Abstract
Antiviral responses induced by double‐stranded RNA (dsRNA) include RNA interference (RNAi) and pattern‐triggered immunity (PTI), but their relative contributions to antiviral defence are not well understood. We aimed at testing the impact of exogenous applied dsRNA on both layers of defence against potato virus X expressing GFP (PVX‐GFP) in Nicotiana benthamiana. Co‐inoculation of PVX‐GFP with either sequence‐specific (RNAi) or nonspecific dsRNA (PTI) showed that nonspecific dsRNA reduced virus accumulation in both inoculated and systemic leaves. However, nonspecific dsRNA was a poor inducer of antiviral immunity compared to a sequence‐specific dsRNA capable of triggering the RNAi response, and plants became susceptible to systemic infection. Studies with a PVX mutant unable to move from cell to cell indicated that the interference with PVX‐GFP triggered by nonspecific dsRNA operated at the single‐cell level. Next, we performed RNA‐seq analysis to examine similarities and differences in the transcriptome triggered by dsRNA alone or in combination with viruses harbouring sequences targeted or not by dsRNA. Enrichment analysis showed an over‐representation of plant‐pathogen signalling pathways, such as calcium, ethylene and MAPK signalling, which are typical of antimicrobial PTI. Moreover, the transcriptomic response to the virus targeted by dsRNA had a greater impact on defence than the non‐targeted virus, highlighting qualitative differences between sequence‐specific RNAi and nonspecific PTI responses. Together, these results further our understanding of plant antiviral defence, particularly the contribution of nonspecific dsRNA‐mediated PTI. We envisage that both sequence‐specific RNAi and nonspecific PTI pathways may be triggered via topical application of dsRNA, contributing cumulatively to plant protection against viruses.
Keywords: antiviral defence, pattern‐triggered immunity, potato virus X, RNA interference, transcriptomic response to PTI, transcriptomic response to RNAi
Innate immunity against PVX‐GFP conferred by nonspecific dsRNA is weaker than the RNAi response triggered by sequence‐specific dsRNA, which correlates with a differential impact on defence responses at the transcriptome level.
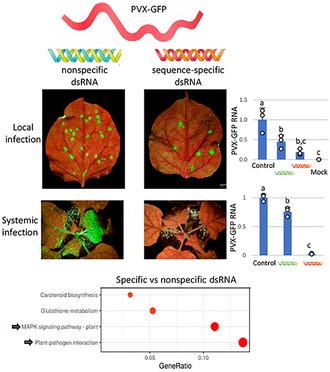
1. INTRODUCTION
Viruses are obligate intracellular pathogens that depend on a living cell to multiply and proliferate by hijacking the host cell metabolism and replication machinery. To protect themselves, plants have evolved a multilayered immune system for defence against viruses, including innate immunity and RNA silencing, among others (Ngou et al., 2022; Soosaar et al., 2005). RNA silencing or RNA interference (RNAi) is a regulatory mechanism evolutionarily conserved in most eukaryotes that relies on the sequence‐specific degradation of targeted RNAs guided by complementary small RNAs (sRNAs) (Baulcombe, 2004; D'Ario et al., 2017). In addition to its crucial activity in regulating plant development and growth, RNAi plays a main role in adaptive immunity against pathogens, including plant viruses (Ding & Voinnet, 2007; Guo et al., 2019). RNAi against viruses is triggered by double‐stranded RNA (dsRNA) molecules that derive either from viral replication intermediates or from hairpin RNAs. The dsRNA is recognized by specific RNase type III‐like enzymes designated in plants as DICER‐like proteins (DCL), which cleave the dsRNA into sRNA duplexes of 21–24 nucleotides long (Zhang et al., 2015). Then, the sRNAs bind to Argonaute (AGO) protein family members that constitute the core of the RNA‐induced silencing complexes (Carbonell & Carrington, 2015).
Innate immunity largely relies on plant perception of the microorganism through the recognition of conserved pathogen‐associated molecular patterns (PAMPs) by cellular pattern recognition receptors (PRRs), thus initiating the so‐called pattern‐triggered immunity (PTI) (Yuan et al., 2021). Despite the broad knowledge on PTI against other pathogens, much less is known about PTI against plant viruses. Nonetheless, several findings suggest that PTI also plays an important role in plant–virus interplay. Firstly, the exogenous application of dsRNA, a well‐known PAMP in animal antiviral immunity (Wang & He, 2019), has been reported to be an elicitor of PTI against plant virus infection (Niehl et al., 2016; Samarskaya et al., 2022). In contrast to sequence‐specific RNAi, antiviral PTI triggered by dsRNA is independent of RNA sequence and may also be activated by non‐viral sequences, for example, green fluorescent protein (GFP)‐derived dsRNA and the synthetic dsRNA analogue polyinosinic‐polycytidilic acid (poly[I:C]) (Niehl et al., 2016). However, plant PRRs involved in dsRNA recognition remain to be identified. Secondly, single and double Arabidopsis mutants in the PRR coreceptor kinases Somatic Embryogenesis Receptor‐like Kinase (SERK)1, SERK3 and SERK4 exhibit increased susceptibility to different RNA viruses (Huang et al., 2023; Kørner et al., 2013; Nicaise & Candresse, 2017; Yang et al., 2010). Thirdly, plant viruses have acquired the ability to suppress nonspecific PTI mechanisms via the action of viral effectors (Kong et al., 2018; Nicaise & Candresse, 2017; Zvereva et al., 2016). In particular, tobamovirus movement proteins suppress the dsRNA‐induced defence response leading to callose deposition at plasmodesmata (PD), thus enabling virus movement (Huang et al., 2023).
The antiviral PTI‐like responses induced by nonspecific dsRNA are somehow similar to those of antimicrobial PTI and include the induction of ethylene (ET) production, the activation of mitogen‐activated protein kinases (MAPKs), and the triggering of defence gene expression (Kørner et al., 2013; Niehl et al., 2016; Samarskaya et al., 2022; Yuan et al., 2021). However, the perception of poly(I:C) did not lead to the production of reactive oxygen species (ROS), indicating differences between dsRNA‐induced and microbial‐induced PTI signalling pathways (Huang et al., 2023; Niehl et al., 2016). Antimicrobial PTI is regulated by phytohormones, with salicylic acid (SA), jasmonic acid (JA) and ET acting as central players in triggering the immune signalling network (Pieterse et al., 2012). For instance, it has been reported that SA and JA signalling are required for activation of PTI against Pseudomonas syringae induced by exogenous application of bacterial RNA in Arabidopsis (Lee et al., 2016). Nevertheless, the contribution of phytohormone signalling to nonspecific dsRNA‐mediated PTI against infecting viruses remains largely uncharacterized.
Plant viruses represent a serious threat to agricultural production and food supply (Savary et al., 2019). Conventional breeding for introgression of major resistance genes even with new genetic marker technologies is a time‐consuming and laborious process (Boiteux et al., 2012). Moreover, the heavy use of chemicals to control vectors transmitting plant viruses has a significant negative impact on human health and environment (Chagnon et al., 2015). Thus, alternative biotechnological approaches are needed to confer protection against virus diseases, which are more sustainable, environmentally friendly and positively perceived by society. Since the first example describing that exogenous application of dsRNA‐induced RNAi‐based protection against RNA viruses (Tenllado & Díaz‐Ruíz, 2001), many studies have been documented reporting the topical application of dsRNA to confer protection against pests and pathogens in numerous plant species (reviewed in Voloudakis et al., 2022; Hoang et al., 2022). Given that dsRNA is the trigger of antiviral RNA silencing and can also act as a potent PTI elicitor, we envisage that both sequence‐specific RNAi and nonspecific PTI pathways may be triggered via topical application of dsRNA, contributing synergistically to plant protection against viruses.
Plant antiviral responses include sequence‐specific RNAi and nonspecific PTI pathways, but their relative contributions to antiviral defence are not thoroughly understood. In this study, we have compared the impact of exogenously applied dsRNA on both these layers of defence and explore its effect on the local and systemic accumulation of potato virus X (PVX) expressing GFP (PVX‐GFP) in Nicotiana benthamiana. We found that while sequence‐specific dsRNA halted virus spread throughout the plant, nonspecific dsRNA reduced virus accumulation locally but was unable to prevent systemic infection in N. benthamiana. Previous findings on PTI‐responsive genes elicited by nonspecific dsRNA were based on a limited number of candidate genes altered by microbial elicitors, which were selected from publicly available gene expression data (Niehl et al., 2016; Samarskaya et al., 2022). To further investigate the dsRNA‐triggered antiviral defence at trancriptome‐wide level, we performed next‐generation sequencing to examine similarities and differences in the alteration of the whole transcriptome triggered by dsRNA alone or in combination with viruses harbouring sequences targeted or not by dsRNA. After examining the RNA‐seq data, the transcriptomic response to the virus targeted by dsRNA had a greater impact on defence than the nontargeted virus, highlighting qualitative differences between sequence‐specific RNAi and nonspecific PTI immune responses.
2. RESULTS
2.1. Differential reduction in virus abundance in sequence‐specific and nonspecific dsRNA‐treated plants
Nicotiana benthamiana plants were inoculated with mixtures of PVX‐GFP and dsRNA molecules homologous to either GFP (dsGFP, 717 bp) or the Potato virus Y (PVY) coat protein (CP) gene (dsPVY, 902 bp) (Figure 1a,b). Both dsRNAs were produced in the RNAse III‐deficient strain Escherichia coli HT115(DE3) upon IPTG induction. The combination of PVX‐GFP plus nucleic acid extracted from bacterial cells not expressing the dsPVY was used as negative control. The dsPVY and PVX CP gene share low overall sequence identity (<50%), with no stretches of nucleotide identity longer than seven bases. A total of 12 plants from each treatment were tested in each of two independent experiments, and the subsequent viral infection was monitored by detecting GFP foci in the inoculated leaves at 7 days post‐inoculation (dpi). There were fewer fluorescent spots or infection foci in the inoculated leaves treated with dsGFP than with the negative control, implying that sequence‐specific dsRNA partially interfered with PVX‐GFP infection (Figure 1c). However, there was no significant difference in the number of GFP foci between nonspecific dsRNA (dsPVY)‐ and control‐treated leaves. Treatments with either dsGFP or dsPVY did not cause a significant change in the size of GFP foci (Figure 1d). To examine the effect of dsRNA treatments on the accumulation level of PVX‐GFP, we measured the relative amount of virus in inoculated leaves from dsGFP‐, dsPVY‐ and control‐treated plants at 7 dpi. Comparative analysis by reverse transcription‐quantitative PCR (RT‐qPCR) revealed that the level of PVX‐GFP RNA in dsGFP‐ and dsPVY‐treated leaves was reduced 5.4‐ and 2.2‐fold, respectively, compared to the control (Figure 1e).
