Abstract
Black Creek Canal virus (BCCV) is a New World hantavirus which is associated with hantavirus pulmonary syndrome. We have examined the site of expression of the BCCV nucleocapsid protein (NBCCV) in the absence of BCCV glycoproteins and found that the majority of the protein is localized to the Golgi region. Immunofluorescence analysis of BHK21 cells expressing the NBCCV and La Crosse virus nucleocapsid protein (NLACV) showed different intracellular localization patterns of these proteins within the same cell: NLACV is cytoplasmic, whereas NBCCV is perinuclear. NBCCV was found to be colocalized with α-mannosidase II, a marker for the Golgi complex. Also, NBCCV was found to be associated with microsomal membranes following cell fractionation. Sedimentation analysis in density gradients revealed that the membrane association of NBCCV is sensitive to treatments with high-salt and high-pH solutions, which indicates that NBCCV is a peripheral membrane protein. Analysis of NBCCV truncation mutants revealed that the 141-amino-acid C-terminal portion of this protein was capable of targeting green fluorescent protein to the perinuclear region. The difference in the intracellular localization between the NBCCV and NLACV proteins suggests that the mechanisms involved in the morphogenesis of New World hantaviruses are distinct from that documented for other members of the Bunyaviridae family.
The members of the Bunyaviridae family are a diverse group of viruses that infect animals, plants, humans, and insects and are distributed worldwide (4, 9). The viruses share a similar genetic organization in which three RNA segments of negative sense encode three structural proteins: nucleocapsid (N); a glycoprotein precursor (GPC), which is processed into G1 and G2 proteins; and RNA-dependent RNA polymerase (L). In addition, some family members encode nonstructural proteins (4, 9). It has generally been accepted that maturation of all the members of this family, except for plant viruses and Rift Valley fever virus, occurs intracellularly by budding into the cisternae in the Golgi apparatus (1, 5, 11, 13). Along with the overall genetic organization and morphology of virions, this feature of virus assembly has been considered as a criterion for classification of these viruses (4, 10, 15).
The process of virus assembly for the family Bunyaviridae has been previously investigated by electron microscopy, immunofluorescence analysis (IFA), and studies of the expression of viral glycoproteins (1, 11, 14). The general conclusions about the assembly mechanism drawn from these studies are as follows. Once cleaved cotranslationally in the endoplasmic reticulum (ER), the glycoproteins G1 and G2 undergo glycosylation, folding, and heterodimerization in the Golgi complex, where they are retained and gradually accumulated. The nucleocapsid protein is expressed as a cytoplasmic protein. After its interaction with the viral RNA segments and subsequent assembly into the ribonucleoprotein (RNP), it is thought to be targeted to the Golgi complex via a specific recognition of the cytoplasmic portion of either G1 or G2. This specific interaction is thought to consequently trigger the budding of virions into the Golgi cisternae.
Recent studies with representatives of the Hantavirus genus, particularly with those designated as New World hantaviruses, challenge the idea that the intracellular mode of virus assembly is the only mechanism utilized by the Bunyaviridae (8, 17). Electron microscopy of Vero E6 cells infected with Sin Nombre virus, a hantavirus found in the southwestern United States, showed accumulation of the virus particles on the cell surface and their absence in the Golgi complex and other intracellular compartments (8). Similar findings were obtained in studies with Black Creek Canal virus (BCCV), another representative of the New World hantaviruses (16). Studies with polarized epithelial cells using electron microscopy and immunofluorescence have shown that BCCV assembly and release occur at the apical cell surface (16). In addition, we have shown that significant amounts of the BCCV nucleocapsid protein (NBCCV) are capable of interacting with actin filaments, and this interaction appears to be important for viral morphogenesis (17).
The hantavirus nucleocapsid protein, which is in the range of 428 to 433 amino acids, is larger than those found in most other members of the family by approximately 160 to 200 amino acids, except for nairoviruses, which also have an N protein of approximately the same length (4, 18, 20, 22). The functional implications of this observation in the cell biology of hantaviruses are unclear. In this study, we have investigated the intracellular localization of the NBCCV in the absence of the viral glycoproteins. Our data show that unlike the nucleocapsid proteins of other members of the Bunyaviridae, this protein demonstrates the unusual property of being expressed as a peripheral membrane-associated protein in the perinuclear region.
MATERIALS AND METHODS
Cell culture and antibodies
The BHK21 cell line was obtained from the American Type Culture Collection (ATCC). The cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin antibiotic solution (Sigma, St. Louis, Mo.). Maintenance of confluent cell cultures was carried out in medium containing 5% FBS.
