Summary
Levodopa-induced dyskinesia (LID) refers to involuntary motor movements of chronic use of levodopa in Parkinson’s disease (PD) that negatively impact the overall well-being of people with this disease. The molecular mechanisms involved in LID were investigated through whole-blood transcriptomic analysis for differential gene expression and identification of new co-expression and differential co-expression networks. We found six differentially expressed genes in patients with LID, and 13 in patients without LID. We also identified 12 co-expressed genes exclusive to LID, and six exclusive hub genes involved in 23 gene-gene interactions in patients with LID. Convergently, we identified novel genes associated with PD and LID that play roles in mitochondrial dysfunction, dysregulation of lipid metabolism, and neuroinflammation. We observed significant changes in disease progression, consistent with previous findings of maladaptive plastic changes in the basal ganglia leading to the development of LID, including a chronic pro-inflammatory state in the brain.
Subject areas: Clinical genetics, Molecular neuroscience, Clinical neuroscience
Graphical abstract
Highlights
-
•
Differential expression analysis revealed ten new transcripts altered in PD patients
-
•
Dyskinesia patients showed an exclusive gene co-expression network
-
•
Alterations in mitochondrial genes may contribute to the pro-inflammatory state in PD
-
•
Inflammatory pathways may contribute to the onset of levodopa-induced dyskinesia
Clinical genetics; Molecular neuroscience; Clinical neuroscience
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disease and the one among movement disorders, affecting 1–2% of the population over the age of 60. PD is the fastest-growing neurological condition and may become the primary neurodegenerative disease in the next two decades.1,2 PD is a complex and multisystemic disease, characterized by widespread alpha-synuclein aggregates, mitochondrial and lysosomal dysfunction, synaptic transport, and neuroinflammation, which lead to the death of neurons, especially dopaminergic.3,4 There are currently no specific diagnostic biomarkers to detect its early stages, and diagnosis occurs decades after the onset of neuronal degeneration.3
The available treatments for PD are effective in alleviating the symptoms, moreover, does not modify the neurodegenerative process. The main treatment with levodopa significantly improves motor symptoms of people with PD.5,6 Levodopa can cross the blood-brain barrier and restores the dopamine levels. Despite its effectiveness in the early stages of treatment, disease progression coupled with the need for increased levodopa dosage can lead to the development of levodopa-induced dyskinesia (LID), characterized by involuntary movements.5,7 LID can affect approximately 30–51.2% of people with PD after only five years of levodopa treatment, becoming more prevalent as the disease progresses, particularly in female individuals and those with a younger onset age of PD.5,8,9
Several known molecular mechanisms contribute to the development of LID, such as the dysregulation of dopamine storage and release control in the synaptic cleft, increased D1 receptors in post-synaptic neurons, release of dopamine by serotonergic and noradrenergic neurons as a replacement of the nigrostriatal degeneration, and greater spontaneous glutamate release, all of which lead to overstimulation of the direct pathway in the basal ganglia, resulting in further movement dysregulation.7,10,11,12,13 Shifts on gene expression at various levels may affect neurotransmitters, but there is still much to explore about the underlying molecular processes involved in the onset and pathophysiology of LID to seek better treatment strategies.5,10
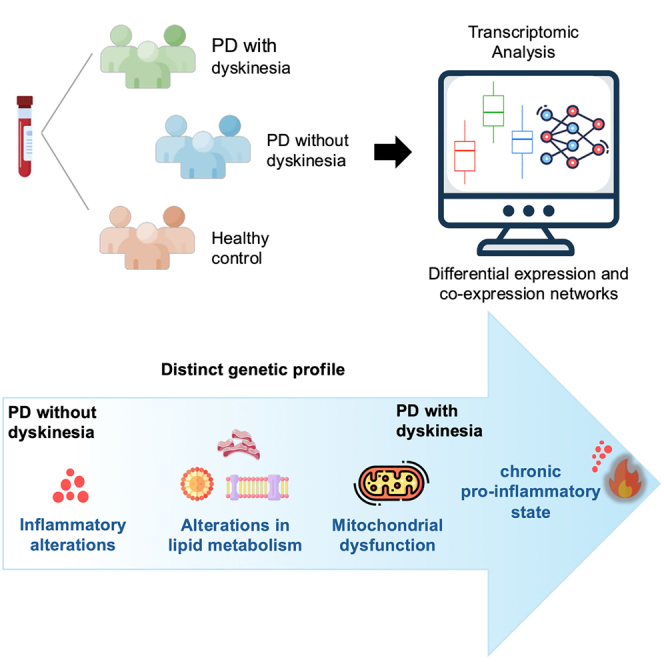
System biology approaches such as differentially expressed genes (DEGs) and differentially co-expressed gene network (DCGNs) in LID is still unexplored, that may help identifying altered gene-gene interactions between conditions.14 The present study aimed at investigating the gene expression, co-expression, and differential gene co-expression based on the transcriptome analysis of peripheral blood in controls (CT) and people with PD with LID (PD-D group) and absence of LID (PD-ND group).
Results
Epidemiological and clinical data
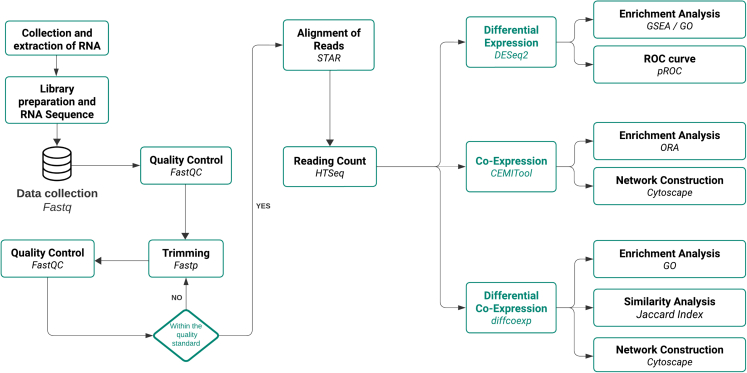
Our data analysis flowchart is depicted in Figure 1. A comprehensive analysis of clinical and epidemiological data revealed that the PD-D group exhibited lower age at disease onset, longer disease and levodopa use durations, and higher levodopa doses when compared to the PD-ND group. Sex and age at evaluation were matched between groups and the analysis is represented in the Table 1. (See also the Table S1; Figure S1 for general data of samples and RNA-sequencing).
Figure 1.
Flowchart for RNA-Seq data analysis to investigate differential expression, co-expression, and differential co-expression
Table 1.
Epidemiological and clinical data and inclusion/exclusion criteria of patients with PD and control group
| General Characteristics | CT (n = 20) | PD-ND (n = 11) | PD-D (n = 15) | p value |
|---|---|---|---|---|
| Sex (Male/Female) | 13/7 | 8/3 | 11/4 | 0.839a |
| Age at clinical evaluation | 59.50 (53–64) | 54 (49.5–60) | 53 (50–60) | 0.228c |
| Age at onset of PDb | – | 50 (46–55) | 41 (38.5–49) | 0.042d |
| Disease duration (years)b | – | 4 (2–7) | 10 (6–11.5) | 0.0006d |
| LEDD (mg/day)b | – | 550 (300–600) | 1200 (737.5–1562) | 0.001d |
| Dose of levodopa (mg/day)b | – | 300 (250–475) | 700 (450–1188) | 0.003d |
| Levodopa duration (years) | – | 3 (1–4.5) | 10 (6.5–12) | 0.0001d |
CT, control group; NA, not applied; PD-D, people with Parkinson’s disease and levodopa-induced dyskinesia; PD-ND, people with Parkinson’s disease without levodopa-induced dyskinesia.
Chi-squared test comparing frequencies between CT, PD-ND and PD-D groups.
Values in median (interquartile range).
Kruskal-Wallis test comparing medians between CT, PD-ND and PD-D groups.
Mann-Whitney test comparing medians between PD-ND and PD-D groups.
Differential gene expression reveals new blood biomarkers in levodopa-induced dyskinesia
We conducted individual comparative analysis of the PD-D and PD-ND groups among themselves and with the CT group. No DEG was observed between PD-D vs. PD-ND samples. When comparing each PD group individually with the CT, we found 13 DEGs in PD-ND and six DEGs in PD-D group. We conducted an exclusion analysis considering all the results and identified ten exclusive DEGs in PD-ND and three exclusive DEGs in PD-D. (DEGs are represented in Table 2; Figures 2A and 2B).
Table 2.
Characterization of differentially expressed genes between people with Parkinson’s disease and control subjects, according to the presence of levodopa-induced dyskinesia
| PD–ND vs. CT | ||||
|---|---|---|---|---|
| Gene | Log2 Fold-Change | p value | p. adjusted (FDR) | Regulation |
| MT-CYB | −0.74 | <0.001 | 0.005 | Downregulated |
| CWC27a | −0.77 | <0.001 | 0.018 | Downregulated |
| MT-ND5 | −0.85 | <0.001 | 0.005 | Downregulated |
| RAPH1a | −1,11 | <0.001 | 0.003 | Downregulated |
| HBA1 | −2,10 | <0.001 | 0.005 | Downregulated |
| MT-ND6 | −2,15 | <0.001 | 0.008 | Downregulated |
| MT-TYa | −2,38 | <0.001 | 0.040 | Downregulated |
| MT-TFa | −2,55 | <0.001 | 0.002 | Downregulated |
| MT-TQa | −2,62 | <0.001 | 0.012 | Downregulated |
| MT-TEa | −2,71 | <0.001 | 0.040 | Downregulated |
| HBA2 | −2,75 | <0.001 | <0.001 | Downregulated |
| HBBP1a | −3,89 | <0.001 | <0.001 | Downregulated |
| ENSG00000281383a | −4,23 | <0.001 | <0.001 | Downregulated |
| PD-D vs. CT | ||||
| BTNL8a | 0.98 | <0.001 | 0.005 | Upregulated |
| TK2 | 0.84 | <0.001 | 0.009 | Upregulated |
| ACAT1a | −0.65 | <0.001 | 0.045 | Downregulated |
| HBA2 | −1.84 | <0.001 | 0.011 | Downregulated |
| ENSG00000281383a | −2.66 | <0.001 | 0.003 | Downregulated |
| HBBP1a | −2.84 | <0.001 | 0.003 | Downregulated |
CT, control group; PD-D, people with Parkinson’s disease with levodopa-induced dyskinesia; PD-ND, people with Parkinson’s disease without levodopa-induced dyskinesia; FDR, false discovery rate.
