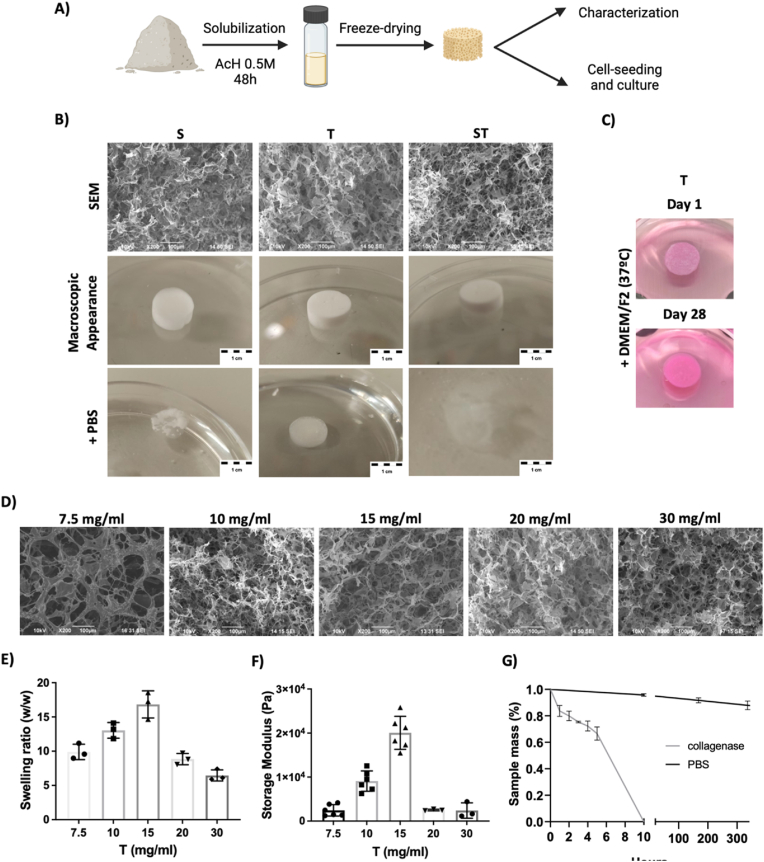
The further step was defining potential approaches to generate biocompatible and potentially implantable 3D structures. For this aim, DCC powder suspension was created using acetic acid and samples were freeze-dried. As a result, porous solid foam scaffolds were created (Fig. 6A and B). Similar to the outcome observed at film formation strategy, when these scaffolds were incubated in DPBS, only the samples from Trypsin obtained DCC (T) retained the mechanical integrity, while those scaffolds made from SDS obtained DCC samples (S and ST) tend to fully dissolve (Fig. 6B). The stability of T scaffolds was also confirmed in cell culture media and 37 °C for 28 days (Fig. 6C), so T samples were used for further studies. First, we assessed porous structure of the materials fabricated using different concentrations. It could be expected increased porosity at low material concentration and more packed structure at higher concentrations, but SEM images were not conclusive in this sense (Fig. 6D) so we proceed to perform further testing with materials of different concentrations.
Fig. 6.
Stable solid foam porous scaffolds can be obtained by freeze-drying the Trypsin-obtained (T) DCC. A) Schematic of DCC powder solubilization and freeze-drying. B-C) SEM and gross morphology of porous solid foam scaffolds (15 mg/ml) and their appearance after being in contact with DPBS, or C) Cell culture media. D-G) Trypsin-obtained (T) DCC-based scaffold material characterization, D) SEM images of scaffolds fabricated from DCC at different concentrations. E) swelling ratio depending on the composition of the freeze-dried solution (n = 3), E) Rheology of hydrated samples on the composition of the freeze-dried solution (n = 3 to 6). F) Stability of scaffolds (15 mg/ml) in DPBS and at treatment with collagenase (n = 3). Data were similar when cell culture media was used.
The swelling ratio is defined as the fractional increase in the weight of samples due to water absorption. Fig. 6E shows at low collagen concentrations, 15 mg/mL or less, the water absorption capacity increases as the collagen concentration increases. This is because the triple-helix structure of collagen allows it to form a three-dimensional network that can trap and retain water molecules. The higher the collagen concentration, the more extensive and dense this network becomes, which can enhance the scaffold's ability to absorb and retain water. However, at collagen concentrations higher than 15 mg/mL, the water absorption capacity decreases, likely due to the difficulty in creating a homogeneous porous scaffold with well-defined and uniform pores because of the low solubility of collagen at high concentrations in acetic acid.
The storage modulus is a parameter used to describe the elastic behavior of scaffolds, particularly in the context of viscoelastic materials. It provides insight into the scaffold's ability to store and release energy during deformation, reflecting its stiffness and elastic properties. It can be observed in Fig. 6F, that at low concentrations, water absorption helps maintain the elasticity and flexibility of the samples. However, the storage modulus value decreases dramatically at higher collagen concentrations, likely due to the low swelling ratio or water absorption capacity of the samples.
Therefore, 15 mg/mL T scaffolds were selected to further studies and incubated with or without collagenase (Fig. 6G). Solid foam scaffolds were stable at DPBS, but show a rapid digestion due to collagenase, suggesting that the 3D structure is maintained by collagens (Fig. 6G).
The next research step was testing materials in cell culture context, and therefore, assays were first corroborated in cell culture media context. Specifically, 15 mg/mL T scaffolds were tested. As previously mentioned, they retain the mechanical integrity during 28 days in cell culture media and conditions (37 °C and 5 % CO2) (Fig. 6C). Interestingly, and compared to data shown in Fig. 6G, the swelling ratio in cell culture media was slightly lower (8,75 SD 2,4), which induces a slower collagenase digestion kinetic due to lower penetration of media at scaffold structure. In any case, materials in culture media were also fully digested by collagenase after 10h of digestion testing.
