Summary
Early bolting is a major breeding objective for globe artichoke (Cynara cardunculus var. scolymus L.). It has been suggested that globe artichoke bolting time is linked to a vernalization requirement, although environmental conditions under which vernalized plants and controls have been grown may not always allow for proper comparison. Here, we defined morphological markers to monitor the vegetative-to-reproductive phase transition at the shoot apex and linked these to expression changes of homologs of key Arabidopsis flowering regulators SOC1, FUL, and AP1. Importantly, we developed an experimental setup where control and vernalized plants grow under comparable conditions. These tools together allowed for comparison of the vegetative-to-reproductive phase transition between early- and late-bolting genotypes and how they respond to vernalization. Our results show that vernalization requirement is significantly lower in early-bolting genotypes, supporting the hypothesis that the early-bolting trait is at least partly underlain by alterations in the network controlling vernalization response.
Subject areas: Biological sciences, Plant biology, Plant development
Graphical abstract
Highlights
-
•
Early bolting is a key trait for globe artichoke
-
•
The trait is thought to be linked to a lower vernalization requirement
-
•
Effect of vernalization was assessed under controlled vernalization conditions
-
•
Result: vernalization requirement is lower in early- than in late-bolting genotypes
Biological sciences; Plant biology; Plant development
Introduction
Globe artichoke (Cynara cardunculus L. var. scolymus [L.] Fiori) is thought to be a domesticated form of wild cardoon (C. cardunculus var. sylvestris [Lamk] Fiori) and cultivated for its sizable, closed, and compact inflorescences, which are colloquially referred to as “heads”. In Mediterranean climate zones, cultivation cycles run from transplanting in summer till harvest between late autumn and late winter. Traditional varieties are often vegetatively propagated whereas modern hybrids are propagated by seeds and designed to be sown and harvested within a single cultivation cycle. Although seeded hybrids have demonstrated their potential for superior yield, quality, and uniformity over traditional varieties, they tend to produce later than many early producing traditional varieties1,2,3,4. For that reason, combining the superior quality and production of hybrids with the earliness of traditional varieties is a major objective for globe artichoke breeders.
Globe artichoke has a seasonal life cycle. At the start of the season, one or more rosettes develop from the roots. These embody the vegetative phase which is characterized by the continuous production of leaves by the shoot apical meristem (SAM), without elongation of the stem internodes. During the vegetative-to-reproductive phase transition, which is also known as the floral transition, the hitherto leaf-producing SAM is converted into an inflorescence meristem (IM) that gives rise to inflorescence stems and capitula (heads). Developmental stages in cardoon and globe artichoke have been described and classified5,6,7,8. These scales however do not describe in detail the changes that occur in the shoot or inflorescence apex between the vegetative-to-reproductive phase change and bolting. Moreover, molecular markers for the vegetative-to-reproductive phase transition, potentially useful for studies on the genetic architecture of flowering, are not available for globe artichoke.
To enable successful reproduction in plants, the vegetative-to-reproductive phase needs to be precisely timed in accordance with a variety of endogenous and environmental cues such as age, day length, cold exposure, ambient temperature, or drought.9,10,11,12 These flowering cues are perceived by flowering regulatory pathways that converge upon a set of flowering integrators that finally direct the vegetative-to-reproductive phase transition. These pathways form an intricate genetic network, which has been extensively studied and reviewed in the model species Arabidopsis (Arabidopsis thaliana) and cereals such as rice, barley, and wheat,9,13,14,15 although the relevance of the different pathways and mechanisms controlling flowering in non-model species are often poorly understood.
In Arabidopsis the major flowering pathways are the vernalization (predetermined requirement for cold exposure to acquire flowering competence), photoperiod, age, gibberellin, and autonomous ones.16 These pathways converge on specific floral integrators. The photoperiod pathway enhances the expression of the floral integrator FLOWERING LOCUS T (FT), a phosphatidylethanolamine binding protein (PEBP) family member17,18 that positively controls the expression of MADS box floral integrator gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1)19,20 and FRUITFULL (FUL).21,22 SOC1 also functions independently from FT, being activated directly by the vernalization and autonomous pathways through repression of the MADS box gene FLOWERING LOCUS C (FLC).23 Moreover, SOC1 and FUL are activated by the age pathway through the microRNA 156 (miR156)-mediated control of several SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes (SPL3, SPL9, and others)24 and by the gibberellin pathway that promotes SOC1 and FUL expression through the degradation of DELLA proteins and the release of active SPL proteins, among others.25,26,27 Finally, SOC1 and FUL function in a partially redundant manner to control the expression of the meristem identity gene LFY.28,29,30
Different studies have indicated that individual genes, or gene families, that constitute this network are largely conserved between Arabidopsis and members of the Asteraceae family, like lettuce (Lactuca sativa L.), chicory (Cichorium intybus L.), safflower (Carthamus tinctorius L.), Chrysanthemum (Chrysanthemum sp.), and gerbera (Gerbera hybrida L.), although gene orthology and function cannot always be inferred directly31,32,33,34,35,36,37. Due to their central role in floral signal integration and floral meristem and organ identity, members of the MADS box gene family have been studied in Asteraceae and homologs of SOC1, FUL, and AP1 have been reported for chrysanthemum, gerbera, lettuce, safflower, and sunflower31,35,37,38,39,40,41. On the contrary, knowledge about the identity of the floral integrators in globe artichoke is scarce, as well as about cues that trigger the floral transition process.
In general, the genetic control of bolting time in globe artichoke is poorly known. Several works have described different factors that modulate bolting but usually associated with specific varieties or cultivation methods and focusing on agronomic and quality traits42,43,44. With respect to photoperiod, different authors have considered globe artichoke to be either an obligate long-day plant45 or a short-day plant46 or considered it to flower independent of photoperiod,47 suggesting a genotype-dependent response. More consistent links have been found with gibberellic acid (GA) and vernalization. Application of GA3 is known to advance the moment of bolting in globe artichoke48,49,50 while vernalization has been proposed to be a major determinant of flowering in globe artichoke.47,51 Moreover, it has been suggested that the presence of a vernalization requirement is linked to late-bolting genotypes, bolting after the winter, whereas the absence of such a requirement might explain the existence of genotypes that can bolt before winter.45,46 However, most studies on the effect of, or requirement for, vernalization in globe artichoke either have been performed under non-controlled field conditions,46,51 by exposing plants to vernalizing temperatures only during part of their life cycle,52,53,54 or do not consider the time of bolting in individual plants.51 This means that, although these studies provide valuable information on the role of vernalization regimes for improved cultivation practices, their results do not allow to quantify the precise effect of vernalization on the time of bolting and/or to compare results between studies.
In order to better understand the floral transition in globe artichoke, we have characterized the morphological changes that occur at the macroscopic and microscopic level in the shoot apex that are associated with the vegetative-to-reproductive phase transition. We also established an experimental procedure that enables study of the vernalization requirement of globe artichoke under controlled and comparable conditions, as well as the identification of globe artichoke homologs of key regulators of the floral transition in Arabidopsis such as SOC1, FUL, and AP1, which can be used as molecular markers for floral transition. These tools have allowed us to compare the vernalization requirement between early- and late-bolting genotypes. This comparison has shown that all genotypes under study are able to respond to vernalization. However, late-bolting genotypes display a stronger vernalization response than early-bolting genotypes, having a higher requirement for vernalization than early-bolting ones.
Results
Novel morphological markers of the shoot apex associated with the transition to the reproductive phase
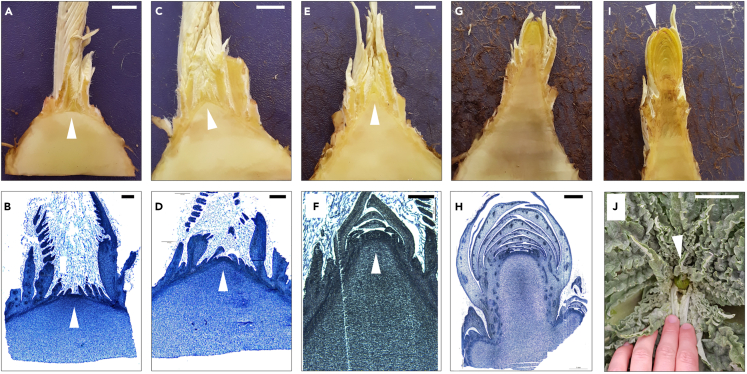
Different developmental stages during the vegetative-to-reproductive phase transition in artichoke have been previously described at the macroscopic level. Bolting stage 0 has been proposed to define plants with no signs of bolting, whereas bolting stage A is assigned to the time when the primary inflorescence is palpable in the center of the basal leaf rosette.5,8 In order to determine the moment of initiation of vegetative-to-reproductive phase transition in artichoke, we decided to complement the previously described stages5,8 by performing a morphological characterization of the SAM at both the macroscopic and microscopic levels. For this, we dissected inflorescence apices in weekly intervals. We defined five stages that describe the morphological changes in the apex during the time before the primary inflorescence becomes palpable inside the rosette, so previous to the described bolting stage A (Table S1). We named these five stages as “pre-bolting stage 0” to “pre-bolting stage 4.” The first detectable sign of the vegetative-to-reproductive phase transition having been initiated is a change in the shape of the hitherto globose or flat vegetative apex, i.e., pre-bolting stage 0 (Figures 1A and 1B), toward a domed, and later pointed or triangular form, which we designated “pre-bolting stage 1” (Figures 1C and 1D). Pre-bolting stage 2 starts with the elongation of the IM and the formation of the first bracts of the primary inflorescence at the tip of the inflorescence apex (Figures 1E and 1F). Subsequently, pre-bolting stage 3 is characterized by an increased number of bracts and further elongation of the primary inflorescence, which now reaches about 1 cm in size (Figure 1G). Pre-bolting stage 4 (Figure 1H) commences with the initiation of second-order inflorescences and coincides with bolting stage A, during which the primary inflorescence becomes palpable in the center of the rosette (Figure 1I).8
Figure 1.
Morphological changes in the SAM around the moment of vegetative-to-reproductive phase transition and a classification of developmental stages
(A) pre-bolting stage 0 (c20, 87 days post transplanting [d.p.t.]), (B) pre-bolting stage 0 (CARI, 127 days d.p.t.), (C) pre-bolting stage 1 (c20, 123 d.p.t.), (D) pre-bolting stage 1 (CARI, 126 d.p.t), (E) pre-bolting stage 2 (c20, 130 d.p.t.), (F) pre-bolting stage 2 (“Green Queen F1,” 140 d.p.t.), (G) pre-bolting stage 3 (c20, 144 d.p.t.), (H) pre-bolting stage 4 (CARI, 147 d.p.t.), (I) bolting stage A (CARI, 144 d.p.t.), (J) bolting stage “B” for comparison. Triangles point to SAM (A–D), the primary inflorescence primordium (E and F), or emerging primary capitulum (I and J). Scale bars: 500 μm (A–G), 1,000 μm (H and I), 4 cm (J).
