Abstract
Inteins (intervening proteins), mobile genetic elements removed through protein splicing, often interrupt proteins required for DNA replication, recombination, and repair. An abundance of in vitro evidence implies that inteins may act as regulatory elements, whereby reduced splicing inhibits production of the mature protein lacking the intein, but in vivo evidence of regulatory intein excision in the native host is absent. The model archaeon Thermococcus kodakarensis encodes 15 inteins, and we establish the impacts of intein splicing inhibition on host physiology and replication in vivo. We report that a decrease in intein splicing efficiency of the recombinase RadA, a Rad51/RecA homolog, has widespread physiological consequences, including a general growth defect, increased sensitivity to DNA damage, and a switch in the mode of DNA replication from recombination-dependent replication toward origin-dependent replication.
In vivo intein splicing efficiency dictates archaeal DNA replication strategies.
INTRODUCTION
The replisome is a collection of proteins that carries out DNA replication. Its functions and concentrations are regulated, intertwined, and coordinated to facilitate synthesis of both leading and lagging strands. Perhaps unexpectedly, critical replication activities are often performed by domain-specific nonorthologous proteins (1, 2) and are best epitomized by the use of evolutionary-unrelated DNA polymerase families (Pol C, Pol D, and Pol B) for replication of bacterial, archaeal, and eukaryotic genomes, respectively (3, 4). The complex systems that regulate the initiation of DNA replication are especially diverse and complex (3, 5–13). DNA replication in most species is controlled by domain-specific initiator protein complexes that assemble at specific locations in the genome, termed origins of replication (ori). Decades of work demonstrate that origin-dependent replication (ODR) is the dominant mechanism of replication in each domain (11).
In some archaea, origin sequences are necessary for replication, and origin recognition proteins (typically Cdc6) initiate replisome assembly by recruiting and helping load the minichromosome maintenance protein (MCM) helicase (14, 15). ODR facilitates accurate replication of the genome, but the sophisticated regulatory strategies used to control the initiation of DNA replication were unlikely to be present in the first cells. Recombination-dependent replication (RDR) initiation is an alternative and effective strategy to replicate archaeal genomes (16). Thermococcus kodakarensis, a hyperthermophilic archaeon, is naturally polyploid (17) and can replicate its genome independently of any ori and the initiator protein Cdc6 (3, 13), demonstrating that some modern species may preferentially initiate replication via recombination at many stochastic sites distributed around the genome. The evolutionary retention of predicted origin sequences and Cdc6 argues for selective usage of ODR versus RDR, but how ODR or RDR is selected as the replicative mechanism is unknown.
Under optimal growth conditions T. kodakarensis predominately uses RDR, but once removed from these conditions, a more judicious use of resources may dictate that an entirely different replication strategy (e.g., ODR) be used to ensure long-term survival (13). RDR requires the retention of multiple genomes and the activity of a recombinase; therefore, control of ploidy and/or the activities of recombinase proteins provide a plausible mechanism to switch between RDR and ODR. Archaea encode RadA, a Rad51/RecA, initiates nucleoprotein filament formation and strand invasion (18–20) that can support DNA synthesis through recombination intermediates in vitro (21). Abundant, active RadA is predicted to be necessary to establish the recombination intermediates that would permit RDR in vivo. Mechanisms that control the active levels of RadA in archaeal cells are critical, as changes to RadA protein levels are likely to alter replication, recombination, and repair (RRR) mechanisms in vivo.
Archaeal loci encoding RadA are often interrupted by intein (intervening protein)–encoding sequences that provide a potential mechanism—via conditional intein splicing (22–35)—to control active RadA levels in vivo. Inteins are mobile genetic elements (MGEs) spliced from host proteins following translation from a precursor protein to yield the isolated intein protein and ligated exteins (LEs) that form mature protein (22, 34–38).
Given that protein splicing (i.e., the totality of reactions involved in intein excision and extein ligation) is known to be affected by environmental conditions (22–35), we sought to understand the mechanism(s) controlling the production of mature RadA (mRadA) in T. kodakarensis and whether intein splicing efficiency could dictate a foundational change in replicative strategy, with high RadA levels supporting RDR and low RadA levels supporting the use of ODR. Given the volatile nature of hydrothermal environments, using an environmentally controlled, intein-based protein switch to regulate DNA replication strategies might prove advantageous. Using otherwise isogenic strains of T. kodakarensis with either mutation to or deletion of the chromosomal intein sequence within RadA (TK1899), we demonstrate that differential RadA protein splicing can influence the physiology of an intein-containing host organism. Quantification of protein splicing efficiency in vitro and mRadA levels in vivo, combined with marker frequency analysis (MFA) to monitor potential origin usage at multiple growth temperatures that alter intein splicing efficiency, demonstrates that the degree of RadA protein splicing directs the choice of ODR versus RDR in T. kodakarensis. While RDR dominates under idealized growth conditions, mutations that result in inefficient Tk-RadA-intein splicing and, in turn, diminished mature Tk-RadA (mTk-RadA) levels, reduce recombination frequencies to tip replication preferences toward ODR.
Our results imply that conditional protein splicing (CPS) of RadA—and, by inference, other intein-invaded replicative machinery components—is likely to shift the in vivo concentrations of mature, spliced factors that drive DNA synthesis, recombination, and repair throughout much of microbial life. An evolutionary shift from intein excision as an initial burden due to the parasitic MGE invasion to an exapted, regulated, and fitness-relevant controlled excision event offers regulatory functions to inteins beyond the originally evolved functions of spreading and splicing. It is thus possible that many intein-encoding species have exapted spontaneous-parasitic intein splicing events to regulatory splicing events that assist in the complex mechanisms controlling key features of RRR strategies.
RESULTS
Tk-RadA-intein houses an active homing endonuclease
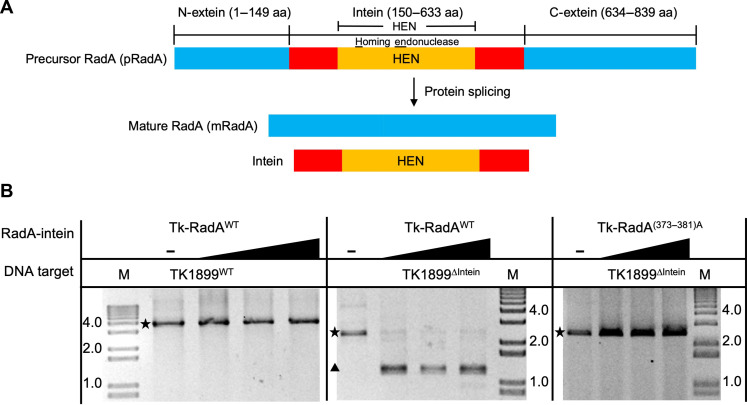
Exhaustive attempts to delete Tk-RadA (TK1899) from the T. kodakarensis genome repeatedly failed, as did initial attempts to delete the intein-encoding sequences (amino acids 150 to 633) within wild-type Tk-RadA (Tk-RadAWT). So-called full-length inteins, such as the intein sequence within Tk-RadA, encode autonomous homing endonuclease (HEN) domains between conserved sequences required for protein splicing; inteins without HEN domains are often termed mini-inteins (Fig. 1A). The HEN domain allows for intein mobility at the DNA level by generating double-stranded DNA breaks in inteinless alleles to drive intein spreading via allelic conversion repair with intein-containing alleles.
Fig. 1. Tk-RadA encodes an intein with an active HEN domain.
(A) Schematic representations of precursor RadA (pRadA; top) splicing to form both intein and mRadA. The precursor RadA is composed of N-extein, intein, and C-extein denoted in blue, red/orange, and blue, respectively. The region of the intein containing the HEN domain is shown in orange. aa, amino acids. (B) In vitro RadA-mediated DNA cleavage demonstrates that HEN activity targets inteinless alleles of TK1899. Tk-RadAWT does not cut its own sequence (TK1899WT) (left) but can cut inteinless allele of RadA from T. kodakarensis (TK1899ΔIntein-encoding sequences) (middle). HEN activities are compromised in the Tk-RadA(373–381)A variant wherein HEN active site residues are altered to alanines (right). The intact DNA target and resultant cleavage product(s) are represented by a star and a triangle, respectively.
We reasoned that our failure to delete the sequences encoding the RadA-intein from the T. kodakarensis chromosome was due to an active HEN domain cleaving the intein-minus allele before recombination. As expected, preparations of Tk-RadAWT did not cleave the TK1899WT target DNA as the intein sequence was intact; however, it demonstrated robust HEN activity against a polymerase chain reaction (PCR)–amplified DNA fragment containing the inteinless allele of TK1899Δintein (Fig. 1B) and the closely related inteinless RadA–encoding sequence from Pyrococcus furiosus (Pf1926) (fig. S1C). The active site of the putative HEN domain within Tk-RadAWT was predicted on the basis of homology with other HEN-containing inteins (39) at residues 373 to 381. When the presumptive HEN active site residues were either changed to alanine [Tk-RadAA(373–381)] or deleted (Tk-RadAΔ373–381), all HEN activity was lost (Fig. 1B and fig. S1C). In addition, Tk-RadA lacking the entire HEN domain was prepared (Tk-RadAΔ286–585) and, as expected, failed to cleave TK1899ΔIntein DNA (fig. S1C). The exact cleavage site, sequence requirements for HEN-mediated DNA cleavage, and HEN active site residues were determined for Tk-RadAWT (figs. S1, A and B, and S2, A to F). We found that Tk-RadA-intein HEN domain recognizes and cleaves long asymmetrical DNA sequence, >23 base pairs (bp), encoding for an inteinless allele of RadA-producing DNA fragments with 3′-hydroxyl overhangs congruent with previous finding of similar HEN domain in T. kodakarensis DNA polymerase B inteins (39).
