Abstract
Production of the human immunodeficiency virus type 1 (HIV-1) Gag-Pol precursor protein results from a −1 ribosomal frameshifting event. In infected cells, this generates Gag and Gag-Pol in a ratio that is estimated to be 20:1, a ratio that is conserved among retroviruses. To examine the impact of this ratio on HIV-1 replication and viral assembly, we altered the Gag/Gag-Pol ratio in virus-producing cells by cotransfecting HIV-1 proviral DNA with an HIV-1 Gag-Pol expression vector. Two versions of the Gag-Pol expression vector were used; one contains an active protease [PR(+)], and the other contains an inactive protease [PR(−)]. In an attempt to produce viral particles with Gag/Gag-Pol ratios ranging from 20:21 to 20:1 (wild type), 293T cells were cotransfected with various ratios of wild-type proviral DNA and proviral DNA from either Gag-Pol expression vector. Viral particles derived from cells with altered Gag/Gag-Pol ratios via overexpression of PR(−) Gag-Pol showed a ratio-dependent defect in their virion protein profiles. However, the defects in virion infectivity were independent of the nature of the Gag-Pol expression vector, i.e., PR(+) or PR(−). Based on equivalent input of reverse transcriptase activity, we estimated that HIV-1 infectivity was reduced 250- to 1,000-fold when the Gag/Gag-Pol ratio in the virion-producing cells was altered from 20:1 to 20:21. Although virion RNA packaging was not affected by altering Gag/Gag-Pol ratios, changing the ratio from 20:1 to 20:21 progressively reduced virion RNA dimer stability. The impact of the Gag/Gag-Pol ratio on virion RNA dimerization was amplified when the Gag-Pol PR(−) expression vector was expressed in virion-producing cells. Virions produced from cells expressing Gag and Gag-Pol PR(−) in a 20:21 ratio contained mainly monomeric RNA. Our observations provide the first direct evidence that, in addition to proteolytic processing, the ratio of Gag/Gag-Pol proteins is also important for RNA dimerization and that stable RNA dimers are not required for encapsidation of genomic RNA in HIV-1.
The genome of the human immunodeficiency virus type 1 (HIV-1), like that of other retroviruses, contains three major structural genes, gag, pol, and env. The Gag and Pol proteins are encoded by overlapping open reading frames. Gag has its own initiation and termination codons, while the synthesis of the HIV-1 Gag-Pol precursor results from a −1 frameshifting event that occurs at a frequency of 5 to 10% during translation of the unspliced Gag or Gag-Pol mRNA (18). Other retroviruses also use similar frameshifting mechanisms (17, 19, 20) or a readthrough suppression mechanism (8–10) to regulate the expression of Gag-Pol proteins. Thus, intracellular Gag/Gag-Pol ratios of around 20:1 are found during the replication of all retroviruses. The HIV frameshift site (a heptanucleotide AU-rich sequence) is found at the 3′ end of the nucleocapsid (NC) coding sequence. This site and a stem structure immediately downstream stall the ribosome during the synthesis of Gag, allowing the ribosome to slip back 1 nucleotide to enable the infrequent (5%) synthesis of the Gag-Pol fusion protein (18). Multimerization of the Gag protein gives rise to viral particles, while expression of polymerase as a Gag-Pol precursor protein ensures that viral enzymes are incorporated into viral particles during viral assembly (for a review, see reference 27). During and after release of virions from cells, the Gag precursor protein is cleaved by viral protease (PR) into mature proteins: matrix, capsid (CA), NC, p6, and two spacer peptides, p2 and p1. Gag-Pol fusion is cleaved to yield matrix, CA, p2, and NC, as well as transframe protein, PR, reverse transcriptase (RT), and integrase (IN).
The synthesis of Gag precursor protein alone has been reported to be sufficient for the assembly and release of virus-like particles (for a review, see reference 45). Incorporation of Gag-Pol or its mature products into virions is required for infectivity, as they mediate the synthesis and integration of viral cDNA in infected cells (46). In addition, cleavage of the precursor proteins by PR is required for morphological maturation of the virion core (15, 23, 24). Viral genomic RNA is also packaged into virions during assembly, driven by the genomic RNA packaging sequence (Ψ) found near the 5′ end of the genome (for a review, see references 3 and 42).
Like other retroviruses, HIV-1 has a dimeric RNA genome. In vitro dimerization analysis of HIV-1 viral RNA has mapped a 50- to 60-nucleotide sequence, termed the dimer initiation sequence, that is important for the formation of the dimeric RNA complex (26, 31, 40). Mutations in the dimer initiation sequence hinder genomic RNA dimerization and virion RNA packaging and result in the production of noninfectious viral particles. It is thought that RNA dimerization is a prerequisite for RNA packaging in HIV-1 (2, 6), and virion packaging of genomic RNA and RNA dimerization are also linked in other retroviruses (5, 38, 44). RNA dimers from PR-defective HIV-1 virions are less heat stable than dimers from wild-type mature HIV-1 (11). Similar observations about Moloney murine leukemia virus have also been reported (12).
Although it is clear that expression of the Gag-Pol precursor alone is insufficient for production of infectious retroviral particles (7, 32), the influence of the Gag/Gag-Pol ratio (20:1) on the viral replication cycle and RNA dimerization is unknown. Our data show that the Gag/Gag-Pol ratio in virion-producing cells is important for the generation of infectious viral particles and the stability of the virion RNA dimer.
MATERIALS AND METHODS
Construction of DNA plasmids.
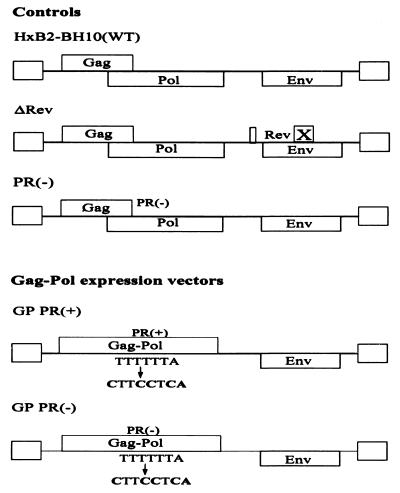
The full-length wild-type HIV-1 plasmid, referred to as HxB2-BH10, has been described previously (43). The ΔRev plasmid differs from HxB2-BH10 by the absence of the full second half of exon 2 of the rev sequence, which was removed to inactivate Rev function. The Rev deletion was achieved by BamHI and XhoI endonuclease digestion, followed by ligation in the presence of a double-stranded DNA adapter which is complementary to both BamHI and XhoI. The GP PR(+) plasmid was constructed using stitch PCR mutagenesis as previously described (30) (Fig. 1). Briefly, GP sense f1 primer 5′ggcaaagaagggcacacagcc3′ and antisense f1 primer 5′cccTGAGGAAGttagcctgtctctcagtac3′ were used to amplify a 130-bp GP f1 fragment. GP sense f2 primer 5′ggctaaCTTCCTCAgggaagatctggccttcc3′ and GP antisense f2 primer 5′gttgacaggtgtaggtcctac3′ were used to amplify a 400-bp GP f2 fragment. The GP f1 and GP f2 fragments were joined by PCR extension. The resulting PCR-amplified fragment was cloned into the HxB2-BH10 proviral DNA via restriction sites ApaI and BclI. The GP f1 antisense and GP f2 sense primers contain mutations that eliminate the five-T heptanucleotide stretch which is responsible for the −1 ribosomal frameshifting during the translation of Gag (18). This GP mutation allows continuous expression of Gag-Pol and bypasses the Gag termination codon. The GP PR(−) plasmid was constructed in the same fashion as GP PR(+), with the exception that a PR-defective full-length HIV-1 plasmid PR(−) (15) was used as the DNA template for GP PR(−) PCR mutagenesis (15). PR(−) was used for the production of PR-defective immature HIV-1 virions as a control.
