Abstract
Replication of Alfalfa mosaic virus (AMV) RNAs depends on the virus-encoded proteins P1 and P2. P1 contains methyltransferase- and helicase-like domains, and P2 contains a polymerase-like domain. Coimmunoprecipitation experiments revealed an interaction between in vitro translated-P1 and P2 and showed that these proteins are present together in fractions with RNA-dependent RNA polymerase activity. A deletion analysis in the yeast two-hybrid system showed that in P1 the C-terminal sequence of 509 amino acids with the helicase domain was necessary for the interaction. In P2, the sequence of the N-terminal 241 aa was required for the interaction. In infected protoplasts, P1 and P2 colocalized at a membrane structure that was identified as the tonoplast (i.e., the membrane that surrounds the vacuoles) by using a tonoplast intrinsic protein as a marker in immunofluorescence studies. While P1 was exclusively localized on the tonoplast, P2 was found both at the tonoplast and at other locations in the cell. As Brome mosaic virus replication complexes have been found to be associated with the endoplasmic reticulum (M. A. Restrepo-Hartwig and P. Ahlquist, J. Virol. 70:8908–8916, 1996), viruses in the family Bromoviridae apparently select different cellular membranes for the assembly of their replication complexes.
The alphavirus-like superfamily comprises a large number of animal and plant viruses sharing the properties of a capped, positive-stranded RNA genome and homologies in their RNA replication proteins. Despite homologies between the guanylytransferase/methyltransferase-like (MT), helicase-like (HEL), and polymerase-like (POL) domains, they may be expressed as parts of a single protein or as separate entities distributed over two or three proteins. Little is known about the nature of the interactions between these polypeptides and the ratio in which they are present in the viral replication complex, although purified RNA-dependent RNA polymerases (RdRp) have been used extensively to study viral replication in vitro. We have studied the interactions between the replicase proteins of Alfalfa mosaic virus (AMV) and their in situ localization to gain insight in the assembly of the replication complex.
AMV is the type species of the genus Alfamovirus. It belongs to the Bromoviridae family of plant viruses, all having a tripartite RNA genome of messenger-sense polarity. RNA 3 encodes the movement protein (MP) and, via a subgenomic mRNA 4, the coat protein (CP). RNAs 1 and 2 encode the P1 and P2 replicase proteins, respectively. The N terminus of P1 contains a domain with sequence homology to the domains involved in RNA capping of the alphavirus Semliki Forest virus (SFV) nsP1 protein and the more closely related Brome mosaic virus (BMV) 1a protein (1, 2, 41). The C-terminal end of P1 has homology to the alphavirus-like supergroup of HEL domains (17, 22), including the SFV nsP2 protein, for which helicase activity was recently shown in vitro (16). The P2 protein contains a central domain with sequence motifs conserved among many polymerases, including the presence of the Mg2+-binding GDD motif (3, 26). The N terminus of P2 does not contain conserved sequence elements. The corresponding sequence of the BMV 2a polymerase protein has been shown to interact with the HEL domain of the BMV 1a protein (23).
In addition to P1 and P2, CP is involved in AMV replication (reviewed in reference 4). CP in the inoculum is required in a step prior to viral minus-strand RNA synthesis, possibly translation of the inoculum RNAs (31, 32). Subsequently, CP expressed from RNA 3 is required for viral plus-strand RNA accumulation in vivo (31, 48) and in vitro (11).
The replication complexes of all eukaryotic positive-stranded RNA viruses studied so far are associated with intracellular membranes. Alphavirus RNA replicase proteins are localized on the cytoplasmic surface of endosomes and lysosomes, probably connected with the 50-nm membrane-associated vesicles that occupy these organelles (14). Endoplasmic reticulum (ER)-derived 50- to 100-nm cytoplasmic vesicles detected in bromovirus-infected plants localize with the site of replication of BMV (39). Replication complexes of other plant viruses are localized in vesicular structures derived from chloroplast membranes or believed to be associated with vesicles on the tonoplast, i.e., the membrane surrounding the vacuoles (19, 29).
In this paper we provide in vitro and in vivo evidence for the interaction between the AMV P1 and P2 replication proteins and show that the HEL domain of P1 and the nonconserved N terminus of P2 are involved in this interaction. In addition, we found a colocalization of P1 and P2 at the tonoplast of infected cowpea protoplasts.
MATERIALS AND METHODS
Construction of two-hybrid plasmids.
AMV P1 and P2 genes were fused to the binding and activation domains of GAL4 encoded by the yeast expression vectors pGBT9, pGAD424, pAS2-1, and pACT2 (Clontech) as follows. The P1 coding sequence plus 3′ untranslated region (3′-UTR) and nos-terminator were excised from pCa17T-Nco (52) with NcoI and PvuII. This sequence was ligated into a NcoI-SmaI digested pUC21 plasmid to give pUC-P1. The P1 sequence was removed using XhoI and PstI and ligated into SalI-PstI digested pGBT9, giving clone pBP1 (1–1126). Similarly, a XhoI-BamHI fragment was cloned in pGAD424 digested with SalI and BglII, to yield plasmid pAP1(1–1126). The resulting fusion proteins contain a 14-amino-acid (aa) linker peptide at the junction with P1.
N- and C-terminal deletions of P1 were made by digesting pBP1(1–1126) or pAP1(1–1126) with the appropriate endonucleases. T4 DNA polymerase was used to generate blunt ends. The following enzymes were used (P1 amino acids encoded by the restriction fragments are given in parentheses): SmaI-NdeI (135 to 1126), BamHI-HpaI (340 to 1126), BamHI-PinAI (509 to 1126), BamHI-EcoRV (618 to 1126), BamHI-BspEI (654 to 1126), EcoRI (684 to 1126), and PinAI-PstI (1 to 509). Additional C-terminal deletions were obtained by BglII-PstI digestion (509 to 997 and 618 to 997). Ligation of an NcoI-EcoRI P1-encoding fragment from pCa17T-Nco in pAS2-1 and pACT2 resulted in clones pBP1(1–686) and pAP1(1–686).
Part of P2 was amplified by PCR from pUT27A (infectious clone of RNA 2) (31) using a primer that introduces an NdeI restriction site around the start codon and a primer complementary to nucleotides 789 to 806 of RNA 2. The PCR product was digested with NdeI and SspI (position 612) and ligated in NdeI-SmaI-digested pUC21. An XhoI (position 262)-StuI fragment of the resulting clone was replaced by an XhoI-SmaI sequence excised from pUT27A, yielding pUC-P2. This plasmid was digested by NdeI, treated with T4 DNA polymerase, and digested with PstI. The fragment containing the P2 open reading frame and RNA 2 3′-UTR was ligated in pGAD424 and pGBT9, each digested with SmaI and PstI, yielding AP2 and BP2, respectively. This resulted in the addition of a 4-aa linker peptide at the junction of the open reading frames.
