Abstract
We have previously examined the transcription and splicing of open reading frames (ORFS) 71 (K13), 72, and 73 of Kaposi's sarcoma-associated herpesvirus (KSHV) in the primary effusion lymphoma cell line BCP-1 (latently infected with KSHV) (45). The three genes encoded by these ORFs (for vFLIP, vCyclin, and latency-associated nuclear antigen [LANA]) are transcribed from a common transcription start site in BCP-1 cells during both latency and the lytic cycles. The resulting transcript is spliced to yield a 5.32-kb message encoding LANA, vCyclin, and vFLIP and a 1.7-kb bicistronic message encoding vCyclin and vFLIP. To investigate whether the vFLIP protein could be expressed from this vCyclin/vFLIP message, we utilized a bicistronic luciferase reporter system. The genes for Renilla and firefly luciferases (which utilize different substrates) were cloned in tandem downstream from a T7 RNA polymerase promoter. Fragments of DNA immediately upstream from the initiating codon of vFLIP were cloned between the two luciferase genes. The relative expression of the two luciferases, one directed by the putative internal ribosome entry site (IRES) sequences and the other by cap-dependent ribosome scanning, was used to compare the activities of the different DNA fragments. A minimum fragment of 233 bp within the coding region of vCyclin was found to direct efficient expression of the downstream cistron (firefly luciferase). The activity of this IRES was orientation dependent and unaffected by methods used to inhibit cap-dependent translation. This is the first demonstration of an IRES element encoded by a DNA virus and may represent a novel mechanism through which KSHV controls protein expression.
Kaposi's sarcoma (KS) is a vascular tumor occurring most commonly in patients with AIDS (3, 4). KS lesions are histologically complex and contain proliferating spindle-shaped cells considered to be of endothelial origin, infiltrating mononuclear cells, plasma cells, and abundant neovascular spaces (7). The recently identified KS-associated herpesvirus (KSHV) (10) is implicated in the etiology of all epidemiological forms of KS, i.e., Mediterranean classic, African endemic, posttransplant or iatrogenic, and the most commonly occurring AIDS-associated form (8). KSHV sequences have also been identified in several rare lymphomas, such as multicentric Castleman's disease and primary effusion lymphoma (PEL), also known as body cavity-based large-cell lymphoma (9, 42). The seroprevalence of KSHV in the general population exhibits variations with geographic distribution. Very low rates of prevalence have been reported for populations in Britain and North America, whereas high rates prevail in Africa and southern Europe (19, 31, 41). However, antibody kinetic studies have shown that a strong correlation exists between conversion to seropositivity and the risk for development of KS (19, 29). Thus, KSHV has been proposed as the etiologic agent for KS and other KSHV-associated malignancies.
KSHV is a gammaherpesvirus that is closely related to three other herpesviruses with oncogenic potential: herpesvirus saimiri, the murine gammaherpesvirus (MHV-68) and, more distantly, Epstein-Barr virus (34, 38). The complete nucleotide sequence of KSHV DNA has revealed several genes, which were probably captured from the host cell during viral evolution and whose products could also play a role in cellular transformation and tumor induction. These include a cyclin D homolog (vCyclin) (11, 21), a Bcl-2 homolog (13), a viral FLICE (FADD [Fas-associated death domain]-like interleukin-1 beta-converting enzyme)-inhibitory protein (vFLIP) (17, 46), and a G-protein coupled receptor homolog (1).
The three genes encoded by open reading frames (ORFs) 71, 72, and 73 (for vFLIP, vCyclin, and latency-associated nuclear antigen [LANA]) are transcribed from a common transcription start site in BCP-1 cells that are uninduced (latent) or induced (lytic) with n-butyrate. The resulting transcript is spliced to yield a 5.32-kb message encoding LANA, vCyclin, vFLIP, and a 1.7-kb bicistronic message encoding vCyclin and vFLIP (16, 45). Since a monocistronic transcript coding for vFLIP alone has never been detected, some other unconventional mechanism involving translational reinitiation, internal ribosomal entry, or leaky ribosomal scanning may be implicated in the expression of vFLIP.
