Abstract
The early steps of adeno-associated virus (AAV) infection involve attachment to a variety of cell surface receptors (heparan sulfate, integrins, and fibroblast growth factor receptor 1) followed by clathrin-dependent or independent internalization. Here we have studied the subsequent intracellular trafficking of AAV particles from the endosomal compartment to the nucleus. Human cell lines were transduced with a recombinant AAV (rAAV) carrying a reporter gene (luciferase or green fluorescent protein) in the presence of agents that affect trafficking. The effects of bafilomycin A1, brefeldin A, and MG-132 were measured. These drugs act at the level of endosome acidification, early-to-late endosome transition, and proteasome activity, respectively. We observed that the transducing virions needed to be routed as far as the late endosomal compartment. This behavior was markedly different from that observed with adenovirus particles. Antiproteasome treatments with MG-132 led to a 50-fold enhancement in transduction efficiency. This effect was accompanied by a 10-fold intracellular accumulation of single-stranded DNA AAV genomes, suggesting that the mechanism of transduction enhancement was different from the one mediated by a helper adenovirus, which facilitates the conversion of the rAAV single-stranded DNA genome into its replicative form. MG-132, a drug currently in clinical use, could be of practical use for potentializing rAAV-mediated delivery of therapeutic genes.
Adeno-associated virus (AAV) is a nonpathogenic human parvovirus with a 4,679-nucleotide single-stranded DNA (ssDNA) genome. It is attractive as a vehicle for therapeutic gene transfer due to its innocuousness and high resistance to extreme conditions (4). Although AAV-based vectors have been shown to be efficient in gene transfer, allowing high and persistent levels of transferred gene expression, little is known about the molecular determinants of cell and tissue permissiveness for transduction. Gene transfer efficiency is limited by the conversion of the ssDNA genome into a double-stranded form suitable for gene expression. Genes encoded by adenovirus (a natural helper of AAV replication) facilitate this genome conversion (9, 10). In the absence of helper virus functions, cellular factors are believed to directly mediate second-strand synthesis (27), and their effect can be enhanced by a variety of genotoxic agents (39).
Distribution of receptors and coreceptors also accounts for differences in susceptibility to AAV transduction between cell types. Heparan sulfate proteoglycans act as primary receptors for AAV and mediate viral attachment and infection of target cells (36). Although heparan sulfate proteoglycans are widely distributed on the surface of many cell types, they may not be sufficient to allow efficient AAV entry. Qing et al. have shown previously that human fibroblast growth factor receptor 1 is needed to confer full AAV infectivity (28). In this context, fibroblast growth factor receptor 1 is used as a coreceptor for AAV entry into the host cell. The second coreceptor described is the αvβ5 integrin (35), which is also a secondary determinant for adenovirus tropism.
As a consequence of the interactions with their receptors, viruses can follow a variety of pathways for entry into cells. A primary one, used by most retroviruses, involves the fusion of the plasma membrane and the viral envelope at the cell surface and the release of the core viral particle into the cytosol (29). Other viruses such as adenoviruses or rhabdoviruses are internalized into clathrin-coated endocytic vesicles (21). A third category, exemplified by polyomaviruses, involves entry into the cell via the clathrin-independent caveola system (1). Different mechanisms are then used to release the viral material from the endosomal compartment. The maturation of endosomes involves a progressive decrease of their internal pH. This low-pH milieu triggers conformational changes in key viral proteins, exposing domains which either facilitate membrane fusion (rhabdoviruses) or disrupt the endosomal membrane (adenoviruses) (21). Depending on the virus, this endosome lysis occurs at different pHs and, consequently, at different stages of endosome maturation (21). It is suggested elsewhere that, once in the cytosol, viral nucleoprotein complexes reach the nuclei by interacting with the cytoskeleton and the nuclear import machinery (37). Degradation of the complexes in the proteasome may occur before they complete their transit and therefore limit the efficiency of infection (31).
AAV is known to enter into the cell in clathrin-coated vesicles (2), but the mechanisms of release of the particle into the cytosol and import into the nucleus are yet to be identified. Here, we have set out to elucidate the intracellular fate of the viral particles by monitoring the effect on AAV-mediated gene transfer efficiency of drugs acting at defined points along the intracellular route. Using drugs that act upon endosome acidification, early-to-late endosome transition, and proteasome activity, we show that AAV particles reach the late endosomal compartment before release and that a significant proportion of them are degraded by the proteasome. As a consequence, pharmacological manipulation of proteasome activity could be an effective means to enhance AAV-mediated gene transfer.
MATERIALS AND METHODS
rAAV preparation.
Recombinant AAV (rAAV) was prepared as described previously (33) except for the following modifications. The adenovirus helper functions were supplied by cotransfection of the pXX6 plasmid, a kind gift of R. J. Samulski (38). The AAV rep and cap genes were supplied by cotransfection by pACG2.1 (18) or PspRC (30). The plasmids encoding the rAAV genomes pSMD2-Luc and pSMD2-eGFP were derived from pSMD2 (34) and contain the luciferase and the enhanced green fluorescent protein (eGFP) genes, respectively, under the transcriptional control of the cytomegalovirus IE1 promoter-enhancer, the intervening sequence, and the polyadenylation site of the human β-globin gene. Transfections were performed with polyethyleneimine (PEI; 25 kDa; Aldrich) as described previously (5). The virus was purified by one round of ultracentrifugation on an isopycnic CsCl2 gradient, followed by dialysis (33). Physical particles were estimated by dot blotting. Infectious particles and pseudo-wild-type particles were quantified by the replication center assay (33) with further modification (30). Briefly, HeLa cells and the rep-cap-expressing cell line HeLa RC (30) were infected in 24-well plates with serial dilutions of the rAAV preparation in the presence of wild-type adenovirus type 5 (wtAd5) at a multiplicity of infection (MOI) of 50. Twenty-four hours later, cells were harvested and transferred onto 0.45-μm-pore-size nylon membranes and lysed under alkaline conditions. The membranes were then hybridized with a probe corresponding to the transgene sequence. A positive signal indicates transduction events at the corresponding dilution. Titers ranged between 1011 and 1012 physical particles per ml and 108 and 109 infectious particles per ml (about 2 ml per preparation) for 25 150-mm plates (109 cells).