FIGURE 1.
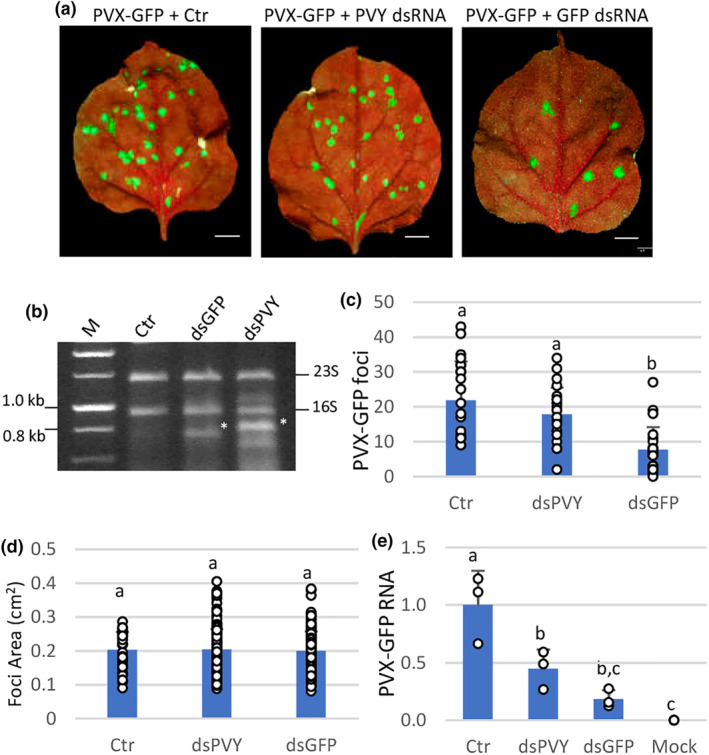
Sequence‐specific and nonspecific dsRNAs interfere differently with PVX‐GFP local infection. Nicotiana benthamiana plants were inoculated with mixtures of PVX‐GFP combined with nucleic acid extracts prepared from Escherichia coli accumulating the dsPVY or dsGFP. (a) Representative inoculated leaves were examined under UV light at 7 days post‐inoculation (dpi). Scale bar denotes 1 cm. (b) E. coli HT115(DE3) cultures transformed with L4440 encoding either a dsRNA consisting of 902 bp of the PVY coat protein (CP) gene (dsPVY) or GFP (dsGFP) were induced with IPTG and processed for total nucleic acid. The total nucleic acid extracted from bacterial cells not expressing dsRNA was used as a control (Crt). Samples were resolved by electrophoresis on 1% agarose gel. The positions of 23S and 16S rRNA are indicated on the right. dsRNA bands are indicated by asterisks. NZYDNA Ladder III was used as dsDNA markers (M). (c) Mean numbers ± SD of infection foci on inoculated leaves of 12 plants (two leaves per plant) with the different treatments at 7 dpi. (d) Mean sizes ± SD of infection foci on inoculated leaves of six plants with the different treatments. (e) Reverse transcription‐quantitative PCR was used to analyse the accumulation of PVX‐GFP genomic RNA levels in the inoculated leaves at 7 dpi. Mock, mock‐inoculated plants. Expression of the 18S rRNA gene served as a control. Data represent the means ± SD of three replicates, each consisting of a pool of 12 plants that received the same treatment. Different letters indicate significant differences determined by employing Scheffé's multiple range test (p < 0.05). Experiments were repeated once more with similar results.
In independent experiments, the effectiveness of sequence‐specific dsRNA (dsGFP) or nonspecific dsRNA (dsPVY) in providing protection against PVX‐GFP was tested by systemic infection assays. All of the 12 N. benthamiana plants inoculated with PVX‐GFP plus control extract showed typical symptoms of mild mottling and GFP fluorescence in systemic leaves at 7 dpi. At the same time point, only 3 of 12 and 10 of 12 of plants treated with PVX‐GFP plus either dsGFP or dsPVY showed viral symptoms and GFP fluorescence, respectively (Figure 2a). At later observation dates (10 dpi), whereas dsGFP‐treated plants remained free of symptoms, all the plants treated with dsPVY displayed viral symptoms and GFP fluorescence in upper leaves, indicating that treatment with nonspecific dsRNA slowed down PVX‐GFP infection. Northern blot analysis from RNA extracted from a pool of the upper leaf tissue from each treatment at 7 dpi revealed that accumulation of PVX‐GFP genomic RNA was slightly lower in plants treated with PVX‐GFP plus dsPVY than in control plants (Figure 2b), whereas PVX‐GFP accumulated below detection limits in dsGFP‐treated plants displaying no symptoms of infection. This was corroborated by RT‐qPCR analysis, where a 98% and 25% reduction in PVX‐GFP RNA levels was quantified in plants treated with the sequence‐specific and nonspecific dsRNAs, respectively (Figure 2c). Taken together, our findings show that nonspecific dsRNA induces a reduced inhibition of PVX‐GFP accumulation in local and systemic leaves compared to sequence‐specific dsRNA.
FIGURE 2.
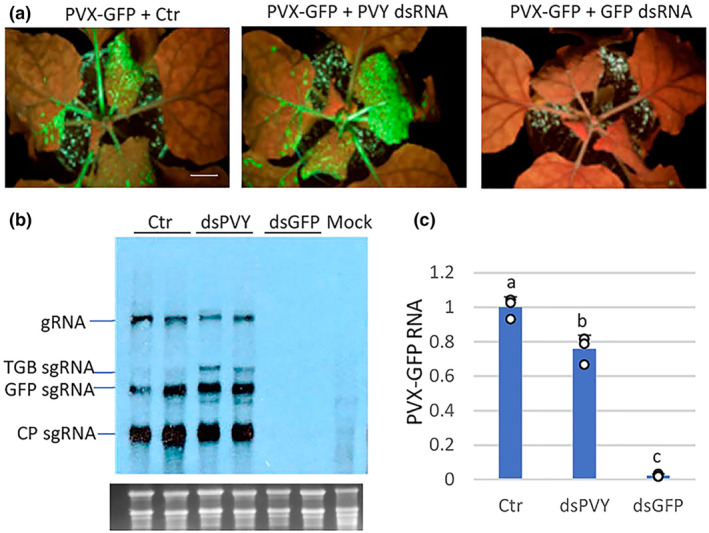
Systemic response of Nicotiana benthamiana plants to inoculation with mixtures of PVX‐GFP combined with dsPVY, dsGFP or control (Ctr) extract. (a) Representative plants were photographed under UV light at 8 days post‐inoculation (dpi). (b) Northern blot analysis of total RNA extracted from upper leaf tissues at 7 dpi. Two independent pooled samples were analysed for each combination. Total RNA (5 μg) was hybridized with a probe complementary to PVX CP. PVX genomic (g)RNA and the major subgenomic (sg)RNAs, triple‐gene‐block (TGB) sgRNA, GFP sgRNA and CP sgRNA, are indicated. Ethidium bromide staining of rRNA is shown as loading control. (c) Reverse transcription‐quantitative PCR was used to analyse the accumulation of PVX‐GFP genomic RNA levels in the systemic leaves at 7 dpi. Expression of the 18S rRNA gene served as a control. Data represent the means ± SD of three replicates, each consisting of a pool of 12 plants that received the same treatment. Different letters indicate significant differences determined by employing Scheffé's multiple range test (p < 0.05). Experiments were repeated once more with similar results.
2.2. Nonspecific dsRNA causes inhibition of PVX‐GFP replication
We next investigated the particular step of the viral replication cycle that was affected by nonspecific dsRNA. N. benthamiana plants were inoculated with mixtures of PVX‐GFP plus either dsPVY or control extracts and with mixtures of PVX‐GFP plus either a bacterial PTI elicitor derived from flagellin (flg22) or water as a control, and samples were taken at an early time point after inoculation. At 4 dpi, there were no significant differences in the number and the size of GFP foci between the leaves treated with dsPVY or flg22 compared to control‐treated leaves (Figure S1). Both PVX‐GFP RNA and CP accumulated less in the inoculated leaves of dsPVY and flg22‐treated plants than in the controls, as assayed by RT‐qPCR and western blot analyses at 4 dpi (Figure 3a,b).
FIGURE 3.