All nuclear eukaryotic mRNAs possess a cap structure (m7GpppN, where N is any nucleotide) at their 5′ end. The cap plays a central role in promoting ribosome binding to the mRNA and controlling the rate of translation initiation (20, 40). Ribosomes can, however, access a eukaryotic mRNA by binding to an internal ribosome entry site (IRES). IRESs were first identified in picornavirus RNA, which does not have a 5′ cap structure (27, 36), and have since been characterized in other viruses, such as hepatitis C virus (47), and also in cellular mRNAs involved in cell proliferation and apoptosis (fibroblast growth factor-2 [48], vascular endothelial growth factor [43], X-linked inhibitor of apoptosis [24], and the protooncogene c-myc [33, 44]). Recently, IRES elements in two cellular mRNAs (encoding omithine decarboxylase [ODC] and PITSLRE protein kinase) have been identified which are regulated in a cell-cycle-dependent manner (15, 37). These data reveal a novel role for IRES elements in the translational regulation of protein expression during cell cycle progression.
In this paper we describe the identification of an IRES element within the KSHV cyclin ORF. This 233-nucleotide sequence could direct the translation of the downstream vFLIP ORF from the vCyclin/vFLIP bicistronic transcript.
MATERIALS AND METHODS
Cells.
The KSHV-positive PEL B-cell line, BCP-1 (6), was grown in RPMI (Gibco) supplemented with 20% (vol/vol) fetal calf serum (FCS), 2 mM glutamine, 60 μg of penicillin/ml, and 100 μg of streptomycin/ml. The endothelial cell line KS-IMM, derived from a KS lesion (2), was grown in MCDB131 media (Gibco) supplemented with 10% (vol/vol) FCS, 10 mM glutamine, 20 ng of endothelial cell growth supplement (Sigma)/ml, 60 μg of penicillin/ml, and 100 μg of streptomycin/ml. HEK293 cells (22) were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% (vol/vol) FCS, 2 mM glutamine, 60 μg of penicillin/ml, and 100 μg of streptomycin/ml. Cells were incubated at 37°C under 4% CO2.
Plasmids.
The plasmid pdLUC was constructed by cloning the firefly luciferase gene (from pGL3-basic; Promega) as an NheI-XbaI fragment downstream of the Renilla luciferase gene at the XbaI site in the plasmid pRL-CMV (Promega). Plasmid pdLUC-SL is identical to pdLUC except for a 60-nucleotide sequence (5′GCTAGCGGTACGGCAGTGCCGTACGACGAATTCGTCGTACGGC ACTGCCGTACCGCTAGC3′), capable of forming a stable 28-bp stem-loop (ΔG = −62 kcal/mol), cloned at the NheI site immediately upstream from the Renilla luciferase start codon (see Fig. 1b).
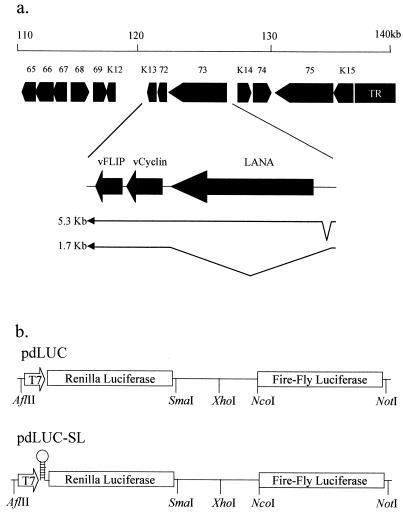
FIG. 1.