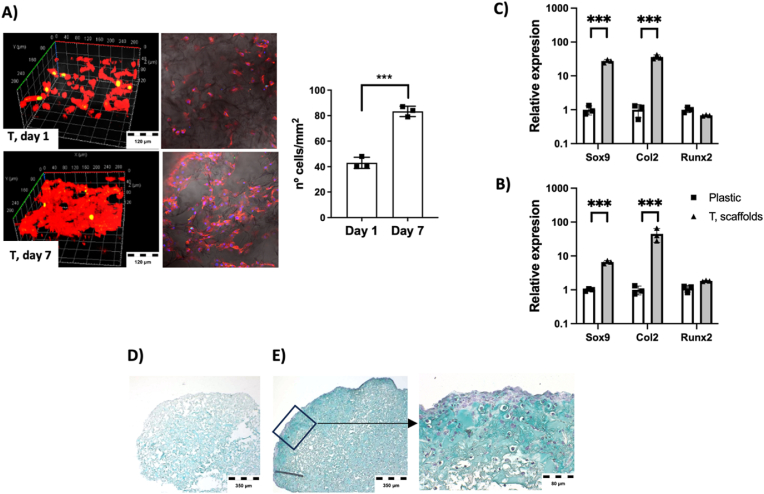
Cell culture studies show cells attached and proliferating on Trypsin-obtained DCC scaffolds (Fig. 7A). At day 1, individual cells were spread at material inner and outer surfaces, while at day 7 cells covered the whole material. This data indicate that the material components are biocompatible, corroborating the biocompatibility observed at material films (see Fig. 5E–G). In scaffolds, cells not only adhere and grow at material surface, but also colonize the inner part of the structure (Fig. 7A–D-E) due to the ingrowth of the cells through the porous scaffolds (See SEM images at Fig. 6). Therefore, the outcome is a 3D culture of cells on, and inside, the solid-foam material (Fig. 7A–D-E). Similar to experimental performed at film formation studies (Fig. 5), here cell-seeded scaffolds were incubated in control media or chondrogenic differentiation media for 21 days, and gene expression data were compared to cells grown on plastic control surface (Fig. 7B and C). In this case, at any condition cells at scaffolds expressed higher amount of Sox9, which is the master regulator of chondrogenesis, and collagen 2, which is a cartilage-specific ECM component. Histology study of samples (Fig. 7D and E) indicate that, at control media (Fig. 7D), cells were colonizing the whole material structure, but they did not undergo terminal cartilage differentiation, as observed by mild blue color staining corresponding to cartilage tissue GAGs. That suggests that signals at DCC scaffolds induced early cell differentiation, but they are not enough to induce proper terminal differentiation. Conversely, scaffolds cultured in differentiation media (Fig. 7E) not only showed cells at the whole material structure, but also their conversion to chondrogenic phenotype with rounded shape and bigger size. They generated high amount of GAG-containing extracellular matrix, observable as deep blue GAG staining at the whole 3D structure, and more evident at surfaces, all it compatible with terminal cartilage differentiation at DCC scaffolds.
Fig. 7.
Freeze-dried Trypsin-obtained (T) DCC scaffolds support in vitro chondrogenic differentiation. A) Confocal microscope imaging of Actin cytoskeleton staining (red fluorescence) at day 1 and day 7 after cell-seeding on T scaffolds, and quantification of cell density. B-C) Gene expression study of selected cartilage markers at cells cultured on control plastic surface or at Trypsin-obtained DCC scaffolds (Data normalized to plastic grown cells) (n = 3), B) Control media, C) Cartilage differentiation media. D-E) Alcian blue histology staining of cell-seeded Trypsin-obtained DCC scaffolds after 21 days of culture in, D) control media or E) chondrogenic differentiation media. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
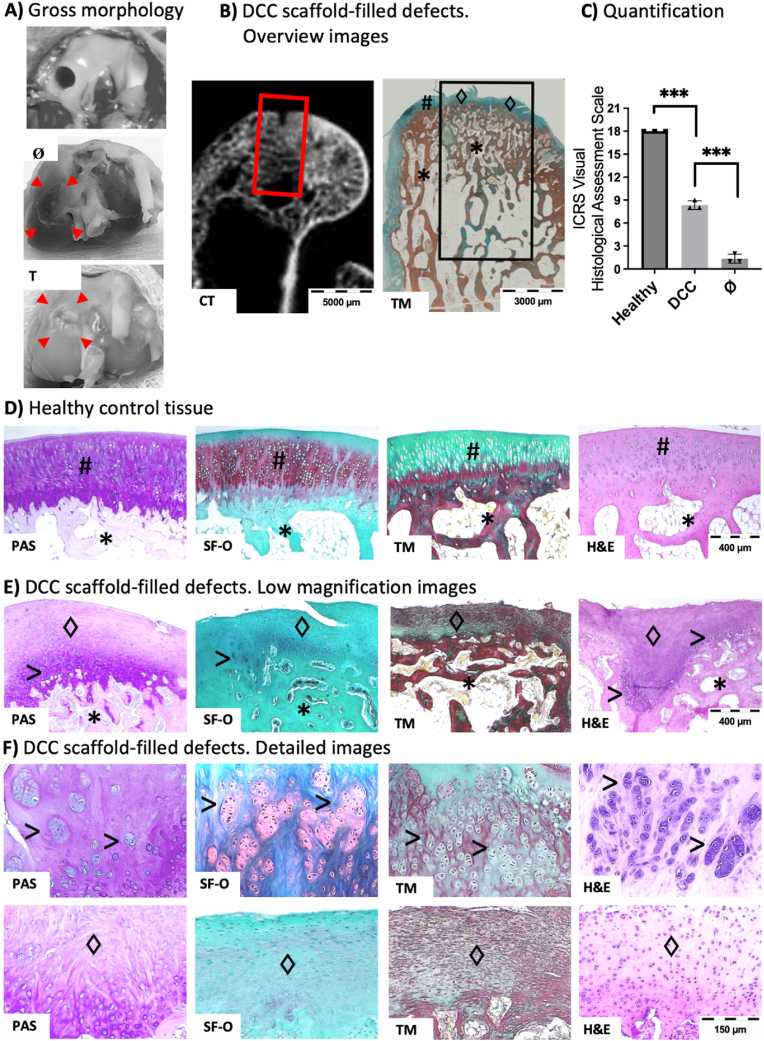
These data encouraged us to test the behavior of scaffolds in an in vivo regenerative approach. For this aim, rabbit osteochondral critical-size defect was selected as implantation model (Fig. 8A). Samples were implanted and then harvested after 3 months. There were no complications during implantation period, and at sample harvest, it was observed no abrasions on the surrounding articular cartilage surfaces and no inflammation of the synovial membranes. At endpoint, the implantation area was filled and opaque, suggesting tissue formation within the defect, while the surface was smooth but irregular (Fig. 8A). Computed tomography (CT) images showed trabecular bone formation at defect area and again irregular tissue structure at bone surface (Fig. 8B), which was confirmed at histology (Fig. 8C–F).
Fig. 8.
DCC scaffold implantation at rabbit osteochondral defect yields subchondral regeneration and fibrocartilage formation. A) Empty defect created at surgery, and gross morphology of tissue at sample harvest (3 months from surgery, red arrowheads indicate the limits and location of the defect), B) Overview images of DCC-implanted defects. At left, a computed tomography (CT) image of a sample after being harvested (Red square indicates the defect area). At right, histology image (Black square indicates the defect area), (TM, Massons Trichrome collagen staining. Green corresponds to normal cartilage or fibrocartilage, Red corresponds to bone tissue) C-F) Histology assessment using different staining (PAS, Periodic acid–Schiff staining. Purple indicates proteoglycans staining; SF-O, Safranin-O staining. Red indicates proteoglycans staining; TM, Massons Trichrome collagen staining; H&E, Hematoxilin and eosin staining. pink for ECM and purple for cell nuclei). C) Quantification of visual assessments by ICRS assessment scale (Healthy, Healthy control tissue samples; DCC, DCC scaffold-implanted samples; Ø, samples from empty defects group). D) representative images of staining at healthy control tissue samples E-F) Representative images of staining at DCC-filled samples. (#, hyaline cartilage; * Subchondral bone; ♢ Fibrocartilage; > chondrocyte clusters). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Histology shows proper trabecular subchondral bone formation at implanted samples, suggesting the implanted DCC scaffolds drive proper bone formation (Fig. 8E and F). Bone tissue regeneration relays mainly on endochondral ossification, a physiological bone formation process in which an initial cartilage callus formation leads to further formation of bone tissue. It has been previously suggested that the implantation of DCC may help to bone formation by mimicking the initial steps of bone regeneration [52]. In this case, the DCC implantations provided cartilage ECM at defect area available for tissue regeneration, and therefore, it would explain how the implanted material boosted endochondral ossification and therefore facilitated bone formation.