Anti-green fluorescent protein (GFP) monoclonal antibody (MAb) was purchased from Invitrogen (Carlsbad, Calif.). The MAb GB04-BF07, which recognizes NBCCV, and anti-BCCV rabbit sera raised against all the BCCV structural proteins were generously provided by Sue Ruo and Thomas Ksiazek (Special Pathogens Branch, Centers for Disease Control and Prevention, Atlanta, Ga.). A rabbit serum against La Crosse virus (LACV) nucleocapsid protein (NLACV) was a gift from Ramaswamy Raju (Meharry Medical College, Nashville, Tenn.). A polyclonal rabbit antibody against α-mannosidase II was kindly provided by Marilyn G. Farquhar (University of California, San Diego, Calif.). Anti-myc MAb was purchased from Clontech (Palo Alto, Calif.).
Construction of recombinant plasmids.
The plasmid pcNBCCV was generated by inserting the NBCCV coding sequence, which was amplified by PCR with primers carrying adapters for XbaI and XhoI restrictases, into pcDNA3.1(−) digested with XbaI and XhoI. The source of the NBCCV gene was the plasmid pNBCCV/rep5 (17). The plasmid pNLAC-myc was constructed by excision of the VP22 coding sequence from the pVP22-mycHis vector (Invitrogen) with HindIII and KpnI and inserting the NLACV coding sequence, which was amplified from plasmid R-108/2 (obtained from Ramaswamy Raju, Meharry Medical College) with primers carrying HindIII and KpnI adapters. The NLACV in this plasmid lacks its own TGA stop codon and is fused with the c-Myc coding sequence. Therefore, it can be detected by both anti-NLACV and anti-Myc antibodies.
The plasmid pGFP/NBCCV was constructed by inserting the NBCCV coding sequence, which was amplified from pNBCCV/rep5 with primers tagged with XhoI and KpnI adapters, into the pEGFP-c3 vector (Clontech) digested with XhoI and KpnI restrictases. pEGFP is a mammalian expression vector designed for construction of GFP-chimeric fusion proteins. The NBCCV coding sequence was fused with the C-terminal portion of GFP in this construct.
The C-terminally truncated NBCCV mutants d119C, d246C, d348C, d401C, and d417C were assembled on the pcDNA3.1(−) vector by inserting PCR-amplified NBCCV fragments into a polylinker site in the vector. The positive chain primer for the truncated sequences was common and contained the first initiating ATG codon of NBCCV and an XbaI adapter upstream of it. The negative chain primers corresponded to the nucleotides 381 to 399, 761 to 780, 1064 to 1086, 1227 to 1245, and 1275 to 1293 of the S segment, followed by a TGA stop codon. Each primer contained an XhoI adapter at the 5′ end. The coding sequences of the amplified DNA fragments corresponded to amino acids 1 to 119, 1 to 246, 1 to 348, 1 to 401, and 1 to 417 of NBCCV. The N-terminally truncated mutants d67N-GFP, d219N-GFP, d287-GFP, d342N-GFP, and d400N-GFP were constructed in the pEGFP-c3 vector. The positive chain primers corresponded to nucleotides 243 to 261, 699 to 718, 903 to 921, 1068 to 1086, and 1242 to 1260 with an XhoI adapter placed upstream. The coding sequence of the amplified DNA fragments corresponded to amino acids 67 to 428, 219 to 428, 287 to 428, 342 to 428, and 400 to 428 of NBCCV. The negative chain primer was common for these truncated mutants, containing a TGA stop codon of NBCCV followed by a KpnI adapter. The sequences were amplified by PCR, digested with XhoI and KpnI, and placed into the pEGFP-c3 linker in frame with the C terminus of the GFP coding sequence.
The pcNPUUV and pcNSEOV plasmids were generated by inserting the N protein coding sequences of Puumala-Sotkamo virus (PUUV) (25) and Seoul-SR virus (SEOV) (2) into the pcDNA(−) vector, which was digested with XbaI and XhoI restrictases. The NPUUV and NSEOV coding sequences were amplified by reverse transcription PCR (RT-PCR) with primers carrying the initiating ATG and stop TGA codons and adapter sequences for the XbaI and XhoI restrictases. The RNA samples, which were used in the RT-PCRs, were prepared from lysates of Vero E6 cells infected with PUUV and SEOV (C. Spiropoulou, Centers for Disease Control and Prevention, Atlanta, Ga.) by using TriPure isolation reagent (Roche Molecular Biochemicals, Indianapolis, Ind.). The size and orientation of inserted DNA fragments were examined by restriction digestion analysis.
IFA.