(a) Map of the left end of the KSHV genome showing the transcription of the latent genes ORF 71, 72, and 73 encoding vFLIP, vCyclin, and LANA, respectively. Two spliced transcripts are observed in PEL cell lines: one tricistronic transcript encoding LANA, vCyclin, and vFLIP and one bicistronic transcript encoding vCyclin and vFLIP. (b) Schematic diagram of the plasmids pdLUC and pdLUC-SL. Both plasmids have the coding sequence for the Renilla and firefly luciferase enzymes cloned downstream of a T7 RNA polymerase promoter. The plasmid pdLUC-SL has a stable 28-bp stem-loop cloned immediately upstream of the Renilla luciferase start codon.
The IRES sequence from encephalomyocarditis virus (EMCV) or fragments of KSHV vCyclin/vFLIP were cloned into the SmaI-NcoI sites of pdLUC and pdLUC-SL.
Transfection of cells.
BCP-1 cells (105 cells per well), HEK293 (5 × 104 cells per well), or KS-IMM (5 × 104 cells per well) were seeded in 24-well trays and incubated overnight. The cells were infected with vTF7-3 (18), a recombinant vaccinia virus expressing T7 RNA polymerase, at 5 PFU per cell in 200 μl of serum-free medium (OptiMEM; Gibco-BRL) for 60 min at 37°C. The inoculum was removed, and the cells were washed once with OptiMEM. The cells were then transfected with 0.5 μg of linearized (AflII and NotI) plasmid DNA and 1.5 μl of Transfast transfection reagent per well according to the instructions of the manufacturer (Promega). Following a 1-h incubation at 37°C, 1 ml of growth medium was added to the wells. The cells were assayed for luciferase activity 24 h later as described below.
Dual luciferase assays.
Transfected cells were washed twice in phosphate-buffered saline and then lysed by addition of 200 μl of passive lysis buffer (Promega). After incubation for 15 min at room temperature, the cell lysates were transferred to Eppendorf tubes and snap frozen on dry ice. The lysates were then thawed and vortexed for 1 min, and the cell debris was removed by spinning at 10,000 × g for 1 min. The activities of Renilla and firefly luciferases were assayed using the dual luciferase system as described by the manufacturer (Promega). Luciferase activities were measured using a Labsystems benchtop luminometer, and the ratio of firefly luciferase activity to Renilla luciferase activity was calculated and used as a measure of IRES function.
Northern blots.
BCP-1 cells (2 × 106) were infected with vaccinia vTF7-3 (18) (multiplicity of infection = 5) and then transfected with 2 μg of linearized plasmid DNA as described above. Twenty-four hours posttransfection, Poly(A)+ RNA was isolated (Sigma) from the BCP-1 cells. The mRNA (equivalent to 106 cells per lane) was separated on formaldehyde agarose gels as described previously (39) and transferred onto a Genescreen-plus membrane by capillary blotting according to the manufacturer's instructions (NEN Research products). The filters were baked at 80°C for 2 h and then hybridized to Renilla or firefly luciferase DNA probes randomly labeled with [α-32P]dCTP. Filters were prehybridized for 20 min and hybridized for 60 min at 60°C in QuickHyb solution (Stratagene). Blots were washed twice for 15 min at 20°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) sodium dodecyl sulfate and once for 30 min at 60°C in 0.1× SSC–0.1% sodium dodecyl sulfate before exposure to X-ray film (X-OMAT-AR-Kodak) at −80°C with an intensifying screen.
