ABSTRACT
Temperate P335 phage TP901-1 represents one of the best-characterized Gram-positive phages regarding its structure and host interactions. Following its reversible adsorption to the polysaccharidic side-chain of the cell wall polysaccharide of its host Lactococcus cremoris 3107, TP901-1 requires a glucosylated cell envelope moiety to trigger its genome delivery into the host cytoplasm. Here, we demonstrate that three distinct single amino acid substitutions in the Tal protein of TP901-1 baseplate are sufficient to overcome the TP901-1 resistance of three L. cremoris 3107 derivatives, whose resistance is due to impaired DNA release of the phage. All of these Tal alterations are located in the N-terminally located gp27-like domain of the protein, conserved in many tailed phages. AlphaFold2 predictions of the Tal mutant proteins suggest that these mutations favor conformational changes necessary to reposition the Tal fiber and thus facilitate release of the tape measure protein from the tail tube and subsequent DNA ejection in the absence of the trigger otherwise required for phage genome release.
IMPORTANCE
Understanding the molecular mechanisms involved in phage-host interactions is essential to develop phage-based applications in the food and probiotic industries, yet also to reduce the risk of phage infections in fermentations. Lactococcus, extensively used in dairy fermentations, has been widely employed to unravel such interactions. Phage infection commences with the recognition of a suitable host followed by the release of its DNA into the bacterial cytoplasm. Details on this latter, irreversible step are still very scarce in lactococci and other Gram-positive bacteria. We demonstrate that a component of the baseplate of the lactococcal phage TP901-1, the tail-associated lysin (Tal), is involved in the DNA delivery into its host, L. cremoris 3107. Specifically, we have found that three amino acid changes in Tal appear to facilitate structural rearrangements in the baseplate necessary for the DNA release process, even in the absence of an otherwise required host trigger.
KEYWORDS: Lactococcus cremoris, TP901-1 phage, Tal, DNA release, recombineering
INTRODUCTION
Lactococcus lactis and Lactococcus cremoris are among the most frequently used microorganisms in global dairy fermentations. Bacteriophages (phages), which are viruses that infect bacteria, represent a major economic concern to the dairy industry as they regularly delay or disrupt the fermentation process. Therefore, phages infecting Lactococcus have been extensively studied to investigate their prevalence, diversity, and host range-determining factors. All known lactococcal phages possess a tail, and 10 distinct groups have been distinguished based on their morphology and genetic relatedness (1). Members of the diverse P335 group are included among the most frequently encountered phages in dairy fermentations.
P335 group phages, which comprise both virulent and temperate phages (2), exhibit a narrow host range, which is believed to reflect a highly specific interaction with their cognate host (3). This interaction is mediated by the so-called “adhesion device,” which is located at the distal end of the phage tail, and is responsible for the recognition of, and reversible binding to, a suitable host-encoded receptor (3). The primary receptor for P335 phages, as well as the majority of lactococcal phages, is known to be (a component of) a specific cell wall polysaccharide (CWPS), which is composed of a peptidoglycan-embedded rhamnan and a surface-exposed oligo- or poly-saccharidic decoration or side-chain (4–6). Furthermore, these CWPS structures may contain additional sugar decorations due to the activity of three-component glycosylation systems (TGSs) (7). These TGSs typically consist of (i) a membrane-associated undecaprenyl-P-sugar (und-P) activating glycosyltransferase, (ii) a flippase that transports the Und-P-sugar moiety to the extracytoplasmic side of the membrane, and (iii) a polytopic GT that catalyzes attachment of the sugar to its final acceptor (8).
The P335 phage TP901-1 is one of the most thoroughly characterized Gram-positive phages and, along with Tuc2009, the best studied lactococcal phage with respect to their structural characteristics (9–11). The structure of the adhesion device or base plate of TP901-1 has previously been partially determined by electron microscopy (9, 12) and X-ray crystallography (11). Recently, AlphaFold2 (13) was used to further assess details of the base plate structure. It consists of six BppU (baseplate protein, upper component) trimers arranged around a central Dit (distal tail protein) hexamer. Each BppU trimer projects three RBP (receptor-binding protein) trimers (11, 14). At the distal extremity of the tail, a trimeric Tal (tail-associated lysin) protrudes, forming the core of the baseplate. It is known that TP901-1 Tal can undergo proteolytic processing, resulting in the removal of its C-terminal domain, which exhibits cell wall-degrading activity (15). The resulting heterogeneous phage population serves to facilitate the phage to infect bacteria most effectively where levels of cell wall cross-linkage may differ (15). The central channel of the tail is proposed to be filled by the TMP (tape measure protein) (14, 16).
TP901-1 recognizes and reversibly adsorbs to the polysaccharidic side-chain of the CWPS of L. cremoris 3107 via its RBPs. This CWPS side-chain (previously termed the polysaccharide pellicle or PSP) of the lactococcal 3107 strain is also the primary receptor of the P335 phage LC3 (17, 18). Following the RBP-mediated adsorption, the phage infection process continues with an irreversible event which involves phage DNA delivery into the bacterial cytoplasm. To date, information pertaining to this DNA delivery or release event in Gram-positive infecting phages remains scarce. Based on the characterization of three TP901-1-resistant derivatives of L. cremoris 3107, it has previously been proposed that TP901-1 and LC3 utilize distinct DNA release trigger(s) or different DNA entry pathways (18). Both phages were shown to be capable of adsorbing these lactococcal derivatives (18), whereas TP901-1 DNA release was blocked causing phage resistance (18, 19). Recently, we demonstrated through comparative genome analysis of these resistant derivatives and subsequent complementation experiments that two distinct GT-encoding genes from L. cremoris 3107 are required for TP901-1 DNA internalization (19). These two GTs form part of a TGS believed to be involved in glucosylation of an unknown cell envelope-associated moiety, which is currently proposed to represent the secondary receptor or molecular trigger for TP901-1 DNA release. The resistant derivative designated as E121 encodes a truncated GT, CsdC3107, which in the parent strain 3107 is the first GT of the system. Mutants E119 and E126 encode a truncated GT termed as CsdG3107, which in strain 3107 catalyzes the last step of the substrate glucosylation process (19). This is the first sugar decoration that has been implicated as a trigger of the DNA delivery step of a Gram-positive phage, prompting us to investigate the phage structure/s implicated in such interaction or process. In the current study, spontaneous TP901-1 mutants able to infect the E119, E121, and E126 mutant strains were isolated. Their comparative genome analysis shows that particular spontaneous mutations in the TP901-1 tal gene allow bypass of the DNA release trigger, which was corroborated by targeted mutational analysis. Finally, the effect of such mutations on Tal structure was predicted using AlphaFold2.
MATERIALS AND METHODS
Bacterial strains and growth conditions
L. cremoris 3107 (20) wild type (WT) and its derived TP901-1-resistant mutants E119, E121, and E126 (18) were grown at 30°C in M17 broth/agar (Oxoid, UK) supplemented with 0.5% glucose (GM17). L. cremoris NZ9000_TP901-1erm carrying TP901-1erm prophage and plasmid pJP005 (15) was grown at 30°C in GM17 supplemented with 2 µg mL−1 of Erythromycin (Ery) and 5 µg mL−1 of Chloramphenicol (Cm).
Recombineering and oligonucleotides
All oligonucleotides used in this study are shown in Table 1. Recombineering was performed as previously described (15, 21, 22). Briefly, competent cells of L. cremoris NZ9000_TP901-1erm harboring pJP005 were transformed with 500 µg of the appropriate recombineering oligonucleotide, and after recovery, serial dilutions were plated on GM17 agar plates containing 2 µg mL−1 Ery and 5 µg mL−1 Cm. Colonies were screened by mismatch amplification mutation analysis-PCR (22), and those containing the desired mutation were further purified. Recombineering oligonucleotides were ordered from Integrated DNA Technologies (Belgium), whereas all other oligonucleotides were ordered from Eurofins Genomics (Germany).
TABLE 1.