FIG. 1.
Schematic representation of proviral DNA constructs used in this study. The wild-type (WT) plasmid is HxB2-BH10 and has been previously described (43). To obtain the GP PR(+) construct, a PCR-amplified fragment was cloned into the HxB2-BH10 proviral DNA via ApaI and BclI restriction sites in order to eliminate the five-T heptanucleotide stretch which is responsible for the −1 ribosomal frameshifting during the translation of Gag and allow continuous expression of Gag-Pol by bypassing the Gag termination codon. The GP PR(−) plasmid was constructed in the same fashion as GP PR(+), with the exception that a PR-defective full-length HIV-1 PR(−) plasmid was used as the DNA template for GP PR(−) PCR mutagenesis (15).
Cotransfection and virus production.
The production of HIV-1 viral particles from cells with an altered Gag/Gag-Pol ratio was achieved by cotransfecting 10 μg of proviral DNA (either HxB2-BH10 or ΔRev) with one of the two Gag-Pol expression vectors [GP PR(+) or GP PR(−)] using the indicated amounts of proviral DNA (see Table 1). Appropriate amounts of a long terminal repeat promotor-driven luciferase reporter plasmid were used to normalize the level of DNA used in cotransfection. HxB2-BH10 or ΔRev was responsible for the 20:1 Gag/Gag-Pol expression ratio in the virus-producing cells, while GP PR(+) and GP PR(−) supplemented the excess of Gag-Pol protein expression in these cells. Supplementation of 10, 5, 2.5, and 1.25 μg of the Gag-Pol expression vectors altered the Gag/Gag-Pol ratio in the HIV-1-producing cells from 20:1 to 20:21, 20:11, 20:6, and 20:3.5, respectively. HxB2-BH10 and the Gag-Pol expression vector GP PR(+) or GP PR(−) were used in cotransfections to produce viral particles for virus infectivity analysis and viral protein profiles within both cellular and virion lysates. ΔRev and a Gag-Pol expression vector, GP PR(+) or GP PR(−), were used in cotransfection experiments to produce mutant viral particles for virion RNA packaging and dimerization analysis. A green fluorescent protein (EGFP; Clontech) reporter plasmid (2 μg) was added to the DNA mixture to determine transfection efficiency.
TABLE 1.
Production of HIV-1 particles from cells with altered Gag/Gag-Pol ratios
| Proviral DNAs used in cotransfections (quantities [-/μg]) | Predicted Gag/ Gag-Pol ratio in virus-producing cells | Level of virion production (ng of p24/ml)a |
|---|---|---|
| WT (10) | 20:1 | 1,176 |
| WT, GP PR(+) (10, 10) | 20:21 | 99 |
| WT, GP PR(+) (10, 5) | 20:11 | 111 |
| WT, GP PR(+) (10, 2.5) | 20:6 | 189 |
| WT, GP PR(+) (10, 1.25) | 20:3.5 | 713 |
| WT, GP PR(−) (10, 10) | 20:21 | 90 |
| WT, GP PR(−) (10, 5) | 20:11 | 120 |
| WT, GP PR(−) (10, 2.5) | 20:6 | 207 |
| WT, GP PR(−) (10, 1.25) | 20:3.5 | 386 |
The levels of virion production (as determined by quantitative p24 assay) from a set of representative experiments are included.
293T cells (1.2 × 106 cells per plate) were plated onto 10-cm-diameter plates (Nunc) and maintained for 24 h in 7 ml of Dulbecco's modified Eagle medium (GIBCO) containing 10% fetal bovine serum (P. A. Biological Co.), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. DNA mixtures were introduced into 293T cells using a previously described calcium phosphate transfection method (29). Cells were washed twice with phosphate-buffered saline 12 h posttransfection and maintained in fresh Dulbecco's modified Eagle medium. Supernatants were collected 36 h posttransfection and centrifuged for 30 min at 3,000 rpm at 4°C (Beckman) to remove cellular debris. The clarified supernatants were either frozen at −70°C or used immediately for further analysis. Cells were washed twice with either phosphate-buffered saline or 1× Tris-buffered saline (TBS buffer, i.e., 50 mM Tris [pH 7.4], 150 mM NaCl), followed by protein extraction using lysis buffer containing 1× TBS, Nonidet P-40 at 10 μl/ml, 20 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, and 1 μM leupeptin. Cell lysates were collected and stored at −20°C for later use.
Assessment of virus replication kinetics. (i) Preparation of viral particles for infectivity assay.
Supernatants obtained from cotransfection of 293T cells were filtered immediately after centrifugation using 0.2-μm-pore-size filters (Schleicher & Schüll). Small volumes of samples were stored at −70°C. Samples were thawed immediately before use for RT and infectivity assays.
(ii) RT activity of clarified virus supernatants.
To determine the RT levels of each virus, 10 μl of supernatant from each sample was mixed with 10 μl of Nonidet P-40. Samples were incubated for 30 min at room temperature for inactivation of the virus, and the RT activity of the viruses in the supernatant was measured using an RT microassay as previously described (13).
(iii) p24 assay.
p24 levels in culture supernatant were quantified via a commercially available p24 antigen detection kit (Abbott Laboratories) used in accordance with the manufacturer's instructions.
(iv) Infectivity assay.