New transcripts associated with Parkinson’s disease.
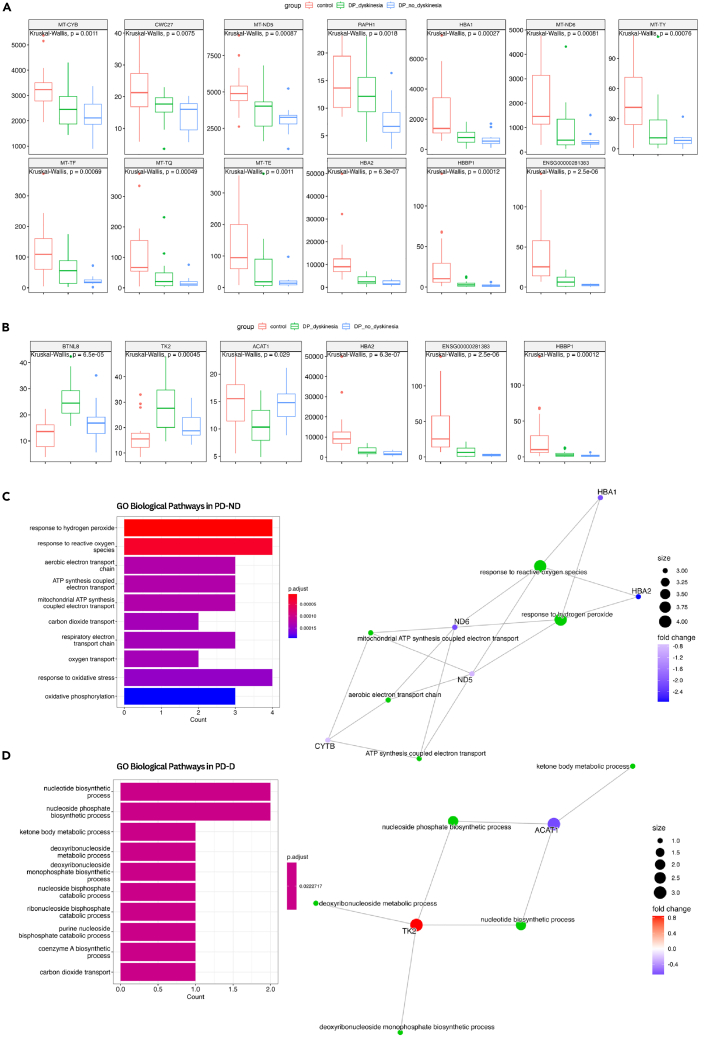
Figure 2.
Differentially expressed genes and enrichment analysis
(A) Boxplot of DEG in PD-ND.
(B) Boxplot of DEG in PD-D.
(C) GO for differentially expressed genes of PD-ND group, with enriched pathways (left) and representative genes for the main pathways (right).
(D) GO for differentially expressed genes of PD-D group, with enriched pathways (left) and representative genes for the main pathways (right). CT, control group; PD-D, people with Parkinson’s disease with levodopa-induced dyskinesia; PD-ND, people with Parkinson’s disease without levodopa-induced dyskinesia; DEG, differentially expressed genes; GO, Gene Ontology. p.adjust, p-value adjusted by FDR.
Gene Ontology analysis of the 6 DEGs between PD-D vs. CT primarily enriched for nucleotide biosynthesis and energy metabolism, highlighting the genes ACAT1 and TK2 (Figure 2C). When investigating the 13 DEGs between PD-ND vs. CT, the genes HBA1, HBA2, CYTB, ND5, and ND6 were related to gas transport, response to reactive oxygen species (ROS), and mitochondrial metabolism (Figure 2D).
Our research identified ten new transcripts associated with PD and two of these new transcripts (HBBP1 and ENSG00000281383) have not been thoroughly studied in terms of their function. We also accessed the potential of prediction of PD for these genes through the ROC curve. The HBA2, HBBP1, and ENSG00000281383 genes were commonly altered in both PD-ND and PD-D groups and exhibited an area under the curve (AUC) over 0.8 (Figure S2A).
The gene set enrichment analysis (GSEA) for PD-ND group revealed that the main exclusive enriched pathways for genes with high expression levels were DNA methylation, RNA polymerase I promoter opening, assembly of the ORC complex at the origin of replication, SIRT1 negatively regulates rRNA expression and HDMs demethylate histones. Pathways with lower enrichment score were RUNX1 regulates transcription of genes involved in differentiation of hematopoietic stem cells, DNA double-strand break repair, metabolism of carbohydrates, heme signaling, and binding and uptake of ligands by scavenger receptors (Figure S2B).
In PD-D group, GSEA revealed that the set of over-expressed genes was particularly associated with the fatty acid metabolism pathways, NCAM1 interactions, integrin cell surface interactions, extracellular matrix organization, and neutrophil degranulation. The main pathways enriched for the under-expressed genes were PD-1 signaling, mitochondrial translation, mitochondrial metabolism, RNA processing, and immune system. Details of these pathways and the involved genes are represented in Figure S2C.
Distinct gene co-expression patterns in levodopa-induced dyskinesia
Gene co-expression analysis was performed for 15,372 transcripts regarding CT, PD-ND, PD-D groups, and a pool of samples (CT + PD-ND + PD-D). Six modules of co-expressed genes were identified in the PD-D group and three modules in the PD-ND group. In the overall analysis by exclusion gene modules and pathways we identified 92 gene-gene interactions in PD-D group and 98 in PD-ND group (all results of co-expression analysis can be observed in the Table S2). The complete network of co-expressed genes in PD-ND and PD-D groups can be found in Figures S3A and S3B.
To focus on exclusive interactions, we systematically compared the interaction results across different groups (Figure S3C). Notably, 23 interactions involving 25 genes were presented in the PD-ND group and 12 exclusive interactions involving 12 genes were found in the PD-D group (Figures 3D and 3E).
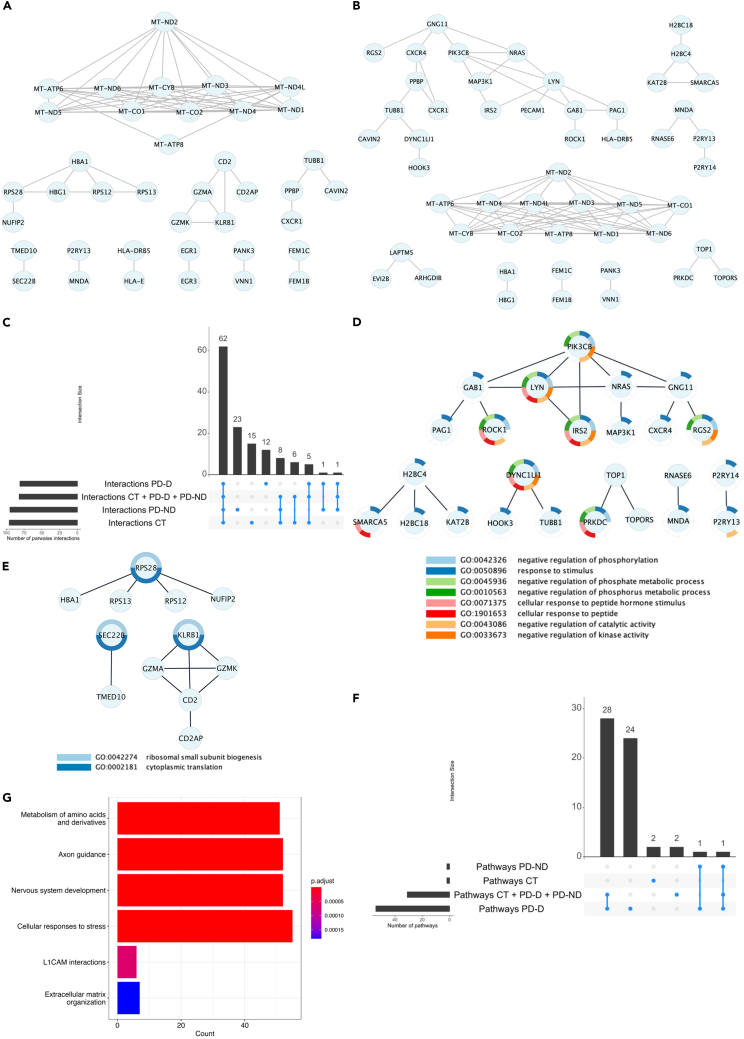
Figure 3.
Networks of co-expressed genes
(A) Complete Network of co-expressed genes in PD-ND.
(B) Complete Network of co-expressed genes in PD-D. Network of exclusive co-expressed genes in PD-ND group.
(C) Number of intersections of gene-gene interactions across all groups.
(D) Network of exclusive co-expressed genes in PD-ND group and biological process enriched to each gene.
(E) Network of exclusive co-expressed genes in PD-D group and biological process enriched to each gene.
(F) Number of intersections of enriched pathways for co-expressed genes.
(G) The main pathways enriched in the group of patients with dyskinesia. CT, control group; PD-D, people with Parkinson’s disease with levodopa-induced dyskinesia; PD-ND, people with Parkinson’s disease without levodopa-induced dyskinesia.
The over representation analysis (ORA) results revealed significantly enriched pathways for the co-expressed gene modules in all investigated groups (FDR-adjusted p value <0.05). Specifically, 54 enriched pathways were found in the PD-D modules, two in PD-ND, two in CT, and 31 in the pooled sample group. The analysis of exclusive pathways was conducted to identify those specifically associated with PD-ND or PD-D groups. We identified 24 exclusive pathways enriched with PD-D (Figure 3F; Table S3) and the main of these pathways associated with LID were showed in Figure 3G. No metabolic pathway was exclusively represented in the PD-ND group.
Exclusive dyskinesia-specific differential co-expression network
We conducted a comprehensive analysis of differential co-expression patterns in people with PD with and without LID. An analysis of 15,372 transcripts highlighted differential co-expressed links (DCLs) and differentially co-expressed genes (DCGs). In the PD-D group, we identified 79 gene-gene interactions, while in the PD-ND group, 3,055 interactions were detected. All interactions were specific to each investigated group, and six DCGs were identified in the PD-D group and 200 in the PD-ND group. Upon examining the similarity between the two investigated groups using the Jaccard index, we confirmed that the gene sets represented in the DCLs of PD-D and PD-ND do not show similarity (Jaccard = 0.0), implicating in distinct molecular mechanisms in each condition.