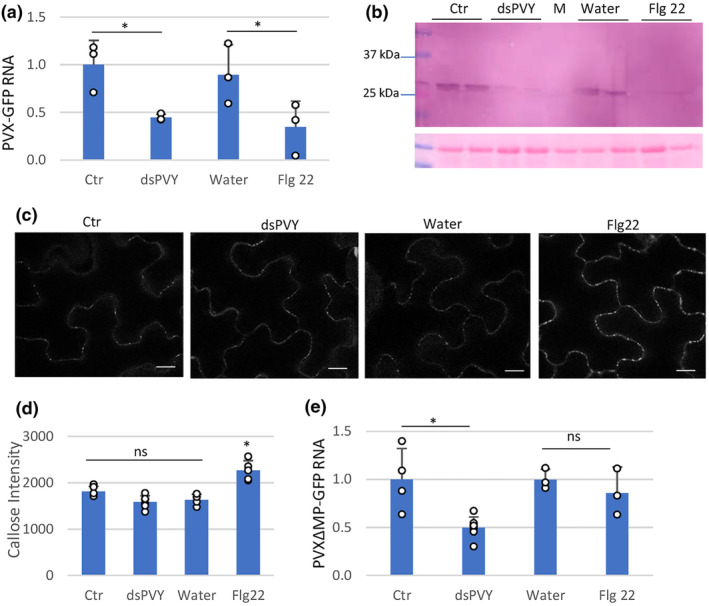
Nonspecific dsRNA elicits inhibition of PVX‐GFP replication. Plants were inoculated with mixtures of PVX‐GFP and either flagellin (flg22), dsPVY, control (Ctr) extracts or water. (a) Reverse transcription‐quantitative PCR (RT‐qPCR) was used to analyse the accumulation of PVX‐GFP genomic RNA levels in the inoculated leaves at 4 days post‐inoculation (dpi). Expression of the 18S rRNA gene served as a control. Data represent the means ± SD of three replicates, each consisting of a pool of nine plants that received the same treatment. (b) Western blot analysis of plant extracts derived from inoculated leaves at 4 dpi, using antibodies against PVX CP. The panel below the blot is the membrane stained with Ponceau S as control of loading. M, mock‐inoculated leaf. (c) Callose spots at plasmodesmata (PD) were visualized upon aniline blue staining of epidermal cells in response to water, bacterial flg22, dsPVY or Ctr extracts. Photographs were taken 30 min after treatment with water, 1 μM flg22, 30 ng/μL of dsPVY or Ctr extracts. Scale bar, 10 μm. (d) Relative PD callose content in water‐, flg22‐, dsPVY‐ and Ctr‐treated leaves. The mean values of callose intensities in individual PD (>200) measured in three leaf discs taken from two independent biological replicates per each treatment. (e) Accumulation of a frameshift mutant in the P25 movement protein of PVX‐GFP (PVXΔMP‐GFP), as assayed by RT‐qPCR at 4 dpi. Asterisks indicate significant differences between treatments (Student's t test, *p < 0.05); ns, not significant. Experiments were repeated once more with similar results.
It has been reported that nonspecific dsRNA‐induced immunity against tobacco mosaic virus (TMV) restricts the progression of virus movement by triggering callose deposition at PD (Huang et al., 2023). To investigate whether PD callose deposition is involved in inhibition of PVX‐GFP accumulation mediated by dsPVY in inoculated leaves, we used in vivo aniline blue staining to quantify PD‐associated callose in dsPVY‐ and flg22‐treated leaves. Treatment with flg22 caused an increase in PD callose intensity compared to leaves treated with water (Figure 3c,d). By contrast, in our experimental conditions PD‐associated callose in dsPVY‐treated leaves remained at control levels, indicating that callose accumulation was not responsible for the inhibition of PVX‐GFP accumulation caused by nonspecific dsRNA.
To test whether virus replication is affected by nonspecific dsRNA, we measured the accumulation of PVX‐GFP RNA in leaves inoculated with a frameshift mutant in the P25 movement protein (PVXΔMP‐GFP) that renders the virus unable to move between cells (Bayne et al., 2005). As shown in Figure 3e, treatment of the leaves with dsPVY elicited a significant reduction in PVXΔMP‐GFP accumulation compared to control‐treated leaves at 4 dpi. Treatment with flg22 did not have a significant impact on viral accumulation in PVXΔMP‐GFP‐inoculated leaves. Altogether, the unaltered size and number of infection sites together with the lack of callose accumulation in dsPVY‐treated leaves suggested that the nonspecific dsRNA‐triggered immunity against PVX‐GFP was not linked to the reduced cell‐to‐cell movement of the virus, but to inhibition of virus replication.
2.3. dsRNA causes transcriptome reprogramming in N. benthamiana
To identify early biological processes and pathways associated with dsRNA‐based immunity, we conducted a global transcriptome RNA‐seq assay with RNA extracted at 4 dpi from N. benthamiana leaves inoculated with four different treatments, that is, dsGFP alone (dsG), PVX‐GFP combined with dsGFP (targeted virus/dsRNA combination, dsG_VG), wild‐type (WT) PVX combined with dsGFP (non‐targeted virus/dsRNAcombination, dsG_V) and bacterial nucleic acid extracts not expressing dsRNA as a control (Ctr). Sequencing of 12 transcriptome libraries generated over 968 million mapped reads. On average, 88.76% of the clean reads had quality scores at the Q30 level. The sequencing data are summarized in Table S1.
Principal component analysis (PCA) was used to sort RNA‐seq‐based transcriptomic data according to gene expression levels. Two principal components explained 61.5% of the overall variance of gene expression profiles (35.8% and 25.7% for principal component 1 (PC1) and principal component 2 (PC2), respectively; Figure 4a). PC1 seems to highlight a shift in gene expression between virus‐infected samples, dsG_V and dsG_VG, and noninfected ones, dsG and Ctr, which suggests a differential transcriptional status in virus‐infected leaves. In addition, the PCA score plot revealed a clear separation between Ctr and samples treated with dsGFP, that is, dsG, dsG_V and dsG_VG, indicating that treatment with dsRNA was the main source of variance underlying PC2.
FIGURE 4.
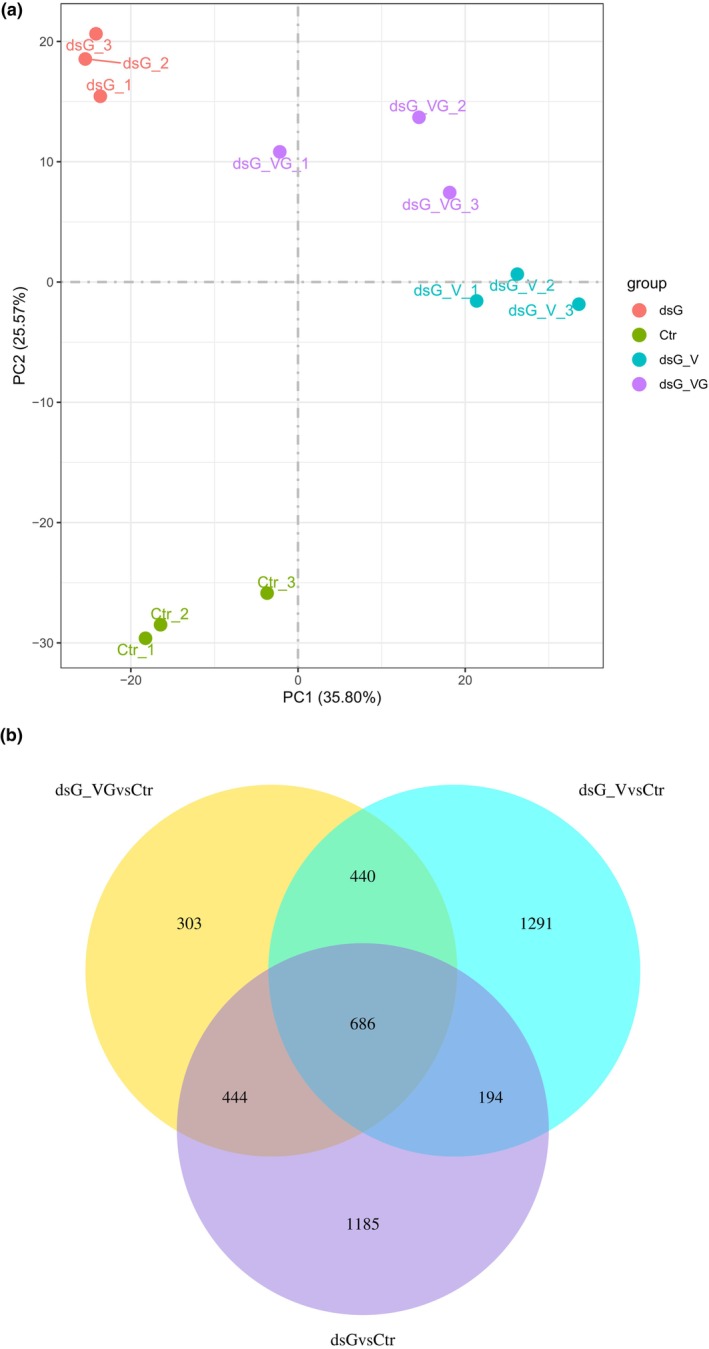
Transcriptional reprogramming associated with either dsRNA alone or virus infections targeted or not by dsRNA. (a) Principal component analysis (PCA) of RNA‐seq data. The PCA was performed using normalized RNA‐seq data of differentially expressed genes (DEGs) between treatments at 4 days post‐inoculation. Each biological replicate is represented in the score plot. The variance explained by each component (%) is given in parentheses. Treatments were as follows: dsGFP alone (dsG), PVX‐GFP combined with dsGFP (dsG_VG), wild‐type PVX combined with dsGFP (dsG_V) and bacterial nucleic acid extracts not expressing dsRNA as a control (Ctr). PCA was performed using the R package. (b) Venn diagrams displaying the number of DEGs with a log2(fold‐change) ≥|1| and adjusted p‐value ≤0.05 in the dsG vs. Ctr, dsG_VG vs. Ctr and dsG_V vs. Ctr pairwise comparisons.