Oligonucleotidesa
| Oligo | Sequence (5′−3′) |
|---|---|
| 218Rec | A*A*A*T*T*CAATTCTTCGCCCATAACCGTCACCAACTTCTTCTACTTTGCCCCTTCCAATTAGGGCAGTAACAATATTTGTACGGTCTTGTTG |
| 226Rec | T*T*A*C*C*ATTTGACTTTCTCCACTCGACATCTGAAAATTCGATGCGCCTTCTATAACCGTCACCAACTTCTTCACCCTTCCCACGTCCAA |
| 381Rec | C*T*C*G*A*ATGATATCAGATTGATACTTCCCAATTTCTGTACTATCGTAGCGCGTCATTTTATTATTATCAAGGTCAGAGATATTGCTTTG |
| 603Rec | T*G*A*C*T*TCTGGTGGGTATTGACCATTCCAACCTGTATAGATCTAGCCAGAGTTTCCGCCACCATTAGTATCAATTTTTGTT |
| 381ScFw | GGTGACGGTTATGGGCGAAGA |
| 381ScRv | GAATTGCATTAATCCCTTGGAG |
| 381MAMA | CCAATTTCTGTACTATCGTAGCGC |
| 200ScFw | GATGAATGACACACTTTCAG |
| 200ScRv | GCTTTGAACTTGTGACAGTTG |
| 218MAMA | CCAACTTCTTCTACTTTGCCCCT |
| 226MAMA | CTGAAAATTCGATGCGCCTTCT |
| 603ScFw | ATTGAAGAATCTGATCATTTTCTTGTAGC |
| 603ScRv | TTGGAGCAATATAACCTCCGCC |
| 603MAMA | CAACCTGTGTCTCCATCTCCACTA |
Asterisk denotes phosphorothioate linkages of recombineering oligonucleotides.
Bacteriophage assays and isolation of TP901-1erm escape mutants
TP901-1erm, a derivative of phage TP901-1 harboring an Ery resistance marker (23), was used for all assays. TP901-1erm prophage was induced from its lysogenic host L. cremoris NZ9000_TP901-1erm when it reached an optical density at 600 nm (OD600nm) of approximately 0.2, using 0.5 µg mL−1 mitomycin C (MitC). The TP901-1erm-derived prophages generated by recombineering were induced in the same way. Where necessary, phages were propagated in 10 mL of GM17 broth cultured with L. cremoris 3107 (or the relevant mutant) at an approximate OD600nm of 0.2. Either plaques, 100–200 µL of a lysate or the resulting lysate of MitC induction were used for propagation. SM buffer (10 mM CaCl2, 100 mM NaCl, 10 mM MgSO4, and 50 mM Tris-HCl at pH 7.5) was employed as the diluent in all phage assays.
Spontaneous TP901-1erm escape mutants (capable of infecting L. cremoris 3107 E-derivatives) were isolated from standard plaque assays (24). Specifically, 200-µL aliquots from overnight propagations of each L. cremoris 3107 E-derivative (E119, E121, and E126) were mixed with 108-9 PFU of TP901-1erm from a fresh propagation, and this mixture was plated as described (24) and incubated at 30°C overnight. This procedure was independently performed four times in order to increase the likelihood of isolating plaques arising from independent events. Isolated visible plaques were propagated once or twice on the specific E-mutant used for the bacterial lawn. The efficiency of plating (EOP) was determined as the ratio of a specific phage titer on a L. cremoris 3107 E-derivative strain relative to that of the parent strain L. cremoris 3107 WT.
Lysogenization assays were performed as previously described (19). The required strain was grown until an approximate OD600nm of 0.2 was reached. Equal volumes of phages and bacteria were mixed at an MOI of 0.05 and incubated at 30°C for 1 hour to allow one round of infection to occur. Following incubation, cells were diluted as necessary and plated on GM17 with and without 2 µg mL−1 Ery. Plates were incubated anaerobically for 48 hours at 30°C. The frequency of lysogenization was determined by calculating the total number of obtained Ery resistant lysogens per mL, divided by the total colony forming units per ml (CFU mL−1) obtained.
Phage DNA extraction, genome sequencing, assembly, and bioinformatic analysis
Phage genomic DNA was extracted from the isolated TP901-1erm escape mutants and TP901-1erm WT using a Phage DNA Isolation Kit (Norgen Biotek, Canada). Moreover, TP901-1erm WT genomic DNA was extracted and sequenced in each batch of experiments. Phage genomic DNA sequencing was performed using an Illumina MiSeq Sequencing System. Genome assemblies were performed with SPAdes v3.14.0 via the MEGAnnotator2 pipeline (25). Open reading frame (ORF) prediction was performed with Prodigal v2.6 (26), and automatic ORF annotation was performed with DIAMOND against NCBI RefSeq database and INTERPRO against PFAM database (27). Subsequently, comparative genome and single nucleotide polymorphism (SNP) analysis was performed between the phage genome sequence of TP901-1erm escape mutants and that of TP901-1erm WT to identify mutations that may have caused these mutant phages to overcome the TP901-1 phage resistance of 3107 mutant strains E119, E121, and E126. Bowtie2 alignment (28), followed by the application of SAMtools (29) to extract base variants, was employed.
Statistical analysis
Statistical analyses were performed using JASP software version (0.18.3) (30). For evaluation of differences in the lysogenization frequency and the EOPs of phages at different temperatures, first, we performed the Shapiro-Wilk test to assess whether the data were normally distributed and the Levene’s test to test the homogeneity of variances. If the data were normally distributed and the variances were equal, we performed one-way analysis of variance (ANOVA), followed by Tukey’s test. If not, we conducted the Kruskal-Wallis test followed by Dunn’s test. In all cases, differences are considered significant when the P value < 0.05.
AlphaFold2 predictions and analysis
AlphaFold2 (13) predictions were performed using either a Colab notebook running AlphaFold v2.3.1 (https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb) or HPC resources from GENCI-IDRIS running AlphaFold v2.3.1. The pLDDT values of any predicted structure, stored in the pdb file as B-factors, as well as the PAE, were plotted and are shown in Fig. S1. The final predicted protein or domain structures were submitted to the Dali server (31) to identify the closest structural homologs in the PDB. Dali provides a root mean square deviation value in Å, as well as an aggregated factor called Z-value. A Z-score above 20 means the two structures are definitely homologous, between 8 and 20 means the two are probably homologous, between 2 and 8 is a gray area, and a Z-Score below 2 is not significant. Visual representations of the structures were prepared with ChimeraX (32). Coot (33) was used to assemble predictions and visually analyze the predictions.
RESULTS
Isolation of TP901-1 escape mutants able to overcome the resistance of L. cremoris 3107 E-derivatives
Three distinct TP901-1-resistant (but LC3-sensitive) mutants of L. cremoris 3107, named E119, E121, and E126, had previously been isolated following chemical mutagenesis with ethyl methanesulfonate (18). We recently demonstrated that the resistance phenotype observed in those derivatives is due to the presence of mutations within two distinct GT-encoding genes, which form part of an L. cremoris 3107 TGS, involved in the glucosylation of a cell-envelope moiety, the latter believed to be involved in triggering TP901-1 DNA release (19). In all three TP901-1-resistant mutants, the mutations led to the introduction of a premature stop codon and, therefore, non-functional truncated products (19). Mutant E121 carries an insertion within csdC3107, which encodes a GT expected to catalyze the transfer of glucose (Glc) from uracil-diphosphate glucose (UDP-Glc) to Und-P, as shown by heterologous enzyme complementation (19). Mutants E119 and E126 each carry a distinct mutation within csdG3107, predicted to encode a GT that transfers this glucose to the final cell-envelope acceptor (19).
Since our attempts to identify the suspected glucosylated cell envelope-associated moiety have so far been unsuccessful (19, 34), we decided to investigate whether it was possible to isolate TP901-1 escape mutants capable of overcoming the phage-resistance of E121, E119, and E126 derivatives. To this end, 108-9 PFU of TP901-1erm WT was mixed with grown cultures of each of the three L. cremoris 3107 E-mutants, after which plaque assays were performed. This procedure was independently executed four times, using a freshly prepared lysate of TP901-1erm WT. Escape mutants were observed at a very low frequency (usually less than five plaques per attempt representing an approximate frequency of <5 × 10−8), albeit slightly more frequently on the L. cremoris 3107 E119 strain relative to E121 and E126. Plaques were propagated once or twice on growing liquid cultures of the corresponding lactococcal mutant used to isolate them, and the presumed mutant phage lysate was tested by standard spot assays using L. cremoris 3107 and the three TP901-1-resistant E-derivatives. However, either the phages present in some of the visible plaques failed to propagate or the phage lysate did not produce discernible zones of lysis on the lawns formed by any of the strains tested. At the same time, apparent bona fide TP901-1erm escape mutants produced clearing in lawns of L. cremoris 3107 WT and the three E-derivatives, irrespective of the mutant strain used to isolate them. This finding indicates that these TP901-1erm phage mutants do not require the presence of the glucosylated moiety on the cell surface of the host to release their DNA. TP901-1erm mutants were named after the strain used to isolate them followed by a letter, in alphabetical order of isolation. A total of 14 genetically distinct escape mutants were isolated (see Table 2 and section below): 8 were isolated on E119, 3 on E121, and 3 on E126.