We used the DCGs from each group to filter their respective DCLs to investigate specific interactions involving these hub genes. In PD-ND group, 200 hub genes are involved in 48 gene-gene interactions among themselves (Figure 4A), and in PD-D we observed that six hub genes are involved in 23 gene-gene interactions (Figure 4B). Gene Ontology analysis for the biological processes in PD-D group was conducted, but no enriched processes were identified through the STRING, g:Profiler, or ToppGene platforms.
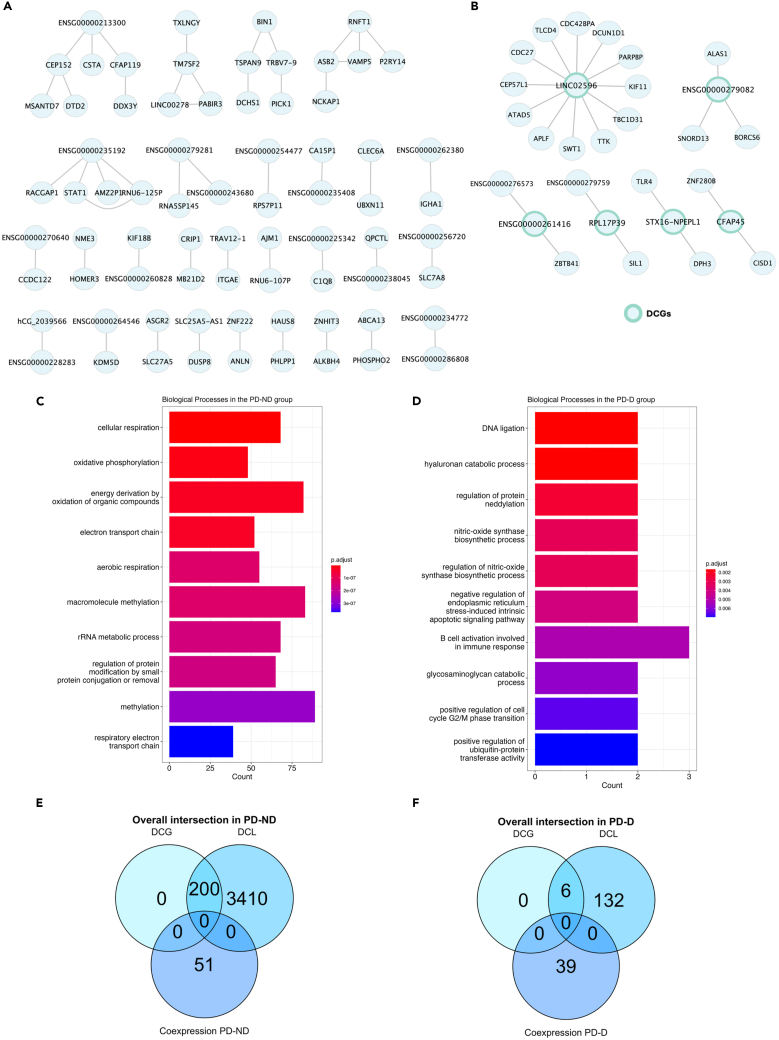
Figure 4.
Networks of differentially co-expressed genes
(A) Network of DCLs of hub genes involved in 48 gene-gene interactions among themselves in PD-ND group, filtered by DCGs.
(B) Network of DCLs of hub genes involved in 23 gene-gene interactions exclusive co-expressed genes in PD-D group, filtered by DCGs.
(C) Top 10 most significant biological processes enriched for differentially co-expressed genes in the PD-ND.
(D) Top 10 most significant biological processes enriched for differentially co-expressed genes in the PD-D.
(E) Overall intersection of co-expression and differential co-expression results in PD-ND group.
(F) Overall intersection of focused LID co-expression and differential co-expression results. PD-D, people with Parkinson’s disease with levodopa-induced dyskinesia; PD-ND, people with Parkinson’s disease without levodopa-induced dyskinesia; DCGs, differentially co-expressed genes; DCLs, differentially co-expressed links.
The Gene Ontology analysis was performed for the complete DCLs of the PD-D and PD-ND groups and found that all enriched pathways were also exclusive to each condition. We identified 152 enriched biological processes in the PD-D group and 220 in the PD-ND group. Subsequently, we performed the exclusion analysis, revealing 110 biological processes exclusive to PD-D group and 178 exclusives to PD-ND group. With a focus on LID, the top 10 most significant processes enriched in the PD-ND and PD-D groups were represented in Figures 4C and 4D, respectively.
Finally, we analyzed the overall distribution of genes identified in the DCLs and DCGs and intersected with the results of co-expression. No overlap was observed between the co-expressed and DCGs in the PD-ND group (Figure 4E) and the PD-D group (Figure 4F). Overall, the analysis revealed a network of DCGs specific to the PD-D group, which helps elucidate the genetic background involved in the LID.
Discussion
In our study, we detected DEGs, gene co-expression networks, and differential co-expression in the blood of people with PD, indicating subsequent molecular adaptations throughout the disease progression. To better discuss our results, we divide into sections offering new insights into molecular changes in PD development and dyskinesia.
Differential gene expression analysis reveals novel biomarkers in Parkinson’s disease
Our results showed no significant difference in gene expression between the PD-D and PD-ND groups. However, as observed in the co-expression analysis results, a composition of different genes was co-expressed within their respective groups. This finding suggests that the fundamental molecular mechanisms distinguishing these groups may involve complex interaction and regulation of multiple genes, rather than isolated changes in gene expression.
DEG analysis revealed ten new genes associated with PD. This finding highlights the complexity of the genetic architecture of PD, which is still not well understood, particularly in admixed populations from the Amazon.15 Of these, three were downregulated in PD overall with AUC > 80% (HBA2, HBBP1, and ENSG00000281383).
Currently, there is no available information regarding the function of the long non-coding RNAs (lncRNA) ENSG00000281383 and its interactions with other genes. There is also no information on the role of HBBP1 in PD, but studies have associated its downregulation with reduced hemoglobin levels and the development of beta-thalassemia.16,17 There are no previous data associating these genes with PD, and a potential regulatory role in neurodegenerative mechanisms of PD needs further clarification through functional investigations.
HBA1 and HBA2 are genes encoding the alpha subunit of hemoglobin, which is expressed in blood cells and dopaminergic neurons of the mesencephalon, where its under-expression is linked to disease severity in postmortem brain tissue from people with PD.18,19,20,21 Furthermore, the downregulation of HBA2 in the blood observed in our results may indicate a reduction in oxygen transport in the body and, consequently, a depletion of its availability in neurons, contributing to oxidative stress and mitochondrial damage, a hallmark characteristic in PD. Another evidence of mitochondrial damage observed in our results is the reduction in the heme signaling metabolic pathway, a molecule produced in the mitochondria that is essential for gas transport in hemoglobin in blood cells and in dopaminergic neurons susceptible to degeneration in PD, as well as in other protein complexes such as the cytochrome family and peroxidase, which are essential for the cellular response to oxidative stress.22
Dysregulation of mitochondrial function and stress response occurs in early and intermediate stages of PD
Mitochondria are organelles that play a crucial role in cellular homeostasis and survival. Mitochondrial dysfunction is described in PD, where genetic and environmental risk factors contribute to the dysregulation of its bioenergetics, biogenesis, transport, and internal quality control.23 Exclusive to people with PD before the onset of motor complications, such as LID (PD-ND group), we observed downregulated mitochondrial genes involved in the electron transport chain (MT-CYB, MT-ND5, and MT-ND6) and mitochondrial transfer RNAs, as MT-TY, MT-TF, MT-TQ, and MT-TE. Alterations in MT-CYB, MT-ND5, and MT-ND6 have been linked to the dysregulation of ATP synthesis and an increase in ROS, which contribute to the elevated damage to both mitochondrial DNA and nuclear DNA.24,25,26,27 Furthermore, the disruption of tRNA metabolism becomes critical for ATP production and protein synthesis in people with PD.28
The gene co-expression analysis for the PD-ND group resulted in a network of genes involved in mitochondrial functions, such as the regulation of apoptosis, response to stress (LYN, PAG1, and MAP3K1), regulation of glucose levels (GAB1 and IRS2), and neurodegeneration (ROCK1 and CXCR4).29,30,31,32,33,34,35 Also, the PIK3CB gene showed a direct or secondary interaction with all genes involved in these processes. A GWAS study conducted in multi-ethnic populations identified new genes associated with PD, including IRS2 and a gene from the PIK3CA family (related to the PIK3CB gene).36 The PIK3CB gene generates a kinase protein primarily associated with immune signaling and apoptosis regulation.37,38 Regarding IRS2, there is an increase in phosphorylation and a simultaneous reduction in the IRS2 gene, along with a decrease in insulin signaling in the basal ganglia of an animal model of PD.31 Furthermore, studies have shown that the dysregulation of insulin pathways, as seen in type II diabetes mellitus, may be a risk factor for PD. These pathways are typically altered in the brains of people with PD, contributing to neurodegeneration and affecting various mitochondrial and autophagy processes.39
The differential gene co-expression analysis revealed over 3,000 statistically significant interactions and 178 unique metabolic processes for the PD-ND group, converging toward mitochondrial functions such as cellular respiration, oxidative phosphorylation, and the electron transport chain, along with methylation and ribosomal RNA metabolic processes. All these factors contribute to the persistent oxidative stress and mitochondrial energy crisis observed in PD, which suggest a series of responses involving the activation of the immune response, response to ROS, DNA repair systems, and selective mitophagy. As mitochondrial damage and cellular stress persist, a cascade of molecular signals leads to programmed neuronal death.40
Dysregulation of lipid homeostasis, endoplasmic reticulum stress, and immune signaling may be related to LID
DEG analysis demonstrated an increased expression of TK2, which is produced in the nucleus and plays role in nucleotide synthesis and the maintenance of mitochondrial DNA. Considering LID usually emerges in advanced stages of the disease, this overexpression is likely to occur in response to frequent oxidative stress and mitochondrial damage.41 Variants in this gene have already been identified in individuals with mitochondrial maintenance disorders, and a comprehensive proteomic analysis observed high expression of this protein in synaptosomes of the substantia nigra in the brains of people with PD.42,43
ACAT1 gene was under expressed and is linked to alterations in other neurological phenotypes, and its inhibition led to the stimulation of autophagy and lysosomal activity in an Alzheimer’s animal model.44,45,46,47,48,49 This gene is typically enriched in the mitochondria-associated endoplasmic reticulum membrane, regulating the connection between these organelles.45
In the co-expression analyses of the PD-D group, SEC22B and TMED10 are hub genes identified in our analysis. These genes are responsible for the regulation of protein and vesicle trafficking associated with the endoplasmic reticulum and the Golgi complex.50,51 Other co-expressed genes, such as HBA1 and RPS/RPL family genes involved in oxygen transport and protein synthesis regulation, respectively, were observed in the PD-D group. A module containing the genes KLRB1, GZMA, GZMK, CD2, and CD2AP, which mediate immune response, neuronal survival, and membrane trafficking, was also observed.52,53,54,55 Furthermore, the enrichment analysis of the co-expression network in LID pointed to important pathways that may be involved in the uptake and metabolism of levodopa, such as amino acid metabolism, axon guidance, L1CAM interactions, and extracellular matrix organization. This is noteworthy since these processes could lead to increased BBB permeability,56,57 potentially influencing the bioavailability of levodopa in the brain and further contributing to dopamine imbalance.