Differentially expressed genes (DEGs) from paired comparisons are presented in Table S1 (adjusted p‐value ≤0.05; log2 ratio ≥|1|). We focused on comparisons between the expression profiles resulting from response to either dsRNA alone (dsG vs. Ctr), the targeted interaction between PVX‐GFP and dsGFP (dsG_VG vs. Ctr), or the non‐targeted interaction between PVX and dsGFP (dsG_V vs. Ctr), to highlight qualitative differences attributable to responses to dsRNA itself, sequence‐specific RNAi and non‐sequence‐specific PTI, respectively. In the dsG vs. Ctr comparison, 2509 DEGs, including 1441 upregulated and 1068 downregulated genes, were identified, whereas 2611 (1962 upregulated and 649 downregulated) and 1873 (1514 upregulated and 359 downregulated) DEGs showed significant changes in the dsG_V vs. Ctr and dsG_VG vs. Ctr comparisons, respectively (Figure 4b). There was substantial overlap in the DEGs altered in each comparison, with 1126, 1130 and 880 genes in common between dsG_VG vs. Ctr and dsG_V vs. Ctr, dsG_VG vs. Ctr and dsG vs. Ctr, and dsG_V vs. Ctr and dsG vs. Ctr comparisons, respectively. A comparison of DEGs between dsG_VG vs. dsG and dsG_V vs. dsG dataset is shown in Figure S2.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to infer pathways significantly associated with DEGs in each dataset (Table S2). KEGG enrichment analysis (p < 0.05) allowed the identification of several terms including photosynthesis, plant–pathogen interaction, porphyrin and chlorophyll metabolism, cutin, suberine and wax biosynthesis, pentose and glucuronate interconversions, and MAPK signalling pathways, among others, over‐represented in the dsG vs. Ctr comparison (Figure 5a). It is noteworthy that, among others, the KEGG terms plant–pathogen interaction and MAPK signalling pathway were also over‐represented in the set of DEGs from dsG_V vs. Ctr and dsG_VG vs. Ctr comparisons, but not in dsG_V vs. dsG and dsG_VG vs. dsG comparisons (Figure 5b–e). This suggests that contribution of virus infection itself to the enrichment of genes related to these KEGG terms was not significant in our experimental conditions. Furthermore, the plant–pathogen interaction and MAPK signalling pathways were still over‐represented in the dsG_VG vs. dsG_V comparison (Figure 5f) albeit with a reduced number of DEGs, suggesting that the interaction of dsGFP with PVX‐GFP (RNAi) altered gene expression differentially compared to the interaction of dsGFP with WT PVX (PTI). Indeed, the over‐representation of the plant–pathogen interaction and MAPK signalling pathway terms in the dsG_VG vs. Ctr comparison was greater than in dsG_V vs. Ctr (the ratio of the number of DEGs annotated to the plant–pathogen interaction pathway to the total number of annotated DEGs was 51/328 [dsG_VG vs. Ctr] and 59/504 [dsG_V vs. Ctr]; and 35/328 [dsG_VG vs. Ctr] and 48/504 [dsG_V vs. Ctr] for MAPK signalling pathway), indicating a deeper impact of the sequence‐specific RNAi response on the transcriptome (Table S2). The KEGG term plant hormone signal transduction was uniquely over‐represented in the dsG_V vs. Ctr comparison, and includes genes involved in auxin metabolism and signalling, abscisic acid (ABA), cytokinin, gibberellin and ET signalling, and SA response (Table S3).
FIGURE 5.
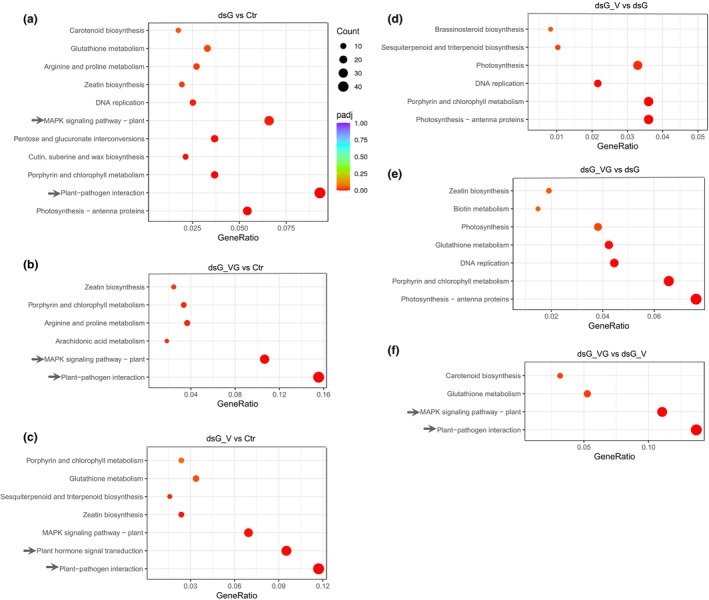
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed genes (DEGs). KEGG terms enriched (adjusted p‐value <0.05) in the (a) dsG vs. Ctr, (b) dsG_VG vs. Ctr, (c) dsG_V vs. Ctr, (d) dsG_V vs. dsG, (e) dsG_VG vs. dsG and (f) dsG_VG vs. dsG_V comparisons are shown. Gene ratio is the percentage of total DEGs in the given KEGG term. Dot size represents the number of genes annotated to a specific KEGG term. The KEGG terms plant–pathogen interaction, MAPK signalling and plant hormone signal transduction are indicated by an arrow.
2.4. PTI‐related gene expression induced by nonspecific dsRNA in N. benthamiana
The dsG vs. Ctr comparison differentially altered the expression of 48 (38 upregulated and 10 downregulated) and 34 (21 upregulated and 13 downregulated) DEGs classified in the plant–pathogen interaction and MAPK signalling pathways terms, respectively (Table S3). Many of these DEGs were also altered in the datasets from dsG_V vs. Ctr and dsG_VG vs. Ctr comparisons. DEGs in the plant–pathogen interaction term comprise genes involved in calcium (Ca2+) signalling, WRKY transcription factors, acyltransferases, mitogen‐activated protein kinase (MAPK) 3 and MAPK kinase 5, pathogenesis‐related (PR) genes and associated transcriptional factors, LRR receptor‐like serine/threonine‐protein kinase FLS2, NBS‐LRR resistance genes RPM1 and RPS2, EDS1L‐like protein, and heat shock protein 82, among others. DEGs in the MAPK signalling pathway term include genes involved in ET signalling, Ca2+ signalling, WRKY transcription factors, abscisic acid (ABA) signalling, serine/threonine‐protein kinases, among others. A pictorial representation of the nonredundant list of genes associated with the plant–pathogen interaction and MAPK signalling pathways in the dsG vs. Ctr comparison is shown in Figure 6.
FIGURE 6.
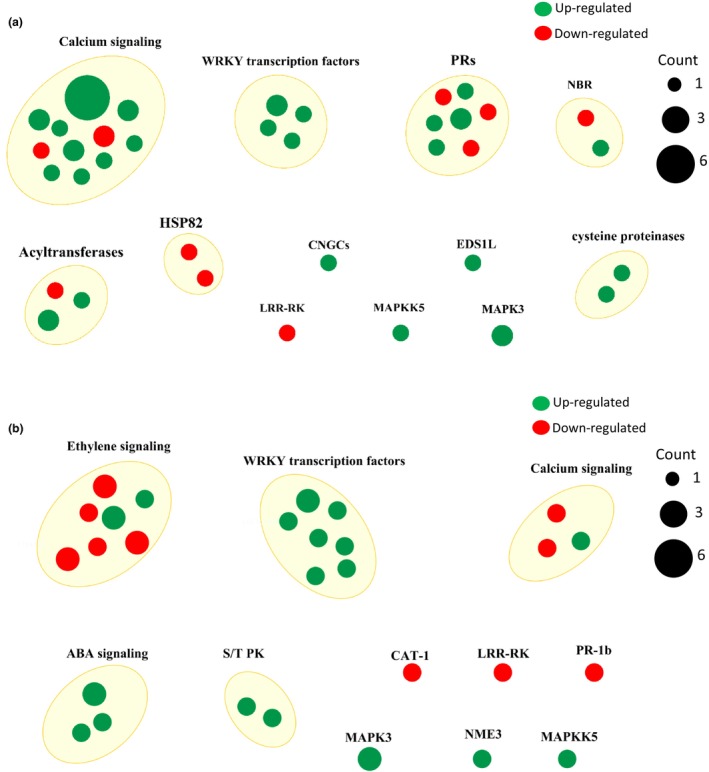
Pictorial representation of the nonredundant list of differentially expressed genes (DEGs) associated with the KEGG term (a) plant–pathogen interaction and (b) MAPK signalling in the dsG vs. Ctr comparison. DEGs were grouped according to their biological function. Dot size represents the number of DEGs annotated to a specific biological function.
Because treatment with dsGFP alone induces defence‐related gene expression, its interfering effect on PVX‐GFP accumulation (Figures 1 and 2) could be interpreted as independent of whether the dsRNA shares sequence homology with the virus. To address this, we monitored PVX accumulation in inoculated leaves of plants treated with dsGFP plus either the non‐targeted virus, wild‐type (WT) potato virus X (PVX), or targeted virus, PVX‐GFP, by RT‐qPCR and western blot analyses at 4 dpi. PVX‐GFP accumulated substantially less (4.4‐fold reduction) in the inoculated leaves of dsGFP‐treated plants than in control, whereas a slight but significant reduction (1.8‐fold) in WT PVX accumulation was detected in dsGFP‐treated leaves compared to the control (Figure S3). Thus, the highly effective control of PVX‐GFP accumulation observed in plants treated with dsGFP was probably due to sequence‐specific dsRNA‐triggered immunity.
2.5. Validation of RNA‐seq analysis
To independently validate the RNA‐seq results, differential expression of several upregulated genes classified in the plant–pathogen interaction and MAPK signalling pathways (MAPK3 [Niben101Scf02171g00008], WRKY transcription factor6 [WRKY6, Niben101Scf02430g03006], Calcium‐binding EF‐hand family protein [CaEF, Niben101Scf13289g00008], 1‐aminocyclopropane‐1‐carboxylate synthase2 [ACS2, Niben101Scf02334g00004] and the NBS‐LRR resistance gene RPS2 [Niben101Scf10333g00018]) was determined by RT‐qPCR using RNA preparations extracted from a new set of samples (not used for RNA‐seq) derived from dsG, dsG_VG, dsG_V and Ctr treatments (Figure 7). These candidate genes were selected for their predicted biological functions associated with pathogen interaction, potentially contributing to the sequence‐specific RNAi and nonspecific PTI responses. In general, the gene expression levels measured by RT‐qPCR in the different treatments confirmed the pattern observed by RNA‐seq analyses. While fold‐change patterns correlated, discrepancies in magnitude between the RT‐qPCR and RNA‐seq platforms is not uncommon and could be attributed to differences in the normalization methods used. The relative accumulation of the mRNAs was greater in dsG_VG compared to both dsG_V and dsG treatments. In addition, samples treated with dsG expressed significantly higher levels of ACS2, MAPK3 and CaEF mRNAs compared to controls.