In terms of cartilage tissue, and compared to staining of healthy control hyaline cartilage tissue (Fig. 8D), the staining of implanted samples show mainly fibrocartilage tissue with reduced tissue-specific staining and no hyaline cartilage morphology (Fig. 8E and F). The newly formed tissue showed some areas of chondrocyte clusters formation (Fig. 8E and F). These clusters are usually observed in pathological cartilage diseases at experimental osteochondral implantation approaches, and they have been previously reported as areas of initial cartilage regeneration [53]. The ISCR scoring method provides values from 0 to 3 to the mentioned visual appreciations (‘surface’, ‘matrix’, ‘cell distribution’, ‘cell population’, ‘subchondral bone’ and ‘cartilage mineralization’). Therefore, the extend of obtained regeneration was quantified by sample scoring (Fig. 8C).
Clinically routinely used cartilage reparative and restorative surgeries, including microfracture, drilling, abrasion arthroscopy, and even autologous chondrocyte implantation, are techniques to restore knee articular lining by forming fibrocartilage tissue covering the damaged area [54]. Fibrocartilage possesses limited mechanical loading properties and degrades with time, and that is why the field requires new cartilage regeneration procedures which ideally, should lead to the regeneration of articular cartilage. Altogether, our results indicate that the implanted DCC material induces subchondral bone formation but failed to regenerate hyaline cartilage, while it formed fibrocartilage, which is a reparative tissue already achievable with conventional clinical techniques.
3. Conclusions
In this work we were aiming to generate DCC potentially useful in biomedical context, and therefore, with biological activity and functional properties in terms of induction of differentiation and regeneration. First, we tested multiple cartilage decellularization methods to define the best approach to obtain DCC. Enzymatic and detergents-based decellularization methods differentially affect ECM components, and this had consequences in further biological behavior. In our research, SDS-treated DCC powder are not useful to be further processed in 2D or 3D structures, because these structures tend to rapidly solubilize, or disaggregate, in physiologic media conditions. Conversely, Trypsin not only decellularized but also partially digested tissue collagens, which may be related with the ability of the resultant DCC to be further processed to mechanically stable 2D films and 3D porous scaffolds. In vitro, these structures are biocompatible and induce and potentiate chondrogenic differentiation, while in vivo, the 3D porous scaffolds induce subchondral bone regeneration and fibrocartilage tissue formation after implantation in rabbit osteochondral defects. Therefore, this work defines an optimal cartilage tissue decellularization protocol able to generate DCC powders processable to biocompatible-bioactive 2D and 3D structures. These structures are useful for in vitro cartilage research and in vivo subchondral bone regeneration, while hyaline cartilage regeneration with DCC as implantable material remains elusive. As potential further steps to ameliorate these results, we envision that adding to the structure some cartilage and bone inducing growth factors during material fabrication step may potentiate its tissue regeneration ability. Alternatively, improvements to the 3D fabrication technique are also feasible, including the assessment of hydrogel forming ability of DCC.
4. Materials and methods
4.1. Cartilage tissue and decellularization
Porcine articular cartilage was extracted postmortem from Dalland Hybrid males/female pigs, weighing between 18 and 22 kg, and previously used in teaching purposes at Biogipuzkoa HRI animal house. The Porcine articular cartilage was maintained frozen at −20 °C until needed.
For decellularizations, first the cartilage was cut into small pieces of around 1 cm3. Then the material was subject to 7 cycles of deep in liquid nitrogen and thaw. At this point, samples were divided to be treated with different compounds. For EDTA (E9884, Sigma-Aldrich, USA) treatment, (0.25 % w/v) was incubated for 30 min at RT in agitation. For Trypsin-EDTA treatment (25050-014, Sigma-Aldrich, USA), 30 min a 37 °C in agitation. For SDS treatment, a 2 % SDS (AM9822, Thermo Fisher, USA) was used, 4h at RT in agitation. For Triton X100 treatment, 1 % Triton X100 (T9284, Sigma Aldrich, USA) diluted in PBS and treatment was 4 h at RT in agitation. For DNase treatment, 300unit/ml of DNAsa I (11284932001, Sigma-Aldrich, USA) were used, 24 h at 37 °C in agitation. Each protocol was followed by 3 PBS (524650, Merk life science, SL) washes. These protocols were tested individually or sequentially in different combinations following this order. At this point, some decellularized tissue pieces were kept for histology studies and the rest were lyophilized for 24 h, micronized using a mixer mill (Retsch MM400, Haan, Germany), the samples were introduced in the cartridges containing a metal ball and submerged in liquid nitrogen for 3 min, this was followed by 4 min of 30 agitations/second. In each step, sample weight was recorded using a precision conventional balance. Micronized powders were stored in a desiccator to avoid moisture until be used.
4.2. Characterization of DNA, GAG and collagen at decellularized samples
Decellularized tissue pieces were fixed in formalin 10 % (HT501128-4L, Sigma-Aldrich, USA) overnight and washed thrice in PBS. For DNA staining, samples were embedded in OCT tissue freezing medium and blocks were frozen at −20 °C for storage. Frozen samples were cut into 6 μm slides (Leica CM3050 cryostat) and then slides were stained with 1 μg/mL DAPI for 10 min, followed by 2 PBS washes and imaging at fluorescence microscope (Leica DMI6000 B). For GAG and collagen staining, samples were embedded in paraffin and cut into 4 μm sections following standard protocols [55]. Then slides were stained for GAG visualization with Alcian blue and hematoxylin (ab150662, Abcam, USA and 05-M06015, Bio-optica, It) and for collagen visualization with Pico Sirius red and fast green (9046, Chondrex, USA), following standard protocols [56]. Images were obtained using an optical microscope.
Decellularized micronized powders were used to quantify multiple parameters in the samples. For DNA quantification, 10 mg of powder were used, and DNA was extracted from using The PureLink Genomic DNA Mini Kit (K182001, Thermo Fisher, USA) and resulting DNA was quantified using the NanoPhotometer N60 (Nanodrop). For the GAG quantification the biocolor Sulfated glycosaminoglycan assay kit was used (B1000, Biocolor, UK); For soluble collagen quantification the Sircol soluble collagen assay kit was used (S1000, Biocolor, UK) and for the insoluble collagen quantification the Sircol insoluble collagen assay kit was used (S2000, Biocolor, UK), in all cases following the manufacturer instructions. All data are provided normalized to mg of powder tissue.