Adenoviruses.
AdΔE1ΔE3-cytomegalovirus-eGFP-simian virus 40-poly(A) (AdGFP) is a kind gift from E. J. Kremer (13); wtAd5 is a kind gift from P. Moullier.
Cell lines and treatments.
HeLa (human cervical carcinoma), 293 (human Ad5 DNA-transformed embryonic kidney), and HepG2 (human hepatoblastoma) cells were seeded in 24-well plates at 5 × 104 cells per well in 10% fetal calf serum-supplemented Dulbecco's modified Eagle's medium and incubated at 37°C in a 5% CO2 atmosphere. Twenty-four hours later, cells were infected in the presence of the inhibitors. In all experiments, cells were infected in Dulbecco's modified Eagle's medium supplemented with either 1 or 10% fetal calf serum for 2 h at an MOI ranging from 1 to 50 infectious particles/cell. Treatments with bafilomycin A1 (Sigma) and MG-132 (Calbiochem) were done by addition of the drug to the culture medium at the time of infection and maintained during infection (1 h 30 min to 2 h). Brefeldin A (Fluka) was applied 5 min prior to infection and kept in the medium for 1 h 30 min to 2 h, concomitantly with the infection.
Luciferase assay.
For transduction analysis, reporter gene expression was measured as follows. Cells were lysed in 250 μl of lysis buffer (25 mM Tris-phosphate, 1 mM dithiothreitol, 1 mM EDTA, 15% glycerol, 8 mM MgCl2, 0.2% Triton X-100) for 10 min. Cell membranes and debris were pelleted by spinning at 10,000 × g. Fifty microliters of the supernatant was mixed with 100 μl of assay buffer (25 mM Tris-phosphate, 1 mM dithiothreitol, 1 mM EDTA, 15% glycerol, 8 mM MgCl2, 2 mM ATP) and 100 μl of 167 μM luciferin (Molecular Probes), and the relative light units (RLU) were measured with a luminometer (Mediators Diagnostika) for 10 s. Each transfection within an assay was done in triplicate. Each assay was performed at least two times. The level of expression is expressed as light units (1 RLU = 10 photons) per second and per well. All the values are given as RLU per second per well and are standardized by the protein content as measured by optical density (see below).
Cell viability assay.
Cell viability was assessed by measuring either the mitochondrial dehydrogenase activity or the total protein content of each point of the assay. Briefly, for mitochondrial dehydrogenase activity, active cells are able to reduce MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; Sigma), which reduction can be measured spectrophotometrically (23), and proteins were quantified with a Bradford-based assay (Bio-Rad).
Fluorescence-activated cell sorting.
For eGFP detection, cells were resuspended in phosphate-buffered saline and sorted for fluorescence expression with a FACScalibur (Becton Dickinson). Data were further analyzed with Cellquest software (Becton Dickinson).
Viral DNA analysis.
Low-molecular-weight (LMW) DNA was extracted according to the Hirt method (11). One-half of the total well LMW DNA content was loaded undigested on a 1% agarose gel. The migration time was 20 h in 1× Tris-borate-EDTA buffer. DNA was transferred onto a positively charged nylon membrane under alkaline conditions. DNA probes were labeled with [α-32P]dCTP by random priming. Membranes were hybridized with DNA probes in Church buffer (6). After two washes in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (each for 15 min at 65°C) and 24 h of exposure, membranes were scanned on a PhosphorImager (Molecular Dynamics). Specific signals were quantified with Image Quant software (Molecular Dynamics).
Real-time PCR.
Oligonucleotide primers and Taqman probes were designed using Primer Express (Perkin-Elmer Applied Biosystems Inc.) and Oligo 4.0 (primer analysis software). The sequences used were the luciferase gene (GenBank accession no. M15077) and the human cytochrome b gene (GenBank accession no. J01415) for standardization. The sequences were as follows: Luc forward (1878luc.f), 5′-GGCGCGTTATTTATCGGAGTT-3′; Luc reverse (1950luc.r), 5′-TACTGTTGAGCAATTCACGTTCATT-3′; human cytochrome b forward (h83cytb), 5′-CCGCATGATGAAACTTCGG-3′; and human cytochrome b reverse (h126cytb), 5′-ATAGTCCTGTGGTGATTTGGAGG-3′.
Quantitative PCR was used to estimate relative values of specific DNA sequences. It is based on real-time detection of PCR products by measuring the increase of fluorescence due to the presence of SYBRgreen. This dye has the property of being fluorescent only in the presence of double-stranded DNA. The increase of fluorescence is proportional to the amount of PCR product. It is also related to the initial number of copies through a particular parameter, the threshold cycle. It is defined as the PCR cycle at which the fluorescence signal rises above a predetermined baseline (threshold) value. The threshold value must be low enough to correspond to the exponential phase and is related to the initial number of template copies. The PCR amplifications were performed using 10 ng of Hirt DNA diluted in a reaction buffer containing 1× Taqman buffer, 5 mM MgCl2, 2.5 U of ampliTaq Gold DNA polymerase, and 200 nM primers (forward and reverse) in a final volume of 25 μl. Cycling conditions consisted of an ampliTaq Gold activation step at 95°C for 10 min followed by 40 cycles of two steps, 15 s of denaturation at 95°C and 60 s of annealing at 60°C. The PCR was performed on an ABI PRISM 7700 sequence detector (Perkin-Elmer Applied Biosystems) allowing automatic collection of the fluorescence emission data. The Luc DNA level of each sample was determined as an average from data obtained from two independent PCRs, each including duplicates.