Similarly, the results of differential co-expression between the PD-D group and control subjects also converged on modifications in genes associated with the following functions: endoplasmic reticulum stress and lipid and protein homeostasis (SIL1, BORCS6, SWT1, TTK, TBC1D1, CDC42BPA, and DCUN1D1)58,59,60,61; DNA repair and cell survival (ATAD5, APLF, PARPBP, and ZNF280B)62,63,64,65; mitochondrial metabolism, regulation of cell death, immune signaling, and neurogenesis (CISD1, ALAS1, TLR4, and CDC27)66,67,68,69; and reorganization of the cytoskeleton and formation of the mitotic spindle, cilia and flagella, microtubules, and centrioles (CFAP45, CEP57L, KIF11, and TBC1D31).70,71,72,73,74 Except for CFAP45 and RPL17P39, the remaining DCGs do not have a described function. They are classified as long intergenic non-coding RNAs (lincRNA) such as LINC02596 and ENSG00000279082, or lncRNAs like ENSG00000261416 and STX16-NPEPL1. Functional studies aiming to identify the regulatory role of these non-coding RNAs should be conducted for a better understanding of their involvement in PD.
Supporting our results, the dysregulation of lipid metabolism and neuroinflammation have also been identified in the plasma and cerebrospinal fluid metabolic profiles of patients with LID.75 Furthermore, a recent study conducted a transcriptomic analysis in the peripheral blood of PD patients and observed several DEGs involved in protein synthesis and ribosomal function.76 Finally, some studies have demonstrated that the use of tetracyclines has significant anti-inflammatory therapeutic potential. For instance, doxycycline has been shown to protect dopaminergic neurons from 6-hydroxydopamine (6-OHDA) induced degeneration and reduce LID in hemi parkinsonian rats, indicating that there may be a pro-inflammatory state contributing to the onset of LID.77,78,79
Transcriptome reveals altered molecular plasticity in the developmental progression of PD and LID
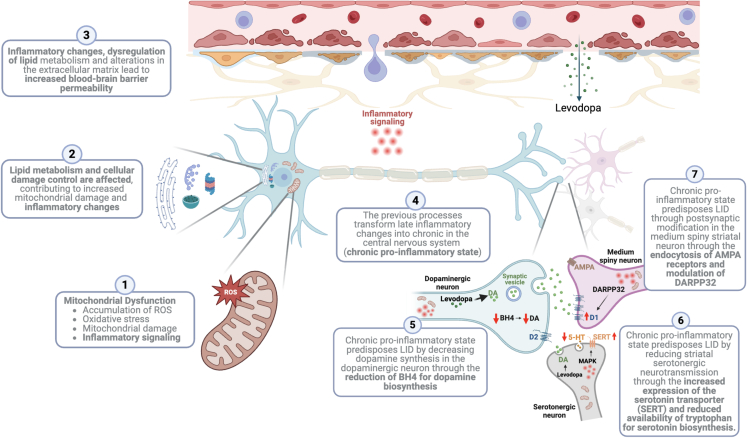
According to these results, we suggest that the disease progression, from the early to the advanced stage when the onset of LID occurs, is crucially influenced by mitochondrial damage. Initial mitochondrial changes promote a series of immune and cellular survival responses that enhances oxidative stress, leading to a cycle of mitochondrial dysfunction across the PD neurodegenerative process. Simultaneously, other mechanisms (alterations in nucleotide and lipid metabolism, inflammation, protein synthesis, endoplasmic reticulum stress, modifications in the membrane, and extracellular matrix) are influenced by multiple dysfunctions in mitochondria and enhances mitochondrial damage. Together, these pathogenic processes may lead to a chronic pro-inflammatory state in the basal ganglia, which has been associated with the development of LID.80,81,82,83,84,85,86,87 See the details of these processes in the Figure 5.
Figure 5.
Mitochondrial damage leads to levodopa-induced dyskinesia (LID) through the chronic pro-inflammatory stage and other processes
(1) Mitochondrial dysfunction is observed during Parkinson’s disease (PD), reinforcing a pro-inflammatory state; (2) initial mitochondrial damage and inflammation contribute to the dysregulation of lipid metabolism and protein synthesis/regulation; (3) these changes affect the permeability of the blood-brain barrier, leading to increased flow of levodopa and immune cells and molecules; (4) the ongoing dysregulation in these systems increasingly reinforces inflammation to a chronic pro-inflammatory stage; (5) chronic neuroinflammation can negatively regulate dopamine (DA) biosynthesis through the reduction of tetrahydrobiopterin (BH4), which is essential for DA synthesis81,82; (6) chronic neuroinflammation reduces serotonin (5-HT) levels in the striatum and contributes to LID by dysregulating DA release through stimulation of the MAPK pathway83 and increasing of serotonin transporter (SERT) expression84,85; (7) Chronic neuroinflammation induces modifications in α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and 32 kDa cyclic AMP-regulated phosphoprotein (DARPP32), affecting corticostriatal glutamatergic synapses and post-synaptic D1 dopamine receptor stimulation.86,87 Created with BioRender.com. Abbreviations: DA, Dopamine; BH4, Tetrahydrobiopterin; 5-HT, Serotonin; MAPK, Mitogen-Activated Protein Kinase; SERT, Serotonin Transporter; AMPA, α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid; DARPP32, 32 kDa Cyclic AMP-Regulated Phosphoprotein.
In conclusion, we have identified novel genes and gene-gene interaction networks associated with the progression of PD and the onset of LID. Our findings align with previous research on the pathophysiology of LID, indicating that dynamic maladaptive plastic changes contribute to motor complications. Furthermore, genes such as HBA2, HBBP1, and ENSG00000281383 are of particular interest, as they seem to play a significant role in PD progression and the development of LID. In individuals with PD but without motor complications, we noticed downregulation of mitochondrial genes involved in the electron transport chain (MT-CYB, MT-ND5, and MT-ND6). These alterations are associated with dysregulation of ATP synthesis and an increase in reactive oxygen species (ROS). Furthermore, gene co-expression network analysis in the PD-ND group revealed a network of genes associated with mitochondrial functions, apoptosis regulation, stress glucose level regulation, and neurodegeneration. Our findings suggest that mitochondrial damage plays a crucial role in the progression of PD and neuroinflammatory chronic stages in PD. Our study highlights the importance of addressing mitochondrial dysfunction as a potential therapeutic target in managing PD progression and the development of LID.
Limitations of the study
We acknowledge certain limitations that could be addressed in future research. While blood serves as an excellent tissue for investigating systemic alterations in individuals, it is known that the administration of levodopa can cause changes in gene expression in inflammatory pathways in the central nervous system (CNS) and blood, as dopamine participates in the regulation of the immune system through its binding to DAR receptors, which are also present in the blood cells.88,89 Thus, our findings related to inflammatory pathways may be more linked to peripheral events than to CNS events. To study this more precisely, cerebrospinal fluid (CSF) samples or the isolation of extracellular vesicles from blood derived from the CNS are good strategies, representing more specifically molecular changes in the CNS that occur during neurodegeneration in PD.
Other limitations include the absence of experimental validations through in vitro or in vivo models of the identified genes and pathways; the lack of a prodromal PD group, which would be crucial for better distinguishing the CT; the small number of samples, which could be expanded to improve the study’s statistical power; and the lack of functional studies to confirm the role of the identified transcripts in PD and LID.
Among the strengths, we highlight that by pairing the groups for age and gender, controlling for various other variables, we present a controlled analysis of gene expression. Also, we explored the data using various methodologies and identified a convergence of results, confirming the pathways and mechanisms involved in PD and LID. We emphasize the importance of genetically characterizing a highly diverse and underrepresented population on the global stage, so that the molecular mechanisms described here can serve as a foundation for further studies aiming to improve the quality of life for patients and explore new therapeutic strategies.
Resource availability
Lead contact
Any additional information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ândrea Ribeiro-dos-Santos (akelyufpa@gmail.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All raw sequences are deposited at the European Nucleotide Archive (ENA), with the accession number PRJEB72411 and are publicly available as of the date of publication.
-
•
All data analysis codes are available in the GitHub repository: https://github.com/tati-souz/RNA-Seq-data-analysis.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We are immensely grateful to everyone who participated in this research. And we emphasize the importance of characterizing the genetic mechanisms present in a highly diverse and underrepresented population on the global scene, such as the Amazon. We hope that the results can serve to improve patients’ quality of life and explore new therapeutic strategies.
We thank the Movement Disorders Clinic of the Hospital Ophir Loyola for enabling the recruitment of patients for the research; lastly, we extend our thanks to doctors Amanda Vidal, Ricardo Vialle, and Elaine Del-Bel for their invaluable contributions and insightful suggestions for improvement.