Early bolting is linked to a reduced vernalization requirement
Some studies have proposed a relevant role for vernalization as a determinant of bolting time in different genotypes.45,51 As mentioned in the introduction, experiments carried out to characterize the vernalization response in globe artichoke did not allow direct comparisons, and conclusions from these studies require assessment because of non-controlled conditions, i.e., comparing genotypes grown in different time frames and climate zones or at different latitudes.
To overcome these limitations and determine the effect of vernalization on the time of bolting in early- and late-bolting genotypes under appropriate conditions, we designed an experimental setup where half of the plants were grown in a net house in a standard growing cycle (transplanting by the end of September and anthesis by the end of April), which naturally includes a vernalization period. The remainder plants were grown under the same conditions, in a net house adjacent to the aforementioned one, but where vernalizing temperatures (<12°C) were prevented by means of a thermostat-controlled heating system (Figure S1). The genotypes used in the experiments were two early-bolting clones (“c1” and “c70”), two late-bolting clones (“c20” and “c154”), two inbred early-bolting lines (“VESB” and “VER”), and one inbred late-bolting line (“CARI”).
The experiments were carried out during three seasons, between 2019 and 2022, comparing plants grown with or without vernalization. The genotypes studied were characterized during at least two seasons, except the inbred line “VER” which was only characterized during the first season. For the vernalized plants, the number of chilling hours (i.e., hours < 10°C) was 698, 837, and 832 in the three seasons, respectively (Table S2). Plants were observed weekly, and their developmental stage scored by eye. A plant was considered to have bolted upon reaching bolting stage B (Figure 1J; Table S1).
The results of the experiments in the three seasons are summarized in Table 1. Out of the 1,170 plants analyzed, a total of 55 did not bolt at the end of the season, 44 that were non-vernalized and 11 that were vernalized. Out of the 44 non-vernalized plants that remained in the vegetative phase, 6 were from early-bolting genotypes and the remainder 38 from late-bolting genotypes, more specifically c20 (7), c154 (7), and CARI (24). This suggests that, in the absence of vernalizing temperatures, late-bolting genotypes have a higher propensity toward remaining vegetative at the end of the season than the early-bolting ones. Under vernalization conditions, at the moment of bolting, early-bolting genotypes c1, c70, VESB, and VER had accumulated an average of 604.0, 403.8, 493.1, and 476.8 h below 10°C, respectively, while the late-bolting genotypes (c20, c154, and CARI) accumulated considerably more hours below 10°C (777.5, 773.3, and 737.0, respectively) (Table 1). Interestingly, late-bolting genotypes each accumulated similar amounts of hours below10°C.
Table 1.
Proportion of plants bolted in time per genotype, season, and treatment
| eEarliness | Genotype | Treatment | Season | n_obsa | Not boltedb | Mean date bolting stage Bc | DTB |
Mean day lengthd | Accumulated hours < 10°C |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mean | SD | per season | meane | ||||||||
| Early | c1 | no vernalization | 2019–2020 | 19 | 0 | 2020/02/15 | 144.9 | 7.8 | 10.8 | 1.5 | |
| Early | c1 | no vernalization | 2020–2021 | 15 | 1 | 2021/02/08 | 137.1 | 3.4 | 10.5 | 0.7 | 0.9 |
| Early | c1 | no vernalization | 2021–2022 | 57 | 2 | 2022/02/14 | 137.2 | 6.5 | 10.8 | 0.5 | |
| Early | c1 | vernalization | 2019–2020 | 0 | 0 | NA | NA | NA | NA | NA | |
| Early | c1 | vernalization | 2020–2021 | 18 | 0 | 2021/02/09 | 138.8 | 3.0 | 10.6 | 617.8 | 604.0 |
| Early | c1 | vernalization | 2021–2022 | 59 | 0 | 2022/02/06 | 129.9 | 4.6 | 10.5 | 590.3 | |
| Early | c70 | no vernalization | 2019–2020 | 32 | 0 | 2020/01/09 | 107.8 | 8.5 | 9.8 | 1.5 | |
| Early | c70 | no vernalization | 2020–2021 | 18 | 0 | 2021/01/19 | 117.1 | 10.8 | 10.0 | 0.1 | 0.7 |
| Early | c70 | no vernalization | 2021–2022 | 57 | 3 | 2022/01/16 | 108.1 | 10.4 | 9.9 | 0.5 | |
| Early | c70 | vernalization | 2019–2020 | 0 | 0 | NA | NA | NA | NA | NA | |
| Early | c70 | vernalization | 2020–2021 | 13 | 1 | 2021/01/23 | 121.4 | 10.8 | 10.1 | 540.8 | 403.8 |
| Early | c70 | vernalization | 2021–2022 | 52 | 0 | 2022/01/05 | 97.1 | 10.0 | 9.7 | 266.7 | |
| Early | VESB | no vernalization | 2019–2020 | 57 | 0 | 2020/02/05 | 134.5 | 12.3 | 10.5 | 1.5 | |
| Early | VESB | no vernalization | 2020–2021 | 0 | 0 | NA | NA | NA | NA | NA | 1.0 |
| Early | VESB | no vernalization | 2021–2022 | 65 | 0 | 2022/02/17 | 140.4 | 7.4 | 10.9 | 0.5 | |
| Early | VESB | vernalization | 2019–2020 | 51 | 0 | 2020/01/21 | 119.4 | 12.6 | 10.0 | 431.6 | |
| Early | VESB | vernalization | 2020–2021 | 0 | 0 | NA | NA | NA | NA | NA | 493.1 |
| Early | VESB | vernalization | 2021–2022 | 49 | 0 | 2022/02/02 | 125.4 | 5.8 | 10.4 | 554.7 | |
| Early | VER | no vernalization | 2019–2020 | 53 | 0 | 2020/02/12 | 141.4 | 7.7 | 10.7 | 1.5 | |
| Early | VER | no vernalization | 2020–2021 | 0 | 0 | NA | NA | NA | NA | NA | 1.5 |
| Early | VER | no vernalization | 2021–2022 | 0 | 0 | NA | NA | NA | NA | NA | |
| Early | VER | vernalization | 2019–2020 | 51 | 0 | 2020/01/26 | 124.5 | 14.3 | 10.2 | 476.8 | |
| Early | VER | vernalization | 2020–2021 | 0 | 0 | NA | NA | NA | NA | NA | 476.8 |
| Early | VER | vernalization | 2021–2022 | 0 | 0 | NA | NA | NA | NA | NA | |
| Late | c20 | no vernalization | 2019–2020 | 0 | 0 | NaN | NA | NA | NA | NA | |
| Late | c20 | no vernalization | 2020–2021 | 8 | 0 | 2021/03/19 | 176.6 | 9.1 | 12.1 | 28.0 | 48.2 |
| Late | c20 | no vernalization | 2021–2022 | 5 | 7 | 2022/04/12 | 199.6 | 3.1 | 13.0 | 68.5 | |
| Late | c20 | vernalization | 2019–2020 | 0 | 0 | NA | NA | NA | NA | NA | |
| Late | c20 | vernalization | 2020–2021 | 10 | 4 | 2021/03/15 | 172.0 | 0.0 | 11.9 | 723.5 | 777.5 |
| Late | c20 | vernalization | 2021–2022 | 12 | 0 | 2022/03/15 | 168.9 | 3.6 | 11.9 | 831.5 | |
| Late | c154 | no vernalization | 2019–2020 | 0 | 1 | NA | NA | NA | NA | NA | |
| Late | c154 | no vernalization | 2020–2021 | 10 | 1 | 2021/03/29 | 191.8 | 12.6 | 12.5 | 51.9 | 57.4 |
| Late | c154 | no vernalization | 2021–2022 | 25 | 5 | 2022/04/10 | 194.3 | 5.1 | 12.9 | 62.9 | |
| Late | c154 | vernalization | 2019–2020 | 0 | 0 | NA | NA | NA | NA | NA | |
| Late | c154 | vernalization | 2020–2021 | 10 | 0 | 2021/03/12 | 169.9 | 3.4 | 11.8 | 715.0 | 773.3 |
| Late | c154 | vernalization | 2021–2022 | 26 | 4 | 2022/03/15 | 171.4 | 3.0 | 11.9 | 831.5 | |
| Late | CARI | no vernalization | 2019–2020 | 16 | 15 | 2020/03/29 | 211.4 | 20.6 | 12.5 | 1.5 | |
| Late | CARI | no vernalization | 2020–2021 | 0 | 0 | NA | NA | NA | NA | NA | 20.8 |
| Late | CARI | no vernalization | 2021–2022 | 39 | 9 | 2022/04/04 | 189.7 | 10.0 | 12.7 | 40.0 | |
| Late | CARI | vernalization | 2019–2020 | 32 | 0 | 2020/03/03 | 161.1 | 3.6 | 11.5 | 642.4 | |
| Late | CARI | vernalization | 2020–2021 | 0 | 0 | NA | NA | NA | NA | NA | 737.0 |
| Late | CARI | vernalization | 2021–2022 | 43 | 2 | 2022/03/15 | 171.9 | 5.9 | 11.9 | 831.5 | |
numberr of observations.
number of individuals that had not bolted at the end of the experiment.
mean date of reaching bolting stage B.
day length refers to the mean calculated number of hours that the sun is above the horizon on a given day upon reaching bolting stage B.
mean of cumulative hours over three seasons.
We also observed that the plants from the early-bolting genotypes, both vernalized and non-vernalized, bolted between January and February, a time frame where nights are still longer than days and usually considered as short-day conditions (Table 1). In contrast, the late-bolting genotypes, under no-vernalization conditions, bolted around the end of March and early April, whereas under vernalization conditions bolting occurred around the middle of March. This time frame indicates that bolting in these genotypes is not strictly confined to short-day conditions (Table 1).