FIGURE 7.
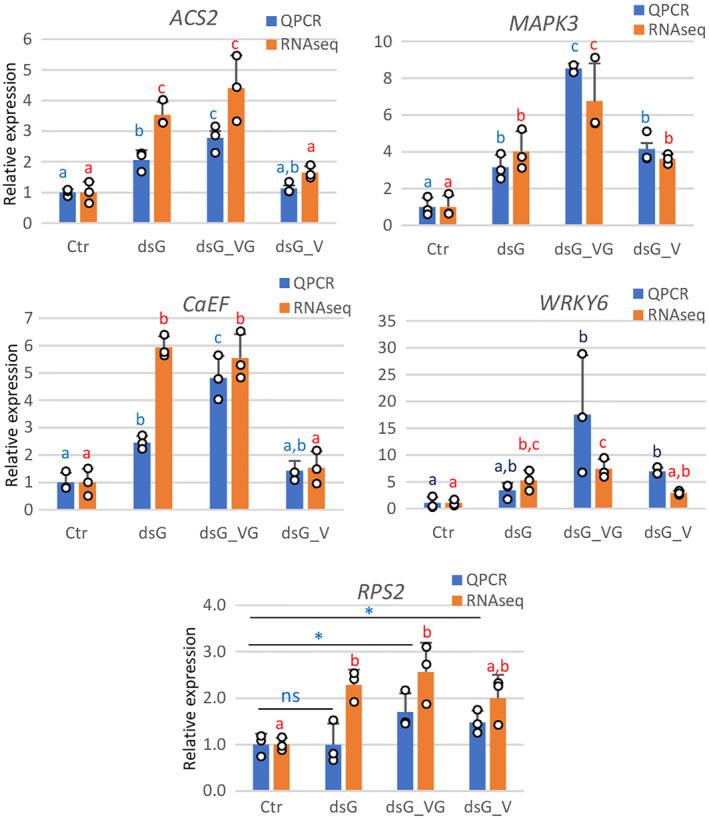
Validation of RNA‐seq data of representative genes from the KEGG terms plant–pathogen interaction and MAPK signalling by reverse transcription‐quantitative PCR (RT‐qPCR). The relative expression levels of Mitogen‐activated protein kinase3 (MAPK3), WRKY transcription factor6 (WRKY6), Calcium‐binding EF‐hand family protein (CaEF), 1‐aminocyclopropane‐1‐carboxylate synthase2 (ACS2) and the NBS‐LRR resistance gene RPS2 was determined by RT‐qPCR using RNA preparations extracted from dsG, dsG_VG, dsG_V and Ctr treatments. The relative expression levels of selected genes based on the number of fragments per kilobase of transcript sequence per millions of base‐pairs sequenced (FPKM) in the three independent samples per treatment used for RNA‐seq are shown for comparison. Different letters indicate significant differences determined by employing Scheffé's multiple range test for between‐group comparisons (p < 0.05). Asterisks indicate significant differences between treatments (Student's t test, p < 0.05); ns, not significant.
3. DISCUSSION
The main goal of this study was to compare the differential efficiency of the antiviral activity triggered by externally delivered, sequence‐specific and nonspecific dsRNAs in N. benthamiana. RNAi mediated by sequence‐specific dsRNA is the main antiviral defence mechanism in plants, but nonspecific dsRNA‐triggered responses have been documented to play a role in antiviral defence (Huang et al., 2023; Niehl et al., 2016). Using PVX‐GFP as a model, we have determined that co‐inoculation with either sequence‐specific or nonspecific dsRNA reduced virus accumulation in both inoculated and systemic leaves, although at different extents (Figures 1 and 2). While the administration of dsRNA specific for the targeted virus induced a potent RNAi‐based antiviral response that resulted in highly effective control of viral disease, the degree of interference with PVX‐GFP infection afforded by nonspecific dsRNA (PTI) was limited; viral titres in the leaves treated with nonspecific dsRNA were several‐fold higher than in the leaves treated with sequence‐specific dsRNA, and the plants became susceptible to systemic infection. Thus, our data point out that nonspecific dsRNA is a poor inducer of antiviral immunity compared to sequence‐specific dsRNA capable of triggering the RNAi response. The coexistence of non‐sequence‐specific immunity and sequence‐specific RNAi as two distinct antiviral mechanisms induced by dsRNA has also been reported in invertebrates, albeit nonspecific dsRNA was shown to evoke an antiviral response much lower in potency than that induced by sequence‐specific dsRNA (Robalino et al., 2005; Wang & He, 2019).
It has been reported that nonspecific dsRNA‐induced immunity against TMV mainly restricted the cell‐to‐cell movement of the virus by callose deposition at PD, but did not affect TMV replication (Huang et al., 2023). Deposition of callose at PD regulates symplastic transport, limiting the ability of the invading pathogen to spread throughout the plant (Chowdhury et al., 2020; Wang, Li, et al., 2021). We show here that cell‐to‐cell movement of PVX‐GFP was not significantly inhibited by nonspecific dsRNA, as judged by the unaltered size of PVX‐GFP infection sites in dsPVY‐treated leaves. Remarkably, studies with a frameshift mutant in the P25 movement protein of PVX indicated that nonspecific dsRNA caused a significant reduction in viral replication (Figure 3). It has been previously reported that PVX mutants defective in the P25 protein do not move out of the initially infected cell, although they do accumulate at WT levels in individual cells (Angell et al., 1996; Bayne et al., 2005). Overall, our data support a model where interference with PVX‐GFP accumulation triggered by nonspecific dsRNA operates at the single‐cell level, and lead to reduce accumulation of the virus in the inoculated leaves. Thus, effects of PTI triggered by nonspecific dsRNA on virus infection may vary with the specific virus–host combination (Huang et al., 2023; this study). Additionally, it cannot be excluded that proteins encoded by PVX‐GFP act as effectors to suppress dsRNA‐induced immunity based on restriction of viral cell‐to‐cell movement, as has been reported previously for other viral proteins (Huang et al., 2023; Nicaise & Candresse, 2017). The bacterial PTI elicitor derived from flagellin, flg22, is known to trigger deposition of callose at PD and restricts TMV cell‐to cell movement in N. benthamiana (Huang et al., 2023). Our results showed that PD callose deposition elicited by flg22 was correlated with a diminished accumulation of PVX‐GFP in the inoculated leaves but did not affect virus replication, suggesting that different PTI elicitors, that is, nonspecific dsRNA and flg22, may restrict PVX‐GFP at distinct steps of virus infection. In this sense, unlike for nonspecific dsRNA, PTI triggered by flg22 has been shown to induce the production of ROS species in N. benthamiana and Arabidopsis thaliana (Huang et al., 2023). However, it was not the intention of this study to compare the amplitude of the responses elicited by flg22 and nonspecific dsRNA, among other reasons because concentrations of the elicitors were quite different.
Previous findings showed that PVX infection was not affected by nonspecific dsRNA despite the induction of PTI‐like responses (Samarskaya et al., 2022). However, careful examination of the experimental conditions revealed differences between the work presented here, which examined the effect of co‐inoculation of dsRNA on the accumulation of the virus in the inoculated leaves, and the other study, where potato plants were challenged with PVX 24 h after dsRNA application and accumulation of PVX RNA was examined in systemically infected leaves. Our observation that accumulation of PVX‐GFP in systemically infected leaves of symptomatic plants was only slightly diminished by nonspecific dsRNAs is consistent with this discrepancy. In a broader sense, our findings could explain other examples in the literature where dsRNA did not confer systemic antiviral protection against non‐targeted viruses (Nilon et al., 2021; Rego‐Machado et al., 2020).
Before this study, no attempt had been reported to examine the whole transcriptomic response to dsRNA either alone or in the context of infections with viruses harbouring sequences targeted or not by dsRNA to shed light on differences between these responses. It is noteworthy that the experimental approach implemented in this work, with samples taken at an early stage of local infection, allowed us to investigate the outputs associated with early events in dsRNA signalling in the absence of the robust transcriptomic response triggered by PVX at later stages of infection (Garcia‐Marcos et al., 2009). PCA revealed that treatment with dsRNA alone had an ample effect on host gene expression, which could be responsible in part for the antiviral responses triggered by dsRNA (Figure 4). KEGG analysis showed a significant enrichment of terms related to plant–pathogen signalling pathways (KEGG terms plant–pathogen interaction and MAPK signalling) in all the three treatments that included dsRNA (Figure 5). Our results further indicated that the transcriptomic response triggered by dsRNA alone included canonical immune pathways or genes known to be involved in defence responses, that is, Ca+2 signalling, ET signalling, MAPK signalling, WRKY transcription factors, PR‐associated transcriptional factors, NBS‐LRR resistance genes, EDS1, and LRR receptor‐like kinases, many of which are typical of antimicrobial PTI (Figure 6) (Li et al., 2020; Niehl et al., 2016; Pruitt, Locci, et al., 2021; Yuan et al., 2021). Moreover, the transcriptomic response to the targeted virus/dsRNA combination (dsGFP plus PVX‐GFP) had a greater over‐representation of genes involved in plant–pathogen signalling pathways than the non‐targeted virus/dsRNA combination (dsGFP plus WT PVX), highlighting qualitative differences between sequence‐specific RNAi and nonspecific PTI immune responses (Table S2). These findings are in accordance with the differential efficiency in antiviral immunity triggered by sequence‐specific vs. nonspecific dsRNA. Moreover, this observation is somehow reminiscent of the intimate relationships between PTI and effector‐triggered immunity (ETI) (Pruitt, Gust, & Nurnberger, 2021; Yuan et al., 2021). Although these two types of immune pathways involve different activation circuits, several downstream outputs (Ca2+ flux, ROS burst, MAPK cascades, transcriptional reprograming and phytohormone signalling) usually converge, albeit with differences in specificity, amplitude and duration.