4.3. Characterization of proteins at decellularized samples
Proteins were extracted from decellularized powders using a mixture of 7 M urea, 2 M thiourea, 4 % CHAPS, and 5 mM DTT and then digested following the filter aided FASP protocol described by Wisniewski et al. with minor modifications [57]. Briefly, Trypsin was added to a Trypsin: protein ratio of 1:20, and the mixture was incubated overnight at 37 °C, dried out in a RVC2 25 speedvac concentrator (Christ), and resuspended in 0.1 % formic acid (FA). Peptides were desalted and resuspended in 0.1 % FA using C18 stage tips (Millipore).
Samples were analyzed on a timsTOF Pro with PASEF (Bruker Daltonics) coupled online to an Evosep ONE liquid chromatograph (Evosep). A total of 200 ng was directly loaded onto the Evosep ONE and resolved using the 30 sample-per-day protocol.
Protein identification and quantification were carried out using MaxQuant software [58] using default settings except for the match between runs (match time window of 5 min, alignment tie window of 20 min) and an LFQ min. ratio count of 1. Searches were carried out against a database consisting of human and mouse protein entries (Uniprot/Swissprot), with precursor and fragment tolerances of 20 ppm and 0.05 Da. Only proteins identified with at least two peptides at FDR<1 % were considered for further analysis. LFQ intensities were used for further analyses and loaded onto the Perseus platform [59]. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD050583.
Data were loaded onto the ClustVis web tool for the visualization and clustering of multivariate data using principal component analysis (PCA) [60]. Data were loaded to MetaboAnalyst [61] to be normalized and statistically analyzed. Partial least squares discriminant analysis (PLS-DA) was used to get the variable importance in projection (VIP) and differentiate the groups. It was used to select and extract data of the relevant specific proteins from the whole database. Venny diagrams was generated using online the tool (https://bioinformatics.psb.ugent.be/webtools/Venn/)All the results were represented using Graphpad Prism software.
4.4. Characterization of presence of biologically active adhesion molecules at decellularized samples
Decellularized powders were diluted in acetic acid 0.5 M (1 mg DCC/ml of Acetic Acid 0.5 M), stirring 24 h at RT. Fibronectin (F0895-1 MG, Merk, USA), collagen I P0245, Merk, USA) and collagen II (C7806-5 MG, Merk, USA) powders were prepared in acetic acid 0.5 M (1 mg/mL). 100 μl of each dilution was added at different wells of glass surfaces (81817, μ-Slide 18 Well Glass Bottom slides, ibidi, Germany) for microscopy studies or 16 well-electronic plates (6465382001, Agilent, USA) for RTCA studies, and they were incubated for 1 h at RT before being removed and wells washed with PBS.
For imaging cell morphology, ATDC5 cells were seeded at 1,5 × 105 cell/cm2 on 12-well chamber slides (154534, Nunc) previously treated with adhesion molecules as described above, and cultured in DMEM/F-12 Complete warm media. At 24 h of incubation at 37 °C and 5%CO2, cells were fixed 30 min using formalin 10 % followed by a PBS wash. Cytoskeleton was stained using ActinRed 555 (R37112, thermos fisher, USA) following the kits protocol. DAPI was used by adding 1 μg/mL DAPI for 10 min, followed by 2 PBS washes. Then, confocal images were recorded in a Zeiss LSM 880 confocal laser scanning microscope, equipped Plan-Apochromat 10 × (0.45 N. A.) and Plan-Apochromat 20 × (0.8 N. A.) objectives. Z-stacks (ca. 50 μm thick) were obtained when required.
The RTCA (Real Time Cell Assay) equipment detects the electron flow between the electrodes located at the bottom surface of the microwells. The adhesion of the cells at the bottom of the wells impedes the electron flow and an impedance signal is given as a cell index data (CI). The magnitude of the signal depends on the cell number, cell size/morphology, and cell-substrate attachment quality. Thus, the RTCA provides a real-time sensitive detection of cell adhesion and cell proliferation. In order to perform RTCA assays. For RTCA assays, 100 μL of DMEM/F-12 (10565018, Thermo Fisher, USA) cell culture media was added to each well, the 16 well-electronic plates were introduced in the xCELLigence RTCA DP equipment (00380601050, Agilent, USA) and an initial background reading was performed. 100 μL of ATDC5 cells (CVCL_3894) suspension at 50.000 cell/mL was added to each well and plates were incubated at room temperature for 30 min before starting the continuous recording of impedance as previously described [62]. All the measurements by the RT-CES were performed inside an incubator at 37C and in a humidified atmosphere of 5%CO2. The impedance was recorded every 5 min for a duration of 8 h to capture the short-term response, followed by measurements every 15 min for 24 h to capture the long-term response. The Cell Index (CI) was derived according to the following equation:
where Rcell (fi) and Rb (fi) are the frequency-dependent electrode resistances with or without cells, respectively, and N is the number of the frequency points at which impedance is measured. The normalized cell index at a given time point was calculated by dividing the cell index at the time point by the cell index at a reference time point. Cell index data were plotted related to cell culture time.
4.5. Generation and characterization of 2D film structures
This procedure was adapted from previous reports, in which coatings were formed from adipose derived ECM [63]. Here, DCC powders were dissolved in acetic acid 0.5 M for 24 h at RT in various concentrations (7.5 mg/ml, 10 mg/ml and 15 mg/ml). Then, each solution was used to create films of different thickness by layering a 500 μl of each solution on wells of 12 well cell culture plates (CLS356500, Sigma Aldrich, USA) or glass surfaces (BPD014, Auxilab, France) and drying it a RT, followed by overnight sterilization inside a cell culture hood using UV light.
For measuring thickness, after the dehydration, a line was scratched on the material. The atomic force microscopy (AFM, JPK NanoWizard II, JPK Instruments AG, Bruker) was used to map the topography of the coating and analyze the thickness of the layer in different material concentrations. Thickness data were plotted related to the amount of decellularized material used to form each film. Based on AFM images, Roughness Average was calculated by Roughness Analysis routine of the WSxM freeware scanning probe microscopy software.
The coating degradation was studied by adding DPBS (14040, Gibco, UK) and keeping the material at 37 °C and 5 % CO2, for 28 days. Then, samples were cleaned in water and allowed to dry before being processed for AFM study as described above. Data are represented normalized to the thickness of the initial dry films.
4.6. Cell culture studies on 2D films
ATDC5 cells were seeded on DCC films formed on glass surfaces (81817, μ-Slide 18 Well Glass Bottom slides, ibidi, Germany) at 15,000 cell/cm2 and cultured in DMEM/F-12 Complete warm media. At 24 h of culture, cells were processed for Actin-Nuclei staining and confocal image recording as described above.
For cell viability imaging, cells were treated with two reagents. Calcein-AM is a non-fluorescent molecule, which is converted to green-fluorescent calcein inside viable cells due to the activity of intracellular esterases. Besides, Propidium Iodide is a red-fluorescent nuclei staining dye which cannot pass through a viable cell membrane. Films were formed on glass surface 12 well plates (CLS356500, Sigma Aldrich, USA) dried at RT and sterilized by UV irradiation, followed by seeding of 4 × 105 ATDC5 cells/well. Then, they were cultured for 1, 4 and 7 days at 37 °C and 5%CO2. The media was replaced by warm DMEM:F12 media containing 10 μg/ml Propidium iodide (V13242, Invitrogen) and 2.5 μg/ml Calcein-AM (56496, Merck) and incubated for 30min at 37 °C and 5%CO2. Then, live images with a 20× objective were acquired in a Zeiss lsm880 confocal microscope.