Briefly, peripheral blood mononuclear cells (PBMCs) were isolated from buffy pack (supplied by the Red Cross Blood Bank, Melbourne, Victoria, Australia) as described previously (4). PBMCs were then stimulated with phytohemagglutinin (10 μg/ml; Murex Diagnostics) for 3 days and cultured in Iscove's medium containing 10% fetal calf serum (PA Biologicals) and 5% interleukin-2 (Boehringer, Mannheim, Germany). The infectivity of the viruses derived from cells with altered Gag/Gag-Pol ratios was measured using a 50% tissue culture infective dose (TCID50) method previously described (4). Two hundred microliters of virus supernatant derived from cells with a predicted Gag/Gag-Pol ratio of 20:21, 20:11, 20:6, or 20:3.5 was mixed with 105 PBMCs in a 96-well tissue culture plate. Four 10-fold dilutions of each virus were tested; each dilution was tested in triplicate. Supernatants were collected on days 3, 7, 10, and 14 postinfection and frozen immediately at −70°C. Viral infectivity was measured by monitoring activity using an RT microassay (13).
Analysis of viral protein within virion-producing cells and virion particles. (i) Intracellular viral protein analysis.
Cell lysates were rapidly freeze-thawed three times to weaken the cellular membrane. Cell debris was subsequently removed by centrifugation for 30 min at 4°C and 14,000 rpm (Beckman). The transfection efficiency of the samples was determined by measuring the level of EGFP from the reporter plasmid using a Bio Imaging Analyzer (Fuji Photo Film Co.). Cellular protein from each sample normalized for an equivalent level of EGFP was mixed with 3 μl of sample buffer (100 mM Tris [pH 6.8], 3% sodium dodecyl sulfate [SDS], 33% glycerol, 0.03% bromophenol blue), denatured for 10 min in 95°C, and resolved by SDS–10% polyacrylamide gel electrophoresis (PAGE). Resolved proteins were transferred to a nitrocellulose membrane (Amersham, Amersham, England). The membrane was blocked for 2 h in 3% casein dissolved in 1× TBS containing 0.3% Tween 20 (TBST) and probed overnight with pooled HIV-1-seropositive patient sera. After three washes with 1× TBST buffer, the membrane was incubated with anti-human horseradish peroxidase-conjugated secondary antibody (DAKO) for 2 h at room temperature. An enhanced chemiluminescence technique was used for visualization of HIV-1 proteins present in the cellular lysates (Amersham). Results were visualized by autoradiography.
(ii) Assessment of Gag/Gag-Pol ratios in virus-producing cells following transfection.
To determine the Gag/Gag-Pol ratios in virus-producing cells, cellular proteins standardized by transfection efficiency were resolved by SDS-PAGE and probed with monoclonal antibodies to RT and p24 antigen for detection and quantitation of Pr160gag-pol, p66/51 RT, and p24 CA, respectively. Data obtained by autoradiography were scanned, and expression levels of these proteins for each sample were quantified using laser densitometry analysis.
(iii) Virion purification and protein analysis.
Supernatants from transfected cells were purified and concentrated by ultracentrifugation through a 20% sucrose cushion using a Beckman L-90 ultracentrifuge (SW 41 rotor) at 35,000 rpm for 1 h at 4°C (29). Pellets were resuspended in 50 μl of TBS lysis buffer.
(iv) Analysis of virion protein profile.
Equal amounts of virion protein (normalized by the transfection efficiency of EGFP) from each sample were mixed with 3 μl of sample buffer containing 5 mM β-mercaptoethanol and heated for 10 min at 95°C. Virion proteins were then resolved by SDS–10% PAGE as described above. The resolved virion protein samples were transferred onto nitrocellulose membranes by electrophoresis using a Bio-Rad transfer apparatus. Virion HIV-1 protein profiles of the samples were determined by Western analysis as described above.
(v) Quantification of virion protein by dot blot assay.
The total virion protein recovered was standardized by a protein dot blot technique. Briefly, twofold dilutions of each protein sample were loaded directly onto a nitrocellulose membrane (Hybond-C extra; Amersham). The membrane was air dried for 30 min at room temperature and blocked for 2 h in 3% casein in 1× TBST. Samples were probed for total HIV-1 virion protein production as described above. Results were analyzed either by autoradiography or by phosphorimaging for quantification of total virion protein.
(vi) Analysis of virion RNA packaging.
For virion RNA analysis, HxB2-BH10 proviral DNA was replaced with ΔRev proviral DNA in a cotransfection. This was done to ensure that virion particles were produced only when the Gag-Pol-expressing vector was supplemented in the virus-producing cell, as both Gag (from the ΔRev plasmid) and Rev (from either of the Gag-Pol expression plasmids) are required for successful viral particle production. Pelleted virions were prepared by ultracentrifugation as described above and resuspended in 500 μl of Trizol (GIBCO) for genomic RNA extraction. Samples were subsequently incubated for 30 min in ice to allow complete lysis to occur, and 200 μl of chloroform was added to remove virion proteins. Samples were mixed well and centrifuged for 5 min at 14,000 rpm and 4°C. The aqueous layer containing the RNA was transferred to a fresh Eppendorf tube and precipitated with 1/10 volume of 3 M sodium acetate and 1 ml of ice-cold 100% ethanol at −20°C overnight. Precipitated RNA samples were centrifugated for 30 min at 14,000 rpm (Beckman) and 4°C. The RNA pellets were washed with 200 μl of 70% ethanol and then air dried for 1 h at room temperature. Each pellet was resuspended in 10 μl of RNase-free water and stored at −20°C until use.
The impact of the Gag/Gag-Pol ratio on genomic packaging of virion RNA was assessed by RNA dot blot assay (39) using a Bio-Rad dot blot apparatus. Serial 10-fold dilutions of virion RNA samples that were normalized by virion protein were used to construct a standard curve. Each sample was mixed with 29 μl of RNA denaturation buffer (37% formaldehyde, 60% deionized formamide, 3% 20× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) and incubated at 65°C for 10 min for RNA denaturation. After incubation, samples were immediately chilled in ice. To each tube, 78 μl of 20× SSC buffer was added and samples were loaded onto a Hybond membrane (Amersham) via a dot blot apparatus (Bio-Rad). The wells were washed twice with 10× SSC, and the membrane was air dried overnight at room temperature. The membrane was exposed to UV light for 90 s to cross-link the RNA onto the membrane and blocked for 1 h at 42°C with 10 ml of hybridization buffer (40 ml of the hybridization buffer contains 8 ml of 5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA at pH 7.7]; 20 ml of deionized formamide, 4 g of dextran sulfate, 400 μl of salmon sperm DNA [10 mg/ml], 3 ml of 20% sodium dodecyl sulfate, and 5 ml of double-distilled H2O).