This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Biocomputacional Protocol no. 3381/2013/CAPES; and Pró-Reitoria de Pesquisa e Pós-Graduação, Universidade Federal do Pará. A.R.S. was supported by CNPq/Productivity (312916/2021-3). The funders had no role in study design, data collection, analysis, interpretation or writing of the report.
Author contributions
Recruitment of participants and collection of biological material., T.P.S., C.S.S., and G.L.E.; clinical assessment of participants., A.F.E. and B.L.S.L.; definition of the methodology, A.R.S., T.P.S., G.S.A., and G.B.S-S; extraction, quality control, library preparation, and sequencing, T.P.S., C.S.S., G.C.C., and L.M.; article writing, T.P.S.; data analysis, T.P.S., A.R.S., G.B.S.S., and G.S.A.; illustration, T.P.S. and B.L.S-L.; review and coordination, A.R.S., G.S.A., and B.L.S-L.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Peripheral blood (whole blood) from patients with Parkinson`s disease with dyskinesia | Movement Disorders Clinic of the Hospital Ophir Loyola | N/A |
| Peripheral blood (whole blood) from patients with Parkinson`s disease without dyskinesia | Movement Disorders Clinic of the Hospital Ophir Loyola | N/A |
| Peripheral blood (whole blood) from heathy donors | Movement Disorders Clinic of the Hospital Ophir Loyola | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol LS Reagent | Thermo Fisher Scientific by Invitrogen™ | Cat#10296010 |
| Critical commercial assays | ||
| TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold kit | Illumina Inc., Chicago, IL, USA | Cat# RS-122-2301, RS-122-2302, and RS-122-2303 |
| Deposited data | ||
| Raw sequences of RNA-Seq | This study | ENA: PRJEB72411 |
| Software and algorithms | ||
| Code of RNA-Seq Data analysis | This study | https://github.com/tati-souz/RNA-Seq-data-analysis. |
| Fastp tool | Chen et al.90 | |
| Star Aligner Software | Dobin et al.91 | https://doi.org/10.1093/bioinformatics/bts635 |
| HTSeq software | Anders et al.92 | https://academic.oup.com/bioinformatics/article/31/2/166/2366196 |
| DESeq2 package | Love et al.93 | https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| RStudio 4.2.3 software | R Core team (2023)94 | https://www.R-project.org/ |
| pROC package | Robin et al.95 | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-77 |
| ClusterProfiller package | Yu et al.96 | https://www.liebertpub.com/doi/10.1089/omi.2011.0118 |
| Ggplot2 package | Wilkinson et al.97 | https://academic.oup.com/biometrics/article/67/2/678/7381027?login=false |
| CEMitool package | Russo et al.98 | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2053-1 |
| Cytoscape software | Shannon et al.99 | https://genome.cshlp.org/content/13/11/2498 |
| Diffcoexp package | Wei et al.100 | https://github.com/hidelab/diffcoexp |
| Other | ||
| Qubit 2.0 Fluorometer | Thermo Fisher Scientific | Cat#32866 |
| NanoDrop ND1000 | Thermo Fisher Scientific | Cat# ND2000CLAPTOP |
| 2200 TapeStation System | Agilent | Cat#G2964AA |
| NextSeq Sequencing 550 System | Illumina | Cat# SY-415-1002 |
Experimental model and study participant details
Human participants
The use of human subjects was approved by the Hospital Ophir Loyola Ethics Committee (Number 82848118.5.0000.5550), and all participants provided written informed consent. We conducted an observational cross-sectional study to profile gene and gene-gene co-expression networks in people with PD with LID (PD-D), people with PD without LID (PD-ND), and non-PD controls (CT). Participants with PD diagnosed according to the UK Parkinson’s Disease Society Brain Bank were recruited in the Movement Disorders Clinic of the Hospital Ophir Loyola, in Belém, Brazil, between January 2018 and December 2021. People with PD and control subjects were recruited using a 1:1 ratio matching for sex and age within four years (See the detailed information of clinical characteristics is Table S1).
Brazilian populations have a high rate of miscegenation, reflecting the different ethnic contributions throughout our history. Consequently, our genomes and/or biological markers represent mosaics of European, African, indigenous, and other ancestries. In the Amazon region, we observed an average contribution of European ancestry genes at 60%, indigenous (original peoples) at 28%, and African ancestry at 12%.101 It is worth noting that the Amazon region has the largest contribution of genes from indigenous peoples. Additionally, previous work by our group on Parkinson’s patients did not observe statistically significant differences in the average contribution of genomic ancestry genes (59% European, 26% indigenous, and 15% African).102
Inclusion and exclusion criteria for participants
As inclusion criteria, we enrolled participants with age at evaluation over 40 years and, for people with PD, a stable levodopa dose in the previous month. CT samples were obtained from volunteers who were invited to participate in the research. Participants with a history of autoimmune, inflammatory, cancer and other neurological diseases were excluded. In addition, PD patients with severe psychotic symptoms (score >2 in item 1.2 of the Movement Disorder Society – Unified Parkinson’s Disease Rating Scale, MDS-UPDRS) and clinical diagnosis of dementia were removed or control subjects with a family history of PD.
Method details
Clinical evaluations
All participants with PD were evaluated by the same movement disorders specialist (B.L.S.-L.) using the MDS-UPDRS in the ON-stage. We defined the presence of LID by a score ≥1 on item 4.1 of the MDS-UPDRS Part IV (time spent with dyskinesia) and confirmed if the patient presented abnormal movements at evaluation. Blood samples were collected on the same day of clinical evaluation.
Blood collection and total RNA extraction
Peripheral blood was collected on the same day and period, between 09:00 and 11:00 a.m., without fasting. All people with PD were instructed to take their regular morning antiparkinsonian drugs before peripheral blood intravenous puncture. Blood samples were stored with TRIzol LS (Thermo Fisher Scientific) following the manufacturer’s instructions at −20°C until further use. Total RNA extraction from the blood samples followed the nucleic acid extraction protocol using TRIzol LS (Thermo Fisher Scientific), in accordance with the manufacturer’s instructions. After extraction, the total RNA was assessed for integrity, purity, and concentration using the Qubit 2.0 Fluorometer (Thermo Fisher Scientific), NanoDrop ND1000 (Thermo Fisher Scientific), and 2200 TapeStation System (Agilent).
Library preparation, sequencing and quality control
For library preparation (total transcriptome), 1 μg of total RNA per sample was used. A total of 46 libraries were constructed (20 controls, 15 PD-D, 11 PD-ND) using the TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold kit (Illumina Inc., Chicago, IL, USA). The libraries were normalized and sequenced on the NextSeq Sequencing System (Illumina) using the NextSeq High Output 150 cycles (Illumina). After sequencing, quality control was performed, where low-quality reads (phred-score = Q30) were discarded, the 3′ ends of sequences were trimmed, and contaminants were removed using the fastp tool.90 For alignment, the STAR software91 was used with the human genome reference GRCh38. Counting of messenger RNAs (mRNAs) and long non-coding RNAs (lncRNAs) was performed using HTSeq.92
Differential gene expression analysis
Differential expression analysis was performed for the PD-D, PD-ND, and CT groups using the DESeq293 package in RStudio software (v.4.2.3).66,67,68,69 DESeq2 normalizes counts by library size and adjusts gene expression, taking gene size into consideration. To compare CT group vs. general PD group (PD-D + PD-ND) and the subgroups with and without dyskinesia, the controlled covariates were age, sex, and sequencing batch. Furthermore, when we compared PD-D vs. PD-ND, the covariates controlled were age, sex, sequencing batch, duration of disease, and levodopa equivalent daily dose (LEDD) to avoid confounding factors and false-positive results. Adjusted p-values (FDR) < 0.05 were considered statistically significant. To identify potential biomarkers associated with PD, we investigated the sensitivity and specificity of DEGs using the ROC curve through the pROC package in software (v.4.2.1).66,67,68,69 Thus, we selected those genes with an area under the curve (AUC) > 80% as potential disease biomarkers. We conducted Gene Set Enrichment Analysis (GSEA) to identify enriched metabolic pathways for all genes in PD-D and PD-ND groups. Consequently, pathways with high and positive enrichment scores are presumed to be associated with over-expressed genes, while those with high and negative scores are linked to under-expressed genes. Furthermore, Gene Ontology analyses for DEGs were carried out using the ClusterProfiler package96 in R software (v.4.2.1). The graphics are constructed with ggplot2 package97 in RStudio.
Gene co-expression analysis
Gene co-expression analysis was performed using the CEMitool98 package in RStudio, for PD-D, PD-ND and CT groups. Correlation was computed to assess the similarity of gene expression within each group. This method allows us to infer modules of co-expressed genes that may play the same biological process roles. We also performed the joint co-expression analysis of CT + PD-D + PD-ND to select genes exclusively co-expressed in the PD-D and PD-ND groups. Finally, co-expression networks were drawn using Cytoscape software (v.3.10).99 The CEMitool package also performs Over Representation Analysis (ORA) to assess the pathways enrichment.
Differential co-expression gene network analysis
The differential co-expression gene network analysis aims to identify modules of Differential Co-expressed Links (DCLs), representing pairs of genes that exhibit statistically differences on correlations between two distinct conditions. Moreover, it identifies Differentially Co-expressed Genes (DCGs) referring to genes with a higher number of DCLs than expected by chance (gene hubs). We employed the diffcoexp package100 in the RStudio, setting for spearman correlation method for gene pairwise similarity between the groups (PD-D x CT and PD-ND x CT). Subsequently, we constructed differential co-expression networks using Cytoscape software (v.3.10). Significance was determined by adjusted p-values (FDR) < 0.05, and genes with a correlation coefficient (r) > 0.8 were selected. To assess network specificity for each group, a similarity analysis utilizing the Jaccard index was conducted. This helped to determine whether the networks of DCGs were exclusive to each group. For insights into the biological processes involving DCGs and genes in DCLs, GO analysis was performed through the ClusterProfiler package96 in RStudio.
Quantification and statistical analysis
All statistical analyses detailed in the results (Table 1) were performed using RStudio. The selection of tests was based on the data types, number of samples, and data distribution pattern (see Table S1). To compare the frequency of each sex between groups, the chi-square test was used. The Kruskal-Wallis test was employed to compare the age at clinical evaluation, and the Mann-Whitney test was conducted to compare the means of age at disease onset, levodopa dosage, and treatment duration.