TABLE 2.
| TP901-1erm spontaneous mutant | Number of SNPs | SNP position in tal | Nucleotide change | Amino acid change | Location/domain | ||
|---|---|---|---|---|---|---|---|
| E119A | 174 | 653 | G | to | C | G218A | Structural domain |
| E119B/ E121Ba/ E121Cb | 13/1/1 | 653 | G | to | T | G218V | Structural domain |
| E119A | 174 | 654–56 | Deletion | E219del | Structural domain | ||
| E119Db/ E119E/E119Fb, c/E119Gd | 1/3/1/136 | 676 | G | to | A | G226R | Structural domain |
| E119C/ E121A/E126A/E126B/E126C | 76/43/74/109/87 | 1,141 | T | to | C | W381R | Structural domain |
| E119Hb | 1 | 1,809 | G | to | A | G603D | Proteolytic processing site |
Only one SNP but query coverage 92%.
Only one SNP.
E119F is genetically identical to E119D, but it was included in the table because it was recovered in a different batch of experiments.
E119G has 94 mutations in tal that results in 20 aa changes.
Position and nucleotide change found in the tal gene, as well as the resulting mutation in the encoded aa and its domain location.
El19I has no mutations in tal. Its genome contains 21 SNPs, which results in 3 aa changes in the terminase large subunit (TerL), 8 aa substitutions in the tail-acting protein (Tap), and 1 aa change in tail terminator protein (Ttp).
Most TP901-1erm escape mutants exhibit a single missense substitution in Tal
The genomes of TP901-1erm escape mutants as well as the genome of TP901-1erm WT were sequenced to identify the mutated gene(s) that is (are) responsible for allowing the phage to overcome the phage resistance phenotype. The genome of TP901-1 WT is 37.6 kb in length, has a 35.38% G + C content, and carries 56 predicted protein-coding regions (35). Following comparative genome analysis between the Illumina-sequenced TP901-1erm WT and phage derivatives, major deletion or insertion events were not identified. Therefore, a SNP analysis was performed to identify mutation events that may have caused the TP901-1erm escape mutants to overcome phage resistance. A SNP analysis was performed using the published TP901-1 data and the Illumina TP901-1erm WT data to rule out sequencing errors. To discard SNPs not involved in TP901-1 DNA release, TP901-1erm escape mutants were compared with the genomic sequence of the TP901-1erm WT obtained from the lysate used to isolate them. Overall, 15 TP901-1erm-independent escape mutants (Table 2) were obtained, of which two, i.e., mutants E119D and E119F, were shown to be genetically identical, though obtained in two separate attempts (in total, 14 genetically distinct mutants). Furthermore, several mutant phages were excluded as they were isolated in the same batch of experiments and their genome sequence turned out to be genetically identical and thus were considered to have originated from a single mutant. The genomes of the TP901-1erm escape mutants exhibit similar characteristics to that of the parent phage. Using as threshold an allelic variation frequency of 80%, the number of SNPs identified on the spontaneous phage mutants ranged between 1 and 174 SNPs (Table 2). Some of these SNPs occurred in more than one of the TP901-1erm escape mutant genomes and with some genes containing more than one mutation in a given genome. Interestingly, the 2,757-bp gene annotated as tal (tail-associated lysin/ORF47), which encodes a 918 amino acid (aa) product, was mutated in 14 out of the 15 TP901-1erm mutant genomes (Table 2). Among the escape mutants, E119A and E119G contain more than one mutation in tal, while the remaining mutants were shown to carry a single missense substitution. Remarkably, five of the phage mutants were shown to carry just a single SNP within their genomes: in mutants E119D/F, this mutation causes a glycine (G) to valine (V) substitution at aa residue 218 of Tal; the SNP in mutants E121B/C brings about a G to arginine (R) change at aa position 226 of Tal, while the SNP in mutant E119H produces a G to aspartic acid (D) alteration at aa position 603 of Tal. G218V and G226R substitutions also appear in other spontaneous TP901-1erm mutants, being the more frequently found mutations in Tal, along with a tryptophan (W) substitution to R at aa position 381 of the Tal protein. To determine how efficiently TP901-1erm spontaneous mutants are able to infect L. cremoris 3107 E-derivatives, quantitative plaque assays were performed using representative mutant phages possessing one of the most frequently obtained mutations (G218V, G226R, and W381R) or the substitution G603D (Table 3). Additionally, two phages containing the same mutation in tal were tested, i.e., E119B and E121C, to analyze whether having one SNP or more, and the strain used to isolate them, had any effect on the EOP. Interestingly, each phage mutant tested exhibited a higher EOP on L. cremoris 3107 E119 when compared with that of E121 and E126. Mutant phages E119B and E121C showed similar EOPs on each host, while E119H exhibited the lowest EOP on all E-derivatives (Table 3). Phages E119B, E121C, and E119F produced smaller plaques on the E-mutants than on L. cremoris 3107 WT, being in some cases pinpoint plaques. The ability of these mutants to overcome the phage insensitivity of L. cremoris 3107 E119, E121, and E126 mutants suggests that either Tal is interacting with the cell envelope component glucosylated by the TGS CsdCG3107 from L. cremoris 3107 or TP901-1 escape mutants bypass the need for this trigger to “activate” Tal.
TABLE 3.
EOPsa of representative spontaneous TP901-1erm mutants on L. cremoris 3107 E-derivatives
| Phage | L. cremoris 3107 E-derivatives | ||
|---|---|---|---|
| E119 | E121 | E126 | |
| TP901-1erm | ≤10−8 | ≤10−8 | ≤10−8 |
| E119B (Mut G218V) | 0.14 ± 0.03 | 0.10 ± 0.03 | 0.06 ± 0.03 |
| E121C (Mut G218V) | 0.14 ± 0.02 | 0.07 ± 0.03 | 0.07 ± 0.06 |
| E119F (Mut G226R) | 0.14 ± 0.09 | 0.07 ± 0.04 | 0.01 ± 4.7 x 10−3 |
| E119D (Mut W381R) | 0.08 ± 0.02 | 0.08 ± 0.01 | 0.05 ± 0.01 |
| E119H (Mut G603D) | 0.03 ± 0.01 | 4.1 × 10−4 ± 7.0 x 10−5 | 8.7 × 10−4 ± 5.0 x 10−4 |
The EOP was determined as the ratio of a specific phage titer on a derivative strain to that of the parent strain L. cremoris 3107 WT.
Three single amino acid substitutions in Tal are sufficient to overcome TP901-1 resistance of L. cremoris 3107 E-derivatives
The structure of TP901-1 Tal (residues 1–918) was predicted by AlphaFold2 and was assembled within the Dit/BppU/RBP heteromeric protein complex to complete the adhesion device structure (3, 14). The TP901-1 Tal N-terminus possesses a T4 phage gp27 fold (36), followed by two linker domains (residues 401–589) and two catalytic domains (residues 618–918). The first catalytic domain (residues 618–772) resembles a domain of a cell wall-degrading enzyme of Bacillus phage phi29, structurally related to lysozymes (3, 37), while the second one (residues 782–918) is a D,D-endopeptidase domain, shown to be able to hydrolyze peptidoglycan cross-bridges (15). To corroborate our indications that certain TP901-1 Tal alterations are essential to allow infection of L. cremoris 3107 E-derivatives, four specific mutations were incorporated in tal of TP901-1erm prophage by recombineering (22). These phages produce a Tal protein carrying the following single aa substitutions, which are also found in the spontaneous TP901-1erm escape mutants: G218V, G226R, W381R, and G603D (Table 2). The first three mutations are located within the T4 gp27-like N-terminal domain. The last mentioned (G603D) mutation is located in a Glycine-rich motif (NGGGNSGGGD; the first and last G of this sequence corresponding to G597 and G604, respectively) located before the first catalytic domain (16, 38). This motif can undergo proteolytic processing, resulting in a heterogeneous population of two phage types (15). It has been shown that a mutant TP901-1erm phage, TP901-1ermGly>Arg, engineered to contain three R instead of G residues in the proteolytic site of Tal, is unable to undergo proteolysis and therefore possesses a full-length tail fiber. This phage was shown to be better adapted to efficiently infect cells with a higher degree of cross-linkage in their cell wall, such as cells in the stationary phase (15). Since we expected a similar effect with the Tal G603D mutation, TP901-1ermGly>Arg was also tested in our experiments.