N- and C-terminal deletions of P2 were made by digesting pAP2(1–790) or pBP2(1–790) with the appropriate endonucleases, and T4 DNA polymerase was used to generate blunt ends. The following enzyme combinations were used (P2 amino acids encoded by the restriction fragments are given in parentheses): PmlI-PstI (1 to 608), EcoRV-PstI (1 to 478), PinAI-PstI (1 to 352), XmnI-PstI (1 to 241), Eco47III-PstI (1 to 156), BamHI-PinAI (1 to 44 and 352 to 790), and EcoRI-ClaI (516 to 790). EcoRI (76 to 478, 76–352, and 76–241) or BamHI-EcoNI (118 to 478) was used to make additional N-terminal deletions. pBP2(352–790) was constructed by ligation of a P2 fragment, isolated from pUC-P2 by digestion with PinAI and PstI, to pAS2-1 previously cut with NdeI and PstI.
All enzymes used were from GIBCO/BRL, and all enzymatic incubations were performed under conditions recommended by the supplier. All recombinant DNAs were checked by restriction mapping and nucleotide sequence analysis.
Construction of other plasmids.
pCaHA17T gives Cauliflower mosaic virus 35S promoter-driven expression of the P1 protein with an N-terminal fusion to a hemagglutinin (HA) tag. Plasmid pCa17T-Nco (52) was digested with NcoI. The HA sequence was inserted by ligation of a synthetic DNA fragment, obtained by hybridization of two oligonucleotides, CATGTACCCATACGATGTTCCAGATTACGC and CATGGCGTAATCTGGAACATCGTATGGGTA.
For the construction of pMON-HA-SITIP, pPE1000 (18) was first modified to allow in-frame fusion of the HA epitope-coding sequence with the cDNA of a tonoplast intrinsic protein (γ-TIP) (20). The QuickChange site-directed mutagenesis method (Stratagene) was used to create an XbaI site upstream and to remove four bases downstream of the HA epitope, using suitable primers. From the resulting plasmid, an XbaI-EcoRI fragment was removed and ligated in the multiple cloning site of pMON999 (46), giving pMON-HA. The complete coding sequence of γ-TIP was released as an EcoRI fragment from pGAD10-TIP, a clone isolated from an Arabidopsis cDNA library (J. E. Carette, unpublished results). This fragment was ligated in pMON-HA digested with the same enzyme.
Plasmids pT7L4P1 and pT7L4P2 encode RNAs 1 and 2, respectively, with the 5′-UTRs replaced by the 5′-UTR of RNA 4. They were constructed by replacing the CP gene and 3′-UTR in the cDNA 4 clone pT72-42NcoCP (49) (NcoI-EcoRI fragment) by a cDNA 1 sequence with the P1 gene and 3′-UTR (to yield pT7L4P1) or a cDNA 2 sequence with the P2 gene and 3′-UTR (to yield pT7L4P2). The cDNA 1 sequence was derived from plasmids pUT17A (31) (NcoI-EcoRI fragment) and pCa17T-Nco (52) (NcoI-NcoI fragment). The cDNA 2 fragment was derived from plasmid pCa27T-Nco (52) (NcoI-SphI fragment). The sequence downstream of the initiation codon in pT7L4P2 was restored to the wild-type P2 sequence using the Promega Altered Sites in vitro mutagenesis system with primer AATACTTCCATCATGTTCACTCTTTTG.
pGEX-NP2 encodes a protein with aa 43 to 349 of P2 fused to the C terminus of glutathione S-transferase (GST). Vector pGEX-2T (Amersham Pharmacia Biotech) was digested with EcoRI. A cDNA 2 fragment was cut from pCa27T using BamHI and DraI. Vector and insert were treated with T4 DNA polymerase and ligated together.
Antibodies.
The P1-specific rabbit polyclonal antibody was directed against the C-terminal aa 1100 to 1120 of P1 (51). For detection of CP, we used a rabbit polyclonal CP-specific antibody. An N-terminal fragment of P2 was overexpressed as a GST fusion protein from pGEX-NP2 in Escherichia coli strain BL21, cleaved with thrombin to remove the GST sequence, and used to immunize rabbits for the production of anti-P2 polyclonal antiserum (Eurogentec). The antibody against HA was a monoclonal mouse antibody (12CA5; Roche). The fluorescein isothiocyanate (FITC)-labeled secondary antibodies were from Jackson Laboratories; the Alexa-568- labeled secondary antibodies were from Molecular Probes.
In vitro RNA transcription and translation.
AMV P1, P2, MP, and CP were translated from RNA transcripts of plasmids pT7L4P1, pT7L4P2, pAL3, and pT72-42, respectively. Transcription of linearized plasmids with T7 RNA polymerase was done as described elsewhere (31, 47). In vitro translation was done in a rabbit reticulocyte lysate according to the protocol provided by the manufacturer (Promega), with minor modifications.
Immunoprecipitation of in vitro translated proteins.
Precipitation was performed with 10 μl of in vitro-translated protein, in the presence of 400 μl of NET-gel buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.2% Igepal CA630, 1 mM EDTA, 0.25% teleostean gelatin) and 2.5 μl of antibody. The sample was incubated at 4°C under continuous rotation. After 60 min, 50 μl of a 50% protein A–Sepharose CL-4B (Amersham Pharmacia Biotech) solution in NET-gel was added, and incubation was continued for another 60 min. The beads were washed three times with 1-ml portions of NET-gel buffer and washed once with NET buffer (which contains no gelatin). The beads were resuspended in 30 μl of protein loading buffer (27), and the samples were boiled for 3 min and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the gel was fixed, dried, and exposed to X-ray film.
Immunoprecipitation of RdRp complexes.
Proteins were precipitated from 50 μl of RdRp glycerol fractions by 5 μl of antibody in the presence of 800 μl of NET-gel buffer. The samples were incubated and washed as described above. After electrophoresis, the gel was analyzed by the Western blot technique (45).
Protoplast infection and immunofluorescent labeling.