Variant intein sequences affect RadA splicing efficiency
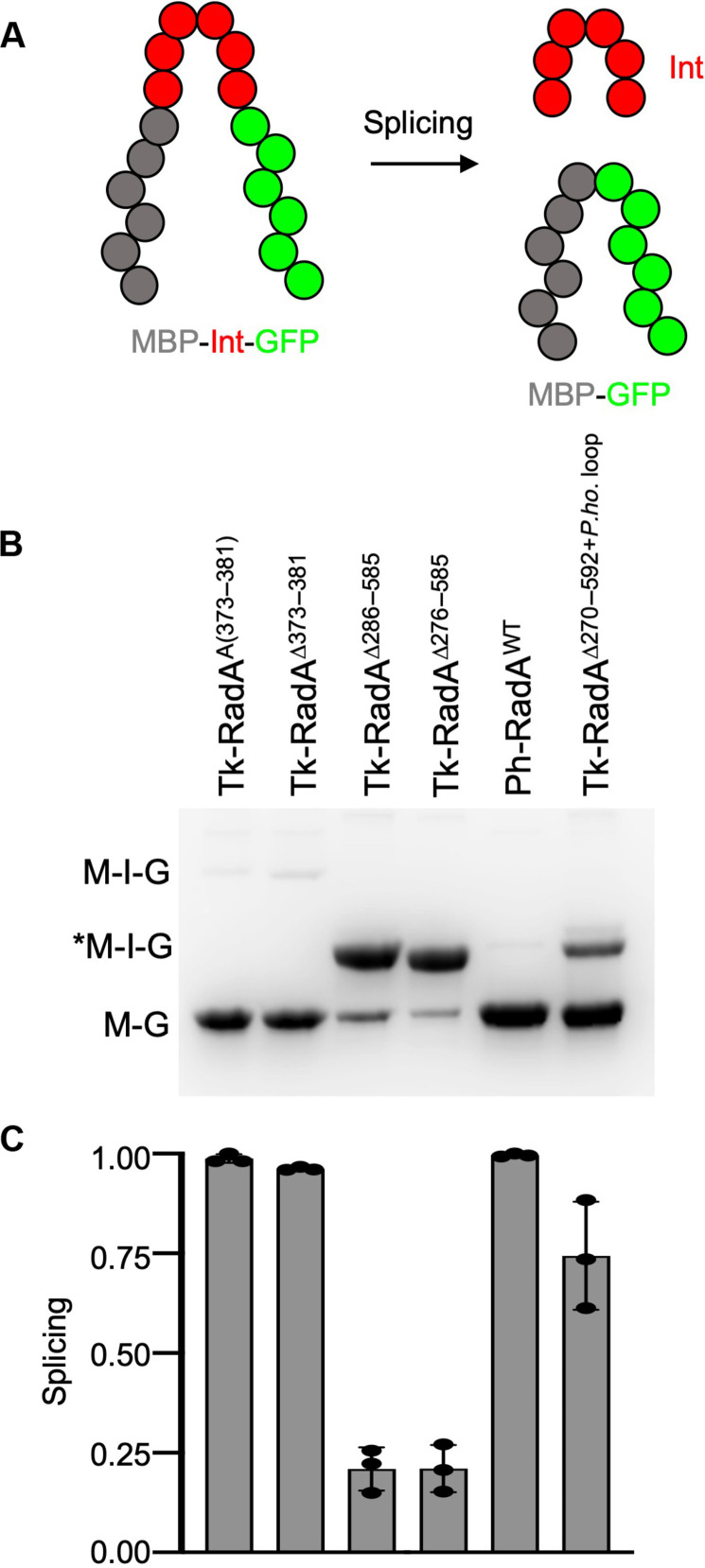
Intein splicing accuracy and efficiency are often quantified using reporters with fully or partially substituted exteins (Fig. 2A). Placing the Tk-RadA-intein between the artificial exteins maltose-binding protein [N-terminal extein (N-extein)] and green fluorescent protein (GFP) [C-terminal extein (C-extein)] generates a splicing reporter we refer to as MIG. MIG constructs provide a rapid mechanism to detail the impacts of intein composition and solution conditions on the accuracy and efficiency of protein splicing determined by the relative fluorescence of unspliced precursor to LEs in total cell lysates. Precursor and ligated extein species are monitored in-gel under conditions [i.e., samples are not boiled before SDS–polyacrylamide gel electrophoresis (PAGE)] where GFP fluorescence is maintained (Fig. 2B) (40).
Fig. 2. Intein-extein partnerships and intein composition affect the efficiency of intein splicing.
(A) Schematic of the maltose-binding protein (MBP)–intein (Int)–GFP (MIG) reporter constructs wherein intein-containing fluorescent precursor proteins must excise the intein and ligate the exteins to generate a matured, smaller fluorescent product (MBP-GFP). (B and C) Native PAGE and quantification of fluorescent signals reveal the impact of intein composition and the presence of the HEN domain on the efficiency of intein excision.
Once the Tk-RadA-inteinA(373–381) and Tk-RadA-inteinΔ373–381 were placed within a MIG reporter, splicing was highly efficient and proceeded to completion during expression in Escherichia coli, even at 15°C (Fig. 2, B and C). Tk-RadA-inteinWT with the active HEN was toxic to E. coli within the MIG construct, and, thus, the relative efficiency of splicing was determined with constructs that lack HEN activity. Internal components of the Tk-RadA-intein, excluding those residues known to be directly involved in intein-mediated catalysis, can influence the efficiency of splicing (41, 42). While HEN inactivation does not fully inhibit splicing, removal of the entire HEN domain (either Δ276 to 585 or Δ286 to 585) from the Tk-RadA-intein markedly lowers splicing efficiency from ~100% to only ~20% during expression at 15°C (Fig. 2, B and C). By comparison, the closely related Pyrococcus horikoshii RadA mini-intein (Ph-RadA-intein), nearly identical to the Tk-RadA-intein but naturally lacking the HEN domain (fig. S3, A and B), splices very efficiently within the MIG splicing reporter (Fig. 2, B and C), as previously observed (22–24).
When comparing the sequences of the Ph-RadA-intein and the Tk-RadA-inteinΔ276–585, which are identical in length and are highly conserved (fig. S3A), one region of the sequence displays minimal conservation (red; fig. S3A). This poorly conserved region (Tk-RadA amino acids 270 to 275 and 586 to 592 compared to Ph-RadA amino acids 273 to 285; note that the Tk sequence is split because of Δ276 to 585), where the HEN domain of Tk-RadA is found, is where Ph-RadA once presumably housed a HEN domain that was lost during evolution (22). Substitution of the corresponding P. horikoshii intein amino acid residues 273 to 285 (referred to as P.ho. loop) for the T. kodakarensis intein amino acids into an otherwise HEN-deleted Tk-RadA-intein sequence (Tk-RadA-inteinΔ270–592+P.ho. loop) largely restored the splicing defect of Tk-RadA-intein variants with large HEN domain deletions (Fig. 2, B and C).
Native RadA exteins block intein splicing following HEN deletion
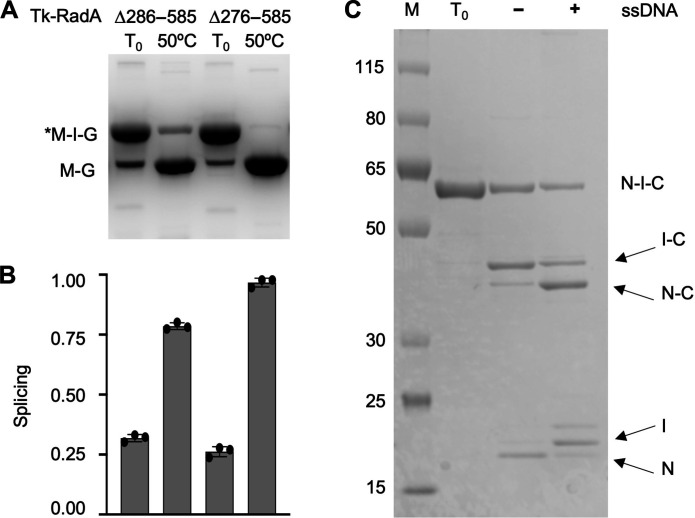
Given that Tk-RadA is an essential protein, the construction of T. kodakarensis strains wherein severe intein splicing defects would sufficiently limit the production of mTk-RadA was predicted to be problematic. Inefficient splicing of Tk-RadA-intein variants lacking the HEN domain (Δ276 to 585 or Δ286 to 585) can be rescued by incubation at elevated temperature within the MIG reporter (Fig. 3, A and B). The splicing efficiencies of Tk-RadA-inteinΔ286–585 and Tk-RadA-inteinΔ276–585 increased from only ~20 to ~80 and ~100%, respectively, within MIG constructs upon incubation at only 50°C. We carried out additional in vitro and MIG reporter assays to develop an intein splicing efficiency, which might translate in vivo, to generate a HEN-deleted Tk-RadA-intein that would reduce, but not eliminate, splicing (Fig. 3).
Fig. 3. The efficiency and accuracy of pTk-RadA splicing is radically improved at high temperature or through addition of ssDNA in vitro.
(A and B) Native PAGE and quantification of fluorescent signals reveal the impact of temperature on the efficiency of intein excision of Tk-RadAΔ286–858. (C) SDS-PAGE of purified pTk-RadAΔ286–585 before incubation (T0) without (−) and with (+) an ssDNA cofactor that markedly increases the efficiency and accuracy of intein splicing and extein ligation. The lane labeled M contains proteins of known molecular weights measured in kilodaltons.
As anticipated, we were unable to obtain large HEN domain deletions of the Tk-RadA-intein within the natural extein context of T. kodakarensis, presumably due to the low splicing efficiency and thus insufficient in vivo levels of mTk-RadA to support growth. We were, however, successful in generating T. kodakarensis strains encoding large HEN domain deletions within the Tk-RadA-intein when accompanied by the addition of the P.ho. loop that was demonstrated (Fig. 2, B and C) to assist splicing of HEN-deleted RadA inteins in vitro. The failure to recover strains at 85°C wherein the HEN-encoding sequences were selectively deleted demonstrates that temperature alone was unable to restore sufficient splicing of the HEN-deleted Tk-RadA-intein within T. kodakarensis to support growth when expressed in the native extein context, despite temperature having a large impact on splicing efficiency within the MIG reporter (Fig. 3A); Ph-RadA-intein splicing efficiencies within the native exteins, rather than the MIG reporter, are also grossly affected by intein-extein interactions (24). Upon expression of the Tk-RadA-inteinΔ286–585 in a construct containing the native exteins, no splicing is observed during expression in E. coli and subsequent purification (Fig. 3C). Attempts to increase splicing of the Tk-RadA-intein lacking the HEN domain by incubating at 65°C only modestly increased splicing, and while the levels of precursor protein are reduced, splicing of this variant was inefficient (~10%), and off-pathway products resulting from cleavage of the N-extein from the intein-C-extein before ligation dominate (Fig. 3C).