RESULTS
The major latently expressed loci within the KSHV genome contain three genes: those for LANA, vCyclin (Cyclin D homolog), and vFLIP (Flice inhibitory protein homolog) (30). Two mRNA transcripts from this region are seen in cells latently infected with KSHV. Both are transcribed from a common transcription start site and are differentially spliced to yield a tricistronic message encoding LANA, vCyclin, and vFLIP and a bicistronic message encoding vCyclin and vFLIP (Fig. 1a) (16, 45). There have been no reports of further splicing events resulting in a monocistronic transcript encoding for vFLIP alone, suggesting that this ORF may be translated via an unconventional mechanism. To test the hypothesis that this bicistronic transcript could contain an IRES allowing the translation of the downstream vFLIP ORF, we constructed a bicistronic luciferase reporter plasmid (pdLUC) (Fig. 1b). In this plasmid the downstream cistron (firefly luciferase) would ordinarily be accessed inefficiently by ribosomes which have completed translation of the upstream cistron (Renilla luciferase). However, if an IRES sequence (e.g., EMCV IRES) (Fig. 2a) is inserted before the downstream ORF, translation is considerably enhanced (up to 100-fold). This system offers the advantage that both reporter enzymes can be assayed in the same cell lysate preparation, with the activity of the two luciferase enzymes (which utilize different substrates) determined using a benchtop luminometer. In addition, the upstream reporter (Renilla luciferase) acts as an internal control to account for differences in transfection efficiency. The plasmids were constructed such that transcription of the bicistronic reporter was driven from a T7 RNA polymerase promoter. The constructs were transfected into cells infected with a recombinant vaccinia virus (vTF7-3) (18) which expresses T7 RNA polymerase, therefore allowing cytoplasmic transcription of the bicistronic mRNAs. Since this system bypasses the nuclear transcription pathway, the possibility of splicing accounting for expression of the downstream cistron is eliminated.
FIG. 2.
Assay for IRES activity in BCP-1 cells. The indicated restriction fragments from the vCyclin gene were cloned between the SmaI and NcoI sites of pdLUC (a) or pdLUC-SL (b). The activities of firefly (FF) and Renilla (RL) luciferase (in relative light units) and the ratio (%) of the two from a representative experiment are shown. The restriction sites shown on the schematic diagram of vCyclin and vFLIP are as follows: NcoI, BamHI, SacII, AatII, Eco47III, PvuII, HindIII, NcoI.
The KSHV vCyclin coding sequence contains an IRES element.
Restriction enzyme fragments of the vCyclin coding sequence immediately upstream from the vFLIP start codon were cloned into the pdLUC plasmid as shown in Fig. 2a. These plasmids were transfected into the BCP-1 cell line (latently infected with KSHV), which had been infected with vaccinia virus vTF7-3 (18) at a multiplicity of infection of 5. The Renilla luciferase and firefly luciferase activities were measured in cell lysates 24 h posttransfection. The ratio of firefly luciferase activity to Renilla luciferase activity was calculated and used as a measure of IRES function. These data (Fig. 2a) show that a 233-nucleotide sequence between the SacII and Eco47III restriction sites (nucleotides 123206 to 122973; GenBank accession no. U75698) (38) in the vCyclin coding sequence is capable of directing efficient translation of the firefly luciferase. Reducing the size of this fragment further using the AatII restriction site resulted in a complete loss of IRES activity. Although this putative IRES sequence was less efficient (10.6 versus 30%) than the well-characterized IRES from EMCV, it still promoted translation well above background levels. We also note that the expression of firefly luciferase from pdLuc8 (SacII-Eco47III) was more efficient (10.6 versus 6.2%) than that from pdLuc3 (SacII-NcoI) (Fig. 2a). This may reflect the smaller nucleotide distance from the IRES element to the downstream cistron, or alternatively, the increased activity could be due to the removal of two start codons that could potentially compete for scanning ribosomes. The function of this vCyclin IRES was orientation dependent (Fig. 2a).