Published: August 26, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110835.
Supplemental information
References
- 1.Dorsey E.R., Sherer T., Okun M.S., Bloem B.R. The Emerging Evidence of the Parkinson Pandemic. J. Parkinsons Dis. 2018;8:S3–S8. doi: 10.3233/JPD-181474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalia L.V., Lang A.E. Parkinson’s disease. Lancet. 2015;386:896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- 3.Bloem B.R., Okun M.S., Klein C. Parkinson’s disease. Lancet. 2021;397:2284–2303. doi: 10.1016/S0140-6736(21)00218-X. [DOI] [PubMed] [Google Scholar]
- 4.Chahine L.M., Beach T.G., Brumm M.C., Adler C.H., Coffey C.S., Mosovsky S., Caspell-Garcia C., Serrano G.E., Munoz D.G., White C.L., et al. In vivo distribution of α-synuclein in multiple tissues and biofluids in Parkinson disease. Neurology. 2020;95:e1267–e1284. doi: 10.1212/WNL.0000000000010404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon D.K., Kwatra M., Wang J., Ko H.S. Levodopa-Induced Dyskinesia in Parkinson’s Disease: Pathogenesis and Emerging Treatment Strategies. Cells. 2022;11:3736. doi: 10.3390/cells11233736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang A.E., Espay A.J. Disease Modification in Parkinson’s Disease: Current Approaches, Challenges, and Future Considerations: Disease Modification in PD. Mov. Disord. 2018;33:660–677. doi: 10.1002/mds.27360. [DOI] [PubMed] [Google Scholar]
- 7.Bezard E. Experimental reappraisal of continuous dopaminergic stimulation against L-dopa-induced dyskinesia. Mov. Disord. 2013;28:1021–1022. doi: 10.1002/mds.25251. [DOI] [PubMed] [Google Scholar]
- 8.López I.C., Ruiz P.J.G., del Pozo S.V.F., Bernardos V.S. Motor complications in Parkinson’s disease: Ten year follow-up study. Movement Disorders. 2010;25:2735–2739. doi: 10.1002/mds.23219. [DOI] [PubMed] [Google Scholar]
- 9.Turcano P., Mielke M.M., Bower J.H., Parisi J.E., Cutsforth-Gregory J.K., Ahlskog J.E., Savica R. Levodopa-induced dyskinesia in Parkinson disease. Neurology. 2018;91:e2238–e2243. doi: 10.1212/WNL.0000000000006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandopadhyay R., Mishra N., Rana R., Kaur G., Ghoneim M.M., Alshehri S., Mustafa G., Ahmad J., Alhakamy N.A., Mishra A. Molecular Mechanisms and Therapeutic Strategies for Levodopa-Induced Dyskinesia in Parkinson’s Disease: A Perspective Through Preclinical and Clinical Evidence. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.805388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calabresi P., Centonze D., Gubellini P., Marfia G.A., Pisani A., Sancesario G., Bernardi G. Synaptic transmission in the striatum: from plasticity to neurodegeneration. Prog. Neurobiol. 2000;61:231–265. doi: 10.1016/S0301-0082(99)00030-1. [DOI] [PubMed] [Google Scholar]
- 12.Lindgren H.S., Andersson D.R., Lagerkvist S., Nissbrandt H., Cenci M.A. l-DOPA-induced dopamine efflux in the striatum and the substantia nigra in a rat model of Parkinson’s disease: temporal and quantitative relationship to the expression of dyskinesia. J. Neurochem. 2010;112:1465–1476. doi: 10.1111/j.1471-4159.2009.06556.x. [DOI] [PubMed] [Google Scholar]
- 13.Nutt J.G. Pharmacokinetics and pharmacodynamics of levodopa. Mov. Disord. 2008;23:S580–S584. doi: 10.1002/mds.22037. [DOI] [PubMed] [Google Scholar]
- 14.Chowdhury H.A., Bhattacharyya D.K., Kalita J.K. (Differential) Co-Expression Analysis of Gene Expression: A Survey of Best Practices. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020;17:1154–1173. doi: 10.1109/TCBB.2019.2893170. [DOI] [PubMed] [Google Scholar]
- 15.Schumacher-Schuh A.F., Bieger A., Okunoye O., Mok K.Y., Lim S.-Y., Bardien S., Ahmad-Annuar A., Santos-Lobato B.L., Strelow M.Z., Salama M., et al. Underrepresented Populations in Parkinson’s Genetics Research: Current Landscape and Future Directions. Mov. Disord. 2022;37:1593–1604. doi: 10.1002/mds.29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma S.-P., Xi H.-R., Gao X.-X., Yang J.-M., Kurita R., Nakamura Y., Song X.-M., Chen H.-Y., Lu D.-R. Long noncoding RNA HBBP1 enhances γ-globin expression through the ETS transcription factor ELK1. Biochem. Biophys. Res. Commun. 2021;552:157–163. doi: 10.1016/j.bbrc.2021.03.051. [DOI] [PubMed] [Google Scholar]
- 17.Ma Y., Liu S., Gao J., Chen C., Zhang X., Yuan H., Chen Z., Yin X., Sun C., Mao Y., et al. Genome-wide analysis of pseudogenes reveals HBBP1’s human-specific essentiality in erythropoiesis and implication in β-thalassemia. Dev. Cell. 2021;56:478–493.e11. doi: 10.1016/j.devcel.2020.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Biagioli M., Pinto M., Cesselli D., Zaninello M., Lazarevic D., Roncaglia P., Simone R., Vlachouli C., Plessy C., Bertin N., et al. Unexpected expression of alpha- and beta-globin in mesencephalic dopaminergic neurons and glial cells. Proc. Natl. Acad. Sci. USA. 2009;106:15454–15459. doi: 10.1073/pnas.0813216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng Q., Zhou X., Chen J., Pan M., Gao H., Zhou J., Wang D., Chen Q., Zhang X., Wang Q., Xu Y. Lower hemoglobin levels in patients with parkinson’s disease are associated with disease severity and iron metabolism. Brain Res. 2017;1655:145–151. doi: 10.1016/j.brainres.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Ferrer I., Gómez A., Carmona M., Huesa G., Porta S., Riera-Codina M., Biagioli M., Gustincich S., Aso E. Neuronal Hemoglobin is Reduced in Alzheimer’s Disease, Argyrophilic Grain Disease, Parkinson’s Disease, and Dementia with Lewy Bodies. J. Alzheimers Dis. 2011;23:537–550. doi: 10.3233/JAD-2010-101485. [DOI] [PubMed] [Google Scholar]
- 21.Freed J., Chakrabarti L. Defining a role for hemoglobin in Parkinson’s disease. NPJ Parkinsons Dis. 2016;2 doi: 10.1038/npjparkd.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latunde-Dada G.O. Academic Press; 2015. Encyclopedia of Food and Health. [Google Scholar]
- 23.Chen Z., Rasheed M., Deng Y. The epigenetic mechanisms involved in mitochondrial dysfunction: Implication for Parkinson’s disease. Brain Pathol. 2022;32 doi: 10.1111/bpa.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown M.D., Shoffner J.M., Kim Y.L., Jun A.S., Graham B.H., Cabell M.F., Gurley D.S., Wallace D.C. Mitochondrial DNA sequence analysis of four Alzheimer’s and Parkinson’s disease patients. Am. J. Med. Genet. 1996;61:283–289. doi: 10.1002/(SICI)1096-8628(19960122)61:3<283::AID-AJMG15>3.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 25.Petruzzella V., Sardanelli A.M., Scacco S., Panelli D., Papa F., Trentadue R., Papa S. Dysfunction of mitochondrial respiratory chain complex I in neurological disorders: genetics and pathogenetic mechanisms. Adv. Exp. Med. Biol. 2012;942:371–384. doi: 10.1007/978-94-007-2869-1_17. [DOI] [PubMed] [Google Scholar]
- 26.Pignataro D., Francia S., Zanetta F., Brenna G., Brandini S., Olivieri A., Torroni A., Biamonti G., Montecucco A. A missense MT-ND5 mutation in differentiated Parkinson Disease cytoplasmic hybrid induces ROS-dependent DNA Damage Response amplified by DROSHA. Sci. Rep. 2017;7:9528. doi: 10.1038/s41598-017-09910-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Eitzen U., Kösel S., Grasbon-Frodl E.M., Egensperger R., Graeber M.B. Sequence analysis of the MTCYB gene in Parkinson disease. Neurogenetics. 2000;3:47–48. doi: 10.1007/PL00022980. [DOI] [PubMed] [Google Scholar]
- 28.Venkatesan D., Iyer M., Raj N., Gopalakrishnan A.V., Narayanasamy A., Kumar N.S., Vellingiri B. Assessment of tRNAThr and tRNAGln Variants and Mitochondrial Functionality in Parkinson’s Disease (PD) Patients of Tamil Nadu Population. J. Mol. Neurosci. 2023;73:912–920. doi: 10.1007/s12031-023-02154-7. [DOI] [PubMed] [Google Scholar]
- 29.Cassarino D.S., Halvorsen E.M., Swerdlow R.H., Abramova N.N., Parker W.D., Sturgill T.W., Bennett J.P. Interaction among mitochondria, mitogen-activated protein kinases, and nuclear factor-kappaB in cellular models of Parkinson’s disease. J. Neurochem. 2000;74:1384–1392. doi: 10.1046/j.1471-4159.2000.0741384.x. [DOI] [PubMed] [Google Scholar]
- 30.Ma J., Dong L., Chang Q., Chen S., Zheng J., Li D., Wu S., Yang H., Li X. CXCR4 knockout induces neuropathological changes in the MPTP-lesioned model of Parkinson’s disease. Biochim. Biophys. Acta, Mol. Basis Dis. 2023;1869 doi: 10.1016/j.bbadis.2022.166597. [DOI] [PubMed] [Google Scholar]