RadA drives recombination by forming nucleoprotein filaments on single-stranded DNA (ssDNA) as a first step in homologous recombination. As the addition of substrates (ssDNA) can rescue Ph-RadA-intein splicing within native exteins (22, 23, 34), we tested whether the addition of ssDNA might similarly increase the splicing efficiency of Tk-RadA-inteinΔ286–585 within the native extein context (Fig. 3C). As predicted, addition of ssDNA increased the accuracy of Tk-RadA-inteinΔ286–585 splicing within native exteins from only ~10% to ~70% in vitro (Fig. 3C). While it is speculative as to any potential role for ssDNA in Tk-RadA splicing, our results indicate that Tk-RadAΔ286–585 retains at least partial ssDNA binding capacity in the precursor form. Further, these results suggest a possible means by which some mini-inteins might rely on cellular signals or substrates (e.g., ssDNA) to compensate for splicing defects following HEN domain loss.
These marked splicing defects within native exteins (Fig. 3C) likely explain our inability to obtain HEN deletions without the addition of the P.ho. loop in vivo in T. kodakarensis. While inviability associated with an inability to efficiently splice an essential protein such as RadA may appear an obvious result, it is the first such description within an intein-containing organism. Hence, our results have implications for other intein-containing microbes and support the hypothesis that targeted approaches to impairing protein splicing in human pathogens, particularly for the RecA intein of Mycobacterium tuberculosis, represent a viable antimicrobial strategy.
Variable protein splicing within T. kodakarensis
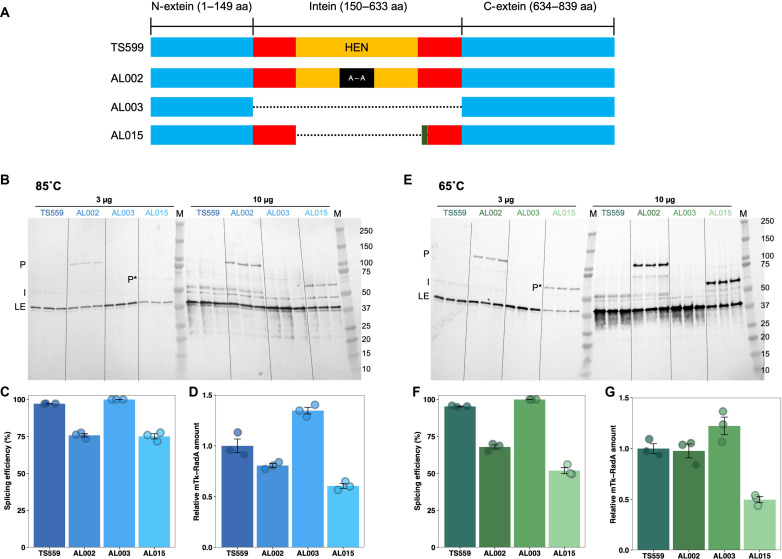
Following an allelic exchange of genomic sequences to inactivate the Tk-RadAWT-intein HEN active site (amino acids 373 to 381), sequences encoding the entire Tk-RadAWT-intein could then be easily deleted or allelically exchanged on the T. kodakarensis genome (Fig. 4A). To evaluate the impacts of variable protein splicing on the viability, fitness, and potential replicative strategies used in vivo because of altered levels of mTk-RadA, we generated a series of otherwise isogenic strains of T. kodakarensis wherein TK1899 sequences (encoding RadA) were modified to generate alleles that encode Tk-RadA variants, with predicted alteration in intein splicing efficiency (Fig. 4A). All strains retain the native promoter and genomic locus and encode the same extein sequences, ensuring that mTk-RadA in each strain is identical at the primary sequence level. Intein sequence variants, however, are likely to affect the efficiency of intein excision and thus steady-state mTk-RadA protein levels in vivo. Whole-genome sequencing (WGS), at greater than 100× coverage for each strain, revealed that the newly constructed strains contained only the desired allelic modifications to TK1899 and were otherwise isogenic throughout the remainder of the >2-Mbp genome compared to the parental strain TS559.
Fig. 4. Intein splicing defects affect in vivo levels of mTk-RadA.
(A) Tk-RadA allelic variants introduced into the T. kodakarensis genome at the native locus retain native extein and promoter sequences. Strains differ solely in the sequences encoding the Tk-RadA-intein. The full-length precursor of Tk-RadAWT from the parental strain TS559 contains the unspliced intein (red and orange; amino acids 150 to 633) with an active HEN domain (orange; amino acids 286 to 585) embedded within N-extein and C-extein (blue; amino acids 1 to 149 and 634 to 839, respectively). AL002 was constructed from TS559 and encodes an inactive HEN (red, orange, and black; amino acids 373 to 381) with alanine substitution of amino acids 373 to 381 [Tk-RadAA(373–381)]. AL003 (Tk-RadAΔIntein) encodes a Tk-RadA variant lacking the entire intein sequence (amino acids 150 to 633) introduced into AL002. Reintroduction of the allele lacking the HEN domain with a small loop derived from the mini-intein of P. horikoshii RadA protein into AL003 generated AL015 (Tk-RadAΔ270–592+P.ho. loop). (B and E) In vivo Tk-RadA protein and intein levels were monitored by Western blotting using polyclonal antibodies against the pTk-RadAWT. Strains were grown to mid-exponential phase in biological triplicate at 85°C (B) and 65°C (E). All Tk-RadAWT isoforms were detected, including pTk-RadA (P), pTk-RadA lacking the HEN domain (P*), mature, ligated exteins (LEs) of Tk-RadA, and the spliced intein (I). (C and F) Tk-RadA splicing efficiencies at 85°C (C) and 65°C (F) were quantified from the ratio of LE/P or LE/P* intensities. (D and G) Relative mTk-RadA levels at 85°C (D) and 65°C (G) were quantified from the ratio of LE (variant strains)/LE (TS559). Error bars represent SE from the mean of a minimum of three biological replicates.
The efficiency of intein splicing in vivo, as well as the steady-state abundance of mTk-RadA (resultant from properly LEs) and the Tk-RadA-intein, was quantified via Western blotting using polyclonal antibodies raised against the unspliced precursor Tk-RadAWT (pTk-RadAWT; Fig. 4, B and E; each strain was evaluated with triplicate biological replicates). Total cell lysates derived from the T. kodakarensis parental strain (TS559) were compared with lysates from otherwise isogenic strains encoding variant sequences of TK1899 that result in (i) direct production of mTk-RadAWT without the need for splicing due to the removal of all intein-encoding sequences (strain AL003), (ii) a pTk-RadAA(373–381) variant wherein the full intein-encoding sequences are retained but modified such that the HEN active site is compromised because of replacement of residues 373 to 381 with alanines (strain AL002), or (iii) a pTk-RadAΔ270–592+P.ho. loop variant wherein the entire HEN domain was removed and a small sequence derived from the Ph-RadA-intein that improves splicing (Fig. 2, B and C) was added (strain AL015). When total cellular lysates were resolved and probed with polyclonal antibodies raised against pTk-RadAWT, we were readily able to detect the anticipated dominant protein products of TK1899, namely, pTk-RadAWT, mTk-RadAWT, and the Tk-RadA-intein (Fig. 4, B and E).
In the parental strain TS559 wherein the native extein-intein partnerships of Tk-RadA are retained, in vivo protein splicing is efficient (~95% of pTk-RadAWT is processed to mTk-RadA and Tk-RadA-intein) and accurate (no evidence of off-pathway reactions) (Fig. 4, B and C). The retention of Tk-RadA-intein as a stand-alone protein outlines potential biological roles, including HEN activity. When the intein-encoding sequences of TK1899 are fully removed from the genome (strain AL003), mTk-RadAWT is the direct product of translation; Western blotting reveals only a single band corresponding to mTk-RadAWT, and, as anticipated Tk-RadA-intein signal is completely absent (Fig. 4, B and E). mTk-RadAWT levels in strain AL003 increase to ~135% compared to those in the parental strain TS559 (Fig. 4, D and G). We rationalize that this difference in mTk-RadAWT levels results from some native pTk-RadAWT misfolding, improperly splicing, or degrading in TS559.
The importance of intein sequences beyond those immediately engaged in the chemistry of protein splicing is revealed in strain AL002, where nine alanine substitutions in the active center of the HEN domain of the Tk-RadA-intein diminish the efficiency of splicing by ~25% and obvious levels of nonspliced pTk-RadAA(373–381) are retained at steady state (Fig. 4, B, C, E, and F). In strain AL015, removal of sequences encoding the entire HEN domain and addition of the corresponding sequences from the Ph-RadA-intein, generating Tk-RadA Δ270–592+P.ho. loop, result in a smaller precursor protein (P*) that is also less efficiently spliced compared to pTk-RadAWT (~75%) due to changes within intein residues not involved in the chemistry of protein splicing (Fig. 4, B, D, E, and F). The in vivo splicing deficiencies for pTk-RadAA(373–381) and pTk-RadAΔ270–592+P.ho. loop were substantial from 85°C grown strains (Fig. 4, B and C) and were further exacerbated when strains were grown at only 65°C (Fig. 4, E and F), demonstrating that the environmental conditions can affect splicing efficiencies and thus regulate production of intein-free proteins in vivo.
Impaired splicing affects mRadA protein levels
Changes to the efficiency of pTk-RadA splicing in vivo directly affect the steady-state levels of mTk-RadA available for cellular activities, including formation of nucleoprotein filaments necessary for initiation of homologous recombination. In both strains AL002 and AL015, wherein the efficiency of Tk-RadA-intein splicing was reduced, the impact on steady-state protein levels was notable, with HEN-inactivation and HEN-deletion P.ho. loop addition within the Tk-RadA-intein resulting in ~20 and ~40% less of the identical mTk-RadA product, respectively (Fig. 4, B and D) when strains were grown at the optimal temperature of 85°C.