Cowpea (Vigna unguiculata cv. Blackeye) mesophyl protoplasts were isolated and transfected with plasmid DNA as described elsewhere (46). The same protocol was used for infection of 5 × 105 protoplasts with 10 μg of phenol-extracted AMV RNA. Protoplasts were incubated for 42 h at 25°C under continuous illumination. Fixation and antibody incubations were performed as described previously (50). Primary antibodies were diluted 1:100 (HA and P2 antibodies) or 1:1,000 (CP antibody) in blocking buffer, which is phosphate-buffered saline containing 0.1% acetylated bovine serum albumin (Aurion) and 0.02% teleostean gelatin (Sigma). Goat anti-mouse or goat anti-rabbit antibodies conjugated to FITC or Alexa-568 were diluted 1:40 in blocking buffer. For nuclear staining, cells were incubated for 15 min with a 5-ng/ml solution of propidium iodide in phosphate-buffered saline-before the last washing step.
Confocal laser scanning microscopy (CLSM).
Immunofluorescence images were obtained with a Bio-Rad MRC-1024 ES confocal microscope. Images with FITC-labeled antibodies were acquired with an excitation wavelength of 488 nm and a 522 DF 32-nm band-pass filter. The Alexa-568 antibody and propidium iodide nuclear stain were detected with a 568-nm laserline and 585-nm long-pass filter. Images were acquired using one laserline at a time to minimize background and merged later using the Confocal Assistant 4.02 software. Each experiment was performed at least three times to ensure the reproducibility of the results.
Yeast two-hybrid system.
All yeast experiments were performed with Saccharomyces cerevisiae strain PJ69-4A (21), which carries the adenine (A) and histidine (H) reporter genes. Yeast cells were grown on minimal synthetic medium plates containing methionine and uracil (MU plates) plus other selective components if required. Binding domain (pB) and activation domain (pA) plasmids carry tryptophan (T) and leucine (L) auxotrophic markers, respectively. Yeast transformations were performed as described before (15). All pB and pA constructs were separately transformed to yeast and plated on MUAHL or MUAHT medium. Single transformants were maintained on these media, and colonies were replated to medium lacking A and H to test for activation of the reporter genes. Combinations of pB and pA constructs were cotransformed and plated on MUAH medium. After 2 to 3 days on MUAH plates, single colonies were transferred to both MUAH and MU plates to test for interaction. Growth on selective medium was monitored over 6 days. Double transformations were performed at least three times.
RdRp isolation and activity assay.
The RdRp was purified from Nicotiana benthamiana plants infected with AMV strain 425 as described previously (37), omitting the DEAE ion-exchange chromatography step. The glycerol gradient was subdivided into 16 fractions that were assayed for in vitro RdRp activity (37).
RESULTS
Interaction of P1 and P2 in the yeast two-hybrid system.
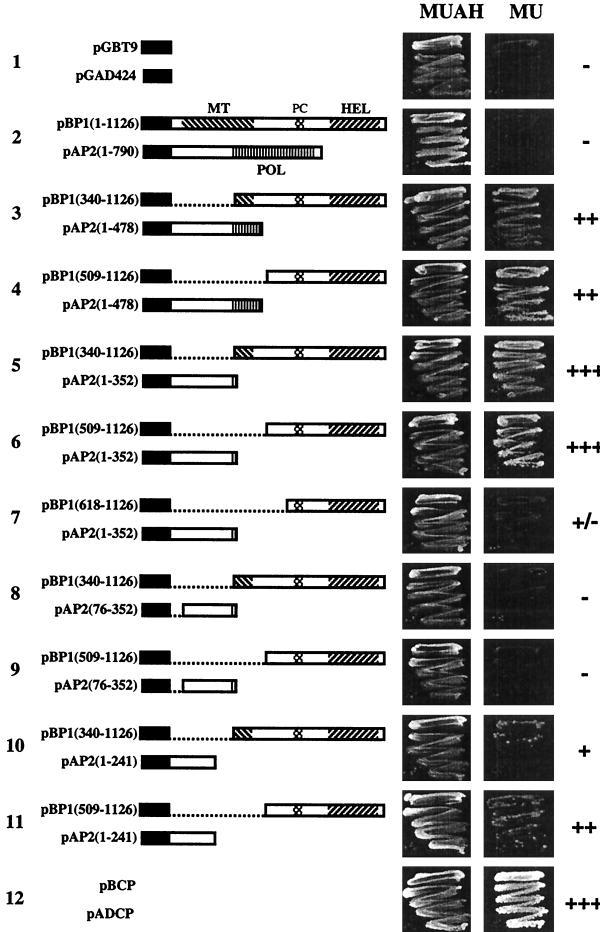
Viral sequences were fused to the C terminus of the activation domain or DNA binding domain of GAL4. When the N-terminal 352 aa of P2 were fused to the binding domain, the construct showed activation of the ADE and HIS3 genes in the absence of a P1 construct. Therefore, most of the P1 derivatives were fused to the binding domain (pBP1 constructs), whereas the P2 derivatives were mainly fused to the activation domain (pAP2 constructs). In addition, we tested a few constructs with P1 or P2 sequences fused to the activation or DNA binding domain, respectively (pAP1 and pBP2 constructs). Figure 1 shows schematic representations of the full-length P1 and P2 proteins and a selection of pBP1 and pAP2 constructs that were used in two-hybrid assays. Table 1 summarizes all P1-P2 interactions tested. None of the constructs shown in Fig. 1 or Table 1 was able to self-activate the ADE and HIS3 selectable marker genes. Western blot analysis revealed stable expression in yeast of all fusion proteins shown in Fig. 1 (data not shown). Growth of yeast on nonselective (MUAH) or selective (MU) medium is shown in Fig. 1. No growth was observed using full-length P1 and P2 (Fig. 1, row 2). However, cotransformation with several constructs encoding C-terminal P1 and N-terminal P2 sequences resulted in growth of the yeast cells. Growth of colonies within 1 to 2 days was found with the C-terminal 618 aa of P1 [pBP1 (509–1126)] and the N-terminal 352 aa of P2 [pAP2 (1–352)] (Fig. 1, row 6; Table 1). N-terminal extension of this P1 sequence in pBP1(340–1126) did not affect this growth rate (Fig. 1, row 5), but truncation to a sequence corresponding to the C-terminal 509 aa of P1 [pBP1(618–1126)] strongly reduced the growth rate (Fig. 1, row 7). When the C terminus of the P1 sequence in pBP1(509–1126) was truncated by deleting 129 aa [pBP1(509–997)], the growth of yeast colonies was abolished (Table 1). When the P2 sequence in pAP2(1–352) was reduced to a sequence corresponding to the N-terminal 241 aa of P2 [pAP2(1–241)], growth was reduced (Fig. 1, compare rows 6 and 11). Further deletions at the N terminus [pAP2(75–242) and pAP2(75–351)] or C terminus [pAP2(1–157)] of this P2 sequence abolished growth (Table 1). With the N-terminal sequence of P1 (aa 1 to 509 or 1 to 686), no interaction with any P2 sequence was detectable (Table 1). Similarly, no interaction was found between C-terminal domains of P2 and any P1 sequence tested (Table 1).