The amount of genomic RNA in viral RNA samples was quantified using a radioactive antisense oligonucleotide probe, 790H (5′CTGACGCTCTCGCACCC3′), that has been previously described (30). Briefly, the 790H oligomer binds specifically to HIV-1 genomic RNA sequence positions 338 to 354 (DNA sequence positions 791 to 807). The 790H oligomer was 5′ end labeled using T4 polynucleotide kinase and γ-32P-labeled ATP (3,000 Ci/mmol; Amersham). The membrane containing the RNA samples was incubated overnight at 42°C with the radioactively labeled probe. The membrane was washed once for 30 min with 1× SSC–0.1% SDS and twice with 0.2× SSC–0.1× SDS for 30 min. The results were visualized by autoradiography. The amount of genomic RNA packaged by wild-type virions was used as a control to determine the impact of the Gag/Gag-Pol ratio on virion RNA packaging.
Virion RNAs that were normalized by virion protein concentration were also analyzed by Northern blot assay using three serial 10-fold dilutions to determine the stability of the 9-kb unspliced genomic RNA. Samples were mixed with 10% 5× formaldehyde running buffer, 17.5% formaldehyde, and 50% formamide and incubated for 15 min at 65°C and then quickly chilled in ice for 15 min. A 2-μl sample of sterile formaldehyde gel loading buffer (50% glycerol, 1 mM EDTA, 0.25% bromophenol blue, 0.25% xylene cyanol FF) was added to each tube, and virion RNAs were separated on a 1% denaturing agarose gel. RNA samples were transferred onto a Hybond membrane, UV cross-linked, and probed for 9-kb HIV-1 genomic RNA using an HIV-specific PCR radioactive probe as previously described (14). Results were visualized by autoradiography.
(vii) Analysis of virion RNA dimerization.
Virion pellets were resuspended in 500 μl of dimerization buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 1% SDS, 50 mM NaCl, yeast tRNA at 50 μg/ml, proteinase K at 100 μg/ml), phenol-chloroform extracted, and isolated for melting curve analysis as previously described (11, 12).
Similar amounts of genomic RNA were used to analyze the stability of the virion RNA dimer in each preparation by heating the samples at the indicated temperatures for 10 min and then quickly chilling them in ice. Heat-denatured dimeric and monomeric RNAs were separated by electrophoresis in a 1% native agarose gel in 0.5× Tris-borate-EDTA buffer. Samples were transferred overnight onto a Hybond N membrane (Amersham). The membrane containing the RNA samples was air dried for 2 h at room temperature and exposed to UV light for 90 s for cross-linking. The membrane was blocked for 1 h at 42°C with 10 ml of hybridization buffer as described above.
Dimeric and monomeric RNAs were subjected to an overnight incubation with a radioactive riboprobe (pGEM7zHIV-1) which is complementary to the 5′ end of the HIV-1 genomic RNA sequences. Briefly, a 1,390-bp HindIII-SphI HIV-1 DNA fragment was cloned into an in vitro transcription vector, pGEM7z, via restriction sites HindIII and SphI. An 800-bp DNA fragment corresponding to the Gag-encoding region was subsequently removed by PstI and BssHII restriction digestion and S1 nuclease treatment, followed by self-religation to generate pGEM7z HIV-1. The removal of the 800-bp Gag coding region shortens the length of the in vitro HIV-1 RNA transcript and reduces the nonspecific binding of the riboprobe. The radioactive riboprobe was synthesized by linearization of pGEM7z HIV-1 with BamHI, followed by T7 RNA polymerase-directed in vitro transcription (Promega) in the presence of [α-32P]CTP (NEN). After probing, the membrane was washed once for 30 min with 1× SSC–0.1% SDS buffer and twice for 30 min (each time) with 0.2× SSC–0.1% SDS buffer, and the results were visualized by autoradiography. Migration of wild-type mature and PR-defective immature dimeric and monomeric RNA at the respective heating temperatures was used as a control to determine the effect of the Gag/Gag-Pol ratio on RNA dimerization for viruses derived from cells with altered ratios.
RESULTS
Effects of altered Gag/Gag-Pol ratios on HIV-1 protein profiles in virus-producing cells.
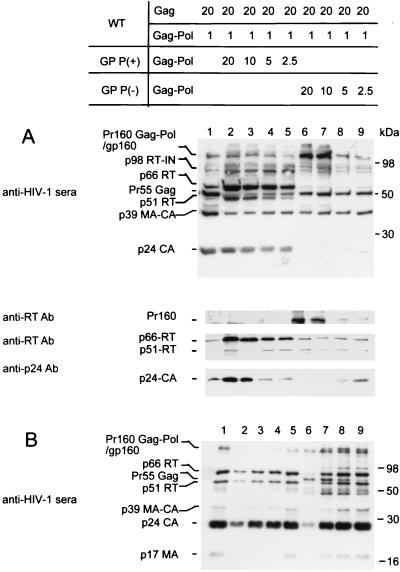
293T cells were transfected with wild-type DNA (HxB2-BH10; producing a 20:1 Gag/Gag-Pol ratio as a control) or cotransfected with either a Gag-Pol expression vector encoding an active PR site, GP PR(+), or a PR-defective Gag-Pol expression vector, GP PR(−), to progressively alter this ratio from 20:1 to 20:21. Following transfection, cells were lysed and expression of EGFP was quantified to standardize transfection efficiency for each sample (data not shown). Cellular proteins from samples normalized for transfection efficiency were resolved by SDS–10% PAGE and probed with human HIV antisera. Compared with wild-type DNA alone (Fig. 2A, lane 1), cotransfection with the GP PR(+) plasmid increased intracellular levels of Gag-Pol products, e.g., p66 RT and p51 RT (Fig. 2A, lanes 2 to 5). In contrast, cotransfection of the GP PR(−) plasmid resulted in higher levels of precursor Pr160 Gag-Pol and reduced levels of Gag-Pol products (e.g., p24-CA) (Fig. 2A, lanes 6 to 9). Intermediate Gag-Pol processing products corresponding to p98 RT-IN, p110 PR-RT-IN, and p119 p2-p7-PR-RT-IN were also detected when either GP PR(+) or GP PR(−) was cotransfected. The wild-type HIV-1 pattern was gradually restored as the ratio approached 20:1.
FIG. 2.
Impact of altered Gag/Gag-Pol ratios on HIV-1 protein profiles of infected cells and virions. Viral proteins were semiquantified by Western blot analysis. Cell lysates (A) and purified virions (see Materials and Methods) (B) were resolved by SDS–10% PAGE. Resolved proteins were probed using sera from HIV-1-infected individuals and monoclonal antibodies (Ab) to RT and p24 as described in Materials and Methods. Lane 1 (A and B) shows wild-type (WT) HIV-1 protein profiles in cell lysates and virions, respectively; lanes 2 to 5 (A and B) show cell lysate and virion protein profiles with estimated HIV-1 Gag/Gag-Pol ratios of 20:21, 20:11, 20:6, and 20:3.5, respectively. A GP PR(+) expression vector was used to produce excessive levels of Gag-Pol in lanes 2 to 5 (see Materials and Methods). Similarly, lanes 6 to 9 show cell lysate and virion protein profiles with estimated HIV-1 Gag/Gag-Pol ratios of 20:21, 20:11, 20:6, and 20:3.5, respectively. Expression vector GP PR(−) was used to produce an excess of Gag-Pol protein that is not processed by PR in lanes 6 to 9 (see Materials and Methods). MA, matrix.