It is worth mentioning that Ca2+ signalling has been found to be involved in antiviral immunity against TMV triggered by nonspecific dsRNA (Huang et al., 2023). Furthermore, it has been reported that a wound‐induced Ca2+ signalling cascade stabilizes mRNAs encoding key components of RNAi machinery, notably AGO1/2, DCL1 and RNA‐dependent RNA polymerase 6, enhancing plant defence against virus infection (Wang, Gong, et al., 2021). Thus, it is tempting to speculate that the strong Ca2+ signalling evoked by dsRNA in this study activated RNAi‐related gene expression and resulted in highly effective control of viral disease when the dsRNA shares sequence homology with the cognate virus. This was somewhat corroborated in our transcriptomic analysis, where a significant induction of AGO2 and DCL2 mRNAs was detected by RNA‐seq analysis (Table S1).
The altered expression of genes classified in the KEGG term plant hormone signal transduction detected by RNA‐seq analysis suggested an important contribution of hormone signalling in the nonspecific PTI response to virus infection. There are precedents arguing for a role of the phytohormones SA and JA in PTI‐based defences induced by exogenous application of bacterial RNA derived from P. syringae in Arabidopsis (Lee et al., 2016). In addition, SA was required for both local and systemic resistance induced by bacterial PAMPs like flg22 and lipopolysaccharides against P. syringae (Mishina & Zeier, 2007). Besides SA‐responsive genes, we identified several DEGs involved in other hormone signalling pathways in the dataset from plants treated with the non‐targeted virus/dsRNA combination (dsGFP plus WT PVX), suggesting that other phytohormones might interact with SA in the PTI response to nonspecific dsRNA (Table S3). In this scenario, ET enhanced SA‐responsive PR‐1 expression in Arabidopsis, and in Nicotiana it was crucial for the onset of systemic acquired resistance (De Vos et al., 2006; Verberne et al., 2003).
In summary, we show here that although PTI‐based defences triggered by nonspecific dsRNA reduced virus accumulation in both local and systemic tissues, antiviral immunity conferred by nonspecific dsRNA was weaker compared to the RNAi response triggered by sequence‐specific dsRNA. Such differential efficiency in virus reduction was correlated with a deeper impact on defence responses in plants treated with the targeted virus/dsRNA combination compared to those elicited by the non‐targeted virus/dsRNA combination. However, given that comparisons were made between virus variants, that is, PVX‐GFP and WT PVX, we cannot discount that some of the differences observed on defence responses were derived from viral particularities. Furthermore, we demonstrated that unlike other examples of nonspecific dsRNA‐based PTI, which restricted virus cell‐to cell movement, PTI induced by nonspecific dsRNA partially inhibited PVX‐GFP accumulation at the single‐cell level.
4. EXPERIMENTAL PROCEDURES
4.1. Plasmid constructs
The complete CP coding sequence and flanking regions of PVY (902 bp) was cloned into L4440 as described (Necira et al., 2021). L4440 is a plasmid vector that has two convergent T7 promoters flanking the multiple cloning sites (Timmons et al., 2001). The complete GFP coding sequence (717 bp) was amplified by PCR using pSLJ‐GFP (Johansen & Carrington, 2001) and cloned into the SacI and PstI sites of L4440. The upstream primer was 5′‐GAGCTCATGGCAAGTAAAGGAGAAGAAC‐3′ (italicized sequence corresponds to the SacI restriction site). The downstream primer was 5′‐CTGCAGTTTGTATAGTTCATCCATGCCAT‐3′ (italicized sequence corresponds to the PstI restriction site). Plasmids were transformed into Escherichia coli HT115(DE3) using standard CaCl2 transformation protocols. HT115(DE3) is an RNAase III‐deficient E. coli strain that was modified to express T7 RNA polymerase from an isopropyl β‐D‐1‐thiogalactopyranoside (IPTG)‐inducible promoter (Timmons et al., 2001).
The binary vector pGR107 expressing the infectious cDNA of PVX has been previously described (Lu et al., 2003). The PVX‐GFP binary vector and its derivative PVXΔMP‐GFP were previously described (Aguilar et al., 2015; Schwach et al., 2005).
4.2. dsRNA production
Single colonies of HT115(DE3) containing the L4440 plasmid derivatives were grown at 37°C for 16 h in Luria‐Bertani (LB) broth with ampicillin and tetracycline at a final concentration of 500 and 12.5 μg/mL, respectively. The culture was diluted 75‐fold in the same medium and allowed to grow to OD595 = 0.5. T7 RNA polymerase was induced by the addition of 10 μM IPTG, and the culture was incubated further with shaking for 2 h at 37°C. After that, the culture broths were centrifuged (2468 g, 15 min) to collect the cells, and bacterial pellets were resuspended in 1 M ammonium acetate (Tenllado et al., 2003). Total nucleic acid was extracted after a phenol‐chloroform step prior to ethanol precipitation. Concentration of total nucleic acid was adjusted with distilled water to 0.5 μg/μL. The nucleic acids prepared using non‐induced cultures of HT115(DE3) containing the L4440 derivatives were used as negative control in all the experiments. The concentration of dsRNA in different preparations was estimated to be approximately 60 ng/μL, as judged by comparison with dsDNA markers (NZYDNA Ladder III) with each band corresponding to a precise quantity of dsDNA.
4.3. Virus inoculation and topical application of dsRNA
In order to ensure the uniformity of the viral inocula in all the experiments, inoculum stocks were prepared from local N. benthamiana leaves agro‐infiltrated with either WT PVX, PVX‐GFP or PVXΔMP‐GFP. For this, agroinfiltrated leaf tissue was cut in small slices (1 cm2), homogenized and collected into 500 mg aliquots that were stored at −80°C until use. Inoculation with WT PVX or PVX‐GFP on 3‐ to 4‐week‐old plantlets was performed by grinding each aliquot in sodium phosphate buffer (0.02 M, pH 7) at 1:5 (wt/vol). Inoculation with PVXΔMP‐GFP was performed by grinding agroinfiltrated leaf tissue at 1:2 (wt/vol) to increase the number of infection foci. A 20 μL‐dose of infected sap combined with an equal volume of dsRNA extract (1:1 vol/vol sap:dsRNA; 1.2 μg dsRNA) was applied to two leaves of each plant previously dusted with carborundum as abrasive (Carlo Erba). Plants were kept in environment‐controlled growth chambers with 16/8 h day/night photoperiod, about 2500 lux of daylight intensity and 60% relative humidity. The number of PVX‐GFP‐derived foci was assessed with a Black Ray long‐wave UV lamp (UVP). For GFP foci measurements, six leaves from three plants for each treatment were scanned with image‐analysis software ImageJ (http://rsbweb.nih.gov/ij/).
4.4. RNA and protein gel blot analysis
To minimize the effects of interleaf variability, whole leaf tissue from all plants corresponding to the same treatment type were pooled. Total RNA was extracted from inoculated leaves at 4 and 7 dpi and from upper leaves 7 dpi as described (García‐Marcos et al., 2013). RNA samples were separated on 1% agarose formaldehyde gels and transferred to Hybond‐N membranes (Roche Molecular Biochemicals). Membrane hybridization was carried out overnight at 65°C using digoxigenin‐labelled riboprobes corresponding to PVX CP sequences.
Total proteins were extracted by grinding leaf disks as described (Tena‐Fernández et al., 2013). Samples were boiled and fractionated in 15% SDS‐PAGE gels. PVX CP was detected with a commercial rabbit antibody (1:300 dilution) (070375/500; Loewe Biochemica GmbH) using an appropriate secondary antibody conjugated with alkaline phosphatase (Sigma‐Aldrich). Detection was performed using BCIP/NBT substrate solutions (Duchefa).
4.5. RT‐qPCR analysis
RT‐qPCR for the analysis of gene expression was performed with gene‐specific primers (Table S3). The relative quantification of PCR products was calculated by the comparative cycle threshold (ΔΔC t) method as described (García‐Marcos et al., 2013). Virus detection was performed using primers that amplify a region from nucleotides 2621 to 2753 of the PVX sequence. Amplification of 18S rRNA was chosen for normalization because of its similar level of expression across all treatments. All RT‐qPCR experiments were performed in triplicate.
4.6. Callose staining
Callose staining was carried out as described (Huang et al., 2022). Briefly, individual leaf disks were soaked with 0.1% aniline blue solution (in 50 mM potassium phosphate buffer, pH 8.0) containing either water, 1 μM flg22 (MedChemExpress), 30 ng/μL dsPVY or control extracts. Aniline blue fluorescence was imaged 30 min after dsPVY/flg22 or control treatment using a TCS SP8 STED 3X confocal microscope (Leica) with Application Suite X software (Leica) and using a 405 nm diode laser for excitation and filtering the emission at 430–490 nm. Eight‐bit images were acquired with a HC PL APO 40×/1.30 oil CS2 objective. Callose fluorescence intensity was quantified with ImageJ software using the plug‐in calloseQuant (Huang et al., 2022). Callose spots were measured in five or six images taken from three leaf discs per plant from two different plants for each treatment.
4.7. Library preparation for transcriptome sequencing
Three independent biological replicates were used to monitor differences in gene expression between treatments, each replicate consisting of a pool of 10 treated leaves. The integrity and quality of the total RNA were checked using NanoDrop 2000 spectrophotometer (Thermo Scientific). The transcriptome libraries, sequencing and bioinformatics analysis were performed at Novogene, UK; mRNA was purified from total RNA using poly‐T oligo‐attached magnetic beads. After fragmentation, the first‐strand cDNA was synthesized using random hexamer primers, followed by the second‐strand cDNA synthesis. The library was checked with Qubit and real‐time PCR (Rotor‐Gene Q thermal cycler; Qiagen) for quantification and bioanalyser for size distribution detection. Quantified libraries were pooled and sequenced on Illumina NovaSeq PE150 platform according to effective library concentration and data amount. The clustering of the index‐coded samples was performed according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced and paired‐end reads were generated.
Raw reads were firstly processed through in‐house Perl scripts. In this step, clean reads were obtained by removing reads containing adapter, reads containing poly‐N and low‐quality reads from raw data. At the same time, Q30 and GC content of the clean data were calculated (Table S1). All the downstream analyses were based on the clean data with high quality.