Given the variable splicing efficiencies due to temperature changes observed in vitro and in E. coli, we grew T. kodakarensis strains at 65°C to determine the impact of environmental conditions on the efficiency of Tk-RadA protein splicing and the steady-state abundance of mTk-RadA in vivo in the native host (Fig. 4, E to G). A 20°C reduction in temperature had minimal and no impact on the splicing efficiency of Tk-RadA in strains TS559 and AL003, respectively. The noted temperature dependence of thermococcal RadA proteins in vitro (22) is thus largely compensated for in vivo. In strains AL002 and AL015, where splicing efficiency was compromised at 85°C, the reduction in growth temperature to 65°C resulted in even greater splicing deficiencies, with protein splicing reduced to only ~66 and ~53%, respectively (Fig. 4, E and F). The deficiency in splicing at 65°C also substantially affects the steady-state protein levels of mTk-RadAWT. As observed at 85°C, the RadA-inteinless strain AL003 retains a greater (~120%) amount of mTk-RadAWT than observed in TS559. Unexpectedly, the reduction in splicing efficiency in strain AL002 at all temperatures only very modestly reduces (~95%) the steady-state level of mTk-RadAWT at 65°C, implying that a combination of splicing and degradation rates affects protein levels in strain AL002. In line with the increased splicing deficiencies in strain AL015, the steady abundance of mTk-RadAWT is halved (~50%) compared to the otherwise isogenic parental strain with the native Tk-RadA-intein sequence (Fig. 4, E and G).
Intein splicing defects compromise growth and response to DNA damage
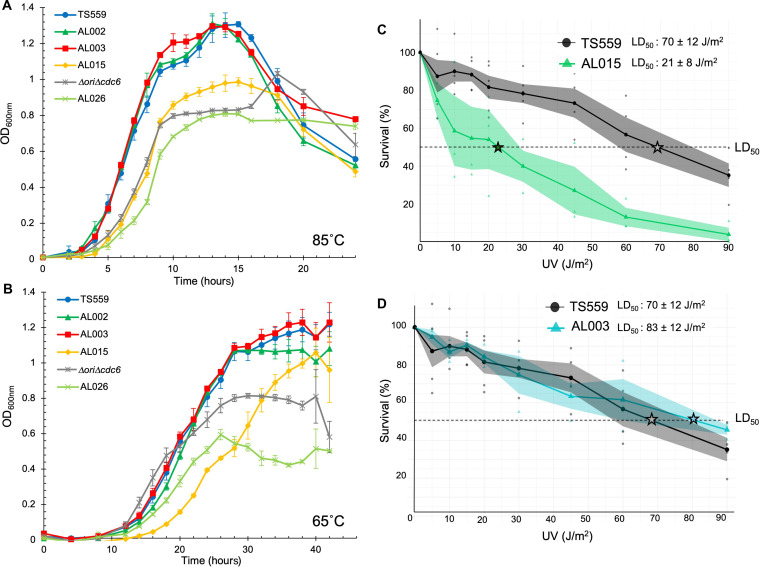
Deletion of the sequences encoding the RadA-intein in strain AL003 or inactivation of the HEN activities within the RadA intein in strain AL002 maintained mRadA protein levels in vivo at both 65° and 85°C (Fig. 4, D and G) and expectedly resulted in negligible impacts on growth compared to the parental strain TS559 at both temperatures (Fig. 5, A and B). These results suggest no negative or positive fitness impact due to the absence of a functional HEN-containing intein nor the complete loss of the intein from the cytoplasm of T. kodakarensis. In contrast, strain AL015, wherein RadA-intein splicing is compromised and mRadAWT levels are reduced compared to the parental strain (Fig. 4, D and G), demonstrated a consistent growth lag, and reduced culture density was observed (Fig. 5, A and B). Given that the parental (TS559) and AL015 strains are otherwise isogenic, the most parsimonious cause of the growth defect is a direct association with RadA-intein excision defects. While the reduction in splicing efficiency is moderate and may not have been predicted to manifest a phenotypic consequence or fitness impact, reduced splicing efficiency and reduced mRadAWT protein levels in AL015 have a demonstrated fitness cost for T. kodakarensis (Fig. 5, A to C).
Fig. 5. Impaired intein excision and reduced mTk-RadA protein levels impair cellular fitness.
(A and B) Growth of triplicate biological replicates of strains of T. kodakarensis monitored by changes in optical density at 600 nm (OD600nm) at 85°C over 24 hours (A) and 65°C over 42 hours (B) reveals that impaired intein splicing negatively affects growth rate and final culture densities. Deletion of the Tk-RadA-intein (AL003) or inactivation of the HEN domain (AL002) does not obviously affect growth at either temperature. Deletion of the HEN domain in AL015 hinders growth at both temperatures, and when combined with the deletion of the ori and Cdc6 in AL026, it synergistically reduces growth. (C) Reduced splicing efficiencies and reduced mTk-RadA levels in strain AL015 result in decreased survival upon UV exposure, adumbrating impacts to RadA-mediated DNA repair mechanisms in vivo. (D) Deletion of the Tk-RadA-intein–encoding sequences in strain AL003 results in a modest increase in survival upon UV exposure. The median lethal dose (LD50) denoted with a star was calculated for both strains based on the death curve data using Finney’s probit analysis calculator.
RadA oligomerizes and forms nucleoprotein filaments that drive strand invasion; these invasions can initiate homology-directed DNA repair, including the repair of bulky DNA lesions resultant from exposure to ultraviolet (UV) light. To ascertain whether the modest defects in RadA-intein splicing and associated approximately twofold reductions in mRadAWT steady-state protein levels affected RadA-mediated DNA repair mechanisms, we used UV sensitivity assays to compare the ability of parental (TS559) and AL015 strains to repair bulky DNA damage. We detected a notable and substantial (~3.5-fold) UV-sensitive phenotype within AL015 cells following UV damage compared to the parental strain, showing that a ~50% reduction in mTk-RadA abundance greatly affected RadA-mediated DNA repair mechanisms and compromises cellular fitness (Fig. 5C). To complement evidence that reduced levels of mTk-RadA directly correlate with an increased sensitivity to UV damage in strain AL015, we also examined the UV sensitivity of strain AL003 (encoding an inteinless RadA, thereby directly producing mTk-RadA without a pTk-RadA intermediate), in which a modest increase (~20%) in steady-state levels of mTk-RadA was observed (Fig. 5D). Although the increase in mTk-RadA levels in AL003 is modest, the UV median lethal dose (LD50) increases, in full support of mTk-RadA levels playing a critical role in initiating efforts to repair bulky lesions in vivo. These results also suggest that intein, as a single protein, has no effect on the organism’s UV sensitivity.
Reduced RadA-intein splicing can control the dominant mode of DNA replication
The defects in DNA repair due to reduced mRadAWT levels (Fig. 5C) suggested that other RadA-mediated processes, such as RDR, could be similarly affected by defects in RadA-intein excision proficiency. The dominant mode of replication in T. kodakarensis cells is easily determined by MFA (Fig. 6A) (13, 43). In rapidly growing but unsynchronized planktonic T. kodakarensis cultures that are using an origin(s) of replication, DNA harvested during exponential growth should retain an overrepresentation of sequences adjacent to the origin compared to sequences located further away from the origin(s). When sequence abundances recovered from (i) growing and replicating cultures and (ii) nongrowing, nonreplicating stationary phase cultures are compared, and any regional overabundance defines an origin(s) of replication and would be consistent with ODR [e.g., a peak(s) in the plotted frequency of sequence abundance defines an origin, with a smooth wave of regional overabundance extending bidirectionally away from the origin providing confidence that at least some of the cells in culture are using ODR as the dominant mode). When a relatively equal abundance of all genomic sequences is returned in MFA, all sites on the genome are equally likely to serve as replication origins and would be consistent with RDR.
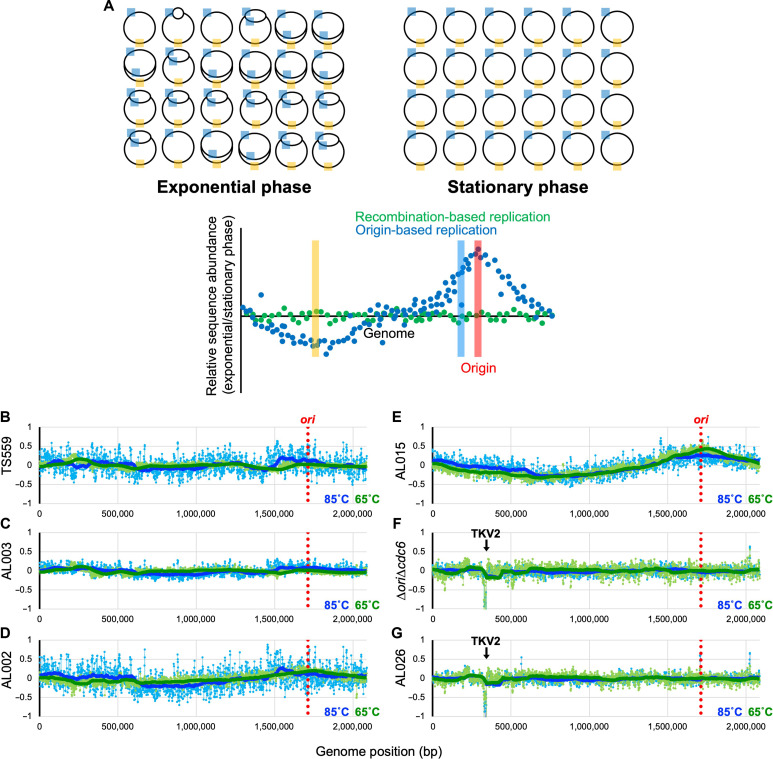
Fig. 6. Impaired intein excision elicits a radical shift of replication strategies.