For cell viability quantification, resazuring-based viability assay was conducted. In this assay the intracellular reduction of the non-fluorescent blue resazurin by the mitochondrial respiratory chain in live cells to pink-fluorescent resorufin is measured. Films were formed on 96 well plates (153596, Teknovas), dried at RT and sterilized by UV irradiation, followed by seeding of 1 × 105 ATDC5 cells/well. Then, they were cultured for 1, 4 and 7 days at 37 °C and 5%CO2. The media was replaced by warm DMEM:F12 media containing 1/10 of the Deep Blue Cell Viability reagent (Biolegend, 424701) and incubated for 4h at 37 °C and 5%CO2. Then, 100 μl of supernatant was transferred to triplicate wells in a clear bottom-black 96 well plate (3603, Corning) and the fluorescence was measured in a plate spectrophotometer (GeniosPro, Tecan) at the ex/em wavelength of 535/590 nm. Results were normalized to the measurement of the control sample (wells without films).
For cell differentiation studies, cells were seeded as described above on 2D films or control cell culture wells and cultured in control media or Mesencult ACF chondrogenic differentiation medium for 21 days. The Monarch total RNA miniprep kit for RNA extraction and Nanodrop was used to test the quality. For Quantitative Real-Time PCR (qPCR) assay, primers for target genes were purchased at Merck, Custom DNA Oligos website. RNA was reverse transcribed to cDNA and prepared for RT-PCR reaction using the GoTaq(R) 2-Step RT-qPCR kit (A6010, Promega Biotech Iberica, SP) and based on the manufacturer's indications. RT-PCR reaction was performed using a CFX Connect Real-time PCR detection system (Biorad, USA) and differential gene expression was calculated by using the 2−ΔΔCt method. Gene expression levels of target genes were measured by normalization to the housekeeping gene GAPDH exploiting control samples as calibrators. Data are provided normalized to data of cells seeded in control cell culture standard plastic surface. All the reactions were conducted in triplicates.
| NAME | SEQUENCE 5'--3′ | Tm | GC Content (%) |
|---|---|---|---|
| Gapdh-F | ATGACATCAAGAAGGTGGTG | 60 | 45.00 |
| Gapdh-R | CATACCAGGAAATGAGCTTG | 59,7 | 45.00 |
| Col2a1-F | ACTGGTAAGTGGGGCAAGAC | 62,9 | 55.00 |
| Col2a1-R | CCACACCAAATTCCTGTTCA | 63,4 | 45.00 |
| Runx2-F | GGACTGTGGTTACCGTCAT | 66,7 | 52.63 |
| Runx2-R | GGAGGATTTGTGAAGACTGTT | 66,6 | 42.86 |
| Sox9-F | GAGGAAGTCGGTGAAGAACG | 63,8 | 55.00 |
| Sox9-R | CTGAGATTGCCCAGAGTGCT | 64,6 | 55.00 |
4.7. Freeze dried 3D scaffold formation and characterization
DCC powders were dissolved in acetic acid 0.5 M for 24 h at RT at 15 mg/mL concentration. The solution was freeze-dried for 24 h. Pictures were obtained from dry samples and samples immersed in PBS for 2 h.
A sputter coater (Alto 1000, Galan) was used to coat the sample's surface with gold/palladium before their visualization in JSM-6490LV (JEOL) scanning electron microscope (SEM) equipment with a 10/15 kV acceleration voltage.
Rheological experiments were carried out in a parallel-plate geometry (200 mm diameter with a 1,2 mm gap) of a Discovery HR20 rheometer (TA Instruments, New Castle, Denver, CO USA). Samples were immersed in water for 10 min for hydration and then deposited on the rheology equipment. Oscillatory rheology (n = 5) was used to measure the rheological properties of scaffold after hydration. Measurements were performed at room temperature in constant deformation control mode over a range frequency from 0,01–10 Hz [64].
Swelling properties of the solid foam scaffolds were determined by water absorption (n = 3). Lyophilized foams weight was recorded (Wd) before adding distillate water for 24 h until maximum hydration. The excess liquid was carefully removed, and wet weight recorded. The mass swelling ratio (S) was determined.
In vitro degradation test of the scaffold was performed by measured weight of the materials (n = 5) before and after the incubation. The samples were incubated at 37 °C in DPBS or DMEM/F-12, and with or without 0,01 % collagenase (C0130, Sigma Aldrich, USA). After the indicated incubation periods, samples were removed from de solutions, dried in vacuum and weighed.
4.8. Cell culture studies on 3D freeze-dried scaffolds
For cell culture studies, DCC freeze-dried scaffold pieces was transferred to a 48-well plate well (3548, Costar, USA) and injected with 30 μL of a ATDC5 cell dilution, trying to seed evenly all around the sample, resulting in 250,000 cells seeded per sample. Cell-seeded samples were incubated at 37 °C and 5 % CO2 for 1 h in a Forma Steri-cycle CO2 incubator (Model 381 Thermo Scientific, USA), then covered with cell culture media for further studies.
Imaging of seeded scaffolds was carried out as described above, by fixing the samples in formalin 10 %, staining of the cytoskeleton and nuclei and confocal Z-stacks (ca. 50 μm thick) imaging.
For long-term cell cultures, cell-seeded samples were cultured for 21 days in complete warm media or Mesencult ACF chondrogenic differentiation medium. Gene expression studies were conducted as described above. Samples were paraffin embedded and processed for GAG staining by Alcian blue as described above.
4.9. In vivo implantation studies with 3D freeze-dried scaffolds
This manuscript follows the ARRIVE guidelines for full and transparent reporting of research involving animals. For in vivo studies, all animal handling and experimental procedures were previously approved by the Animal Care and Use Committee of Hospital Clínico San Carlos and competent authority at Comunidad Autonoma de Madrid (Authorized animal experimental project reference: PROEX 013.6/22; approval date 18–11–2021; title “development of in vivo implantable experimental approaches for regenerative medicine, stem cell biology and disease modelling purposes”), according to the EC guidelines for ethical care of experimental animals. The selected surgical procedure was the critical size osteochondral defect in rabbit femoral condyle. The animals were randomly allocated to the following groups: 1- Critical size defect empty, 2- Critical size defect filled with DCC scaffold. The sample size was established at n = 3 per group according to the primary outcome measure (histology). A total of 3 animals were used, with 2 implants per animal (right and left femur heads). New Zealand White rabbits (weight 3–3.5 kg) were treated with an intramuscular injection of xylazine (RompumVR) (3 mg/kg), ketamine (KetolarVR) (80 mg/kg) and atropin sulfate (Atropina BraunVR) (1 mg/kg). The knee regions were then shaved and disinfected. A medial parapatellar incision was made on the bilateral knee joint. The dissection continued until the medial femoral condyle was exposed. A 4-mm diameter defect to a depth of 5 mm centered on the medial femoral condyle was created using a surgical drill bit. Free-dried scaffold samples were reconstituted in PBS and were placed tightly within the defect site to ensure a perfect filling of the defect. After scaffold implantation, knee joint synovium, capsule and skin were carefully sutured separately. Animals were allowed free movement and fed a standard diet ad libitum after surgical procedure. Animals were euthanized by intravenous injection of 100 mg/kg of sodium pentobarbital. Knee joints were exposed, samples were harvested, images were obtained, and samples were fixed in 4 % formalin.