RESULTS
Our studies were performed using human cell lines which exhibit different levels of permissiveness for rAAV transduction, high for 293 and HeLa and low for HepG2. This was designed in order to avoid a possible bias associated with different susceptibilities to AAV attachment or genome conversion. In all the experiments described, cells were seeded in 24-well plates and infected 24 h later with rAAV-Luc (1 to 50 infectious particles/cell) in the presence of increasing concentrations of the inhibitor. Luciferase expression and cell viability were quantified 24 to 48 h later. At an MOI of 50 and in the absence of any drug treatment, the luciferase expression was between 106 and 107 RLU per well for 293 and HeLa cells and between 104 and 105 RLU for HepG2 cells (see untreated transduced control cells in Fig. 1, 2, and 3).
FIG. 1.
Effect of bafilomycin A1 on AAV-mediated transduction. Cells were infected with rAAV-Luc at an MOI of 50, in the presence of increasing doses of bafilomycin A1. Luciferase activity and cell viability were assessed 24 h postinfection. (A) 293 cells. (B) HepG2 cells. (C) HeLa cells. (D) HeLa cells were infected for 2 h with rAAV-Luc at an MOI of 2, 25, or 100, in the presence of increasing doses of bafilomycin A1. Values are given with the standard deviations (n = 3). All the values are standardized with the protein content of the sample. ni, noninfected cells.
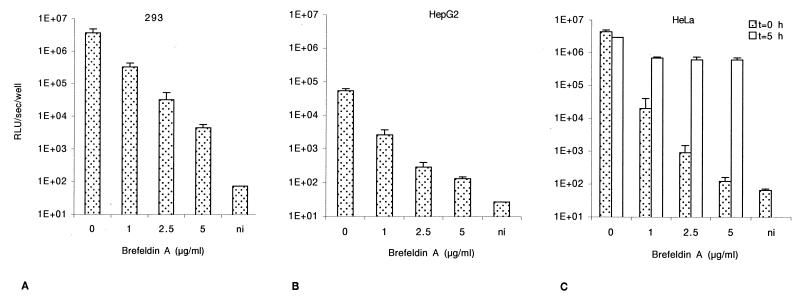
FIG. 2.
Effect of brefeldin A on AAV-mediated cell transduction. Cells were infected with rAAV-Luc at an MOI of 50 for 1 h 30 min in the presence of increasing amounts of brefeldin A. They were then washed to remove both unattached virus and the drug and further incubated in fresh complete medium. Luciferase activity was assessed at 24 h postinfection. (A) 293 cells. (B) HepG2 cells. (C) HeLa cells. In panel C, open bars correspond to postinfection pulse treatment with brefeldin A. Cells were infected with rAAV-Luc at an MOI of 50 at t = 0 h for 1 h 30 min, washed, and incubated in fresh medium for 3 h 30 min followed by an additional treatment (1 h 30 min) with brefeldin A (from t = 5 h to 6 h 30 min). After washing, cells were incubated in fresh medium until luciferase assay at 24 h postinfection. All the values are standardized with the protein content and shown with the standard deviations (n = 3). (D) Comparison of effects of brefeldin A treatment on adenovirus and rAAV transduction. HeLa cells were infected either with AdGFP (dotted bars, MOI of 1) or rAAV-eGFP (hatched bars, MOI of 10), in the presence of increasing doses of brefeldin A for 2 h. Cells were analyzed by flow cytometry at 24 h postinfection. The left vertical axis indicates the percentages of GFP-positive cells, and the right vertical axis indicates the mean fluorescence per cell. ni, noninfected cells.
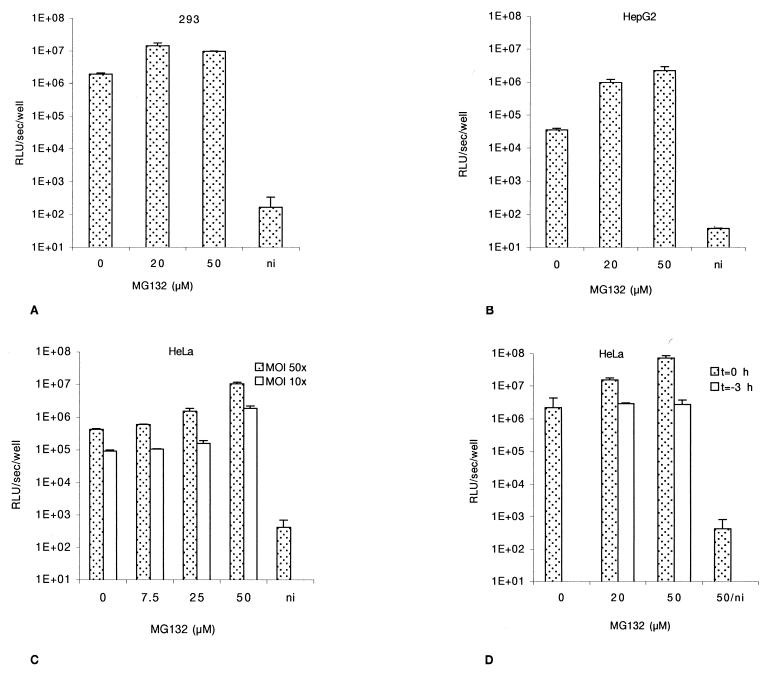
FIG. 3.