(A) Overview of MFA. The T. kodakarensis genome is shown by a black circle, with cyan and orange boxes denoting generic regions in the genome proximal and distal, respectively, to the ori. When ODR prevails as the dominant replicative strategy, DNA isolated from actively replicating cultures during exponential phase retains an overrepresentation of the DNA regions closest to the ori (cyan) and an underrepresentation of regions of DNA located distantly from the ori (orange). When the frequencies of DNA sequences from replicating cultures are compared to the frequencies of DNA sequences from nonreplicating cultures of the same strain in stationary phase, MFA permits identification of any areas of the genome that are overrepresented in replicating cells, with the peak defining a replication origin. The hypothetical blue scatter plot displays the anticipated wave pattern with a single apex at the predicted origin that would be anticipated from cultures wherein ODR dominates. In contrast, when RDR dominates within a growing culture, MFA will not reveal any sequence enrichment and will be consistent with the hypothetical green scatter plot. (B to G) Strains were grown at 85°C (blue) and 65°C (green), and DNAs were purified from cells harvested at both mid-exponential and stationary phase. MFA reports the log2 ratio of the mapped reads from exponentially growing cells divided by reads from stationary cultures. A 100-period moving average is plotted using the thick blue/green solid lines to show the trend from the MFA. The genome of T. kodakarensis was segregated into 1000-bp bins. The location of the ori-cdc6 region is indicated by the red dotted line. The downward arrow in (F) and (G) represents the location of the TKV2-excised region.
The sole T. kodakarensis replication origin, the hallmark of ODR, is located not far from the RadA (TK1899) locus, between TK1900 (predicted voltage-gated potassium channel) and TK1901 (Cdc6; the origin recognition protein) at ~1.75 Mbp of the chromosome (Fig. 6; red dotted line defines the location of the ori). MFA of TS559 cultures grown at 85° and 65°C reaffirmed (13) that an obvious single origin is not present and, therefore, is not necessary for normal growth of T. kodakarensis. These results are consistent with the preponderance of DNA replication initiation through RDR at every position of the genome with equal preference instead of a clear peak at the origin or anywhere else on the genome (Fig. 6B). The loss of the Tk-RadA-intein in strain AL003 did not affect the use of RDR as the dominant mode of replication at either 85° or 65°C (Fig. 6C). AL003 appeared to generate a flatter MFA profile compared to TS559 (Fig. 6, B and C), suggesting a greater propensity for RDR due to the increased levels of mTk-RadA (Fig. 4, B and E). For AL002, particularly at 65°C, MFA indicates a modest shift in the population toward ODR, although the change was subtle (Fig. 6D). However, a pronounced switch in the mode of DNA replication from RDR to ODR is observed for AL0015, particularly at 65°C (Fig. 6, D and E). This switch from RDR to ODR correlated with deficiencies in Tk-RadA splicing (Fig. 4, B, C, E, and F), a resultant decrease in mTk-RadAWT (Fig. 4, B, D, E, and G), an increase in pTk-RadA (Fig. 4, B and E), a reduced overall fitness (Fig. 5, A and B), and a UV hypersensitivity phenotype (Fig. 5C).
AL015, when compared to TS559 (parental), is completely isogenic besides changes to the Tk-RadA-intein, and it still encodes mTk-RadAWT. When grown at either 65° or 85°C, there was clear evidence of origin usage, and, thus, a foundational switch in replicative strategies was manifested by a maximally twofold change in steady-state mTk-RadA levels in vivo. MFA of the AL002 strain, wherein splicing efficiency is decreased to only ~80 and ~66% at 85° and 65°C, respectively, displayed a more subtle transition to ODR from the RDR default; MFA provides a population average; thus, some cells in strain AL002 are likely to prefer RDR, whereas others prefer ODR. This shift correlated with accumulation of pTk-RadA rather than steady-state protein levels of mTk-RadAWT (Fig. 4C). Unexpectedly, it appeared that the splicing efficiency decrease was responsible for changes in the prominence of ODR versus RDR, as total steady-state protein levels of mTk-RadAWT in AL002 were only reduced by ~20 or ~5% when compared to TS559 at 85° and 65°C, respectively. Evidence for origin utilization in strain AL002 at 65°C, wherein mTk-RadAWT levels were effectively unchanged, suggested that the very act of splicing, or the retention of pTk-RadAA(373–381), tipped the ratio of T. kodakarensis cells that rely on RDR versus ODR.
ΔoriΔcdc6 strains must use RDR
The preference for RDR over ODR in T. kodakarensis permits deletion of the origin (ori) sequence and adjacently encoded origin recognition protein Cdc6 (TK1901) without substantial growth defects (13). ΔoriΔcdc6 strains rely entirely on RDR (Fig. 6F), and deletion of the natural origin and initiator protein does not reveal evidence of a secondary or cryptic origin of replication (13). Construction of the ΔoriΔcdc6 strain was coincident with the spontaneous deletion of TKV2, a prophage genome known to be dispensable for growth of T. kodakarensis (44). Mapping of MFA results to the TS559 genome details this loss through a precise peak at ~0.32 Mbp of the genome (Fig. 6, F and G). We rationalized that introduction of the Tk-RadA Δ270–592+P.ho. loop allele into a ΔoriΔcdc6 strain incapable of ODR (generating strain AL026) would challenge both ODR and RDR mechanisms in T. kodakarensis. The reduced efficiency of intein splicing and reduced mTk-RadA levels favor ODR, but the deletion of the origin and initiator protein precludes use of ODR. As predicted, growth of AL026 strains is compromised, more markedly at 65°C, wherein intein excision is more impaired (Fig. 5, A and B), but AL026 strains are incapable of ODR as revealed by MFA (Fig. 6G). Thus, although reduced RadA splicing (e.g., strain AL015) would normally dictate a switch in replication strategies favoring ODR when intein excision is compromised, ODR is not permitted, and, therefore, AL026 must use RDR. RDR is required for the growth of AL026 but leads to markedly reduced growth at 65°C due to suboptimal concentrations of mTk-RadA. This represents the first demonstration of a growth defect due to impaired splicing and the potential power of regulated intein splicing to regulate microbial physiology.
mRadA activities are unaffected by precursor RadA
The massive phenotypic impacts resultant from modestly decreased intein excision and a twofold reduction in mRadAWT protein levels suggest that even small changes in intein excision efficiencies can manifest large physiological responses in vivo, supporting continued efforts to develop antimicrobials that control intein splicing in essential genes. Given that RadA functions as an oligomer, the precursor RadA (pRadA) retains a response to addition of substrate (Fig. 3) and is likely at least partially folded. We rationalized that increased steady-state levels of pRadA could affect on mRadA function. Work with the closely related P. horikoshii RadA demonstrated that while the pRadA cannot hydrolyze adenosine 5′-triphosphate (ATP), it can bind DNA (23, 24), hinting that precursor forms of intein-containing proteins can perform some but not all functions. Therefore, if elevated steady-state pRadA levels permit pRadA to interrupt mRadA oligomerization and function, then the true impact of deficiencies in RadA-intein excision may reflect a reduced mRadA level and inhibition of mRadA function due to unfavorable and interfering interactions between pRadA and mRadA.
RadA activity can be reconstituted and quantified through an in vitro recombinase assay that monitors the efficiency of purified recombinant mTk-RadA to promote strand invasion of an ssDNA oligonucleotide into a supercoiled plasmid (fig. S4) (21). Nucleoprotein filament formation was facilitated by incubating the purified recombinase with the 5′-Fluorescein amidites (FAM) deoxyoligonucleotide (L93) before the addition of supercoiled plasmid DNA (pUC19), which contains a sequence complementary to the L93 oligo. RadA mediates strand invasion of the L93 oligo into pUC19, forming a displacement loop (D loop) in a RadA protein concentration and ATP-dependent manner (fig. S4A). The D-loop formation can subsequently be resolved and quantified using native gel electrophoresis, revealing a shift in fluorescent signal caused by the 5′-FAM L93 oligo traveling slower with the supercoiled pUC19.
Purified mTk-RadA, as expected, functions as a recombinase and successfully catalyzes the strand invasion of the 5′-FAM L93 oligo into the supercoiled pUC19, resulting in the D loop (fig. S4A). The optimal temperature for RadA-mediated invasion was established to be 65°C, comparable with prior results using RadA from Pyrococcus abysii (fig. S4D) (21). Incubating the recombinase reaction with increasing amounts of pTk-RadA, in an attempt to inhibit mTk-RadA, did not affect the efficiency of D-loop formation (fig. S4B), suggesting that the unspliced pRadAs were not interacting with nor impairing the capacity of mRadA to drive D-loop formation even when present at equal molar concentrations. Note that while we added pTk-RadA preparations that contained almost no-spliced proteins into the recombinase reactions, the in vitro recombinase assay conditions appeared to promote the splicing of pTk-RadA, as observed by SDS-PAGE (fig. S4C); given that the bulk of the added precursor protein does not splice, we remain confident that the excess pTk-RadA does affect mTk-RadA function in vitro. While we did generate a pTk-RadA variant wherein we mutated the first cysteine residue on the intein splicing junctions to completely prevent pTk-RadA from splicing (and thus eliminate any remaining concerns of splicing of pTk-RadA in the recombinase reactions), the resulting variant was unstable and repeatedly precipitated out of solution during heat treatment. The results obtained thus argue that increased pTk-RadA levels do not negatively affect the functions of mTk-RadA and that the massive phenotypic impacts result directly from reduced splicing efficiencies and reduced mTk-RadA protein levels in vivo.
DISCUSSION
Inteins can be vertically inherited, transferred via endosymbiosis, move between closely related strains during mating, propagate via intragenomic transfer, or be horizontally transferred between species or even domains, often as a result of viral-mediated events (35, 45). T. kodakarensis encodes the most inteins relative to genome size reported, with at least 15 inteins distributed in 12 genes (Table 1) (36). About 1% of the T. kodakarensis genome is composed of inteins, with these enigmatic elements representing over 20 kbp of sequence. While inteins might be traditionally viewed as molecular parasites and fitness-negative MGEs, the past decade has yielded compelling work to demonstrate that the rate and accuracy of protein splicing for dozens of inteins are highly dependent on environment, suggesting that CPS may provide regulatory benefits to the intein-containing organism (26).