Quantification of Gag/Gag-Pol ratios of mutant viruses in virus-producing cells.
As assessed by laser densitometry, the expression of the p66/51 RT in virus-producing cell correlated with the input levels of GP PR(+) used during transfection. An approximately twofold increase in RT protein was detected for every subsequent twofold increase in GP PR(+) plasmid DNA used, with the p66/RT densitometry units being 150,000, 87,000, 35,000, and 15,000 (Fig. 2A, lanes 2, 3, 4, and 5, respectively). A similar, Gag/Gag-Pol ratio-dependent, p24 CA level was also detected when an anti-p24 monoclonal antibody was used (Fig. 2A, lanes 2 to 5). Densitometry units of p24 CA expression were 268,000, 160,000, 50,320, and 25,770, respectively. No detectable difference in p66/51 RT was observed in cells transfected with an excess of GP PR(−) compared to the wild-type control, while intracellular p24 CA expression levels showed reduction in a dose-responsive manner (Fig. 2A, lanes 6 to 9). The intensity of intracellular p24 CA signals was inversely proportional to the detectable levels of Pr160gag-pol (Fig. 2A, lanes 6 to 9).
Effects of altered Gag/Gag-Pol ratios on virion protein patterns.
Purified viral particles derived from cotransfected cells (as described above) were analyzed by Western blot assay. Virions derived from cells overexpressing GP PR(+) showed slightly low levels of Pr160gag-pol, p66 RT, and p24 CA but otherwise maintained virion protein profiles similar to those of wild-type virions (Fig. 2B, lanes 1 and 2). The virion proteins returned toward wild-type levels as the Gag/Gag-Pol ratio approached 20:1 (Fig. 2B, lanes 1 to 5). Cells with a Gag/Gag-Pol ratio of 20:21 produced 10 times less supernatant p24 than cells producing only wild-type virus (Table 1). Overexpression of GP PR(−), however, altered both the levels and profiles of virion proteins in a ratio-dependent fashion (Fig. 2B, lanes 6, 7, 8, and 9). Notably, the p66 RT band disappeared and only low levels of p24 CA were detected when the Gag/Gag-Pol ratio was altered from 20:1 to 20:21 (Fig. 2B, lanes 1 and 6). The levels of mature and intermediate virion proteins, e.g., p66 RT, p51 RT, p24 CA, and p39 MA-CA, were inversely related to the presence of Pr55gag (Fig. 2B, lanes 6 to 9).
Assessment of the effect of an altered Gag/Gag-Pol ratio on virus infectivity.
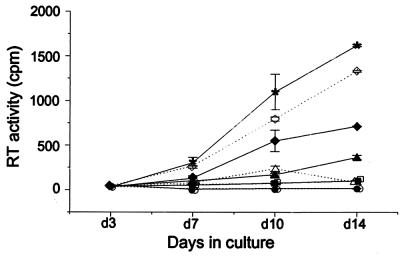
Wild-type or mutant virus suspensions derived from harvested cells were assayed for infectivity, RT activity, and p24 antigen concentration. End point dilution infectivity assay results (TCID50) were normalized to RT or p24. Progeny viruses derived from cells with a Gag/Gag-Pol ratio of 20:21 were 250 to 1,000 times less infectious than the wild-type virus (Fig. 3 and Table 2). Similar decreases in infectivity, in a clearly ratio-dependent fashion, were seen in virions derived from cells with excess GP PR(+) or GP PR(−).
FIG. 3.
Infectivity of viral particles produced by cells with a wild-type or altered Gag/Gag-Pol ratio. Freshly isolated PBMCs were phytohemagglutinin stimulated for 3 days and then infected with either wild-type or mutant virus. Supernatants were collected 3, 7, 10, and 14 days (d) after infection, and the RT activity in each sample was measured. The results shown represent the mean and standard deviation of triplicate samples. Open symbols represent viruses derived from cells with an excess of PR(+) Gag-Pol, and closed symbols (except asterisks) represent viruses derived from cells with excess of PR(−) Gag-Pol. Symbols: ∗, wild-type virus; ⧫ and ◊, progeny viruses of cells with a predicted 20:3.5 Gag/Gag-Pol ratio; ▴ and ▵, predicted 20:6 Gag/Gag-Pol ratio; ■ and □, predicted 20:11 Gag/Gag-Pol ratio; ● and ○, predicted 20:21 Gag/Gag-Pol ratio.
TABLE 2.
Infectivity of virus particles derived from cells with altered Gag/Gag-Pol ratiosa
| Proviral DNA(s) used | Gag/Gag-Pol ratio in virus-producing cells | Gag and Gag- Pol processing phenotyped | Virion infectivity (TCID50)a standardized for:
|
|
|---|---|---|---|---|
| RT activityb | p24 levelc | |||
| WT | 20:1 | ++++ | 1,300.0 | 480.0 |
| WT, GP PR(+) | 20:21 | ++++ | 1.4 | 5.6 |
| WT, GP PR(+) | 20:11 | ++++ | 1.5 | 5.0 |
| WT, GP PR(+) | 20:6 | ++++ | 13.0 | 17.0 |
| WT, GP PR(+) | 20:3.5 | ++++ | 130.0 | 79.0 |
| WT, GP PR(−) | 20:21 | ± | 5.2 | 3.5 |
| WT, GP PR(−) | 20:11 | + | 5.0 | 4.7 |
| WT, GP PR(−) | 20:6 | ++ | 28.0 | 27.0 |
| WT, GP PR(−) | 20:3.5 | +++ | 150.0 | 140.0 |
TCID50 was measured as described in Materials and Methods.
The values shown are TCID50s (10−4) per count per minute of RT activity.
The values shown are TCID50s (10−1) per nanogram of p24 antigen.
++++, very strong; +++, strong; ++, medium; +, weak; ±, very weak.
Relationship of intracellular Gag/Gag-Pol ratio, virion RNA packaging, and RNA dimerization.