- 31.Morris J.K., Zhang H., Gupte A.A., Bomhoff G.L., Stanford J.A., Geiger P.C. Measures of striatal insulin resistance in a 6-hydroxydopamine model of Parkinson’s disease. Brain Res. 2008;1240:185–195. doi: 10.1016/j.brainres.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Napolitano M., Picconi B., Centonze D., Bernardi G., Calabresi P., Gulino A. L-DOPA treatment of parkinsonian rats changes the expression of Src, Lyn and PKC kinases. Neurosci. Lett. 2006;398:211–214. doi: 10.1016/j.neulet.2005.12.077. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Araujo G., Nakagami H., Takami Y., Katsuya T., Akasaka H., Saitoh S., Shimamoto K., Morishita R., Rakugi H., Kaneda Y. Low alpha-synuclein levels in the blood are associated with insulin resistance. Sci. Rep. 2015;5 doi: 10.1038/srep12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y., Yang X., Guan S., Ma T., Jiang Z., Gao M., Xu Y., Cong B. The role of phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (PAG1) in regulating the progression of oral squamous cell carcinoma. Arch. Oral Biol. 2023;156 doi: 10.1016/j.archoralbio.2023.105810. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q., Hu C., Huang J., Liu W., Lai W., Leng F., Tang Q., Liu Y., Wang Q., Zhou M., et al. ROCK1 induces dopaminergic nerve cell apoptosis via the activation of Drp1-mediated aberrant mitochondrial fission in Parkinson’s disease. Exp. Mol. Med. 2019;51:1–13. doi: 10.1038/s12276-019-0318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J.J., Vitale D., Otani D.V., Lian M.M., Heilbron K., 23andMe Research Team. Iwaki H., Lake J., Solsberg C.W., Leonard H., et al. Multi-ancestry genome-wide association meta-analysis of Parkinson’s disease. Nat. Genet. 2024;56:27–36. doi: 10.1038/s41588-023-01584-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bommarito D., Martin A., Forcade E., Nastke M.-D., Ritz J., Bellucci R. Enhancement of tumor cell susceptibility to natural killer cell activity through inhibition of the PI3K signaling pathway. Cancer Immunol. Immunother. 2016;65:355–366. doi: 10.1007/s00262-016-1804-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu W., Wang Z., Zhang Z., Xu J., Jiang Y. PIK3CB promotes oesophageal cancer proliferation through the PI3K/AKT/mTOR signalling axis. Cell Biol. Int. 2022;46:1399–1408. doi: 10.1002/cbin.11847. [DOI] [PubMed] [Google Scholar]
- 39.Cullinane P.W., de Pablo Fernandez E., König A., Outeiro T.F., Jaunmuktane Z., Warner T.T. Type 2 Diabetes and Parkinson’s Disease: A Focused Review of Current Concepts. Mov. Disord. 2023;38:162–177. doi: 10.1002/mds.29298. [DOI] [PubMed] [Google Scholar]
- 40.Dionísio P.A., Amaral J.D., Rodrigues C.M.P. Oxidative stress and regulated cell death in Parkinson’s disease. Ageing Res. Rev. 2021;67 doi: 10.1016/j.arr.2021.101263. [DOI] [PubMed] [Google Scholar]
- 41.Pérez-Pérez M.-J., Priego E.-M., Hernández A.-I., Familiar O., Camarasa M.-J., Negri A., Gago F., Balzarini J. Structure, physiological role, and specific inhibitors of human thymidine kinase 2 (TK2): present and future. Med. Res. Rev. 2008;28:797–820. doi: 10.1002/med.20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bychkov I.O., Itkis Y.S., Tsygankova P.G., Krylova T.D., Mikhaylova S.V., Klyushnikov S.A., Pechatnikova N.L., Degtyareva A.V., Nikolaeva E.A., Seliverstov Y.A., et al. Mitochondrial DNA maintenance disorders in 102 patients from different parts of Russia: Mutational spectrum and phenotypes. Mitochondrion. 2021;57:205–212. doi: 10.1016/j.mito.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Plum S., Eggers B., Helling S., Stepath M., Theiss C., Leite R.E.P., Molina M., Grinberg L.T., Riederer P., Gerlach M., et al. Proteomic Characterization of Synaptosomes from Human Substantia Nigra Indicates Altered Mitochondrial Translation in Parkinson’s Disease. Cells. 2020;9:2580. doi: 10.3390/cells9122580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De La Torre A.L., Huynh T.N., Chang C.C.Y., Pooler D.B., Ness D.B., Lewis L.D., Pannem S., Feng Y., Samkoe K.S., Hickey W.F., Chang T.Y. Stealth Liposomes Encapsulating a Potent ACAT1/SOAT1 Inhibitor F12511: Pharmacokinetic, Biodistribution, and Toxicity Studies in Wild-Type Mice and Efficacy Studies in Triple Transgenic Alzheimer’s Disease Mice. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms241311013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harned T.C., Stan R.V., Cao Z., Chakrabarti R., Higgs H.N., Chang C.C.Y., Chang T.Y. Acute ACAT1/SOAT1 Blockade Increases MAM Cholesterol and Strengthens ER-Mitochondria Connectivity. Int. J. Mol. Sci. 2023;24:5525. doi: 10.3390/ijms24065525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shibuya Y., Chang C.C.Y., Huang L.-H., Bryleva E.Y., Chang T.-Y. Inhibiting ACAT1/SOAT1 in microglia stimulates autophagy-mediated lysosomal proteolysis and increases Aβ1-42 clearance. J. Neurosci. 2014;34:14484–14501. doi: 10.1523/JNEUROSCI.2567-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vance J.E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 48.Xu Y., Wan W. Acetylation in the regulation of autophagy. Autophagy. 2023;19:379–387. doi: 10.1080/15548627.2022.2062112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J., Wang J., Zhou Z., Park J.-E., Wang L., Wu S., Sun X., Lu L., Wang T., Lin Q., et al. Importance of TFEB acetylation in control of its transcriptional activity and lysosomal function in response to histone deacetylase inhibitors. Autophagy. 2018;14:1043–1059. doi: 10.1080/15548627.2018.1447290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anwar M.U., Sergeeva O.A., Abrami L., Mesquita F.S., Lukonin I., Amen T., Chuat A., Capolupo L., Liberali P., D’Angelo G., van der Goot F.G. ER-Golgi-localized proteins TMED2 and TMED10 control the formation of plasma membrane lipid nanodomains. Dev. Cell. 2022;57:2334–2346.e8. doi: 10.1016/j.devcel.2022.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Bonaud A., Gargowitsch L., Gilbert S.M., Rajan E., Canales-Herrerias P., Stockholm D., Rahman N.F., Collins M.O., Taskiran H., Hill D.L., et al. Sec22b is a critical and nonredundant regulator of plasma cell maintenance. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2213056120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang C., Chen Y., Chen S., She J., Shi Q., Wang P. KLRB1 is a novel prognostic biomarker in endometrial cancer and is associated with immune infiltration. Transl. Cancer Res. 2023;12:3641–3652. doi: 10.21037/tcr-23-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Springer T.A., Dustin M.L., Kishimoto T.K., Marlin S.D. The lymphocyte function-associated LFA-1, CD2, and LFA-3 molecules: cell adhesion receptors of the immune system. Annu. Rev. Immunol. 1987;5:223–252. doi: 10.1146/annurev.iy.05.040187.001255. [DOI] [PubMed] [Google Scholar]
- 54.Tao Q.-Q., Chen Y.-C., Wu Z.-Y. The role of CD2AP in the Pathogenesis of Alzheimer’s Disease. Aging Dis. 2019;10:901–907. doi: 10.14336/AD.2018.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Z., He H., Wang K., Shi X., Wang Y., Su Y., Wang Y., Li D., Liu W., Zhang Y., et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368 doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 56.Cenci M.A., Ohlin K.E., Rylander D. Plastic effects of L-DOPA treatment in the basal ganglia and their relevance to the development of dyskinesia. Parkinsonism Relat. Disord. 2009;15:S59–S63. doi: 10.1016/S1353-8020(09)70782-5. [DOI] [PubMed] [Google Scholar]
- 57.Ohlin K.E., Sebastianutto I., Adkins C.E., Lundblad C., Lockman P.R., Cenci M.A. Impact of L-DOPA treatment on regional cerebral blood flow and metabolism in the basal ganglia in a rat model of Parkinson’s disease. Neuroimage. 2012;61:228–239. doi: 10.1016/j.neuroimage.2012.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bell C.J., Gupta N., Tremblay K.D., Mager J. Borcs6 is required for endo-lysosomal degradation during early development. Mol. Reprod. Dev. 2022;89:337–350. doi: 10.1002/mrd.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S., Sun L., Hao M., Liu X. Circ-SWT1 Ameliorates H2O2-Induced Apoptosis, Oxidative Stress and Endoplasmic Reticulum Stress in Cardiomyocytes via miR-192-5p/SOD2 Axis. Cardiovasc. Toxicol. 2022;22:378–389. doi: 10.1007/s12012-022-09720-2. [DOI] [PubMed] [Google Scholar]
- 60.Kumar A., Karuppagounder S.S., Chen Y., Corona C., Kawaguchi R., Cheng Y., Balkaya M., Sagdullaev B.T., Wen Z., Stuart C., et al. 2-Deoxyglucose drives plasticity via an adaptive ER stress-ATF4 pathway and elicits stroke recovery and Alzheimer’s resilience. Neuron. 2023;111:2831–2846.e10. doi: 10.1016/j.neuron.2023.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao L., Longo-Guess C., Harris B.S., Lee J.-W., Ackerman S.L. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat. Genet. 2005;37:974–979. doi: 10.1038/ng1620. [DOI] [PubMed] [Google Scholar]
- 62.Mehrotra P.V., Ahel D., Ryan D.P., Weston R., Wiechens N., Kraehenbuehl R., Owen-Hughes T., Ahel I. DNA repair factor APLF is a histone chaperone. Mol. Cell. 2011;41:46–55. doi: 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mochizuki A.L., Katanaya A., Hayashi E., Hosokawa M., Moribe E., Motegi A., Ishiai M., Takata M., Kondoh G., Watanabe H., et al. PARI Regulates Stalled Replication Fork Processing To Maintain Genome Stability upon Replication Stress in Mice. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00117-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park S.H., Kim Y., Ra J.S., Wie M.W., Kang M.-S., Kang S., Myung K., Lee K.-Y. Timely termination of repair DNA synthesis by ATAD5 is important in oxidative DNA damage-induced single-strand break repair. Nucleic Acids Res. 2021;49:11746–11764. doi: 10.1093/nar/gkab999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhai J., Yang Z., Cai X., Yao G., An Y., Wang W., Fan Y., Zeng C., Liu K. ZNF280B promotes the growth of gastric cancer in vitro and in vivo. Oncol. Lett. 2018;15:5819–5824. doi: 10.3892/ol.2018.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abe K., Ikeda M., Ide T., Tadokoro T., Miyamoto H.D., Furusawa S., Tsutsui Y., Miyake R., Ishimaru K., Watanabe M., et al. Doxorubicin causes ferroptosis and cardiotoxicity by intercalating into mitochondrial DNA and disrupting Alas1-dependent heme synthesis. Sci. Signal. 2022;15 doi: 10.1126/scisignal.abn8017. [DOI] [PubMed] [Google Scholar]