Table 1. Inteins present in the T. kodakarensis genome.
| Gene | Protein | Function | RRR | Number of Intein | Intein Length | Extein Length |
|---|---|---|---|---|---|---|
| TK0001 | Pol B | DNA polymerase | Yes | 2 | 360/536 | 760 |
| TK0470 | Rgy | Reverse gyrase | Yes | 1 | 489 | 1222 |
| TK0764 | LHR | Large helicase–related protein | Yes | 1 | 525 | 865 |
| TK1091 | TopA | DNA topoisomerase 1 | Yes | 1 | 511 | 718 |
| TK1305 | IF2 | Initiation factor 2 | No | 1 | 546 | 598 |
| TK1332 | Ski2-like | DNA repair and recombination | Yes | 1 | 403 | 722 |
| TK1620 | MCM-3 | Replicative DNA helicase | Yes | 2 | 140/335 | 682 |
| TK1736 | RNR | Ribonucleotide reductase | No | 2 | 454/382 | 910 |
| TK1853 | KlbA | Type II/IV secretion system adenosine triphosphatase | No | 1 | 523 | 675 |
| TK1899 | RadA | Homologous recombinase | Yes | 1 | 482 | 354 |
| TK1903 | Pol D | DNA polymerase II | Yes | 1 | 474 | 1324 |
| TK2218 | RFC | Clamp loader | Yes | 1 | 540 | 322 |
The preponderance of inteins in RRR-related genes in T. kodakarensis (11 of 15; Table 1) is in line with the prevalence of inteins in RRR-related gene across both prokaryotic domains (~65%) (36). Given the nonrandom distribution and retention of inteins in RRR-related genes, coupled with the wealth of in vitro work demonstrating CPS in response to environmental signals acutely relevant to the intein-containing organism (22–35), argue that some inteins have been exapted from one-time parasites to beneficial regulatory elements. However, the major criticism of this hypothesis was the complete absence of work demonstrating a connection between RRR-related protein function and intein splicing efficiencies within an organism that naturally houses inteins. This study provides a compelling connection between intein activity and RRR-protein activity in vivo. In this case, we show that the growth rates, response to fitness challenges, and the dominant mode of DNA replication can be regulated by differences in the intein splicing efficiency or RadA levels.
While the reduction in splicing we observe is based on intein-specific mutations in otherwise isogenic strains, we convincingly demonstrate that a reduction in mTk-RadA levels due to variable intein splicing results in T. kodakarensis favoring ODR over RDR (Fig. 6), a reduction in growth rate, and an increase in UV sensitivity (Fig. 5). In addition, we show for the splicing-deficient AL015 strain that the switch from RDR to ODR becomes more apparent at lower temperature, which correlates with intein splicing accuracy and efficiency both in vivo and in vitro (Figs. 2 to 4). Although our findings rely on an artificial down-regulation of intein activity by intein-specific mutations on the T. kodakarensis chromosome, our work suggests that CPS could potentially be a prevalent mechanism of regulating the efficiency, rate, or mechanisms of DNA RRR under distinct environmental conditions. These results represent seminal findings in an emerging field that will lay the groundwork for the investigation of CPS within T. kodakarensis and other intein-containing organisms.
We demonstrate that a relatively modest reduction (~50%) in the splicing of a single intein can radically affect microbial physiology. Future work will be required to determine whether this is specific to homologous recombinases such as RadA that form nucleoprotein filaments where activity can be highly sensitive to protein concentration or more generalizable to most intein-containing proteins. Nevertheless, while it has been long hypothesized that splicing inhibition would result in diminished growth, this work represents the first in vivo demonstration and shows the potential power of protein splicing inhibition to control microbial growth. It remains plausible that variant intein sequences within identical extein sequences can influence the folding of sequence-identical exteins and thus affect the function of the mature protein. Regardless, knowing that differential intein excision can radically affect total cellular physiology and fitness, combined with (i) the retention of inteins in several devastating human pathogens, including M. tuberculosis and Cryptococcus neoformans, and (ii) the complete absence of inteins in metazoans, lends support to the ongoing search for targeted protein splicing inhibitors as previously unidentified antimicrobials (46, 47). The ability for modest changes in the splicing of a single intein to radically affect microbial growth, combined with the retention of inteins in many pathogens, but the near absence (only 1%) or complete loss of inteins in eukaryotic genomes and metazoans, respectively, offers intriguing opportunities to target intein splicing with therapeutics (46, 47).
The integration of inteins into the active sites or critical regulatory regions of RRR-related proteins may reflect a selection for intein integrations that retain the greatest likelihood of impacting function of the RRR-related protein in the precursor form. The Tk-RadA-intein is located within the ATP-binding P loop. Alternatively, inteins may cluster to these RRR proteins due to their high degree of genetic conservation and, thus, HEN target site conservation, which would promote horizontal intein transfer. We reason therefore that in some, but not all, cases, retention within RRR proteins is due to exaptation. It will thus be critical to determine both the efficiency of splicing and the impact on steady-state mature protein concentrations in vivo for inteins in different classes of genes and across each domain to fully understand the biological role inteins play in regulating RRR mechanisms. Given that even very subtle changes to in vivo protein concentrations and splicing efficiencies may result in marked changes in cellular physiology, it will be equally critical to evaluate the fitness importance of intein splicing efficiency—an ancient selectable characteristic of inteins—to the fitness importance of regulated intein splicing—an evolutionarily recent exaptation of intein biology.
MATERIALS AND METHODS
Microbial growth and medium conditions
Parental (TS559) and newly constructed T. kodakarensis strains were anaerobically maintained in an artificial sea water–based medium supplemented with tryptone (5 g/liter), yeast extract (5 g/liter), pyruvate (5 g/liter), elemental sulfur (S°; 2 g/liter), a KOD1-vitamin mixture, and 1 mM agmatine sulfate at 85° or 65°C (48). Growth rates were monitored via optical density measurements at 600 nm in liquid cultures. Cultures were prepared with 1:100 inoculums from overnight cultures grown in the same medium, and the growth rates of a minimum of three independent biological replicates were monitored and reported (Figs. 4 and 5).
T. kodakarensis strain constructions
The genomic sequences encoding Tk-RadA (TK1899) were targeted for allelic modification using standard markerless modification protocols in T. kodakarensis strain TS559 (48) and ΔoriΔcdc6 (13, 48). Briefly, nonreplicative plasmids containing sequences homologous to the flanking regions of TK1899 along with the desired allelic change(s) were temporarily integrated and then subsequently excised from the TS559 or ΔoriΔcdc6 genome in the region surrounding TK1899 based on restoration of agmatine prototrophy and resistance to 6-methylpurine, respectively. Sanger sequencing of amplicons generated via diagnostic PCR using primers adjacent to TK1899 with genomic DNA purified from newly constructed strains confirmed the exact end points of deletions and any allelic modifications. Preparations of genomic DNA from strains presumed to be modified at TK1899 were sequenced at >100× coverage via MinION Nanopore sequencing (Oxford Nanopore Technologies, Oxford, UK) to finalize WGS. Analyses of WGS results confirmed the introduction of sequences encoding allelic variants at TK1899 and the absence of any secondary mutations throughout the remainder of the genome. WGS also confirmed the loss of TKV2 in strains AL026 and ΔoriΔcdc6. HEN activity of native Tk-RadA necessitated a unique path to strain constructions to ensure that intein invasion of inteinless alleles of TK1899 did not prohibit generation of desired alleles. TS559 was used to first generate AL002 [Tk-RadAA(373–381)], the inactive HEN activity lacking variant of Tk-RadA. AL002 was used to generate AL003 (Tk-RadAΔIntein), the inteinless variant of Tk-RadA. AL003 was used to generate AL015 (Tk-RadAΔ270–592+P.ho. loop), and the mini-intein variant of Tk-RadA. ΔoriΔcdc6 was used to generate AL026.
Cloning, expression, and purification of Tk-RadA variants
The wild-type, intein-containing sequences of Tk-RadA (TK1899) was amplified from purified TS559 genomic DNA, incorporating sequences encoding a C-terminal 6×His tag during amplification and cloned into the Sal I site of pQE-80L via In-Fusion Snap Assembly (Takara Bio USA Inc.). Sequence variants, used to produce proteins variants of Tk-RadA, were introduced via QuikChange mutagenesis on the Tk-RadAWT expression plasmid. Sanger sequencing confirmed the full sequence of the entire insert for all constructs. Each expression plasmid was transformed into Rosetta 2 competent cells (Novagen). Transformants were grown at 37°C in LB medium containing sodium ampicillin salt (100 μg/ml) and chloramphenicol (25 μg/ml). Protein expression was induced at optical density at 600 nm (OD600nm) of ~0.4 through addition of isopropyl-β-d-thiogalactopyranoside to 0.5 mM final concentration, and protein production was permitted for an additional 4 hours of 37°C growth. Cells were harvested via centrifugation (15,000g for 15 min at 4°C), the supernatant was discarded, and cell pellets were frozen at −20°C until protein purifications. Cell pellets were thawed, resuspended in 20 mM tris-HCl (pH 8.3), 500 mM NaCl, 10% glycerol, and 30 mM imidazole (buffer A), and lysed using sonication, and cellular debris were removed through centrifugation (75,000g at 4°C for 20 min). Clarified lysates were passed through a 5-ml HiTrap Chelating HP column (Cytiva) charged with nickel, washed extensively with buffer A, and then eluted using a linear, 20-column volume gradient from 100% buffer A to 100% buffer B [20 mM tris-HCl (pH 8.3), 500 mM NaCl, 10% glycerol, and 500 mM imidazole]. Fractions containing Tk-RadA were identified via SDS-PAGE, pooled, and dialyzed into 20 mM tris-HCl (pH 8.3), 200 mM NaCl, and 50% glycerol for long-term storage.