FIG. 1.
Examples of interactions between AMV proteins in the yeast two-hybrid system, 4 days after plating. Yeast cells were grown on medium that does not select for interaction (MUAH) and medium that selects for adenine and histidine auxotrophy (MU). In the schematic representations of P1 and P2 segments fused to the GAL4 binding domain (B) or activation domain (A) that were used to transform yeast cells, GAL4 sequences are represented by black bars, MT, HEL, and POL, domains and the proline cluster (PC) are hatched, and deletions are shown as dotted lines. Typical interactions are shown along with positive and negative controls (rows 12 and 1, respectively). Growth rates of yeast cells are indicated by plus and minus signs as explained in the footnote to Table 1.
TABLE 1.
Summary of two-hybrid interactions tested for selective growth
| Binding domain fusion | Interaction with indicated activation domain fusiona
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pAP2
|
pAP1
|
Other
|
||||||||||||||
| 1–790 | 1–608 | 1–478 | 76–478 | 1–352 | 76–352 | 1–241 | 76–241 | 1–156 | 118–478 | 1–44/352–790 | 1–1126 | 1–686 | 509–1126 | pAD CP | pGAD 424 | |
| pBP1 | ||||||||||||||||
| 1–126 | − | − | − | − | − | |||||||||||
| 135–1126 | − | − | − | − | − | |||||||||||
| 340–1126 | − | − | ++ | − | +++ | − | + | − | − | − | ||||||
| 509–1126 | − | − | ++ | − | +++ | − | ++ | − | − | − | − | − | − | − | − | − |
| 509–997 | − | − | − | − | − | |||||||||||
| 618–1126 | − | − | − | − | +/− | − | − | − | − | − | ||||||
| 618–997 | − | − | − | − | − | |||||||||||
| 654–1126 | − | − | ||||||||||||||
| 684–1126 | − | − | ||||||||||||||
| 1–509 | − | − | − | − | − | − | − | |||||||||
| 1–686 | − | − | − | − | − | − | ||||||||||
| pBP2 | ||||||||||||||||
| 1–790 | − | − | − | − | ||||||||||||
| 1–44/352–790 | − | − | − | − | − | |||||||||||
| 352–790 | − | − | − | − | − | |||||||||||
| 516–790 | − | − | − | − | − | |||||||||||
| Other | ||||||||||||||||
| pBCP | − | − | − | − | − | +++ | ||||||||||
| pGBT9 | − | − | ||||||||||||||
Blank, interaction not tested; −, no growth on selective medium within 6 days; +/−, growth within 5 to 6 days; +, growth within 4 days; ++, growth within 3 days; +++, growth within 1 to 2 days.
CP is known to be present in purified AMV RdRp preparations (37). In the two-hybrid system, a strong CP-CP interaction is detectable (44), resulting in fast growth on selective medium (Fig. 1, row 12; constructs pADCP and pBCP in Table 1). However, no interaction of CP could be detected with any of the P1 or P2 domains tested in the two-hybrid system (Table 1). In addition, in yeast transformed with several combinations of two P1 constructs or two P2 constructs, no evidence for a P1-P1 or P2-P2 interaction was obtained (Table 1).
Coimmunoprecipitation of in vitro-translated P1 and P2.
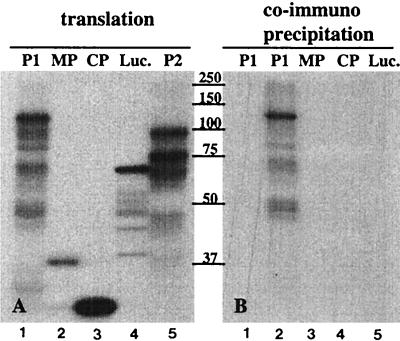
To confirm the interaction observed in the two-hybrid system, P1 and P2 were translated in a cell-free system and interaction between the translation products was analyzed by coimmunoprecipitation. This assay was also used to analyze a possible interaction of P1 or P2 with other AMV proteins. Figure 2A shows the products obtained by translation of the AMV proteins P1, P2, MP, and CP, along with luciferase as a control: in Fig. 2B, the radiolabeled translation products were mixed with unlabeled P2 and subjected to precipitation with a P2 antiserum. Labeled P1 coprecipitated with P2 (Fig. 2B, lane 2), but MP, CP, and luciferase did not. The P2 antiserum did not precipitate P1 in the absence of P2 (Fig. 2B, lane 1). Addition of RNase A (0.1 μg ml−1) to the translated proteins before binding had no affect on precipitation (data not shown). These results corroborate the interactions observed in the two-hybrid system between P1 and P2 and the finding that an interaction between replicase proteins and CP was not detectable. The fact that interaction between full-length P1 and P2 was detectable by coimmunoprecipitation but not in the two-hybrid system may be due to steric hindrance caused by the activation or DNA binding domains in the yeast system or impaired nuclear import of these large fusion proteins.
FIG. 2.
Immunoprecipitation of in vitro-translated proteins. AMV P1, P2, MP, and CP and luciferase (Luc.) were translated in a rabbit reticulocyte lysate in the presence of [35S]methionine, and samples of the translation mixtures were analyzed by SDS-PAGE (A). The remaining parts of the translation mixtures were incubated with nonlabeled P2 (except the lane 1, to which no P2 was added) (B) and subjected to immunoprecipitation with a polyclonal anti-P2 serum followed by SDS-PAGE. The labeled proteins in the translation mixtures are indicated above the lanes. Positions of molecular size markers (in kilodaltons) are indicated between the panels.
Coimmunoprecipitation of P1 and P2 from purified RdRp.