First, to analyze if any of the engineered mutations affect the phage’s ability to infect the L. cremoris 3107 WT strain, following prophage induction from the lysogenic host and propagation on L. cremoris 3107, the titer of each mutant phage was determined on L. cremoris 3107 (Table 4). Lysates resulting from inductions contained 1–2 × 107 PFU mL−1, except for TP901-1ermW381R, which produced a lysate containing 1 × 106 PFU mL−1. The propagations were performed in parallel, under the same conditions, using 100 µL of each mutant “induction” per 10 mL of culture of L. cremoris 3107. All phages increased their initial titer by at least 10-fold, indicating that all are able to produce new infective virions (Table 4), although TP901-1ermG218V seemed to produce a smaller number of infective virions compared with the other phages.
TABLE 4.
Engineered TP901-1 mutants after propagation on L. cremoris 3107 and EOP on L. cremoris 3107 E-derivatives
| Titer (PFU mL−1) | EOP L. cremoris 3107 E-derivative | |||
|---|---|---|---|---|
| Phage | L. cremoris 3107 | E119 | E121 | E126 |
| TP901-1erm | 2.4 × 108 ± 1.7 × 108 |
≤10−8 | ≤10–8 | ≤10–8 |
| TP901-1ermG218V | 2.6 × 106 ± 3.6 × 105 |
1.5 × 10−2 ± 7.0 × 10−3 |
6.4 × 10−3 ± 2.7 × 10−3 |
1.1 × 10−3 ± 8.1 × 10−4 |
| TP901-1ermG226R | 5.6 × 107 ± 4.3 × 107 |
1.0 × 10−1 ± 5.4 × 10−2 |
5.9 × 10−2 ± 3.8 × 10−2 |
9.8 × 10−3 ± 7.5 × 10−3 |
| TP901-1ermW381R | 1.6 × 106 ± 1.1 × 106 |
1.5 × 10−1 ± 5.0 × 10−2 |
1.1 × 10−1 ± 4.2 × 10−2 |
2.0 × 10−2 ± 1.1 × 10−2 |
| TP901-1ermG603D | 1.1 × 108 ± 3.1 × 107 |
N.D.a | N.D. | N.D. |
| TP901-1ermGlyArg | 4.6 × 108 ± 3.6 × 108 |
N.D. | N.D. | N.D. |
N.D., not determined.
Next, to investigate the effect of the mentioned Tal mutations on the ability of its corresponding mutant phages to infect lactococcal 3107-resistant derivatives, plaque assays were performed and their EOP was determined (Table 4). TP901-1erm mutants harboring mutations in the N-terminal domain of Tal (substitutions G218V, G226R, and W381R), formed visible plaques on each of the L. cremoris 3107 E-derivatives as well as on the WT strain, exhibiting the highest EOPs on E119 and the lowest EOP on E126. TP901-1ermG226R and TP901-1ermW381Rwere shown to exhibit similar EOPs on the three lactococcal mutants, while TP901-1ermG218V yielded EOPs around 10 times lower than the former two TP901-1erm mutants. In all cases, plaques formed on the WT host had a regular size, while they were tiny on the three lactococcal E-derivatives, suggesting that these hosts could affect either the phage latent period or the burst size, apart from the rate constants for phage binding to the specific host (39). The reduction of plaque size on the E-derivatives was also observed with the spontaneous mutant phages E119B, E121C, and E119F, as mentioned above. Double phage mutants were also constructed by recombineering (TP901-1ermG218V,W381R and TP901-1ermG226R,W381R). However, when they were induced from their lysogenic host by MitC, either the lysates were unable to infect L. cremoris 3107 WT and its derivatives, or no intact virions were formed. The other two single mutant phages tested, TP901-1ermG603D and TP901-1ermGly>Arg, seemed to overcome TP901-1 resistance of L. cremoris 3107 E-derivatives, but their EOP could not be accurately determined, as they produced very hazy plaques which were observed only after 48 hours of incubation. Plaque assays were also performed using an overlay containing agarose instead of agar, but no improvement in the plaque visualization was achieved. These findings reinforce the notion that TP901-1 Tal is implicated in the DNA release process triggered by the glucosylated moiety of L. cremoris 3107 and that three aa, G218, G226, and W381, and possibly G603, are involved in this interaction and/or in the genome delivery process.
To support the previous results and considering that lysogeny assays reflect the capacity of a phage to adsorb to its host and to deliver its DNA followed by integration, rather than its ability to form a plaque, we determined the lysogenization frequencies of the engineered mutant phages. An MOI of 0.05 was used for these experiments since some of the phage mutants did not reach a high titer. TP901-1erm lysogenization frequencies of the three L. cremoris 3107 E-derivatives were lower than those of the WT strain (Fig. 1; Table S1). All tested engineered mutant phages exhibited a similar lysogenization frequency of L. cremoris 3107 (10−4), except for TP901-1ermG226R, whose lysogenization frequency was significantly higher than TP901-1erm (1.5-fold higher) (Fig. 1). The lysogenization frequencies obtained for L. cremoris E119 when tested using mutant phages harboring the Tal substitutions G218V, G226R, and W381R were significantly different compared with TP901-1erm WT and approximately the same frequency as of L. cremoris 3107 WT. The frequency of lysogenization of E119 with the other two phages was similar to TP901-1erm (Fig. 1). In the case of L. cremoris E121, a considerable increase in the frequency of lysogeny compared with TP901-1erm was observed with all phage mutants, but the statistical analysis was not performed as the frequency of lysogeny of TP901-1erm was below the detection limits (Table S1). The lysogenization frequencies of L. cremoris E119 and E121 by the phages carrying Tal substitutions G218V, G226, and W381R support the hypothesis that these three aa play an important role in Tal’s function. In contrast, L. cremoris E126 did not reveal any substantial increase in the frequency of lysogenization with any of the tested phages. Although this was unexpected, L. cremoris 3107 E-mutants carry additional mutations (19) which may affect growth and/or lysogeny establishment. Consistent with this, the EOP on L. cremoris E126 of most mutant phages (spontaneous or engineered; Tables 2 and 4) was lower compared with the EOP observed for the other two E-derivatives.
Fig 1.
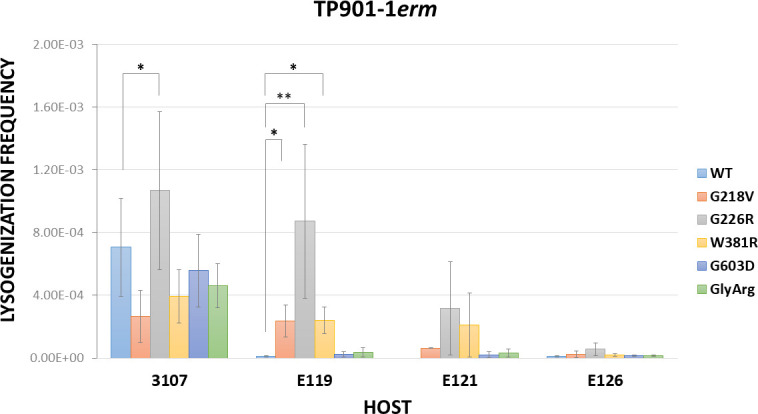
Engineered TP901-1erm mutant lysogenization frequency of L. cremoris 3107 and E-derivatives. Data are the mean of three biological replicates ± standard deviation. One-way ANOVA or Kruskal-Wallis test were performed to compare the lysogenization frequency of L. cremoris 3107, depending on the data distribution and homogeneity of variance. They were followed by Tukey’s test or Dunn’s test, respectively, to determine if there are significant differences compared with TP901-1erm WT. The lysogenization frequency of E121 was not statistically analyzed as TP901-1erm WT frequency was below the limits of detection of our assay ( Table S1). A P value ≤ 0.05 was considered significant and is represented by an asterisk “*”and a P value ≤ 0.01 was represented by two asterisks “∗∗.”