HEN assays
DNA amplicons were generated via two rounds of PCR, each followed by gel extraction and PCR clean-up, respectively, using Nucleospin Gel and PCR Clean-Up kit (MACHEREY-NAGEL Inc.). HEN activity was evaluated at 75°C for 1 hour in 20 μl of reactions containing 0 to 0.5 nM of the Tk-RadAWT or Tk-RadA variant proteins and 15 nM of purified DNA substrate in 50 mM tris-HCl (pH 8.3), 100 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol. The reactions were stopped by addition of 100 μl of 0.6 M tris-HCl (pH 8.0) and 12 mM EDTA and extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, v/v/v). Precipitation of the aqueous phase was facilitated with 2.6× volumes of 100% ethanol and GlycoBlue Coprecipitant (50 μg/ml; Invitrogen). Purified DNAs were resolved through either 1% (Fig. 1 and fig. S1) or 3% (fig. S2) 1× tris-borate EDTA (TBE) agarose gels run in TBE and visualized by EtBr staining. The cleaved fragments generated from TK1899ΔIntein-encoding sequences were Sanger sequenced by Azenta Life Sciences (fig. S1B) to map the exact cut sites.
D-loop formation assay
The protocol to monitor D-loop formation was adapted from Hogrel et al. (21) with a few modifications. Supercoiled pUC19 plasmids were purified using a QIAGEN plasmid purification kit following a low-temperature modified protocol intended to increase isolation of supercoiled plasmids from E. coli cells (49). The 5′-FAM L93 single-stranded fluorescently labeled DNA substrates (5′-[FAM]-AAA-GGC-GGT-AAT-ACG-GTT-ATC-CAC-AGA-ATC-AGG-GGA-TAA-CGC-AGG-AAA-GAA-CAT-GTG-AGC-AAA-AGG-CCA-GCA-AAA-GGC-CAG-GAA-CCG-TAA-AAA-3′) was obtained from Eurofins Genomics LLC. Briefly, 25 nM 5′-FAM L93 was mixed with purified mTk-RadA or pTk-RadA at various concentrations from 0 to 1600 nM or 0 to 800 nM, respectively (detailed in fig. S4) in 20 mM tris-HCl (pH 8.0), 125 mM NaCl, 10 mM dithiothreitol, bovine serum albumin (50 μg/ml), 10 mM MgCl2, and 2.5 mM ATP (when indicated), followed by 10 min of incubation at 65°C (note that incubation temperatures were varied in fig. S4C). Following the initial incubation, 25 nM supercoiled pUC19 was added, and the reactions were incubated again for 10 min at 65°C (unless noted otherwise). The reactions were terminated by the addition of Proteinase K (50 μg/ml), 0.5% SDS, and 40 mM EDTA, followed by 15 min of incubation at 37°C. Reactions were resolved through 1.2% TBE agarose gel following the addition of an equal volume of 20% Ficoll. The unincorporated and D loop–incorporated 5′-FAM L93 oligos were visualized with a Typhoon FLA 9500 (GE Healthcare). The percentage of oligo complexed with the plasmid through the activities of Tk-RadA was quantified with ImageQuant software and plotted in Excel.
Western blot analysis
Purified pTk-RadAWT was used as an antigen to generate polyclonal antibodies in guinea pigs (Cocalico Biologicals Inc.); test and terminal bleeds were confirmed against purified Tk-RadAWT and lysates from T. kodakarensis for specificity. T. kodakarensis cultures were grown at either 85° or 65°C until optical density measurements of 0.4 to 0.5 were achieved, representing mid-log phase growth, and then rapidly chilled on ice. Chilled cells were harvested via centrifugation (15,000g for 10 min at 4°C) and resuspended in 100 μl of 25 mM tris-HCl (pH 8.0), 500 mM NaCl, 10% glycerol, and 2% SDS. Protein concentrations were quantified using the Qubit Protein Assay (Thermo Fisher Scientific). Proteins were resolved through 4 to 20% acrylamide gels, blotted onto polyvinylidene difluoride (PVDF) membranes, blocked with 5% bovine serum albumin, and probed using the primary anti–Tk-RadA antibody (1:10,000 dilution). Blots were washed, then probed with immunoglobulin G–alkaline phosphatase–conjugated goat anti–guinea pig secondary antibodies (1:1000 dilution), and visualized using the 1-Step NBT/BCIP Substrate Solution (Thermo Fisher Scientific). Western blot bands were quantified with ImageQuant software and plotted in Excel.
UV sensitivity assay
Parental (TS559) and AL015 T. kodakarensis strains were grown at 85°C in rich medium supplemented with 1 mM agmatine sulfate and KOD1-vitamin mixture as previously described to an OD600nm of ~0.4 to 0.5 before being rapidly harvested via centrifugation (8000g for 15 min at 4°C), the supernatant was discarded, and the cell pellets were resuspended in 150 ml of 1× artificial sea water. The resuspended cells were anaerobically irradiated by exposure to a UV light in 10 ml of aliquots at 100 μW/cm2 for 0, 5, 10, 15, 20, 30, 45, 60, and 90 s (resulting in total exposures of 0, 5, 10, 15, 20, 30, 45, 60, and 90 J/m2, respectively) and immediately put on ice. Irradiated cultures were serially diluted 10-fold from 10−1 to 10−7, and 10 μl from each dilution was plated onto rich medium–solidified plates supplemented with polysulfides, agmatine sulfate, and KOD1-vitamin mixture. The plates were incubated anaerobically at 85°C for 48 hours. Given that T. kodakarensis colonies are flat, pale, and difficult to image conventionally, colony-forming unit counts were determined after transferring colonies to PVDF (0.2 μm) and staining the proteins lysed from transferred cells. PVDF membranes were prerinsed with 100% methanol for 10 min and pressed onto the colony-containing plates to facilitate colony transfer. The colony-containing membranes were flash frozen with liquid nitrogen to facilitate cell lysis and protein release and then stained with Coomassie Brilliant Blue G-250 for 20 min with gentle rocking. The membranes were destained twice in 100% methanol for 5 min and allowed to air dry. The fraction of viable cells at each UV dose was determined by comparing the number of colony-forming units from countable spots at each UV dose to that of the same dilution spot from the no UV control for each strain. Assays were done in minimally triplicate for each strain.
Marker frequency analysis
Genomic DNAs were isolated at mid-exponential (0.4 to 0.5 OD600nm) and late stationary phase (after the OD600nm reading peaked and modestly decreased). Illumina libraries were prepared using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs) as directed by the manufacturer. The quality of each library was assessed using an Agilent Bioanalyzer 2100 using an Agilent High Sensitivity Kit (Agilent Technologies). Libraries were pooled in equimolar proportion and sequenced on an Illumina NextSeq instrument, using a NextSeq 1000/2000 P2 reagent kit. Raw sequencing data were uploaded into the Galaxy platform and processed via fastp for adaptor trimming and filtering of low-quality reads, HISAT2 for aligning the processed data to the TS559 reference genome, and bamCoverage to bin the TS559 reference genome into nonoverlapping 1000-bp bins for generating a bedGraph file that classifies the sequencing reads into the corresponding bin normalized to reads per kilobase per million mapped reads. Graphs plotting the log2 (exponential/stationary) ratios were generated using Excel.
In vitro splicing assays
All MIG splicing reporter constructs (Figs. 2B and 3A) containing either Tk-RadA-inteinA(373–381), Tk-RadA-inteinD373–381, Tk-RadA-inteinΔ286–585, Tk-RadA-inteinΔ276–585, or Tk-RadA-inteinΔ270–592+P.ho. loop were commercially synthesized and sequenced (GenScript, USA) based on previous MIG reporters in the pACYC vector backbone (24). Construction of the Pho-RadA-inteinWT was previously described (24). All inteins within the MIG reporter are flanked by 10 residues on both the N and C termini from the natural RadA exteins, which are identical in all constructs.
For native extein splicing assays with Tk-RadA-inteinΔ286–585 (Fig. 3C), this construct was commercially synthesized and subcloned into the pET45b(+) vector backbone in-frame with an N-terminal His tag (GenScript, USA). This construct contains the entire natural C-extein, which forms interactions with the intein that inhibits splicing (23, 24), and a deletion in the N-extein (residues 1 to 112) previously shown to not influence extein-intein interactions or response to ssDNA (23).
Plasmids were transformed into E. coli BL21(DE3), and protein expression was induced in mid-log phase by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (GoldBio). Proteins were expressed for ~20 hours at 15°C, and cells were harvested at 4000g.
For MIG splicing assays, cell pellets were resuspended in 50 mM tris-HCl (pH 8.0) and 10% glycerol and lysed by sonication. Insoluble material was removed by centrifugation, and clarified lysates were either examined immediately for splicing during expression or incubated at 50°C as described in Figs. 2 and 3. To measure splicing efficiencies, lysates were mixed with Laemmli sample buffer and resolved using 8 to 16% Tris-Glycine eXtended gels (Bio-Rad). Samples were not heated in Laemmli sample buffer to maintain GFP fluorescence. GFP fluorescence was measured in-gel using an Amersham Imager 680 (GE Healthcare).
For the Tk-RadA-inteinΔ286–585 in native exteins (Fig. 3), cell pellets were resuspended in 20 mM tris-HCl (pH 8.0), 500 mM NaCl, and 30 mM imidazole and lysed by sonication, and insoluble material was removed by centrifugation. Clarified lysates were purified using Ni-charged MagBeads (GenScript) and dialyzed into 20 mM tris-HCl (pH 8.5), 200 mM NaCl, and 10% glycerol. Purified protein was mixed with ssDNA (187.5 ng/μl; Bayou Biolabs) or tris-EDTA buffer and incubated as described in Fig. 3C. Reactions were mixed with SDS sample buffer, resolved through 8 to 16% bis-tris gels (GenScript, USA), and stained with Coomassie Brilliant Blue dye.
Relative levels of precursor, LE, and other species were quantified by densitometry using ImageJ (imagej.nih.gov) based on three biological replicates. Average and SD are shown in the bar graphs.
Acknowledgments
We thank M. Belfort and Z. Kelman for comments.
Funding: This work was supported by funding from the US NASA awards 80NSSC20K0613 and 80NSSC23K1354 (to T.J.S.), US National Science Foundation award MCB-2016857 (to T.J.S.), and the US National Institutes of Health awards R15-GM143662 (to C.W.L.) and R35-GM143963 (to T.J.S.). This study was also privately funded from New England Biolabs Inc. (to K.M.Z. and A.F.G.).