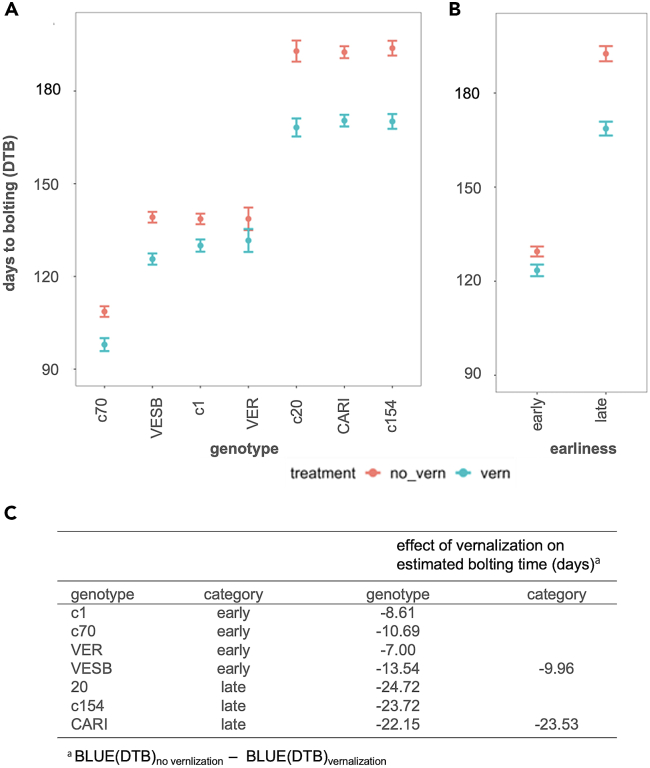
To measure the level of earliness, we considered the number of days to bolting stage B (DTBs). Then DTBs were modeled using a linear mixed model to account for both fixed effects and random effects such as between-plot variation between different seasons (random effects). The analysis revealed that the terms “genotype” (p value = 1.16 × 10−80), “treatment” (p value = 3.01 × 10−30), and “genotype:treatment” (p value = 9.09 × 10−6) significantly affected bolting time, whereas “season” did not (p value = 0.23) (Table S3). The estimated means were extracted from the model for each “genotype:treatment” combination, and the significance of differences between best linear unbiased estimates (BLUEs) was assessed by whether the difference between BLUEs was larger or smaller than the value of the least significant difference (LSD, 6.86 days) (Figure 2A). The statistical model showed that vernalization had a significant effect on bolting time in all genotypes that were observed in multiple seasons, indicating that both early- and late-bolting genotypes were sensitive to vernalization (Figure 2A).
Figure 2.
Model predictions and effects of vernalization on DTB in early- and late-bolting genotypes
(A) BLUEs (best linear unbiased estimates) for each genotype:treatment combination. Non bolters were not included in the analysis. Error bars indicate the predicted interval.
(B) BLUEs categorized by early- vs. late-bolting habit. Error bars indicate the predicted interval.
(C) Effect of vernalization on bolting time BLUEs per genotype and genotype category. LSD (least significant difference) for both models is 6.86.
To determine whether the effect of vernalization was different between the early- and the late-bolting genotypes, the modeling was repeated substituting explanatory variable “genotype” with “earliness,” a dichotomous variable distinguishing early-bolting genotypes (c1, c70, VESB, and VER) from late-bolting ones (c20, c154, and CARI). In this model, the term “earliness” was significant while “season” was not (Table S3). Differences in estimated means were not significant for the effect of vernalization on early-bolting genotypes (6.02 < 6.86), but they were significant for late-bolting genotypes (23.89 > 6.86) (Figures 2B and 2C). These results indicate that the late-bolting genotypes significantly respond to vernalization whereas the early-bolting genotypes, as a category, probably due to different vernalization responses inside the group, apparently do not respond.
Whereas in the late-bolting genotypes, vernalization advanced the estimated mean bolting time by 23.53 days in the model, in the early-bolting ones vernalization advanced mean bolting time by 9.96 days (Figure 2C). More specifically, in the early-bolting genotypes, the effect of vernalization was largest for VESB (13.54 days advance), followed by c70 (10.69 days), c1 (8.61 days), and VER (7.00 days), while in the late-bolting genotypes the effect was largest for c20 (24.72 days), followed by c154 (23.72 days) and CARI (22.15 days) (Figure 2C). The larger effect of vernalization on late-bolting genotypes is in agreement with the latter having accumulated more hours below 10°C, as registered during the vernalization experiment (Table S2; Figure S2), which indicates that early-bolting genotypes have a lower vernalization requirement for bolting than late-bolting genotypes. This analysis clearly indicates that our experimental setup allows for measuring the effect of vernalization in the tested genotypes under comparable conditions, making it feasible to extend the results of this study to the behavior of plants under field conditions.
Vernalization triggers the vegetative-to-reproductive phase change only in late-bolting genotype
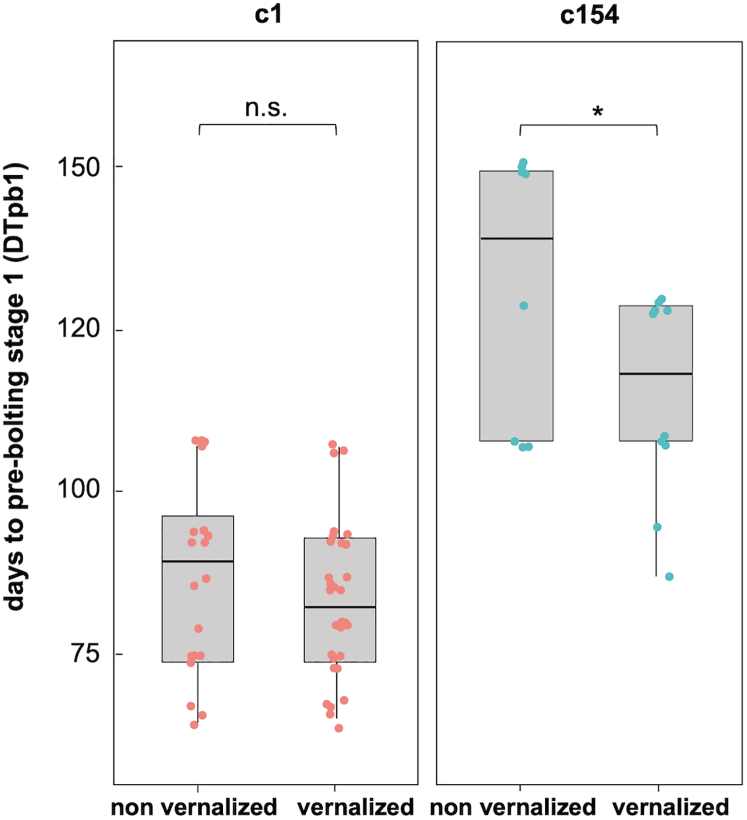
Bolting is a late and advanced phase of the floral transition, which occurs days before the vegetative-to-reproductive phase change is detected. To better understand the effect of vernalization on the timing of the vegetative-to-reproductive phase transition in these plants, we decided to monitor the transition in selected early- and late-bolting genotypes by direct observation of the plant apices. For this purpose, we selected early-bolting genotype c1 and late-bolting genotype c154. During the 2020–2021 season, we performed an additional vernalization experiment wherein shoot apices were collected and analyzed at different time points before bolting. The vegetative-to-reproductive phase transition was considered to have commenced when the SAM reached pre-bolting stage 1 (Figures 1A and 1B; Table S1). This experiment showed that vernalized plants from genotype c1 reached this stage on average at 88.3 days after transplanting whereas non-vernalized plants reached this stage at 84.1 days after transplanting, having accumulated only 148.7 h below 10°C (Table 2; Figure 3). This analysis indicates that vernalization apparently does not affect the time of floral transition of early-bolting genotype c1. In contrast, non-vernalized plants from late-bolting genotype c154 reached pre-bolting stage 1 later than plants from genotype c1, at an average of 131.6 days after transplanting (Table 2; Figure 3). Under vernalization conditions, plants from genotype c154 showed a significant advancement of the vegetative-to-reproductive phase transition with respect to non-vernalized plants (115.0 days, p = 0.043), having accumulated 459.4 h below 10°C (Table 2; Figure 3). This analysis also reflected that the two different genotypes characterized initiate the floral transition in short-day conditions (third week of December for both vernalized and non-vernalized c1 plants, last days of January for non-vernalized c154 plants, and middle January for vernalized ones) (Table 2).
Table 2.
Proportion of plants bolted in time per genotype, season, and treatment
| Earliness | Genotype | Treatment | Season | n_obsa | Mean date bolting stage Bb | DTpb1 |
Mean daylengthc | Acc. hours < 10°Cd | |
|---|---|---|---|---|---|---|---|---|---|
| mean | SD | ||||||||
| Early | c1 | no vernalization | 2020–2021 | 20 | 2020/12/21 | 88.3 | 15.0 | 9.7 | 0.0 |
| Early | c1 | vernalization | 2020–2021 | 30 | 2020/12/17 | 84.1 | 12.0 | 9.6 | 148.7 |
| Late | c154 | no vernalization | 2020–2021 | 9 | 2021/01/29 | 127.4 | 23.2 | 10.4 | 0.3 |
| Late | c154 | vernalization | 2020–2021 | 10 | 2021/01/17 | 115.0 | 16.2 | 10.0 | 459.4 |
Number of observations.
Mean date of reaching pre-bolting stage 1.
Day length refers to the calculated number of hours that the sun is above the horizon on a given day.
Number of accumulated hours below 10°C.
Figure 3.
Days to pre-bolting stage 1 (DTpb1) in early-bolting genotype c1 and late-bolting genotype c154
Data from 2020 to 2021 season. Red and blue dots represent individual samples. Significance levels: n.s., not significant; ∗, p < 0.05.