TP901-1ermW381R infectivity is temperature dependent
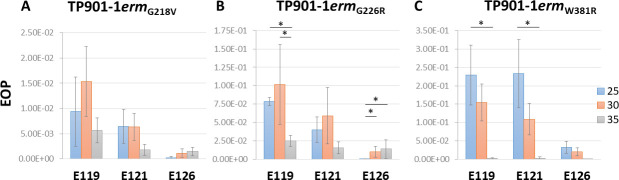
Temperature critically influences phage infection as it can affect host growth and its gene expression, the phage infection stages and type of cycle; and the phage per se, since temperature may affect the function of its structural components. We thus investigated the temperature effect on the infection of L. cremoris 3107 WT and E-derivatives by the engineered mutants. Lactococcal cells were grown at 30°C, while the plates were incubated overnight at the maximum temperature that allows growth of L. cremoris 3107, 35°C, as well as at a lower than optimal growth temperature for this strain, i.e., 25°C. First, we assessed that the titer of each phage mutant originating from the same lysate on L. cremoris 3107 WT plates incubated at the different temperatures was similar to the value obtained at 30°C. Then, we calculated the EOP values of three lactococcal E-derivatives (Fig. 2; Table S2). EOPs obtained at 30°C were also included in the table for comparison. TP901-1ermG218V EOPs were similar on each mutant at each temperature (Fig. 2A; Table S2). TP901-1ermG226R exhibited similar EOPs on E121 independently of the temperature used. EOP values on E119 were significantly different, exhibiting slightly higher EOPs when compared with 35°C. In contrast, on E126, the EOP significantly increased with temperature, being nearly 40-fold higher at 35°C compared with 25°C (Fig. 2B; Table S2). Regarding TP901-1ermW381R, the increase in temperature had the same effect on all mutants, and the EOPs on the three mutants decreased more than 100-fold as the temperature of incubation increased, being significantly different on E119 and E121 when compared with EOPs at 25°C to 35°C. Surprisingly, in both cases, the differences between 30°C and 35°C were not significant (Fig. 2C; Table S2). In general, the plaque size of TP901-1ermG218V and TP901-1ermG226R did not vary much with temperature. In the case of TP901-1ermW381R, plaques observed on E119 and E121 at 25°C were bigger compared with the other temperatures and had a similar size to the regular-size plaques observed on L. cremoris 3107 at 30°C.
Fig 2.
Engineered TP901-1erm mutants EOP on L. cremoris 3107 and E-derivatives at different temperatures. Data are the mean of three biological replicates ± standard deviation. One-way ANOVA or Kruskal-Wallis test were performed to compare the EOPs at different temperatures, depending on the data distribution and homogeneity of variance, followed by Tukey’s test or Dunn’s test, respectively, to determine differences between two temperatures. A P value ≤ 0.05 was considered significant and is represented by an asterisk “*.”
Modelling of Tal
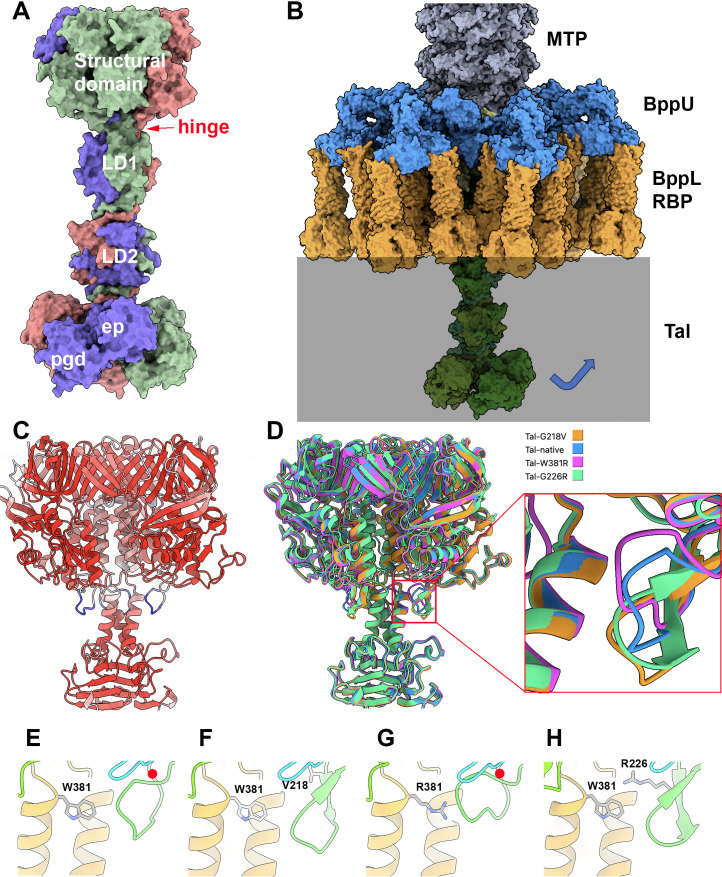
The release of AlphaFold2, a machine-learning, based model to predict protein structures developed by DeepMind, has represented a milestone advance in structural biology (40), due to its atomic accuracy even in cases in which no similar structure is known (13). It synergizes with experimental methods including X-ray crystallography and cryo-electron microscopy and enables structural analyses of large and flexible assemblies resistant to experimental approaches (41). The X-ray structure of the TP901-1 baseplate has previously been reported (PDB id 4v96), though without its Tal component (11). Recently, the full-length Tal trimer was predicted with AlphaFold2 (Fig. 3A) and added to the X-ray-determined structure of the baseplate (14) using Coot (33) (Fig. 3B). The central channel of Tal’s structural N-terminal domain (1–390) is filled by three α-helices that link this domain to the functional C-terminal domain. These helices and the C-terminal domain are believed to dramatically rearrange in order to allow TMP exit and subsequent DNA ejection in the early infection stages (14). Such a rearrangement has recently been reported for phage T5 (42, 43). Based on this, a similar mechanism is proposed to occur for TP901-1 upon baseplate/glucosylated moiety binding to allow consecutive TMP/DNA exit, involving the expulsion of the three helices within the Tal channel and rotation of Tal’s middle and C-terminal domain (Fig. 3B). These structural changes may be induced or facilitated by the impact of the Tal C-terminus on the cell wall and/or its peptidoglycan affinity due to the two cell wall-degradative enzymatic activities adjacently specified by this Tal portion (Fig. 3B). No saccharide-binding domains were found running Dali and Foldseek (31, 44) on linker domains, consistent with previous predictions (3). If such rotation and structural rearrangement occur in TP901-1 Tal, the rotation hinge should be located around residues 375–385 (Fig. 3A, C, and D). Mutations at positions 381, 226, and 218 may play an important role as W381 is located in the putative hinge area and faces G218 and G226. AlphaFold2 predictions of mutants G218V, G226R, and W381R suggest that a conformational change occurs in the loop 217–227 (Fig. 3E through H). These mutations may weaken the Tal rotation area and therefore favor the conformational changes necessary to reposition the Tal fiber and thus facilitate TMP/DNA ejection.
Fig 3.
Structural analysis of TP901-1 Tal. (A) Surface representation of the structural prediction of trimeric Tal; structural domain (residues 1–380); LD1 and LD2, linker domains 1 and 2; pgd, peptidoglycan depolymerase domain; ep, endopeptidase domain. (B) Surface representation of the TP901-1 complete baseplate; MTP, major tail protein (gray); Dit, distal tail protein (yellow, hidden); BppU (blue); RBP/BppL (orange); Tal (green). The cell wall is symbolized by the gray area. (C) Ribbon representation of trimeric Tal N-terminus (1–463) colored by pLDDT. Note the flexible loops 217–227 in blue. (D) Superposition of native and mutated Tal [same orientation as (C)]; inset; close-up of helices 380–396 and loops 217–227 colored by monomer. (E) Ribbon and side-chain view of native Tal W381. (F) Ribbon and side-chain view of Tal W381 and mutant G218V. (G) Ribbon and side-chain view of W381R Tal. (H) Ribbon and side-chain view of Tal W381 and mutant G226R. (E–H) Ribbon is rainbow colored. (E, G) The red dots identify G218 and G226. Figure made with ChimeraX (32). The pLDDT values of any predicted structure, stored in the pdb file as B-factors, as well as the PAE were plotted and are shown in Fig. S1.
DISCUSSION
Phage infection commences with an initial reversible adsorption to the host, followed by an irreversible step that coincides with DNA release into the bacterial cytoplasm. Knowledge on and understanding of this latter step in phages that infect Gram-positive bacteria is still very limited. It is known that Lactococcus Ceduovirus and TP901-1 require two distinct host receptors for reversible and irreversible adsorption, and in both cases, the primary receptor is part of the lactococcal CWPS. The Ceduovirus secondary receptor is the membrane‐associated protein YjaE (45, 46) or PIP (phage infection protein) (47, 48). It has been reported that the phage genes corresponding to l14-15-16 from phage c2 and ORF34-35-36 from phage bIL67 correlate to host lactococcal phage determinants Pip and YjaE, respectively (45). The annotation of the adhesion device of these phages is ambiguous in publicly available databases, but it has been suggested that l13-l14 represent functionally equivalent Dit-Tal components (3). Based on previous work, it has been suggested that TP901-1 irreversibly binds to an as yet unknown glucosylated cell envelope moiety, triggering DNA release (19). In the present study, we provide evidence to support the notion that the Tal protein of TP901-1, which forms part of its baseplate, is implicated in DNA release. Of note, the lactococcal-purified cell wall (without the plasma membrane) has been shown to be necessary and sufficient for adsorption and irreversible inactivation of (some) Skunaviruses (49).