For histological evaluation, specimens were decalcified, paraffin embedded, and sectioned at 4 μm thick sections. Conventional staining was conducted with hematoxylin-eosin staining; collagen staining (Masson's trichrome); and Proteoglycan staining (Safranin O and PAS). All histological processing was performed by “Micros veterinaria SL” histological services (https://microsvet.es/). Images were obtained using a routine inverted microscope coupled with camera.
Images were evaluated following the ISCR scoring method [65] which provides values from 0 to 3 to the following visual appreciations: ‘surface’, ‘matrix’, ‘cell distribution’, ‘cell population’, ‘subchondral bone’ and ‘cartilage mineralization’.
4.10. Statistics
All data were plotted and statistically analyzed using the GraphPad Prism software (La Jolla, CA, USA). In all cases, data normality was verified using the Shapiro–Wilk test and a normal QQ plot was assessed before statistical analysis using parametric tests (t-test, ANOVA) and Tukey's multiple comparisons tests. Data are presented as mean ± SD, and each individual point is provided in the plots. Additionally, the n number of each assay is provided in the figure legends. P values of less than 0.05 were considered statistically significant (ns p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001).
Funding
Grants PID2021-127191OB-I00, RTI2018-101708-A-I00 and PRE2022-102680 funded by MCIN/AEI/10.13039/501100011033 and by “ERDF A way of making Europe”. Grant RYC2018-025502-I and PRE2018-084542 funded by MCIN/AEI/10.13039/501100011033 and by “ESF Investing in your future”. Grant MDM-2017-0720 Maria de Maeztu Units of Excellence Program was funded by the Spanish State Research Agency.
CRediT authorship contribution statement
Unai Mendibil: Software, Investigation, Formal analysis, Data curation. Yaiza Lópiz-Morales: Writing – review & editing, Visualization, Validation, Project administration, Investigation. Blanca Arnaiz: Visualization, Software, Methodology, Investigation, Formal analysis. Raquel Ruiz-Hernández: Investigation. Pablo Martín: Investigation. Desiré Di-Silvio: Investigation, Formal analysis. Nerea Garcia-Urquia: Investigation. Felix Elortza: Resources, Methodology, Investigation. Mikel Azkargorta: Methodology, Investigation, Formal analysis, Data curation. Beatriz Olalde: Writing – review & editing, Visualization, Supervision, Resources, Project administration, Funding acquisition, Conceptualization. Ander Abarrategi: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank the staff at CIC biomaGUNE core facility platforms. We would like to thank the animal facility staff at Hospital Clinico San Carlos Madrid.
Contributor Information
Beatriz Olalde, Email: beatriz.olalde@tecnalia.com.
Ander Abarrategi, Email: aabarrategi@cicbiomagune.es.
Data availability
We have shared the link to data at "data availability" section
References
- 1.Barui S., Ghosh D., Laurencin C.T. Osteochondral regenerative engineering: challenges, state-of-the-art and translational perspectives. Regen Biomater. 2023;10 doi: 10.1093/rb/rbac109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei W., Dai H. Articular cartilage and osteochondral tissue engineering techniques: recent advances and challenges. Bioact. Mater. 2021;6:4830–4855. doi: 10.1016/j.bioactmat.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendibil U., Ruiz-Hernandez R., Retegi-Carrion S., Garcia-Urquia N., Olalde-Graells B., Abarrategi A. Tissue-specific decellularization methods: rationale and strategies to achieve regenerative compounds. International Journal of Molecular Sciences 2020. 2020;21:5447. doi: 10.3390/ijms21155447. Page 5447 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendibil U., Ruiz-Hernández R., Arnaiz B., Khatami N., Abarrategi A. Handbook of the Extracellular Matrix. 2023. Extracellular matrix isolation: sources and methods; pp. 1–33. [Google Scholar]
- 5.Iazzolino G., Mendibil U., Arnaiz B., Ruiz-de-Angulo A., Azkargorta M., Uribe K.B., Khatami N., Elortza F., Olalde B., Gomez-Vallejo V., Llop J., Abarrategi A. Decellularization of xenografted tumors provides cell-specific in vitro 3D environment. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.956940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia C., Mei S., Gu C., Zheng L., Fang C., Shi Y., Wu K., Lu T., Jin Y., Lin X., Chen P. Decellularized cartilage as a prospective scaffold for cartilage repair. Mater. Sci. Eng. C. 2019;101:588–595. doi: 10.1016/j.msec.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y., Yan L., Chen S., Pei M. Functionality of decellularized matrix in cartilage regeneration: a comparison of tissue versus cell sources. Acta Biomater. 2018;74:56–73. doi: 10.1016/j.actbio.2018.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiyotake E.A., Beck E.C., Detamore M.S. Cartilage extracellular matrix as a biomaterial for cartilage regeneration. Ann. N. Y. Acad. Sci. 2016;1383:139–159. doi: 10.1111/nyas.13278. [DOI] [PubMed] [Google Scholar]
- 9.Monibi F.A., Cook J.L. Tissue-derived extracellular matrix bioscaffolds: emerging applications in cartilage and meniscus repair. Tissue Eng Part B Rev. 2017;23:386–398. doi: 10.1089/ten.TEB.2016.0431. [DOI] [PubMed] [Google Scholar]
- 10.Huang Z., Godkin O., Schulze-Tanzil G. The challenge in using mesenchymal stromal cells for recellularization of decellularized cartilage. Stem Cell Reviews and Reports. 2016;13(1):50–67. doi: 10.1007/s12015-016-9699-8. [DOI] [PubMed] [Google Scholar]
- 11.Monzavi S.M., Kajbafzadeh A.M., Sabetkish S., Seifalian A. Extracellular matrix scaffold using decellularized cartilage for hyaline cartilage regeneration. Adv. Exp. Med. Biol. 2021;1345:209–223. doi: 10.1007/978-3-030-82735-9_17. [DOI] [PubMed] [Google Scholar]
- 12.Benders K.E.M., Boot W., Cokelaere S.M., Van Weeren P.R., Gawlitta D., Bergman H.J., Saris D.B.F., Dhert W.J.A., Malda J. Multipotent stromal cells outperform chondrocytes on cartilage-derived matrix scaffolds. Cartilage. 2014;5:221–230. doi: 10.1177/1947603514535245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elder B.D., Kim D.H., Athanasiou K.A. Developing an articular cartilage decellularization process toward facet joint cartilage replacement. Neurosurgery. 2010;66:722–727. doi: 10.1227/01.NEU.0000367616.49291.9F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinheiro A., Cooley A., Liao J., Prabhu R., Elder S. Comparison of natural crosslinking agents for the stabilization of xenogenic articular cartilage. J. Orthop. Res. 2016;34:1037–1046. doi: 10.1002/jor.23121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider C., Lehmann J., Van Osch G.J.V.M., Hildner F., Teuschl A., Monforte X., Miosga D., Heimel P., Priglinger E., Redl H., Wolbank S., Nürnberger S. Systematic comparison of protocols for the preparation of human articular cartilage for use as scaffold material in cartilage tissue engineering. Tissue Eng Part C Methods. 2016;22:1095–1107. doi: 10.1089/ten.TEC.2016.0380. [DOI] [PubMed] [Google Scholar]