Effect of MG-132 on AAV-mediated transduction. Cells were infected with rAAV-Luc for 2 h at an MOI of 50 and then washed and further incubated in fresh complete medium. Luciferase activity was assessed at 24 h postinfection. (A) 293 cells. (B) HepG2 cells. (C) HeLa cells infected at an MOI of 50 or 10. (D) MG-132 reversion. Dotted bars represent cells infected with rAAV-Luc and treated at t = 0 for 2 h with MG-132. Open bars represent cells pulse treated for 2 h with MG-132 3 h prior to infection with rAAV-Luc for 2 h. ni, noninfected cells. Values are given with the standard deviations (n = 3). All the values are standardized with the protein content.
To rule out the possibility of an alteration of virus binding and internalization following drug addition, cells were exposed to rAAV (MOI of 1,000) in the presence of drug and the internalized vector genomes were visualized by fluorescent in situ hybridization after 10 and 60 min. The results (not shown) indicated that rAAV was internalized normally following bafilomycin A1, brefeldin A, and MG-132 treatments.
Low endosomal pH favors release of AAV particles.
We determined the initial fate of the internalized virus. Once in the endosomal compartment, viruses may require acidification to escape the vesicle and achieve successful infection. To demonstrate a requirement for low endosomal pH, cells were treated with bafilomycin A1, an inhibitor of vacuolar proton ATPases (3). 293 cells were incubated for 2 h with the rAAV at an MOI of 50 with increasing doses of bafilomycin A1 in the medium. At 50 nM, luciferase activity was 15- to 50-fold lower than in untreated cells (Fig. 1A). Higher doses did not further affect the level of reporter gene expression (data not shown). The susceptibility of HepG2 cells receiving the same treatment was more limited. A slight increase or no effect in luciferase expression was observed at 50 and 100 nM, respectively (Fig. 1B). A limited effect of bafilomycin A1 was detected at higher doses, reaching a 10-fold decrease at 300 nM (Fig. 1B), without significant toxicity. Using HeLa cells, bafilomycin A1 treatment (50 to 150 nM) led to a dose-dependent decrease of luciferase activity, reaching 43-fold at a dose of 150 nM (Fig. 1C). At this concentration of bafilomycin A1, the decrease of luciferase activity was 16, 14, and 19-fold at MOIs of 2, 25 and 100, respectively (Fig. 1D). Therefore, the rate of inhibition was independent of the MOI. Higher doses of bafilomycin A1 did not induce a higher decrease of the luciferase activity, and no significant toxic effect was observed.
AAV particles are processed further than the early endocytic vesicle.
Once in early endosomes, viral particles can be further transported to the late endosomal compartment or released in the cytosol (32). To test whether AAV particles are trafficked further than the early endosomal compartment, cells were treated with brefeldin A, a fungal antibiotic which causes early endosomes to form a tubular network and prevents early-to-late endosome transition (19, 25). Cells were incubated for 2 h with the AAV vector at an MOI of 50, and brefeldin A was added in the infection mixture at doses ranging from 0.5 to 5 μg/ml. Treatment with brefeldin A led to a decrease of 2 to 3 orders of magnitude in the efficiency of gene transfer, as measured by the level of luciferase activity in the three cell lines. The diminution of luciferase activity at 5 μg of brefeldin A per ml was above 800-fold for 293 cells and 400-fold for HepG2 cells as shown in Fig. 2A and B. In HeLa cells, at an MOI of 50, the diminution reached more than 3 × 104-fold upon treatment with 5 μg of brefeldin A per ml (Fig. 2C, dotted bar). A dose-dependent diminution of gene delivery was also observed at MOIs of 5 and 20 (data not shown). At the highest dose of brefeldin A, a toxic effect was observed.
Brefeldin A displays a pleiotropic effect, and the decrease of luciferase activity could have been due to an alteration in the synthesis of the luciferase protein rather than an inhibition of gene transfer. Since the effect of brefeldin A is readily reversible when the drug is removed (25), and because the treatments were simultaneous with infection and 24 h before the beginning of transgene expression, we believe that the observed effect was not artifactual. In order to confirm that point, we performed a postinfection pulse treatment with brefeldin A. HeLa cells were incubated with the AAV-Luc at an MOI of 50 for 2 h. Five hours postinfection, cells were treated for 1.5 h with increasing doses of brefeldin A. The medium was then changed, and cells were further incubated until 24 h postinfection. Figure 2C shows that, when the treatment was applied 5 h postinfection, a less-than-fivefold decrease of luciferase activity was observed (open bars), showing that the drug is effective only when present in the cell at the same time as the virus.
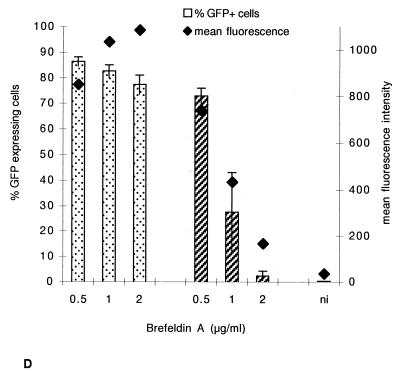
Finally, to assess the specificity of brefeldin A for AAV trafficking, we compared the effect of the drug on HeLa cells treated under the same conditions and infected either with an rAAV encoding GFP driven by the cytomegalovirus promoter (MOI of 10) or with an adenovirus bearing the same expression cassette (MOI of 1). Adenovirus is known to readily escape the early endosome and to be released into the cytosol (17) and is therefore not expected to be sensitive to brefeldin A. Indeed, we did not observe any effect of brefeldin A on adenovirus-mediated gene transfer, whereas a strong diminution was seen with rAAV (Fig. 2D). Altogether, these results indicated that a high proportion of incoming AAV particles must be routed toward the late endosomal compartment.
AAV transduction efficiency is enhanced by the peptide aldehyde MG-132.