Analysis of artichoke MADS box genes to develop expression markers for the vegetative-to-reproductive phase transition
We have identified morphological markers in the shoot apex to assess the developmental stages during the transition from the vegetative to the reproductive phases in globe artichoke (Figure 1). In order to generate gene-expression markers for the vegetative-to-reproductive phase transition, which might signal developmental changes not revealed by morphological markers, we identified homologs of genes with known roles as flowering pathway integrators.55 MADS box genes SOC1 and FUL have been described as flowering integrators in multiple plant species.34,56 In Arabidopsis, an increase in expression levels of these genes accompanies the vegetative-to-reproductive phase transition.14 Thus, we looked for homologs of the SOC1 and FUL genes in the globe artichoke genome.57
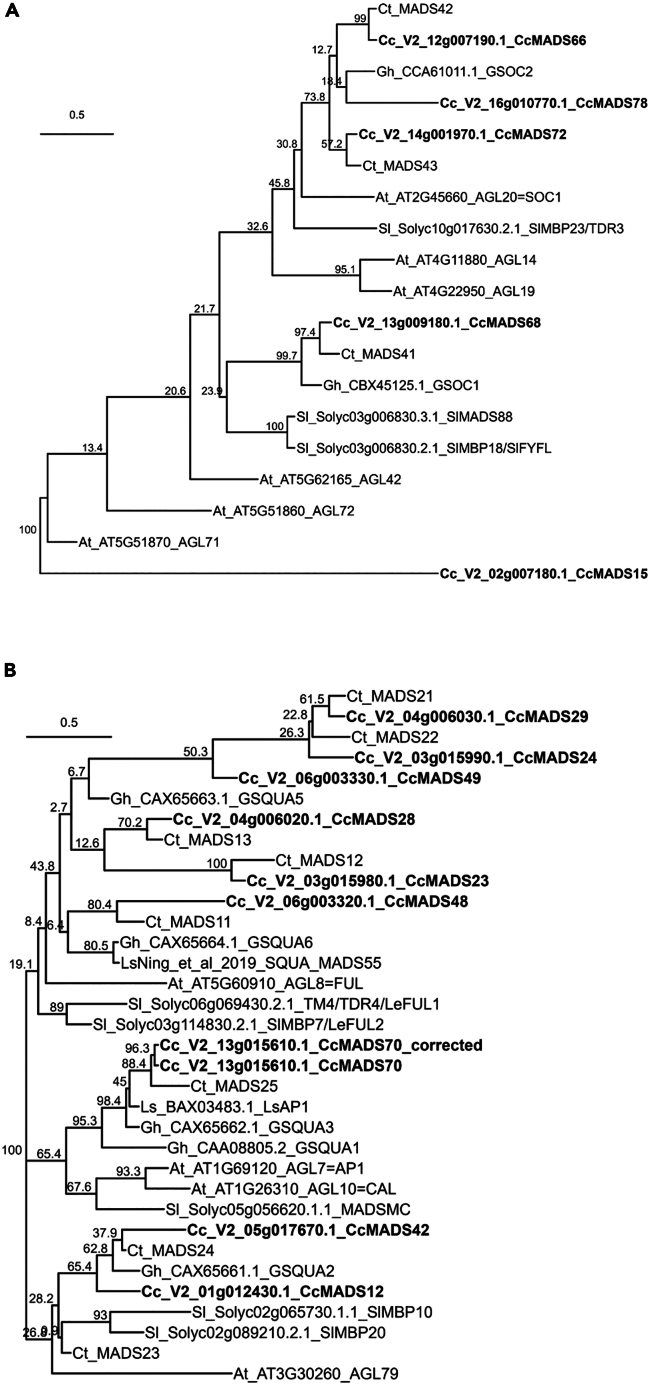
A basic local alignment search tool of MADS box proteins (BLASTp) from Arabidopsis, tomato, and lettuce on the globe artichoke genome yielded a total of 82 hits. In plants, two major lineages of MADS box genes can be distinguished by phylogenetic studies, being Type I and Type II (MIKC-like). Type I genes contain an MADS domain, whereas Type II genes possess an additional Keratin-like domain (K-domain).58 A Pfam domain scan of the 82 proteins identified significant SRF/MADS domains (PF00319.21) in 67 proteins and significant K-domains (PF01486.20) in 33 proteins. The genes were named CcMADS1 till CcMADS81 (Table S4). After clustering the proteins with Arabidopsis, tomato, and lettuce homologs, they were divided into 28 Type I and 53 Type II MADS box genes (Figure S3). In a total of 53 Type II MADS box genes, we identified 3 MIKC∗-type (Mδ subfamily) genes. The remainder 50 MIKCC genes were divided into 13 subfamilies. In brief, we identified 82 MADS box genes in the globe artichoke genome that represent the main clades found in other angiosperms. In a more detailed phylogenetic study of the SOC1 subfamily, we clustered globe artichoke proteins with SOC1 homologs and subfamily members from Arabidopsis, tomato, lettuce, safflower, and gerbera (Figure 4A). Of the five globe artichoke SOC1 subclade members, CcMADS66, CcMADS68, and CcMADS72 contain both an SRF and a K-box Pfam domain (Table S4). Moreover, when considering both gene structure and protein sequence similarity to AtSOC1, CcMADS66 and CcMADS72 were most similar (61.2% and 63.9%, respectively). These two genes also contain the canonical C-terminal “eVETeLvIGpP” TM3/SOC1 motif,59 which was not found in the remainder genes except for CcMADS66. Taken together, these data suggest that the SOC1 subfamily contains at least two true SOC1 homologs, CcMADS72 and CcMADS66, which we designated CcSOC1a and CcSOC1b. Remainder homologs, CcMADS78, CcMADS68, and CcMADS15, were designated CcSOC1Like-A, CcSOC1Like-B, and CcSOC1Like-C.
Figure 4.
ML phylogenetic tree of SOC1 and AP1 proteins including globe artichoke
(A) SOC1 proteins.
(B) AP1 proteins. Species are Cc, globe artichoke; Sl, tomato; At, Arabidopsis; Ct, safflower; Gh, gerbera. Nodes annotated with 1,000-bootstrap values. Proteins from globe artichoke in bold italics.
To study the AP1/FUL subfamily in globe artichoke in more detail, proteins belonging to this subfamily, CcMADS23, CcMADS24, CcMADS28, CcMADS29, CcMADS42, CcMADS48, CcMADS49, and CcMADS70, were clustered with AP1 homologs from Arabidopsis, tomato, lettuce, and safflower (Figure 4B). The globe artichoke proteins, ranging from 74 aa to 205 aa (Figure S4) are all shorter than AP1 from Arabidopsis (256 aa) or LsAP1 from lettuce (253 aa),41 albeit short proteins in the AP1 family are encountered in the lettuce as well.35 The number of exons in the globe artichoke AP1 genes ranged from one exon (CcMADS24, CcMADS29, and CcMADS49) to nine exons (CcMADS23), compared to the eight exons in AtAP1. CcMADS70 (V2_13g015610.1) was annotated with seven exons, although we identified an eighth-exon for this gene, leading to “CcMADS70_corrected,” coding a 211 aa protein with high sequence similarity to LsAP141 (Figure S4B). With respect to protein domains, only CcMADS12 contained significant SRF and K-domains, whereas the remainder had only one of the two domains. The euAPETALA1 (euAP1) motif60,61 was identified in CcMADS70_corrected, supporting an AP1 identity for this gene (Figure S4B), which was hence named “CcAP1.” The euFUL and AGL79 lineages could not be clearly distinguished from each other in the phylogenetic study. With respect to motifs, complete C-terminal euFUL motifs were only identified in CcMADS23 and CcMADS42 and absent in the remainder proteins. Taken together, these results indicate that there are two FUL-like genes in globe artichoke that code the correct motif although without complete MADS and K-domains. CcMADS42 was designated “CcFUL like-A” and CcMADS23 “CcFUL like-B.”
CcSOC1b and CcFUL like-B genes are molecular markers for floral transition in artichoke
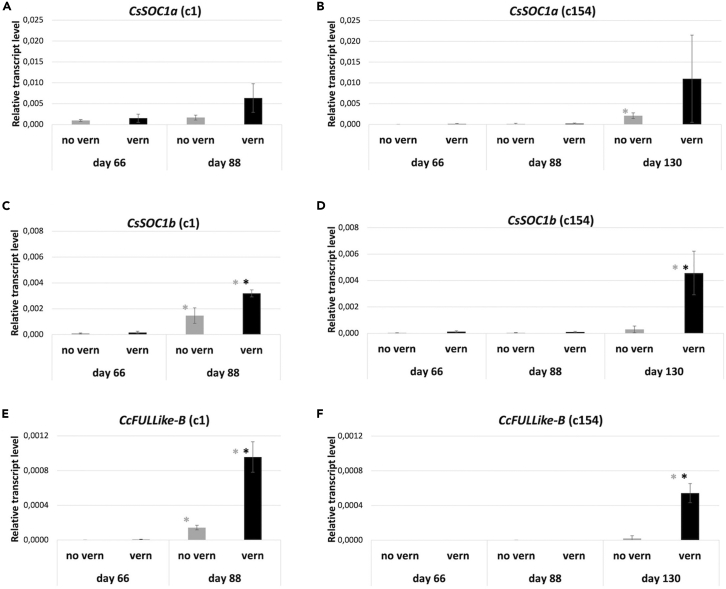
After this analysis we decided to use quantitative real-time PCR to assess the usefulness of the CcSOC1a, CcSOC1b, and CcFUL like-B MADS box genes as markers for the process of transition to the reproductive phase in globe artichoke. We again used plants from early-bolting genotype c1, and late-bolting one c154, grown under vernalization and no-vernalization conditions. Shoot apices were sampled at three different time points, before floral transition (day 67 after transplanting), day 88 after transplanting (average time for floral transition in non-vernalized c1 plants [Table 2]), and for c154 at a third time point at day 130 post transplanting (average time for floral transition in non-vernalized c154 plants [Table 2]) (Figure 5).
Figure 5.
Expression of CcSOC1a, CcSOC1b, and CcFUL like-B genes between early- and late-bolting genotypes
Expression level for CcSOC1a (A and B), CcSOC1b (C and D), and CcFUL like-B (E and F) genes was determined in samples taken before the floral transition (day 66) and at pre-bolting stage 1 in both early-bolting genotype c1 (day 88) (A, C, and E) and late-bolting genotype c154 (B, D, and F) (day 130). Analysis was performed on plants that have gone through vernalization (black bars) or not (gray bars). Expression level was represented as the 2-ΔCt average of three biological samples. Error bars represent the standard deviations of the three biological samples. Black asterisks indicate significant differences (p ≤ 0.05) between treatments. Gray asterisks indicate significant differences (p ≤ 0.05) with respect to the previous time point. Genotype c1 was not sampled at day 130.
In agreement with the inferred roles for these genes, expression of CcSOC1a, CcSOC1b, and CcFUL like-B was very low or absent in apices in pre-bolting stage 0 before the floral transition (day 67), independent of growing conditions (Figures 5A–5F). At 88 days, when the floral transition had started in genotype c1 under both vernalization and no-vernalization conditions, we detected a clear up-regulation of CcSOC1b and CcFUL like-B genes with respect to the values of day 67, being stronger in the plants that were in vernalization conditions (Figures 5C and 5E), while the CcSOC1a expression did not change. On the other hand, at this same time point, no change in gene expression was detected in genotype c154 for none of the three tested genes, independent of growing conditions (Figures 5B, 5D, and 5F). At 130 days, expression of CcSOC1b and CcFUL like-B genes was clearly detected in shoot apices of vernalized c154 plants, whereas expression did not change with respect to the previous measurements of day 67 and 88 in the non-vernalized ones (Figures 5D and 5F). Again, no clear changes in expression of CcSOC1a in the genotype c154 at day 130 were observed. These results confirm that CcSOC1b and CcFUL like-B are strongly up-regulated during the floral transition in globe artichoke, being even more evident in the late flowering genotype c154. This indicates that CcSOC1b and CcFUL like-B up-regulation and probably the floral transition are accelerated by the vernalization treatment in both early- and late-bolting genotypes. Finally, our results also support the use of CcSOC1b and CcFUL like-B genes as expression markers for early transition to bolting in globe artichoke.