BHK21 cells were seeded in 12-well plates on coverslips, and once they achieved 60% confluence they were transfected with plasmids with a Fugene 6 transfection kit (BMB, Indianapolis, Ind.). For visualization of the expressed GFP and the GFP-chimeric proteins, cells were washed twice with phosphate-buffered saline (PBS) and examined with an inverted fluorescence microscope. For indirect IFA, the cells were fixed in 3.7% paraformaldehyde for 30 min at room temperature and then permeabilized in 0.1% Triton X-100 for 5 min. To eliminate nonspecific binding, the cells were preincubated with PBS containing 3% bovine serum albumin for 15 min at room temperature in a humidity chamber. Antibody dilutions of 1:100 for anti-BCCV sera (1947), 1:500 for anti-N protein MAb, 1:50 for anti-GFP MAb (GB04-BF07), 1:50 for anti-NLACV, 1:50 for anti-α-mannosidase II polyclonal antibody, and 1:200 for anti-Myc MAb were used. The first round of incubations was carried out for 30 min at room temperature. After washing, the first antibody was detected with either anti-rabbit fluorescein isothiocyanate (FITC)-conjugated secondary antibody or anti-mouse tetramethyl rhodamine isothiocyanate (TRITC)-conjugated antibody. Coverslips were washed in PBS, mounted with Vectashield (Vector Laboratories, Burlingame, Calif.), and examined with a Nikon Optiphot microscope.
Preparation of microsomal membranes
BHK21 cells were grown in Ti 150 flasks until 60% confluent and transfected with a plasmid. At 24 h posttransfection, the cells were harvested and washed in 10 ml of cold PBS twice. The pellet was resuspended in 1 ml of PBS, and the cells were counted. Approximately 5 × 107 cells/ml were used per sample. The cells were transferred into an Eppendorf tube, pelleted, and resuspended in 200 μl of isosmotic medium (5 mM Tris-HCl [pH 7.4], 0.5 mM MgCl2, and protein inhibitor cocktail in a concentration recommended by the manufacturer [BMB]). The sample was incubated on ice for 10 min and then transferred to a Dounce homogenizer, and homogenization was carried out until at least 90% of the cells were visibly disrupted. To establish an isosmotic environment, the sample with homogenized cells was adjusted to 0.25 M sucrose by adding 22.2 μl of 2.5 M sucrose. The cell debris and nuclei were removed by centrifugation at low speed (2,000 × g, 10 min, 4°C) twice. The supernatant was collected and transferred into a 500-μl centrifuge tube (Beckman, Palo Alto, Calif.). The total volume in the tube was adjusted to 450 μl with mineral oil, and the sample was centrifuged at 32,500 rpm in an SW55 Ti rotor (Beckman) for 1 h. The supernatant represented the cytosolic fraction, while the pellet, which was dissolved in 200 μl of a combination of 10 mM Tris-HCl (pH 6.8), protease inhibitors, and 1% sodium dodecyl sulfate (SDS) overnight at 4°C, corresponded to the membrane fraction. Both samples were mixed with equal volumes of 2× Laemmli buffer (12), separated in a 10% polyacrylamide gel with SDS, and analyzed by Western blotting.
Sucrose gradient analysis.
Prior to analysis, NBCCV-microsomes were subjected to treatments with 1 M NaCl, 4 M urea, or 15 mM sodium carbonate (pH 11). Untreated NBCCV-microsomes were also examined. For each treatment, an aliquot of NBCCV-microsomes (∼20 μl) was constituted to the desired concentration in the presence of peptide inhibitors in a total volume of 50 μl. The treatment was carried out on ice for 45 min. Following the treatment, the protein samples were combined with 1.1 ml of 30% sucrose and layered on a 60%–45%–40% (wt/wt) sucrose gradient (24). Thirty percent sucrose (1.2 ml) with diluted protein samples was overlaid with 200 μl of PBS. The tubes were centrifuged in an SW55 Ti rotor (Beckman) at 86,000 × g for 18 h at 4°C. The gradient was fractionated from the bottom into nine fractions. Each fraction (∼450 μl) was precipitated with either ethanol or acetone. The proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and examined by Western blotting with anti-N MAb.
RESULTS
NBCCV is localized in the Golgi apparatus.
In our previous studies, we investigated the interaction of NBCCV with the cytoskeleton (17) and observed that, in BCCV-infected cells, significant amounts of NBCCV were associated with actin filaments. However, the majority of the nucleocapsid antigen in the infected cells was localized in the perinuclear region. To further investigate the intracellular localization of NBCCV, we have examined its site of expression in the absence of the BCCV glycoproteins. The NBCCV coding sequence was inserted into the pcDNA3.1(−) plasmid under the cytomegalovirus promoter, the resulting plasmid was introduced into BHK21 cells, and the expression of NBCCV was analyzed. NBCCV migrated at 43 kDa and had the same electrophoretic mobility as the protein derived from lysates of BCCV-infected cells (not shown). The transfected cells were examined by IFA with anti-N MAb. Figure 1A shows that in BHK21 cells expressing NBCCV alone, the majority of this protein is localized to the perinuclear region. This distinctive site of intracellular localization of the NBCCV was also observed in Vero, Vero E6, HeLa, and primary human umbilical endothelial cells, using either anti-N MAb or anti-BCCV polyclonal antibody.