The N. benthamiana reference genome and gene model annotation files were downloaded from the SGN ftp site (https://solgenomics.net/ftp/genomes/Nicotiana_benthamiana/assemblies/) directly. An index of the reference genome was built using Hisat2 v. 2.0.5 and paired‐end clean reads were aligned to the reference genome using Hisat2 v. 2.0.5. FeatureCounts v. 1.5.0‐p3 was used to count the reads numbers mapped to each gene. The expected number of fragments per kilobase of transcript sequence per millions of base‐pairs sequenced (FPKM) of each gene was calculated based on the length of the gene and reads count mapped to this gene.
4.8. KEGG pathway enrichment analysis of differentially expressed genes
Differential expression analysis of the two assayed treatments (three replicates per treatment) was performed using the DESeq2 R package (v. 1.20.0) (Anders & Huber, 2010). The resulting p‐values were adjusted using the Benjamini and Hochberg's correction for controlling the false discovery rate. Genes with a log2(fold‐change) ≥|1| and adjusted p‐value ≤0.05 were assigned as differentially expressed. Fold‐change calculations were performed for paired‐comparisons made between treatments.
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a collection of manually curated databases containing resources on genomic, biological‐pathway and disease information (Kanehisa & Goto, 2000). KEGG pathway enrichment analysis of DEGs was implemented by the clusterProfiler R package (v. 3.8.1), in which gene length bias was corrected. KEGG pathways with adjusted p value less than 0.05 were considered significantly enriched. Gene ontology (GO) enrichment analysis rendered highly generic GO terms and was not used for subsequent analyses.
4.9. Statistical analysis
All statistical analyses were performed using the statistical software SPSS Statistics v. 25 (IBM Corp.). For each experiment, samples were assessed for normality via the Shapiro–Wilk test and for equality of variances using Levene's test. For experiments with normally distributed samples of equal variance, one‐way analysis of variance (ANOVA) followed by Scheffé's post hoc test was performed. Otherwise, a nonparametric Mann–Whitney U test was employed, with the Bonferroni correction for multiple comparisons between samples applied. For comparisons between pairs of means (pairwise comparisons), Student's t tests were employed.
CONFLICT OF INTEREST STATEMENT
Authors have no conflict of interest to declare.
Supporting information
Figure S1. Unaltered size of PVX‐GFP infection sites in dsPVY‐treated leaves. Plants were inoculated with mixtures of PVX‐GFP and either flagellin (flg22), dsPVY or control (Ctr) extracts. (A) Representative inoculated leaves were examined under UV light at 4 days post‐inoculation (dpi). Scale bar denotes 1 cm. (B) Mean numbers ± SD of infection foci on inoculated leaves of six plants (two leaves per plant) with the different treatments at 7 dpi. (C) Mean sizes ± SD of infection foci on inoculated leaves of six plants with the different treatments. Different letters indicate significant differences determined by employing Scheffé’s multiple range test (p < 0.05). Experiments were repeated once more with similar results.
Figure S2. Venn diagrams displaying the number of differentially expressed genes with a log2(fold‐change) ≥|1| and adjusted p‐value ≤0.05 in the Ctr vs. dsG, dsG_VG vs. dsG and dsG_V vs. dsG pairwise comparisons.
Figure S3. GFP dsRNA interferes differently with PVX‐GFP and wild‐type (WT) PVX infection. Nicotiana benthamiana plants were inoculated with mixtures of dsGFP extracts and either PVX‐GFP or wild‐type (WT) PVX and with mixtures of control (Ctr) extracts plus either PVX‐GFP or WT PVX. Reverse transcription‐quantitative PCR was used to analyse the accumulation of PVX‐GFP (A) and WT PVX (B) genomic RNA levels in the inoculated leaves at 4 days post‐inoculation (dpi). Expression of the 18S rRNA gene served as a control. Data represent the means ± SD of three replicates, each consisting of a pool of four plants that received the same treatment. Asterisks indicate significant differences between treatments (Student’s t test, p < 0.05). (C) Western blot analysis of plant extracts derived from inoculated leaves at 4 dpi, using antibodies against PVX coat protein (CP). The panel below the blot is the membrane stained with Ponceau S as control of loading. M, mock‐inoculated leaf.
Table S1. Differentially expressed Nicotiana benthamiana genes performed for paired‐comparisons made between treatments.
Table S2. KEGG analysis of differentially expressed Nicotiana benthamiana genes performed for paired‐comparisons made between treatments.
Table S3. Differentially expressed Nicotiana benthamiana genes classified in MAPK signalling, plant–pathogen interaction and plant hormone signal transduction in the different comparisons.
ACKNOWLEDGEMENTS
This work is dedicated to the memory of mentor and friend, José Ramón Díaz‐Ruiz Alba. The research was funded by grants PID2019‐109304RB‐I00 and PID2022‐137691OB‐I00 from the Spanish Ministry of Science and Innovation (MCIN/AEI/10.13039/501100011033), and by “ERDF A way of making Europe”. Khouloud Necira was funded by the Ministry of Higher Education and Scientific Research of Tunisia, grant numbers 2019‐BALT‐1077 and 2019‐BALT‐1079. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Necira, K. , Contreras, L. , Kamargiakis, E. , Kamoun, M.S. , Canto, T. & Tenllado, F. (2024) Comparative analysis of RNA interference and pattern‐triggered immunity induced by dsRNA reveals different efficiencies in the antiviral response to potato virus X. Molecular Plant Pathology, 25, e70008. Available from: 10.1111/mpp.70008
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/ under accession number GSE253964.
REFERENCES
- Aguilar, E. , Almendral, D. , Allende, L. , Pacheco, R. , Chung, B.N. , Canto, T. et al. (2015) The P25 protein of potato virus X (PVX) is the main pathogenicity determinant responsible for systemic necrosis in PVX‐associated synergisms. Journal of Virology, 89, 2090–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. & Huber, W. (2010) Differential expression analysis for sequence count data. Genome Biology, 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angell, S.M. , Davies, C. & Baulcombe, D.C. (1996) Cell‐to‐cell movement of potato virus X is associated with a change in the size exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii . Virology, 215, 197–201. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Bayne, E.H. , Rakitina, D.V. , Morozov, S.Y. & Baulcombe, D.C. (2005) Cell‐to‐cell movement of potato potexvirus X is dependent on suppression of RNA silencing. The Plant Journal, 44, 471–482. [DOI] [PubMed] [Google Scholar]
- Boiteux, L.S. , de Noronha Fonseca, M.E. , Vieira, J.V. & de Cássia Pereira‐Carvalho, R. (2012) Breeding for resistance to viral diseases. In: Fritsche‐Neto, R. & Borém, A. (Eds.) Plant breeding for biotic stress resistance. Berlin/Heidelberg: Springer, pp. 57–79. [Google Scholar]
- Carbonell, A. & Carrington, J.C. (2015) Antiviral roles of plant ARGONAUTES. Current Opinion in Plant Biology, 27, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnon, M. , Kreutzweiser, D. , Mitchell, E.A. , Morrissey, C.A. , Noome, D.A. & Van der Sluijs, J.P. (2015) Risks of large‐scale use of systemic insecticides to ecosystem functioning and services. Environmental Science and Pollution Research, 22, 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, R.N. , Lasky, D. , Karki, H. , Zhang, Z. , Goyer, A. , Halterman, D. et al. (2020) HCPro suppression of callose deposition contributes to strain‐specific resistance against potato virus Y. Phytopathology, 110, 164–173. [DOI] [PubMed] [Google Scholar]
- D'Ario, M. , Griffiths‐Jones, S. & Kim, M. (2017) Small RNAs: big impact on plant development. Trends in Plant Science, 22, 1056–1068. [DOI] [PubMed] [Google Scholar]
- De Vos, M. , Van Zaanen, W. , Koornneef, A. , Korzelius, J.P. , Dicke, M. , Van Loon, L.C. et al. (2006) Herbivore‐induced resistance against microbial pathogens in Arabidopsis . Plant Physiology, 142, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W. & Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Marcos, A. , Pacheco, R. , Manzano, A. , Aguilar, E. & Tenllado, F. (2013) Oxylipin biosynthesis genes positively regulate programmed cell death during compatible infections with the synergistic pair potato virus X‐potato virus Y and tomato spotted wilt virus. Journal of Virology, 87, 5769–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Marcos, A. , Pacheco, R. , Martiáñez, J. , González‐Jara, P. , Díaz‐Ruíz, J.R. & Tenllado, F. (2009) Transcriptional changes and oxidative stress associated with the synergistic interaction between Potato virus X and Potato virus Y and their relationship with symptom expression. Molecular Plant–Microbe Interactions, 22, 1431–1444. [DOI] [PubMed] [Google Scholar]
- Guo, Z. , Li, Y. & Ding, S.W. (2019) Small RNA‐based antimicrobial immunity. Nature Reviews Immunology, 19, 31–44. [DOI] [PubMed] [Google Scholar]
- Hoang, B.T.L. , Fletcher, S.J. , Brosnan, C.A. , Ghodke, A.B. , Manzie, N. & Mitter, N. (2022) RNAi as a foliar spray: efficiency and challenges to field applications. International Journal of Molecular Sciences, 23, 6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Mutterer, J. & Heinlein, M. (2022) In vivo aniline blue staining and semi‐automated quantification of callose deposition at plasmodesmata. Methods in Molecular Biology, 2457, 151–165. [DOI] [PubMed] [Google Scholar]
- Huang, C. , Sede, A.R. , Elvira‐González, L. , Yan, Y. , Rodriguez, M. , Mutterer, J. et al. (2023) dsRNA‐induced immunity targets plasmodesmata and is suppressed by viral movement proteins. The Plant Cell, 35, 3845–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, L.K. & Carrington, J.C. (2001) Silencing on the spot: induction and suppression of RNA silencing in the agrobacterium‐mediated transient expression system. Plant Physiology, 126, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. & Goto, S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, J. , Wei, M. , Li, G. , Lei, R. , Qiu, Y. , Wang, C. et al. (2018) The cucumber mosaic virus movement protein suppresses PAMP‐triggered immune responses in Arabidopsis and tobacco. Biochemical and Biophysical Research Communications, 498, 395–401. [DOI] [PubMed] [Google Scholar]