Early virion degradation may limit the efficiency of AAV-mediated transduction. We examined whether incoming AAV virions could be eliminated by the proteasome, which degrades ubiquitin-conjugated proteins through an ATP-dependent mechanism and is present in both cytosol and nucleus (16). This was assessed by treating cells with the peptide aldehyde MG-132, a strong inhibitor of the chymotrypsin-like activity of the proteasome (16).
Cells were infected for 2 h at an MOI of 50 with rAAV in the presence of increasing doses of MG-132 (up to 50 μM), and luciferase activity was assessed 24 h later. Transduction efficiency in 293 cells was increased sevenfold at 20 μM MG-132. Higher doses did not further enhance the transduction level (Fig. 3A). HepG2 cells showed higher sensitivity to MG-132, with a 60-fold transduction increase at 50 μM (Fig. 3B). Similarly, luciferase activity in HeLa cells was also enhanced in a dose-dependent fashion, increasing 20- and 25-fold at MOIs of 10 and 50, respectively (Fig. 3C). At the highest dose, MG-132 showed some toxicity.
The action of MG-132 on the proteasome can be fully reversed within 1 h after withdrawal of the compound (31). HeLa cells were pulsed for 2 h with 20 and 50 μM MG-132 3 h before rAAV infection. The luciferase activity was quantified 24 h postinfection. As seen in Fig. 3D, under coincubation of the cells with rAAV and MG-132 the luciferase activity increased as expected (33-fold at 50 μM, dotted bars), whereas both doses were ineffective when the cells were pulsed prior to infection (Fig. 3D, solid bars).
Lactacystin also affects the chymotrypsin-like and trypsin-like activities of the proteasome, although in an irreversible manner. A similar enhancement of rAAV transduction was observed when cells were treated with this inhibitor (20 μM). In contrast, calpain inhibitors I and II (up to 100 μM) had no effect on transduction efficiency (data not shown).
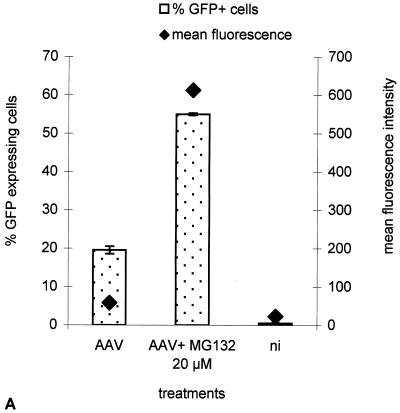
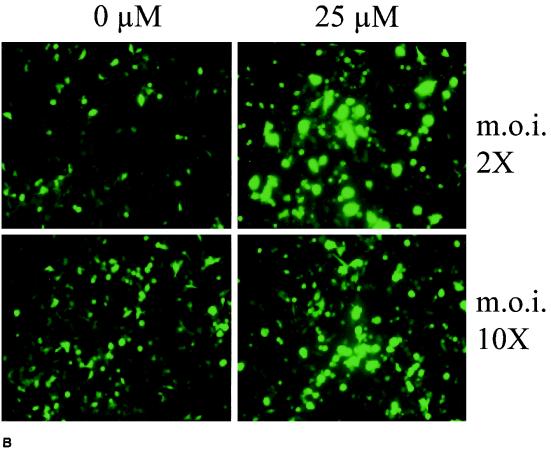
The enhancement effect of MG-132 was also analyzed by flow cytometry, following transduction with rAAV-eGFP (Fig. 4). Cells were infected for 2 h at an MOI of 2 or 10 with the vector in the presence of increasing doses of MG-132 (up to 50 μM). GFP expression was assessed 24 to 48 h later. We observed that the amount of cells expressing the reporter gene increased from 19 to 55% (2.9-fold increase) under 20 μM MG-132 treatment, whereas cellular expression, measured as the mean fluorescence per cells (FL1-H), was increased 10.5-fold (Fig. 4A), leading to an overall increase of 30-fold, consistent with our observation in the luciferase assay. The effect of MG-132 on rAAV transduction was further confirmed by direct microscopic observation showing a strong enhancement of the GFP signal upon 25 μM MG-132 treatment (Fig. 4B). The specificity of MG-132 for the viral infection mechanism and not for a subsequent step like transgene expression was assessed by comparing its effects on a transfected plasmid. The pSMD2-eGFP plasmid, which was used to generate rAAV-Luc, was transfected into HeLa cells with PEI (25 kDa), and luciferase expression was measured 24 h later. The results showed that PEI-complexed plasmid was not significantly affected by MG-132 (data not shown).
FIG. 4.
Effect of MG-132 on the activity of AAV-eGFP. (A) GFP expression was measured by flow cytometry in HeLa cells 24 h following infection with rAAV-eGFP at an MOI of 10, in the presence or absence of 20 μM MG-132. The left vertical axis indicates the percentages of GFP-positive cells, and the right vertical axis indicates the mean fluorescence per cell. (B) HeLa cells were infected with rAAV-eGFP at MOIs of 2 and 10 in the presence or absence of 25 μM MG-132. Magnification, 20× (Zeiss Axiovert 135 epifluorescence microscope). The difference of intensity between the upper and lower panels is due to different exposure times at the MOI of 2 (2 s) and the MOI of 10 (1 s). ni, noninfected cells.