- 16.Vas W.J., Shah M., Blacker T.S., Duchen M.R., Sibbons P., Roberts S.J. Decellularized cartilage directs chondrogenic differentiation: creation of a fracture callus mimetic. Tissue Eng Part A. 2018;24:1364–1376. doi: 10.1089/ten.TEA.2017.0450. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z., Shi Y., Wei X., He J., Yang S., Dickson G., Tang J., Xiang J., Song C., Li G. Fabrication and repair of cartilage defects with a novel acellular cartilage matrix scaffold. Tissue Eng Part C Methods. 2009;16:865–876. doi: 10.1089/ten.TEC.2009.0444. [DOI] [PubMed] [Google Scholar]
- 18.Yin H., Wang Y., Sun X., Cui G., Sun Z., Chen P., Xu Y., Yuan X., Meng H., Xu W., Wang A., Guo Q., Lu S., Peng J. Functional tissue-engineered microtissue derived from cartilage extracellular matrix for articular cartilage regeneration. Acta Biomater. 2018;77:127–141. doi: 10.1016/j.actbio.2018.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z., Li Z., Li Z., Wu B., Liu Y., Wu W. Cartilaginous extracellular matrix derived from decellularized chondrocyte sheets for the reconstruction of osteochondral defects in rabbits. Acta Biomater. 2018;81:129–145. doi: 10.1016/j.actbio.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Sutherland A.J., Detamore M.S. Bioactive microsphere-based scaffolds containing decellularized cartilage. Macromol. Biosci. 2015;15:979–989. doi: 10.1002/mabi.201400472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothrauff B.B., Yang G., Tuan R.S. Tissue-specific bioactivity of soluble tendon-derived and cartilage-derived extracellular matrices on adult mesenchymal stem cells. Stem Cell Res. Ther. 2017;8:1–17. doi: 10.1186/s13287-017-0580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.High-precision bioactive scaffold with dECM and extracellular vesicles targeting 4E-BP inhibition for cartilage injury repair. Mater Today Bio. 2024 doi: 10.1016/j.mtbio.2024.101114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Z., Yao H., Sun H., Gu Z., Hu X., Yang J., Shi J., Yang H., Dai J., Chong H., Wang D.A., Lin L., Zhang W. Enhanced hyaline cartilage formation and continuous osteochondral regeneration via 3D-Printed heterogeneous hydrogel with multi-crosslinking inks. Mater Today Bio. 2024;26 doi: 10.1016/j.mtbio.2024.101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H., Xing F., Yu P., Lu R., Ma S., Shakya S., Zhou X., Peng K., Zhang D., Liu M. Biomimetic fabrication bioprinting strategies based on decellularized extracellular matrix for musculoskeletal tissue regeneration: current status and future perspectives. Mater. Des. 2024;243 [Google Scholar]
- 25.Golebiowska A.A., Intravaia J.T., Sathe V.M., Kumbar S.G., Nukavarapu S.P. Decellularized extracellular matrix biomaterials for regenerative therapies: advances, challenges and clinical prospects. Bioact. Mater. 2024;32:98–123. doi: 10.1016/j.bioactmat.2023.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itoh M., Itou J., Imai S., Okazaki K., Iwasaki K. A survey on the usage of decellularized tissues in orthopaedic clinical trials. Bone Joint Res. 2023;12:179–188. doi: 10.1302/2046-3758.123.BJR-2022-0383.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farr J., Gracitelli G.C., Shah N., Chang E.Y., Gomoll A.H. High failure rate of a decellularized osteochondral allograft for the treatment of cartilage lesions. Am. J. Sports Med. 2016;44:2015–2022. doi: 10.1177/0363546516645086. [DOI] [PubMed] [Google Scholar]
- 28.Nie X., Chuah Y.J., Zhu W., He P., Peck Y., Wang D.A. Decellularized tissue engineered hyaline cartilage graft for articular cartilage repair. Biomaterials. 2020;235 doi: 10.1016/j.biomaterials.2020.119821. [DOI] [PubMed] [Google Scholar]
- 29.at ASTM Volume 13.02: Medical And Surgical Materials And Devices (II) ASTM International; West Conshohocken, PA, USA: 2019. Standard Guide for Evaluating Extracellular Matrix Decellularization Processes. [DOI] [Google Scholar]
- 30.Cederlund A.A., Aspden R.M. Walking on water: revisiting the role of water in articular cartilage biomechanics in relation to tissue engineering and regenerative medicine. J R Soc Interface. 2022;19 doi: 10.1098/rsif.2022.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rathnayake M.S.B., Farrugia B.L., Kulakova K., ter Voert C.E.M., van Osch G.J.V.M., Stok K.S. Macromolecular interactions in cartilage extracellular matrix vary according to the cartilage type and location. Cartilage. 2021;13:476S–485S. doi: 10.1177/19476035211000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffin D.J., Vicari J., Buckley M.R., Silverberg J.L., Cohen I., Bonassar L.J. Effects of enzymatic treatments on the depth-dependent viscoelastic shear properties of articular cartilage. J. Orthop. Res. 2014;32:1652–1657. doi: 10.1002/jor.22713. [DOI] [PubMed] [Google Scholar]
- 33.DiDomenico C.D., Kaghazchi A., Bonassar L.J. Measurement of local diffusion and composition in degraded articular cartilage reveals the unique role of surface structure in controlling macromolecular transport. J. Biomech. 2019;82:38–45. doi: 10.1016/j.jbiomech.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 34.Gahlawat S., Nanda V., Shreiber D.I. Purification of recombinant bacterial collagens containing structural perturbations. PLoS One. 2023;18 doi: 10.1371/journal.pone.0285864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuboki Y., Tsuzaki M., Sasaki S., Liu C.F., Mechanic G.L. Location of the intermolecular cross-links in bovine dentin collagen, solubilization with trypsin and isolation of cross-link peptides containing dihydroxylysinonorleucine and pyridinoline. Biochem. Biophys. Res. Commun. 1981;102:119–126. doi: 10.1016/0006-291x(81)91497-2. [DOI] [PubMed] [Google Scholar]
- 36.Ma H., Zou T., Li H., Cheng H. The interaction of sodium dodecyl sulfate with trypsin: multi-spectroscopic analysis, molecular docking, and molecular dynamics simulation. Int. J. Biol. Macromol. 2020;162:1546–1554. doi: 10.1016/j.ijbiomac.2020.08.020. [DOI] [PubMed] [Google Scholar]