Tal proteins from most phages are longer than ~400 residues and in some cases reach up to 2,000 residues or more. Typically, these proteins possess a conserved N-terminal structural domain, followed by an extension exhibiting large structural and functional diversity (3). The involvement of Tal in the early stages of infection has been experimentally demonstrated in other Gram-positive infecting phages. Bacillus subtilis phage SPP1, following recognition of glycosylated teichoic acids present on the host cell surface, irreversibly binds through Tal to the ectodomain of a membrane protein, YueB, an interaction that triggers genome delivery into the cytoplasm (50, 51). This interaction was shown by the isolation of phage mutants specifically affected in YueB binding and the use of antibodies against the C-terminal region of Tal, consisting of 1,108 residues, which interfered with its interaction with YueB and triggered DNA ejection (51). In the case of the Listeria phage A118, it has been shown that antibodies raised against gp18, the Tal protein, can neutralize adsorption of A118 (52). Tal protein from A118 possesses an extension of ~300 aa following the N-terminal structural domain, harboring a glycosidase domain at its extremity. Interestingly, the first 404 residues of SPP1 Tal fold as the A118 Tal N-terminal domain (53). Currently, we cannot affirm that TP901-1 Tal is directly interacting with the secondary receptor based on our results and structural predictions. Firstly, the mutations found here to be sufficient to overcome TP901-1 resistance are not part of a carbohydrate binding domain (CBM). In fact, no CBMs are predicted within Tal (3), although it is expected to possess peptidoglycan affinity due to its enzymatic activity (15). Nevertheless, it cannot be excluded that there may be, as yet, undiscovered alternative triggers for this process. Secondly, TP901-1 recognition of the secondary receptor seems to induce Tal conformational changes leading to TMP exit which is necessary for DNA release (Fig. 4). Tal mutations G218V, G226R, and W381R, which are either located in the putative Tal rotation hinge or facing it, appear to weaken this Tal rotation area, facilitating Tal rearrangements without the requirement of the glucosylated trigger. This mechanism is based on the one described for coliphage T5. Upon contact of the tail tip with T5’s receptor, the membrane protein FhuA, the Tal-like protein pb3, which obstructs the tail exit channel, opens and rotates on the tail side, thus allowing the TMP (pb2) to insert into the membrane (42, 43).
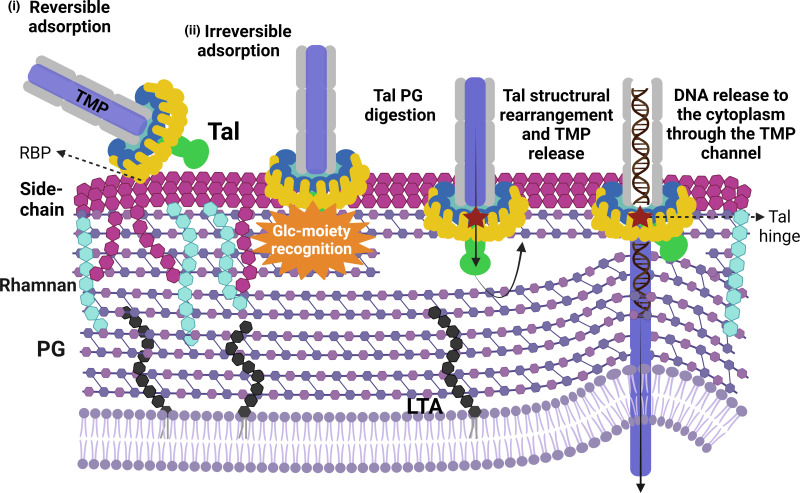
Fig 4.
Schematic proposed model of TP901-1 first stages of infection. TP901-1 baseplate is depicted: MTP (gray); TMP (blue); Dit (turquoise); BppU (dark-blue); RBPs (yellow); Tal (green). The cell wall of L. cremoris 3107 WT contains a thick peptidoglycan (PG) layer; lipoteichoic acids (LTA) anchored to cytoplasmic membrane; CWPS, consisting of the rhamnan (light-blue) and the polysaccharidic side-chain (purple) and cell wall proteins (not shown). (i) TP901-1 recognizes and reversibly adsorbs to the side-chain of the CWPS of L. cremoris 3107 via its RBPs. (ii) TP901-1 irreversibly adsorbs to a glucosylated cell envelope-associated moiety, triggering conformational changes in Tal that facilitate release of the TMP from the tail tube, the formation of a channel, and the concomitant release of the genome into the cytoplasm. These structural changes may be induced or facilitated by the impact of the Tal C-terminus on the cell wall and its enzymatic activity, which may allow the membrane to “bulge” closer to this TMP channel. Tal mutations in the putative Tal rotation hinge or facing it, appear to weaken this Tal rotation area (red star), facilitating Tal rearrangements without the requirement of the glucosylated trigger. Created with BioRender.com.
We have observed that the Tal mutations G226R and W381R have an effect on the EOPs at different temperatures (Fig. 2), although the effect of G226R is milder and contradictory depending on the host. The impact of temperature on phage infection can be attributable to different factors, i.e., the host gene expression or physical properties of the cell surface receptors (54, 55); the phage lifestyle (56) and its gene expression (57); and/or phage particle stiffness (58). Considering that our results show that the infectivity of only one of the mutants, TP901-1ermW381R, is greatly affected by temperature and shows the same tendency in the three E-derivatives, it seems that it is not a consequence of the aforementioned factors, as they may be expected to equally affect each mutant infection. Even though Tal substitutions G218V, G226R, and W381R are predicted to cause a similar structural impact, it could be that this specific mutation, W381R, leads to a temperature-sensitive phenotype. For instance, it has been described that some mutations in the tailspike-encoding gene of Salmonella phage P22 are critical for its proper folding when it is produced at restrictive temperatures. If the tail spike of such much mutants is formed at permissive temperature, they have similar thermal stability and biological activity properties to the wild type (59, 60).
The phenotypic effect of Tal mutation G603D is unclear. The spontaneous mutant harboring this single mutation formed plaques on the E-derivatives after overnight incubation (although it exhibited the lowest EOP compared with the other spontaneous mutants), whereas the recombineering mutant formed very hazy plaques after 48 hours. Given that the aa substitution is located in the Glycine-rich motif, in the junction between the second structural linker domain and the first catalytic domain, we presume that it cannot undergo proteolysis, as is the case for TP901-1ermGly>Arg (15). Therefore, the entire mutant population is expected to possess a full-length tail fiber, maintaining the cell wall-degrading domains, an adaptation that has been shown to promote TP901-1ermGly>Arg infection of stationary phase cells. The delay in the appearance of plaques is consistent with this hypothesis.
The latter mechanism for infection optimization under particular conditions was also described for the P335 phage Tuc2009 (15). The aa sequence identity of TP901-1 and Tuc2009 Tal’s is 96% (3, 15). In fact, as their baseplates are very similar, they have been classified in the same group (II) of adhesion devices stablished for P335 phages, that is based on comparative genome and morphological analyses (38). The main difference between the baseplates of TP901-1 and Tuc2009 is the presence of an additional protein termed BppA in the Tuc2009 baseplate (10, 11). Furthermore, their RBPs differ significantly, and accordingly, they target different lactococcal hosts (3, 38, 53). It would be interesting to define if, following initial attachment of Tuc2009 to its host L. cremoris UC509.9, DNA release to its host is similar to that observed for TP901-1, requiring the participation of TalTuc2009 and the presence of a glucosylated secondary receptor. Curiously, TalTuc2009 contains G218 and G226, while the residue 381 is a phenylalanine (F). This aa, together with G218 and G226, are also present in the putative Tal of an intact prophage found in the genome of L. cremoris 3107. The 3107 prophage is related to TP901-1 (93% nucleotide identity/47% coverage) (61). The putative Tal protein shares 97% and 95% aa identity with TalTP901-1 and TalTuc2009, respectively. To our knowledge, it is not known if there is any interaction between TP901-1 and this prophage.
In the case of the P335 phage LC3, aa sequence identity between the LC3 RBP C-terminal domain and that of TP901-1 is 95%, with conserved receptor-binding residues, reflecting the fact that TP901-1 and LC3 are able to infect the same lactococcal strain (3, 38). However, their DNA release mechanism differ (18) and it has been suggested that this could be a consequence of different interactions with the host through their distinct adhesion devices (19). LC3 belongs to the group III of P335 phages, which encompass a “stubby” baseplate with a short Tal, the latter consisting of only the N-terminal structural domain (3, 38). Our results, which implicate TP901-1 Tal in the DNA release mechanism and the differences between both Tals, support the latter hypothesis of two distinct DNA release mechanisms.