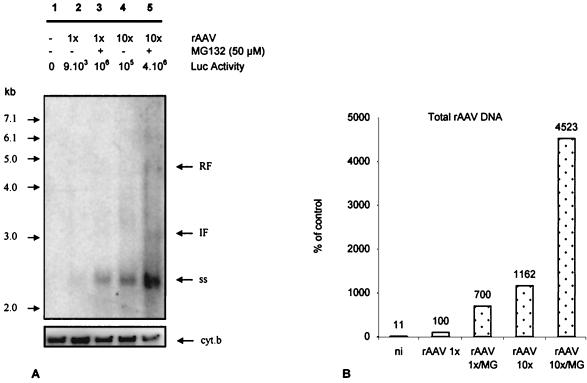
We then reasoned that, if the virus was targeted to the proteasome before it had delivered its genome to the nucleus, most of the undelivered genome would be readily degraded in the cytoplasm after 24 h, since the half-life of naked DNA in the cytosol is 50 to 90 min (15). On the other hand, when antiproteasome treatment is applied, viral capsid would not be degraded and the viral genome would still be protected. Therefore, we should be able to detect more viral genome per cell. HeLa cells were infected at MOIs of 1 and 10 in the presence or absence of 50 μM MG-132 for 2 h. Twenty-four hours postinfection, the luciferase expression was measured and the LMW DNA was extracted, blotted, and hybridized with a luciferase probe (Fig. 5A). The amounts of DNA loaded per lane were standardized using a cytochrome b probe detecting the mitochondrial genome. A major hybridizing signal was detected in each sample, with an apparent molecular size of 2.4 kb, and was identified as AAV ssDNA (10). This signal increased in the presence of MG-132. At the highest MOI and in the presence of MG-132 (lane 5), the expected intermediate (3-kb) and replicative (4.4-kb) forms of the AAV genome were detected. The total amounts of signal were determined by PhosphorImager analysis (see Materials and Methods) and compared (Fig. 5B). In the presence of MG-132, the amounts of vector genome were seven- and fourfold higher at MOIs of 1 and 10, respectively. The increased amount of vector genome present in the cell paralleled the enhancement of luciferase activity in the presence of MG-132 (105- and 36-fold at MOIs of 1 and 10, respectively [Fig. 5A]). In lane 5 (MOI of 10, 50 μM MG-132), ssDNA represents more than 95% of the hybridizing signal, and most of the increase in the overall signal was attributed to the accumulation of ssDNA.
FIG. 5.
Accumulation of viral DNA following MG-132 treatment. HeLa cells were infected with rAAV-Luc at an MOI of 1 or 10 in the presence or absence of 50 μM MG-132 for 2 h and cultivated for 24 h. Luciferase expression was monitored and standardized with the protein content. LMW DNA was extracted, and undigested DNA was resolved on a 1% agarose gel, transferred to a nylon membrane, and hybridized against a radiolabeled luciferase DNA probe. (A) Hybridizing LMW DNA against the luciferase gene probe reveals rAAV genome. Lane 1, uninfected cells; lanes 2 and 3, infected cells at an MOI of 1 in the absence and presence of MG-132, respectively; lanes 4 and 5, infected cells at an MOI of 10 in the absence and presence of MG-132, respectively. Luciferase expression (background subtracted) is indicated above each lane as RLU per well per microgram of protein. The DNA molecular size ladder is shown on the left of the membrane. ss, single-stranded rAAV genome; RF, rAAV replicative (4.4-kb) form; IF, intermediate form. The lower part of the gel shows the same membrane hybridized against a cytochrome b probe. (B) DNA quantification of rAAV genome. The intensity of the signal corresponding to the ssAAV genome was normalized using the intensity of the cytochrome b signal with Image Quant software. Lane 2 of panel A is taken as the 100% signal for cytochrome b and rAAV. MG, MG-132; ni, noninfected cells.
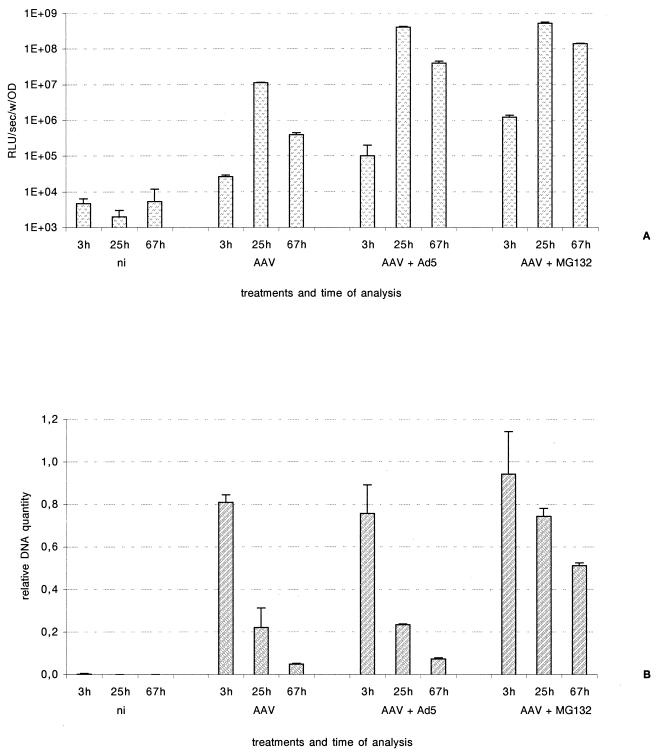
Adenovirus coinfection enhances rAAV transduction by increasing the rate of conversion of ssDNA to double-stranded DNA. The global amount of intracellular rAAV DNA is not expected to change significantly upon adenovirus enhancement, in contrast to what we had observed with MG-132. To verify this point, we performed the following experiment. HeLa cells were infected with rAAV-Luc at an MOI of 50 and either coinfected with an adenovirus (MOI of 1) or treated with 50 μM MG-132. The transduction efficiency was monitored through luciferase activity, and the total amount of vector DNA was measured using real-time PCR (see Materials and Methods) (Fig. 6). At 25 h postinfection, luciferase expression was increased 40-fold following adenovirus coinfection and 50-fold after MG-132 treatment. These values reached 100- and 350-fold, respectively, at 67 h postinfection (Fig. 6A). The total amounts of rAAV genome detected in MG-132-treated samples were 3.7-fold (25 h) and 10-fold (67 h) higher than those in untreated cells, whereas coinfection with adenovirus did not lead to a significant increase of amounts of rAAV genome in the cell (Fig. 6B). These results confirmed that MG-132 treatment allows the incoming rAAV genomes to accumulate in the cells.