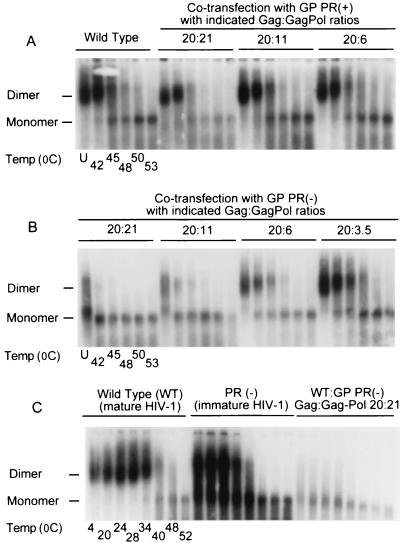
To investigate the impact of the Gag/Gag-Pol ratio on RNA dimerization and dimer stability, virus suspensions obtained using the transfection methods described above were studied. Similar amounts of virion RNA were treated at different temperatures, and dimeric and monomeric virion RNAs were resolved by electrophoresis (Fig. 4). Dimeric RNA was observed in all viruses derived from cells in which GP PR(+) was overexpressed. These RNA dimers were only slightly less stable than wild-type dimers (Fig. 4A, compare the wild-type ratio with 20:21 at 45, 48, and 50°C), and the degree of instability correlated with the level of excess PR(+) Gag-Pol proteins in virion-producing cells (Fig. 4A). In contrast to the results obtained with GP PR(+), supplementing virus-producing cells with a PR-inactive Gag-Pol expression vector [GP PR(−)] markedly reduced RNA dimerization in a concentration-dependent fashion (Fig. 4B). When the Gag/Gag-Pol ratio of the virion-producing cells was altered from 20:1 to 20:21 with the GP PR(−) protein expression vector, progeny virions contained mainly monomeric RNA (Fig. 4B). The few RNA dimers in these aberrant viral particles dissociated at temperatures 15 to 20°C lower than the dimeric RNA found in PR-defective immature and wild-type mature HIV-1 virions, respectively (Fig. 4C).
FIG. 4.
Effect of altered Gag/Gag-Pol ratios on virion RNA dimerization. The impact of various Gag/Gag-Pol ratios on genomic RNA dimerization was determined using melting curve and electrophoretic analysis of wild-type (WT) and mutant dimers. Virion RNA was resuspended in RNA dimerization buffer and heat denatured for 10 min at the indicated temperatures. Dimers and monomers were electrophoresed in a 1% native agarose gel and probed with an HIV-1 riboprobe as described in Materials and Methods. RNA dimerization analysis involved wild-type HIV-1 and mutant viruses derived from cells cotransfected with wild-type and GP PR(+) expression vectors to achieve Gag/Gag-Pol ratios of 20:1 to 20:21 (A), cells cotransfected with wild-type and GP PR(−) expression vectors to achieve Gag/Gag-Pol ratios of 20:3.5 to 20:21 (B), and cells transfected with wild-type proviral DNA, transfected with a PR-defective full-length HIV-1 Pr-defective plasmid [PR(−)] for the production of PR-defective immature HIV-1 particles, and cotransfected with wild-type GP PR(−) to achieve a Gag/Gag-Pol ratio of 20:21 (C). U, unheated.
Recent work on Rous sarcoma virus (RSV) has shown that a 50% reduction in virion RNA packaging in a mutant RSV is associated with a defect in virion RNA dimer formation (35). To rule out this possibility, the level of packaged genomic RNA in our monomeric RNA virions was compared with wild-type HIV-1 and PR-defective immature HIV-1 virions [via transfection of PR(−) alone]. Wild-type and mutant viruses were examined by RNA dot blot assay to assess the influence of the Gag/Gag-Pol ratio on RNA packaging (Fig. 5B), and the stability of the full-length genomic RNA was analyzed by Northern blotting (Fig. 5C). Equivalent amounts of genomic RNA packaging were found in wild-type HIV-1 (Fig. 5B, row 1), PR-defective immature HIV-1 virions (Fig. 5B, row 3), and the monomeric RNA virions (Fig. 5B, row 4) when normalized for HIV-1 protein concentration. Transfection of GP PR(−) alone into 293T cells results in expression of the Gag-Pol precursor protein. Expression of the Gag-Pol precursor protein without Gag yielded low but detectable levels of HIV-1 proteins (8- to 10-fold lower than those seen following wild-type plasmid transfection as determined by Fuji FLA 2000 phosphorimaging) (Fig. 5A, row 2) but negligible levels of virion genomic RNA (at least 100-fold lower than the wild type) when normalized for HIV-1 protein concentration (Fig. 5B, row 2) compared to that from the wild-type virus (Fig. 5B, row 1). These results show that the monomeric RNA packaged into virions was not contaminated with pelletable Gag-Pol precursor protein. Similar levels of genomic RNA packaging were found in all of the other viruses tested, and the level of the viral RNA packaged was not affected by the stability of RNA dimers or the nature of the Gag-Pol expression vector [i.e., GP PR(+) or GP PR (−)] (data not shown).
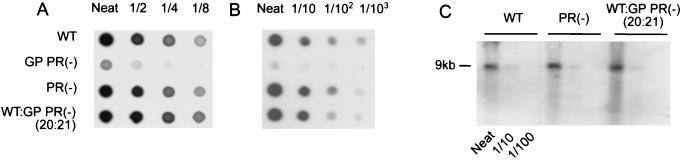
FIG. 5.
Effect of altered Gag/Gag-Pol ratios on packaging of genomic RNA. Three types of HIV-1 virions were used in this analysis; they were wild-type HIV-1 (WT), Pr-defective immature HIV-1 [PR(−)], and virions containing mainly monomeric RNA [WT:GP PR(−) (20:21)]. Supernatant collected from cells expressing only Gag-Pol precursor protein was also used as a control. Quantitation of pelleted viral protein was performed by protein dot immunoblot assay (A). Samples for RNA analysis (B and C) were standardized for protein concentration (A). Virion RNA encapsidation efficiency of wild-type and mutant viruses was analyzed by RNA dot blot hybridization assay (B). A series of 10-fold dilutions was done for quantitation. The stability of the virion-packaged RNA genome was determined by Northern blot analysis (see Materials and Methods for details of procedures) (C).
Northern analysis of the packaged genomic RNA showed that the 9-kb genomic RNA derived from our monomeric RNA virions was intact in comparison to the wild-type mature and PR-defective immature HIV-1 (Fig. 5C). Alteration of the Gag/Gag-Pol ratio neither affected RNA packaging nor promoted degradation of the 9-kb genomic RNA.
DISCUSSION
Using a cotransfection system, we have demonstrated that alteration of the Gag/Gag-Pol ratio in virus-producing cells reduces the infectivity of progeny viruses and hinders the formation of stable virion RNA dimers without impairing virion packaging of genomic RNA. This study shows for the first time that maintenance of the normal Gag/Gag-Pol ratio is important for HIV-1 replication. Although both the proteolytic processing of HIV-1 proteins and a normal intracellular Gag/Gag-Pol ratio are required for RNA dimerization, the formation of stable RNA dimers is not essential for the packaging of HIV-1 genomic RNA.