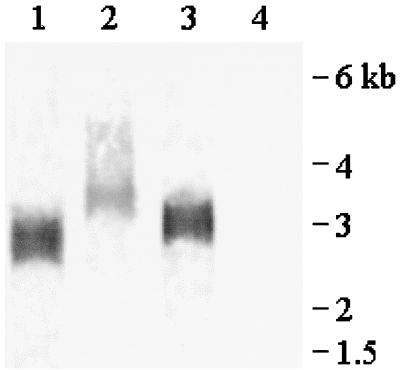
A Northern blot analysis of poly(A)+ RNA isolated from BCP-1 cells infected with vaccinia vTF7-3 (18) and transfected with pdLuc, pdLuc-EMCV, or pdLuc8 and probed with firefly luciferase DNA showed that the cells expressed bicistronic mRNA of the predicted size (Fig. 3). We did not observe any smaller transcripts representing alternative transcription start sites within the KSHV sequences, even on overexposed blots (data not shown). These data suggest that the enhanced expression of the downstream firefly luciferase gene resulted from IRES-driven translation rather than from unusual splicing events, RNA fragmentation, or a cryptic vaccinia virus promoter.
FIG. 3.
Northern blot of poly(A)+ RNA purified from BCP-1 cells infected with vaccinia vTF73 (18) and transfected with pdLUC (lane 1), pdLUC-EMCV (lane 2), or pdLUC-8 (lane 3) or untransfected (lane 4). The blot was probed with DNA corresponding to the firefly luciferase gene. The approximate size predicted for each transcript is 2.7 kb (pdLUC), 3.3 kb (pdLUC-EMCV), or 2.9 kb (pdLUC-8). Size markers (kb) are indicated.
The function of the vCyclin IRES element was also tested in both HEK293 (22) and KSIMM cells (2) (a KSHV-negative endothelial cell line derived from a KS biopsy). Although the EMCV IRES directed efficient translation of the second cistron in these cells, none of the vCyclin sequences appeared to function as an IRES (Table 1). These data suggest that specific KSHV or cellular factors may be required for efficient functioning of the vCyclin IRES.
TABLE 1.
IRES activity in HEK293 cells and KSIMM cellsa
| Cell line | Plasmid | Activity
|
||
|---|---|---|---|---|
| RL | FF | FF:RL (%) | ||
| HEK293 | pdLuc | 4,122 | 6 | 1.4 |
| pdLuc-EMCV | 4,122 | 731 | 18 | |
| pdLuc8 | 5,595 | 5 | 0.9 | |
| KSIMM | pdLuc | 1,969 | 4 | 0.2 |
| pdLuc-EMCV | 1,194 | 299 | 25 | |
| pdLuc8 | 1,120 | 8 | 0.7 | |
The activities of firefly (FF) and Renilla (RL) luciferases (in relative light units) and the ratio (%) of the two from a representative experiment are shown. The structures of the plasmids are as shown in Fig. 2.
Inhibition of translation of the upstream cistron does not affect the activity of the IRES.
To show that the enhanced expression of the firefly luciferase cistron was due to an IRES sequence within the vCyclin gene and was not dependent on the translation of the upstream Renilla luciferase cistron, we constructed a plasmid that inhibited translation of the first cistron. This was achieved by using an equivalent reporter construct (pdLUC-SL; Fig. 1b) that contained an inverted repeat, with the potential to form a stable 28-bp stem-loop structure in the 5′ untranslated region (UTR) immediately upstream from the Renilla luciferase start codon. The data shown in Fig. 2b show that translation of the first cistron is efficiently inhibited by the presence of the stable stem-loop structure. However, translation of the second cistron via the IRES sequence within the vCyclin gene was unaffected by the presence of the stem-loop. This confirms that translation of the second cistron is dependent not on translation of the first cistron but rather on the presence of specific sequence elements within the IRES sequence.
The predicted RNA secondary structure of the vCyclin IRES.