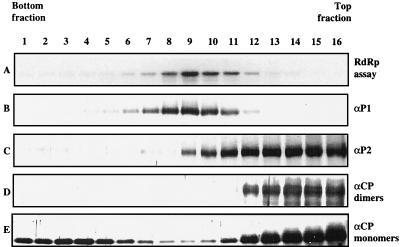
AMV RdRp was solubilized from membrane structures sedimenting at 30,000 × g and centrifuged in a glycerol gradient (37). The gradient was subdivided into 16 fractions, each of which was analyzed for polymerase activity by an in vitro RdRp assay and for the presence of P1, P2, and CP by Western blotting. Over 80% of the total RdRp activity was present in fractions 8 to 11, with a peak of 37% in fraction 9 (Fig. 3A). The sedimentation pattern of P1 (Fig. 3B) closely corresponded to the distribution of the RdRp activity. P2 was present in all fractions that showed RdRp activity, but the majority of P2 was found in the top fractions of the gradient that showed little or no RdRp activity (Fig. 3C). The antiserum against CP detected CP monomers as well as CP dimers. CP monomers were found in all fractions of the gradient, particularly in the top and botton fractions, with relatively little CP in the fractions that contained RdRp activity (Fig. 3E). CP dimers were found exclusively in the top fractions and did not cosediment with RdRp activity. We do not know why these dimers do not dissociate in the presence of SDS and reducing agents in the gel loading buffer.
FIG. 3.
Analysis of AMV RdRp by glycerol gradient centrifugation. Samples from each gradient fraction were assayed for RdRp activity (A) and analyzed by Western blotting (B to E). RdRp activity was measured in vitro, using plus-strand AMV RNA 3 as the template; a gel run with the radiolabeled minus-strand RNA 3 products is shown. Western blots were analyzed using polyclonal antibodies against AMV proteins P1 (B), P2 (C), and CP (D and E). The antiserum against CP detected CP dimers and monomers, which are separately shown in panels D and E, respectively. Sedimentation is from right to left.
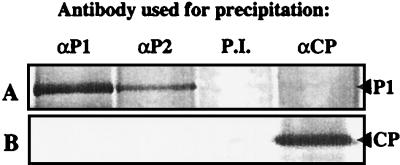
Fraction 9 of the gradient shown in Fig. 3 was incubated with antiserum against P1, P2, or CP or with preimmune serum. After incubation with protein A-Sepharose, the immunoprecipitates were run on an SDS-gel and analyzed by Western blotting using the P1 (Fig. 4A) or CP (Fig. 4B) antiserum. P1 was detectable in immunoprecipitates obtained with the P1 and P2 antisera but not in precipitates obtained with the CP or preimmune serum (Fig. 4A). CP was detectable in the precipitate obtained with the CP antiserum but not in any of the other three precipitates (Fig. 4B). These results support the notion that P1 and P2 are both subunits of the RdRp complex, while no interaction of CP with this complex was detectable.
FIG. 4.
Immunoprecipitation of RdRp complexes. RdRp purified by glycerol gradient centrifugation (Fig. 3, fraction 9) was subjected to immunoprecipitation with a polyclonal antiserum against P1, P2, or CP or with preimmune (P.I.) serum, as indicated. The immunoprecipitates were analyzed by Western blotting using antiserum against P1 (A) or CP (B). The positions of P1 and CP are indicated at the right.
Localization of P1 and P2 in infected cowpea protoplasts.
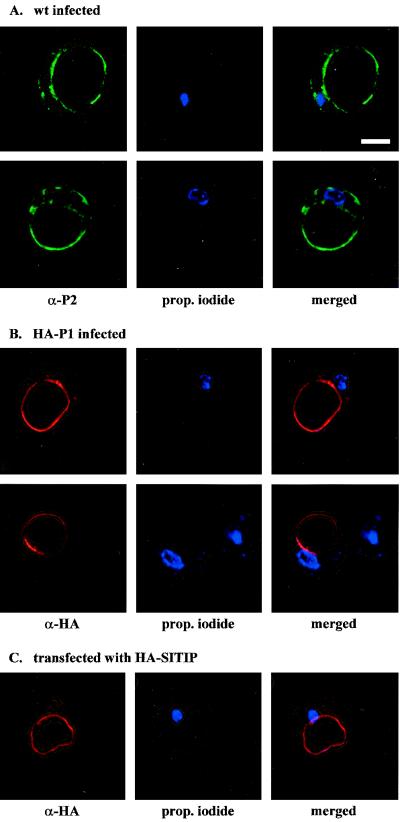
A possible colocalization of P1 and P2 in AMV-infected cowpea protoplasts was analyzed by immunofluorescence CLSM. Using antibodies against CP, it was determined by immunofluorescence that 10 to 50% of the protoplasts were infected. The P1 antibody did not give a specific signal in any of the cells (results not shown). The P2 antibody gave a faint signal throughout the cytoplasmic part of the cell and distinct labeling of the tonoplast (Fig. 5A). Labeling was not concentrated around or near the nucleus (propidium iodide staining shown in blue).
FIG. 5.
In situ localization of P1 and P2 in AMV-infected cowpea protoplasts by immunofluorescence microscopy. Each column of images shows the analysis of a single protoplast, stained with a specific antiserum (left) and with propidium iodide to reveal the nucleus (middle); merge of the two stained images are shown at the right (A) Two different protoplasts infected with wild-type (wt) AMV that were analyzed with a polyclonal antiserum against P2. (B) Two different protoplasts infected with AMV that was engineered to express HA-tagged P1. The protoplasts were analyzed with an antiserum against the HA epitope. (C) Protoplast transfected with a plasmid expressing γ-TIP fused to an HA epitope. The protoplast was analyzed with an antiserum against the HA epitope. Bar = 10 μm.
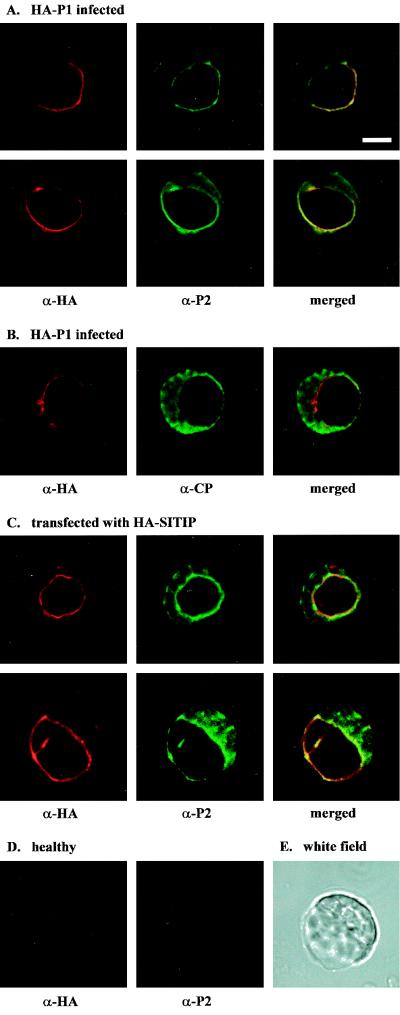
N. benthamiana plants were inoculated with wild-type AMV cDNA 2 and 3 clones and a cDNA 1 clone encoding a P1 protein with the N terminus extended with an HA epitope. The modified virus accumulated at wild-type levels and expressed the HA-P1 protein (results not shown). RNA extracted from the purified chimeric virus was used to inoculate cowpea protoplasts. Analysis of the protoplasts by immunofluorescence CLSM with an HA antibody showed a clear signal of tonoplast-associated HA-P1 in 10 to 50% of the cells (Fig. 5B). This percentage is similar to the percentage of fluorescent protoplasts obtained with antibodies against P2 or CP. Double labeling with HA and P2 antibodies showed a clear colocalization of P1 and P2 around the vacuole (Fig. 6A). CP was detected throughout the cytoplasm, with no distinct pattern around the vacuole (Fig. 6B). The HA and P2 antibodies did not show background labeling of healthy protoplasts (Fig. 6D).