The identification of translational suppression in retroviruses has provided crucial information regarding retroviral gene expression. With the exception of foamy viruses, the expression of pol gene products in all retroviruses requires translational suppression (28) to maintain a Gag/Gag-Pol expression ratio of 20:1 in virion-producing cells. The evolutionary conservation of the Gag/Gag-Pol ratio in retroviruses supports its importance in the retroviral replication cycle (17–20, 34). In our study, we have examined the relevance of this ratio by altering the Gag/Gag-Pol ratio in the virus-producing cell via supplementing Gag-Pol expression vectors in HIV-1-transfected cells.
Gag/Gag-Pol ratio and viral protein expression.
The impact of overexpression of PR(+) Gag-Pol was reflected in intracellular viral protein concentrations. The reduction in intracellular mature Gag and Gag-Pol products in cells containing an excess of PR(−) Gag-Pol is considered to be due to the trans-dominant negative effect of PR-defective Gag-Pol proteins (1, 33). The reduction of viral particle production in cells containing excess GP PR(+) can, in part, be explained by overexpression of viral PR. Intracellular overexpression of HIV-1 PR inhibits HIV-1 replication (21), while overexpression of intracellular Gag-Pol precursor proteins interferes with HIV-1 assembly (22, 37). The reduced levels of p24 and RT in the supernatant of cells containing excess GP PR(−) is possibly due to inactivation of the viral PR function, as our p24 immunoassay used for p24 antigen detection does not recognize Pr55gag as efficiently as p24 CA. Therefore, it is also likely that we have underestimated the number of viral particles being produced from cells overexpressing PR-defective Gag-Pol, hence overestimating the infectivity of the mutant viral particles produced via overexpression of PR(−) Gag-Pol.
Gag/Gag-Pol ratio, virion RNA packaging, and dimerization.
Our data show that alteration of the Gag/Gag-Pol ratio did not influence HIV-1 RNA packaging. In contrast, expression of Gag-Pol precursor protein alone yielded eightfold less virion protein than that found in the wild type and packaged negligible levels (100 times less than the wild type) of genomic RNA for equivalent amounts of virion proteins. The latter observation is in agreement with the study of Kaye and Lever, which showed that expression of Gag-Pol protein alone yields pelletable virion proteins that do not encapsidate virion RNA (25). Others have also reported that expression of Gag-Pol protein alone is insufficient for the production of virus-like particles (36, 41) and that the formation of viral particles requires the expression of Gag precursor proteins (for a review, see reference 45). We have previously shown that expression of HIV-1 Gag-Pol [either GP PR(+) or GP PR(−)] is important for the virion packaging of primer tRNALys3 in HIV-1 (29) and that overexpression of primer tRNALys3 enhances virion packaging of primer tRNA (16). Using a converse approach, our experiments also suggest that overexpression of Gag-Pol does not affect the virion packaging of primer tRNALys3 (M. Shehu-Xhilaga et al., unpublished data).
These studies show that overexpression of PR(+) Gag-Pol protein has a minor effect on virion RNA dimerization and that this impairment of virion RNA dimerization can be amplified via overexpression of a trans-dominant PR-negative Gag-Pol expression vector. Fu et al. (11, 12) have shown that proteolytic processing of retroviral precursor proteins is important for the maturation of virion RNA dimers to a more heat-stable form. In addition to the importance of precursor protein processing in virion RNA dimer maturation, we have found that a Gag-Gag-Pol ratio reduction also yields viral particles with unstable (highly heat-sensitive) RNA dimers, a defect which is clearly not due to the inactivation of the viral PR alone.
Furthermore, our experiments provide direct evidence that the formation of stable virion RNA dimers is not required for virion RNA packaging. Previous work has shown that in retroviruses, genomic RNA packaging and virion RNA dimerization are closely linked (for a review, see reference 3). A recent publication by Parent et al. (35) showed that a mutation in the RS matrix sequence interfered with the formation of RNA dimers and reduced virion RNA packaging by 50%. With our work, we were able to produce viral particles containing mainly monomeric RNA merely by increasing the concentration of PR(−) Gag-Pol in HIV-1-producing cells. This was achieved without introducing mutations into the HIV-1 RNA packaging region or the 5′ end of the Gag coding sequences.
HIV-1 virions produced from cells containing altered Gag/Gag-Pol ratios showed dramatically reduced infectivity (Table 2). The mechanism of this effect is unclear, as virions derived from cells with a Gag/Gag-Pol ratio of 20:21 [due to an excess of GP PR(+)] had protein profiles and levels of virion-encapsidated RNA equivalent to those of wild-type HIV-1, despite having ∼1,000-fold decreased infectivity. As the mRNAs for both GP PR(+) and GP PR(−) contain wild-type RNA packaging sequences, it is conceivable that the packaging of GP PR(+) or GP PR(−) mRNA can, in part, contribute to the reduction in viral infectivity observed here. The highest intracellular level of GP PR(+) or GP PR(−), however, has never exceeded the mRNA levels of wild-type HxB2-BH10 (Table 1). If the viral infectivity of the progeny viruses with altered Gag/Gag-Pol ratios is a function of the encapsidated genome, a maximum twofold reduction in viral infectivity would be anticipated. Clearly, the maintenance of virion infectivity is at least one explanation for the conserved Gag/Gag-Pol ratio of 20:1 in all retroviruses. The maintenance of this evolutionarily conserved Gag/Gag-Pol ratio in retroviruses may represent a previously unexplored mechanism for regulation of retroviral assembly, and ultrastructural study of retroviral particles produced in cells with altered Gag/Gag-Pol ratios may provide further insight into the functional importance of this ratio in the retroviral replication cycle.
ACKNOWLEDGMENTS
We thank John Mills for helpful criticism and review of the manuscript. We also thank Melissa Hill and Katherine Kedzierska for assistance with the infectivity assay.
M.S.-X. is a recipient of an NHMRC Dora Lush Ph.D. scholarship and is also supported by the Monash University Postgraduate Research Fund. S.M.C. is supported by NCHVR and the MBC research fund. J.M. is a recipient of an NHMRC Peter Doherty postdoctoral fellowship.