Author contributions: Conceptualization: G.L.S.L., C.W.L., A.M.G., and T.J.S. Methodology: G.L.S.L., C.W.L., A.M.G., and T.J.S. Investigation: G.L.S.L., C.W.L., J.L.M., A.M.G., and K.M.Z. Visualization: G.L.S.L., C.W.L., J.L.M., A.M.G., and K.M.Z. Supervision: G.L.S.L., C.W.L., A.M.G., and T.J.S. Writing—original draft: G.L.S.L., C.W.L., and T.J.S. Writing—review and editing: All authors.
Competing interests: K.M.Z. and A.F.G. are employees of New England Biolabs Inc. This affiliation does not affect the authors’ impartiality, objectivity of data generation, or its interpretation, adherence to journal standards and policies or availability of data. All other authors declare that they have no competing interests.
Data and materials availability: Datasets from WGS and MFA analyses were deposited to the NCBI Sequence Read Archive (SRA) and can be accessed via PRJNA941823. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
REFERENCES AND NOTES
- 1.Kelman L. M., Kelman Z., Archaeal DNA replication. Annu. Rev. Genet. 48, 71–97 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Raymann K., Forterre P., Brochier-Armanet C., Gribaldo S., Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in archaea. Genome Biol. Evol. 6, 192–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausiannikava D., Allers T., Diversity of DNA replication in the archaea. Genes 8, 56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cubonová L., Richardson T., Burkhart B. W., Kelman Z., Connolly B. A., Reeve J. N., Santangelo T. J., Archaeal DNA polymerase D but not DNA polymerase B is required for genome replication in Thermococcus kodakarensis. J. Bacteriol. 195, 2322–2328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell S. D., Archaeal orc1/cdc6 proteins. Subcell. Biochem. 62, 59–69 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Beattie T. R., Bell S. D., Molecular machines in archaeal DNA replication. Curr. Opin. Chem. Biol. 15, 614–619 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Kunkel T. A., Burgers P. M. J., Arranging eukaryotic nuclear DNA polymerases for replication: Specific interactions with accessory proteins arrange Pols α, δ, and ϵ in the replisome for leading-strand and lagging-strand DNA replication. Bioessays 39, 1700070 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riera A., Barbon M., Noguchi Y., Reuter L. M., Schneider S., Speck C., From structure to mechanism-understanding initiation of DNA replication. Genes Dev. 31, 1073–1088 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bleichert F., Botchan M. R., Berger J. M., Mechanisms for initiating cellular DNA replication. Science 355, eaah6317 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Samson R. Y. Y., Xu Y., Gadelha C., Stone T. A. A., Faqiri J. N. N., Li D., Qin N., Pu F., Liang Y. X. X., She Q., Bell S. D. D., Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 3, 485–496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa A., Hood I. V., Berger J. M., Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 82, 25–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forterre P., Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 33, 457–465 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Gehring A. M., Astling D. P., Matsumi R., Burkhart B. W., Kelman Z., Reeve J. N., Jones K. L., Santangelo T. J., Genome replication in Thermococcus kodakarensis independent of Cdc6 and an origin of replication. Front Microbiol. 8, 2084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudier M., Schuwirth B. S., Westcott S. L., Wigley D. B., Structural basis of DNA replication origin recognition by an ORC protein. Science 317, 1213–1216 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Samson R. Y., Abeyrathne P. D., Bell S. D., Mechanism of archaeal MCM helicase recruitment to DNA replication origins. Mol. Cell 61, 287–296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawkins M., Malla S., Blythe M. J., Nieduszynski C. A., Allers T., Accelerated growth in the absence of DNA replication origins. Nature 503, 544–547 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spaans S. K., van der Oost J., Kengen S. W. M., The chromosome copy number of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Extremophiles 19, 741–750 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seitz E. M., Brockman J. P., Sandler S. J., Clark A. J., Kowalczykowski S. C., RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev. 12, 1248–1253 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wardell K., Haldenby S., Jones N., Liddell S., Ngo G. H. P., Allers T., RadB acts in homologous recombination in the archaeon Haloferax volcanii, consistent with a role as recombination mediator. DNA Repair 55, 7–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova K. S., Koonin E. V., Archaeology of eukaryotic DNA replication. Cold Spring Harb. Perspect. Biol. 5, a012963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogrel G., Lu Y., Alexandre N., Bossé A., Dulermo R., Ishino S., Ishino Y., Flament D., Role of RadA and DNA polymerases in recombination-associated DNA synthesis in hyperthermophilic archaea. Biomolecules 10, 1045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lennon C. W., Stanger M., Banavali N. K., Belfort M., Conditional protein splicing switch in hyperthermophiles through an intein-extein partnership. mBio 9, e02304–e02317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lennon C. W., Stanger M., Belfort M., Protein splicing of a recombinase intein induced by ssDNA and DNA damage. Genes Dev. 30, 2663–2668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Topilina N. I., Novikova O., Stanger M., Banavali N. K., Belfort M., Post-translational environmental switch of RadA activity by extein-intein interactions in protein splicing. Nucleic Acids Res. 43, 6631–6648 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yalala V. R., Lynch A. K., Mills K. V., Conditional alternative protein splicing promoted by inteins from Haloquadratum walsbyi. Biochemistry 61, 294–302 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood D. W., Belfort M., Lennon C. W., Inteins-mechanism of protein splicing, emerging regulatory roles, and applications in protein engineering. Front. Microbiol 14, 1305848 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lennon C. W., Wahl D., Goetz J. R., Weinberger J., Reactive chlorine species reversibly inhibit DnaB protein splicing in mycobacteria. Microbiol. Spectr. 9, e0030121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green C. M., Li Z., Smith A. D., Novikova O., Bacot-Davis V. R., Gao F., Hu S., Banavali N. K., Thiele D. J., Li H., Belfort M., Spliceosomal Prp8 intein at the crossroads of protein and RNA splicing. PLOS Biol. 17, e3000104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topilina N. I., Green C. M., Jayachandran P., Kelley D. S., Stanger M. J., Piazza C. L., Nayak S., Belfort M., SufB intein of Mycobacterium tuberculosis as a sensor for oxidative and nitrosative stresses. Proc. Natl. Acad. Sci. U.S.A. 112, 10348–10353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills K. V., Paulus H., Reversible inhibition of protein splicing by zinc ion. J. Biol. Chem. 276, 10832–10838 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Reitter J. N., Cousin C. E., Nicastri M. C., Jaramillo M. V., Mills K. V., Salt-dependent conditional protein splicing of an intein from Halobacterium salinarum. Biochemistry 55, 1279–1282 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Woods D., Vangaveti S., Egbanum I., Sweeney A. M., Li Z., Bacot-Davis V., Lesassier D. S., Stanger M., Hardison G. E., Li H., Belfort M., Lennon C. W., Conditional DnaB protein splicing is reversibly inhibited by zinc in mycobacteria. mBio 11, e01403–e01420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callahan B. P., Topilina N. I., Stanger M. J., Van Roey P., Belfort M., Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat. Struct. Mol. Biol. 18, 630–633 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lennon C. W., Stanger M. J., Belfort M., Mechanism of single-stranded DNA activation of recombinase intein splicing. Biochemistry 58, 3335–3339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lennon C. W., Belfort M., Inteins. Curr. Biol. 27, R204–R206 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Novikova O., Jayachandran P., Kelley D. S., Morton Z., Merwin S., Topilina N. I., Belfort M., Intein clustering suggests functional importance in different domains of life. Mol. Biol. Evol. 33, 783–799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavankumar T. L., Inteins: Localized distribution, gene regulation, and protein engineering for biological applications. Microorganisms 6, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeder D. L., Weiss R. B., Dunn D. M., Cherry J. L., González J. M., Diruggiero J., Robb F. T., Niederhausern V., Aoyagi A., Mahmoud M., Hannenhalli S., Lupas A. N., Koretke K. K., Diruggiero J., Divergence of the hyperthermophilic archaea Pyrococcus furiosus and P. horikoshii inferred from complete genomic sequences. Genetics 152, 1299 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishioka M., Fujiwara S., Takagi M., Imanaka T., Characterization of two intein homing endonucleases encoded in the DNA polymerase gene of Pyrococcus kodakaraensis strain KOD1. Nucleic Acids Res. 26, 4409–4412 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinberger Ii J., Lennon C. W., Monitoring protein splicing using in-gel fluorescence immediately following SDS-PAGE. Bio. Protoc. 11, e4121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naor A., Altman-Price N., Soucy S. M., Green A. G., Mitiagina Y., Turgeman-Grotta I., Davidovich N., Gogarten J. P., Gophna U., Impact of a homing intein on recombination frequency and organismal fitness. Proc. Natl. Acad. Sci. U.S.A. 113, E4654–E4661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinzon S., Cawood A. R., Ruiz M. A., Gophna U., Altman-Price N., Mills K. V., Protein splicing activity of the Haloferax volcanii PolB-c intein is sensitive to homing endonuclease domain mutations. Biochemistry 59, 3359–3367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andersson A. F., Pelve E. A., Lindeberg S., Lundgren M., Nilsson P., Bernander R., Replication-biased genome organisation in the crenarchaeon Sulfolobus. BMC Genomics 11, 454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tagashira K., Fukuda W., Matsubara M., Kanai T., Atomi H., Imanaka T., Genetic studies on the virus-like regions in the genome of hyperthermophilic archaeon, Thermococcus kodakarensis. Extremophiles 17, 153–160 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Green C. M., Novikova O., Belfort M., The dynamic intein landscape of eukaryotes. Mob. DNA 9, 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tharappel A. M., Li Z., Li H., Inteins as drug targets and therapeutic tools. Front. Mol. Biosci. 9, 821146 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wall D. A., Tarrant S. P., Wang C., Mills K. V., Lennon C. W., Intein inhibitors as novel antimicrobials: Protein splicing in human pathogens, screening methods, and off-target considerations. Front. Mol. Biosci. 8, 752824 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liman G. L. S., Stettler M. E., Santangelo T. J., Transformation techniques for the anaerobic hyperthermophile Thermococcus kodakarensis. Methods Mol. Biol. 2522, 87–104 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carbone A., Fioretti F. M., Fucci L., Ausió J., Piscopo M., High efficiency method to obtain supercoiled DNA with a commercial plasmid purification kit. Acta Biochim. Pol. 59, 275–278 (2012). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4