Discussion
Earliness is a key trait for globe artichoke breeders. Despite multiple studies having addressed the timing of flowering in model plant species, reports on the genetic architecture of this trait in crop species remain relatively scarce. Among other factors, this can likely be attributed to the complexity of growing and handling plants of large size, such as globe artichoke, in controlled environmental conditions. Some studies have proposed a relevant role for vernalization as a determinant of bolting time in different genotypes.45,51 However, conclusions from these studies require assessment because of non-controlled conditions, i.e., comparing genotypes grown in different time frames and climate zones or at different latitudes. Thus, most experiments have been performed in the field under conditions in which the individual contributions of vernalization and photoperiod cannot easily be assayed as separate variables.46 In other studies, an artificial vernalization treatment was applied exclusively during an early developmental phase, after which plants were grown and observed under field conditions.52,53,54 In such cases, it is not possible to rule out the possible contribution of other factors, such as devernalization, referring to a possible reversal of the vernalized state after prolonged exposure to temperatures above a certain threshold.62
In this study we aimed to answer the question of whether bolting in globe artichoke is controlled by vernalization, and how cold exposure under natural growth conditions affects development of different artichoke genotypes selected by their early- or late-bolting phenotypes. To our knowledge, this is the first time that an experiment has been performed in which globe artichoke plants have been observed under controlled vernalization temperatures during their whole life cycle. This enabled the study of bolting time between vernalization and no-vernalization treatments under comparable conditions. Moreover, our setting allows vernalization to follow a natural cycle, making the results and experimental design applicable for breeding programs as well.
In addition to observing bolting visually in the field, we characterized morphological changes in the apex that are associated with the vegetative-to-reproductive phase transition and linked this transition to the expression of homologs of key flowering regulators SOC1 and FUL. The results of our vernalization experiments indicate that early-bolting genotypes have a significantly lower vernalization response than late-bolting genotypes, suggesting that a reduced requirement for vernalization is a relevant component of earliness in globe artichoke.
Vernalization response is an important determinant of bolting time in globe artichoke
Regarding the effect of vernalization, we found that the 698–935 h below 10°C advanced the time of bolting on average by 10.0 and 23.5 days for early- and late-bolting genotypes, respectively. To our knowledge, this is the first study that allows direct comparison on the vernalization response of early- and late-bolting genotypes. However, previous reports have identified general trends influencing this response in globe artichoke. Artificial vernalization prior to transplanting, equivalating to about 500 chilling hours (hours < 10°C) reduced the time to bolting by 11 days in the late-bolting variety “Orlando F1” but had no significant effect on three other commercial hybrids.63 Therefore, in that study, the effect of the artificial vernalization treatment on bolting time is lower than the 23.5 days we found for late-bolting genotypes. Another study on the effect of artificially vernalizing varieties prior to transplanting has been described by Rangarajan et al. (2000),52 who reported a significant effect on early yield after treating plantlets for about 450 h at 13°C in combination with late planting in spring, which meant that little to no natural vernalization was present. The effect was larger for the late-bolting variety “Green Globe Improved” than for the early-bolting variety “Imperial Star.” Another study by García and Cointry (2010)64 failed to find an effect of vernalizing seeds, seedlings, and plantlets of the early-bolting variety “Imperial Star” for 240 h at 3°C, perhaps because of a lack of vernalization requirement in this variety.
The results from these studies are in line with our results insofar that the effect of vernalization is largely genotype dependent. Although complete control of environmental conditions was out of the scope of this study, the experimental design that we developed addresses some of the challenges associated with field experiments or artificial vernalization, allowing for direct comparisons between genotypes of the effect of vernalization on bolting time. Moreover, the use of a heated net house with plastic insulation potentially offers an effective and affordable way for breeders of large-sized field crops to apply selection pressure against vernalization requirement in their programs.
Vernalization requirement could be necessary for both floral transition and bolting in globe artichoke
Vernalization has been reported to be a major factor determining flowering time in globe artichoke.47,51 Similarly, genotypes with an early-bolting phenotype could reflect the absence of a vernalization requirement.45,46 In species like Arabidopsis, vernalization is a gradual and quantitative process that prepares the plant to respond to other cues (environmental or endogenous) that promote the floral transition.9,14 In our experiments, cold depended on the prevailing weather in each season, which dictated not only the amount of accumulated chilling hours but also when cold started to be perceived by the plants. Despite observing a clear vernalization response in terms of bolting in all the genotypes tested, probably we are not detecting a saturated vernalization response. Interestingly, when we monitor the floral transition by morphological changes in the SAM, we observed that early-bolting genotype c1 initiates the floral transition independently of vernalization whereas the same transition is significantly advanced in late-bolting genotype c154 under vernalization conditions. This observation suggests that early-bolting genotypes do not require vernalization to initiate the floral transition but that somehow low temperatures are able to modulate bolting, as differences in bolting time were observed for this genotype. At the same time, our analysis supports the view that vernalizing temperatures can advance the floral transition in late-bolting genotypes, as well as modulate bolting progression. As commented, our microscopic observations indicate that early-bolting genotypes, or at least the c1 genotype, initiate the floral transition independently of chilling temperatures. Interestingly, at the molecular level, we clearly detected a stronger activation of homologs of floral integrators CcSOC1b and CcFUL like-B, in the vernalized c1 plants than in non-vernalized ones at the floral transition/pre-bolting stage 1, suggesting an early floral transition or a stronger activation of those genes, and hence vernalization response.
Effect of photoperiod on bolting
When DTB is compared between genotypes and seasons, it is apparent that late-bolting genotypes c20, c154, and CARI, under vernalization conditions, bolt within a relatively narrow time frame of 161–172 days in each season. Contrarily, when observed in no-vernalization conditions, DTB is more variable in these genotypes with a range of 177–211 days (Table 1). This suggests that vernalized plants react more uniformly to a stable bolting cue, such as photoperiod or age, while non-vernalized plants present a later and more variable response to those possible cues. In terms of photoperiod, the daily light hours measured for vernalized late-bolting genotypes were around 11.5–11.9 h at the moment of bolting. This may support the hypothesis that late-bolting genotypes are long-day plants. However, early-bolting genotypes c1 and c70, independently of the vernalization treatment, presented mean DTB values that varied 13–15 days between seasons (Table 1) and with an average of 9.6 daily light hours, rendering the early-bolting genotypes short-day plants. These observations are in line with reports that suggest critical photoperiod in globe artichoke being genotype dependent,45,46 hence putting emphasis on the genotype dimension when dealing with this trait. Our morphological analyses of the apex around the moment of the floral transition suggested that, at least for the early- and late-bolting genotypes tested, the floral transition took place in short-day conditions. These observations render globe artichoke a short-day plant in terms of floral transition, despite bolting occurring in both long- and short-day conditions. Interestingly it has been shown that the artificial extension of the day length in globe artichoke also accelerates bolting.42 Thus, we could speculate that globe artichokes are short-day plants that initiate the floral transition in response to vernalization. Once floral transition happens, inflorescence development and bolting could be promoted by cold temperatures as well as by the natural day length extension associated with the change of season. To determine the putative contribution of each factor in inflorescence development and bolting, additional experiment would be required.
No indications for devernalization or epigenetic memory of the vernalized state
In globe artichoke there have been mentions of the concept of devernalization as a heat-induced reversal of the vernalized state. Critical temperatures for this to occur, however, appear to vary widely according to different reports, with mentions being >18.3°C (65°F),62 >26°C,51 >32°C,52 and >33°C.63 If temperatures above 28°C were considered as a putative measure for devernalization, these conditions were met for 170–240 h in both the vernalization and non-vernalization compartments during our experiments. Most of these hours were, however, accumulated in the first month of the experiment, when vernalizing temperatures still had to occur, and in the last month, when most plants had already bolted.
Four of the varieties in this study were clones, and this might raise the question of whether an epigenetic memory of vernalization65 could have existed that explains the earliness in early-bolting clonal varieties. According to our results, this appears not to be the case. Both early- and late-bolting clones bolted later under the no-vernalization treatment, rendering a fully vernalized state prior to the experiments unlikely. Likewise, plants that have been successfully vernalized in the first season do not bolt earlier after resprouting in the next season and require being vernalized again.3,66
Morphological and molecular markers for the vegetative-to-reproductive phase transition
Different scales that describe phenological or developmental stages in artichoke have been described. A macroscopic 8-stage scale of the apex that spans the vegetative-to-reproductive phase change till anthesis of peripheral flowers was described by Foury (1967).5,8 Another scale with 15 stages by Baggio et al. (2011)7 is specifically tailored to the development of the head from the moment it reaches about 1.3 cm in size until the end of the reproductive phase. A scale describing phenological stages in Cyncara cardunculus according to the Biologische Bundesanstalt, Bundessortenamt und Chemische Industrie scale has been developed as well.6 None of the scales specifically conveys the changes in the vegetative or inflorescence apex that occur between the vegetative-to-reproductive phase transition and the start of bolting, when the primary inflorescence becomes visible in the rosette. We addressed this by the addition of five pre-bolting stages to the scale originally published by Foury (1967).5
The morphological markers developed in this study allow us to place the beginning of the vegetative-to-reproductive phase transition in globe artichoke, either just before or during pre-bolting stage 1. This is the stage where SAM doming occurs, which is also an indicator of the vegetative-to-reproductive phase transition in other plant species.67 In addition, we observed that pre-bolting stage 1 coincides with an up-regulation of expression of CcSOC1b and CcFUL like-B, globe artichoke homologs of SOC1 and FUL. This further supports the placement of the vegetative-to-reproductive phase transition at the start of pre-bolting stage 1. The connection between doming and the vegetative-to-reproductive phase transition, however, may not be universal for Asteraceae, and instead in lettuce this transition has been placed after doming and at the start of the elongation phase.68
MADS box genes in globe artichoke
Phylogenetic analysis allowed us to identify 82 putative MADS box genes in the artichoke genome and determine as well putative orthologs of genes in this family that are key elements in the control of flowering time in other plant species. In that regard, we have focused on the identification of FUL and SOC1 orthologs. FUL is known to act redundantly with SOC1 in promoting flowering, increasing their expression levels at the time of vegetative-to-reproductive phase transition in Arabidopsis.69,70 This is in line with the marked increase in expression levels observed for CcSOC1b and CcFUL like-B around the estimated time of floral transition in globe artichoke. SOC1 homologs with increased expression around the time of vegetative-to-reproductive transition have also been reported for other Asteraceae species such as chrysanthemum39,71 and lettuce.41
Research on the role of FUL genes in Asteraceae is limited. Ectopic expression of two homologs of FUL from Chrysanthemum morifolium in tobacco led to early flowering.72 Overexpression of FUL ortholog GSQUA2 from gerbera led to accelerated flowering in the same species.73 These results suggest that the function of FUL orthologs as flowering promotors is conserved in Asteraceae.
FLC is a central gene in the control of vernalization in Arabidopsis and other Brassicaceae.74,75 Our phylogenetic study revealed that the FLC subfamily in globe artichoke contains two members (Table S4). Of the two, CcMADS65 is most likely a closer homolog of Arabidopsis FLM and FLC genes than CcMADS10, although both have low protein similarity to FLC from Arabidopsis (30.3% and 25.7%, respectively). Furthermore, there exist few reports of FLC orthologs in Asteraceae. An MADS box gene in chicory (Cichorium intybus), with high protein sequence similarity to FLC from Arabidopsis, is repressed by vernalization and acts as a flowering repressor.32 Puglia et al., 2016,76 report on the isolation of the partial sequence of ccMFL, an FLC homolog from cultivated cardoon that was, however, not further characterized. It is possible that FLC genes have been subject to neofunctionalization in Asteraceae, as suggested by a recent study of MIKCC genes in Chrysanthemum lavandulifolium, where FLC genes might rather be involved in flower or capitulum development.77 The absence of a clear homolog of Arabidopsis FLC in globe artichoke could mean that vernalization requirement and response in this species are underlain by a different genetic architecture.