REFERENCES
- 1.Babé L M, Ros J, Craik C S. Trans-dominant inhibitory human immunodeficiency virus type 1 protease monomers prevent protease activation and virion maturation. Proc Natl Acad Sci USA. 1995;92:10069–10073. doi: 10.1073/pnas.92.22.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Crowe S M, et al. HIV infection of monocyte-derived macrophages in vitro reduces phagocytosis of Candida albicans. J Leukoc Biol. 1994;56:318–327. doi: 10.1002/jlb.56.3.318. [DOI] [PubMed] [Google Scholar]
- 5.Darlix J-L, Gabus C, Allain B. Analytical study of avian reticuloendotheliosis virus dimeric RNA generated in vivo and in vitro. J Virol. 1992;66:7245–7252. doi: 10.1128/jvi.66.12.7245-7252.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darlix J-L, Gabus C, Nugeyre M T, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein K M, Goff S P. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J Virol. 1988;62:2179–2182. doi: 10.1128/jvi.62.6.2179-2182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein K M, Goff S P. Mutational analysis of the gag-pol junction of Moloney murine leukemia virus: requirements for expression of the gag-pol fusion protein. J Virol. 1992;66:6601–6608. doi: 10.1128/jvi.66.11.6601-6608.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Y-X, Hatfield D L, Rein A, Levin J G. Translational readthrough of the murine leukemia virus gag gene amber codon does not require virus-induced alteration of tRNA. J Virol. 1989;63:2405–2410. doi: 10.1128/jvi.63.5.2405-2410.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y-X, Levin J G, Hatfield D L, Schaefer T S, Gorelick R J, Rein A. Suppression of UAA and UGA termination codons in mutant murine leukemia viruses. J Virol. 1989;63:2870–2873. doi: 10.1128/jvi.63.6.2870-2873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goff S, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorry P R, Howard J L, Churchill M J, Anderson J L, Cunningham A, Adrian D, McPhee D A, Purcell D F J. Diminished production of human immunodeficiency virus type 1 in astrocytes results from inefficient translation of gag, env, and nef mRNAs despite efficient expression of Tat and Rev. J Virol. 1999;73:352–361. doi: 10.1128/jvi.73.1.352-361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Mak J, Cao Q, Li Z, Wainberg M A, Kleiman L. Incorporation of excess wild type and mutant tRNALys3 into HIV-1. J Virol. 1994;68:7676–7683. doi: 10.1128/jvi.68.12.7676-7683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacks T, Madhani H D, Masiarz F R, Varmus H. Signals for ribosomal framshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 19.Jacks T, Townsley K, Varmus H E, Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci USA. 1987;84:4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacks T, Varmus H E. Expression of the Rous sarcoma virus Pol gene by ribosomal frameshifting. Science. 1985;232:1237. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 21.Junker U, Escaich S, Plavec I, Baker J, McPhee F, Ros J R, Craik C S, Böhnlein E. Intracellular expression of human immunodeficiency virus type 1 (HIV-1) protease variants inhibits replication of wild-type and protease inhibitor-resistant HIV-1 strains in human T-cell lines. J Virol. 1996;70:7765–7772. doi: 10.1128/jvi.70.11.7765-7772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karacostas V, Wolffe E J, Nagashima K, Gonda M A, Moss B. Overexpression of the HIV-1 Gag-Pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 23.Katoh I, Yasunaga T, Ikawa Y, Yoshinaka Y. Inhibition of retroviral protease activity by an aspartyl proteinase inhibitor. Nature. 1987;329:654–656. doi: 10.1038/329654a0. [DOI] [PubMed] [Google Scholar]
- 24.Katoh I, Yoshinaka Y, Rein A, Shibuya M, Odaka T, Oroszlan S. Murine leukemia virus mutation: protease region required for conversion from “immature” to “mature” core form and for virus infectivity. Virology. 1985;145:280–292. doi: 10.1016/0042-6822(85)90161-8. [DOI] [PubMed] [Google Scholar]
- 25.Kaye J F, Lever A M L. trans-acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J Virol. 1996;70:880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 27.Levin J G, Hatfield D L, Oroszlan S, Rein A. Mechanisms of translational suppression used in the biosynthesis of reverse transcriptase. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 5–31. [Google Scholar]
- 28.Linial M L. Foamy viruses are unconventional retroviruses. J Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mak J, Jiang M, Wainberg M A, Hammarskjöld M-L, Rekosh D, Kleiman L. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J Virol. 1994;68:2065–2072. doi: 10.1128/jvi.68.4.2065-2072.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak J, Khorchid A, Cao Q, Huang Y, Lowy I, Prasad V R, Parniak M A, Wainberg M A, Kleiman L. Effects of mutations in Pr160gag-pol upon tRNALys3 and Pr160gag-pol incorporation into HIV-1. J Mol Biol. 1997;265:419–431. doi: 10.1006/jmbi.1996.0742. [DOI] [PubMed] [Google Scholar]
- 31.Marquet R, Paillart J-C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mergener K, Facke M, Welker R, Brinkmann V, Gelderblom H R, Kräusslich H G. Analysis of HIV particle formation using transient expression of subviral constructs in mammalian cells. Virology. 1992;186:25–39. doi: 10.1016/0042-6822(92)90058-w. [DOI] [PubMed] [Google Scholar]
- 33.Morin N, Cherry E, Wainberg M A. Cotransfection of mutated forms of human immunodeficiency virus type 1 Gag-Pol with wild-type constructs can interfere with processing and viral replication. J Hum Virol. 1998;1:240–247. [PubMed] [Google Scholar]
- 34.Panganiban A T. Retroviral gag gene amber codon suppression is caused by an intrinsic cis-acting component of the viral mRNA. J Virol. 1988;62:3574–3580. doi: 10.1128/jvi.62.10.3574-3580.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parent L J, Cairns T M, Albert J A, Wilson C B, Wills J W, Craven R C. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J Virol. 2000;74:164–172. doi: 10.1128/jvi.74.1.164-172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J, Morrow C D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J, Morrow C D. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol. 1991;65:5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prats A-C, Roy C, Wang P, Erard M, Housset V, Gabus C, Paoletti C, Darlix J-L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 1.74–1.84. [Google Scholar]
- 40.Skripkin E, Paillart J-C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith A J, Cho M-I, Hammarskjöld M-L, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanstrom R, Wills J W. Retroviral gene expression. II. Synthesis, processing, and assembly of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 43.Terwilliger E F, Cohen E A, Lu Y C, Sodroski J G, Haseltine W A. Functional role of human immunodeficiency virus type 1 Vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torrent C, Bordet T, Darlix J L. Analytical study of rat retrotransposon VL30 RNA dimerization in vitro and packaging in murine leukemia virus. J Mol Biol. 1994;240:434–444. doi: 10.1006/jmbi.1994.1459. [DOI] [PubMed] [Google Scholar]
- 45.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 46.Wu X, Liu H, Xiao H, Conway J A, Hunter E, Kappes J C. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 1997;16:5113–5122. doi: 10.1093/emboj/16.16.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]