FIG. 1.
Intracellular localization of NBCCV. BHK21 cells were seeded onto coverslips in a 12-well plate, grown to 60% confluency, and transfected with pcNBCCV. The cells were fixed at 24 h posttransfection, and the NBCCV antigen was identified with IFA by using anti-N MAb (A). The cells were costained with phalloidin to show cellular morphology (B). The NBCCV antigen staining is concentrated in the perinuclear region of each transfected cell. A few cells also exhibit a filamentous pattern of the antigen. Panels C and D show BHK21 cells coexpressing the NBCCV and NLACV proteins. The cells were transfected with the pcNBCCV and pcNLACV plasmids and examined by double immunofluorescence staining at 24 h posttransfection. Panel C shows staining with anti-BCCV N MAb, and panel D shows staining with anti-NLACV rabbit polyclonal antibody. While the NLACV antigen is localized diffusely in the cytoplasm, the NBCCV antigen is found in the perinuclear region in the same cells.
BHK21 cells expressing NBCCV were also cotransfected with pcNLAC-myc, a plasmid that directs expression of NLACV. LACV is a representative of the Bunyavirus genus of the family Bunyaviridae, and its N protein is reported to be localized throughout the cytoplasm (19). NLACV is thought to migrate to the Golgi complex, the site of LACV assembly, only in the presence of the LACV glycoproteins as infection progresses (5, 9). At 24 h posttransfection, the cells were analyzed by double immunofluorescence staining with anti-N MAb and anti-NLACV rabbit sera. Figure 1C and D show that, in the same cell, NLACV is diffusely localized in the cytoplasm, whereas NBCCV is localized exclusively in the perinuclear region.
To ascertain whether NBCCV is associated with the Golgi complex, we performed double immunofluorescence staining with α-mannosidase II antibody, a marker for the Golgi complex, and with anti-N MAb. Figure 2A and B show that both antigens are colocalized in the perinuclear region. This result indicates that NBCCV is localized to the perinuclear region and is likely to be associated with the Golgi membranes.
FIG. 2.
NBCCV is localized in the Golgi region. Panels A and B show BHK21 cells that were dispersedly seeded and transfected with the pcNBCCV plasmid. The cells were examined by double immunofluorescence staining at 24 h posttransfection. Panel A represents staining with anti-N MAb, and panel B shows staining with anti-α-mannosidase II polyclonal antibody. Panels C and D show cells that were separately transfected with the plasmid pGFP/NBCCV (C), driving expression of NBCCV fused with GFP. Panel D represents cells transfected with pEGFP-c3, driving expression of GFP alone. The cells were examined at 24 h posttransfection by direct GFP fluorescence.
We next determined whether this unusual characteristic of the NBCCV is observed if the protein is fused with a heterologous cytoplasmic protein, GFP. GFP is uniformly distributed throughout the cytoplasm, and its detection in the living cell is possible by direct visualization under UV light (3). We have placed the GFP coding sequence in frame with the coding sequence for the N-terminal end of NBCCV in the context of the pEGFP-c3 vector. The resulting plasmid, pGFP/NBCCV, was introduced into BHK21 cells for transient expression of the GFP/BCCV-chimeric protein. The GFP/NBCCV protein migrated at 75 kDa, which is in accordance with its predicted size. When we examined the live cells expressing the chimeric protein, we found that fluorescence was localized to the perinuclear region (Fig. 2C and D). Every one of 500 GFP/NBCCV-expressing cells examined exhibited prominent perinuclear localization. Thus, the ability of NBCCV to be transported to the Golgi-like region is not affected by adding a foreign sequence and is not an artifact in fixation of cells.
The C-terminal 141 amino acids of NBCCV are sufficient for NBCCV perinuclear localization.
The observed targeting of the NBCCV to the Golgi region suggests that this protein might possess a specific sequence or sequences responsible for its unusual subcellular localization. To identify the NBCCV amino acid region important for its perinuclear localization, we constructed two series of N- and C-terminal-truncated mutants of NBCCV (Fig. 3A), and expressed them in BHK21 cells. Although the C-terminal-truncated proteins were detected easily, the signals for the N-terminally truncated mutants were barely detectable by either Western blotting or IFA. This differential detection between the N- and C-terminally truncated proteins was thought to be likely due to a polarized distribution of the NBCCV antigenic domains to the N-terminal end of the protein. To overcome this obstacle, we added the GFP coding sequence in front of each of the N-terminally truncated coding sequences. Western blotting revealed that the C- and the GFP-tagged N-terminally truncated mutants were synthesized and were observed to have the predicted molecular weights. The C-terminally truncated mutant proteins (d119C, d246C, d348C, d401C, and d417C) were found to be uniformly distributed throughout the cytoplasm, in contrast to the full-length N protein observed in the perinuclear region (Fig. 3B). The removal of as few as 11 amino acids from the C-terminal end of the NBCCV protein was found to cause this change in localization (Fig. 3B). The intracellular localization of the N-terminally truncated proteins varied from a Golgi-like pattern to a cytoplasmic pattern, depending on the length of the deletions introduced at the N terminus of NBCCV (Fig. 3B). The d67N-GFP, d219N-GFP, and d287-GFP mutant proteins, lacking amino acids 67, 219, and 287, respectively, showed typical perinuclear localization patterns. In contrast, the d342N-GFP and d400N-GFP mutant proteins, encoding 86- and 28-amino-acid C-terminal portions of NBCCV, respectively, were localized exclusively in the cytoplasm.