It is well established that modular shuffling within phage structures, as happens with TP901-1, along with the diversity of host cell wall structures involved in phage sensitivity, results in a vast repertoire of possible phage-host combinations. However, it is still remarkable to note that a single aa substitution has such a profound effect on a complex, multi-stage process as phage infection, broadening the host range or completely changing phage susceptibility. This study has opened avenues for mutational analysis of the tal gene of TP901-1, as well as different phage assays with cell wall extracts. Moreover, the noteworthy effect of temperature on some phage mutants needs further detailed investigation in order to draw firm mechanistic conclusions. Indeed, shedding light on the molecular characteristics and precise function of the Tal protein is essential to understand the TP901-1 DNA release process. This demonstrates the value of acquiring a thorough understanding of the molecular mechanisms involved in phage-host interactions, as this information is essential not only to combat infections in undesirable contexts such as food fermentations but also to develop rationale phage-based applications such as therapeutic or biocontrol agents and biosensors.
ACKNOWLEDGMENTS
This publication has emanated from research conducted with the financial support of the Science Foundation Ireland under grant number 12/RC/2273-P2. J.M. is supported under the Science Foundation Ireland Frontiers of the Future grant number 20/FFP-P/8664. This work was performed in part using HPC resources from GENCI-IDRIS (Grant 2023 AD010714075).
For the purpose of open access, we have applied a CC BY public copyright license to any author-accepted manuscript version arising from this submission.
Contributor Information
Jennifer Mahony, Email: j.mahony@ucc.ie.
Douwe van Sinderen, Email: d.vansinderen@ucc.ie.
Edward G. Dudley, The Pennsylvania State University, University Park, Pennsylvania, USA
DATA AVAILABILITY
The Illumina raw reads of TP901-1erm mutants were deposited in SRA repository under BioProject no. PRJNA1097360.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00694-24.
Fig. S1; Tables S1 and S2.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Deveau H, Labrie SJ, Chopin M-C, Moineau S. 2006. Biodiversity and classification of lactococcal phages. Appl Environ Microbiol 72:4338–4346. doi: 10.1128/AEM.02517-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Labrie SJ, Josephsen J, Neve H, Vogensen FK, Moineau S. 2008. Morphology, genome sequence, and structural proteome of type phage P335 from Lactococcus lactis. Appl Environ Microbiol 74:4636–4644. doi: 10.1128/AEM.00118-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goulet A, Mahony J, Cambillau C, van Sinderen D. 2022. Exploring structural diversity among adhesion devices encoded by lactococcal P335 phages with AlphaFold2. Microorganisms 10:2278. doi: 10.3390/microorganisms10112278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mahony J, Frantzen C, Vinogradov E, Sadovskaya I, Theodorou I, Kelleher P, Chapot-Chartier M-P, Cambillau C, Holo H, van Sinderen D. 2020. The CWPS Rubik’s cube: linking diversity of cell wall polysaccharide structures with the encoded biosynthetic machinery of selected Lactococcus lactis strains. Mol Microbiol 114:582–596. doi: 10.1111/mmi.14561 [DOI] [PubMed] [Google Scholar]
- 5. Sadovskaya I, Vinogradov E, Courtin P, Armalyte J, Meyrand M, Giaouris E, Palussière S, Furlan S, Péchoux C, Ainsworth S, Mahony J, van Sinderen D, Kulakauskas S, Guérardel Y, Chapot-Chartier M-P. 2017. Another bick in the wall: a rhamnan polysaccharide trapped inside Peptidoglycan of Lactococcus lactis. mBio 8:e01303-17. doi: 10.1128/mBio.01303-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Theodorou I, Courtin P, Palussière S, Kulakauskas S, Bidnenko E, Péchoux C, Fenaille F, Penno C, Mahony J, van Sinderen D, Chapot-Chartier M-P. 2019. A dual-chain assembly pathway generates the high structural diversity of cell-wall polysaccharides in Lactococcus lactis. J Biol Chem 294:17612–17625. doi: 10.1074/jbc.RA119.009957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Theodorou I, Courtin P, Sadovskaya I, Palussière S, Fenaille F, Mahony J, Chapot-Chartier M-P, van Sinderen D. 2020. Three distinct glycosylation pathways are involved in the decoration of Lactococcus lactis cell wall glycopolymers. J Biol Chem 295:5519–5532. doi: 10.1074/jbc.RA119.010844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mann E, Whitfield C. 2016. A widespread three-component mechanism for the periplasmic modification of bacterial glycoconjugates. Can. J. Chem 94:883–893. doi: 10.1139/cjc-2015-0594 [DOI] [Google Scholar]
- 9. Bebeacua C, Lai L, Vegge CS, Brøndsted L, van Heel M, Veesler D, Cambillau C. 2013. Visualizing a complete Siphoviridae member by single-particle electron microscopy: the structure of lactococcal phage TP901-1. J Virol 87:1061–1068. doi: 10.1128/JVI.02836-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Legrand P, Collins B, Blangy S, Murphy J, Spinelli S, Gutierrez C, Richet N, Kellenberger C, Desmyter A, Mahony J, van Sinderen D, Cambillau C. 2016. The atomic structure of the phage Tuc2009 baseplate tripod suggests that host recognition involves two different carbohydrate binding modules. mBio 7:e01781-15. doi: 10.1128/mBio.01781-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Veesler D, Spinelli S, Mahony J, Lichière J, Blangy S, Bricogne G, Legrand P, Ortiz-Lombardia M, Campanacci V, van Sinderen D, Cambillau C. 2012. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci USA 109:8954–8958. doi: 10.1073/pnas.1200966109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bebeacua C, Bron P, Lai L, Vegge CS, Brøndsted L, Spinelli S, Campanacci V, Veesler D, van Heel M, Cambillau C. 2010. Structure and molecular assignment of lactococcal phage TP901-1 baseplate. J Biol Chem 285:39079–39086. doi: 10.1074/jbc.M110.175646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahony J, Goulet A, van Sinderen D, Cambillau C. 2023. Partial atomic model of the tailed lactococcal phage TP901-1 as predicted by AlphaFold2: revelations and limitations. Viruses 15:2440. doi: 10.3390/v15122440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stockdale SR, Mahony J, Courtin P, Chapot-Chartier M-P, van Pijkeren J-P, Britton RA, Neve H, Heller KJ, Aideh B, Vogensen FK, van Sinderen D. 2013. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J Biol Chem 288:5581–5590. doi: 10.1074/jbc.M112.444901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mahony J, Alqarni M, Stockdale S, Spinelli S, Feyereisen M, Cambillau C, Sinderen D van. 2016. Functional and structural dissection of the tape measure protein of lactococcal phage TP901-1. Sci Rep 6:36667. doi: 10.1038/srep36667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ainsworth S, Sadovskaya I, Vinogradov E, Courtin P, Guerardel Y, Mahony J, Grard T, Cambillau C, Chapot-Chartier M-P, van Sinderen D. 2014. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio 5:e00880-14. doi: 10.1128/mBio.00880-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ostergaard Breum S, Neve H, Heller KJ, Vogensen FK. 2007. Temperate phages TP901-1 and ΦLC3, belonging to the P335 species, apparently use different pathways for DNA injection in Lactococcus lactis subsp. cremoris 3107. FEMS Microbiol Lett 276:156–164. doi: 10.1111/j.1574-6968.2007.00928.x [DOI] [PubMed] [Google Scholar]
- 19. Ruiz-Cruz S, Erazo Garzon A, Kelleher P, Bottacini F, Breum SØ, Neve H, Heller KJ, Vogensen FK, Palussière S, Courtin P, Chapot-Chartier M-P, Vinogradov E, Sadovskaya I, Mahony J, van Sinderen D. 2022. Host genetic requirements for DNA release of lactococcal phage TP901-1. Microb Biotechnol 15:2875–2889. doi: 10.1111/1751-7915.14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Braun V, Hertwig S, Neve H, Geis A, Teuber M. 1989. Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. Microbiology 135:2551–2560. doi: 10.1099/00221287-135-9-2551 [DOI] [Google Scholar]
- 21. van Pijkeren J-P, Neoh KM, Sirias D, Findley AS, Britton RA. 2012. Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered 3:209–217. doi: 10.4161/bioe.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Pijkeren J-P, Britton RA. 2012. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res 40:e76–e76. doi: 10.1093/nar/gks147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koch B, Christiansen B, Evison T, Vogensen FK, Hammer K. 1997. Construction of specific erythromycin resistance mutations in the temperate lactococcal bacteriophage TP901-1 and their use in studies of phage biology. Appl Environ Microbiol 63:2439–2441. doi: 10.1128/aem.63.6.2439-2441.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lillehaug D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol 83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x [DOI] [PubMed] [Google Scholar]
- 25. Lugli GA, Fontana F, Tarracchini C, Milani C, Mancabelli L, Turroni F, Ventura M. 2023. MEGAnnotator2: a pipeline for the assembly and annotation of microbial genomes. Microbiome Res Rep 2:15. doi: 10.20517/mrr.2022.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sonnhammer ELL, Eddy SR, Durbin R. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405–420. doi: [DOI] [PubMed] [Google Scholar]
- 28. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. JASP Team . 2024. JASP (Version 0.18.3)[Computer software]. https://jasp-stats.org/faq/how-do-i-cite-jasp/.