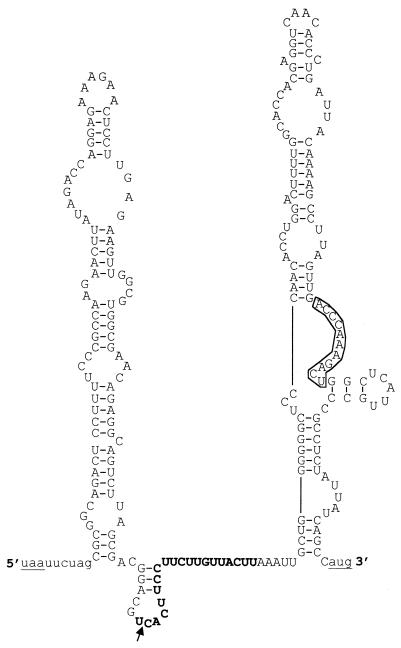
An RNA secondary model was determined for the minimum vCyclin IRES sequence. The program RNA mfold (32, 49) was used to produce a structure with the lowest free energy (ΔG = −61.4 kcal/mol), as shown in Fig. 4. The model predicts that this RNA sequence folds into two stable stem-loop structures separated by a short non-base-paired region. Sequence comparisons revealed a stretch of 11 nucleotides that is 100% complementary to a sequence in 18S rRNA (nucleotides 1150 to 1160). The secondary structure model predicts that this sequence is in a single-stranded loop and therefore has the potential for base-pairing interactions with 18S rRNA. A recent report (12) described an IRES element within the 5′ UTR of the mRNA encoding the homeodomain protein Gtx, which contained a nine-nucleotide segment 100% complementary to 18S rRNA (nucleotides 1124 to 1132). This 9-nucleotide fragment still functioned as an IRES even when taken out of the context of the rest of the 5′ UTR, suggesting that this IRES functioned at least in part via base-pairing between rRNA and the mRNA. To test if this was also the case for the vCyclin IRES, we cloned the 11-nucleotide sequence into the pdLUC and pdLUC-SL reporter plasmids. However, this sequence was unable to promote the translation of the second luciferase cistron on its own (data not shown). Although these data do not rule out the possibility of direct base pairing between the IRES sequence and 18S rRNA, there must be additional elements within the vCyclin IRES that are necessary for its correct function. One such element could be the polypyrimidine-rich sequence found at nucleotides 123199 to 123183. The presence of polypyrimidine-rich sequences in several other IRES elements in viruses and cellular mRNAs has been noted (37). The fact that IRES activity is lost when the two stem-loop structures are separated at the AatII site suggests that multiple structural and sequence elements are essential for correct function of the vCyclin IRES.
FIG. 4.
Predicted secondary structure of the shortest functional vCyclin IRES. The sequence between the SacII and Eco47III sites (nucleotides 123206 to 122973; GenBank accession no. U75698) (38) is shown in upper case. Vector sequences are shown in lower case. The stop codon for the Renilla luciferase gene and the start codon for the firefly luciferase gene are underlined. The 11-nucleotide sequence complementary to 18S rRNA (nucleotides 1150 to 1160) is boxed, and the polypyrimidine-rich sequence is shown in bold. The arrow shows the cleavage position of the AatII site. The RNA was folded using RNA mfold version 3.0 (32, 49). ΔG = −61.4 kcal/mol.
DISCUSSION
Analysis of KSHV gene expression has revealed a cluster of latently expressed transcripts encoding LANA (ORF 73), vCyclin (ORF 72), and vFLIP (ORF K13). The vCyclin and vFLIP coding sequences are present on a bicistronic transcript (30, 45). We have identified an IRES within the vCyclin coding sequence (nucleotides 123206 to 122973) (38) that has the potential to direct the translation of vFLIP from this bicistronic transcript. IRES elements were first identified in the 5′ UTR of picornaviruses (27, 36) and are essential for the cap-independent translation of the viral polyprotein. More recently, IRES elements have been characterized for several cellular genes (14, 15, 23, 24, 28, 33, 37, 44).
It is unusual to find an IRES driving the translation of a gene within the coding sequence of an upstream cistron (vCyclin). Most IRESs examined to date are located within 5′ UTR of mRNAs. Although we have not yet formally shown that this IRES actually controls the expression of vFLIP in vivo, we assume that the data presented here using a dual luciferase reporter system will parallel the situation in KSHV-infected cells.