FIG. 3.
Identification of the NBCCV perinuclear localization sequence. Panel A shows a diagram of the NBCCV C- and N-terminally truncated mutants. The numbers indicate the position of ATG and TGA codons in the constructs relative to the amino acid sequence of the NBCCV protein. Panel B shows IFA of BHK21 cells expressing the NBCCV truncated proteins. The cells were fixed in 4% paraformaldehyde at 24 h posttransfection, permeabilized, and stained with either anti-N MAb (the upper row) or anti-GFP MAb (the lower row).
These results indicate that the sequence responsible for the NBCCV Golgi-like localization is contained within the last 141 C-terminal amino acids.
NBCCV is associated with microsomal membranes.
To confirm the IFA results suggesting membrane association of the NBCCV, we examined BHK21 cells expressing NBCCV or its truncated variants by subcellular fractionation. The transfected cells were disrupted with a Dounce homogenizer and subjected to differential centrifugation to obtain microsomal and cytosolic fractions. In addition to NBCCV and its truncated mutants, we also examined the cells expressing GFP, which is a cytosolic protein, and the beta subunit of cytoplasmic coat protein (β-COP), which is associated with the Golgi apparatus (21).
The protein samples prepared from the cytosolic and microsomal fractions were analyzed by Western blotting with specific antibodies. Figure 4 shows that NBCCV, whether expressed alone or as a fusion protein with GFP, is primarily associated with the microsomal fraction just like β-COP. In contrast the C-terminally truncated mutants and the GFP were detected only in the cytosolic fraction. The GFP-N-terminally truncated chimeric proteins are seen in both the microsomal and the cytosolic fractions. As observed with IFA, the difference in the fractionation patterns of these proteins depends on the size of the deletions at the N terminus of NBCCV. The d67N-GFP, d219N-GFP, and d287-GFP mutants are found in the microsomal fraction, whereas d342N-GFP and d400N-GFP are found in the cytosol. Based on these results and the IFA localization patterns, we conclude that NBCCV is a membrane-associated protein.
FIG. 4.
NBCCV is associated with microsomal membranes. BHK21 cells were harvested at 24 h posttransfection, disrupted in a homogenizer, and subjected to differential centrifugation to obtain microsomal (M) and cytosolic (C) fractions. The proteins of both fractions were separated by SDS-PAGE and analyzed by Western blotting with a specific antibody. The first row shows detection of β-COP (expressed endogenously), GFP, NBCCV, and GFP-NBCCV with anti-β-COP, anti-GFP, and anti-N MAbs, respectively. The second row represents subcellular fractionation of BHK21 cells expressing the C-terminally truncated proteins, which were detected with anti-N MAb. The third row shows detection of GFP-N-terminally truncated mutants with anti-GFP MAb.
NBCCV is a peripheral membrane-associated protein.
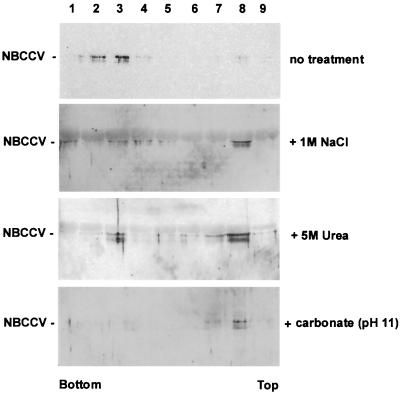
We further examined the membrane association properties of NBCCV (Fig. 5). NBCCV-microsomes were treated with 1 M NaCl, 4 M urea, or 15 mM sodium carbonate (pH 11) solutions and then examined by sedimentation analysis in sucrose gradients. In parallel, we have carried out the same analysis for untreated NBCCV microsomes. It is expected that the membrane-associated form of NBCCV should sediment toward the bottom portion of the gradient, whereas the soluble form of NBCCV would remain at the top of the gradient. Following the fractionation, the protein samples were separated by SDS-PAGE and examined by Western blotting. Figure 5 shows that when the NBCCV-microsomes were not subjected to the treatments with the high-salt or high-pH solutions, the majority of the protein sedimented in fractions 2 and 3 (lanes 2 and 3). The densities of the fractions containing NBCCV were in the range from 1.27 to 1.20 g/ml, which is consistent with data for other membrane-associated proteins analyzed in this system (24). In the presence of either 1 M NaCl, 4 M urea, or 15 mM sodium carbonate (pH 11), the NBCCV protein was found at the top of the gradient (lane 8), as expected for a soluble protein (24). Taken together, these results indicate that NBCCV is a peripheral membrane-associated protein, which is oriented to the cytosol and is associated with the membranes via electrostatic interactions.