- 31. Holm L, Kääriäinen S, Rosenström P, Schenkel A. 2008. Searching protein structure databases with DaliLite v.3. Bioinformatics 24:2780–2781. doi: 10.1093/bioinformatics/btn507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruiz-Cruz S, Sadovskaya I, Mahony J, Grard T, Chapot-Chartier M-P, van Sinderen D, Vinogradov E. 2023. Structural studies of the deacylated glycolipids and lipoteichoic acid of Lactococcus cremoris 3107. Carbohydr Res 531:108898. doi: 10.1016/j.carres.2023.108898 [DOI] [PubMed] [Google Scholar]
- 35. Brøndsted L, Ostergaard S, Pedersen M, Hammer K, Vogensen FK. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93–109. doi: 10.1006/viro.2001.0871 [DOI] [PubMed] [Google Scholar]
- 36. Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415:553–557. doi: 10.1038/415553a [DOI] [PubMed] [Google Scholar]
- 37. Xiang Y, Morais MC, Cohen DN, Bowman VD, Anderson DL, Rossmann MG. 2008. Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage Φ29 tail. Proc Natl Acad Sci USA 105:9552–9557. doi: 10.1073/pnas.0803787105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mahony J, Oliveira J, Collins B, Hanemaaijer L, Lugli GA, Neve H, Ventura M, Kouwen TR, Cambillau C, van Sinderen D. 2017. Genetic and functional characterisation of the lactococcal p335 phage-host interactions. BMC Genomics 18:146. doi: 10.1186/s12864-017-3537-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abedon ST, Yin J. 2009. Bacteriophage plaques: theory and analysis. Methods Mol Biol 501:161–174. doi: 10.1007/978-1-60327-164-6_17 [DOI] [PubMed] [Google Scholar]
- 40. Yang Z, Zeng X, Zhao Y, Chen R. 2023. AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct Target Ther 8:115. doi: 10.1038/s41392-023-01381-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goulet A, Cambillau C. 2022. Present Impact of AlphaFold2 revolution on structural biology, and an illustration with the structure prediction of the bacteriophage J-1 Host adhesion device. Front Mol Biosci 9:907452. doi: 10.3389/fmolb.2022.907452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Degroux S, Effantin G, Linares R, Schoehn G, Breyton C. 2023. Deciphering bacteriophage T5 host recognition mechanism and infection trigger. J Virol 97:e0158422. doi: 10.1128/jvi.01584-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Linares R, Arnaud C-A, Effantin G, Darnault C, Epalle NH, Boeri Erba E, Schoehn G, Breyton C. 2023. Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation. Sci Adv 9:eade9674. doi: 10.1126/sciadv.ade9674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Kempen M, Kim SS, Tumescheit C, Mirdita M, Lee J, Gilchrist CLM, Söding J, Steinegger M. 2024. Fast and accurate protein structure search with Foldseek. Nat Biotechnol 42:243–246. doi: 10.1038/s41587-023-01773-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Millen AM, Romero DA. 2016. Genetic determinants of lactococcal C2viruses for host infection and their role in phage evolution. J Gen Virol 97:1998–2007. doi: 10.1099/jgv.0.000499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stuer-Lauridsen B, Janzen T, Schnabl J, Johansen E. 2003. Identification of the host determinant of two prolate-headed phages infecting Lactococcus lactis. Virology 309:10–17. doi: 10.1016/s0042-6822(03)00012-6 [DOI] [PubMed] [Google Scholar]
- 47. Monteville MR, Ardestani B, Geller BL. 1994. Lactococcal bacteriophages require a host cell wall carbohydrate and a plasma membrane protein for adsorption and ejection of DNA. Appl Environ Microbiol 60:3204–3211. doi: 10.1128/aem.60.9.3204-3211.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Valyasevi R, Sandine WE, Geller BL. 1991. A membrane protein is required for bacteriophage c2 infection of Lactococcus lactis subsp. lactis C2. J Bacteriol 173:6095–6100. doi: 10.1128/jb.173.19.6095-6100.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Geller BL, Ngo HT, Mooney DT, Su P, Dunn N. 2005. Lactococcal 936-species phage attachment to surface of Lactococcus lactis. J Dairy Sci 88:900–907. doi: 10.3168/jds.S0022-0302(05)72756-9 [DOI] [PubMed] [Google Scholar]
- 50. São-José C, Baptista C, Santos MA. 2004. Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J Bacteriol 186:8337–8346. doi: 10.1128/JB.186.24.8337-8346.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vinga I, Baptista C, Auzat I, Petipas I, Lurz R, Tavares P, Santos MA, São-José C. 2012. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol Microbiol 83:289–303. doi: 10.1111/j.1365-2958.2011.07931.x [DOI] [PubMed] [Google Scholar]
- 52. Bielmann R, Habann M, Eugster MR, Lurz R, Calendar R, Klumpp J, Loessner MJ. 2015. Receptor binding proteins of Listeria monocytogenes bacteriophages A118 and P35 recognize serovar-specific teichoic acids. Virology 477:110–118. doi: 10.1016/j.virol.2014.12.035 [DOI] [PubMed] [Google Scholar]
- 53. Goulet A, Spinelli S, Mahony J, Cambillau C. 2020. Conserved and diverse traits of adhesion devices from Siphoviridae recognizing proteinaceous or saccharidic receptors. Viruses 12:512. doi: 10.3390/v12050512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niu C, Shang N, Liao X, Feng E, Liu X, Wang D, Wang J, Huang P, Hua Y, Zhu L, Wang H. 2013. Analysis of soluble protein complexes in Shigella flexneri reveals the influence of temperature on the amount of lipopolysaccharide. Mol Cell Proteomics 12:1250–1258. doi: 10.1074/mcp.M112.025270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tokman JI, Kent DJ, Wiedmann M, Denes T. 2016. Temperature significantly affects the plaquing and adsorption efficiencies of Listeria phages. Front Microbiol 7:631. doi: 10.3389/fmicb.2016.00631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meng B, Qi Z, Li X, Peng H, Bi S, Wei X, Li Y, Zhang Q, Xu X, Zhao H, Yang X, Wang C, Zhao X. 2023. Characterization of Mu-Like Yersinia phages exhibiting temperature dependent infection. Microbiol Spectr 11:e0020323. doi: 10.1128/spectrum.00203-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ghosh S, Shaw R, Sarkar A, Gupta SKD. 2020. Evidence of positive regulation of mycobacteriophage D29 early gene expression obtained from an investigation using a temperature-sensitive mutant of the phage. FEMS Microbiol Lett 367:fnaa176. doi: 10.1093/femsle/fnaa176 [DOI] [PubMed] [Google Scholar]
- 58. Sae-Ueng U, Bhunchoth A, Phironrit N, Treetong A, Sapcharoenkun C, Chatchawankanphanich O, Leartsakulpanich U, Chitnumsub P. 2022. Thermoresponsive C22 phage stiffness modulates the phage infectivity. Sci Rep 12:13001. doi: 10.1038/s41598-022-16795-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mitraki A, King J. 1992. Amino acid substitutions influencing intracellular protein folding pathways. FEBS Lett 307:20–25. doi: 10.1016/0014-5793(92)80894-m [DOI] [PubMed] [Google Scholar]
- 60. Yu MH, King J. 1984. Single amino acid substitutions influencing the folding pathway of the phage P22 tail spike endorhamnosidase. Proc Natl Acad Sci USA 81:6584–6588. doi: 10.1073/pnas.81.21.6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ruiz-Cruz S, Parlindungan E, Erazo Garzon A, Alqarni M, Lugli GA, Ventura M, van Sinderen D, Mahony J. 2020. Lysogenization of a Lactococcal host with three distinct temperate phages provides homologous and heterologous phage resistance. Microorganisms 8:1685. doi: 10.3390/microorganisms8111685 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1; Tables S1 and S2.
Data Availability Statement
The Illumina raw reads of TP901-1erm mutants were deposited in SRA repository under BioProject no. PRJNA1097360.