- Kørner, C.J. , Klauser, D. , Niehl, A. , Domínguez‐Ferreras, A. , Chinchilla, D. , Boller, T. et al. (2013) The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Molecular Plant–Microbe Interactions, 26, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Lee, B. , Park, Y.S. , Lee, S. , Song, G.C. & Ryu, C.M. (2016) Bacterial RNAs activate innate immunity in Arabidopsis . New Phytologist, 209, 785–797. [DOI] [PubMed] [Google Scholar]
- Li, P. , Lu, Y.J. , Chen, H. & Day, B. (2020) The lifecycle of the plant immune system. CRC Critical Reviews in Plant Sciences, 39, 72–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Malcuit, I. , Moffett, P. , Ruiz, M.T. , Peart, J. , Wu, A.J. et al. (2003) High throughput virus‐induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO Journal, 22, 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina, T.E. & Zeier, J. (2007) Pathogen‐associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis . The Plant Journal, 50, 500–513. [DOI] [PubMed] [Google Scholar]
- Necira, K. , Makki, M. , Sanz‐García, E. , Canto, T. , Djilani‐Khouadja, F. & Tenllado, F. (2021) Topical application of Escherichia coli‐encapsulated dsRNA induces resistance in Nicotiana benthamiana to potato viruses and involves RDR6 and combined activities of DCL2 and DCL4. Plants, 10, 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou, B.P.M. , Ding, P. & Jones, J.D.G. (2022) Thirty years of resistance: zig‐zag through the plant immune system. The Plant Cell, 34, 1447–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise, V. & Candresse, T. (2017) Plum pox virus capsid protein suppresses plant pathogen‐associated molecular pattern (PAMP)‐triggered immunity. Molecular Plant Pathology, 18, 878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehl, A. , Wyrsch, I. , Boller, T. & Heinlein, M. (2016) Double‐stranded RNAs induce a pattern‐triggered immune signaling pathway in plants. New Phytologist, 211, 1008–1019. [DOI] [PubMed] [Google Scholar]
- Nilon, A. , Robinson, K. , Pappu, H.R. & Mitter, N. (2021) Current status and potential of RNA interference for the management of tomato spotted wilt virus and thrips vectors. Pathogens, 10, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Van‐der‐Does, D. , Zamioudis, C. , Leon‐Reyes, A. & Van‐Wees, S.C.M. (2012) Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology, 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Pruitt, R.N. , Gust, A.A. & Nurnberger, T. (2021) Plant immunity unified. Nature Plants, 7, 382–383. [DOI] [PubMed] [Google Scholar]
- Pruitt, R.N. , Locci, F. , Wanke, F. , Zhang, L. , Saile, S.C. , Joe, A. et al. (2021) The EDS1‐PAD4‐ADR1 node mediates Arabidopsis pattern‐triggered immunity. Nature, 598, 495–499. [DOI] [PubMed] [Google Scholar]
- Rego‐Machado, C.M. , Nakasu, E.Y.T. , Silva, J.M.F. , Natalia, L. , Tatsuya, N. & Alice, K.I.N. (2020) siRNA biogenesis and advances in topically applied dsRNA for controlling virus infections in tomato plants. Scientific Reports, 10, 22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robalino, J. , Bartlett, T. , Shepard, E. , Prior, S. , Jaramillo, G. , Scura, E. et al. (2005) Double‐stranded RNA induces sequence‐specific antiviral silencing in addition to nonspecific immunity in a marine shrimp: convergence of RNA interference and innate immunity in the invertebrate antiviral response? Journal of Virology, 79, 13561–13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarskaya, V.O. , Spechenkova, N. , Markin, N. , Suprunova, T.P. , Zavriev, S.K. , Love, A.J. et al. (2022) Impact of exogenous application of Potato virus Y‐specific dsRNA on RNA interference, pattern‐triggered immunity and poly(ADP‐ribose) metabolism. International Journal of Molecular Sciences, 23, 7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary, S. , Willocquet, L. , Pethybridge, S.J. , Esker, P. , McRoberts, N. & Nelson, A. (2019) The global burden of pathogens and pests on major food crops. Nature Ecology & Evolution, 3, 430–439. [DOI] [PubMed] [Google Scholar]
- Schwach, F. , Vaistij, F.E. , Jones, L. & Baulcombe, D.C. (2005) An RNA‐dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiology, 138, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soosaar, J.L.M. , Burch‐Smith, T.M. & Dinesh‐Kumar, S.P. (2005) Mechanisms of plant resistance to viruses. Nature Reviews Microbiology, 3, 789–798. [DOI] [PubMed] [Google Scholar]
- Tena‐Fernández, F. , González, I. , Doblas, P. , Rodríguez, C. , Sahana, N. , Kaur, H. et al. (2013) The influence of cis‐acting P1 protein and translational elements on the expression of Potato virus Y helper‐component proteinase (HCPro) in heterologous systems and its suppression of silencing activity. Molecular Plant Pathology, 14, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado, F. & Díaz‐Ruíz, J.R. (2001) Double‐stranded RNA‐mediated interference with plant virus infection. Journal of Virology, 75, 12288–12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado, F. , Martínez‐García, B. , Vargas, M. & Díaz‐Ruíz, J.R. (2003) Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnology, 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L. , Court, D.L. & Fire, A. (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans . Gene, 263, 103–112. [DOI] [PubMed] [Google Scholar]
- Verberne, M.C. , Hoekstra, J. , Bol, J.F. & Linthorst, H.J.M. (2003) Signaling of systemic acquired resistance in tobacco depends on ethylene perception. The Plant Journal, 35, 27–32. [DOI] [PubMed] [Google Scholar]
- Voloudakis, A.E. , Kaldis, A. & Patil, B.L. (2022) RNA‐based vaccination of plants for control of viruses. Annual Review of Virology, 9, 521–548. [DOI] [PubMed] [Google Scholar]
- Wang, P.H. & He, J.G. (2019) Nucleic acid sensing in invertebrate antiviral immunity. International Review of Cell and Molecular Biology, 345, 287–360. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Gong, Q. , Wu, Y. , Huang, F. , Ismayil, A. , Zhang, D. et al. (2021) A calmodulin‐binding transcription factor links calcium signaling to antiviral RNAi defense in plants. Cell, Host & Microbe, 29, 1393–1406. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Li, X. , Fan, B. , Zhu, C. & Chen, Z. (2021) Regulation and function of defense‐related callose deposition in plants. International Journal of Molecular Sciences, 22, 2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Gou, X. , He, K. , Xi, D. , Du, J. , Lin, H. et al. (2010) BAK1 and BKK1 in Arabidopsis thaliana confer reduced susceptibility to turnip crinkle virus. European Journal of Plant Pathology, 127, 149–156. [Google Scholar]
- Yuan, M. , Ngou, B.P.M. , Ding, P. & Xin, X.F. (2021) PTI–ETI crosstalk: an integrative view of plant immunity. Current Opinion in Plant Biology, 62, 102030. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Wu, Z. , Li, Y. & Wu, J. (2015) Biogenesis, function, and applications of virus‐derived small RNAs in plants. Frontiers in Microbiology, 6, 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvereva, A.S. , Golyaev, V. , Turco, S. , Gubaeva, E.G. , Rajeswaran, R. , Schepetilnikov, M.V. et al. (2016) Viral protein suppresses oxidative burst and salicylic acid‐dependent autophagy and facilitates bacterial growth on virus‐infected plants. New Phytologist, 211, 1020–1034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Unaltered size of PVX‐GFP infection sites in dsPVY‐treated leaves. Plants were inoculated with mixtures of PVX‐GFP and either flagellin (flg22), dsPVY or control (Ctr) extracts. (A) Representative inoculated leaves were examined under UV light at 4 days post‐inoculation (dpi). Scale bar denotes 1 cm. (B) Mean numbers ± SD of infection foci on inoculated leaves of six plants (two leaves per plant) with the different treatments at 7 dpi. (C) Mean sizes ± SD of infection foci on inoculated leaves of six plants with the different treatments. Different letters indicate significant differences determined by employing Scheffé’s multiple range test (p < 0.05). Experiments were repeated once more with similar results.
Figure S2. Venn diagrams displaying the number of differentially expressed genes with a log2(fold‐change) ≥|1| and adjusted p‐value ≤0.05 in the Ctr vs. dsG, dsG_VG vs. dsG and dsG_V vs. dsG pairwise comparisons.
Figure S3. GFP dsRNA interferes differently with PVX‐GFP and wild‐type (WT) PVX infection. Nicotiana benthamiana plants were inoculated with mixtures of dsGFP extracts and either PVX‐GFP or wild‐type (WT) PVX and with mixtures of control (Ctr) extracts plus either PVX‐GFP or WT PVX. Reverse transcription‐quantitative PCR was used to analyse the accumulation of PVX‐GFP (A) and WT PVX (B) genomic RNA levels in the inoculated leaves at 4 days post‐inoculation (dpi). Expression of the 18S rRNA gene served as a control. Data represent the means ± SD of three replicates, each consisting of a pool of four plants that received the same treatment. Asterisks indicate significant differences between treatments (Student’s t test, p < 0.05). (C) Western blot analysis of plant extracts derived from inoculated leaves at 4 dpi, using antibodies against PVX coat protein (CP). The panel below the blot is the membrane stained with Ponceau S as control of loading. M, mock‐inoculated leaf.
Table S1. Differentially expressed Nicotiana benthamiana genes performed for paired‐comparisons made between treatments.
Table S2. KEGG analysis of differentially expressed Nicotiana benthamiana genes performed for paired‐comparisons made between treatments.
Table S3. Differentially expressed Nicotiana benthamiana genes classified in MAPK signalling, plant–pathogen interaction and plant hormone signal transduction in the different comparisons.
Data Availability Statement
The data that support the findings of this study are openly available in Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/ under accession number GSE253964.