FIG. 5.
Membrane association properties of NBCCV. NBCCV-microsomes that were either untreated (the first gel from top) or treated with 1 M NaCl (second gel), 4 M urea (third gel), or 15 mM sodium carbonate (pH 11) (fourth gel) were analyzed in sucrose density gradients. The fractions (∼450 μl each) were collected from the bottom of the gradient, precipitated with either ethanol or acetone, and examined by Western blotting with anti-N MAb. Lane numbers correspond to fractions collected from the gradients.
Intracellular localization of other hantaviral nucleocapsid proteins.
We next addressed the question whether the property of perinuclear localization is also observed for other hantaviral N proteins. Nucleocapsid proteins of PUUV and SEOV (NPUUV and NSEOV, respectively) were expressed in BHK21 cells that were transfected with the pcNPUUV and pcNSEOV plasmids. At 24 h posttransfection, the cells were examined by IFA with an anti-N MAb that is cross-reactive with a broad range of hantaviruses. Figure 6A and B show that, in many cells, both NPUUV and NSEOV are also localized in the perinuclear region, as observed for NBCCV. However, the percentage of cells showing this pattern was somewhat lower than that observed with NBCCV: 50 to 60% of antigen-positive cells were found to show Golgi-type localization for NPUUV, and 65 to 70% did so for NSEOV. A fraction of cells (approximately 5%) also exhibited filamentous staining of the antigen, suggesting actin-filament association.
FIG. 6.
NPUUV and NSEOV are localized to the perinuclear region. The intracellular localization of these proteins was examined by IFA. BHK21 cells were grown on coverslips and transfected either with pcNPUUV or pcNSEOV plasmids. At 24 h posttransfection, the cells were examined by IFA with anti-N MAb as the first antibody. Panel A shows expression of NPUUV, and panel B shows intracellular localization of NSEOV.
DISCUSSION
We have demonstrated that NBCCV is expressed as a peripheral membrane-associated protein in the perinuclear region. The evidence that supports this conclusion is presented at both the cellular and biochemical levels. We found that the NBCCV expressed in BHK21 cells is colocalized with anti-α-mannosidase II, a marker for the Golgi apparatus. Centrifugal separation of subcellular components demonstrated that NBCCV is associated with microsomal membranes. In addition, analysis of NBCCV truncation mutants revealed that a 141-amino-acid C-terminal portion of this protein directs that GFP (3) be localized in a Golgi-like region. In our previous studies (17), we observed that the majority of N protein in BCCV-infected cells was localized to the perinuclear region. Therefore, the absence of viral RNA in cells expressing the N protein alone, examined in this study, is not required for its perinuclear localization. The unusual localization of a hantavirus N protein does not appear to be restricted only to BCCV. Studies with Sin Nombre virus nucleocapsid protein expressed in a variety of permissive cell types, including human endothelial cells, also show a perinuclear pattern for this protein (C. Spiropoulou, personal communication). Our results indicate that this characteristic is not limited only to the group of New World hantaviruses, since PUUV and SEOV, representatives of Old World hantaviruses, also demonstrate a distinct perinuclear localization of the nucleocapsid antigens.
The genetic structure of the hantavirus N genes and their counterparts in viruses comprising the other genera of the Bunyaviridae family, such as Bunyavirus, Phlebovirus, and Tospovirus, differs as well (4, 18, 20, 22). The hantavirus nucleocapsid genes encode proteins that are in range of 428 to 433 amino acids, whereas in all the other members of the family, except for nairoviruses, this sequence is shorter by 160 to 200 amino acids. The observed size difference may reflect the acquisition of a sequence by an ancestor of the hantaviruses during evolution. Alternatively, the ancestral virus of the family Bunyaviridae may also have had this characteristic, but it was lost in some genera, therefore distinguishing the hantaviruses and nairoviruses from the rest of the family.