The KSHV cyclin complexed with cyclin-dependent kinase-6 (cdk6) has been shown to inactivate the retinoblastoma (Rb) protein and thereby promote cell cycle progression and proliferation (11, 21). It has been demonstrated that ectopic expression of vCyclin in cells with elevated levels of cdk6 leads to apoptotic cell death after the cells enter S phase. Studies on the mechanisms involved in this caspase-3-mediated apoptosis indicated that it was independent of cellular p53 or pRb status, and it was not suppressed by Bcl-2. In contrast, the KSHV-encoded v-Bcl-2 efficiently suppressed vCyclin-/cdk6-induced apoptosis (35). Abnormally proliferating cells are also known to be a target for cytotoxic T-cell (CTL)-mediated cell killing through FasL or tumor necrosis factor death receptors. Cellular FLIP (cFLIP) and KSHV vFLIP have been shown to inhibit FasL-mediated apoptosis by binding to and blocking the death-effector domain on the cytoplasmic C terminus of the Fas receptor (CD95) (26, 35, 46). KSHV vFLIP protects cells from Fas-mediated apoptosis by inhibiting caspase activation and permits clonal growth in the presence of death stimuli in vitro. KSHV vFLIP also mediates a tumor-progressive activity by prevention of death receptor-induced apoptosis triggered by CTLs in vivo (17). It therefore seems likely that the function of vFLIP is to prevent CTL-mediated killing of proliferating cells infected with KSHV, and this may explain why the expression of vFLIP is so intimately linked to the expression of vCyclin.
Correct progression of the cell cycle requires that certain proteins be present or active at specific times. It has been shown that cells arrested in G2/M phase or in mitosis translate mRNAs at about 25% of the rate of interphase cells. This is in part due to an inhibition of the translation initiation step due to a loss of the cap binding protein's ability to bind the 5′ m7GpppN cap structure present on eukaryotic mRNAs (5). This results in a general loss of cap-dependent translation in this phase of the cell cycle (5, 25). Recently, two reports (15, 37) have shown that two cellular mRNAs encoding ODC and PITSLRE protein kinase are expressed from IRES elements specifically during the G2/M phase of the cell cycle. These mRNAs bypass the general inhibition of cap-dependent translation in G2/M-arrested cells by utilizing a cap-independent IRES during this period of the cell cycle. These data may have parallels with the expression of KSHV vCyclin and vFLIP. If vFLIP is to protect KSHV-infected cells from CTL-mediated apoptosis, then the levels of this inhibitor of FasL-mediated apoptosis must remain at an appropriate level throughout the cell cycle. One way of achieving this would be to ensure that vFLIP is expressed via an IRES which is not susceptible to cap-dependent inhibition of translation at G2/M.
It has been previously noted that the few eukaryotic mRNAs that possess IRESs encode growth factors (fibroblast growth factor 2 and vascular endothelial growth factor), oncogenes (c-myc and ODC), and an inhibitor of apoptosis (XIAP) (37). IRES-dependent translation of these mRNAs may be essential for the survival and proliferation of cells under stressful conditions. In this paper we have identified an IRES which potentially controls the expression of a virally encoded anti apoptotic protein, vFLIP, that is intimately linked to the expression of a cell growth-promoting protein, vCyclin. Therefore, it is conceivable that the expression of vFLIP from an IRES represents a novel mechanism whereby KSHV ensures continued protection against CTL-induced apoptosis throughout the cell cycle.
ACKNOWLEDGMENTS
This work was supported by a Career Establishment Grant from the Medical Research Council.
We thank James Stewart for reagents and useful discussions.