Since earliness is a key breeding target for commercial hybrids, gaining an understanding of the genetic architecture of this trait supports the development of tools for efficient selection and introgression into the most market-relevant elite genetic backgrounds. With the advent of affordable mass sequencing and data analysis, characterization of transcriptome changes in response to vernalization is a promising approach toward identification of genes involved in earliness. Using this approach would allow the identification of additional molecular markers for the floral transition and/or earliness, such as the ones developed in this study, that could be helpful tools in globe artichoke breeding programs.
Limitations of the study
The genotypes that were used for this study were selected from a commercial breeding program in South-Eastern Spain, meaning that they represent a gene pool that is optimized for the prevailing climate and cultivation cycle in that region. For this reason, we had to design the experiments to match these conditions, leaving other varieties and cultivation practices out of scope. We do believe that the results from this study are representative for globe artichoke in general, predominantly because of the wide genetic diversity included in the germplasm under study. We do, however, acknowledge the possibility of different genetic architectures yet to be discovered existing in the species, leaving open the possibility to make gains in earliness under different cultivation cycles and practices. Moreover, we did not consider differences in developmental speed that occur between the onset of bolting and the moment of harvest.
Another limitation of our study was that only minimum temperatures were controlled, but not mean or maximum temperatures. This is due to the impracticality of raising large plants such as globe artichoke in the fully controlled environment of a phytotron. Since the net house compartment in which the non-vernalization treatment was applied required the construction of plastic walls for the heating to be effective, control of daytime temperature was limited to manually raising or lowering the internal and/or outward-facing compartment walls to manage air exchange. This made temperature control challenging under high irradiance, high outside temperature, and low wind conditions and might have induced stress in the non-vernalized plants that could not be accounted for. A final limitation that we like to mention is the destructive nature of sampling shoot apices, which precluded multiple observations of gene expression in individual plants.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Vicente Balanzà (vbalanza@ibmcp.upv.es).
Materials availability
This study did not generate new or unique reagents.
Data and code availability
-
•
This paper does not report original code.
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
The authors like to thank BASF|Nunhems for the support and facilities that made the experiments possible. In particular, the artichoke breeding team, the Statistics team, and external contactors have been crucial to this work. Michael Dobres (Head of Breeding EMEA II at BASF|Nunhems at the time) and Peter Visser (R&D Crop Lead for leafies, artichoke, okra, celeriac at BASF|Nunhems) facilitated the collaboration project between BASF|Nunhems and CSIC-IBMCP. This project was financially supported by BASF|Nunhems.
Author contributions
Conceptualization and methodology: P.V., F.M., V.B., R. Benlloch, and R. Berentsen. Investigation: R. Berentsen, R. Benlloch, and V.B. Writing – original draft: R. Berentsen. Writing – review and editing: R. Berentsen, V.B., and F.M. Funding acquisition: F.M. and P.V. Supervision, F.M., V.B., and R. Benlloch.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Early- and late- bolting globe artichoke genotypes | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RNAlater™ solution | Thermo Fisher Scientific | Cat#AM7021 |
| EZNA® Plant RNA kit | Omega Bio-tek | Cat# R6827-02 |
| RNase-free DNase I Set | Omega Bio-tek | Cat# E1091-02 |
| SuperScript IV First-Strand Synthesis System | Thermo Fisher Scientific | Cat#18091050 |
| Oligonucleotides | ||
| qPCR primers | This paper | N/A |
| Software and algorithms | ||
| R-Studio | Posit PBC | https://posit.co/downloads/ |
| R-package: Phangorn v2.10 | Schliep et al.78 | https://cran.r-project.org/web/packages/phangorn/index.html |
| R-package: ggtree | Yu et al.79 | https://www.bioconductor.org/packages/release/bioc/html/ggtree.html |
| CLC Genomics Workbench v21.0.3 | Qiagen | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-clc-genomics-workbench/ |
| Other | ||
| data loggers | Onset Computer Corp | Cat# MX2302A |
| solar radiation shields | Onset Computer Corp | Cat# RS-3B |
Experimental model and study participant details
Plant material
A total of seven genotypes were selected from the BASF Vegetable Seeds artichoke breeding program. These constituted two early-bolting clones, “c1” and “c70”, two late-bolting clones, “c20” and “c154”, two early-bolting lines “VER” and “VESB” and one late-bolting line “CARI”. For microscopy, samples from the commercial hybrid ‘Green Queen’ were taken when certain pre-bolting stages were insufficient. Clones were produced at the BASF|Nunhems Cell Biology Services lab in Haelen, the Netherlands, according to internal standard protocol. Care was taken to avoid temperatures <13°C during production and transport. Clones were delivered at the 3–4 leaf stage between 20 August and 31 August in the years 2019, 2020 and 2021. Lines were sown in the third week of July in a commercial nursery and allowed to germinate for seven days at 16°C. Both clones and lines were kept in pots under shade cloth for 1–2 weeks between delivery and the start of the experiments at the 4–5 leaf stage.
Method details
Experimental design and treatment
The experiments took place in a net house in vicinity of Águilas, Spain (230 m MSL). The limestone soil was solarized, tilled, and fertilized, with compost prior to transplanting, which took place on 25 September 2019, 25 September 2020 and 01 October 2021. Plants were assigned to plots of 10 individuals each, which were subsequently randomized, with experimental groups containing 30–70 individuals in accordance with availability. Planting distance was 0.8 m within the row and 1.5 m between the rows. A border row made up of commercial hybrids surrounded the perimeter (Figure S1).
Along the external wall of the net house, two compartments were created. In one compartment minimum temperatures were controlled by a thermostat-controlled oil heater set to 13°C. To allow the system to heat effectively, a double ceiling, an external wall, and internal walls were constructed from thin transparent plastic. The internal and external walls were raised in the morning by the grower and lowered in the late afternoon or early evening to allow for ventilation and to moderate daytime temperatures. The other compartment was identical, but with no heating system, double ceiling nor outward facing plastic wall, allowing circulation of cool air during the night. The temperature in each compartment was monitored with a HOBO MX2302A data logger with RS-3B solar radiation shields (both from Onset Computer Corp, Bourne, USA) mounted in the center of the compartment at a height of 1.5 m. Daylength was calculated with the R-package chillR80 for a latitude of 37.49°N.
Macroscopic characterization
Plants were observed weekly for symptoms of bolting according to the developmental scale we designed (Table S1). Plants with multiple rosettes or suspected disease symptoms were removed from the experiment. Bolting observation campaigns lasted from 2019 to 05–10 until 2020-05-08 (150 days) during the 2019–2020 season, from 2020 to 11–30 till 2021-05-19 (170 days) and from 2021 to 12–13 till 2022-04-20 (128 days). The end dates for the bolting observation campaigns coincided with rising temperatures in the net house becoming unconducive to plant growth and development. Plants were considered to have bolted upon reaching bolting stage B and the number of days-to-bolting (DTB) was calculated for statistical analysis.
Tissue fixation, sectioning and staining
At predetermined intervals, individuals randomly selected from genotypes were dissected and their apices were scored according to the developmental scale (Figure 1; Table S1). During the 2021–2022 season, apices were preserved for microscopy by fixation for 24h in a buffer consisting of 50% (v/v) ethanol, 10% formaldehyde and 5% (v/v) acetic acid, followed by dehydration in ethanol and storage at 4°C. For microscopy, the samples were infiltrated and embedded in paraffin and cut with a microtome to 8 μm sections. Toluidine staining was performed according to the protocol described by.81 Microscopy was performed using a Leica 5000 microscope (Leica, Germany).
Identification of MADS box genes in globe artichoke
Protein sequences from MADS box genes were obtained for Arabidopsis (The Arabidopsis Information Resource82 and Parenicova et al., 200383), tomato (Sol Genomics Network84), and lettuce.35 For lettuce, an additional AP1 homologue was included from Fukuda et al., 2017.41 Arabidopsis, lettuce and tomato MADS box protein sequences were BLASTed against the 28.632 predicted proteins in globe artichoke57 using CLC Genomics Workbench v21.0.3 (QIAGEN, Aarhus, Denmark) with default settings, an E-value cutoff of 10−5, and no masking of low-complexity regions. The resulting list was subsequently filtered for unique sequences. In the resulting 82 hits, Pfam SRF (PF00319.21) and K-box (PF01486.20) domains were predicted by a Hidden Markov model using HMMER 3.1b1. Protein sequences were aligned with MUSCLE.85 Maximum likelihood (ML) phylogenies were inferred with the Phangorn v2.1078 package from the aforementioned protein sequences, to which additional protein sequences were added from the Asteraceae safflower37 and gerbera.71 The optimal model was selected with modelTest, followed by 1000 iterations of Nearest Neighbor Interchange (NNI) bootstraps. Phylograms were constructed and annotated with ggtree.79 To ensure legibility of the resulting dendrograms, each was optimized manually to reflect the resolved phylogeny with the lowest number of different species. These were globe artichoke and Arabidopsis (Type I MADS-box genes, Figure S3A), globe artichoke, Aradibopsis, safflower, and tomato (Type II MADS-box genes, Figure S3B), Arabidopsis, tomato, safflower, and gerbera (SOC1, Figure 4A), Arbidopsis, tomato, safflower, gerbera, and lettuce (FUL, Figure 4B).
RNA sampling and Q-PCR analysis
SAMs dissected in the net house were immediately transferred to 300 μL RNAlater solution (Thermo Fisher Scientific, USA) for storage at −18°C. RNA extraction was performed with the EZNA Plant RNA kit (Omega Bio-tek, USA) with on-column DNAse treatment with an RNase-free DNase I Set (Omega BioTek, USA) and converted to cDNA with a SuperScript IV First-Strand Synthesis System (Thermo Fisher, USA). Primers were designed for CcSOC1a, CcSOC1b, CcAP1 and CcFUL like-A and CcFUL like-B with CLC Genomics Workbench v21.0.3 (QIAGEN, Aarhus, Denmark) (Table S5). Amplicons were checked with high resolution melting prior to qPCR on a magnetic induction cycler (Mic) qPCR system (Bio Molecular Systems, Australia). Relative expression for three biological replicates was calculated according to the method. From CcAP1 and CcFUL like-A no pure amplicon could be obtained precluding quantification of these two genes. CcElf1α was used as reference gene.86
Quantification and statistical analysis
Statistical analysis
Bolting data were modeled using a linear mixed model with the main effects (genotype, treatment and season) and the interaction terms (genotype: treatment and treatment:season) set as fixed effects. The term “plot” was set as a random factor and nested within “season” to account for the between plot variation within each season. The estimated means were extracted from the model for each “genotype:treatment” combination, and the significance of differences between best linear unbiased estimators (BLUEs) was assessed by calculating the LSD and determining whether the difference between BLUEs was larger or smaller than the value of the LSD (6.86 days). In an additional modeling to determine the effect of the treatment on early- versus late-bolting genotypes, the explanatory variable “genotype” was substituted with “earliness”, a dichotomous variable distinguishing early (genotypes c1, c70, VESB and VER) from late (genotypes c20, c154 and CARI). DTpb1values for genotypes c1 and c154 were analyzed with a one-sided Student’s t-test and qPCR results with a two-sided t-test, both with α = 0.05.