Membrane-associated proteins are classified as integral or peripheral proteins. Typical examples of integral proteins are viral glycoproteins, which interact with the lipid bilayer directly via their transmembrane domains. Peripheral membrane proteins are soluble proteins by their nature and are associated with membranes via electrostatic interactions. Our studies showed that association of NBCCV with microsomal membranes is sensitive to treatments with high-salt or high-pH solutions, indicating that NBCCV employs electrostatic interactions in its association with membranes and is therefore a peripheral protein. This is consistent with a computer analysis of the NBCCV amino acid sequence, which failed to reveal the presence of NH2-terminal signal motifs or transmembrane sequences (data not shown).
The fact that NBCCV is expressed as a membrane-associated protein in the perinuclear region might suggest that BCCV assembly occurs at the Golgi complex. However, there is evidence that would seriously challenge this suggestion. First, since NBCCV is associated with membranes in the absence of other viral components, an interaction of NBCCV with the cytoplasmic portions of the viral glycoproteins is not required for localization in this cellular compartment. Second, examination of BCCV-infected cells by electron microscopy has not revealed the presence of virus particles inside the Golgi stacks (16), and in BCCV-infected polarized epithelial cells, the viral glycoprotein was expressed at the apical cell surfaces, from which the virus was released (16). Third, double immunofluorescent staining of endothelial cells infected with Sin Nombre virus by using anti-G1, anti-G2, and anti-N antibodies has shown different localization patterns of the hantavirus glycoproteins relative to the nucleocapsid protein within the perinuclear region (C. Spiropoulou, unpublished data). These results indicate that these proteins probably are not physically associated in this organelle, as would be expected for interactions that occur during virus assembly.
Our data raise the question how a membrane-associated form of NBCCV could be utilized in the assembly of the RNP and how the RNP is targeted to the cell surface. It is possible that the RNA replication process for hantaviruses could occur on membranes in a manner similar to that of polioviruses (6). Membranes might serve as a physical matrix for assembly of hantaviral RNPs. The targeting of the newly assembled RNPs to the cell surface could be mediated through actin filaments, which might also be involved in the release of virions from the plasma membrane. Recent findings have shown that actin and actin-associated proteins, such as nonmuscle myosin II, tropomyosin 5, centeractin, neurabin I, and β-actin itself, are localized in a Golgi-like region (7, 23). This model fits with our previous findings that BCCV assembly occurs at the plasma membrane; NBCCV interacts with both G (soluble)- and F (filamentous)-actins; and depolymerization of actin filaments by cytochalasin D in infected cells dramatically reduces virus release. In contrast, representatives of other genera of the Bunyaviridae are resistant to cytochalasin D treatment and do not exhibit association of their N protein with either actin or Golgi-like membranes (17).
Although the assembly of New World hantaviruses appears to occur at the plasma membrane, the assembly of Old World hantaviruses has been reported to occur intracellularly (26). Thus, it may be the case that viruses in this genus employ a dual mode of virus assembly.
ACKNOWLEDGMENTS
We thank Marilyn G. Farquhar, Thomas Ksiazek, Ramaswamy Raju, and Sue Ruo for providing us with important reagents. We also thank Christina Spiropoulou and Stuart T. Nichol for helpful discussions, L. R. Melsen for assistance with figures, and Tanya Cassingham for assistance in preparing the manuscript.
This study was supported by NIH grant AI 12680.
REFERENCES
- 1.Anderson G W J, Smith J F. Immunoelectron microscopy of rift valley fever viral morphogenesis in primary rat hepatocytes. Virology. 1987;161:91–100. doi: 10.1016/0042-6822(87)90174-7. [DOI] [PubMed] [Google Scholar]
- 2.Arikawa J, Lapenotiere H F, Iacono-Connors L, Wang M L, Schmaljohn C S. Coding properties of the S and the M genome segments of Sapporo rat virus: comparison to other causative agents of hemorrhagic fever with renal syndrome. Virology. 1990;176:114–125. doi: 10.1016/0042-6822(90)90236-k. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1996. [Google Scholar]
- 4.Bishop D H L. Biology and molecular biology of bunyaviruses. In: Elliott R M, editor. The Bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 19–61. [Google Scholar]