- 67.Geldenhuys W.J., Benkovic S.A., Lin L., Yonutas H.M., Crish S.D., Sullivan P.G., Darvesh A.S., Brown C.M., Richardson J.R. MitoNEET (CISD1) Knockout Mice Show Signs of Striatal Mitochondrial Dysfunction and a Parkinson’s Disease Phenotype. ACS Chem. Neurosci. 2017;8:2759–2765. doi: 10.1021/acschemneuro.7b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mishra M., Akatsu H., Heese K. The novel protein MANI modulates neurogenesis and neurite-cone growth. J. Cell Mol. Med. 2011;15:1713–1725. doi: 10.1111/j.1582-4934.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Z., Ning J., Bao X.-Q., Shang M., Ma J., Li G., Zhang D. Fecal microbiota transplantation protects rotenone-induced Parkinson’s disease mice via suppressing inflammation mediated by the lipopolysaccharide-TLR4 signaling pathway through the microbiota-gut-brain axis. Microbiome. 2021;9:226. doi: 10.1186/s40168-021-01107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dougherty G.W., Mizuno K., Nöthe-Menchen T., Ikawa Y., Boldt K., Ta-Shma A., Aprea I., Minegishi K., Pang Y.-P., Pennekamp P., et al. CFAP45 deficiency causes situs abnormalities and asthenospermia by disrupting an axonemal adenine nucleotide homeostasis module. Nat. Commun. 2020;11:5520. doi: 10.1038/s41467-020-19113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ito K.K., Watanabe K., Ishida H., Matsuhashi K., Chinen T., Hata S., Kitagawa D. Cep57 and Cep57L1 maintain centriole engagement in interphase to ensure centriole duplication cycle. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202005153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu M., Aneja R., Sun X., Xie S., Wang H., Wu X., Dong J.-T., Li M., Joshi H.C., Zhou J. Parkin Regulates Eg5 Expression by Hsp70 Ubiquitination-dependent Inactivation of c-Jun NH2-terminal Kinase. J. Biol. Chem. 2008;283:35783–35788. doi: 10.1074/jbc.M806860200. [DOI] [PubMed] [Google Scholar]
- 73.Senatore E., Chiuso F., Rinaldi L., Intartaglia D., Delle Donne R., Pedone E., Catalanotti B., Pirone L., Fiorillo B., Moraca F., et al. The TBC1D31/praja2 complex controls primary ciliogenesis through PKA-directed OFD1 ubiquitylation. EMBO J. 2021;40 doi: 10.15252/embj.2020106503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valentine M.T., Gilbert S.P. To step or not to step? How biochemistry and mechanics influence processivity in Kinesin and Eg5. Curr. Opin. Cell Biol. 2007;19:75–81. doi: 10.1016/j.ceb.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santos-Lobato B.L., Gardinassi L.G., Bortolanza M., Peti A.P.F., Pimentel Â.V., Faccioli L.H., Del-Bel E.A., Tumas V. Metabolic Profile in Plasma AND CSF of LEVODOPA-induced Dyskinesia in Parkinson’s Disease: Focus on Neuroinflammation. Mol. Neurobiol. 2022;59:1140–1150. doi: 10.1007/s12035-021-02625-1. [DOI] [PubMed] [Google Scholar]
- 76.D’Angiolini S., Lui M., Mazzon E., Calabrò M. Network Analysis Performed on Transcriptomes of Parkinson’s Disease Patients Reveals Dysfunction in Protein Translation. Int. J. Mol. Sci. 2024;25:1299. doi: 10.3390/ijms25021299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bortolanza M., Do Nascimento G.C., Raisman-Vozari R., Del-Bel E. Doxycycline and its derivative, COL-3, decrease dyskinesia induced by l -DOPA in hemiparkinsonian rats. Br. J. Pharmacol. 2021;178:2595–2616. doi: 10.1111/bph.15439. [DOI] [PubMed] [Google Scholar]
- 78.Bortolanza M., Nascimento G.C., Socias S.B., Ploper D., Chehín R.N., Raisman-Vozari R., Del-Bel E. Tetracycline repurposing in neurodegeneration: focus on Parkinson’s disease. J. Neural. Transm. 2018;125:1403–1415. doi: 10.1007/s00702-018-1913-1. [DOI] [PubMed] [Google Scholar]
- 79.Lazzarini M., Martin S., Mitkovski M., Vozari R.R., Stühmer W., Bel E.D. Doxycycline restrains glia and confers neuroprotection in a 6-OHDA Parkinson model. Glia. 2013;61:1084–1100. doi: 10.1002/glia.22496. [DOI] [PubMed] [Google Scholar]
- 80.Pisanu A., Boi L., Mulas G., Spiga S., Fenu S., Carta A.R. Neuroinflammation in l-DOPA-induced dyskinesia: beyond the immune function. J. Neural. Transm. 2018;125:1287–1297. doi: 10.1007/s00702-018-1874-4. [DOI] [PubMed] [Google Scholar]
- 81.Miller A.H., Haroon E., Raison C.L., Felger J.C. Cytokine targets in the brain: impact on neurotransmitters and neurocircuits. Depress. Anxiety. 2013;30:297–306. doi: 10.1002/da.22084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bortolanza M., Padovan-Neto F.E., Cavalcanti-Kiwiatkoski R., dos Santos-Pereira M., Mitkovski M., Raisman-Vozari R., Del-Bel E. Are cyclooxygenase-2 and nitric oxide involved in the dyskinesia of Parkinson’s disease induced by l-DOPA? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2014.0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carta M., Carlsson T., Kirik D., Björklund A. Dopamine released from 5-HT terminals is the cause of L-DOPA-induced dyskinesia in parkinsonian rats. Brain. 2007;130:1819–1833. doi: 10.1093/brain/awm082. [DOI] [PubMed] [Google Scholar]
- 84.Wu Y., Dissing-Olesen L., MacVicar B.A., Stevens B. Microglia: Dynamic Mediators of Synapse Development and Plasticity. Trends Immunol. 2015;36:605–613. doi: 10.1016/j.it.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu C.-B., Blakely R.D., Hewlett W.A. The proinflammatory cytokines interleukin-1beta and tumor necrosis factor-alpha activate serotonin transporters. Neuropsychopharmacology. 2006;31:2121–2131. doi: 10.1038/sj.npp.1301029. [DOI] [PubMed] [Google Scholar]
- 86.Feyder M., Bonito-Oliva A., Fisone G. L-DOPA-Induced Dyskinesia and Abnormal Signaling in Striatal Medium Spiny Neurons: Focus on Dopamine D1 Receptor-Mediated Transmission. Front. Behav. Neurosci. 2011;5:71. doi: 10.3389/fnbeh.2011.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Silverdale M.A., Kobylecki C., Hallett P.J., Li Q., Dunah A.W., Ravenscroft P., Bezard E., Brotchie J.M. Synaptic recruitment of AMPA glutamate receptor subunits in levodopa-induced dyskinesia in the MPTP-lesioned nonhuman primate. Synapse. 2010;64:177–180. doi: 10.1002/syn.20739. [DOI] [PubMed] [Google Scholar]
- 88.Buttarelli F.R., Fanciulli A., Pellicano C., Pontieri F.E. The Dopaminergic System in Peripheral Blood Lymphocytes: From Physiology to Pharmacology and Potential Applications to Neuropsychiatric Disorders. Curr. Neuropharmacol. 2011;9:278–288. doi: 10.2174/157015911795596612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Penedo M.A., Rivera-Baltanás T., Pérez-Rodríguez D., Allen J., Borrajo A., Alonso-Crespo D., Fernández-Pereira C., Nieto-Araujo M., Ramos-García S., Barreiro-Villar C., et al. The role of dopamine receptors in lymphocytes and their changes in schizophrenia. Brain Behav. Immun. Health. 2021;12 doi: 10.1016/j.bbih.2021.100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2023. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- 95.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilkinson L. ggplot2: Elegant Graphics for Data Analysis by WICKHAM, H. Biometrics. 2011;67:678–679. doi: 10.1111/j.1541-0420.2011.01616.x. [DOI] [Google Scholar]
- 98.Russo P.S.T., Ferreira G.R., Cardozo L.E., Bürger M.C., Arias-Carrasco R., Maruyama S.R., Hirata T.D.C., Lima D.S., Passos F.M., Fukutani K.F., et al. CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinf. 2018;19:56. doi: 10.1186/s12859-018-2053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wei W., Amberkar S., Hide W. Bioconductor. Diffcoexp, Differential Co-expression Analysis. https://bioconductor.org/packages/release/bioc/html/diffcoexp.html
- 101.Santos N.P.C., Ribeiro-Rodrigues E.M., Ribeiro-Dos-Santos A.K.C., Pereira R., Gusmão L., Amorim A., Guerreiro J.F., Zago M.A., Matte C., Hutz M.H., Santos S.E.B. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum. Mutat. 2010;31:184–190. doi: 10.1002/humu.21159. [DOI] [PubMed] [Google Scholar]
- 102.Bispo A.G., Silva C.S., Sena-dos-Santos C., Dalledone Moura D., Koshimoto B.H.B., Santos-Lobato B.L., Ribeiro-Dos-Santos Â., Cavalcante G.C. Investigation of PRKN Mutations in Levodopa-Induced Dyskinesia in Parkinson’s Disease Treatment. Biomedicines. 2023;11:2230. doi: 10.3390/biomedicines11082230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All raw sequences are deposited at the European Nucleotide Archive (ENA), with the accession number PRJEB72411 and are publicly available as of the date of publication.
-
•
All data analysis codes are available in the GitHub repository: https://github.com/tati-souz/RNA-Seq-data-analysis.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.