FIG. 6.
Analysis of the colocalization of P1, P2, and CP in AMV-infected cowpea protoplasts by immunofluorescence microscopy. Each column of images shows the analysis of a single protoplast: left and middle, protoplasts analyzed with specific antisera; right, the left and middle images merged. (A) Two protoplasts infected with AMV that was engineered to express HA-tagged P1. The protoplasts were analyzed with antisera against the HA epitope (left) and P2 (middle). (B) Protoplast infected with AMV that was engineered to express HA-tagged P1. The protoplast was analyzed with antisera against the HA epitope (left) and CP (middle). (C) Two protoplasts transfected with a plasmid expressing γ-TIP fused to an HA epitope. The protoplasts were analyzed with antisera against HA (left) and P2 (middle). (D) Healthy protoplasts analyzed with antisera against the HA epitope (left) and P2 (middle). (E) White-field picture of the lower protoplast shown in panel C. Bar = 10 μm.
Colocalization with γ-TIP.
When transfected to cowpea protoplasts, plasmid pMON-HA-SITIP expressed γ-TIP fused N-terminally to the HA tag. The HA antibody detected the protein exclusively at the tonoplast and sometimes around small vesicles (Fig. 5C). Cowpea protoplasts cotransfected with wild-type AMV RNAs and pMON-HA-SITIP were double labeled with antibodies against HA and P2. The γ-TIP and P2 proteins were found to colocalize at the tonoplast (Fig. 6C, merged pictures). The white-field picture shown in Fig. 6E corresponds to the protoplast analyzed in the lower panels of Fig. 6C. These results confirm that the colocalization of P1 and P2 (Fig. 6A) occurs at the tonoplast.
DISCUSSION
P1 and P2 replication proteins interact in vitro and in vivo.
The AMV P1 and P2 proteins show similarities with their BMV 1a and 2a counterparts in amino acid sequence and in the organization of the MT, HEL, and POL domains. Thus, it was expected that AMV P1 and P2 would interact in the same way as reported for BMV 1a and 2a (23, 24). The AMV P1-P2 interaction was demonstrated by co immunoprecipitation of full-length in vitro-translated proteins and of RdRp complexes purified from infected plants. Application of the yeast two-hybrid system permitted the identification of C-terminal sequences of P1 and N-terminal sequences of P2 that are involved in the P1-P2 interaction.
The HEL domains of BMV 1a and AMV P1 are part of protease-resistant domains, which start with a proline cluster and end at the C termini of the proteins (34) (domains indicated in Fig. 1). For the 1a protein, this protease resistance was destroyed by a small C-terminal deletion or by N-terminal deletions that remove the proline cluster. The study by O'Reilly et al. (34) showed a good correlation between the presence of the protease-resistant domain and the ability of 1a to interact with 2a in the LexA two-hybrid system. In the AMV P1 protein, the protease-resistant domain and proline cluster are present between aa 654 and 1126 (34), but no interaction of this sequence with the N terminus of P2 was detectable in our GAL4 two-hybrid system. The slightly longer P1 sequence of aa 618 to 1126 showed a relatively weak interaction with P2, as suggested by the growth rate of the yeast cells. Extension of the P1 sequence to aa 509 to 1126 was required in our assays to obtain maximum growth. The P1 sequence upstream of the proline cluster may act as a spacer preventing the GAL4 DNA binding domain from interfering with proper folding of the protease-resistant domain, or it may directly be required for interaction with P2.
The (putative) polymerases of alphavirus-like viruses, which express their HEL and POL domains in separate proteins, have N termini with very low sequence similarity (ProDom 2000.1 database) (8). The finding that for BMV this N terminus links the POL protein to the MT/HEL protein led to the hypothesis that this N terminus coordinates the assembly of a balanced number of MT/HEL and POL units in replication complexes (23, 33). Our finding that the N-terminal 242 aa of AMV P2 are required for interaction with the HEL domain of P1 further supports the proposed role of N termini of POL proteins. The 1a proteins of BMV, Cowpea chlorotic mottle virus and Cucumber mosaic virus (CMV) are able to form homodimers, and it has been suggested that the 2a protein interacts with a 1a dimer in the replication complex (33, 35). A similar stoichiometry of two MT and HEL domains to one POL domain has been observed in the complex of the 126K and 183K replicase proteins of Tobacco mosaic virus (TMV) (53). For BMV, 1a-1a dimerization involved MT-MT as well as MT-HEL interactions (35). We did not observe an interaction between full-length AMV P1 proteins in the two-hybrid system, although we could detect the protein on Western blots of yeast extracts (data not shown). This may be due to the relatively large size of the protein (13). However, we were also unable to detect dimer formation of the MT domain present in the N-terminal P1 sequence of aa 1 to 686 [Table 1, vectors pBP1(1–686) and pAP1(1–686)]. Moreover, no interaction of this sequence was observed with C-terminal sequences of P1 that contain the HEL domain (Table 1). In addition, we did not find evidence for a P2-P2 interaction. It is interesting that fusions of the N-terminal sequences of AMV P2 (this study) and BMV 2a (33) to a DNA binding domain both resulted in self-activation of the selectable marker in yeast although the two sequences show very little similarity. The possibility that P2 and 2a act as transcriptional activators cannot be ruled out.
It has been proposed that the stimulatory effect of CP on plus-strand AMV RNA synthesis in an in vitro assay is triggered by association of CP with the viral RdRp (11). We did not observe an interaction of CP with P1 or P2 in the two-hybrid system or by coimmunoprecipitation assays of in vitro-translated proteins. Also, our inability to precipitate the P1 present in the purified RdRp by a CP antiserum, or CP by a P1 or P2 antiserum, suggests that CP does not interact with the enzyme through a P1-P2 complex or via a putative host subunit of the enzyme. Possibly, CP stimulates plus-strand RNA synthesis by its RNA binding activity.