- 37.Lee J.M., Lee E.H., Kim I.S., Kim J.E. Tgfbi deficiency leads to a reduction in skeletal size and degradation of the bone matrix. Calcif. Tissue Int. 2015;96:56–64. doi: 10.1007/s00223-014-9938-4. [DOI] [PubMed] [Google Scholar]
- 38.Kou I., Ikegawa S. SOX9-dependent and -independent transcriptional regulation of human cartilage link protein. J. Biol. Chem. 2004;279:50942–50948. doi: 10.1074/jbc.M406786200. [DOI] [PubMed] [Google Scholar]
- 39.Foldager C.B., Toh W.S., Gomoll A.H., Olsen B.R., Spector M. Distribution of basement membrane molecules, Laminin and collagen type IV. Cartilage. 2014;5:123–132. doi: 10.1177/1947603513518217. Normal and Degenerated Cartilage Tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamill K.J., Kligys K., Hopkinson S.B., Jones J.C.R. Laminin deposition in the extracellular matrix: a complex picture emerges. J. Cell Sci. 2009;122:4409–4417. doi: 10.1242/jcs.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harvey S.J., Thorner P.S. Type IV collagen: a network for development, differentiation, and disease. Adv. Dev. Biol. 2005;15:1–64. [Google Scholar]
- 42.Foldager C.B., Toh W.S., Christensen B.B., Lind M., Gomoll A.H., Spector M. Collagen type IV and Laminin expressions during cartilage repair and in late clinically failed repair tissues from human subjects. Cartilage. 2016;7:52–61. doi: 10.1177/1947603515604022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stechschulte D.J., Wu J.J., Eyre D.R. Fibronectin lacking the ED-B domain is a major structural component of tracheal cartilage. J. Biol. Chem. 1997;272:4783–4786. doi: 10.1074/jbc.272.8.4783. [DOI] [PubMed] [Google Scholar]
- 44.Chevalier X. Fibronectin, cartilage, and osteoarthritis. Semin. Arthritis Rheum. 1993;22:307–318. doi: 10.1016/s0049-0172(05)80010-1. [DOI] [PubMed] [Google Scholar]
- 45.Bernardtgy B.A., Yamadag K.M., K. Olden@ Carbohydrates selectively protect a specific domain of fibronectin against proteases. OGIFAL CHEMISTRY. 1982;257:49–8554. [PubMed] [Google Scholar]
- 46.Yao Y., Wang Y. ATDC5: an excellent in vitro model cell line for skeletal development. J. Cell. Biochem. 2013;114:1223–1229. doi: 10.1002/jcb.24467. [DOI] [PubMed] [Google Scholar]
- 47.Frahs S.M., Reeck J.C., Yocham K.M., Frederiksen A., Fujimoto K., Scott C.M., Beard R.S., Brown R.J., Lujan T.J., Solov’yov I.A., Estrada D., Oxford J.T. Prechondrogenic ATDC5 cell attachment and differentiation on graphene foam; modulation by surface functionalization with fibronectin. ACS Appl. Mater. Interfaces. 2019;11:41906–41924. doi: 10.1021/acsami.9b14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loeser R.F. Integrin-mediated attachment of articular chondrocytes to extracellular matrix proteins. Arthritis Rheum. 1993;36:1103–1110. doi: 10.1002/art.1780360811. [DOI] [PubMed] [Google Scholar]
- 49.Fujita T., Azuma Y., Fukuyama R., Hattori Y., Yoshida C., Koida M., Ogita K., Komori T. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. JCB (J. Cell Biol.) 2004;166:85–95. doi: 10.1083/jcb.200401138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dy P., Wang W., Bhattaram P., Wang Q., Wang L., Ballock R.T., Lefebvre V. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev. Cell. 2012;22:597–609. doi: 10.1016/j.devcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng A., Genever P.G. SOX9 determines RUNX2 transactivity by directing intracellular degradation. J. Bone Miner. Res. 2010;25:2680–2689. doi: 10.1002/jbmr.174. [DOI] [PubMed] [Google Scholar]
- 52.Dennis S.C., Berkland C.J., Bonewald L.F., Detamore M.S. Endochondral ossification for enhancing bone regeneration: converging native extracellular matrix biomaterials and developmental engineering in vivo. Tissue Eng Part B Rev. 2015;21:247–266. doi: 10.1089/ten.teb.2014.0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lotz M.K., Otsuki S., Grogan S.P., Sah R., Terkeltaub R., D'Lima D. Cartilage cell clusters. Arthritis Rheum. 2010;62:2206–2218. doi: 10.1002/art.27528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armiento A.R., Alini M., Stoddart M.J. Articular fibrocartilage - why does hyaline cartilage fail to repair? Adv. Drug Deliv. Rev. 2019;146:289–305. doi: 10.1016/j.addr.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 55.Fischer A.H., Jacobson K.A., Rose J., Zeller R. Paraffin embedding tissue samples for sectioning. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot4989. pdb.prot4989-pdb.prot4989. [DOI] [PubMed] [Google Scholar]
- 56.Schmitz N., Laverty S., Kraus V.B., Aigner T. Basic methods in histopathology of joint tissues. Osteoarthritis Cartilage. 2010;18(Suppl 3) doi: 10.1016/j.joca.2010.05.026. [DOI] [PubMed] [Google Scholar]
- 57.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 58.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 59.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 60.Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ewald J.D., Zhou G., Lu Y., Kolic J., Ellis C., Johnson J.D., Macdonald P.E., Xia J. Web-based multi-omics integration using the Analyst software suite. Nat. Protoc. 2024;2024:1–31. doi: 10.1038/s41596-023-00950-4. [DOI] [PubMed] [Google Scholar]
- 62.Gomez I.J., Arnaiz B., Cacioppo M., Arcudi F., Prato M. Nitrogen-doped carbon nanodots for bioimaging and delivery of paclitaxel. J. Mater. Chem. B. 2018;6:5540–5548. doi: 10.1039/c8tb01796d. [DOI] [PubMed] [Google Scholar]
- 63.Cicuéndez M., Casarrubios L., Feito M.J., Madarieta I., Garcia-Urkia N., Murua O., Olalde B., Briz N., Diez-Orejas R., Portolés M.T. Effects of human and porcine adipose extracellular matrices decellularized by enzymatic or chemical methods on macrophage polarization and immunocompetence. Int. J. Mol. Sci. 2021;22:3847. doi: 10.3390/ijms22083847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia-Urkia N., Luzuriaga J., Uribe-Etxebarria V., Irastorza I., Fernandez-San-Argimiro F.J., Olalde B., Briz N., Unda F., Ibarretxe G., Madarieta I., Pineda J.R. Enhanced adipogenic differentiation of human dental pulp stem cells in enzymatically decellularized adipose tissue solid foams. Biology. 2022;11 doi: 10.3390/biology11081099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mainil-Varlet P., Aigner T., Brittberg M., Bullough P., Hollander A., Hunziker E E., Kandel R., Nehrer S., Pritzker K., Roberts S., Stauffer E. Histological assessment of cartilage repair: a report by the histology endpoint committee of the international cartilage repair society (ICRS) J Bone Joint Surg Am. 2003;85-A:45e57. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We have shared the link to data at "data availability" section