Floral transition data were analyzed with a two samples t-test, one tail, considering p-value<0.05 as significant.
Published: August 27, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110829.
Contributor Information
Francisco Madueño, Email: madueno@ibmcp.upv.es.
Vicente Balanzà, Email: vbalanza@ibmcp.upv.es.
Supplemental information
References
- 1.Welbaum G.E., Warfield S.C. Growing globe artichokes from seed. Acta Hortic. 1992;318:111–116. doi: 10.17660/ActaHortic.1992.318.13. [DOI] [Google Scholar]
- 2.Mauromicale G., Ierna A. Effects of gibberellic acid and sowing date on harvest time and yields of seed-grown globe artichoke (Cynara scolymus L) Agronomie. 1995;15:527–538. doi: 10.1051/agro:19950902. [DOI] [Google Scholar]
- 3.Macua J., Lahoz I., Bozal J. Evolution in Seed-Propagated Artichoke Growing in Cold Zones of Spain. Acta Hortic. 2012;942:331–336. doi: 10.17660/ActaHortic.2012.942.48. [DOI] [Google Scholar]
- 4.Baixauli C., Giner A., Aguilar J., Nájera I., Maroto J., Pascual B., Torres J., López-Galarza S., San Bautista A. Respuesta productiva y agronómica de diferentes cvs de alcachofa multiplicados por semilla. Agrícola vergel: Fruticultura, horticultura, floricultura. 2014;33:184–188. [Google Scholar]
- 5.Foury C. Étude de la biologie florale de l'artichaut (Cynara scolymus L.); application à la sélection (1ere partie) Ann. Amelior. Plant. (Paris) 1967;17:357–373. [Google Scholar]
- 6.Archontoulis S., Struik P., Vos J., Danalatos N. Phenological growth stages of Cynara cardunculus: codification and description according to the BBCH scale. Ann. Appl. Biol. 2010;156:253–270. doi: 10.1111/j.1744-7348.2009.00384.x. [DOI] [Google Scholar]
- 7.Baggio M., Palla F., Boscardin D., Mantovani N., Grando M., Augustin L., Suzin M., Donida B., Lombardo S., Mauromicale G. Floral Biology of Globe Artichoke (Cynara cardunculus var. scolymus) Acta Hortic. 2012;942:297–302. doi: 10.17660/ActaHortic.2012.942.42. [DOI] [Google Scholar]
- 8.Pesce G., Mauromicale G. In: The Globe Artichoke Genome. Compendium of Plant Genomes. Portis E., Acquadro A., Lanteri S., editors. Springer; Cham: 2019. Cynara Cardunculus L.: Historical and Economic Importance, Botanical Descriptions, Genetic Resources and Traditional Uses. [DOI] [Google Scholar]
- 9.Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61:1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- 10.Cho L.-H., Yoon J., An G. The control of flowering time by environmental factors. Plant J. 2017;90:708–719. doi: 10.1111/tpj.13461. [DOI] [PubMed] [Google Scholar]
- 11.Fudge J.B., Lee R.H., Laurie R.E., Mysore K.S., Wen J., Weller J.L., Macknight R.C. Medicago truncatula SOC1 genes are up-regulated by environmental cues that promote flowering. Front. Plant Sci. 2018;9:496. doi: 10.3389/fpls.2018.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weigmann M., Maurer A., Pham A. Barley yield formation under abiotic stress depends on the interplay between flowering time genes and environmental cues. Sci. Rep. 2019;9:6397. doi: 10.1038/s41598-019-42673-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huijser P., Schmid M. The control of developmental phase transitions in plants. Development. 2011;138:4117–4129. doi: 10.1242/dev.063511. [DOI] [PubMed] [Google Scholar]
- 14.Andrés F., Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 15.Trevaskis B., Hemming M.N., Dennis E.S., Peacock W.J. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007;12:352–357. doi: 10.1016/j.tplants.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Srikanth A., Schmid M. Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 2011;68:2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Putterill J., Robson F., Lee K., Simon R., Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 18.Kardialsky I., Shukla V., Ahn J., Dagenais N., Christensen S., Nguyen J., Chory J., Harrison M., Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 19.Lee H., Suh S.S., Park E., Cho E., Ahn J.H., Kim S.G., Lee J.S., Kwon Y.M., Lee I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14:2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michaels S.D., Himelblau E., Kim S.Y., Schomburg F.M., Amasino R.M. Integration of Flowering Signals in Winter-Annual Arabidopsis. Plant Physiol. 2005;137:149–156. doi: 10.1104/pp.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandel M.A., Yanofsky M.F. A gene triggering flower formation in Arabidopsis. Nature. 1995;377:522–524. doi: 10.1038/377522a0. [DOI] [PubMed] [Google Scholar]
- 22.Teper-Bamnolker P., Samach A. The Flowering Integrator FT Regulates SEPALLATA3 and FRUITFULL Accumulation in Arabidopsis Leaves. Plant Cell. 2005;17:2661–2675. doi: 10.1105/tpc.105.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helliwell C.A., Wood C.C., Robertson M., James Peacock W., Dennis E.S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang J.W., Czech B., Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Yu S., Galvão V.C., Zhang Y.C., Horrer D., Zhang T.Q., Hao Y.H., Feng Y.Q., Wang S., Schmid M., Wang J.W. Gibberellin Regulates the Arabidopsis Floral Transition through miR156-Targeted SQUAMOSA PROMOTER BINDING–LIKE Transcription Factors. Plant Cell. 2012;24:3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyun Y., Richter R., Vincent C., Martinez-Gallegos R., Porri A., Coupland G. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell. 2016;37:254–266. doi: 10.1016/j.devcel.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Bao S., Hue C., Shen L., Yu H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integrated Plant Biol. 2020;62:118–131. doi: 10.1111/jipb.12892. [DOI] [PubMed] [Google Scholar]
- 28.Ferrándiz C., Gu Q., Martienssen R., Yanofsky M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- 29.Melzer S., Lens F., Gennen J., Vanneste S., Rohde A., Beeckman T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 2008;40:1489–1492. doi: 10.1038/ng.253. [DOI] [PubMed] [Google Scholar]
- 30.Balanzá V., Martínez-Fernández I., Ferrándiz C. Sequential action of FRUITFULL as a modulator of the activity of the floral regulators SVP and SOC1. J. Exper. Bot. 2014;65:1193–1203. doi: 10.1093/jxb/ert482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruokolainen S., Ng Y.P., Albert V.A., Elomaa P., Teeri T.H. Over-expression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration. Ann. Bot. 2011;107:1491–1499. doi: 10.1093/aob/mcr112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Périlleux C., Pieltain A., Juacquemin G., Bouché F., Detry N., D'Aloia M., Thiry L., Aljochim P., Delansnay M., Mathiey A.-S., et al. A root chicory MADS box sequence and the Arabidopsis flowering repressor FLC share common features that suggest conserved function in vernalization and de-vernalization responses. Plant J. 2013;75:390–402. doi: 10.1111/tpj.12208. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Huang H., Ma Y., Fu J., Wang L., Dai S. Construction and de novo characterization of a transcriptome of Chrysanthemum lavandulifolium: analysis of gene expression patterns in floral bud emergence. Plant Cell Tissue Organ Cult. 2014;116:297–309. doi: 10.1007/s11240-013-0404-1. [DOI] [Google Scholar]
- 34.Leijten W., Koes R., Roobeek I., Frugis G. Translating Flowering Time from Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species. Plants. 2018;7:111. doi: 10.3390/plants7040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ning K., Han Y., Chen Z., Luo C., Wang S., Zhang W., Li L., Zhang X., Fan S., Wang Q. Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 2019;42:1868–1881. doi: 10.1111/pce.13523. [DOI] [PubMed] [Google Scholar]
- 36.Han R., Truco M.J., Lavelle D.O., Michelmore R.W. A composite analysis of flowering time regulation in lettuce. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Ge H., Ahmad N., Li J., Wang Y., Liu X., Liu W., Li X., Wang N., Wang F., Dong Y. Genome-Wide Identification of MADS-box Family Genes in Safflower (Carthamus tinctorius L.) and Functional Analysis of CtMADS24 during Flowering. Int. J. Mol. Sci. 2023;24:1026. doi: 10.3390/ijms24021026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu D., Kotilainen M., Mehto E., Elomaa P., Helariutta Y., Albert V., Teeri T. Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae) Plant J. 1999;17:51–62. doi: 10.1046/j.1365-313X.1999.00351.x. [DOI] [PubMed] [Google Scholar]
- 39.Li T., Niki T., Nishijima T., Douzono M., Koshioka M., Hisamatsu T. Roles of CmFL, CmAFL1, and CmSOC1 in the transition from vegetative to reproductive growth in Chrysanthemum morifolium Ramat. J. Hortic. Sci. Biotechnol. 2009;84:447–453. doi: 10.1080/14620316.2009.11512547. [DOI] [Google Scholar]
- 40.Blackman B.K., Rasmussen D.A., Strasburg J.L., Raduski A.R., Burke J.M., Knapp S.J., Michaels S.D., Rieseberg L.H. Contributions of Flowering Time Genes to Sunflower Domestication and Improvement. Genetics. 2011;187:271–287. doi: 10.1534/genetics.110.121327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda M., Yanai Y., Nakano Y., Sasaki H., Uragami A., Okada K. Isolation and Gene Expression Analysis of Flowering-related Genes in Lettuce (Lactuca sativa L.) Hort. J. 2017;86:340–348. doi: 10.2503/hortj.OKD-036. [DOI] [Google Scholar]
- 42.Ledda L., Mameli M.G., Milia M., Marras G.F. Influence of plant shading and ovoli typology on globe artichoke development, early production and head atrophy: preliminary results. Acta Hortic. 2004;660:365–371. doi: 10.17660/ActaHortic.2004.660.53. [DOI] [Google Scholar]
- 43.Lanteri S., Ledda L., Di Leo I., Mameli M.G., Portis E. Molecular and Morphological Variation among and within Populations of Cynara scolymus L. cv. ‘Spinoso sardo. Acta Hortic. 2005;681:333–340. doi: 10.17660/ActaHortic.2005.681.44. [DOI] [Google Scholar]