FIG. 6.
Real-time PCR quantification of rAAV-Luc genomes following infection in the presence or absence of wtAd and MG-132. HeLa cells were infected with rAAV-Luc at an MOI of 50 and either coinfected with an adenovirus at an MOI of 1 or treated with 50 μM MG-132. (A) The luciferase activity was measured at the indicated times postinfection. The values were standardized with the protein content. (B) LMW DNA was extracted at the indicated times following infection. The amount of vector DNA was measured using real-time PCR with a luciferase-specific probe. Human cytochrome b sequences were amplified for standardization. The results are expressed as relative DNA signal (AAV) in arbitrary units. ni, noninfected cells; OD, optical density.
DISCUSSION
It has been shown previously that AAV internalization requires dynamin, indicating that clathrin-dependent endocytosis is the main pathway for rAAV entry into cells (7). Yet, it was suggested elsewhere that a dynamin-independent pathway could also be involved in rAAV infection (7). In our hands, internalization of rAAV was unaffected by cytochalasin B, an actin microfilament network-disrupting agent (24), which excluded phagocytosis and macropinocytosis as primary entry routes (data not shown). In addition, the cell lines studied showed none (HepG2) or partial (HeLa and 293) susceptibility to potassium depletion (A.-M. Douar, unpublished results), which specifically blocks the clathrin-dependent endocytic pathway (14). It is likely that AAV enters cells via both clathrin-dependent and -independent pathways, whose respective contributions may vary from cell to cell.
It was recently shown that the trafficking of AAV depends on acidification of the endosome (2). In this respect, AAV behaves like many other viruses, which require a low endosomal pH to escape the endocytic vesicle. Our studies confirm this observation and further extend it. The requirement of an acidic pH in endosomes was more or less stringent depending on the cell type: transduction of HeLa and 293 cells could be inhibited at low doses of bafilomycin A1, whereas HepG2 exhibited a weaker sensitivity to the antibiotic.
AAV and its natural helper, adenovirus, follow very different pathways for leaving the endosomal compartment. The adenovirus capsid has a strong endosomolytic activity and is released into the cytoplasm early after entry. This process is insensitive to bafilomycin A1 and therefore independent of the V-ATPase proton pump (26). Interestingly, coinfections with early-region-mutant adenoviruses do not enhance rAAV transduction, indicating that the presence of adenovirus capsid does not potentiate AAV entry (10). We show here with brefeldin A treatment that, in contrast to adenovirus, AAV particles must remain in the endosomal compartment up to a later stage in order to be efficiently released. Acidification is required for early-to-late endosomal transition, and this may explain why AAV entry strongly depends on low endosomal pH. Other viruses (Semliki Forest virus and influenza virus) are also routed toward the late endosomal compartment (21). This pathway may be advantageous for the infectious process since the particles will be directly delivered to the perinuclear region.
Finally, we observe that the proteasome inhibitor MG-132 increases rAAV transduction efficiency up to 60-fold. We show that the less permissive the cell line, the higher the enhancement, and that the drug must be present early during infection in order to mediate its augmenting effect. Duan et al. have recently reported an enhancing effect of antiproteasome treatments on rAAV-mediated transduction (8). Minor differences with our data must be pointed out. First, they found that the calpain inhibitor I (MG101) was enhancing transduction, whereas it was inactive in our system. Second, the effect of MG-132 was observed at doses 10 times lower than the one that we used. This may reflect differences in the physiological status of the transduced cells (differentiated or primary cultures versus established cell lines).
Our analysis of viral DNA following MG-132 treatment indicates that protection against proteasome-mediated degradation leads to an intracellular accumulation of ssDNA viral genomes which in turn may increase their chances to become converted into transcriptionally active double-stranded DNA templates. This accumulation of vector ssDNA was not observed in the presence of helper adenovirus, indicating different mechanisms of transduction enhancement. MG-132 can modify the cell cycle by inducing p53 expression (20) and activate stress kinases and induce heat shock gene transcription (12, 22), all of which could eventually affect cellular permissiveness for AAV transduction by promoting ssDNA genome conversion (39). However, the fact that the antiproteasome treatment is effective only when given at the time of exposure to the vector and is without effect 5 h later argues against a major contribution of these indirect effects of MG-132 on transduction enhancement. This effect of MG-132, a drug currently used with human patients, could be of interest for potentiating the gene transfer capability of AAV vectors in the clinic.
ACKNOWLEDGMENTS
We thank Antoine Kichler and Eric Kremer for helpful discussions and critical reading of the manuscript.
This work was performed with the financial support of the Association Française contre les Myopathies (AFM).