REFERENCES
- 1.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 2.Benelli R, Repetto L, Carlone S, Parravicini C, Albini A. Establishment and characterization of two new Kaposi's sarcoma cell cultures from an AIDS and a non-AIDS patient. Res Virol. 1994;145:251–259. doi: 10.1016/s0923-2516(07)80030-6. [DOI] [PubMed] [Google Scholar]
- 3.Beral V. Epidemiology of Kaposi's sarcoma. In: Beral V, Jaffe H W, Weiss R A, editors. Cancer, HIV and AIDS. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 5–22. [Google Scholar]
- 4.Beral V, Peterman T A, Berkelman R L, Jaffe H W. Kaposi's sarcoma among persons with AIDS: a sexually transmitted infection? Lancet. 1990;335:123–128. doi: 10.1016/0140-6736(90)90001-l. [DOI] [PubMed] [Google Scholar]
- 5.Bonneau A M, Sonenberg N. Involvement of the 24 kDa capbinding protein in regulation of protein synthesis and mitosis. J Biol Chem. 1987;262:11134–11139. [PubMed] [Google Scholar]
- 6.Boshoff C, Gao S J, Healy L E, Matthews S, Thomas A J, Coignet L, Warnke R A, Strauchen J A, Matutes E, Kamel O W, Moore P S, Weiss R A, Chang Y. Establishing a KSHV+ cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood. 1998;91:1671–1679. [PubMed] [Google Scholar]
- 7.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 8.Boshoff C, Weiss R A. Kaposi's sarcoma-associated herpesvirus. Adv Cancer Res. 1998;75:57–86. doi: 10.1016/s0065-230x(08)60739-3. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman E, Nador R G, Aozasa K, Delsol G, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus in non-AIDS related lymphomas occurring in body cavities. Am J Pathol. 1996;149:53–57. [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 11.Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden K, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 12.Chappell S A, Edelman G M, Mauro V P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng E H, Nicholas J, Bellows D S, Hayward G S, Guo H G, Reitz M S, Hardwick J M. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coldwell M J, Mitchell S A, Stoneley M, MacFarlane M, Willis A E. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- 15.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 16.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djerbi M, Screpanti V, Catrina A I, Bogen B, Biberfeld P, Grandien A. The inhibitor of death receptor signaling, FLICE-inhibitory protein defines a new class of tumor progression factors. J Exp Med. 1999;190:1025–1032. doi: 10.1084/jem.190.7.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuerst T R, Niles E G, Studier F W, Moses B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesises bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 20.Gingras A C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 21.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 23.Henis-Korenblit S, Strumpf N L, Goldstaub D, Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holcik M, Korneluk R G. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J T, Schneider R J. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell. 1991;65:271–280. doi: 10.1016/0092-8674(91)90161-q. [DOI] [PubMed] [Google Scholar]
- 26.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer J L, Schroter M, Burns K, Mattmann C, Rimoldi D, French L E, Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 27.Jang S K, Kräusslich H G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 30.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 31.Lennette E T, Blackbourn D J, Levy J A. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet. 1996;348:858–861. doi: 10.1016/S0140-6736(96)03240-0. [DOI] [PubMed] [Google Scholar]
- 32.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 33.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 34.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ojala P M, Tiainen M, Salven P, Veikkola T, Castanos-Velez E, Sarid R, Biberfeld P, Makela T P. Kaposi's sarcoma-associated herpesvirus-encoded v-cyclin triggers apoptosis in cells with high levels of cyclin-dependent kinase 6. Cancer Res. 1999;59:4984–4989. [PubMed] [Google Scholar]
- 36.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 37.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 38.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Shatkin A J. Capping of eukaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 41.Simpson G R, Schulz T F, Whitby D, Cook P M, Boshoff C, Rainbow L, Howard M R, Gao S J, Bohenzky R A, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder R S, Weller I V, Weiss R A, Moore P S. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 42.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 43.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoneley M, Paulin F E, Le Quesne J P, Chappell S A, Willis A E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 45.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 46.Thome M, Schneider P, Hofmann K, Fickenscher H, Meini E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidl C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 47.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A C. Alternative translation of human FGF-2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. Norwell, Mass: Kluwer Academic; 1999. pp. 11–43. [Google Scholar]