Replication proteins localize at the tonoplast.
Infection of plant cells by many positive-strand RNA viruses is accompanied with the appearance of vesicular structures that originate from various cellular membranes. For a few viruses it has been shown that the viral RdRp and synthesis of viral RNA are associated with these structures; for others, the accumulation of these structures has been observed only by electron microscopy, and their role in RNA replication is unproven (reviewed in references 10 and 28). Induction of vesicular structures by Cowpea mosaic virus (CPMV) is due to proliferation of the ER membrane and requires de novo biosynthesis of lipids (6). TMV also replicates at the ER membrane, but replication is not dependent on de novo lipid synthesis and the membrane aggregates appear to be derived from preexisting ER (6, 38). Replication of Turnip yellow mosaic virus is associated with small virus-induced invaginations of the chloroplast membrane (29). Similar 50- to 70-nm vesicles are abundant at the tonoplast of umbravirus-infected cells (12, 30).
The family Bromoviridae consists of the genera Alfamovirus, Ilarvirus, Bromovirus, Cucumovirus, and Oleavirus (36, 43). So far, replication proteins have been localized only in bromovirus-infected cells. Many ER-derived vesicles have been detected adjacent to the nuclei of bromovirus-infected cells, and the BMV 1a and 2a proteins colocalize with these structures (5, 25, 39, 40). Here, we showed that the AMV replicase proteins P1 and P2 colocalize at the tonoplast of infected cells. This in situ localization was verified by using an intrinsic tonoplast protein as marker. In sucrose gradients of homogenates of AMV-infected cells, RdRp activity has been reported to cosediment with chloroplasts and chloroplast membranes (9), but these fractions may have contained tonoplast structures as well. Although P1 was found exclusively at the tonoplast of infected protoplasts, P2 was detectable both at the tonoplast and in the cytoplasm. The cytoplasmic fraction of P2 may correspond to P2 that was detectable in the top fractions of the gradient shown in Fig. 3. Alternatively, this slow-sedimenting P2 may have been released from replication complexes during solubilization of the RdRp. The BMV 1a protein contains the sorting signal for targeting of 1a and 2a to the ER (7, 40). By transient expression of P1 and P2 fused to the green fluorescent protein, we are currently investigating the signals responsible for targeting of the AMV replicase to the tonoplast.
The generation of ER-derived vesicles in CPMV- and TMV-infected cells is accompanied by drastic alterations of the endomembrane system (6, 38). No structural changes of the ER were observed in N. benthamiana plants infected with AMV (Carette, unpublished). By electron microscopy, tonoplast-associated vesicles have been observed in AMV-infected leaf tissue, appearing before cytoplasmic virus particles could be detected (28, 42; J. van Lent, personal communication). Similarly, the formation of tonoplast-associated vesicles protruding into the vacuole has been reported for cucumovirus-infected cells (19, 28) and was occasionally seen in cells infected with the ilarvirus Tobacco streak virus (28). The CMV-induced vesicles were found to contain double-stranded RNA, pointing to a role of these vesicles in RNA replication (19). Our finding that AMV P1 and P2 co-localize at the tonoplast corroborates the assumption that this membrane is the site of replication of alfamo-, ilar-, and cucumoviruses. Replication at the tonoplast or ER may be used as a diagnostic tool to further clarify the taxonomy of viruses in the family of Bromoviridae.
ACKNOWLEDGMENTS
We thank P. James for providing yeast strain PJ69-4A, P. Ealing for the pPE1000 plasmid, Gerda Lamers for help with the confocal microscope, Corrie Houwing for growing the cowpea plants, and Jan van Lent and Joan Wellink for helpful discussions.
REFERENCES
- 1.Ahola T, Ahlquist P. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J Virol. 1999;73:10061–10069. doi: 10.1128/jvi.73.12.10061-10069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bol J F. Alfalfa mosaic virus and ilarviruses: involvement of coat protein in multiple steps of the replication cycle. J Gen Virol. 1999;80:1089–1102. doi: 10.1099/0022-1317-80-5-1089. [DOI] [PubMed] [Google Scholar]
- 5.Burgess J, Motoyoshi F, Fleming E N. Structural changes accompanying infection of tobacco protoplasts with two spherical viruses. Planta. 1974;117:133–144. doi: 10.1007/BF00390795. [DOI] [PubMed] [Google Scholar]
- 6.Carette J E, Stuiver M, van Lent J W M, Wellink J, van Kammen A. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J Virol. 2000;74:6556–6563. doi: 10.1128/jvi.74.14.6556-6563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Ahlquist P. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J Virol. 2000;74:4310–4318. doi: 10.1128/jvi.74.9.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corpet F, Gouzy J, Kahn D. Recent improvements of the ProDom database of protein domain families. Nucleic Acids Res. 1999;27:263–267. doi: 10.1093/nar/27.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Graaff M, Coscoy L, Jaspars E M J. Localization and biochemical characterization of alfalfa mosaic virus replication complexes. Virology. 1993;194:878–881. doi: 10.1006/viro.1993.1335. [DOI] [PubMed] [Google Scholar]
- 10.De Graaff M, Jaspars E M J. Plant viral RNA synthesis in cell-free systems (a literature review) Annu Rev Phytopathol. 1994;32:311–335. [Google Scholar]
- 11.De Graaff M, Man in 't Veld M R, Jaspars E M J. In vitro evidence that the coat protein of alfalfa mosaic virus plays a direct role in the regulation of plus and minus RNA synthesis: implications for the life cycle of alfalfa mosaic virus. Virology. 1995;208:583–589. doi: 10.1006/viro.1995.1189. [DOI] [PubMed] [Google Scholar]
- 12.Falk B W, Morris T J, Duffus J E. Unstable infectivity and sedimentable ds-RNA associated with lettuce speckles mottle virus. Virology. 1979;96:239–248. doi: 10.1016/0042-6822(79)90187-9. [DOI] [PubMed] [Google Scholar]
- 13.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 14.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz R D, Woods R A. High efficiency transformation in yeast. In: Johnston J A, editor. Molecular genetics of yeast: practical approaches. Oxford, England: Oxford University Press; 1994. pp. 121–134. [Google Scholar]
- 16.Gomez de Cedrón M, Ehsani N, Mikkola M L, García J A, Kääriäinen L. RNA helicase activity of Semliki Forest virus replicase protein nsP2. FEBS Lett. 1999;448:19–22. doi: 10.1016/s0014-5793(99)00321-x. [DOI] [PubMed] [Google Scholar]