REFERENCES
- 1.Anderson H A, Chen Y, Norkin L C. Bound simian virus 40 translocates to caveolin-enriched membrane domains, and its entry is inhibited by drugs that selectively disrupt caveolae. Mol Biol Cell. 1996;7:1825–1834. doi: 10.1091/mbc.7.11.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett J S, Wilcher R, Samulski R J. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer N, Schober D, Prchla E, Murphy R F, Blaas D, Fuchs R. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J Virol. 1998;72:9645–9655. doi: 10.1128/jvi.72.12.9645-9655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berns K I. Parvoviridae: the virus and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2173–2197. [Google Scholar]
- 5.Boussif O, Lezoualc'h F, Zanta M A, Mergny M D, Scherman D, Demeneix B, Behr J P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan D, Li Q, Kao A W, Yue Y, Pessin J E, Engelhardt J F. Dynamin is required for recombinant adeno-associated virus type 2 infection. J Virol. 1999;73:10371–10376. doi: 10.1128/jvi.73.12.10371-10376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan D, Yue Y, Yan Z, Yang J, Engelhardt J F. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J Clin Investig. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari F K, Samulski T, Shenk T, Samulski R J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher K J, Gao G P, Weitzman M D, DeMatteo R, Burda J F, Wilson J M. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 12.Kim D, Kim S H, Li G C. Proteasome inhibitors MG132 and lactacystin hyperphosphorylate HSF1 and induce hsp70 and hsp27 expression. Biochem Biophys Res Commun. 1999;254:264–268. doi: 10.1006/bbrc.1998.9840. [DOI] [PubMed] [Google Scholar]
- 13.Kremer E J, Boutin S, Chillon M, Danos O. Canine adenovirus vectors: an alternative for adenovirus-mediated gene transfer. J Virol. 2000;74:505–512. doi: 10.1128/jvi.74.1.505-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin J M, Brown M S, Goldstein J L, Anderson R G. Depletion of intracellular potassium arrests coated pit formation and receptor-mediated endocytosis in fibroblasts. Cell. 1983;33:273–285. doi: 10.1016/0092-8674(83)90356-2. [DOI] [PubMed] [Google Scholar]
- 15.Lechardeur D, Sohn K J, Haardt M, Joshi P B, Monck M, Graham R W, Beatty B, Squire J, O'Brodovich H, Lukacs G L. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- 16.Lee D H, Goldberg A L. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 17.Leopold P L, Ferris B, Grinberg I, Worgall S, Hackett N R, Crystal R G. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum Gene Ther. 1998;9:367–378. doi: 10.1089/hum.1998.9.3-367. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Samulski R J, Xiao X. Role for highly regulated rep gene expression in adeno-associated virus vector production. J Virol. 1997;71:5236–5243. doi: 10.1128/jvi.71.7.5236-5243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, Klausner R D. Brefeldin A's effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell. 1991;67:601–616. doi: 10.1016/0092-8674(91)90534-6. [DOI] [PubMed] [Google Scholar]
- 20.Magae J, Illenye S, Tejima T, Chang Y C, Mitsui Y, Tanaka K, Omura S, Heintz N H. Transcriptional squelching by ectopic expression of E2F-1 and p53 is alleviated by proteasome inhibitors MG-132 and lactacystin. Oncogene. 1997;15:759–769. doi: 10.1038/sj.onc.1201251. [DOI] [PubMed] [Google Scholar]
- 21.Marsh M, Helenius A. Virus entry into animal cells. Adv Virus Res. 1989;36:107–151. doi: 10.1016/S0065-3527(08)60583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meriin A B, Gabai V L, Yaglom J, Shifrin V I, Sherman M Y. Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J Biol Chem. 1998;273:6373–6379. doi: 10.1074/jbc.273.11.6373. [DOI] [PubMed] [Google Scholar]
- 23.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 24.Paccaud J P, Siddle K, Carpentier J L. Internalization of the human insulin receptor. The insulin-independent pathway. J Biol Chem. 1992;267:13101–13106. [PubMed] [Google Scholar]
- 25.Pelham H R. Multiple targets for brefeldin A. Cell. 1991;67:449–451. doi: 10.1016/0092-8674(91)90517-3. [DOI] [PubMed] [Google Scholar]
- 26.Perez L, Carrasco L. Involvement of the vacuolar H(+)-ATPase in animal virus entry. J Gen Virol. 1994;75:2595–2606. doi: 10.1099/0022-1317-75-10-2595. [DOI] [PubMed] [Google Scholar]
- 27.Qing K, Khuntirat B, Mah C, Kube D M, Wang X S, Ponnazhagan S, Zhou S, Dwarki V J, Yoder M C, Srivastava A. Adeno-associated virus type 2-mediated gene transfer: correlation of tyrosine phosphorylation of the cellular single-stranded D sequence-binding protein with transgene expression in human cells in vitro and murine tissues in vivo. J Virol. 1998;72:1593–1599. doi: 10.1128/jvi.72.2.1593-1599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 29.Russell S J, Cosset F L. Modifying the host range properties of retroviral vectors. J Gene Med. 1999;1:300–311. doi: 10.1002/(SICI)1521-2254(199909/10)1:5<300::AID-JGM59>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 30.Salvetti A, Oreve S, Chadeuf G, Favre D, Cherel Y, Champion A P, David A J, Moullier P. Factors influencing recombinant adeno-associated virus production. Hum Gene Ther. 1998;9:695–706. doi: 10.1089/hum.1998.9.5-695. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard J M. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slepnev V I, De Camilli P. Endocytosis: an overview. In: Kabanov A V, Felgner P L, Seymou L W, editors. Self-assembling complexes for gene delivery: from laboratory to clinical trial. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1998. pp. 71–88. [Google Scholar]
- 33.Snyder R O, Xiao X, Samulski R J. Production of recombinant adeno-associated viral vectors. In: Dracopoli J H N, Krof B, Moir D, Morton C, Seidman C, Seidman J, Smith D, editors. Current protocols in human genetics. New York, N.Y: John Wiley and Sons, Inc.; 1996. pp. 12.1.1–12.1.24. [Google Scholar]
- 34.Snyder R O, Spratt S K, Lagarde C, Bohl D, Kaspar B, Sloan B, Cohen L K, Danos O. Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum Gene Ther. 1997;8:1891–1900. doi: 10.1089/hum.1997.8.16-1891. [DOI] [PubMed] [Google Scholar]
- 35.Summerford C, Bartlett J S, Samulski R J. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 36.Summerford C, Samulski R J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suomalainen M, Nakano M Y, Keller S, Boucke K, Stidwill R P, Greber U F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao X, Li J, Samulski R J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakinoglu A O, Heilbronn R, Burkle A, Schlehofer J R, zur Hausen H. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 1988;48:3123–3129. [PubMed] [Google Scholar]