- 44.Fadda A., Virdis A., Barberis A., Ledda L., Melito S. Impact of different photoperiodic treatments on Spinoso Sardo globe artichoke (Cynara cardunculus L. var. scolymus Fiori) head traits and elementary composition. Acta Hortic. 2020;1284:131–136. doi: 10.17660/ActaHortic.2020.1284.17. [DOI] [Google Scholar]
- 45.Basnizki J., Zohary D. Plant Breeding Reviews; 1994. Breeding of Seed-Planted Artichoke; pp. 253–269. [DOI] [Google Scholar]
- 46.Virdis A., Motzo R., Giunta F. Key phenological events in globe artichoke (Cynara cardunculus var. scolymus) development. Ann. Appl. Biol. 2009;155:419–429. doi: 10.1111/j.1744-7348.2009.00354.x. [DOI] [Google Scholar]
- 47.Foury C., Pécaut P. Quelques aspects du developpement de l'artichaut (Cynara scolymus L.) : Problemes poses par la substitution de la reproduction sexuee a la multiplication vegetative. C. R. Acad. Agric. Fr. 1988;74:85–92. [Google Scholar]
- 48.Snyder M.J., Welch N.C., Rubatzky V.E. Influence of Gibberellin on Time of Bud Development in Globe Artichoke. Hortscience. 1971;6:484–485. doi: 10.21273/HORTSCI.6.5.484. [DOI] [Google Scholar]
- 49.Calabrese N., De Palma E., Bianco V. Yield and Quality of New Commercial Seed Grown Artichoke Hybrids. Acta Hortic. 2004;660:77–82. doi: 10.17660/ActaHortic.2004.660.8. [DOI] [Google Scholar]
- 50.Maroto J. Effects of gibberellic acid (GA3) applications on globe artichoke production. Acta Hortic. 2007;730:137–142. doi: 10.17660/ActaHortic.2007.730.15. [DOI] [Google Scholar]
- 51.Welbaum G.E. Annual Culture of Globe Artichoke from Seed in Virginia. HortTechnology. 1994;4:147–150. doi: 10.21273/HORTTECH.4.2.147. [DOI] [Google Scholar]
- 52.Rangarajan A., Ingall B.A., Zeppelin V.C. Vernalization Strategies to Enhance Production of Annual Globe Artichoke. HortTechnology. 2000;10:585–588. doi: 10.21273/HORTTECH.10.3.585. [DOI] [Google Scholar]
- 53.García S., Cointry E., Firpo I., López-Anido F., Cravero V., Asprelli P. Vernalization of seed-grown artichoke. Acta Hortic. 2004;660:443–447. doi: 10.17660/ActaHortic.2004.660.66. [DOI] [Google Scholar]
- 54.Riahi J., Nicoletto C., Bouzaein G., Arfaoui K., Ghezal I., Sambo P., Kouki Khalfallah K. Effect of artificial vernalization on the production of Tunisian globe artichoke derived from nursery's ovoli: earliness, yield and quality traits. Acta Hortic. 2020;1284:101–108. doi: 10.17660/ActaHortic.2020.1284.13. [DOI] [Google Scholar]
- 55.Simpson G.G., Dean C. Arabidopsis, the Rosetta Stone of Flowering Time? Science. 2002;296:285–289. doi: 10.1126/science.296.5566.2. [DOI] [PubMed] [Google Scholar]
- 56.Parcy F. Flowering: a time for integration. Int. J. Dev. Biol. 2005;49:585–593. doi: 10.1387/ijdb.041930fp. [DOI] [PubMed] [Google Scholar]
- 57.Acquadro A., Portis E., Valentino D., Barchi L., Lanteri S. “Mind the Gap”: Hi-C Technology Boosts Contiguity of the Globe Artichoke Genome in Low-Recombination Regions. G3 (Bethesda). 2020;10:3557–3564. doi: 10.1534/g3.120.401446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alvarez-Buylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas de Pouplana L., Martínez-Castilla L., Yanofsky M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vandenbussche M., Theissen G., Van de Peer Y., Gerats T. Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 2003;31:4401–4409. doi: 10.1093/nar/gkg642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Litt A., Irish V.F. Duplication and Diversification in the APETALA1/FRUITFULL Floral Homeotic Gene Lineage: Implications for the Evolution of Floral Development. Genetics. 2003;165:821–833. doi: 10.1093/genetics/165.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shan H., Zhang N., Liu C., Xu G., Zhang J., Chen Z., Kong H. Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol. Phylogenet. Evol. 2007;44:26–41. doi: 10.1016/j.ympev.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 62.Gerakis P.A., Markarian D., Honma S. Vernalization of globe artichoke, Cynara scolymus L. J. Am. Soc. Hortic. Sci. 1969;94:254–258. doi: 10.21273/JASHS.94.3.254. [DOI] [Google Scholar]
- 63.Mauromicale G., Ierna A., Cavallaro V. Effects of vernalization and gibberellic acid on bolting, harvest time and yield of seed-grown Globe artichoke. Acta Hortic. 2005;681:243–250. doi: 10.17660/ActaHortic.2005.681.31. [DOI] [Google Scholar]
- 64.García S.M., Cointry E.L. Vernalization of seed and plantlets and development of globe artichoke. Int. J. Veg. Sci. 2010;16:184–190. doi: 10.1080/19315260903396885. [DOI] [Google Scholar]
- 65.Michaels S.D., Amasino R.M. Memories of winter: vernalization and the competence to flower. Plant Cell Environ. 2000;23:1145–1153. doi: 10.1046/j.1365-3040.2000.00643.x. [DOI] [Google Scholar]
- 66.Virdis A., Motzo R., Giunta F. The phenology of seed-propagated globe artichoke. Ann. Appl. Biol. 2014;164:128–137. doi: 10.1111/aab.12087. [DOI] [Google Scholar]
- 67.Tal L., Friedlander G., Gilboa N.S., Unger T., Gilad S., Eshed Y. Coordination of Meristem Doming and the Floral Transition by Late Termination, a Kelch Repeat Protein. Plant Cell. 2017;29:681–696. doi: 10.1105/tpc.17.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Z., Han Y., Ning K., Ding Y., Zhao W., Yan S., Luo C., Jiang X., Ge D., Liu R., et al. Inflorescence Development and the Role of LsFT in Regulating Bolting in Lettuce (Lactuca sativa L.) Front. Plant Sci. 2017;8:2248. doi: 10.3389/fpls.2017.02248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hempel F.D., Weigel D., Mandel M.A., Ditta G., Zambryski P.C., Feldman L.J., Yanofsky M.F. Floral determination and expression of floral regulatory genes in Arabidopsis. Development. 1997;124:3845–3853. doi: 10.1242/dev.124.19.3845. [DOI] [PubMed] [Google Scholar]
- 70.Borner R., Kampmann G., Chandler J., Gleißner R., Wisman E., Apel K., Melzer S. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 2000;24:591–599. doi: 10.1046/j.1365-313x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- 71.Fu J., Qi S., Yang L., Dai Y., Dai S. Characterization of Chrysanthemum ClSOC1-1 and ClSOC1-2, homologous genes of SOC1. Plant Mol. Biol. Report. 2014;32:740–749. doi: 10.1007/s11105-013-0679-8. [DOI] [Google Scholar]
- 72.Goloveshkina E.N., Shchennikova A.V., Kamionskaya A.M., Skryabin K.G., Shulga O.A. Influence of ectopic expression of Asteraceae MADS box genes on plant ontogeny in tobacco. Plant Cell Tissue Organ Cult. 2012;109:61–71. doi: 10.1007/s11240-011-0074-9. [DOI] [Google Scholar]
- 73.Ruokolainen S., Ng Y.P., Broholm S.K., Albert V.A., Elomaa P., Teeri T.H. Characterization of SQUAMOSA-like genes in Gerbera hybrida, including one involved in reproductive transition. BMC Plant Biol. 2010;10:128. doi: 10.1186/1471-2229-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sheldon C.C., Rouse D.T., Finnegan E.J., Peacock W.J., Dennis E.S. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC) Proc. Natl. Acad. Sci. USA. 2000;97:3753–3758. doi: 10.1073/pnas.97.7.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raman H., Raman R., Coombes N., Song J., Prangnell R., Bandaranayake C., Tahira R., Sundaramoorthi V., Killian A., Meng J., et al. Genome-wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 2016;39:1228–1239. doi: 10.1111/pce.12644. [DOI] [PubMed] [Google Scholar]
- 76.Puglia G., Raccuia S., Pappalardo H., Genovese C., Argento S., Melilli M. Characterization of a MADS Flowering Locus C - like (MFL) in Cynara cardunculus var. altilis under different sowing and planting density. Acta Hortic. 2016;1147:301–308. doi: 10.17660/ActaHortic.2016.1147.42. [DOI] [Google Scholar]
- 77.Li J., Zhang Q., Kong D., Pu Y., Wen X., Dai S. Genome-wide identification of the MIKCc-type MADS-box gene family in Chrysanthemum lavandulifolium reveals their roles in the capitulum development. Front. Plant Sci. 2023;14:1153490. doi: 10.3389/fpls.2023.1153490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schliep K.P. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinformatics. 2020;69:e96. doi: 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
- 80.Luedeling E., Fernández E. 2022. chillR: Statistical Methods for Phenology Analysis in Temperate Fruit Trees. [Google Scholar]
- 81.Balanzá V., Ballester P., Colombo M., Fourquin C., Martinez-Fernández I., Ferrándiz C. In: Riechmann J., Wellmer F., editors. Vol. 1110. Humana Press; 2014. Genetic and Phenotypic Analyses of Carpel Development in Arabidopsis. (Flower Development. Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 82.TAIR . Carnegie Institution; 2022. The Arabidopsis Information Resource.http://arabidopsis.org [Google Scholar]
- 83.Parenicova L., de Folter S., Kieffer M., Horner D., Favalli C., Busscher J., Cook H., Ingram R., Kater M., Davies B., Angenent G. Molecular and Phylogenetic Analyses of the Complete MADS-Box Transcription Factor Family in Arabidopsis: New Openings to the MADS World. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandez-Pozo N., Menda N., Edwards J.D., Saha S., Tecle I.Y., Strickler S.R., Bombarely A., Fisher-York T., Pujar A., Foerster H., et al. The Sol Genomics Network (SGN) - from genotype to phenotype to breeding. Nucleic Acids Res. 2015;43:D1036–D1041. doi: 10.1093/nar/gku1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sonnante G., D'Amore R., Blanco E., Pierri C.L., De Palma M., Luo J., Tucci M., Martin C. Novel hydroxycinnamoyl-coenzyme A quinate transferase genes from artichoke are involved in the synthesis of chlorogenic acid. Plant Physiol. 2010;153:1224–1238. doi: 10.1104/pp.109.150144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report original code.
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.