- 17.Gorbalenya A E, Koonin E V. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 1989;17:8413–8440. doi: 10.1093/nar/17.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock K R, Phillips L D, White D W R, Ealing P M. pPE1000: a versatile vector for the expression of epitope-tagged foreign proteins in transgenic plants. BioTechniques. 1997;22:861–865. doi: 10.2144/97225bm16. [DOI] [PubMed] [Google Scholar]
- 19.Hatta T, Francki R I B. Cytopathic structures associated with tonoplasts of plant cells infected with cucumber mosaic and tomato aspermy viruses. J Gen Virol. 1981;53:343–346. [Google Scholar]
- 20.Höfte H, Hubbard L, Reizer J, Ludevid D, Herman E M, Chrispeels M J. Vegetative and seed-specific forms of tonoplast intrinsic protein in the vacuolar membrane of Arabidopsis thaliana. Plant Physiol. 1992;99:561–570. doi: 10.1104/pp.99.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadaré G, Haenni A-L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao C C, Ahlquist P. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao C C, Quadt R, Hershberger R P, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a form a complex in vitro. J Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim K S. An ultrastructural study of inclusions and disease development in plant cells infected by cowpea chlorotic mottle virus. J Gen Virol. 1977;35:535–543. [Google Scholar]
- 26.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Martelli G P, Russo M. Virus-host relationships, symptomatological and ultrastructural aspects. In: Francki R I B, editor. Polyhedral virions with tripartite genomes. New York, N.Y: Plenum Press; 1985. pp. 163–205. [Google Scholar]
- 29.Matthews R E F. Plant virology. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 30.Murant A F, Goold R A, Roberts I M, Cathro J. Carrot mottle—a persistent aphid-borne virus with unusual properties and particles. J Gen Virol. 1969;4:329–341. [Google Scholar]
- 31.Neeleman L, Bol J F. Cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology. 1999;254:324–333. doi: 10.1006/viro.1998.9568. [DOI] [PubMed] [Google Scholar]
- 32.Olsthoorn R C L, Mertens S, Brederode F Th, Bol J F. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 1999;18:4856–4864. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Reilly E K, Paul J D, Kao C C. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J Virol. 1997;71:7526–7532. doi: 10.1128/jvi.71.10.7526-7532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Reilly E K, Tang N, Ahlquist P, Kao C C. Biochemical and genetic analysis of the interaction between the helicase-like and polymerase-like proteins of the brome mosaic virus. Virology. 1995;214:59–71. doi: 10.1006/viro.1995.9954. [DOI] [PubMed] [Google Scholar]
- 35.O'Reilly E K, Wang Z, French R, Kao C C. Interactions between the structural domains of the RNA replication proteins of plant-infecting RNA viruses. J Virol. 1998;72:7160–7169. doi: 10.1128/jvi.72.9.7160-7169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pringle C R. Virus taxonomy—1999. Arch Virol. 1999;144:421–429. doi: 10.1007/s007050050515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quadt R, Rosdorff H J M, Hunt T W, Jaspars E M J. Analysis of the protein composition of alfalfa mosaic virus RNA-dependent RNA polymerase. Virology. 1991;182:309–315. doi: 10.1016/0042-6822(91)90674-z. [DOI] [PubMed] [Google Scholar]
- 38.Reichel C, Beachy R N. Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc Natl Acad Sci USA. 1998;95:11169–11174. doi: 10.1073/pnas.95.19.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Restrepo-Hartwig M A, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Restrepo-Hartwig M A, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol. 1999;73:10303–10309. doi: 10.1128/jvi.73.12.10303-10309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozanov M N, Koonin E V, Gorbalenya A E. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J Gen Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- 42.Rubio-Huertos M. Atlas on ultrastructure of plant tissues infected with viruses. Madrid, Spain: Consejo Superior de Investigaciones Científicas; 1978. [Google Scholar]
- 43.Rybicki E P. Bromoviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Sixth Report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 450–457. [Google Scholar]
- 44.Tenllado F, Bol J F. Genetic dissection of the multiple functions of alfalfa mosaic virus coat protein in viral RNA replication, encapsidation, and movement. Virology. 2000;268:29–40. doi: 10.1006/viro.1999.0170. [DOI] [PubMed] [Google Scholar]
- 45.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Bokhoven H, Verver J, Wellink J, van Kammen A. Protoplasts transiently expressing the 200K coding sequence of cowpea mosaic virus B-RNA support replication of M-RNA. J Gen Virol. 1993;74:2233–2241. doi: 10.1099/0022-1317-74-10-2233. [DOI] [PubMed] [Google Scholar]
- 47.van der Kuyl A C, Neeleman L, Bol J F. Deletion analysis of cis- and trans-acting elements involved in replication of alfalfa mosaic virus RNA 3 in vivo. Virology. 1991;183:687–694. doi: 10.1016/0042-6822(91)90997-p. [DOI] [PubMed] [Google Scholar]
- 48.van der Kuyl A C, Neeleman L, Bol J F. Role of alfalfa mosaic virus coat protein in regulation of the balance between viral plus and minus strand RNA synthesis. Virology. 1991;185:496–499. doi: 10.1016/0042-6822(91)90807-n. [DOI] [PubMed] [Google Scholar]
- 49.van der Vossen E A G, Neeleman L, Bol J F. Early and late functions of alfalfa mosaic virus coat protein can be mutated separately. Virology. 1994;202:891–903. doi: 10.1006/viro.1994.1411. [DOI] [PubMed] [Google Scholar]
- 50.van Lent J W M, Storms M, van der Meer F, Wellink J, Goldbach R W. Tubular structures involved in movent of cowpea mosaic virus are also formed in infected cowpea protoplasts. J Gen Virol. 1991;72:2615–2623. doi: 10.1099/0022-1317-72-11-2615. [DOI] [PubMed] [Google Scholar]
- 51.van Pelt-Heerschap H. Immunochemical analysis of the alfalfa mosaic virus gene products. Ph.D. thesis. Leiden, The Netherlands: Leiden University; 1987. [Google Scholar]
- 52.van Rossum C M A, Garcia M L, Bol J F. Accumulation of alfalfa mosaic virus RNAs 1 and 2 requires the encoded proteins in cis. J Virol. 1996;70:5100–5105. doi: 10.1128/jvi.70.8.5100-5105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe T, Honda A, Iwata A, Ueda S, Hibi T, Ishihama A. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126/183K protein heterodimer. J Virol. 1999;73:2633–2640. doi: 10.1128/jvi.73.4.2633-2640.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]