- 5.Bupp K, Stillmock K, Gonzalez-Scarano F. Analysis of the intracellular transport properties of recombinant La Crosse virus glycoproteins. Virology. 1996;220:485–490. doi: 10.1006/viro.1996.0336. [DOI] [PubMed] [Google Scholar]
- 6.Caliguiri L A, Tamm I. Characterization of poliovirus-specific structures associated with cytoplasmic membranes. Virology. 1970;42:112–122. doi: 10.1016/0042-6822(70)90243-6. [DOI] [PubMed] [Google Scholar]
- 7.Fath K R, Trimbur G M, Burgess D R. Molecular motors are differentially distributed on Golgi membranes from polarized epithelial cells. J Cell Biol. 1994;126:661–675. doi: 10.1083/jcb.126.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldsmith C S, Elliott L H, Peters C J, Zaki S R. Ultrastructural characteristics of Sin Nombre virus, causative agent of hantavirus pulmonary syndrome. Arch Virol. 1995;140:2107–2122. doi: 10.1007/BF01323234. [DOI] [PubMed] [Google Scholar]
- 9.Gonzales-Scarano F, Nathanson N. Bunyaviruses. In: Fields B N, Knipe D M, editors. Virology. Vol. 2. New York, N.Y: Raven Press; 1990. pp. 1195–1228. [Google Scholar]
- 10.Karabatsos N, editor. International catalogue of arboviruses including certain other viruses. San Antonio, Tex: American Society of Tropical Medicine and Hygiene; 1985. [Google Scholar]
- 11.Kuismanen E, Hedman K, Saraste J, Pettersson R F. Uukuniemi virus maturation: accumulation of virus particles and viral antigens in the Golgi complex. Mol Cell Biol. 1982;2:1444–1458. doi: 10.1128/mcb.2.11.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Matsuoka Y, Chen S-Y, Compans R W. Bunyavirus protein transport and assembly. Curr Top Microbiol Immunol. 1991;169:161–180. doi: 10.1007/978-3-642-76018-1_6. [DOI] [PubMed] [Google Scholar]
- 14.Matsuoka Y, Chen S-Y, Compans R W. A signal for Golgi retention in the bunyavirus G1 glycoprotein. J Biol Chem. 1994;269:22565–22573. [PubMed] [Google Scholar]
- 15.Murphy F A, Harrison A K, Whitfield S G. Bunyaviridae: morphologic and morphogenetic similarities of Bunyamwera serologic supergroup viruses and several other anthropod-borne viruses. Intervirology. 1973;1:297–316. doi: 10.1159/000148858. [DOI] [PubMed] [Google Scholar]
- 16.Ravkov E V, Nichol S T, Compans R W. Polarized entry and release in epithelial cells of Black Creek Canal virus, a New World hantavirus. J Virol. 1997;71:1147–1154. doi: 10.1128/jvi.71.2.1147-1154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravkov E V, Nichol S T, Peters C J, Compans R W. Role of actin microfilaments in Black Creek Canal virus morphogenesis. J Virol. 1998;72:2865–2870. doi: 10.1128/jvi.72.4.2865-2870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravkov E V, Rollin P E, Ksiazek T G, Peters C J, Nichol S T. Genetic and serologic analysis of Black Creek Canal virus and its association with human disease and Sigmodon hispidus infection. Virology. 1995;210:482–489. doi: 10.1006/viro.1995.1366. [DOI] [PubMed] [Google Scholar]
- 19.Rossier C, Patterson J, Kolakofsky D. La Crosse virus small genome mRNA is made in the cytoplasm. J Virol. 1986;58:647–650. doi: 10.1128/jvi.58.2.647-650.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmaljohn C S. Molecular biology of hantaviruses. In: Elliott R M, editor. The Bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 63–90. [Google Scholar]
- 21.Serafini T, Stenbeck G, Brecht A, Lottspeich F, Orci L, Rothman J E, Wieland F T. A coat subunit of Golgi-derived non-clathrin-coated vesicles with homology to the clathrin-coated vesicle coat protein beta-adaptin. Nature. 1991;349:215–220. doi: 10.1038/349215a0. [DOI] [PubMed] [Google Scholar]
- 22.Spiropoulou C F, Morzunov S, Feldmann H, Sanchez A, Peters C J, Nichol S T. Genome structure and variability of a virus causing hantavirus pulmonary syndrome. Virology. 1994;200:715–723. doi: 10.1006/viro.1994.1235. [DOI] [PubMed] [Google Scholar]
- 23.Stephens D J, Banting G. Direct interaction of the trans-Golgi network membrane protein, TGN38, with the F-actin binding protein, neurabin. J Biol Chem. 1999;274:30080–30086. doi: 10.1074/jbc.274.42.30080. [DOI] [PubMed] [Google Scholar]
- 24.Towner J S, Ho T V, Semler B L. Determinants of membrane association for poliovirus protein 3AB. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 25.Vapalahti O, Kallio-Kokko H, Salonen E M, Brummer-Korvenkontio M, Vaheri A. Cloning and sequencing of Puumala virus Sotkamo strain S and M RNA segments: evidence for strain variation in hantaviruses and expression of the nucleocapsid protein. J Gen Virol. 1992;73:829–838. doi: 10.1099/0022-1317-73-4-829. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Zang L, Feng M, Liang Z, Wang S, Zheng S, Zhang L, Jiang Z, Chen D. Transmission electron microscope study of the hemorrhagic spots in patients with epidemic hemorrhagic fever in the early stage. Ultrastruct Pathol. 1997;21:281–287. doi: 10.3109/01913129709021924. [DOI] [PubMed] [Google Scholar]